
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 29 June 2011
Sec. Microbial Physiology and Metabolism
volume 2 - 2011 | https://doi.org/10.3389/fmicb.2011.00135
This article is part of the Research Topic The microbial sulfur cycle View all 21 articles
 Kimberly L. Keller1,2
Kimberly L. Keller1,2 Judy D. Wall1,2,3*
Judy D. Wall1,2,3*Progress in the genetic manipulation of the Desulfovibrio strains has provided an opportunity to explore electron flow pathways during sulfate respiration. Most bacteria in this genus couple the oxidation of organic acids or ethanol with the reduction of sulfate, sulfite, or thiosulfate. Both fermentation of pyruvate in the absence of an alternative terminal electron acceptor, disproportionation of fumarate and growth on H2 with CO2 during sulfate reduction are exhibited by some strains. The ability to produce or consume H2 provides Desulfovibrio strains the capacity to participate as either partner in interspecies H2 transfer. Interestingly the mechanisms of energy conversion, pathways of electron flow and the parameters determining the pathways used remain to be elucidated. Recent application of molecular genetic tools for the exploration of the metabolism of Desulfovibrio vulgaris Hildenborough has provided several new datasets that might provide insights and constraints to the electron flow pathways. These datasets include (1) gene expression changes measured in microarrays for cells cultured with different electron donors and acceptors, (2) relative mRNA abundances for cells growing exponentially in defined medium with lactate as carbon source and electron donor plus sulfate as terminal electron acceptor, and (3) a random transposon mutant library selected on medium containing lactate plus sulfate supplemented with yeast extract. Studies of directed mutations eliminating apparent key components, the quinone-interacting membrane-bound oxidoreductase (Qmo) complex, the Type 1 tetraheme cytochrome c3 (Tp1-c3), or the Type 1 cytochrome c3:menaquinone oxidoreductase (Qrc) complex, suggest a greater flexibility in electron flow than previously considered. The new datasets revealed the absence of random transposons in the genes encoding an enzyme with homology to Coo membrane-bound hydrogenase. From this result, we infer that Coo hydrogenase plays an important role in D. vulgaris growth on lactate plus sulfate. These observations along with those reported previously have been combined in a model showing dual pathways of electrons from the oxidation of both lactate and pyruvate during sulfate respiration. Continuing genetic and biochemical analyses of key genes in Desulfovibrio strains will allow further clarification of a general model for sulfate respiration.
The ability to obtain energy from substrate oxidation coupled with sulfate reduction is a unique mechanism that is shared by a heterogeneous group of microbes that include proteobacteria, firmicutes and archaea; mesophiles, thermophiles, and psychrophiles. While this means of energy generation is restricted to anaerobes, these sulfate-reducing microbes (SRM) are found in a wide variety of environments including oxic waters and soils. Whereas oxygen at high concentrations is toxic to SRM, large variations in sensitivity are observed (Sass and Cypionka, 2007). Oxygen is reduced by many SRM perhaps as a protective mechanism because sustained growth supported through oxygenic respiration has not been demonstrated (Marschall et al., 1993; Hansen, 1994a).
Of this group of microbes, members of the δ-proteobacterial genus Desulfovibrio have been most intensively studied because of their rapid growth and ease of manipulation. Desulfovibrio vulgaris Hildenborough was the first of the sulfate-reducing bacteria to have its genome sequenced (Heidelberg et al., 2004). D. vulgaris was also the first to be genetically manipulated through conjugation (Powell et al., 1989; van den Berg et al., 1989) and the second to be modified by marker exchange mutagenesis (Fu and Voordouw, 1997), after Desulfovibrio fructosovorans (Rousset et al., 1991). Genome sequences of many more SRM are now available.1 Improved genetic tools have been developed (Wall et al., 1993; Rousset et al., 1998; Bender et al., 2007; Keller et al., 2009; Zane et al., 2010), transposon mutant libraries have been generated (Groh et al., 2005; Deutschbauer, personal communication, and this lab) and systems biology tools have been applied (Mukhopadhyay et al., 2006; Redding et al., 2006; Tang et al., 2007; Pereira et al., 2008; He et al., 2010). Therefore this chapter will focus on the contributions of molecular genetics to our understanding of energy conversion in the Desulfovibrio strains. Confirmation that these insights might be applicable to other SRM genera remain to be established.
Our discussion will consider energy conversion by Desulfovibrio strains grown with lactate as carbon and electron source and sulfate as electron acceptor. Because electron flow from lactate oxidation with sulfate reduction provides robust growth of Desulfovibrio, data from systems biology approaches are now available from cells grown on these substrates. Strengthening relevance, the released fermentation products, lactate and H2, are likely to be the sources of electrons found in natural environments where these bacteria flourish. Currently many transcriptomic data and some proteomic data have been generated2 and transposon libraries have been selected with cells grown on these substrates (Deutschbauer, personal communication, and Wall lab).
As a terminal electron acceptor, sulfate has four properties that together give the SRM access to growth niches unavailable to other microbes. First, sulfate is a thermodynamically stable oxidized form of sulfur and must be activated prior to being reduced. Second, sulfate is reduced by soluble enzymes in the cytoplasm making active transport of the sulfate necessary because it is an ionized species at physiological pHs (H2SO4; pKa1, −3; pKa2, 2). Third, sulfide, HS− (H2S, pKa 6.9) the reduced product, is toxic at elevated concentrations (Caffrey and Voordouw, 2010) and must be transported out of the cell or defense systems invoked. Fourth, the fact that the reductive enzymes are soluble and not membrane-bound suggests that reduction of sulfate may not be uniquely or tightly associated with an electron transport system in the membrane.
The common sources of carbon and electrons used by SRM are fermentation products from facultative anaerobes. Most Desulfovibrio strains are incomplete oxidizers of organic acids and alcohols producing acetate in quantities nearly stoichiometric to the added substrate (Postgate, 1984a; Rabus et al., 2006). Therefore, about 95% of the carbon substrate oxidized by Desulfovibrio strains is used for energy generation and the residual used to produce cell material (Noguera et al., 1998; Rabus et al., 2006). The inability of these strains to oxidize acetate has been suggested to result from the high reduction potential of the fumarate/succinate couple (E0′ = 33 mV, Eq. 1, from Thauer et al., 1977) in the standard TCA cycle (Thauer, 1989). Therefore, reduction of the activated form of sulfate, adenosine 5′-phosphosulfate (APS, Eq. 2) or bisulfite (Eq. 3) cannot be coupled with succinate oxidation.
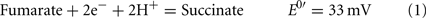
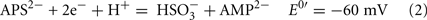
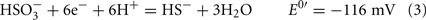
For those strains of the genus Desulfobacter or Desulfobacterium that do completely oxidize organic acids to CO2 with sulfate as electron acceptor, the standard TCA cycle is not used (Hansen, 1994a).
The Desulfovibrio pathway for lactate oxidation is through pyruvate to acetyl-CoA and from acetyl-CoA to acetate via acetylphosphate which is of sufficient energy to generate ATP by substrate-level phosphorylation (See Eqs 4–8). Early researchers believed that the SRM were restricted to substrate-level phosphorylation for ATP generation. However, the demonstration that Desulfovibrio strains could grow with H2 oxidation, with carbon supplied by CO2 and acetate, and sulfate as terminal electron acceptor (Butlin et al., 1949; Peck, 1960) firmly established the capacity for electron transport-linked phosphorylation. In addition, Peck (1966) convincingly argued that a mechanism, in addition to substrate-level phosphorylation, was essential during lactate plus sulfate growth. Peck pointed out that the two moles of ATP generated during the oxidation of two moles of lactate would be consumed during sulfate activation. Therefore, energy to support growth must be provided by an additional mechanism.
Odom and Peck (1981a) proposed an unusual and controversial model for increasing the energy budget called the “hydrogen cycle.” This model was proposed, in part, as an explanation for the observation of a H2 transient seen in cultures of Desulfovibrio inoculated into medium with lactate or pyruvate as electron donors and sulfate as electron acceptor. They proposed that electrons and protons generated from organic acid oxidation could serve as substrates for a cytoplasmically located hydrogenase. The resulting H2 could diffuse through the cytoplasmic membrane and be reoxidized by periplasmic hydrogenases. The electrons produced by the H2 oxidation would be delivered to the c-type cytochrome pool for return to the cytoplasm through transmembrane protein complexes and used for sulfate reduction. The protons released would contribute to the chemiosmotic potential.
In contrast, Lupton et al. (1984) proposed that trace H2 was produced and consumed in the initial growth phases of Desulfovibrio cultures on organic acids plus sulfate to control redox state of electron carriers. These researchers also provided evidence that they interpreted as eliminating the argument that the purpose of the transient H2 production was to allow fermentation to make ATP for sulfate activation and reduction in growth initiation. They showed that with thiosulfate as terminal electron acceptor, a substrate that does not require activation prior to reduction, H2 was still produced and consumed when cells were introduced into fresh medium.
Evidence both for and against the hydrogen cycling model has been published. Odom and Peck (1981a) demonstrated that spheroplasts produced from Desulfovibrio gigas were unable to oxidize lactate and reduce sulfate without the restoration of the periplasmic proteins, hydrogenase and the type I cytochrome c3 (Tp1-c3). From this result it was inferred that electrons were routed from lactate through the periplasm and back to sulfate. In addition, Peck et al. (1987) were able to trap and measure H2 released by D. vulgaris from pyruvate oxidation coupled to sulfate reduction with membrane-inlet mass spectrometry. On the other hand, the existence of free H2, predicted by the model to be a periplasmic intermediate in the oxidation of lactate or pyruvate, would lead to the expectation that a H2 atmosphere would strongly inhibit oxidation of these substrates. Lupton et al. (1984) showed that there was no inhibition of lactate or pyruvate oxidation by H2 during sulfate respiration. This lack of inhibition of substrate oxidation by H2 was confirmed by Pankhania et al. (1986) for cells with lactate. Also, a mutant of Desulfovibrio desulfuricans ATCC27774 was isolated that could grow on lactate plus sulfate but not H2 plus sulfate (Odom and Wall, 1987). This result is incongruent with periplasmic H2 as an intermediate in lactate oxidation during sulfate reduction. In addition, if the cycling process – H2 production, membrane diffusion and H2 oxidation – were essential for sulfate respiration with organic acids, SRM should all have hydrogenases. Desulfovibrio sapovorans is one example that lacks hydrogenase (Postgate, 1984b).
More recent reviews have diminished the putative role of hydrogen cycling in the metabolism of the SRM. Widdel and Hansen (1991) suggested that H2 production is simply an accident of having constitutive hydrogenases active in the cell. Rabus et al. (2006) proposed that constitutive hydrogenases would produce hydrogen whenever there was an imbalance between reductant produced or consumed by the cells. An alternative suggestion of the latter researchers derived from the postulation that in natural environments the SRM participate routinely in interspecies H2 transfer when sulfate is absent. As a result, SRM might not be able to redirect electrons completely when sulfate was available, thus driving the release of electrons as H2 (Rabus et al., 2006). In contrast to these theories, Noguera et al. (1998) elegantly generated and tested a model for H2 production and consumption by D. vulgaris growing on lactate plus sulfate. These researchers proposed that dual pathways were operating, simultaneously, for electron flow and that the free energy available determined the distribution of electron flow. Their model predicted 52% of the electron flow would move from lactate through H2 and finally to sulfate; whereas, 48% would move to sulfate without H2 formation. After subtraction of the reductant moving to biomass, their experiments showed that 47% of the remaining electrons were directly coupled to sulfate reduction and 53% formed H2 before being used for sulfate reduction (Noguera et al., 1998).
The predicted pathways of sulfate reduction and lactate oxidation are presented in the equations below (Eqs 4–16). Dissimilatory sulfate reduction occurs by a two step process requiring a minimum of four enzymes (Table 1). First sulfate is activated to form APS at the expense of two ATP equivalents by sulfate adenylyltransferase. Two electrons are then used by APS reductase to reduce APS to 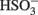 (Eqs 10 and 12). The activation reaction is pulled by the hydrolysis of the released pyrophosphate (Eq. 11). Then bisulfite reductase reduces
(Eqs 10 and 12). The activation reaction is pulled by the hydrolysis of the released pyrophosphate (Eq. 11). Then bisulfite reductase reduces 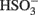 with six electrons to HS−, in one step (Eq. 13). The equations show protonated species considering the published pKa’s of the compounds3 and assuming that the internal pH is ca. 7.5 (Cypionka, 1995). It should be noted that the pKa’s of
with six electrons to HS−, in one step (Eq. 13). The equations show protonated species considering the published pKa’s of the compounds3 and assuming that the internal pH is ca. 7.5 (Cypionka, 1995). It should be noted that the pKa’s of 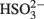 , H2S, and
, H2S, and 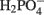 are 7.2, 7.0, and 7.2, respectively, and therefore the protonated and deprotonated forms were considered to be present in about equal quantities. The net result for sulfate reduction (Eq. 14) indicates a consumption of eight moles of electrons with eight and a half moles of protons.
are 7.2, 7.0, and 7.2, respectively, and therefore the protonated and deprotonated forms were considered to be present in about equal quantities. The net result for sulfate reduction (Eq. 14) indicates a consumption of eight moles of electrons with eight and a half moles of protons.
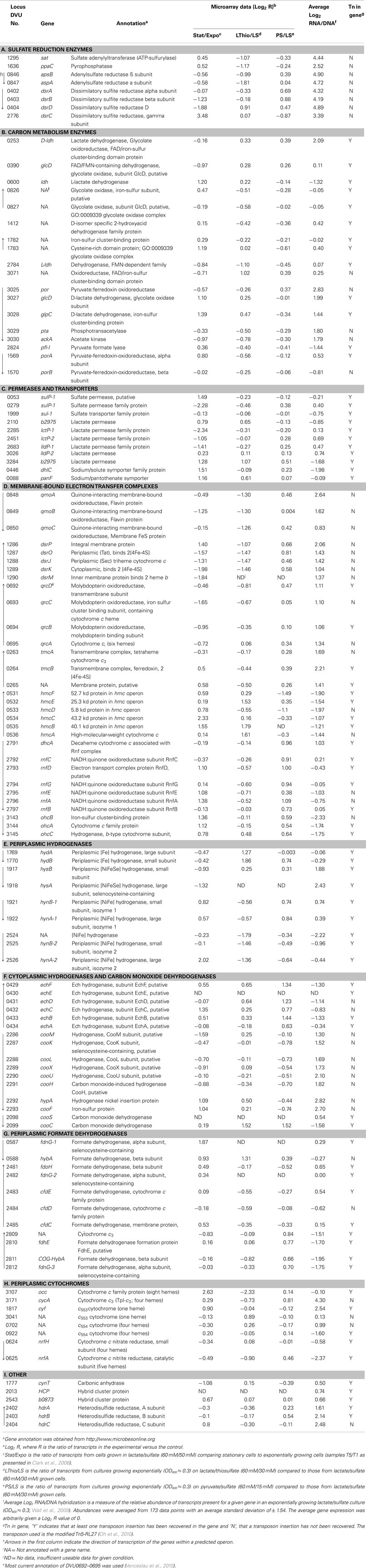
Table 1. Expression of genes encoding enzymes putatively involved in Lactate/Sulfate growth of Desulfovibrio vulgaris Hildenborough.
Lactate oxidation by the incomplete oxidizers in the Desulfovibrio genus, proceeds with pyruvate and acetyl-CoA as intermediates. Reductant is generated from lactate through lactate dehydrogenase and pyruvate:ferredoxin oxidoreductase (Eqs 4 and 5), while acetyl-CoA is the source for substrate-level phosphorylation producing ATP (Eqs 6 and 7). Seven protons were produced in the reactions as shown (Eq. 8). By interconversion of the adenylate nucleotides (Eq. 15) and summing Eqs 8, 9, 14, and 15, the combined oxidation/reduction reaction for growth with lactate plus sulfate is shown as Eq. 16. With the assumptions made here, 0.5 mole of protons are produced in excess of those consumed during reduction of one mole of sulfate. Clearly nitrogen metabolism, biomass production, transport functions and/or small variations in internal pH that affect the concentrations of protonated species will all affect the actual balance of electrons and protons (Hooper and DiSpirito, 1985).
Lactate oxidation:
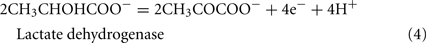
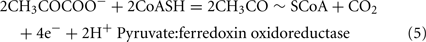
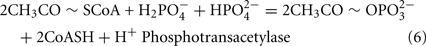
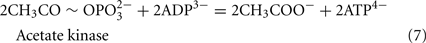
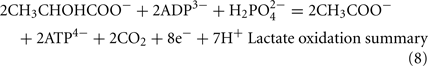
Dissolved CO2:
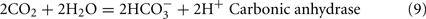
Sulfate reduction:
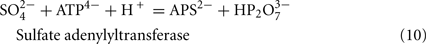
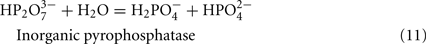
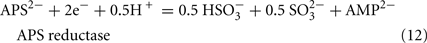
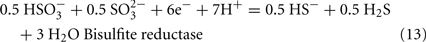
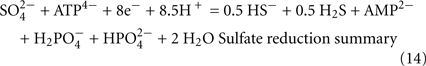
Adenylate nucleotide interchange:
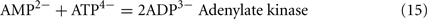
Summary lactate plus sulfate metabolism:
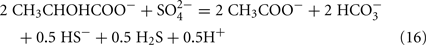
Transport functions of substrates and products have been addressed experimentally in variable detail. Cypionka (1995) has summarized his detailed work on sulfate transport in SRM. In freshwater SRM strains, sulfate was shown to be symported with protons, two if sulfate were in excess and three electrogenically if sulfate were limited. Since the genetic experiments to be described here with D. vulgaris were carried out with excess sulfate, transport of sulfate likely occurs with the electroneutral symport of two electrons. Energy for the sulfate transport, i.e., the two protons used, is compensated by the diffusion of H2S from the cell. However, in the slightly alkaline pH of the cell much of the sulfide would be ionized, HS−. Little is known about the export of this charged ion. Cypionka (1995) suggests that it may be impermeable or symported with one proton. If true, energy spent transporting sulfate into the cell would be balanced by the electroneutral export of sulfide or diffusion of the protonated form.
Transport of the monocarboxylates, lactate and acetate, into and out of the cell, respectively, is likely to be by symport with one proton (Dong et al., 1993). Thus, the reciprocal transport of these compounds will not affect the proton balance of the cell. Although acetic acid in its protonated form can apparently diffuse across the cell membrane (Thauer and Morris, 1984), its continued production against a chemical gradient of acetate during oxidation of lactate or pyruvate would suggest an energy linked export process. Six genes have been annotated as lactate permeases in D. vulgaris, five for L-lactate and one for D-lactate (Table 1); whereas, no genes were annotated as acetate transporters. BLAST analysis4 of the D. vulgaris genome showed that DVU0446 is 61% identical and DVU0088 is 23% identical to the acetate permease (actP, b4067) from Escherichia coli str. K-12 substr. MG1655. Further experimentation will be needed to establish whether these genes encode transporters for acetate and whether they might be essential.
The model of Noguera et al. (1998) is predicated on the existence of two routes for electron flow during lactate plus sulfate growth and controlled by a thermodynamic switch that responds to free energy availability. This is consistent with the differences in reduction potential for the two oxidation steps generating electrons from lactate. For the first oxidation step, the E0′ (mV) of the pyruvate/lactate couple is -190 mV; whereas, for pyruvate oxidation, the acetyl-CoA + CO2/pyruvate couple is -498 mV. When sulfate is terminal electron acceptor, the E0′ (mV) of 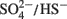 is -200 (calculated for [HS−] of 0.1 mM; Thauer et al., 2007). The standard reduction potential of the lactate dehydrogenases is above that of the H+/H2 couple (E0′ of -414 mV), even if H2 at more environmental concentrations is considered (E0′ of -270 mV at 1 Pa:H2), and would require energy-driven reverse electron flow to accomplish H2 generation. In contrast, pyruvate oxidation with ferredoxin reduction has sufficient redox potential to generate H2. Thus it is tempting to suggest that the two pathways for electron transport coincide with electrons from lactate and those from pyruvate.
is -200 (calculated for [HS−] of 0.1 mM; Thauer et al., 2007). The standard reduction potential of the lactate dehydrogenases is above that of the H+/H2 couple (E0′ of -414 mV), even if H2 at more environmental concentrations is considered (E0′ of -270 mV at 1 Pa:H2), and would require energy-driven reverse electron flow to accomplish H2 generation. In contrast, pyruvate oxidation with ferredoxin reduction has sufficient redox potential to generate H2. Thus it is tempting to suggest that the two pathways for electron transport coincide with electrons from lactate and those from pyruvate.
It is clear that the lactate dehydrogenases are membrane-bound flavoproteins capable of delivering electrons directly to the menaquinone pool in the membrane (Reed and Hartzell, 1999; Thauer et al., 2007). In contrast, the pyruvate:ferredoxin oxidoreductase is a soluble enzyme in the cytoplasm that delivers electrons to the soluble electron carrier, ferredoxin (Kletzin and Adams, 1996; Garczarek et al., 2007). Indeed, these electron pools must mix when the SRM grow on lactate in coculture with hydrogenotrophic methanogens, since lactate can be completely oxidized without the production of detectable pyruvate (Walker et al., 2009). During this syntrophic growth by D. vulgaris, the Coo membrane-bound hydrogenase has been shown to be needed, although it was not essential for monoculture on lactate plus sulfate (Walker et al., 2009). Interestingly, this protein complex belongs to a family of transmembrane complexes that function as proton (or sodium) – translocating hydrogenases and that use reduced ferredoxin as electron donors (Welte et al., 2010; Figure 1). The recently proposed mechanism of flavin-based electron bifurcation (Herrmann et al., 2008) could offer a possible solution to the necessity of lactate supported hydrogen production. This mechanism would suggest that, when concentrations of H2 are maintained at very low levels, the exergonic reduction of protons to form H2 from the reduced ferredoxin (E0′ ≤ 420 mV) could be coupled with the endergonic formation of hydrogen with electrons from menaquinone (E0′ -75 mV). The Coo membrane-bound hydrogenase complex is a flavoprotein that could allow bifurcation of electrons from the two sources, reduced ferredoxin and menaquinone. This model (Figure 1) would also provide an explanation for the decrease or cessation of electron flow in this pathway as hydrogen concentrations increased. Currently no experiments have been conducted to explore electron bifurcation during lactate oxidation and sulfate reduction in Desulfovibrio.
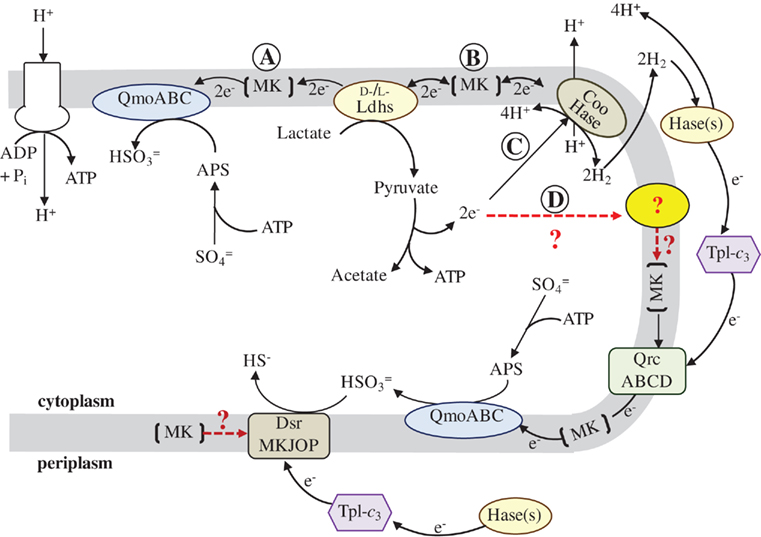
Figure 1. Proposed model for the flow of electrons during sulfate respiration in D. vulgaris Hildenborough. Abbreviations: QmoABC, Quinone-interacting membrane-bound oxidoreductase (DVU0848–0850); Ldhs, lactate dehydrogenases (nine annotated); Coo Hase, CO-induced membrane-bound hydrogenase (DVU2286–2293); Hase(s), periplasmic hydrogenases (four annotated); TpI-c3, Type-1 tetraheme cytochrome c3 (DVU3171); QrcABCD, Type-1 cytochrome c3:menaquinone oxidoreductase, formerly molybdopterin oxidoreductase (DVU0692–0695); DsrMKJOP, (DVU1290–1286); and MK, Menaquinone pool. Red, dashed lines and (?) indicate metabolic pathways for which less evidence is available. The reaction arrows were drawn as unidirectional for clarity of the model and electron flow. Pathway description is given in text.
In 2004, the genome sequence of D. vulgaris (Heidelberg et al., 2004) became available. In the ensuing few years, several additional SRM genomes have become accessible. If hydrogen cycling were a major mechanism for augmenting the energy budget of the SRM, one might predict that conserved hydrogenases would be found among these bacteria for this process. By making the simple comparison of the genomes of two species that from physiological analyses were thought to be closely related, D. vulgaris and Desulfovibrio alaskensis G20 (Hauser et al., 2011), it can be seen that the candidate cytoplasmic hydrogenases from D. vulgaris (Ech hydrogenase, DVU0429–0434, and Coo hydrogenase, DVU2286–2293) are not conserved in the G20 strain. However, that does not preclude the existence of multiple electron pathways to sulfate in the SRM.
Here we examine D. vulgaris gene expression data from microarrays of cells grown under different nutrient conditions, the relative abundance of transcripts from cells grown on lactate plus sulfate, and a random transposon mutant library to determine whether genes important for enzymes needed for energy generation in this growth mode become evident (Table 1).
Microarray data provide information about the changes in expression of genes in response to environmental changes but do not allow absolute levels of gene expression to be determined. Changes in gene expression for D. vulgaris have been generated through the Virtual Institute for Microbial Stress and Survival (VIMSS) program5 supported by the US Department of Energy and are available at www.microbesonline.org (Dehal et al., 2010). These data are log2 ratios of experimental versus control transcript abundances from gene probe microarrays with 12 replicates for each ratio. All transcript preparations were made from cells that were in the mid-exponential growth phase (OD600 of ca. 0.3) with the exception of the experiment following gene expression changes throughout the growth curve (Clark et al., 2006).
Our simple interpretations of the differential expression of genes in microarrays (Table 1) are as follows. Comparison of transcripts from cultures in stationary phase versus exponential phase for cells growing with lactate (60 mM) plus sulfate (50 mM; Clark et al., 2006) represent a lactate-limiting condition and preparation for a non-growing state. We have assumed that an indicator for gene importance in the metabolism of lactate plus sulfate as cells enter stationary phase, at the substrate concentrations given, would be an increase in expression of carbon acquisition genes. A decrease in those genes needed for sulfate reduction and energy generation might also be expected since the cells are slowing growth and sulfate is plentiful. For cultures grown to mid-exponential phase with thiosulfate versus sulfate as electron acceptor, we would predict that transcript changes would reflect a decrease for the sulfate-reducing enzymes but that sulfite reduction and carbon and electron metabolism would remain unchanged. For cultures growing exponentially with pyruvate as carbon and electron source compared to lactate, the genes differentially transcribed should include those required for lactate transport and oxidation. Clearly a direct connection between transcript changes and enzyme levels is often not evident (Torres-García et al., 2009). However, because energy conversion is such a large proportion of the metabolism of these bacteria, the possibility of correlated changes was tacitly assumed.
All microarray experiments in the database of the VIMSS program have been normalized to DNA signals on the microarrays. As a result, a robust measure of the relative abundance of mRNA for each gene has been accumulated during microarray experiments, with 173 data points in the calculation for each (Wall et al., 2009). We determined an average expression level from the mRNA/DNA signals and arbitrarily set the average for all genes to 1 or log2 = 0 ± 1.54. Thus a transcript value that is one SD above the average is a gene expressed approximately three fold more highly than average and is in the top 16% expression level of all genes. Because so much of the metabolic capacity of anaerobes is devoted to energy conversion, we assumed that the genes for the proteins and enzymes responsible would likely be among those above the average in expression.
To reveal genes needed for growth of D. vulgaris on lactate plus sulfate, the final information source comes from mutation analysis. The creation of a random transposon library in this bacterium with a modified Tn5-RL27 transposon (Larsen et al., 2002; Oh et al., 2010) has been initiated. To date, the Wall lab has 8,869 D. vulgaris mutants in which the insertion site of the transposon has been identified by sequencing that can be seen on the following website http://desulfovibriomaps.biochem.missouri.edu/mutants/. Most mutants were selected on lactate (60 mM) plus sulfate (30 mM) medium supplemented with yeast extract to avoid loss of auxotrophs. The library currently has mutations in 2208 unique genes or 62% of the ca. 3500 genes predicted in the genome. Our assumption for interpreting these data was that if a gene were essential for this growth mode, no mutations would be found in the gene. Of course, essential genes not specific to lactate/sulfate growth would also be identified among those not mutated. Conversely, genes actually needed, but whose loss could be compensated by expression of others, would likely be found to be mutated. As yet the library is not saturating for non-essential genes and the data must be interpreted with that caveat in mind.
Figure 2 provides two examples of the transposon mutant analysis. The enzymes for sulfate reduction have been biochemically confirmed (Peck and LeGall, 1994) and, as a result, the gene annotations are firm (Table 1). There are apparently single copies of these genes in the genome and we would not expect mutations to be found in cells growing by sulfate respiration. In Figure 2A, a section of the genome with the genes for the bisulfite reductase is illustrated with the insertion sites of the transposon indicated. There are no mutations in dsrABD while the flanking genes were successfully mutated. In the second example (Figure 2B), the genes putatively encoding the cytoplasmic hydrogenase associated with carbon monoxide dehydrogenase, cooMKLXUH, also lack insertions of the transposon, a result not entirely expected. This is a region is about 8.2 kb. With 8869 transposon insertions, on average we should expect an insertion about every 425 bp (with the conservative assumption that the entire genome is an appropriate target). Interestingly the region in which transposon mutations have not been recovered is precisely congruent with the Coo hydrogenase operon. It is probable that this region is needed for growth on lactate/sulfate or to survive the manipulations of the mutagenesis process.
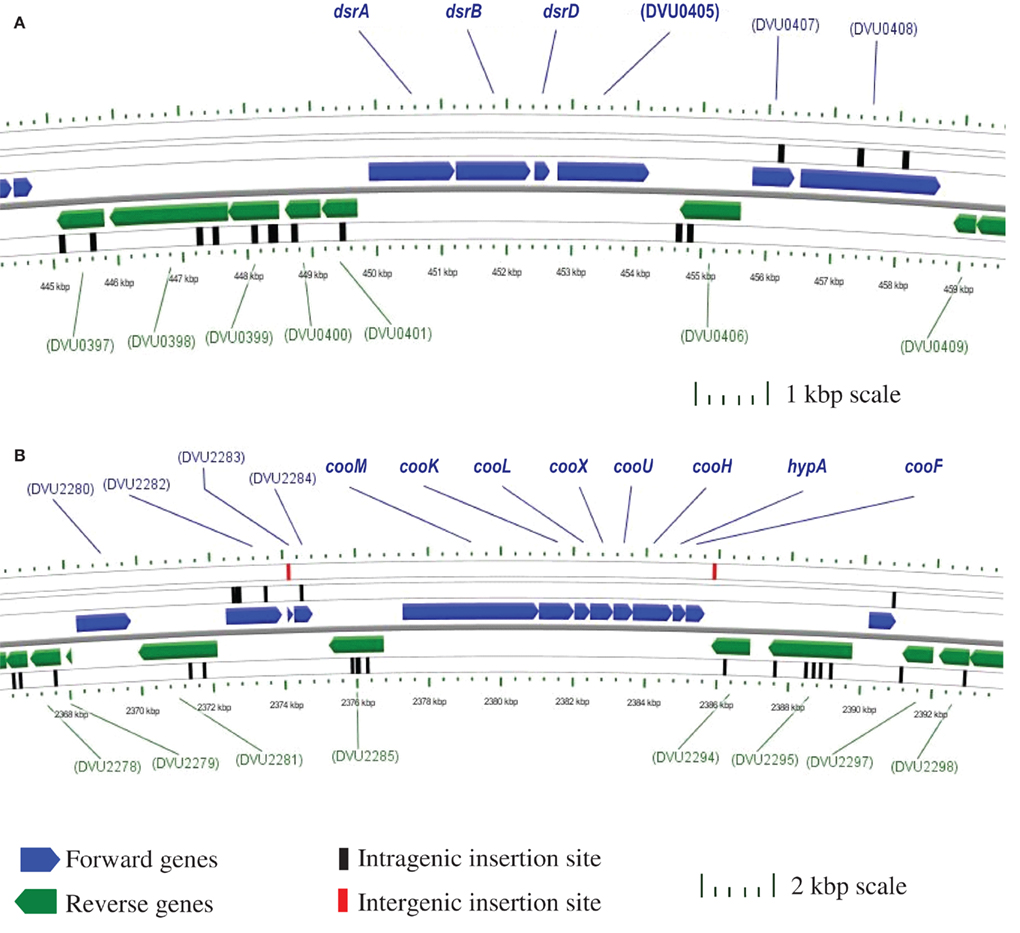
Figure 2. Chromosomal maps generated with CGView (Stothard and Wishart, 2004) indicating the location of transposon insertions in the Desulfovibrio vulgaris Hildenborough genome. Open reading frames are designated by TIGR DVU numbers. (A) No intragenic transposons are within the 4.2 kb four gene dissimilatory bisulfite reductase (dsr) operon; however within the 5 kb regions flanking the operon there are 14 intragenic insertions. (B) No intragenic transposons are within the 8.2 kb eight gene putative CO-induced membrane-bound hydrogenase (coo) operon. Also, there are 21 intragenic transposon insertions within the 10 kb regions up- and down-stream of the operon, as well as two intergenic transposon insertions.
The molecular information about genes potentially involved in lactate/sulfate growth of D. vulgaris – differential expression in defined culture conditions, relative abundance of mRNA and essentiality during growth with lactate/sulfate – are considered for the different categories of functions (Table 1). First, the genes known to encode APS reductase (apsBA) and bisulfite reductase (dsrABD), tended to decrease in expression in stationary phase cells limited for electron donor where growth rate was decreasing (Table 1). The expression levels of apsBA were also decreased when thiosulfate replaced sulfate. This result might be predicted since the substrate for the APS reductase was absent. Also as expected, dsrABD mRNA did not decrease with thiosulfate, since the initial two electron reduction of thiosulfate generates sulfide and bisulfite (Thauer et al., 2007). Measurements of the dissimilatory bisulfite reductase genes in Desulfobacterium autotrophicum grown in lactate/sulfate compared to lactate/thiosulfate (lactate limiting in both media) showed that early exponential phase expression of the dsrAB genes with sulfate was slightly higher than that observed with bisulfite (Neretin et al., 2003). As these D. autotrophicum cells entered stationary phase, the dsrAB expression in bisulfite medium exceeded that with sulfate. Major changes were not seen in the production of dsrAB transcripts in D. vulgaris with alternative electron acceptors consistent with the report of constitutive expression of these genes (Brusseau, 1998, as reported in Neretin et al., 2003).
Neither apsBA nor dsrABD was responsive to the change in electron donor from lactate to pyruvate. The predictions of abundant mRNAs for these genes (Table 1) and the absence of transposon mutations in the library (Figure 2A) were accurate and serve as the proof of principle for the use of these features as predictive measures. The transcript abundances for these genes were among the top 1% of all D. vulgaris genes.
For the highly expressed genes encoding sulfate adenylyltransferase and pyrophosphatase, sat and ppaC, the microarray data showed that transcription decreased with thiosulfate as terminal electron acceptor (Table 1). Because these enzymes function primarily for sulfate utilization, the mRNA decrease for the encoding genes might reflect a control by sulfate concentrations. No transposon mutations in these genes were isolated from cells grown on lactate/sulfate medium consistent with an essential function.
Because sulfate carries two charges at physiological pHs, transport functions would be expected. However, all growth conditions from which molecular and genetic information were obtained had plentiful sulfate, where electroneutral uptake with two protons might be expected (Cypionka, 1995). Three candidate sulfate permease genes were annotated, but no single one was apparently essential since transposon mutants lacking each were recovered (Table 1). The abundance level of the mRNA of each was near that of the average for D. vulgaris genes. Microarray changes also did not indicate a unique ortholog among the genes, although DVU0279 was strongly decreased in expression in stationary phase when carbon and electron flow was limited.
For genes encoding possible enzymes for lactate transport and oxidation, the story is not clear. While some biochemical analysis for lactate dehydrogenase has been accomplished (Odom and Peck, 1981b; Ogata et al., 1981; Stams and Hansen, 1982), the genes encoding the enzyme(s) responsible for either oxidation or transport have not been identified. Of the six genes annotated as putative lactate permeases, only two showed increases in transcription as the cells were being limited for carbon, DVU2110 and DVU3284. Neither of these was essential alone since mutations were found in each in lactate/sulfate grown cells and the mRNA abundance of both was well below average (Table 1). The two most highly transcribed orthologs were still only 1.6–1.7 times the average gene in expression.
The number of candidate genes encoding the lactate dehydrogenase (LDH) activities that have been annotated is about nine (Table 1), including those annotated as glycolate oxidases. Because many Desulfovibrio strains can readily use both L- and D-lactate (Stams and Hansen, 1982), at least two enzymes would seem to be necessary. Biochemical analysis of the LDHs showed that all are membrane-bound and NAD(P)+ -independent (Hansen, 1994b). However, the L- and D-LDH differ in their sensitivity to oxygen with the L-LDH from Desulfovibrio baculatus HL21 DSM 2555 (formerly, Desulfovibrio desulfuricans HL21) being extremely unstable (Stams and Hansen, 1982; Hansen, 1994a); whereas, the D-LDH from D. vulgaris Miyazaki is apparently much more stable when exposed to oxygen (Ogata et al., 1981).
Of the nine ldh orthologs and paralogs in D. vulgaris, none was increased in response to a change in electron donor from lactate to pyruvate (Table 1). Genes for three enzymes were increased from 2.1 to 2.6 fold upon entering stationary phase presumably because of carbon and electron donor limitation, DVU0600, DVU1783, and the complex DVU3027–3028. Of the three, only the latter complex had more abundant transcripts than the average gene. This complex is also part of a nine gene operon that includes por, encoding the primary pyruvate:ferredoxin oxidoreductase; pta, phosphotransacetylase; and ackA, the acetate kinase; and is preceded by a gene annotated as a lactate permease, DVU3026. Interestingly, por, pta, and ackA have not been mutated by the insertion of the transposon; whereas, seven transposon insertion sites have been identified within the coding sequences of the putative permease and lactate dehydrogenase, DVU3026, 3027, and 3028. We suggest that the transposon data mean that these genes are either not involved in lactate metabolism or that there are other enzymes able to compensate for the mutations in these genes.
The genes encoding the transmembrane electron transport complexes (TMC) that are proposed to function with APS reductase and bisulfite reductase are qmoABC and dsrMKJOP, respectively (Table 1; Matias et al., 2005; Pereira et al. (2007)). They are strictly conserved among sulfate reducers, underscoring their importance for sulfate reduction. The assignment of QmoABC and DsrMKJOP as electron carriers to their respective reductases also receives support from the location of their encoding genes (Pereira et al., 2007). qmoABC are immediately adjacent to and downstream of apsBA. Although the dsrMKJOP operon is not in the proximity of the structural genes for bisulfite reductase in D. vulgaris, it is contiguous with orthologs of the structural genes in the sulfur oxidizer, Allochromatium vinosum (Dahl et al., 2005). The genes for both of these TMCs were down regulated when D. vulgaris entered stationary growth phase caused by electron donor limitation (Table 1), as suggested for genes encoding functions involved in energy generation. Curiously both are also decreased in expression when thiosulfate replaced sulfate as terminal electron acceptor, even though DsrMKJOP would be predicted to be essential for thiosulfate use (Thauer et al., 2007). A change in electron donor did not appear to impact the transcription of the genes for either of the TMCs importantly. The relative mRNA abundances for these genes were among the top 20% of all genes in the bacterium and no transposon mutants were identified, supporting the supposition that their function is essential for growth on lactate plus sulfate.
A deletion of the qmoABC-DVU0851 was recently constructed by marker exchange mutagenesis in D. vulgaris (Zane et al., 2010). This mutant confirmed the essential role of the encoded complex in sulfate reduction. The deletion mutant was unable to grow with sulfate as terminal electron acceptor with any electron donor including H2 or formate, although growth with sulfite or thiosulfate was unimpaired. The hypothetical gene, DVU0851, was shown not to be necessary for sulfate reduction. Interestingly no other TMC of D. vulgaris was able to compensate for the loss of QmoABC.
Recently an additional four protein transmembrane complex, QrcDCBA or Type 1 cytochrome c3:menaquinone oxidoreductase, has been proposed to be essential to carry electrons coming from H2 or formate to the cytoplasmic membrane for delivery to APS reductase in D. vulgaris (Venceslau et al., 2010). The Qrc complex was suggested to be necessary because the QmoABC complex apparently has no periplasmic interface that would allow electrons from the soluble cytochrome c matrix to traverse the membrane to reach the sulfate-reducing enzymes. Curiously 10 of 12 fully sequenced Desulfovibrio strains have highly conserved qrc operons2; however, D. desulfuricans ATCC 27774 and Desulfovibrio piger do not. Because these two species still have a conserved Qmo complex, another complex may be needed to serve the Qrc function of delivery of periplasmic electrons to the membrane for sulfate reduction (Venceslau et al., 2010).
A transposon mutation in the D. alaskensis G20 gene Dde_2933 corresponding to qrcB, was unable to grow with H2 or formate as electron donor (Li et al., 2009). In D. vulgaris, QrcDCBA was also decreased in expression during stationary phase when energy generation was decreased (Table 1) and was found to have above average transcript abundance. Surprisingly, transposon insertions in the genes for this complex were isolated in cells growing on lactate plus sulfate, showing that either this complex was not required for electrons from all donors or that compensation for this complex was possible.
A deletion of DVU0692–0694, ΔqrcBCD, has been constructed in D. vulgaris (Wall lab, unpublished). Preliminary experiments showed that this mutant was capable of growing on lactate plus sulfate but the rate was slower and the yield was about 40% less than that of wild-type cells. As for the D. alaskensis G20 mutant, the D. vulgaris deletion could not grow with H2 or formate with sulfate as electron acceptor, consistent with the predicted role for this complex by Venceslau et al. (2010). A need for the Qrc complex should be bypassed if electrons from the periplasmic cytochrome pool could be transferred to bisulfite reductase through a cytochrome c containing complex. Because DsrJ of the DsrMKJOP complex is a periplasmic cytochrome c and the complex is predicted to be the electron conduit to bisulfite reductase, this complex would be predicted to be able to accept periplasmically generated electrons from H2 or formate carried by the abundant Tp1-c3. Sulfite does support growth of the qrc mutant on formate. The inability of the ΔqmoABC mutant to grow on lactate, H2 or formate during sulfate respiration would suggest that the Qrc complex acts prior to Qmo. The possible role of QrcABCD must be further investigated.
Of the remaining TMCs characterized or recognized in the D. vulgaris genome, the High molecular weight cytochrome (Hmc) complex has been proposed by many researchers to be a conduit for electrons from H2 to sulfate (Peck, 1993; Dolla et al., 2000; Pereira et al., 2007). A mutant deleted for hmcBCDE was impaired in syntrophic growth with Methanococcus maripaludis (Walker et al., 2009). The low level of transcript abundance for this complex during growth on lactate/sulfate (Table 1) would not appear compatible with a primary role for Hmc for energy conversion on these substrates. In addition, the presence of multiple transposon insertions within the Hmc coding sequences in the mutant library casts doubt on its importance. The inference that a functional Hmc is not essential for D. vulgaris in lactate plus sulfate medium is consistent with monoculture growth, albeit slower, of the deletion mutant (Dolla et al., 2000; Walker et al., 2009). These observations support a role for this TMC when H2 must be maintained at a low level.
A role for the decaheme cytochrome c and the RnfCDGEAB complex in Desulfovibrio has not been established. This complex was first identified in Rhodobacter capsulatus as complex required for nitrogen fixation (Schmehl et al., 1993). It is also in the family of electron bifurcating flavoproteins that are NADH ferredoxin oxidoreductases (Herrmann et al., 2008).Transcript changes and abundances were not particularly informative. However, several transposon mutations have been identified in the encoding genes indicating that the complex is not essential to D. vulgaris cells growing on lactate plus sulfate. A deletion of rnfC constructed by marker replacement confirmed a role for this complex in the ability of D. vulgaris to grow with molecular nitrogen as the sole nitrogen source (Wall lab, unpublished).
Finally, tmcABCD and ohcABC (Pereira et al., 2007) have been identified as encoding TMCs but a role for each is not yet known. The tmc genes are expressed at about the same level as the dsrMKJOP operon suggesting a role in the metabolism (Table 1), but preliminary characterization of a mutant deleted for the genes had no apparent phenotype (Wall lab, unpublished). This was consistent with finding transposon mutations within the structural genes for the complex. In contrast, ohcABC transcripts were in the lower 15% abundance of all D. vulgaris genes, perhaps indicating a non-critical role in energy metabolism in cells growing with lactate plus sulfate.
The presence and activity of cytoplasmic hydrogenases were predicted to be necessary for energy augmentation by hydrogen cycling in sulfate reducers (Odom and Peck, 1981a). Although the distribution of cytoplasmic hydrogenases in Desulfovibrio strains is not conserved, there are candidate enzymes that could function in most cases. D. vulgaris has two putative energy-converting hydrogenases with subunits related to the proton-pumping NADH:ubiquinone oxidoreductase (complex I), Ech hydrogenase (Friedrich and Scheide, 2000) and the Coo hydrogenase, CooH, originally described in Rhodospirillum rubrum (Kerby et al., 1992). These enzyme complexes are reversible, oxidizing reduced ferredoxin with the production of H2 while generating an electrochemical proton potential or using reversed electron transport to catalyze reduced ferredoxin from H2 (Thauer et al., 2007). These complexes are encoded in DVU0429–0434 and DVU2286–2293, respectively (Table 1). Transcription expression changes were not dramatic for these complexes, but the transcript abundance difference between the two was striking. The cooMKLZUHhypAcooF operon is among the top 15% of expressed genes; whereas, the echABCDEF operon is well below average in expression. As seen earlier in Figure 1, mutations in this coo operon were not recovered in cells grown with lactate/sulfate, suggesting that this cytoplasmic hydrogenase is needed for this growth mode. It should be noted that the putative structural genes for the CO dehydrogenase enzyme, cooSC, were found to carry transposon insertions, as did the ech operon.
Through examination of a mutation in cooL, the Coo hydrogenase has also been shown to be essential for syntrophic growth of D. vulgaris with Methanococcus on lactate but not pyruvate (Walker et al., 2009). However, there was no requirement for this hydrogenase for respiration of lactate with sulfate. This observation is curiously in conflict with the absence of transposon mutations in these genes when selection was carried out on lactate/sulfate medium. Interestingly, Coo hydrogenase belongs to a family of transmembrane complexes that function as proton (or sodium) – translocating hydrogenases and that use reduced ferredoxin as electron donors (Welte et al., 2010; Figure 1). The recently proposed mechanism of flavin-based electron bifurcation (Herrmann et al., 2008) could offer a possible solution to the necessity of lactate supported hydrogen production. This mechanism would suggest that, when concentrations of H2 are maintained at very low levels, the exergonic reduction of protons to form H2 from the reduced ferredoxin (E0′ ≤ 420 mV) could be coupled with the endergonic formation of hydrogen with electrons from menaquinone (E0′ -75 mV). The Coo membrane-bound hydrogenase complex is a flavoprotein potentially allowing bifurcation of electrons from the two sources, reduced ferredoxin and menaquinone. While the energy available for pumping a proton (or sodium ion) might be dissipated through the bifurcation, the overall contribution to the proton gradient from the oxidation of the additional H2 might compensate. This model (Figure 1) would also provide an explanation for the cessation of electron flow in this pathway as hydrogen concentrations increased. Currently no experiments have been conducted to explore electron bifurcation function during lactate oxidation and sulfate reduction in Desulfovibrio.
While the four periplasmic hydrogenases have been well characterized (Fontecilla-Camps et al., 1997; Vignais and Billoud, 2007), no single enzyme has been shown to be essential for uptake or production of H2 (Casalot et al., 2002; Caffrey et al., 2007). While the transcript abundance of the [NiFeSe] hydrogenase appears to be the highest of the four, this may reflect the inclusion of selenium in the medium trace elements which strongly increases the production of the [NiFeSe] hydrogenase while repressing the [FeFe] and [NiFe]-1 hydrogenases (Valente et al., 2006). Multiple deletions and various combinations will be needed to determine the roles of these enzymes.
Of the annotated c-type cytochromes that are not apparently associated with large TMCs, the Type I tetraheme cytochrome c3 (Tp1-c3, DVU3171) and the monoheme c553 cytochrome (DVU1817) have highly abundant transcripts (Table 1) suggesting a major role in electron flow during energy conversion. The high expression levels are congruent with the abundance of the cytochromes as well (Moura et al., 1991; Pollock and Voordouw, 1994; Aubert et al., 1998). Tp1-c3 has been suggested to be the electron acceptor for each of the periplasmic hydrogenases and to serve as a capacitor for electrons in the periplasm of the SRM (Heidelberg et al., 2004). Interestingly, there is little expression modulation of the gene encoding, Tp1-c3, cycA, upon entry into stationary phase or in response to altered electron acceptors or donors as has previously been reported (Payne et al., 2002). No transposon mutations have been recovered that would eliminate Tp1-c3, although the small size of cycA may be a factor in mutant isolation. However, the successful construction of a marker exchange deletion of the encoding gene (Semkiw et al., 2010), showed that Tp1-c3 is not essential for growth on lactate/sulfate. Similar mutants have been generated in D. alaskensis G20 (Rapp-Giles et al., 2000; Li et al., 2009). The D. vulgaris ΔcycA mutant was unable to grow with H2 or formate with sulfate or sulfite as electron acceptor (Semkiw et al., 2010). Growth was slowed with lactate plus sulfate or sulfite but the biomass produced was not different from wild-type. For cells growing with sulfate respiration, the observations mirror those for the Δqrc mutant of this bacterium or transposon interruptions in the homologs found in D. alaskensis G20 (Li et al., 2009). These results suggest a pathway for periplasmically generated electrons through Tp1-c3 and the Qrc complex to the Qmo. Whether both the cytochrome and the Qrc complex are needed to transfer electrons for bisulfite reduction remains to be established.
The gene for the abundant cytochrome c553 (cyf, DVU1817) is located just upstream of the cytochrome c oxidase, a putative heme-copper oxygen reductase. It has been suggested that the cytochrome c553 might serve as an electron donor to that membrane-bound complex (Dolla et al., 2007). No major differences in expression were apparent in the microarray data. It might be predicted that constitutive expression could be advantageous if this cytochrome plays a role in oxygen protection. A deletion of cyf grew well on lactate plus sulfate (Wall lab, unpublished) confirming that this cytochrome is not a key electron carrier for energy conversion under these conditions. A possible role in oxygen tolerance will require further experimentation.
The function of other c-type cytochromes is still undetermined and will likely require multiple deletions and biochemical analysis to gain insight. The lack of transposon mutations in a number of the encoding genes may simply reflect the small size of the genes and the lack of saturation in the library of mutants.
In summary, the data in Table 1 show that the transcriptional changes in the microarray data suggest that genes for both sulfate and sulfite reduction are not constitutively expressed but respond to the energy demands of the cell, decreasing in expression in stationary phase and when thiosulfate is provided as electron acceptor. The operons for the proposed transmembrane complexes for donating electrons to APS reductase and that for bisulfite reductase were similarly decreased under these conditions; whereas, operons for other putative transmembrane complexes were not uniformly changed in transcription in these conditions. The genes and operons for sulfate and sulfite reduction were among the most highly transcribed genes in the cells and, as would be predicted while growing with lactate plus sulfate, transposon insertions in these genes were not recovered. Transposons were recovered in almost all other operons and genes listed with the interesting exception of the Coo hydrogenase complex which showed a surprisingly low number of insertions. This complex has been shown not to be essential for growth on lactate plus sulfate but it appears to be needed for syntrophic growth with Methanococcus maripaludis with lactate as sole source of carbon and reductant (Walker et al., 2009 and for the efficient recovery of transposon mutants. Because this complex is a member of the energy-converting [NiFe] hydrogenases (Hedderich, 2004), it is predicted that this complex is integral to the energy budget at some stage of culturing for single colony formation on medium with lactate as electron donor and sulfate as electron acceptor.
Important information about electron carriers has been gained from constructed deletions and from transposon insertions. However, the genes encoding a number of the enzymes believed to be needed for electron flow in D. vulgaris have not been specifically identified in part because multiple ORFs have been similarly annotated. Examples include the periplasmic hydrogenases, formate dehydrogenases, lactate permeases, lactate dehydrogenases, and others. When multiple genes encode isozymes, the loss of one enzyme by mutation of its encoding gene may be compensated by others. This compensation or suppression could explain why transposon mutations are observed in genes for what would be predicted to be key enzymes (Table 1). To elucidate the function of individual members of protein families, multiple genes will likely need to be eliminated simultaneously. Until recently, the number of sequential deletions in the same strain of D. vulgaris was limited by the number of reliable antibiotic resistant markers available to use for marker exchange deletion construction. A markerless deletion system has now been developed based on the counterselection of the gene for uracil phosphoribosyltransferase, upp, by resistance to the toxic uracil analog 5-fluorouracil (Keller et al., 2009). This method allows the generation of in-frame deletions without a residual antibiotic resistance gene. The application of this technique may contribute critical information about any unique roles of the four periplasmic hydrogenases. The flexible electron flow pathways and mechanisms of energy conversion in D. vulgaris will require the combined application of genetic, biochemical and systems biology approaches.
To provide a framework for organizing the information obtained to date for electron flow from lactate to sulfate, a model is presented in Figure 1. This scheme draws heavily from the models presented by Noguera et al. (1998) and Walker et al. (2009) and is driven by the observations of two proteins putatively involved in the electron pathways, the Tp1-c3 and the Coo hydrogenase. First, the phenotype of a deletion of the cycA gene encoding the Tp1-c3 indicates that electrons from periplasmic H2 (or formate) are exclusively dependent on this cytochrome for reaching sulfate. Thus the ability of the ΔcycA mutant to grow on organic acids with sulfate and produce sulfide means that some proportion of the electrons generated from substrate oxidation must not cycle as molecular H2. Thus the hydrogen cycling hypothesis may contribute but is not essential to the energy budget of SRM grown with organic acids. Second, a mutant in the Coo hydrogenase complex cooL::Tn5-RL27 was unable to grow by syntrophy with a methanogen on lactate but could grow in monoculture with lactate plus sulfate (Walker et al., 2009). An interpretation of these observations is that the mutation in the Coo hydrogenase complex prevented H2 generation from lactate but did not block electron flow to sulfate. With pyruvate as electron donor, syntrophy was established as well as growth of monocultures (Walker et al., 2009). Thus a path for H2 production is available for electrons from pyruvate that is not available to electrons from lactate. In addition, the apparent lack of recovery of transposon mutations in the cooMKLXUHhypAcooF genes in the recently generated library, points to Coo hydrogenase as potentially important, but not necessarily essential, player in electron flow from lactate to sulfate.
The model accommodates these seemingly contradictory observations by suggesting two pathways for the electrons from lactate (Figure 1A and B) and from pyruvate (Figure 1C, and D). In each case, one pathway has the potential for transfer of electrons into the menaquinone pool for delivery to the membrane associated complex, Qmo, that is dedicated for the first step in sulfate reduction (pathways A and D). Whereas the DsrMKJOP complex is thought to be the transmembrane complex for electron donation to bisulfite reductase, it may also receive electrons directly from the menaquinone pool. The second path in each case, B and C, provides a route for the electrons to generate H2. The model is already too simple for electron flow from pyruvate since H2 generation in the absence of a functional Coo hydrogenase is documented (Walker et al., 2009). The question marks in the model indicate the components that have a high uncertainty, including the membrane associated component(s) accepting electrons from pyruvate oxidation.
A future effort will be to determine the gatekeepers for directing electrons into one path or another. Will this be just the availability of acceptors or will the rate of flux through the pathways determine the choices? The complexity of this simple system continues to impress. It is already evident that there will not be a single wiring circuit for electron flow from these substrates in the SRM as it does not extend to the physiologically similar D. alaskensis G20 strain that lacks orthologs encoding the cytoplasmic hydrogenases of D. vulgaris.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work, conducted by ENIGMA, was supported by the Office of Science, Office of Biological and Environmental Research, of the US Department of Energy under Contract No. DE-AC02-05CH11231. A portion of the work was supported by the Office of Science, Office of Biological and Environmental Research, Genomics Program:GTL BioHydrogen Production and BioEthanol contract DE-FG02-083464691. In addition, some of this work was supported as part of the Virtual Institute for Microbial Stress and Survival (http://VIMSS.lbl.gov) supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomics Program:GTL through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy. We thank Barbara J. Rapp-Giles, Elizabeth S. Semkiw, and Grant M. Zane from the Wall lab for graciously sharing their unpublished data and Adam Deutschbauer and Jennifer Kuehl for their contributions to the transposon library.
Aubert, C., Leroy, G., Bianco, P., Forest, E., Bruschi, M., and Dolla, A. (1998). Characterization of the cytochromes c from Desulfovibrio desulfuricans G201. Biochem. Biophys. Res. Commun. 242, 213–218.
Bender, K. S., Yen, H.-C. B., Hemme, C. L., Yang, Z., He, Z., He, Q., Zhou, J., Huang, K. H., Alm, E. J., Hazen, T. C., Arkin, A. P., and Wall, J. D. (2007). Analysis of a ferric uptake regulator (fur) mutant of Desulfovibrio vulgaris Hildenborough. Appl. Enivron. Microbiol. 73, 5389–5400.
Brusseau, G. A. (1998). The Role of Syntrophy in the Expression of key Metabolic Enzymes in Sulfate-Reducing Bacteria. Ph.D. dissertation, Northwestern University, Evanston, IL.
Butlin, K. R., Adams, M. E., and Thomas, M. (1949). The isolation and cultivation of sulphate-reducing bacteria. J. Gen. Microbiol. 3, 46–59.
Caffrey, S. M., Park, H. S., Voordouw, J. K., He, Z., Zhou, J., and Voordouw, G. (2007). Function of periplasmic hydrogenases in the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 189, 6159–6167.
Caffrey, S. M., and Voordouw, G. (2010). Effect of sulfide on growth physiology and gene expression of Desulfovibrio vulgaris Hildenborough. Antonie van Leeuwenhoek 97, 11–20.
Casalot, L., Valette, O., De Luca, G., Dermoun, Z., Rousset, M., and de Philip, P. (2002). Construction and physiological studies of hydrogenase depleted mutants of Desulfovibrio fructosovorans. FEMS Microbiol. Lett. 214, 107–112.
Clark, M. E., He, Q., He, Z., Huang, K. H., Alm, E. J., Wan, X. F., Hazen, T. C., Arkin, A. P., Wall, J. D., Zhou, J. Z., and Fields, M.W. (2006). Temporal transcriptomic analysis as Desulfovibrio vulgaris Hildenborough transitions into stationary phase during electron donor depletion. Appl. Environ. Microbiol. 72, 5578–5588.
Cypionka, H. (1995). “Solute transport and cell energetic,” in Sulfate-Reducing Bacteria, ed. L. L. Barton (New York, NY: Plenum Press), 151–184.
Dahl, C., Engels, S., Pott-Sperling, A. S., Schulte, A., Sander, J., Lübbe, Y., Deuster, O., and Brune, D. C. (2005). Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J. Bacteriol. 187, 1392–1404.
Dehal, P. S., Joachimiak, M. P., Price, M. N., Bates, J. T., Baumohl, J. K., Chivian, D., Friedland, G. D., Huang, K. H., Keller, K., Novichkov, P. S., Dubchak, I. L., Alm, E. J., and Arkin, A. P. (2010). MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 38(Suppl. 1), D396–D400.
Dolla, A., Kurtz, D. M. Jr., Teixeira, M., and Voordouw, G. (2007). “Biochemical, proteomic and genetic characterization of oxygen survival mechanisms in sulphate-reducing bacteria of the genus Desulfovibrio,” in Sulphate-Reducing Bacteria: Environmental and Engineered Systems, eds L. L. Barton, and W. A. Hamilton (Cambridge: Cambridge University Press), 185–213.
Dolla, A., Pohorelic, B. K., Voordouw, J. K., and Voordouw, G. (2000). Deletion of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox-potential niche establishment. Arch. Microbiol. 174, 143–151.
Dong, J. M., Taylor, J. S., Latour, D. J., Iuchi, S., and Lin, E. C. C. (1993). Three overlapping ict genes involved in L-lactate utilization by Escherichia coli. J. Bacteriol. 175, 6671–6678.
Fontecilla-Camps, J. C., Frey, M., Garcin, E., Hatchikian, C., Montet, Y., Piras, C., Vernède, X., and Volbeda, A. (1997). Hydrogenase: a hydrogen-metabolizing enzyme. What do the crystal structures tell us about its mode of action? Biochimie 79, 661–666.
Friedrich, T., and Scheide, D. (2000). The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 479, 1–5.
Fu, R., and Voordouw, G. (1997). Targeted gene-replacement mutagenesis of dcrA, encoding an oxygen sensor of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Microbiology 143, 1815–1826.
Garczarek, F., Dong, M., Typke, D., Witkowska, H. E., Hazen, T. C., Nogales, E., Biggin, M. D., and Glaeser, R. M. (2007). Octomeric pyruvate:ferredoxin oxidoreductase from Desulfovibrio vulgaris. J. Struct. Biol. 159, 9–18.
Groh, J. L., Luo, Q., Ballard, J. D., and Krumholz, L. R. (2005). A method adapting microarray technology for signature-tagged mutagenesis of Desulfovibrio desulfuricans G20 and Shewanella oneidensis MR-1 in anaerobic sediment survival experiments. Appl. Environ. Microbiol. 71, 7064–7074.
Hansen, T. A. (1994a). Metabolism of sulfate-reducing prokaryotes. Antonie Van Leeuwenhoek 66, 165–185.
Hansen, T. A. (1994b). “NAD(P)-independent lactate dehydrogenase from sulfate-reducing prokaryotes,” in Methods in Enzymology: Inorganic Microbial Sulfur Metabolism, eds H. D. Jr. Peck, and J. LeGall (New York: Academic Press), 21–23.
Hauser, L. J., Land, M. L., Brown, S. D., Larimer, F., Keller, K. L., Rapp-Giles, B. J., Price, M. N., Lin, M., Bruce, D. C., Detter, J. C., Tapia, R., Han, C. S., Goodwin, L. A., Cheng, J.-F., Pitluck, S., Copeland, A., Lucas, S., Nolan, M., . Lapidus, A. L, Palumbo, A. V., and Wall, J. D. (2011). The complete genome sequence and updated annotation of Desulfovibrio alaskensis G20. J. Bacteriol.. [Epub ahead of print].
He, Z., Zhou, A., Baidoo, E., He, Q., Joachimiak, M. P., Benke, P., Phan, R., Mukhopadhyay, A., Hemme, C. L., Huang, K., Alm, E. J., Fields, M. W., Wall, J., Stahl, D., Hazen, T. C., Keasling, J. D., Arkin, A. P., and Zhou, J. (2010). Global transcriptional, physiological, and metabolite analyses of the responses of Desulfovibrio vulgaris Hildenborough to salt adaptation. Appl. Environ. Microbiol. 76, 1574–1586.
Hedderich, R. (2004). Energy-converting [NiFe] hydrogenases from Archaea and extremophiles: ancestors of complex I. J. Bioenerg. Biomembr. 36, 65–75.
Heidelberg, J. F., Seshadri, R., Haveman, S. A., Hemme, C. L., Paulsen, I. T., Kolonay, J. F., Eisen, J. A., Ward, N., Methe, B., Brinkac, L. M., Daugherty, S. C., Deboy, R. T., Dodson, R. J., Durkin, A. S., Madupu, R., Nelson, W. C., Sullivan, S. A., Fouts, D., Haft, D. H., Delengut, J., Peterson, J. D., Davidsen, T. M., Zafar, N., Zhou, L., Radune, D., Dimitrov, G., Hance, M., Tran, K., Khouri, H., Gill, J., Utterback, T. R., Feldblyum, T. V., Wall, J. D., Voordouw, G., and Fraser, C. M. (2004). The genome sequence of the anaerobic sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22, 554–559.
Herrmann, G., Jayamani, E., Mai, G., and Buckel, W. (2008). Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 190, 784–791.
Hooper, A. B., and DiSpirito, A. A. (1985). In bacteria which grow on simple reductants, generation of a proton gradient involves extracytoplasmic oxidation of substrate. Microbiol. Rev. 49, 140–157.
Keller, K. L., Bender, K. S., and Wall, J. D. (2009). Development of a markerless genetic exchange system in Desulfovibrio vulgaris Hildenborough and its use in generating a strain with increased transformation efficiency. Appl. Environ. Microbiol. 75, 7682–7691.
Kerby, R. L., Hong, S. S., Ensign, S. A., Coppoc, L. J., Ludden, P. W., and Roberts, G. P. (1992). Genetic and physiological characterization of the Rhodospirillum rubrum carbon monoxide dehydrogenase system. J. Bacteriol. 174, 5284–5294.
Kletzin, A., and Adams, M. W. (1996). Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J. Bacteriol. 178, 248–257.
Larsen, R. A., Wilson, M. M., Guss, A. M., and Metcalf, W. W. (2002). Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178, 193–201.
Li, X., Luo, Q., Wofford, N. Q., Keller, K. L., McInerney, M. J., Wall, J. D., and Krumholz, L. R. (2009). A molybdopterin oxidoreductase is involved in H2 oxidation in Desulfovibrio desulfuricans G20. J. Bacteriol. 191, 2675–2682.
Lupton, F. S., Conrad, R., and Zeikus, J. G. (1984). Physiological function of hydrogen metabolism during growth of sulfidogenic bacteria on organic substrates. J. Bacteriol. 159, 843–849.
Marschall, C., Frenzel, P., and Cypionka, H. (1993). Influence of oxygen on sulfate reduction and growth of sulfate-reducing bacteria. Arch. Microbiol. 159, 168–173.
Matias, P. M., Pereira, I. A., Soares, C. M., and Carrondo, M. A. (2005). Sulphate respiration from hydrogen in Desulfovibrio bacteria: a structural biology overview. Prog. Biophys. Mol. Biol. 89, 292–329.
Moura, J. J., Costa, C., Liu, M. Y., Moura, I., and LeGall, J. (1991). Structural and functional approach toward a classification of the complex cytochrome c system found in sulfate-reducing bacteria. Biochim. Biophys. Acta 1058, 61–66.
Mukhopadhyay, A., He, Z., Alm, E. J., Arkin, A. P., Baidoo, E. E., Borglin, S. C., Chen, W., Hazen, T. C., He, Q., Holman, H. Y., Huang, K., Huang, R., Joyner, D. C., Katz, N., Keller, M., Oeller, P., Redding, A., Sun, J., Wall, J., Wei, J., Yang, Z., Yen, H. C., Zhou, J., and Keasling, J. D. (2006). Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J. Bacteriol. 188, 4068–4078.
Neretin, L. N., Schippers, A., Pernthaler, A., Hamann, K., Amann, R., and Jørgensen, B. B. (2003). Quantification of dissimilatory (bi)sulphite reductase gene expression in Desulfobacterium autotrophicum using real-time RT-PCR. Environ. Microbiol. 5, 660–671.
Noguera, D. R., Brusseau, G. A., Rittmann, B. E., and Stahl, D. A. (1998). A unified model describing the role of hydrogen in the growth of Desulfovibrio vulgaris under different environmental conditions. Biotechnol. Bioeng. 59, 732–746.
Odom, J. M., and Peck, H. D. Jr. (1981a). Hydrogen cycling as a general mechanism for energy coupling in the sulfate reducing bacteria, Desulfovibrio sp. FEMS Microbiol. Lett. 12, 47–50.
Odom, J. M., and Peck, H. D. Jr. (1981b). Localization of dehydrogenases, reductases, and electron transfer components in the sulfate-reducing bacterium Desulfovibrio gigas. J. Bacteriol. 147, 161–169.
Odom, J. M., and Wall, J. D. (1987). Properties of a hydrogen-inhibited mutant of Desulfovibrio desulfuricans ATCC 27774. J. Bacteriol. 169, 1335–1337.
Ogata, M., Arihara, K., and Yagi, T. (1981). D-lactate dehydrogenase of Desulfovibrio vulgaris. J. Biochem. 89, 1423–1431.
Oh, J., Fung, E., Price, M. N., Dehal, P. S., Davis, R.W., Giaever, G., Nislow, C., Arkin, A. P., and Deutschbauer, A. (2010). A universal TagModule collection for parallel genetic analysis of microorganisms. Nuc. Acids Res. 38, e146.
Pankhania, I. P., Gow, L. A., and Hamilton, W. A. (1986). The effect of hydrogen on the growth of Desulfovibrio vulgaris Hildenborough on lactate. J. Gen. Microbiol. 132, 3349–3356.
Payne, R. B., Gentry, D. M., Rapp-Giles, B. J., Casalot, L., and Wall, J. D. (2002). Uranium reduction by Desulfovibrio desulfuricans strain G20 and a cytochrome c3 mutant. Appl. Environ. Microbiol. 268, 3129–3132.
Peck, H. D. Jr. (1960). Evidence for oxidative phosphorylation during the reduction of sulfate with hydrogen by Desulfovibrio desulfuricans. J. Biol. Chem. 235, 2734–2738.
Peck, H. D. Jr. (1966). Phosphorylation coupled with electron transfer in extracts of the sulfate reducing bacterium, Desulfovibrio gigas. Biochem. Biophys. Res. Comm. 22, 112–118.
Peck, H. D. Jr. (1993). “Bioenergetics strategies of the sulfate-reducing bacteria,” in The Sulfate-Reducing Bacteria: Contemporary Perspectives, eds J. M. Odom, and R. Jr. Singleton (New York, NY: Springer), 41–76.
Peck, H. D. Jr., and LeGall, J. (1994). Methods in Enzymology: Inorganic Microbial Sulfur Metabolism. New York: Academic Press.
Peck, H. D. Jr., LeGall, J., Lespinat, P. A., Berlier, Y., and Fauque, G. (1987). A direct demonstration of hydrogen cycling by Desulfovibrio vulgaris employing membrane-inlet mass spectrometry. FEMS Microbiol. Lett. 40, 295–299.
Pereira, I. A. C., Haveman, S. A., and Voordouw, G. (2007). “Biochemical, genetic and genomic characterization of anaerobic electron transport pathways in sulphate-reducing “ Deltaproteobacteria,” in Sulphate-Reducing Bacteria, eds L. L. Barton, and W. A. Hamilton (Cambridge: Cambridge University Press), 215–240.
Pereira, P. M., He, Q., Valente, F. M., Xavier, A. V., Zhou, J., Pereira, I. A., and Louro, R. O. (2008). Energy metabolism in Desulfovibrio vulgaris Hildenborough: insights from transcriptome analysis. Antonie Van Leeuwenhoek 93, 347–362.
Pollock, W. B., and Voordouw, G. (1994). Molecular biology of c-type cytochromes from Desulfovibrio vulgaris Hildenborough. Biochimie 76, 554–560.
Postgate, J. R. (1984a). The Sulphate-Reducing Bacteria, 2nd Edn. New York: Cambridge University Press.
Postgate, J. R. (1984b). “Genus Desulfovibrio,” in Bergey’s Manual of Systematic Bacteriology, eds N. R. Krieg, and J. G. Holt (Baltimore, MD: Williams & Wilkins), 666–672.
Powell, B., Mergeay, M., and Christofi, N. (1989). Transfer of broad host-range plasmids to sulphate-reducing bacteria. FEMS Microbiol. Lett. 59, 269–273.
Rabus, R., Hansen, T. A., and Widdel, F. (2006). “Dissimilatory sulfate- and sulfur-reducing prokaryotes,” in The Prokaryotes, eds M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (New York, NY: Springer-Verlag), 659–768.
Rapp-Giles, B. J., Casalot, L., English, R. S., Ringbauer, J. A. Jr., Dolla, A., and Wall, J. D. (2000). Cytochrome c3 mutants of Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 66, 671–677.
Redding, A. M., Mukhopadhyay, A., Joyner, D. C., Hazen, T. C., and Keasling, J. D. (2006). Study of nitrate stress in Desulfovibrio vulgaris Hildenborough using iTRAQ proteomics. Brief Funct. Genomic. Proteomic. 2, 133–143.
Reed, D. W., and Hartzell, P. L. (1999). The Archaeoglobus fulgidus D-lactate dehydrogenase is a Zn2+ flavoprotein. J. Bacteriol. 181, 7580–7587.
Rousset, M., Casalot, L., Rapp-Giles, B. J., Dermoun, Z., de Philip, P., Bélaich, J-P., and Wall, J. D. (1998). New shuttle vectors for the introduction of cloned DNA in Desulfovibrio. Plasmid 39, 114–122.
Rousset, M., Dermoun, Z., Chippaux, M., and Bélaich, J. P. (1991). Marker exchange mutagenesis of the hydN genes in Desulfovibrio fructosovorans. Mol. Microbiol. 7, 1735–1740.
Sass, H., and Cypionka, H. (2007). “Response of sulphate-reducing bacteria to oxygen,” in Sulphate-Reducing Bacteria, eds L. L. Barton, and W. A. Hamilton (Cambridge: Cambridge University Press), 167–183.
Schmehl, M., Jahn, A., Meyer zu Vilsendorf, A., Hennecke, S., Masepohl, B., Schuppler, M., Marxer, M., Oelze, J., and Klipp, W. (1993). Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 241, 602–615.
Semkiw, E. S., Zane, G. M., and Wall, J. D. (2010). “The role of the type-1 tetraheme cytochrome c3 in Desulfovibrio vulgaris Hildenborough metabolism (Abstract),” in 110th General Meeting of American Society For Microbiology, San Diego, CA, K-773.
Stams, A. J. M., and Hansen, T. A. (1982). Oxygen-labile L(+) lactate dehydrogenase activity in Desulfovibrio desulfuricans. FEMS Microbiol. Lett. 13, 389–394.
Stothard, P., and Wishart, D. S. (2004). Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539.
Tang, Y., Pingitore, F., Mukhopadhyay, A., Phan, R., Hazen, T. C., and Keasling, J. D. (2007). Pathway confirmation and flux analysis of central metabolic pathways in Desulfovibrio vulgaris Hildenborough using gas chromatography-mass spectrometry and Fourier transform-ion cyclotron resonance mass spectrometry. J. Bacteriol. 189, 940–949.
Thauer, R. K. (1989). “Energy metabolism of sulfate-reducing bacteria,” in Autotrophic Bacteria, eds H. G. Schegel, and B. Bowien (Berlin: Springer-Verlag), 397–413.
Thauer, R.K., Jungermann, K., and Decker, K. (1977). Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41, 100–180.
Thauer, R. K., and Morris, J. G. (1984). “Metabolism of chemotrophic anaerobes: old views and new aspects,” in The Microbe 1984. Part II. Prokaryotes and Eukaryotes, eds D. P. Kelly, and N. G. Carr (Cambridge: Cambridge University Press), 123–168.
Thauer, R. K., Stackebrandt, E., and Hamilton, W. A. (2007). “Energy metabolism of sulphate reducing bacteria,” in Sulphate-Reducing Bacteria, eds L. L. Barton, and W. A. Hamilton (Cambridge: Cambridge University Press), 1–37.
Torres-García, W., Zhang, W., Runger, G. C., Johnson, R. H., and Meldrum, D. R. (2009). Integrative analysis of transcriptomic and proteomic data of Desulfovibrio vulgaris: a non-linear model to predict abundance of undetected proteins. Bioinformatics 25, 1905–1914.
Valente, F. M., Almeida, C. C., Pacheco, I., Carita, J., Saraiva, L. M., and Pereira, I. A. (2006). Selenium is involved in regulation of periplasmic hydrogenase gene expression in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 188, 3228–3235.
van den Berg, W. A. M., Stevens, A. A. M., Verhagen, M. F. J. M., van Dongen, W. M. A. M., and Hagen, W. R. (1989). Overproduction of the prismane protein from Desulfovibrio desulfuricans ATCC 27774 in Desulfovibrio vulgaris (Hildenborough) and EPR spectroscopy of the [6Fe-6S] cluster in different redox states. Biochim. Biophys. Acta-Protein Struct. M. 1206, 240–246.
Venceslau, S. S., Lino, R. R., and Pereira, I. A. (2010). The Qrc membrane complex, related to the alternative complex III, is a menaquinone reductase involved in sulfate respiration. J. Biol. Chem. 285, 22774–22783.
Vignais, P. M., and Billoud, B. (2007). Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–42072.
Walker, C. B., He, Z., Yang, Z. K., Ringbauer, J. A. Jr., He, Q., Zhou, J., Voordouw, G., Wall, J. D., Arkin, A. P., Hazen, T. C., Stoylar, S., and Stahl, D. A. (2009). The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J. Bacteriol. 191, 5793–5801.
Wall, J. D., Arkin, A. P., Balci, N. C., and Rapp-Giles, B. (2009). “Genetics and genomics of sulfate respiration in Desulfovibrio,” in Microbial Sulfur Metabolism, eds C. Dahl, and C. G. Friedrich (Berlin: Springer-Verlag.), 1–12.
Wall, J. D., Rapp-Giles, B. J., and Rousset, M. (1993). Characterization of a small plasmid from Desulfovibrio desulfuricans and its use for shuttle vector construction. J. Bacteriol. 175, 4121–4128.
Welte, C., Krätzer, C., and Deppenmeier, U. (2010). Involvement of Ech hydrogenase in energy conservation of Methanosarcina mazei. FEBS J. 277, 3396–3403.
Widdel, F., and Hansen, T. A. (1991). “The dissimilatory sulfate- and sulfur-reducing bacteria,” in The Prokaryotes, eds A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (New York, NY: Springer-Verlag), 659–768.
Keywords: Desulfovibrio, sulfate respiration, sulfite reduction, Coo hydrogenase, cytochrome c3
Citation: Keller KL, and Wall JD (2011) Genetics and molecular biology of the electron flow for sulfate respiration in Desulfovibrio. Front. Microbio. 2:135. doi: 10.3389/fmicb.2011.00135
Received: 22 January 2011;
Accepted: 10 June 2011;
Published online: 29 June 2011.
Edited by:
Thomas E. Hanson, University of Delaware, USAReviewed by:
Larry L. Barton, University of New Mexico, USACopyright: © 2011 Keller and Wall. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Judy D. Wall, Department of Biochemistry, University of Missouri, Columbia, MO 65211, USA. e-mail:d2FsbGpAbWlzc291cmkuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.