- Winogradsky Institute of Microbiology, Russian Academy of Sciences, Moscow, Russia
Northern wetlands play a key role in the global carbon budget, particularly in the budgets of the greenhouse gas methane. These ecosystems also determine the hydrology of northern rivers and represent one of the largest reservoirs of fresh water in the Northern Hemisphere. Sphagnum-dominated peat bogs and fens are the most extensive types of northern wetlands. In comparison to many other terrestrial ecosystems, the bacterial diversity in Sphagnum-dominated wetlands remains largely unexplored. As demonstrated by cultivation-independent studies, a large proportion of the indigenous microbial communities in these acidic, cold, nutrient-poor, and water-saturated environments is composed of as-yet-uncultivated bacteria with unknown physiologies. Most of them are slow-growing, oligotrophic microorganisms that are difficult to isolate and to manipulate in the laboratory. Yet, significant breakthroughs in cultivation of these elusive organisms have been made during the last decade. This article describes the major prerequisites for successful cultivation of peat-inhabiting microbes, gives an overview of the currently captured bacterial diversity from northern wetlands and discusses the unique characteristics of the newly discovered organisms.
Northern Wetlands: Global Functions, Key Characteristics and Major Biogeochemical Processes
Wetlands are ecosystems in which the water table is permanently or periodically close to the soil surface. The global area of natural wetlands is about 5.3–5.7 × 106 km2 (Matthews and Fung, 1987; Aselmann and Crutzen, 1989). More than half of this area is located between 50 and 70°N and is therefore referred as northern wetland. Northern wetlands are peat-accumulating ecosystems in which the amount of carbon sequestered in net primary production exceeds the amount of carbon lost to the atmosphere by decomposition of organic matter (Moore and Bellamy, 1974; Clymo, 1984). Hence, northern peatlands are recognized as a persistent sink for atmospheric CO2, with carbon accumulation rates of 10–30 g C m−2 year−1 and a global carbon pool of 200–450 Pg of carbon, which is about one-third of the global soil C pool (Gorham, 1991). Carbon in peat is roughly equivalent to an atmospheric carbon pool of 100–200 ppmv CO2 (∼25–50% of the current atmospheric burden). Decomposition of organic matter in deep, anoxic peat layers generates methane (CH4), which diffuses to the surface and is then partially emitted to the atmosphere, making northern wetlands a globally important source of CH4 (Matthews and Fung, 1987; Barlett and Harris, 1993; Hein et al., 1997; Panikov, 1999; Smith et al., 2004). The climate impact of peatlands, therefore, is the net result of reduced radiative forcing due to CO2 uptake and storage as peat, and enhanced radiative forcing due to CH4 emissions (Frolking and Roulet, 2007).
Apart of their importance in the terrestrial carbon cycle, northern wetlands hold a key role in the global water balance. These wetlands determine the hydrology of northern rivers and represent one of the largest reservoirs of fresh water in the Northern Hemisphere. Despite the carbon sequestering properties of peatlands, they are major sources of dissolved organic carbon (DOC), which is exported to aquatic ecosystems draining peatland catchments (Freeman et al., 2004; Fenner et al., 2007). Currently observed intensified DOC export from peatlands is becoming an increasing cause for concern in the water industry.
Sphagnum-dominated peatlands represent one of the most extensive types of northern wetlands. Sphagnum moss is characteristic of peat bogs and poor fens. Bogs are ombrotrophic ecosystems that are decoupled from the groundwater of the surrounding watershed and receive all water and nutrient inputs from the atmosphere. These peatlands typically have pH values below 4.0 and negligible concentrations of basic cations in surface waters. Poor fens also are acidic, but have somewhat higher surface water pH (4.0–5.5) than bogs, because they receive some minerotrophic drainage in addition to precipitation (Bedford et al., 1999). Both bogs and fens are nutrient-poor by nature. The total concentration of mineral nutrients in these wetlands is usually in the range of 5–50 mg L−1. N and S cycling is dominated by organic forms and transformations (Moore et al., 2004). Peat water usually contains very low concentrations of  and
and  (3–100 μM or several mg L−1; Lamers et al., 2000; Kravchenko, 2002; Moore et al., 2004; Kip et al., 2011). Sulfate concentrations are in the range of 10–300 μM, with the highest values measured in fens or in polluted wetlands (Nedwell and Watson, 1995; Blodau et al., 2007; Pester et al., 2010; Kip et al., 2011). Iron concentrations are also very low. Microbial Fe(III) reduction in an upland fen was shown to account for <7% of the anaerobic organic carbon mineralization (Küsel et al., 2008). Therefore, transformations of mineral N, S, and Fe are of minor importance in oligotrophic wetlands.
(3–100 μM or several mg L−1; Lamers et al., 2000; Kravchenko, 2002; Moore et al., 2004; Kip et al., 2011). Sulfate concentrations are in the range of 10–300 μM, with the highest values measured in fens or in polluted wetlands (Nedwell and Watson, 1995; Blodau et al., 2007; Pester et al., 2010; Kip et al., 2011). Iron concentrations are also very low. Microbial Fe(III) reduction in an upland fen was shown to account for <7% of the anaerobic organic carbon mineralization (Küsel et al., 2008). Therefore, transformations of mineral N, S, and Fe are of minor importance in oligotrophic wetlands.
Degradation of plant litter is the basis of the microbial food chain in these ecosystems. Since Sphagnum species are the major primary producers in ombrotrophic peatlands, the litter produced here originates largely from Sphagnum, and peat consists primarily of bryophyte remains. Sphagnum-derived litter is known to decompose very slowly, with 0.1–25% first-year mass loss (Clymo, 1965; Aerts et al., 2001; Scheffer et al., 2001), which is due to the presence of various highly decay-resistant phenolic compounds and waxes (Verhoeven and Liefveld, 1997) and also due to the low nutrient (particularly, N and P) concentrations in Sphagnum litter. Other factors that inhibit decomposition are high acidity, low temperatures and anoxic conditions prevailing within the peat profile. The end-products of anaerobic plant debris degradation are then transformed into methane, which diffuses into the aerobic part of the bog profile.
In summary, the key biogeochemical processes driven by microorganisms in acidic northern wetlands are (i) degradation of plant-derived organic matter, (ii) methanogenesis, (iii) methanotrophy, and (iv) N2 fixation since available forms of nitrogen are mostly at very low or undetectable levels. Of these, only the microorganisms involved in CH4 cycle, i.e., methanogenic archaea (Galand et al., 2003; Sizova et al., 2003; Kotsyurbenko et al., 2004, 2007; Juottonen et al., 2005; Bräuer et al., 2006, 2011; Cadillo-Quiroz et al., 2006, 2009, 2010) and methanotrophic bacteria (Dedysh et al., 1998, 2000, 2001, 2002; Dedysh et al. 2007; Morris et al., 2002; Raghoebarsing et al., 2005; Chen et al., 2008a,b; Dedysh, 2009; Kip et al., 2010, 2011) have received considerable research attention. Several representatives of these microbial groups are now available in pure cultures and are described taxonomically. Many of them display unusual characteristics, which are reviewed below. Knowledge about microorganisms responsible for degradation of plant litter in these ecosystems is much more limited. Most of this information is limited to fungi (Thormann et al., 2002, 2004; Rice et al., 2006), which are not addressed in this review, while reports on hydrolytic capabilities of peat-inhabiting prokaryotes and their role in decomposition processes are very rare (Pankratov et al., 2011). The same is true for the research on nitrogen-fixing microorganisms (Kravchenko and Doroshenko, 2003; Dedysh et al., 2004b; Doroshenko et al., 2007; Zadorina et al., 2009). Finally, the functional role of many other microbial inhabitants of northern wetlands remains completely unknown.
“Cultured Versus Uncultured” in Sphagnum-Dominated Wetlands
Despite the importance of northern wetlands in global carbon and water cycles, the microbial community composition in these ecosystems remains insufficiently described. Only a few studies have addressed the overall diversity of bacterial 16S rRNA genes in acidic northern peatlands (Juottonen et al., 2005; Dedysh et al., 2006; Morales et al., 2006; Hartman et al., 2008; Ausec et al., 2009; Pankratov et al., 2011). Peat material used in these studies was collected mostly from the surface layers (0–40 cm depth), around the oxic–anoxic interface, i.e., from the region of highest biological activity.
Figure 1 gives an overview of the 16S rRNA gene diversity retrieved in six cultivation-independent studies of acidic northern peatlands in different geographic locations. An additional dataset obtained for tropical acidic peatland in Thailand (Kanokratana et al., 2011) was included for comparison (right column in Figure 1). Interestingly, northern and tropical acidic peatlands appear to display similar patterns of bacterial diversity. These habitats are usually dominated by members of the phyla Acidobacteria and Proteobacteria. Peat-inhabiting acidobacteria belong to subdivisions 1, 3, 4, and 8 of this phylum. Of these, only subdivision 1 is relatively well represented by cultured and characterized strains, many of which were isolated from Sphagnum-dominated wetlands. Subdivisions 3 and 8 contain only a few described members, while subdivision 4 does not include taxonomically characterized representatives. Proteobacteria found in peat bogs most commonly belong to the Alpha- or Delta- classes. Alphaproteobacteria usually prevail in methane-emitting wetlands, and a large proportion of these bacteria is composed of methanotrophs and methylotrophs from the families Methylocystaceae and Beijerinckiaceae (Dedysh et al., 2006; Dedysh, 2009). Chemo-heterotrophs from the families Bradyrhizobiaceae, Acetobacteraceae, Hyphomicrobiaceae, and Caulobacteraceae, and phototrophs of the genera Rhodoblastus, Rhodomicrobium, and Rhodopseudomonas are also common. Today, much of this diversity is available in culture (Figure 2). Peat-inhabiting Deltaproteobacteria belong to phylogenetic lineages represented by the genera Syntrophobacter, Syntrophus, Smithella, Geobacter, and Anaeromyxobacter. These bacteria seem to prevail in wetlands with somewhat increased sulfate levels (Morales et al., 2006).
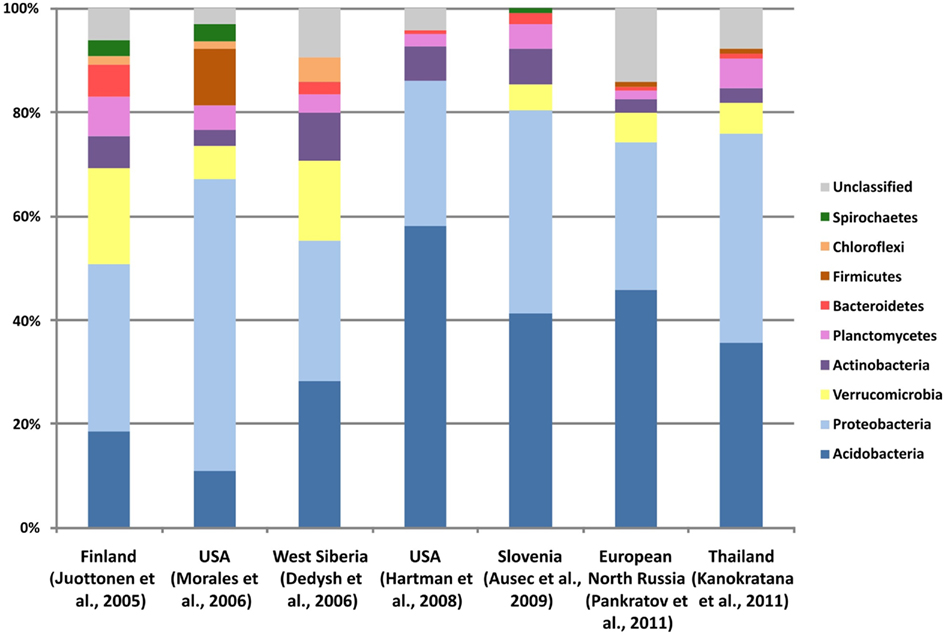
Figure 1. Taxonomic composition of bacterial communities in northern Sphagnum-dominated wetlands of various geographic locations, determined in different cultivation-independent studies (columns 1–6 from left to right). A dataset obtained for a tropical acidic peatland in Thailand (column 7) is included for comparison.
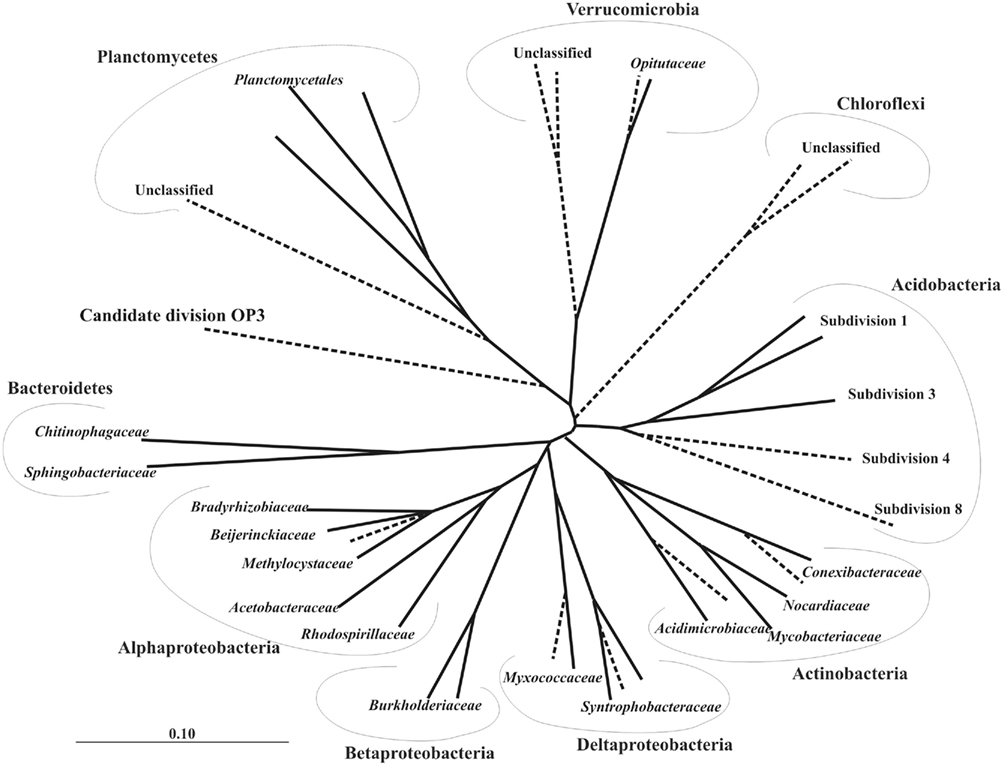
Figure 2. Unrooted 16S rRNA gene-based neighbor-joining tree showing the major phylogenetic lineages of bacteria that have been detected in Sphagnum-dominated wetlands by means of cultivation-independent techniques. The lineages lacking cultivated representatives are indicated by dashed lines. Bar, 0.1 substitutions per nucleotide position.
Less abundant but numerically significant groups of sequences in clone libraries made from acidic peat affiliate with the phyla Verrucomicrobia, Actinobacteria, and Planctomycetes (Figure 1). With the sole exception of the Opitutus-like group, most peat-inhabiting verrucomicrobia belong to taxonomically uncharacterized groups, for which isolates are not available. Surprisingly little is known about Actinobacteria that thrive in wetlands. In contrast to well-characterized soil Actinobacteria, peat-inhabiting members of this phylum belong to poorly described sub-groups. Significant proportion of Actinobacteria-related 16S rRNA gene sequences retrieved from Sphagnum-dominated wetlands affiliate with the Acidimicrobidae, a family containing only two characterized members, Acidimicrobium ferrooxidans and Ferrimicrobium acidiphilum. Another group of commonly detected sequences shows a distant relationship to Conexibacter woesei, a deep-rooting member of the phylum Actinobacteria.
The Planctomycetes is one of the bacterial groups that are strongly underrepresented in clone libraries obtained with the widely used Bacteria-specific PCR primer 9-27f. Yet, 16S rRNA gene sequences of planctomycetes are commonly found in these clone libraries made from acidic peat. They are highly diverse and represent nearly all currently known major lineages of this phylum. Many Planctomycetes-related 16S rRNA gene sequences display only a distant relationship (≤90% similarity) to those of taxonomically characterized organisms, although a large proportion of peat-derived sequences can now be classified as belonging to the lineages defined by the recently described acidophilic genera Schlesneria, Singulisphaera, and Zavarzinella.
Finally, minor groups of 16S rRNA gene sequences in the clone libraries affiliate with the Beta- and Gammaproteobacteria, Chloroflexi, Bacteroidetes, Spirochaetes, Firmicutes, and several candidate divisions such as Obsidian Pool 3 (OP3), OD1, and OP8. Of these, sequences related to the Firmicutes as well as to the Beta- and Gammaproteobacteria usually belong to well-characterized microbial groups.
As evidenced by the results of cultivation-independent studies, a large proportion of the indigenous bacteria populations in northern acidic wetlands is represented by as-yet-uncultivated organisms with unknown physiologies and metabolic potentials (Figure 2). Most of these bacteria cannot be cultured using conventional cultivation approaches.
Major Prerequisites for Successful Cultivation of Peat-Inhabiting Microbes
Direct counts of microbial cells in acidic wetlands are typically about 108–109 g−1 of wet peat (Williams and Crawford, 1983; Dedysh et al., 2001, 2006; Kotsyurbenko et al., 2004). Twenty to 70% of these cells are detectable by fluorescence in situ hybridization (FISH) with the bacteria-specific probe EUB338-mix, while up to 10% of total cells are targeted with archaeal probes ARCH915 and ARC344. The DAPI-stained objects that are not detected by any of these domain-specific probes are represented by cells of a very small size, i.e., ≤0.5 μm in length (Dedysh et al., 2006). Their nature and metabolic status remain poorly understood.
Only a minor part of peat-inhabiting bacteria can be cultured using a surface plating technique and conventional media such as nutrient agar, R2A and others. The numbers of colony forming units (CFUs) obtained by this conventional approach are low and usually range between 105 and 106 CFUs g−1 of wet peat (Golovchenko et al., 2005; Dedysh et al., 2006). This corresponds to approximately 0.01–1.0% of total DAPI cell counts.
Nearly one half of the colonies that develop on nutrient agar plates after 1–2 weeks of incubation are formed by fast growing Betaproteobacteria, particularly members of the genus Burkholderia (Opelt and Berg, 2004; Belova et al., 2006; Dedysh et al., 2006; Opelt et al., 2007; Vandamme et al., 2007). Other prokaryotes that most commonly develop on these plates affiliate with the Alphaproteobacteria (genera Bradyrhizobium, Mesorhizobium, Sphingomonas, Brevundimonas, Caulobacter, Hyphomicrobium), Actinobacteria (genera Mycobacterium, Rhodococcus, Streptomyces, Micromonospora), Gammaproteobacteria (genera Pseudomonas, Serratia, Rahnella), Firmicutes (genera Paenibacillus, Bacillus), and Bacteroidetes (genera Pedobacter, Dyadobacter, Chryseobacterium). These easily cultured isolates are closely related (98–100% 16S rRNA gene sequence similarity) to taxonomically described organisms and, with a very few exceptions, none of them represent the major 16S rRNA gene sequence groups detected in peatlands by means of cultivation-independent techniques. In other words, those who intend exploring the unknown bacterial diversity in northern wetlands cannot rely on a routine approach using standard media and short incubation times. Patience is one of the major virtues required in hunting for novel microbes. Other prerequisites for the successful cultivation of peat-inhabiting microbes are given below.
The Use of Dilute Acidic Media
Most commonly used nutrient media have a near-neutral pH and a salt content of 1–3 g L−1. This contrasts dramatically to acidic (pH 3.5–5.5) peat water with a mineral salt content of 5–50 mg L−1 and explains why most peat-inhabiting bacteria do not develop on conventional media. Therefore, a more accurate simulation of the peat bog environment in the laboratory requires dilute (1:10–1:100) and acidic (pH 4.0–5.5) media. High potential of this strategy was demonstrated by isolation of novel acidophilic methane-oxidizing bacteria from northern wetlands (Dedysh et al., 1998). Later, a number of peat-inhabiting methanotrophs were isolated on mildly acidic, low-ionic-strength media (Dedysh, 2009; Kip et al., 2011). Though methanogenic archaea are beyond the scope of this review, they have also been successfully retrieved from acidic wetlands using strongly dilute, acidic media (Sizova et al., 2003; Bräuer et al., 2006; Kotsyurbenko et al., 2007; Cadillo-Quiroz et al., 2009). The same strategy was applied for isolation of heterotrophic bacteria from poorly studied phyla. For example, peat-inhabiting members of the Acidobacteria were cultured using low-ionic-strength, low-nutrient media MM, MM1, or 10-fold diluted R2A (Dedysh et al., 2006; Pankratov et al., 2008). One of these media, MM1, does not contain phosphates since they were shown to inhibit growth of some acidobacteria (Pankratov and Dedysh, 2010). In this case, medium pH was adjusted to 4.0–5.0 with alginic acid, which offers a unique possibility of decreasing medium pH without increasing its ionic strength.
Extended Incubation Time
Most microbes that thrive in cold and nutrient-poor northern wetlands are slow-growing organisms. Even under optimal growth conditions, isolation of these microbes requires long incubation. Colonies of peat-inhabiting methanotrophs, acidobacteria, planctomycetes, and other fastidious bacteria appear on solid media only after 4–8 weeks of incubation (Dedysh et al., 2000; Pankratov et al., 2008, in press; Kulichevskaya et al., 2009). In the case of the acidotolerant facultative anaerobe Telmatospirillum sibiriense, development of colonies was observed after 5 months of anaerobic incubation (Sizova et al., 2007).
Most of these difficult-to-culture bacteria produce very small (0.1–0.5 mm in diameter) colonies, which can be observed and picked with the use of a dissecting microscope only. The same phenomenon was reported for several rarely cultured groups of soil bacteria, which were most abundant among the mini-colonies that developed after >12 weeks of incubation (Davis et al., 2011). The use of gellan gum (phytagel, gelrite), which produces a very clear medium, allows for easy discrimination of these mini-colonies (Janssen et al., 2002; Stott et al., 2008). This polysaccharide is also free of contaminants, which may inhibit growth of some microbes. Gellan gum was successfully applied for isolation of diverse peat-inhabiting bacteria, including methanotrophs and acidobacteria (Dedysh et al., 2002, in press; Pankratov et al., 2008, in press). Notably, many of these organisms were unable to grow on agar-containing media. It should be kept in mind, however, that polymerization of gellan gum occurs in the presence of bivalent cations, which are normally present in very low concentrations in ombrotrophic wetlands. The possibility that this may negatively effect isolation of some particular bacterial groups cannot be excluded.
A Biofilm-Mediated Approach Combined with Fish-Based Monitoring of the Enrichment/Isolation Procedure
As discussed above, most colonies that develop rapidly on the surface of various solid media are formed by bacteria which are not predominant in the peat bog environment. Many of these colonies, however, represent not a pure bacterial culture but a co-culture of two or more microorganisms. Examination of such mixed colonies obtained from acidic peat revealed that many of them contained cells of rarely cultured organisms, such as acidobacteria or planctomycetes (Dedysh et al., 2006). This observation suggested that an isolation approach based on enrichment of mixed cultures or microbial biofilms might in some cases be more efficient for isolating these elusive bacteria than a routine “single-colony pick-up” strategy. The two simplest versions of this alternative approach are given below.
Microbial biofilms consisting of cells of peat-inhabiting bacteria can be obtained by using a simple technique invented by Schlesner (1994) for isolation of planctomycetes. Briefly, the bottom of a Petri dish is covered with a layer of sterilized water agar, which contains cycloheximide to inhibit growth of fungi. Up to 20 sterilized cover slides are placed vertically and partially into the agar layer. These plates are then inoculated with 10–15 ml of peat water, which serves as the only source of nutrients for developing microorganisms, and incubated for 1–2 months. Every 2–3 weeks, a few slides are taken out and examined for the presence of target cells. In our experience, the best screening tool to recognize target cells, as opposed to easily culturable cells, is FISH. It allows direct visualization of the target bacteria within the entire population of cells present in the biofilm (Figure 3). This greatly simplifies all further isolation/purification procedures. Various PCR-based techniques can also be used for screening purposes, but FISH offers a clear advantage of seeing the target. The examples of successful application of this biofilm-mediated enrichment approach include isolation of peat-inhabiting subdivision 3 acidobacterium Bryobacter aggregatus (Kulichevskaya et al., 2010) as well as cultivation of several mildly acidophilic planctomycetes (Kulichevskaya et al., 2007, 2008, 2009, in press).
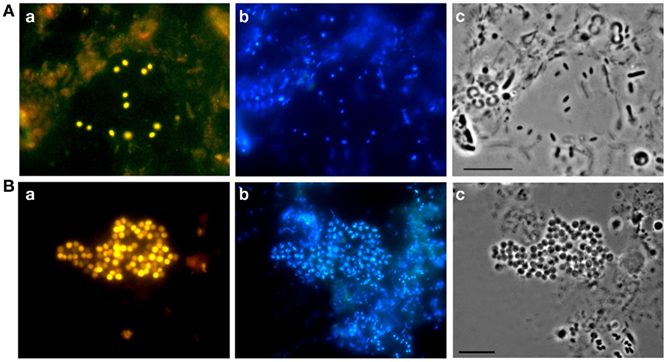
Figure 3. Specific detection of acidobacteria (A) and planctomycetes (B) in microbial biofilms by FISH: epifluorescence micrographs of in situ hybridization with probes HoAc1402 (Aa) and Pla46 + Pla886 (Ba), DAPI staining (b), and the respective phase-contrast images (c). Scale bars, 10 μm.
Careful inspection of impure cultures that are commonly discarded in routine cultivation practice is another way of obtaining an unusual organism. A good example is the isolation of a difficult-to-culture acidobacterium, Telmatobacter bradus (Pankratov et al., in press). This microorganism is a facultative anaerobe which can grow only under reduced oxygen tension. Despite this fact, T. bradus was recovered from aerobically incubated plates where it developed in a co-culture with an unidentified rapidly growing aerobic alphaproteobacterium. The alphaproteobacterium created a microhabitat with reduced oxygen concentration through its rapid growth. Application of FISH with the Acidobacteria-specific probe HoAc1402 was again the main tool to identify one of the organisms in this co-culture as a target for further isolation work.
Some Tips for Isolation of Anaerobes
So far, only a very few anaerobic prokaryotes have been cultivated from acidic wetlands. One of the factors that may have hampered isolation of anaerobes is the proper choice of a reducing agent for anaerobic media preparation. As shown by Sizova et al. (2003), many conventional reducing agents such as Na2S + L-cysteine-HCl and ascorbic acid did not support growth of methanogenic consortia from Sphagnum-derived peat, but success was achieved with titanium(III)citrate. Later, a similar approach was successfully applied for isolation of several methanogens from acidic peatlands (Bräuer et al., 2006; Cadillo-Quiroz et al., 2009) as well as for isolation and cultivation of some facultatively anaerobic peat-inhabiting bacteria (Sizova et al., 2007; Pankratov et al., in press).
In summary, the major tools for culturing microbes from acidic northern wetlands are readily available, although they need to be carefully adjusted for each particular target bacterium and site. Nevertheless, the bacterial diversity in acidic northern wetlands remains largely unexplored and should benefit from further development of improved cultivation techniques.
Currently Captured Bacterial Diversity
This chapter gives a brief overview of the currently cultured diversity of peat-inhabiting bacteria that have been characterized, named, and deposited in the public culture collections. At present, the list of these prokaryotic organisms does not exceed 30 species (Table 1), which compares poorly to the hundreds and thousands of species isolated and described from other ecosystems. Most bacterial taxa characteristic for northern wetlands have been described only during the last decade. Some representatives of the most abundant bacterial groups in acidic wetlands are discussed below.
Members of the Alphaproteobacteria
Three different physiological groups of alphaproteobacteria commonly occur in acidic wetlands: (i) methanotrophs, (ii) chemo-organotrophs, and (iii) phototrophs. Of these, methanotrophic alphaproteobacteria have received the most research attention due to their important role in reducing methane emission from northern wetlands (reviewed in Dedysh, 2009).
Methanotrophs
Methanotrophic bacteria that have been cultivated from these ecosystems are members of the families Beijerinckiaceae (the genera Methylocella, Methylocapsa, and Methyloferula) and Methylocystaceae (the genus Methylocystis). All of them are cold-tolerant, N2-fixing organisms that grow between pH 3.5–4.2 and 7.0–7.5. Methylocella palustris was the first methanotrophic bacterium isolated from acidic Sphagnum-dominated wetlands (Dedysh et al., 1998, 2000). Another species of this genus, Methylocella tundrae, was later isolated from tundra wetlands (Dedysh et al., 2004a). Members of the genus Methylocella have many unique features compared to other extant methanotrophs. Notably, they lack an extensive intracellular membrane (ICM) system, use only soluble methane monooxygenase MMO (sMMO) for methane oxidation, and are able to grow on some substrates containing C–C bonds (see below). Another unusual acidophilic methanotroph that possesses only sMMO is Methyloferula stellata (Vorobev et al., in press). In contrast to Methylocella spp., it is unable to utilize multicarbon compounds. Obligate methanotrophy is also characteristic of the peat-inhabiting methanotroph Methylocapsa acidiphila (Dedysh et al., 2002). Despite its close phylogenetic relationship to Methylocella and Methyloferula, Methylocapsa possesses a particulate MMO (pMMO) enzyme and an extensive ICM system. All methanotrophs from the Beijerinckiaceae are highly sensitive to salt stress and prefer dilute media with a low salt content (0.2–0.5 g L−1). One-carbon substrates are utilized via the serine pathway, although Methyloferula also displays activity of ribulose-1.5-bisphosphate carboxylase (RubisCO).
Methylocystis-like methanotrophs are one of the numerically dominant and metabolically active methanotroph populations in northern wetlands (Dedysh et al., 2001; Chen et al., 2008a,b; Belova et al., 2011). Currently, two peat-inhabiting members of this genus are available in cultures – Methylocystis heyeri H2 (Dedysh et al., 2007) and Methylocystis sp. H2s (Belova et al., 2011). These are mesophilic and mildly acidophilic methanotrophs which possess both a particulate and a soluble MMO and a well-developed ICM system. Methane and methanol are utilized via the serine pathway. However, in the absence of C1 substrates, these methanotrophs are able to grow slowly on acetate (Belova et al., 2011). A unique characteristic of M. heyeri is the phospholipid fatty acid profile. In addition to the signature fatty acid of alphaproteobacterial methanotrophs (18:1ω8c), cells also contain large amounts of what was previously considered a signature fatty acid of gammaproteobacterial methanotrophs, 16:1ω8c (Dedysh et al., 2007).
Chemo-organotrophs
Members of the family Rhodospirillaceae are common inhabitants of acidic wetlands. An interesting, non-phototrophic representative of this family, the facultatively anaerobic, acidotolerant bacterium Telmatospirillum sibiriense, was isolated by Sizova et al. (2007 from a Siberian fen. These motile spirilla grow chemoorganotrophically on several organic acids and glucose under anoxic and micro-oxic conditions, which span the largest part of the bog profile. At low oxygen partial pressure in the dark, they are also capable of lithotrophic growth on H2:CO2. Oligotrophy, metabolic versatility, ability to fix N2 and cell motility make these bacteria perfectly adapted to the peat bog environment.
Two aerobic, acidophilic, peat-inhabiting chemo-organotrophs, Acidisoma sibiricum and Acidisoma tundrae, were described by Belova et al. 2009). These psychrotolerant bacteria develop in the pH range of 3.0–7.5 and utilize most sugars, polyalcohols, some organic acids and several polysaccharides, including xylan and starch.
Phototrophs
Stagnant water bodies with large amounts of organic matter and low oxygen tensions are the preferred habitats for the purple non-sulfur bacteria (PNSB). They also develop in acidic freshwater wetlands. These metabolically flexible microorganisms grow phototrophically under anoxic conditions in the light or chemoorganotrophically under micro-oxic to oxic conditions in the dark. The examples of taxonomically characterized PNSB from acidic wetlands are Rhodoblastus acidophilus (Imhoff, 2001), which was originally described as “Rhodopseudomonas acidophila” (Pfennig, 1969), and Rhodoblastus sphagnicola (Kulichevskaya et al., 2006b). The latter bacterium was isolated from anaerobic cellulolytic enrichments made with peat suspension incubated under light. Several other phototrophs, such as Rhodopseudomonas palustris and Rhodomicrobium vannielii, are also often obtained from acidic wetlands. The isolates usually display 99–100% 16S rRNA gene identity to the type strains of these species.
Members of the Betaproteobacteria
The most typical representatives of this bacterial group in Sphagnum-dominated wetlands are members of the genus Burkholderia (Opelt and Berg, 2004; Belova et al., 2006; Opelt et al., 2007). The species Burkholderia bryophila was described based on a characterization of a number of strains isolated from Sphagnum mosses (Vandamme et al., 2007). These acidotolerant bacteria utilize various carbon substrates including aromatic compounds, fix N2 and show high antagonistic potential against fungal pathogens.
The Phylum Acidobacteria
The Acidobacteria is one of the most diverse groups of prokaryotes in acidic wetlands (Dedysh et al., 2006). Despite this fact, cultured representatives of peat-inhabiting acidobacteria are now available for subdivisions 1 and 3 only. These are the genera Granulicella (Pankratov and Dedysh, 2010), Telmatobacter (Pankratov et al., in press), and Bryocella (Dedysh et al., in press) in subdivision 1 and the genus Bryobacter in subdivision 3 (Kulichevskaya et al., 2010). All of these characterized acidobacteria are mesophilic or psychrotolerant, acidophilic chemo-organotrophs that grow at pH values between 3.0 and 6.5–7.5. Granulicella spp., Bryocella elongata, and B. aggregatus are strict aerobes. T. bradus is a facultative anaerobe that grows under reduced oxygen tension and under anoxic conditions. The preferred growth substrates are sugars though several organic acids and polyalcohols can also be utilized by some strains. Most peat-inhabiting acidobacteria can utilize glucuronic and galacturonic acids, which are released during decomposition of Sphagnum moss. The ability to degrade various plant-derived polymers, such as pectin, xylan, laminarin, lichenan, and starch varies between different species, but only T. bradus is capable of hydrolyzing cellulose. B. elongata possesses an ability to develop in a co-culture with exopolysaccharide-producing acidophilic methanotrophs, presumably by means of feeding on their capsular material. None of the cultured acidobacteria are capable of C1-metabolism or N2 fixation. However, members of this phylum seem to play an important role in degrading plant-derived polymers in acidic oligotrophic wetlands.
The Phylum Planctomycetes
Planctomycetes represent one of the most abundant bacterial groups detectable by FISH in acidic Sphagnum-dominated wetlands (Dedysh et al., 2006; Kulichevskaya et al., 2006a). Several peat-inhabiting, acidophilic planctomycetes were recently isolated and described as members of the novel genera Schlesneria, Singulisphaera, and Zavarzinella (Kulichevskaya et al., 2007, 2008, 2009, in press). These are mesophilic, moderately acidophilic chemo-organotrophs capable of growth at pH values between 4.2 and 7.5. They grow best in aerobic conditions on media containing carbohydrates or N-acetylglucosamine. However, Schlesneria paludicola is also capable of fermentation, while members of the genus Singulisphaera grow well in micro-oxic conditions. All planctomycetes isolated from peat are able to degrade various heteropolysaccharides, but not cellulose or chitin. None of them is capable of fixing N2. These cultured representatives, however, do not cover all planctomycete diversity detected in acidic wetlands by means of cultivation-independent approaches (Ivanova and Dedysh, unpublished data). The existence of other, still-unknown physiological types of planctomycetes in wetlands cannot be excluded.
The Phylum Actinobacteria
Currently available cultures of peat-inhabiting actinobacteria include several species of the genus Mycobacterium (Kazda and Müller, 1979; Kazda, 1980) and a member of the family Micromonosporaceae, Verrucosispora gifhornensis (Rheims et al., 1998). They utilize sugars, several polyalcohols and organic acids as well as some aromatic compounds, but none of them is capable of cellulose degradation.
The Phylum Bacteroidetes
In contrast to many aquatic and terrestrial habitats, members of the phylum Bacteroidetes are not abundant in acidic wetlands. The species cultured from acidic peat lack the ability to degrade cellulose or chitin. Two examples of such bacteria are Mucilaginibacter spp. (Pankratov et al., 2007) and Chitinophaga arvensicola (Pankratov et al., 2006). They do, however, possess some hydrolytic potential being able to degrade some heteropolysaccharides, such as xylan, laminarin, or pectin, in acidic and cold conditions.
New Knowledge Gained Due to Cultivation Efforts
As outlined in Table 1, cultivation efforts of the last decade have resulted in significant success in uncovering the microbial diversity in acidic northern wetlands. Many characteristics of the newly isolated organisms, such as their acid and salt tolerance as well as temperature preferences, distinguish them from bacteria inhabiting neutral mesophilic habitats. This is not surprising since acidic northern wetlands possess unique physico-chemical conditions. More interesting are several unexpected physiological discoveries made after isolation of novel representatives of the bacterial groups presumed to be already well documented. Some of these findings are discussed in this chapter.
Existence of Methanotrophs Containing Only a Soluble Methane Monooxygenase
Until recently, possession of a particulate methane monooxygenase enzyme and a well-developed intracytoplasmic membrane system in which pMMO is bound was considered a characteristic feature of all extant aerobic methanotrophs. The first methanotrophic bacterium that did not meet these criteria was isolated from acidic Sphagnum-dominated wetlands and named Methylocella palustris (Dedysh et al., 2000). Two other recognized species of this genus, M. silvestris (Dunfield et al., 2003) and M. tundrae (Dedysh et al., 2004a) were later isolated from an acidic forest soil and tundra wetlands, respectively. All members of the genus Methylocella lack extensive intracytoplasmic membrane structures typical of most other methanotrophs. The absence of pMMO in M. palustris was initially suggested by the failure to detect a pmoA gene, which encodes the 27-kDa polypeptide of pMMO, by PCR or by DNA probing with pmoA from Methylococcus capsulatus Bath (Dedysh et al., 2000). Analysis of M. silvestris BL2T by SDS-PAGE detected no pMMO-specific polypeptides, and DNA probing with pmoA gene fragments from two close phylogenetic relatives also found no evidence for pMMO (Theisen et al., 2005). Finally, the absence of any pmo gene homologs in M. silvestris BL2T was conclusively verified by analyzing the genome sequence of this bacterium (Chen et al., 2010).
The absence of pMMO in Methylocella spp. implies that these bacteria cannot be detected using a pmoA-based PCR assay considered universal and specific for all other methanotrophs (Holmes et al., 1995). However, they do possess the mmoX gene encoding the α-subunit of sMMO and can be detected via retrieval of these genes from the environment. A real-time quantitative PCR method was recently developed and validated targeting Methylocella mmoX that allowed detection and quantification of these unusual methanotrophs in a variety of environmental samples (Rahman et al., 2011). Interestingly, Methylocella spp. were detected not only in acidic or neutral habitats, but also in alkaline environments, suggesting that these methanotrophs are not limited to acidic pH in nature.
The diversity of methanotrophs that lack pMMO and use only an sMMO for methane oxidation appears to be not restricted to Methylocella species. Recently, three novel isolates were obtained from Sphagnum-dominated wetlands and an acidic forest soil and were described as representing a novel genus and species, Methyloferula stellata (Vorobev et al., in press). In these bacteria, the mmoX gene could not be amplified with any of the previously known mmoX-targeted primers, which explains why these methanotrophs escaped detection in all previous cultivation-independent studies. The absence of a universal assay for the specific detection of all pMMO-lacking methanotrophs complicates assessment of their abundance and distribution as well as their contribution to the processes of methane oxidation in northern wetlands and other natural environments. We have only recently begun exploring this new group of pMMO-lacking methanotrophs.
Facultative Methanotrophy
For a long time, all methanotrophs were considered to be obligately methylotrophic, i.e., unable to grow on compounds containing C–C bonds. This notion has recently been revised since the ability to grow on methane as well as on some multicarbon substrates, i.e., facultative methanotrophy, was demonstrated in several methanotrophic bacteria. Interestingly, all currently characterized facultative methanotrophs were isolated from acidic environments, such as Sphagnum-dominated wetlands or acidic boreal forest soils. Unusual, pMMO-lacking methanotrophs of the genus Methylocella were the first to be conclusively shown to have a facultative capability (Dedysh et al., 2005a). In addition to methane, they are capable of growth on acetate, pyruvate, succinate, malate, and ethanol. Acetate and methane were used as model substrates to conclusively verify facultative growth in M. silvestris BL2T. The growth rate and carbon conversion efficiency of M. silvestris BL2T was higher on acetate than on methane, and when both substrates were provided in excess acetate was preferably used and methane oxidation shut down. Further detailed experiments demonstrated that transcription of the mmo operon in Methylocella was repressed by acetate (Theisen et al., 2005).
We now know that a facultative lifestyle may also occur in pMMO-possessing methanotrophs. Several members of the genera Methylocapsa and Methylocystis were recently shown to be capable of growing on methane as well as on some multicarbon substrates such as acetate or ethanol (Dunfield et al., 2010; Belova et al., 2011; Im et al., 2011). Unlike Methylocella, these methanotrophs prefer to utilize methane, but growth can also occur on acetate and/or ethanol in the absence of methane. Two of these organisms, M. heyeri H2 and Methylocystis sp. strain H2s, were isolated from acidic Sphagnum peat. The latter was shown to represent a numerically abundant methanotroph population in geographically distinct northern wetlands (Belova et al., 2011).
Metabolic Versatility within the Beijerinckiaceae
Research on acidic wetlands has changed our perception of the family Beijerinckiaceae. Former knowledge of this bacterial group characterized its representatives as aerobic, acidophilic chemo-heterotrophs with the ability to fix N2. During the last decade, a novel sub-group of acidophilic serine pathway methano- and methylotrophs that are phylogenetically closely related to the genus Beijerinckia was discovered in acidic Sphagnum peat bogs and forest soils (Dedysh et al., 1998, 2000, 2002; Dedysh et al., 2004a; Dunfield et al., 2003, 2010; Vorob’ev et al., 2009; Vorobev et al., in press). Moreover, methylotrophic autotrophy was found in one earlier described member of the genus Beijerinckia, Beijerinckia mobilis (Dedysh et al., 2005b). Now this family accommodates bacteria with strikingly different lifestyles, such as chemo-heterotrophs, facultative methylotrophs and also facultative and obligate methanotrophs (Figure 4). The 16S rRNA gene sequence similarity values between these phenotypically distinct bacteria range from 96 to 98%, which makes it impossible to predict the phenotype of any novel member in this family based on 16S rRNA gene sequence information alone. Since Beijerinckiaceae is the only bacterial family containing methanotrophic and non-methanotrophic bacteria, it has been chosen for the ongoing comparative genomic study, which should enable elucidating the genetic and metabolic tradeoffs required for a specialized methanotrophic lifestyle compared to more generalist chemoorganotrophic lifestyles.
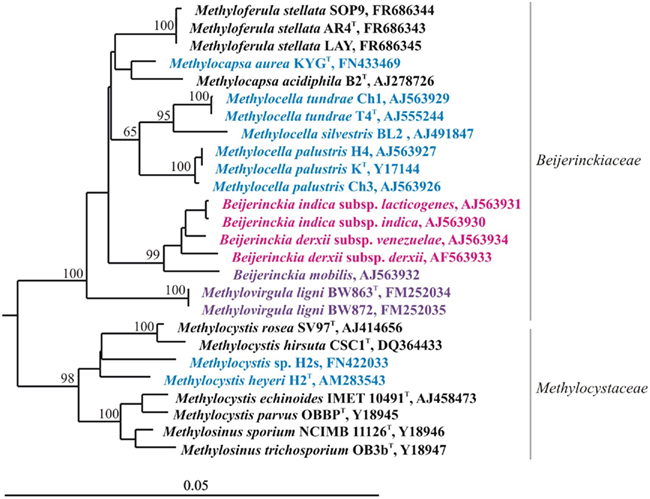
Figure 4. 16S rRNA gene-based neighbor-joining tree showing members of the families Beijerinckiaceae and Methylocystaceae. Black letters indicate obligate methanotrophs, blue indicates facultative methanotrophs, violet indicates facultative methylotrophs, while pink indicates chemo-heterotrophs. Bootstrap values (1000 data resamplings) >60% are shown. Bar, 0.05 substitutions per nucleotide position.
Cellulose Degradation in Members of the Acidobacteria
The Acidobacteria is one of the cosmopolitan but poorly characterized groups of the domain Bacteria. The role of these organisms in natural environments remains poorly understood. Recent analysis of the genomes of three bacteria from the phylum Acidobacteria suggested a potential role of these prokaryotes in degradation of plant, fungal, and insect-derived organic matter (Ward et al., 2009). The genomes were shown to contain the genes encoding candidate cellulases and β-glucosidases, suggesting that acidobacteria are able to degrade cellulose substrates, although experimental confirmation for this ability was missing. Recently, however, the first proofs for cellulose degradation by acidobacteria became available (Pankratov et al., in press). This ability was found in a peat-inhabiting facultative anaerobe Telmatobacter bradus. So far, this is the only characterized cellulolytic acidobacterium, although two other uncharacterized subdivision 1 acidobacteria, strains KBS 83 and CCO287, and subdivision 3 acidobacterium KBS 96 were also recently reported to possess cellulolytic potential (Eichorst et al., 2011; Pankratov et al., 2011). The actual rates of cellulose decomposition by these bacteria are very low and are not comparable to those in well-characterized cellulose degraders. The latter, however, are either absent or present in only very low numbers in cold and acidic northern wetlands. This makes acidobacteria important members of the indigenous hydrolytic microbial community. This may also be true for other acidic water-logged habitats such as a primary tropical peat swamp forest in southern Thailand (Kanokratana et al., 2011). Metagenomic analysis of the microbial community in the surface peat layer revealed a variety of putative genes encoding a range of cellulolytic and hemicellulolytic enzymes from the Acidobacteria, suggesting the key role of these microbes in plant debris degradation.
Intriguing Groups of as-yet-Uncultivated Microbes
Many bacteria that have been detected in acidic northern wetland by means of molecular techniques have so far resisted all cultivation attempts. The reasons behind our failure to culture these microbes are unclear and we also do not have any insights into their phenotypes or ecological functions. Several groups of these elusive bacteria are discussed below.
An Uncultured Group of Alphaproteobacteria Associated with Sphagnum Mosses
A specific and highly abundant group of alphaproteobacteria was shown to colonize the hyaline cells of the outer stem cortex and the surface of stem leaves of Sphagnum cuspidatum (Raghoebarsing et al., 2005). Molecular identification of these bacteria showed their distant affiliation to acidophilic methanotrophs of the genera Methylocella and Methylocapsa (93% 16S rRNA gene sequence identity) and, therefore, these bacteria were assumed to be “symbiotic” methanotrophs that live in and on Sphagnum mosses. A number of similar 16S rRNA gene sequences were later retrieved from an acidic peat bog in West Siberia (Dedysh et al., 2006) as well as from other boreal ecosystems. These sequences form a common cluster, which is distinct from the Methylocystaceae and the Beijerinckiaceae, and does not contain cultivated representatives (Figure 2). Interestingly, members of this bacterial group were also detected in cellulolytic peat enrichment cultures, which were incubated without methane (Pankratov et al., 2011). None of the currently available pmoA or mmoX sequence groups retrieved from Sphagnum mosses or Sphagnum-derived peat can be linked to these bacteria. Therefore, the biology of organisms within this cluster remains obscure.
Uncharacterized Acidobacteria
Despite the recent success in cultivating acidobacteria, most members of this phylum that thrive in northern wetlands resist isolation. This is especially true for subdivision 4 and 8 acidobacteria, which seem to be typical for the peat bog environment. At present, subdivision 4 has no characterized members. Subdivision 8 of the Acidobacteria is currently represented by three neutrophilic organisms that display highly contrasting characteristics. Holophaga foetida is a strictly anaerobic, homoacetogenic bacterium that degrades aromatic compounds (Liesack et al., 1994). Geothrix fermentans is also a strict anaerobe that oxidizes acetate as well as several other simple organic acids and long-chain fatty acids with Fe(III) as the electron acceptor (Coates et al., 1999). By contrast, Acanthopleuribacter pedis is a strictly aerobic chemo-organotroph that utilizes only a very limited number of growth substrates including glucose and several amino-acids (Fukunaga et al., 2008). The 16S rRNA gene sequences commonly retrieved from acidic wetlands affiliate with either Holophaga or Geothrix, but display only a very distant relationship to these characterized bacteria. Further cultivation efforts are needed to elucidate the metabolic potentials of the as-yet-uncultivated acidobacteria and their roles in northern wetlands.
Peat-Inhabiting Verrucomicrobia
Verrucomicrobia-affiliated 16S rRNA gene sequences are abundant in clone libraries obtained from Sphagnum peat (Figure 1). Most of these clones belong to a broad phylogenetic sequence cluster for which cultured representatives have not yet been reported. Therefore, no conclusions can be made concerning the lifestyle of peat-inhabiting verrucomicrobia. Most cultivated members of this phylum are heterotrophs but several recently characterized Verrucomicrobia are thermoacidophilic methanotrophs (Op den Camp et al., 2009). So far, these bacteria were detected in thermal environments only and none of the 16S rRNA gene sequences retrieved from northern wetlands fall close to those of thermoacidophilic methanotrophs. Therefore, the occurrence of methanotrophic Verrucomicrobia in acidic northern wetlands remains a highly intriguing question.
Candidate Division OP3
The candidate division OP3 is one of the primary bacterial groups for which no information is currently available. OP3-related 16S rRNA gene sequences have been recovered from many anoxic environments, such as flooded paddy soils, marine sediments, hypersaline deep sea, freshwater lakes, and methanogenic bioreactors (Glöckner et al., 2010). They were also retrieved from Sphagnum-dominated wetlands (Dedysh et al., 2006; Morales et al., 2006; Ivanova and Dedysh, unpublished results). The analysis of metagenomic fosmid libraries constructed from flooded paddy soils revealed that OP3 bacteria share a high proportion of orthologs with members of the Deltaproteobacteria and may possess an anaerobic respiration mode (Glöckner et al., 2010). They may have a facultatively anaerobic lifestyle since OP3-related 16S rRNA gene sequences were found both in oxic and anoxic peat layers.
Designing novel isolation strategies and cultivating uncultured microbes from northern wetlands remain highly challenging tasks given that most microbial inhabitants of these acidic, cold, and nutrient-poor environments are slow-growing, oligotrophic organisms. Hunting for such microbes is perhaps seen as technologically out of step with the increasingly molecular character of modern microbiology. The newly isolated microorganisms, however, are a unique source of novel, unexpected findings, which may revise many old paradigms in our knowledge. They also provide the means to study cell biology, to verify hypotheses emerging from genome sequence data, and to adjust the currently used molecular detection techniques as well as our ideas about the functional role of these microbes in the environment.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Svetlana N. Dedysh was supported by the Program “Molecular and Cell Biology” of Russian Academy of Sciences and the Russian Fund of Basic Research (grants no. 09-04-00004 and 11-04-91333). The author would like to thank Irina S. Kulichevskaya for providing the images in Figure 3 and Peter F. Dunfield for editing this manuscript.
References
Aerts, R., Walléen, B., Malmer, N., and de Caluwe, H. (2001). Nutritional constraints on Sphagnum-growth and potential decay in northern peatlands. J. Ecol. 89, 292–299.
Albert, R. A., Archambault, J., Rossello-Mora, R., Tindall, B. J., and Matheny, M. (2005). Bacillus acidicola sp. nov., a novel mesophilic acidophilic species isolated from acidic Sphagnum peat bogs in Wisconsin. Int. J. Syst. Evol. Microbiol. 55, 2125–2130.
Aselmann, I., and Crutzen, P. J. (1989). Global distribution of natural freshwater wetlands and rice paddies, their net primary productivity, seasonality and possible methane emissions. J. Atmos. Chem. 8, 307–358.
Ausec, L., Kraigher, B., and Mandic-Mulec, I. (2009). Differences in the activity and bacterial community structure of drained grassland and forest peat soil. Soil Biol. Biochem. 41, 1874–1881.
Barlett, K. B., and Harris, R. C. (1993). Review and assessment of methane emissions from wetlands. Chemosphere 26, 261–320.
Bedford, B. L., Walbridge, M. R., and Aldous, A. (1999). Patterns in nutrient availability and plant diversity of temperate North American wetlands. Ecology 80, 2151–2169.
Belova, S. E., Baani, M., Suzina, N. E., Bodelier, P. L. E., Liesack, W., and Dedysh, S. N. (2011). Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ. Microbiol. Rep. 3, 36–46.
Belova, S. E., Pankratov, T. A., and Dedysh, S. N. (2006). Bacteria of the genus Burkholderia as a typical component of the microbial community of Sphagnum peat bogs. Microbiology 75, 90–96.
Belova, S. E., Pankratov, T. A., Detkova, E. N., Kaparullina, E. N., and Dedysh, S. N. (2009). Acidisoma tundrae gen. nov., sp. nov. and Acidisoma sibiricum sp. nov., two novel acidophilic and psychrotolerant members of the Alphaproteobacteria from acidic northern wetlands. Int. J. Syst. Evol. Microbiol. 59, 2383–2290.
Blodau, C., Mayer, B., Peiffer, S., and Moore, T. R. (2007). Support for an anaerobic sulfur cycle in two Canadian peatland soils. J. Geophys. Res. Biogeosci. 112, 1–10.
Bräuer, S. L., Cadillo-Quiroz, H., Ward, R. J., Yavitt, J. B., and Zinder, S. H. (2011). Methanoregula boonei gen. nov., sp. nov., an acidophilic methanogen isolated from an acidic peat bog. Int. J. Syst. Evol. Microbiol. 61, 45–52.
Bräuer, S. L., Cadillo-Quiroz, H., Yashiro, E., Yavitt, J. B., and Zinder, S. H. (2006). Isolation of a novel acidiphilic methanogen from an acidic peat bog. Nature 442, 192–194.
Cadillo-Quiroz, H., Bräuer, S. L., Yashiro, E., Sun, C., Yavitt, J. B., and Zinder, S. H. (2006). Vertical profiles of methanogenesis and methanogens in two contrasting acidic peatlands in central New York State, USA. Environ. Microbiol. 8, 1428–1440.
Cadillo-Quiroz, H., Yavitt, J. B., and Zinder, S. H. (2009). Methanosphaerula palustris gen. nov., sp. nov., a hydrogenotrophic methanogen isolated from a minerotrophic fen peatland. Int. J. Syst. Evol. Microbiol. 59, 928–935.
Cadillo-Quiroz, H., Yavitt, J. B., Zinder, S. H., and Thies, J. E. (2010). Diversity and community structure of Archaea inhabiting the rhizoplane of two contrasting plants from an acidic bog. Microb. Ecol. 59, 757–767.
Chen, Y., Crombie, A., Rahman, M. T., Dedysh, S. N., Liesack, W., Stott, M. B., Alam, M., Theisen, A. R., Murrell, J. C., and Dunfield, P. F. (2010). Complete genome sequence of the aerobic facultative methanotroph Methylocella silvestris BL2. J. Bacteriol. 192, 3840–3841.
Chen, Y., Dumont, M. G., McNamara, N. P., Chamberlain, P. M., Bodrossy, L., Stralis-Pavese, N., and Murrell, J. C. (2008a). Diversity of the active methanotrophic community in acidic peatlands as assessed by mRNA and SIP-PLFA analyses. Environ. Microbiol. 10, 446–459.
Chen, Y., Dumont, M. G., Neufeld, J. D., Bodrossy, L., Stralis-Pavese, N., McNamara, N. P., Ostle, N., Briones, M. J. I., and Murrell, J. C. (2008b). Revealing the uncultivated majority: combining DNA stable-isotope probing, multiple displacement amplification and metagenomic analyses of uncultivated Methylocystis in acidic peatlands. Environ. Microbiol. 10, 2609–2622.
Clymo, R. S. (1984). The limits to peat bog growth. Philos. Trans. R. Soc. Lond. B Biol. Sci. 303, 605–654.
Coates, J. D., Ellis, D. J., Gaw, C. V., and Lovley, D. R. (1999). Geothrix fermentans gen. nov., sp. nov., a novel Fe(III)-reducing bacterium from a hydrocarbon-contaminated aquifer. Int. J. Syst. Bacteriol. 49, 1615–1622.
Davis, K. E. R., Sangwan, P., and Janssen, P. H. (2011). Acidobacteria, Rubrobacteridae and Chloroflexi are abundant among very slow-growing and mini-colony-forming bacteria. Environ. Microbiol. 13, 798–805.
Dedysh, S. N. (2009). Exploring methanotroph diversity in acidic northern wetlands: molecular and cultivation-based studies. Microbiology 78, 655–669.
Dedysh, S. N., Belova, S. E., Bodelier, P. L. E., Smirnova, K. V., Khmelenina, V. N., Chidthaisong, A., Trotsenko, Y. A., Liesack, W., and Dunfield, P. F. (2007). Methylocystis heyeri sp. nov., a novel type II methanotrophic bacterium possessing “signature” fatty acids of type I methanotrophs. Int. J. Syst. Evol. Microbiol. 57, 472–479.
Dedysh, S. N., Berestovskaja, Y. Y., Vasylieva, L. V., Belova, S. E., Khmelenina, V. N., Suzina, N. E., Trotsenko, Y. A., Liesack, W., and Zavarzin, G. A. (2004a). Methylocella tundrae sp. nov., a novel methanotrophic bacterium from acidic tundra peatlands. Int. J. Syst. Evol. Microbiol. 54, 151–156.
Dedysh, S. N., Ricke, P., and Liesack, W. (2004b). NifH and NifD phylogenies: a molecular basis for understanding nitrogen fixation capabilities of methanotrophic bacteria. Microbiology 150, 1301–1313.
Dedysh, S. N., Derakshani, M., and Liesack, W. (2001). Detection and enumeration of methanotrophs in acidic Sphagnum peat by 16S rRNA fluorescence in situ hybridization, including the use of newly developed oligonucleotide probes for Methylocella palustris. Appl. Environ. Microbiol. 67, 4850–4857.
Dedysh, S. N., Dunfield, P. F., Derakshani, M., Stubner, S., Heyer, J., and Liesack, W. (2003). Differential detection of type II methanotrophic bacteria in acidic peatlands using newly developed 16S rRNA-targeted fluorescent oligonucleotide probes. FEMS Microbiol. Ecol. 43, 299–308.
Dedysh, S. N., Khmelenina, V. N., Suzina, N. E., Trotsenko, Y. A., Semrau, J. D., Liesack, W., and Tiedje, J. M. (2002). Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 52, 251–261.
Dedysh, S. N., Knief, C., and Dunfield, P. (2005a). Methylocella species are facultatively methanotrophic. J. Bacteriol. 187, 4665–4670.
Dedysh, S. N., Smirnova, K. V., Chmelenina, V. N., Suzina, N. E., Liesack, W., and Trotsenko, Y. A. (2005b). Methylotrophic autotrophy in Beijerinckia mobilis. J. Bacteriol. 187, 3884–3888.
Dedysh, S. N., Kulichevskaya, I. S., Serkebaeva, Y. M., Mityaeva, M., Sorokin, V. V., Suzina, N. E., Rijpstra, W. I. C., and Sinninghe Damsté, G. S. (in press). Bryocella elongata gen. nov., sp. nov., a novel member of subdivision 1 of the Acidobacteria isolated from a methanotrophic enrichment culture, and emended description of Edaphobacter aggregans Koch et al. 2008. Int. J. Syst. Evol. Microbiol.
Dedysh, S. N., Liesack, W., Khmelenina, V. N., Suzina, N. E., Trotsenko, Y. A., Semrau, J. D., Bares, A. M., Panikov, N. S., and Tiedje, J. M. (2000). Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs representing a novel sub-type of serine pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50, 955–969.
Dedysh, S. N., Panikov, N. S., Liesack, W., Großkopf, R., Zhou, J., and Tiedje, J. M. (1998). Isolation of acidophilic methane-oxidizing bacteria from northern peat wetlands. Science 282, 281–284.
Dedysh, S. N., Pankratov, T. A., Belova, S. E., Kulichevskaya, I. S., and Liesack, W. (2006). Phylogenetic analysis and in situ identification of bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 72, 2110–2117.
Doroshenko, E. V., Boulygina, E. S., Spiridonova, E. M., Tourova, T. P., and Kravchenko, I. K. (2007). Isolation and characterization of nitrogen-fixing bacteria of the genus Azospirillum from the soil of a Sphagnum peat bog. Microbiology 76, 93–101.
Dunfield, P. F., Belova, S. E., Vorob’ev, A. V., Cornish, S. L., and Dedysh, S. N. (2010). Methylocapsa aurea sp. nov., a facultatively methanotrophic bacterium possessing a particulate methane monooxygenase, and emended description of the genus Methylocapsa. Int. J. Syst. Evol. Microbiol. 60, 2659–2664.
Dunfield, P. F., Khmelenina, V. N., Suzina, N. E., Trotsenko, Y. A., and Dedysh, S. N. (2003). Methylocella silvestris sp. nov., a novel methanotrophic bacterium isolated from an acidic forest cambisol. Int. J. Syst. Evol. Microbiol. 53, 1231–1239.
Eichorst, S. A., Kuske, C. R., and Schmidt, T. M. (2011). Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl. Environ. Microbiol. 77, 586–596.
Fenner, N., Freeman, C., Lock, M. A., Harmens, H., Reynolds, B., and Sparks, T. (2007). Interactions between elevated CO2 and warming could amplify DOC exports from peatland catchments. Environ. Sci. Technol. 41, 3146–3152.
Freeman, C., Fenner, N., Ostle, N. J., Kang, H., Dowrick, D. J., Reynolds, B., Lock, M. A., Sleep, D., Hughes, S., and Hudson, J. (2004). Export of dissolved organic carbon from peatlands under elevated carbon dioxide levels. Nature 430, 195–198.
Frolking, S., and Roulet, N. T. (2007). Holocene radiative forcing impact of northern peatland carbon accumulation and methane emissions. Global Change Biol. 13, 1079–1088.
Fukunaga, Y., Kurahashi, M., Yanagi, K., Yokota, A., and Harayama, S. (2008). Acanthopleuribacter pedis gen. nov., a marine bacterium isolated from a chiton, and description of Acanthopleuribacteraceae fam. nov., Acanthopleuribacterales ord. nov., Holophagaceae fam. nov., Holophagales ord. nov. and Holophagae classis nov. in the phylum “Acidobacteria.” Int. J. Syst. Evol. Microbiol. 58, 2597–2601.
Galand, P. E., Fritze, H., and Yrjälä, K. (2003). Microsite-dependent changes in methanogenic populations in a boreal oligotrophic fen. Environ. Microbiol. 5, 1133–1143.
Glöckner, J., Kube, M., Shrestha, P. M., Weber, M., Glöckner, F. O., Reinhardt, R., and Liesack, W. (2010). Phylogenetic diversity and metagenomics of candidate division OP3. Environ. Microbiol. 12, 1218–1229.
Golovchenko, A. V., Sannikova, Y. V., Dobrovol’skaya, T. G., and Zvyagintsev, D. G. (2005). The saprotrophic bacterial complex in the raised peat bogs of western Siberia. Microbiology 74, 545–551.
Gorham, E. (1991). Northern peatlands: role in carbon cycle and probable responses to climate warming. Ecol. Appl. 1, 182–195.
Hartman, W. H., Richardson, C. J., Vigalys, R., and Bruland, G. L. (2008). Environmental and anthropogenic controls over bacterial communities in wetland soils. Proc. Natl. Acad. Sci. U.S.A. 105, 17842–17847.
Hein, R., Crutzen, P. J., and Heinmann, M. (1997). An inverse modeling approach to investigate the global atmospheric methane cycle. Global Biogeochem. Cycles 11, 43–76.
Holmes, A. J., Costello, A., Lidstrom, M. E., and Murrell, J. C. (1995). Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132, 203–208.
Im, J., Lee, S.-W., Yoon, S., DiSpirito, A. A., and Semrau, J. D. (2011). Characterization of a novel facultative Methylocystis species capable of growth on methane, ethanol, and acetate. Environ. Microbiol. Rep. 3, 174–181.
Imhoff, J. F. (2001). Transfer of Rhodopseudomonas acidophila to the new genus Rhodoblastus as Rhodoblastus acidophilus gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 51, 1863–1866.
Janssen, P. H., Yates, P. S., Grinton, B. E., Taylor, P. M., and Sait, M. (2002). Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68, 2391–2396.
Juottonen, H., Galand, P., Tuittila, E.-S., Laine, J., Fritze, H., and Yrjälä, K. (2005). Methanogen communities and bacteria along an ecohydrological gradient in a northern raised bog. Environ. Microbiol. 7, 1547–1557.
Kanokratana, P., Uengwetwanit, T., Rattanachomsri, U., Bunterngsook, B., Nimchua, T., Tangphatsornruang, S., Plengvidhya, V., Champreda, V., and Eurwilaichitr, L. (2011). Insights into the phylogeny and metabolic potential of a primary tropical peat swamp forest microbial community by metagenomic analysis. Microb. Ecol. 61, 518–528.
Kazda, J., and Müller, K. (1979). Mycobacterium komossense sp. nov. Int. J. Syst. Bacteriol.. 29, 361–365.
Kip, N., Ouyang, W., van Winden, J., Raghoebarsing, A., van Niftrik, L., Pol, A., Pan, Y., Bodrossy, L., van Donselaar, E. G., Reichart, G.-J., Jetten, M. S. M., Sinninghe Damsté, J. S., and Op den Camp, H. J. M. (2011). Detection, isolation and characterization of acidophilic methanotrophs from Sphagnum mosses. Appl. Environ. Microbiol. 77, 5643–5654.
Kip, N., van Winden, J., Pan, Y., Bodrossy, L., Reichart, G.-J., Smolders, A. J. P., Jetten, M. S. M., Sinninghe Damsté, J. S., and Op den Camp, H. J. M. (2010). Global prevalence of methane oxidation by symbiotic bacteria in peat-moss ecosystems. Nat. Geosci. 3, 617–621.
Kotsyurbenko, O. R., Chin, K.-J., Glagolev, M. V., Stubner, S., Simankova, M. V., Nozhevnikova, A. N., and Conrad, R. (2004). Acetoclastic and hydrogenotrophic methane production and methanogenic populations in an acidic West-Siberian peat bog. Environ. Microbiol. 6, 1159–1173.
Kotsyurbenko, O. R., Friedrich, M. W., Simankova, M. V., Nozhevnikova, A. N., Golyshin, P. N., Timmis, K. N., and Conrad, R. (2007). Shift from acetoclastic to H2-dependent methanogenesis in a West Siberian peat bog at low pH values and isolation of an acidophilic Methanobacterium strain. Appl. Environ. Microbiol. 73, 2344–2348.
Kravchenko, I. K. (2002). Methane oxidation in boreal peat soils treated with various nitrogen compounds. Plant Soil 242, 157–162.
Kravchenko, I. K., and Doroshenko, E. V. (2003). Nitrogen-fixing activity in peat soils from a raised bog. Microbiology 72, 98–102.
Kuhner, C. H., Matthies, C., Acker, G., Schmittroth, M., Göner, A.S., and Drake, H. L. (2000). Clostridium akagii sp. nov. and Clostridium acidisoli sp. nov.: acid-tolerant, N2-fixing clostridia isolated from acidic forest soil and litter. Int. J. Syst. Evol. Microbiol. 50, 873–881.
Kulichevskaya, I. S., Baulina, O. I., Bodelier, P. L. E., Rijpstra, W. I. C., Sinninghe Damsté, G.S., and Dedysh, S. N. (2009). Zavarzinella formosa gen. nov., sp. nov., a novel stalked, Gemmata-like planctomycete from a siberian peat bog. Int. J. Syst. Evol. Microbiol. 59, 357–364.
Kulichevskaya, I. S., Detkova, E. N., Bodelier, P. L. E., Rijpstra, W. I. C., Sinninghe Damsté, G. S., and Dedysh, S. N. (in press). Singulisphaera rosea sp. nov., a novel planctomycete from acidic Sphagnum peat. Int. J. Syst. Evol. Microbiol. [Epub ahead of print].
Kulichevskaya, I. S., Ivanova, A. O., Baulina, O. I., Bodelier, P. L. E., Sinninghe Damsté, J. S., and Dedysh, S. N. (2008). Singulisphaera acidiphila gen. nov., sp. nov., a non-filamentous, Isosphaera-like planctomycete from acidic northern wetlands. Int. J. Syst. Evol. Microbiol. 58, 1186–1193.
Kulichevskaya, I. S., Ivanova, A. O., Belova, S. E., Baulina, O. I., Bodelier, P. L. E., Rijpstra, W. I. C., Sinninghe Damsté, J. S., Zavarzin, G. A., and Dedysh, S. N. (2007). Schlesneria paludicola gen. nov., sp. nov., the first acidophilic member of the order Planctomycetales, from Sphagnum-dominated boreal wetlands. Int. J. Syst. Evol. Microbiol. 57, 2680–2687.
Kulichevskaya, I. S., Pankratov, T. A., and Dedysh, S. N. (2006a). Detection of representatives of the Planctomycetes in Sphagnum peat bogs by molecular and cultivation approaches. Microbiology 75, 329–335.
Kulichevskaya, I. S., Guzev, V. S., Gorlenko, V. M., Liesack, W., and Dedysh, S. N. (2006b). Rhodoblastus sphagnicola sp. nov., a novel acidophilic purple non-sulfur bacterium from Sphagnum peat bog. Int. J. Syst. Evol. Microbiol. 56, 1397–1402.
Kulichevskaya, I. S., Suzina, N. E., Liesack, W., and Dedysh, S. N. (2010). Bryobacter aggregatus gen. nov., sp. nov., a peat-inhabiting, aerobic chemoorganotroph from Subdivision 3 of the Acidobacteria. Int. J. Syst. Evol. Microbiol. 60, 301–306.
Küsel, K., Blöthe, M., Schulz, D., Reiche, M., and Drake, H. L. (2008). Microbial reduction of iron and porewater biogeochemistry in acidic peatlands. Biogeosciences 5, 1537–1549.
Lamers, L. P. M., Bobbink, R., and Roelofs, J. G. M. (2000). Natural nitrogen filter fails in polluted raised bogs. Global Change Biol. 6, 583–586.
Liesack, W., Bak, F., Kreft, J. U., and Stackebrandt, E. (1994). Holophaga foetida gen. nov., sp. nov., a new homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch. Microbiol. 162, 85–90.
Matthews, E., and Fung, I. (1987). Methane emission from natural wetlands: global distribution, area, and environmental characteristics of sources. Global Biogeochem. Cycles 1, 61–86.
Moore, T., Blodau, C., Turunen, J., Roulet, N., and Richard, P. J. H. (2004). Patterns of nitrogen and sulfur accumulation and retention in ombrotrophic bogs, eastern Canada. Global Change Biol. 11, 356–367.
Morales, S. E., Mouser, P. J., Ward, N., Hudman, S. P., Gotelli, N. J., Ross, D. S., and Lewis, T. A. (2006). Comparison of bacterial communities in New England Sphagnum bogs using terminal restriction fragment length polymorphism. Microb. Ecol. 52, 34–44.
Morris, S. A., Radajewski, S., Willison, T. W., and Murrell, J. C. (2002). Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68, 1446–1453.
Nedwell, D. B., and Watson, A. (1995). CH4 production, oxidation and emission in a U.K. ombrotrophic peat bog: influence of SO42- from acid rain. Soil Biol. Biochem. 27, 893–903.
Op den Camp, H. J. M., Islam, T., Stott, M. B., Harhangi, H. R., Hynes, A., Schouten, S., Jetten, M. S. M., Birkeland, N.-K., Pol, A., and Dunfield, P. F. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1, 293–306.
Opelt, K., and Berg, G. (2004). Diversity and antagonistic potential of bacteria associated with bryophytes from nutrient-poor habitats of the Baltic Sea Coast. Appl. Environ. Microbiol. 70, 6569–6579.
Opelt, K., Chobot, V., Hadacek, F., Schönmann, S., Eberl, L., and Berg, G. (2007). Investigations of the structure and function of bacterial communities associated with Sphagnum mosses. Environ. Microbiol. 9, 2795–2809.
Panikov, N. S. (1999). Fluxes of CO2 and CH4 in high latitude wetlands: measuring, modeling and predicting response to climate change. Polar Res. 18, 237–244.
Pankratov, T. A., and Dedysh, S. N. (2010). Granulicella paludicola gen. nov., sp. nov., G. pectinivorans sp. nov., G. aggregans sp. nov. and G. rosea sp. nov., novel acidophilic, polymer-degrading acidobacteria from Sphagnum peat bogs. Int. J. Syst. Evol. Microbiol. 60, 2951–2959.
Pankratov, T. A., Ivanova, A. O., Dedysh, S. N., and Liesack, W. (2011). Bacterial populations and environmental factors controlling cellulose degradation in an acidic Sphagnum peat. Environ. Microbiol. 13, 1800–1814.
Pankratov, T. A., Kirsanova, L. A., Kaparullina, E. N., Kevbrin, V. V., and Dedysh, S. N. (in press). Telmatobacter bradus gen. nov., sp. nov., a cellulolytic facultative anaerobe from subdivision 1 of the Acidobacteria, and emended description of Acidobacterium capsulatum Kishimoto et al. 1991. Int. J. Syst. Evol. Microbiol.
Pankratov, T. A., Kulichevskaya, I. S., Liesack, W., and Dedysh, S. N. (2006). Isolation of aerobic, gliding, xylanolytic and laminarinolytic bacteria from acidic Sphagnum peatlands and emended description of Chitinophaga arvensicola Kämpfer et al. Int. J. Syst. Evol. Microbiol. 56, 2761–2764.
Pankratov, T. A., Serkebaeva, Y. M., Kulichevskaya, I. S., Liesack, W., and Dedysh, S. N. (2008). Substrate-induced growth and isolation of Acidobacteria from acidic Sphagnum peat. ISME J. 2, 551–560.
Pankratov, T. A., Tindall, B. J., Liesack, W., and Dedysh, S. N. (2007). Mucilaginibacter paludis gen. nov., sp. nov. and Mucilaginibacter gracilis sp. nov., pectin-, xylan-, and laminarin-degrading members of the family Sphingobacteriaceae from acidic Sphagnum peat bog. Int. J. Syst. Evol. Microbiol. 57, 2349–2354.
Pester, M., Bittner, N., Deevong, P., and Wagner, M. (2010). A “rare biosphere” microorganism contributes to sulfate reduction in a peatland. ISME J. 4, 1–12.
Pfennig, N. (1969). Rhodopseudomonas acidophila, sp. n., a new species of the budding purple nonsulfur bacteria. J. Bacteriol. 99, 597–602.
Raghoebarsing, A. A., Smolders, A. J. P., Schmid, M. C., Rijpstra, W. I. C., Wolters-Arts, M., Derksen, J., Jetten, M. S. M., Schouten, S., Damste, J. S. S., Lamers, L. P. M., Roelofs, J. G. M., Op den Camp, H. J. M., and Strous, M. (2005). Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature 436, 1153–1156.
Rahman, M. T., Crombie, A., Chen, Y., Stralis-Pavese, N., Bodrossy, L., Meir, P., McNamara, N. P., and Murrell, C. J. (2011). Environmental distribution and abundance of the facultative methanotroph Methylocella. ISME J. 5, 1061–1066.
Rheims, H., Schumann, P., Rohde, M., and Stackebrandt, E. (1998). Verrucosispora gifhornensis gen. nov., sp. nov., a new member of the actinobacterial family Micromonosporaceae. Int. J. Syst. Evol. Microbiol. 48, 1119–1127.
Rice, A. V., Tsuneda, A., and Currah, R. S. (2006). In vitro decomposition of Sphagnum by some microfungi resembles white rot of wood. FEMS Microbiol. Ecol. 56, 372–382.
Scheffer, R. A., van Logtestijn, R.S.P., and Verhoeven, J. T. A. (2001). Decomposition of Carex and Sphagnum litter in two mesotrophic fens differing in dominant plant species. Oikos 92, 44–54.
Schlesner, H. (1994). The development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from various aquatic habitats using dilute media. Syst. Appl. Microbiol. 17, 135–145.
Sizova, M. V., Panikov, N. S., Spiridonova, E. M., Slobodova, N. V., and Tourova, T. P. (2007). Novel facultative anaerobic acidotolerant Telmatospirillum sibiriense gen. nov. sp. nov. isolated from mesotrophic fen. Syst. Appl. Microbiol. 30, 213–220.
Sizova, M. V., Panikov, N. S., Tourova, T. P., and Flanagan, P. W. (2003). Isolation and characterization of oligotrophic acido-tolerant methanogenic consortia from a Sphagnum peat bog. FEMS Microbiol. Ecol. 45, 301–315.
Smith, L. C., MacDonald, G. M., Velichko, A. A., Beilman, D. W., Borisova, O. K., Frey, K. E., Kremenetski, K. V., and Sheng, Y. (2004). Siberian peatlands a net carbon sink and global methane source since the early Holocene. Science 303, 353–356.
Stott, M. B., Crowe, M. A., Mountain, B. W., Smirnova, A. V., Hou, S., Alam, M., and Dunfield, P. F. (2008). Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ. Microbiol. 10, 2030–2041.
Theisen, A. R., Ali, M. H., Radajewski, S., Dumont, M. G., Dunfield, P. F., McDonald, I. R., Dedysh, S. N., Miguez, C. B., and Murrell, J. C. (2005). Regulation of methane oxidation in the facultative methanotroph Methylocella silvestris BL2. Mol. Microbiol. 58, 682–692.
Thormann, M. N., Bayley, S. E., and Currah, R. S. (2002). The relative ability of fungi from Sphagnum fuscum to decompose selected carbon sources. Can. J. Microbiol. 48, 204–211.
Thormann, M. N., Bayley, S. E., and Currah, R. S. (2004). Microcosm tests of the effects of temperature and microbial species number on the decomposition of Carex aquatilis and Sphagnum fuscum litter from southern boreal peatlands. Can. J. Microbiol. 50, 793–802.
Vandamme, P., Opelt, K., Knöchel, N., Berg, C., Schönmann, S., De Brandt, E., Eberl, L., Falsen, E., and Berg, G. (2007). Burkholderia bryophila sp. nov. and Burkholderia megapolitana sp. nov., moss-associated species with antifungal and plant-growth-promoting properties. Int. J. Syst. Evol. Microbiol. 57, 2228–2235.
Verhoeven, J. T. A., and Liefveld, W. M. (1997). The ecological significance of organochemical compounds in Sphagnum. Acta Bot. Neerl. 46, 117–130.
Vorobev, A. V., Baani, M., Doronina, N. V., Brady, A. L., Liesack, W., Dunfield, P. F., and Dedysh, S. N. (in press). Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium possessing only a soluble methane monooxygenase. Int. J. Syst. Evol. Microbiol.
Vorob’ev, A. V., de Boer, W., Folman, L. B., Bodelier, P. L. E., Doronina, N. V., Suzina, N. E., Trotsenko, Y. A., and Dedysh, S. N. (2009). Methylovirgula ligni gen. nov., sp. nov., an obligately acidophilic, facultatively methylotrophic bacterium with highly divergent mxaF gene. Int. J. Syst. Evol. Microbiol. 59, 2538–2545.
Ward, N. L., Challacombe, J. F., Janssen, P. H., Henrissat, B., Coutinho, P. M., Wu, M., Xie, G., Haft, D. H., Sait, M., Badger, J., Barabote, R. D., Bradley, B., Brettin, T. S., Brinkac, L. M., Bruce, D., Creasy, T., Daugherty, S. C., Davidsen, T. M., Deboy, R. T., Detter, J. C., Dodson, R. J., Durkin, A. S., Ganapathy, A., Gwinn-Giglio, M., Han, C. S., Khouri, H., Kiss, H., Kothari, S. P., Madupu, R., Nelson, K. E., Nelson, W. C., Paulsen, I., Penn, K., Ren, Q. H., Rosovitz, M. J., Selengut, J. D., Shrivastava, S., Sullivan, S. A., Tapia, R., Thompson, L. S., Watkins, K. L., Yang, Q., Yu, C. H., Zafar, N., Zhou, L. W., and Kuske, C. R. (2009). Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75, 2046–2056.
Williams, R. T., and Crawford, R. L. (1983). Microbial diversity of Minnesota peatlands. Microb. Ecol. 9, 201–214.
Keywords: northern wetlands, as-yet-uncultivated bacteria, isolation approaches, methanotrophs, Acidobacteria, Planctomycetes
Citation: Dedysh SN (2011) Cultivating uncultured bacteria from northern wetlands: knowledge gained and remaining gaps. Front. Microbio. 2:184. doi: 10.3389/fmicb.2011.00184
Received: 23 July 2011; Paper pending published: 05 August 2011;
Accepted: 19 August 2011; Published online: 16 September 2011.
Edited by:
Josh David Neufeld, University of Waterloo, CanadaReviewed by:
Marc Gregory Dumont, Max-Planck-Institute for Terrestrial Microbiology, GermanyLevente Bodrossy, CSIRO Marine and Atmospheric Research, Australia
Copyright: © 2011 Dedysh. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Svetlana N. Dedysh, Winogradsky Institute of Microbiology, Russian Academy of Sciences, Prospect 60-Letya Octyabrya 7/2, Moscow 117312, Russia. e-mail:ZGVkeXNoQG1haWwucnU=