- Department of Microbiology, Institute of Health Biosciences, The University of Tokushima Graduate School, Tokushima, Japan
Human immunodeficiency virus type 1 (HIV-1) is tropic and pathogenic only for humans, and does not replicate in macaque monkeys routinely used for experimental infections. This specially narrow host range (species tropism) has impeded much the progress of HIV-1/acquired immunodeficiency syndrome (AIDS) basic research. Extensive studies on the underlying mechanism have revealed that Vif, one of viral accessory proteins, is critical for the HIV-1 species tropism in addition to Gag-capsid protein. Another auxiliary protein Vpu also has been demonstrated to affect this HIV-1 property. In this review, we focus on functional interactions of these HIV-1 proteins and species specific-restriction factors. In addition, we describe an evolutional viewpoint that is relevant to the species tropism of HIV-1 controlled by the accessory proteins.
Introduction
Human immunodeficiency virus type 1 (HIV-1) is strictly adapted to humans, and cause disease-inducing persistent infection only in humans (Nomaguchi et al., 2008). This property is unique among primate immunodeficiency viruses, and represent one of the most evident and important viral characteristics to understand the biology/molecular biology of HIV-1. Of numerous primate immunodeficiency viruses so far identified (Kirchhoff, 2009; Sharp and Hahn, 2011), HIV-1 with an extremely limited host range exhibits exceptionally high replication ability, transmissibility, and pathogenicity in sensitive host humans. For basic HIV-1 researchers, it would be final goal to realize the basis/mechanism underlying these properties by experimental approaches.
Primate immunodeficiency viruses can be divided into three groups based on their genome structure in the central regions (Kirchhoff, 2009; Fujita et al., 2010; Sharp and Hahn, 2011). While viruses of HIV-1 type contain vpr and vpu genes, viruses of HIV-2 type carry vpx and vpr genes in tandem (Figure 1). The other simian immunodeficiency viruses (SIVs), the prototype virus, have only the vpr gene in the corresponding genomic region. HIV-1 is believed to emerge from the prototype virus via SIVmon/mus/gsn (isolated from the mona, mustached, and greater spot-nosed monkeys), SIVcpz (isolated from the chimpanzees), and SIVgor (isolated from the gorilla) through mutational and recombinational events. SIVmon/mus/gsn is known to recombine with SIVrcm (isolated from the red-capped mangabey monkey) to generate SIVcpz (for genome structures, see, Figure 1). SIVcpz served as parental virus for HIV-1 (M and N) and SIVgor (and finally for HIV-1 P).
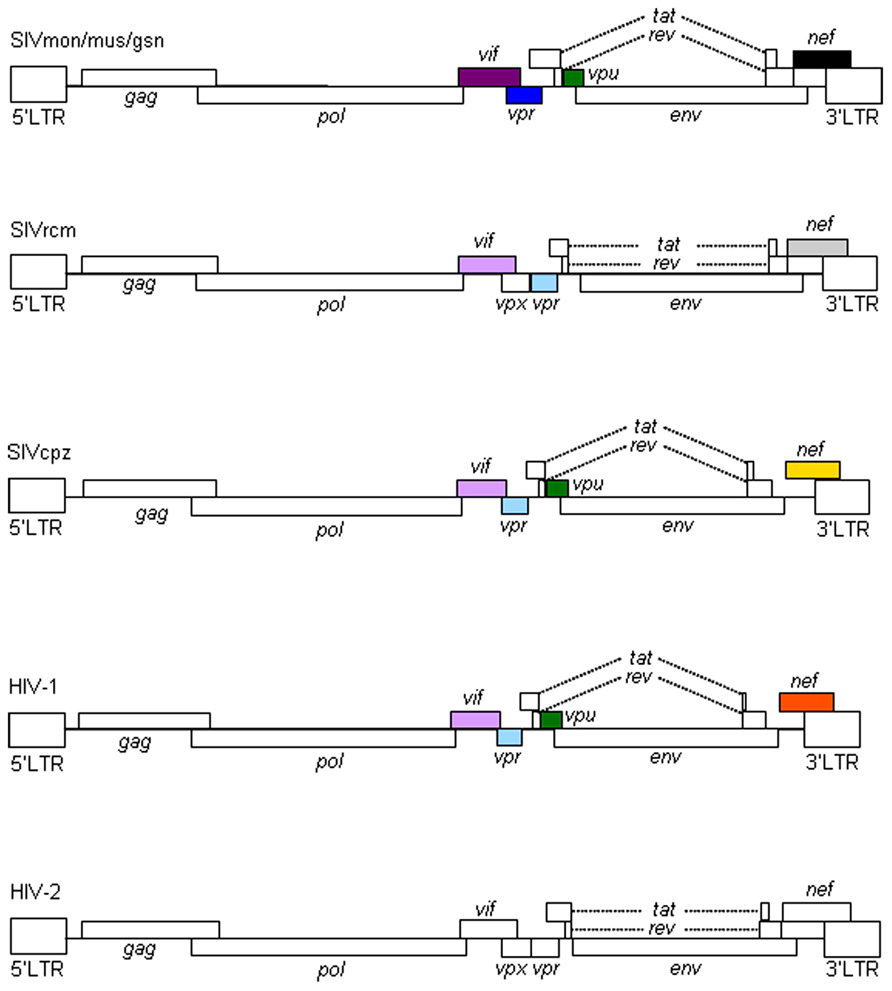
FIGURE 1. Genome organization of primate immunodeficiency viruses. Various proviral genomes are schematically shown. As indicated by colored boxes, the vpr and vpu genes of SIVcpz/HIV-1 came from those of SIVrcm and SIVmon/mus/gsn, respectively. Also, the vif genes of SIVcpz/HIV-1 originated from that of SIVrcm. In addition, as shown by colored boxes, HIV-1 nef gene is similar to but distinct from SIVcpz nef gene. HIV-1 nef gene is different from those of SIVmon/mus/gsn, SIVrcm, and HIV-2 as indicated. For virus designations, see text.
The biological and molecular biological bases for species tropism of HIV-1 should reside in the above outlined evolutional processes. In particular, the so-called accessory proteins encoded by extra genes are important. Each virus group has a unique set of the accessory proteins in terms of their combinations and of their activities. Therefore, studies on viral accessory proteins are also meaningful for understanding viral evolution by cross-species transmission.
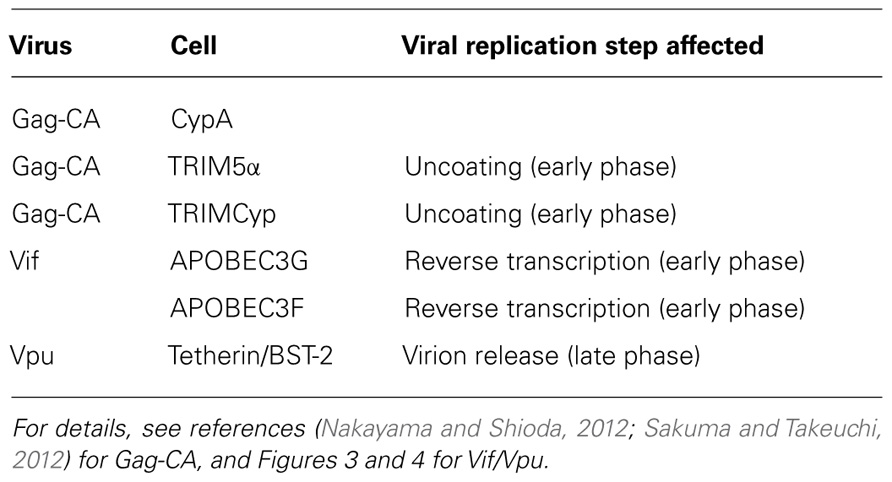
Viral and Cellular Determinants for HIV-1 Species Tropism
Our early studies have already suggested the possible viral determinants and viral replication stage involved in the HIV-1 species tropism described above (Shibata et al., 1991, 1995; Shibata and Adachi, 1992). By the use of numerous chimeric molecular clones between HIV-1 and dual-tropic (tropic for human and monkey cells) SIVmac (isolated from the macaque monkey), we have claimed in essence, together with a work on the cyclophilin A (CypA; Dorfman and Gottlinger, 1996), that Gag-capsid (CA) and a viral protein(s) encoded by the central genomic region of HIV-1 are the determinants. We also have showed that HIV-1 is replication-incompetent in monkey cells because a certain replication step(s) before/during reverse transcription, other than the viral entry into cells, does not proceed normally. Subsequent extensive studies by us and others have clearly indicated that the interactions of Gag-CA/CypA, Gag-CA/tripartite motif (TRIM) proteins, and Vif/apolipoprotein B mRNA-editing enzyme-catalytic (APOBEC) proteins are major determinants for the HIV-1 species tropism (Nomaguchi et al., 2008, 2011; Nakayama and Shioda, 2012; Sakuma and Takeuchi, 2012) as summarized in Table 1. Gag-CA, CypA, and TRIM proteins have been described in detail in two articles in the Research Topic of this journal (Nakayama and Shioda, 2012; Sakuma and Takeuchi, 2012).
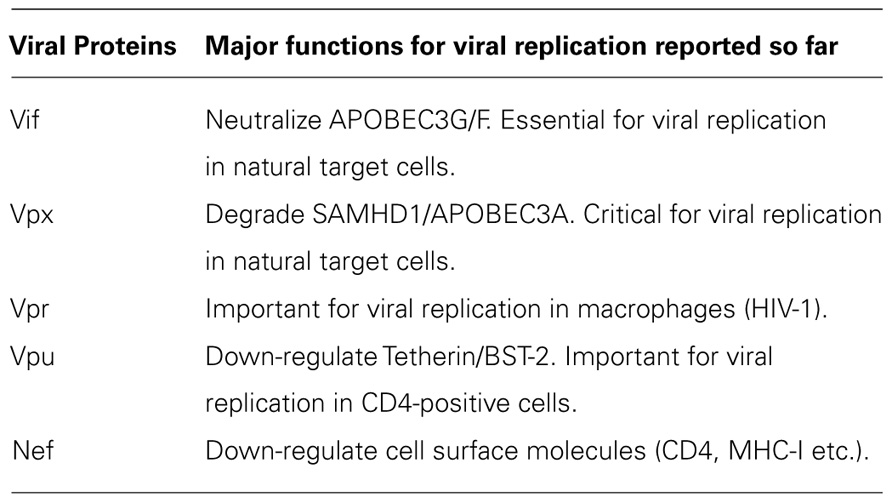
Accessory Proteins of Primate Immunodeficiency Viruses
All primate immunodeficiency viruses encode a number of extra proteins (Vif, Vpx, Vpr, Vpu, and Nef) in addition to regulatory (Tat and Rev) and structural (Gag, Pol, and Env) proteins (Figure 1). Structural proteins are common to all retroviruses, but the regulatory and accessory proteins are unique to the complex primate lentiviruses and not found in the other simple mammalian retroviruses. Regulatory Tat and Rev proteins are trans-activators for transcription and for the expression of late viral proteins, respectively. While the regulatory and structural proteins are essential for viral replication, the extra proteins, unfairly generically called “accessory,” are dispensable under certain circumstances. However, in some cells, some of them are essential and the others are quite critical/important for optimal viral replication as illustrated for ΔVif and ΔVpu viruses (viruses that lack Vif or Vpu) in Figure 2. Another point to be mentioned here is relating to Vpr/Vpx proteins. Although Vpr and Vpx are genetically very similar (Khamsri et al., 2006), some primate immunodeficiency viruses bear two of them as described above (Fujita et al., 2010). Furthermore, the other viruses have Vpr only. What about the functional relationship of the two proteins? At present, the function of Vpr/Vpx is least well understood relative to that of the other accessory proteins (Malim and Emerman, 2008; Fujita et al., 2010). Table 2 summarizes the important information regarding these accessory proteins so far reported. In total, it is fairly reasonable to believe that the accessory proteins are regulators to optimize viral replication and persistence in vivo thereby enhancing viral transmission between individuals.
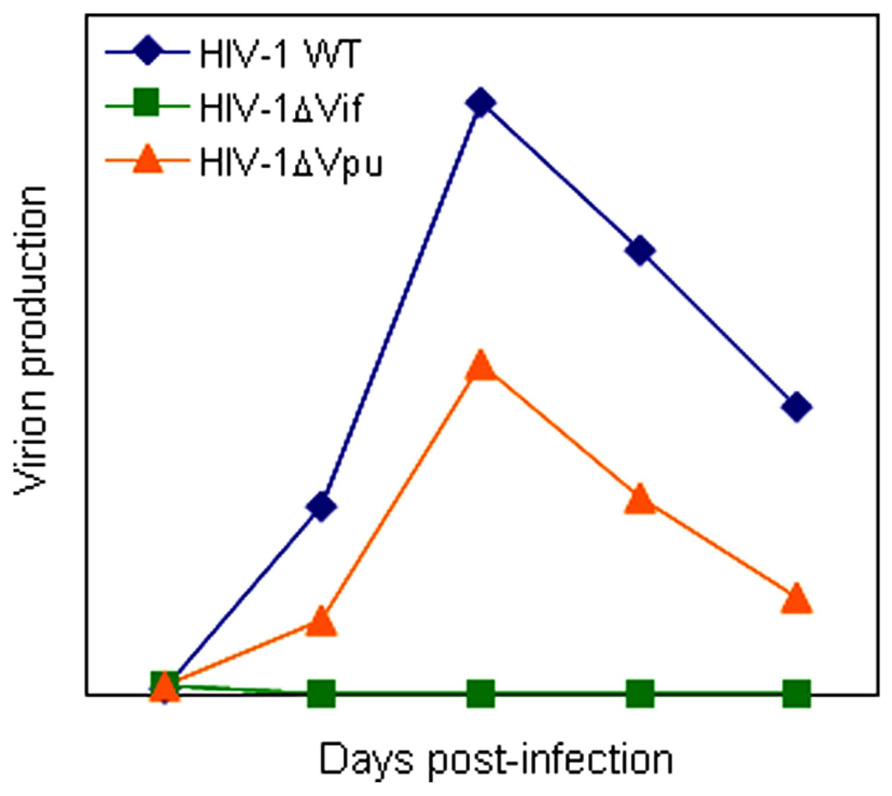
FIGURE 2. A schema of replication kinetics by HIV-1 wild-type and mutant viruses. Viral growth properties in cells are illustrated based on numerous infection experiments in our laboratory. WT, wild-type.
Vif and Vpu Proteins
Vif protein is essential for viral replication in natural target cells such as CD4-positve lymphocytes and macrophages. Recent identification of its cellular object for attack (Sheehy et al., 2002) has clearly revealed the biological and biochemical bases for the growth property of ΔVif virus in natural target cells. This finding (identification of a family of APOBEC3 proteins, cellular cytidine deaminases, as potent inhibitors of HIV-1 replication in primary cells) has also contributed much to establish the concept of “the restriction factor” to well understand virus–cell interaction (Malim and Emerman, 2008; Sato et al., 2012). Of the APOBEC3 family proteins, APOBEC3G and APOBEC3F (Kitamura et al., 2011) strongly inhibit viral replication in the absence of Vif (Figure 3). Although HIV-1 Vif can abrogate the activities of human APOBEC3, it cannot do so against monkey APOBEC3. In contrast, SIVmac Vif can neutralize the anti-viral activity of APOBEC3 of both origins. Finally, it has been demonstrated that Vif and APOBEC3 are the major determinants for the HIV-1 species tropism by constructing macaque-tropic HIV-1 (HIV-1mt) and monitoring the HIV-1mt growth property in various genetic contexts of macaques (Hatziioannou et al., 2006, 2009; Kamada et al., 2006; Igarashi et al., 2007; Thippeshappa et al., 2011).
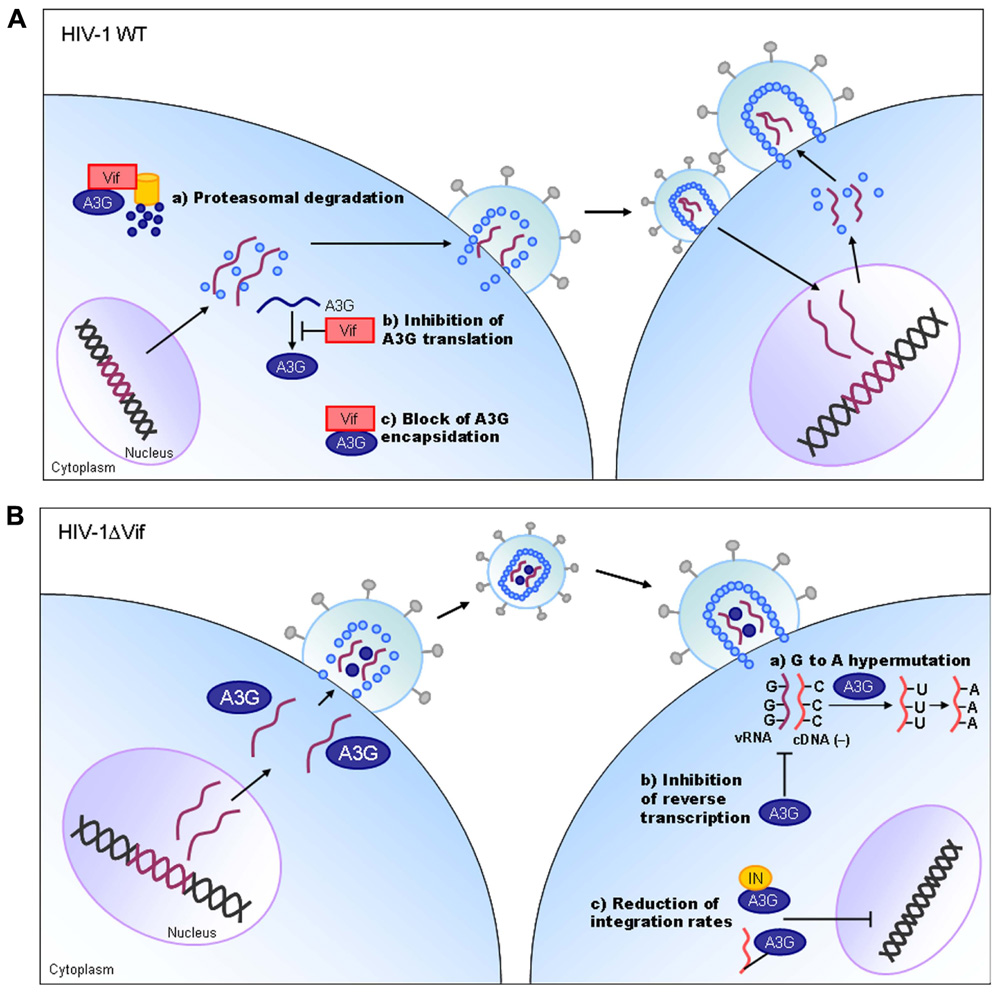
FIGURE 3. HIV-1 replication and APOBEC3G. On the basis of results reported so far, the action mechanism of Vif is depicted. Replication process for wild-type (WT) and ΔVif mutant viruses are schematically shown on the basis of previously reported review articles (Holmes et al., 2007; Huthoff and Towers, 2008; Strebel et al., 2009). A3G, APOBEC3G; IN, viral integrase protein.
Vpu protein, unique to viruses of the HIV-1 group (Figure 1), modulates viral replication in human CD4-positive cell lines and primary cells. Mutant HIV-1 without Vpu (ΔVpu virus) grows poorly relative to wild-type virus. Recently, a cellular protein named Tetherin (also called BST-2) has been identified as a restriction factor against HIV-1 and is antagonized by Vpu (Neil et al., 2008; Van Damme et al., 2008). Vpu down-regulates the Tetherin from cell surface, and thereby promotes extracellular production of progeny virions (Malim and Emerman, 2008; Arias et al., 2011; Sato et al., 2012). The baseline mechanism for this action of Vpu is well studied as shown in Figure 4. Here, it must be attentive that the anti-Tetherin activity of Vpu is host species-specific as observed for Vif. HIV-1 Vpu acts against human but not (or poorly) macaque Tetherins (Sauter et al., 2009, 2010). Although the biological effect of Vpu is much milder than that of Vif as judged by the growth kinetics of mutant viruses (Figure 2), Vpu may be critical for interspecies transmission through mutation/adaptation/recombinations (Kirchhoff, 2009; Sauter et al., 2009, 2010; Sharp and Hahn, 2011). Thus, Vpu and Tetherin affect the HIV-1 species tropism, but the effect may be relatively small.
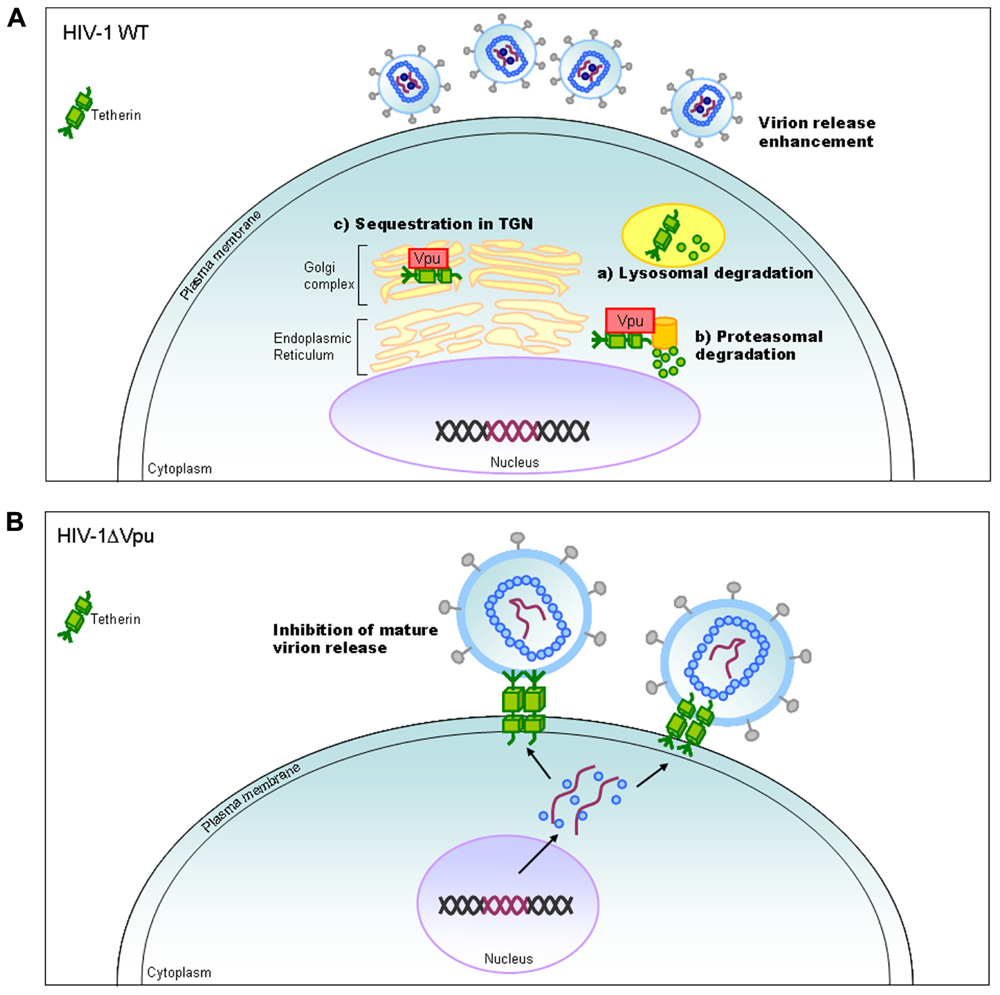
FIGURE 4. HIV-1 replication and Tetherin. On the basis of results reported so far, the action mechanism of Vpu is depicted. Replication process for wild-type (WT) and ΔVpu mutant viruses are schematically shown on the basis of previously reported review articles (Tokarev et al., 2009; Douglas et al., 2010; Evans et al., 2010). TGN, trans-Golgi network.
In sum, Vif and Vpu counteract the major restriction factors APOBEC3 proteins and Tetherin/BST-2, respectively, and represent viral determinants for the host range of HIV-1 (Tables 1 and 2). It is intriguing to note that these factors would have shaped HIV-1 and made it unique among various primate immunodeficiency viruses (Figure 1).
Vpx and Vpr Proteins
Vpx and Vpr proteins are necessary for efficient viral replication (Malim and Emerman, 2008; Fujita et al., 2010). In macrophages, ΔVpx replication is not detectable and this defect has been shown to be present at post-entry and before/during the reverse transcription process (Fujita et al., 2008, 2010; Srivastava et al., 2008). Also in some lymphocyte cell lines and in primary lymphocytes, Vpx protein is critical for viral replication (Ueno et al., 2003; Fujita et al., 2010; Doi et al., 2011). Because ΔVpr virus is somewhat replication-defective in some cells (for both HIV-1 and HIV-2), it is not unreasonable to assume that Vpr may play a role in the viral growth cycle. As such, Vpx and Vpr are important for in vivo viral replication and finally for viral pathogenicity (Fujita et al., 2010).
Very recently, SAMHD1 and APOBEC3A have been reported to be myeloid cell-specific restriction factors against HIV-1 counteracted by Vpx (Berger et al., 2011; Hrecka et al., 2011; Laguette et al., 2011). Whether these proteins are associated with the HIV-1 species tropism described in this review article, and whether they can explain the in vitro and in vivo situation of HIV-2/SIVmac mutant viruses mentioned above remain to be determined (Fujita et al., 2010; Nomaguchi et al., 2011).
Conclusion
In this review, we have described the major determinants for the species tropism of HIV-1. Structural Gag-CA and accessory Vif and Vpu proteins are clearly involved in this host range of HIV-1 as viral factors (Table 1). Cellular proteins that interact with these and contribute to this tropism are definitely the restriction factors (Table 1). In total, interplays between the viral and cellular responsible factors decide this unique and limited tropism of HIV-1. Whether there are the other factors that affect the HIV-1 species tropism is awaiting further investigations. In this regard, the biology of Vpx deserves attention. Because Vpx is present in SIVmac but not in HIV-1 (Figure 1), it may inactivate a cellular anti-viral protein(s) which is not recognized by HIV-1 proteins.
In both basic and applicable points of view, the narrow host range of HIV-1 is burdensome obstacle to overcome. Assuming that HIV-1mt can grow and cause disease similarly with SIVmac in macaques, we would be able to better perform model studies to precisely analyze viral replication and pathogenicity in vivo, and to establish the effective anti-HIV-1/AIDS strategies. To the best of our knowledge, there are no such HIV-1mt clones so far (Hatziioannou et al., 2006, 2009; Kamada et al., 2006; Igarashi et al., 2007; Kuroishi et al., 2009; Saito et al., 2011; Thippeshappa et al., 2011). We may further improve the ability of HIV-1mt by today's powerful methodology if we knew all the cellular determinants for the species tropism of HIV-1. Studies in this direction are in progress in our laboratory.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ms Kazuko Yoshida for excellent editorial assistance. This work was supported by a grant from the Ministry of Health, Labour and Welfare of Japan (Research on HIV/AIDS project no. H22-003).
References
Arias, J. F., Iwabu, Y., and Tokunaga, K. (2011). Structural basis for the antiviral activity of BST-2/tetherin and its viral antagonism. Front. Microbiol. 2:250. doi: 10.3389/fmicb.2011.00250
Berger, G., Durand S., Fargier, G., Nguyen, X.-N., Cordeil, S., Bouaziz, S., Muriaux, D., Darlix, J.-L., and Cimarelli, A. (2011). APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog. 7, e1002221. doi: 10.1371/journal.ppat.1002221
Doi, N., Fujiwara, S., Adachi, A., and Nomaguchi, M. (2011). Rhesus M1.3S cells suitable for biological evaluation of macaque-tropic HIV/SIV clones. Front. Microbiol. 2:115. doi: 10.3389/fmicb.2011.00115
Dorfman, T., and Gottlinger, H. G. (1996). The human immunodeficiency virus type 1 capsid p2 domain confers sensitivity to the cyclophilin-binding drug SDZ NIM 811. J. Virol. 70, 5751–5757.
Douglas, J. L., Gustin, J. K., Viswanathan, K., Mansouri, M., Moses, A. V., and Fruh, K. (2010). The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog. 6, e1000913. doi: 10.1371/journal.ppat.1000913
Evans, D. T., Serra-Moreno, R., Singh, R. K., and Guatelli, J. C. (2010). BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 18, 388–396.
Fujita, M., Otsuka, M., Miyoshi, M., Khamsri, B., Nomaguchi, M., and Adachi, A. (2008). Vpx is critical for reverse transcription of the human immunodeficiency virus type 2 genome in macrophages. J. Virol. 82, 7752–7756.
Fujita, M., Otsuka, M., Nomaguchi, M., and Adachi, A. (2010). Multifaceted activity of HIV Vpr/Vpx proteins: the current view of their virological functions. Rev. Med. Virol. 20, 68–76.
Hatziioannou, T., Ambrose, Z., Chung, N. P. Y., Piatak, M. Jr., Yuan, F., Trubey, C. M., Coalter, V., Kiser, R., Schneider, D., Smedley, J., Pung, R., Gathuka, M., Estes, J. D., Veazey, R. S., KewalRamani, V. N., Lifson, J. D., and Bieniasz, P. D. (2009). A macaque model of HIV-1 infection. Proc. Natl. Acad. Sci. U.S.A. 106, 4425–4429.
Hatziioannou, T., Princiotta, M., Piatak, M. Jr., Yuan, F., Zhang, F., Lifson, J. D., and Bieniasz, P. D. (2006). Generation of siman-tropic HIV-1 by restriction factor evasion. Science 314, 95.
Holmes, R. K., Malim, M. H., and Bishop, K. N. (2007). APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32, 118–128.
Hrecka, K., Hao, C., Gierszewska, M., Swanson, S. K., Kesik-Brodacka, M., Srivastava, S., Florens, L., Washburn, M. P., and Skowronski, J. (2011). Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661.
Huthoff, H., and Towers, G. J. (2008). Restriction of retroviral replication by APOBEC3G/F and TRIM5alpha. Trends Microbiol. 16, 612–619.
Igarashi, T., Iyengar, R., Byrum, R. A., Buckler-White, A., Dewar, R. L., Buckler, C. E., Lane, H. C., Kamada, K., Adachi, A., and Martin, M. A. (2007). Human immunodeficiency virus type 1 derivative with 7% simian immunodeficiency virus genetic content is able to establish infections in pig-tailed macaques. J. Virol. 81, 11549–11552.
Kamada, K., Igarashi, T., Martin, M. A., Khamsri, B., Hatcho, K., Yamashita, T., Fujita, M., Uchiyama, T., and Adachi, A. (2006). Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc. Natl. Acad. Sci. U.S.A.103, 16959–16964.
Khamsri, B., Murao, F., Yoshida, A., Sakurai, A., Uchiyama, T., Shirai, H., Matsuo, Y., Fujita, M., and Adachi, A. (2006). Comparative study on the structure and cytopathogenic activity of HIV Vpx/Vpr proteins. Microbes Infect. 8, 10–15.
Kirchhoff, F. (2009). Is the high virulence of HIV-1 an unfortunate coincidence of primate lentiviral evolution? Nat. Rev. Microbiol. 7, 467–476.
Kitamura, S., Ode, H., and Iwatani, Y. (2011). Structual features of antiviral APOBEC3 proteins are linked to their functional activities. Front. Microbiol. 2:258. doi: 10.3889/fmicb.2011.00258
Kuroishi, A., Saito, A., Shingai, Y., Shioda, T., Nomaguchi, M., Adachi, A., Akari, H., and Nakayama, E. E. (2009). Modification of a loop sequensce between alpha-helices 6 and 7 of virus capsid (CA) protein in a human immunodeficiency virus type 1 (HIV-1) derivative that has simian immunodeficiency virus (SIVmac239) Vif and CA alpha-helices 4 and 5 loop improves replication in cynomolgus monkey cells. Retrovirology 6, 70.
Laguette, N., Sobhian, B., Casartelli, N., Ringeard, M., Chable-Bessia, C., Segeral, E., Yatim, A., Emiliani, S., Schwartz, O., and Benkirane, M. (2011). SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657.
Malim, M. H., and Emerman, M. (2008). HIV-1 accessory proteins-ensuring viral survival in a hostile environment. Cell Host Microbe 3, 388–398.
Nakayama, E. E., and Shioda, T. (2012). TRIM5α and species tropism of HIV/SIV. Front. Microbiol. 3:13. doi: 10.3389/fmicb.2012.00013
Neil, S. J., Zang, T., and Bieniasz, P. D. (2008). Tether ininhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430.
Nomaguchi, M., Doi, N., Kamada, K., and Adachi, A. (2008). Species barrier of HIV-1 and its jumping by virus engineering. Rev. Med. Virol. 18, 261–275.
Nomaguchi, M., Fujita, M., and Adachi, A. (2011). The fourth major restriction factor against HIV/SIV. Front. Microbiol. 2:132. doi: 10.3389/fmicb.2011.00132
Saito, A., Nomaguchi, M., Iijima, S., Kuroishi, A., Yoshida, T., Lee, Y. J., Hayakawa, T., Kono, K., Nakayama, E. E., Shioda, T., Yasutomi, Y., Adachi, A., Matano, T., and Akari, H. (2011). Improved capacity of a monkey-tropic HIV-1 derivative to replicate in cynomolgus monkeys with minimal modifications. Microbes Infect. 13, 58–64.
Sakuma, R., and Takeuchi, H. (2012). SIV replication in human cells. Front. Microbiol. 3:162. doi: 10.3389/fmicb.2012.00162
Sato, K., Gee, P., and Koyanagi, Y. (2012). Vpu and BST-2: still not there yet? Front. Microbiol. 3:131. doi: 10.3389/fmicb.2012.00131
Sauter, D., Schindler, M., Specht, A., Landford, W. N., Munch, J., Kim, K.-A., Votteler, J., Schubert, U., Bibollet-Ruche, F., Keele, B. R. F., Takehisa, J., Ogando, Y., Ochsenbauer, C., Kappes, J. C., Ayouba, A., Peeters, M., Learn, G. H., Shaw, G., Sharp, P. M., Bieniasz, P., Hahn, B. H., Hatziioannou, T., and Kirchhoff, F. (2009). Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6, 409–421.
Sauter, D., Specht, A., and Kirchhoff, F. (2010). Tetherin: holding on and letting go. Cell 141, 392–398.
Sharp, P. M., and Hahn, B. H. (2011). Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 1, a006841.
Sheehy, A. M., Gaddis, N. C., Choi, J. D., and Malim, M. H. (2002). Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650.
Shibata, R., and Adachi, A. (1992). SIV/HIV recombinants and their use in studying biological properties. AIDS Res. Hum. Retroviruses 8, 403–409.
Shibata, R., Kawamura, M., Sakai, H., Hayami, M., Ishimoto, A., and Adachi, A. (1991). Generation of a chimeric human and simian immunodeficiency virus infectious to monkey peripheral blood mononuclear cells. J. Virol. 65, 3514–3520.
Shibata, R., Sakai, H., Kawamura, M., Tokunaga, K., and Adachi, A. (1995). Early replication block of human immunodeficiency type 1 in monkey cells. J. Gen. Virol. 76, 2723–2730.
Srivastava, S., Swanson, S. K., Manel, N., Florens, L., Washburn, M. P., and Skowronski, J. (2008). Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable to enhance macrophage infection. PLoS Pathog. 4, e1000059. doi: 10.1371/journal.ppat.1000059
Strebel, K., Luban, J., and Jeang, K. T. (2009). Human cellular restriction factors that target HIV-1 replication. BMC Med. 7, 48. doi: 10.1186/1741-7015-7-48
Thippeshappa, R., Polacino, P., Kimata, M. T. Y., Siwak, E. B., Anderson, D., Wang, W., Sherwood, L., Arora, R., Wen, M., Zhou, P., Hu, S.-L., and Kimata, J. T. (2011). Vif substitution enables persistent infection of pig-tailed macaques by human immunodeficiency virus type 1. J. Virol. 85, 3767–3779.
Tokarev, A., Skasko, M., Fitzpatrick, K., and Guatelli, J. (2009). Antiviral activity of the interferon-induced cellular protein BST-2/tetherin. AIDS Res. Hum. Retroviruses 25, 1197–1210.
Ueno, F., Shiota, H., Miyaura, M., Yoshida, A., Sakurai, A., Tatsuki, J., Koyama, A. H., Akari, H., Adachi, A., and Fujita, M. (2003). Vpx and Vpr proteins up-regulate the viral infectivity by a distinct mechanism in lymphocytic cells. Microbes Infect. 5, 387–395.
Keywords: HIV-1, species tropism, accessory protein, Vif, Vpu
Citation: Nomaguchi M, Doi N, Matsumoto Y, Sakai Y, Fujiwara S and Adachi A (2012) Species tropism of HIV-1 modulated by viral accessory proteins. Front. Microbio. 3:267. doi: 10.3389/fmicb.2012.00267
Received: 02 July 2012; Paper pending published: 08 July 2012;
Accepted: 09 July 2012; Published online: 26 July 2012.
Edited by:
Mikako Fujita, Kumamoto University, JapanReviewed by:
Mikako Fujita, Kumamoto University, JapanYasuyuki Miyazaki, The University of Tokushima Graduate School, Japan
Copyright: © 2012 Nomaguchi, Doi,Matsumoto, Sakai, Fujiwara and Adachi. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Akio Adachi, Department of Microbiology, Institute of Health Biosciences, The University of Tokushima Graduate School, 3-18-15 Kuramoto-cho, Tokushima-shi, Tokushima 770-8503, Japan. e-mail:YWRhY2hpQGJhc2ljLm1lZC50b2t1c2hpbWEtdS5hYy5qcA==