- Department of Viral Infections, Research Institute for Microbial Diseases, Osaka University, Suita, Osaka, Japan
Human immunodeficiency virus type 1 (HIV-1) infects humans and chimpanzees but not old world monkeys (OWMs) such as the rhesus monkey (Rh) and cynomolgus monkey (CM). HIV-1 efficiently enters cells of OWMs but encounters a block before reverse transcription. This narrow host range is attributed to a barrier in the host cell. In 2004, the screening of a Rh cDNA library identified tripartite motif 5α (TRIM5α) as a cellular antiviral factor. TRIM5α is one of splicing variants produced by TRIM5 gene and TRIM5 proteins are members of the TRIM family containing RING, B-box 2, and coiled-coil domains. The RING domain is frequently found in E3 ubiquitin ligase and TRIM5α is degraded via the ubiquitin–proteasome-dependent pathway. Among TRIM5 splicing variants, TRIM5α alone has an additional C-terminal PRYSPRY (B30.2) domain. Previous studies have shown that sequence variation in variable regions of the PRYSPRY domain among different monkey species affects species-specific retrovirus infection, while amino acid sequence differences in the viral capsid protein determine viral sensitivity to restriction. TRIM5α recognizes the multimerized capsid proteins (viral core) of an incoming virus by its PRYSPRY domain and is thus believed to control retroviral infection. There are significant intraspecies variations in the Rh-TRIM5 gene. It has also been reported that some Rh and CM individuals have retrotransposed cyclophilin A open reading frame in the TRIM5 gene, which produces TRIM5–cyclophilin A fusion protein (TRIMCyp). TRIMCyp, which was originally identified as an anti-HIV-1 factor of New World owl monkeys, is an interesting example of the gain of a new function by retrotransposition. As different TRIM5 genotypes of Rh showed different levels of simian immunodeficiency virus replication in vivo, the TRIM5 genotyping is thought to be important in acquired immunodeficiency syndrome monkey models.
Introduction
Human immunodeficiency virus type 1 (HIV-1) is a causative agent of acquired immunodeficiency syndrome (AIDS). More than two million people are infected with HIV-1 annually around the world. Nevertheless, the host range of HIV-1 is extremely narrow, being limited to humans and chimpanzees (Gao et al., 1999). This narrow host range has hampered the establishment improved animal models of HIV-1 infection that are needed to facilitate the development of an efficacious vaccine against HIV-1 infection. In this review, we summarize current understanding regarding the species barrier of HIV-1 as discussed from the viewpoint of animal model development, focusing on tripartite motif 5α (TRIM5α), a restriction factor in monkeys.
Life Cycle of HIV-1
Human immunodeficiency virus type 1 belongs to the family Retroviridae, subfamily Lentivirus. It is an enveloped virus with a single-stranded RNA genome with positive polarity. HIV-1 enters CD4+ T cells and macrophages through plasma membrane fusion. The virus RNA genome is subsequently reverse transcribed by viral-associated reverse transcriptase and resultant double-strand cDNA is transported to the nucleus. In the nucleus, viral-associated integrase (IN) inserts viral cDNA into the human chromosome. The transcription is enhanced by cellular activation and mRNA and full-length viral genome RNA are exported from the nucleus. Viral proteins assemble beneath the plasma membrane and virus particles bud from plasma membrane (Figure 1).
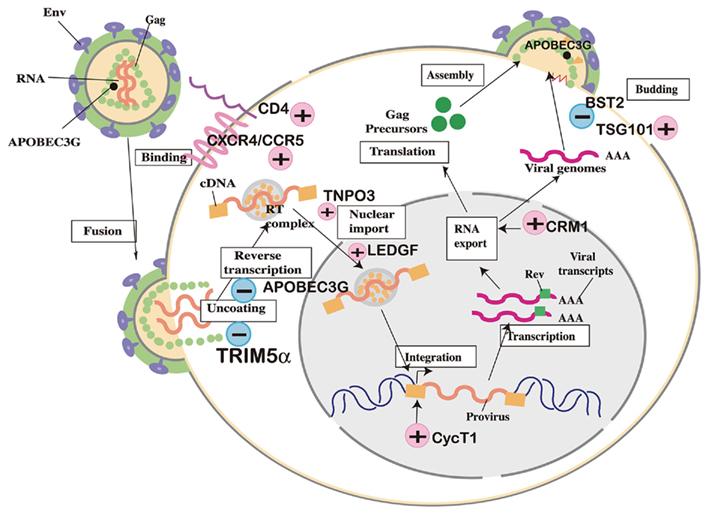
Figure 1. Cellular factors involved in human immunodeficiency virus (HIV) replication cycle. (+) Positive factors required for viral replication. (−) Negative factors that suppress viral replication.
Host Factors Required for HIV-1 Replication in Human and Species-Specific Barrier of HIV-1 in Mice
Many trials of small animal models for HIV-1 infection have failed due to lack of host factors in mice, which are necessary for efficient virus replication (Figure 1). CD4, the cellular receptor for HIV-1, was first identified as a host range barrier because mouse CD4 does not bind to HIV-1 envelope protein (Landau et al., 1988). Human CD4 transgenic mice, however, were not susceptible to HIV-1 infection (Lores et al., 1992). Chemokine receptors were identified as entry co-receptors (Alkhatib et al., 1996; Bleul et al., 1996; Choe et al., 1996; Deng et al., 1996; Doranz et al., 1996; Dragic et al., 1996; Feng et al., 1996), but mice transgenic for human CD4 and either human CXCR4 (Sawada et al., 1998) or CCR5 (Browning et al., 1997) failed to show productive infection, even though murine CXCR4 is a functional co-receptor for CXCR4-tropic HIV-1 (Bieniasz et al., 1997).
Human immunodeficiency virus type 1 pre-integration complex (PIC) containing viral cDNA and viral IN are translocated into the nucleus. Two host cellular proteins have recently been reported to mediate PIC import. The first is a lens epithelium-derived growth factor (LEDGF/p75; Cherepanov et al., 2003; Maertens et al., 2003), a protein implicated in the regulation of gene expression and cellular stress responses. LEDGF interacts with HIV-1 IN and is thought to guide PIC toward sites of active transcription for integration of viral cDNA into the human chromosome. The second is Transportin 3 (TNPO3/Transportin-SR2) identified by two independent screenings of host factors involved in HIV-1 replication (Brass et al., 2008; Christ et al., 2008). TNPO3 also binds to IN, but it is also thought to associate with viral capsid (CA) protein and supports nuclear translocation of PICs. It is currently unclear whether mouse orthologs of either or both of these factors are defective in nuclear transport and integration of HIV-1.
The integrated HIV-1 genome is then transcribed from its promoter in the long terminal repeat (LTR) by using NF-κB and Sp1 (Jones et al., 1986; Staal et al., 1990; Cullen, 1991). HIV-1 non-structural proteins, Tat, Rev, and Nef, are early gene products produced from multiply spliced mRNAs (Feinberg et al., 1986). Tat binds to the 5′ region of nascent HIV-1 transcripts and facilitates the elongation of transcribed RNA (Laspia et al., 1989; Feinberg et al., 1991). Mouse cells do not show Tat-dependent transcriptional activation of HIV-1. Cyclin T1 (CycT1) is responsible for this transcriptional barrier in mice (Newstein et al., 1990; Garber and Jones, 1999). CycT1 protein is a component of the CDK9/pTEFb transcription factor complex (Mancebo et al., 1997; Wei et al., 1998). Human but not mouse CycT1 binds to Tat and activates transcription from HIV-1 LTR (Garber et al., 1998). Nevertheless, triple-transduction of mouse cells with human CD4, CXCR4, and CycT1 was insufficient to induce productive viral infection (Bieniasz and Cullen, 2000). Additional barriers have been reported in the late stages of the viral life cycle (Mariani et al., 2000; Keppler et al., 2001; Koito et al., 2003a,b; Nagai-Fukataki et al., 2011). CRM1, a nuclear export factor that functions in association with Rev, has been suggested to be one of the late-phase factors important for the export of unspliced full-length viral genome from the nucleus (Zheng et al., 2003). Further studies are necessary to identify host cellular factors that are necessary for virus replication in humans but defective in mouse cells in order to establish small animal models of HIV-1 infection.
TRIM5α, One of the Species-Specific Barriers to HIV-1 in Monkeys
Amino acid sequences of CD4, CXCR4, CCR5, CycT1, and CRM1 in Old World monkeys (OWMs) are almost identical to those of the human orthologs, while New World monkeys, such as common marmosets and squirrel monkeys, have less functional CD4 and CCR5 receptors (LaBonte et al., 2002). Nevertheless, HIV-1 fails to replicate in activated CD4+ T lymphocytes obtained from OWM, such as the rhesus monkey (Rh; Macaca mulatta; Shibata et al., 1995; Himathongkham and Luciw, 1996) and cynomolgus monkey (CM; Macaca fascicularis; Akari et al., 1996, 1999). Several studies have suggested that the blockade of HIV-1 replication in OWM cells occurs at a post-entry step (Shibata et al., 1995; Himathongkham and Luciw, 1996; Chackerian et al., 1997) and appears to result from a failure to initiate reverse transcription (Himathongkham and Luciw, 1996). Importantly, resistance against HIV-1 infection was shown to be dominant in heterokaryons between human and OWM cells, suggesting the presence of inhibitory factor(s) against HIV-1 infection but not for simian immunodeficiency virus (SIV) in OWM cells (Munk et al., 2002).
In 2004, the screening of an Rh cDNA library identified TRIM5α as a factor that confers resistance to HIV-1 infection (Stremlau et al., 2004; Figures 1 and 2). Both Rh and CM TRIM5α restrict HIV-1 infection but fail to restrict SIV isolated from a macaque monkey (SIVmac; Stremlau et al., 2004; Nakayama and Shioda, 2010). In contrast, human TRIM5α is almost powerless to restrict the aforementioned viruses, but potently restricts N-tropic murine leukemia viruses (N-MLV) and equine infectious anemia virus (EIAV; Hatziioannou et al., 2004; Figure 3).
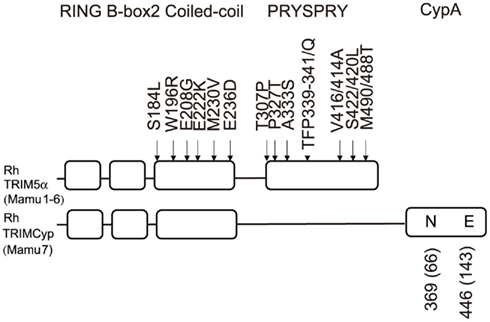
Figure 2. Domains of rhesus monkey (Rh) TRIM5α and TRIMCyp proteins. The RING, B-box2, Coiled-coil, PRYSPRY, or CypA domains of Rh-TRIM5α and TRIMCyp are shown by squares. Polymorphisms are shown outside the squares. The numbers in parentheses show the amino acid positions counting from the initiation methionine codon of the CypA open reading frame.
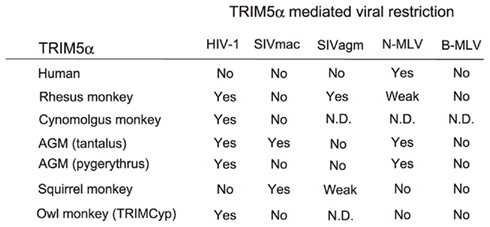
Figure 3. Species-specific restriction by TRIM5α. “Yes” denotes restriction. “Weak” denotes weak restriction. “No” denotes no restriction. “N. D.” denotes no result has yet been published. SIVmac, simian immunodeficiency virus isolated from a macaque (Ohkura et al., 2006). SIVagm, simian immunodeficiency virus isolated from an African green monkey (Song et al., 2005b). N-MLV, N-tropic murine leukemia virus (Ohkura et al., 2006); B-MLV, B-tropic murine leukemia virus (Ohkura et al., 2006). AGM, African green monkey (Nakayama et al., 2005; Kim et al., 2011). Rhesus monkey (Stremlau et al., 2004; Ylinen et al., 2005; Ohkura et al., 2006), cynomolgus monkey (Nakayama et al., 2005), and owl monkey TRIMCyp (Nisole et al., 2004; Sayah et al., 2004) are also included.
Tripartite Motif of TRIM5α
TRIM5α is a member of the TRIM family of proteins, and consists of RING, B-box 2, coiled-coil, and PRYSPRY (B30.2) domains (Reymond et al., 2001; Figure 2). Proteins with the RING domains possess E3 ubiquitin ligase activity (Jackson et al., 2000); therefore, TRIM5α was thought to restrict HIV-1 by proteasome-dependent pathways. However, a proteasome inhibitor MG132 did not rescue HIV-1 infection from TRIM5α-mediated restriction, even though the levels of HIV-1 late reverse-transcription products were recovered (Anderson et al., 2006; Wu et al., 2006; Maegawa et al., 2010). TRIM5α is thus thought to use both proteasome-dependent and -independent pathways to restrict HIV-1. The distinct molecular mechanism of the proteasome-independent pathway has yet to be elucidated. It was shown that incubation of an artificially constructed HIV-1 core structure composed of the capsid–nucleocapsid (CA–NC) fusion protein with the chimeric protein containing the Rh-TRIM5α B-box 2, coiled-coil, and PRYSPRY domains and the RING domain of TRIM21 (TRIM5-21R) caused apparent breaks in the CA structure without any other cellular components (Langelier et al., 2008; Zhao et al., 2011). It is therefore likely that direct binding of Rh-TRIM5α proteins to incoming HIV-1 CA proteins causes CA disassembly, which is observed as proteasome-independent restriction.
The intact B-box 2 domain is also required for TRIM5α-mediated antiviral activity, as TRIM5α restrictive activity is diminished by several amino acid substitutions in the B-box 2 domain (Javanbakht et al., 2005). TRIM5α has been shown to form a dimer (Kar et al., 2008; Langelier et al., 2008), while the B-box 2 domain mediates higher-order self-association of Rh-TRIM5α oligomers (Li and Sodroski, 2008; Diaz-Griffero et al., 2009). The coiled-coil domain of TRIM5α is important for the formation of homo-oligomers (Mische et al., 2005), and the homo-oligomerization of TRIM5α is essential for antiviral activity (Javanbakht et al., 2006; Nakayama et al., 2006).
PRYSPRY Domain of TRIM5α, A Determinant of Species-Specific Restriction of Viruses
The PRYSPRY domain is specific for the α-isoform among at least three splicing variants transcribed from the TRIM5 gene. Soon after the identification of TRIM5α as a restriction factor of Rh, several studies found that differences in the amino acid sequences of the variable region 1 (V1) of TRIM5α PRYSPRY domain of different monkey species affect the species-specific restriction of retrovirus infection (Nakayama et al., 2005; Perez-Caballero et al., 2005; Sawyer et al., 2005; Stremlau et al., 2005; Yap et al., 2005; Ohkura et al., 2006; Perron et al., 2006; Kono et al., 2008, 2009). The PPYSPRY domain is thought to recognize viral cores, as TRIM5α lacking this domain does not show antiviral activity. Overexpression of truncated TRIM5α lacking the PRYSPRY domain shows a dominant negative effect on antiviral activity of full-length TRIM5α (Berthoux et al., 2005; Nakayama et al., 2006). Biochemical studies have shown that TRIM5α associates with CA in detergent-stripped N-MLV virions (Sebastian and Luban, 2005) or with an artificially constituted HIV-1 core structure composed of the CA–NC fusion protein in a PRYSPRY domain-dependent manner (Stremlau et al., 2006). Although the precise three-dimensional crystal structure of the PRYSPRY domain has not been resolved, TRIM5-21R assembled and formed two-dimensional paracrystalline hexagonal arrays in vitro (Ganser-Pornillos et al., 2011). This assembly required RING and B-box 2 domains but was independent of the PRYSPRY domain. However, the hexagonal lattices of HIV-1 CA that mimic the surface of core act as template for stabilization of TRIM5-21R arrays in a PRYSPRY-dependent manner (Ganser-Pornillos et al., 2011). As the interaction between individual CA monomers and TRIM5α is very weak, CA recognition by TRIM5α is thought to be a synergistic combination of direct binding interactions with the PRYSPRY domain, higher-order assembly of TRIM5α, template-based assembly, and lattice complementarity.
Variable Susceptibility of Simian Immunodeficiency Viruses Among Monkey Species
Simian immunodeficiency virus isolated from sooty mangabey (SIVsm) and SIV isolated from African green monkey (SIVagm) replicate in their natural hosts (VandeWoude and Apetrei, 2006) and CD4+ human cells. SIVmac evolved from SIVsm in captive macaques, and replicates efficiently in Rh (Shibata et al., 1995; Himathongkham and Luciw, 1996) and CM (Akari et al., 1996, 1999) as well as in human CD4+ cells but not in African green monkey (AGM) cells. We found that a 37-amino acid residue region including a 20-amino acid duplication in the V1 of AGM TRIM5α determined species-specific restriction against SIVmac239 (Nakayama et al., 2005). However, AGM TRIM5α failed to restrict SIVagm, which naturally infects AGM, while Rh-TRIM5α can restrict SIVagm infection (Song et al., 2005b; Figure 3).
In contrast to HIV-1, AGM TRIM5α restricted SIVmac239 mainly in a proteasome-dependent manner, as SIVmac239 escaped completely from attacks by RING mutants of TRIM5α that could still moderately restrict HIV-1 infection. Kim et al. reported that AGM TRIM5α derived from Chlorocebus tantalus but not Chlorocebus pygerythrus subspecies of AGM restrict SIVmac239, while both potently restrict HIV-1 (Figure 3). Both AGM TRIM5α share the 20-amino acid duplication but a C. pygerythrus-specific leucine residue at the 34th position within the RING domain compromised the ability of C. pygerythrus AGM TRIM5α to restrict SIVmac239 infection (Kim et al., 2011). This result is consistent with the observation of RING-proteasome dependency of SIVmac239 restriction by TRIM5α.
Human immunodeficiency virus type 2 (HIV-2) is assumed to have originated from SIVsm as a result of zoonotic events involving monkeys and humans (Hahn et al., 2000). Previous studies have shown that HIV-2 strains vary widely in their ability to grow in cells of OWM, such as baboons, Rh, and CM (Castro et al., 1990, 1991; Locher et al., 1998, 2003; Fujita et al., 2003). By testing CM and Rh recombinant TRIM5α, three amino acid residues of TFP at the 339th to 341st positions of Rh-TRIM5α V1 were shown to be indispensable for restricting particular HIV-2 strains that are still resistant to CM TRIM5α bearing a single Q instead of TFP at the 339th to 341st positions (Kono et al., 2008; Figure 4). The TFP motif is also critical to restrict SIVsm (Kirmaier et al., 2010). Baboon and sooty mangabey (SM) TRIM5α bearing SFP at the 339th to 341st positions can potently restrict HIV-1, only weakly restrict HIV-2, and failed to restrict SIVmac239 (Newman et al., 2006; Kono et al., 2008, 2009).

Figure 4. HIV-2/SIV capsid sequence variations and restriction patterns of rhesus (Rh) and cynomolgus monkey (CM) TRIM5α/TRIMCyp alleles. “Yes” denotes restriction. “Weak” denotes weak restriction. “No” denotes no restriction. “N. D.” denotes no result has yet been published. The unique QQ sequence at the 89th–90th positions of SIVmac, which is critical for escape from Rh TRIMCyp, RhCypA (Kirmaier et al., 2010), is shown in red. Arginine 97 at the base of the loop between helices 4 and 5, which is important to escape from TFP alleles of Rh-TRIM5α, RhTFP (Kirmaier et al., 2010), is shown in blue. The glutamine and alanine residues at position 120 of GH123 or analogous positions of other HIV-2 strains, which is critical for resistance against Q alleles of Rh-TRIM5α, RhQ (Kirmaier et al., 2010) and CM TRM5α (Song et al., 2007; Kono et al., 2008), are shown in green. CMCypA(NE) and CMCypA(DK) denote the minor and major alleles of CM TRIMCyp, respectively.
Viral Determinant of Sensitivity to Monkey TRIM5α
Tripartite motif 5α is thought to recognize viral cores through its PRYSPRY domain. To determine the region in viral CA that interacts with TRIM5α, we focused on HIV-2, which closely resembles SIVmac (Hahn et al., 2000). Sequence analysis showed that the CM TRIM5α-sensitive viruses had proline (P) at the 119th or 120th position of CA, while the CM TRIM5α-resistant viruses had alanine (A), glutamine (Q), or glycine (G) at the same position (Figure 4). Replacing the proline of a CM TRIM5α-sensitive HIV-2 molecular clone with A, Q, or G changed the phenotype from sensitive to resistant and the mutant viruses replicated well in the presence of CM TRIM5α. The reverse was observed when the glutamine of a resistant SIVmac molecular clone was replaced with proline (Song et al., 2007; Miyamoto et al., 2011). The 119th or 120th position is located in a loop between α-helices 6 and 7 (L6/7; Figure 5).
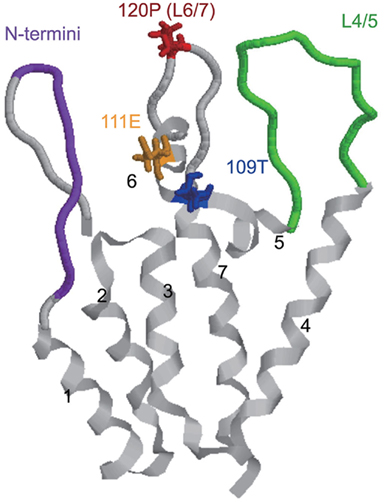
Figure 5. Structure of the N-terminal half of HIV-2 capsid monomer. The ribbons represent the backbone of HIV-2 capsid proteins, and seven α-helices are labeled. The positions important in Rh-TRIM5α recognition are highlighted as N-termini (the 5th to 13th amino acid residues) in purple, the loop between α-helices 4 and 5 (L4/5) in green, the 109th T in blue, the 111th E in orange, and the 120th P in red.
In the case of Rh-TRIM5α, Ylinen et al. replaced a loop between α-helices 4 and 5 (L4/5) of SIVmac239 CA with that of HIV-2 in the SIVmac239 background and found that the resultant mutant virus showed impaired growth ability in Rh cells compared with the parental SIVmac239. However, the reciprocal virus with SIVmac239 CA L4/5 in the HIV-2 background did not gain resistance against Rh-TRIM5α, suggesting that Rh-TRIM5α interacts mainly with L4/5 but other portion(s) of HIV-2 CA are also involved (Ylinen et al., 2005). Lin and Emerman (2008) also reported that SIVagm with HIV-1 L4/5 and L6/7 was susceptible to Rh-TRIM5α restriction. In fact, we found that the 120th amino acid of HIV-2 CA, the determinant of CM TRIM5α sensitivity, also contributes to Rh-TRIM5α susceptibility (Kono et al., 2010). Furthermore, studies on chimeric viruses between Rh-TRIM5α-sensitive HIV-2 and Rh-TRIM5α-resistant SIVmac239 revealed that multiple regions including L4/5 in the N-terminal half of SIVmac239 CA contribute to evasion of SIVmac239 from Rh-TRIM5α (Kono et al., 2010; Figure 5).
To elucidate further details regarding the structure of CA recognized by TRIM5α, we generated mutant HIV-2 viruses each carrying 1 of 20 amino acid residues at position 120, and examined their susceptibilities to CM TRIM5α-mediated restriction. Amino acid residues with hydrophobic side chains or aromatic rings were associated with sensitivity to CM TRIM5α, while those with small side chains or amide groups conferred resistance (Miyamoto et al., 2011). Computer-assisted three-dimensional models showed that the mutations at the 120th position in L6/7 affected the conformation of the neighboring loop L4/5 by a hydrogen bond between aspartic acid 97 in L4/5 and arginine 119 in L6/7 (Figure 6).
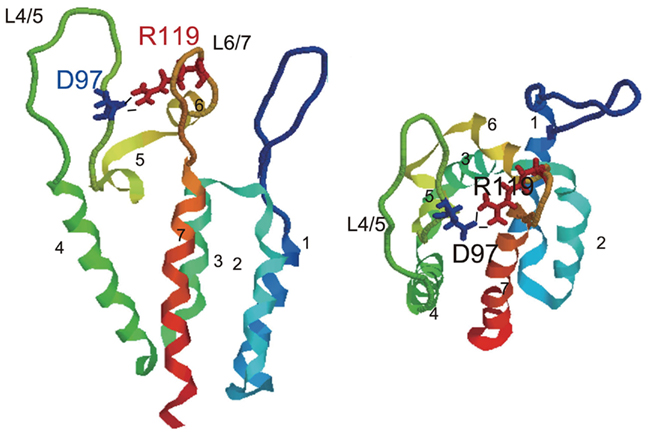
Figure 6. The hydrogen bond between two external loops of HIV-2 capsid. The structures of the N-terminal domain of GH123 are shown, and seven color-coded α-helices are labeled. Blue and red wireframes denote side chains of glutamic acid at the 97th (D97) and arginine at the 119th (R119) positions, respectively. Black lines indicate hydrogen bonds between D97 and R119. Models are shown from two different angles.
Taken together, these observations suggested that TRIM5α recognized the overall outer surface of the N-terminal half of viral CA including L4/5 and L6/7. To determine further details regarding the interaction between CA and TRIM5α, biochemical and structural analyses of the PRYSPRY domain, especially the V1 loop bound with CA, are required. In contrast to SIV/HIV-2, the L4/5 loop of HIV-1 also binds cyclophilin A (CypA). It is not yet clear whether monkey TRIM5α does or does not recognize HIV-1 CA with endogenous CypA.
Intraspecies Genetic Variation of the Rh-TRIM5 Gene
The TRIM5 gene varies considerably among primate species (Sawyer et al., 2005; Song et al., 2005a; Newman et al., 2006). It is not surprising that the PRYSPRY domain is highly variable as TRIM5α interacts with the retroviral core through this region, as described above, and the main pressure for positive selection may be endogenous retroviruses (Kaiser et al., 2007). Interestingly, there is a 339TFP341-to-Q polymorphism in Rh-TRIM5α (Newman et al., 2006; Figure 2), which reduces its anti-HIV-2 activity (Kono et al., 2008). Newman et al. (2006) grouped Rh-TRIM5α into six alleles (Mamu-1 to -6) including rare alleles Mamu-2 and Mamu-6. Wilson et al. (2008a) showed that Mamu-1 and -3 alleles restrict HIV-1, HIV-2, EIAV, and feline immunodeficiency virus (FIV), but not N-MLV, B-MLV, or SIVmac239, while Mamu-4 and -5 alleles restrict HIV-1, EIAV, and FIV but not HIV-2, N-MLV, B-MLV, or SIVmac239 using a TRIM5α-transduced cat cell line (CRFK).
Lim et al. independently reported 11 Rh-TRIM5α alleles in which alleles 1–5 contained 339TFP341. Remaining alleles 6–11 contained 339Q. They established B-lymphoblastoid cell lines (B-LCLs) from Rh and used these B-LCLs for infection with VSV-G pseudo-typed GFP-expressing viruses. They found more GFP-positive cells in B-LCLs with Rh-TRIM5α Q allele than in B-LCLs with Rh TFP allele infected with SIVmac239-, HIV-1-, and SIVsmE543-based GFP-expressing viruses. It should be noted that the anti-HIV-1 activity of the Rh-TRIM5α Q allele is significantly stronger than the anti-SIVmac239 and SIVsmE543 activities of the Rh-TRIM5α TFP allele (Lim et al., 2010b). Lim et al. (2010a,b) retrospectively analyzed plasma viral load of Rh after SIVmac251 challenge by intravenous route and found that Rh with the Q allele was associated with higher levels of plasma viral RNA at the time when the levels of viral RNA stabilized after a period of acute infection (0.6 log median difference at 70 days after infection), more rapid loss of central memory CD4+ T cells, and higher rate of progression to AIDS. These results were consistent with their own in vitro observations described above.
On the other hand, Wilson et al. (2008a) failed to detect anti-SIVmac239 activity of both Rh-TRIM5α Q and TFP alleles. Similarly, Kirmaier et al. (2010) detected virtually no anti-SIVmac239 activity in both Rh-TRIM5α TFP and Q alleles (Figure 4), although numbers of infected cells in Mamu-4 (Rh-TRIM5α Q allele) are slightly higher than those in Mamu-1 (Rh-TRIM5α TFP allele). In contrast, Kirmaier et al. (2010) reported that the Rh-TRIM5α TFP allele restricted SIVsmE543 and SIVsmE041, although the Rh-TRIM5α Q allele did not show any anti-SIVsmE543 or anti-SIVsmE041 activity. SIVmac239 is a molecular clone of a highly adapted, emergent virus of Rh, generated in the 1980s by experimental passage of SIV-positive plasma through several monkeys (Daniel et al., 1985). In contrast, SIVsmE041 is a primary isolate from SM and SIVsmE543 was cloned after experimental passage of SIVsm through two Rh (Hirsch et al., 1997). SIVmac and SIVsm shared Q at the 118th position of CA, corresponding to the 120th position of GH123 (HIV-2), but SIVmac239 and SIVmac251 have an R-to-S change at position 97 at the base of L4/5 of CA that are critical for resistance against Rh-TRIM5α TFP allele (Figure 4).
In the case of SIVsmE543 in vivo, Rh-TRIM5αTFP/TFP homozygotes markedly diminished viral replication compared to Rh-TRIM5αQ/Q homozygotes at peak (2 log reduction) and 8 weeks (3 log reduction) after intravenous or intrarectal infection, consistent with the in vitro results (Kirmaier et al., 2010). It should be noted that the suppression of SIVsmE543 by Rh-TRIM5α TFP is more dramatic than that of SIVmac251. In low-dose repeated mucosal challenge experiments, two groups reported similar results using SIVsmE660, the CA sequence of which closely resembles that of SIVsmE543 (Reynolds et al., 2011; Yeh et al., 2011). Several studies evaluated MHC class I and TRIM5 genotypes in SIV-infected Rh, and concluded that TRIM5 genotype independently affected plasma viral load and survival rate after SIV infection (Lim et al., 2010a; Reynolds et al., 2011; Yeh et al., 2011). Taken together, these observations indicate that it is necessary to perform TRIM5 genotyping of Rh when using SIVsm. It is also better to do so when using SIVmac239 and SIVmac251, although Fenizia et al. (2011) recently reported that there was no difference in SIVmac251 susceptibility among Rh with different TRIM5 genotypes in repeated rectal challenge.
TRIM5 and CypA Fusion Protein in New World Monkey
Cells of the NWM, owl monkey (Aotus trivirgatus), are resistant to HIV-1 infection. Treatment of owl monkey cells with cyclosporin A, an inhibitor of CypA, allowed HIV-1 infection (Towers et al., 2003). In 2004, soon after the discovery of TRIM5α, analysis of the owl monkey TRIM5 gene identified a long interspersed nuclear element (LINE)-1-mediated retrotransposition of CypA between exons 7 and 8, resulting in expression of a fusion protein designated as TRIMCyp (Nisole et al., 2004; Sayah et al., 2004). Owl monkey TRIMCyp contained the N-terminal half of TRIM5α, RING, B-box 2, and coiled-coil, but the PRYSPRY domain was replaced with CypA. As the CypA domain of owl monkey TRIMCyp binds to L4/5 of HIV-1 CA, owl monkey TRIMCyp showed similar antiviral activity to TRIM5α (Figure 3). The interaction between HIV-1 CA and CypA can be inhibited by cyclosporine A. This is a very interesting example of a gain-of-function by retrotransposition. The owl monkey has been shown to express only TRIMCyp, and not TRIM5α.
TRIMCyp in OWMs
The expression of TRIMCyp was thought to be an anomaly unique to owl monkeys, but in 2008 another CypA insertion was found in several species of OWMs belonging to the Genus Macaca, Rh, CM, and the pig-tailed monkey (PM; Macaca nemestrina; Brennan et al., 2008; Newman et al., 2008; Virgen et al., 2008; Wilson et al., 2008b). It is reasonable to assume that the retrotransposition event occurred in the common ancestor of these three macaques. Insertion of the CypA gene was at the 3′ end of the TRIM5 gene, which is different from the owl monkey, indicating that CypA retrotransposition into the TRIM5 gene in OWMs occurred independently from that in owl monkeys. A G-to-T transversion linked with CypA insertion altering the canonical splicing acceptor of TRIM5 exon 7 caused alternative splicing (Brennan et al., 2008). The resultant mRNA lacks exons 7 and 8, and the PRYSPRY domain is replaced with CypA. In PM, TRIM5α mRNA is absent. Instead, TRIM5 isoforms TRIM5θ and TRIM5η were detected. These isoforms are splicing variants of the TRIMCyp (Brennan et al., 2008). TRIM5θ is truncated at the N-terminus of the PRYSPRY domain and TRIM5η lacks nine amino acid residues encoded by exon 7 (Brennan et al., 2007). PM TRIMCyp restricted HIV-2 but not HIV-1 infection (Liao et al., 2007; Brennan et al., 2008).
In Rh, the allele frequency of TRIMCyp (Mamu-7) was 25% in an Indian population but TRIMCyp was completely absent from a Chinese population (Wilson et al., 2008b). Rh TRIMCyp restricted HIV-2 but not HIV-1 infection (Wilson et al., 2008b). In CM, Brennan et al. (2008) initially reported that the amino acid residue at position 357 of CM TRIMCyp, corresponding to position 54 counting from the methionine of CypA, was arginine (R), and CM TRIMCyp with R at this position failed to restrict HIV-1. Subsequently, Ylinen et al. (2010) reported another allele of CM TRIMCyp encoding histidine (H) at this position (Mafa TRIMCyp2) and Mafa TRIMCyp2 protein potently restricted HIV-1 but not HIV-2. Recently, Dietrich et al. (2011) examined 15 CMs from Indonesia, Indochina, Mauritius, and the Philippines carrying TRIMCyp, and did not find R at this position. We also examined 64 CMs from Malaysia, the Philippines, and Indonesia carrying TRIMCyp (34 heterozygotes and 30 homozygotes for TRIMCyp), and found that none of these 94 TRIMCyp genes carried R at this position (Saito et al., 2011a). On the other hand, both Dietrich et al. and our group found that TRIMCyp frequency in CM was apparently higher than that in Rh. TRIMCyp frequency tended to be higher in eastern Asia than in western Asia. Dietrich et al. and our group also found major and minor haplotypes of CM TRIMCyp with single nucleotide polymorphisms in the CypA domain. The major haplotype of CM TRIMCyp bears aspartic acid (D) and lysine (K) at positions 369 and 446, respectively (Brennan et al., 2008; Ylinen et al., 2010). The minor haplotype encodes asparagine (N) and glutamic acid (E) at positions 369 and 446, respectively (Dietrich et al., 2011; Saito et al., 2011a). N369 and E446 were also found in PM and Rh TRIMCyps, and the CypA portion of the NE haplotype of CM TRIMCyp has the same amino acid sequence as that of Rh TRIMCyp. The major CM haplotype of the TRIMCyp suppressed HIV-1 but not HIV-2, while the minor haplotype of TRIMCyp suppressed HIV-2 but not HIV-1 as PM and Rh TRIMCyp did (Saito et al., 2011a; Figure 4).
The original CypA sequence retrotransposed into the macaque TRIM5 locus must have been the authentic macaque CypA. There are two or three amino acid differences between authentic CypA and the CypA portion of TRIMCyp in Rh, CM, and PM, and TRIMCyp with the authentic CypA sequence has been shown to restrict HIV-1 but to only weakly restrict HIV-2 (Virgen et al., 2008; Price et al., 2009). TRIMCyp from all three of these OWM species share H at the 372nd position, corresponding to the 69th position of CypA where the authentic macaque CypA has R. Rh and PM TRIMCyps and the minor haplotype of CM TRIMCyp share N at the 369th position (the 66th position in CypA), where the authentic CypA and major haplotypes of CM TRIMCyp (Mafa TRIMCyp2) have D. Structural analysis of CypA domain revealed that these mutations caused drastic changes in configuration of the active site loop (from the 64th amino acid residue to the 74th residue in CypA) in Rh TRIMCyp, leading to a decreased binding affinity to HIV-1 CA but an increased affinity to HIV-2 CA (Price et al., 2009). Therefore, these mutations enhanced antiviral activity of TRIMCyp against HIV-2 but diminished anti-HIV-1 activity (Price et al., 2009). In the case of the major haplotype of CM TRIMCyp, an additional E-to-K change at the 446th position (the 143rd position in CypA) decreased affinity to HIV-2 CA by its positive charge (Ylinen et al., 2010), and the D at the 369th position (the 66th position in CypA) supported its anti-HIV-1 activity.
How did these interspecies and intraspecies variations occur in TRIMCyp? It is reasonable to assume that the R-to-H mutation at the 372nd position (R69H) together with the D-to-N mutation at the 369th position (D66N), which enhanced antiviral activity of TRIMCyp against HIV-2 but diminished anti-HIV-1 activity (Price et al., 2009), arose early in a macaque common ancestor. After the separation of CM from other species, an additional E-to-K change at the 446th position (E143K) and the N-to-D reversion at the 369th position (N66D) may also have occurred in the major haplotype of CM TRIMCyp. Alternatively, polymorphisms at the 369th and 446th positions may have arisen early in a macaque common ancestor but only CM could transmit these polymorphisms until the present. As described above, CM TRIM5α has Q at amino acid position 339 (Nakayama et al., 2005), where Rh-TRIM5α has a Q-to-TFP polymorphism (Newman et al., 2006; Figure 2). This Q-to-TFP polymorphism in the PRYSPRY domain also altered the spectrum of anti-lentiviral activity of TRIM5α (Kono et al., 2008; Wilson et al., 2008a; Kirmaier et al., 2010; Lim et al., 2010b; Figure 4). Therefore, it is tempting to speculate that the selection pressure in CM drove amplification and diversification in TRIMCyp, while that in Rh drove diversification of the PRYSPRY domain of TRIM5α. It will be of interest to examine what retroviruses have driven the evolution of TRIMCyp and TRIM5 genes (Figure 7).
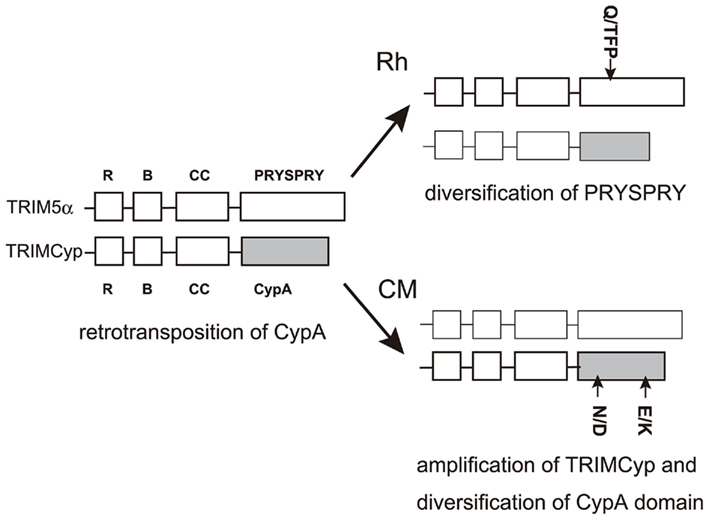
Figure 7. Diversity of Rh and CM TRIM5 genes. The RING, B-box2, Coiled-coil, and PRYSPRY domains of TRIM5α and TRIMCyp are shown by squares. CypA domains in TRIMCyp are shown as filled squares. Major alleles in Rh and CM are shown in bold lines. Polymorphisms are shown outside the squares.
With respect to SIV infection, Rh TRIMCyp failed to restrict SIVmac239 (Brennan et al., 2008; Wilson et al., 2008b; Kirmaier et al., 2010) but could restrict SIVsm (Kirmaier et al., 2010). The unique LPA-to-QQ change at positions 89–91 in L4/5 of SIVmac was critical for escape from Rh TRIMCyp (Figure 4). Rh heterozygous for the TFP allele of TRIM5α and TRIMCyp suppressed viral infection of both SIVsmE543 (Kirmaier et al., 2010) and SIVsm660 (Reynolds et al., 2011) more efficiently than Rh homozygous for either TRIMCyp or TRIM5α. It is possible that Rh heterozygous for the TFP allele of TRIM5α and TRIMCyp express two different molecules that bind distinct regions of CA and eliminate incoming virus more effectively than Rh with TRIM5 molecules targeting only one region of CA.
Other Restriction Factors and Development of Monkey-Tropic HIV-1
To establish a monkey model for the study of HIV-1/AIDS, Kamada et al. (2006) developed an HIV-1 strain with minimal segments of SIVmac239. This virus (NL-ScaVR and DT5R) contains the L4/5 of CA and the entire vif segment of SIVmac239, and was designed to escape restriction mediated by ApoB mRNA editing catalytic subunit (APOBEC) 3G and CypA in OWM cells. APOBEC3G modifies the minus strand viral DNA during reverse transcription, resulting in impairment of viral replication (Sheehy et al., 2002; Harris et al., 2003; Mangeat et al., 2003), but this activity could be counteracted by the viral Vif protein (Mariani et al., 2003; Marin et al., 2003; Sheehy et al., 2003). Although HIV-1 Vif can potently suppress human APOBEC3G, it is not effective against Rh APOBEC3G, which at least partly explains the restriction of HIV-1 replication in monkey cells. CypA binds directly to L4/5 of HIV-1 CA but not to SIVmac CA and augments HIV-1 infection in human cells but inhibits its replication in OWM cells (Kootstra et al., 2003; Berthoux et al., 2004; Nakayama et al., 2008). Although DT5R could replicate in PM primary CD4+ T cells as well as in the CM T cell line HSC-F but not in Rh cells (Kamada et al., 2006), inoculation of this monkey-tropic HIV-1 (HIV-1mt) into PM did not cause CD4+ T cell depletion or any clinical symptoms (Igarashi et al., 2007), probably due to inefficient viral growth in monkeys. In the case of CM, replacement of L6/7 of HIV-1 with that of SIVmac239 greatly enhanced viral replication in PBMC (Kuroishi et al., 2009, 2010) and in animals (Saito et al., 2011b). However, the virus could not escape completely from CM TRIM5α (Kuroishi et al., 2010). Another HIV-1mt carrying 202 amino acid residues of SIVmac239 CA and vif, generated by Hatziioannou et al. (2006), could replicate efficiently in Rh cells, confirming that the N-terminal half of CA is required to be that of SIVmac to escape from Rh-TRIM5α. Unfortunately, this virus has replaced nearly all of CA sequence with that of SIVmac239 and has lost important CTL epitopes of HIV-1, and thus further improvement is required to use Rh as an HIV-1 infection model. H87Q and/or V86M mutations induced by adaptation of HIV-1 to the cells expressing Rh-TRIM5α (Pacheco et al., 2010) would be useful. In contrast, lack of functional TRIM5α expression in PM enabled Hatziioannou et al. (2009) to construct an HIV-1mt strain that differs from HIV-1 only in the vif gene and can efficiently replicate in PM. This is the most promising HIV-1/monkey model at present, if PMs are available in sufficient numbers for research.
Other host factors capable of suppressing HIV-1 replication were recently identified (Figure 1). One is tetherin (also know as BST2 or CD317; Neil et al., 2008; Van Damme et al., 2008). BST2 is an interferon-inducible membrane protein that interferes with the detachment of virus particles from infected cells. HIV-1 overcomes this restriction by expressing an accessory protein, Vpu, which counteracts BST2. BST2 restriction is also counteracted by primate lentiviruses that do not express a Vpu protein. Anti-BST2 functions are provided by the Env protein in HIV-2 and SIVtan (Gupta et al., 2009) or the Nef protein in SIVsm/mac and SIVagm (Jia et al., 2009; Zhang et al., 2009). As chimeric virus containing the tat, rev, vpu, and env of the HXB2 strain of HIV-1 in the genetic background of SIVmac239 is pathogenic in Rh and PM (Joag et al., 1996), BST2 in monkeys can be canceled by HIV-1 Vpu. Another recently identified host factor is SAMHD1 as dendritic and myeloid-cell-specific HIV-1 restriction factor counteracted by HIV-2/SIV Vpx (Laguette et al., 2011; Yeh et al., 2011). As HIV-1 lacks Vpx, it is necessary to elucidate whether monkey SAMHD1 restricts HIV-1.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology, and the Ministry of Health, Labour, and Welfare, Japan.
References
Akari, H., Mori, K., Terao, K., Otani, I., Fukasawa, M., Mukai, R., and Yoshikawa, Y. (1996). In vitro immortalization of old world monkey T lymphocytes with Herpesvirus saimiri: its susceptibility to infection with simian immunodeficiency viruses. Virology 218, 382–388.
Akari, H., Nam, K. H., Mori, K., Otani, I., Shibata, H., Adachi, A., Terao, K., and Yoshikawa, Y. (1999). Effects of SIVmac infection on peripheral blood CD4+CD8+ T lymphocytes in cynomolgus macaques. Clin. Immunol. 91, 321–329.
Alkhatib, G., Combadiere, C., Broder, C. C., Feng, Y., Kennedy, P. E., Murphy, P. M., and Berger, E. A. (1996). CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272, 1955–1958.
Anderson, J. L., Campbell, E. M., Wu, X., Vandegraaff, N., Engelman, A., and Hope, T. J. (2006). Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J. Virol. 80, 9754–9760.
Berthoux, L., Sebastian, S., Sayah, D. M., and Luban, J. (2005). Disruption of human TRIM5alpha antiviral activity by nonhuman primate orthologues. J. Virol. 79, 7883–7888.
Berthoux, L., Sebastian, S., Sokolskaja, E., and Luban, J. (2004). Lv1 inhibition of human immunodeficiency virus type 1 is counteracted by factors that stimulate synthesis or nuclear translocation of viral cDNA. J. Virol. 78, 11739–11750.
Bieniasz, P. D., and Cullen, B. R. (2000). Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol. 74, 9868–9877.
Bieniasz, P. D., Fridell, R. A., Anthony, K., and Cullen, B. R. (1997). Murine CXCR-4 is a functional coreceptor for T-cell-tropic and dual-tropic strains of human immunodeficiency virus type 1. J. Virol. 71, 7097–7100.
Bleul, C. C., Farzan, M., Choe, H., Parolin, C., Clark-Lewis, I., Sodroski, J., and Springer, T. A. (1996). The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 382, 829–833.
Brass, A. L., Dykxhoorn, D. M., Benita, Y., Yan, N., Engelman, A., Xavier, R. J., Lieberman, J., and Elledge, S. J. (2008). Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926.
Brennan, G., Kozyrev, Y., and Hu, S. L. (2008). TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc. Natl. Acad. Sci. U.S.A. 105, 3569–3574.
Brennan, G., Kozyrev, Y., Kodama, T., and Hu, S. L. (2007). Novel TRIM5 isoforms expressed by Macaca nemestrina. J. Virol. 81, 12210–12217.
Browning, J., Horner, J. W., Pettoello-Mantovani, M., Raker, C., Yurasov, S., Depinho, R. A., and Goldstein, H. (1997). Mice transgenic for human CD4 and CCR5 are susceptible to HIV infection. Proc. Natl. Acad. Sci. U.S.A. 94, 14637–14641.
Castro, B. A., Barnett, S. W., Evans, L. A., Moreau, J., Odehouri, K., and Levy, J. A. (1990). Biologic heterogeneity of human immunodeficiency virus type 2 (HIV-2) strains. Virology 178, 527–534.
Castro, B. A., Nepomuceno, M., Lerche, N. W., Eichberg, J. W., and Levy, J. A. (1991). Persistent infection of baboons and rhesus monkeys with different strains of HIV-2. Virology 184, 219–226.
Chackerian, B., Long, E. M., Luciw, P. A., and Overbaugh, J. (1997). Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J. Virol. 71, 3932–3939.
Cherepanov, P., Maertens, G., Proost, P., Devreese, B., Van Beeumen, J., Engelborghs, Y., De Clercq, E., and Debyser, Z. (2003). HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278, 372–381.
Choe, H., Farzan, M., Sun, Y., Sullivan, N., Rollins, B., Ponath, P. D., Wu, L., Mackay, C. R., Larosa, G., Newman, W., Gerard, N., Gerard, C., and Sodroski, J. (1996). The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85, 1135–1148.
Christ, F., Thys, W., De Rijck, J., Gijsbers, R., Albanese, A., Arosio, D., Emiliani, S., Rain, J. C., Benarous, R., Cereseto, A., and Debyser, Z. (2008). Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 18, 1192–1202.
Daniel, M. D., Letvin, N. L., King, N. W., Kannagi, M., Sehgal, P. K., Hunt, R. D., Kanki, P. J., Essex, M., and Desrosiers, R. C. (1985). Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science 228, 1201–1204.
Deng, H., Liu, R., Ellmeier, W., Choe, S., Unutmaz, D., Burkhart, M., Di Marzio, P., Marmon, S., Sutton, R. E., Hill, C. M., Davis, C. B., Peiper, S. C., Schall, T. J., Littman, D. R., and Landau, N. R. (1996). Identification of a major co-receptor for primary isolates of HIV-1. Nature 381, 661–666.
Diaz-Griffero, F., Qin, X. R., Hayashi, F., Kigawa, T., Finzi, A., Sarnak, Z., Lienlaf, M., Yokoyama, S., and Sodroski, J. (2009). A B-box 2 surface patch important for TRIM5alpha self-association, capsid-binding avidity and retrovirus restriction. J. Virol. 83, 10737–10751.
Dietrich, E. A., Brennan, G., Ferguson, B., Wiseman, R. W., O’Connor, D., and Hu, S. L. (2011). Variable prevalence and functional diversity of the antiretroviral restriction factor TRIMCyp in Macaca fascicularis. J. Virol. 85, 9956–9963.
Doranz, B. J., Rucker, J., Yi, Y., Smyth, R. J., Samson, M., Peiper, S. C., Parmentier, M., Collman, R. G., and Doms, R. W. (1996). A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85, 1149–1158.
Dragic, T., Litwin, V., Allaway, G. P., Martin, S. R., Huang, Y., Nagashima, K. A., Cayanan, C., Maddon, P. J., Koup, R. A., Moore, J. P., and Paxton, W. A. (1996). HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381, 667–673.
Feinberg, M. B., Baltimore, D., and Frankel, A. D. (1991). The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. U.S.A. 88, 4045–4049.
Feinberg, M. B., Jarrett, R. F., Aldovini, A., Gallo, R. C., and Wong-Staal, F. (1986). HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell 46, 807–817.
Feng, Y., Broder, C. C., Kennedy, P. E., and Berger, E. A. (1996). HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272, 872–877.
Fenizia, C., Keele, B. F., Nichols, D., Cornara, S., Binello, N., Vaccari, M., Pegu, P., Robert-Guroff, M., Ma, Z. M., Miller, C. J., Venzon, D., Hirsch, V., and Franchini, G. (2011). TRIM5alpha does not affect simian immunodeficiency virus SIV(mac251) replication in vaccinated or unvaccinated Indian rhesus macaques following intrarectal challenge exposure. J. Virol. 85, 12399–12409.
Fujita, M., Yoshida, A., Sakurai, A., Tatsuki, J., Ueno, F., Akari, H., and Adachi, A. (2003). Susceptibility of HVS-immortalized lymphocytic HSC-F cells to various strains and mutants of HIV/SIV. Int. J. Mol. Med. 11, 641–644.
Ganser-Pornillos, B. K., Chandrasekaran, V., Pornillos, O., Sodroski, J. G., Sundquist, W. I., and Yeager, M. (2011). Hexagonal assembly of a restricting TRIM5alpha protein. Proc. Natl. Acad. Sci. U.S.A. 108, 534–539.
Gao, F., Bailes, E., Robertson, D. L., Chen, Y., Rodenburg, C. M., Michael, S. F., Cummins, L. B., Arthur, L. O., Peeters, M., Shaw, G. M., Sharp, P. M., and Hahn, B. H. (1999). Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397, 436–441.
Garber, M. E., and Jones, K. A. (1999). HIV-1 Tat: coping with negative elongation factors. Curr. Opin. Immunol. 11, 460–465.
Garber, M. E., Wei, P., Kewalramani, V. N., Mayall, T. P., Herrmann, C. H., Rice, A. P., Littman, D. R., and Jones, K. A. (1998). The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12, 3512–3527.
Gupta, R. K., Mlcochova, P., Pelchen-Matthews, A., Petit, S. J., Mattiuzzo, G., Pillay, D., Takeuchi, Y., Marsh, M., and Towers, G. J. (2009). Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc. Natl. Acad. Sci. U.S.A. 106, 20889–20894.
Hahn, B. H., Shaw, G. M., De Cock, K. M., and Sharp, P. M. (2000). AIDS as a zoonosis: scientific and public health implications. Science 287, 607–614.
Harris, R. S., Bishop, K. N., Sheehy, A. M., Craig, H. M., Petersen-Mahrt, S. K., Watt, I. N., Neuberger, M. S., and Malim, M. H. (2003). DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809.
Hatziioannou, T., Ambrose, Z., Chung, N. P., Piatak, M. Jr., Yuan, F., Trubey, C. M., Coalter, V., Kiser, R., Schneider, D., Smedley, J., Pung, R., Gathuka, M., Estes, J. D., Veazey, R. S., Kewalramani, V. N., Lifson, J. D., and Bieniasz, P. D. (2009). A macaque model of HIV-1 infection. Proc. Natl. Acad. Sci. U.S.A. 106, 4425–4429.
Hatziioannou, T., Perez-Caballero, D., Yang, A., Cowan, S., and Bieniasz, P. D. (2004). Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. U.S.A. 101, 10774–10779.
Hatziioannou, T., Princiotta, M., Piatak, M. Jr., Yuan, F., Zhang, F., Lifson, J. D., and Bieniasz, P. D. (2006). Generation of simian-tropic HIV-1 by restriction factor evasion. Science 314, 95.
Himathongkham, S., and Luciw, P. A. (1996). Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology 219, 485–488.
Hirsch, V., Adger-Johnson, D., Campbell, B., Goldstein, S., Brown, C., Elkins, W. R., and Montefiori, D. C. (1997). A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J. Virol. 71, 1608–1620.
Igarashi, T., Iyengar, R., Byrum, R. A., Buckler-White, A., Dewar, R. L., Buckler, C. E., Lane, H. C., Kamada, K., Adachi, A., and Martin, M. A. (2007). Human immunodeficiency virus type 1 derivative with 7% simian immunodeficiency virus genetic content is able to establish infections in pig-tailed macaques. J. Virol. 81, 11549–11552.
Jackson, P. K., Eldridge, A. G., Freed, E., Furstenthal, L., Hsu, J. Y., Kaiser, B. K., and Reimann, J. D. (2000). The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10, 429–439.
Javanbakht, H., Diaz-Griffero, F., Stremlau, M., Si, Z., and Sodroski, J. (2005). The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5alpha. J. Biol. Chem. 280, 26933–26940.
Javanbakht, H., Yuan, W., Yeung, D. F., Song, B., Diaz-Griffero, F., Li, Y., Li, X., Stremlau, M., and Sodroski, J. (2006). Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding. Virology 353, 234–246.
Jia, B., Serra-Moreno, R., Neidermyer, W., Rahmberg, A., Mackey, J., Fofana, I. B., Johnson, W. E., Westmoreland, S., and Evans, D. T. (2009). Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 5, e1000429. doi:10.1371/journal.ppat.1000429
Joag, S. V., Li, Z., Foresman, L., Stephens, E. B., Zhao, L. J., Adany, I., Pinson, D. M., Mcclure, H. M., and Narayan, O. (1996). Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J. Virol. 70, 3189–3197.
Jones, K. A., Kadonaga, J. T., Luciw, P. A., and Tjian, R. (1986). Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science 232, 755–759.
Kaiser, S. M., Malik, H. S., and Emerman, M. (2007). Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science 316, 1756–1758.
Kamada, K., Igarashi, T., Martin, M. A., Khamsri, B., Hatcho, K., Yamashita, T., Fujita, M., Uchiyama, T., and Adachi, A. (2006). Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc. Natl. Acad. Sci. U.S.A. 103, 16959–16964.
Kar, A. K., Diaz-Griffero, F., Li, Y., Li, X., and Sodroski, J. (2008). Biochemical and biophysical characterization of a chimeric TRIM21-TRIM5alpha protein. J. Virol. 82, 11669–11681.
Keppler, O. T., Yonemoto, W., Welte, F. J., Patton, K. S., Iacovides, D., Atchison, R. E., Ngo, T., Hirschberg, D. L., Speck, R. F., and Goldsmith, M. A. (2001). Susceptibility of rat-derived cells to replication by human immunodeficiency virus type 1. J. Virol. 75, 8063–8073.
Kim, J., Tipper, C., and Sodroski, J. (2011). Role of TRIM5alpha RING domain E3 ubiquitin ligase activity in capsid disassembly, reverse transcription blockade, and restriction of simian immunodeficiency virus. J. Virol. 85, 8116–8132.
Kirmaier, A., Wu, F., Newman, R. M., Hall, L. R., Morgan, J. S., O’Connor, S., Marx, P. A., Meythaler, M., Goldstein, S., Buckler-White, A., Kaur, A., Hirsch, V. M., and Johnson, W. E. (2010). TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 8, e1000462. doi:10.1371/journal.pbio.1000462
Koito, A., Kameyama, Y., Cheng-Mayer, C., and Matsushita, S. (2003a). Susceptibility of mink (Mustera vision)-derived cells to replication by human immunodeficiency virus type 1. J. Virol. 77, 5109–5117.
Koito, A., Shigekane, H., and Matsushita, S. (2003b). Ability of small animal cells to support the postintegration phase of human immunodeficiency virus type-1 replication. Virology 305, 181–191.
Kono, K., Bozek, K., Domingues, F. S., Shioda, T., and Nakayama, E. E. (2009). Impact of a single amino acid in the variable region 2 of the Old World monkey TRIM5alpha SPRY (B30.2) domain on anti-human immunodeficiency virus type 2 activity. Virology 388, 160–168.
Kono, K., Song, H., Shingai, Y., Shioda, T., and Nakayama, E. E. (2008). Comparison of anti-viral activity of rhesus monkey and cynomolgus monkey TRIM5alphas against human immunodeficiency virus type 2 infection. Virology 373, 447–456.
Kono, K., Song, H., Yokoyama, M., Sato, H., Shioda, T., and Nakayama, E. E. (2010). Multiple sites in the N-terminal half of simian immunodeficiency virus capsid protein contribute to evasion from rhesus monkey TRIM5alpha-mediated restriction. Retrovirology 7, 72.
Kootstra, N. A., Munk, C., Tonnu, N., Landau, N. R., and Verma, I. M. (2003). Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl. Acad. Sci. U.S.A. 100, 1298–1303.
Kuroishi, A., Bozek, K., Shioda, T., and Nakayama, E. E. (2010). A single amino acid substitution of the human immunodeficiency virus type 1 capsid protein affects viral sensitivity to TRIM5 alpha. Retrovirology 7, 58.
Kuroishi, A., Saito, A., Shingai, Y., Shioda, T., Nomaguchi, M., Adachi, A., Akari, H., and Nakayama, E. E. (2009). Modification of a loop sequence between alpha-helices 6 and 7 of virus capsid (CA) protein in a human immunodeficiency virus type 1 (HIV-1) derivative that has simian immunodeficiency virus (SIVmac239) vif and CA alpha-helices 4 and 5 loop improves replication in cynomolgus monkey cells. Retrovirology 6, 70.
LaBonte, J. A., Babcock, G. J., Patel, T., and Sodroski, J. (2002). Blockade of HIV-1 infection of new world monkey cells occurs primarily at the stage of virus entry. J. Exp. Med. 196, 431–445.
Laguette, N., Sobhian, B., Casartelli, N., Ringeard, M., Chable-Bessia, C., Segeral, E., Yatim, A., Emiliani, S., Schwartz, O., and Benkirane, M. (2011). SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657.
Landau, N. R., Warton, M., and Littman, D. R. (1988). The envelope glycoprotein of the human immunodeficiency virus binds to the immunoglobulin-like domain of CD4. Nature 334, 159–162.
Langelier, C. R., Sandrin, V., Eckert, D. M., Christensen, D. E., Chandrasekaran, V., Alam, S. L., Aiken, C., Olsen, J. C., Kar, A. K., Sodroski, J. G., and Sundquist, W. I. (2008). Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J. Virol. 82, 11682–11694.
Laspia, M. F., Rice, A. P., and Mathews, M. B. (1989). HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 59, 283–292.
Li, X., and Sodroski, J. (2008). The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J. Virol. 82, 11495–11502.
Liao, C. H., Kuang, Y. Q., Liu, H. L., Zheng, Y. T., and Su, B. (2007). A novel fusion gene, TRIM5-cyclophilin A in the pig-tailed macaque determines its susceptibility to HIV-1 infection. AIDS 21(Suppl. 8), S19–S26.
Lim, S. Y., Chan, T., Gelman, R. S., Whitney, J. B., O’Brien, K. L., Barouch, D. H., Goldstein, D. B., Haynes, B. F., and Letvin, N. L. (2010a). Contributions of Mamu-A*01 status and TRIM5 allele expression, but not CCL3L copy number variation, to the control of SIVmac251 replication in Indian-origin rhesus monkeys. PLoS Genet. 6, e1000997. doi:10.1371/journal.pgen.1000997
Lim, S. Y., Rogers, T., Chan, T., Whitney, J. B., Kim, J., Sodroski, J., and Letvin, N. L. (2010b). TRIM5alpha modulates immunodeficiency virus control in rhesus monkeys. PLoS Pathog. 6, e1000738. doi:10.1371/journal.ppat.1000738
Lin, T. Y., and Emerman, M. (2008). Determinants of cyclophilin A-dependent TRIM5 alpha restriction against HIV-1. Virology 379, 335–341.
Locher, C. P., Blackbourn, D. J., Herndier, B. G., Reyes-Teran, G., Barnett, S. W., Murthy, K. K., and Levy, J. A. (1998). Transient virus infection and pathogenesis of a new HIV type 2 isolate, UC12, in baboons. AIDS Res. Hum. Retroviruses 14, 79–82.
Locher, C. P., Witt, S. A., Herndier, B. G., Abbey, N. W., Tenner-Racz, K., Racz, P., Kiviat, N. B., Murthy, K. K., Brasky, K., Leland, M., and Levy, J. A. (2003). Increased virus replication and virulence after serial passage of human immunodeficiency virus type 2 in baboons. J. Virol. 77, 77–83.
Lores, P., Boucher, V., Mackay, C., Pla, M., Von Boehmer, H., Jami, J., Barre-Sinoussi, F., and Weill, J. C. (1992). Expression of human CD4 in transgenic mice does not confer sensitivity to human immunodeficiency virus infection. AIDS Res. Hum. Retroviruses 8, 2063–2071.
Maegawa, H., Miyamoto, T., Sakuragi, J., Shioda, T., and Nakayama, E. E. (2010). Contribution of RING domain to retrovirus restriction by TRIM5alpha depends on combination of host and virus. Virology 399, 212–220.
Maertens, G., Cherepanov, P., Pluymers, W., Busschots, K., De Clercq, E., Debyser, Z., and Engelborghs, Y. (2003). LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278, 33528–33539.
Mancebo, H. S., Lee, G., Flygare, J., Tomassini, J., Luu, P., Zhu, Y., Peng, J., Blau, C., Hazuda, D., Price, D., and Flores, O. (1997). P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11, 2633–2644.
Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L., and Trono, D. (2003). Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103.
Mariani, R., Chen, D., Schrofelbauer, B., Navarro, F., Konig, R., Bollman, B., Munk, C., Nymark-Mcmahon, H., and Landau, N. R. (2003). Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114, 21–31.
Mariani, R., Rutter, G., Harris, M. E., Hope, T. J., Krausslich, H. G., and Landau, N. R. (2000). A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 74, 3859–3870.
Marin, M., Rose, K. M., Kozak, S. L., and Kabat, D. (2003). HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9, 1398–1403.
Mische, C. C., Javanbakht, H., Song, B., Diaz-Griffero, F., Stremlau, M., Strack, B., Si, Z., and Sodroski, J. (2005). Retroviral restriction factor TRIM5alpha is a trimer. J. Virol. 79, 14446–14450.
Miyamoto, T., Yokoyama, M., Kono, K., Shioda, T., Sato, H., and Nakayama, E. E. (2011). A single amino acid of human immunodeficiency virus type 2 capsid protein affects conformation of two external loops and viral sensitivity to TRIM5alpha. PLoS ONE 6, e22779. doi:10.1371/journal.pone.0022779
Munk, C., Brandt, S. M., Lucero, G., and Landau, N. R. (2002). A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. U.S.A. 99, 13843–13848.
Nagai-Fukataki, M., Ohashi, T., Hashimoto, I., Kimura, T., Hakata, Y., and Shida, H. (2011). Nuclear and cytoplasmic effects of human CRM1 on HIV-1 production in rat cells. Genes Cells 16, 203–216.
Nakayama, E. E., Maegawa, H., and Shioda, T. (2006). A dominant-negative effect of cynomolgus monkey tripartite motif protein TRIM5alpha on anti-simian immunodeficiency virus SIVmac activity of an African green monkey orthologue. Virology 350, 158–163.
Nakayama, E. E., Miyoshi, H., Nagai, Y., and Shioda, T. (2005). A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5alpha determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J. Virol. 79, 8870–8877.
Nakayama, E. E., Shingai, Y., Kono, K., and Shioda, T. (2008). TRIM5alpha-independent anti-human immunodeficiency virus type 1 activity mediated by cyclophilin A in old world monkey cells. Virology 375, 514–520.
Nakayama, E. E., and Shioda, T. (2010). Anti-retroviral activity of TRIM5 alpha. Rev. Med. Virol. 20, 77–92.
Neil, S. J., Zang, T., and Bieniasz, P. D. (2008). Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430.
Newman, R. M., Hall, L., Connole, M., Chen, G. L., Sato, S., Yuste, E., Diehl, W., Hunter, E., Kaur, A., Miller, G. M., and Johnson, W. E. (2006). Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc. Natl. Acad. Sci. U.S.A. 103, 19134–19139.
Newman, R. M., Hall, L., Kirmaier, A., Pozzi, L. A., Pery, E., Farzan, M., O’Neil, S. P., and Johnson, W. (2008). Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 4, e1000003. doi:10.1371/journal.ppat.1000003
Newstein, M., Stanbridge, E. J., Casey, G., and Shank, P. R. (1990). Human chromosome 12 encodes a species-specific factor which increases human immunodeficiency virus type 1 tat-mediated trans activation in rodent cells. J. Virol. 64, 4565–4567.
Nisole, S., Lynch, C., Stoye, J. P., and Yap, M. W. (2004). A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. U.S.A. 101, 13324–13328.
Ohkura, S., Yap, M. W., Sheldon, T., and Stoye, J. P. (2006). All three variable regions of the TRIM5alpha B30.2 domain can contribute to the specificity of retrovirus restriction. J. Virol. 80, 8554–8565.
Pacheco, B., Finzi, A., Stremlau, M., and Sodroski, J. (2010). Adaptation of HIV-1 to cells expressing rhesus monkey TRIM5alpha. Virology 408, 204–212.
Perez-Caballero, D., Hatziioannou, T., Yang, A., Cowan, S., and Bieniasz, P. D. (2005). Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J. Virol. 79, 8969–8978.
Perron, M. J., Stremlau, M., and Sodroski, J. (2006). Two surface-exposed elements of the B30.2/SPRY domain as potency determinants of N-tropic murine leukemia virus restriction by human TRIM5alpha. J. Virol. 80, 5631–5636.
Price, A. J., Marzetta, F., Lammers, M., Ylinen, L. M., Schaller, T., Wilson, S. J., Towers, G. J., and James, L. C. (2009). Active site remodeling switches HIV specificity of antiretroviral TRIMCyp. Nat. Struct. Mol. Biol. 16, 1036–1042.
Reymond, A., Meroni, G., Fantozzi, A., Merla, G., Cairo, S., Luzi, L., Riganelli, D., Zanaria, E., Messali, S., Cainarca, S., Guffanti, A., Minucci, S., Pelicci, P. G., and Ballabio, A. (2001). The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151.
Reynolds, M. R., Sacha, J. B., Weiler, A. M., Borchardt, G. J., Glidden, C. E., Sheppard, N. C., Norante, F. A., Castrovinci, P. A., Harris, J. J., Robertson, H. T., Friedrich, T. C., Mcdermott, A. B., Wilson, N. A., Allison, D. B., Koff, W. C., Johnson, W. E., and Watkins, D. I. (2011). The TRIM5{alpha} genotype of rhesus macaques affects acquisition of simian immunodeficiency virus SIVsmE660 infection after repeated limiting-dose intrarectal challenge. J. Virol. 85, 9637–9640.
Saito, A., Kono, K., Nomaguchi, M., Yasutomi, Y., Adachi, A., Shioda, T., Akari, H., and Nakayama, E. E. (2011a). Geographic, genetic and functional diversity of antiretroviral host factor TRIMCyp in cynomolgus macaque (Macaca fascicularis). J. Gen. Virol. doi:10.1099/vir.0.038075-0. [Epub ahead of print].
Saito, A., Nomaguchi, M., Iijima, S., Kuroishi, A., Yoshida, T., Lee, Y. J., Hayakawa, T., Kono, K., Nakayama, E. E., Shioda, T., Yasutomi, Y., Adachi, A., Matano, T., and Akari, H. (2011b). Improved capacity of a monkey-tropic HIV-1 derivative to replicate in cynomolgus monkeys with minimal modifications. Microbes Infect. 13, 58–64.
Sawada, S., Gowrishankar, K., Kitamura, R., Suzuki, M., Suzuki, G., Tahara, S., and Koito, A. (1998). Disturbed CD4+ T cell homeostasis and in vitro HIV-1 susceptibility in transgenic mice expressing T cell line-tropic HIV-1 receptors. J. Exp. Med. 187, 1439–1449.
Sawyer, S. L., Wu, L. I., Emerman, M., and Malik, H. S. (2005). Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U.S.A. 102, 2832–2837.
Sayah, D. M., Sokolskaja, E., Berthoux, L., and Luban, J. (2004). Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430, 569–573.
Sebastian, S., and Luban, J. (2005). TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology 2, 40.
Sheehy, A. M., Gaddis, N. C., Choi, J. D., and Malim, M. H. (2002). Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650.
Sheehy, A. M., Gaddis, N. C., and Malim, M. H. (2003). The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9, 1404–1407.
Shibata, R., Sakai, H., Kawamura, M., Tokunaga, K., and Adachi, A. (1995). Early replication block of human immunodeficiency virus type 1 in monkey cells. J. Gen. Virol. 76(Pt 11), 2723–2730.
Song, B., Gold, B., O’Huigin, C., Javanbakht, H., Li, X., Stremlau, M., Winkler, C., Dean, M., and Sodroski, J. (2005a). The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J. Virol. 79, 6111–6121.
Song, B., Javanbakht, H., Perron, M., Park, D. H., Stremlau, M., and Sodroski, J. (2005b). Retrovirus restriction by TRIM5alpha variants from old world and new world primates. J. Virol. 79, 3930–3937.
Song, H., Nakayama, E. E., Yokoyama, M., Sato, H., Levy, J. A., and Shioda, T. (2007). A single amino acid of the human immunodeficiency virus type 2 capsid affects its replication in the presence of cynomolgus monkey and human TRIM5alphas. J. Virol. 81, 7280–7285.
Staal, F. J., Roederer, M., and Herzenberg, L. A. (1990). Intracellular thiols regulate activation of nuclear factor kappa B and transcription of human immunodeficiency virus. Proc. Natl. Acad. Sci. U.S.A. 87, 9943–9947.
Stremlau, M., Owens, C. M., Perron, M. J., Kiessling, M., Autissier, P., and Sodroski, J. (2004). The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in old world monkeys. Nature 427, 848–853.
Stremlau, M., Perron, M., Lee, M., Li, Y., Song, B., Javanbakht, H., Diaz-Griffero, F., Anderson, D. J., Sundquist, W. I., and Sodroski, J. (2006). Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. U.S.A. 103, 5514–5519.
Stremlau, M., Perron, M., Welikala, S., and Sodroski, J. (2005). Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J. Virol. 79, 3139–3145.
Towers, G. J., Hatziioannou, T., Cowan, S., Goff, S. P., Luban, J., and Bieniasz, P. D. (2003). Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9, 1138–1143.
Van Damme, N., Goff, D., Katsura, C., Jorgenson, R. L., Mitchell, R., Johnson, M. C., Stephens, E. B., and Guatelli, J. (2008). The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252.
VandeWoude, S., and Apetrei, C. (2006). Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin. Microbiol. Rev. 19, 728–762.
Virgen, C. A., Kratovac, Z., Bieniasz, P. D., and Hatziioannou, T. (2008). Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc. Natl. Acad. Sci. U.S.A. 105, 3563–3568.
Wei, P., Garber, M. E., Fang, S. M., Fischer, W. H., and Jones, K. A. (1998). A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92, 451–462.
Wilson, S. J., Webb, B. L., Maplanka, C., Newman, R. M., Verschoor, E. J., Heeney, J. L., and Towers, G. J. (2008a). Rhesus macaque TRIM5 alleles have divergent antiretroviral specificities. J. Virol. 82, 7243–7247.
Wilson, S. J., Webb, B. L., Ylinen, L. M., Verschoor, E., Heeney, J. L., and Towers, G. J. (2008b). Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc. Natl. Acad. Sci. U.S.A. 105, 3557–3562.
Wu, X., Anderson, J. L., Campbell, E. M., Joseph, A. M., and Hope, T. J. (2006). Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc. Natl. Acad. Sci. U.S.A. 103, 7465–7470.
Yap, M. W., Nisole, S., and Stoye, J. P. (2005). A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 15, 73–78.
Yeh, W. W., Rao, S. S., Lim, S. Y., Zhang, J., Hraber, P. T., Brassard, L. M., Luedemann, C., Todd, J. P., Dodson, A., Shen, L., Buzby, A. P., Whitney, J. B., Korber, B. T., Nabel, G. J., Mascola, J. R., and Letvin, N. L. (2011). The TRIM5 gene modulates penile mucosal acquisition of simian immunodeficiency virus in rhesus monkeys. J. Virol. 85, 10389–10398.
Ylinen, L. M., Keckesova, Z., Wilson, S. J., Ranasinghe, S., and Towers, G. J. (2005). Differential restriction of human immunodeficiency virus type 2 and simian immunodeficiency virus SIVmac by TRIM5alpha alleles. J. Virol. 79, 11580–11587.
Ylinen, L. M., Price, A. J., Rasaiyaah, J., Hue, S., Rose, N. J., Marzetta, F., James, L. C., and Towers, G. J. (2010). Conformational adaptation of Asian macaque TRIMCyp directs lineage specific antiviral activity. PLoS Pathog. 6, e1001062. doi:10.1371/journal.ppat.1001062
Zhang, F., Wilson, S. J., Landford, W. C., Virgen, B., Gregory, D., Johnson, M. C., Munch, J., Kirchhoff, F., Bieniasz, P. D., and Hatziioannou, T. (2009). Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6, 54–67.
Zhao, G., Ke, D., Vu, T., Ahn, J., Shah, V. B., Yang, R., Aiken, C., Charlton, L. M., Gronenborn, A. M., and Zhang, P. (2011). Rhesus TRIM5alpha disrupts the HIV-1 capsid at the inter-hexamer interfaces. PLoS Pathog. 7, e1002009. doi:10.1371/journal.ppat.1002009
Keywords: TRIM5α, TRIMCyp, HIV-1, HIV-2, SIV, rhesus monkey, cynomolgus monkey
Citation: Nakayama EE and Shioda T (2012) TRIM5α and species tropism of HIV/SIV. Front. Microbio. 3:13. doi: 10.3389/fmicb.2012.00013
Received: 09 December 2011; Paper pending published: 20 December 2011;
Accepted: 09 January 2012; Published online: 24 January 2012.
Edited by:
Masako Nomaguchi, The University of Tokushima Graduate School, JapanReviewed by:
Masaru Yokoyama, National Institute of Infectious Diseases, JapanCecilia Cheng-Mayer, Aaron Diamond AIDS Research Center, USA
Copyright: © 2012 Nakayama and Shioda. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Emi E. Nakayama, Department of Viral Infections, Research Institute for Microbial Diseases, Osaka University, 3-1 Yamada-oka, Suita, Osaka 565-0871, Japan. e-mail:ZW1pZW5AYmlrZW4ub3Nha2EtdS5hYy5qcA==

