
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 03 February 2012
Sec. Microbial Immunology
Volume 3 - 2012 | https://doi.org/10.3389/fmicb.2012.00023
This article is part of the Research TopicSystems Biology of Microbial InfectionView all 12 articles
Listeria monocytogenes is a foodborne human pathogen that can cause invasive infection in susceptible animals and humans. For proliferation within hosts, this facultative intracellular pathogen uses a reservoir of specific metabolic pathways, transporter, and enzymatic functions whose expression requires the coordinated activity of a complex regulatory network. The highly adapted metabolism of L. monocytogenes strongly depends on the nutrient composition of various milieus encountered during infection. Transcriptomic and proteomic studies revealed the spatial–temporal dynamic of gene expression of this pathogen during replication within cultured cells or in vivo. Metabolic clues are the utilization of unusual C2- and C3-bodies, the metabolism of pyruvate, thiamine availability, the uptake of peptides, the acquisition or biosynthesis of certain amino acids, and the degradation of glucose-phosphate via the pentose phosphate pathway. These examples illustrate the interference of in vivo conditions with energy, carbon, and nitrogen metabolism, thus affecting listerial growth. The exploitation, analysis, and modeling of the available data sets served as a first attempt to a systemic understanding of listerial metabolism during infection. L. monocytogenes might serve as a model organism for systems biology of a Gram-positive, facultative intracellular bacterium.
A successful infection by bacterial pathogens requires multiple adaptation processes including adhesion to host tissues, modulation of the immune response, or toxic activity toward the host defense system. Some pathogens enter epithelial cells or are internalized by professional phagocytes, and these steps are often followed by bacterial manipulation of the host cell actin skeleton and the manipulation of the endocytic route. Most of these processes, which often require specific virulence factors that enable the microbes to overcome the various physical and biochemical barriers of the infected host, have been characterized in detail.
In contrast, little attention has been given to the metabolic requirements and the metabolic flexibility of bacteria during infection, in parts due to limitation of analytical tools, and because the bacterial metabolism in vivo and in vitro has erroneously been assumed to be similar (Muñoz-Elías and McKinney, 2006). Therefore, our knowledge about the substrates used by pathogens during infection, and, equally important, the effect of a bacterial infection on the metabolism of the host cell is still fragmentary (Joseph and Goebel, 2007; Fuchs et al., 2011; Rohmer et al., 2011). Furthermore, the structural conservation of metabolic enzymes was considered to prevent the identification of microbe-specific inhibitors. Key metabolic enzymes, however, that are specifically required during growth within host cells could constitute a promising new set of possible targets for antibacterial compounds urgently needed or be used for the development of food formulas that suppress growth of pathogenic bacteria (Boigegrain et al., 2005; Becker et al., 2006; Liautard et al., 2006). Recent progress has been made in determining the major carbon sources used by intracellularly replicating pathogens such as Listeria monocytogenes, Shigella flexneri, and pathogenic Escherichia coli (Lucchini et al., 2005; Eylert et al., 2008; Götz and Goebel, 2010; Götz et al., 2010). These data suggest that pathogens, in order to efficiently replicate within a host or its cells, have to coordinate their metabolism with the availability of nutrients during their life cycle (for review, see Eisenreich et al., 2010).
Listeria monocytogenes is a Gram-positive pathogen that mainly affects immunocompromised individuals, pregnant women, and newborns. Severe infections are characterized by bacteremia, meningoencephalitis, abortion, or neonatal sepsis. The most common vehicles of transmission of this saprophytic bacterium to humans are dairy products and other foods including eggs, seafood, and vegetables. Three hundred eighty-six documented cases of listeriosis were reported for 2010 in Germany (Robert Koch-Institut, 2011), and about 1600 in the USA (Centers for Disease Control and Prevention, 2011). The high lethality rate of up to 20–30% despite early antibiotic treatment resulted in increasing efforts to understand listerial pathogenicity and to find tools against this pathogen (Vázquez-Boland et al., 2001).
Upon uptake by contaminated food, L. monocytogenes enter non-phagocytic cells such as epithelial cells, hepatocytes, or fibroblasts by the activity of the surface-associated internalins A and B. In contrast to other facultative intracellular pathogens like Salmonella, a hallmark of L. monocytogenes is that it is capable to escape from the phagocytic vacuole by disrupting the phagosomal membrane via the expression of listeriolysin (Hly) and phospholipase A (PlcA). Listerial cells thus access the host cell cytoplasm where they are not only able to replicate, but also to actively move by actin polymerization mediated by ActA. Cell-to-cell spreading and subsequent disruption of the vacuolar double-membrane by Hly and PlcB has also been observed. Having passed the gut epithelium, L. monocytogenes is capable to resist killing by professional phagocytes. It might disseminate via the lymph and the blood to the liver and the spleen and even cross the blood–brain or the blood–placenta barrier. All main virulence factors are under control of the positive regulatory factor A (PrfA; for more details, see reviews such as Vázquez-Boland et al., 2001; Dussurget et al., 2004; Hamon et al., 2006; Cossart and Toledo-Arana, 2008; Camejo et al., 2011).
Here, we will summarize recent omic-studies relevant for the topic of listerial metabolism during infection, and introduce isotopolog profiling analysis (IPA) as a technique that allows novel insights in metabolic fluxes during infection. Then, metabolic adaptations and requirement of L. monocytogenes in cultured cells and in vivo, as well as the underlying regulatory factors, will be resumed. Modeling approaches as further tools that pave the way toward a systems level understanding of listerial metabolism during infection will be presented, followed by challenges and future perspectives in this research field.
So far, mainly two strategies have been followed to improve our systemic understanding of metabolic adaptations by listeriae within host compartments: firstly, the differences between the transcriptomes and the proteomes of apathogenic and pathogenic strains, and secondly the analysis of differential gene expression upon the transition from one physiologically relevant condition to another.
Comparative genomics is important to identify factors and pathways contributing to virulence properties, both on the genus level with insights into host specificities and on the species level regarding the biodiversity of certain bacterial lineages (Zhao et al., 2011). Detection of differences between the genomes of L. monocytogenes and the apathogenic species L. innocua gave first insights into the reasons that cause a Listeria species to be pathogenic (Buchrieser et al., 2003). In a triple analysis involving the whole genome sequences of L. monocytogenes, L. welshimeri, and L. innocua, it was shown that genome reduction led to the apathogenic L. welshimeri, the latter being derived from early evolutionary events. This finding points to an ancestor more compact than L. monocytogenes (Hain et al., 2006).
However, the genetic equipment itself does not sufficiently describe differences of more closely related strains with respect to their virulence properties. Several proteomic approaches have been performed in the context of listerial adaption to the host environment, thus contributing to the systemic understanding of metabolism during infection (Cabanes et al., 2011). A comparative in vitro proteomic approach investigated the protein expression profiles of L. monocytogenes and L. innocua with a focus on the secretome of both species (Trost et al., 2005). In vitro comparative transcriptome analysis of L. monocytogenes strains revealed differences of the two major lineages/serovar 1/2a, and serovars 4b and 1/2b including stress-related sigma factor B (see below) and virulence factors (Severino et al., 2007). Related studies analyzed the secreted proteomes of L. monocytogenes strains belonging to serovars 4b, 1/2b, and 1/2a (Dumas et al., 2008, 2009a,b). Due to the identification of factors possibly involved in substrate degradation, those studies might reveal novel insight into the listerial metabolism in vivo. Donaldson et al. (2011) focused on proteome differences between an avirulent and two virulent L. monocytogenes strains representing the two lineages mentioned above. Their data revealed that most proteins of the intermediary metabolism are stronger expressed in pathogenic strains in comparison to apathogenic strains. Comparison between the two virulent serovars also revealed metabolic differences in their intramacrophagic proteome, possibly reflecting unequal proliferation rates (Donaldson et al., 2009).
Two pilot studies had investigated the transcriptome of L. monocytogenes infecting human epithelial (Caco-2) cells and the murine macrophage cell line P388D1. Both studies revealed that up to 19% of the listerial genes are differentially expressed in comparison to their level of transcription in BHI medium (Chatterjee et al., 2006; Joseph et al., 2006). Differences in the results point to cell-specific metabolic adaptations during the intracellular replication of L. monocytogenes, whereas common findings support the assumption that several metabolic traits play a central role for listerial replication in vivo (see below). The analysis of the proteome of L. monocytogenes strain EGDe replicating in macrophages demonstrated the upregulation of specific metabolic pathways (van de Velde et al., 2009). An in vitro proteomic study revealed global changes in gene expression when L. monocytogenes enters stationary phase, a growth condition also relevant in vivo (Weeks et al., 2004). These data clearly indicate that specific metabolic adaptations significantly contribute to the capability of pathogens to replicate within macrophages.
The analysis of the bacterial response to changing conditions in vitro also elucidates the dynamic of metabolism in vivo. Wen et al. (2011) investigated the reversible transition from the bacilli-like to the cocci-morphology in the long-term-survival (LTS) phase of L. monocytogenes serotype 4b strain F2365 at different growth stages in tryptic soy broth with yeast extract (TSBYE). Transcriptome analysis identified 225 differentially expressed genes (≥4-fold; P < 0.05) with the upregulation of metabolic genes including those involved in the synthesis of branched-chain amino acids (BCAAs). Combined treatment with potassium lactate and sodium diacetate led to altered metabolism, including a shift toward fermentative production of acetoin (Stasiewicz et al., 2011).
The study of biosynthetic pathways and fluxes under in vivo conditions is crucial to understand the metabolism and physiology of microorganisms on a systems level (Winder et al., 2011). While many important features on metabolism can be deduced on the basis of genome sequences, RNA transcripts, protein and metabolite profiles, and numeric modeling, the direct observation of metabolic pathways and fluxes during listerial growth in eukaryotic host cells has long been hampered by the lack of an adequate technology. One of the recent methods for the quantitative analysis of metabolite fluxes throughout biological systems is based on growing infected host cells in medium containing stable isotope labeled nutrients (Eylert et al., 2008). For heterotrophic organisms including pathogenic bacteria such as L. monocytogenes and their eukaryotic host cells, simple carbohydrates, amino acids, and glycerol are among the typical carbon sources and, therefore, serve as suitable tracers in these labeling studies.
Any perturbation of the natural isotope equilibrium by the supply of such an isotope enriched compound will naturally spread in the experimental system via a large number of enzyme catalyzed reactions. In other words, a broad spectrum of biosynthetic pathways (if not all) is covered by this experimental approach. As a consequence, comprehensive information about the nature and the dynamics of the metabolic network can be obtained. Analytical methods to monitor the distribution of the stable isotope (e.g., 13C) in biosynthetic products are NMR spectroscopy and/or mass spectrometry typically coupled with gas chromatography (GC/MS). Both methods have their specific advantages and disadvantages. Mass spectrometry is a well-established tool to determine isotopolog patterns in amino acids at adequate sensitivity (Zamboni et al., 2009). Signals with high signal-to-noise ratio can be obtained even with small sample amounts, i.e., 108 bacterial cells or 1 mg of dry cell pellet is sufficient for the measurement of protein bound amino acids (Eisenreich et al., 2010).
After protein hydrolysis, the resulting amino acids are converted into tert-butyldimethylsilyl-derivatives (TBDMS amino acids). On the basis of the mass patterns, the relative fractions of isotopomers (i.e., molecular ions or fragments thereof comprising a defined number of 13C-atoms) can be determined for approximately 50 mass fragments of 15 TBDMS amino acids under realistic conditions. However, from these analytes only 29 fragments of 12 different TBDMS amino acids (glycine, alanine, serine, aspartate, threonine, glutamate, valine, leucine, isoleucine, methionine, tyrosine, and phenylalanine) meet the demands for a reliable isotopomer quantification (Antoniewicz et al., 2007). Only 13 fragments comprise all carbon atoms of the original amino acids (i.e., for glycine, alanine, serine, aspartate, threonine, glutamate, proline, methionine, valine, lysine, histidine, phenylalanine, and tyrosine), whereas all other fragments are observed after loss of one or more carbon atoms from the original amino acid. For most TBDMS amino acids, fragments are detected where C-1 (the carboxylic atom) has been lost during the ionization. Provided that the labeling strategy had generated specific profiles in all of these fragments, the biosynthetic pathways leading to the analyzed amino acids can be identified in typical studies. Moreover, the same data can be used to predict the patterns in the precursors for the respective amino acids as a basis to elucidate the fluxes in the central intermediary metabolisms. For accurate data, the overall 13C-enrichment should be at least 0.2%; better results are obtained with enrichments >1%.
Notably, the measurements can be done in a high throughput manner by automated systems, albeit data processing, and interpretation still need considerable efforts by expert users even when supported by the available software to deconvolute isotopolog enrichments from the original data. Considering the isotope fractions in the molecular masses of the original metabolites and one or more fragments thereof, some limited information can be gleaned about the positional distribution of the 13C-label. However, the positional resolution is still low in comparison with isotopolog profiling by NMR spectroscopy that, in turn, is worse in sensitivity (Eisenreich and Bacher, 2007). Nevertheless, the isotopomer patterns in amino acids detected by GC/MS already reveal considerable information about the carbon fluxes in the bacterial cells and their hosts (see below).
When more than 1012 bacterial cells are available (>10 mg of dry cell pellet), one- and two-dimensional NMR spectroscopy can be used in addition to GC/MS. Indeed, high-resolution 13C-NMR spectroscopy is capable to assess 13C-enrichment for each non-equivalent carbon atom of a metabolite under study. Isotopomers carrying one 13C-atom at different positions display completely different 13C-NMR spectra with singlet signals at the chemical shifts for the respective labeled carbon atoms. Notably, this is in sharp contrast to mass spectrometry, where each of these isotopomers gives rise to identical signals due to the identical molecular masses. The same holds true for multiply 13C-labeled isotopologs. Whereas the mass spectra only show the sum of all isotopologs comprising two or more 13C-atoms, respectively, these isotopologs can be better distinguished by NMR spectroscopy. Due to scalar 13C13C couplings, the 13C-NMR signals of a multiply 13C-labeled metabolite appear as specific multiplets in the spectra.
On the basis of the 13C-NMR coupling signatures observed for every single (non-equivalent) carbon atom in a 13C-labeled compound, information on the abundance of 12C and 13C at the respective neighbored carbon atoms is obtained (i.e., in a framework comprising not more than three to four bonds around the index atom). As a result, a set of isotopolog groups can be identified for each 13C-NMR signal. On the basis of the signal intensities (i.e., by deconvolution of the complex coupling patterns providing integral values for each component in the 13C-NMR multiplets), these sets are quantified. For most very small non-symmetrical molecules (i.e., comprising not more than three carbon atoms), all isotopomers/isotopologs display specific NMR signal patterns and can be clearly assigned and quantified on this basis (Eisenreich and Bacher, 2007).
Due to the fact that many of the potential long-range 13C-couplings cannot be completely resolved in the 13C-NMR spectrum of a more complex metabolite (comprising more than three carbon atoms), the observables are typically not sufficient for the direct observation of all individual isotopologs. However, in combination with GC/MS analysis, a sufficient number of constraints can be determined for amino acids to clearly assign molecules with single and multiple labels at adjacent carbon positions. As described below, isotopolog profiling studies have indeed contributed important and novel insights into the listerial metabolism during infection.
A hallmark of L. monocytogenes is its capability to replicate within host cells. Thus, auxotrophic mutants indicate possible nutrient limitations within a host cell. For example, listerial strains defective in the biosynthesis of aromatic compounds as precursors for menaquinone show a strong attenuation in cell culture and mice infection assays, and a more slightly in vivo attenuation was observed for mutants requiring the three aromatic amino acids, threonine, and adenine (Alexander et al., 1993; Marquis et al., 1993; Stritzker et al., 2004). These studies indicate that hosts provide sufficient organic and inorganic compounds to overcome selected auxotrophies. In addition, proteins involved in nucleotide synthesis (PyrD, PyrE, PyrF, and PyrAB) are expressed at a lower level during replication of L. monocytogenes within human THP-1 monocytes or macrophages, indicating a sufficient supply of nucleotides from the host cell (Klarsfeld et al., 1994; van de Velde et al., 2009). The reduced replication rate of a pyrE mutant in macrophages (Schauer et al., 2010) again might point to specificities of host cell lines with respect to their metabolic status.
The cytosol serves as a compartment that protects pathogens from killing by the host’s immune system. Its composition with respect to metabolites is largely unknown, as well as the intracytosolic availability of nutrients. Typically, the cytosol exhibits low concentrations of magnesium, sodium, iron, and calcium ions at neutral pH (reviewed in Ray et al., 2009). In total, up to 100 suitable carbon as well as nitrogen, phosphorus, and sulfur sources are present within eukaryotic cells (Brown et al., 2008). The intracellular milieu is a reducing environment that contains 10 mM reduced glutathione and only 0.5 mM oxidized glutathione (Hwang et al., 1992), a fact important for growth of L. monocytogenes that depends on the availability of reduced nitrogen and sulfur sources (Joseph and Goebel, 2007).
It was assumed that facultative intracellular bacteria have developed a balanced strategy to exploit just enough nutrients from the host cell to maintain survival and proliferation in the intracellular niche (Joseph and Goebel, 2007). A prerequisite for such a strategy, however, is that the intracellular compartment is rich in nutrients, a hypothesis for which the following pros and cons might be stated:
Pros: Strikingly, non-pathogenic bacteria such as Bacillus subtilis or E. coli expressing Hly are able to replicate within the cytosol following phagocytic uptake and escape from the vacuole (Bielecki et al., 1990; Monack and Theriot, 2001). The vacuolar passage, for example by a low pH, might prime the bacterium for intracellular replication since microinjected non-pathogenic L. innocua, E. coli, Salmonella enterica serovar Typhimurium, Yersinia enterocolitica, or B. subtilis failed to proliferate within the cytosol (Goetz et al., 2001; Slaghuis et al., 2004; Hain et al., 2008). Cons: In case cells are a habitat providing easy access to substrates, much more pathogens beside L. monocytogenes, S. flexneri, Burkholderia pseudomallei, and Francisella tularensis would exploit this niche (Ray et al., 2009). Furthermore, all intracellular pathogens exhibit a preference for certain cell types that might depend on differences in the cytosolic nutrient composition. For example, the pathogenic strains EGDe and F2365 could be more metabolically active in J774.1 than in P388D1 macrophages (Chatterjee et al., 2006).
Occupation of the intracellular niche plays a pivotal role for L. monocytogenes virulence, allowing the pathogen to temporarily escape from the immune system of the host and to initiate systemic infection. The host cell cytosol with cell-specific and dynamic nutrient composition is a complex environment that this intracellular pathogen actively adapts to (Eisenreich et al., 2010). Here, we will resume what is known about the listerial mechanisms of substrate acquisition, and the specific metabolic adaptations of L. monocytogenes that allow its successful transition from the extra- to the intracellular milieu (Figure 1).
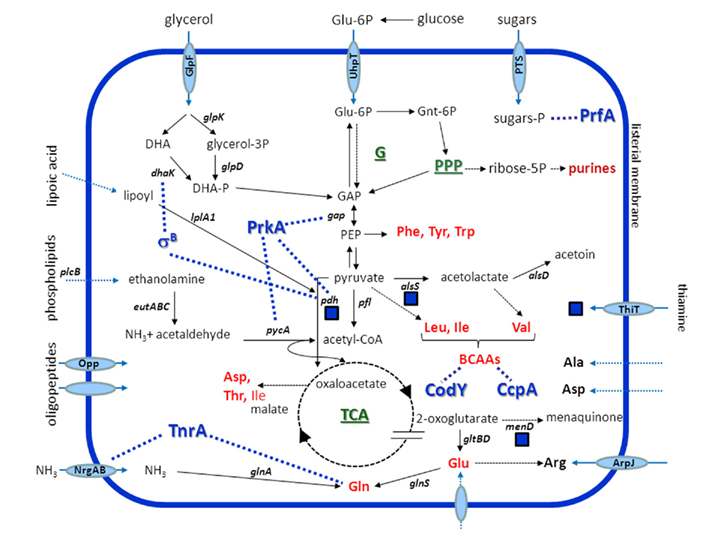
Figure 1. Simplified view of listerial metabolic enzymes, transporters, and pathways relevant during infection. Cofactor thiamine is symbolized by a blue square. Dashed lines indicate putative interactions of master regulators with transporters, enzymes, or metabolites, dashed arrows unknown uptake mechanisms. Blue, shaded genes encode putative interaction partners of PrkA. De novo biosynthesized amino acids are shown in red, complex pathways in green. Abbreviations: G, glycolysis; PEP, phosphoenolpyruvate; P, phosphate. Functions of genes are mentioned in the text.
The intracellular utilization of glucose or glucose-6-phosphate, but also of non-carbohydrate nutrients such as glycerol and amino acids has been demonstrated by IPA studies (Eylert et al., 2008). A multiple mutant unable to take up and catabolize glycerol due to a lack of GlpF, glycerol kinase (GlpK), glycerol-3-phosphate dehydrogenase (GlpD), and DhaK is also attenuated in intracellular growth (own unpublished data). This is in line with the upregulation of these genes within the cytoplasm of host cells (Chatterjee et al., 2006; Joseph et al., 2006) and within the intestine of mice (Toledo-Arana et al., 2009). In vitro, genes encoding glycolysis enzymes and genes involved in the metabolism and the biosynthesis of BCAA are downregulated, and those required for gluconeogenesis are upregulated in the presence of glycerol. All PrfA-dependent genes show a higher level of transcription under these conditions (Joseph et al., 2008). On the other hand, a strain lacking Hpt, a transporter involved in the exploitation of hexose phosphate from the host cell, is attenuated in vivo and within cells, and its gene, uhpT, revealed to be highly upregulated during growth in intracellular strains (Chatterjee et al., 2006; Joseph et al., 2006). In addition, the decreased transcription of hpr encoding the phosphocarrier protein of a phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) during intraepithelial replication reflects the lower amount of Hpr in the absence of glucose (Asanuma and Hino, 2003). These data support the assumption that glucose-6-phosphate rather than glucose is used for bacterial metabolism within hosts (Chico-Calero et al., 2002). This was confirmed by the observation of carbon catabolite derepression during intracellular growth (Joseph et al., 2006), and the finding that a mutant unable to uptake glucose does not show replication attenuation in macrophages or epithelial cells (Stoll and Goebel, 2010). Instead of glycolysis, the oxidative pentose phosphate pathway (PPP) is assumed to be the predominant pathway of sugar metabolism within host cells, since the genes required for glycolysis are down- and those for PPP are upregulated (Chatterjee et al., 2006; Joseph et al., 2006). The non-oxidative branch of PPP is also upregulated in epithelial cells and results in the production of xylulose- and ribose-5-phosphate as precursors of nucleotide biosynthesis (Chatterjee et al., 2006). This was not supported by proteomic analysis that revealed a decreased abundance of two enzymes of this pathway, namely the transaldolase lmo2743 and ribose-5-phosphate epimerase during listerial replication in human THP-1 monocytes (van de Velde et al., 2009). This contradiction might be due to cell-type specific metabolite compositions. Another possible explanation was only recently been provided by a proteomic comparison between two pathogenic strains EGDe and F2365 of L. monocytogenes with the non-pathogenic strain HCC23. The pattern of intermediary metabolism proteins suggests that an initial period of reduced glycolysis during intracellular replication is followed by a resumption of glycolysis (Donaldson et al., 2011).
Glutamine, convertible to glutamate, is the optimal nitrogen source for L. monocytogenes in vitro (Fisher, 1999). Little is known about the in vivo nitrogen metabolism of L. monocytogenes that is assumed to use ammonium, arginine, and/or ethanolamine as nitrogen sources during intracellular replication (Buchrieser et al., 2003; Joseph et al., 2006; Joseph and Goebel, 2007). NrgAB that is activated by TnrA at low concentrations of nitrogen sources is responsible for the uptake of ammonium ions. Gene nrgAB is strongly downregulated within murine macrophages (Chatterjee et al., 2006), but upregulated in human epithelial cells (Joseph et al., 2006). These data again suggest a cell-type dependent availability of nitrogen sources and/or cell-type specific metabolic adaptations of L. monocytogenes.
Ethanolamine lyase EutABC has been demonstrated to be required for wildtype-like intracellular replication of L. monocytogenes (Joseph et al., 2006). Ethanolamine might be derived from phoshatidylethanolamine by the activity of listerial phospholipases. Thus, phospholipids ubiquitous in host environments such as the gut might serve as important nitrogen, carbon, and energy source during infection. This hypothesis is in line with the finding that ethanolamine can be utilized under anaerobic conditions only (Price-Carter et al., 2001; Winter et al., 2010; Srikumar and Fuchs, 2011). The role of host peptides in nitrogen metabolism (see below) remains to be investigated in more detail.
At least six listerial transporters possibly involved in the uptake of amino acids and oligopeptides were recently identified to contribute to intramacrophagic survival and replication (Joseph et al., 2006; Schauer et al., 2010). Induced expression of the oligopeptide transporter lmo0135–0137 associated with cysteine transport has been demonstrated to contribute to proliferation in Caco-2 cells and in vivo (Chatterjee et al., 2006; Schauer et al., 2010). In addition, the oligopeptide permease OppABCDF is not only a prerequisite for intramacrophagic growth, but also for full virulence of L. monocytogenes in mice (Borezee et al., 2000; Port and Freitag, 2007). These findings support the assumption that L. monocytogenes uses intracellular peptides as a source of amino acids (Marquis et al., 1993).
Transcriptomic data showed that genes involved in the biosynthesis of glutamate, glutamine, and arginine belonging to the glutamate family are upregulated during intraepithelial replication of L. monocytogenes (Joseph et al., 2006). Similar findings were made for tryptophan and the BCAAs valine, leucine, and isoleucine. BCAAs are among the most abundant amino acids in proteins, and maintaining their pools is a prerequisite for high-level synthesis of proteins (Sonenshein, 2007). The downregulation of aminoacyl tRNA synthase genes glyS, serS, cysS, alaS, hisS, valS, thrS, ileS, leuS, tyrS, and trpS suggests that the respective amino acids are available for L. monocytogenes within macrophages (Chatterjee et al., 2006). In a pilot study, this hypothesis was at least partially confirmed by 13C-isotopolog profiling experiments, demonstrating that up to 50% of the amino acids alanine, aspartate, and glutamate are recruited from the host cells during intracellular replication of L. monocytogenes (Eylert et al., 2008). Results of the same study indicate a restricted anabolism under these conditions since only 7 of 14 amino acids investigated, namely alanine, aspartate, glutamate, serine, threonine, valine, and glycine ordered by decreased 13C-incorporation, were identified to be de novo biosynthesized (Eylert et al., 2008).
A hallmark of the central metabolism of L. monocytogenes is the incomplete tricarboxylic acid (TCA) cycle due to a lack of 2-oxoglutarate dehydrogenase as demonstrated by genome analysis and 13C-labeling studies (Glaser et al., 2001; Eisenreich et al., 2006). 2-Oxoglutarate as a substrate of glutamate synthase links carbon and nitrogen metabolism. Vice versa, amino acids such as arginine, proline, and histidine can be converted to 2-oxoglutarate.
It could be demonstrated that oxaloacetate, the direct or indirect precursor of aspartate and threonine, is generated by an anaplerotic reaction afforded by pyruvate carboxylation (Eylert et al., 2008). This reaction is catalyzed by the ATP-dependent pyruvate carboxylase PycA (Schär et al., 2010). The critical role for PycA for proliferation of L. monocytogenes in vivo and within epithelial cells or macrophages makes it an interesting candidate for antilisterial compound search (Schindler and Zähner, 1972). It is worth to note that pyruvate is also the precursor for the synthesis of BCAAs that, as demonstrated by several studies cited here, play a pivotal role during intracellular replication of listeriae. Thus, the biosynthesis of BCAAs that is mainly regulated by catabolite control protein A (CcpA) and CodY in an opposite manner also balances the distribution of pyruvate and 2-oxoglutarate (Sonenshein, 2007).
Oxidative decarboxylation of pyruvate results in acetyl-coenzyme A used for the synthesis of valine and leucine, whereas isoleucine is more indirectly derived via pyruvate carboxylation to oxaloacetate as a precursor for aspartate and threonine synthesis. These data suggest a competition of two metabolic pathways important for intracellular replication, namely the biosynthesis of BCAA and the replenishing of the TCA cycle. This metabolic sink might partially be compensated by the increased and decreased expression of several enzymes involved in amino acid metabolism, among them D-alanyl-D-alanine ligase and D-alanine transaminase, an enzyme essential for virulence (Johnson et al., 2004; van de Velde et al., 2009).
The cofactors riboflavin, thiamine, biotin, and lipoate are supplements of defined growth media for L. monocytogenes. The vitamin thiamine is required by enzymes of the central metabolism including Pdh, and the need of its de novo biosynthesis by L. monocytogenes suggests its intracellular limitation (Schauer et al., 2009). Its precursors might be derived from glutamine or from 2-oxoglutarate by overexpressed glutamate dehydrogenase (van de Velde et al., 2009). The same authors also argued that the unexpected induced expression of PurQ and PurM results in the formation of 1-(5′-phosphoribosyl)-5-amino-imidazole that might predominantly be used as a thiamine precursor. Indeed, insertional knockout of purQ attenuated listerial replication inside macrophages (Schauer et al., 2010). Interestingly, Madeo and coworkers recently identified thiamine to play a yet unknown role in listerial tolerance toward low pH as encountered within host compartments. This was explained by the fact that the thiamine-dependent conversion of pyruvate to acetolactate and further to acetoin comprises two proton-consuming steps (Madeo et al., 2012). The first step is catalyzed by the product of alsS induced under acidic conditions (Stasiewicz et al., 2011). The use of host-derived lipoic acid is also relevant for L. monocytogenes infection. Lipoyl modification of Pdh subunit E2 by lipoate protein ligase LplA1 was demonstrated to contribute to intracellular replication and virulence of this pathogen (O’Riordan et al., 2003).
Fundamental shifts in the expression pattern of genes are involved in the adaptation of listerial metabolism to several host compartments. A comprehensive approach was recently performed using tiling arrays to describe the listerial RNome during transition of L. monocytogenes from saprophytism to virulence. To this end, the authors not only studied three physiologically relevant conditions, namely stationary phase, hypoxia, and temperature shifts, but also investigated sigma B (σB)- and PrfA-negative mutants (Toledo-Arana et al., 2009). Two PTS required for mannitol and mannose uptake, and ribose-5-phosphate isomerase as part of the non-oxidative PPP are also upregulated in intestine and stationary phase. A remarkable metabolic adaptation to the intestine and the blood is the increased transcription of the gene cluster involved in ethanolamine and 1,2-propanediol utilization including the cofactor cobalamine (see above), of PTS systems for galactitol, fructose, and cellobiose uptake, and of DhaKs involved in glycerol utilization. Interestingly, the gene clusters lmo0315–lmo0318 responsible for thiamine biosynthesis, and lmo1983–1991 involved in the biosynthesis of BCAAs are also upregulated during listerial replication in blood (Toledo-Arana et al., 2009). This resembles previous findings that these metabolic traits are required for intracellular replication (Joseph et al., 2006). In the host intestinal lumen, the authors observed a σB-mediated activation of genes, whereas in blood, gene transcription was mainly controlled by PrfA.
Listeria monocytogenes also actively replicates within the spleen of infected mice. Following intravenous application of strain EGDe, 30% of all genes differentially regulated in mice spleen were found to be involved in metabolism (Camejo et al., 2009). Upregulated genes include uhpT and, in contrast to epithelial cells and macrophages, genes involved in glycolysis (gap, pgi, fbaA, pgm), whereas those involved in the non-oxidative phase of the PPP appeared to be downregulated. Genes involved in the expression of the Pdh complex including its activator, lipoate protein ligase, were induced in vivo. The upregulation of genes encoding pyruvate–formate lyase indicates an anoxic degradation of pyruvate to formate and acetyl-CoA in the spleen. The upregulation of genes responsible for the biosynthesis of aromatic amino acids (aroA, pheA), of BCAAs and of amino acids of the aspartate and glutamate families is in line with the transcription pattern in epithelial cells and macrophages. An increased expression of mannose-, maltose-, and cellobiose-specific PTS and a decreased expression of PTS responsible for fructose, galactitol, and mannitol uptake was observed. The induction of glnA encoding glutamine synthetase might hint to an inactivation of TnrA within the spleen. Remarkably, listerial genes involved in thiamine synthesis are also upregulated during proliferation in mice spleen.
The human intestine is a yet underinvestigated compartment with respect to its impact on bacterial metabolism. Here, L. monocytogenes not only encounters predominantly anaerobic conditions as a prerequisite for ethanolamine utilization, but also a higher concentration of carbon dioxide known to act as a signal for bacterial metabolism (Neidhardt et al., 1974; Kröger et al., 2011). It has been hypothesized that the biosynthesis of threonine via oxaloacetate and overexpression of threonine synthase is favored by the high concentration of carbon dioxide (van de Velde et al., 2009). Carbon dioxide is also required for oxidative carboxylation of glucose-6-phosphate, the first reaction in the PPP.
Some of the data on the listerial metabolism in cell culture on the one hand and in vivo on the other hand, for example the role of glycolysis, the non-oxidative PPP or the uptake of sugars, are inconsistent. These discrepancies not only indicate to a more active multiplication status in mouse organs compared to cell culture studies using immortal cells (Camejo et al., 2009), but also point to compartment-specific metabolic needs.
During invasive infection, L. monocytogenes not only proliferates within epithelial cells and macrophages, but encounters a frequent change of the surrounding milieu. Therefore, in vivo growth is assumed to require the concerted activity of the following regulators:
Three global regulators of L. monocytogenes, CcpA, TnrA, and CodY sense key metabolites generated in carbon- and nitrogen metabolism (Fisher, 1999; Sonenshein, 2007). CodY, a global transcriptional regulator, seems to play a critical role in the intracellular adaptation of L. monocytogenes. CodY is known to lose its repressing activity when intracellular levels of GTP and BCAAs decrease. As shown by Bennett et al. (2007), the CodY regulon comprises genes involved in amino acid metabolism, nitrogen assimilation, and sugar uptake. Derepression of the CodY operon revealed to be essential for listerial replication in various cell culture models and in mice. The authors therefore hypothesized that CodY senses the metabolic state of the host cell cytoplasm and triggers an adaptive listerial response, which is characterized by the induction of genes involved in the de novo biosynthesis of amino acids such as glutamate, arginine, histidine, tryptophan, and serine, in the intermediary carbon metabolism, in the uptake of sugars or in peptide, and ammonium transport. The induction and requirement of argD encoding N-acetylornithine aminotransferase during intracellular replication simultaneously with the induced expression of the arginine transporter ArpJ might reflect a limited availability of this amino acid under intracellular conditions (Klarsfeld et al., 1994; Joseph et al., 2006). Listerial TnrA, probably encoded by lmo1298, is known as the principal global regulator of nitrogen metabolism in B. subtilis and represses the synthesis of glutamate synthase (Glt) and glutamine synthetase (GlnA). TnrA expression revealed to be downregulated within epithelial cells (Chatterjee et al., 2006), a finding that links to the inactivation of TnrA in the presence of glutamine (Wray et al., 2001). Less is known about the role of the listerial catabolite CcpA. Repression of sugar utilization pathways by glucose is independent of CcpA (Gopal et al., 2010), and CcpA is not involved in carbon source regulation of virulence genes (Behari and Youngman, 1998).
The transition of L. monocytogenes from the saprophytic to the pathogenic lifestyle requires an adequate response to environmental conditions such as low pH, bile stress, carbon starvation, and the presence of reactive oxygen species on the level of transcription and expression (Abram et al., 2008a; Hain et al., 2008; O’Byrne and Karatzas, 2008; Ryan et al., 2008; Soni et al., 2011; Zhang et al., 2011). The regulon controlled by σB plays a major role in this adaptation process. Moreover, σB has been demonstrated to contribute to invasion of epithelial cells and to gastrointestinal infection (Kim et al., 2004; Garner et al., 2006; McGann et al., 2007; Ollinger et al., 2009). Interestingly, the two most highly represented categories of σB-dependent factors were transport and metabolism proteins, among them pyruvate dehydrogenase Pdh, a dihydroxyacetone kinase DhaK, the glycerol transporter GlpF, and a mannose-specific PTS (Kazmierczak et al., 2003). Accordingly, a L. monocytogenes σB mutant lacking σB less efficiently used glycerol as a carbon and energy source (Abram et al., 2008b). An intensive transcriptional reorganization was observed in an in vivo study based on tiling microarrays in which a σB mutant was used for infection (Toledo-Arana et al., 2009).
The main virulence regulator of L. monocytogenes, PrfA, belongs to the family of cAMP receptor protein (Crp)/fumarate nitrate reductase regulators (de las Heras et al., 2011). PrfA not only activates nine key virulence factors including Hpt, but also controls the expression of further 136 factors including those involved in the metabolic activity of this pathogen during infection (Milohanic et al., 2003). The direct or indirect regulatory PrfA function requires an interplay with σB that controls the general stress response of L. monocytogenes (Ollinger et al., 2008, 2009). Interestingly, there is a close link between metabolism and virulence in L. monocytogenes since the carbon sources utilized by the pathogen affect the expression of virulence genes. Especially, glucose, fructose, mannose, or cellobiose transported by PTS downregulated PrfA-dependent genes (Freitag et al., 2009), a finding that points to a low concentration of these sugars within cells. Vice versa, overproduction of PrfA resulted in growth inhibition in glucose-containing media and in an increased de novo biosynthesis of BCAA (Eisenreich et al., 2006; Marr et al., 2006). This sugar-mediated gene repression of PrfA-dependent genes depends rather on inhibition of PrfA than on changes of the PrfA concentration (Renzoni et al., 1997). Taken together, the intracellular availability of carbon sources might act as a key signal for virulence gene expression in L. monocytogenes (de las Heras et al., 2011).
Another mechanism that regulates the metabolic adaptation of pathogens to their hosts is the phosphorylation status of proteins. In L. monocytogenes, a eukaryotic-type serine/threonine-kinase (PrkA, lmo1820) and a serine/threonine-phosphatase (Stp, lmo1821) have been identified that play a role during intracellular replication and in vivo, respectively (Archambaud et al., 2005; Zemansky et al., 2009). In a proteomic approach, 62 proteins were identified as interacting partners of PrkA including 19 involved in carbohydrate and amino acid metabolism (Lima et al., 2011). PrkA directly influences important metabolic enzymes such as Pdh, glyceraldehyde-3-phosphate (GAP) dehydrogenase, pyruvate carboxylase PycA, an aminotransferase involved in BCAA synthesis, and the acetate kinase Acs. The role of universal stress proteins (Usps), which support the adaptation to energy deficiencies, in listerial stress resistance and in virulence has only recently been demonstrated (Seifart Gomes et al., 2011). In E. coli, the activation of uspA is regulated by fructose-6-phosphate (Persson et al., 2007), but a similar mechanism in Gram-positive pathogens remains to be elucidated.
Systems biology approaches of listerial infection use a combination of genome-based bioinformatics tools including sequence and domain analysis, function and structure prediction for proteins, and phylogenetic comparisons. The focus of modeling has been on (intracellular) metabolism, pathogenicity factors, host–pathogen interaction, and the immune response of the host (Schauer et al., 2010; Sauer et al., 2011).
To model listerial metabolism in vitro and within cells, elementary mode analysis was applied (Eisenreich et al., 2006). This method allows to enumerate all possible enzyme combinations or independent pathways within the bacterial cell. Each pathway balances all internal metabolites involved. Furthermore, the metabolic flux models allows to fill in gaps of knowledge from the retrobiosynthetic analysis of the isotopolog compositions, for instance areas where no labeled metabolites were transported by the metabolic flux.
In a more recent study, quantitative literature data on intracellular replication of defined mutants was combined with the replication rates derived from a genome-scale screening for mutants showing reduced intraepithelial survival of L. monocytogenes (Schauer et al., 2010). The application of extreme pathway and elementary mode analysis revealed a critical role of glycerol and purine metabolism, of fucose utilization, and of the synthesis of glutathione, aspartate semialdehyde, serine, and BCAAs during intracellular replication of L. monocytogenes. Thus, new targets for antibiotic intervention became visible by this approach. Furthermore, the modeling demonstrated that degradation of glucose indeed occurs to a large extent via the PPP. To achieve those insights in metabolic traits relevant for listerial infection, numerous all-against-all protein sequence comparisons were performed to establish specific and general protein families.
The number of metabolic pathways to consider in such genome-scale models reaches several thousand and more. Two criteria helped here to zoom in on the key pathways involved: Firstly, only those mutants of L. monocytogenes were considered that showed wildtype-like replication in rich medium, but impaired growth in macrophages.
Secondly, the pathways critical for survival in the macrophage were analyzed to particularly identify enzymes with a role in at least two of these pathways. The list of key enzymes reoccurring in the affected elementary modes by the different affected mutants could then be calculated and ranked. By this procedure, it was possible to quickly identify the most important listerial pathways required for intracellular survival.
Modeling metabolic pathways often results in combinatorial explosion, i.e., an intractable high number of potential flux modes. However, recent advances have the potential to overcome this disadvantage. For instance, flux distributions were decomposed into elementary flux modes in genome-scale metabolic networks of E. coli grown in rich medium containing various carbon sources (Chan and Ji, 2011). Furthermore, software such as EMILiO increases the scope of strain design to include reactions with individually optimized fluxes. Unlike existing approaches experiencing an explosion in complexity to solve this problem, this allowed to generate diverse strain designs regarding production of succinate, L-glutamate, and L-serine (Yang et al., 2011). A third possibility is the application of metabolic flux patterns. These are sets of reactions representing the basic routes through a particular subsystem that are compatible with admissible fluxes in a (possibly) much larger metabolic network (Schuster et al., 2010). The subsystem can be made up by reactions one is interested in, for example production of a certain metabolite. Furthermore, growth, growth boundary, and inactivation models of L. monocytogenes have extensively been developed in food research. Validated approaches exploited available experimental data and combined all three aspects using on different food sources as a medium (Coroller et al., 2012). Stochastic-based examples are multiplicative heteroscedastic models taking into account differing growth variance for heterogeneous populations (Cao et al., 2010), or the dependence of successful growth on inoculum and cell counts (Vermeulen et al., 2009).
Each of the approaches reviewed above has obvious limitations. Interfacing the transcriptome analysis with a mutant library screening underscored that the up- or downregulation of a gene or an operon not necessarily coincides with a phenotype under the same conditions (Joseph et al., 2006), a general drawback of these approaches. On the other hand, a proteomic analysis does not reveal the whole proteome under given conditions. Mutant analysis and IPA, on the other hand, focus on the role of single genes and metabolic traits, thus providing parts of the metabolic puzzle only. Contradictory results derived from different approaches posed the relevance of single metabolic pathways of L. monocytogenes for infection in question. Examples are the overexpression of glutamate dehydrogenase during listerial replication in human THP-1 monocytes in contrast to its decreased expression in strain F2365 (van de Velde et al., 2009; Donaldson et al., 2011), or the induction or repression of the non-oxidative branch of the PPP (see above). These contradictions not only point to host cell-specific or temporary metabolic adaptations of L. monocytogenes, but also to the need of more standardized experimental conditions in omic-approaches.
Another prerequisite for systems biology of listerial metabolism is to quantify and qualify the metabolic state of the bacterial and the host cells during different stages of infection (Zamboni and Sauer, 2009). So far, data sets on metabolites have been derived only from in vitro approaches under non-physiological conditions. For example, L. monocytogenes 10403S cells grown at 37 and 8°C were analyzed (Singh et al., 2011). At low temperature, an increase of solute concentrations such as amino acids, sugars, organic acids, urea cycle intermediates, polyamines, and different compatible solutes was observed in the listerial cytoplasm, thus lowering the freezing point of intracellular water and decreasing ice crystal formation. A statistical analysis (PCA) was instrumental to reveal this system response. Mathematical modeling can consider such effects by adjusting metabolite concentrations and by thermodynamic considerations. Furthermore, compartment models are soon expected to improve such studies (Cheng et al., 2008) including pools and fluxes of the mammalian cell (Lopes et al., 2010). Future research should also consider microcompartments and organelles that both are used to optimize metabolic processes. Compartmentalization is common also in L. monocytogenes including carboxysomes responsible for cobalamine-dependent 1,2-propanediol degradation, thus contributing to functional diversity.
To reveal an even more comprehensive picture of listerial metabolism during infection, cells such as primary macrophages and fibroblasts should be considered for future cell culture experiments. It is also of interest to determine whether and to what extent the host metabolism is modulated during infection, for instance by L. monocytogenes secreting c-di-AMP (Woodward et al., 2010). In addition to in vivo studies using mice and guinea pigs (Cabanes et al., 2008), small animals such as Caenorhabditis elegans or Galleria mellonella might be established as new model organisms. C. elegans is a facile and proven model host for the study of microbial pathogenesis and metabolism (Sifri et al., 2005; Thomsen et al., 2006; Srikumar and Fuchs, 2011). Because both host and pathogen are easily amenable to genetic manipulations, the effect of metabolic perturbations upon infection, changing culture conditions, or gene knockouts can be investigated in this infection model. Notably, the model also reduces costs and ethical constraints in comparison to infection experiments with higher animals. A further advantage of the model is that metabolic fluxes from cells or tissues of an infected host animal to L. monocytogenes and vice versa might be followed, an approach that has long been hampered by the lack of technology to identify and quantify metabolism throughout an appropriate biological system (Spanier et al., 2009). G. mellonella, the greater wax moth, is another relatively simple, non-mammalian host model system that can be used to assess not only the virulence of listerial strains, but also the contribution of metabolic capacities to larvae killing as exemplified by the attenuated phenotype of an EGDe ΔuhpT mutant (Mukherjee et al., 2010). A major disadvantage in contrast to nematodes, however, is the need of subcutaneous instead of oral application of the pathogen.
Listeria monocytogenes is highly adapted to the cytoplasm of mammalian host cells where it is able to multiply with a generation time comparable to that in rich medium. A comprehensive analysis of its metabolism is a prerequisite for a systemic understanding of L. monocytogenes infection. Taking into account all omic-studies and mutant data, certain metabolic enzymes and pathways of L. monocytogenes revealed to have a critical role during infection and might serve as new targets for the development of antilisterial compounds (Figure 1). Metabolic clues for intracellular and in vivo replication are the degradation of phosphorylated glucose via the PPP, the utilization of C3-bodies as alternative carbon- and energy source, pyruvate as a sink for central metabolism including downstream reactions such as pyruvate decarboxylation, biosynthesis of BCAAs, the availability of thiamine, and the acquisition of host cell derived nitrogen sources including ethanolamine. The regulation of these metabolic capabilities involves the activity of factors such as PrfA and σB, resulting in a fine-tuned balance of metabolic flows with the host cell and in vivo. For a deeper understanding especially of host–pathogen interdependencies with respect to metabolism, new technologies such as isotopolog profiling, additional animal models including invertebrates, and novel systems biological approaches have to be combined in future research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Work in the authors’ laboratories is supported by grants of the German Research Foundation (DFG), including the priority program SPP1316 (DA 208/13-1, EI-384/6, and FU-375/5).
σB, sigma factor; BBCAAs, branched-chain amino acids; GAP, glyceraldehyde-3-phosphate; Hly, listeriolysin; IPA, isotopolog profiling analysis; Pdh, pyruvate dehydrogenase; PlcA/PlcB, phospholipase A/B; PPP, pentose phosphate pathway; PrfA, positive regulatory factor A.
Abram, F., Starr, E., Karatzas, K. A., Matlawska-Wasowska, K., Boyd, A., Wiedmann, M., Boor, K. J., Connally, D., and O’Byrne, C. P. (2008a). Identification of components of the sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl. Environ. Microbiol. 74, 6848–6858.
Abram, F., Su, W.-L., Wiedmann, M., Boor, K. J., Coote, P., Botting, C., Karatzas, K. A. G., and O’Byrne, C. P. (2008b). Proteomic analyses of a Listeria monocytogenes mutant lacking sigma B identify new components of the sigma B regulon and highlight a role for sigma B in the utilization of glycerol. Appl. Environ. Microbiol. 74, 594–604.
Alexander, J. E., Andrew, P. W., Jones, D., and Roberts, I. S. (1993). Characterization of an aromatic amino acid-dependent Listeria monocytogenes mutant: attenuation, persistence, and ability to induce protective immunity in mice. Infect. Immun. 61, 2245–2248.
Antoniewicz, M. R., Kelleher, J. K., and Stephanopoulos, G. (2007). Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal. Chem. 79, 7554–7559.
Archambaud, C., Gouin, E., Pizarro-Cerda, J., Cossart, P., and Dussurget, O. (2005). Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Mol. Microbiol. 56, 383–396.
Asanuma, N., and Hino, T. (2003). Molecular characterization of HPr and related enzymes, and regulation of HPr phosphorylation in the ruminal bacterium Streptococcus bovis. Arch. Microbiol. 179, 205–213.
Becker, D., Selbach, M., Rollenhagen, C., Ballmaier, M., Meyer, T. F., Mann, M., and Bumann, D. (2006). Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440, 303–307.
Behari, J., and Youngman, P. (1998). A homolog of CcpA mediates catabolite control in Listeria monocytogenes but not carbon source regulation of virulence genes. J. Bacteriol. 180, 6316–6324.
Bennett, H. J., Pearce, D. M., Glenn, S., Taylor, C. M., Kuhn, M., Sonenshein, A. L., Andrew, P. W., and Roberts, I. S. (2007). Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63, 1453–1467.
Bielecki, J., Youngman, P., Connelly, P., and Portnoy, D. A. (1990). Bacillus subtilis expressing a haemolysin gene from Listeria monocytogenes can grow in mammalian cells. Nature 345, 175–176.
Boigegrain, R.-A., Liautard, J.-P., and Köhler, S. (2005). Targeting of the virulence factor acetohydroxyacid synthase by sulfonylureas results in inhibition of intramacrophagic multiplication of Brucella suis. Antimicrob. Agents Chemother. 49, 3922–3925.
Borezee, E., Pellegrini, E., and Berche, P. (2000). OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68, 7069–7077.
Brown, S. A., Palmer, K. L., and Whiteley, M. (2008). Revisiting the host as a growth medium. Nat. Rev. Microbiol. 6, 657–666.
Buchrieser, C., Rusniok, C., Kunst, F., Cossart, P., and Glaser, P. (2003). Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol. Med. Microbiol. 35, 207–213.
Cabanes, D., Lecuit, M., and Cossart, P. (2008). Animal models of Listeria infection. Curr. Protoc. Microbiol. 9, 9B.1.1–9B.1.17.
Cabanes, D., Sousa, S., and Cossart, P. (2011). “Listeria genomics,” in Genomics of Foodborne Bacterial Pathogens, eds M. Wiedmann and W. Zhang (New York: Springer), 141–70.
Camejo, A., Buchrieser, C., Couvé, E., Carvalho, F., Reis, O., Ferreira, P., Sousa, S., Cossart, P., and Cabanes, D. (2009). In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 5, e1000449. doi:10.1371/journal.ppat.1000449
Camejo, A., Carvalho, F., Reis, O., Leitao, E., Sousa, S., and Cabanes, D. (2011). The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2, 379–394.
Cao, R., Francisco-Fernández, M., and Quinto, E. J. (2010). A random effect multiplicative heteroscedastic model for bacterial growth. BMC Bioinformatics 11, 77. doi:10.1186/1471-2105-11-77
Centers for Disease Control and Prevention. (2011). Statistics of Listeriosis. Atlanta. Available at: http://www.cdc.gov/listeria/statistics.html
Chan, S. H. J., and Ji, P. (2011). Decomposing flux distributions into elementary flux modes in genome-scale metabolic networks. Bioinformatics 27, 2256–2262.
Chatterjee, S. S., Hossain, H., Otten, S., Kuenne, C., Kuchmina, K., Machata, S., Domann, E., Chakraborty, T., and Hain, T. (2006). Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74, 1323–1338.
Cheng, S., Liu, Y., Crowley, C. S., Yeates, T. O., and Bobik, T. A. (2008). Bacterial microcompartments: their properties and paradoxes. Bioessays 30, 1084–1095.
Chico-Calero, I., Suárez, M., González-Zorn, B., Scortti, M., Slaghuis, J., Goebel, W., and Vázquez-Boland, J. A. (2002). Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc. Natl. Acad. Sci. U.S.A. 99, 431–436.
Coroller, L., Kan-King-Yu, D., Leguerinel, I., Mafart, P., and Membré, J.-M. (2012). Modelling of growth, growth/no-growth interface and nonthermal inactivation areas of Listeria in foods. Int. J. Food Microbiol. 152, 139–152.
Cossart, P., and Toledo-Arana, A. (2008). Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect. 10, 1041–1050.
de las Heras, A., Cain, R. J., Bielecka, M. K., and Vazquez-Boland, J. A. (2011). Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 14, 118–127.
Donaldson, J. R., Nanduri, B., Burgess, S. C., and Lawrence, M. L. (2009). Comparative proteomic analysis of Listeria monocytogenes strains F2365 and EGD. Appl. Environ. Microbiol. 75, 366–373.
Donaldson, J. R., Nanduri, B., Pittman, J. R., Givaruangsawat, S., Burgess, S. C., and Lawrence, M. L. (2011). Proteomic expression profiles of virulent and avirulent strains of Listeria monocytogenes isolated from macrophages. J. Proteomics 74, 1906–1917.
Dumas, E., Desvaux, M., Chambon, C., and Hébraud, M. (2009a). Insight into the core and variant exoproteomes of Listeria monocytogenes species by comparative subproteomic analysis. Proteomics 9, 3136–3155.
Dumas, E., Meunier, B., Berdagué, J.-L., Chambon, C., Desvaux, M., and Hébraud, M. (2009b). The origin of Listeria monocytogenes 4b isolates is signified by subproteomic profiling. Biochim. Biophys. Acta 1794, 1530–1536.
Dumas, E., Meunier, B., Berdagué, J.-L., Chambon, C., Desvaux, M., and Hébraud, M. (2008). Comparative analysis of extracellular and intracellular proteomes of Listeria monocytogenes strains reveals a correlation between protein expression and serovar. Appl. Environ. Microbiol. 74, 7399–7409.
Dussurget, O., Pizarro-Cerda, J., and Cossart, P. (2004). Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 58, 587–610.
Eisenreich, W., and Bacher, A. (2007). Advances of high-resolution NMR techniques in the structural and metabolic analysis of plant biochemistry. Phytochemistry 68, 2799–2815.
Eisenreich, W., Dandekar, T., Heesemann, J., and Goebel, W. (2010). Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8, 401–412.
Eisenreich, W., Slaghuis, J., Laupitz, R., Bussemer, J., Stritzker, J., Schwarz, C., Schwarz, R., Dandekar, T., Goebel, W., and Bacher, A. (2006). 13C isotopologue perturbation studies of Listeria monocytogenes carbon metabolism and its modulation by the virulence regulator PrfA. Proc. Natl. Acad. Sci. U.S.A. 103, 2040–2045.
Eylert, E., Schär, J., Mertins, S., Stoll, R., Bacher, A., Goebel, W., and Eisenreich, W. (2008). Carbon metabolism of Listeria monocytogenes growing inside macrophages. Mol. Microbiol. 69, 1008–1017.
Fisher, S. H. (1999). Regulation of nitrogen metabolism in Bacillus subtilis: vive la différence! Mol. Microbiol. 32, 223–232.
Freitag, N. E., Port, G. C., and Miner, M. D. (2009). Listeria monocytogenes – from saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 7, 623–628.
Fuchs, T. M., Eisenreich, W., Heesemann, J., and Goebel, W. (2011). Metabolic adaptation of human pathogenic and related nonpathogenic bacteria to extra- and intracellular habitats. FEMS Microbiol. Rev. doi: 10.1111/j.1574-6976.2011.00301.x. [Epub ahead of print].
Garner, M. R., Njaa, B. L., Wiedmann, M., and Boor, K. J. (2006). Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect. Immun. 74, 876–886.
Glaser, P., Frangeul, L., Buchrieser, C., Rusniok, C., Amend, A., Baquero, F., Berche, P., Bloecker, H., Brandt, P., Chakraborty, T., Charbit, A., Chetouani, F., Couvé, E., de Daruvar, A., Dehoux, P., Domann, E., Domínguez-Bernal, G., Duchaud, E., Durant, L., Dussurget, O., Entian, K. D., Fsihi, H., García-del Portillo, F., Garrido, P., Gautier, L., Goebel, W., Gómez-López, N., Hain, T., Hauf, J., Jackson, D., Jones, L. M., Kaerst, U., Kreft, J., Kuhn, M., Kunst, F., Kurapkat, G., Madueno, E., Maitournam, A., Vicente, J. M., Ng, E., Nedjari, H., Nordsiek, G., Novella, S., de Pablos, B., Pérez-Diaz, J. C., Purcell, R., Remmel, B., Rose, M., Schlueter, T., Simoes, N., Tierrez, A., Vázquez-Boland, J. A., Voss, H., Wehland, J., and Cossart, P. (2001). Comparative genomics of Listeria species. Science 294, 849–852.
Goetz, M., Bubert, A., Wang, G., Chico-Calero, I., Vazquez-Boland, J. A., Beck, M., Slaghuis, J., Szalay, A. A., and Goebel, W. (2001). Microinjection and growth of bacteria in the cytosol of mammalian host cells. Proc. Natl. Acad. Sci. U.S.A. 98, 12221–12226.
Gopal, S., Berg, D., Hagen, N., Schriefer, E.-M., Stoll, R., Goebel, W., and Kreft, J. (2010). Maltose and maltodextrin utilization by Listeria monocytogenes depend on an inducible ABC transporter which is repressed by glucose. PLoS ONE 5, e10349. doi:10.1371/journal.pone.0010349
Götz, A., Eylert, E., Eisenreich, W., and Goebel, W. (2010). Carbon metabolism of enterobacterial human pathogens growing in epithelial colorectal adenocarcinoma (Caco-2) cells. PLoS ONE 5, e10586. doi:10.1371/journal.pone.0010586
Götz, A., and Goebel, W. (2010). Glucose and glucose 6-phosphate as carbon sources in extra- and intracellular growth of enteroinvasive Escherichia coli and Salmonella enterica. Microbiology 156, 1176–1187.
Hain, T., Hossain, H., Chatterjee, S. S., Machata, S., Volk, U., Wagner, S., Brors, B., Haas, S., Kuenne, C. T., Billion, A., Otten, S., Pane-Farre, J., Engelmann, S., and Chakraborty, T. (2008). Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e σB regulon. BMC Microbiol. 8, 20. doi:10.1186/1471-2180-8-20
Hain, T., Steinweg, C., Kuenne, C. T., Billion, A., Ghai, R., Chatterjee, S. S., Domann, E., Kärst, U., Goesmann, A., Bekel, T., Bartels, D., Kaiser, O., Meyer, F., Pühler, A., Weisshaar, B., Wehland, J., Liang, C., Dandekar, T., Lampidis, R., Kreft, J., Goebel, W., and Chakraborty, T. (2006). Whole-genome sequence of Listeria welshimeri reveals common steps in genome reduction with Listeria innocua as compared to Listeria monocytogenes. J. Bacteriol. 188, 7405–7415.
Hamon, M., Bierne, H., and Cossart, P. (2006). Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 4, 423–434.
Hwang, C., Sinskey, A. J., and Lodish, H. F. (1992). Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257, 1496–1502.
Johnson, J., Jinneman, K., Stelma, G., Smith, B. G., Lye, D., Messer, J., Ulaszek, J., Evsen, L., Gendel, S., Bennett, R. W., Swaminathan, B., Pruckler, J., Steigerwalt, A., Kathariou, S., Yildirim, S., Volokhov, D., Rasooly, A., Chizhikov, V., Wiedmann, M., Fortes, E., Duvall, R. E., and Hitchins, A. D. (2004). Natural atypical Listeria innocua strains with Listeria monocytogenes pathogenicity island 1 genes. Appl. Environ. Microbiol. 70, 4256–4266.
Joseph, B., and Goebel, W. (2007). Life of Listeria monocytogenes in the host cells’ cytosol. Microbes Infect. 9, 1188–1195.
Joseph, B., Mertins, S., Stoll, R., Schar, J., Umesha, K. R., Luo, Q., Muller-Altrock, S., and Goebel, W. (2008). Glycerol metabolism and PrfA activity in Listeria monocytogenes. J. Bacteriol. 190, 5412–5430.
Joseph, B., Przybilla, K., Stühler, C., Schauer, K., Slaghuis, J., Fuchs, T. M., and Goebel, W. (2006). Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188, 556–568.
Kazmierczak, M. J., Mithoe, S. C., Boor, K. J., and Wiedmann, M. (2003). Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 185, 5722–5734.
Kim, H., Boor, K. J., and Marquis, H. (2004). Listeria monocytogenes sigma B contributes to invasion of human intestinal epithelial cells. Infect. Immun. 72, 7374–7378.
Klarsfeld, A. D., Goossens, P. L., and Cossart, P. (1994). Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE and an arginine ABC transporter gene, arpJ. Mol. Microbiol. 13, 585–597.
Kröger, C., Srikumar, S., Ellwart, J., and Fuchs, T. M. (2011). Bistability in myo-inositol utilization by Salmonella enterica serovar Typhimurium. J. Bacteriol. 193, 1427–1435.
Liautard, J.-P., Jubier-Maurin, V., Boigegrain, R.-A., and Köhler, S. (2006). Antimicrobials: targeting virulence genes necessary for intracellular multiplication. Trends Microbiol. 14, 109–113.
Lima, A., Duran, R., Schujman, G. E., Marchissio, M. J., Portela, M. M., Obal, G., Pritsch, O., de, M. D., and Cervenansky, C. (2011). Serine/threonine protein kinase PrkA of the human pathogen Listeria monocytogenes: biochemical characterization and identification of interacting partners through proteomic approaches. J. Proteomics 74, 1720–1734.
Lopes, T. J. S., Luganskaja, T., Vujic Spasic, M., Hentze, M. W., Muckenthaler, M. U., Schümann, K., and Reich, J. G. (2010). Systems analysis of iron metabolism: the network of iron pools and fluxes. BMC Syst. Biol. 4, 112. doi:10.1186/1752-0509-4-112
Lucchini, S., Liu, H., Jin, Q., Hinton, J. C. D., and Yu, J. (2005). Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect. Immun. 73, 88–102.
Madeo, M., O’Riordan, N., Fuchs, T. M., Utratna, M., Karatzas, K. A. G., and O’Byrne, C. P. (2012). Thiamine plays a critical role in the acid tolerance of Listeria monocytogenes. FEMS Microbiol. Lett. 326, 137–143.
Marquis, H., Bouwer, H. G., Hinrichs, D. J., and Portnoy, D. A. (1993). Intracytoplasmic growth and virulence of Listeria monocytogenes auxotrophic mutants. Infect. Immun. 61, 3756–3760.
Marr, A. K., Joseph, B., Mertins, S., Ecke, R., Müller-Altrock, S., and Goebel, W. (2006). Overexpression of PrfA leads to growth inhibition of Listeria monocytogenes in glucose-containing culture media by interfering with glucose uptake. J. Bacteriol. 188, 3887–3901.
McGann, P., Wiedmann, M., and Boor, K. J. (2007). The alternative sigma factor sigma B and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins. Appl. Environ. Microbiol. 73, 2919–2930.
Milohanic, E., Glaser, P., Coppée, J.-Y., Frangeul, L., Vega, Y., Vázquez-Boland, J. A., Kunst, F., Cossart, P., and Buchrieser, C. (2003). Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47, 1613–1625.
Monack, D. M., and Theriot, J. A. (2001). Actin-based motility is sufficient for bacterial membrane protrusion formation and host cell uptake. Cell. Microbiol. 3, 633–647.
Mukherjee, K., Altincicek, B., Hain, T., Domann, E., Vilcinskas, A., and Chakraborty, T. (2010). Galleria mellonella as a model system for studying Listeria pathogenesis. Appl. Environ. Microbiol. 76, 310–317.
Muñoz-Elías, E. J., and McKinney, J. D. (2006). Carbon metabolism of intracellular bacteria. Cell. Microbiol. 8, 10–22.
Neidhardt, F. C., Bloch, P. L., and Smith, D. F. (1974). Culture medium for enterobacteria. J. Bacteriol. 119, 736–747.
O’Byrne, C. P., and Karatzas, K. A. G. (2008). The role of sigma B (sigma B) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence. Adv. Appl. Microbiol. 65, 115–140.
Ollinger, J., Bowen, B., Wiedmann, M., Boor, K. J., and Bergholz, T. M. (2009). Listeria monocytogenes sigma B modulates PrfA-mediated virulence factor expression. Infect. Immun. 77, 2113–2124.
Ollinger, J., Wiedmann, M., and Boor, K. J. (2008). Sigma B- and PrfA-dependent transcription of genes previously classified as putative constituents of the Listeria monocytogenes PrfA regulon. Foodborne Pathog. Dis. 5, 281–293.
O’Riordan, M., Moors, M. A., and Portnoy, D. A. (2003). Listeria intracellular growth and virulence require host-derived lipoic acid. Science 302, 462–464.
Persson, O., Valadi, A., Nyström, T., and Farewell, A. (2007). Metabolic control of the Escherichia coli universal stress protein response through fructose-6-phosphate. Mol. Microbiol. 65, 968–978.
Port, G. C., and Freitag, N. E. (2007). Identification of novel Listeria monocytogenes secreted virulence factors following mutational activation of the central virulence regulator, PrfA. Infect. Immun. 75, 5886–5897.
Price-Carter, M., Tingey, J., Bobik, T. A., and Roth, J. R. (2001). The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183, 2463–2475.
Ray, K., Marteyn, B., Sansonetti, P. J., and Tang, C. M. (2009). Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat. Rev. Microbiol. 7, 333–340.
Renzoni, A., Klarsfeld, A., Dramsi, S., and Cossart, P. (1997). Evidence that PrfA, the pleiotropic activator of virulence genes in Listeria monocytogenes, can be present but inactive. Infect. Immun. 65, 1515–1518.
Rohmer, L., Hocquet, D., and Miller, S. I. (2011). Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19, 341–348.
Ryan, S., Hill, C., and Gahan, C. G. M. (2008). Acid stress responses in Listeria monocytogenes. Adv. Appl. Microbiol. 65, 67–91.
Sauer, J.-D., Pereyre, S., Archer, K. A., Burke, T. P., Hanson, B., Lauer, P., and Portnoy, D. A. (2011). Listeria monocytogenes engineered to activate the Nlrc4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proc. Natl. Acad. Sci. U.S.A. 108, 12419–12424.
Schär, J., Stoll, R., Schauer, K., Loeffler, D. I. M., Eylert, E., Joseph, B., Eisenreich, W., Fuchs, T. M., and Goebel, W. (2010). Pyruvate carboxylase plays a crucial role in carbon metabolism of extra- and intracellularly replicating Listeria monocytogenes. J. Bacteriol. 192, 1774–1784.
Schauer, K., Geginat, G., Liang, C., Goebel, W., Dandekar, T., and Fuchs, T. M. (2010). Deciphering the intracellular metabolism of Listeria monocytogenes by mutant screening and modelling. BMC Genomics 11, 573. doi:10.1186/1471-2164-11-573
Schauer, K., Stolz, J., Scherer, S., and Fuchs, T. M. (2009). Both thiamine uptake and biosynthesis of thiamine precursors are required for intracellular replication of Listeria monocytogenes. J. Bacteriol. 191, 2218–2227.
Schindler, P. W., and Zähner, H. (1972). Stoffwechselprodukte von Mikroorganismen. Arch. Mikrobiol. 82, 66–75.
Schuster, S., de Figueiredo, L. F., and Kaleta, C. (2010). Predicting novel pathways in genome-scale metabolic networks. Biochem. Soc. Trans. 38, 1202–1205.
Seifart Gomes, C., Izar, B., Pazan, F., Mohamed, W., Mraheil, M. A., Mukherjee, K., Billion, A., Aharonowitz, Y., Chakraborty, T., and Hain, T. (2011). Universal stress proteins are important for oxidative and acid stress resistance and growth of Listeria monocytogenes EGD-e in vitro and in vivo. PLoS ONE 6, e24965. doi:10.1371/journal.pone.0024965
Severino, P., Dussurget, O., Vêncio, R. Z. N., Dumas, E., Garrido, P., Padilla, G., Piveteau, P., Lemaître, J.-P., Kunst, F., Glaser, P., and Buchrieser, C. (2007). Comparative transcriptome analysis of Listeria monocytogenes strains of the two major lineages reveals differences in virulence, cell wall, and stress response. Appl. Environ. Microbiol. 73, 6078–6088.
Sifri, C. D., Begun, J., and Ausubel, F. M. (2005). The worm has turned - microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 13, 119–127.
Singh, A. K., Ulanov, A. V., Li, Z., Jayaswal, R. K., and Wilkinson, B. J. (2011). Metabolomes of the psychrotolerant bacterium Listeria monocytogenes 10403S grown at 37°C and 8°C. Int. J. Food Microbiol. 148, 107–114.
Slaghuis, J., Goetz, M., Engelbrecht, F., and Goebel, W. (2004). Inefficient replication of Listeria innocua in the cytosol of mammalian cells. J. Infect. Dis. 189, 393–401.
Sonenshein, A. L. (2007). Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5, 917–927.
Soni, K. A., Nannapaneni, R., and Tasara, T. (2011). The contribution of transcriptomic and proteomic analysis in elucidating stress adaptation responses of Listeria monocytogenes. Foodborne Pathog. Dis. 8, 843–852.
Spanier, B., Lasch, K., Marsch, S., Benner, J., Liao, W., Hu, H., Kienberger, H., Eisenreich, W., and Daniel, H. (2009). How the intestinal peptide transporter PEPT-1 contributes to an obesity phenotype in Caenorhabditis elegans. PLoS ONE 4, e6279. doi:10.1371/journal.pone.0006279
Srikumar, S., and Fuchs, T. M. (2011). Ethanolamine utilization contributes to proliferation of Salmonella enterica serovar Typhimurium in food and in nematodes. Appl. Environ. Microbiol. 77, 281–290.
Stasiewicz, M. J., Wiedmann, M., and Bergholz, T. M. (2011). The transcriptional response of Listeria monocytogenes during adaptation to growth on lactate and diacetate includes synergistic changes that increase fermentative acetoin production. Appl. Environ. Microbiol. 77, 5294–5306.
Stoll, R., and Goebel, W. (2010). The major PEP-phosphotransferase systems (PTSs) for glucose, mannose and cellobiose of Listeria monocytogenes, and their significance for extra- and intracellular growth. Microbiology 156, 1069–1083.
Stritzker, J., Janda, J., Schoen, C., Taupp, M., Pilgrim, S., Gentschev, I., Schreier, P., Geginat, G., and Goebel, W. (2004). Growth, virulence, and immunogenicity of Listeria monocytogenes aro mutants. Infect. Immun. 72, 5622–5629.
Thomsen, L. E., Slutz, S. S., Tan, M.-W., and Ingmer, H. (2006). Caenorhabditis elegans is a model host for Listeria monocytogenes. Appl. Environ. Microbiol. 72, 1700–1701.
Toledo-Arana, A., Dussurget, O., Nikitas, G., Sesto, N., Guet-Revillet, H., Balestrino, D., Loh, E., Gripenland, J., Tiensuu, T., Vaitkevicius, K., Barthelemy, M., Vergassola, M., Nahori, M.-A., Soubigou, G., Régnault, B., Coppée, J.-Y., Lecuit, M., Johansson, J., and Cossart, P. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956.
Trost, M., Wehmhöner, D., Kärst, U., Dieterich, G., Wehland, J., and Jänsch, L. (2005). Comparative proteome analysis of secretory proteins from pathogenic and nonpathogenic Listeria species. Proteomics 5, 1544–1557.
van de Velde, S., Delaive, E., Dieu, M., Carryn, S., van Bambeke, F., Devreese, B., Raes, M., and Tulkens, P. M. (2009). Isolation and 2-D-DIGE proteomic analysis of intracellular and extracellular forms of Listeria monocytogenes. Proteomics 9, 5484–5496.
Vázquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., Domínguez-Bernal, G., Goebel, W., González-Zorn, B., Wehland, J., and Kreft, J. (2001). Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640.
Vermeulen, A., Gysemans, K. P. M., Bernaerts, K., Geeraerd, A. H., Debevere, J., Devlieghere, F., and van Impe, J. F. (2009). Modelling the influence of the inoculation level on the growth/no growth interface of Listeria monocytogenes as a function of pH, aw and acetic acid. Int. J. Food Microbiol. 135, 83–89.
Weeks, M. E., James, D. C., Robinson, G. K., and Smales, C. M. (2004). Global changes in gene expression observed at the transition from growth to stationary phase in Listeria monocytogenes ScottA batch culture. Proteomics 4, 123–135.
Wen, J., Deng, X., Li, Z., Dudley, E. G., Anantheswaran, R. C., Knabel, S. J., and Zhang, W. (2011). Transcriptomic response of Listeria monocytogenes during the transition to the long-term-survival phase. Appl. Environ. Microbiol. 77, 5966–5972.
Winder, C. L., Dunn, W. B., and Goodacre, R. (2011). TARDIS-based microbial metabolomics: time and relative differences in systems. Trends Microbiol. 19, 315–322.
Winter, S. E., Thiennimitr, P., Winter, M. G., Butler, B. P., Huseby, D. L., Crawford, R. W., Russell, J. M., Bevins, C. L., Adams, L. G., Tsolis, R. M., Roth, J. R., and Bäumler, A. J. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429.
Woodward, J. J., Iavarone, A. T., and Portnoy, D. A. (2010). c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705.
Wray, L. V., Zalieckas, J. M., and Fisher, S. H. (2001). Bacillus subtilis glutamine synthetase controls gene expression through a protein-protein interaction with transcription factor TnrA. Cell 107, 427–435.
Yang, L., Cluett, W. R., and Mahadevan, R. (2011). EMILiO: a fast algorithm for genome-scale strain design. Metab. Eng. 13, 272–281.
Zamboni, N., Fendt, S.-M., Rühl, M., and Sauer, U. (2009). (13)C-based metabolic flux analysis. Nat. Protoc. 4, 878–892.
Zamboni, N., and Sauer, U. (2009). Novel biological insights through metabolomics and 13C-flux analysis. Curr. Opin. Microbiol. 12, 553–558.
Zemansky, J., Kline, B. C., Woodward, J. J., Leber, J. H., Marquis, H., and Portnoy, D. A. (2009). Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J. Bacteriol. 191, 3950–3964.
Zhang, Q., Feng, Y., Deng, L., Feng, F., Wang, L., Zhou, Q., and Luo, Q. (2011). SigB plays a major role in Listeria monocytogenes tolerance to bile stress. Int. J. Food Microbiol. 145, 238–243.
Keywords: Listeria monocytogenes, infection, metabolism, systems biology, modeling, intracellular
Citation: Fuchs TM, Eisenreich W, Kern T and Dandekar T (2012) Toward a systemic understanding of Listeria monocytogenes metabolism during infection. Front. Microbio. 3:23. doi: 10.3389/fmicb.2012.00023
Received: 12 December 2011;
Paper pending published: 30 December 2011;
Accepted: 13 January 2012;
Published online: 03 February 2012.
Edited by:
Reinhard Guthke, Leibniz-Institute for Natural Product Research and Infection Biology – Hans-Knoell-Institute, GermanyCopyright: © 2012 Fuchs, Eisenreich, Kern and Dandekar. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Thilo M. Fuchs, Abteilung Mikrobiologie, Zentralinstitut für Ernährungs- und Lebensmittelforschung, Technische Universität München, Weihenstephaner Berg 3, 85350 Freising, Germany. e-mail:dGhpbG8uZnVjaHNAd3p3LnR1bS5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.