
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 28 May 2012
Sec. Virology
Volume 3 - 2012 | https://doi.org/10.3389/fmicb.2012.00181
This article is part of the Research Topic Global host proteomic responses to virus infection View all 11 articles
Over the years, a vast array of information concerning the interactions of viruses with their hosts has been collected. However, recent advances in proteomics and other system biology techniques suggest these interactions are far more complex than anticipated. One particularly interesting and novel aspect is the analysis of cellular proteins incorporated into mature virions. Though sometimes considered purification contaminants in the past, their repeated detection by different laboratories suggests that a number of these proteins are bona fide viral components, some of which likely contribute to the viral life cycles. The present mini review focuses on cellular proteins detected in herpesviruses. It highlights the common cellular functions of these proteins, their potential implications for host–pathogen interactions, discusses technical limitations, the need for complementing methods and probes potential future research avenues.
Over the last decades, many host–pathogen interactions have been characterized using genetics, biochemical, and microscopy approaches. These discoveries relied on mutants, pharmacological reagents, immunoprecipitations, immunofluorescence, electron microscopy, cell fractionation, and Western blotting to name a few of the methods employed. These approaches provided much precious information but, given the typical focus of these approaches on individual molecules, likely only revealed a small portion of the proteins involved. Other methods such as high throughput two-hybrid and genetic screens, nucleic acid arrays, RNA interference, and proteomics are now proving essential tools to tackle the complexity of these interactions. The main advantages of mass spectrometry, for instance, are that it is a fast, sensitive and potentially a quantitative approach to identify putative novel players, particularly when coupled to efficient purification schemes. Already, proteomics revealed how viruses modulate the expression of host proteins (Rassmann et al., 2006; Sun et al., 2008; Tong et al., 2008; Antrobus et al., 2009; Pastorino et al., 2009; Thanthrige-Don et al., 2009; Zandi et al., 2009; Zhang et al., 2009, 2010; Coombs et al., 2010; Emmott et al., 2010; Lu et al., 2010, 2012; Munday et al., 2010; Bartel et al., 2011; Lietzen et al., 2011; Ramirez-Boo et al., 2011; Chou et al., 2012). A relatively new and interesting field is the characterization of host–pathogen interactions within mature purified virions. As reviewed on several occasions, several studies reported the presence of individual cellular proteins in viral particles (Bernhard et al., 2005; Maxwell and Frappier, 2007; Viswanathan and Fruh, 2007; Friedel and Haas, 2011; Zheng et al., 2011). This includes vaccinia virus (Krauss et al., 2002), influenza virus (Shaw et al., 2008), HIV (Gurer et al., 2002; Cantin et al., 2005; Ott, 2008), vesicular stomatitis virus (Moerdyk-Schauwecker et al., 2009), and several herpesviruses (see below). Though these cellular components have often been considered purification contaminants, the presence of similar proteins in both related and unrelated viruses suggests that some of them may be biologically relevant. The identification of virion-associated host proteins could thus lead to the discovery of novel therapeutic tools against viruses. The present review focuses on their identification and putative roles with respect to the proteomics of herpesviruses.
Thus far, the protein composition of eight different herpesvirions has been studied by mass spectrometry. These studies include the alphaherpesvirinae herpes simplex virus type 1 (HSV-1) and pseudorabies virus (PRV; Loret et al., 2008; Kramer et al., 2011), the betaherpesvirinae human and murine cytomegaloviruses (HCMV and MCMV, respectively; Kattenhorn et al., 2004; Varnum et al., 2004) and the gammaherpesvirinae Kaposi sarcoma herpesvirus (KSHV), gamma herpesvirus 68 (γHV68), Epstein–Barr virus (EBV), and Alcelaphine (Bortz et al., 2003; Johannsen et al., 2004; Bechtel et al., 2005; Zhu et al., 2005; Dry et al., 2008). Interestingly, host proteins were detected in all herpesvirions analyzed so far, as summarized in Table 1. For instance, our laboratory previously reported the protein composition of mature extracellular HSV-1 viral particles and identified as many as 49 cellular proteins (Loret et al., 2008). Similarly, studies focusing on PRV and EBV reported up to 48 and 43 cellular proteins, respectively (Johannsen et al., 2004; Kramer et al., 2011). Meanwhile, Varnum et al. (2004) found as many as 70 different host proteins in extracellular HCMV virions. While fewer cellular proteins were reported for other viral particles, it is clear that herpesviruses can potentially incorporate many proteins from its host. Moreover, of the 173 different proteins detected in herpesvirions, nine protein groups are present in at least four distinct herpesvirions. This includes 14-3-3, actin, annexins, cofilin, translation factors, GAPDH, heat shock proteins, pyruvate kinase M2, and various Rab GTPases. These results indicate that, first of all, it is common for herpesviruses to incorporate cellular proteins into their viral particles and, secondly, that different viruses share similar host proteins. Most excitingly, it also suggests that these host proteins may play common roles throughout the herpesviral family. This defines an interesting and novel set of host–pathogen interactions taking place within the virus itself, rather than the cell.
It is tempting to speculate that some viruses might have a higher capacity to steal cellular proteins because of their size and symmetry. Herpesviruses are indeed large viruses containing a layer called the tegument between their capsids and envelopes that could accommodate non-viral proteins. Though some host proteins may randomly be incorporated into virions, others may rather be selected to insure the optimal replication of the viruses that carry them.
Bioinformatics databases such as the KEGG, Gene Ontology, or DAVID are useful tools to get an overview of the functional interplay of proteins (Ashburner et al., 2000; Huang Da et al., 2009; Kanehisa et al., 2010). As pointed out by Friedel and Haas (2011), complex statistical tools are available to quantitatively evaluate the implication of proteins in various processes but these are beyond the scope of the present review. Here an analysis of the proteins identified in herpesvirions was instead performed with the Ingenuity Pathways Analysis database (Ingenuity® Systems), which contains all the known physical and functional links among cellular proteins and defines their most significant functions. That analysis indicates that many of the cellular proteins found in herpesvirions normally modulate trafficking, cell proliferation, cell death, cell migration, cell metabolism, or the cytoskeleton (Figure 1, upper pie chart). Though subtle differences between family members are noticeable when looking at individual viruses, similar functions are found (Figure 1, other charts). Immune-related molecules are also important constituents for several viruses, including HSV-1, KSHV, γHV68, Alcelaphine, and MCMV. Altogether, this provides an overall picture whereby herpesviruses, not surprisingly, modulate all of the important aspects of the cell but where each virus might deploy its energies slightly differently. The main surprise is that so many cellular proteins are detected within assembled viral particles, which raises an important question as to their biological significance and mode of action.
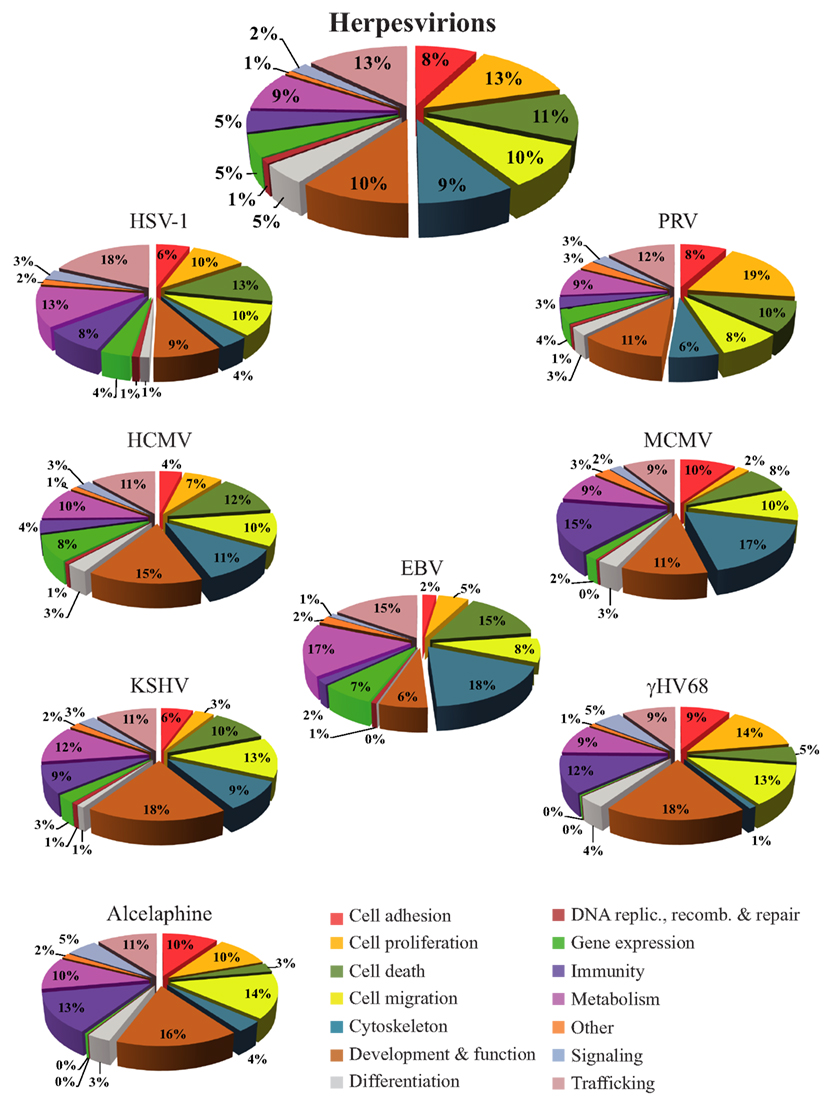
Figure 1. The proteins from Table 1 were analyzed with the Ingenuity database to define their putative functions in the context of an infection. To this end, the protein accession numbers (or GI numbers) were queried from the Ingenuity database. For the purpose of this figure, all known functions associated with these proteins were exported to Microsoft Excel and regrouped. In the top pie chart, the cellular proteins found in all the herpesvirions were analyzed collectively, while the other pie charts depict the host proteins incorporated into each virus. Since each protein can be associated with multiples functions in the database, the results of those analyses are expressed as relative values instead of raw numbers, which consequently exceeds the original number of proteins analyzed. The percentages therefore represent the number of proteins falling into a given category with the total of each pie chart being 100%. A graphical legend of the categories is provided at the bottom right corner of the figure.
The overall picture that several important cellular functions might be modulated by the host proteins incorporated into viral particles is intriguing. This clever strategy is consistent with the parasitic nature of all viruses, including herpesviruses, which would presumably gain some replication advantage from stealing cellular modulators rather than coding for them in their own genomes. The most critical question is the benefit for the viruses to incorporate these cellular proteins in their assembled particles, particularly since these proteins also exist in the cells. While this is open to discussion, one possibility is that some of the incorporated cellular proteins may be remnants of the final capsid envelopment process. Alternatively, this may allow the prompt action of some of these proteins immediately upon viral entry. This could jumpstart the expression and/or duplication of the viral genome, as it is the case for the herpesviral VHS, VP16, ICP0, and ICP4 proteins that are present in virions (Lam et al., 1996; Everett, 2000; Halford and Schaffer, 2001; Ellison et al., 2005; Hancock et al., 2006; Loret et al., 2008; Sarma et al., 2008; Loret and Lippe, 2012). Other early potential sites of action are the process of viral entry itself, intracellular capsid transport, import of the viral genome through the nuclear pore or immune modulation, all common steps among herpesviruses. Whatever the case might be, the question remains as to why the cellular pool of these proteins would not suffice. Several options may be considered. First, it may be that the virions incorporate specific isoforms, splice variants or post-translationally modified proteins that could have properties or functions distinct than their cellular counterparts. Second, the incorporation of a host protein from one cell type might permit the infection of a different cell type that does not express such protein. For example, alpha herpesviruses initially infect mucosal cells and could acquire host proteins that are beneficial to infect dormant neuronal cells. Finally, the host proteins might be in complex with viral proteins and it is those complexes that are active to promote the infection. These possibilities are of course speculative at this point and need to be explored.
One aspect where the incorporation of host proteins in mature virions might be beneficial is molecules involved in intracellular trafficking. Work by numerous laboratories demonstrated that the transport machinery used to move cellular proteins is also employed by viruses (Simons and Warren, 1984; Lodish et al., 2000; Sollner, 2004; Greber and Way, 2006; Mercer et al., 2010). This is essential for their proteins and particles to reach their final destination, for example, the site of viral replication, assembly, and/or envelopment. Along with SNARES proteins, Rab and Arf GTPases are master regulators of molecular trafficking throughout the cell (Sollner and Rothman, 1996; Zerial and McBride, 2001; Mizuno-Yamasaki et al., 2012). So far, VAMP3, a SNARE, was identified in PRV virions (Kramer et al., 2011) but it may only be a matter of time until other SNARES are discovered in other members of the herpes family. This is relevant as another SNARE was reported to facilitate the envelopment of MCMV capsids (Cepeda and Fraile-Ramos, 2011). In contrast, a great number of Rab proteins have been identified in herpesvirions, particularly HSV-1 and PRV (Table 1). One stimulating option is that these proteins regulate the displacement of viral capsids in the cell, which could justify their incorporation in the viral particles. As Rab and Arf proteins collectively modulate several intracellular transport steps within the cell, it is anticipated they may be involved in various stages of the infection. For instance, Rab1, which is present in HSV-1 extracellular virions (Loret et al., 2008), and Rab43 were recently demonstrated to modulate the final envelopment of the virus (Zenner et al., 2011). Similarly, Rab6, found in HSV-1 and PRV (Loret et al., 2008; Kramer et al., 2011), is also necessary for the efficient assembly of the related HCMV (Indran and Britt, 2011). It will now be of interest to determine if the virion-associated pool of these GTPases actively participates in the viral life cycle. Interestingly, several Rab proteins have been implicated in autophagosome formation and maturation (Chua et al., 2011). While it is difficult to consider how virion-incorporated Rab proteins play a role at that stage, they might rather be incorporated into the virions as a consequence of their involvement in autophagosome formation and concomitant viral envelopment. Given the vast impact of Rab proteins on the cell, it will be a major challenge to decipher all their roles in the life cycle of herpesviruses, particularly for the pool present in mature virions.
Molecular trafficking is not only dependent on SNARES, Rab, and Arf proteins, it is also intimately linked to the cytoskeleton. It is thus not surprising that herpesviruses devote some of their resources toward regulating this central cellular machinery. For instance, herpesviruses significantly reorganize both cellular and nuclear actin as well as microtubules (Norrild et al., 1986; Avitabile et al., 1995; Sharma-Walia et al., 2004; Simpson-Holley et al., 2005; De Regge et al., 2006; Saksena et al., 2006). They also travel along microtubules during both entry and egress and interact with several cellular molecular motors (Sodeik et al., 1997; Smith et al., 2001; Dohner et al., 2002; Marozin et al., 2004; Lee et al., 2006; Wolfstein et al., 2006; Radtke et al., 2010) as well as cortical and nuclear actin filaments (Forest et al., 2005; Feierbach et al., 2006; Roberts and Baines, 2011). Furthermore, some members incorporate in their viral particles tubulin or actin-related components (Table 1; Wong and Chen, 1998; Grunewald et al., 2003). Actin has been reported to compensate the loss of various viral tegument proteins in PRV (del Rio et al., 2005; Michael et al., 2006) and may thus act as an abundant filling agent, so its significance in herpesviral particles remains enigmatic. Similarly, the relevance of intermediate filament components vimentin and keratins in some herpes virions (Table 1) is difficult to assess given these filaments are not as well characterized as other cytoskeletal elements. It may nevertheless be important for herpesviruses, particularly since they are not all associated with the common skin or hair contaminants often detected in mass spectrometry (Hertel, 2011).
Viruses tend to monopolize for their own purpose their host expression apparatus, including protein translation (Bushell and Sarnow, 2002). For example, the prototypic HSV-1 ICP27 viral protein regulates all aspect of mRNAs including transcription, splicing, nuclear export, and translation for the benefit of the virus (Rice and Knipe, 1988; Sekulovich et al., 1988; Sandri-Goldin and Mendoza, 1992; Smith et al., 1992; Hardwicke and Sandri-Goldin, 1994; Hardy and Sandri-Goldin, 1994; Brown et al., 1995; Soliman et al., 1997; Chen et al., 2002; Lindberg and Kreivi, 2002; Ellison et al., 2005; Larralde et al., 2006; Fontaine-Rodriguez and Knipe, 2008). As these cellular functions are highly regulated, the inclusion of DDX3X, a multifunctional RNA helicase that also regulates transcription, nuclear export, and translation that is used by several viruses (Schroder, 2010, 2011) may be relevant. Its incorporation into mature virions could thus accelerate viral gene expression in the early stages of the infection. Similarly, the presence of translation initiation or elongation factors in virions (Table 1) may also jumpstart gene expression in favor of the viruses.
Interestingly, HSV-1 does not require cells to be in the S-phase and even arrests the cell cycle at the G1/S transition step (Shadan et al., 1994; Song et al., 2000), which partly explains why it can grow in non-dividing neurons. While the precise mechanism of this arrest is unclear, it is known that the viral ICP0 protein and the VP16 cellular partner HCF modulate the cell cycle (Hobbs and DeLuca, 1999; Lomonte and Everett, 1999; Piluso et al., 2002). Moreover, ICP0 interacts with the host cyclin D3 (Kawaguchi et al., 1997). However, it was recently reported that stress, rather than the cell cycle per se, may be a critical feature (Bringhurst and Schaffer, 2006). Clearly, the interaction of herpesviruses with the cell proliferation apparatus is complex and likely involves several host and viral proteins. Identifying novel players that might be incorporated into mature virions may thus be very useful to clarify this process.
An interesting scenario is the possible regulation of apoptosis by host proteins loaded onto viral particles. Apoptosis is regulated both negatively and positively by several viruses (Teodoro and Branton, 1997; Goodkin et al., 2004), presumably to insure their survival at the early stages of the infection but their efficient release later on. Conceptually, the presence of anti-apoptotic proteins in herpes particles might thus provide a mean to quickly evade death upon entry, while the presence of pro-apoptotic proteins on newly assembled/enveloped viral particles may trigger or stimulate their extracellular release. Only further work will resolve this open question.
Several factors generally contribute to variation among proteomic studies. Hence, the preparation of the samples (e.g., in-gel trypsin digestion versus liquid digestion and chromatography) may lead to the detection of different populations of tryptic peptides. Moreover, the sensitivity of the mass spectrometers and the abundance of the proteins in the samples also impact peptide detection. The relative abundance of a peptide is itself influenced by the complexity of the samples, where some proteins may evade identification. Finally, each protein differs in its properties (ionization, resolution in SDS-PAGE gels), which will be reflected in their detection. This includes SNARES, which are transmembrane proteins resistant to SDS extraction (Yang et al., 1999; Kubista et al., 2004). It is thus likely that some of the proteins in Table 1 are present in more viral particles than reported and that additional proteins are indeed incorporated in herpesvirions. More specific aspects regarding herpesviruses includes the purification schemes employed to enrich the viral particles, which will directly influence the purity of the samples and hence the potential detection of contaminants. One important caveat is that some host proteins may simply stick to the large viral particles. Another one is common contaminants such as some hair/skin associated keratins or as mentioned above actin, which may simply fill the virions. However, even potential contaminants cannot simply be discarded since actin and even some keratins may indeed participate in viral life cycles. Moreover, the relative abundance of all the cellular proteins within the cell is unknown, so it is not possible to rule out potential contaminants on the sole basis of abundance. It is thus critical to orthogonally validate all proteomics hits.
Various tools are available to define the biological relevance of host proteins identified in viral particles, including Western blotting, immuno-electron microscopy or functional screens. One powerful method is RNA interference. However, given the dual presence of the host proteins within the viral particles and the cell itself, this becomes a challenging task. RNA interference also has its own caveats (false positives and negatives). Another common step is the expression of dominant positive or negative mutants. In all cases, one major difficulty is that the host proteins may be essential for the cells and their depletion may lead to cytotoxicity, thus proper controls are needed. In addition, the host proteins might be essential for the virus within the cells but only accessory within the virions. Consequently, depletion of a protein may have limited impact on the virus since complemented by the other pool of that protein in the virus or the cell. Small reduction or stimulation in viral yields may thus result. It such cases, it may be necessary to produce the virus on cells that lack these proteins to see if this makes a difference. One should also consider animal models since tissue culture based screens may miss important players, for instance modulators of the immune system or virulence factors. Clearly, multiple experimental strategies are needed to ultimately insure the biological significance of the host proteins found in viral particles.
The identification and functions of host proteins in viral particles is an important step toward the elucidation of novel host-pathogen interactions. In the case of herpesvirions, this is well under way with eight different family members analyzed so far. One main aspect is to sort biologically relevant cellular proteins from sticky contaminants. The orthogonal validation of the host proteins found in herpesvirions using biologically relevant assays is thus critical. As pointed out above, it will necessary to analyze all these proteins in the background of two pools, one cellular and one virion-associated, which are likely to complement one another. An interesting possibility is that some isoforms or specific post-translationally modified host proteins may be loaded into the capsids. Thus a detailed analysis of the host proteins present in viral particles will be important and a potential way to distinguish them from their cell-associated counterparts. Another issue is the expected variation among cell types. In that respect, it would be most interesting to examine the cellular protein content of HSV-1 produced in neurons in opposition to the virions produced on other cell types. Finally, the mechanisms by which all these host proteins are recruited to the viral particles will also need to be explored. Thus the proteomics of viral particles is only the beginning of the adventure, which should prove most exciting yet challenging.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
I am indebted to the Canadian Institutes of Health Research (grant # MOP82921) for funding our proteomics research. I also wish to thank Kerstin Radtke for excellent suggestions and Daniel Henaff for critical reading of the manuscript.
Antrobus, R., Grant, K., Gangadharan, B., Chittenden, D., Everett, R. D., Zitzmann, N., and Boutell, C. (2009). Proteomic analysis of cells in the early stages of herpes simplex virus type-1 infection reveals widespread changes in the host cell proteome. Proteomics 9, 3913–3927.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J. C., Richardson, J. E., Ringwald, M., Rubin, G. M., and Sherlock, G. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29.
Avitabile, E., Di Gaeta, S., Torrisi, M. R., Ward, P. L., Roizman, B., and Campadelli-Fiume, G. (1995). Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol. 69, 7472–7482.
Bartel, S., Doellinger, J., Darsow, K., Bourquain, D., Buchholz, R., Nitsche, A., and Lange, H. A. (2011). Proteome analysis of vaccinia virus IHD-W-infected HEK 293 cells with 2-dimensional gel electrophoresis and MALDI-PSD-TOF MS of on solid phase support N-terminally sulfonated peptides. Virol. J. 8, 380.
Bechtel, J. T., Winant, R. C., and Ganem, D. (2005). Host and viral proteins in the virion of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 79, 4952–4964.
Bernhard, O. K., Diefenbach, R. J., and Cunningham, A. L. (2005). New insights into viral structure and virus-cell interactions through proteomics. Expert Rev. Proteomics 2, 577–588.
Bortz, E., Whitelegge, J. P., Jia, Q., Zhou, Z. H., Stewart, J. P., Wu, T. T., and Sun, R. (2003). Identification of proteins associated with murine gammaherpesvirus 68 virions. J. Virol. 77, 13425–13432.
Bringhurst, R. M., and Schaffer, P. A. (2006). Cellular stress rather than stage of the cell cycle enhances the replication and plating efficiencies of herpes simplex virus type 1 ICP0- viruses. J. Virol. 80, 4528–4537.
Brown, C. R., Nakamura, M. S., Mosca, J. D., Hayward, G. S., Straus, S. E., and Perera, L. P. (1995). Herpes simplex virus trans-regulatory protein ICP27 stabilizes and binds to 3′ ends of labile mRNA. J. Virol. 69, 7187–7195.
Bushell, M., and Sarnow, P. (2002). Hijacking the translation apparatus by RNA viruses. J. Cell Biol. 158, 395–399.
Cantin, R., Methot, S., and Tremblay, M. J. (2005). Plunder and stowaways: incorporation of cellular proteins by enveloped viruses. J. Virol. 79, 6577–6587.
Cepeda, V., and Fraile-Ramos, A. (2011). A role for the SNARE protein syntaxin 3 in human cytomegalovirus morphogenesis. Cell. Microbiol. 13, 846–858.
Chen, I. H., Sciabica, K. S., and Sandri-Goldin, R. M. (2002). ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76, 12877–12889.
Chou, W., Ngo, T., and Gershon, P. D. (2012). An overview of the vaccinia virus infectome: a survey of the proteins of the poxvirus-infected cell. J. Virol. 86, 1487–1499.
Chua, C. E., Gan, B. Q., and Tang, B. L. (2011). Involvement of members of the Rab family and related small GTPases in autophagosome formation and maturation. Cell. Mol. Life Sci. 68, 3349–3358.
Coombs, K. M., Berard, A., Xu, W., Krokhin, O., Meng, X., Cortens, J. P., Kobasa, D., Wilkins, J., and Brown, E. G. (2010). Quantitative proteomic analyses of influenza virus-infected cultured human lung cells. J. Virol. 84, 10888–10906.
De Regge, N., Nauwynck, H. J., Geenen, K., Krummenacher, C., Cohen, G. H., Eisenberg, R. J., Mettenleiter, T. C., and Favoreel, H. W. (2006). Alpha-herpesvirus glycoprotein D interaction with sensory neurons triggers formation of varicosities that serve as virus exit sites. J. Cell Biol. 174, 267–275.
del Rio, T., Decoste, C. J., and Enquist, L. W. (2005). Actin is a component of the compensation mechanism in pseudorabies virus virions lacking the major tegument protein VP22. J. Virol. 79, 8614–8619.
Dohner, K., Wolfstein, A., Prank, U., Echeverri, C., Dujardin, D., Vallee, R., and Sodeik, B. (2002). Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13, 2795–2809.
Dry, I., Haig, D. M., Inglis, N. F., Imrie, L., Stewart, J. P., and Russell, G. C. (2008). Proteomic analysis of pathogenic and attenuated alcelaphine herpesvirus 1. J. Virol. 82, 5390–5397.
Ellison, K. S., Maranchuk, R. A., Mottet, K. L., and Smiley, J. R. (2005). Control of VP16 translation by the herpes simplex virus type 1 immediate-early protein ICP27. J. Virol. 79, 4120–4131.
Emmott, E., Rodgers, M. A., Macdonald, A., Mccrory, S., Ajuh, P., and Hiscox, J. A. (2010). Quantitative proteomics using stable isotope labeling with amino acids in cell culture reveals changes in the cytoplasmic, nuclear, and nucleolar proteomes in Vero cells infected with the coronavirus infectious bronchitis virus. Mol. Cell Proteomics 9, 1920–1936.
Everett, R. D. (2000). ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22, 761–770.
Feierbach, B., Piccinotti, S., Bisher, M., Denk, W., and Enquist, L. W. (2006). Alpha-herpesvirus infection induces the formation of nuclear actin filaments. PLoS Pathog. 2, e85. doi:10.1371/journal.ppat.0020085
Fontaine-Rodriguez, E. C., and Knipe, D. M. (2008). Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J. Virol. 82, 3538–3545.
Forest, T., Barnard, S., and Baines, J. D. (2005). Active intranuclear movement of herpesvirus capsids. Nat. Cell Biol. 7, 429–431.
Friedel, C. C., and Haas, J. (2011). Virus-host interactomes and global models of virus-infected cells. Trends Microbiol. 19, 501–508.
Goodkin, M. L., Morton, E. R., and Blaho, J. A. (2004). Herpes simplex virus infection and apoptosis. Int. Rev. Immunol. 23, 141–172.
Grunewald, K., Desai, P., Winkler, D., Heymann, J., Belnap, D., Baumeister, W., and Steven, A. (2003). Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302, 1396–1398.
Gurer, C., Cimarelli, A., and Luban, J. (2002). specific incorporation of heat shock protein 70 family members into primate lentiviral virions. J. Virol. 76, 4666–4670.
Halford, W. P., and Schaffer, P. A. (2001). ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 75, 3240–3249.
Hancock, M. H., Corcoran, J. A., and Smiley, J. R. (2006). Herpes simplex virus regulatory proteins VP16 and ICP0 counteract an innate intranuclear barrier to viral gene expression. Virology 352, 237–252.
Hardwicke, M. A., and Sandri-Goldin, R. M. (1994). The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J. Virol. 68, 4797–4810.
Hardy, W. R., and Sandri-Goldin, R. M. (1994). Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68, 7790–7799.
Hertel, L. (2011). Herpesviruses and intermediate filaments: close encounters with the third type. Viruses 3, 1015–1040.
Hobbs, W. E. II, and DeLuca, N. A. (1999). Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 73, 8245–8255.
Huang Da, W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57.
Indran, S. V., and Britt, W. J. (2011). A role for the small GTPase Rab6 in assembly of human cytomegalovirus. J. Virol. 85, 5213–5219.
Johannsen, E., Luftig, M., Chase, M. R., Weicksel, S., Cahir-Mcfarland, E., Illanes, D., Sarracino, D., and Kieff, E. (2004). Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U.S.A. 101, 16286–16291.
Kanehisa, M., Goto, S., Furumichi, M., Tanabe, M., and Hirakawa, M. (2010). KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38, D355–D360.
Kattenhorn, L. M., Mills, R., Wagner, M., Lomsadze, A., Makeev, V., Borodovsky, M., Ploegh, H. L., and Kessler, B. M. (2004). Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 78, 11187–11197.
Kawaguchi, Y., Van Sant, C., and Roizman, B. (1997). Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71, 7328–7336.
Kramer, T., Greco, T. M., Enquist, L. W., and Cristea, I. M. (2011). Proteomic characterization of pseudorabies virus extracellular virions. J. Virol. 85, 6427–6441.
Krauss, O., Hollinshead, R., Hollinshead, M., and Smith, G. L. (2002). An investigation of incorporation of cellular antigens into vaccinia virus particles. J. Gen. Virol. 83, 2347–2359.
Kubista, H., Edelbauer, H., and Boehm, S. (2004). Evidence for structural and functional diversity among SDS-resistant SNARE complexes in neuroendocrine cells. J. Cell. Sci. 117, 955–966.
Lam, Q., Smibert, C. A., Koop, K. E., Lavery, C., Capone, J. P., Weinheimer, S. P., and Smiley, J. R. (1996). Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15, 2575–2581.
Larralde, O., Smith, R. W., Wilkie, G. S., Malik, P., Gray, N. K., and Clements, J. B. (2006). Direct stimulation of translation by the multifunctional herpesvirus ICP27 protein. J. Virol. 80, 1588–1591.
Lee, G. E., Murray, J. W., Wolkoff, A. W., and Wilson, D. W. (2006). Reconstitution of herpes simplex virus microtubule-dependent trafficking in vitro. J. Virol. 80, 4264–4275.
Lietzen, N., Ohman, T., Rintahaka, J., Julkunen, I., Aittokallio, T., Matikainen, S., and Nyman, T. A. (2011). Quantitative subcellular proteome and secretome profiling of influenza A virus-infected human primary macrophages. PLoS Pathog. 7, e1001340. doi:10.1371/journal.ppat.1001340
Lindberg, A., and Kreivi, J. P. (2002). Splicing inhibition at the level of spliceosome assembly in the presence of herpes simplex virus protein ICP27. Virology 294, 189–198.
Lodish, H., Berk, A., Zipursky, S. L., Matsudaira, P., Baltimore, D., and Darnell, J. (2000). Viruses: Structure, Function, and Uses. Molecular Cell Biology, 4th Edn. (New York: W.H. Freeman and Company), 191–207.
Lomonte, P., and Everett, R. D. (1999). Herpes simplex virus type 1 immediate-early protein Vmw110 inhibits progression of cells through mitosis and from G(1) into S phase of the cell cycle. J. Virol. 73, 9456–9467.
Loret, S., Guay, G., and Lippe, R. (2008). Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82, 8605–8618.
Loret, S., and Lippe, R. (2012). Biochemical analysis of ICP0, ICP4, UL7 and UL23 incorporated into extracellular herpes simplex type 1 virions. J. Gen. Virol. 93, 624–634.
Lu, Q., Bai, J., Zhang, L., Liu, J., Jiang, Z., Michal, J. J., He, Q., and Jiang, P. (2012). Two-dimensional liquid chromatography-tandem mass spectrometry coupled with isobaric tags for relative and absolute quantification (iTRAQ) labeling approach revealed first proteome profiles of pulmonary alveolar macrophages infected with porcine reproductive and respiratory syndrome virus. J. Proteome Res. 11, 2890–2903.
Lu, Z., Qin, A., Qian, K., Chen, X., Jin, W., Zhu, Y., and Eltahir, Y. M. (2010). Proteomic analysis of the host response in the bursa of Fabricius of chickens infected with Marek’s disease virus. Virus Res. 153, 250–257.
Marozin, S., Prank, U., and Sodeik, B. (2004). Herpes simplex virus type 1 infection of polarized epithelial cells requires microtubules and access to receptors present at cell-cell contact sites. J. Gen. Virol. 85, 775–786.
Mercer, J., Schelhaas, M., and Helenius, A. (2010). Virus entry by endocytosis. Annu. Rev. Biochem. 79, 803–833.
Michael, K., Klupp, B. G., Mettenleiter, T. C., and Karger, A. (2006). Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 80, 1332–1339.
Mizuno-Yamasaki, E., Rivera-Molina, F., and Novick, P. (2012). GTPase networks in membrane traffic. Annu. Rev. Biochem. PMID: 22463690.[Epub ahead of print]..
Moerdyk-Schauwecker, M., Hwang, S. I., and Grdzelishvili, V. Z. (2009). Analysis of virion associated host proteins in vesicular stomatitis virus using a proteomics approach. Virol. J. 6, 166.
Munday, D. C., Hiscox, J. A., and Barr, J. N. (2010). Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus subgroup B using SILAC coupled to LC-MS/MS. Proteomics 10, 4320–4334.
Norrild, B., Lehto, V. P., and Virtanen, I. (1986). Organization of cytoskeleton elements during herpes simplex virus type 1 infection of human fibroblasts: an immunofluorescence study. J. Gen. Virol. 67(Pt 1), 97–105.
Padula, M. E., Sydnor, M. L., and Wilson, D. W. (2009). Isolation and preliminary characterization of herpes simplex virus 1 primary enveloped virions from the perinuclear space. J. Virol. 83, 4757–4765.
Pastorino, B., Boucomont-Chapeaublanc, E., Peyrefitte, C. N., Belghazi, M., Fusai, T., Rogier, C., Tolou, H. J., and Almeras, L. (2009). Identification of cellular proteome modifications in response to West Nile virus infection. Mol. Cell Proteomics 8, 1623–1637.
Piluso, D., Bilan, P., and Capone, J. P. (2002). Host cell factor-1 interacts with and antagonizes transactivation by the cell cycle regulatory factor Miz-1. J. Biol. Chem. 277, 46799–46808.
Radtke, K., Kieneke, D., Wolfstein, A., Michael, K., Steffen, W., Scholz, T., Karger, A., and Sodeik, B. (2010). Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 6, e1000991. doi:10.1371/journal.ppat.1000991
Ramirez-Boo, M., Nunez, E., Jorge, I., Navarro, P., Fernandes, L. T., Segales, J., Garrido, J. J., Vazquez, J., and Moreno, A. (2011). Quantitative proteomics by 2-DE, 16O/18O labelling and linear ion trap mass spectrometry analysis of lymph nodes from piglets inoculated by porcine circovirus type 2. Proteomics 11, 3452–3469.
Rassmann, A., Henke, A., Zobawa, M., Carlsohn, M., Saluz, H. P., Grabley, S., Lottspeich, F., and Munder, T. (2006). Proteome alterations in human host cells infected with coxsackievirus B3. J. Gen. Virol. 87, 2631–2638.
Rice, S. A., and Knipe, D. M. (1988). Gene-specific transactivation by herpes simplex virus type 1 alpha protein ICP27. J. Virol. 62, 3814–3823.
Saksena, M. M., Wakisaka, H., Tijono, B., Boadle, R. A., Rixon, F., Takahashi, H., and Cunningham, A. L. (2006). Herpes simplex virus type 1 accumulation, envelopment, and exit in growth cones and varicosities in mid-distal regions of axons. J. Virol. 80, 3592–3606.
Sandri-Goldin, R. M., and Mendoza, G. E. (1992). A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 6, 848–863.
Sarma, N., Agarwal, D., Shiflett, L. A., and Read, G. S. (2008). Small interfering RNAs that deplete the cellular translation factor eIF4H impede mRNA degradation by the virion host shutoff protein of herpes simplex virus. J. Virol. 82, 6600–6609.
Schroder, M. (2010). Human DEAD-box protein 3 has multiple functions in gene regulation and cell cycle control and is a prime target for viral manipulation. Biochem. Pharmacol. 79, 297–306.
Schroder, M. (2011). Viruses and the human DEAD-box helicase DDX3: inhibition or exploitation? Biochem. Soc. Trans. 39, 679–683.
Sekulovich, R. E., Leary, K., and Sandri-Goldin, R. M. (1988). The herpes simplex virus type 1 alpha protein ICP27 can act as a trans-repressor or a trans-activator in combination with ICP4 and ICP0. J. Virol. 62, 4510–4522.
Shadan, F. F., Cowsert, L. M., and Villarreal, L. P. (1994). n-Butyrate, a cell cycle blocker, inhibits the replication of polyomaviruses and papillomaviruses but not that of adenoviruses and herpesviruses. J. Virol. 68, 4785–4796.
Sharma-Walia, N., Naranatt, P. P., Krishnan, H. H., Zeng, L., and Chandran, B. (2004). Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 envelope glycoprotein gB induces the integrin-dependent focal adhesion kinase-Src-phosphatidylinositol 3-kinase-rho GTPase signal pathways and cytoskeletal rearrangements. J. Virol. 78, 4207–4223.
Shaw, M. L., Stone, K. L., Colangelo, C. M., Gulcicek, E. E., and Palese, P. (2008). Cellular proteins in influenza virus particles. PLoS Pathog. 4, e1000085. doi:10.1371/journal.ppat.1000085
Simons, K., and Warren, G. (1984). Semliki forest virus: a probe for membrane traffic in the animal cell. Adv. Protein Chem. 36, 79–132.
Simpson-Holley, M., Colgrove, R. C., Nalepa, G., Harper, J. W., and Knipe, D. M. (2005). Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 79, 12840–12851.
Smith, G. A., Gross, S. P., and Enquist, L. W. (2001). Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. U.S.A. 98, 3466–3470.
Smith, I. L., Hardwicke, M. A., and Sandri-Goldin, R. M. (1992). Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology 186, 74–86.
Sodeik, B., Ebersold, M. W., and Helenius, A. (1997). Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136, 1007–1021.
Soliman, T. M., Sandri-Goldin, R. M., and Silverstein, S. J. (1997). Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 71, 9188–9197.
Sollner, T. H. (2004). Intracellular and viral membrane fusion: a uniting mechanism. Curr. Opin. Cell Biol. 16, 429–435.
Sollner, T. H., and Rothman, J. E. (1996). Molecular machinery mediating vesicle budding, docking and fusion. Experientia 52, 1021–1025.
Song, B., Liu, J. J., Yeh, K. C., and Knipe, D. M. (2000). Herpes simplex virus infection blocks events in the G1 phase of the cell cycle. Virology 267, 326–334.
Sun, J., Jiang, Y., Shi, Z., Yan, Y., Guo, H., He, F., and Tu, C. (2008). Proteomic alteration of PK-15 cells after infection by classical swine fever virus. J. Proteome Res. 7, 5263–5269.
Teodoro, J. G., and Branton, P. E. (1997). Regulation of apoptosis by viral gene products. J. Virol. 71, 1739–1746.
Thanthrige-Don, N., Abdul-Careem, M. F., Shack, L. A., Burgess, S. C., and Sharif, S. (2009). Analyses of the spleen proteome of chickens infected with Marek’s disease virus. Virology 390, 356–367.
Tong, A., Wu, L., Lin, Q., Lau, Q. C., Zhao, X., Li, J., Chen, P., Chen, L., Tang, H., Huang, C., and Wei, Y. Q. (2008). Proteomic analysis of cellular protein alterations using a hepatitis B virus-producing cellular model. Proteomics 8, 2012–2023.
Varnum, S. M., Streblow, D. N., Monroe, M. E., Smith, P., Auberry, K. J., Pasa-Tolic, L., Wang, D., Camp, D. G. II, Rodland, K., Wiley, S., Britt, W., Shenk, T., Smith, R. D., and Nelson, J. A. (2004). Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78, 10960–10966.
Viswanathan, K., and Fruh, K. (2007). Viral proteomics: global evaluation of viruses and their interaction with the host. Expert Rev. Proteomics 4, 815–829.
Wolfstein, A., Nagel, C. H., Radtke, K., Dohner, K., Allan, V. J., and Sodeik, B. (2006). The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7, 227–237.
Wong, M. L., and Chen, C. H. (1998). Evidence for the internal location of actin in the pseudorabies virion. Virus Res. 56, 191–197.
Yang, B., Gonzalez, L. Jr., Prekeris, R., Steegmaier, M., Advani, R. J., and Scheller, R. H. (1999). SNARE interactions are not selective. Implications for membrane fusion specificity. J. Biol. Chem. 274, 5649–5653.
Zandi, F., Eslami, N., Soheili, M., Fayaz, A., Gholami, A., and Vaziri, B. (2009). Proteomics analysis of BHK-21 cells infected with a fixed strain of rabies virus. Proteomics 9, 2399–2407.
Zenner, H. L., Yoshimura, S., Barr, F. A., and Crump, C. M. (2011). Analysis of Rab GTPase-activating proteins indicates that Rab1a/b and Rab43 are important for herpes simplex virus 1 secondary envelopment. J. Virol. 85, 8012–8021.
Zerial, M., and McBride, H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2, 107–117.
Zhang, L., Jia, X., Zhang, X., Sun, J., Peng, X., Qi, T., Ma, F., Yin, L., Yao, Y., Qiu, C., and Lu, H. (2010). Proteomic analysis of PBMCs: characterization of potential HIV-associated proteins. Proteome Sci. 8, 12.
Zhang, X., Zhou, J., Wu, Y., Zheng, X., Ma, G., Wang, Z., Jin, Y., He, J., and Yan, Y. (2009). Differential proteome analysis of host cells infected with porcine circovirus type 2. J. Proteome Res. 8, 5111–5119.
Zheng, J., Sugrue, R. J., and Tang, K. (2011). Mass spectrometry based proteomic studies on viruses and hosts – a review. Anal. Chim. Acta 702, 149–159.
Keywords: herpes, virus, host–pathogen interactions, HSV, PRV, HCMV, EBV γHV68, KSHV
Citation: Lippé R (2012) Deciphering novel host–herpesvirus interactions by virion proteomics. Front. Microbio. 3:181. doi: 10.3389/fmicb.2012.00181
Received: 11 April 2012; Accepted: 27 April 2012;
Published online: 28 May 2012.
Edited by:
Kevin Coombs, University of Manitoba, CanadaReviewed by:
Bruce W. Banfield, Queen’s University, CanadaCopyright: © 2012 Lippé. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Roger Lippé, Department of Pathology and Cell Biology, University of Montreal, PO Box 6128, Succursale Centre-Ville, Montreal, QC, Canada H3C 3J7. e-mail:cm9nZXIubGlwcGVAdW1vbnRyZWFsLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.