
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 16 March 2012
Sec. Terrestrial Microbiology
Volume 3 - 2012 | https://doi.org/10.3389/fmicb.2012.00084
This article is part of the Research TopicMicrobiology of wetlandsView all 18 articles
Salt marshes located on the east coast of temperate North America are highly productive, typically nitrogen-limited, and support diverse assemblages of free-living nitrogen fixing (diazotrophic) bacteria. This article reviews and analyzes data from North Inlet estuary (SC, USA), addressing diazotroph assemblage structure and the influence of plant host and environmental conditions on the assemblage. The North Inlet estuary is a salt marsh ecosystem in which anthropogenic influences are minimal and the distributions of diazotrophs are governed by the natural biota and dynamics of the system. Denaturing gradient gel electrophoresis fingerprinting and phylogenetic analyses of recovered sequences demonstrated that the distributions of some diazotrophs reflect plant host specificity and that diazotroph assemblages distributed across marsh gradients are also heavily influenced by edaphic conditions. Broadly distributed diazotrophs that are capable of maintaining populations under all environmental conditions spanning such gradients are also present in these assemblages. Statistical analyses indicate that the structures of diazotroph assemblages in different vegetation zones are significantly (p < 0.01) different. New data presented here demonstrate the heterogeneity of salt marsh rhizosphere microenvironments, and corroborate previous findings from different plant hosts growing at several locations within this estuary. The data from these collected works support the hypothesis that the biogeography of microorganisms is non-random and these biogeographic patterns are predictable.
Numerous studies have revealed the immense diversity of microorganisms and have lent insight into their distributions within and across ecosystems (Ward et al., 1990; Øvreås, 2000; Floyd et al., 2005). A number of recent studies have demonstrated that microorganisms have distinct patterns of distribution (microbial biodiversity over space and time) reminiscent of those of macroorganisms (Hughes Martiny et al., 2006). Biogeography concepts can be used to understand why organisms are distributed as they are within and across ecosystems and why stable, mixed communities exist (Green et al., 2008). On large regional scales, biogeographic patterns of microorganisms are more heavily influenced by edaphic parameters than by the factors that typically dictate the well documented biogeography of plants and animals, such as latitude and mean annual temperature (Fierer and Jackson, 2006). However, on the local scale, microbial biogeography is influenced by both edaphic and biotic parameters such as vegetation type, carbon availability, nutrient availability, and soil moisture. These patterns have been observed in terrestrial, wetland, and aquatic ecosystems (Crump et al., 2004; Dolan, 2005; Fierer and Jackson, 2006).
Salt marsh ecosystems are excellent locations for the investigation of biogeographical patterns due to the small number of dominant macrophytes and to predictable environmental effectors such as tidal inundation and salinity. Niche differentiation patterns are well-established as gradients of these effectors and develop in response to tidal flooding and bioturbation in salt marsh ecosystems. Microbial communities express niche differentiation based on environmental gradients, often responding strongly to one or more specific environmental effectors in the ecosystem (Crump et al., 2004; Macalady et al., 2008; Dumbrell et al., 2010; Davis et al., 2011). For example, microbial diversity in estuarine tidal creeks is strongly influenced by salinity (Hollibaugh et al., 2000; Selje and Simon, 2003; Crump et al., 2004), which reflects both the direct impact of salinity on microbial communities and the influences of advective inputs of marine and freshwater microorganisms. Niche-based theories of ecology help to explain the partitioning of limiting resources between competing species and the differentiation of niche space across all species within a community (Leibold and McPeek, 2008). Particularly with reference to functional assemblages (i.e., assemblages of organisms carrying out specific ecological activities), such theories can give insight into the distribution of organisms throughout ecosystems and their ability to contribute significantly to ecosystem function in differing niche dimensions.
This review will examine recent findings from North American salt marsh ecosystems, focusing specifically on the diverse assemblage of free-living diazotrophic (nitrogen fixing) organisms found there. The salt marsh diazotroph assemblage includes organisms that are quite specific for a given marsh microenvironment as well as for specific marsh plant hosts. This assemblage is also sufficiently stable in composition (determined through field manipulation experiments discussed below in the Section Niche Specialization) to make analysis of geographic patterns feasible.
Coastal salt marshes are characteristically dominated by herbs, low shrubs and grasses bordering saline water bodies and are subjected to periodic flooding as a result of fluctuations (tidal or non-tidal) in the sea/ocean (Adam, 1990). The marshes depend on the tidal cycle to maintain water regimes, flush peat and sediment with water and dissolved nutrients, and to maintain salinity gradients (Gardner, 1973; Hemond and Fifield, 1982; Adam, 1990). Tidally dominated salt marshes are transitional between the mainly submerged sea grass communities and fully exposed terrestrial zones, and have received extensive study due to their importance as nursery grounds for commercially important fisheries species and as sources of carbon and nitrogen to nearshore waters (Long and Long, 1983; Schubauer and Hopkinson, 1984; Dame and Kenny, 1986; Dame et al., 1986; Morris and Haskin, 1990; Morris et al., 1990; King and Lester, 1995; Dai and Wiegert, 1997).
The coastal salt marshes of the south-eastern United States are typically dominated by halophyte perennial graminoids which have extensive underground rhizome systems (Odum, 1988). The number of plant species that are numerically significant in coastal salt marshes is small (Schubauer and Hopkinson, 1984) and a clear zonation of plant species that is based on edaphic environmental conditions, interspecies competition and minor differences in elevation is routinely observed (Bertness and Ellison, 1987; Adam, 1990; Pennings and Callaway, 1992). Key abiotic factors that influence plant zonation include sediment porewater salinity, soluble sulfide concentration, pH and redox potential (Bertness, 1991). Microbial activities have also been thought to influence the development of plant zonation, as expressed through their ability to affect the concentrations of nutrients and other solutes, particularly sulfide (King et al., 1982). The low marsh, which is regularly flooded and where salinities are higher, is dominated by halophyte grasses such as Spartina alterniflora and Spartina patens. The high marsh typically has lower salinities, undergoes irregular flooding (with frequency of flooding based on tidal heights and the elevation gradient), is closer to the terrestrial zone typically fringing the marsh, and is dominated by rushes and grasses having lower salt tolerance, such as Juncus roemerianus. Areas of disturbance in the marsh, such as those experiencing long-term waterlogged conditions or unusually high rates of evapotranspiration, are often dominated by successional species such as Salicornia virginica (Pennings and Callaway, 1992; Levine et al., 1998). These plant zones are easily recognized in the field and some are monotypic with respect to the plant present.
Crab Haul Creek Basin, North Inlet, South Carolina hosts a typical tidally dominated salt marsh with obvious plant zonation, fresh water input primarily from local rainfall, and nitrogen limitation. North Inlet is a high salinity, relatively pristine estuary (Blood and Vernberg, 1992) which serves as a benchmark system in the United States National Estuarine Research Reserve. The Crab Haul Creek Basin has a relatively minor elevation gradient, just over 1 m from mean high tide to the highest points within the basin, with semi-diurnal tides averaging 1.5 m (Kjerfve, 1986). This relatively flat landscape results in the edaphic parameter gradients that have influenced the plant zonation patterns observed in this system (Figures 1 and 2). Porewater salinity and sulfide concentration gradients, in particular, drive the development of macrophyte zonation along this elevation gradient. Monotypic stands of short and tall growth forms of S. alterniflora occur in the low marsh, the successional species S. virginica in the waterlogged region of the midmarsh (as well as in a mixed stand with the short growth form S. alterniflora), and J. roemerianus in the high marsh and in patches of slightly higher elevation within the low marsh zone of short growth form S. alterniflora. This pattern of plant zonation has been stable for decades (e.g., Dame and Kenny, 1986; Morris and Haskin, 1990).
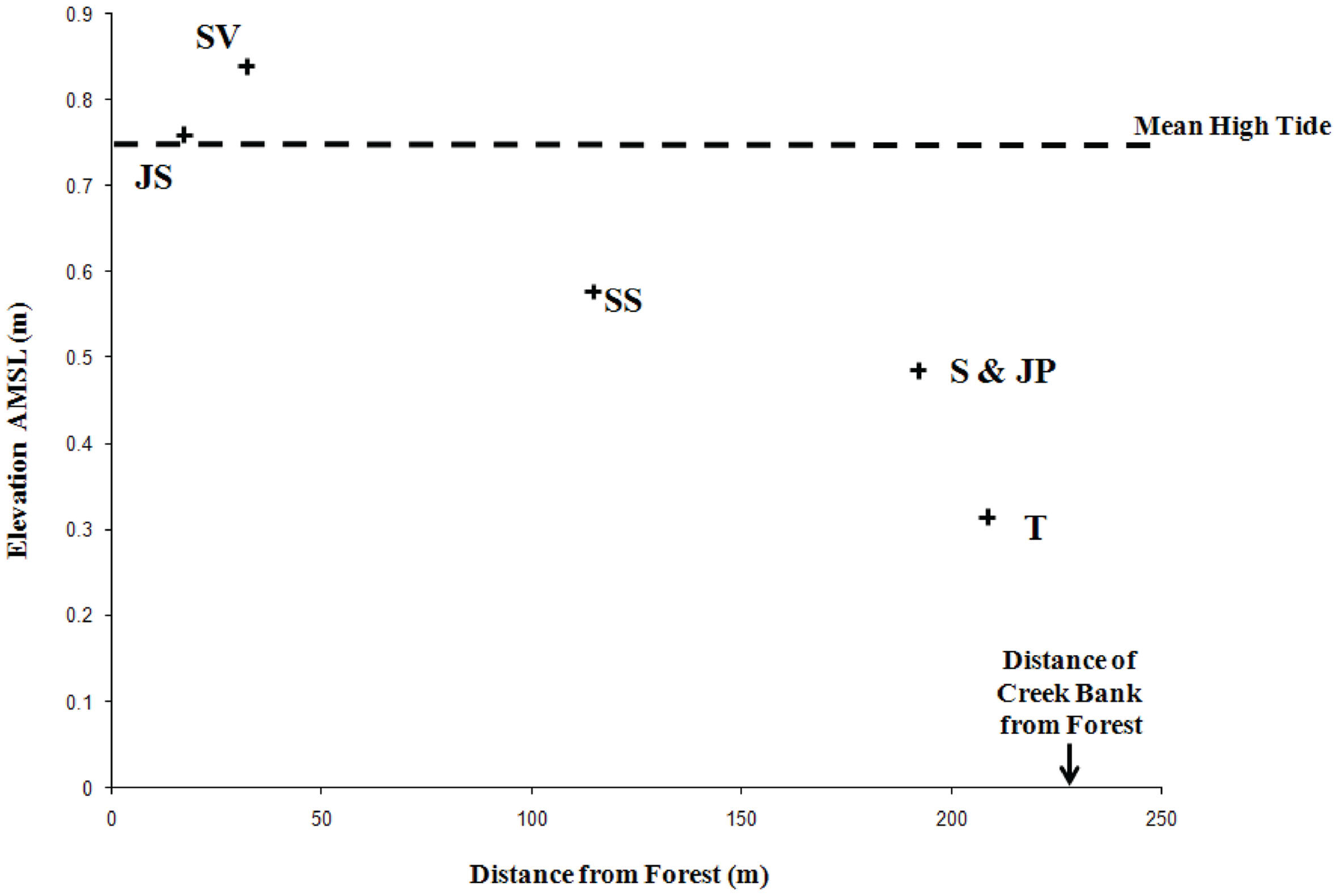
Figure 1. Differences in site elevation above mean sea level (AMSL) and distance of each sampling location from the forest edge. Site designations: Juncus roemerianus stand (JS); Salicornia virginica (SV); mixed plant zone of co-occurring S. virginica and short form Spartina alterniflora (SS); short form S. alterniflora (S); patch of J. roemerianus in the S zone (JP); and tall form S. alterniflora (T). Reproduced with kind permission from Davis et al. (2011), Figure 1.
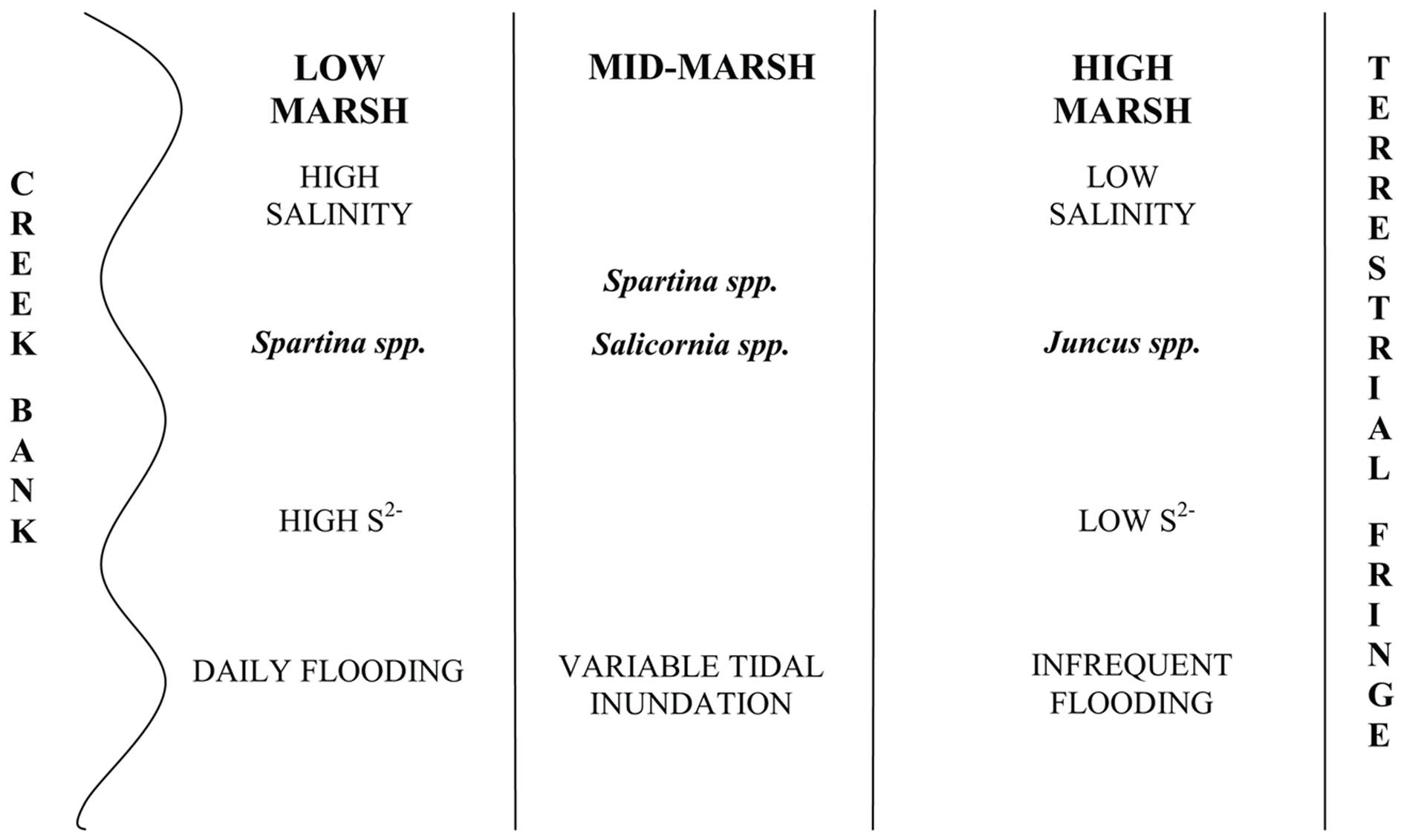
Figure 2. Typical marsh zonation including vegetation zonation and gradients of influential abiotic parameters as observed in Crab Haul Creek Basin, North Inlet.
Productivity in salt marshes has important impacts on coastal processes, and salt marshes are among the most productive ecosystems known (Keefe, 1972; Turner, 1976; Morris and Haskin, 1990). The average annual production ranges from 167 to 938 g m−2 year−1, however this value varies greatly with temporal and local conditions (Kirwan et al., 2009). Despite these high levels of productivity, salt marshes are often nitrogen-limited as combined nitrogen compounds (nitrate, nitrite and ammonia) can be rapidly depleted and sources of “new” nitrogen, such as N2 fixation, must be utilized (Howarth et al., 1988). Phosphorus can also be a limiting factor for productivity of salt marshes and a seasonal switch is sometimes observed between nitrogen limitation and phosphorus limitation (Valiela and Teal, 1974; Howarth et al., 1988; van Wijnen and Bakker, 1999). Estuaries that are not subject to high levels of anthropogenic nutrient inputs are particularly susceptible to the effects of nutrient limitation. Nitrogen limitation has been demonstrated in these relatively pristine systems through nutrient addition experiments (Valiela and Teal, 1974; Marinucci et al., 1983; Hopkinson and Schubauer, 1984; Morris, 1991; Dai and Wiegert, 1997; van Wijnen and Bakker, 1999; Bagwell and Lovell, 2000a; Piceno and Lovell, 2000a,b) and through ecosystem-level mass balance considerations (Hopkinson and Schubauer, 1984; Hopkinson, 1988; Morris, 1995). Biological nitrogen fixation in salt marshes is thought to be more important than in most other ecosystems due to its significant contribution of N to the nitrogen budget in these systems (Capone and Carpenter, 1982).
Only organisms of the domains Bacteria and Archaea are capable of the reduction of gaseous dinitrogen to ammonia (nitrogen fixation or diazotrophy; Postgate, 1998). Ammonium levels in the sediment porewater are much higher than in salt marsh surface waters, demonstrating that the sediments are a net source of nitrogen for the system (Mendelssohn, 1979; Morris and Dacey, 1984; Sundareshwar et al., 2003). The diazotrophs in the sediments of salt marshes are thus the main source of the considerable contribution of fixed nitrogen from salt marshes to the global nitrogen cycle. This was confirmed by 15N2 enrichment studies using microcosms of sediment from a California estuary where nitrogen fixation was the primary method of N transfer to vegetation and other primary producers (Moseman-Valtierra et al., 2010). Diazotrophs associated with salt marsh grasses are present in high numbers, are most active on the roots of these plants (Patriquin, 1978; McClung et al., 1983; Whiting et al., 1986; Yoch and Whiting, 1986) and are supported by root exudates, as well as by products of plant decomposition (Whiting et al., 1986; Bagwell et al., 2001; Bergholz et al., 2001). Diazotrophs associated with salt marsh macrophytes, particularly the cordgrasses (Spartina spp.), have been studied extensively and the remarkable diversity of the salt marsh diazotroph assemblages documented (Bagwell et al., 1998, 2001, 2002; Piceno et al., 1999; Bagwell and Lovell, 2000a; Lovell et al., 2000, 2001a,b; Piceno and Lovell, 2000a,b; Bergholz et al., 2001; Burke et al., 2002, 2003; Brown et al., 2003; LaRocque et al., 2004; Moisander et al., 2005; Welsh et al., 2007; Moseman-Valtierra et al., 2009, 2010; Gamble et al., 2010; Cramer et al., 2011; Davis et al., 2011).
Bacterial diazotroph assemblages associated with salt marsh grasses are primarily composed of organisms taxonomically related to the α-, β-, δ-, ε-, and γ-Proteobacteria (Bagwell et al., 1998, 2001, 2002; Piceno et al., 1999; Bagwell and Lovell, 2000a; Lovell et al., 2000, 2001a,b; Piceno and Lovell, 2000a,b; Burke et al., 2002, 2003; Brown et al., 2003; Moisander et al., 2005; Welsh et al., 2007; Gamble et al., 2010; Davis et al., 2011). Archaeal diazotrophs have not been extensively studied in most ecosystems (Zehr et al., 2003) and have yet to be examined in salt marshes. Salt marsh diazotroph assemblages consist of mainly novel, uncultivated organisms whose physiologies have not yet been determined. This is a common observation of diazotroph assemblages across natural ecosystems and the diversity of these assemblages in combination with their potentially very different physiologies has inspired many studies (Zehr et al., 2003; Bürgmann et al., 2004). Culture-dependent studies have also revealed diverse culturable assemblages, largely composed of Proteobacteria (Bagwell et al., 1998, 2001; Bagwell and Lovell, 2000a; Bergholz et al., 2001; LaRocque et al., 2004). These assemblages are capable of utilizing a host of carbon compounds found in plant root exudates as determined in studies on isolates from washed roots (Bagwell et al., 2001; Bergholz et al., 2001), and of surviving substantial ranges of environmental conditions such as pH and salinity (Cramer et al., 2011).
The development of nitrogenase gene specific polymerase chain reaction (PCR) primers has allowed the investigation of salt marsh diazotroph assemblages via culture-independent methods (Zehr and McReynolds, 1989; Ueda et al., 1995; Olson et al., 1998; Piceno et al., 1999; Widmer et al., 1999; Lovell et al., 2000; Welsh et al., 2007). Briefly, the nitrogenase enzyme is composed of two multi-subunit metalloproteins (reviewed in Postgate, 1998; Zehr et al., 2003). Component I contains the active site for N2 reduction and is composed of two heterodimers encoded by the nifD and nifK genes. Component II couples ATP hydrolysis to inter-protein electron transfer and is composed of two identical subunits encoded by the nifH gene. Primers specific for the nifH gene have been used extensively in culture-independent studies of diazotroph assemblages across many ecosystems resulting in a robust database of gene sequences from uncultivated microorganisms (Zehr et al., 2003). Primers specific for nifD (Ueda et al., 1995) have also been developed but have not been used as broadly or as frequently as the nifH-specific primers (Zehr et al., 2003). The diazotroph assemblages associated with salt marsh macrophytes have been investigated through PCR-amplification using nifH-specific primers and the nifH amplicons have in turn been analyzed using methods such as denaturing gradient gel electrophoresis (DGGE; Bagwell et al., 1998, 2001, 2002; Piceno et al., 1999; Bagwell and Lovell, 2000a; Lovell et al., 2000, 2001a,b; Piceno and Lovell, 2000a,b; Brown et al., 2003; Welsh et al., 2007, 2010; Gamble et al., 2010; Davis et al., 2011), restriction fragment length polymorphism (RFLP; Burke et al., 2002, 2003), and terminal restriction fragment length polymorphism (T-RFLP; Moseman-Valtierra et al., 2009, 2010). PCR-amplification of reverse-transcribed mRNA extracts has also been used to a lesser extent (Brown et al., 2003). In the North Inlet estuary, diazotrophs have been investigated via culture-dependent methods as well as DGGE (Bagwell et al., 1998, 2001, 2002; Piceno et al., 1999; Bagwell and Lovell, 2000a; Lovell et al., 2000, 2001a,b; Piceno and Lovell, 2000a,b; Brown et al., 2003; Gamble et al., 2010; Davis et al., 2011), reverse sample genome probing (Bagwell and Lovell, 2000a), clone library construction and screening (Lovell et al., 2008) and sequencing of reverse-transcribed mRNA extracts (Brown et al., 2003). The composition of the diazotroph assemblages of this ecosystem are similar to those recovered from other salt marsh ecosystems (e.g., Moisander et al., 2005) and other estuaries (e.g., Affourtit et al., 2001; Burns et al., 2002; Moisander et al., 2007). Due to the number of studies that have been done in this area, detailed phylogenetic and statistical analyses can be conducted on the nifH sequence data that have been obtained. Assemblages from the North Inlet estuary have proven to be stable, ubiquitous, functionally versatile, distributed in predictable patterns across the marsh landscape and are characterized by their content of novel and undescribed taxa (Gamble et al., 2010; Davis et al., 2011).
Culture-dependent and culture-independent analyses of diazotroph assemblages associated with the rhizospheres of salt marsh macrophytes of North Inlet have demonstrated the occurrence of plant host specific organisms. This is not uncommon, nor is it unexpected in natural ecosystems. The colonization of plant species by specific microbial populations has been repeatedly observed in terrestrial ecosystems (reviewed in Berg and Smalla, 2009). Soil quality and soil type heavily influences the root colonizing microorganisms, however it has been proven repeatedly that plant species also exerts a direct influence (Berg and Smalla, 2009). The biogeography of soil microbial communities on a large scale (i.e., continental) is controlled primarily by soil abiotic parameters, particularly soil pH (Fierer and Jackson, 2006), and it is assumed that this premise holds true for salt marsh sediment microbial communities. However, as previously mentioned, parameters such as vegetation type, carbon and nutrient availability, and soil moisture have a more direct influence on the diversity of the local microbial community (Fierer and Jackson, 2006). Plant type and the root exudates that individual plant species produce affect the relative abundance and diversity of indigenous microbial communities at the root and in the rhizosphere (Berg and Smalla, 2009) resulting in plant host specific phylotypes.
LaRocque et al. (2004) examined diazotrophs isolated from J. roemerianus rhizoplanes from two habitats differing in abiotic parameters. The two monotypic Juncus stands were in different marsh zones having different salinities (29 and 40 parts per thousand), different porewater chemistries and experiencing different extents and durations of tidal inundation (LaRocque et al., 2004). The higher salinity Juncus habitat is regularly inundated during daily tidal cycles, whereas the lower salinity stand is in the high marsh and not as frequently inundated. Diazotrophs from both habitats were analyzed in parallel using the same protocols (including nifH PCR-amplification). The presence of nifH was confirmed in each isolate and isolates were grouped based on their carbon substrate utilization patterns. Three major substrate utilization clusters were identified and each cluster contained isolates from both monotypic stands. DGGE analysis of rhizoplane (sediment free roots) and rhizosphere (sediments influenced by the roots) nifH amplicons from both Juncus stands confirmed the presence of host specific diazotrophs – diazotrophs that were present in both assemblages despite differences in salinity, sulfide, and pH. The use of both culture-dependent and independent methods in this study reduced the impact of biases that could have been introduced had only data from a single method been examined.
Bergholz et al. (2001) isolated diazotrophs from the rhizoplanes of S. patens growing in two physically different habitats of North Inlet estuary, along with diazotrophs from other macrophytes. The diazotrophs were grouped based on substrate utilization and strains from S. patens formed two distinct clusters each containing strains from both habitats. Of the 16 physiologically different groups, only one strain isolated from S. patens was similar to diazotroph strains isolated from other macrophytes in the same salt marsh ecosystem. Diazotroph strains from S. virginica were different from strains from other plants growing in other marsh zones, but were more similar to strains from host plants growing in the same marsh zone (Bagwell et al., 2001). As described above and in Figure 2, marsh zones are defined by edaphic parameters (Bertness and Ellison, 1987; Adam, 1990; Pennings and Callaway, 1992). Diazotrophs isolated from plant hosts in the same zone are presumed to be heavily influenced by the edaphic parameters of the marsh zone.
nifH DGGE analysis of diazotroph assemblages recovered from the rhizosphere and rhizoplane of host plants in North Inlet also confirm the plant host specificity observed in the culture-dependent studies (Gamble et al., 2010; Davis et al., 2011). Assemblages from S. alterniflora growing in two differing tidal drainage systems within North Inlet estuary showed seasonal variability but no significant site-influenced variability (Gamble et al., 2010). The host plants were the same in both locations (short and tall growth forms of S. alterniflora) and the subtle changes in edaphic parameters with changes in seasons shaped the seasonally responsive segments of the diazotroph assemblages in these two locations. Assemblages from the rhizospheres of plant hosts in the Crab Haul Creek Basin that exhibit zonation due to elevation differences, contained host specific diazotrophs as well as diazotrophs having broad distributions across the landscape (Davis et al., 2011). As described above, plant zones contain specific macrophytes due to the differences in marsh environments. A clear response of diazotroph assemblages to plant host zonation and seasonal changes in edaphic parameters was observed from principal component analysis (PCA) of the recovered nifH amplicons (Gamble et al., 2010; Davis et al., 2011). The results obtained by Davis et al. (2011) confirmed the seasonal responses (Figure 3) and plant host specificity (Figure 4) observed by Gamble et al. (2010), and also included diazotroph assemblages associated with multiple plant hosts.
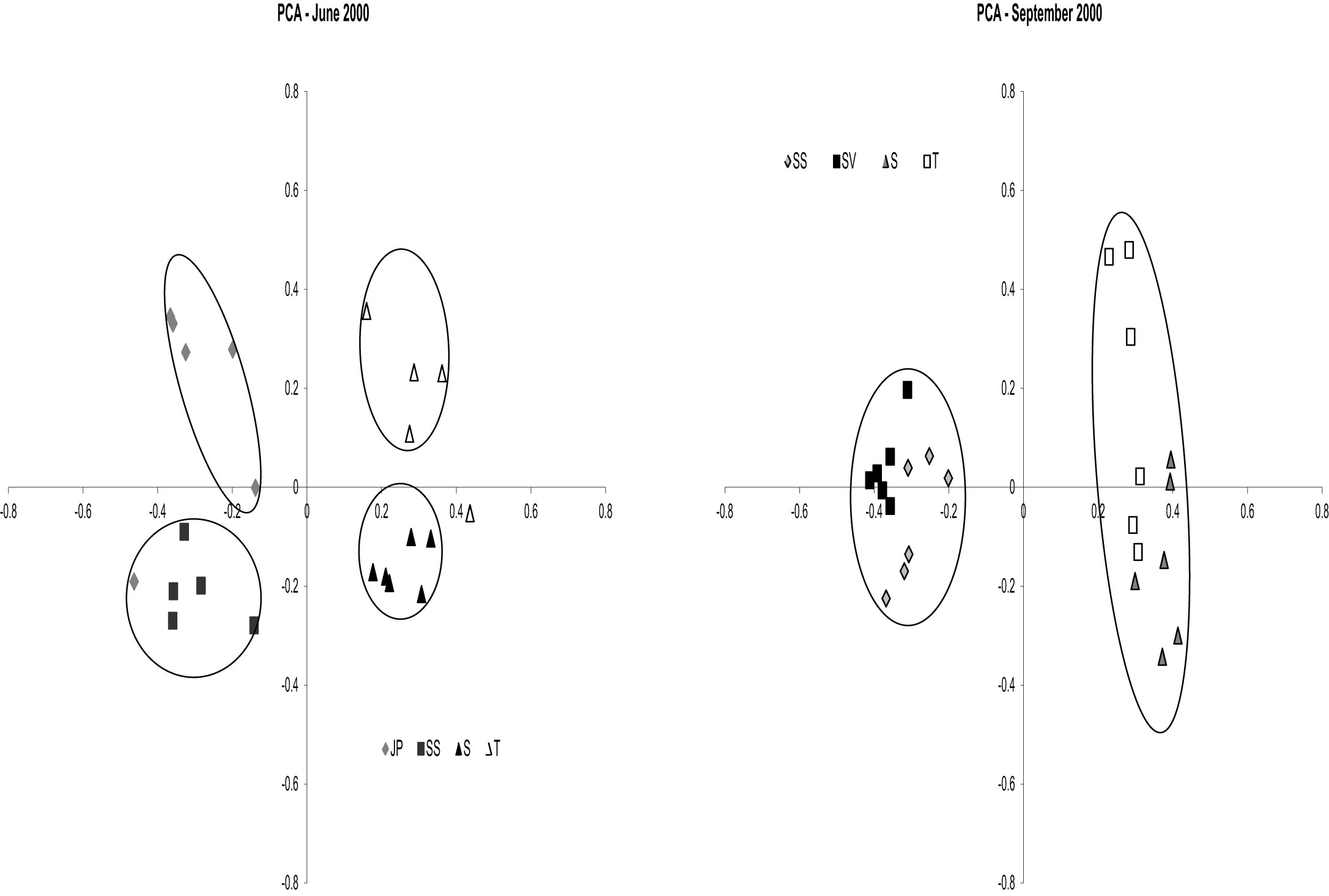
Figure 3. Principal Components Analysis results for dates June (SS, JP, S, and T) and September 2000 (SV, SS, S, and T). Circles denote significance (p < 0.001) for clustering. For June 2000 Axis 1 represents 28.4% of the variance and Axis 2 represents 14.7% of the variance. For September 2000 Axis 1 represents 31.5% of the variance and Axis 2 represents 11.5% of the variance. Reproduced with kind permission from Davis et al. (2011), Figure 3.
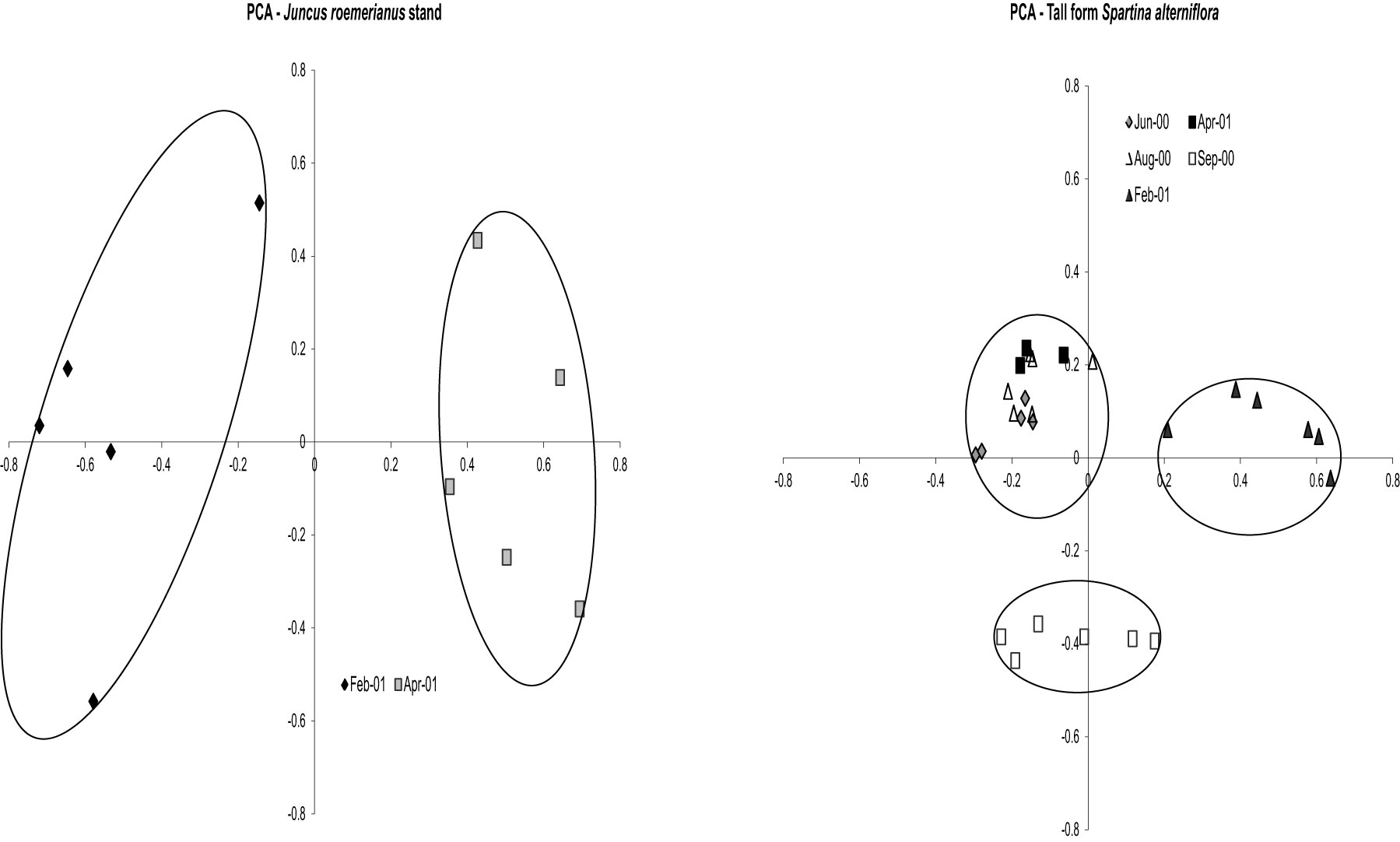
Figure 4. Principal Components Analysis results for vegetation zones Juncus roemerianus stand and tall form Spartina alterniflora on all dates. Circles denote significance (p < 0.001) for clustering. For J. roemerianus stand Axis 1 represents 31.2% of the variance and Axis 2 represents 20.8% of the variance. For tall form S. alterniflora Axis 1 represents 25.8% of the variance and Axis 2 represents 15.5% of the variance. Reproduced with kind permission from Davis et al. (2011), Supplementary Figure 1A.
To confirm the results from PCA analysis, mothur (Schloss et al., 2009) was used to conduct parsimony tests on neighbor-joining trees created using nifH sequences (400–420 bp). These sequences were recovered from DGGE banding patterns of rhizoplane and rhizosphere samples from all plant zones except the mixed zone of co-occurring S. virginica and short form S. alterniflora (Davis, 2010). Included in the phylogenetic analysis were plant zones (with designations): J. roemerianus stand (JS), S. virginica (SV), short form S. alterniflora (SS), patch of J. roemerianus in the SS zone (JP), and tall form S. alterniflora (TS). Using a significance threshold of p < 0.01 it was confirmed that diazotroph assemblages in the same marsh zone (based on edaphic parameters) had similar structures (Table 1). Phylogenetic analyses of the sequences recovered from these assemblages also revealed clusters of plant host specific sequences that were considered identical (sensu Venter et al., 2004; see also Lovell et al., 2008) throughout the nifH phylogenetic trees (Figure 5; Table A2 in Appendix, 19 clusters in the high marsh; Figures 6 and 7; Table A4 in Appendix, 39 clusters in the low marsh). These results provide additional and extensive evidence of diazotroph specificity for the plant host.
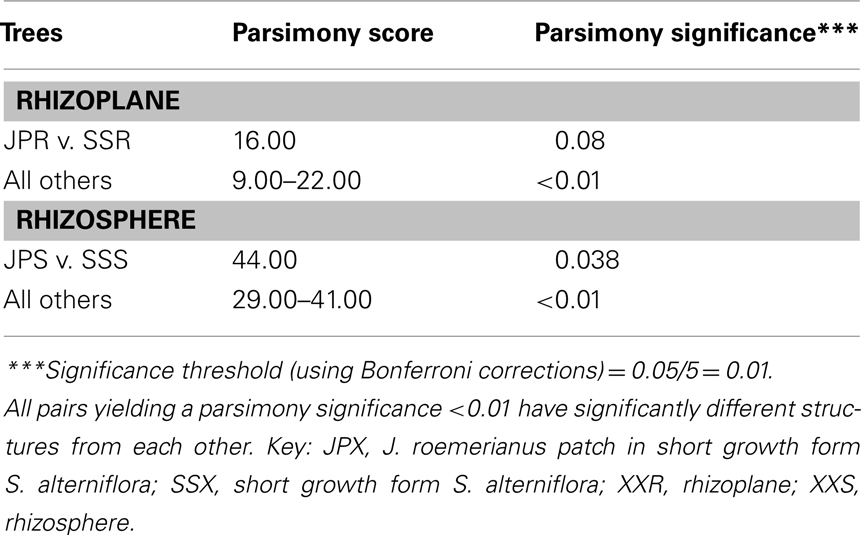
Table 1. Results from parsimony tests (mothur) of neighbor-joining trees containing diazotroph sequences recovered from band stabs of DGGE gels of PCR-amplified nifH amplicons from rhizoplane (roots) or rhizosphere (sediments) of different plant zones.
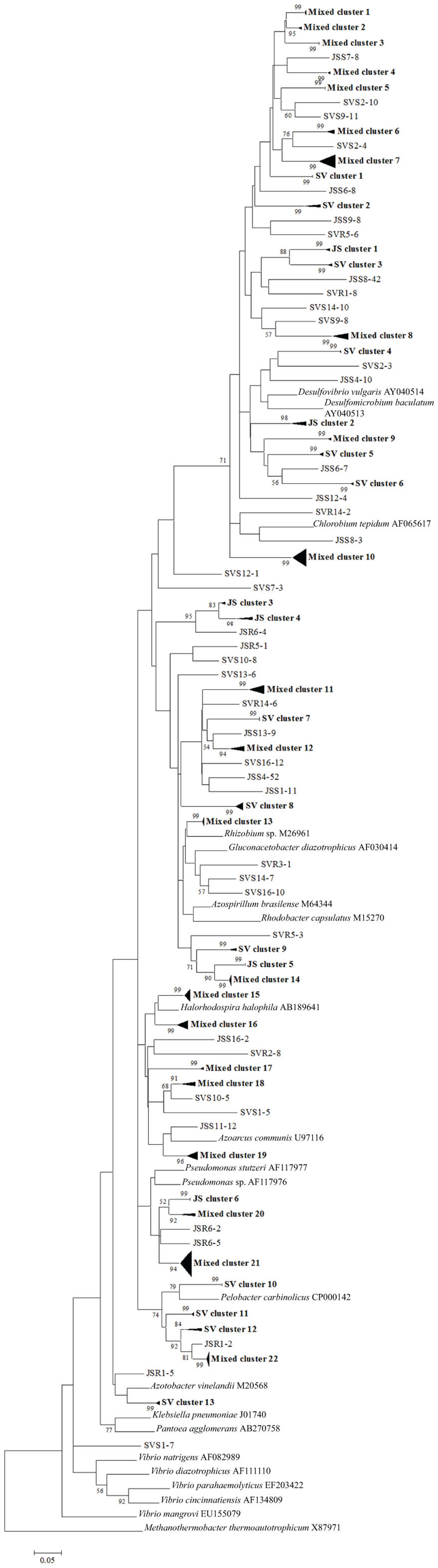
Figure 5. Phylogenetic analysis of all nifH sequences from DGGE bands of rhizoplane (R) and rhizosphere (S) samples from the high marsh plant zones (nucleotide sequences, Neighbor-Joining, Jukes Cantor correction, complete deletion of gaps and missing data, 1000 bootstrap replicates). Boot strap values below 50% are not shown. Abbreviations: JS, Juncus roemerianus stand; SV, Salicornia virginica.
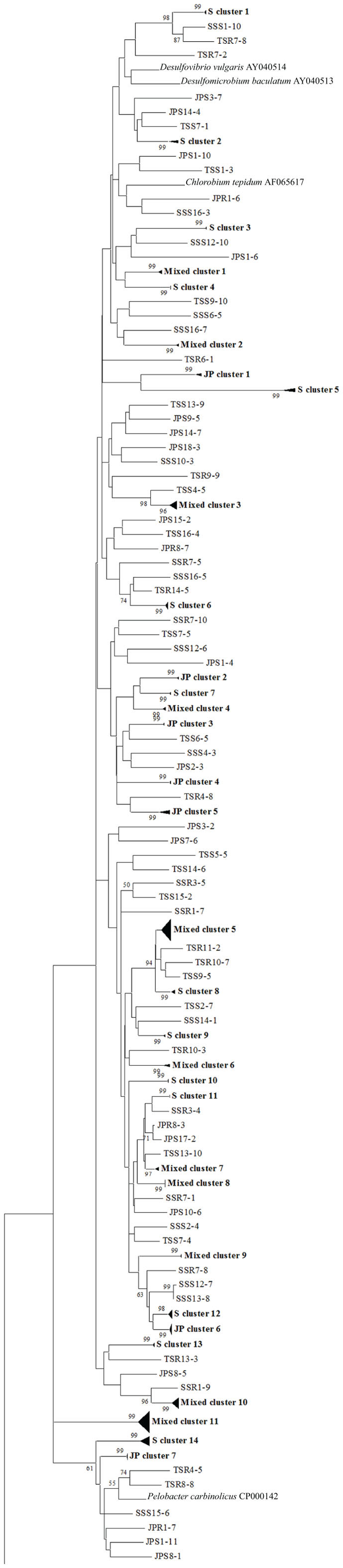
Figure 6. Phylogenetic analysis of all nifH sequences from DGGE bands of rhizoplane (R) and rhizosphere (S) samples from the low marsh plant zones (nucleotide sequences, Neighbor-Joining, Jukes Cantor correction, complete deletion of gaps and missing data, 1000 bootstrap replicates). Boot strap values below 50% are not shown. Abbreviations: JP, J. roemerianus patch; SS, short form S. alterniflora; TS, tall form S. alterniflora. The phylogram is continued in Figure 7.
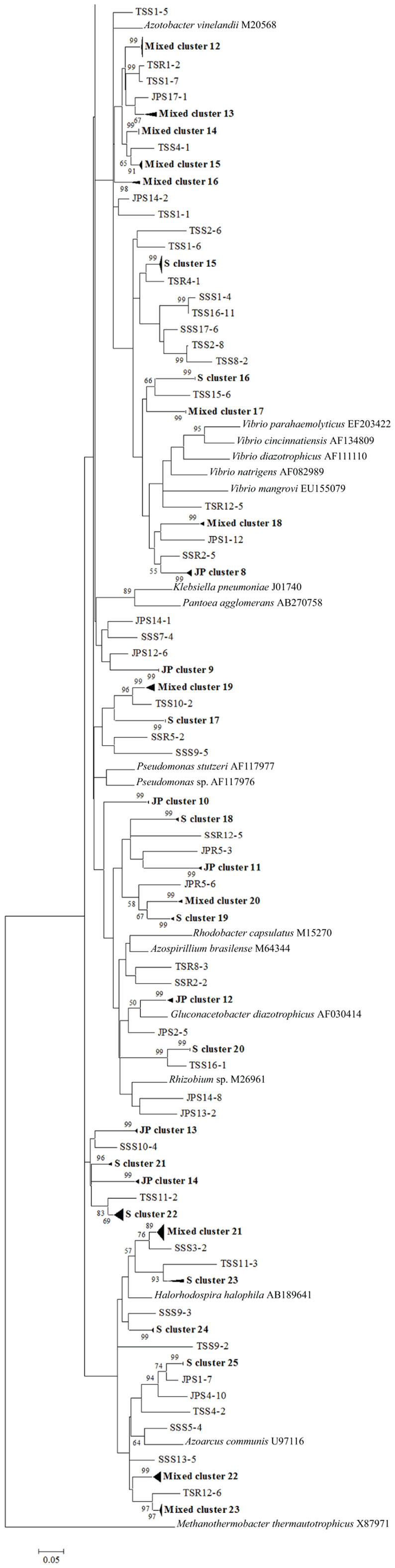
Figure 7. Continuation of Figure 6 – phylogenetic analysis of all nifH sequences recovered from the low marsh plant zones (nucleotide sequences, Neighbor-Joining, Jukes Cantor correction, complete deletion of gaps and missing data, 1000 bootstrap replicates). Abbreviations: JP, J. roemerianus patch; SS, short form S. alterniflora; TS, tall form S. alterniflora.
Multiple analyses of datasets obtained in several different studies provide strong evidence of plant host specific diazotroph assemblages associated with the rhizospheres of salt marsh plants. These organisms have been detected through both culture-dependent and culture-independent methods, in different sites throughout North Inlet estuary, and over the course of more than a decade. It is very clear that salt marsh diazotrophs have a distinct biogeography driven by both biotic (host plant) and abiotic environmental parameters (i.e., salinity, tidal inundation, elevation, etc.). The results of these studies also indicate substantial resilience of some members of the diazotroph assemblages that are capable of withstanding wide ranges of environmental conditions and are present in multiple marsh zones.
Studies of diazotroph assemblages associated with the salt marsh plants in North Inlet have demonstrated a diverse yet stable assemblage. Bagwell and Lovell (2000b) conducted a series of field fertilization experiments and through reverse sample genome probing revealed a highly resilient, niche specialized diazotroph assemblage. The assemblage responded to environmental manipulations through increases and decreases in quantities of specific taxa, but not through loss of taxa (Bagwell and Lovell, 2000b). DGGE analysis of diazotroph assemblages in long- and short-term fertilization experiments (fertilized with 16.3 g N m−2 NH4NO3 and 18.0 g P m−2 P2O5) confirmed the stability of these assemblages (Piceno and Lovell, 2000a,b). The results of these experiments indicate that salt marsh diazotrophs are quite competitive with non-diazotrophs and are able to maintain their populations despite the energy expense of nitrogen fixation. The resilience of diazotrophs associated with salt marsh plants has also been observed during periods of extreme environmental conditions (Davis, 2010).
In stark contrast to the diversity of diazotrophs associated with the rhizospheres of salt marsh plants, nifH sequences recovered from standing dead S. alterniflora biomass from North Inlet and Sapelo Island (GA, USA) salt marshes revealed a diazotroph assemblage with markedly lower diversity (Lovell et al., 2001b). These assemblages can produce significant levels of diazotrophy (based on rates of nitrogen fixation) despite the highly mineralized and frequently dry microenvironments they occupy (Newell et al., 1985, 1992). The analysis methods (including nifH-specific PCR), which were also utilized in other studies, revealed that approximately 90% of nifH sequences from these assemblages were similar to those from α-Proteobacteria. This demonstrates that the methods of nifH sequence recovery employed in these studies are sound, as the α-Proteobacteria are poorly represented in most salt marsh rhizosphere diazotroph assemblages (Lovell et al., 2000, 2008; Gamble et al., 2010; Davis et al., 2011). For example, Figures 5–7 show the small number of nifH sequences that are similar to those from α-Proteobacteria (*-labeled clusters in Tables A1 and A4 in Appendix) in comparison to the large number of recovered sequences related to those from the γ-Proteobacteria. The majority of the α-Proteobacteria-like salt marsh sequences have been recovered from the high marsh, particularly in the J. roemerianus plant zone. In addition, approximately half of the α-Proteobacteria-like sequences that were recovered from the low marsh were from the patch of J. roemerianus growing in the short form S. alterniflora zone. This analysis clearly demonstrates specificity of diazotroph assemblages for specific niches.
Principal component analysis and phylogenetic analyses by Gamble et al. (2010) revealed that changes in edaphic parameters associated with plant senescence, tidal intrusion, and nutrient cycling produced different responses from the seasonally (growing season/summer vs. plant senescence/winter) responsive groups within the diazotroph assemblages (Gamble et al., 2010). A seasonally responsive γ-Proteobacteria clade was detected during the Spartina growing season while, an anaerobic clade was only detected during plant senescence. Some clades were also recovered throughout all seasons, indicating a resilient portion of the assemblage. These diazotrophs are capable of competing for resources and maintaining populations over seasonal changes in both biotic and abiotic environmental variables.
Phylogenetic analyses of the recovered sequences from differing plant zones by Davis et al. (2011) demonstrated broadly distributed (and mostly novel), frequently detected (FD) diazotroph sequences that did not respond detectably to seasonal changes (Davis et al., 2011). These sequences were not detected in studies employing the same methods to examine diazotrophic assemblages associated with standing dead Spartina (Lovell et al., 2001b) or seagrasses (Bagwell et al., 2002). This indicates that they were unlikely to derive from sample contamination. Sequences were also identified (via statistical tests in PCA) as contributing significantly to the structure of the diazotroph assemblages (LV – sequences from bands producing long eigenvectors in Euclidean bi-plot analysis of PCA results; for full explanation see Davis et al., 2011). LV sequences drive the formation of these assemblages throughout the ecosystem. Phylogenetic analysis revealed several groups of FD and LV sequences that were virtually identical (sensu Venter et al., 2004; see also Lovell et al., 2008) to each other and to previously recovered “seasonally non-responsive” (detected throughout all seasons) sequences from Gamble et al. (2010). This work demonstrates that there are diazotrophs that have become specialized for this ecosystem. They have been recovered from multiple sites in North Inlet estuary and differ from those recovered from other ecosystems. Most importantly, these organisms have maintained ubiquity and strong influence on ecosystem function in an ecosystem characterized by distinct plant zones resulting from varying edaphic parameters. It has been determined in terrestrial ecosystems that free-living diazotrophs are functionally significant (Bürgmann et al., 2004; Hsu and Buckley, 2009). It is reasonable to assume, based on the results of these studies, that these organisms are equally important in salt marsh ecosystems. Phylogenetic analysis of sequences recovered from high marsh and low marsh zones (Figures 5–7) also confirm the presence of diazotrophs specific to marsh zones. Throughout the phylograms are clades containing virtually identical sequences recovered from host plants in the same marsh zone (plant host labeled clusters in bolded font in Figures 5–7, and sequences listed in Tables A2 and A4 in Appendix), indicating niche specificity of these diazotrophs for the edaphic parameters present in these zones.
These data have given insight into the distributions of diazotroph populations throughout the nitrogen-limited salt marsh ecosystem of North Inlet, SC, USA. These diazotrophs are specialized for the niches within this ecosystem, whether the niches are defined by the microenvironment of the plant host, or by marsh zone, or by both. They maintain populations that are readily detectable by methods employed in multiple studies (particularly nifH based PCR and DGGE fingerprinting) and influence ecosystem function as well as diazotroph assemblage structure.
Studies of this salt marsh ecosystem have revealed that the structure of a functional microbial assemblage can be predicted, based on specific environmental parameters. Plant hosts and edaphic factors that vary in response to elevation are the key parameters that predict the biogeography of these organisms. On a continental scale, the biogeography of microbial communities in terrestrial ecosystems is predicted by abiotic parameters, particularly soil pH, rather than by geographic distance or vegetation type (Fierer and Jackson, 2006). However, on a local scale, environmental parameters and vegetation type more heavily influence the diversity of microbial populations. These patterns have been observed in this estuarine system and can be used to predict biogeographical patterns of diazotroph assemblages associated with salt marsh grass rhizospheres. Diazotroph assemblages of salt marsh ecosystems are also capable of resisting relatively large changes in environmental parameters to maintain detectable and functionally important populations (Davis, 2010) and can be understood in the context of biogeographical concepts that have been applied to macroorganisms.
Strong evidence of plant host specificity and niche specialization in these assemblages has been compiled in this review; however the methods used to obtain the data to support these hypotheses are not perfect. Limitations of DGGE include the co-migration of DNA fragments which will result in a loss of information (Gelsomino et al., 1999; Maarit-Niemi et al., 2001; Kirk et al., 2004). Also, the detection of nifH gene sequences by PCR does not confirm the expression of this gene. Methods of quantification such as quantitative reverse transcriptase-PCR would be useful as this would identify the dominant diazotrophic phylotypes that actively fix nitrogen and could help to quantify the sizes of their populations. Continuing difficulties in the cultivation of these organisms results in a very limited understanding of their physiology, with much of that understanding inferred from known physiologies of related organisms. Finally, the evolutionary distances separating most salt marsh diazotrophs from their nearest characterized relatives limits even inference of ecologically significant characteristics of many of these organisms. A great deal remains to be done in identifying the taxa that contribute substantially to diazotrophy in salt marsh ecosystems, though a small start has been made (Brown et al., 2003). Greater knowledge of these organisms may allow us to refine our cultivation strategies and finally isolate the key organisms into pure culture for characterization. Perhaps metagenomic approaches could be employed, but the great diversity of distinct strains within even well studied lineages may limit the gains from these methods as chimeric genomic fragments may be encountered. Clearly, many challenges remain to be overcome.
Based on current climate models, coastal salt marshes are in danger of being completely submerged by the end of the century (Nicholls et al., 2007; Craft et al., 2009). This is due to the accelerating rate of sea level rise. Currently, it is expected that the ecogeomorphic feedbacks of these dynamic coastal ecosystems [i.e., rapid vegetation growth during years of low and high sea level (Morris et al., 2002); sediment trapping, organic matter accretion and limiting erosion (Fagherazzi et al., 2004)] that have allowed the survival of intertidal wetlands over time will continue to do so. However, the ever increasing number of completely submerged wetlands indicates that the current rate of sea level rise may exceed the abilities of ecogeomorphic feedbacks to preserve tidal ecosystems, particularly salt marshes (Kirwan et al., 2010). Continuous submersion of coastal salt marshes will result in losses of vegetation (Penland et al., 2001; Hartig et al., 2002; Kearnery et al., 2002; Kirwan et al., 2010) and consequently loss of microorganisms restricted to a particular plant host or microenvironment (Berg and Smalla, 2009). Loss of microorganisms critical to ecosystem function would clearly be detrimental to the ecosystem that is dependent on their activity and interactions with macrophytes (primary producers). These events will eventually interrupt food webs and littoral biogeochemical cycles affecting the quality of estuarine water due to eutrophication and increasing the export of particulates (Kearnery et al., 2002).
The ability to predict biogeographical distributions of microorganisms as critical to ecosystem function as diazotrophs are to salt marshes will be advantageous to ecosystem modelers, especially those involved in determining the effects of global climate change. These data clearly show that the distribution of diazotrophs is non-random and is driven directly or indirectly by edaphic parameters in their habitat. The assemblages contain microorganisms that appear to be restricted in their distributions, as well as broadly distributed microorganisms. Each group is clearly engaged in ecosystem function as evidenced by their resilience and stability within the assemblages. However, even with this apparent ability to prosper over wide ranges of environmental parameters, there are limits to their survival. The flooding of coastal wetlands as sea level continues to rise could result in large losses from these assemblages. The biogeographical patterns observed in North Inlet estuary can be applied to tidally dominated coastal wetlands when the edaphic parameters of the system in question are taken into consideration.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge the Belle W. Baruch Institute for Coastal and Marine Sciences for access to sampling sites and for logistical support. Six hundred twenty-seven sequences recovered from DGGE bands (Figures 5–7; Tables A1– A4 in Appendix) were deposited with the National Center for Biotechnology Information (NCBI) GenBank (accession numbers HM750282 – HM750907). We thank Paul Bodelier for his invitation to contribute to this special topics volume on the Microbiology of Wetlands. This is contribution 1645 of the Belle W. Baruch Institute for Coastal and Marine Sciences.
Affourtit, J., Zehr, J. P., and Pearl, H. W. (2001). Distribution of nitrogen-fixing microorganisms along the Neuse River Estuary, North Carolina. Microb. Ecol. 41, 114–123.
Bagwell, C. E., Dantzler, M., Bergholz, P. W., and Lovell, C. R. (2001). Host-specific ecotype diversity of rhizoplane diazotrophs of the perennial glasswort Salicornia virginica and selected salt marsh grasses. Aquat. Microb. Ecol. 23, 293–300.
Bagwell, C. E., LaRocque, J. R., Smith, G. W., Polson, S. W., Friez, M. J., Longshore, J. W., and Lovell, C. R. (2002). Molecular diversity of diazotrophs in oligotrophic tropical seagrass bed communities. FEMS Microbiol. Ecol. 39, 113–119.
Bagwell, C. E., and Lovell, C. R. (2000a). Microdiversity of culturable diazotrophs from the rhizoplanes of the salt marsh grasses Spartina alterniflora and Juncus roemerianus. Microb. Ecol. 39, 128–136.
Bagwell, C. E., and Lovell, C. R. (2000b). Persistence of selected Spartina alterniflora rhizoplane diazotrophs exposed to natural and manipulated environmental variability. Appl. Environ. Microbiol. 66, 4625–4633.
Bagwell, C. E., Piceno, Y. M., Ashburne-Lucas, A., and Lovell, C. R. (1998). Physiological diversity of the rhizosphere diazotroph assemblages of selected salt marsh grasses. Appl. Environ. Microbiol. 64, 4276–4282.
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13.
Bergholz, P. W., Bagwell, C. W., and Lovell, C. R. (2001). Physiological diversity of rhizoplane diazotrophs of the saltmeadow cordgrass, Spartina patens: implications for host specific ecotypes. Microb. Ecol. 42, 466–476.
Bertness, M. D. (1991). Zonation of Spartina patens and Spartina alterniflora in New England salt marsh. Ecology 72, 138–148.
Bertness, M. D., and Ellison, A. M. (1987). Determinants of pattern in a New England salt marsh plant community. Ecol. Monogr. 57, 129–147.
Blood, E. R., and Vernberg, F. J. (1992). “Characterization of the physical, chemical, and biological conditions and trends in Winyah Bay and North Inlet estuaries: 1970-1985,” in Characterization of the Physical, Chemical, and Biological Conditions and Trends in Three South Carolina Estuaries. Winyah Bay and North Inlet Estuaries Vol II. Charleston, SC: Sea Grant Consortium, 162.
Brown, M. M., Friez, M. J., and Lovell, C. R. (2003). Expression of nifH genes by diazotrophic bacteria in the rhizosphere of short form Spartina alterniflora. FEMS Microbiol. Ecol. 43, 411–417.
Bürgmann, H., Widmer, F., Von Sigler, W., and Zeyer, J. (2004). New molecular screening tools for analysis of free-living diazotrophs in soil. Appl. Environ. Microbiol. 70, 240–247.
Burke, D. J., Hamerlynck, E. P., and Hahn, D. (2002). Interactions among plant species and microorganisms in salt marsh sediments. Appl. Environ. Microbiol. 68, 1157–1164.
Burke, D. J., Hamerlynck, E. P., and Hahn, D. (2003). Interactions between the salt marsh grass Spartina patens, arbuscular mycorrhizal fungi and sediment bacteria during the growing season. Soil Biol. Biochem. 35, 501–511.
Burns, J. A., Zehr, J. P., and Capone, D. G. (2002). Nitrogen-fixing phylotypes of Chesapeake Bay and Neuse River Estuary sediments. Microb. Ecol. 44, 336–343.
Capone, D. G., and Carpenter, E. J. (1982). Nitrogen fixation in the environment. Science 217, 1140–1142.
Craft, C., Clough, J., Ehman, J., Joye, S., Park, R., Pennings, S., Guo, H., and Machmuller, M. (2009). Forecasting the effects of accelerated sea-level rise on tidal marsh ecosystem services. Front. Ecol. Environ. 7, 73–78.
Cramer, M. J., Haghshenas, N., Bagwell, C. E., Matsui, G. Y., and Lovell, C. R. (2011). Celerinatantimonas diazotrophica gen. nov., sp nov., a nitrogen-fixing bacterium representing a new family in the Gammaproteobacteria, Celerinatantimonadaceae fam. nov. Int. J. Syst. Evol. Microbiol. 61, 1053–1060.
Crump, B. C., Hopkinson, C. S., Sogin, M. L., and Hobbie, J. E. (2004). Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 70, 1494–1505.
Dai, T., and Wiegert, R. G. (1997). A field study of photosynthetic capacity and its response to nitrogen fertilization in Spartina alterniflora. Estuar. Coast. Shelf Sci. 45, 273–283.
Dame, R., Chrzanowski, T., Bildstein, K., Kjerfve, B., Mckellar, H., Nelson, D., Spurrier, J., Stancyk, S., Stevenson, H., Vernberg, J., and Zingmark, R. (1986). The outwelling hypothesis and North Inlet, South Carolina. Mar. Ecol. Prog. Ser. 33, 217–229.
Dame, R. F., and Kenny, P. D. (1986). Variability of Spartina alterniflora primary production in the euhaline North Inlet estuary. Mar. Ecol. Prog. Ser. 32, 71–80.
Davis, D. A. (2010). Diazotroph Assemblages of Salt Marsh Plants Growing Along an Environmental Gradient: Assemblage Composition and Responses to Environmental Conditions. Ph.D. thesis, University of South Carolina, Columbia, SC.
Davis, D. A., Gamble, M. D., Bagwell, C. E., Bergholz, P. W., and Lovell, C. R. (2011). Responses of salt marsh plant rhizosphere diazotroph assemblages to changes in marsh elevation, edaphic conditions and plant host species. Microb. Ecol. 61, 386–398.
Dolan, J. R. (2005). An introduction to the biogeography of aquatic microbes. Aquat. Microb. Ecol. 41, 39–48.
Dumbrell, A. J., Nelson, M., Helgason, T., Dytham, C., and Fitter, A. H. (2010). Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 4, 337–345.
Fagherazzi, S., Marani, M., and Blum, L. K. (ed.). (2004). The Ecogeomorphology of Tidal Marshes. Washington, DC: AGU.
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631.
Floyd, M. M., Tang, J., Kane, M., and Emerson, D. (2005). Captured diversity in a culture collection: case study of the geographic and habitat distributions of environmental isolates held at the American type culture collection. Appl. Environ. Microbiol. 71, 2813–2823.
Gamble, M. D., Bagwell, C. E., LaRocque, J., Bergholz, P. W., and Lovell, C. R. (2010). Seasonal variability of diazotroph assemblages associated with the rhizosphere of the salt marsh cordgrass, Spartina alterniflora. Microb. Ecol. 59, 253–265.
Gardner, L. (1973). The effect of hydrologic factors on the pore water chemistry of intertidal marsh sediments. SouthEastern Geol. 15, 17–28.
Gelsomino, A., Keijzer-Wolters, A. C., Cacco, G., and Van Elsas, J. D. (1999). Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gradient gel electrophoresis. J. Microbiol. Methods 38, 1–15.
Green, J. L., Bohannon, B. J. M., and Whitaker, R. J. (2008). Microbial biogeography: from taxonomy to traits. Science 320, 1039–1043.
Hartig, E. K., Gornitz, V., Kolker, A., Mushacke, F., and Fallon, D. (2002). Anthropogenic and climate-change impacts on salt marshes of Jamaica Bay, New York City. Wetlands 22, 71–89.
Hemond, H. F., and Fifield, J. L. (1982). Subsurface flow in a salt marsh peat: a model and field study. Limnol. Oceanogr. 27, 126–136.
Hollibaugh, J., Wong, P. S., and Murrell, M. C. (2000). Similarity of particle-associated and free-living bacterial communities in northern San Francisco Bay, California. Aquat. Microb. Ecol. 21, 103–114.
Hopkinson, C. S. (1988). “Patterns of organic carbon exchange between coastal ecosystems: the mass balance approach in salt marsh ecosystems,” in Coastal-Offshore Ecosystem Interactions (Lecture Notes on Coastal and Estuarine Studies), ed. B.-O. Jansson (New York: Springer-Verlag), 122–154.
Hopkinson, C. S., and Schubauer, J. P. (1984). Static and dynamic aspects of nitrogen cycling in the salt marsh graminoid Spartina alterniflora. Ecology 65, 961–969.
Howarth, R. W., Marino, R., and Lane, J. (1988). Nitrogen fixation in freshwater, estuarine, and marine ecosystems. 1. Rates and importance. Limnol. Oceanogr. 33, 669–687.
Hsu, S. F., and Buckley, D. H. (2009). Evidence for the functional significance of diazotroph community structure in soil. ISME J. 3, 124–136.
Hughes Martiny, J. B., Bohannan, B. J. M., Brown, J. H., Colwell, R. K., Fuhrman, J. A., Green, J. L., Horner-Devine, M. C., Kane, M., Adams-Krumins, J., Kuske, C. R., Morin, P. J., Naeem, S., Øvreås, L., Reysenbach, A.-L., Smith, V. L., and Staley, J. T. (2006). Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112.
Kearnery, M. S., Rogers, A. S., Townshend, J. R. G., Rizzo, E., and Stutzer, D. (2002). Landsat imagery shows decline of coastal marshes in Chesapeake and Delaware Bays. Eos (Washington DC) 83, 173–178.
King, G. M., Klug, M. J., Wiegert, R. G., and Chalmers, A. G. (1982). Relation of soil water movement and sulfide concentration to Spartina alterniflora production in a Georgia salt marsh. Science 218, 61–63.
King, S. E., and Lester, J. N. (1995). The value of salt-marsh as a sea defense. Mar. Pollut. Bull. 30, 180–189.
Kirk, J. L., Beaudete, L. A., Hart, M., Moutoglis, P., Klironomos, J. N., Lee, H., and Trevors, J. T. (2004). Methods of studying soil microbial diversity. J. Microbiol. Methods 58, 169–188.
Kirwan, M. L., Guntenspergen, G. R., D’Alpaos, A., Morris, J. T., Mudd, S. M., and Temmerman, S. (2010). Limits on the adaptability of coastal marshes to rising sea level. Geophys. Res. Lett. 37, L23401.
Kirwan, M. L., Guntenspergen, G. R., and Morris, J. T. (2009). Latitudinal trends in Spartina alterniflora productivity and the response of coastal marshes to global change. Glob. Chang. Biol. 15, 1982–1989.
Kjerfve, B. (1986). “Circulation and salt flux in a well mixed estuary,” in Physics of Shallow Estuaries and Bays, ed. J. van De Kreeke (New York: Springer-Verlag), 22–29.
LaRocque, J. R., Bergholz, P. W., Bagwell, C. E., and Lovell, C. R. (2004). Influence of host plant-derived and abiotic environmental parameters on the composition of the diazotroph assemblage associated with roots of Juncus roemerianus. Antonie Van Leeuwenhoek 86, 249–261.
Leibold, M. A., and McPeek, M. A. (2008). Coexistence of niche and neutral prespectives in community ecology. Ecology 87, 1399–1410.
Levine, J. M., Brewer, J. S., and Bertness, M. D. (1998). Nutrients, competition and plant zonation in a New England salt marsh. J. Ecol. 86, 285–292.
Lovell, C. R., Bagwell, C. E., Czako, M., Marton, L., Piceno, Y. M., and Ringelberg, D. B. (2001a). Stability of a rhizosphere microbial community exposed to natural and manipulated environmental variability. FEMS Microbiol. Ecol. 38, 69–76.
Lovell, C. R., Friez, M. J., Longshore, J. W., and Bagwell, C. E. (2001b). Recovery and phylogenetic analysis of nifH sequences from diazotrophic bacteria associated with dead aboveground biomass of Spartina alterniflora. Appl. Environ. Microbiol. 67, 5308–5314.
Lovell, C. R., Decker, P. V., Bagwell, C. E., Thompson, S., and Matsui, G. Y. (2008). Analysis of a diverse assemblage of diazotrophic bacteria from Spartina alterniflora using DGGE and clone library screening. J. Microbiol. Methods 73, 160–171.
Lovell, C. R., Piceno, Y. M., Quattro, J. M., and Bagwell, C. E. (2000). Molecular analysis of diazotroph diversity in the rhizosphere of the smooth cordgrass, Spartina alterniflora. Appl. Environ. Microbiol. 66, 3814–3822.
Maarit-Niemi, R., Heiskanen, I., Wallenius, K., and Lindstrom, K. (2001). Extraction and purification of DNA in rhizosphere soil samples for PCR-DGGE analysis of bacterial consortia. J. Microbiol. Methods 45, 155–165.
Macalady, J. L., Dattagupta, S., Schaperdoth, I., Jones, D. S., Druschel, G. K., and Eastman, D. (2008). Niche differentiation among sulfur-oxidizing bacterial populations in cave waters. ISME J. 2, 590–601.
Marinucci, A. C., Hobbie, J. E., and Helfrich, J. V. K. (1983). Effect of litter nitrogen on decomposition and microbial biomass in Spartina alterniflora. Microb. Ecol. 9, 27–40.
McClung, C. R., Van Berkum, P., Davis, R. E., and Sloger, C. (1983). Enumeration and localization of N2-fixing bacteria associated with roots of Spartina alterniflora Loisel. Appl. Environ. Microbiol. 45, 1914–1920.
Mendelssohn, I. A. (1979). Nitrogen metabolism in the height forms of Spartina alterniflora in North Carolina. Ecology 60, 574–584.
Moisander, P. H., Morrison, A. E., Ward, B. B., Jenkins, B. D., and Zehr, J. P. (2007). Spatial-temporal variability in diazotroph assemblages in Chesapeake Bay using an oligonucleotide nifH microarray. Environ. Microbiol. 9, 1823–1835.
Moisander, P. H., Piehler, M. F., and Paerl, H. W. (2005). Diversity and activity of epiphytic nitrogen-fixers on standing dead stems of the salt marsh grass Spartina alterniflora. Aquat. Microb. Ecol. 39, 271–279.
Morris, J. T. (1991). Effects of nitrogen loading on wetland ecosystems in particular reference to atmospheric deposition. Annu. Rev. Ecol. Syst. 22, 257–279.
Morris, J. T. (1995). The mass balance of salt and water in intertidal sediments: results from North Inlet, South Carolina. Estuaries 18, 556–567.
Morris, J. T., and Dacey, J. W. H. (1984). Effects of O2 on ammonium uptake and root respiration by Spartina alterniflora. Am. J. Bot. 71, 979–985.
Morris, J. T., and Haskin, B. (1990). A 5-yr record of aerial primary production and stand characteristics of Spartina alterniflora. Ecology 71, 2209–2217.
Morris, J. T., Kjerfve, B., and Dean, J. M. (1990). Dependence of estuarine productivity on anomalies in mean sea level. Limnol. Oceanogr. 35, 926–930.
Morris, J. T., Sundareshwar, P. V., Nietch, C. T., Kjerfve, B., and Cahoon, D. R. (2002). Responses of coastal wetlands to rising sea level. Ecology 83, 2869–2877.
Moseman-Valtierra, S. M., Armaiz-Nolla, K., and Levin, L. A. (2010). Wetland response to sedimentation and nitrogen loading: diversification and inhibition of nitrogen-fixing microbes. Ecol. Appl. 20, 1556–1568.
Moseman-Valtierra, S. M., Johnson, R., Zhang, R., and Qian, P. Y. (2009). Differences in cordgrass structure between a mature and developing marsh reflect distinct N2-fixing communities. Wetlands 29, 919–930.
Newell, S. Y., Fallon, R. D., Cal Rodriguez, R. M., and Groene, L. C. (1985). Influence of rain, tidal wetting and relative humidity on release of carbon dioxide by standing-dead salt-marsh plants. Oecologia 68, 73–79.
Newell, S. Y., Hopkinson, C. S., and Scott, L. A. (1992). Patterns of nitrogenase activity (acetylene reduction) associated with standing, decaying shoots of Spartina alterniflora. Estuar. Coast Shelf Sci. 35, 127–140.
Nicholls, R. J., Wong, P. P., Burkett, V. R., Codignotto, J. O., Hay, J. E., Mclean, R. F., Ragoonaden, S., and Woodroffe, C. D. (2007). “Coastal systems and low-lying areas,” in Climate Change 2007: Impacts, Adaptation and Vulnerability. Contribution of Working Group II to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, eds M. L. Parry, O. F. Canziani, J. P. Palutikof, P. J. Van Der Linden, and C. E. Hanson (Cambridge: Cambridge University Press), 315–356.
Odum, W. E. (1988). Comparative ecology of tidal freshwater and salt marshes. Annu. Rev. Ecol. Syst. 19, 147–176.
Olson, J. B., Steppe, T. F., Litaker, R. W., and Paerl, H. W. (1998). N2-fixing microbial consortia associated with the ice cover of Lake Bonney, Antarctica. Microb. Ecol. 36, 231–238.
#x000D8;vreås, L. (2000). Population and community level approaches for analysing microbial diversity in natural environments. Ecol. Lett. 3, 236–251.
Patriquin, D. G. (1978). Factors affecting nitrogenase activity (acetylene-reducing activity) associated with excised roots of the emergent halophyte Spartina alterniflora Loisel. Aquat. Bot. 4, 193–210.
Penland, S., Wayne, L., Britsch, L. D., Williams, S. J., Beall, A. D., and Butterworth, V. C. (2001). Geomorphic Classification of Coastal Land Loss Between 1932 and 1990 in the Mississippi River Delta Plain, Southeastern Louisiana. USGS Open-file Report 00–417, Woods Hole, MA.
Pennings, S. C., and Callaway, R. M. (1992). Salt marsh zonation: the relative importance of competition and physical factors. Ecology 73, 681–690.
Piceno, Y. M., and Lovell, C. R. (2000a). Stability in natural bacterial communities: I. Nutrient addition effects on rhizosphere diazotroph assemblage composition. Microb. Ecol. 39, 32–40.
Piceno, Y. M., and Lovell, C. R. (2000b). Stability in natural bacterial communities: II. Plant resource allocation effects on rhizosphere diazotroph assemblage composition. Microb. Ecol. 39, 41–48.
Piceno, Y. M., Noble, P. A., and Lovell, C. R. (1999). Spatial and temporal assessment of diazotroph assemblage composition in vegetated salt marsh sediments using denaturing gradient gel electrophoresis. Microb. Ecol. 38, 157–167.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., Lesniewski, R. A., Oakley, B. B., Parks, D. H., Robinson, C. J., Sahl, J. W., Stres, B., Thallinger, G. G., Van Horn, D. J., and Weber, C. F. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541.
Schubauer, J. P., and Hopkinson, C. S. (1984). Above- and belowground emergent macrophyte production and turnover in a coastal marsh ecosystem, Georgia. Limnol. Oceanogr. 29, 1052–1065.
Selje, N., and Simon, M. (2003). Composition and dynamics of particle-associated and free-living communities in the Weser estuary, Germany. Aquat. Microb. Ecol. 30, 221–237.
Sundareshwar, P. V., Morris, J. T., Koepfler, E. K., and Fornwalt, B. (2003). Phosphorus limitation of coastal ecosystem processes. Science 299, 563–565.
Turner, R. E. (1976). Geographic variations in salt marsh macrophyte production: a review. Contrib. Mar. Sci. 20, 47–68.
Ueda, T., Suga, Y., Yahiro, N., and Matsuguchi, T. (1995). Genetic diversity of N2- fixing bacteria associated with rice roots by molecular analysis of a nifD library. Can. J. Microbiol. 41, 235–240.
Valiela, I., and Teal, J. M. (1974). “Nutrient limitation in salt marsh vegetation,” in Ecology of Halophytes, eds R. J. Reimold and W. H. Queen (New York: Academic Press, Inc.), 547–563.
van Wijnen, H. J., and Bakker, J. P. (1999). Nitrogen and phosphorous limitation in a coastal barrier salt marsh: the implications for vegetation succession. J. Ecol. 87, 265–272.
Venter, J. C., Remington, K., Heidelberg, J. F., Halpern, A. L., Rusch, D., Eisen, J. A., Wu, D., Paulsen, I., Nelson, K. E., Nelson, W., Fouts, D. E., Levy, S., Knap, A. H., Lomas, M. W., Nealson, K., White, O., Peterson, J., Hoffman, J., Parsons, R., Baden-Tillson, H., Pfannkoch, C., Rogers, Y.-H., and Smith, H. O. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74.
Ward, D. M., Weller, R., and Bateson, M. M. (1990). 16S ribosomal-RNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345, 63–65.
Welsh, A., Burke, D. J., and Hahn, D. (2007). Analysis of nitrogen-fixing members of the ε-subclass of Proteobacteria in salt marsh sediments. Appl. Environ. Microbiol. 73, 7747–7752.
Welsh, A. K., Burke, D. J., Hamerlynck, E. P., and Hahn, D. (2010). Seasonal analyses of arbuscular mycorrhizae, nitrogen-fixing bacteria and growth performance of the salt marsh grass Spartina patens. Plant Soil 330, 251–266.
Whiting, G. J., Gandy, E. L., and Yoch, D. C. (1986). Tight coupling of root-associated nitrogen fixation and plant photosynthesis in the salt marsh grass Spartina alterniflora and carbon dioxide enhancement of nitrogenase activity. Appl. Environ. Microbiol. 52, 108–113.
Widmer, F., Shaffer, B. T., Porteous, L. A., and Seidler, R. J. (1999). Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site int eh Oregon Cascade mountain range. Appl. Environ. Microbiol. 65, 374–380.
Yoch, D. C., and Whiting, G. Y. (1986). Evidence for 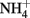 switch-off regulation of nitrogenase activity by bacteria in salt marsh sediments and roots of the grass Spartina alterniflora. Appl. Environ. Microbiol. 51, 143–149.
switch-off regulation of nitrogenase activity by bacteria in salt marsh sediments and roots of the grass Spartina alterniflora. Appl. Environ. Microbiol. 51, 143–149.
Zehr, J. P., Jenkins, B. D., Short, S. M., and Steward, G. F. (2003). Nitrogenase gene diversity and microbial community structure: a cross-system diversity. Environ. Microbiol. 5, 539–554.
Zehr, J. P., and McReynolds, L. A. (1989). Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl. Environ. Microbiol. 55, 2522–2526.

Table A1. Sequences included in the mixed clusters designated in Figure 5 (*α-Proteobacteria clusters).
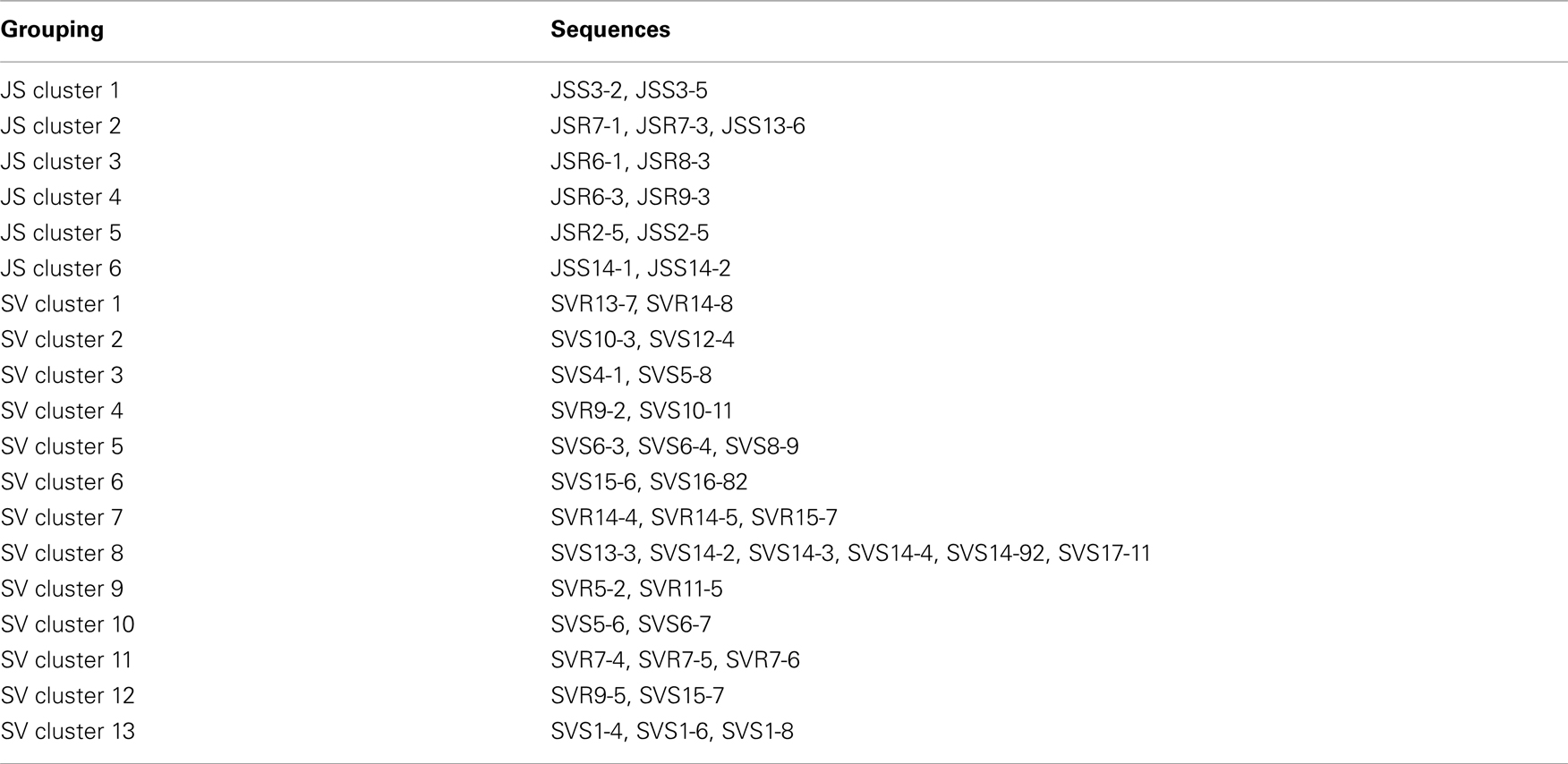
Table A2. Sequences included in the plant host clusters designated in Figure 5.
Keywords: diazotroph, salt marsh, plant host, vegetation zones, biogeography
Citation: Lovell CR and Davis DA (2012) Specificity of salt marsh diazotrophs for vegetation zones and plant hosts: results from a North American marsh. Front. Microbio. 3:84. doi: 10.3389/fmicb.2012.00084
Received: 23 November 2011; Accepted: 19 February 2012;
Published online: 16 March 2012.
Edited by:
Paul Bodelier, Netherlands Institute of Ecology, NetherlandsReviewed by:
Eric Boyd, Montana State University, USACopyright: © 2012 Lovell and Davis. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Charles R. Lovell, Department of Biological Sciences, University of South Carolina, 715 Sumter Street, Columbia, SC 29208, USA. e-mail:bG92ZWxsQGJpb2wuc2MuZWR1
†Present address: Debra A. Davis, Department of Biological Sciences, University of Alabama, P.O. Box 870344, Tuscaloosa, AL 35487, USA.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.