- 1 Research Institute for Microbial Diseases, Osaka University, Osaka, Japan
- 2 Graduate School of Frontier Biosciences, Osaka University, Osaka, Japan
Type IV secretion systems (T4SSs) play a central role in the pathogenicity of many important pathogens, including Agrobacterium tumefaciens, Helicobacter pylori, and Legionella pneumophila. The T4SSs are related to bacterial conjugation systems, and are classified into two subgroups, type IVA (T4ASS) and type IVB (T4BSS). The T4BSS, which is closely related to conjugation systems of IncI plasmids, was originally found in human pathogen L. pneumophila; pathogenesis by L. pneumophila infection requires functional Dot/Icm T4BSS. A zoonotic pathogen, Coxiella burnetii, and an arthropod pathogen, Rickettsiella grylli – both of which carry T4BSSs highly similar to the Legionella Dot/Icm system – are evolutionarily closely related and comprise a monophyletic group. A growing body of bacterial genomic information now suggests that T4BSSs are not limited to Legionella and related bacteria and IncI plasmids. Here, we review the current knowledge on T4BSS apparatus and component proteins, gained mainly from studies on L. pneumophila Dot/Icm T4BSS. Recent structural studies, along with previous findings, suggest that the Dot/Icm T4BSS contains components with primary or higher-order structures similar to those in other types of secretion systems – types II, III, IVA, and VI, thus highlighting the mosaic nature of T4BSS architecture.
Introduction
Legionella pneumophila is the causative agent of the acute pneumonia known as legionellosis or Legionnaires’ disease. The genus Legionella was established in 1979 after a large outbreak at the American Legion convention in Philadelphia 3 years earlier (Brenner et al., 1979). L. pneumophila enters eukaryotic host cells using the host cells’ own mechanisms: phagocytosis or macropinocytosis (Horwitz, 1984; Watarai et al., 2001b; Peracino et al., 2010). Early studies by Marcus Horwitz and his colleagues revealed that the Legionella-containing vacuoles (LCVs) escape from endocytic maturation processes, including the acidification of LCVs and LCV–lysosome fusion (Horwitz, 1983; Horwitz and Maxfield, 1984). The LCVs acquire endoplasmic reticulum (ER)-like properties over time and L. pneumophila multiply within the resulting replicative niche (Horwitz and Silverstein, 1980; Swanson and Isberg, 1995; Tilney et al., 2001).
By 1998, taking advantage of the available forward genetic approach, Ralph Isberg’s and Howard Shuman’s laboratories independently discovered ∼20 L. pneumophila genes that are required for the establishment of the replicative niche, intracellular replication, or macrophage killing (Berger et al., 1994; Brand et al., 1994; Segal and Shuman, 1997; Andrews et al., 1998; Purcell and Shuman, 1998; Segal et al., 1998; Vogel et al., 1998). These genes have been named independently by the two groups: dot (for defect in organelle trafficking) or icm (for intracellular multiplication). The dot/icm genes were believed to encode a type IV secretion system (T4SSs) – defined as bacterial macromolecular transport systems closely related to conjugation systems – because (a) several Dot/Icm proteins have limited sequence-level similarity to components of conjugation systems; and (b) L. pneumophila has the Dot/Icm-dependent ability to mediate the conjugal transfer of IncQ plasmids (Segal and Shuman, 1997; Segal et al., 1998; Vogel et al., 1998). When the nucleotide sequences of IncI plasmids colIb-P9 and R64 became available to the public in 1999–2000, it became obvious that the dot/icm genes are closely related to the tra/trb genes of these IncI plasmids (Segal and Shuman, 1999b; Komano et al., 2000; Wilkins and Thomas, 2000). It had been known that T4SSs play central roles as DNA or protein transporters in the pathogenicity of many important pathogens, including Agrobacterium tumefaciens, Bordetella pertussis, Brucella species, and Helicobacter pylori. In 2002, L. pneumophila was shown to deliver a protein substrate RalF to the host cell cytosol using the Dot/Icm system; this established that the Dot/Icm system can translocate effector proteins (Nagai et al., 2002). Since then, over a hundred L. pneumophila proteins have been experimentally shown to be translocated via the Dot/Icm system (Hubber and Roy, 2010 as a review). Together, the dot/icm genes encode a T4SS classified as type IVB, which is closely related to the I-type conjugation systems (Lawley et al., 2003), but is distinct from the conventional T4SSs now classified as type IVA (Christie and Vogel, 2000; Sexton and Vogel, 2002). Hereafter in this article, we use the term “type IVB secretion system (T4BSS)” to mean the secretion/conjugation system family closely related to the Legionella Dot/Icm system and the I-type conjugation system of IncI conjugal plasmids, unless otherwise indicated.
Transport substrate proteins, including VirE2 and VirF of the Agrobacterium VirB/D type IVA secretion system (T4ASS), have C-terminal translocation signals (Vergunst et al., 2000, 2005). Likewise, RalF carries a C-terminal disordered region necessary for translocation via the Dot/Icm system (Amor et al., 2005; Nagai et al., 2005). The C-terminal signal hypothesis has been confirmed by studies on a number of L. pneumophila effector proteins.
Coxiella burnetii is a zoonotic pathogen and the causative agent of human Q-fever. Like L. pneumophila, C. burnetii establishes a specialized replicative compartment within host cells; the properties of lysosome-derived Coxiella-containing vacuoles are distinct from those of ER-derived LCVs. C. burnetii carries genes closely related to the L. pneumophila dot/icm genes (Segal and Shuman, 1999b; Sexton and Vogel, 2002; Seshadri et al., 2003). The C. burnetii Dot/Icm proteins, including DotB, IcmW, and IcmS, have been shown to be able to functionally substitute for their L. pneumophila counterparts in intracellular replication; this implies that the Coxiella Dot/Icm system is functional and plays a critical role in interactions with its host cells (Zamboni et al., 2003; Zusman et al., 2003). Until recently, C. burnetii was believed to be an obligate pathogen (Omsland et al., 2009). L. pneumophila has been successfully employed as a surrogate host to identify and analyze C. burnetii effector proteins (Pan et al., 2008; Voth et al., 2009; Chen et al., 2010).
Bacteria of the genus Rickettsiella are obligate intracellular pathogens of a wide variety of arthropods. A phylogenetic study of Rickettsiella popilliae and Rickettsiella grylli demonstrated that these bacteria carry genes orthologous to dotB, dotO/icmB, and icmQ (Leclerque and Kleespies, 2008). Analysis of the published draft genome sequence of R. grylli (GenBank accession no. NZ_AAQJ02000001) demonstrated that R. grylli encodes a nearly full set of the dot/icm genes on its chromosome (Figure 1). A phylogenetic analysis using 16S rRNA sequences placed R. grylli as the nearest neighbor of C. burnetii, under the family Coxiellaceae (Roux et al., 1997). The families Legionellaceae and Coxiellaceae are the only members of the order Legionellales, showing the intracellular pathogens Legionella, Coxiella, and Rickettsiella to be evolutionarily closely related bacteria, and comprise a monophyletic group.
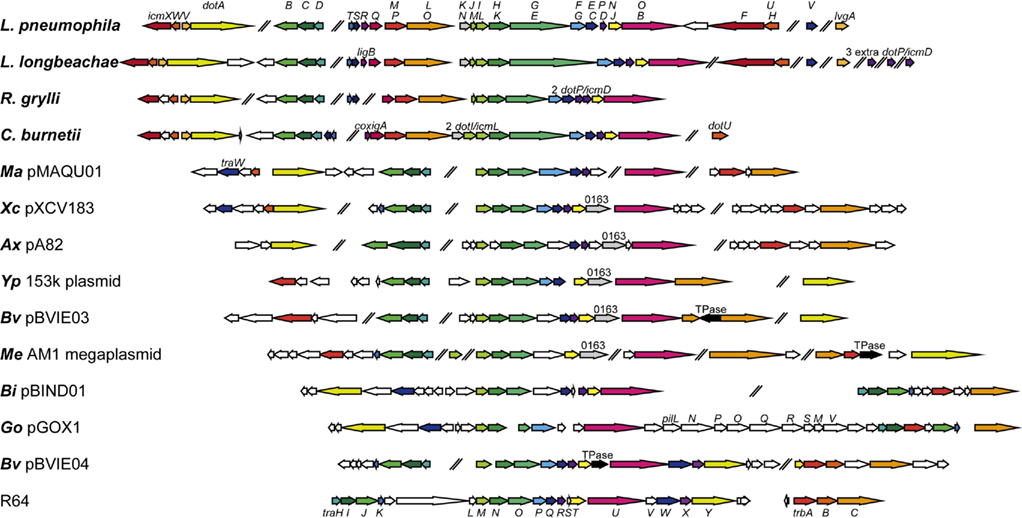
Figure 1. Genetic organizations of selected T4BSSs. Genetic organizations of T4BSSs from the following bacteria or plasmids are illustrated. Legionella pneumophila strain Philadelphia 1 (GenBank accession no. NC_002942); Legionella longbeachae NSW150 (NC_013861); Rickettsiella grylli (NZ_AAQJ02000001); Coxiella burnetii RSA 493 (NC_002971); Marinobacter aquaeolei VT8 pMAQU01 (NC_008738); Xanthomonas campestris pv. vesicatoria str. 85-10 pXCV183 (NC_007507); Achromobacter xylosoxidans A8 pA82 (NC_014642); Yersinia pseudotuberculosis IP 31758 153 kbp plasmid (NC_009705); Burkholderia vietnamiensis G4 pBVIE03 (NC_009229); Methylobacterium extorquens AM1 megaplasmid (NC_012811); Beijerinckia indica subsp. indica ATCC 9039 pBIND01 (NC_012811); Gluconobacter oxydans 621H pGOX1 (NC_006672); Burkholderia vietnamiensis G4 pBVIE04 (NC_009228), and IncI plasmid R64 (NC_005014). ORFs designated as “0163” are conserved in several T4BSSs but not in Legionella, Coxiella, Rickettsiella, or R64. ORFs designated as “TPase” are putative transposase derivatives. Notably, B. vietnamiensis G4 harbors multiple plasmids that carry distinct T4BSSs.
Genetic Organizations of T4BSSs
We noticed that the current release of the BLAST non-redundant protein database (nr) contains quite a few proteins from various pathogenic and environmental bacteria that have significant similarity to L. pneumophila Dot/Icm proteins. This tempted us to conduct phylogenetic analyses of these Dot/Icm-related proteins. We constructed a phylogenetic tree of a C-terminal domain of DotG/IcmE (862–1046; Figure 2), because DotG is supposed to be a central core component of the T4BSS, and the DotG domain is conserved in T4BSSs and T4ASSs (see below for details). Legionella species and their plasmids have been shown to encode T4ASSs (Lvh and Trb), genomic-island associated T4SSs (GI or LGI) which may function as integrative and conjugative elements (ICEs), and/or F-type conjugation systems (Segal et al., 1999; Cazalet et al., 2004, 2010; Chien et al., 2004; Glockner et al., 2008; D’Auria et al., 2010; Kozak et al., 2010; Schroeder et al., 2010). The proteins of these distinct systems are distantly homologous to DotG and are suitable as outgroups in the phylogenetic analysis. DotG of Legionella, Coxiella, and Rickettsiella, as well as DotG-homolog TraO of various plasmids, form distinct monophyletic clades. In addition to these, a dozen proteins from a wide variety of proteobacteria are placed in the major clade of DotG/TraO, distinct from outgroups. Similar results were obtained from a phylogenetic analysis using secretion ATPase DotB (data not shown). We looked closely at the genomic sequences of bacteria that encode DotG- and DotB-like proteins and whose genome projects have been completed, to identify other T4BSS components; in Figure 1, we have illustrated their genetic organizations.

Figure 2. A phylogenetic tree of DotG/IcmE862–1046. Proteins that have regions homologous to DotG/IcmE862–1046 were selected by multiple rounds of PSIBLAST (Altschul et al., 1997) using the non-redundant protein database (nr), as of November 30, 2010. Legionella proteins homologous to Ti plasmid VirB10, RP4 plasmid TrbI and F plasmid TraB were incorporated in the analysis as outgroups. The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The bootstrap consensus tree inferred from 500 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site. Evolutionary analyses were conducted in MEGA4 (Tamura et al., 2007).
Interestingly, nearly all the T4BSSs found in the analysis are encoded on plasmids. Notable exceptions include Legionella, Coxiella, and Rickettsiella Dot/Icm systems. It is most likely that a common ancestor of these closely related bacteria acquired a T4BSS on its chromosome, and that the T4BSS played a critical role in survival of the ancestor. The T4BSS acquisition on chromosome might be related to the alteration of life style as intracellular bacterium. Genes encoding T4BSS tend to gather in several conserved gene clusters; it appears that there is little pressure to keep them in a single locus. The conserved gene clusters include (a) dotD–dotC–dotB (traH–traI–traJ in I-type conjugation systems), (b) dotM/icmP–dotL/icmO (trbA–trbC), and (c) dotI/icmL–dotH/icmK–dotG/icmE (traM–traN–traO). Together with other genes found in all T4BSSs, including dotA (traY) and dotO/icmB (traU), these conserved genes are expected to encode core components that play fundamental roles in transport activity. On the other hand, the genes found only in the Dot/Icm systems of Legionella and related bacteria may encode components that are important for life as intracellular pathogens. In the following section, we discuss T4BSS component proteins in detail, taking advantages of developing genomic information and structural insights. We do not intend to thoroughly review the type IVB effector proteins and the regulation of T4BSSs here. Please refer to excellent reviews recently published on these subjects (Segal et al., 2005; Ninio and Roy, 2007; Shin and Roy, 2008; Franco et al., 2009; Isberg et al., 2009; Hubber and Roy, 2010).
Core Components of T4BSSs
The Putative Core Complex of the L. pneumophila Dot/Icm System
In 2006, Joseph Vogel and his colleague reported an excellent systematic study on component proteins of the Dot/Icm system (Vincent et al., 2006b). Notably, they demonstrated that DotC, DotD, DotF/IcmG, DotG/IcmE, and DotH/IcmK were fractionated into outer membrane fractions. DotC and DotD are lipoproteins and sorted to outer membranes even in the absence of other Dot/Icm components. DotH is localized to the outer membranes, which requires the lipoproteins DotC and DotD. Thus, DotC and DotD appear to behave as a pilotin for DotH. DotF and DotG are intrinsically inner membrane proteins with single transmembrane helices. In wild-type L. pneumophila, DotF and DotG are fractionated into both inner and outer membrane fractions. The outer membrane fractionation of DotF and DotG requires the DotC, DotD, and DotH. These results suggest the presence of a complex spanning both inner and outer membranes that contains these five proteins, DotC, DotD, DotF, DotG, and DotH (Figure 3).
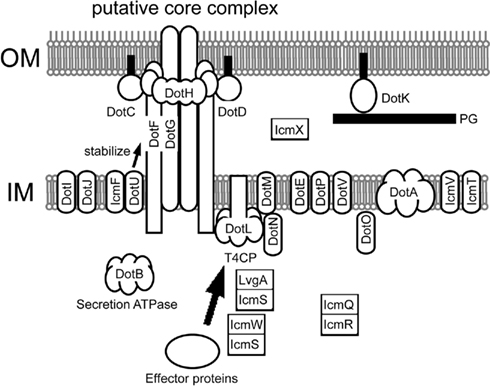
Figure 3. Legionella pneumophila Dot/Icm T4BSS. The putative core complex containing DotC, DotD, DotH, DotG, and DotF was suggested by Vincent et al. (2006b). A possible scenario of its assembly is as follows: (1) outer membrane lipoproteins DotC and DotD recruit intrinsic periplasmic protein DotH to the outer membrane, thus forming a DotC–DotD–DotH outer membrane complex; (2) the C-terminal domain of DotG participates in the outer membrane complex, resulting in a complex spanning both inner and outer membranes; and (3) DotF participates in the core complex by binding to DotG and/or the DotC–DotD–DotH complex. Subcellular localization of Dot/Icm proteins are depicted based on lines of experimental evidence (Roy and Isberg, 1997; Zuckman et al., 1999; Coers et al., 2000; Matthews and Roy, 2000; Sexton et al., 2004a,2004b; Vincent et al., 2006b), or prediction from amino acid sequences (DotE, DotJ, DotV, and IcmT).
DotG/IcmE: The Central Component of the Core Complex?
In 2009, major advances in structural research on T4ASS were achieved by Gabriel Waksman and his colleagues. They biochemically isolated the core complex spanning both inner and outer membranes from the conjugation system of an IncN plasmid pKM101 (Figure 4A; Fronzes et al., 2009). Furthermore, they isolated the outer membrane complex from the core complex treated with trypsin, and solved the crystal structure of the outer membrane complex (Figure 4B; Chandran et al., 2009). The core complex has 14-fold rotation symmetry and contains three proteins, VirB7, VirB9, and VirB10, at a 1:1:1 molar ratio. In the outer membrane complex, VirB10 faces to the central cavity and two alpha helices from each monomer are inserted into the outer membrane. VirB9 constitutes the outer lobe. VirB7 takes an extended form and wraps around the complex.
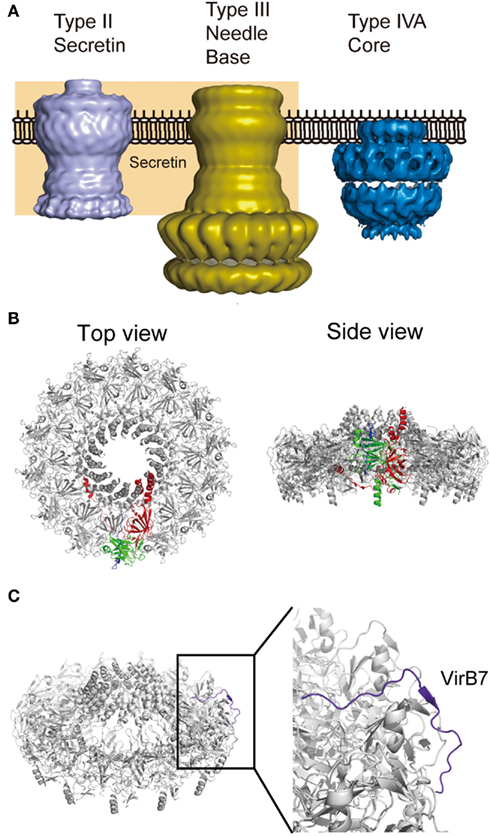
Figure 4. Core complex of pKM101 T4ASS. (A) Comparison of electron micrographic structures of Vibrio cholera secretin GspD (type II secretin; EMDB accession EMD-1763; Reichow et al., 2010), type III injectisome isolated from ΔinvJ Salmonella typhimurium (type III needle base; EMD-1224; Marlovits et al., 2006), and T4ASS core complex of pKM101 conjugal plasmid (type IVA core; EMD-5031; Fronzes et al., 2009). (B) Top and side views of pKM101 outer membrane complex (PDB accession 3JQO; Chandran et al., 2009). One of each protomer in the complex is shown in color: VirB7 (blue), VirB9 (green), and VirB10 (red). (C) VirB7 takes an extended form in the complex. Figures are generated using PyMol (Schrodinger, 2010) and resources deposited to indicated databases.
DotG is an integral membrane protein with single transmembrane helix in an N-terminal region. As described above, the C-terminal region of DotG (862–1046) is well conserved in T4BSSs including I-type conjugation systems (TraO). However, the sizes of DotG family proteins significantly vary: Legionella, Coxiella and Rickettsiella DotG proteins are significantly larger than other siblings because of a variable region, which often contains penta-peptide repeats (Segal et al., 1998), between the N-terminal transmembrane and the C-terminal conserved regions. As previously suggested (Segal et al., 1998; Vogel et al., 1998), the C-terminal conserved region of DotG is significantly similar to the TrbI domain (Pfam PF03743) found in VirB10 family proteins of T4ASSs. Interestingly, the VirB10 region in the outer membrane complex of the pKM101 conjugation system corresponds nicely to the Pfam TrbI domain. The size variation of VirB10 family proteins is documented as well; for instance, Cag7/CagY of the Helicobacter cagPAI-associated T4ASS is far larger than the Agrobacterium VirB10 (Liu et al., 1999). These suggest that T4BSS has an outer membrane complex similar to that of T4ASS, and DotG is the counterpart of VirB10 in the outer membrane complex.
DotC, DotD, and DotH/IcmK
Besides VirB10, the core complex of T4ASS also contains VirB7 and VirB9. Agrobacterium VirB7 is an outer membrane lipoprotein; it forms a heterodimer with VirB9 and stabilizes several VirB proteins, including VirB9 (Fernandez et al., 1996). None of T4BSS proteins has detectable sequence-level similarity to T4ASS VirB7 and VirB9. Possible candidates for T4BSS counterparts of these VirB proteins include DotC, DotD, and DotH. Like DotG, DotC, DotD, and DotH are well conserved in T4BSSs, including I-type conjugation systems (TraI, TraH, and TraN, respectively). However, the genes encoding DotH and DotG, and the genes encoding DotC and DotD are often found in separate gene clusters. In contrast, the genes encoding VirB7, VirB9, and VirB10 are typically found in single gene clusters of T4ASSs.
DotC and DotD are outer membrane lipoproteins required for the outer membrane targeting of DotH (Vincent et al., 2006b). L. pneumophila strains that produce lipidation-site cysteine mutant of DotC or DotD are partially defective in intracellular growth (Yerushalmi et al., 2005). The defects due to these mutations are additive, which suggest a genetic interaction between DotC and DotD. DotD consists of a disordered N-terminal domain and a globular C-terminal domain (Nakano et al., 2010). The crystal structure of the C-terminal domain is remarkably similar to the N-terminal subdomain of secretins, whereas these domain/subdomains are poorly related to each other at the amino acid sequence-level (Nakano et al., 2010; Figure 5). Secretins form a protein family that participates in several macromolecule translocation processes across bacterial outer membranes, notably type II and type III secretion (Genin and Boucher, 1994; Hardie et al., 1996). Secretins are integral outer membrane proteins that form substrate conduits. The protease-resistant C-terminal domain of secretins forms rings with 12- or 14-fold rotation symmetry (Opalka et al., 2003; Collins et al., 2004; Chami et al., 2005), and is embedded into the outer membrane. The N-terminal region of secretins extends into the periplasm and may interact with inner membrane partners as well as substrates. The periplasmic domain of secretins contains a most N-terminal DotD-like subdomain, followed by often-repeated Secretin_N domain(s) (Pfam 03958). Crystal structures of periplasmic domains of enterotoxigenic Escherichia coli (ETEC) GspD and enteropathogenic E. coli (EPEC) EscC secretins, from type II and type III secretion systems, respectively, containing the N-terminal DotD-like subdomains, were not captured as multimers of cylindrical shape (Korotkov et al., 2009; Spreter et al., 2009). Recently cryo-EM structure of dodecameric full-length type II secretin GspD from Vibrio cholera has been reported (Figures 4A and 5D; Reichow et al., 2010). The periplasmic domain of V. cholera GspD forms a vestibule, which binds to the substrate cholera toxin and tip of pseudopilins. These findings imply that the C-terminal domain of DotD may form a periplasmic ring that is a part of the T4BSS core complex (Figure 5D). It should be noted that there is no counterpart of the putative DotD ring in the pKM101 core complex.
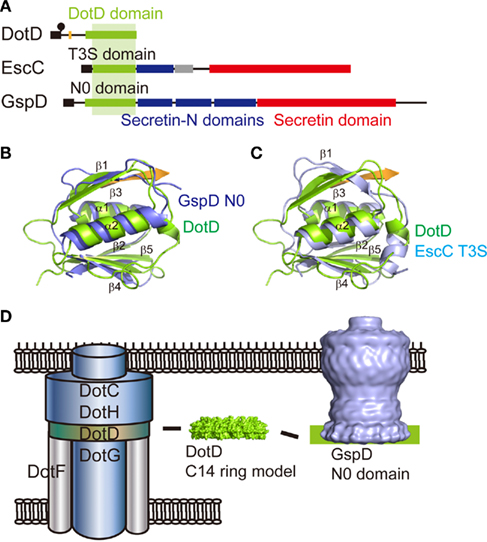
Figure 5. Comparison of the C-terminal domain of DotD with secretin periplasmic subdomains. (A) Domain organizations of L. pneumophila DotD (Nakano et al., 2010), EPEC secretin EscC (Spreter et al., 2009), and ETEC secretin GspD (Korotkov et al., 2009). (B) DotD (green, PDB accession 3ADY) superimposed onto the N0 domain of ETEC secretin GspD (blue, PDB 3EZJ). (C) DotD (green) superimposed onto the T3S domain of EPEC secretin EscC (light blue, PDB 3GR5). (D) A model of T4BSS core complex. DotD may form a periplasmic ring, like the N0 domain of type II secretin.
Assuming that the core complex of T4BSS carries the VirB9 counterpart, DotH is the strongest candidate. Both proteins are recruited to outer membranes, probably through interaction with cognate outer membrane lipoproteins. Structural analyses indicated that VirB9 is rich in beta-strands (Bayliss et al., 2007; Chandran et al., 2009). Similarly, the protease-resistant C-terminal domain, representing about two thirds of DotH, is predicted to be rich in beta-strands, using PHDsec (our unpublished results). Surface exposure of residues Asn-226, Pro-227, and Asp-228 of Agrobacterium VirB9 has been reported (Bayliss et al., 2007). Likewise, the surface exposure of DotH in L. pneumophila treated in certain conditions has been reported (Watarai et al., 2001a). Along these lines, the N-terminal disordered domain of DotD may serve as the VirB7 counterpart. The VirB7 in the pKM101 outer membrane complex takes an extended conformation and wraps around the complex (Figure 4C; Chandran et al., 2009). Mature VirB7 of pKM101 is a small peptide of 33 amino acids long, comparable in size to the N-terminal disordered region of DotD (46 amino acids), which may interact with outer membrane components such as DotH. Notably, some VirB7 family T4ASS proteins including Helicobacter CagT are significantly larger than Agrobacterium VirB7, having an extra C-terminal region whose function is not known (Alvarez-Martinez and Christie, 2009).
Alternatively, DotC and DotH may structurally correspond to the Secretin_N and Secretin domains, respectively. In this case, DotD, DotC, and DotH form an outer membrane complex functionally equivalent to secretins. The DotG outer membrane complex may associate with the secretin-like DotC–DotD–DotH complex. Clearly, further studies are called to clarify the nature of the T4BSS core complex.
DotF/IcmG
DotF is a ∼30 kDa protein composed of a small cytoplasmic domain, a transmembrane domain, and a large periplasmic domain. The periplasmic domain contains a putative coiled-coil region that is potentially responsible for protein–protein interaction, such as self-association and interaction with DotG (Vincent et al., 2006b). The DotF homologs can be found in T4BSSs, including I-type conjugation systems (TraP), whereas some T4BSSs have no protein significantly similar to DotF. Furthermore, even among homologous proteins, the region of similarity is limited mostly to C-terminal regions. Consistently, unlike other components of the putative core complex, DotF appears to be not essential for full activities of the Dot/Icm system. An internal deletion/kanamycin-cassette insertion dotF mutant (icmG635::Kan) was shown to be partially cytotoxic to HL-60 cells (Purcell and Shuman, 1998). This dotF mutant is not able to replicate within a protozoan host, Acanthamoeba castellanii, but shows only a partial defect in growth within HL-60 cells (Segal and Shuman, 1999a). Similar results were obtained using an in-frame dotF deletion strain (our unpublished results). Two-hybrid interactions of DotF and several effector proteins, including RalF, LidA, and Sid proteins, have been reported (Luo and Isberg, 2004). This raises the possibility that DotF is somehow involved in substrate recognition or signal transduction from the transport substrates to the T4BSS machinery.
DotI/IcmL and DotJ/IcmM
DotI and DotJ are closely related integral inner membrane proteins, essential to Dot/Icm-dependent activities. Both proteins carry single transmembrane helices in N-terminal conserved regions. DotJ consists only of the N-terminal conserved region. DotI has an extra periplasmic domain. DotI is conserved in all T4BSSs including I-type conjugation systems (TraM). Legionella species encode multiple additional DotI-related proteins whose functions are not known. C. burnetii appears to have only the DotI-type protein but in two copies. R. grylli has both DotI- and DotJ-types of proteins in terms of domain organization, but the N-terminal conserved regions of both proteins are more closely related to that of DotI than that of DotJ. Other T4BSSs have only the DotI-type proteins. These suggest that the gene duplication of dotI occurred in a common ancestor of Legionella, Coxiella, and Rickettsiella, and the DotJ-type protein was evolved after species differentiation.
The gene encoding DotI is associated with the genes encoding core components DotH and DotG, and the gene order dotI–dotH–dotG appears to be well conserved in T4BSSs. These suggest pivotal roles of the DotI in T4BSS activities. DotI (and DotJ) may form an inner membrane complex that associates with the core complex.
DotE/IcmC, DotV, and DotP/IcmD
DotE and DotV are closely related small integral inner membrane proteins having four transmembrane helices. TraQ, of the I-type conjugation system, has the same domain organization as DotE and DotV, but the sequence-level similarity, if any, between them is difficult to detect by homology search programs (BLAST, e.g.). Proteins with domain organization similar to DotE/DotV are found in most T4BSSs; the genes are typically located immediately downstream of dotF. DotV is only found in Legionella species.
DotP, which has sequence-level similarity to DotE/DotV, appears to be a shorter version of DotE/DotV, having two transmembrane helices. As with TraQ, TraR of the I-type conjugation system has the same domain organization as DotP, but poor sequence-level similarity. Proteins having similar domain organization to DotP are found in most T4BSSs; the genes are typically located immediately downstream of dotE. In addition to the two transmembrane helices, DotP is predicted to have a cleavable signal sequence at the N-terminus (Purcell and Shuman, 1998). Interestingly, multiplication of the gene encoding DotP is found in L. longbeachae and R. grylli genomes.
DotO/IcmB
DotO is a large protein associated with all T4BSSs, including I-like conjugation systems (TraU). The DotO family protein is distantly homologous to VirB4 of T4ASS. Like VirB4, DotO has conserved Walker motifs for nucleotide binding (Purcell and Shuman, 1998). A cellular localization study showed L. pneumophila DotO to be targeted to the inner membrane (Vincent et al., 2006b). The surface exposure of DotO was reported in L. pneumophila that had been treated under the same conditions as those in which DotH was surface-exposed (Watarai et al., 2001a). The specific function of DotO, however, remains unclarified.
DotL/IcmO, DotM/IcmP, and DotN/IcmJ: T4CP and Its Partners
DotL is a member of the type IV coupling protein family (T4CP). T4CPs are associated with nearly all type IV secretion and conjugation systems, and are related to the FtsK/SpoIIIJ family DNA motor proteins (Errington et al., 2001; Aussel et al., 2002; Massey et al., 2006). The typical T4CP has transmembrane helices at its N-terminus, followed by a large cytoplasmic domain. The cytoplasmic domain carries conserved Walker motifs and forms a hexamer ring, which is anchored to the inner membranes via the N-terminal transmembrane helices (Gomis-Ruth et al., 2001, 2002). Studies on conjugation systems indicate that T4CP interacts with a nucleoprotein complex called relaxosome (de la Cruz et al., 2010). The relaxosome contains a protein called relaxase, which is a bona fide protein substrate; relaxase is translocated into recipient cells even in the absence of trailing DNA (Draper et al., 2005; Lang et al., 2010). These suggest that T4CP links protein substrates as well as DNA substrates to membrane-embedded transport apparatus of T4SSs.
T4BSSs are also associated with T4CPs DotL (TrbC in I-type conjugation systems). The genes encoding DotL are often coupled with the genes encoding DotM (TrbA), whereas the homolog of TrbB of I-type conjugation systems is missing in the Dot/Icm systems of Legionella and related bacteria. DotL is essential for viability of L. pneumophila strain Lp02 (Buscher et al., 2005). This phenomenon is strain-dependent: DotL is not essential for the viability of L. pneumophila strain JR32, a derivative of strain Philadelphia 1, as Lp02 is (Buscher et al., 2005). Transposon-inserted suppressor mutations of lethality through DotL disruption were mapped in several dot/icm genes, suggesting that lethality requires a functional Dot/Icm system (Buscher et al., 2005). Insertion mutants of DjlA (DnaJ-like protein) were also identified as suppressors for the lethality phenotype (Vincent et al., 2006a). DjlA mutants of Legionella species have been shown to be severely defective in intracellular growth (Ohnishi et al., 2004; Vincent et al., 2006a). These suggest involvement of the DnaK chaperone system in the assembly/quality control of the Dot/Icm apparatus. Interestingly, DotM and a cytoplasmic/inner membrane protein, DotN, showed the same essentiality as DotL, suggesting genetic interactions between these proteins (Buscher et al., 2005). Furthermore, DotL and DotM proteins are destabilized in Lp02-derived strains lacking DotL, DotM, or DotN, suggesting biochemical interactions between these proteins (Vincent et al., 2006b).
DotB: Secretion ATPase Possibly Originated from the Type IV Pilus Biogenesis System
DotB is another protein in T4BSS that carries conserved Walker motifs. Purified DotB is a hexametric ATPase in a ring shape, the activity of which is essential to Dot/Icm-dependent activities (Sexton et al., 2004b, 2005). The formation of the ring structure does not require ATP binding/hydrolysis. In L. pneumophila cells the majority of DotB was found to be cytoplasmic, while small amounts of DotB were recovered in inner membrane fractions.
Secretion ATPases from type II and type IV secretion systems are well conserved at the sequence-level. DotB is no exception; DotB orthologs are well conserved in T4BSSs, including I-type conjugation systems (TraJ). Interestingly, phylogenetic analyses clearly indicate that DotB has a closer relationship to PilT, the ATPase involved in the retraction of type IV pili, than to VirB11 of T4ASS (Planet et al., 2001). Type IV pilus biogenesis systems are closely related to the type II secretion system, and DotB is found in the major clade to which ATPases from type II secretion and type IV pilus biogenesis systems belong. In this connection, it is notable that many plasmids harboring T4BSSs carry type IV pilus biogenesis systems as well. Of particular interest, the gene encoding DotO and the pil genes encoding a type IV pilus biogenesis system of Gluconobacter oxydans pGOX1 comprise a single transcription unit (Figure 1). The common ancestor of the genes encoding DotB might originate from the co-existing type IV pilus biogenesis system. In summary, phylogenetic analyses clearly indicate that at least one component of T4BSS has a distinct origin from T4ASS, which highlights the mosaic nature of T4BSS architecture.
IcmT
IcmT is a small integral inner membrane protein. IcmT orthologs can be found in most T4BSSs, including I-type conjugation systems (TraK). The gene encoding IcmT is often associated with the gene cluster dotD–dotC–dotB. IcmT is essential for Dot/Icm-dependent activities, but its specific function remains to be clarified.
DotA
DotA is one of the most mysterious components of T4BSSs. DotA is required for Dot/Icm-dependent activities; historically, defective DotA mutants have been frequently used in studies of the Dot/Icm system with regard to pathogenicity of L. pneumophila. The DotA in L. pneumophila is an integral inner membrane protein composed of a cleavable signal sequence, seven transmembrane helices, a large periplasmic domain and a small cytoplasmic C-terminal domain (Roy and Isberg, 1997). DotA is well conserved at sequence-level in T4BSSs, including I-like conjugation systems (TraY), but the large periplasmic domain appears to be specific to DotA orthologs in Legionella species. It should be emphasized that cleavable signal sequences of integral inner membrane proteins is rarely found in prokaryotes. More surprisingly, DotA is somehow secreted into the extracellular milieu from culture-grown L. pneumophila in a Dot/Icm-dependent fashion (Nagai and Roy, 2001). The extracellular DotA forms ring-like oligomers with unknown 46 kDa protein. The elucidation of the specific function of DotA awaits future studies.
Components Only Found in Dot/Icm Systems of Legionella and Closely Related Bacteria
IcmF and DotU/IcmH
IcmF and DotU were originally identified as components of the L. pneumophila Dot/Icm system; most T4BSSs lack them, with the notable exception of those in Legionella species. Homologous proteins to IcmF and DotU are prevalent in a wide variety of proteobacteria, while genes encoding them are associated with gene clusters encoding the conserved IcmF-associated homologous proteins (IAHPs). Now it has been well established that these IcmF and DotU homologs are components of the type VI secretion system (Cascales, 2008). L. pneumophila IcmF and DotU are partially required for Dot/Icm-dependent activities (Sexton et al., 2004a; VanRheenen et al., 2004; Zusman et al., 2004). The loss of IcmF or DotU results in decreased amounts of core components, most notably DotH and DotG (Sexton et al., 2004a). Moreover, the overexpression of DotH was shown to suppress defects in intracellular replication of the double-deletion mutant of IcmF and DotU. This suggests that IcmF and DotU work together to stabilize the core complex of the Dot/Icm system (Sexton et al., 2004a).
IcmW, IcmS, and LvgA
IcmW, IcmS, and LvgA are small acidic cytoplasmic proteins partially required for Dot/Icm-dependent activities (Zuckman et al., 1999; Coers et al., 2000; Vincent and Vogel, 2006; Vincent et al., 2006b). These proteins are destabilized in L. pneumophila mutant strains lacking IcmW, IcmS, or LvgA, suggesting interactions between them (Vincent and Vogel, 2006; Vincent et al., 2006b). Binary complexes of IcmW–IcmS and of IcmS–LvgA have been consistently reported (Ninio et al., 2005; Vincent and Vogel, 2006; Cambronne and Roy, 2007). It is not clear if the ternary complex exists. The physical properties of these proteins mimic those of transport chaperones of type III secretion systems, which are often associated with cognate type III effector proteins and required for their translocation and/or stability in bacterial cells. This prompted speculation that these proteins may interact with effector proteins like the type III chaperones; several effector proteins, including WipA, SdeA, SidH, and SidG, were identified as proteins that interact with IcmW and/or IcmS (Bardill et al., 2005; Ninio et al., 2005; Cambronne and Roy, 2007). A detailed study on the effector protein SidG showed that the IcmW–IcmS complex binds to a distinct region of SidG from its C-terminal translocation signal, which probably results in a conformational change that facilitates recognition of the C-terminal translocation signal by the Dot/Icm machinery (Cambronne and Roy, 2007). Decreased levels of DotL and DotM have been seen in IcmW or IcmS mutants, which suggest potential interactions between DotL–DotM and IcmW–IcmS (Vincent et al., 2006b). The IcmW–IcmS complex may therefore play a role in recruitment of effector proteins to the transport apparatus (Figure 3).
Interestingly, IcmS is only found in Legionella, Coxiella, and Rickettsiella Dot/Icm systems, whereas IcmW is distributed beyond this scope; IcmW orthologs are found in Marinobacter aquaeolei VT8 pMAQU01 and Xanthomonas campestris pv. vesicatoria str. 85-10 pXCV183 as well. LvgA orthologs are found only in Legionella species. Taken into account their possible function facilitating substrate translocation, and the large numbers of effector proteins that L. pneumophila are believed to translocate, it is tempting to speculate that these proteins evolved to meet the increasing demands to translocate a wide variety of effector proteins as intracellular pathogens.
IcmQ and IcmR
IcmQ and IcmR are cytoplasmic proteins required for Dot/Icm-dependent activities (Coers et al., 2000). IcmQ is composed of an N-terminal domain, a short linker domain, and a large C-terminal domain. Purified IcmQ tends to aggregate, which can be prevented by the addition of purified IcmR (Dumenil and Isberg, 2001). This suggests a chaperone–substrate kind of relationship between IcmR–IcmQ. Purified IcmQ associates with synthetic lipid vesicles, leading to vesicle disruption, as evidenced by the release of preloaded calcein dye (Dumenil et al., 2004). The C-terminal domain plays a primary role in membrane targeting mediated by electrostatic interactions, while the N-terminal domain may be inserted into lipid bilayers and disrupts membranes (Dumenil et al., 2004; Raychaudhury et al., 2009). The N-terminal domain also binds to IcmR, which prevents IcmQ from the stable association with lipid vesicles. Consistently in L. pneumophila lacking IcmR, significant amounts of IcmQ are localized to membrane fractions (Dumenil et al., 2004). Thus IcmR may have a regulatory function on IcmQ. The site of function as well as the specific function of IcmQ is unclear.
While IcmQ orthologs are found in Legionella, Coxiella, and Rickettsiella Dot/Icm systems, the situation regarding IcmR is more complicated. Proteins homologous to IcmR are found only in L. pneumophila. In other bacteria closely related to L. pneumophila, the genes located at immediately upstream of the genes encoding IcmQ encode proteins functionally equivalent, but poorly related, to IcmR (so-called FIRs). It has been reported that various Legionella species (other than L. pneumophila) and C. burnetii carry FIRs which bind to cognate IcmQs (Feldman and Segal, 2004; Feldman et al., 2005). Structural analysis of the complex of the N-terminal domain of IcmQ (Qn) and IcmR provides insights into the molecular basis of the IcmQ–FIR interaction (Raychaudhury et al., 2009). The Qn–IcmR complex forms a four-helix bundle – two helices each from IcmQ and IcmR. Two alpha helices of IcmQ are amphipathic and the formation of the Qn–IcmR complex is mediated by hydrophobic interactions. The hydrophobic nature of residues participating in the interaction with IcmQ is conserved among IcmR and FIRs, although they are poorly related at sequence-level in general. It still remains unclear why IcmR/FIRs are so divergent compared to other Dot/Icm components.
IcmX
IcmX is a primarily periplasmic protein conserved in Legionella, Coxiella, and Rickettsiella Dot/Icm systems (Matthews and Roy, 2000). It has been suggested that IcmX is a distant homolog of TraW of I-type conjugation systems (Segal et al., 2005), but we are unable to follow the similarity, if any, between IcmX and TraW by homology search and phylogenetic analyses. A truncated form of IcmX was found in culture supernatant in a Dot/Icm-dependent fashion (Matthews and Roy, 2000). Its physiological meaning, as well as the specific function of IcmX, remains unclear.
IcmV
IcmV is an integral inner membrane protein conserved in Legionella, Coxiella, and Rickettsiella Dot/Icm systems, whose specific function remains unknown.
DotK/IcmN
DotK is an outer membrane lipoprotein found in Legionella species and C. burnetii Dot/Icm systems. Two L. pneumophila mutants, which carry a transposon insertion in the coding region of DotK (icmN3007::Kan) or a transposon insertion accompanying a partial deletion of the promoter and the coding regions of DotK (icmN3006::Kan), were reported to be partially defective in growth within a protozoan host, A. castellanii (Segal et al., 1998; Segal and Shuman, 1999a). However, the defect was not complemented with plasmids carrying DotK or DotKJIHG, which makes it difficult to interpret the data on these mutations. DotK carries the OmpA family domain (Pfam PF00691; Morozova et al., 2004), which is conserved in bacterial peptidoglycan-binding proteins – notably in an outer membrane porin OmpA, a flagellar stator MotB and peptidoglycan-associated lipoproteins (PALs). The OmpA family domain functions as a peptidoglycan-binding domain, suggesting that DotK anchors the Dot/Icm apparatus to the peptidoglycan layer.
Perspectives
Genomic and metagenomic analyses have revealed that T4BSSs are widely prevalent in bacterial realm (Figure 1). It is well recognized that Legionella and Coxiella Dot/Icm T4BSSs play pivotal roles in infection, while most of other T4BSSs may represent conjugation systems. Rickettsiella is a facultative arthropod pathogen and phylogenetically closely related to Legionella and Coxiella. Recently some pea aphids were reported to carry Rickettsiella species as endosymbionts which modify insect body color (Tsuchida et al., 2010). Rickettsiella infection appears to up-regulate production of green pigments by host insects. Although the molecular mechanisms underlying the phenomenon have not been clarified, it is tempting to guess that Rickettsiella T4BSS may play a role in endosymbiosis. Future studies on T4BSSs of Legionella and related bacteria might shed lights not only on the molecular basis of bacterial pathogenesis but also on evolutionary history from intracellular pathogens to mutualistic endosymbionts.
Structural studies on secretion systems of pathogenic bacteria including T4SSs are rapidly advancing in recent years. Together with pioneer works on characterizations of L. pneumophila Dot/Icm proteins, we are getting a grip on the T4BSS core complex containing DotC, DotD, DotH, DotG, and DotF (Figures 3 and 5D). Intriguingly, the T4BSS core complex appears to be considerably different from the core complex of pKM101 T4ASS. Sequence-level similarity is only found between C-terminal domains of DotG and VirB10. T4BSS appears to be more complicated than T4ASS; T4BSSs contain roughly twice the number of component proteins than T4ASSs. Lines of evidence now suggest the mosaic nature of T4BSS architecture: (a) secretion ATPase DotB is phylogenetically related to ATPases from type II secretion and related systems; (b) L. pneumophila Dot/Icm T4BSS contains the genes encoding icmF and dotU, which are now recognized to encode components of the type VI secretion system; and (c) The C-terminal domain of DotD is structurally similar to a N-terminal subdomain of secretins of type II and type III secretion systems. The nature of the T4BSS core complex as well as the roles of component proteins in type IVB secretion and I-type conjugation remain as major challenges of future studies.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge many authors of related works, which because of constraints of space were not cited here. Research in the Nagai Laboratory is supported in part by the Targeted Proteins Research Program (TPRP) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Mitsubishi Foundation.
References
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Alvarez-Martinez, C. E., and Christie, P. J. (2009). Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73, 775–808.
Amor, J. C., Swails, J., Zhu, X., Roy, C. R., Nagai, H., Ingmundson, A., Cheng, X., and Kahn, R. A. (2005). The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site. J. Biol. Chem. 280, 1392–1400.
Andrews, H. L., Vogel, J. P., and Isberg, R. R. (1998). Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect. Immun. 66, 950–958.
Aussel, L., Barre, F. X., Aroyo, M., Stasiak, A., Stasiak, A. Z., and Sherratt, D. (2002). FtsK is a DNA motor protein that activates chromosomedimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108, 195–205.
Bardill, J. P., Miller, J. L., and Vogel, J. P. (2005). IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol. Microbiol. 56, 90–103.
Bayliss, R., Harris, R., Coutte, L., Monier, A., Fronzes, R., Christie, P. J., Driscoll, P. C., and Waksman, G. (2007). NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system. Proc. Natl. Acad. Sci. U.S.A. 104, 1673–1678.
Berger, K. H., Merriam, J. J., and Isberg, R. R. (1994). Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol. Microbiol. 14, 809–822.
Brand, B. C., Sadosky, A. B., and Shuman, H. A. (1994). The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol. Microbiol. 14, 797–808.
Brenner, D. J., Steigerwalt, A. G., and McDade, J. E. (1979). Classification of the Legionnaires’ disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, family nova. Ann. Intern. Med. 90, 656–658.
Buscher, B. A., Conover, G. M., Miller, J. L., Vogel, S. A., Meyers, S. N., Isberg, R. R., and Vogel, J. P. (2005). The DotL protein, a member of the TraG-coupling protein family, is essential for Viability of Legionella pneumophila strain Lp02. J. Bacteriol. 187, 2927–2938.
Cambronne, E. D., and Roy, C. R. (2007). The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 3, e188. doi:10.1371/journal.ppat.0030188
Cazalet, C., Gomez-Valero, L., Rusniok, C., Lomma, M., Dervins-Ravault, D., Newton, H. J., Sansom, F. M., Jarraud, S., Zidane, N., Ma, L., Bouchier, C., Etienne, J., Hartland, E. L., and Buchrieser, C. (2010). Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires’ disease. PLoS Genet. 6, e1000851. doi: 10.1371/journal.pgen.1000851
Cazalet, C., Rusniok, C., Bruggemann, H., Zidane, N., Magnier, A., Ma, L., Tichit, M., Jarraud, S., Bouchier, C., Vandenesch, F., Kunst, F., Etienne, J., Glaser, P., and Buchrieser, C. (2004). Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36, 1165–1173.
Chami, M., Guilvout, I., Gregorini, M., Remigy, H. W., Muller, S. A., Valerio, M., Engel, A., Pugsley, A. P., and Bayan, N. (2005). Structural insights into the secretin PulD and its trypsin-resistant core. J. Biol. Chem. 280, 37732–37741.
Chandran, V., Fronzes, R., Duquerroy, S., Cronin, N., Navaza, J., and Waksman, G. (2009). Structure of the outer membrane complex of a type IV secretion system. Nature 462, 1011–1015.
Chen, C., Banga, S., Mertens, K., Weber, M. M., Gorbaslieva, I., Tan, Y., Luo, Z. Q., and Samuel, J. E. (2010). Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc. Natl. Acad. Sci. U.S.A. 107, 21755–21760.
Chien, M., Morozova, I., Shi, S., Sheng, H., Chen, J., Gomez, S. M., Asamani, G., Hill, K., Nuara, J., Feder, M., Rineer, J., Greenberg, J. J., Steshenko, V., Park, S. H., Zhao, B., Teplitskaya, E., Edwards, J. R., Pampou, S., Georghiou, A., Chou, I. C., Iannuccilli, W., Ulz, M. E., Kim, D. H., Geringer-Sameth, A., Goldsberry, C., Morozov, P., Fischer, S. G., Segal, G., Qu, X., Rzhetsky, A., Zhang, P., Cayanis, E., De Jong, P. J., Ju, J., Kalachikov, S., Shuman, H. A., and Russo, J. J. (2004). The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305, 1966–1968.
Christie, P. J., and Vogel, J. P. (2000). Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8, 354–360.
Coers, J., Kagan, J. C., Matthews, M., Nagai, H., Zuckman, D. M., and Roy, C. R. (2000). Identification of icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38, 719–736.
Collins, R. F., Frye, S. A., Kitmitto, A., Ford, R. C., Tonjum, T., and Derrick, J. P. (2004). Structure of the Neisseria meningitidis outer membrane PilQ secretin complex at 12 A resolution. J. Biol. Chem. 279, 39750–39756.
D’Auria, G., Jimenez-Hernandez, N., Peris-Bondia, F., Moya, A., and Latorre, A. (2010). Legionella pneumophila pan genome reveals strain-specific virulence factors. BMC Genomics 11, 181. doi: 10.1186/1471-2164-11-181
de la Cruz, F., Frost, L. S., Meyer, R. J., and Zechner, E. L. (2010). Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 34, 18–40.
Draper, O., Cesar, C. E., Machon, C., de la Cruz, F., and Llosa, M. (2005). Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc. Natl. Acad. Sci. U.S.A. 102, 16385–16390.
Dumenil, G., and Isberg, R. R. (2001). The Legionella pneumophila IcmR protein exhibits chaperone activity for IcmQ by preventing its participation in high-molecular-weight complexes. Mol. Microbiol. 40, 1113–1127.
Dumenil, G., Montminy, T. P., Tang, M., and Isberg, R. R. (2004). IcmR-regulated membrane insertion and efflux by the Legionella pneumophila IcmQ protein. J. Biol. Chem. 279, 4686–4695.
Errington, J., Bath, J., and Wu, L. J. (2001). DNA transport in bacteria. Nat. Rev. Mol. Cell Biol. 2, 538–545.
Feldman, M., and Segal, G. (2004). A specific genomic location within the icm/dot pathogenesis region of different Legionella species encodes functionally similar but nonhomologous virulence proteins. Infect. Immun. 72, 4503–4511.
Feldman, M., Zusman, T., Hagag, S., and Segal, G. (2005). Coevolution between nonhomologous but functionally similar proteins and their conserved partners in the Legionella pathogenesis system. Proc. Natl. Acad. Sci. U.S.A. 102, 12206–12211.
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Fernandez, D., Spudich, G. M., Zhou, X. R., and Christie, P. J. (1996). The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J. Bacteriol. 178, 3168–3176.
Franco, I. S., Shuman, H. A., and Charpentier, X. (2009). The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell. Microbiol. 11, 1435–1443.
Fronzes, R., Schafer, E., Wang, L., Saibil, H. R., Orlova, E. V., and Waksman, G. (2009). Structure of a type IV secretion system core complex. Science 323, 266–268.
Genin, S., and Boucher, C. A. (1994). A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol. Gen. Genet. 243, 112–118.
Glockner, G., Albert-Weissenberger, C., Weinmann, E., Jacobi, S., Schunder, E., Steinert, M., Hacker, J., and Heuner, K. (2008). Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol. 298, 411–428.
Gomis-Ruth, F. X., Moncalian, G., de la Cruz, F., and Coll, M. (2002). Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein. Detailed structural features and mapping of the active site cleft. J. Biol. Chem. 277, 7556–7566.
Gomis-Ruth, F. X., Moncalian, G., Perez-Luque, R., Gonzalez, A., Cabezon, E., de la Cruz, F., and Coll, M. (2001). The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409, 637–641.
Hardie, K. R., Seydel, A., Guilvout, I., and Pugsley, A. P. (1996). The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol. Microbiol. 22, 967–976.
Horwitz, M. A. (1983). The Legionnaires’ disease bacterium (Legionella pneumophila) inhibits phagosome lysosome fusion in human monocytes. J. Exp. Med. 158, 2108–2126.
Horwitz, M. A. (1984). Phagocytosis of the Legionnaires’ disease bacterium (Legionella pneumophila) occurs by a novel mechanism: engulfment within a pseudopod coil. Cell 36, 27–33.
Horwitz, M. A., and Maxfield, F. R. (1984). Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J. Cell Biol. 99, 1936–1943.
Horwitz, M. A., and Silverstein, S. C. (1980). Legionnaires’ disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Invest. 66, 441–450.
Hubber, A., and Roy, C. R. (2010). Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26, 261–283.
Isberg, R. R., O’Connor, T. J., and Heidtman, M. (2009). The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24.
Komano, T., Yoshida, T., Narahara, K., and Furuya, N. (2000). The transfer region of IncI1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol. Microbiol. 35, 1348–1359.
Korotkov, K. V., Pardon, E., Steyaert, J., and Hol, W. G. (2009). Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure 17, 255–265.
Kozak, N. A., Buss, M., Lucas, C. E., Frace, M., Govil, D., Travis, T., Olsen-Rasmussen, M., Benson, R. F., and Fields, B. S. (2010). Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968. J. Bacteriol. 192, 1030–1044.
Lang, S., Gruber, K., Mihajlovic, S., Arnold, R., Gruber, C. J., Steinlechner, S., Jehl, M. A., Rattei, T., Frohlich, K. U., and Zechner, E. L. (2010). Molecular recognition determinants for type IV secretion of diverse families of conjugative relaxases. Mol. Microbiol. 78, 1539–1555.
Lawley, T. D., Klimke, W. A., Gubbins, M. J., and Frost, L. S. (2003). F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224, 1–15.
Leclerque, A., and Kleespies, R. G. (2008). Type IV secretion system components as phylogenetic markers of entomopathogenic bacteria of the genus Rickettsiella. FEMS Microbiol. Lett. 279, 167–173.
Liu, G., McDaniel, T. K., Falkow, S., and Karlin, S. (1999). Sequence anomalies in the Cag7 gene of the Helicobacter pylori pathogenicity island. Proc. Natl. Acad. Sci. U.S.A. 96, 7011–7016.
Luo, Z. Q., and Isberg, R. R. (2004). Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. U.S.A. 101, 841–846.
Marlovits, T. C., Kubori, T., Lara-Tejero, M., Thomas, D., Unger, V. M., and Galan, J. E. (2006). Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441, 637–640.
Massey, T. H., Mercogliano, C. P., Yates, J., Sherratt, D. J., and Lowe, J. (2006). Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell 23, 457–469.
Matthews, M., and Roy, C. R. (2000). Identification and subcellular localization of the Legionella pneumophila IcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect. Immun. 68, 3971–3982.
Morozova, I., Qu, X., Shi, S., Asamani, G., Greenberg, J. E., Shuman, H. A., and Russo, J. J. (2004). Comparative sequence analysis of the icm/dot genes in Legionella. Plasmid 51, 127–147.
Nagai, H., Cambronne, E. D., Kagan, J. C., Amor, J. C., Kahn, R. A., and Roy, C. R. (2005). A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. U.S.A. 102, 826–831.
Nagai, H., Kagan, J. C., Zhu, J., Kahn, R. A., and Roy, C. R. (2002). A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295, 679–682.
Nagai, H., and Roy, C. R. (2001). The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J. 20, 5962–5970.
Nakano, N., Kubori, T., Kinoshita, M., Imada, K., and Nagai, H. (2010). Crystal Structure of Legionella DotD: insights into the relationship between type IVB and type II/III secretion systems. PLoS Pathog. 6, e1001129. doi:10.1371/journal.ppat.1001129
Ninio, S., and Roy, C. R. (2007). Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 15, 372–380.
Ninio, S., Zuckman-Cholon, D. M., Cambronne, E. D., and Roy, C. R. (2005). The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol. Microbiol. 55, 912–926.
Ohnishi, H., Mizunoe, Y., Takade, A., Tanaka, Y., Miyamoto, H., Harada, M., and Yoshida, S. (2004). Legionella dumoffii DjlA, a member of the DnaJ family, is required for intracellular growth. Infect. Immun. 72, 3592–3603.
Omsland, A., Cockrell, D. C., Howe, D., Fischer, E. R., Virtaneva, K., Sturdevant, D. E., Porcella, S. F., and Heinzen, R. A. (2009). Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc. Natl. Acad. Sci. U.S.A. 106, 4430–4434.
Opalka, N., Beckmann, R., Boisset, N., Simon, M. N., Russel, M., and Darst, S. A. (2003). Structure of the filamentous phage pIV multimer by cryo-electron microscopy. J. Mol. Biol. 325, 461–470.
Pan, X., Luhrmann, A., Satoh, A., Laskowski-Arce, M. A., and Roy, C. R. (2008). Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320, 1651–1654.
Peracino, B., Balest, A., and Bozzaro, S. (2010). Phosphoinositides differentially regulate bacterial uptake and Nramp1-induced resistance to Legionella infection in Dictyostelium. J. Cell. Sci. 123, 4039–4051.
Planet, P. J., Kachlany, S. C., DeSalle, R., and Figurski, D. H. (2001). Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. U.S.A. 98, 2503–2508.
Purcell, M., and Shuman, H. A. (1998). The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect. Immun. 66, 2245–2255.
Raychaudhury, S., Farelli, J. D., Montminy, T. P., Matthews, M., Menetret, J. F., Dumenil, G., Roy, C. R., Head, J. F., Isberg, R. R., and Akey, C. W. (2009). Structure and function of interacting IcmR-IcmQ domains from a type IVb secretion system in Legionella pneumophila. Structure 17, 590–601.
Reichow, S. L., Korotkov, K. V., Hol, W. G., and Gonen, T. (2010). Structure of the cholera toxin secretion channel in its closed state. Nat. Struct. Mol. Biol. 17, 1226–1232.
Roux, V., Bergoin, M., Lamaze, N., and Raoult, D. (1997). Reassessment of the taxonomic position of Rickettsiella grylli. Int. J. Syst. Bacteriol. 47, 1255–1257.
Roy, C. R., and Isberg, R. I. (1997). Topology of Legionella pneumophila DotA: an inner membrane protein required for replication in macrophages. Infect. Immun. 65, 571–578.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Schroeder, G. N., Petty, N. K., Mousnier, A., Harding, C. R., Vogrin, A. J., Wee, B., Fry, N. K., Harrison, T. G., Newton, H. J., Thomson, N. R., Beatson, S. A., Dougan, G., Hartland, E. L., and Frankel, G. (2010). Legionella pneumophila strain 130b possesses a unique combination of type IV secretion systems and novel Dot/Icm secretion system effector proteins. J. Bacteriol. 192, 6001–6016.
Segal, G., Feldman, M., and Zusman, T. (2005). The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 29, 65–81.
Segal, G., Purcell, M., and Shuman, H. A. (1998). Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U.S.A. 95, 1669–1674.
Segal, G., Russo, J. J., and Shuman, H. A. (1999). Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34, 799–809.
Segal, G., and Shuman, H. A. (1997). Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 65, 5057–5066.
Segal, G., and Shuman, H. A. (1999a). Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67, 2117–2124.
Segal, G., and Shuman, H. A. (1999b). Possible origin of the Legionella pneumophila virulence genes and their relation to Coxiella burnetii [letter]. Mol. Microbiol. 33, 669–670.
Seshadri, R., Paulsen, I. T., Eisen, J. A., Read, T. D., Nelson, K. E., Nelson, W. C., Ward, N. L., Tettelin, H., Davidsen, T. M., Beanan, M. J., Deboy, R. T., Daugherty, S. C., Brinkac, L. M., Madupu, R., Dodson, R. J., Khouri, H. M., Lee, K. H., Carty, H. A., Scanlan, D., Heinzen, R. A., Thompson, H. A., Samuel, J. E., Fraser, C. M., and Heidelberg, J. F. (2003). Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. U.S.A. 100, 5455–5460.
Sexton, J. A., Miller, J. L., Yoneda, A., Kehl-Fie, T. E., and Vogel, J. P. (2004a). Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect. Immun. 72, 5983–5992.
Sexton, J. A., Pinkner, J. S., Roth, R., Heuser, J. E., Hultgren, S. J., and Vogel, J. P. (2004b). The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J. Bacteriol. 186, 1658–1666.
Sexton, J. A., and Vogel, J. P. (2002). Type IVB secretion by intracellular pathogens. Traffic 3, 178–185.
Sexton, J. A., Yeo, H. J., and Vogel, J. P. (2005). Genetic analysis of the Legionella pneumophila DotB ATPase reveals a role in type IV secretion system protein export. Mol. Microbiol. 57, 70–84.
Shin, S., and Roy, C. R. (2008). Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell. Microbiol. 10, 1209–1220.
Spreter, T., Yip, C. K., Sanowar, S., Andre, I., Kimbrough, T. G., Vuckovic, M., Pfuetzner, R. A., Deng, W., Yu, A. C., Finlay, B. B., Baker, D., Miller, S. I., and Strynadka, N. C. (2009). A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat. Struct. Mol. Biol. 16, 468–476.
Swanson, M. S., and Isberg, R. R. (1995). Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63, 3609–3620.
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599.
Tilney, L. G., Harb, O. S., Connelly, P. S., Robinson, C. G., and Roy, C. R. (2001). How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J. Cell. Sci. 114, 4637–4650.
Tsuchida, T., Koga, R., Horikawa, M., Tsunoda, T., Maoka, T., Matsumoto, S., Simon, J. C., and Fukatsu, T. (2010). Symbiotic bacterium modifies aphid body color. Science 330, 1102–1104.
VanRheenen, S. M., Dumenil, G., and Isberg, R. R. (2004). IcmF and DotU are required for optimal effector translocation and trafficking of the Legionella pneumophila vacuole. Infect. Immun. 72, 5972–5982.
Vergunst, A. C., Schrammeijer, B., den Dulk-Ras, A., de Vlaam, C. M., Regensburg-Tuink, T. J., and Hooykaas, P. J. (2000). VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290, 979–982.
Vergunst, A. C., van Lier, M. C., den Dulk-Ras, A., Stuve, T. A., Ouwehand, A., and Hooykaas, P. J. (2005). Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 102, 832–837.
Vincent, C. D., Buscher, B. A., Friedman, J. R., Williams, L. A., Bardill, P., and Vogel, J. P. (2006a). Identification of non-dot/icm suppressors of the Legionella pneumophila DeltadotL lethality phenotype. J. Bacteriol. 188, 8231–8243.
Vincent, C. D., Friedman, J. R., Jeong, K. C., Buford, E. C., Miller, J. L., and Vogel, J. P. (2006b). Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 62, 1278–1291.
Vincent, C. D., and Vogel, J. P. (2006). The Legionella pneumophila IcmS-LvgA protein complex is important for Dot/Icm-dependent intracellular growth. Mol. Microbiol. 61, 596–613.
Vogel, J. P., Andrews, H. L., Wong, S. K., and Isberg, R. R. (1998). Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876.
Voth, D. E., Howe, D., Beare, P. A., Vogel, J. P., Unsworth, N., Samuel, J. E., and Heinzen, R. A. (2009). The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J. Bacteriol. 191, 4232–4242.
Watarai, M., Andrews, H. L., and Isberg, R. R. (2001a). Formation of a fibrous structure on the surface of Legionella pneumophila associated with exposure of DotH and DotO proteins after intracellular growth. Mol. Microbiol. 39, 313–329.
Watarai, M., Derre, I., Kirby, J., Growney, J. D., Dietrich, W. F., and Isberg, R. R. (2001b). Legionella pneumophila is internalized by a macropinocytotic uptake pathway controlled by the Dot/Icm system and the mouse Lgn1 locus. J. Exp. Med. 194, 1081–1096.
Wilkins, B. M., and Thomas, A. T. (2000). DNA-independent transport of plasmid primase protein between bacteria by the I1 conjugation system. Mol. Microbiol. 38, 650–657.
Yerushalmi, G., Zusman, T., and Segal, G. (2005). Additive effect on intracellular growth by Legionella pneumophila Icm/Dot proteins containing a lipobox motif. Infect. Immun. 73, 7578–7587.
Zamboni, D. S., McGrath, S., Rabinovitch, M., and Roy, C. R. (2003). Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 49, 965–976.
Zuckerkandl, E., and Pauling, L. (1965). “Evolutionary divergence and convergence in proteins,” in Evolving Genes and Proteins, eds V. Bryson, and H. J. Vogel (New York: Academic Press), 97–166.
Zuckman, D. M., Hung, J. B., and Roy, C. R. (1999). Pore-forming activity is not sufficient for Legionella pneumophila phagosome trafficking and intracellular growth. Mol. Microbiol. 32, 990–1001.
Zusman, T., Feldman, M., Halperin, E., and Segal, G. (2004). Characterization of the icmH and icmF genes required for Legionella pneumophila intracellular growth, genes that are present in many bacteria associated with eukaryotic cells. Infect. Immun. 72, 3398–3409.
Keywords: Legionella, Coxiella, Rickettsiella, Dot/Icm, type IVB secretion, type IV secretion, protein secretion, conjugation
Citation: Nagai H and Kubori T (2011) Type IVB secretion systems of Legionella and other Gram-negative bacteria. Front. Microbio. 2:136. doi: 10.3389/fmicb.2011.00136
Received: 15 January 2011; Paper pending published: 04 March 2011;
Accepted: 11 June 2011; Published online: 27 June 2011.
Edited by:
Carmen Buchrieser, Pasteur Institute, FranceReviewed by:
Christoph Dehio, Biozentrum University of Basel, SwitzerlandRémi Fronzes, CNRS/Pasteur Institute, France
Copyright: © 2011 Nagai and Kubori. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Hiroki Nagai, Research Institute for Microbial Diseases, Osaka University, 3-1 Yamadaoka, Suita, Osaka 565-0871, Japan. e-mail:aG5hZ2FpQGJpa2VuLm9zYWthLXUuYWMuanA=

