
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 04 May 2012
Sec. Evolutionary and Genomic Microbiology
Volume 3 - 2012 | https://doi.org/10.3389/fmicb.2012.00167
This article is part of the Research Topic Recent advances in the biology of planctomycetes and verrucomicrobia View all 13 articles
 John A. Fuerst* and Evgeny Sagulenko
John A. Fuerst* and Evgeny Sagulenko
Planctomycetes are known to display compartmentalization via internal membranes, thus resembling eukaryotes. Significantly, the planctomycete Gemmata obscuriglobus has not only a nuclear region surrounded by a double-membrane, but is also capable of protein uptake via endocytosis. In order to clearly analyze implications for homology of their characters with eukaryotes, a correct understanding of planctomycete structure is an essential starting point. Here we outline the major features of such structure necessary for assessing the case for or against homology with eukaryote cell complexity. We consider an evolutionary model for cell organization involving reductive evolution of Planctomycetes from a complex proto-eukaryote-like last universal common ancestor, and evaluate alternative models for origins of the unique planctomycete cell plan. Overall, the structural and molecular evidence is not consistent with convergent evolution of eukaryote-like features in a bacterium and favors a homologous relationship of Planctomycetes and eukaryotes.
The phylum Planctomycetes is a distinctive one situated within the domain Bacteria with characteristics of immense significance for understanding cell biology and the evolution of cell organization (Fuerst and Sagulenko, 2011). Their most dramatic feature is a shared cell compartmentalization correlated with intracellular membranes (Fuerst, 2005). In at least one genus, Gemmata, this is displayed by an even more complex compartment containing nucleoid DNA and surrounded by an envelope of two closely apposed membranes. Other members of the PVC superphylum (in phyla Planctomycetes, Verrucomicrobia, and Chlamydia), e.g. Verrucomicrobia, also display at least simpler forms of this cell compartmentalization (Lee et al., 2009b). Functional complexity accompanies this structural complexity, since Gemmata obscuriglobus has an endocytosis-like ability to incorporate large proteins from the extracellular medium (Lonhienne et al., 2010). This feature correlates with possession unique within the Bacteria of homologs of the coatomer MC class of eukaryote proteins associated with endocytosis (Lonhienne et al., 2010; Santarella-Mellwig et al., 2010). These characters pose puzzles regarding the exact relevance of Planctomycetes and the PVC superphylum to the evolution of eukaryotes (Devos and Reynaud, 2010; Koonin, 2010; McInerney et al., 2011; Reynaud and Devos, 2011). What might Planctomycetes tell us about evolution of eukaryote cell biology? We will endeavur here to clarify cell structural features of Planctomycetes of special significance to the question of potential homologies with eukaryotes and their endomembrane systems.
For a deeper appreciation of features of PVC/Planctomycetes members leading to a concept of eukaryote homology, we need to make clear the evidence underlying interpretations of their distinctiveness. This applies to evidence for internal membranes of G. obscuriglobus and the distinction of the protein-rich wall of Planctomycetes from the cell wall of Gram-negative Bacteria (Cavalier-Smith, 2010). Eukaryotic features such as compartmentalized cells and true tubulins capable of forming microtubules in the related Verrucomicrobia of the PVC superphylum pose similar problems (Schlieper et al., 2005; Martin-Galiano et al., 2011; Pilhofer et al., 2011).
We can compare cell plan features of G. obscuriglobus as a representative planctomycete to those of other bacteria with internal membranes and to those of Escherichia coli as typical of others without such membranes (Figure 1). These plans can be contrasted also with simplest representative microbial eukaryotes Saccharomyces and Giardia (Figure 1). This clearly illustrates the distinctive nature of the Gemmata ICM and other internal membranes compared with that of the plasma membrane of other bacteria, the uniqueness of the paryphoplasm, and the way in which Gemmata paryphoplasm and nuclear envelope pericisternal space are topologically similar to the endoplasmic reticulum (ER) lumen in eukaryotes. In Gemmata, in addition to the two major compartments present in other planctomycete species, there is a third compartment, the “nuclear body,” enclosing a condensed nucleoid and some of the cell’s ribosomes. 3D reconstruction from serial sectioning of freeze-substituted G. obscuriglobus has shown that the nuclear body envelope is largely continuous and is compatible with the view of the nuclear body as a separate compartment of the cell (Lindsay et al., 2001). This nuclear body compartment resembles the nucleus of eukaryote cells with its double-membraned nuclear envelope, with the exception of apparent presence of ribosomes inside the nuclear region. Among Planctomycetes, such structures, at least in their complete form, are not so far found outside this genus. In G. obscuriglobus the outer membrane of the nuclear envelope is continuous with membranes originating from the ICM. This means that the nuclear envelope and membranes continuous with it form an endomembrane system comparable to the ER of eukaryotes including rough ER. The pericisternal space between membranes of the eukaryote nuclear envelope and continuous with the ER lumen (see Saccharomyces, Giardia in Figure 1) is analogous to the space between membranes of the Gemmata nuclear envelope and perhaps also with paryphoplasm. In rough ER of eukaryotes, ribosomes are found bound to ER membrane and the outer membrane of the nuclear envelope (see Saccharomyces, Giardia, in Figure 1). In G. obscuriglobus and cyanobacterium Synechococcus, ribosomes can be found bound to internal membranes (Figure 1). In G. obscuriglobus, like eukaryotes, some ribosomes are found bound to the nuclear envelope’s outer membrane, but G. obscuriglobus also has ribosomes lining the inner side of the nuclear envelope, as well as free ribosomes both inside and outside the nuclear compartment. In G. obscuriglobus, ribosomes are also seen bound to ICM on its inner side, but ribosomes are never seen in the paryphoplasm or bound to the cytoplasmic membrane (in complete contrast to E. coli, where ribosomes are normally bound to cytoplasmic membrane as well as free in the cytoplasm; Bibi and Herskovits, 2000). We predict that internal membrane-bounded ribosomes of Gemmata are involved in co-translational secretion across bound membrane. At a molecular level, in ribosomal proteins such as L17 in Planctomycetes, indels involving alpha-helices unique within domain Bacteria have been reported (Kamneva et al., 2010). These may be correlated with changes from typical bacterial translation. Note that no internal membranes are found in wild-type E. coli, and internal membranes such as the chromatophores of Rhodopseudomonas viridis (Konorty et al., 2008) and the magnetosomes of magnetotactic bacteria originate from the plasma membrane (Remsen et al., 1968; Komeili et al., 2006; Schuler, 2008). Magnetosomes of the alpha-proteobacterium Magnetospirillum in Figure 1 represent the class of structures where plasma membrane invaginates to form internal membranes (Komeili et al., 2006). However the thylakoid membranes of cyanobacteria functioning in oxygen-evolving photosynthesis may well bear comparison with those of Gemmata internal membranes. Cryo-electron tomography has demonstrated that thylakoid membranes seem not to show clear continuity with plasma membrane in Prochlorococcus (Ting et al., 2007), Synechococcus (van de Meene et al., 2006), and Synechocystis (Liberton et al., 2006), as well as other cyanobacteria (Nevo et al., 2007; Konorty et al., 2008). For Synechocystis fluorescence microscopy of live cells is consistent with this conclusion (Schneider et al., 2007).
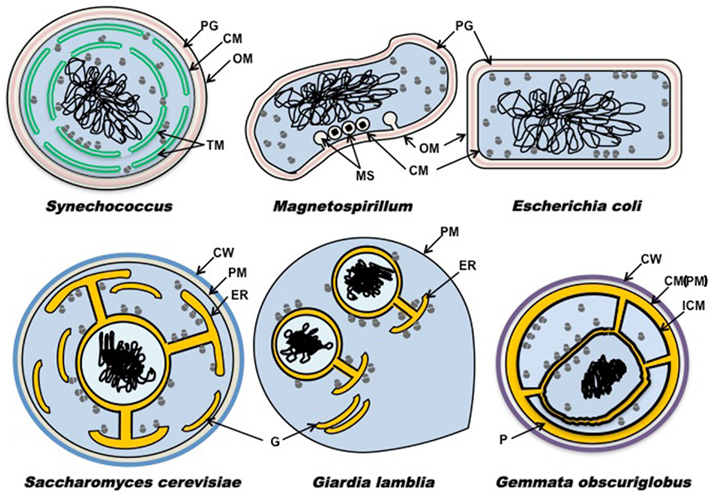
Figure 1. Schematic diagram showing plans of membrane distribution. Membrane topology is in representative Bacteria Gemmata obscuriglobus, Synechococcus, Escherichia coli, and Magnetospirillum relative to representative microbial members of the Eucarya Saccharomyces cerevisiae and Giardia lamblia. Cytoplasm containing ribosomes is shown in blue, nucleus is shown in light-blue, DNA in black, and ribosome-free spaces (ER lumen in the eukaryotes, paryphoplasm in G. obscuriglobus) in yellow orange, peptidoglycan in pink, mannoprotein-glucan (Klis et al., 2006; yeast) walls or protein (planctomycete) walls in blue or purple respectively. Magnetite particles are shown as black diamonds (Magnetospirillum only). CW, cell wall; CM, cytoplasmic membrane; ER, endoplasmic reticulum; G, Golgi apparatus; ICM, intracytoplasmic membrane; MS, magnetosome; OM, outer membrane; P, paryphoplasm; PM, plasma membrane; PG, peptidoglycan; TM, thylakoid membrane.
It is important not to confuse the characteristically planctomycete- and PVC superphylum-specific paryphoplasm with the periplasm found in non-planctomycete and non-PVC bacteria. Planctomycete internal membranes are not mere invaginations of the plasma membrane (PM; in bacteria also called the cytoplasmic membrane (CM), in the present context not used in the text to avoid confusion of two terms for the same structure). There is no existing ultrastructural data known to us suggesting any direct connection between PM of planctomycete cells and the ICM defining the inner boundary of the paryphoplasm, as there is none relating any further internal membranes in the cytoplasm to the PM.
It is clear that the “paryphoplasm” compartment in G. obscuriglobus plays a major role in endocytotic protein uptake in this planctomycete bacterium (Lonhienne et al., 2010), confirming its functional reality. Those proteins are taken up by the cell into the paryphoplasm but such incorporated proteins have never been detected in other cell compartments, e.g., the nuclear region and ribosome-containing compartments, confirming the separate functional nature of paryphoplasm. It is illuminating to consider what we know of the mechanism of protein uptake in G. obscuriglobus in relation to the question of the relation of PM to ICM and the topological nature of the paryphoplasm to any periplasm that might occur in this planctomycete. During uptake of protein from the external milieu, vesicles form from infolding of PM, and such vesicles form separate compartments within the paryphoplasm (Lonhienne et al., 2010). It is the contents of these vesicles which are equivalent topologically to external milieu (or to periplasm in contact with such), not the paryphoplasm itself, which is a space in which the vesicles are located. The paryphoplasm is thus not equivalent to periplasm topologically. A separate membrane-trafficking compartment might be a result of specialization for efficient endocytosis-based nutrition.
Once we clearly understand the unique nature of the cell plan in Planctomycetes, we can start to approach the question of their potential eukaryote homology. The consilience of such planctomycete features as presence of endocytosis abilities and eukaryote membrane-deforming MC-protein homologs, the absence of FtsZ and other divisome proteins other than FtsK, presence of protein cell walls, absence of peptidoglycan, presence of sterols accompanied by sterol-synthesizing enzymes with eukaryote homology (Frickey and Kannenberg, 2009), presence of some dnaK (Hsp70) genes with eukaryote homology (Ward-Rainey et al., 1997), C1 transfer enzymes falling between Bacteria and Archaea in phylogenetic trees (Chistoserdova et al., 2004), and in anammox Planctomycetes the presence of ATP synthase in internal membranes (van Niftrik et al., 2010) points to some dramatically unusual Bacteria. And these features may also be most parsimoniously consistent with some perhaps quite heretical evolutionary models. Such models include one where a last common ancestor of the PVC group has some properties of all three Domains including the Eucarya and a complex eukaryote-like cell plan – i.e., a PVC ancestor with properties of a compartmentalized progenote last universal common ancestor (LUCA). Occurrence of a simple compartmentalized cell plan throughout the PVC group combined with occurrence of now established eukaryotic tubulin homologs of ancient origin in at least Prosthecobacter among the Verrucomicrobia (Martin-Galiano et al., 2011) is consistent with such a concept. An alternative model may derive key eukaryote features from a planctomycete or other PVC members (or their ancestors), their cells acting as a kind of tool kit for eukaryality – where genes and cell organizational plans central to eukaryote abilities may have evolved to be later transferred laterally to a eucaryal lineage.
Endocytosis is considered a key innovation in the origin of the eukaryotic cell (Cavalier-Smith, 2009), and is linked to evolution of the primordial endomembrane system of the last eukaryotic common ancestor (LECA; Dacks et al., 2008). The endocytosis system in a simple eukaryote such as Giardia (albeit of secondary simplicity) is illuminating in terms of what homology might encompass in terms of simplicity. In Giardia, endocytosis is performed via contiguous and communicating peripheral vesicles (PV) and the ER-like tubulovesicular network (TVN), which appear to combine the functions of the separate early and late endosomes and lysosomes of the endocytic system of more complex organisms (Lanfredi-Rangel et al., 1998; Abodeely et al., 2009). Very interestingly in relation to the paryphoplasm in Gemmata and its role as harboring endocytotic vesicles, Giardia endocytosis and degradation of exogenous proteins occurs in the ER (TVN) or in a compartment communicating with ER – Giardia differs from other eukaryotes in that the endocytotic system is not excluded from the ER, so that “lysosomal” protein degradation occurs in the ER. So too in Gemmata both endocytotic vesicle formation from PM and protein degradation occur in the same paryphoplasm region (Lonhienne et al., 2010). In both Gemmata and Giardia reductive evolution may have occurred via distinct selective forces but resulting in similarly simple endocytosis organelles. In such a meeting point of pro- and eukaryote cell structure and function we may find clues to the minimal eukaryote and maximally complex prokaryote and the evolutionary connections between them via potential common ancestors.
What does the most complex of the PVC superphylum, G. obscuriglobus, imply for homologies with eukaryotes? If a distinct nuclear body compartment exists separated by a double-membrane envelope from other ribosome-containing cytoplasm, then some translation must occur in a non-DNA containing compartment as occurs in eukaryotes. Transcription must be confined to the nuclear body with its condensed nucleoid, and at least some mRNA and possibly proteins must be transported through the nuclear envelope. Secondly, separation of the nuclear region from the remainder of the ribosome-rich cytoplasm implies a nuclear transport system and even nuclear pore complexes (NPCs). The MC-protein homolog appearing to operate in endocytosis in Gemmata might also form the basis for an NPC, since MC proteins and some nucleoporins share underlying structure (Devos et al., 2004; Devos, 2012). Like endomembrane systems and membrane-deforming proteins needed for such vesicle-rich systems (Field and Dacks, 2009; Field et al., 2011), NPCs seem to have been a feature of the LECA (Wilson and Dawson, 2011). This is no surprise considering the shared membrane-deforming domains of the MC-like proteins underlying these structures, shared also with the ancestral “protocoatomer” (Devos et al., 2004). These domains are also shared, exclusively within the Bacteria, with MC proteins of PVC superphylum members (Santarella-Mellwig et al., 2010), including those correlated with endocytosis in Gemmata (Lonhienne et al., 2010). In G. obscuriglobus the ribosomes bound to nuclear envelope membrane on both sides in linear arrays are consistent with polysomes and imply co-translational protein secretion into the NE lumen, making this an analog of the eukaryotic ER lumen. This further implies ER signal sequences in proteins destined for the lumen, chaperone proteins such as calreticulin and calnexin assisting folding of lumen proteins (Johnson et al., 2001) and even the ERAD system for degrading incorrectly folded lumen proteins (Meusser et al., 2005). Absence of a clear Golgi system is not critical to proto-eukaryality, as Giardia cell biology indicates – Giardia has no clear morphologically recognizable Golgi, but encystation-specific vesicles perform some Golgi functions and a primordial or a least simple secretory apparatus is present (Marti et al., 2003; no matter whether the simplicity of any Golgi-like function in Giardia is a result of secondary loss). The ability of Gemmata to endocytose proteins implies return of membrane-forming endocytotic vesicles back to the PM and thus some form of exocytosis and even secretion of vesicle contents. In molecular terms, a receptor- and clathrin-mediated endocytosis mechanism implies potential homologs of accessory eukaryotic proteins needed for endocystosis. These include SNAREs, Rab GTPases, actins and myosins, and dynamins (Fuerst and Sagulenko, 2010), though it should be noted that yeast seems able to dispense with the Arp2 adaptin operating in the clathrin system of mammals (Conibear, 2010). Other elements of the system may be missing or replaced non-orthologously in Gemmata. Actins may be crucial especially in organisms which experience significant turgor pressure affecting endocytotic vesicle formation (Conibear, 2010). SNARE-like motifs have been found in bacterial proteins including IncA of species of the PVC group member Chlamydia (where they interact with SNAREs of hosts of these intracellular pathogens; Delevoye et al., 2008) and IncA of another intracellular pathogen Legionella (Paumet et al., 2009b). G. obscuriglobus and several other Planctomycetes have proteins with SNARE-associated domains (as do Bacteria from other phyla (Delevoye et al., 2008; Paumet et al., 2009a). Planctomycetes may not be the only organisms displaying homologies in their cell biology to eukaryotes. Several members of the Crenarcheota and Thaumarcheota within the Archaea have now been shown to possess an alternative cell division machinery. This machinery shares elements with the ESCRT-III system of eukaryotes involved in endocytosis via the late endosome multivesicular body and in formation of intraluminal vesicles as well as cytokinesis (Lindas et al., 2008; Ettema and Bernander, 2009; Pelve et al., 2011).
Significant to the question of homology of Planctomycetes with eukaryotes, bioinformatic analysis of the predicted subcellular proteome of anammox planctomycete Kuenenia stuttgartiensis indicates that signal peptides of anammox proteins are more similar to those of eukaryotes than to those of any model bacteria whether Gram-positive or Gram-negative (Medema et al., 2010). The specialized ammonium-oxidizing anammoxosome organelle in Kuenenia appears to correlate with a distinct subproteome of hundreds of proteins, and the Tat translocation system seems confined to the anammoxosome membrane. This study also found that signal peptide prediction algorithms trained on eukaryotes gave better yield of true positives, and that use of Gram-negative predictors trained with proteobacteria signals could be deceptive – this type of problem may be a wider one applying also to genome annotation for Planctomycetes, we would suggest.
The whole question of signal peptides in Planctomycetes is significant for considering homology with eukaryotes since protein transport through membranes in all Planctomycetes must occur across internal membranes such as ICM as well as across the PM. Rhodopirellula baltica has an exceptionally high number of genes with signal peptides, and an exceptionally high number of Tat signal peptides (Glockner et al., 2003).
The presently understood evolutionary relationships of Planctomycetes and the PVC group place them firmly within the Bacteria, at least as based on 16S rRNA and sequences such as multiple ribosomal proteins derived from genomes (Fuerst and Sagulenko, 2011). There are also a number of molecular features which link Planctomycetes with Bacteria, e.g., Shine-Dalgarno sequence in mRNA (Leary et al., 1998), promoters (Liesack and Stackebrandt, 1989), common conserved oligonucleotides in 16S rRNA (Woese, 1987), presence of some or many peptidoglycan synthesis enzymes (Pilhofer et al., 2008; Bernander and Ettema, 2010), presence of genes for lipid A (Sutcliffe, 2010), bacterial secretion systems (Glockner et al., 2003), and bacterial flagellar systems (Pallen et al., 2009). They also have some distinctive eukaryotic molecular features, such as discoidin domains (Studholme et al., 2004), an integrin homolog (Fuerst et al., 2002), and MC (membrane coat) proteins homologous to clathrins (Santarella-Mellwig et al., 2010). Other features include novel cytochrome domains, solute-binding proteins, and N-terminal export signal peptides containing eukaryote-homologous domains such as calx-β, cadherin and thrombospondin Type 3 (Studholme et al., 2004). The phylogenetic position of Planctomycetes and PVC superphylum within the Bacteria is important to understand in relation to possible retention of features from a postulated eukaryote-like LUCA on the one hand, and potential of these organisms to form an intermediate between either LUCA or Bacteria and the eukaryotes on the other (Reynaud and Devos, 2011). Conversion from a bacterium to a eukaryote poses problems connected with the distinctive nature of their ribosomes, so that retention of eukaryote features from a LUCA is a simpler hypothesis to explain PVC compartments and other eukaryote features. Evidence consistent with a deep branching of Planctomycetes comes from slowly evolving 16SrRNA position trees (Brochier and Philippe, 2002) and proteome phylogenetics (Jun et al., 2010). Some phylogenetic analyses deduce that Planctomycetes are relatively shallow, based on analysis of concatenated ribosomal proteins combined with clustering with phylum Chlamydiae (Glockner et al., 2004). At least one genome-based analysis separates Chlamydiae as a deep phylum (Huson et al., 2005), and the whole PVC superphylum could conceivably also be as deep.
Some features of plantomycetes superficially resemble those of some archaea, e.g., cell walls dominated by protein. However, the proteins comprising the cell walls of at least the planctomycete R. baltica appear unique to Planctomycetes (with the distinctive YTV signature; Hieu et al., 2008) rather than converging to the archaeal S-layer glycoprotein structure. In anammox Planctomycetes ether-linked lipids occur as in Archaeal membranes, but these occur within the unique ladderane lipids, not known elsewhere in nature including the Archaea (Sinninghe Damste et al., 2004)
Our present knowledge of genomes of Planctomycetes is not sufficient to assume absence of any homology at gene level between Planctomycetes and eukaryotes. Such a conclusion would be premature considering the limited amount of analysis of planctomycete genomes and the very large percentages of the deduced proteome of Planctomycetes consisting of hypothetical proteins relative to proteomes of other bacteria deduced using identical algorithms (Table 1).
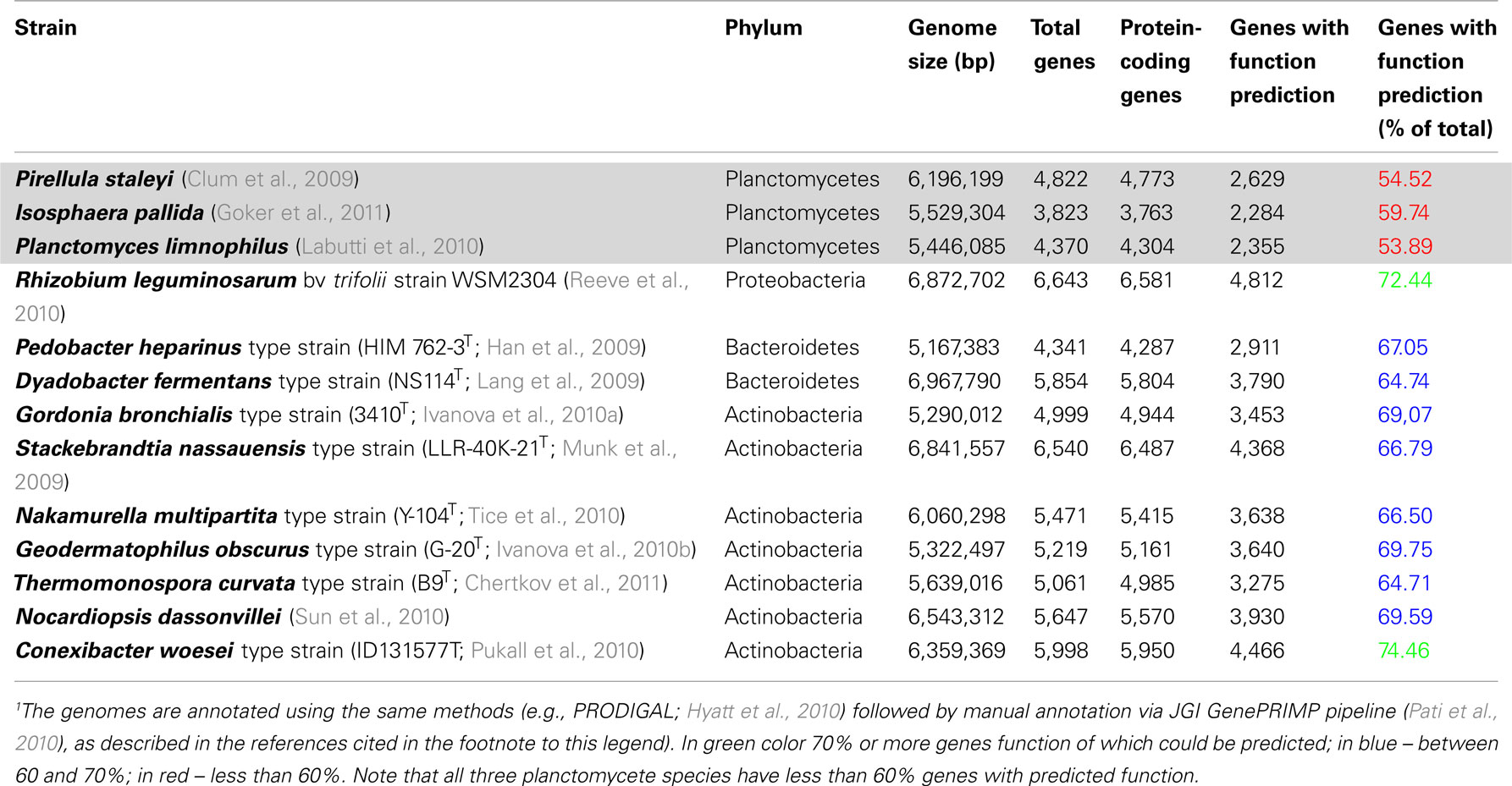
Table 1. Comparison of fraction of genes annotated with function prediction in planctomycete genomes relative to other recently sequenced genomes1.
Such large proportions of divergent hypothetical proteins difficult to relate to those of other organisms would be expected if Planctomycetes retain many ancestral proteins. The homology between planctomycete and eukaryote genes is in any case likely to be only detectable at the secondary structure level, as seems the case for many of the MC clathrin-like proteins of Planctomycetes (Santarella-Mellwig et al., 2010). For example, gp4978 was originally annotated as a hypothetical protein GobsU_11075 of G. obscuriglobus, before secondary structure analysis indicated presence of the combination of α-solenoid and β propeller domains characteristic for coatomer proteins such as clathrin (Santarella-Mellwig et al., 2010). However, an alignment of gp4978 with clathrin heavy-chain from yeast in fact displays many identities occurring in homologous positions of the primary amino acid sequence (see Supplementary Figure S8 in Lonhienne et al., 2010). Even α-solenoids or stacked pair α-helix (SPAH) structures as they may also be sometimes termed, are difficult to detect due to limits of present structure determination methods (Field et al., 2011). The homology may well be a very ancient one, much more so than homologies detectable between genes of Eucarya and Archaea for example. Perhaps it even goes back as far as the progenote pre-Domain gene pool (Woese, 1998) or a eukaryote-like LUCA. Even the apparent homologies between archaeal and eukaryal genes detectable may be remnants of very ancient HGT events or even vertical connections to a progenote pre-Domain with resemblances to all three Domains, but presumably in this case not enough of the original signal has been lost to eliminate detection via simple Blast searches based on primary sequence similarity.
Serious consideration should be given to the possibility that complex features have been retained by Planctomycetes from a eukaryote-like ancestor such as a eukaryote-like LUCA (i.e., that reductive evolution has occurred; Forterre and Gribaldo, 2010; Forterre, 2011). Such reductive evolution from an ancestor with complex cell structure may have resulted in retention of some eukaryote-like features such as compartmentalization and loss/modification of others. The prediction can be made that an ancient link between a proto-eukaryote and PVCs would give rise to exactly the kind of remote sequence similarities and difficulties in confirming gene homologies which are found with Planctomycetes.
The concept of a compartmentalized last bacterial common ancestor (LBCA) derived from the eukaryote-like LUCA has been proposed partly on the basis of the shared compartmentalization among the PVC superphylum phyla, and the occurrence of eukaryote-like tubulins and MC proteins in that group (Forterre, 2011). If there were also a complex last archaeal common ancestor (LACA), then we might expect evidence for compartmentalized ancestors of Archaea. Such evidence has been found from phylogenomics, the functionally complex membranes of Ignicoccus (Kuper et al., 2010) and the ESCRT cell division and actin-homologous proteins in some archaea (Ettema and Bernander, 2009; Yutin et al., 2009; Makarova et al., 2010; Forterre, 2011). Combining these concepts yields the conclusion of a eukaryote-like or proto-eukaryote compartmentalized LUCA. Reductive evolution and gene loss from such an organism may have proceeded more extensively in most Bacteria and Archaea. However, LBCA and LACA as well as some more lately evolved members of both domains may have retained proto-eukaryote features. In Figure 2A, we have illustrated such a complex “synkaryotic” (Forterre and Gribaldo, 2010) LUCA cell. This cell would have possessed an endomembrane system ER communicating with both PM and periplasm of the cell and with the nuclear cisterna, and endocytosis via MC-protein-associated vesicles infolded from the PM. Ribosomes are illustrated both within a nuclear envelope-bounded nuclear region and in other cytoplasm, in a similar manner to their distribution in Gemmata. The controversial indication of at least some translation occurring within nuclei of even advanced eukaryote cells (Iborra et al., 2001, 2004) could be a remnant of such an early distribution where transcription and translation remained coupled to some extent.
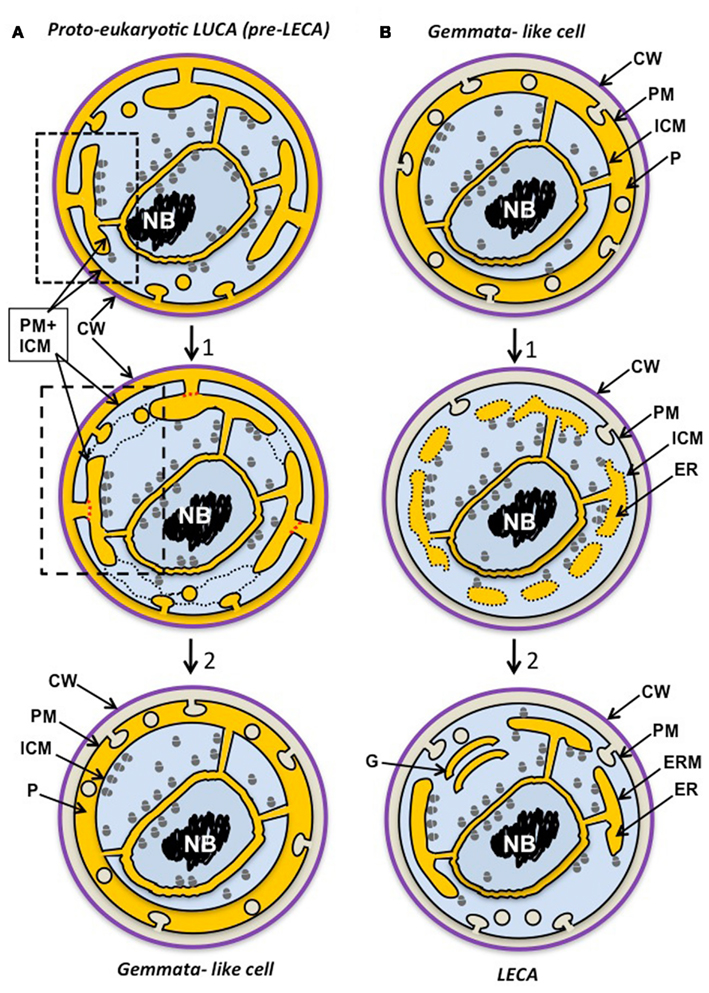
Figure 2. Possible scenarios for evolution of Gemmata obscuriglobus. (A) The planctomycete cell plan originates via reductive evolution from a proto-eukaryote, either a eukaryote-like LUCA ancestor of the three domains itself or a later precursor to the last eukaryotic common ancestor (LECA). This proto-eukaryote lineage has evolved endocytotic protein uptake via PM vesicle budding as its main form of nutrition (following protocoatomer evolution) and this has allowed bound ribosomes initially on a PM to become attached to internal ER-like membranes and a nuclear compartment. The proto-eukaryote has a simplified mitochondrion-less (“archaezoan”) eukaryote cell plan where ER endomembrane is continuous with PM (dotted box), endocytotic vesicles bud from PM, and ribosomes are bound only to endomembranes, including inner and outer nuclear envelope membranes (unlike modern eukaryotes); translation is not compartmented, and there may be no introns or pre-mRNA splicesosomal processing. In a second stage of differentiation (1), the cell plan first becomes simplified such that ER membrane becomes separated from PM (red dotted lines), ER vescicles fuse (black dotted lines), and endocytotic vesicle formation occurs in the now specialized ER-like paryphoplasm compartment. This results in increased endocytosis efficiency via macromolecular crowding in a confined compartment. Finally a Gemmata-like cell evolves capable of endocytosis by PM infolding into the paryphoplasm compartment which can be specialized for endocytotic nutrient acquisition and expansion of lysosome-like nutrient degradation in low osmolarity, high turgor pressure habitats. The nucleus preserves the eukaryote-like advantages of macromolecular crowding, replication and transcription factories and efficient chromosomal segregation without DNA-plasma membrane contact. Note that topologically equivalent compartments or cell components appear in the same color in different stages and scenarios – ER and paryphoplasm (yellow), ribosome-containing cytoplasm (blue), chromosomal DNA (black) and cell wall (purple). CW, cell wall; PM, plasma membrane; ICM, intracytoplasmic membrane; P, paryphoplasm. (B) In this second scenario a eukaryotic cell plan originates from the cell plan of a complex Gemmata obscuriglobus-like planctomycete bacterium (an ancestor of modern Gemmata) possessing a nuclear envelope, endocytosis and endocytotic vesicles in the paryphoplasm (yellow) and a cell wall, an organism capable of protein uptake and thus with a competitive novel nutrition relative to other bacteria. This ancestral form passes through a stage of differentiation (1) where aspects of planctomycete cell plan are conserved – such as ribosomes inside and outside of the nucleus and accompanying incomplete compartmentation of translation – but there are significant changes to membrane–bounded ER-like compartments such that, starting with paryphoplasm, separate compartments are walled off (forming some isolated vesicles that disperse – indicated by dotted lines), making possible greater RER surface for protein synthesis and complex processing of proteins through differentiated compartments. Vesicle transport is now used for wall protein addition and PM is generated via exocytotic vesicle transport from ER; finally in the mitochondrion-less LECA cell, endomembrane is differentiated such that some form of Golgi apparatus (similar to the simple form in Giardia) allows protein processing via trafficking. Translation is now predominantly in the cytoplasm outside the nucleus, allowing splicesosomal pre-mRNA splicing in the separate nuclear compartment. A stage after this would involve a loss of the cell wall correlated with development of phagocytosis. Cell wall loss via mutation in an endocytosis-capable ancestral eukaryote would lead to selection for phagocytic ability and a new form of nutrition via particle (particulate organic matter, POM) acquisition – e.g., dead bacterial cells and degradation products – and later, predation of other cells in a microbial community. ER and paryphoplasm (yellow), ribosome-containing cytoplasm (blue), chromosomal DNA (black), and cell wall (purple). CW, cell wall; CM, cytoplasmic membrane; G. Golgi apparatus; ICM, intracytoplasmic membrane; ER, endoplasmic reticulum; ERM, endoplasmic reticulum membrane; P, paryphoplasm.
The idea of such a compartmentalized LUCA implies that organisms similar to PVC group compartmentalized cells might be found among the deep-branching Archaea. The features of the archaeon Ignicoccus hospitalis are consistent with features of a comparmetalized Archaeal ancestor. The energized ATP synthase-containing outermost membrane in Ignicoccus (Kuper et al., 2010) implies that the inner membrane of that organism may be equivalent to the ICM of PVC bacteria. A compartmentalized LBCA could even encompass a full peptidoglycan wall, and could also possess some outer membrane synthesis genes. An even more radical concept would propose that LUCA was a progenote (Woese, 1990, p. 310) with a genome reflecting a stage before “annealing” of the Domains (Woese, 1998, p. 313). This progenote would possess some genes ancestral to and homologous with each Domain including those of Eucarya, making this also a proto-eukaryote with the potential for compartmentalization. This proto-eukaryote LUCA would have an endomembrane system and endocytosis abilities as outlined below. This has the advantage of explaining retention of both bacterial and eucaryal features in the PVC group – they are effectively pre-Bacterial and pre-Eucaryal characters (see Figure 2A) and the retention of compartmentalization. A compartmentalized LBCA or LUCA ancestor to the LBCA could explain the occurrence of some properties in members of the cyanobacteria such as internal membranes unconnected in a clear way to the PM, and ribosomes bound to internal membrane. In relation to how internal membranes in cyanobacteria might be formed, it is of interest that the only clear bacterial homolog of dynamin important in eukaryote endocytotic vesicle formation via membrane bending is from cyanobacteria (Lowe and Low, 2006). Perhaps in organisms where compartmentalization by internal membranes performed a useful function, including the diverged phyla Planctomycetes and Cyanobacteria, LBCA compartmentalization was retained, but in most Bacteria complete reductive loss of internal membranes occurred.
Alternatively to a reductive scenario, the Planctomycetes as well as some groups of archaea may have acted as the foundaries of early eukaryote-homologous genes. They effectively and independently assembled tool-kits ready to provide separately developed Eucarya domain members (with 18S rRNA and 80S ribosomes) with the elements needed for full eukaryality (e.g., endocytosis, cytoskeleton, nuclear envelope assembly nuclear transport), for example via lateral gene transfer. So, Planctomycetes or a PVC ancestor might donate endocytosis-like MC proteins, true tubulins, sterols and nuclear envelope assembly, while archaea would donate actins, the ESCRT system for advanced endocytotic late endosome function, and eukaryotic histones (Forterre, 2011). Supplementary elements (e.g., for spliceosome origins) could be supplied from virus genomes. Advantages of this model are that compartmentalized eukaryote first eukaryotic common ancestor (FECA) or LECA evolution could occur quite late after bacterial and archaeal domain development, and yet a simple Eucarya ribosomal RNA-defined domain lineage could have developed very early.
Even cell compartmentalization as such could have been supplied to an early eukaryote such as LECA or its ancestor (Figure 2B) from a Gemmata-like cell. This would require a remodeling of the paryphoplasm and ICM such that a more dispersed ER and endomembrane system would be evolved in the proto-eukaryotic ancestor, but this could also have happened via lateral transfer of gene sets coding for compartment membranes.
To extend the idea of a complex LUCA in a radical way, there seems no reason why endocytosis could not have been a relatively primary form of heterotrophic nutrition in LUCA or a primordial progenote. A protocoatomer capable of allowing the sort of protein uptake we see in Gemmata could have evolved from duplication of existing motifs to form an MC-like protein able to stabilize endocytotic vesicles. This could have been selected for due to the competitive advantage supplied by a powerful form of nutrition for LUCA, which could by this means have utilized polypeptides present abiotically in a complex primordial soup or released from dead progenote cells. During reductive evolution perhaps following a thermal crisis precipitating the annealing of the distinct Domains, most Bacteria and Archaea could have lost this ability and the associated membrane compartmentalization. This compartmentalization was retained along with MC homologs in the PVC superphylum, while being modified for specialized metabolism in organisms like anammox planctomycete ammonium oxidizers and the photosynthetic cyanobacteria. Alternatively, in a eukaryotes-late scenario, planctomycete ancestors could have invented simple endocytosis and associated necessary proteins and supplied FECA or LECA with this machinery by lateral transfer.
Occurrence of endocytosis and endocytotic components in this planctomycete species is completely consistent with recent analyses showing that the endomembrane system of eukaryotes evolved before LECA. All major vesicle coats and organelles associated with these coats appear from phylogenetic analysis to have evolved before LECA, even though some endocytosis proteins may have evolved later (Dacks and Field, 2007; Dacks et al., 2008, 2009). Mitochondria do not appear to have underpinned the origin and expansion of the endomembrane system and endocytosis- related gene families of eukaryotes.
Planctomycete wall structure and budding reproduction are an indication of their distinctiveness relative to other Bacteria, and these do not seem likely to be homologous with eukaryote characters without further molecular evidence. However, such features may prove to be relevant to possible evolutionary pathways to or from eukaryotes, e.g., loss of peptidoglycan and new sets of cell division proteins (Bernander and Ettema, 2010), new types of membrane reorganization during division (Lee et al., 2009a), and even the prediction of a wall-less phagocytic planctomycete similar to forms postulated in some early models for eukaryogenesis (Cavalier-Smith, 1981).
The problem of eukaryote origins and the nature of LUCA is too significant to superficially dismiss the relevance of Planctomycetes on the basis of anything less than the clearest understanding of available data on their structure and function. This includes molecular level interpretation of protein structure, as in the recent controversy regarding MC proteins (McInerney et al., 2011; Devos, 2012). We may not have sufficient evidence concerning molecular homologies of planctomycete genes with those of eukaryotes to exclude analogy and convergent evolution of similar structures and functions, but we also have insufficient evidence to exclude homology, especially since models for eukaryote evolution including eukaryote-like common ancestry need to be considered in judging such homology.
Analogy is inherently difficult to test and ambiguous conceptually. It implies (1) similarity of structure without an explanation of common ancestry and thus selection for similar function in absence of similar genes (convergence) or (2) constraint to develop similar structure and function due to common gene ancestry (parallelism). The latter case is very difficult to distinguish from homology (Gould, 2002). To reject homology in favor of analogy one needs very clear alternative models for how similar structures might arise from different gene lineages. In this case, one needs to explain how endomembranes and endocytosis correlated with clathrin-like proteins can evolve in a cell lineage with no ancestral tree or net connections to Eucarya. In the absence of such models analogy as an explanation for the remarkable eukaryote features of Planctomycetes remains unconvincing. Parallelism and homology understood in classical macrobiologically derived senses both imply homologous genes, the central issue for argument here and for future tests.
The questions arising regarding planctomycete relevance to eukaryote evolution and planctomycete homology with eukaryotes will rely for their answers on experimental data concerning actual abilities of Planctomycetes as well as on genomics and in silico analysis. The key to the evolutionary significance of Planctomycetes is to be found in a “top-down” approach starting with cell biology phenomena and structures such as endocytosis and the membrane-bounded nuclear body of Gemmata, proteomics of organelles and membranes, as well as in silico evidence. Considering the large proportion of hypothetical proteins in planctomycete proteomes, a “bottom-up” approach starting with the genome and deduced proteome alone would be likely to end in an impasse. Interactive feedback between top-down and bottom-up approaches will instead be needed, combined with an absence of too firm an adherence to existing dogma relating to eukaryote and Domain evolution. The discovery of endocytotic protein uptake in G. obscuriglobus is a good example of that approach, and there will most probably be other important examples. Such experimental data concerning Planctomycetes and their relatives must be taken into account in formulating and evaluating future hypotheses concerning origins and evolution of eukaryote cell organization. All the consequences of assuming homology of the planctomycete Gemmata to eukaryotes in structure and function make testable experimental predictions, especially after genetic systems for Planctomycetes are developed (Jogler et al., 2011), regardless of detectable sequence similarity of proposed homologs. Assumption of only analogy with eukaryote endocytosis is on the other hand unproductive concerning experimental predictions.
Attempted rejections of the hypothesis of homology of planctomycete features with those of eukaryotes requires more than alternative scenarios of eukaryote origins involving the less and less tenable (Poole and Penny, 2007; Forterre, 2011) fusion of Bacteria and Archaea domains. The concept of shared ancestry between eukaryotes and PVC group members is a key one for testing the hypothesis of homology. To test only the hypothesis of Planctomycetes as intermediates in a linear progression to eukaryotes is to ignore forms of homology which may lead to deeper understanding of the meaning of Planctomycetes for cell organization evolution. If we can clear the intellectual air to allow more illuminating evolutionary models, we may be able to see so well we can even make out the landscape of the mythical land of LUCA itself.
Anammox: anaerobic ammonium oxidation; a process whereby ammonium is oxidized to N2 with nitrite as electron acceptor, performed by some autotrophic Planctomycetes such as “Candidatus Brocadia anammoxidans” and “Candidatus Kuenenia stuttgartiensis.” Anammox Planctomycetes contain a special anammoxosome organelle bounded by a single membrane separating this ribosome-free organelle from the ribosome-rich pirellulosome in which it is contained.
Domains: in context other than referral to protein structural and folding domains, the three Domains of life, Bacteria, Archaea, and Eucarya, as originally defined on the basis of ribosomal RNA sequence analysis, but since largely confirmed via genome analysis (Woese et al., 1990).
FECA: the first eukaryotic common ancestor, the first type of cell on the stem lineage leading to the last ancestor shared by all extant eukaryotes – this organism may have existed quite early in evolution of the Domains, and some characteristics of FECA may have been lost by the time LECA evolved. If Eucarya is the root Domain, then LUCA may have been equivalent to FECA.
ICM: intracytoplasmic membrane of Planctomycetes; a single membrane surrounding the major pirellulosome compartment with ribosomes and nucleoid in Planctomycetes – it separates the outer ribosome-free paryphoplasm region from the inner ribosome-rich pirellulosome region of cytoplasm.
Ladderane: a type of lipid unique to anammox Planctomycetes containing a ladder-like arrangement of up to five fused cyclobutane rings in a linearly concatenated chain.
LACA: last archaeal common ancestor.
LBCA: last bacterial common ancestor.
LECA: last eukaryotic common ancestor – the last ancestor shared by all branches of the domain Eucarya.
LGT: lateral gene transfer; such transfer of course accompanies any endosymbiotic acquisition or engulfment of one type of cell by another, but could also occur via other less extensive mechanisms, e.g., virus infection. Care is needed in interpreting genome data to invoke LGT where it is not the only explanation for apparently mixed gene origins (Kurland et al., 2003).
LUCA: last universal common ancestor of all three domains of life.
MC protein: membrane-coating protein, a type of protein with characteristic domains such as alpha-solenoids and/or beta-propellors, and consistent with formation of regions of membrane curvature; such proteins encompass diverse proteins such as those homologous with clathrins associated with receptor-mediated endocytosis in eukaryotes, COPI, COPII, and some nuclear pore proteins of eukaryotes. Within Bacteria only limited species contain homologs of MC proteins, including many members of the PVC superphylum (Devos et al., 2004; Devos, 2012)
Nuclear body: a membrane-bounded organelle of the planctomycete G. obscuriglobus in which all the DNA of the cell is bound by an envelope of two closely apposed membranes.
Nucleoid: the region of the bacterial cell that contains the genomic DNA, usually seen in thin sections as a fibrillar region; in electron micrographs of cryosubstituted Escherichia coli cell sections, the nucleoid seems to occupy much of the cell, whereas it forms a highly condensed fibrillar structure in Planctomycetes.
Paryphoplasm: a region of the cytoplasm of the planctomycete cell between the cytoplasmic membrane and the ICM that contains no detectable ribosome-like particles but appears to contain RNA.
Pirellulosome: a region of planctomycete cell cytoplasm bound by the ICM that contains the fibrillar nucleoid and all the DNA of the cell.
Progenote: an early type of “organism” consisting of a community of cells rather than a single organismal lineage, occurring before separation of the three Domains of life, where extensive lateral gene transfer in a gene pool is postulated to have occurred, before “annealing” of genomes into Domain characteristics (Woese, 1990, 1998).
PVC: a superphylum consisting of the Bacteria phyla Planctomycetes, Verrucomicrobia, and Chlamydia (Wagner and Horn, 2006), as well as phyla such as the uncultured marine sponge species in phylum Poribacteria, the marine phylum Lentisphaerae, and several other phyla such as the uncultured OP3 phylum with its original member from a hot spring Obsidian Pool at Yellowstone National Park (Glockner et al., 2010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Australian Research Council for support of Planctomycete research in John A. Fuerst’s laboratory.
Abodeely, M., Dubois, K. N., Hehl, A., Stefanic, S., Sajid, M., Desouza, W., Attias, M., Engel, J. C., Hsieh, I., Fetter, R. D., and Mckerrow, J. H. (2009). A contiguous compartment functions as endoplasmic reticulum and endosome/lysosome in Giardia lamblia. Eukaryotic Cell 8, 1665–1676.
Bernander, R., and Ettema, T. J. G. (2010). FtsZ-less cell division in archaea and bacteria. Curr. Opin. Microbiol. 13, 747–752.
Bibi, E., and Herskovits, A. A. (2000). Association of Escherichia coli ribosomes with the inner membrane requires the signal recognition particle receptor but is independent of the signal recognition particle. Proc. Natl. Acad. Sci. U.S.A. 97, 4621–4626.
Brochier, C., and Philippe, H. (2002). Phylogeny: a non-hyperthermophilic ancestor for bacteria. Nature 417, 244.
Cavalier-Smith, T. (1981). “The origin and early evolution of the eukaryotic cell,” in Molecular and Cellular Aspects of Microbial Evolution, 32 Edn, eds M. J. Carlile, J. F. Collins, and B. E. B. Mosley (Cambridge: Cambridge University Press), 33–84.
Cavalier-Smith, T. (2009). Predation and eukaryote cell origins: a coevolutionary perspective. Int. J. Biochem. Cell Biol. 41, 307–322.
Cavalier-Smith, T. (2010). Origin of the cell nucleus, mitosis and sex: roles of intracellular coevolution. Biol. Direct 5, 7.
Chertkov, O., Sikorski, J., Nolan, M., Lapidus, A., Lucas, S., Del Rio, T. G., Tice, H., Cheng, J. F., Goodwin, L., Pitluck, S., Liolios, K., Ivanova, N., Mavromatis, K., Mikhailova, N., Ovchinnikova, G., Pati, A., Chen, A., Palaniappan, K., Djao, O. D., Land, M., Hauser, L., Chang, Y. J., Jeffries, C. D., Brettin, T., Han, C., Detter, J. C., Rohde, M., Goker, M., Woyke, T., Bristow, J., Eisen, J. A., Markowitz, V., Hugenholtz, P., Klenk, H. P., and Kyrpides, N. C. (2011). Complete genome sequence of Thermomonospora curvata type strain (B9). Stand. Genomic Sci. 4, 13–22.
Chistoserdova, L., Jenkins, C., Kalyuzhnaya, M. G., Marx, C. J., Lapidus, A., Vorholt, J. A., Staley, J. T., and Lidstrom, M. E. (2004). The enigmatic Planctomycetes may hold a key to the origins of methanogenesis and methylotrophy. Mol. Biol. Evol. 21, 1234–1241.
Clum, A., Tindall, B. J., Sikorski, J., Ivanova, N., Mavrommatis, K., Lucas, S., Glavina, T., Del, R., Nolan, M., Chen, F., Tice, H., Pitluck, S., Cheng, J. F., Chertkov, O., Brettin, T., Han, C., Detter, J. C., Kuske, C., Bruce, D., Goodwin, L., Ovchinikova, G., Pati, A., Mikhailova, N., Chen, A., Palaniappan, K., Land, M., Hauser, L., Chang, Y. J., Jeffries, C. D., Chain, P., Rohde, M., Goker, M., Bristow, J., Eisen, J. A., Markowitz, V., Hugenholtz, P., Kyrpides, N. C., Klenk, H. P., and Lapidus, A. (2009). Complete genome sequence of Pirellula staleyi type strain (ATCC 27377). Stand. Genomic Sci. 1, 308–316.
Conibear, E. (2010). Converging views of endocytosis in yeast and mammals. Curr. Opin. Cell Biol. 22, 513–518.
Dacks, J. B., and Field, M. C. (2007). Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J. Cell. Sci. 120, 2977–2985.
Dacks, J. B., Peden, A. A., and Field, M. C. (2009). Evolution of specificity in the eukaryotic endomembrane system. Int. J. Biochem. Cell Biol. 41, 330–340.
Dacks, J. B., Poon, P. P., and Field, M. C. (2008). Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc. Natl. Acad. Sci. U.S.A. 105, 588–593.
Delevoye, C., Nilges, M., Dehoux, P., Paumet, F., Perrinet, S., Dautry-Varsat, A., and Subtil, A. (2008). SNARE protein mimicry by an intracellular bacterium. PLoS Pathog. 4, e1000022. doi:10.1371/journal.ppat.1000022
Devos, D., Dokudovskaya, S., Alber, F., Williams, R., Chait, B. T., Sali, A., and Rout, M. P. (2004). Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2, e380. doi:10.1371/journal.pbio.0020380
Devos, D. P. (2012). Regarding the presence of membrane coat proteins in bacteria: confusion? What confusion? Bioessays 34, 38–39.
Ettema, T. J., and Bernander, R. (2009). Cell division and the ESCRT complex: asurprise from the archaea. Commun. Integr. Biol. 2, 86–88.
Field, M. C., and Dacks, J. B. (2009). First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes. Curr. Opin. Cell Biol. 21, 4–13.
Field, M. C., Sali, A., and Rout, M. P. (2011). On a bender-BARs, ESCRTs, COPs, and finally getting your coat. J. Cell Biol. 193, 963–972.
Forterre, P. (2011). A new fusion hypothesis for the origin of eukarya: better than previous ones, but probably also wrong. Res. Microbiol. 162, 77–91.
Forterre, P., and Gribaldo, S. (2010). Bacteria with a eukaryotic touch: a glimpse of ancient evolution? Proc. Natl. Acad. Sci. U.S.A. 107, 12739–12740.
Frickey, T., and Kannenberg, E. (2009). Phylogenetic analysis of the triterpene cyclase protein family in prokaryotes and eukaryotes suggests bidirectional lateral gene transfer. Environ. Microbiol. 11, 1224–1241.
Fuerst, J. A. (2005). Intracellular compartmentation in Planctomycetes. Annu. Rev. Microbiol. 59, 299–328.
Fuerst, J. A., Jenkins, C., and Kedar, V. (2002). Gene discovery within the planctomycete division of the domain bacteria using sequence tags from genomic DNA libraries. Genome Biol. 3, research0031–research0031.11.
Fuerst, J. A., and Sagulenko, E. (2010). Protein uptake by bacteria: an endocytosis-like process in the planctomycete Gemmata obscuriglobus. Commun. Integr. Biol. 3, 572–575.
Fuerst, J. A., and Sagulenko, E. (2011). Beyond the bacterium: Planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 9, 403–413.
Glockner, F. O., Kube, M., Bauer, M., Teeling, H., Lombardot, T., Ludwig, W., Gade, D., Beck, A., Borzym, K., Heitmann, K., Rabus, R., Schlesner, H., Amann, R., and Reinhardt, R. (2003). Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. U.S.A. 100, 8298–8303.
Glockner, F. O., Teeling, H., Lombardot, T., Bauer, M., and Ludwig, W. (2004). Evaluation of the phylogenetic position of the planctomycete ‘Rhodopirellula baltica’ SH 1 by means of concatenated ribosomal protein sequences, DNA-directed RNA polymerase subunit sequences and whole genome trees. Int. J. Syst. Evol. Microbiol. 54, 791–801.
Glockner, J., Kube, M., Shrestha, P. M., Weber, M., Glockner, F. O., Reinhardt, R., and Liesack, W. (2010). Phylogenetic diversity and metagenomics of candidate division OP3. Environ. Microbiol. 12, 1218–1229.
Goker, M., Cleland, D., Saunders, E., Lapidus, A., Nolan, M., Lucas, S., Hammon, N., Deshpande, S., Cheng, J. F., Tapia, R., Han, C., Goodwin, L., Pitluck, S., Liolios, K., Pagani, I., Ivanova, N., Mavromatis, K., Pati, A., Chen, A., Palaniappan, K., Land, M., Hauser, L., Chang, Y. J., Jeffries, C. D., Detter, J. C., Beck, B., Woyke, T., Bristow, J., Eisen, J. A., Markowitz, V., Hugenholtz, P., Kyrpides, N. C., and Klenk, H. P. (2011). Complete genome sequence of Isosphaera pallida type strain (IS1B). Stand. Genomic Sci. 4, 63–71.
Gould, S. J. (2002). The Structure of Evolutionary Theory. Cambridge, MA: The Belknap Press of Harvard University Press.
Han, C., Spring, S., Lapidus, A., Del Rio, T. G., Tice, H., Copeland, A., Cheng, J. F., Lucas, S., Chen, F., Nolan, M., Bruce, D., Goodwin, L., Pitluck, S., Ivanova, N., Mavromatis, K., Mikhailova, N., Pati, A., Chen, A., Palaniappan, K., Land, M., Hauser, L., Chang, Y. J., Jeffries, C. C., Saunders, E., Chertkov, O., Brettin, T., Goker, M., Rohde, M., Bristow, J., Eisen, J. A., Markowitz, V., Hugenholtz, P., Kyrpides, N. C., Klenk, H. P., and Detter, J. C. (2009). Complete genome sequence of Pedobacter heparinus type strain (HIM 762-3). Stand. Genomic Sci. 1, 54–62.
Hieu, C. X., Voigt, B., Albrecht, D., Becher, D., Lombardot, T., Glockner, F. O., Amann, R., Hecker, M., and Schweder, T. (2008). Detailed proteome analysis of growing cells of the planctomycete Rhodopirellula baltica SH1T. Proteomics 8, 1608–1623.
Huson, D. H., Henz, S. R., Auch, A. F., Nieselt-Struwe, K., and Schuster, S. C. (2005). Whole-genome prokaryotic phylogeny. Bioinformatics 21, 2329–2335.
Hyatt, D., Chen, G. L., Locascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119. doi:10.1186/1471-2105-11-119
Iborra, F. J., Jackson, D. A., and Cook, P. R. (2001). Coupled transcription and translation within nuclei of mammalian cells. Science 293, 1139–1142.
Iborra, F. J., Jackson, D. A., and Cook, P. R. (2004). The case for nuclear translation. J. Cell. Sci. 117, 5713–5720.
Ivanova, N., Sikorski, J., Jando, M., Lapidus, A., Nolan, M., Lucas, S., Del Rio, T. G., Tice, H., Copeland, A., Cheng, J. F., Chen, F., Bruce, D., Goodwin, L., Pitluck, S., Mavromatis, K., Ovchinnikova, G., Pati, A., Chen, A., Palaniappan, K., Land, M., Hauser, L., Chang, Y. J., Jeffries, C. D., Chain, P., Saunders, E., Han, C., Detter, J. C., Brettin, T., Rohde, M., Goker, M., Bristow, J., Eisen, J. A., Markowitz, V., Hugenholtz, P., Klenk, H. P., and Kyrpides, N. C. (2010a). Complete genome sequence of Gordonia bronchialis type strain (3410). Stand. Genomic Sci. 2, 19–28.
Ivanova, N., Sikorski, J., Jando, M., Munk, C., Lapidus, A., Glavina Del Rio, T., Copeland, A., Tice, H., Cheng, J. F., Lucas, S., Chen, F., Nolan, M., Bruce, D., Goodwin, L., Pitluck, S., Mavromatis, K., Mikhailova, N., Pati, A., Chen, A., Palaniappan, K., Land, M., Hauser, L., Chang, Y. J., Jeffries, C. D., Meincke, L., Brettin, T., Detter, J. C., Rohde, M., Goker, M., Bristow, J., Eisen, J. A., Markowitz, V., Hugenholtz, P., Kyrpides, N. C., and Klenk, H. P. (2010b). Complete genome sequence of Geodermatophilus obscurus type strain (G-20). Stand. Genomic Sci. 2, 158–167.
Jogler, C., Glockner, F. O., and Kolter, R. (2011). Characterization of Planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum Planctomycetes. Appl. Environ. Microbiol. 77, 5826–5829.
Johnson, S., Michalak, M., Opas, M., and Eggleton, P. (2001). The ins and outs of calreticulin: from the ER lumen to the extracellular space. Trends Cell Biol. 11, 122–129.
Jun, S. R., Sims, G. E., Wu, G. A., and Kim, S. H. (2010). Whole-proteome phylogeny of prokaryotes by feature frequency profiles: Aa alignment-free method with optimal feature resolution. Proc. Natl. Acad. Sci. U.S.A. 107, 133–138.
Kamneva, O. K., Liberles, D. A., and Ward, N. L. (2010). Genome-wide influence of indel substitutions on evolution of bacteria of the PVC superphylum, revealed using a novel computational method. Genome Biol. Evol. 2, 870–886.
Klis, F. M., Boorsma, A., and De Groot, P. W. J. (2006). Cell wall construction in Saccharomyces cerevisiae. Yeast 23, 185–202.
Komeili, A., Li, Z., Newman, D. K., and Jensen, G. J. (2006). Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311, 242–245.
Konorty, M., Kahana, N., Linaroudis, A., Minsky, A., and Medalia, O. (2008). Structural analysis of photosynthetic membranes by cryo-electron tomography of intact Rhodopseudomonas viridis cells. J. Struct. Biol. 161, 393–400.
Koonin, E. V. (2010). The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 11, 209.
Kuper, U., Meyer, C., Muller, V., Rachel, R., and Huber, H. (2010). Energized outer membrane and spatial separation of metabolic processes in the hyperthermophilic Archaeon Ignicoccus hospitalis. Proc. Natl. Acad. Sci. U.S.A. 107, 3152–3156.
Kurland, C. G., Canback, B., and Berg, O. G. (2003). Horizontal gene transfer: a critical view. Proc. Natl. Acad. Sci. U.S.A. 100, 9658–9662.
Labutti, K., Sikorski, J., Schneider, S., Nolan, M., Lucas, S., Glavina Del Rio, T., Tice, H., Cheng, J. F., Goodwin, L., Pitluck, S., Liolios, K., Ivanova, N., Mavromatis, K., Mikhailova, N., Pati, A., Chen, A., Palaniappan, K., Land, M., Hauser, L., Chang, Y. J., Jeffries, C. D., Tindall, B. J., Rohde, M., Goker, M., Woyke, T., Bristow, J., Eisen, J. A., Markowitz, V., Hugenholtz, P., Kyrpides, N. C., Klenk, H. P., and Lapidus, A. (2010). Complete genome sequence of Planctomyces limnophilus type strain (Mu 290). Stand. Genomic Sci. 3, 47–56.
Lanfredi-Rangel, A., Attias, M., De Carvalho, T. M., Kattenbach, W. M., and De Souza, W. (1998). The peripheral vesicles of trophozoites of the primitive protozoan Giardia lamblia may correspond to early and late endosomes and to lysosomes. J. Struct. Biol. 123, 225–235.
Lang, E., Lapidus, A., Chertkov, O., Brettin, T., Detter, J. C., Han, C., Copeland, A., Glavina Del Rio, T., Nolan, M., Chen, F., Lucas, S., Tice, H., Cheng, J. F., Land, M., Hauser, L., Chang, Y. J., Jeffries, C. D., Kopitz, M., Bruce, D., Goodwin, L., Pitluck, S., Ovchinnikova, G., Pati, A., Ivanova, N., Mavrommatis, K., Chen, A., Palaniappan, K., Chain, P., Bristow, J., Eisen, J. A., Markowitz, V., Hugenholtz, P., Goker, M., Rohde, M., Kyrpides, N. C., and Klenk, H. P. (2009). Complete genome sequence of Dyadobacter fermentans type strain (NS114). Stand. Genomic Sci. 1, 133–140.
Leary, B. A., Ward-Rainey, N., and Hoover, T. R. (1998). Cloning and characterization of Planctomyces limnophilus rpoN: complementation of a Salmonella typhimurium rpoN mutant strain. Gene 221, 151–157.
Lee, K. C., Webb, R. I., and Fuerst, J. A. (2009a). The cell cycle of the planctomycete Gemmata obscuriglobus with respect to cell compartmentalization. BMC Cell Biol. 10, 4. doi:10.1186/1471-2121-10-4
Lee, K. C., Webb, R. I., Janssen, P. H., Sangwan, P., Romeo, T., Staley, J. T., and Fuerst, J. A. (2009b). Phylum Verrucomicrobia representatives share a compartmentalized cell plan with members of bacterial phylum Planctomycetes. BMC Microbiol. 9, 5. doi:10.1186/1471-2180-9-5
Liberton, M., Howard Berg, R., Heuser, J., Roth, R., and Pakrasi, H. B. (2006). Ultrastructure of the membrane systems in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. Protoplasma 227, 129–138.
Liesack, W., and Stackebrandt, E. (1989). Evidence for unlinked rrn operons in the planctomycete Pirellula marina. J. Bacteriol. 171, 5025–5030.
Lindas, A. C., Karlsson, E. A., Lindgren, M. T., Ettema, T. J., and Bernander, R. (2008). A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. U.S.A. 105, 18942–18946.
Lindsay, M. R., Webb, R. I., Strous, M., Jetten, M. S., Butler, M. K., Forde, R. J., and Fuerst, J. A. (2001). Cell compartmentalisation in Planctomycetes: novel types of structural organisation for the bacterial cell. Arch. Microbiol. 175, 413–429.
Lonhienne, T. G., Sagulenko, E., Webb, R. I., Lee, K. C., Franke, J., Devos, D. P., Nouwens, A., Carroll, B. J., and Fuerst, J. A. (2010). Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. U.S.A. 107, 12883–12888.
Makarova, K. S., Yutin, N., Bell, S. D., and Koonin, E. V. (2010). Evolution of diverse cell division and vesicle formation systems in archaea. Nat. Rev. Microbiol. 8, 731–741.
Marti, M., Regos, A., Li, Y., Schraner, E. M., Wild, P., Muller, N., Knopf, L. G., and Hehl, A. B. (2003). An ancestral secretory apparatus in the protozoan parasite Giardia intestinalis. J. Biol. Chem. 278, 24837–24848.
Martin-Galiano, A. J., Oliva, M. A., Sanz, L., Bhattacharyya, A., Serna, M., Yebenes, H., Valpuesta, J. M., and Andreu, J. M. (2011). Bacterial tubulin distinct loop sequences and primitive assembly properties support its origin from a eukaryotic tubulin ancestor. J.Biol. Chem. 286, 19789–19803.
McInerney, J. O., Martin, W. F., Koonin, E. V., Allen, J. F., Galperin, M. Y., Lane, N., Archibald, J. M., and Embley, T. M. (2011). Planctomycetes and eukaryotes: a case of analogy not homology. Bioessays 33, 810–817.
Medema, M. H., Zhou, M., Van Hijum, S. A., Gloerich, J., Wessels, H. J., Siezen, R. J., and Strous, M. (2010). A predicted physicochemically distinct sub-proteome associated with the intracellular organelle of the anammox bacterium Kuenenia stuttgartiensis. BMC Genomics 11, 299. doi:10.1186/1471-2164-11-299
Meusser, B., Hirsch, C., Jarosch, E., and Sommer, T. (2005). ERAD: the long road to destruction. Nat. Cell Biol. 7, 766–772.
Munk, C., Lapidus, A., Copeland, A., Jando, M., Mayilraj, S., Glavina Del Rio, T., Nolan, M., Chen, F., Lucas, S., Tice, H., Cheng, J. F., Han, C., Detter, J. C., Bruce, D., Goodwin, L., Chain, P., Pitluck, S., Goker, M., Ovchinikova, G., Pati, A., Ivanova, N., Mavromatis, K., Chen, A., Palaniappan, K., Land, M., Hauser, L., Chang, Y. J., Jeffries, C. D., Bristow, J., Eisen, J. A., Markowitz, V., Hugenholtz, P., Kyrpides, N. C., and Klenk, H. P. (2009). Complete genome sequence of Stackebrandtia nassauensis type strain (LLR-40K-21). Stand. Genomic Sci. 1, 234–241.
Nevo, R., Charuvi, D., Shimoni, E., Schwarz, R., Kaplan, A., Ohad, I., and Reich, Z. (2007). Thylakoid membrane perforations and connectivity enable intracellular traffic in cyanobacteria. EMBO J. 26, 1467–1473.
Pallen, M. J., Snyder, L. a. S., Lornan, N. J., and Futterer, K. (2009). Bacterial flagellar diversity and evolution: seek simplicity and distrust it? Trends Microbiol. 17, 1–5.
Pati, A., Ivanova, N. N., Mikhailova, N., Ovchinnikova, G., Hooper, S. D., Lykidis, A., and Kyrpides, N. C. (2010). GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat. Methods 7, 455–457.
Paumet, F., Wesolowski, J., Garcia-Diaz, A., Delevoye, C., Aulner, N., Shuman, H. A., Subtil, A., and Rothman, J. E. (2009a). Intracellular bacteria encode inhibitory SNARE-like proteins. PLoS ONE 4, e7375. doi:10.1371/journal.pone.0007375
Paumet, F., Wesolowski, J., Garcia-Diaz, A., Delevoye, C., Aulner, N., Shuman, H. A., Subtil, A., and Rothman, J. E. (2009b). Intracellular bacteria encode inhibitory SNARE-like proteins. PLoS ONE 4, e7375. doi:10.1371/journal.pone.0007375
Pelve, E. A., Lindas, A. C., Martens-Habbena, W., De La Torre, J. R., Stahl, D. A., and Bernander, R. (2011). Cdv-based cell division and cell cycle organization in the thaumarchaeon Nitrosopumilus maritimus. Mol. Microbiol. 82, 555–566.
Pilhofer, M., Ladinsky, M. S., Mcdowall, A. W., Petroni, G., and Jensen, G. J. (2011). Microtubules in bacteria: ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol. 9, e1001213. doi:10.1371/journal.pbio.1001213
Pilhofer, M., Rappl, K., Eckl, C., Bauer, A. P., Ludwig, W., Schleifer, K. H., and Petroni, G. (2008). Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J. Bacteriol. 190, 3192–3202.
Poole, A. M., and Penny, D. (2007). Evaluating hypotheses for the origin of eukaryotes. Bioessays 29, 74–84.
Pukall, R., Lapidus, A., Glavina Del Rio, T., Copeland, A., Tice, H., Cheng, J. F., Lucas, S., Chen, F., Nolan, M., Bruce, D., Goodwin, L., Pitluck, S., Mavromatis, K., Ivanova, N., Ovchinnikova, G., Pati, A., Chen, A., Palaniappan, K., Land, M., Hauser, L., Chang, Y. J., Jeffries, C. D., Chain, P., Meincke, L., Sims, D., Brettin, T., Detter, J. C., Rohde, M., Goker, M., Bristow, J., Eisen, J. A., Markowitz, V., Kyrpides, N. C., Klenk, H. P., and Hugenholtz, P. (2010). Complete genome sequence of Conexibacter woesei type strain (ID131577). Stand. Genomic Sci. 2, 212–219.
Reeve, W., O’Hara, G., Chain, P., Ardley, J., Brau, L., Nandesena, K., Tiwari, R., Malfatti, S., Kiss, H., Lapidus, A., Copeland, A., Nolan, M., Land, M., Ivanova, N., Mavromatis, K., Markowitz, V., Kyrpides, N., Melino, V., Denton, M., Yates, R., and Howieson, J. (2010). Complete genome sequence of Rhizobium leguminosarum bv trifolii strain WSM2304, an effective microsymbiont of the South American clover Trifolium polymorphum. Stand. Genomic Sci. 2, 66–76.
Remsen, C. C., Watson, S. W., Waterbury, J. B., and Truper, H. G. (1968). Fine structure of Ectothiorhodospira mobilis Pelsh. J. Bacteriol. 95, 2374–2392.
Reynaud, E. G., and Devos, D. P. (2011). Transitional forms between the three domains of life and evolutionary implications. Proc Biol Sci. 278, 3321–3328.
Santarella-Mellwig, R., Franke, J., Jaedicke, A., Gorjanacz, M., Bauer, U., Budd, A., Mattaj, I. W., and Devos, D. P. (2010). The compartmentalized bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum have membrane coat-like proteins. PLoS Biol. 8, e1000281. doi:10.1371/journal.pbio.1000281
Schlieper, D., Oliva, M. A., Andreu, J. M., and Lowe, J. (2005). Structure of bacterial tubulin BtubA/B: evidence for horizontal gene transfer. Proc. Natl. Acad. Sci. U.S.A. 102, 9170–9175.
Schneider, D., Fuhrmann, E., Scholz, I., Hess, W. R., and Graumann, P. L. (2007). Fluorescence staining of live cyanobacterial cells suggest non-stringent chromosome segregation and absence of a connection between cytoplasmic and thylakoid membranes. BMC Cell Biol. 8, 39. doi:10.1186/1471-2121-8-39
Schuler, D. (2008). Genetics and cell biology of magnetosome formation in magnetotactic bacteria. FEMS Microbiol. Rev. 32, 654–672.
Sinninghe Damste, J. S., Rijpstra, W. I., Strous, M., Jetten, M. S., David, O. R., Geenevasen, J. A., and Van Maarseveen, J. H. (2004). A mixed ladderane/n-alkyl glycerol diether membrane lipid in an anaerobic ammonium-oxidizing bacterium. Chem. Commun. (Camb.) 2590–2591.
Studholme, D. J., Fuerst, J. A., and Bateman, A. (2004). Novel protein domains and motifs in the marine planctomycete Rhodopirellula baltica. FEMS Microbiol. Lett. 236, 333–340.
Sun, H., Lapidus, A., Nolan, M., Lucas, S., Del Rio, T. G., Tice, H., Cheng, J. F., Tapia, R., Han, C., Goodwin, L., Pitluck, S., Pagani, I., Ivanova, N., Mavromatis, K., Mikhailova, N., Pati, A., Chen, A., Palaniappan, K., Land, M., Hauser, L., Chang, Y. J., Jeffries, C. D., Djao, O. D., Rohde, M., Sikorski, J., Goker, M., Woyke, T., Bristow, J., Eisen, J. A., Markowitz, V., Hugenholtz, P., Kyrpides, N. C., and Klenk, H. P. (2010). Complete genome sequence of Nocardiopsis dassonvillei type strain (IMRU 509). Stand. Genomic Sci. 3, 325–336.
Sutcliffe, I. C. (2010). A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 18, 464–470.
Tice, H., Mayilraj, S., Sims, D., Lapidus, A., Nolan, M., Lucas, S., Glavina Del Rio, T., Copeland, A., Cheng, J. F., Meincke, L., Bruce, D., Goodwin, L., Pitluck, S., Ivanova, N., Mavromatis, K., Ovchinnikova, G., Pati, A., Chen, A., Palaniappan, K., Land, M., Hauser, L., Chang, Y. J., Jeffries, C. D., Detter, J. C., Brettin, T., Rohde, M., Goker, M., Bristow, J., Eisen, J. A., Markowitz, V., Hugenholtz, P., Kyrpides, N. C., Klenk, H. P., and Chen, F. (2010). Complete genome sequence of Nakamurella multipartita type strain (Y-104). Stand. Genomic Sci. 2, 168–175.
Ting, C. S., Hsieh, C., Sundararaman, S., Mannella, C., and Marko, M. (2007). Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium. J. Bacteriol. 189, 4485–4493.
van de Meene, A. M., Hohmann-Marriott, M. F., Vermaas, W. F., and Roberson, R. W. (2006). The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch. Microbiol. 184, 259–270.
van Niftrik, L., Van Helden, M., Kirchen, S., Van Donselaar, E. G., Harhangi, H. R., Webb, R. I., Fuerst, J. A., Op Den Camp, H. J., Jetten, M. S., and Strous, M. (2010). Intracellular localization of membrane-bound ATPases in the compartmentalized anammox bacterium ‘Candidatus Kuenenia stuttgartiensis’. Mol. Microbiol. 77, 701–715.
Wagner, M., and Horn, M. (2006). The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 17, 241–249.
Ward-Rainey, N., Rainey, F. A., and Stackebrandt, E. (1997). The presence of a dnaK (HSP70) multigene family in members of the orders Planctomycetales and Verrucomicrobiales. J. Bacteriol. 179, 6360–6366.
Wilson, K. L., and Dawson, S. C. (2011). Evolution: functional evolution of nuclear structure. J. Cell Biol. 195, 171–181.
Woese, C. R., Kandler, O., and Wheelis, M. L. (1990). Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U.S.A. 87, 4576–4579.
Keywords: Planctomycetes, evolution, compartmentalization, cell complexity, LUCA, eukaryote evolution, origin of nucleus, Gemmata obscuriglobus
Citation: Fuerst JA and Sagulenko E (2012) Keys to eukaryality: Planctomycetes and ancestral evolution of cellular complexity. Front. Microbio. 3:167. doi: 10.3389/fmicb.2012.00167
Received: 19 January 2012; Accepted: 13 April 2012;
Published online: 04 May 2012.
Edited by:
Naomi L. Ward, University of Wyoming, USAReviewed by:
Jonathan H. Badger, J. Craig Venter Institute, USACopyright: © 2012 Fuerst and Sagulenko. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: John A. Fuerst, School of Chemistry and Molecular Biosciences, The University of Queensland, St Lucia 4072, QLD, Australia e-mail:ai5mdWVyc3RAdXEuZWR1LmF1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.