
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 02 July 2012
Sec. Evolutionary and Genomic Microbiology
Volume 3 - 2012 | https://doi.org/10.3389/fmicb.2012.00207
This article is part of the Research Topic Recent advances in genomic and genetic studies in the Archaea View all 12 articles
Secreted proteins make up a significant percentage of a prokaryotic proteome and play critical roles in important cellular processes such as polymer degradation, nutrient uptake, signal transduction, cell wall biosynthesis, and motility. The majority of archaeal proteins are believed to be secreted either in an unfolded conformation via the universally conserved Sec pathway or in a folded conformation via the Twin arginine transport (Tat) pathway. Extensive in vivo and in silico analyses of N-terminal signal peptides that target proteins to these pathways have led to the development of computational tools that not only predict Sec and Tat substrates with high accuracy but also provide information about signal peptide processing and targeting. Predictions therefore include indications as to whether a substrate is a soluble secreted protein, a membrane or cell wall anchored protein, or a surface structure subunit, and whether it is targeted for post-translational modification such as glycosylation or the addition of a lipid. The use of these in silico tools, in combination with biochemical and genetic analyses of transport pathways and their substrates, has resulted in improved predictions of the subcellular localization of archaeal secreted proteins, allowing for a more accurate annotation of archaeal proteomes, and has led to the identification of potential adaptations to extreme environments, as well as phyla-specific pathways among the archaea. A more comprehensive understanding of the transport pathways used and post-translational modifications of secreted archaeal proteins will also facilitate the identification and heterologous expression of commercially valuable archaeal enzymes.
During protein translation at the ribosome, the presence of an N-terminal signal sequence or a transmembrane helix can target the corresponding protein for translocation into or across the cytoplasmic membrane (Driessen and Nouwen, 2008; Natale et al., 2008; Ellen et al., 2010b; Yuan et al., 2010; Calo and Eichler, 2011). Archaeal proteins targeted for secretion, including polymer degrading enzymes, antimicrobial porteins, or proteins involved in intercellular signaling may be released into the extracellular environment. However, most proteins remain associated with the cell, requiring either a direct interaction with the membrane, an indirect association with membrane-anchored proteins, or, as recently predicted, attachment to the cell wall (Figure 1; Gimenez et al., 2007; Albers and Pohlschroder, 2009; Storf et al., 2010; Haft et al., 2012). Proteins that remain associated with the cell surface are involved in a wide variety of cellular processes, including nutrient uptake, motility, and surface adhesion (Ellen et al., 2010b; Albers and Meyer, 2011).
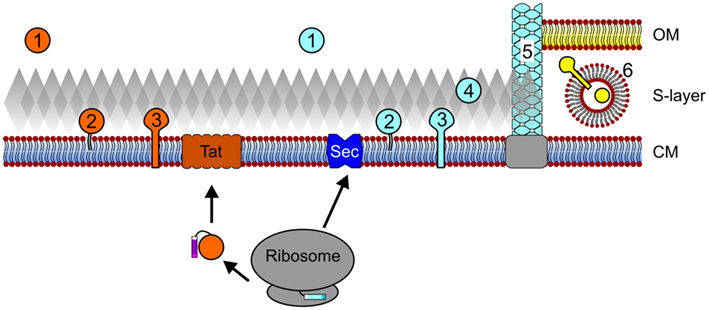
Figure 1. Archaeal protein secretion and subcellular localization. Proteins that contain Tat signal peptides pass the cytoplasmic membrane (CM) through the Tat translocon after translation and folding. Conversely, protein translocation through the Sec pore can occur co- and possibly post-translationally. Upon secretion and signal peptide processing, Sec and Tat substrates can be released into the extracellular milieu (1), be embedded into the membrane via a lipid anchor (2) or a C-terminal transmembrane segment (3). In silico data also suggest that Sec substrates may be anchored to the cell wall in an archaeosortase-dependent manner (4) and a number of type IV pilin-like proteins have been shown to assemble into cell appendages (5). In I. hospitalis, an additional outer membrane (OM) is present and periplasmic vesicles (6) are thought to play a role in the trafficking of secreted and outer membrane proteins across the periplasmic space. See text for details. Cell components are not drawn to scale.
In bacteria and archaea, the universally conserved Sec pathway is commonly used to transport secreted proteins to the cytoplasmic membrane (Nather and Rachel, 2004; Pohlschroder et al., 2005b; Driessen and Nouwen, 2008). Since these proteins are secreted in an unfolded conformation through a narrow proteinaceous pore, protein folding, post-translational modifications, such as glycosylation, and associations with cofactors, must occur in an extracytoplasmic environment devoid of ATP, and hence ATP-dependent chaperones. These complications are circumvented by substrates of the twin arginine transport (Tat) pathway, at least in part, because protein folding, cofactor incorporation, and possibly protein modifications, are accomplished in the cytoplasm (Pohlschroder et al., 2005a; Natale et al., 2008). Regardless of the transport pathway used, archaeal secreted proteins that are not released into the extracellular environment must be anchored to the cell, the only exception known to date being proteins of the archaeal genus Ignicoccus, which has an outer membrane that provides a periplasmic space that can retain secreted proteins (Huber et al., 2002; Nather and Rachel, 2004).
The results generated by several in vivo studies of archaeal secreted proteins and their transport pathways has allowed for the identification of the signal peptides required to target secreted proteins to a specific transport pathway as well as amino- or carboxy-terminal motifs within the protein that mediate substrate interactions with extracytoplasmic structures (Kobayashi et al., 1994; O’Connor and Shand, 2002; Rose et al., 2002; Dilks et al., 2005; Gimenez et al., 2007; De Castro et al., 2008; Albers and Pohlschroder, 2009; Ng et al., 2009; Kwan and Bolhuis, 2010; Storf et al., 2010; Calo and Eichler, 2011). In turn, the identification of these motifs in a large number of substrates has allowed for the development of software programs that facilitate in silico analyses of secreted protein sequences encoded by a large number and variety of archaeal genomes (Rose et al., 2002; Szabo et al., 2007a; Bagos et al., 2009; Storf et al., 2010). These in silico analyses have already lead to important insights into the strategies used to secrete and anchor proteins to the cell surface, which vary in part depending on the function of the secreted protein. Ultimately, the trends revealed by in silico analyses may clarify how an organism adapts to the selective pressures imposed on it, and may make clear which aspects of the environment had the greatest impact on the evolution of the organism.
Besides the well-studied protein transport pathways, archaea may use additional, poorly understood or currently unknown, means by which to facilitate the egress of proteins across the membrane. For example, some archaeal species produce vesicles that are released into the extracellular environment (Soler et al., 2008; Ellen et al., 2010b), or in the case of Ignicoccus, are present in the periplasmic space (Nather and Rachel, 2004). Future studies may clarify the role of these mechanisms in protein transport.
In this review, we describe the highly diverse array of archaeal secreted proteins. We consider the pathways used to transport these proteins to the cytoplasmic membrane, the ultimate destinations of substrates, and whether these proteins are secreted into the extracellular environment or are anchored to the cytoplasmic membrane or cell wall. We also explore the mechanisms that facilitate the tethering of secreted substrates to the cell surface, and discuss the types of functions displayed by cell-associated and secreted proteins. Finally, we describe the in silico approaches used to predict the subcellular localization of substrates and the post-translational modifications that these substrates undergo and identify potential trends in the use of these pathways and modifications in various microorganisms.
In the following section we briefly describe the two main routes for archaeal protein transport across the cytoplasmic membrane, the Sec and the Tat pathways, while focusing on the processing and modification of the substrates transported by these pathways. These systems have recently been more extensively reviewed elsewhere (Pohlschroder et al., 2005b; Ellen et al., 2010b; Yuan et al., 2010; Calo and Eichler, 2011).
In silico and in vivo analyses indicate that all species, whether eukaryotic or prokaryotic, transport proteins by way of the universally conserved Sec pathway, which acts as a conduit for inserting proteins into the cytoplasmic membrane or secreting them into the extracytoplasmic environment (Yuan et al., 2010; Calo and Eichler, 2011). Proteins are targeted to the Sec pathway by conserved amino-terminal signal peptides that have a tripartite structure consisting of a charged amino-terminus, a hydrophobic stretch and a signal peptidase recognition motif (Bardy et al., 2003; Ng et al., 2007; Zimmermann et al., 2011; Table 1). The Sec pathway consists of several components, including the signal recognition particle (SRP). The SRP recognizes either the signal peptide or transmembrane segments in the substrate as the nascent peptide chain emerges from the ribosome, which results in a translational arrest. Subsequently, the SRP–ribosome nascent chain complex is targeted to the proteinaceous Sec pore where the substrate is translocated across the membrane co-translationally (Grudnik et al., 2009). Conversely, SRP-independent post-translational Sec transport requires chaperones to maintain the precursor in an unfolded conformation to allow for the secretion through the approximately 20 Å Sec pore – a pore just large enough to allow for transport of an unfolded polypeptide (Van den Berg et al., 2004; Mori et al., 2010; Yuan et al., 2010). While the key components required for co-translational transport are universally conserved, post-translational transport across the ER membrane depends on a luminal ATPase, Bip/Kar2, whereas bacterial post-translational transport requires the cytoplasmic ATPase, SecA. There is evidence for archaeal post-translational translocation of Sec substrates (Dale et al., 2000; Ortenberg and Mevarech, 2000; Irihimovitch and Eichler, 2003). However, as archaea lack homologs of either of the two aforementioned ATPases, the energetics of archaeal post-translational transport is not understood to date.
The Tat pathway, which unlike the Sec pathway, is not universally conserved, being limited to prokaryotes, chloroplasts, and a few protists, transports proteins in a folded conformation (Pohlschroder et al., 2005a; Natale et al., 2008; Yuan et al., 2010; Robinson et al., 2011). Because they are folded prior to transport, Tat substrates are too large for the Sec pore to accommodate. Therefore, it is not surprising that the Tat pathway consists of components and mechanisms that are fundamentally different from those employed by the Sec pathway. However, just as Sec substrates are targeted to the Sec pathway by a signal peptide, a Tat signal peptide targets proteins to the Tat pathway (Berks et al., 2005; Robinson et al., 2011). Organisms containing a functional Tat pathway require at least TatA and TatC, which play particularly pivotal roles, with TatC (and TatB in many organisms) apparently being involved in substrate targeting, while multimers of TatA, a single transmembrane spanning protein, allow for the formation of pores of varying sizes to accommodate the diverse sizes of secreted substrates (Dilks et al., 2005; Gohlke et al., 2005; Leake et al., 2008; Robinson et al., 2011).
While the Sec and Tat pathways are mechanistically very different, the tripartite structures of the signal sequences that target substrates to these systems are surprisingly similar (Table 1). However, there are some subtle differences between Sec and Tat signal peptides. Among the most significant of these differences is the highly conserved twin arginine motif that lies within the charged region of the Tat signal peptide. The Tat signal peptide also has a hydrophobic stretch that is generally less hydrophobic than that of the Sec signal sequence (Rose et al., 2002; Bendtsen et al., 2005).
In addition to the regions of signal peptides that are critical to targeting secreted proteins to a protein transport pathway, Sec and Tat signal peptides also usually contain a recognition sequence that targets it for cleavage by a signal peptidase, although unprocessed Sec substrate signal peptides occasionally serve as amino-terminal membrane anchors (Eichler, 2002; Paetzel et al., 2002; Tuteja, 2005; Ng et al., 2007). Substrates having signal peptides that are processed by the universally conserved signal peptidase I (SPase I) are either released from the membrane or anchored to the cell by way of a carboxy-terminal membrane anchor or protein–protein interaction (Rose et al., 2002; Dilks et al., 2005; Tuteja, 2005; Gimenez et al., 2007; De Castro et al., 2008; Uthandi et al., 2010).
Recent in silico data suggest that several euryarchaeal SPase I processed Sec substrates have an additional carboxy-terminal sorting signal that targets these proteins to the cell wall or to another cell surface structure (Haft et al., 2012). Originally identified in Gram-positive bacteria, an enzyme known as a sortase processes these sorting signals, which are characterized by a signature motif containing a hydrophobic transmembrane α-helix and a cluster of basic amino acids (Spirig et al., 2011; Table 1). Upon cleavage, substrate proteins are typically covalently linked to the peptidoglycan cell wall by the sortase. Several types of sortases, each processing distinct motifs within the conserved tripartite structure have now been identified in bacteria, and similar C-terminal tripartite sorting signals have also been found in archaea. Furthermore, using comparative genomics, potential archaeosortases have been identified (Haft et al., 2012). However, sorting signal processing and modification in archaea awaits in vitro and in vivo confirmation.
Bacteria and archaea, but to the best of our knowledge not eukaryotes, express proteins that have signal peptides processed by signal peptidases distinct from the universally conserved SPase I. Bacterial signal peptides recognized by SPase II are largely similar to those processed by SPase I, containing a charged amino-terminus and a hydrophobic stretch; however, they also contain a short motif known as a lipobox that lies subsequent to the hydrophobic stretch (Table 1). The lipobox contains an invariant cysteine residue that is acylated by a prolipoprotein diacylglyceryl transferase prior to signal peptidase processing by SPase II, which then cleaves the precursor immediately upstream of the lipid-modified cysteine. The acylated cysteine residue anchors the processed substrate to the cytoplasmic membrane (Sankaran and Wu, 1994; Hutchings et al., 2009; Thompson et al., 2010; Okuda and Tokuda, 2011). Although archaea, particularly euryarchaea, express Sec and Tat substrates containing lipobox motifs, neither an archaeal SPase II homolog nor a prolipoprotein diacylglyceryl transferase homolog has yet been identified (Mattar et al., 1994; Dilks et al., 2005; Falb et al., 2005; Gimenez et al., 2007; Storf et al., 2010).
The highly conserved nature of SPase II processing site sequences across prokaryotic domains makes the apparent absence of an archaeal SPase II homolog even more intriguing (Storf et al., 2010). Perhaps the mechanisms involved in archaeal lipoprotein biosynthesis are unique to archaea, possibly owing to the distinct membrane lipid composition in the two prokaryotic domains. Studies performed on bacterial and archaeal lipoprotein mutants in which the conserved lipobox cysteine was replaced with a serine lend credence to the hypothesis that this cysteine is critical in both domains. Mutant lipoproteins are not processed in either archaea or bacteria, and like in bacteria, unprocessed archaeal substrates often remain cell-associated while a subset of similar mutant lipoprotein precursors are secreted into the supernatant (Hutchings et al., 2009; Storf et al., 2010).
SPase III specifically processes the subunits of a wide array of type IV pilus-like surface structures; included among these are the flagella of archaea (see below; Pohlschroder et al., 2011). Similar to SPase I and SPase II, SPase III recognizes a processing site located adjacent to the hydrophobic stretch of the N-terminal signal peptide that targets the substrate to the transport pathway (Arts et al., 2007; Francetic et al., 2007). However, unlike signal peptides processed by SPase I or SPase II, the processing site is N-terminal to the hydrophobic stretch, which hence remains part of the mature protein where it is essential for subunit–subunit interactions that allow the formation of an α-helical scaffold facilitating the assembly of type IV pili and pilus-like structures (Bardy and Jarrell, 2002; Albers et al., 2003; Table 1). Although the translocation of archaeal type IV pilin-like proteins into the cytoplasmic membrane has not been studied, in Gram-negative bacteria, they were shown to be secreted by the Sec pathway in a signal particle-dependent fashion (Arts et al., 2007; Francetic et al., 2007).
Additional post-translational modifications of archaeal secreted proteins, other than signal peptide processing, include both O- and N-glycosylation (Eichler and Adams, 2005; Calo et al., 2010; Jarrell et al., 2010). The first non-eukaryotic N-glycosylated protein reported was the surface S-layer glycoprotein of the haloarchaeon Halobacterium salinarum, a protein that was subsequently also shown to be O-glycosylated (Mescher et al., 1974; Sumper et al., 1990). While little is known about the O-linked glycosylation process, genetic and biochemical studies of the S-layer, as well as of the flagella subunits in two methanogenic archaea and a haloarchaeon, revealed a set of archaeal glycosylation (Agl) enzymes that catalyze the assembly and attachment of N-linked glycans to target proteins (Calo et al., 2010; Jarrell et al., 2010). As in eukaryotes, in the final step of glycosylation, an oligosaccharyltransferase homolog covalently links an assembled oligosaccharide onto the asparagine residue of a target motif (N-X-T/S) in the protein. Interestingly, aside from the oligosaccharyltransferase, the Agl components of haloarchaea and methanogens, or even closely related methanogens, show no homology to each other, indicating a wide diversity in the specifics of post-translational modification by N-glycosylation across the archaea (Calo et al., 2010; Jarrell et al., 2010). Initial studies on N-glycosylated proteins in crenarchaea support this hypothesis; however, much less is known about the components required for N-glycosylation in these organisms (Peyfoon et al., 2010; Meyer et al., 2011).
Secreted proteins play a crucial role in the interaction of cells with their environment. They fulfill a variety of functions such as degradation of complex polymeric substances (e.g., carbohydrates, proteins), nutrient uptake, signal transduction, or formation of surface complexes such as the cell wall or pili (for examples see Table 2). While for the majority of secreted proteins no function can be predicted, many secreted proteins encoded in model archaea or heterologously expressed in a model archaeon have been characterized, leading to a better understanding of the physiology of the organisms, as well as the transport pathways and post-translational modification (i.e., some have been used as reporters). In the following section, several examples are given for the variety of tasks that secreted archaeal proteins fulfill and what is known about their secretion and localization.
Several archaea produce peptides or proteins with antimicrobial activity that are collectively termed archaeocins. Thus far they have been identified in halophiles (halocins) and crenarchaea from the genus Sulfolobus (sulfolobicins; Ellen et al., 2011). Halocins are plasmid-encoded, secreted antimicrobials that can be subdivided into protein or peptide (micro) halocins (O’Connor and Shand, 2002).
HalH4 from Haloferax mediterranei is the best-characterized protein halocin. It is a 35 kDa protein predicted to be secreted by the Tat pathway that is active against other haloarchaea (Cheung et al., 1997). Microhalocins are peptides ranging from about 3 to 8 kDa in mass that are released from a larger prepro-protein that also contains a predicted Tat signal peptide (Price and Shand, 2000; O’Connor and Shand, 2002). They are effective against other haloarchaea, and in some cases, against thermophilic crenarchaea (Haseltine et al., 2001). In the case of halocin 8, the C-terminal 76 amino acid peptide carries antimicrobial activity while the N-terminal protein portion termed HalI, which remains membrane-associated, confers immunity against the halocin, probably by sequestration (Sun et al., 2005).
Sulfolobicins, proteins (about 20 kDa) associated with the membrane and extracellular vesicles, were first identified in Sulfolobus islandicus (Prangishvili et al., 2000). The genes encoding sulfolobicins were recently identified in S. acidocladarius and S. tokodaii, sulA and sulB, both of which are necessary for antimicrobial activity (Figure 2A). The gene products carry putative SPase I processed Sec signal peptides and were suggested to be peripheral membrane proteins (Ellen et al., 2011).
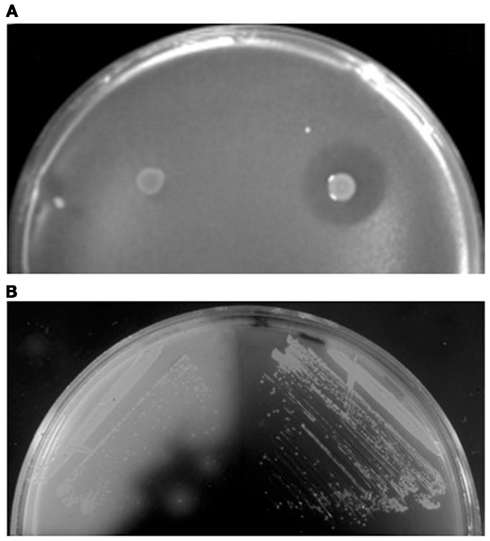
Figure 2. Detection of extracellular protein activities. (A) Overlay assay with colonies of S. acidocaldarius wt (right) and ΔsulAB mutant (left) grown on a lawn of S. solfataricus strain P2. Clear halos surrounding the colonies indicate killing of archaea by the sulfolobicins. Image reproduced with permission from Ellen et al. (2011). (B) Iodine vapor staining of H. volcanii expressing wild-type (left) or signal sequence mutated (right) α-amylase grown on rich medium supplemented with 0.2% soluble starch. Clear halos surrounding colonies indicate starch hydrolysis by extracytoplasmic α-amylase (Rose et al., 2002).
Nutrient sources in the environment are often in a form that cannot be directly taken up by cells. Polymeric molecules such as carbohydrates must be cleaved into smaller subunits, proteins degraded into peptides, and inorganic phosphate liberated from organic compounds (Worthington et al., 2003; Wende et al., 2010; Xu et al., 2011). Many archaeal enzymes that perform these tasks have been identified and characterized, either by sequence homology search and heterologous expression, or by purification of the native enzymes from cell culture supernatants of the host (Moracci et al., 2007). In the case of native enzymes, protein identification by N-terminal sequencing provided valuable information regarding the signal peptidase cleavage site and in some cases post-translational modifications by glycosylation or lipid modification. In addition, functional assays can also be used as reporters for in vivo studies on secretion, using either plate assays or measuring enzyme activity in cell culture supernatant (Figure 2B; Rose et al., 2002; Worthington et al., 2003; Hutcheon et al., 2005; Wende et al., 2010; Xu et al., 2011).
Examples of secreted enzymes include proteases such as halolysins and pyrolysins (Kamekura et al., 1992; de Vos et al., 2001; Shi et al., 2006; De Castro et al., 2008), proteins involved in starch degradation such as α-amylase and cyclodextrin glycosyltransferase (Kobayashi et al., 1994; Hutcheon et al., 2005; Bautista et al., 2012), and a copper-containing oxidase (laccase; Uthandi et al., 2010). In some cases, secretion and localization have been studied in more detail. These studies showed that not all secreted enzymes are soluble extracellular proteins, with some being anchored to the membrane via a lipid anchor (Mattar et al., 1994; Gimenez et al., 2007; Storf et al., 2010) or a C-terminal transmembrane segment (Gimenez et al., 2007). Interestingly, an α-amylase from S. solfataricus that contains a C-terminal membrane anchor in addition to an SPase I processed signal sequence was detected in both membrane and extracellular fractions (Worthington et al., 2003; Ellen et al., 2010a). Apparently, part of the protein pool is released by an unknown mechanism, which may contribute to more efficient substrate degradation. Direct attachment to the S-layer was demonstrated for a protease from Staphylothermus marinus (Mayr et al., 1996), and tetrathionate hydrolase from Acidianus ambivalens was suggested to be localized to the pseudo-periplasmic space, i.e., between the cytoplasmic membrane and the S-layer (Protze et al., 2011). More recently, another tetrathionate hydrolase from A. hospitalis was found to be assembled into “zipper-like particles” on the cell surface and extracellular space (Krupovic et al., 2012) (Figure 3A). The secretion mechanism of enzymes has only been addressed in haloarchaea. Secretion by the Tat pathway was demonstrated for the protease SptA from Natrinema sp. J7 (Shi et al., 2006), arabinanase, halocyanin 2 and 3, a DsbA-like protein, and two hypothetical proteins from Haloferax volcanii (Gimenez et al., 2007; Storf et al., 2010) as well as α-amylases from Natronococcus sp. strain Ah36 and Haloarcula hispanica (Rose et al., 2002; Hutcheon et al., 2005).
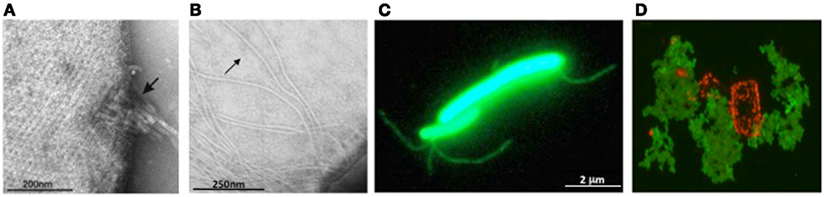
Figure 3. Archaeal surface structures. (A) Tetrathionate hydrolase from A. hospitalis YS8 assembles into zipper-like particles on the cell surface. Image reproduced with permission from Krupovic et al. (2012). (B) Electron micrographs of M. maripaludis cells expressing type IV pilus-like structures. Arrows indicate EppA-processed pili, while additional, thicker structures, are PibD-processed flagella. Samples were negatively stained with 2% phosphotungstic acid. Image reproduced with permission from (VanDyke et al., 2008). (C) Non-type IV Mth60 fimbriae of planktonic M. thermoautotrophicus cells by staining with AlexaFluor®488 (Thoma et al., 2008). Image courtesy of R. Wirth, University of Regensburg, Germany. (D) Secreted halomucin complexes (stained green with specific antibody coupled to fluorescein) surrounds quadratic Haloarcula marismortui cell (stained red by Nile blue for polyhydroxy butyrate). Unpublished image, courtesy of D. Oesterhelt, Max Planck Institute of Biochemistry, Martinsried, Germany.
Enzymes, especially those of archaeal hyperthermophiles, have potential industrial applications, in particular hydrolytic enzymes that degrade complex polymers. At high temperatures, starch and cellulose are more accessible and the risk of microbial contamination is low. Most known hyperthermophiles (cells that grow at 80°C and above) are archaea and their enzymes can withstand the harsh conditions employed in industrial processes (Egorova and Antranikian, 2005). For example, an α-amylase with high homology to a protein from Thermococcus kodakarensis, which was identified in an environmental DNA library, is stable and highly active under the conditions applied for starch liquefaction (Richardson et al., 2002).
Nutrients such as peptides and oligosaccharides produced by extracellular enzymes are taken up by dedicated transporters. Archaea make extensive use of binding protein-dependent ATP binding cassette (ABC) transporters. Binding proteins have high affinity for their substrates. Although substrate binding proteins of Gram-negative bacteria reside in the periplasmic space, they are usually associated with the cytoplasmic membrane in archaea. Different mechanisms are employed for anchoring.
The class of oligopeptide-binding proteins characterized in the crenarchaeal species S. solfataricus and Aeropyrum pernix contains an N-terminal secretory signal sequence with an SPase I cleavage site and an additional C-terminal transmembrane segment (Elferink et al., 2001; Gogliettino et al., 2010; Balestrieri et al., 2011). The membrane anchor is preceded by a stretch of glycosylated serine and threonine residues. Several euryarchaeal binding proteins were shown to be lipid anchored after processing by the yet to be identified SPase II. These lipobox-containing proteins are associated with transport of maltose in Thermococcus litoralis (Horlacher et al., 1998), or predicted to bind iron and maltose, respectively, in H. volcanii (Gimenez et al., 2007). In addition, a putative phosphate binding protein from Hfx. volcanii was predicted to be processed by an archaeosortase and may therefore have a C-terminal lipid anchor (Haft et al., 2012). Interestingly, BasA and CosA from Hbt. salinarum are two lipid anchored binding proteins shown to be involved in chemotaxis rather than substrate uptake despite their homology to ABC transporter solute binding proteins (Kokoeva et al., 2002).
A third class of binding proteins contains a prepilin-like signal sequence that is cleaved by SPase III. Removal of the short positively charged leader peptide has been shown for S. solfataricus glucose and arabinose binding proteins (Albers et al., 1999, 2003; Elferink et al., 2001). In silico analyses predicted substrate binding proteins with SPase III processing sites in many Eury- and Crenarchaea, however these predictions await experimental confirmation (Szabo et al., 2007b). Similar to pilins and archaeal flagellins that are assembled into cell appendages, sugar binding proteins are thought to be incorporated into a macromolecular structure called the bindosome (see below).
Most archaeal cells are surrounded by a membrane-anchored, crystalline S-layer that provides stability and osmoprotection to the cells (Engelhardt, 2007a,b; Albers and Meyer, 2011). Known S-layers are composed of a single protein or two subunits that are secreted by the Sec pathway and whose signal peptides are processed by SPase I. In Sulfolobales, anchoring to the cell membrane occurs via a C-terminal transmembrane segment, which is preceded by a stalk-like structure creates a pseudo-periplasmic space. While haloarchaeal S-layer proteins also contain a C-terminal hydrophobic stretch, it has recently been proposed that it is part of a C-terminal conserved tripartite sorting signal that is cleaved and lipid-modified by a yet uncharacterized system possibly involving the predicted archaeosortase enzyme (Haft et al., 2012). Consistent with this observation, Hfx. volcanii and Hbt. salinarum S-layer glycoproteins contain lipid modifications (Kikuchi et al., 1999; Konrad and Eichler, 2002). Conversely, the S-layer glycoprotein of Methanothermus fervidus does not contain a C-terminal transmembrane segment and the mechanism of cell surface attachment is elusive (Brockl et al., 1991).
Archaea also produce a diverse array of cell appendages that provide functions such as motility and surface adhesion. Most characterized cell appendage subunits have an N-terminal type IV pilin-like signal peptide (see above; Albers et al., 2003; Bardy et al., 2003; Szabo et al., 2007b; Tripepi et al., 2010).
The best-characterized archaeal cell surface structure is the flagellum, which is required for swimming motility. In contrast to bacterial flagellar biogenesis, which uses a type III secretion-like mechanism, the archaeal flagellum biosynthesis machinery includes homologs of the bacterial type IV pilus biogenesis components, including the archaeal prepilin-peptidase homolog PibD/FlaK (Journet et al., 2005; Ghosh and Albers, 2011). In addition to conferring swimming motility, flagella also play a role in surface adhesion in S. solfataricus, M. maripaludis, and Pyrococcus furiosus (Nather et al., 2006; Zolghadr et al., 2010; Jarrell et al., 2011). In fact, the thermal stability and adhesive properties led to the suggestion that Pyrococcus flagella be used as “molecular glue” (US Patent US 2008/0305524). Finally, motility assays developed for several flagellated archaea have been used to monitor successful secretion and post-translational modification of flagellins (Chaban et al., 2007; Tripepi et al., 2010; Lassak et al., 2012).
Additional genes encoding archaeal type IV pilin-like proteins were identified, using a program trained on flagellin subunits (see below), ultimately leading to the identification and characterization of non-flagellar pilus-like structures. For example, S. acidocaldarius, use Aap and Ups-pili, along with flagella, for surface adhesion and biofilm formation (Henche et al., 2012). DNA damage such as that caused by UV light irradiation strongly induces expression of the subunits of the Ups-pili, cell surface structures that subsequently facilitate cell aggregation. Once aggregated, these cells are thought to exchange chromosomal DNA, allowing efficient repair of the damaged DNA (Frols et al., 2008; Ajon et al., 2011). Interestingly, as noted above, S. solfataricus also contains sugar binding proteins that are processed by the same SPase III as the flagellin/pilin subunits. It is believed that these proteins are also assembled into a large pilus-like structure, the bindosome, since homologs of pilus-biosynthesis genes are also required for bindosome function, i.e., the growth of cells on certain sugars (Zolghadr et al., 2007).
Most archaea appear to only have one SPase III homolog; some, however, contain additional paralogs with specialized functions. M. maripaludis expresses a prepilin-peptidase, EppA, that recognizes type IV pilins with a specific SPase III processing site (previously designated as domain of unknown function 361), along with FlaK/PibD (Szabo et al., 2007b; Wang et al., 2008; Ng et al., 2011) (Figure 3B).
The only known archaeal pili that do not contain subunits with a type IV pilin signal peptide are the Mth60 fimbriae produced by Methanothermobacter thermoautotrophicus, which are involved in adhesion to solid surfaces (Thoma et al., 2008; Figure 3C). Mth60 is a small (16 kDa) glycoprotein with no known homologs in any other species. It contains a predicted SPase I processed Sec signal peptide and while it is not known how Mth60 fimbriae are assembled, the recombinant protein was shown to polymerize into fibers at the optimal growth temperature of M. thermoautotrophicus.
Halomucin is the largest known archaeal protein, produced by the extremely halophilic archaeon Haloquadratum walsbyi (Figure 3D). The 9195 amino acid protein with homology to eukaryotic mucin, is predicted to be secreted via the Sec pathway and its likely function is to provide a hydration shell in their low water activity environment (Bolhuis et al., 2006; Sublimi Saponetti et al., 2011).
Other unusual cell surface structures have been described, but are not yet characterized in molecular detail. One fascinating example is the hamus produced by the euryarchaeon SM1, which grows in cold sulfidic springs (Rudolph et al., 2001; Moissl et al., 2005). Each cell of strain SM1 produces about 100 hami. The hami are highly complex structures having a helical basic structure from which prickles emanate at regular distances, forming a barbwire-like appearance. These structures consist of large subunits of about 120 kDa. The N-terminal sequence of this protein is not known and therefore the mode of secretion cannot be assessed (Moissl et al., 2005). Finally, cannulae from the crenarchaeaon Pyrodictium abyssi are tube-like extracellular structures that cross the S-layer and the pseudo-periplasmic space, but do not protrude into the cytoplasm. Cannulae are composed of at least three related glycoproteins, but their sequences have not been published (Mai et al., 1998). While most currently characterized archaeal surface appendages appear to follow the type IV pilin assembly route, a more diverse variety of specialized secretion systems can be expected from the molecular analysis of structures that have so far only been studied on a morphological level.
As described above, the fate of a secretory protein upon translocation is determined by specific sequence motifs within the signal peptide, such as those for signal peptidase processing and lipid modification (Bardy et al., 2003; Ng et al., 2007). Additionally, the presence of a C-terminal membrane anchor or a putative sorting signal is a determinant for the localization of the mature protein. Because most of these motifs are relatively well conserved, as has been confirmed by numerous in vivo studies, it was possible to develop in silico programs that can provide more detailed information indicating whether a substrate is secreted into the extracellular environment, is tethered to a membrane by an amino-terminal transmembrane or lipid anchor, or is incorporated into a surface structure. Predictions such as these, in conjunction with in vivo and in vitro confirmations, can also deepen our understanding of the extent to which specific transport pathways and signal peptidases are used, which may point to the evolutionary pressures encountered by a particular species (see below).
A variety of software programs have been developed to predict specific signal peptide classes. Phobius, for example, is a combined transmembrane helix and SPase I processed Sec signal peptide prediction program (Kall et al., 2004). As laid out above, prokaryotic Sec substrates can contain signal peptides cleaved by either SPase I, SPase II, or SPase III, and in many prokaryotes structurally similar signal peptides can differentially target proteins to the Sec or Tat pathway (Bardy et al., 2003; Ng et al., 2007; Calo et al., 2010). Hence, to accurately predict the pathways employed to transport specific prokaryotic proteins, and their ultimate destinations, Phobius needs to be used in conjunction with programs that predict whether signal peptides contain a lipobox or a prepilin-peptidase processing site, as well as programs that can distinguish between Tat and Sec substrates.
TatFind and TatP are two programs that were specifically trained to identify Tat substrates (Rose et al., 2002; Bendtsen et al., 2005). Although structurally conserved, Tat signal peptide sequences vary somewhat across prokaryotic species. In some species, a small fraction of Tat substrates tolerate replacement of one of the arginines in the twin arginine motif with a lysine, while in others replacing either arginine disrupts transport (Berks et al., 2005). There is also some variability in the specificity of the amino acid residues allowed near the twin arginines, both among bacterial species and between prokaryotic domains (McDonough et al., 2008). However, TatFind, which was trained on a large set of predicted haloarchaeal Tat substrates, and TatP, have been validated by in vivo verification of a large number of substrates in archaea as well as bacteria, suggesting that Tat signal peptide sequences are reasonably similar across bacterial and archaeal species (Rose et al., 2002; Dilks et al., 2003, 2005; Gimenez et al., 2007; McDonough et al., 2008; Bagos et al., 2009; Storf et al., 2010; Uthandi et al., 2010). Consistent with this finding, Coulthurst et al. (2012) identified a Tat signal peptide recognition system in the euryarchaeon Archaeoglobus fulgidus resembling that of Escherichia coli.
Lipoprotein prediction programs such as LipoP (Juncker et al., 2003) and pred-lipo (Bagos et al., 2008), the latter being trained specifically on Gram-positive bacterial lipoproteins, were developed to identify bacterial Sec signal peptides containing SPase II processing sites. In silico and limited in vivo analyses suggest that the bacterial and archaeal lipoboxes are similar, and therefore, these programs can be used to identify archaeal lipoproteins (Storf et al., 2010). Using these programs in tandem gives the best results since although both programs identify most putative lipoproteins, each misses a subset. When used consecutively, TatFind and lipoprotein prediction programs can also predict Tat lipoproteins. However, the only prediction program trained on Tat substrates processed by SPase II is TatLipo, which was trained on haloarchaeal Tat substrates and is not yet fully tested on non-haloarchaeal proteins (Storf et al., 2010). Tat substrates not identified by LipoP or pred-lipo are assumed to be processed by SPase I.
Unlike the other types of signal peptides discussed here, signal peptides processed by SPase III show readily apparent differences in bacteria and archaea, requiring independent prediction programs be developed to specifically detect archaeal (FlaFind; Szabo et al., 2007b) and bacterial type IV pilin-like proteins (PilFind; Imam et al., 2011). In archaeal SPase III substrates, the most important residues for cleavage to occur are the amino acids at positions −2, −1, and +1, relative to the cleavage site. In particular, a positively charged amino acid residue at position −2 appears to be specific to archaeal SPase III recognition sites. Conversely, many archaeal SPase III substrates do not contain a glutamate residue at the +5 position, which appears to be a nearly universal feature of the signal peptides of type IV pilin-like proteins in bacteria (Imam et al., 2011). As noted above, Tat signal peptides containing SPase III processing sites have not been identified. Clearly, applying prediction programs in concert can clarify subcellular localization of processed substrates by ensuring more accurate predictions of the class of the signal peptide that each substrate contains.
When applied to the large number of genome sequences completed in recent years, the prediction tools discussed above have enabled us to confirm trends first observed several years ago, and have also helped identify patterns in the usage of secretion pathways and protein anchoring mechanisms. These patterns may have significant implications for the evolutionary history or relationships of the organisms in which they are observed.
Genomic analyses indicate that the Sec protein transport pathway is conserved, and very likely essential, in all organisms (Pohlschroder et al., 2005b). The requirement of the Sec pathway for protein transport across the membrane as well as for the insertion of membrane proteins, strongly suggests that it evolved early in the history of living organisms with the insertion of membrane proteins preceding the transport of proteins with a SPase I-cleavable signal peptide. The Tat pathway likely evolved later, rerouting Sec substrates to this pathway, hence the similarities of their signal peptides. The fact that a highly diverse set of bacterial and archaeal species use the Tat pathway to transport secreted proteins suggests that this pathway was also present before the divergence of bacteria and archaea from the common ancestor (Dilks et al., 2003; Storf et al., 2010). The lack of this pathway in most eukaryotes is likely due to the transport of proteins into the lumen of the ER, an ATP-containing compartment that lacks many of the challenges of the extracytoplasmic environment prokaryotic secreted proteins face. Most prokaryotic species are predicted to use the Sec pathway to transport a large majority of their secreted proteins (Dilks et al., 2003; Storf et al., 2010). Interestingly, in vivo studies strongly support in silico analyses of nearly 20 haloarchaeal genomes showing that the species of this euryarchaeal class transport nearly half of their secreted proteins through the Tat pathway (Bolhuis, 2002; Rose et al., 2002; Dilks et al., 2005; Falb et al., 2005; Gimenez et al., 2007; Storf et al., 2010). Similar analyses performed on over a 100 non-haloarchaeal genomes indicated that the use of the Tat pathway to transport such a large percentage of secreted proteins may be unique to the halophilic archaea (Dilks et al., 2003). Perhaps, the extensive use of the Tat pathway is an adaptation to the high salt environments that haloarchaea inhabit. Highly negatively charged surfaces, providing them with a hydration shell, are a prerequisite for the stability of proteins exposed to high salt concentration and hence require rapid, efficient protein folding, a process that may be best accomplished in the cytoplasm, which contains ATP-dependent chaperones (Frolow et al., 1996; Kennedy et al., 2001). It should be noted that halophilic bacteria appear to use the Tat pathway for transporting only a very limited number of substrates. However, although they face the same high salt concentration challenges as haloarchaea, as noted above, halophilic bacteria express a Sec transport designated ATPase (SecA), possibly resulting in fundamental differences in Sec transport in these two prokaryotic domains that might have lead to distinctly different adaptations to high salt (Mongodin et al., 2005).
Analysis of the signal peptidase processing sites also suggest that the vast majority of haloarchaeal Tat substrates are lipoproteins, possibly because lipid anchoring avoids the need to insert a hydrophobic stretch of the secreted protein into the cytoplasmic membrane (Storf et al., 2010). For haloarchaea, transporting a carboxy-terminal hydrophobic anchor across a membrane and inserting it into that membrane in an unfolded conformation is problematic since the solubility of hydrophobic amino acids is low in a high salt environment – it is unlikely that the Tat pathway is able to laterally insert the transmembrane segments of their substrates as is observed for Sec substrates. Under these conditions, using chaperones to maintain the solubility of the long hydrophobic stretch of amino acids might be counterproductive since hydrophobic interactions between the carboxy-terminal anchor and the chaperones may be enhanced, ultimately resulting in the chaperones becoming affixed to the secreted substrate. The observation that only a small subset of non-haloarchaeal Tat substrates contain a predicted lipobox also supports the notion that the extensive use of lipid anchors by Tat substrates is likely an adaptation to high salt. Conversely, similar portions of non-haloarchaeal and haloarchaeal Sec substrates appear to have signal peptides that contain lipoboxes.
The genomes of most euryarchaeal species encode proteins having lipoboxes, again indicating the presence of lipid anchoring in the common ancestor of bacteria and archaea. The absence of archaeal homologs of the bacterial lipoprotein biosynthesis components may be due to the distinct cytoplasmic membrane compositions of bacteria and archaea. Interestingly, to date, in silico analyses of crenarchaeal genomes suggest that the crenarchaea lack lipoproteins (Storf et al., 2010). Perhaps crenarchaeal secreted proteins, compared to their euryarchaeal counterparts, are more frequently anchored to the membrane via amino- or carboxy-terminal transmembrane segments, or perhaps crenarchaea use as yet unidentified mechanisms to anchor or retain proteins, possibly by forming a periplasmic space (Ellen et al., 2010a). This is consistent with the fact that crenarchaea also appear to lack both archaeosortase homologs and substrates that contain the carboxy-terminal motifs targeted by an archaeosortase (Haft et al., 2012).
Conversely, species in nearly all archaeal kingdoms express type IV pilin-like proteins, although not every species within a given kingdom necessarily does, strongly suggesting that type IV pili are ancient surface structures (Szabo et al., 2007b; Pohlschroder et al., 2011). Interestingly, even the extraordinarily small genome of the symbiont Nanoarchaeum equitans, a species that cannot even produce its own lipids, encodes homologs of type IV pilus-biosynthesis pathway as well as FlaFind positive substrates (Huber et al., 2002; Szabo et al., 2007b). It is possible that these proteins play an integral role in their attachment to their symbiotic host I. hospitalis (Moissl-Eichinger and Huber, 2011). The lack of both lipobox-containing proteins and type IV pilins in eukaryotes is not surprising, again because proteins are transported into the ER-lumen, rather than directly across the cytoplasmic membrane.
To date, archaea adapted to extreme conditions other than high salt, such as high temperature or low pH, have not shown a preference for either of the known protein transport pathways, nor do they appear to have a preferred mechanism for anchoring secreted proteins to the cell surface (Storf et al., 2010).
The combination of in vivo and in vitro experiments with in silico predictions has greatly extended our understanding of the diversity of archaeal protein secretion and the localization of Sec and Tat substrates. However, additional strategies may have evolved for the export of proteins and their targeting. One possible approach toward the exploration of alternative secretion routes involves the proteomics of secreted proteins. Proteomic studies aim to generate a global view of all proteins present in a sample, and because quantitative methods are available, the relative expression of certain gene products under a variety of conditions can also be evaluated.
In recent years, there has been some progress on the proteomics of archaeal extracellular, surface exposed and membrane proteins (Chong and Wright, 2005; Saunders et al., 2006; Burghardt et al., 2008; Ellen et al., 2009, 2010b; Williams et al., 2010a,b). One typical observation was that proteins expected to be cytoplasmic, based on functional homology to cytoplasmic proteins, or the lack of a predicted Tat or Sec signal peptide are frequently recovered from the extracellular fractions. These proteins may originate from lysed cells, but in some cases could indicate alternative secretion strategies. Difficulties in determining which secreted proteins are transported through as yet undefined secretion pathways include an inability to identify motifs that target proteins to these pathways. Furthermore, although many predicted proteins are homologous to proteins having known functions, making the qualifier “hypothetical” obsolete, their functions have not yet been characterized. While computational analyses do allow us to make predictions, based on homology, concerning the general functions of these proteins, thereby indicating new directions for further investigations of their physiological function, homologous proteins sometimes have very different functions and hence predictions based on homology may be misleading with regard to subcellular localization. Most significantly though, the majority of predicted proteins show no homology to known proteins, making investigations of their functions and subcellular localization much more complicated.
Vesicles may be one possible mechanism for transporting proteins across the hydrophobic membrane in a Sec and Tat-independent manner. Extracellular vesicles have been detected in various archaea, including Thermococcales and Sulfolobus species as well as I. hospitalis (Prangishvili et al., 2000; Reysenbach et al., 2006; Soler et al., 2008; Ellen et al., 2009). Proteomic analysis of Sulfolobus derived S-layer coated vesicles revealed the presence of homologs of the eukaryotic endosomal sorting complex (ESCRT-III) proteins (Lindas et al., 2008; Samson et al., 2008). These proteins play an important role in cell division of Sulfolobus (Samson and Bell, 2011) and may thus have an additional role in eukaryotic-like vesicle budding and release. Unfortunately, the physiological roles of extracellular vesicles in archaea are not yet clear, although sulfolobicins are associated with these structures (Prangishvili et al., 2000; Ellen et al., 2011). Interestingly, Sulfolobus turreted icosahedral virus (STIV) particles released from the host cells contain ESCRT-III protein homologs, and the expression of these proteins in the host cell is up-regulated during viral infection. It is tempting to speculate that STIV hijacks the cellular vesicle release mechanism for its own biogenesis (Snyder and Young, 2011; Maaty et al., 2012).
The vesicles present in the periplasmic space of I. hospitalis bud off the inner membrane and probably fuse with the outer membrane (Figure 4A). Whether this is a strategy by which I. hospitalis outer membrane proteins are transported across the periplasmic space is not yet known. In fact, I. hospitalis requires the transport of many outer membrane proteins as this crenarchaeon is the only reported organism with an energized outer membrane and ATP synthesis within the periplasmic space (Kuper et al., 2010; Figure 4A). The presence of the outer membrane of the only diderm archaeon identified to date raises numerous intriguing questions such as why the type IV Iho670 pili required for I. hospitalis surface adhesion are secreted across the outer membrane (Muller et al., 2009), or how secreted outer membrane proteins that are likely to be required for cell–cell adhesion with the I. hospitalis symbiont N. equitans are being targeted to and transported across this outer membrane (Junglas et al., 2008; Figure 4B).
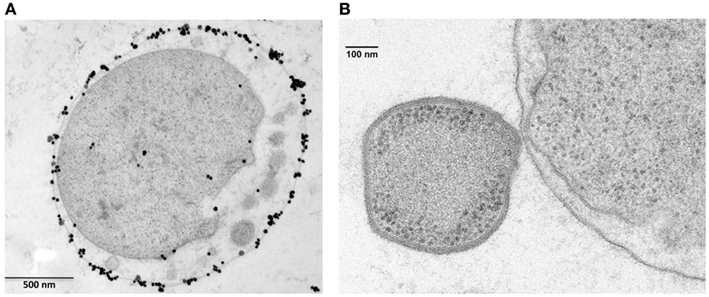
Figure 4. Ignicoccus hospitalis protein transport into and across the outer membrane. (A) Localization of A1AO ATP synthase on I. hospitalis outer membrane by EM of ultrathin sections and labeled with antibodies specifically raised against the purified 440-kDa ATPase complex (Kuper et al., 2010). Image courtesy of T. Heimerl, H. Huber, and R. Rachel, University of Regensburg, Germany. (B) Electron micrographs of I. hospitalis ultrathin sections. Outer membrane of I. hospitalis and the cell surface of N. equitans in direct contact (Junglas et al., 2008). Image courtesy of T. Heimerl, H. Huber, and R. Rachel, and image (B) courtesy of H. Huber and R. Rachel, University of Regensburg, Germany.
In conclusion, further experimental studies, including proteomic analyses, will lead to the improvement of existing prediction programs and may allow the identification of phyla-specific sequences. However, along with a better understanding of “classical” secretory routes via the Tat and Sec pathways, the recent development of several eury- and crenarchaeal models (Leigh et al., 2011) combined with in silico analyses, such as the recent comparative genomics that identified the putative archaeosortases (Haft et al., 2012), may lead to the discovery of novel protein transport and anchoring strategies.
Exciting new trends have emerged from recent analyses of archaeal genomes that are shaping studies designed to identify and characterize the secreted proteins and the mechanisms underlying the transport, processing, and modification of secreted proteins. The most promising of these approaches combine in vivo and in silico analyses. Finally, researchers have identified a plethora of prokaryotic proteins having useful, commercially valuable enzymatic activities. Many of these enzymes are secreted proteins. Knowledge about the secretion pathways as well as signal peptide processing and other post-translational modifications will be invaluable in designing strategies for efficiently producing these proteins.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Research at Microdish is in part supported by the EU Seventh Framework Large Scale Integrated Project “HotZyme”. Mechthild Pohlschroder was supported by a National Science Foundation grant MCB02-MT and by a National Aeronautics and Space Administration grant NNX10AR84G.
Ajon, M., Frols, S., Van Wolferen, M., Stoecker, K., Teichmann, D., Driessen, A. J., Grogan, D. W., Albers, S. V., and Schleper, C. (2011). UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol. Microbiol. 82, 807–817.
Albers, S. V., Konings, W. N., and Driessen, A. J. M. (1999). A unique short signal sequence in membrane-anchored proteins of archaea. Mol. Microbiol. 31, 1595–1596.
Albers, S. V., and Meyer, B. H. (2011). The archaeal cell envelope. Nat. Rev. Microbiol. 9, 414–426.
Albers, S. V., and Pohlschroder, M. (2009). Diversity of archaeal type IV pilin-like structures. Extremophiles 13, 403–410.
Albers, S. V., Szabo, Z., and Driessen, A. J. (2003). Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J. Bacteriol. 185, 3918–3925.
Arts, J., van Boxtel, R., Filloux, A., Tommassen, J., and Koster, M. (2007). Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J. Bacteriol. 189, 2069–2076.
Bagos, P. G., Tsirigos, K. D., Liakopoulos, T. D., and Hamodrakas, S. J. (2008). Prediction of lipoprotein signal peptides in Gram-positive bacteria with a hidden Markov model. J. Proteome Res. 7, 5082–5093.
Bagos, P. G., Tsirigos, K. D., Plessas, S. K., Liakopoulos, T. D., and Hamodrakas, S. J. (2009). Prediction of signal peptides in archaea. Protein Eng. Des. Sel. 1, 27–35.
Balestrieri, M., Gogliettino, M., Fiume, I., Pocsfalvi, G., Catara, G., Rossi, M., and Palmieri, G. (2011). Structural and functional insights into Aeropyrum pernix OppA, a member of a novel archaeal OppA subfamily. J. Bacteriol. 193, 620–630.
Bardy, S. L., Eichler, J., and Jarrell, K. F. (2003). Archaeal signal peptides – a comparative survey at the genome level. Protein Sci. 12, 1833–1843.
Bardy, S. L., and Jarrell, K. F. (2002). FlaK of the archaeon Methanococcus maripaludis possesses preflagellin peptidase activity. FEMS Microbiol. Lett. 208, 53–59.
Bautista, V., Esclapez, J., Perez-Pomares, F., Martinez-Espinosa, R. M., Camacho, M., and Bonete, M. J. (2012). Cyclodextrin glycosyltransferase: a key enzyme in the assimilation of starch by the halophilic archaeon Haloferax mediterranei. Extremophiles 16, 147–159.
Bendtsen, J. D., Nielsen, H., Widdick, D., Palmer, T., and Brunak, S. (2005). Prediction of twin-arginine signal peptides. BMC Bioinformatics 6, 167. doi:10.1186/1471-2105-6-167
Berks, B. C., Palmer, T., and Sargent, F. (2005). Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr. Opin. Microbiol. 8, 174–181.
Bolhuis, A. (2002). Protein transport in the halophilic archaeon Halobacterium sp. NRC-1: a major role for the twin-arginine translocation pathway? Microbiology 148, 3335–3346.
Bolhuis, H., Palm, P., Wende, A., Falb, M., Rampp, M., Rodriguez-Valera, F., Pfeiffer, F., and Oesterhelt, D. (2006). The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics 7, 169. doi:10.1186/1471-2164-7-169
Brockl, G., Behr, M., Fabry, S., Hensel, R., Kaudewitz, H., Biendl, E., and Konig, H. (1991). Analysis and nucleotide sequence of the genes encoding the surface-layer glycoproteins of the hyperthermophilic methanogens Methanothermus fervidus and Methanothermus sociabilis. Eur. J. Biochem. 199, 147–152.
Burghardt, T., Saller, M., Gurster, S., Muller, D., Meyer, C., Jahn, U., Hochmuth, E., Deutzmann, R., Siedler, F., Babinger, P., Wirth, R., Huber, H., and Rachel, R. (2008). Insight into the proteome of the hyperthermophilic crenarchaeon Ignicoccus hospitalis: the major cytosolic and membrane proteins. Arch. Microbiol. 190, 379–394.
Calo, D., and Eichler, J. (2011). Crossing the membrane in archaea, the third domain of life. Biochim. Biophys. Acta 1808, 885–891.
Calo, D., Kaminski, L., and Eichler, J. (2010). Protein glycosylation in archaea: sweet and extreme. Glycobiology 20, 1065–1076.
Chaban, B., Ng, S. Y. M., Kanbe, M., Saltzman, L., Nimmo, G., Aizawa, S. I., and Jarrell, K. F. (2007). Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol. Microbiol. 66, 596–609.
Cheung, J., Danna, K. J., O’Connor, E. M., Price, L. B., and Shand, R. F. (1997). Isolation, sequence, and expression of the gene encoding halocin H4, a bacteriocin from the halophilic archaeon Haloferax mediterranei R4. J. Bacteriol. 179, 548–551.
Chong, P. K., and Wright, P. C. (2005). Identification and characterization of the Sulfolobus solfataricus P2 proteome. J. Proteome Res. 4, 1789–1798.
Coulthurst, S. J., Dawson, A., Hunter, W. N., and Sargent, F. (2012). Conserved signal peptide recognition systems across the prokaryotic domains. Biochemistry 51, 1678–1686.
Dale, H., Angevine, C. M., and Krebs, M. P. (2000). Ordered membrane insertion of an archaeal opsin in vivo. Proc. Natl. Acad. Sci. U.S.A. 97, 7847–7852.
De Castro, R. E., Ruiz, D. M., Gimenez, M. I., Silveyra, M. X., Paggi, R. A., and Maupin-Furlow, J. A. (2008). Gene cloning and heterologous synthesis of a haloalkaliphilic extracellular protease of Natrialba magadii (Nep). Extremophiles 12, 677–687.
de Vos, W. M., Voorhorst, W. G. B., Dijkgraaf, M., Kluskens, L. D., Van Der Oost, J., and Siezen, R. J. (2001). Purification, characterization, and molecular modeling of pyrolysin and other extracellular thermostable serine proteases from hyperthermophilic microorganisms. Methods Enzymol. 330, 383–393.
Dilks, K., Gimenez, M. I., and Pohlschroder, M. (2005). Genetic and biochemical analysis of the twin-arginine translocation pathway in halophilic archaea. J. Bacteriol. 187, 8104–8113.
Dilks, K., Rose, R. W., Hartmann, E., and Pohlschroder, M. (2003). Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185, 1478–1483.
Driessen, A. J., and Nouwen, N. (2008). Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77, 643–667.
Egorova, K., and Antranikian, G. (2005). Industrial relevance of thermophilic archaea. Curr. Opin. Microbiol. 8, 649–655.
Eichler, J. (2002). Archaeal signal peptidases from the genus Thermoplasma: structural and mechanistic hybrids of the bacterial and eukaryal enzymes. J. Mol. Evol. 54, 411–415.
Eichler, J., and Adams, M. W. (2005). Posttranslational protein modification in archaea. Microbiol. Mol. Biol. Rev. 69, 393–425.
Elferink, M. G. L., Albers, S. V., Konings, W. N., and Driessen, A. J. M. (2001). Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol. Microbiol. 39, 1494–1503.
Ellen, A. F., Albers, S. V., and Driessen, A. J. (2010a). Comparative study of the extracellular proteome of Sulfolobus species reveals limited secretion. Extremophiles 14, 87–98.
Ellen, A. F., Zolghadr, B., Driessen, A. M., and Albers, S. V. (2010b). Shaping the archaeal cell envelope. Archaea 2010, 608243.
Ellen, A. F., Albers, S. V., Huibers, W., Pitcher, A., Hobel, C. F. V., Schwarz, H., Folea, M., Schouten, S., Boekema, E. J., Poolman, B., and Driessen, A. J. M. (2009). Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 13, 67–79.
Ellen, A. F., Rohulya, O. V., Fusetti, F., Wagner, M., Albers, S. V., and Driessen, A. J. (2011). The sulfolobicin genes of Sulfolobus acidocaldarius encode novel antimicrobial proteins. J. Bacteriol. 193, 4380–4387.
Engelhardt, H. (2007a). Are S-layers exoskeletons? The basic function of protein surface layers revisited. J. Struct. Biol. 160, 115–124.
Engelhardt, H. (2007b). Mechanism of osmoprotection by archaeal S-layers: a theoretical study. J. Struct. Biol. 160, 190–199.
Falb, M., Pfeiffer, F., Palm, P., Rodewald, K., Hickmann, V., Tittor, J., and Oesterhelt, D. (2005). Living with two extremes: conclusions from the genome sequence of Natronomonas pharaonis. Genome Res. 15, 1336–1343.
Francetic, O., Buddelmeijer, N., Lewenza, S., Kumamoto, C. A., and Pugsley, A. P. (2007). Signal recognition particle-dependent inner membrane targeting of the PulG Pseudopilin component of a type II secretion system. J. Bacteriol. 189, 1783–1793.
Frolow, F., Harel, M., Sussman, J. L., Mevarech, M., and Shoham, M. (1996). Insights into protein adaptation to a saturated salt environment from the crystal structure of a halophilic 2Fe-2S ferredoxin. Nat. Struct. Biol. 3, 452–458.
Frols, S., Ajon, M., Wagner, M., Teichmann, D., Zolghadr, B., Folea, M., Boekema, E. J., Driessen, A. J., Schleper, C., and Albers, S. V. (2008). UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol. Microbiol. 70, 938–952.
Ghosh, A., and Albers, S. V. (2011). Assembly and function of the archaeal flagellum. Biochem. Soc. Trans. 39, 64–69.
Gimenez, M. I., Dilks, K., and Pohlschroder, M. (2007). Haloferax volcanii twin-arginine translocation substates include secreted soluble, C-terminally anchored and lipoproteins. Mol. Microbiol. 66, 1597–1606.
Gogliettino, M., Balestrieri, M., Pocsfalvi, G., Fiume, I., Natale, L., Rossi, M., and Palmieri, G. (2010). A highly selective oligopeptide binding protein from the archaeon Sulfolobus solfataricus. J. Bacteriol. 192, 3123–3131.
Gohlke, U., Pullan, L., Mcdevitt, C. A., Porcelli, I., De Leeuw, E., Palmer, T., Saibil, H. R., and Berks, B. C. (2005). The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc. Natl. Acad. Sci. U.S.A. 102, 10482–10486.
Grudnik, P., Bange, G., and Sinning, I. (2009). Protein targeting by the signal recognition particle. Biol. Chem. 390, 775–782.
Haft, D. H., Payne, S. H., and Selengut, J. D. (2012). Archaeosortases and exosortases are widely distributed systems linking membrane transit with posttranslational modification. J. Bacteriol. 194, 36–48.
Haseltine, C., Hill, T., Montalvo-Rodriguez, R., Kemper, S. K., Shand, R. F., and Blum, P. (2001). Secreted euryarchaeal microhalocins kill hyperthermophilic crenarchaea. J. Bacteriol. 183, 287–291.
Henche, A. L., Koerdt, A., Ghosh, A., and Albers, S. V. (2012). Influence of cell surface structures on crenarchaeal biofilm formation using a thermostable green fluorescent protein. Environ. Microbiol. 14, 779–793.
Horlacher, R., Xavier, K. B., Santos, H., Diruggiero, J., Kossmann, M., and Boos, W. (1998). Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J. Bacteriol. 180, 680–689.
Huber, H., Hohn, M. J., Rachel, R., Fuchs, T., Wimmer, V. C., and Stetter, K. O. (2002). A new phylum of archaea represented by a nanosized hyperthermophilic symbiont. Nature 417, 63–67.
Hutcheon, G. W., Vasisht, N., and Bolhuis, A. (2005). Characterisation of a highly stable alpha-amylase from the halophilic archaeon Haloarcula hispanica. Extremophiles 9, 487–495.
Hutchings, M. I., Palmer, T., Harrington, D. J., and Sutcliffe, I. C. (2009). Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold ‘em, knowing when to fold ‘em. Trends Microbiol. 17, 13–21.
Imam, S., Chen, Z., Roos, D. S., and Pohlschroder, M. (2011). Identification of surprisingly diverse type IV pili, across a broad range of Gram-positive bacteria. PLoS ONE 6, e28919. doi:10.1371/journal.pone.0028919
Irihimovitch, V., and Eichler, J. (2003). Post-translational secretion of fusion proteins in the halophilic archaea Haloferax volcanii. J. Biol. Chem. 278, 12881–12887.
Jarrell, K. F., Jones, G. M., and Nair, D. B. (2010). Biosynthesis and role of N-linked glycosylation in cell surface structures of archaea with a focus on flagella and s layers. Int. J. Microbiol. 2010, 470138.
Jarrell, K. F., Stark, M., Nair, D. B., and Chong, J. P. (2011). Flagella and pili are both necessary for efficient attachment of Methanococcus maripaludis to surfaces. FEMS Microbiol. Lett. 319, 44–50.
Journet, L., Hughes, K. T., and Cornelis, G. R. (2005). Type III secretion: a secretory pathway serving both motility and virulence (review). Mol. Membr. Biol. 22, 41–50.
Juncker, A. S., Willenbrock, H., Von Heijne, G., Brunak, S., Nielsen, H., and Krogh, A. (2003). Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12, 1652–1662.
Junglas, B., Briegel, A., Burghardt, T., Walther, P., Wirth, R., Huber, H., and Rachel, R. (2008). Ignicoccus hospitalis and Nanoarchaeum equitans: ultrastructure, cell-cell interaction, and 3D reconstruction from serial sections of freeze-substituted cells and by electron cryotomography. Arch. Microbiol. 190, 395–408.
Kall, L., Krogh, A., and Sonnhammer, E. L. (2004). A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036.
Kamekura, M., Seno, Y., Holmes, M. L., and Dyallsmith, M. L. (1992). Molecular cloning and sequencing of the gene for halophilic alkaline serine protease (halolysin) from an unidentified halophilic strain (172P1) and expression of the gene in Haloferax volcanii. J. Bacteriol. 174, 736–742.
Kennedy, S. P., Ng, W. V., Salzberg, S. L., Hood, L., and Dassarma, S. (2001). Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 11, 1641–1650.
Kikuchi, A., Sagami, H., and Ogura, K. (1999). Evidence for covalent attachment of diphytanylglyceryl phosphate to the cell-surface glycoprotein of Halobacterium halobium. J. Biol. Chem. 274, 18011–18016.
Kobayashi, T., Kanai, H., Aono, R., Horikoshi, K., and Kudo, T. (1994). Cloning, expression, and nucleotide sequence of the α-amylase gene from the haloalkaliphilic archaeon Natronococcus sp. strain Ah-36. J. Bacteriol. 176, 5131–5134.
Kokoeva, M. V., Storch, K. F., Klein, C., and Oesterhelt, D. (2002). A novel mode of sensory transduction in archaea: binding protein-mediated chemotaxis towards osmoprotectants and amino acids. EMBO J. 21, 2312–2322.
Konrad, Z., and Eichler, J. (2002). Lipid modification of proteins in archaea: attachment of a mevalonic acid-based lipid moiety to the surface-layer glycoprotein of Haloferax vokanii follows protein translocation. Biochem. J. 366, 959–964.
Krupovic, M., Peixeiro, N., Bettstetter, M., Rachel, R., and Prangishvili, D. (2012). Archaeal tetrathionate hydrolase goes viral: secretion of a sulfur metabolism enzyme in the form of virus-like particles. Appl. Environ. Microbiol. doi: 10.1128/AEM.01186-12. [Epub ahead of print].
Kuper, U., Meyer, C., Muller, V., Rachel, R., and Huber, H. (2010). Energized outer membrane and spatial separation of metabolic processes in the hyperthermophilic archaeon Ignicoccus hospitalis. Proc. Natl. Acad. Sci. U.S.A. 107, 3152–3156.
Kwan, D., and Bolhuis, A. (2010). Analysis of the twin-arginine motif of a haloarchaeal Tat substrate. FEMS Microbiol. Lett. 308, 138–143.
Lassak, K., Neiner, T., Ghosh, A., Klingl, A., Wirth, R., and Albers, S. V. (2012). Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 83, 110–124.
Leake, M. C., Greene, N. P., Godun, R. M., Granjon, T., Buchanan, G., Chen, S., Berry, R. M., Palmer, T., and Berks, B. C. (2008). Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc. Natl. Acad. Sci. U.S.A. 105, 15376–15381.
Leigh, J. A., Albers, S. V., Atomi, H., and Allers, T. (2011). Model organisms for genetics in the domain archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol. Rev. 35, 577–608.
Lindas, A. C., Karlsson, E. A., Lindgren, M. T., Ettema, T. J., and Bernander, R. (2008). A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. U.S.A. 105, 18942–18946.
Maaty, W. S., Selvig, K., Ryder, S., Tarlykov, P., Hilmer, J. K., Heinemann, J., Steffens, J., Snyder, J. C., Ortmann, A. C., Movahed, N., Spicka, K., Chetia, L., Grieco, P. A., Dratz, E. A., Douglas, T., Young, M. J., and Bothner, B. (2012). Proteomic analysis of Sulfolobus solfataricus during Sulfolobus turreted icosahedral virus infection. J. Proteome Res. 11, 1420–1432.
Mai, B., Frey, G., Swanson, R. V., Mathur, E. J., and Stetter, K. O. (1998). Molecular cloning and functional expression of a protein-serine/threonine phosphatase from the hyperthermophilic archaeon Pyrodictium abyssi TAG11. J. Bacteriol. 180, 4030–4035.
Mattar, S., Scharf, B., Kent, S. B., Rodewald, K., Oesterhelt, D., and Engelhard, M. (1994). The primary structure of halocyanin, an archaeal blue copper protein, predicts a lipid anchor for membrane fixation. J. Biol. Chem. 269, 14939–14945.
Mayr, J., Lupas, A., Kellermann, J., Eckerskorn, C., Baumeister, W., and Peters, J. (1996). A hyperthermostable protease of the subtilisin family bound to the surface layer of the Archaeon Staphylothermus marinus. Curr. Biol. 6, 739–749.
McDonough, J. A., Mccann, J. R., Tekippe, E. M., Silverman, J. S., Rigel, N. W., and Braunstein, M. (2008). Identification of functional Tat signal sequences in Mycobacterium tuberculosis proteins. J. Bacteriol. 190, 6428–6438.
Mescher, M. F., Strominger, J. L., and Watson, S. W. (1974). Protein and carbohydrate composition of the cell envelope of Halobacterium salinarium. J. Bacteriol. 120, 945–954.
Meyer, B. H., Zolghadr, B., Peyfoon, E., Pabst, M., Panico, M., Morris, H. R., Haslam, S. M., Messner, P., Schaffer, C., Dell, A., and Albers, S. V. (2011). Sulfoquinovose synthase – an important enzyme in the N-glycosylation pathway of Sulfolobus acidocaldarius. Mol. Microbiol. 82, 1150–1163.
Moissl, C., Rachel, R., Briegel, A., Engelhardt, H., and Huber, R. (2005). The unique structure of archaeal “hami,” highly complex cell appendages with nano-grappling hooks. Mol. Microbiol. 56, 361–370.
Moissl-Eichinger, C., and Huber, H. (2011). Archaeal symbionts and parasites. Curr. Opin. Microbiol. 14, 364–370.
Mongodin, E. F., Nelson, K. E., Daugherty, S., Deboy, R. T., Wister, J., Khouri, H., Weidman, J., Walsh, D. A., Papke, R. T., Sanchez Perez, G., Sharma, A. K., Nesbo, C. L., Macleod, D., Bapteste, E., Doolittle, W. F., Charlebois, R. L., Legault, B., and Rodriguez-Valera, F. (2005). The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl. Acad. Sci. U.S.A. 102, 18147–18152.
Moracci, M., Cobucci-Ponzano, B., Perugino, G., and Rossi, M. (2007). “Biotechnology,” in Archaea: Molecular and Cellular Biology, ed. R. Cavicchioli (Washington: ASM Press), 478–495.
Mori, T., Ishitani, R., Tsukazaki, T., Nureki, O., and Sugita, Y. (2010). Molecular mechanisms underlying the early stage of protein translocation through the Sec translocon. Biochemistry 49, 945–950.
Muller, D. W., Meyer, C., Gurster, S., Kuper, U., Huber, H., Rachel, R., Wanner, G., Wirth, R., and Bellack, A. (2009). The Iho670 fibers of Ignicoccus hospitalis: a new type of archaeal cell surface appendage. J. Bacteriol. 191, 6465–6468.
Natale, P., Bruser, T., and Driessen, A. J. (2008). Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane – distinct translocases and mechanisms. Biochim. Biophys. Acta 1778, 1735–1756.
Nather, D. J., and Rachel, R. (2004). The outer membrane of the hyperthermophilic archaeon Ignicoccus: dynamics, ultrastructure and composition. Biochem. Soc. Trans. 32, 199–203.
Nather, D. J., Rachel, R., Wanner, G., and Wirth, R. (2006). Flagella of Pyrococcus furiosus: multifunctional organelles, made for swimming, adhesion to various surfaces, and cell-cell contacts. J. Bacteriol. 188, 6915–6923.
Ng, S. Y., Chaban, B., Vandyke, D. J., and Jarrell, K. F. (2007). Archaeal signal peptidases. Microbiology 153, 305–314.
Ng, S. Y., Vandyke, D. J., Chaban, B., Wu, J., Nosaka, Y., Aizawa, S., and Jarrell, K. F. (2009). Different minimal signal peptide lengths recognized by the archaeal prepilin-like peptidases FlaK and PibD. J. Bacteriol. 191, 6732–6740.
Ng, S. Y., Wu, J., Nair, D. B., Logan, S. M., Robotham, A., Tessier, L., Kelly, J. F., Uchida, K., Aizawa, S., and Jarrell, K. F. (2011). Genetic and mass spectrometry analyses of the unusual type IV-like pili of the archaeon Methanococcus maripaludis. J. Bacteriol. 193, 804–814.
O’Connor, E. M., and Shand, R. F. (2002). Halocins and sulfolobicins: the emerging story of archaeal protein and peptide antibiotics. J. Ind. Microbiol. Biotechnol. 28, 23–31.
Okuda, S., and Tokuda, H. (2011). Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65, 239–259.
Ortenberg, R., and Mevarech, M. (2000). Evidence for post-translational membrane insertion of the integral membrane protein bacterioopsin expressed in the heterologous halophilic archaeon Haloferax volcanii. J. Biol. Chem. 275, 22839–22846.
Paetzel, M., Karla, A., Strynadka, N. C. J., and Dalbey, R. E. (2002). Signal peptidases. Chem. Rev. 102, 4549–4580.
Peyfoon, E., Meyer, B., Hitchen, P. G., Panico, M., Morris, H. R., Haslam, S. M., Albers, S. V., and Dell, A. (2010). The S-layer glycoprotein of the crenarchaeote Sulfolobus acidocaldarius is glycosylated at multiple sites with chitobiose-linked N-glycans. Archaea. PMID: 20936123.
Pohlschroder, M., Ghosh, A., Tripepi, M., and Albers, S. V. (2011). Archaeal type IV pilus-like structures – evolutionarily conserved prokaryotic surface organelles. Curr. Opin. Microbiol. 14, 357–363.
Pohlschroder, M., Gimenez, M. I., and Jarrell, K. F. (2005a). Protein transport in archaea: Sec and twin arginine translocation pathways. Curr. Opin. Microbiol. 8, 713–719.
Pohlschroder, M., Hartmann, E., Hand, N. J., Dilks, K., and Haddad, A. (2005b). Diversity and evolution of protein translocation. Annu. Rev. Microbiol. 59, 91–111.
Prangishvili, D., Holz, I., Stieger, E., Nickell, S., Kristjansson, J. K., and Zillig, W. (2000). Sulfolobicins, specific proteinaceous toxins produced by strains of the extremely thermophilic archaeal genus Sulfolobus. J. Bacteriol. 182, 2985–2988.
Price, L. B., and Shand, R. F. (2000). Halocin S8: a 36-amino-acid microhalocin from the haloarchaeal strain S8a. J. Bacteriol. 182, 4951–4958.
Protze, J., Muller, F., Lauber, K., Nass, B., Mentele, R., Lottspeich, F., and Kletzin, A. (2011). An extracellular tetrathionate hydrolase from the thermoacidophilic archaeon Acidianus ambivalens with an activity optimum at pH 1. Front. Microbiol. 2:68. doi:10.3389/fmicb.2011.00068
Reysenbach, A. L., Liu, Y. T., Banta, A. B., Beveridge, T. J., Kirshtein, J. D., Schouten, S., Tivey, M. K., Von Damm, K. L., and Voytek, M. A. (2006). A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature 442, 444–447.
Richardson, T. H., Tan, X. Q., Frey, G., Callen, W., Cabell, M., Lam, D., Macomber, J., Short, J. M., Robertson, D. E., and Miller, C. (2002). A novel, high performance enzyme for starch liquefaction – discovery and optimization of a low pH, thermostable α-amylase. J. Biol. Chem. 277, 26501–26507.
Robinson, C., Matos, C. F., Beck, D., Ren, C., Lawrence, J., Vasisht, N., and Mendel, S. (2011). Transport and proofreading of proteins by the twin-arginine translocation (Tat) system in bacteria. Biochim. Biophys. Acta 1808, 876–884.
Rose, R. W., Bruser, T., Kissinger, J. C., and Pohlschroder, M. (2002). Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45, 943–950.
Rudolph, C., Wanner, G., and Huber, R. (2001). Natural communities of novel archaea and bacteria growing in cold sulfurous springs with a string-of-pearls-like morphology. Appl. Environ. Microbiol. 67, 2336–2344.
Samson, R. Y., and Bell, S. D. (2011). Cell cycles and cell division in the archaea. Curr. Opin. Microbiol. 14, 350–356.
Samson, R. Y., Obita, T., Freund, S. M., Williams, R. L., and Bell, S. D. (2008). A role for the ESCRT system in cell division in archaea. Science 322, 1710–1713.
Sankaran, K., and Wu, H. C. (1994). Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269, 19701–19706.
Saunders, N. F. W., Ng, C., Raftery, M., Guilhaus, M., Goodchild, A., and Cavicchioli, R. (2006). Proteomic and computational analysis of secreted proteins with type 1 signal peptides from the antarctic archaeon Methanococcoides burtonii. J. Proteome Res. 5, 2457–2464.
Shi, W. L., Tang, X. F., Huang, Y. P., Gan, F., Tang, B., and Shen, P. (2006). An extracellular halophilic protease SptA from a halophilic archaeon Natrinema sp J7: gene cloning, expression and characterization. Extremophiles 10, 599–606.
Snyder, J. C., and Young, M. J. (2011). Potential role of cellular ESCRT proteins in the STIV life cycle. Biochem. Soc. Trans. 39, 107–110.
Soler, N., Marguet, E., Verbavatz, J. M., and Forterre, P. (2008). Virus-like vesicles and extracellular DNA produced by hyperthermophilic archaea of the order Thermococcales. Res. Microbiol. 159, 390–399.
Spirig, T., Weiner, E. M., and Clubb, R. T. (2011). Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 82, 1044–1059.
Storf, S., Pfeiffer, F., Dilks, K., Chen, Z. Q., Imam, S., and Pohlschroder, M. (2010). Mutational and bioinformatic analysis of haloarchaeal lipobox-containing proteins. Archaea 2010, 11.
Sublimi Saponetti, M., Bobba, F., Salerno, G., Scarfato, A., Corcelli, A., and Cucolo, A. (2011). Morphological and structural aspects of the extremely halophilic archaeon Haloquadratum walsbyi. PLoS ONE 6, e18653. doi:10.1371/journal.pone.0018653
Sumper, M., Berg, E., Mengele, R., and Strobel, I. (1990). Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J. Bacteriol. 172, 7111–7118.
Sun, C., Li, Y., Mei, S., Lu, Q., Zhou, L., and Xiang, H. (2005). A single gene directs both production and immunity of halocin C8 in a haloarchaeal strain AS7092. Mol. Microbiol. 57, 537–549.
Szabo, Z., Sani, M., Groeneveld, M., Zolghadr, B., Schelert, J., Albers, S. V., Blum, P., Boekema, E. J., and Driessen, A. J. (2007a). Flagellar motility and structure in the hyperthermoacidophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 189, 4305–4309.
Szabo, Z., Stahl, A. O., Albers, S. V., Kissinger, J. C., Driessen, A. J., and Pohlschroder, M. (2007b). Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J. Bacteriol. 189, 772–778.
Thoma, C., Frank, M., Rachel, R., Schmid, S., Nather, D., Wanner, G., and Wirth, R. (2008). The Mth60 fimbriae of Methanothermobacter thermoautotrophicus are functional adhesins. Environ. Microbiol. 10, 2785–2795.
Thompson, B. J., Widdick, D. A., Hicks, M. G., Chandra, G., Sutcliffe, I. C., Palmer, T., and Hutchings, M. I. (2010). Investigating lipoprotein biogenesis and function in the model Gram-positive bacterium Streptomyces coelicolor. Mol. Microbiol. 77, 943–957.
Tripepi, M., Imam, S., and Pohlschroder, M. (2010). Haloferax volcanii flagella are required for motility but are not involved in PibD-dependent surface adhesion. J. Bacteriol. 192, 3093–3102.
Uthandi, S., Saad, B., Humbard, M. A., and Maupin-Furlow, J. A. (2010). LccA, an archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax volcanii. Appl. Environ. Microbiol. 76, 733–743.
Van den Berg, B., Clemons, W. M. Jr., Collinson, I., Modis, Y., Hartmann, E., Harrison, S. C., and Rapoport, T. A. (2004). X-ray structure of a protein-conducting channel. Nature 427, 36–44.
VanDyke, D. J., Wu, J., Ng, S. Y., Kanbe, M., Chaban, B., Aizawa, S., and Jarrell, K. F. (2008). Identification of a putative acetyltransferase gene, MMP0350, which affects proper assembly of both flagella and pili in the archaeon Methanococcus maripaludis. J. Bacteriol. 190, 5300–5307.
Wang, Y. A., Yu, X., Ng, S. Y., Jarrell, K. F., and Egelman, E. H. (2008). The structure of an archaeal pilus. J. Mol. Biol. 381, 456–466.
Wende, A., Johansson, P., Vollrath, R., Dyall-Smith, M., Oesterhelt, D., and Grininger, M. (2010). Structural and biochemical characterization of a halophilic archaeal alkaline phosphatase. J. Mol. Biol. 400, 52–62.
Williams, T. J., Burg, D. W., Ertan, H., Raftery, M. J., Poljak, A., Guilhaus, M., and Cavicchioli, R. (2010a). Global proteomic analysis of the insoluble, soluble, and supernatant fractions of the psychrophilic archaeon Methanococcoides burtonii. Part II: the effect of different methylated growth substrates. J. Proteome Res. 9, 653–663.
Williams, T. J., Burg, D. W., Raftery, M. J., Poljak, A., Guilhaus, M., Pilak, O., and Cavicchioli, R. (2010b). Global proteomic analysis of the insoluble, soluble, and supernatant fractions of the psychrophilic archaeon Methanococcoides burtonii. Part I: the effect of growth temperature. J. Proteome Res. 9, 640–652.
Worthington, P., Hoang, V., Perez-Pomares, F., and Blum, P. (2003). Targeted disruption of the alpha-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataficus. J. Bacteriol. 185, 482–488.
Xu, Z., Du, X., Li, T., Gan, F., Tang, B., and Tang, X.-F. (2011). Functional insight into the C-terminal extension of halolysin SptA from haloarchaeon Natrinema sp J7. PLoS ONE 6, e23562. doi:10.1371/journal.pone.0023562
Yuan, J., Zweers, J. C., Van Dijl, J. M., and Dalbey, R. E. (2010). Protein transport across and into cell membranes in bacteria and archaea. Cell. Mol. Life Sci. 67, 179–199.
Zimmermann, R., Eyrisch, S., Ahmad, M., and Helms, V. (2011). Protein translocation across the ER membrane. Biochim. Biophys. Acta 1808, 912–924.
Zolghadr, B., Klingl, A., Koerdt, A., Driessen, A. J., Rachel, R., and Albers, S. V. (2010). Appendage-mediated surface adherence of Sulfolobus solfataricus. J. Bacteriol. 192, 104–110.
Keywords: Tat transport, Sec transport, protein transport, archaea, lipoprotein, pili, cell surface structures, archaeosortase
Citation: Szabo Z and Pohlschroder M (2012) Diversity and subcellular distribution of archaeal secreted proteins. Front. Microbio. 3:207. doi: 10.3389/fmicb.2012.00207
Received: 16 March 2012; Accepted: 21 May 2012;
Published online: 02 July 2012.
Edited by:
Frank T. Robb, University of California, USAReviewed by:
Thijs Ettema, Uppsala University, SwedenCopyright: © 2012 Szabo and Pohlschroder. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Mechthild Pohlschroder, Department of Biology, University of Pennsylvania, 201 Leidy Laboratories, Philadelphia, PA 19104, USA. e-mail:cG9obHNjaHJAc2FzLnVwZW5uLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.