- 1 Instituto de Biología Molecular y Celular de Plantas (UPV-CSIC), Valencia, Spain
- 2 Istituto di Virologia Vegetale (CNR), Unità Organizzativa di Bari, Bari, Italy
As a consequence of two unique physical properties, small size and circularity, viroid RNAs do not code for proteins and thus depend on RNA sequence/structural motifs for interacting with host proteins that mediate their invasion, replication, spread, and circumvention of defensive barriers. Viroid genomes fold up on themselves adopting collapsed secondary structures wherein stretches of nucleotides stabilized by Watson–Crick pairs are flanked by apparently unstructured loops. However, compelling data show that they are instead stabilized by alternative non-canonical pairs and that specific loops in the rod-like secondary structure, characteristic of Potato spindle tuber viroid and most other members of the family Pospiviroidae, are critical for replication and systemic trafficking. In contrast, rather than folding into a rod-like secondary structure, most members of the family Avsunviroidae adopt multibranched conformations occasionally stabilized by kissing-loop interactions critical for viroid viability in vivo. Besides these most stable secondary structures, viroid RNAs alternatively adopt during replication transient metastable conformations containing elements of local higher-order structure, prominent among which are the hammerhead ribozymes catalyzing a key replicative step in the family Avsunviroidae, and certain conserved hairpins that also mediate replication steps in the family Pospiviroidae. Therefore, different RNA structures – either global or local – determine different functions, thus highlighting the need for in-depth structural studies on viroid RNAs.
Introduction
Since their discovery some 50 years ago (Diener and Raymer, 1967; Diener, 1972, 2003) viroids have exerted a particular fascination because they are the lowest step of the biological scale: simple RNA genomes of just ∼250–400 nucleotides (nt). How such tiny molecules manipulate the machinery of the organisms they parasitize to ensure their replication and systemic invasion and, possibly as a fortuitous consequence, often incite disease? How they select their specific hosts, hitherto restricted to higher plants, and evade the defensive response they mount? Moreover, given that viroids do not encode proteins, these remarkable goals must be achieved by direct interaction of the genomic RNAs (or derivatives thereof) with host components. Which are the RNA sequence/structural motifs involved in these interactions? Viroids have also caught much attention because some “code” for ribozymes (of the hammerhead class, the simplest described so far) that play a key role in their replication. In other words, they are catalytic RNAs possessing a genotype and expressing a phenotype without recurring to protein intermediation. Altogether, these astonishing properties make viroids unique systems to investigate how cells decipher signals contained in foreign (and most likely endogenous) RNAs or, considered from the reverse perspective, how RNA structure determines function. Here we present an overview of the ideas on this topic – since the inception of the viroid concept to recent developments – showing that this research field has been and continues to be fertile and promising. For in-depth treatment of other aspects on viroid biology the reader is referred to previous reviews (Hadidi et al., 2003; Flores et al., 2005, 2011a; Tsagris et al., 2008; Ding, 2009).
Circularity and Rolling-Circle Replication
Besides their minimal size, viroid genomes display another peculiar structural property with deep functional connotations: they are circular RNAs. Strong hints in this respect were obtained even before Potato spindle tuber viroid (PSTVd), the type member of the group, was identified as a physical entity. While treatments with exonucleases abolished infectivity of the RNA from Tobacco mosaic virus (TMV), no detectable effect was observed on PSTVd infectivity, which also remained unaffected following co-treatment with alkaline phosphatase to remove hypothetical terminal phosphoryl groups that might impede exonuclease attack (Diener, 1971). Electron microscopy under denaturing conditions confirmed this point (Sänger et al., 1976; McClements and Kaesberg, 1977) that was further corroborated by direct sequencing of PSTVd (Gross et al., 1978). Because circular RNAs cannot be translated by ribosome scanning from their 5′-end and, additionally, PSTVd lacked typical AUG initiation codons (Gross et al., 1978), it was concluded that this was a non-protein-coding RNA. All subsequent work with different viroids – in which no internal ribosomal entry site has been reported and attempts to verify their translation in vitro and in vivo have failed (Davies et al., 1974; Semancik et al., 1977) – sustain this notion. Therefore, to complete their biological cycle viroids are significantly more host-dependent than viruses (that encode one or more proteins).
The circular nature of the genomic viroid RNA has another key consequence in replication. When entering an appropriate host, this RNA – to which the (+) polarity is assigned by convention – is reiteratively transcribed by a cellular RNA polymerase into oligomeric (−) strands that, by themselves or after cleavage and circularization, serve as templates for a second RNA–RNA transcription round leading to oligomeric (+) strands that are cleaved and ligated into the final circular product. This rolling-circle mechanism with only RNA intermediates (Grill and Semancik, 1978) has two variants. In PSTVd and members of its family (Pospiviroidae) replication takes place in the nucleus and all steps are catalyzed by host enzymes (Branch and Robertson, 1984; Branch et al., 1988), while in Avocado sunblotch viroid (ASBVd; Symons, 1981) and members of its family (Avsunviroidae), replication occurs in plastid (mostly chloroplasts) with cleavage of oligomeric strands being mediated by hammerhead ribozymes (Hutchins et al., 1986; Daròs et al., 1994). A more detailed account of viroid replication can be found in a recent review (Flores et al., 2009).
Rod-Like Versus Branched Secondary Structure: in silico and in vitro Evidence
Viroid genomes fold up on themselves adopting collapsed secondary structures wherein stretches of nucleotides paired through canonical Watson–Crick interactions are flanked by loops of apparently unpaired nucleotides. Circularity facilitates intramolecular folding, and together, the two features afford protection against both exonucleases (demanding free termini), and endonucleases (acting preferentially on single-stranded regions). This peculiar secondary structure became evident upon sequencing PSTVd: thermodynamics-based predictions, RNase, and bisulfite probing in vitro, and electron microscopy revealed that the 359-nt PSTVd RNA adopts a rod-like secondary structure with a width roughly similar to that of a double-stranded DNA (Sogo et al., 1973; Sänger et al., 1976; Gross et al., 1978; Riesner et al., 1979; Figure 1). Additional support for this view was provided when similar structures of maximal base-pairing were predicted for two other viroids related to PSTVd but different in size and with just 60–73% nucleotide sequence identity: Chrysanthemum Stunt viroid (CSVd), and Citrus exocortis viroid (CEVd; Haseloff and Symons, 1981; Gross et al., 1982). Although probing with nucleases and dimethyl sulfate supplied evidence for a bifurcation in the left-terminal domain of the PSTVd rod-like structure (Gast et al., 1996), subsequent nuclear magnetic resonance (NMR) analysis of the 69-nt forming this domain, confirmed by temperature-gradient gel electrophoresis, and UV melting experiments, sustain the elongated rod form as the thermodynamically favored conformation (Dingley et al., 2003). Further supporting this notion, recent application of SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) – a relatively novel technique that interrogates local backbone RNA flexibility in solution at single-nucleotide resolution (Merino et al., 2005; Weeks and Mauger, 2011) and permits coupling these data to computer-assisted structure prediction – has failed to reveal the presence of Y-shaped, or cruciform structures in CEVd and some other members of the family Pospiviroidae (Xu et al., 2012). However, despite this evidence, the universality of the rod-like structure of viroids should not be taken for granted within this family, as illustrated by early results indicating that the most stable secondary structure predicted for Pear blister canker viroid (PBCVd) is cruciform and that none of the other energetically close conformations are elongated (Hernández et al., 1992).
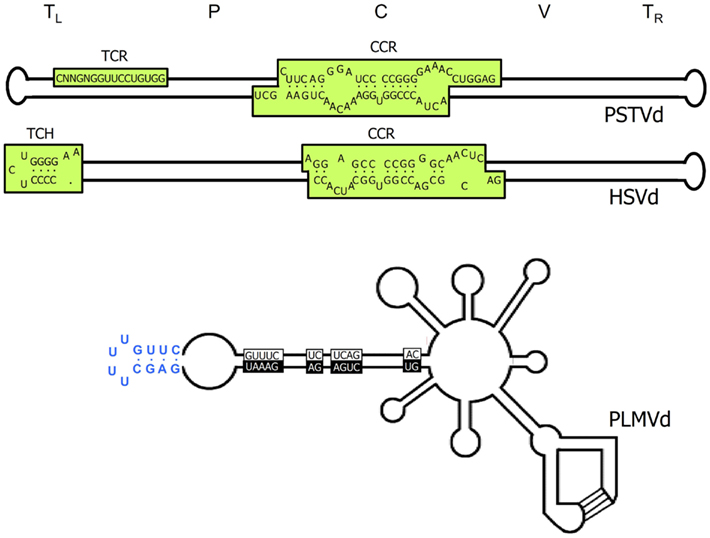
Figure 1. Structural features of viroids. Upper and middle panels, schemes of the characteristic rod-like secondary structures of the genomic RNAs of Potato spindle tuber viroid (PSTVd) and Hop stunt viroid (HSVd) respectively (family Pospiviroidae). The approximate location of the five structural domains – terminal left (TL), pathogenic (P), central (C), variable (V), and terminal right (TR) – is indicated, as well as that of the central conserved region (CCR), the terminal conserved region (TCR), and the terminal conserved hairpin (TCH). Lower panel, scheme of the multibranched secondary structure of the genomic RNA of Peach latent mosaic viroid (PLMVd; family Avsunviroidae), in which the sequences conserved in most natural hammerhead ribozymes are boxed with black and white backgrounds for the (+) and (−) polarities, respectively; the kissing-loop interaction is indicated with lines, and the characteristic 12-nt hairpin insertion of the reference variant containing the pathogenicity determinant of an extreme chlorosis (peach calico) is highlighted with blue color. For a more detailed representation of the PSTVd secondary structure see Figure 5.
Within the family Avsunviroidae embracing chloroplastic viroids (Flores et al., 2000, 2005), the rod-like structure is the exception rather than the rule. Even if ASBVd, the type member of this family, was initially proposed to fold into an elongated conformation (Symons, 1981), other data point to a bifurcated left-terminal domain (Gast et al., 1996; Navarro and Flores, 2000). Moreover, molecular characterization of Peach latent mosaic viroid (PLMVd; Hernández and Flores, 1992), and Chrysanthemum chlorotic mottle viroid (CChMVd; Navarro and Flores, 1997) predicted multibranched most stable conformations, and subsequent sequencing of many natural variants of PLMVd (Ambrós et al., 1998, 1999; Pelchat et al., 2000; Malfitano et al., 2003; Rodio et al., 2006; Fekih Hassen et al., 2007; Yazarlou et al., 2012), and CChMVd (De la Peña et al., 1999; De la Peña and Flores, 2002) not only corroborated this view, but also provided evidence for the significance of the multibranched conformations in vivo. Additional credence for such a PLMVd secondary structure was obtained by in vitro nuclease mapping and oligonucleotide binding shift assays (Bussière et al., 2000); this work also indicated the existence of an element of tertiary structure, specifically a pseudoknot interaction between two hairpin loops stabilizing the proposed branched conformation (see below), and suggested the possibility of a similar kissing-loop interaction in CChMVd (Bussière et al., 2000). SHAPE examination of several PLMVd variants is also consistent with the proposed branched conformation or with variations thereof (Dubé et al., 2011). Finally, computer-assisted analysis of the sequence of the fourth member of this family, Eggplant latent viroid (ELVd), has predicted a secondary structure with two bifurcations (Fadda et al., 2003a).
Rod-Like Versus Branched Secondary Structure: in vivo Evidence
Is the elongated conformation obtained for PSTVd and related viroids by in silico and in vitro approaches physiologically relevant? This issue is not trivial because structural inferences derived from thermodynamics-based studies and probing analyses of RNA in solution should not be interpreted as directly reflecting the situation in vivo. While predictions of RNA secondary structure by free energy minimization (Nussinov and Jacobson, 1980; Zuker and Stiegler, 1981) are usually correct for small structural motifs like hairpins, they must be considered just tentative for larger RNAs because of the lack of accurate thermodynamic estimates for all sequence motifs and higher-order interactions (Mathews and Turner, 2006). On the other hand, nucleotides non-reacting in chemical probing in vitro do not necessarily reflect base-pairing in vivo, because they might be involved in tertiary non-canonical interactions or in RNA-protein contacts. Moreover, in its physiological habitat the genomic viroid RNA must interact with different host proteins mediating replication and movement, and these interactions may impact deeply on its structure: for instance, the rod-like conformation proposed for most members of the family Potato spindle tuber viroid must be transiently unfolded during replication by the enzymatic complex catalyzing transcription. In this same context, since RNA folding occurs during transcription, the specific initiation site of viroid strands can have functional implications.
Nonetheless, data from three independent lines support that this elongated conformation is biologically significant: (i) a less-than-unit (341 nt) infectious PSTVd variant, identified in plants agrotransformed with the dimeric form of an in vitro-deleted 350-bp (non-infectious) PSTVd-cDNA unit, results from an additional 9-nt deletion in vivo that restores the rod-like secondary structure (Wassenegger et al., 1994), (ii) the longer-than-unit natural variants of Coconut cadang–cadang viroid (CCCVd; Haseloff et al., 1982) and CEVd (Semancik et al., 1994; Fadda et al., 2003b) accumulating in infected tissues result from repetitions of the right terminal domain that also preserve the rod-like secondary structure, and (iii) replication and directional PSTVd trafficking across specific cellular boundaries is mediated or influenced by RNA motifs, particularly loops, of the rod-like secondary structure (see below).
What about in the family Avsunviroidae? Evidence for the physiological relevance of the branched conformations proposed for PLMVd and CChMVd is particularly strong. Sequencing of many natural variants has shown that they display an extreme sequence heterogeneity (Ambrós et al., 1998, 1999; De la Peña et al., 1999; Pelchat et al., 2000; De la Peña and Flores, 2002; Malfitano et al., 2003; Rodio et al., 2006; Fekih Hassen et al., 2007; Yazarlou et al., 2012), most likely resulting from the high mutation rate of these viroids during replication catalyzed by a single-subunit, nuclear-encoded chloroplastic RNA polymerase without proof-reading ability (Navarro et al., 2000; Rodio et al., 2007); this mutation rate has been measured for CChMVd and is the highest reported for any biological entity (Gago et al., 2009). Remarkably, however, the sequence heterogeneity occurs in such a manner that the changes map at loops or, when affecting a base-pair, they are mostly co-variations or substitutions of Watson–Crick by wobble base-pairs (or vice-versa) that do not distort the computer-predicted branched conformations. Co-variations or compensatory mutations are regarded the most compelling feature for testing computer-predicted structures in RNA (Gutell et al., 1994).
The CChMVd natural variability also supports that the branched conformation is further stabilized by a kissing-loop interaction (Gago et al., 2005) resembling another one proposed in PLMVd from in vitro assays. Specifically, site-directed mutagenesis, bioassays, and progeny analyses showed that: (i) single CChMVd mutants partially disrupting the kissing-loops display low or no infectivity, which was recovered in double mutants restoring the interaction, (ii) mutations affecting regions adjacent to the kissing-loops reverted to wild-type or led to stem rearrangements preserving this interaction, and (iii) the interchange between four nucleotides from each of the two kissing-loops generated a viable CChMVd variant with eight mutations. Moreover, denaturing and non-denaturing PAGE revealed that the kissing-loop interaction determines proper in vitro folding of CChMVd RNA, and that this interaction only exists in the (+) polarity strand (Gago et al., 2005), as most likely also occurs in PLMVd (Dubé et al., 2010). Conservation of a similar loop-loop interaction in two viroids with low sequence similarity strongly suggests a role in the adoption and stabilization of a compact folding critical for viroid viability in vivo (Gago et al., 2005), although what may be this specific role remains unknown. Interestingly, a similar kissing-loop interaction has been predicted in the (+) strand of a recently described viroid-like RNA from grapevine with hammerhead ribozymes in both polarity strands (Wu et al., 2012). Even if preliminary bioassays have failed to detect autonomous replication for this small grapevine RNA, it is most likely a new viroid species of the genus Pelamoviroid formed so far by PLMVd and CChMVd (Navarro and Flores, 1997).
Another structural aspect of interest in PLMVd is that its secondary structure contains a long double-stranded region, interspersed with small loops, in which the nucleotides forming part of both hammerhead structures face each other (Hernández and Flores, 1992). Yet, in the most stable secondary structures predicted for some sequence variants, this region (the so-called hammerhead arm) adopts an alternative cruciform conformation that is also supported by co-variations in natural variants (a solid argument for relevance in vivo; Ambrós et al., 1998, 1999), and by recent SHAPE probing in vitro (Dubé et al., 2011). Intriguingly, a similar cruciform domain is formed in the hammerhead arm of CChMVd (Navarro and Flores, 1997; De la Peña et al., 1999), but with an extra-helical A in the junction between two of the stems that is indispensable for infectivity, possibly because this singular location induces a deformation of the cruciform domain critical for interacting with other RNA regions or host factors involved in CChMVd replication, transport, or accumulation (De la Peña and Flores, 2001). This nucleotide also forms part of the (+) hammerhead structure, an alternative folding transiently adopted during replication in vivo, and causes in vitro a moderate decrease of the trans-cleaving rate constant with respect to the same ribozyme without this residue. Therefore, certain natural hammerheads appear to deviate from the optimal catalytic format due to the involvement of some of their nucleotides in critical function(s) other than self-cleavage, a consequence of the need to compress genetic information in very small genomes like those of viroids (De la Peña and Flores, 2001).
The secondary structure with two bifurcations predicted for ELVd is also partly supported by co-variations (Fadda et al., 2003a). However, the limited sequence heterogeneity detected in members of the family Pospiviroidae (Góra et al., 1994), the replication of which is mediated by the multi-subunit nuclear RNA polymerase II (Mühlbach and Sänger, 1979; Flores and Semancik, 1982; Schindler and Mühlbach, 1992), has precluded the implementation of this approach for validating their proposed secondary structure.
Conserved Domains, Sequence Motifs, and Hairpins in the Family Pospiviroidae: Functional Significance
Early pair-wise comparisons looking for local similarities between members of the family Pospiviroidae, mostly of the genus Pospiviroid whose type species is PSTVd, led to the proposal that the rod-like secondary structure could be splitted into five domains: TL (terminal left), P (pathogenic), C (central), V (variable), and TR (terminal right; Figure 1; Keese and Symons, 1985). These domains, besides playing a role in intermolecular RNA rearrangements contributing to viroid evolution (Keese and Symons, 1985; Koltunow and Rezaian, 1989), were associated with specific functions; for instance, the P domain was related to symptom expression, although subsequent work unveiled a more complex scenario controlled by discrete determinants located in the five domains (Sano et al., 1992; Qi and Ding, 2003).
Within the C domain it is found the central conserved region (CCR), formed by two stretches of conserved nucleotides in the upper and lower strands flanked by an imperfect inverted repeat in the upper strand (McInnes and Symons, 1991), and within the TL domain are found the terminal conserved region (TCR) or the terminal conserved hairpin (TCH), because both appear mutually exclusive (Koltunow and Rezaian, 1988; Puchta et al., 1988; Flores et al., 1997; Figure 1). These elements, the conservation of which in sequence and secondary structure or location strongly suggests their involvement in key functional roles, serve as one of the main criteria for viroid classification (Flores et al., 1997). However, data for such a role are only available for the CCR so far. Superimposed on the CCR primary structure, there are other elements of higher-order structure. In particular, studies using a set of biochemical and biophysical techniques showed that, during thermal denaturation, PSTVd and some closely related viroids assume metastable branched conformations that are trapped by quick cooling, while slow cooling renaturation favors adoption of the native rod-like structure (Henco et al., 1979; Riesner et al., 1979). From these studies it was proposed that thermal denaturation might recapitulate unfolding (or transient folding) during replication in vivo (Riesner, 1991).
The metastable conformations contain hairpins, prominent among which is hairpin I (HP I) formed by the upper CCR strand and the flanking imperfect repeats. The finding that the nucleotide variability between members of the family Pospiviroidae preserves HP I (Riesner et al., 1979; Visvader et al., 1985; Polivka et al., 1996) – including the capping palindromic tetraloop with the two central residues phylogenetically conserved, the adjacent 3-bp stem with its central pair also phylogenetically conserved, an internal symmetric loop of 1–3 nt in each strand presumably stabilized by non-canonical interactions (Gast et al., 1998), and a stem of 9–10 bp occasionally interrupted by a 1-nt symmetric or asymmetric internal loop (Visvader et al., 1985; Flores et al., 1997; Figure 2) – adds further support to this structural element serving as the basis for an important function in vivo. In agreement with this notion, results obtained with an in vivo system based in transgenic Arabidopsis thaliana lines expressing dimeric transcripts of CEVd, Apple scar skin viroid (ASSVd), and Hop stunt viroid (HSVd) – that mimic the replicative intermediates – have mapped the cleavage site of the (+) strands of these three viroids at the upper CCR strand (Daròs and Flores, 2004; Gas et al., 2007), in a position homologous to that proposed for PSTVd with an in vitro system (Baumstark et al., 1997). Moreover, as a consequence of the peculiar features of HP I (Figure 2), the corresponding sequence of di- or oligomeric RNAs can alternative form a long double-stranded structure with a GC-rich central region of 10 bp containing the cleavage sites. The adoption in vivo of this double-stranded structure, which would be the actual substrate for cleavage in line with an early proposal (Diener, 1986), could be facilitated by hairpin I. More specifically, during transcription of oligomeric (+) RNAs of the family Pospiviroidae, a kissing-loop interaction between the palindromic tetraloops of two consecutive hairpin I motifs (Figure 3A), paralleling the situation observed in retroviruses (Paillart et al., 2004), might start intramolecular dimerization and their stems then form a longer interstrand duplex (Gas et al., 2007; Figure 3B). Furthermore, the cleavage sites in the double-stranded structure leave two 3′-protruding nucleotides in each strand (Figure 3C), the hallmark of class III RNases that display a clear preference for substrates with a compact secondary structure, like viroids, and generate products with 5′-phosphomonoester and free 3′-hydroxyl termini (MacRae and Doudna, 2007). The monomeric linear CEV (+) strands that accumulate in A. thaliana transgenically expressing the corresponding dimeric transcripts have indeed these termini (Gas et al., 2008), the ligation of which, interestingly, occurs through a novel pathway.
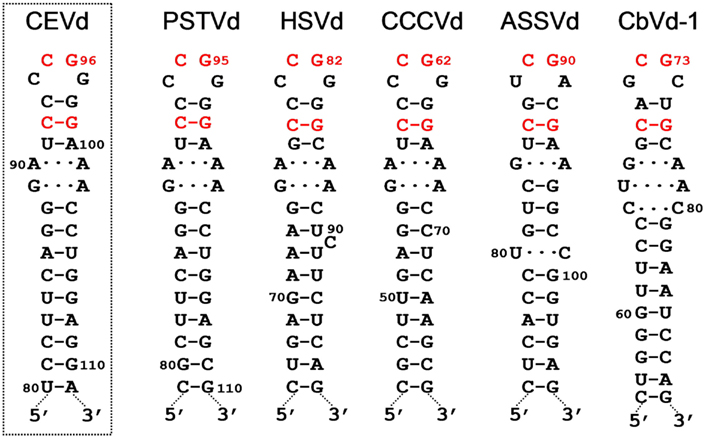
Figure 2. Hairpin I structures of the five type species of the family Pospiviroidae. This element of secondary structure is formed by the upper CCR strand and a flanking inverted repeat of PSTVd, HSVd, CCCVd, ASSVd, and CbVd-1 (Coleus blumei viroid 1). Red fonts indicate conserved nucleotides in structurally similar positions. Continuous and broken lines represent Watson–Crick and non-canonical base-pairs, respectively. Notice that the variability preserves the overall structure of hairpin I, including the terminal palindromic tetraloop, the adjacent 3-bp stem, and the long stem. Left inset, hairpin I of the wild-type CEVd variant used to transform A. thaliana (notice two co-variations with respect to PSTVd at the basis of the long stem). Reproduced with permission from Gas et al. (2007).
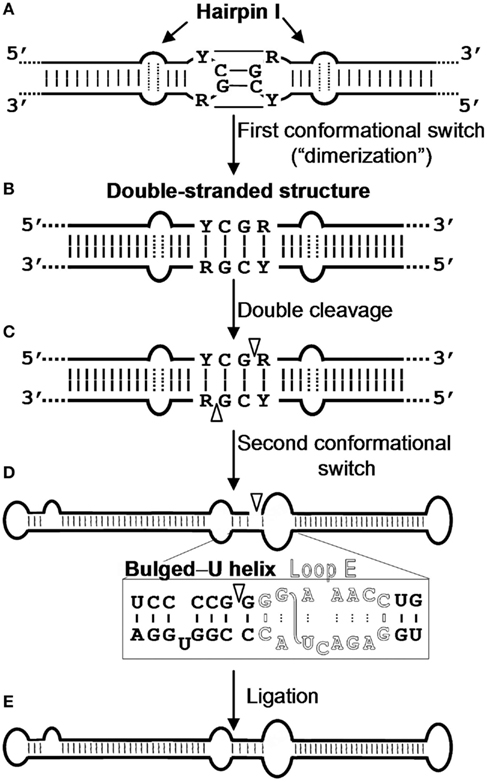
Figure 3. Model for processing in vivo of the oligomeric (+) replicative intermediates of the family Pospiviroidae. The model predicts a kissing-loop interaction between the palindromic tetraloops of two consecutive hairpin I motifs (A), with their stems then forming a longer interstrand duplex (B). This double-stranded structure is the substrate for cleavage at specific positions in both strands (C). Following a second conformational switch, the resulting unit-length strands adopt the extended rod-like structure with loop E (in outlined fonts) and the adjacent bulged-U helix (D), which is the substrate for ligation (E). R and Y refer to purines and pyrimidines, respectively, the S-shaped line denotes the UV-induced cross-link, and white arrowheads mark the cleavage sites in the double-stranded structure and the ligation site in the extended conformation. Reproduced with permission from Gas et al. (2007).
The ensuing ligation of these termini appears to be facilitated by an alternative extended conformation similar, if not identical, to the rod-like secondary structure containing an element of local tertiary structure in the CCR stabilized by non-canonical interactions. Early UV irradiation and enzymatic and chemical probing of purified PSTVd circular RNA unveiled the existence of such an element with high sequence and structural similarity to loop E of eukaryotic 5S rRNA (Branch et al., 1985; Gast et al., 1996), and direct UV irradiation of PSTVd-infected tomato leaves and subsequent RNA analysis showed the same UV-induced cross-linking, thus indicating that loop E is also formed in vivo (Eiras et al., 2007; Wang et al., 2007). Moreover, phylogenetic dissection supports that this element is conserved in other members of the genus Pospiviroid, a notion further confirmed by a structural model for PSTVd loop E – based on isostericity matrix and mutagenic analyses (see below) – that can be extended to other closely related viroids like CEVd (Zhong et al., 2006). In vitro evidence for the involvement of loop E in ligation was derived from incubation with a potato nuclear extract of a monomeric (+) PSTVd RNA with a short repeat of the upper CCR strand: detection of the infectious monomeric circular form led to the proposal that the enzymatic cleavage and ligation of the PSTVd (+) strand results from a switch from a branched structure (containing a GAAA-capped hairpin) to an extended conformation containing loop E (Baumstark et al., 1997). In vivo evidence obtained with 16 CEVd mutants expressed transgenically in A. thaliana corroborates this view with some modifications: processing is driven by an interplay between RNA conformations involving the hairpin I/double-stranded structure formed by the upper CCR strand and flanking nucleotides in cleavage, and loop E and flanking nucleotides of both strands in ligation (Gas et al., 2007, 2008; Figure 3D). In the latter conformation, the 5′-phosphomonoester and free 3′-hydroxyl termini to be joined must lie in close proximity and proper orientation.
Remarkably, loop E has been also involved in host specificity (Wassenegger et al., 1996), pathogenesis (Qi and Ding, 2003), and accumulation (Zhong et al., 2006) of PSTVd. Furthermore, because loop E is part of the binding site of at least two proteins – transcription factor IIIA that activates 5S rRNA transcription, and the ribosomal protein L5 implicated in post-transcriptional delivery of this RNA from the nucleoplasm to the nucleolus – the possibility that loop E could also act in PSTVd as a binding site for recruiting proteins involved in its replication and/or intranuclear transport was advanced (Flores et al., 1997). In support of this proposal, recent data show that L5 and TFIIIA from A. thaliana bind PSTVd (+) RNA in vitro with the same affinity as they bind their cognate 5S rRNA, whereas the affinity for a chloroplastic viroid is significantly lower (Eiras et al., 2011). Because loop E is not conserved in other members of the family Pospiviroidae apart from those forming the genus Pospiviroid, it remains to be determined whether alternative elements of local tertiary structure, different in sequence but functionally equivalent, play a role similar to that of loop E. Altogether these results highlight that a small structural element, like loop E, can mediate multiple functions. This is also the case for HP I, which either itself or its conserved sequence, has been additionally involved in driving the import of PSTVd into the nucleus (Zhao et al., 2001; Abraitiene et al., 2008). In permeabilized tobacco protoplasts, nuclear transport of fluorescein-labeled PSTVd RNA is cytoskeleton-independent, uncoupled to the Ran GTPase cycle, and facilitated by a specific and saturable receptor (Woo et al., 1999). In a related context, a small purine/pyrimidine motif in the TR domain, conserved in all members of the genus Pospiviroid, has been proposed to mediate systemic transport (Gozmanova et al., 2003; Maniataki et al., 2003).
In addition to HP I, the metastable conformations unveiled by thermal denaturation analyses also contain HP II, formed by sequences from the lower strand of the rod-like structure positioned at both sides of the CCR (Henco et al., 1979; Riesner et al., 1979). The functional relevance of HP II is supported by its conservation in the genus Pospiviroid, and by the critical role played by its core region in infectivity (Loss et al., 1991; Qu et al., 1995; Candresse et al., 2001). More specifically, HP II is adopted by sequential folding in the (−) strand replicative intermediate, and is essential for its template activity in (+) strand synthesis (Qu et al., 1995; Schroder and Riesner, 2002). Other hairpins have been only found in specific viroids, like HP III in PSTVd (Riesner et al., 1979), and HP IV in Columnea latent viroid (CLVd; Owens et al., 2003), but their functional role, if any, is not known.
There is an increasing appreciation that the small RNA motifs seemingly unstructured due to the absence of Watson–Crick base-pairs – one example being the loops in the rod-like secondary structure of PSTVd – are instead stabilized by alternative interactions. Actually, each RNA base can form hydrogen bonds with another base via one of the three edges (Watson–Crick, Hoogsteen, and sugar), with their glycosidic bonds being oriented cis or trans relative to each other (Figure 4; Leontis and Westhof, 2001). Isosteric (that is, similar in shape) relationships for each base-pairing family are compiled in isostericity matrices that provide the rationale for explaining and predicting recurrent three-dimensional (3D) motifs in non-homologous RNAs, wherein they are more conserved in structure (their nucleotides adopt similar spatial arrangements) than in sequence (Leontis et al., 2002). This approach has been used to validate a 3D model of PSTVd loop E inferred from comparative sequence analysis as well as from NMR and X-ray crystal structures of similar motifs in other RNAs and, besides, it has allowed the design of disruptive and compensatory mutations; functional analyses of such mutants in vitro and in vivo has shown that the structural integrity of this element of tertiary structure is critical for accumulation (Zhong et al., 2006). RNA signatures also regulate short (cell-to-cell) and long distance movement of viroids through the plasmodesmata and phloem, respectively. More specifically: (i) unidirectional PSTVd trafficking from the bundle sheath to mesophyll in young tobacco leaves demands a bipartite RNA motif (Qi et al., 2004), (ii) entry of PSTVd from non-vascular into phloem tissue to initiate systemic infection is mediated by an U/C motif (forming a water-inserted cis Watson–Crick/Watson–Crick base-pair flanked by short conventional helices), a 3D model based on comparisons with X-ray crystal structures of similar motifs in rRNAs and supported by combined mutagenesis and co-variation analyses (Zhong et al., 2007), and (iii) trafficking from palisade mesophyll to spongy mesophyll requires an RNA motif called loop 6 (consisting of the sequence 5′-CGA-3′…5′-GAC-3′ flanked on both sides by cis Watson–Crick G/C and G/U wobble base-pairs) the 3D model of which, describing all non-Watson–Crick base-pairs, has been derived by isostericity-based sequence comparisons with 3D RNA motifs from the RNA X-ray crystal structure database (Takeda et al., 2011). Finally, extending this approach, a genome-wide mutational analysis has identified loops/bulges in PSTVd that are essential or important for autonomous replication in single cells (protoplasts) or for systemic trafficking, thus providing a framework for future studies on RNA motifs that regulate these two functional properties in PSTVd and perhaps in other RNAs (Zhong et al., 2008; Figure 5). In conclusion, significant advances in understanding viroid-host relationships should be expected from a comprehensive dissection of viroid RNA structural motifs and, particularly, on how they interact with their cognate cellular factors (Takeda and Ding, 2009; Ding, 2010).
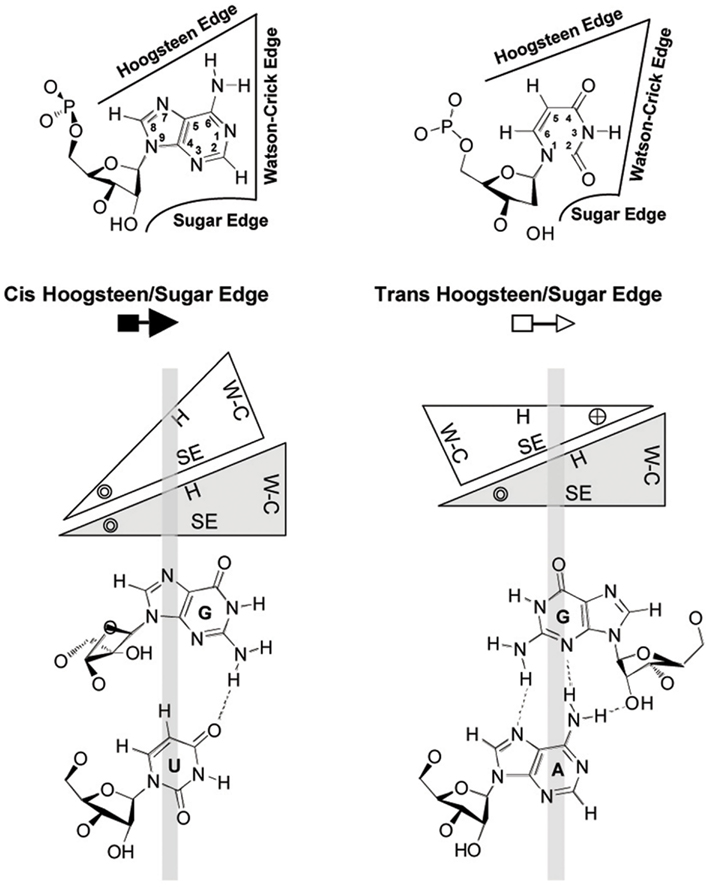
Figure 4. Geometric classification of RNA base-pairing. The upper panel shows that each nucleotide base has three edges (Watson–Crick, Hoogsteen, and sugar) that can potentially form hydrogen bonds with one of the three edges of another base. Thus, each base is represented by a triangle and can potentially pair with up to three other bases. The interacting bases can pair with a cis or trans relative orientation of their glycosidic bonds; this is illustrated in the lower panels for the cis and trans orientations of nucleotides pairing at the Hoogsteen edge of one base and the sugar edge of the second base. In these base-pairs, the Watson–Crick edges of the interacting bases are available for further interactions – with other RNAs, proteins, or small molecules. The cross and circle in the triangle where the Hoogsteen and sugar edges meet indicate 5′ → 3′ and 3′ → 5′ orientations, respectively, of the sugar-phosphodiester backbones relative to the plane of the page. W–C, Watson–Crick edge; H, Hoogsteen edge; SE, sugar edge. Reproduced with permission from Zhong et al. (2006).
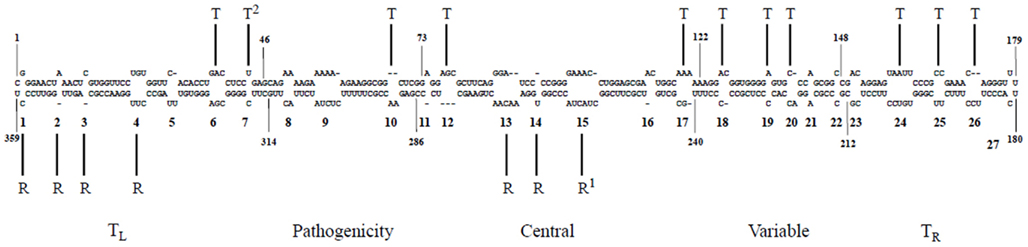
Figure 5. A genomic map of PSTVd loop motifs that are essential/critical for replication and systemic trafficking. Superscripts 1 and 2 refer to data from Zhong et al. (2006) and Zhong et al. (2007), respectively. R, replication; T, trafficking. Reproduced with permission from Zhong et al. (2008).
Hammerhead Ribozymes and Other Elements of Higher-Order Structure in the Family Avsunviroidae: Functional Significance
As indicated above, the finding that members of the family Avsunviroidae are catalytic RNAs is one of the most striking features of viroids. The catalytic activity of these RNAs resides in the ability of their both polarity strands to fold into hammerhead structures, which facilitate splitting of a specific phosphodiester bond through a transesterification that produces 5′-hydroxyl and 2′,3′ cyclic phosphodiester termini (Hutchins et al., 1986; Prody et al., 1986). An account of the data supporting the functional role of hammerhead ribozymes in promoting self-excision into unit-length strands of the multimeric intermediates, generated in replication through a rolling-circle mechanism, has been presented previously (Flores et al., 2000, 2001). From a structural perspective, X-ray crystallography (and to a lower extent NMR, see below), have provided key insights on the actual shape of two of these ribozymes in their natural (full-length) version, and reconciled conflicting interpretations of previous structural and biochemical data obtained with minimal (incomplete) versions thereof. Rather than being composed of a central conserved core flanked by three double-stranded helices (I, II, and III) usually capped by short loops (1, 2, and 3), altogether looking like a hammerhead, the actual shape instead resembles a misshapen Y in which helices II and III are virtually colinear (Martick and Scott, 2006; Chi et al., 2008; Figures 6A,B). Importantly, these studies confirmed physical contacts between loops 1 and 2 predicted by in vitro and in vivo analyses of natural hammerheads showing that this tertiary interaction is critical for catalysis at the low physiological levels of Mg2+ (De la Peña et al., 2003; Khvorova et al., 2003), most likely because it facilitates sampling of the active conformation. Dissection by NMR spectroscopy, site-directed mutagenesis, and kinetic and infectivity analyses of another two natural hammerheads (those of CChMVd) has resulted in a detailed picture of the loop–loop contacts (Figure 6C), the key aspects of which – base-pairing between the pyrimidine at the 5′ side of loop 1 and the purine at the 3′ side of loop 2, and interaction of an extra-helical pyrimidine of loop 1 with loop 2 – appear to exist in other natural hammerheads as well (Dufour et al., 2009). Incorporation of these tertiary stabilization motifs has resulted in a second generation of hammerheads catalyzing trans-cleavage of specific RNAs at the low concentrations of Mg2+ existing in vivo (Carbonell et al., 2011 and references therein). Comparative studies on the effects of mutations in co- and post-transcriptional self-cleavage have revealed an additional feature of natural hammerheads: they have been evolutionarily selected to operate co-transcriptionally, in other words, concurrently with RNA folding during replication (Carbonell et al., 2006). This finding, together with the existence of alternative non-self-cleaving conformations that involve the nucleotides conserved in hammerheads (Hernández and Flores, 1992; Flores et al., 2000), explain why a fraction of the viroid molecules remains uncleaved and can thus serve as templates for replication through a rolling-circle mechanism. Analysis of ELVd RNAs expressed in chloroplasts of Chlamydomonas reinhardtii has shown that deletion mutants able to self-cleave efficiently display ligation defects, indicating that additional nucleotides – apart those forming the hammerhead – are involved in the conformation promoting ligation (Martínez et al., 2009). Moreover, this conformation should favor the physical proximity and positioning of the 5′-hydroxyl and 2′,3′ cyclic phosphodiester termini (resulting from self-cleavage) for their joining most likely catalyzed by a chloroplastic tRNA ligase (Nohales et al., 2012).
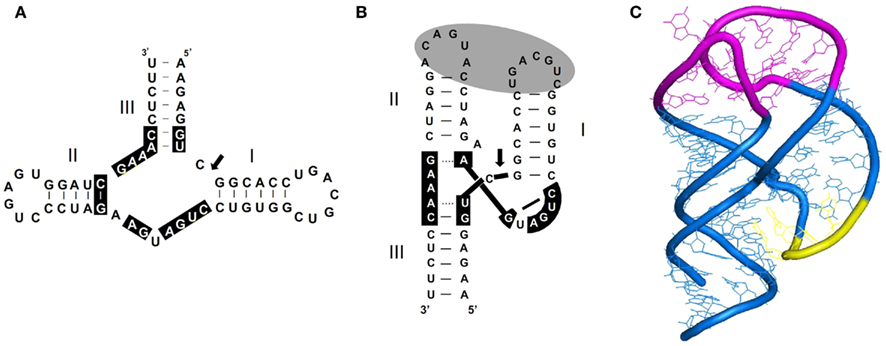
Figure 6. Secondary structure and tridimensional model proposed for most natural hammerhead ribozymes. (A) Schematic representation of a typical hammerhead structure (that of the plus strand of PLMVd) as originally formulated. Residues strictly or highly conserved in most natural hammerheads are on a black background. Arrow marks the self-cleavage site and dashes indicate Watson–Crick (and wobble) base-pairs. (B) Schematic representation of the same hammerhead structure according to X-ray crystallography and NMR data. The proposed tertiary interaction between loops 1 and 2 that facilitates catalytic activity in vivo, is denoted with a gray oval. Dashes indicate Watson–Crick (and wobble) base-pairs and dots non-canonical interactions. (C) Detailed 3D model of this hammerhead structure showing the interactions between loops 1 and 2 (in magenta). Residues in yellow form a local element of higher-order structure (the uridine turn).
Apart from the kissing-loop interaction stabilizing the branched conformation of PLMVd, CChMVd, and the viroid-like RNA from grapevine, the existence of other elements of tertiary structure has been proposed. In particular, co-variations between the hairpin loops A and B of the most stable secondary structure predicted for PLMVd preserve a potential base-pairing interaction between these two hairpin loops, thus suggesting the adoption – at least in some variants and perhaps only transiently – of another kissing-loop interaction (Ambrós et al., 1998). Recent in vitro probing supports the existence of this interaction (Dubé et al., 2011). On the other hand, application of an experimental approach similar to that leading to discovery of loop E in PSTVd – UV irradiation, denaturing PAGE, and Northern blot hybridization – has disclosed another element of tertiary structure between two conserved nucleotides located far apart in the sequence of the PLMVd (+) strand. Moreover, the same cross-linked species was also observed when PLMVd-infected leaves were irradiated prior to RNA extraction, indicating that the UV-sensitive element of tertiary structure exists also in vivo (Hernández et al., 2006). Its functional role, however, remains unknown. Finally, according to new data, the monomeric linear (+) form of ELVd has the potential for trafficking from the cytoplasm into the nucleus and subsequently from this organelle into chloroplasts, where replication occurs; this trafficking appears to be mediated by a RNA sequence and/or structural motif restricted to the left-terminal domain of the ELVd RNA (Gómez and Pallás, 2012).
Concluding Reflections and Prospects
The hypothesis that single-stranded RNA chains fold into a series of hairpin-like helices in which base-pairing by hydrogen bonds takes place, being extruded as loops the bases with no partner available, was proposed more than 50 years ago (Fresco et al., 1960). This first RNA secondary structure model has been fully validated and is now at the core of all studies on RNA structure including predictions with algorithms that search for secondary structures of minimal free energy. These in silico approaches have been complemented by analyses in vitro, with SHAPE playing a prominent role in recent years. Besides, X-ray crystallography and NMR have provided a 3D view of small RNA motifs as well as of RNAs of increasing size, showing that most loops – initially regarded as unstructured motifs – are instead stabilized by complex arrays of non-canonical interactions. Databases storing detailed structural descriptions of these RNA motifs, some of which occur recurrently in many RNAs, have become irreplaceable tools when examining new RNAs, specially if combined with other approaches like isostericity matrices.
In the previous sections we have presented a series of RNA motifs/domains to exemplify the key functions they mediate in viroids. This series, however, is not exhaustive. The initiation sites of ASBVd (+) and (−) RNAs, and of PSTVd (−) RNA, are contained within terminal hairpin loops of their predicted quasi-rod- or rod-like secondary structures, respectively (Navarro and Flores, 2000; Kolonko et al., 2006), with the hairpin loop in PSTVd also binding in vitro, directly or indirectly, the RNA polymerase II involved in replication (Bojic et al., 2012). In contrast, both initiation sites have been mapped in PLMVd at short double-stranded RNA motifs that also include the hammerhead self-cleavage sites (Delgado et al., 2005; Motard et al., 2008), thus illustrating that two quite different structural motifs may play similar functional roles and that even a single motif may be involved in more than one function. Furthermore, given that RNA folds co-transcriptionally, the initiation sites of nascent viroid strands may influence the adoption of transient metastable structures that are functionally important (Delgado et al., 2005).
The collapsed folding of viroid strands makes them potential substrates for the RNA silencing defensive response that they elicit in their hosts, because this response is specifically triggered by double-stranded or highly structured single-stranded RNAs (see for reviews Carthew and Sontheimer, 2009; Llave, 2010). The evidence supporting this notion is compelling and includes detection and characterization in plants infected by members of both families of viroid-derived small RNAs (vd-sRNAs) with the typical properties of the small interfering (si) and micro (mi) RNAs, which by loading the RNA-induced silencing complex (RISC), guide it against specific targets (see for a review Flores et al., 2011a). Some of the vd-sRNAs may be relevant for pathogenesis. Pertinent to this context is an extreme albinism termed peach calico (PC) induced by PLMVd variants of 348–351 nt containing a specific insertion of 12–14 nt, which interestingly, folds into a hairpin capped by a U-rich tetraloop (Malfitano et al., 2003; Rodio et al., 2006; Figure 1). Recent evidence indicates that PC – manifested at the cell level in altered plastids with irregular shape and size, and with rudimentary thylakoids resembling proplastids (Rodio et al., 2007) – is possibly induced via RNA silencing by two 21-nt vd-sRNAs that contain the specific insertion and prime RISC for cleaving the mRNA coding for the chloroplastic heat-shock protein 90 (Navarro et al., 2012a). Therefore, PC appears incited by the primary rather than by the secondary structure of the specific insertion, with its folding into a hairpin perhaps affording additional stability to the motif. These results support a previous proposal (Papaefthimiou et al., 2001; Wang et al., 2004), but do not exclude alternative mechanisms of viroid pathogenesis based on structural motifs of the genomic RNA directly interacting with still unidentified host factors (Owens et al., 1996; Schmitz and Riesner, 1998; De la Peña et al., 1999; see for reviews Owens and Hammond, 2009; Navarro et al., 2012b).
To conclude, the two most prominent structural features of viroids – circularity and compact folding – are also displayed by the RNA of human hepatitis delta virus (HDV), thus illustrating a remarkable example of convergent evolution between plant and animal systems. Like in the family Pospiviroidae, HDV RNA adopts a rod-like secondary structure and replicates in the nucleus through a rolling-circle mechanism involving RNA polymerase II, and like in the family Avsunviroidae, replication is also mediated by ribozymes (although of a non-hammerhead class; see for reviews Taylor and Pelchat, 2010; Flores et al., 2011b). Therefore, structural similarity has a functional correlate, highlighting that additional efforts focusing in the ultimate goal of elucidating RNA structure in vivo should certainly pay off.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. J. A. Daròs for critical reading and suggestions, Dr. B. Ding for kindly providing Figures 4 and 5, and Dr. M. de la Peña for kindly providing Figure 6. Research in Ricardo FLores laboratory is presently supported by grant BFU2011-28443 from the Ministerio de Educación y Ciencia (MEC) of Spain. During this work Pedro Serra has been supported by postdoctoral contracts from the Generalitat Valenciana (APOSTD/2010, program VALi+d) and the MEC (program Juan de la Cierva), and Sofia Minoia by a predoctoral fellowship from the MEC.
References
Abraitiene, A., Zhao, Y., and Hammond, R. (2008). Nuclear targeting by fragmentation of the potato spindle tuber viroid genome. Biochem. Biophys. Res. Commun. 368, 470–475.
Ambrós, S., Hernández, C., Desvignes, J. C., and Flores, R. (1998). Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: implications of the existence of constraints limiting the heterogeneity of viroid quasi-species. J. Virol. 72, 7397–7406.
Ambrós, S., Hernández, C., and Flores, R. (1999). Rapid generation of genetic heterogeneity in progenies from individual cDNA clones of peach latent mosaic viroid in its natural host. J. Gen. Virol. 80, 2239–2252.
Baumstark, T., Schröder, A. R., and Riesner, D. (1997). Viroid processing: switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J. 16, 599–610.
Bojic, T., Beeharry, Y., Zhang, D. J., and Pelchat, M. (2012). Tomato RNA polymerase II interacts with the rod-like conformation of the left terminal domain of the potato spindle tuber viroid positive RNA genome. J. Gen. Virol. (in press). doi:10.1099/vir.0.041574–41570
Branch, A. D., Benenfeld, B. J., and Robertson, H. D. (1985). Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc. Natl. Acad. Sci. U.S.A. 82, 6590–6594.
Branch, A. D., Benenfeld, B. J., and Robertson, H. D. (1988). Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc. Natl. Acad. Sci. U.S.A. 85, 9128–9132.
Branch, A. D., and Robertson, H. D. (1984). A replication cycle for viroids and other small infectious RNAs. Science 223, 450–454.
Bussière, F., Ouellet, J., Côté, F., Lévesque, D., and Perreault, J. P. (2000). Mapping in solution shows the peach latent mosaic viroid to possess a new pseudoknot in a complex, branched secondary structure. J. Virol. 74, 2647–2654.
Candresse, T., Góra-Sochackab, A., and Zagórski, W. (2001). Restoration of secondary hairpin II is associated with restoration of infectivity of a non-viable recombinant viroid. Virus Res. 75, 29–34.
Carbonell, A., De la Peña, M., Flores, R., and Gago, S. (2006). Effects of the trinucleotide preceding the self-cleavage site on eggplant latent viroid hammerheads: differences in co- and post-transcriptional self-cleavage may explain the lack of trinucleotide AUC in most natural hammerheads. Nucleic Acids Res. 34, 5613–5622.
Carbonell, A., Flores, R., and Gago, S. (2011). Trans-cleaving hammerhead ribozymes with tertiary stabilizing motifs: in vitro and in vivo activity against a structured viroid RNA. Nucleic Acids Res. 39, 2432–2444.
Carthew, R. W., and Sontheimer, E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655.
Chi, Y. I., Martick, M., Lares, M., Kim, R., Scott, W. G., and Kim, S. H. (2008). Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol. 6, e234. doi:10.1371/journal.pbio.0060234
Daròs, J. A., and Flores, R. (2004). Arabidopsis thaliana has the enzymatic machinery for replicating representative viroid species of the family Pospiviroidae. Proc. Natl. Acad. Sci. U.S.A. 101, 6792–6797.
Daròs, J. A., Marcos, J. F., Hernández, C., and Flores, R. (1994). Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. U.S.A. 91, 12813–12817.
Davies, J. W., Kaesberg, P., and Diener, T. O. (1974). Potato spindle tuber viroid. XII. An investigation of viroid RNA as a messenger for protein synthesis. Virology 61, 281–286.
De la Peña, M., and Flores, R. (2001). An extra nucleotide in the consensus catalytic core of a viroid hammerhead ribozyme: implications for the design of more efficient ribozymes. J. Biol. Chem. 276, 34586–34593.
De la Peña, M., and Flores, R. (2002). Chrysanthemum chlorotic mottle viroid RNA: dissection of the pathogenicity determinant and comparative fitness of symptomatic and non-symptomatic variants. J. Mol. Biol. 321, 411–421.
De la Peña, M., Gago, S., and Flores, R. (2003). Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 22, 5561–5570.
De la Peña, M., Navarro, B., and Flores, R. (1999). Mapping the molecular determinant of pathogenicity in a hammerhead viroid: a tetraloop within the in vivo branched RNA conformation. Proc. Natl. Acad. Sci. U.S.A. 96, 9960–9965.
Delgado, S., Martínez de Alba, E., Hernández, C., and Flores, R. (2005). A short double-stranded RNA motif of peach latent mosaic viroid contains the initiation and the self-cleavage sites of both polarity strands. J. Virol. 79, 12934–12943.
Diener, T. O. (1971). “A plant virus with properties of a free nucleic acid: potato spindle tuber viroid,” in Comparative Virology, eds K. Maramorosch and E. Kurstak (New York: Academic Press), 433–478.
Diener, T. O. (1972). Potato spindle tuber viroid VIII. Correlation of infectivity with a UV-absorbing component and thermal denaturation properties of the RNA. Virology 50, 606–609.
Diener, T. O. (1986). Viroid processing: a model involving the central conserved region and hairpin I. Proc. Natl. Acad. Sci. U.S.A. 83, 58–62.
Diener, T. O., and Raymer, W. B. (1967). Potato spindle tuber virus: a plant virus with properties of a free nucleic acid. Science 158, 378–381.
Ding, B. (2010). Viroids: self-replicating, mobile, and fast-evolving noncoding regulatory RNAs. Wiley Interdiscip. Rev. RNA 1, 362–375.
Dingley, A. J., Steger, G., Esters, B., Riesner, D., and Grzesiek, S. (2003). Structural characterization of the 69 nucleotide potato spindle tuber viroid left-terminal domain by NMR and thermodynamic analysis. J. Mol. Biol. 334, 751–767.
Dubé, A., Baumstark, T., Bisaillon, M., and Perreault, J. P. (2010). The RNA strands of the plus and minus polarities of peach latent mosaic viroid fold into different structures. RNA 16, 463–473.
Dubé, A., Bolduc, F., Bisaillon, M., and Perreault, J. P. (2011). Mapping studies of the peach latent mosaic viroid reveal novel structural features. Mol. Plant Pathol. 12, 688–701.
Dufour, D., De la Peña, M., Gago, S., Flores, R., and Gallego, J. (2009). Structure-function analysis of the ribozymes of chrysanthemum chlorotic mottle viroid: a loop-loop interaction motif conserved in most natural hammerheads. Nucleic Acids Res. 37, 368–381.
Eiras, M., Kitajima, E. W., Flores, R., and Daròs, J. A. (2007). Existence in vivo of the loop E motif in potato spindle tuber viroid RNA. Arch. Virol. 152, 1389–1393.
Eiras, M., Nohales, M. A., Kitajima, E. W., Flores, R., and Daròs, J. A. (2011). Ribosomal protein L5 and transcription factor IIIA from Arabidopsis thaliana bind in vitro specifically potato spindle tuber viroid RNA. Arch. Virol. 156, 529–533.
Fadda, Z., Daròs, J. A., Fagoaga, C., Flores, R., and Duran-Vila, N. (2003a). Eggplant latent viroid (ELVd): candidate type species for a new genus within family Avsunviroidae (hammerhead viroids). J. Virol. 77, 6528–6532.
Fadda, Z., Daròs, J. A., Flores, R., and Durán-Vila, N. (2003b). Identification in eggplant of a variant of citrus exocortis viroid (CEVd) with a 96 nucleotide duplication in the right terminal region of the rod-like secondary structure. Virus Res. 97, 145–149.
Fekih Hassen, I., Massart, S., Motard, J., Roussel, S., Parisi, O., Kummert, J., Fakhfakh, H., Marrakchi, M., Perreault, J. P., and Jijakli, M. H. (2007). Molecular features of new peach latent mosaic viroid variants suggest that recombination may have contributed to the evolution of this infectious RNA. Virology 360, 50–57.
Flores, R., Daròs, J. A., and Hernández, C. (2000). The Avsunviroidae family: viroids with hammerhead ribozymes. Adv. Virus Res. 55, 271–323.
Flores, R., Di Serio, F., and Hernández, C. (1997). Viroids: the noncoding genomes. Semin. Virol. 8, 65–73.
Flores, R., Di Serio, F., Navarro, B., Duran-Vila, N., and Owens, R. A. (2011a). “Viroids and viroid diseases of plants,” in Studies in Viral Ecology, Vol. 1, Microbial and Botanical Host Systems, ed. C. J. Hurst (Hoboken, NJ: John Wiley & Sons), 311–346.
Flores, R., Grubb, D., Elleuch, A., Nohales, M. A., Delgado, S., and Gago, S. (2011b). Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: variations on a theme. RNA Biol. 8, 200–206.
Flores, R., Gas, M. E., Molina-Serrano, D., Nohales, M. A., Carbonell, A., Gago, S., De la Peña, M., and Daròs, J. A. (2009). Viroid replication: rolling-circles, enzymes and ribozymes. Viruses 1, 317–334.
Flores, R., Hernández, C., De la Peña, M., Vera, A., and Daròs, J. A. (2001). Hammerhead ribozyme structure and function in plant RNA replication. Meth. Enzymol. 341, 540–552.
Flores, R., Hernández, C., Martínez de Alba, E., Daròs, J. A., and Di Serio, F. (2005). Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43, 117–139.
Flores, R., and Semancik, J. S. (1982). Properties of a cell-free system for synthesis of citrus exocortis viroid. Proc. Natl. Acad. Sci. U.S.A. 79, 6285–6288.
Fresco, J. R., Alberts, B. M., and Doty, P. (1960). Some molecular details of the secondary structure of ribonucleic acid. Nature 188, 98–101.
Gago, S., De la Peña, M., and Flores, R. (2005). A kissing-loop interaction in a hammerhead viroid RNA critical for its in vitro folding and in vivo viability. RNA 11, 1073–1083.
Gago, S., Elena, S. F., Flores, R., and Sanjuán, R. (2009). Extremely high variability of a hammerhead viroid. Science 323, 1308.
Gas, M. E., Hernández, C., Flores, R., and Daròs, J. A. (2007). Processing of nuclear viroids in vivo: an interplay between RNA conformations. PLoS Pathog. 3, e182. doi:10.1371/journal.ppat.0030182
Gas, M. E., Molina-Serrano, D., Hernández, C., Flores, R., and Daròs, J. A. (2008). Monomeric linear RNA of citrus exocortis viroid resulting from processing in vivo has 5′-phosphomonoester and 3′-hydroxyl termini: implications for the ribonuclease and RNA ligase involved in replication. J. Virol. 82, 10321–10325.
Gast, F. U., Kempe, D., and Sänger, H. L. (1998). The dimerization domain of potato spindle tuber viroid, a possible hallmark for infectious RNA. Biochemistry 37, 14098–14107.
Gast, F. U., Kempe, D., Spieker, R. L., and Sänger, H. L. (1996). Secondary structure probing of potato spindle tuber viroid (PSTVd) and sequence comparison with other small pathogenic RNA replicons provides evidence for central non-canonical base-pairs, large A-rich loops, and a terminal branch. J. Mol. Biol. 262, 652–670.
Gómez, G., and Pallás, V. (2012). Studies on subcellular compartmentalization of plant pathogenic non coding RNAs give new insights into the intracellular RNA-traffic mechanisms. Plant Physiol. 159, 558–564.
Góra, A., Candresse, T., and Zagórski, W. (1994). Analysis of the population structure of three phenotypically different PSTVd isolates. Arch. Virol. 138, 233–245.
Gozmanova, M., Denti, M. A., Minkov, I. N., Tsagris, M., and Tabler, M. (2003). Characterization of the RNA motif responsible for the specific interaction of potato spindle tuber viroid RNA (PSTVd) and the tomato protein Virp1. Nucleic Acids Res. 31, 5534–5543.
Grill, L. K., and Semancik, J. S. (1978). RNA sequences complementary to citrus exocortis viroid in nucleic acid preparations from infected Gynura aurantiaca. Proc. Natl. Acad. Sci. U.S.A. 75, 896–900.
Gross, H. J., Domdey, H., Lossow, C., Jank, P., Raba, M., Alberty, H., and Sänger, H. L. (1978). Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 273, 203–208.
Gross, H. J., Krupp, G., Domdey, H., Raba, M., Jank, P., Lossow, C., Alberty, H., Ramm, K., and Sanger, H. L. (1982). Nucleotide sequence and secondary structure of citrus exocortis and chrysanthemum stunt viroid. Eur. J. Biochem. 121, 249–257.
Gutell, R. R., Larsen, N., and Woese, C. R. (1994). Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev. 58, 10–26.
Hadidi, A., Flores, R., Randles, J. W., and Semancik, J. S. (2003). Viroids. Collingwood: CSIRO Publishing.
Haseloff, J., Mohamed, N. A., and Symons, R. H. (1982). Viroid RNAs of the cadang-cadang disease of coconuts. Nature 229, 316–321.
Haseloff, J., and Symons, R. H. (1981). Chrysanthemum stunt viroid: primary sequence and secondary structure. Nucleic Acids Res. 9, 2741–2752.
Henco, K., Sänger, H. L., and Riesner, D. (1979). Fine structure melting of viroids as studied by kinetic methods. Nucleic Acids Res. 6, 3041–3059.
Hernández, C., Di Serio, F., Ambrós, S., Daròs, J. A., and Flores, R. (2006). An element of the tertiary structure of peach latent mosaic viroid RNA revealed by UV irradiation. J. Virol. 80, 9336–9340.
Hernández, C., Elena, S. F., Moya, A., and Flores, R. (1992). Pear blister canker viroid is a member of the apple scar skin viroid subgroup (apscaviroids) and also has sequence homologies with viroids from other subgroups. J. Gen. Virol. 73, 2503–2507.
Hernández, C., and Flores, R. (1992). Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. U.S.A. 89, 3711–3715.
Hutchins, C., Rathjen, P. D., Forster, A. C., and Symons, R. H. (1986). Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 14, 3627–3640.
Keese, P., and Symons, R. H. (1985). Domains in viroids: evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl. Acad. Sci. U.S.A. 82, 4582–4586.
Khvorova, A., Lescoute, A., Westhof, E., and Jayasena, S. D. (2003). Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 10, 708–712.
Kolonko, N., Bannach, O., Aschermann, K., Hu, K. H., Moors, M, Schmitz, M., Steger, G., and Riesner, D. (2006). Transcription of potato spindle tuber viroid by RNA polymerase II starts in the left terminal loop. Virology 347, 392–404.
Koltunow, A. M., and Rezaian, M. A. (1988). Grapevine yellow speckle viroid – structural features of a new viroid group. Nucleic Acids Res. 16, 849–864.
Koltunow, A. M., and Rezaian, M. A. (1989). Grapevine viroid-1B, a new member of the apple scar skin viroid group contains the left terminal region of tomato planta macho viroid. Virology 170, 575–578.
Leontis, N. B., Stombaugh, J., and Westhof, E. (2002). Motif prediction in ribosomal RNAs: lessons and prospects for automated motif prediction in homologous RNA molecules. Biochimie 84, 961–973.
Leontis, N. B., and Westhof, E. (2001). Geometric nomenclature and classification of RNA base pairs. RNA 7, 499–512.
Llave, C. (2010). Virus-derived small interfering RNAs at the core of plant-virus interactions. Trends Plant Sci. 15, 701–707.
Loss, P., Schmitz, M., Steger, G., and Riesner, D. (1991). Formation of a thermodynamically metastable structure containing hairpin II is critical for infectivity of potato spindle tuber viroid RNA. EMBO J. 10, 719–727.
MacRae, I. J., and Doudna, J. A. (2007). Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 17, 1–8.
Malfitano, M., Di Serio, F., Covelli, L., Ragozzino, A., Hernández, C., and Flores, R. (2003). Peach latent mosaic viroid variants inducing peach calico contain a characteristic insertion that is responsible for this symptomatology. Virology 313, 492–501.
Maniataki, E., Tabler, M., and Tsagris, M. (2003). Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hairpin with the host protein VirP1. RNA 9, 346–354.
Martick, M., and Scott, W. G. (2006). Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126, 309–320.
Martínez, F., Marqués, J., Salvador, M. L., and Daròs, J. A. (2009). Mutational analysis of eggplant latent viroid RNA processing in Chlamydomonas reinhardtii chloroplast. J. Gen. Virol. 90, 3057–3065.
Mathews, D. H., and Turner, D. H. (2006). Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol. 16, 270–278.
McClements, W. L., and Kaesberg, P. (1977). Size and secondary structure of potato spindle tuber viroid. Virology 76, 477–484.
McInnes, J. L., and Symons, R. H. (1991). “Comparative structure of viroids and their rapid detection using radioactive and nonradioactive nucleic acid probes,” in Viroids and Satellites: Molecular Parasites at the Frontier of Life, ed. K. Maramorosch (Boca Raton, FL: CRC Press), 21–58.
Merino, E. J., Wilkinson, K. A., Coughlan, J. L., and Weeks, K. M. (2005). RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127, 4223–4231.
Motard, J., Bolduc, F., Thompson, D., and Perreault, J. P. (2008). The peach latent mosaic viroid replication initiation site is located at a universal position that appears to be defined by a conserved sequence. Virology 373, 362–375.
Mühlbach, H. P., and Sänger, H. L. (1979). Viroid replication is inhibited by α-amanitin. Nature 278, 185–188.
Navarro, B., and Flores, R. (1997). Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of viroids with hammerhead ribozymes. Proc. Natl. Acad. Sci. U.S.A. 94, 11262–11267.
Navarro, B., Gisel, A., Rodio, M. E., Degado, S., Flores, R., and Di Serio, F. (2012a). Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. (in press). doi:10.1111/j.1365-2313X.2012.04940.x
Navarro, B., Gisel, A., Rodio, M. E., Degado, S., Flores, R., and Di Serio, F. (2012b). Viroids: how to infect a host and cause disease without encoding proteins. Biochimie 94, 1474–1480.
Navarro, J. A., and Flores, R. (2000). Characterization of the initiation sites of both polarity strands of a viroid RNA reveals a motif conserved in sequence and structure. EMBO J. 19, 2662–2670.
Navarro, J. A., Vera, A., and Flores, R. (2000). A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of avocado sunblotch viroid. Virology 268, 218–225.
Nohales, M. A., Molina-Serrano, D., Flores, R., and Daròs, J. A. (2012). Involvement of the chloroplastic isoform of tRNA ligase in the replication of the viroids belonging to the family Avsunviroidae. J. Virol. [Epub ahead of print]. doi:10.1128/JVI.00629-12.
Nussinov, R., and Jacobson, A. B. (1980). Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc. Natl. Acad. Sci. U.S.A. 77, 6309–6313.
Owens, R. A., and Hammond, R. W. (2009). Viroid pathogenicity: one process, many faces. Viruses 1, 298–316.
Owens, R. A., Sano, T., Feldstein, P. A., Hu, Y., and Steger, G. (2003). Identification of a novel structural interaction in columnea latent viroid. Virology 313, 604–614.
Owens, R. A., Steger, G., Hu, Y., Fels, A., Hammond, R. W., and Riesner, D. (1996). RNA structural features responsible for potato spindle tuber viroid pathogenicity. Virology 222, 144–158.
Paillart, J. C., Shehu-Xhilaga, M., Marquet, R., and Mak, J. (2004). Dimerization of retroviral RNA genomes: an inseparable pair. Nat. Rev. Microbiol. 2, 461–472.
Papaefthimiou, I., Hamilton, A. J., Denti, M. A., Baulcombe, D. C., Tsagris, M., and Tabler, M. (2001). Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 29, 2395–2400.
Pelchat, M., Levesque, D., Ouellet, J., Laurendeau, S., Levesque, S., Lehoux, J., Thompson, D. A., Eastwell, K. C., Skrzeczkowski, L. J., and Perreault, J. P. (2000). Sequencing of peach latent mosaic viroid variants from nine North American peach cultivars shows that this RNA folds into a complex secondary structure. Virology 271, 37–45.
Polivka, H., Staub, U., and Gross, H. J. (1996). Variation of viroid profiles in individual grapevine plants: novel grapevine, yellow speckle viroid 1 mutants show alterations of hairpin I. J. Gen. Virol. 77, 155–161.
Prody, G. A., Bakos, J. T., Buzayan, J. M., Schneider, I. R., and Bruening, G. (1986). Autolytic processing of dimeric plant virus satellite RNA. Science 231, 1577–1580.
Puchta, H., Ramm, K., and Sänger, H. L. (1988). The molecular structure of hop latent viroid (HLV), a new viroid occurring worldwide in hops. Nucleic Acids Res. 16, 4197–4216.
Qi, Y., and Ding, B. (2003). Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 15, 1360–1374.
Qi, Y., Pélissier, T., Itaya, A., Hunt, E., Wassenegger, M., and Ding, B. (2004). Direct role of a viroid RNA motif in mediating directional RNA trafficking across a specific cellular boundary. Plant Cell 16, 1741–1752.
Qu, F., Heinrich, C., Loss, P., Steger, G., Tien, P., and Riesner, D. (1995). Multiple pathways of reversion in viroids for conservation of structural domains. EMBO J. 12, 2129–2139.
Riesner, D. (1991). Viroids: from thermodynamics to cellular structure and function. Mol. Plant Microbe Interact. 4, 122–131.
Riesner, D., Henco, K., Rokohl, U., Klotz, G., Kleinschmidt, A. K., Domdey, H., Jank, P., Gross, H. J., and Sänger, H. L. (1979). Structure and structure formation of viroids. J. Mol. Biol. 133, 85–115.
Rodio, M. E., Delgado, S., De Stradis, A. E., Gómez, M. D., Flores, R., and Di Serio, F. (2007). A viroid RNA with a specific structural motif inhibits chloroplast development. Plant Cell 19, 3610–3626.
Rodio, M. E., Delgado, S., Flores, R., and Di Serio, F. (2006). Variants of peach latent mosaic viroid inducing peach calico: uneven distribution in infected plants and requirements of the insertion containing the pathogenicity determinant. J. Gen. Virol. 87, 231–240.
Sänger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. (1976). Viroids are single-stranded covalently-closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 73, 3852–3856.
Sano, T., Candresse, T., Hammond, R. W., Diener, T. O., and Owens, R. A. (1992). Identification of multiple structural domains regulating viroid pathogenicity. Proc. Natl. Acad. Sci. U.S.A. 89, 10104–10108.
Schindler, I. M., and Mühlbach, H. P. (1992). Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci. 84, 221–229.
Schmitz, A., and Riesner, D. (1998). Correlation between bending of the VM region and pathogenicity of different potato spindle tuber viroid strains. RNA 4, 1295–1303.
Schroder, A. R. W., and Riesner, D. (2002). Detection and analysis of hairpin II, an essential metastable structural element in viroid replication intermediates. Nucleic Acids Res. 30, 3349–3359.
Semancik, J. S., Conejero, V., and Gerhart, J. (1977). Citrus exocortis viroid: survey of protein synthesis in Xenopus laevis oocytes following addition of viroid RNA. Virology 80, 218–221.
Semancik, J. S., Szychowski, J. A., Rakowski, A. G., and Symons, R. H. (1994). A stable 463 nucleotide variant of citrus exocortis viroid produced by terminal repeats. J. Gen. Virol. 75, 727–732.
Sogo, J. M., Koller, T., and Diener, T. O. (1973). Potato spindle tuber viroid X. Visualization and size determination by electron microscopy. Virology 55, 70–80.
Symons, R. H. (1981). Avocado sunblotch viroid: primary sequence and proposed secondary structure. Nucleic Acids Res. 9, 6527–6537.
Takeda, R., and Ding, B. (2009). Viroid intercellular trafficking: RNA motifs, cellular factors and broad impacts. Viruses 1, 210–221.
Takeda, R., Petrov, A. I., Leontis, N. B., and Ding, B. (2011). A three-dimensional RNA motif in potato spindle tuber viroid mediates trafficking from palisade mesophyll to spongy mesophyll in Nicotiana benthamiana. Plant Cell 23, 258–272.
Tsagris, E. M., Martínez de Alba, A. E., Gozmanova, M., and Kalantidis, K. (2008). Viroids. Cell. Microbiol. 10, 2168–2179.
Visvader, J. E., Forster, A. C., and Symons, R. H. (1985). Infectivity and in vitro mutagenesis of monomeric cDNA clones of citrus exocortis viroid indicates the site of processing of viroid precursors. Nucleic Acids Res. 13, 5843–5856.
Wang, M. B., Bian, X. Y., Wu, L. M., Liu, L. X., Smith, N. A., Isenegger, D., Wu, R. M., Masuta, C., Vance, V. B., Watson, J. M., Rezaian, A., Dennis, E. S., and Waterhouse, P. M. (2004). On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. U.S.A. 101, 3275–3280.
Wang, Y., Zhong, X., Itaya, A., and Ding, B. (2007). Evidence for the existence of the loop E motif of potato spindle tuber viroid in vivo. J. Virol. 81, 2074–2077.
Wassenegger, M., Heimes, S., and Sänger, H. L. (1994). An infectious viroid RNA replicon evolved from an in vitro-generated non-infectious viroid deletion mutant via a complementary deletion in vivo. EMBO J. 13, 6172–6177.
Wassenegger, M., Spieker, R. L., Thalmeir, S., Gast, F. U., Riedel, L., and Sänger, H. L. (1996). A single nucleotide substitution converts potato spindle tuber viroid (PSTVd) from a noninfectious to an infectious RNA for Nicotiana tabacum. Virology 226, 191–197.
Weeks, K. M., and Mauger, D. M. (2011). Exploring RNA structural codes with SHAPE chemistry. Acc. Chem. Res. 44, 1280–1291.
Woo, Y.-M., Itaya, A., Owens, R. A., Tang, L., Hammond, R. W., Chou, H.-C., Lai, M. M. C., and Ding, B. (1999). Characterization of nuclear import of potato spindle tuber viroid RNA in permeabilized protoplasts. Plant J. 17, 627–635.
Wu, Q., Wang, Y., Cao, M., Pantaleo, V., Burgyan, J., Lib, W. X., and Ding, S. W. (2012). Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc. Natl. Acad. Sci. U.S.A. 109, 3938–3943.
Xu, W., Bolduc, F., Hong, N., and Perreault, J. P. (2012). The use of a combination of computer-assisted structure prediction and SHAPE probing to elucidate the secondary structures of five viroids. Mol. Plant Pathol. (in press). doi:10.1111/j.1364-3703.2011.00776.x
Yazarlou, A., Jafarpour, B., Tarighi, S., Habili, N., and Randles, J. W. (2012). New Iranian and Australian peach latent mosaic viroid variants and evidence for rapid sequence evolution. Arch. Virol. 157, 343–347.
Zhao, Y., Owens, R. A., and Hammond, R. W. (2001). Use of a vector based on potato virus X in a whole plant assay to demonstrate nuclear targeting of potato spindle tuber viroid. J. Gen. Virol. 82, 1491–1497.
Zhong, X., Archual, A. J., Amin, A. A., and Ding, B. (2008). A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell 20, 35–47.
Zhong, X., Leontis, N., Qiang, S., Itaya, A., Qi, Y., Boris-Lawrie, K., and Ding, B. (2006). Tertiary structural and functional analysis of a viroid RNA motif by isostericity matrix and mutagenesis reveal its essential role in replication. J. Virol. 80, 8566–8581.
Zhong, X., Tao, X., Stombaugh, J., Leontis, N., and Ding, B. (2007). Tertiary structure and function of an RNA motif required for plant vascular entry to initiate systemic trafficking. EMBO J. 26, 3836–3846.
Keywords: catalytic RNAs, hammerhead ribozyme, RNA silencing, small non-coding RNAs
Citation: Flores R, Serra P, Minoia S, Di Serio F and Navarro B (2012) Viroids: from genotype to phenotype just relying on RNA sequence and structural motifs. Front. Microbio. 3:217. doi: 10.3389/fmicb.2012.00217
Received: 02 May 2012; Paper pending published: 16 May 2012;
Accepted: 28 May 2012; Published online: 18 June 2012.
Edited by:
K. Andrew White, York University, CanadaReviewed by:
Martin Pelchat, University of Ottawa, Canada; Biao Ding, The Ohio State University, USACopyright: © 2012 Flores, Serra, Minoia, Di Serio and Navarro. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Ricardo Flores, Instituto de Biología Molecular y Celular de Plantas (UPV-CSIC), Universidad Politécnica, 46022 Valencia, Spain. e-mail:cmZsb3Jlc0BpYm1jcC51cHYuZXM=

 Pedro Serra1
Pedro Serra1
