
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 02 March 2012
Sec. Terrestrial Microbiology
Volume 3 - 2012 | https://doi.org/10.3389/fmicb.2012.00078
This article is part of the Research TopicMicrobiology of wetlandsView all 18 articles
Acidic wetlands are global sources of the atmospheric greenhouse gases methane (CH4), and nitrous oxide (N2O). Consumption of both atmospheric gases has been observed in various acidic wetlands, but information on the microbial mechanisms underlying these phenomena is scarce. A substantial amount of CH4 is consumed in sub soil by aerobic methanotrophs at anoxic–oxic interfaces (e.g., tissues of Sphagnum mosses, rhizosphere of vascular plant roots). Methylocystis-related species are likely candidates that are involved in the consumption of atmospheric CH4 in acidic wetlands. Oxygen availability regulates the activity of methanotrophs of acidic wetlands. Other parameters impacting on the methanotroph-mediated CH4 consumption have not been systematically evaluated. N2O is produced and consumed by microbial denitrification, thus rendering acidic wetlands as temporary sources or sinks for N2O. Denitrifier communities in such ecosystems are diverse, and largely uncultured and/or new, and environmental factors that control their consumption activity are unresolved. Analyses of the composition of N2O reductase genes in acidic wetlands suggest that acid-tolerant Proteobacteria have the potential to mediate N2O consumption in such soils. Thus, the fragmented current state of knowledge raises open questions concerning methanotrophs and denitrifiers that consume atmospheric CH4 and N2O in acidic wetlands.
Microbial communities of acidic wetlands are responsible for emissions of the potent greenhouse gases methane (CH4), and nitrous oxide (N2O; Conrad, 1996; Whalen, 2005; Denman et al., 2007). The largest single global source of atmospheric CH4 (100–237 Gt year−1) are acidic wetlands, such as peat bogs and fens that cover 3.5% of the terrestrial surface, but store about 30% of global carbon of terrestrial ecosystems (Gore, 1983; Denman et al., 2007). Largest sources of atmospheric N2O are soils under natural vegetation and agricultural use; nonetheless, certain acidic wetlands that have a high nitrogen content were recently identified as significant sources of N2O displaying N2O emissions in the range of those observed for agricultural soils (Denman et al., 2007; Repo et al., 2009; Marushchak et al., 2011). Conservative estimates suggest that 0.6% of the global annual emission of N2O might be attributed to such acidic wetlands (Repo et al., 2009).
Acidic wetlands are covered by water-adapted vascular plants, and mosses of the genus Sphagnum (Gorham, 1991). Microbes that produce CH4 are methanogenic Archaea, and those that are the main source of N2O are denitrifiers in anoxic habitats like wetlands (Conrad, 1996; Ferry, 1999; Drake et al., 2009). CH4 produced by methanogens is further oxidized by aerobic methanotrophs, which thrive as a significant part of wetland microbial communities and mitigate the net CH4 flux into the atmosphere (Hanson and Hanson, 1996; Shannon et al., 1996; Whalen, 2005; Knorr et al., 2008; Hornibrook et al., 2009). Atmospheric oxygen may penetrate soil due to water table draw downs, and gas transport through the aerenchyma of vascular plants into the rhizosphere (Moog and Bruggemann, 1998; Kettunen et al., 1999; Mainiero and Kazda, 2005; Paul et al., 2006). Acidic wetlands may temporarily turn into sinks of atmospheric CH4, or at least have the potential to consume atmospheric CH4 (≤1.75 ppmv; Table 1; Whalen and Reeburgh, 2000; Jauhiainen et al., 2005; Elberling et al., 2011), a fact that is currently underappreciated, and poorly understood on the level of microbial communities.
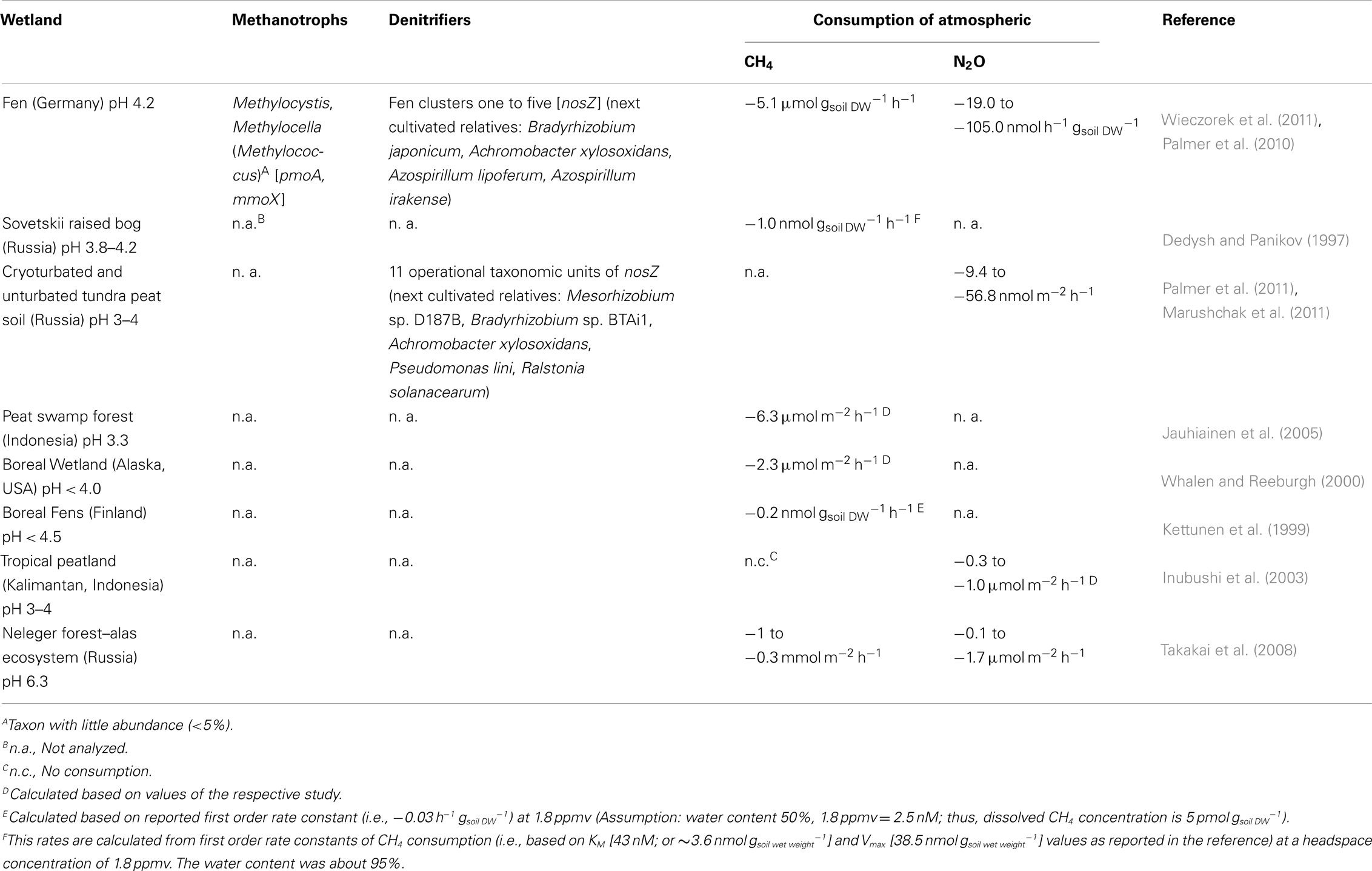
Table 1. Acidic wetlands exhibiting consumption of atmospheric CH4 or N2O, and associated methanotroph and denitrifier communities if such data were available.
Association of denitrifiers with N2O production in nitrogen-rich acidic wetlands is indicated by process-orientated studies of water-logged acidic soils, and denitrification is suggested as the primary N2O producing process in such systems where the water filled pore space exceeds 70% (Davidson, 1991). Denitrifiers catalyze the sequential reduction of N-oxides to nitrous oxide (N2O) and/or dinitrogen (N2) via the sequential action of nitrate, nitrite, nitric oxide, and nitrous oxide reductases (Zumft, 1997). Subunits that contain the catalytic center are encoded by narG/napA, nirK/nirS, norB, and nosZ. Dissimilatory reduction of nitrate via nitrite to ammonium (DNRA) might produce N2O under oxygen limited conditions via the unspecific interaction of nitrite with nitrate reductase (Smith and Zimmermann, 1981; Smith, 1983). However, DNRA potentials are low in acidic wetland soils, and free nitrite is virtually absent from such soils, suggesting that DNRA is negligible with regards to N2O production (Anderson and Leine, 1986; Bowden, 1987). Significant products of chemodenitrification are NO and NO2 rather than N2O (van Cleemput, 1998; Kappelmeyer et al., 2003; Kresovic et al., 2009). N2O is also produced as a by-product during the aerobic conversion of ammonia to nitrate by nitrifiers, but nitrification is the primary source of N2O in oxic rather than water-logged soils (Davidson, 1991; Conrad, 1996). Despite the importance of denitrifiers for N2O production in water-logged soils and the wealth of information on denitrifiers of agricultural and pH-neutral upland soils, studies addressing acid-tolerant denitrifiers in acidic wetlands are just emerging and under-represented in current literature. Evidence is accumulating that soils including acidic wetlands can act as temporary sinks for N2O (Inubushi et al., 2003; Hadi et al., 2005; Chapuis-Lardy et al., 2007; Goldberg et al., 2010). Indeed, negative fluxes of up to −1.7 μmol N2O m−2 h−1 were observed in situ for acidic wetlands (Table 1). Thus, the review intents to delineate the current understanding of microbial communities in acidic wetlands for CH4 and N2O consumption and to highlight research needs.
CH4 fluxes of acidic wetlands are highly variable due to non-linear interactions of environmental factors that control methanogenesis, methanotrophy, and CH4 gas transport (Whalen, 2005; Spahni et al., 2011). One reason of high spatial and temporal variability of CH4 exchange with the atmosphere might be a mitigation of net flux by consumption of atmospheric CH4 (Wieczorek et al., 2011). Hot spots of activity of methanotrophs in acidic wetlands are anoxic–oxic interfaces, where CH4 and oxygen gradients overlap (Sundh et al., 1995; Kettunen et al., 1999). Thus changes in oxygen availability alter both CH4 consumption, and production, and hence, reduce or increase net flux of CH4 from soil into the atmosphere (Kettunen et al., 1999; Whalen, 2005).
Prevalent methanotrophs of acidic wetlands belong to Alphaproteobacteria (Chen et al., 2008a; Dedysh, 2009; Wieczorek et al., 2011). Total cell numbers of methanotrophs of acidic wetlands range from 106 to 108 cells per gram of wet weight (Sundh et al., 1995; Dedysh et al., 2001, 2003; Dedysh, 2002, 2009; Belova et al., 2011). Methanotrophic pure cultures have primarily been isolated from Sphagnum-covered wetlands, and are acid-tolerant strains of Methylocystis (Methylocystaceae), Methylocella, and Methylocapsa (both Beijerinckiaceae; Dedysh et al., 2001, 2003; Dedysh, 2002, 2009; Kip et al., 2011). Methylocella species, and some species of Methylocystis and Methylocapsa utilize also simple multicarbon compounds, such as acetate (Dedysh et al., 2005; Theisen et al., 2005; Dunfield et al., 2010; Belova et al., 2011). Fluctuations of soil-internal methanogenesis may be caused by varying exudation of plants and redox conditions due to water table changes that can be consequences of drought periods, or heavy rainfall (Knorr et al., 2008; Knorr and Blodau, 2009, Dorodnikov et al., 2011). Acetate-utilizing methanotrophs are largely independent of such CH4 deficits in acidic wetlands.
Methylocystis strain H2s apparently represents a key methanotroph in acidic wetlands (Belova et al., 2011; Wieczorek et al., 2011). Strain H2s-related 16S rRNA genes ubiquitously occur in northern wetlands, and contribute with 18–58% (i.e., 0.4 × 107 to 3.4 × 107 cells g−1 of wet weight) to the alphaproteobacterial methanotrophs in such wetlands (Belova et al., 2011). Uncultivated species that are phylogenetically related to Methylocystis, and Methylocapsa are associated with hyaline cells of submerged mosses of Sphagnum. These Sphagnum methanotroph associations have been found in various northern wetlands (Kip et al., 2010). Methanotrophs apparently contribute up to 30% to carbon assimilation of the moss (Raghoebarsing et al., 2005; Kip et al., 2010; Larmola et al., 2010; Parmentier et al., 2011). The association is, based on current observations, not specific since different methanotroph-free Sphagnum species can be colonized by methanotrophs that were provided from the same Sphagnum individual (Larmola et al., 2010). The physiological basis of the interaction between methanotroph and moss has not been fully resolved.
Wetland methanotroph communities are usually dominated by Alphaproteobacteria, but gammaproteobacterial methanotrophs may also be present (Morris et al., 2002; Chen et al., 2008a,b; Wieczorek et al., 2011). Some of them (Methylomonas-, Methylosoma-related strains) have been isolated, which represent the first acid-tolerant members of the Methylococcaceae (Kip et al., 2011). All cultured methanotrophs of acidic wetlands are acid-tolerant and have growth optima below pH6 (Dedysh, 2009; Kip et al., 2011). Most information on the identities and physiologies of methanotrophs of acidic wetlands are derived from Sphagnum-associated microbial communities, but there is a lack of information on vascular plant-associated methanotrophs in acidic wetlands.
Acidic wetlands may temporally turn into net sinks of atmospheric CH4, which has been documented in various studies, or have the capability to utilize atmospheric CH4 concentrations (Table 1; Figure 1; Dedysh and Panikov, 1997; Kettunen et al., 1999; Whalen and Reeburgh, 2000; Jauhiainen et al., 2005; Elberling et al., 2011; Wieczorek et al., 2011). Many wetland sites have annual emission rates of less than 3.8 mol CH4 ha−2 year−1 (Dalal and Allen, 2008) indicating not necessarily only low CH4 production, but also a certain proportion of consumption of atmospheric CH4 over the year.
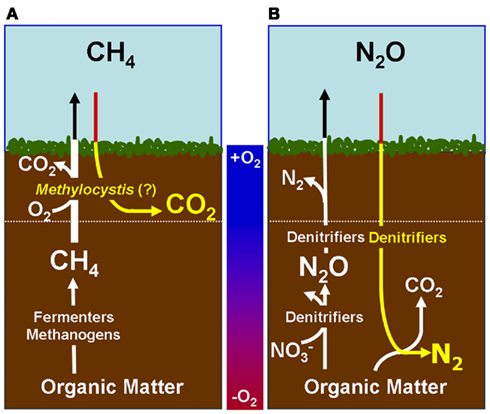
Figure 1. Proposed Model of Microbes that are responsible of atmospheric sink function of CH4 and N2O in natural wetlands. The quantitative contribution to global net fluxes of CH4 and N2O into the atmosphere has not been evaluated in acidic wetlands. (A) Acidic wetland covered by Sphagnum plants with low nitrogen input and mineralization rates. White Pathway: Plant-derived organic matter is degraded by microbial fermentation, acetogenesis, and methanogenesis to CH4 that is transported to oxygen-saturated zones close to the surface. The thickness of the oxygen-saturated layer is dependent of water table level (white dotted line). Methanotrophs (such as Methylocystyceae, Beijerinkiaceae, but as well Methylococcaceae) consume up to 90% of methanogenic CH4. Products are microbial biomass and CO2. The not-consumed proportion of CH4 is released into the atmosphere. Methanotrophs convert CH4 to carbon dioxide (CO2) which is released into the atmosphere. Yellow pathway: Methylocystis species likely consume atmospheric CH4 when CH4 supply from anoxic zones is limited. (B) Natural wetland with high nitrogen input and mineralization rates (e.g., minerotrophic fens, Palsa peats). White pathway: In nitrogen-rich acidic wetlands (e.g., Palsa peats) a part of anaerobic organic matter degradation intermediates are consumed by denitrifiers that reduce 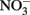 to N2O and N2. N2O and N2 is released into the atmosphere. Ammonia-oxidizing bacteria (AOB) are known to produce small amounts of N2O during oxidation of ammonia (Schmidt et al., 2004; Braker and Conrad, 2011). Yellow pathway: Under as-yet unresolved environmental conditions atmospheric N2O may be further reduced to N2 by to date not cultured denitrifiers that use electrons from organic matter.
to N2O and N2. N2O and N2 is released into the atmosphere. Ammonia-oxidizing bacteria (AOB) are known to produce small amounts of N2O during oxidation of ammonia (Schmidt et al., 2004; Braker and Conrad, 2011). Yellow pathway: Under as-yet unresolved environmental conditions atmospheric N2O may be further reduced to N2 by to date not cultured denitrifiers that use electrons from organic matter.
A very limited number of studies analyzed the composition of methanotroph communities of acidic wetlands, and measured the potential of the respective wetland to consume atmospheric CH4 (Table 1). Methylocystis species that were frequently detected in acidic wetlands are closely related to the solely known methanotroph strains that are capable of consumption of atmospheric CH4, i.e., strains LR1, SC2, and DWT (Dunfield et al., 1999, 2002; Kolb et al., 2005; Baani and Liesack, 2008; Dedysh, 2009; Wieczorek et al., 2011). Nonetheless, pmoA2 genes (that encode for the hydroxylase subunit of a high affinity CH4 mono-oxygenase) that would indicate the capability of consumption of atmospheric CH4 (Dunfield et al., 2002; Yimga et al., 2003; Ricke et al., 2004; Baani and Liesack, 2008) have only been detected at low relative abundances (<5%; Wieczorek et al., 2011). To warrant consumption of atmospheric CH4 in wetlands a small fraction (about 107 cells per gram dry weight in forest soils that consume atmospheric CH4; Kolb et al., 2005; Degelmann et al., 2010) of the total cell number of present methanotrophs (about 108 cells per gram dry weight) need presumably be active to allow for consumption of atmospheric CH4 at rates that were observed in acidic wetland sites. The major proportion of detectable pmoA (encodes the beta-subunit of a low affinity membrane bound CH4 mono-oxygenase) and mmoX (encodes the hydroxylase subunit of a low affinity cytoplasmic bound CH4 mono-oxygenase) genes in an acidic wetland soil that exhibited the capability of consumption of atmospheric levels of CH4 (i.e., 82 and 64%, respectively), affiliated with genes of Methylocystis (Wieczorek et al., 2011). Some Methylocystis species have the capability to utilize atmospheric CH4 concentrations. The abundant Methylocystis strain H2s harbors as well pmoA2 indicating that this methanotroph may consume atmospheric CH4 (Dedysh, personal communication). Thus, Methylocystis species of acidic wetlands likely are capable of atmospheric CH4 consumption (Figure 1), and may have the advantage to survive more likely periods of CH4 deficiency, may be periodically the case in minerotrophic fens, and at oxygen-saturated surface-near soil layers.
Recently described thermoacidophilic methanotrophs of the Methylacidiphilaceae (Op den Camp et al., 2009) as well as anaerobic methanotrophs Candidatus “Methylomirabilis oxyfera” (Ettwig et al., 2010) have not been detected in acidic wetlands. In summary, Methylocystis are likely involved for consumption of atmospheric CH4 in acidic wetlands. Nonetheless, an unequivocal experimental proof is missing. The role of other methanotrophs in acidic wetlands (i.e., Beijerinckiaceae, and Methylococcaceae) in consumption of atmospheric CH4 is not fully unresolved, but not very likely based on their low affinities for CH4. It is not likely that “M. oxyfera” is involved in N2O cycling since it lacks nitric oxide reductase.
One of the most remarkable features of denitrifiers is their capacity to produce and consume N2O (Table 1; Figure 1; Zumft, 1997). Such organisms are widely distributed in various environments and highly diverse (e.g., Zumft, 1997; Braker and Conrad, 2011). Not all denitrifiers known to date are capable of N2O consumption (Zumft and Kroneck, 2007). However, N2O is a common intermediate during denitrification and serves as sole electron acceptor for supporting growth of many denitrifiers (Zumft, 1997; Strohm et al., 2007). DNA based stable isotope probing with 13C-labeled succinate as electron donor and N2O as electron acceptor and cultivation-based studies demonstrated growth of putative denitrifiers by N2O reduction in rice field soil (Ishii et al., 2011). These findings are in line with a 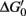 of N2O reduction to N2 that approximates −340 kJ mol−1 under standard conditions, indicating a highly exergonic process (Zumft and Kroneck, 2007). Thus, it might not be surprising that N2O consumption by soil communities is a more common phenomenon than previously thought (Table 1; reviewed in Holtan-Hartwig et al., 2000; Chapuis-Lardy et al., 2007).
of N2O reduction to N2 that approximates −340 kJ mol−1 under standard conditions, indicating a highly exergonic process (Zumft and Kroneck, 2007). Thus, it might not be surprising that N2O consumption by soil communities is a more common phenomenon than previously thought (Table 1; reviewed in Holtan-Hartwig et al., 2000; Chapuis-Lardy et al., 2007).
N2O reductases of denitrifiers catalyze the efficient reduction of N2O to N2 and are copper-dependent metallo-enzymes that exhibit KM values in the lower micromolar range for N2O, highlighting the potential of denitrifiers to consume low concentrations of N2O (Zumft, 1997). Abiotic conversion of N2O at ambient temperatures might occur with (reduced) transition-metal complexes (which occur in the active site of N2O converting enzymes) and metal amides (Banks et al., 1968; Zumft and Kroneck, 2007). Metallo-enzymes like carbon monoxide dehydrogenase and cobalamin-dependent methionine synthase are also capable of reducing N2O to N2, while nitrogenase converts N2O to NH3; bacterial nitrite cytochrome c reductase transforms N2O to an unknown product (Jensen and Burris, 1986; Bannerjee and Matthews, 1990; Lu and Ragsdale, 1991; Drummond and Matthews, 1994; Stach et al., 2000). Such enzymes are hosted by diverse organisms including non-denitrifiers (see references in Zumft, 1997; Zumft and Kroneck, 2007). However, the conversion of N2O by these aforementioned enzymes is regarded as a non-physiological reaction. Low KM values and high specific activities for N2O (as were demonstrated for N2O reductases of denitrifiers) have not been demonstrated for such enzymes, suggesting a minor role, if any, for the consumption of atmospheric N2O concentrations (319 ppb, equivalent to app. 8 nmol N2O per liter soil solution; Conrad, 1996) in soils (Zumft, 1997; Stach et al., 2000; Zumft and Kroneck, 2007). In this context, dissimilatory reducers of nitrate producing ammonia of the Enterobacteriaceae that were shown to consume N2O in the laboratory displayed a KM value of 3 mM for N2O, likewise suggesting that the N2O reduction is a non-physiological reaction and probably of minor importance in situ (Kaldorf et al., 1993). Moreover, assimilatory reduction of N2O that might theoretically proceed via nitrogenase action was not detected in soils (Vieten et al., 2008). Thus, denitrifiers hosting N2O reductases deserve primary attention in the context of N2O consumption in soils.
Denitrification rates and the ratio of N2O to total nitrogen gas products are regulated by oxygen availability, pH, temperature, carbon to nitrogen ratio, the availability of substrates, and electron acceptors, as well as by the denitrifier community composition (van Cleemput, 1998; Dörsch et al., 2011). Anoxia (as indicated by a water filled pore space of greater than 60%), a carbon to nitrogen ratio greater than 30, and pH-neutral conditions favor complete denitrification to N2 in soils including acidic wetlands (Conrad, 1995, 1996; Klemedtsson et al., 2005; Cuhel et al., 2010; Braker and Conrad, 2011). A pH value below 5 is often encountered in peatlands, and is hypothesized to impair N2O reduction and to increase the product ratio of N2O to N2 (Simek and Cooper, 2002; Cuhel et al., 2010). However, acidic wetlands host acid-tolerant denitrifiers capable of N2O reduction and complete denitrification (Palmer et al., 2010, 2011). Moreover, evidence of active denitrification-associated N2O production and consumption, (including consumption of atmospheric N2O concentrations) was obtained by compound specific stable isotope analyses of N2O in one acidic wetland (Goldberg et al., 2008, 2010). In that specific wetland, δ15N values indicated that N2O produced in deep soil layers was partially consumed upon its diffusion to the soil surface (Goldberg et al., 2008, 2010), suggesting that denitrifiers might contribute to the mitigation of N2O emission from acidic wetlands. Denitrifiers that exhibit the genetic potential to consume N2O were detected in acidic wetlands by the analysis of nosZ genes. Detected nosZ genes in acidic wetlands mostly affiliated with nosZ from uncultured soil bacteria or were only distantly related to known genes, suggesting a wealth of uncultured denitrifier diversity in acidic wetlands (Palmer et al., 2010, 2011). Phylogenies of nosZ and 16S rRNA genes from the same organisms are basically congruent (Jones et al., 2008; Palmer et al., 2009), allowing to speculate on the identity of organisms hosting the detected nosZ. Thus, putative N2O reducers in acidic wetlands include Alphaproteobacteria (Azospirillum, Brady-, and Mesorhizobium), Betaproteobacteria (Achromobacter, Ralstonia), and Gammaproteobacteria (Pseudomonas). However, it remains to be determined whether the detected N2O reductase genes were expressed, i.e., the detected organisms reduce N2O in situ (Figure 1).
Analyses of other genes associated with denitrification corroborate that acidic wetlands harbor diverse and new denitrifiers (Palmer et al., 2010, 2011). However, given the low number of studies addressing N2O-consuming denitrifier communities in acidic wetlands, and the selectivity of PCR-based gene marker analyses, the present studies have to be regarded as providing a minimal estimate of denitrifier diversity capable of N2O consumption (Throback et al., 2004; Green et al., 2010; Heylen et al., 2011; Jones et al., 2011). The fact that high affinity respiratory N2O reduction occurs in non-denitrifying organisms that reduce nitrate but lack nitrite reductase activity, and that nitrifiers were shown to have the capability to consume N2O and produce N2 further complicates matters (Yoshinari, 1980; McEwan et al., 1985; Zumft, 1997; Schmidt et al., 2004). Nevertheless, the relevance of N2O consumption for mitigation of N2O emissions, and eventually a temporary sink function of acidic wetlands for N2O is emerging, although the physiology and taxonomic identity of “microbial catalysts” have not been sufficiently resolved. Furthermore, the regulation of N2O reduction in situ and on cellular level, and thus the potential of the soil to reduce N2O to N2, and the diffusivity of N2O within the soil profile will impact the sink functions of wetland soils for N2O but are likewise only poorly resolved.
Few studies addressed the role of methanotrophs and denitrifiers in consumption of atmospheric CH4 and N2O in acidic wetlands. Potential consumption activities have been measured in laboratory-scale experiments, and first evidence for denitrification-associated N2O consumption in situ is available. Systematic in situ flux measurements combined with measurements of environmental parameters, consumption potentials, stable isotope signatures of carbon and nitrogen in CH4 and N2O pools, and community analyses are needed to improve our understanding of environmental factors that trigger CH4 and N2O consumption in acidic wetland soils.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Anderson, I. C., and Leine, J. S. (1986). Relative rates of nitric oxide and nitrous oxide production by nitrifiers, denitrifiers, and nitrate respires. Appl. Environ. Microbiol. 51, 938–945.
Baani, M., and Liesack, W. (2008). Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc. Natl. Acad. Sci. U.S.A. 105, 10203–10208.
Banks, R. G. S., Henderson, R. J., and Pratt, J. M. (1968). Reactions of gases in solution. Part III. Some reactions of nitrous oxide with transition-metal complexes. J. Chem. Soc. A 2886–2889. doi: 10.1039/J19680002886
Bannerjee, R. V., and Matthews, R. G. (1990). Cobalamin-dependent methionine synthase. FASEB J. 4, 1450–1459.
Belova, S. E., Baani, M., Suzina, N. E., Bodelier, P. L. E., Liesack, W., and Dedysh, S. N. (2011). Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ. Microbiol. Rep. 3, 36–46.
Bowden, W. B. (1987). The biogeochemistry of nitrogen in freshwater wetlands. Biogeochemistry 4, 313–348.
Braker, G., and Conrad, R. (2011). “Diversity, structure, and size of N2O-producing microbial communities in soils – what matters for their functioning?” in Advances in Applied Microbiology, Vol. 75, eds A. I. Laskin, S. Sariaslani, and G. M. Gadd (San Diego: Elsevier Academic Press Inc.), 33–70.
Chapuis-Lardy, L., Wrage, N., Metay, A., Chotte, J. L., and Bernoux, M. (2007). Soils, a sink for N2O? A review. Glob. Chang. Biol. 13, 1–17.
Chen, Y., Dumont, M. G., Mcnamara, N. P., Chamberlain, P. M., Bodrossy, L., Stralis-Pavese, N., and Murrell, J. C. (2008a). Diversity of the active methanotrophic community in acidic peatlands as assessed by mRNA and SIP-PLFA analyses. Environ. Microbiol. 10, 446–459.
Chen, Y., Dumont, M. G., Neufeld, J. D., Bodrossy, L., Stralis-Pavese, N., Mcnamara, N. P., Ostle, N., Briones, M. J., and Murrell, J. C. (2008b). Revealing the uncultivated majority: combining DNA stable-isotope probing, multiple displacement amplification and metagenomic analyses of uncultivated Methylocystis in acidic peatlands. Environ. Microbiol. 10, 2609–2622.
Conrad, R. (1995). “Soil microbial processes involved in production and consumption of atmospheric trace gases,” in Advances in Applied Microbiology, ed. J. G. Jones (New York: Plenum Press), 207–250.
Conrad, R. (1996). Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60, 609–642.
Cuhel, J., Simek, M., Laughlin, R. J., Bru, D., Cheneby, D., Watson, C. J., and Philippot, L. (2010). Insights into the effect of soil pH on N2O and N2 emissions and denitrifier community size and activity. Appl. Environ. Microbiol. 76, 1870–1878.
Dalal, R. C., and Allen, D. E. (2008). Greenhouse gas fluxes from natural ecosystems. Aust. J. Bot. 56, 369–407.
Davidson, E. A. (1991). “Fluxes of nitrous oxide and nitric oxide from terrestrial ecosystems,” in Microbial Production and Consumption of Greenhouse Gases, eds J. E. Rogers and W. B. Whitman (Washington, DC: American Society for Microbiology), 219–235.
Dedysh, S. N. (2002). Methanotrophic bacteria of acidic Sphagnum peat bogs. Microbiology 71, 638–650.
Dedysh, S. N. (2009). Exploring methanotroph diversity in acidic northern wetlands: molecular and cultivation-based studies. Microbiology 78, 655–669.
Dedysh, S. N., Derakshani, M., and Liesack, W. (2001). Detection and enumeration of methanotrophs in acidic Sphagnum peat by 16S rRNA fluorescence in situ hybridization, including the use of newly developed oligonucleotide probes for Methylocella palustris. Appl. Environ. Microbiol. 67, 4850–4857.
Dedysh, S. N., Dunfield, P. F., Derakshani, M., Stubner, S., Heyer, J., and Liesack, W. (2003). Differential detection of type II methanotrophic bacteria in acidic peatlands using newly developed 16S rRNA-targeted fluorescent oligonucleotide probes. FEMS Microbiol. Ecol. 43, 299–308.
Dedysh, S. N., Knief, C., and Dunfield, P. F. (2005). Methylocella species are facultatively methanotrophic. J. Bacteriol. 187, 4665–4670.
Dedysh, S. N., and Panikov, N. S. (1997). Effect of methane concentration on the rate of its oxidation by bacteria in Sphagnum peat. Microbiology 66, 470–475.
Degelmann, D. M., Borken, W., Drake, H. L., and Kolb, S. (2010). Different atmospheric methane-oxidizing communities in European Beech and Norway Spruce soils. Appl. Environ. Microbiol. 76, 3228–3235.
Denman, K. L., Brasseur, G., Chidthaisong, A., Ciais, P., Cox, P. M., Dickinson, R. E., Hauglustaine, D., Heinze, C., Holland, E., Jacob, D., Lohmann, U., Ramachandran, S., Da Silva Dias, P. L., Wofsy, S. C., and Zhang, X. (2007). “Couplings between changes in the climate system and biogeochemistry,” in Climate Change 2007: The Physical Science Basis (Cambridge: Cambridge University Press), 499–587.
Dorodnikov, M., Knorr, K.-H., Kuzyakov, Y., and Wilmking, M. (2011). Plant-mediated CH4 transport and contribution of photosynthates to methanogenesis at a boreal mire: a 14C pulse study. Biogeosciences 8, 2365–2375.
Dörsch, P., Braker, G., and Bakken, L. (2011). Community specific pH response of denitrification: experi-ments with cells extracted from org-anic soils. FEMS Microbiol. Ecol. 79, 530–541.
Drake, H. L., Horn, M. A., and Wüst, P. K. (2009). Intermediary ecosystem metabolism as a main driver of methanogenesis in acidic wetland soil. Environ. Microbiol. Rep. 1, 307–318.
Drummond, J. T., and Matthews, R. G. (1994). Nitrous oxide degradation by cobalamin-dependent methionine synthase: characterization of the reactants and products in the inactivation reaction. Biochemistry 33, 3732–3741.
Dunfield, P. F., Belova, S. E., Vorob’Ev, A. V., Cornish, S. L., and Dedysh, S. N. (2010). Methylocapsa aurea sp. nov., a facultative methanotroph possessing a particulate methane monooxygenase, and emended description of the genus Methylocapsa. Int. J. Syst. Evol. Microbiol. 60, 2659–2664.
Dunfield, P. F., Liesack, W., Henckel, T., Knowles, R., and Conrad, R. (1999). High-affinity methane oxidation by a soil enrichment culture containing a type II methanotroph. Appl. Environ. Microbiol. 65, 1009–1014.
Dunfield, P. F., Yimga, M. T., Dedysh, S. N., Berger, U., Liesack, W., and Heyer, J. (2002). Isolation of a Methylocystis strain containing a novel pmoA-like gene. FEMS Microbiol. Ecol. 41, 17–26.
Elberling, B., Askaer, L., Jorgensen, C. J., Joensen, H. P., Kuhl, M., Glud, R. N., and Lauritsen, F. R. (2011). Linking soil O2, CO2, and CH4 concentrations in a wetland soil: implications for CO2 and CH4 fluxes. Env. Sci. Tech. 45, 3393–3399.
Ettwig, K. F., Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M. M., Schreiber, F., Dutilh, B. E., Zedelius, J., de Beer, D., Gloerich, J., Wessels, H. J. C. T., van Alen, T., Luesken, F., Wu, M. L., van de Pas-Schoonen, K. T., Op den Camp, H. J. M., Janssen-Megens, E. M., Francoijs, K.-J., Stunnenberg, H., Weissenbach, J., Jetten, M. S. M., and Strous, M. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548.
Ferry, J. G. (1999). Enzymology of one-carbon metabolism in methanogenic pathways. FEMS Microbiol. Rev. 23, 13–38.
Goldberg, S. D., Knorr, K.-H., Blodau, C., Lischeid, G., and Gebauer, G. (2010). Impact of altering the water table height of an acidic fen on N2O and NO fluxes and soil concentrations. Glob. Chang. Biol. 16, 220–233.
Goldberg, S. D., Knorr, K.-H., and Gebauer, G. (2008). N2O concentration and isotope signature along profiles provide deeper insight into the fate of N2O in soils. Isotopes Environ. Health Stud. 44, 377–391.
Gorham, E. (1991). Northern peatlands – role in the carbon-cycle and probable responses to climatic warming. Ecol. Appl. 1, 182–195.
Green, S. J., Prakash, O., Gihring, T. M., Akob, D. M., Jasrotia, P., Jardine, P. M., Watson, D. B., Brown, S. D., Palumbo, A. V., and Kostka, J. E. (2010). Denitrifying bacteria isolated from terrestrial subsurface sediments exposed to mixed-waste contamination. Appl. Environ. Microbiol. 76, 3244–3254.
Hadi, A., Inubushi, K., Furukawa, Y., Purnomo, E., Rasmadi, M., and Tsuruta, H. (2005). Greenhouse gas emissions from tropical peatlands of Kalimantan, Indonesia. Nutr. Cycl. Agroecosys. 71, 73–80.
Heylen, K., Verbaendert, I., De Vos, P., and Boon, N. (2011). Denitrification in gram-positive bacteria: an underexplored trait. Biochem. Soc. Trans. 39, 254–258.
Holtan-Hartwig, L., Dorsch, P., and Bakken, L. R. (2000). Comparison of denitrifying communities in organic soils: kinetics of NO3- and N2O reduction. Soil Biol. Biochem. 32, 833–843.
Hornibrook, E. R. C., Bowes, H. L., Culbert, A., and Gallego-Sala, A. V. (2009). Methanotrophy potential versus methane supply by pore water diffusion in peatlands. Biogeosciences 6, 1490–1504.
Inubushi, K., Furukawa, Y., Hadi, A., Purnomo, E., and Tsuruta, H. (2003). Seasonal changes of CO2, CH4 and N2O fluxes in relation to land-use change in tropical peatlands located in coastal area of South Kalimantan. Chemosphere 52, 603–608.
Ishii, S., Ohno, H., Tsuboi, M., Otsuka, S., and Senoo, K. (2011). Identification and isolation of active N2O reducers in rice paddy soil. ISME J. 5, 1936–1945.
Jauhiainen, J., Takahashi, H., Heikkinen, J. E. P., Martikainen, P. J., and Vasander, H. (2005). Carbon fluxes from a tropical peat swamp forest floor. Glob. Chang. Biol. 11, 1788–1797.
Jensen, B. B., and Burris, R. H. (1986). Nitrogenase. N2O as a substrate and as a competitive inhibitor of nitrogenase. Biochemistry 25, 1083–1088.
Jones, C. M., Stres, B., Rosenquist, M., and Hallin, S. (2008). Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 25, 1955–1966.
Jones, C. M., Welsh, A., Throback, I. N., Dorsch, P., Bakken, L. R., and Hallin, S. (2011). Phenotypic and genotypic heterogeneity among closely related soil-borne N2- and N2O-producing Bacillus isolates harboring the nosZ gene. FEMS Microbiol. Ecol. 76, 541–552.
Kaldorf, M., Linne yon Berg, K.-H., Meier, U., Servos, U., and Bothe, H. (1993). The reduction of nitrous oxide to dinitrogen by Escherichia coli. Arch. Microbiol. 160, 432–439.
Kappelmeyer, U., Kuschk, P., and Stottmeister, U. (2003). Model experiments on the influence of artificial humic compounds on chemodenitrification. Water Air Soil Pollut. 147, 317–330.
Kettunen, A., Kaitala, V., Lehtinen, A., Lohila, A., Alm, J., Silvola, J., and Martikainen, P. J. (1999). Methane production and oxidation potentials in relation to water table fluctuations in two boreal mires. Soil Biol. Biochem. 31, 1741–1749.
Kip, N., Ouyang, W. J., Van Winden, J., Raghoebarsing, A., Van Niftrik, L., Pol, A., Pan, Y., Bodrossy, L., Van Donselaar, E. G., Reichart, G. J., Jetten, M. S. M., Damste, J. S. S., and Den Camp, H. J. M. O. (2011). Detection, isolation, and characterization of acidophilic methanotrophs from Sphagnum mosses. Appl. Environ. Microbiol. 77, 5643–5654.
Kip, N., Van Winden, J. F., Pan, Y., Bodrossy, L., Reichart, G. J., Smolders, A. J. P., Jetten, M. S. M., Damste, J. S. S., and Op Den Camp, H. J. M. (2010). Global prevalence of methane oxidation by symbiotic bacteria in peat-moss ecosystems. Nat. Geosci. 3, 617–621.
Klemedtsson, L., Von Arnold, K., Weslien, P., and Gundersen, P. (2005). Soil CN ratio as a scalar parameter to predict nitrous oxide emissions. Glob. Chang. Biol. 11, 1142–1147.
Knorr, K. H., and Blodau, C. (2009). Impact of experimental drought and rewetting on redox transformations and methanogenesis in mesocosms of a northern fen soil. Soil Biol. Biochem. 41, 1187–1198.
Knorr, K. H., Glaser, B., and Blodau, C. (2008). Fluxes and 13C isotopic composition of dissolved carbon and pathways of methanogenesis in a fen soil exposed to experimental drought. Biogeosciences 5, 1457–1473.
Kolb, S., Knief, C., Dunfield, P. F., and Conrad, R. (2005). Abundance and activity of uncultured methanotrophic bacteria involved in the consumption of atmospheric methane in two forest soils. Environ. Microbiol. 7, 1150–1161.
Kresovic, M., Jakovljevic, M., Blagojevic, S., and Maksimovic, S. (2009). Specific transformations of mineral forms of nitrogen in acid soils. J. Serbian Chem. Soc. 74, 93–102.
Larmola, T., Tuittila, E. S., Tiirola, M., Nykanen, H., Martikainen, P. J., Yrjala, K., Tuomivirta, T., and Fritze, H. (2010). The role of Sphagnum mosses in the methane cycling of a boreal mire. Ecology 91, 2356–2365.
Lu, W.-P., and Ragsdale, S. W. (1991). Reductive activation of the coenzyme A/Acetyl-CoA isotopic exchange reaction catalyzed by carbon monoxide dehydrogenase from Clostridium thermoaceticum and its inhibition by nitrous oxide and carbon monoxide. J. Biol. Chem. 266, 3554–3564.
Mainiero, R., and Kazda, M. (2005). Effects of Carex rostrata on soil oxygen in relation to soil moisture. Plant Soil 270, 311–320.
Marushchak, M. E., Pitkämäki, A., Koponen, H., Biasi, C., Seppälä, M., and Martikainen, P. J. (2011). Hot spots for nitrous oxide emissions found in different types of permafrost peatlands. Glob. Chang. Biol. 17, 2601–2614.
McEwan, A. G., Greenfield, A. J., Wetzstein, H. G., Jackson, J. B., and Ferguson, S. J. (1985). Nitrous oxide reduction by members of the family Rhodospirillaceae and the nitrous oxide reductase of Rhodopseudomonas capsulata. J. Bacteriol. 164, 823–830.
Moog, P. R., and Bruggemann, W. (1998). Flooding tolerance of Carex species. II. Root gas-exchange capacity. Planta 207, 199–206.
Morris, S. A., Radajewski, S., Willison, T. W., and Murrell, J. C. (2002). Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68, 1446–1453.
Op den Camp, H. J. H., Islam, T., Stott, M. B., Harhangi, H. R., Hynes, A., Schouten, S., Jetten, M. S. M., Birkeland, N.-K., Pol, A., and Dunfield, P. F. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1, 239–306.
Palmer, K., Biasi, C., and Horn, M. A. (2011). Contrasting denitrifier communities relate to contrasting N2O emission patterns from acidic peat soils in arctic tundra. ISME J. doi: ISMEJ.2011.172
Palmer, K., Drake, H. L., and Horn, M. A. (2009). Genome-derived criteria for assigning environmental narG and nosZ sequences to operational taxonomic units of nitrate reducers. Appl. Environ. Microbiol. 75, 5170–5174.
Palmer, K., Drake, H. L., and Horn, M. A. (2010). Association of novel and highly diverse acid-tolerant denitrifiers with N2O fluxes of an acidic fen. Appl. Environ. Microbiol. 76, 1125–1134.
Parmentier, F. J. W., Van Huissteden, J., Kip, N., Op Den Camp, H. J. M., Jetten, M. S. M., Maximov, T. C., and Dolman, A. J. (2011). The role of endophytic methane-oxidizing bacteria in submerged Sphagnum in determining methane emissions of Northeastern Siberian tundra. Biogeosciences 8, 1267–1278.
Paul, S., Küsel, K., and Alewell, C. (2006). Reduction processes in forest wetlands: tracking down heterogeneity of source/sink functions with a combination of methods. Soil Biol. Biochem. 38, 1028–1039.
Raghoebarsing, A. A., Smolders, A. J. P., Schmid, M. C., Rijpstra, W. I. C., Wolters-Arts, M., Derksen, J., Jetten, M. S. M., Schouten, S., Damste, J. S. S., Lamers, L. P. M., Roelofs, J. G. M., Den Camp, H. J. M. O., and Strous, M. (2005). Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature 436, 1153–1156.
Repo, M. E., Susiluoto, S., Lind, S. E., Jokinen, S., Elsakov, V., Biasi, C., Virtanen, T., and Martikainen, P. J. (2009). Large N2O emissions from cryoturbated peat soil in tundra. Nat. Geosci. 2, 189–192.
Ricke, P., Erkel, C., Kube, M., Reinhardt, R., and Liesack, W. (2004). Comparative analysis of the conventional and novel pmo (particulate methane monooxygenase) operons from Methylocystis strain SC2. Appl. Environ. Microbiol. 70, 3055–3063.
Schmidt, I., vanSpanning, R. J. M., and Jetten, M. S. M. (2004). Denitrification and ammonia oxidation by Nitrosomonas europaea wild-type, and NirK and NorB-deficient mutants. Microbiology 150, 4107–4114.
Shannon, R. D., White, J. R., Lawson, J. E., and Gilmour, B. S. (1996). Methane efflux from emergent vegetation in peatlands. J. Ecol. 84, 239–246.
Simek, M., and Cooper, J. E. (2002). The influence of soil pH on denitrification: progress towards the understanding of this interaction over the last 50 years. Eur. J. Soil Sci. 53, 345–354.
Smith, M. S. (1983). Nitrous-oxide production by Escherichia coli is correlated with nitrate reductase-activity. Appl. Environ. Microbiol. 45, 1545–1547.
Smith, M. S., and Zimmermann, K. (1981). Nitrous oxide production by nondenitrifying soil nitrate reducers. Soil Sci. Soc. Am. J. 45, 865–871.
Spahni, R., Wania, R., Neef, L., Van Weele, M., Pison, I., Bousquet, P., Frankenberg, C., Foster, P. N., Joos, F., Prentice, I. C., and Van Velthoven, P. (2011). Constraining global methane emissions and uptake by ecosystems. Biogeosciences 8, 1643–1665.
Stach, P., Einsle, O., Schumacher, W., Kurun, E., and Kroneck, P. M. H. (2000). Bacterial cytochrome c nitrite reductase: new structural and functional aspects. J. Inorg. Biochem. 79, 381–385.
Strohm, T. O., Griffin, B., Zumft, W. G., and Schink, B. (2007). Growth yields in bacterial denitrification and nitrate ammonification. Appl. Environ. Microbiol. 73, 1420–1424.
Sundh, I., Borga, P., Nilsson, M., and Svensson, B. H. (1995). Estimation of cell numbers of methanotrophic bacteria in boreal peatlands based on analysis of specific phospholipid fatty-acids. FEMS Microbiol. Ecol. 18, 103–112.
Takakai, F., Desyatkin, A. R., Lopez, C. M. L., Fedorov, A. N., Desyatkin, R. V., and Hatano, R. (2008). CH4 and N2O emissions from a forest-alas ecosystem in the permafrost taiga forest region, eastern Siberia, Russia. J. Geophys. Res. 113, G02002.
Theisen, A. R., Ali, M. H., Radajewski, S., Dumont, M. G., Dunfield, P. F., Mcdonald, I. R., Dedysh, S. N., Miguez, C. B., and Murrell, J. C. (2005). Regulation of methane oxidation in the facultative methanotroph Methylocella silvestris BL2. Mol. Microbiol. 58, 682–692.
Throback, I. N., Enwall, K., Jarvis, A., and Hallin, S. (2004). Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49, 401–417.
van Cleemput, O. (1998). Subsoils: chemo- and biological denitrification, N2O and N2 emissions. Nutr. Cycl. Agroecosys. 52, 187–194.
Vieten, B., Conen, F., Seth, B., and Alewell, C. (2008). The fate of N2O consumed in soils. Biogeosciences 5, 129–132.
Whalen, S. C. (2005). Biogeochemistry of methane exchange between natural wetlands and the atmosphere. Environ. Eng. Sci. 22, 73–94.
Whalen, S. C., and Reeburgh, W. S. (2000). Methane oxidation, production, and emission at contrasting sites in a boreal bog. Geomicrobiol. J. 17, 237–251.
Wieczorek, A. S., Drake, H. L., and Kolb, S. (2011). Organic acids and ethanol inhibit the oxidation of methane by mire methanotrophs. FEMS Microbiol. Ecol. 77, 28–39.
Yimga, M. T., Dunfield, P. F., Ricke, P., Heyer, H., and Liesack, W. (2003). Wide distribution of a novel pmoA-like gene copy among type II methanotrophs, and its expression in Methylocystis strain SC2. Appl. Environ. Microbiol. 69, 5593–5602.
Keywords: Peat, bog, fen, CH4 cycle, nitrogen cycle, greenhouse gas sink, soil microbiology
Citation: Kolb S and Horn MA (2012) Microbial CH4 and N2O consumption in acidic wetlands. Front. Microbio. 3:78. doi: 10.3389/fmicb.2012.00078
Received: 16 December 2011; Accepted: 15 February 2012;
Published online: 02 March 2012.
Edited by:
Svetlana N. Dedysh, Winogradsky Institute of Microbiology, Russian Academy of Sciences, RussiaReviewed by:
Gesche Braker, Max Planck Institute for Terrestrial Microbiology, GermanyCopyright: © 2012 Kolb and Horn. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Steffen Kolb, Department of Ecological Microbiology, University of Bayreuth, Dr.-Hans-Frisch-Str. 1-3, 95448 Bayreuth, Germany. e-mail:c3RlZmZlbi5rb2xiQHVuaS1iYXlyZXV0aC5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.