
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 19 April 2011
Sec. Microbial Physiology and Metabolism
volume 2 - 2011 | https://doi.org/10.3389/fmicb.2011.00069
This article is part of the Research Topic The microbial sulfur cycle View all 21 articles
 Inês A. Cardoso Pereira*
Inês A. Cardoso Pereira* Ana Raquel Ramos
Ana Raquel Ramos Fabian Grein
Fabian Grein Marta Coimbra Marques
Marta Coimbra Marques Sofia Marques da Silva and Sofia Santos Venceslau
Sofia Marques da Silva and Sofia Santos Venceslau
The number of sequenced genomes of sulfate reducing organisms (SRO) has increased significantly in the recent years, providing an opportunity for a broader perspective into their energy metabolism. In this work we carried out a comparative survey of energy metabolism genes found in 25 available genomes of SRO. This analysis revealed a higher diversity of possible energy conserving pathways than classically considered to be present in these organisms, and permitted the identification of new proteins not known to be present in this group. The Deltaproteobacteria (and Thermodesulfovibrio yellowstonii) are characterized by a large number of cytochromes c and cytochrome c-associated membrane redox complexes, indicating that periplasmic electron transfer pathways are important in these bacteria. The Archaea and Clostridia groups contain practically no cytochromes c or associated membrane complexes. However, despite the absence of a periplasmic space, a few extracytoplasmic membrane redox proteins were detected in the Gram-positive bacteria. Several ion-translocating complexes were detected in SRO including H+-pyrophosphatases, complex I homologs, Rnf, and Ech/Coo hydrogenases. Furthermore, we found evidence that cytoplasmic electron bifurcating mechanisms, recently described for other anaerobes, are also likely to play an important role in energy metabolism of SRO. A number of cytoplasmic [NiFe] and [FeFe] hydrogenases, formate dehydrogenases, and heterodisulfide reductase-related proteins are likely candidates to be involved in energy coupling through electron bifurcation, from diverse electron donors such as H2, formate, pyruvate, NAD(P)H, β-oxidation, and others. In conclusion, this analysis indicates that energy metabolism of SRO is far more versatile than previously considered, and that both chemiosmotic and flavin-based electron bifurcating mechanisms provide alternative strategies for energy conservation.
Sulfate reducing organisms (SRO) are anaerobic prokaryotes found ubiquitously in nature (Rabus et al., 2007; Muyzer and Stams, 2008). They employ a respiratory mechanism with sulfate as the terminal electron acceptor giving rise to sulfide as the major metabolic end-product. These organisms play an important role in global cycling of sulfur and carbon in anaerobic environments, particularly in marine habitats due to the high sulfate concentration, where they are responsible for up to 50% of carbon remineralization (Jørgensen, 1982). Sulfate reduction is a true respiratory process, which leads to oxidative phosphorylation through a still incompletely understood electron-transfer pathway. This electron transport chain links dehydrogenases to the terminal reductases, which are located in the cytoplasm, and therefore, not directly involved in charge translocation across the membrane and generation of transmembrane electrochemical potential. In recent years, the advent of genomic information coupled with biochemical and genetic studies has provided significant advances in our understanding of sulfate respiration, but several important questions remain to be answered including the sites and mechanisms of energy conservation. These studies revealed that sulfate reduction is associated with a set of unique proteins. Some of these proteins are also present in sulfur-oxidizing organisms, whereas others are shared with anaerobes like methanogens. Most biochemical studies have focused on mesophilic sulfate reducers of the Deltaproteobacteria, mostly Desulfovibrio spp. (Matias et al., 2005; Rabus et al., 2007), but previous analyses indicated that the composition of energy metabolism proteins can vary significantly between different SRO (Pereira et al., 2007; Rabus et al., 2007; Junier et al., 2010). The increasing number of SRO genomes available from different classes of both Bacteria and Archaea prompted us to perform a comparative analysis of energy metabolism proteins. In this work we report the analysis of 25 genomes of SRO available at the Integrated Microbial Genomes website. This includes 3 Archaea, 17 Deltaproteobacteria (of the Desulfovibrionacae, Desulfomicrobiacae, Desulfobacteraceae, Desulfohalobiacae, Desulfobulbaceae, and Syntrophobacteraceae families), 4 Clostridia (of the Peptococcaceae and Thermoanaerobacterales families), and T. yellowstonii DSM 11347 of the Nitrospira phylum (Table 1). This analysis extends a previous one in which only the Deltaproteobacteria Desulfovibrio vulgaris Hildenborough, Desulfovibrio desulfuricans G20, and Desulfotalea psychrophila were considered (Pereira et al., 2007). Genes/proteins involved in carbon metabolism are not discussed, with the exception of lactate and formate dehydrogenases. The loci for all genes analyzed can be found in Supplementary Material. A general scheme depicting most of the proteins discussed is presented in Figure 1.
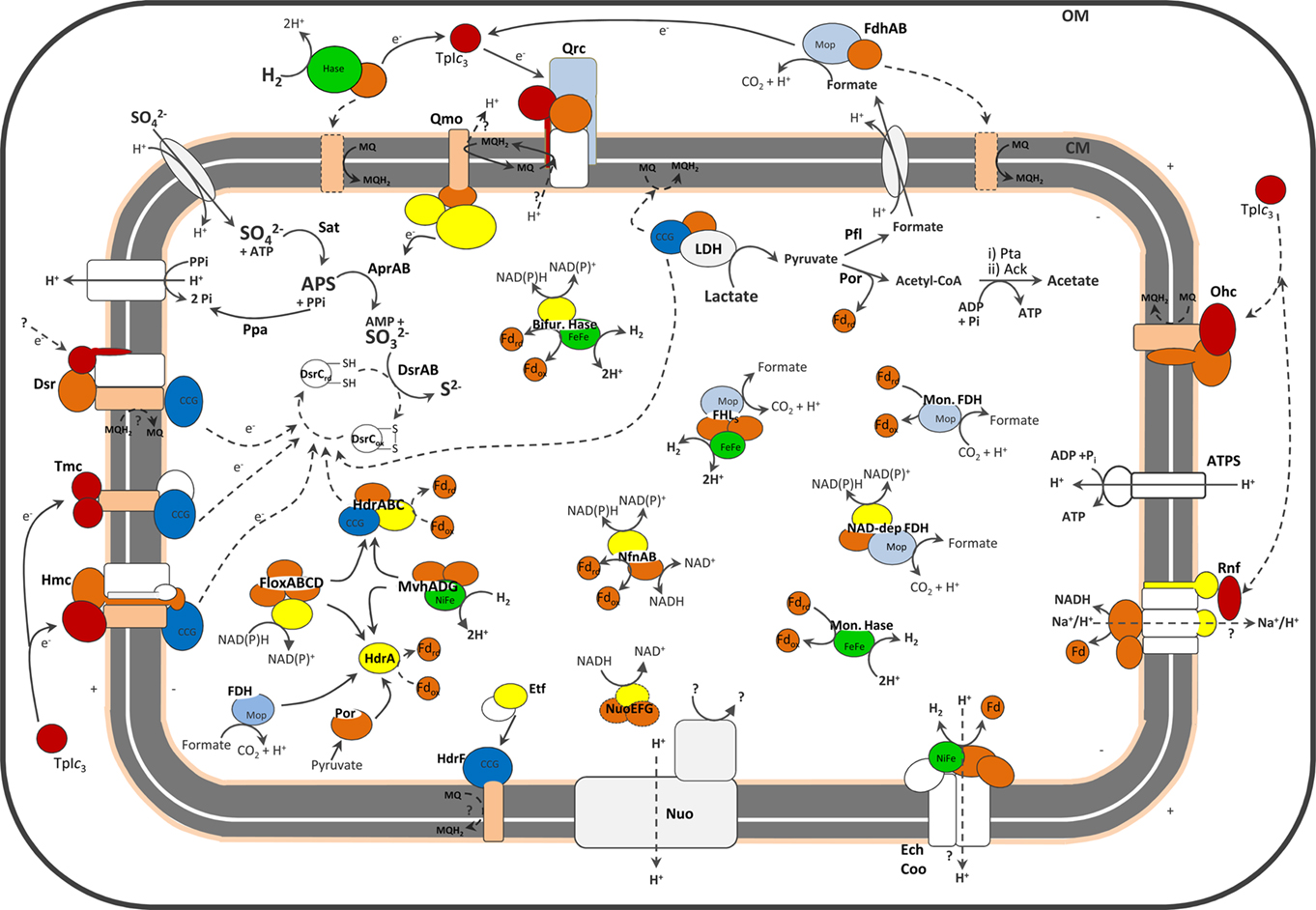
Figure 1. Schematic representation of the cellular location of SRO main energy metabolism proteins. No single organism is represented. For the exact distribution of proteins in each organism please refer to the Tables. The dashed lines represent hypothetical pathways, or (in the case of periplasmic Hases and FDHs) pathways present in only a few organisms. For the sake of clarity a few proteins discussed are not represented. Color code is red for cytochromes c, pale orange for cytochromes b, yellow for flavoproteins, dark orange for FeS proteins, light blue for proteins of molybdopterin family, dark blue for CCG proteins and green for catalytic subunits of Hases.
As expected, all organisms analyzed contain genes for those proteins long known to be directly involved in sulfate reduction (Rabus et al., 2007), including sulfate transporters, ATP sulfurylase (sat), APS reductase (aprAB), and dissimilatory sulfite reductase (dsrAB; Supplementary Material). The hydrolysis of pyrophosphate is carried out by soluble inorganic pyrophosphatases in most cases, but in a few organisms a membrane-associated proton-translocating pyrophosphatase (Serrano et al., 2007) is present, which may allow energy conservation from hydrolysis of pyrophosphate. These include the Gram-positive bacteria (Junier et al., 2010), Syntrophobacter fumaroxidans, Desulfococcus oleovorans, Desulfatibacillum alkenivorans, and Caldivirga maquilingensis. F1F0-ATP synthases are also present in all the SRO analyzed. Other strictly conserved proteins include ferredoxins, which are very abundant proteins in sulfate reducers (Moura et al., 1994). Their crucial role in anaerobic metabolism has gained increasing evidence in recent years (Meuer et al., 2002; Herrmann et al., 2008; Thauer et al., 2008; see Cytoplasmic Electron Transfer section below). All organisms analyzed contain ferredoxin I, which in some cases is present in multiple copies, and most contain also ferredoxin II.
One of the remaining important questions about sulfate reduction is the nature of the electron donors to the terminal reductases AprAB and DsrAB. Two membrane complexes, QmoABC and DsrMKJOP (Figures 1 and 2) have been proposed to perform this function (Pereira, 2008).
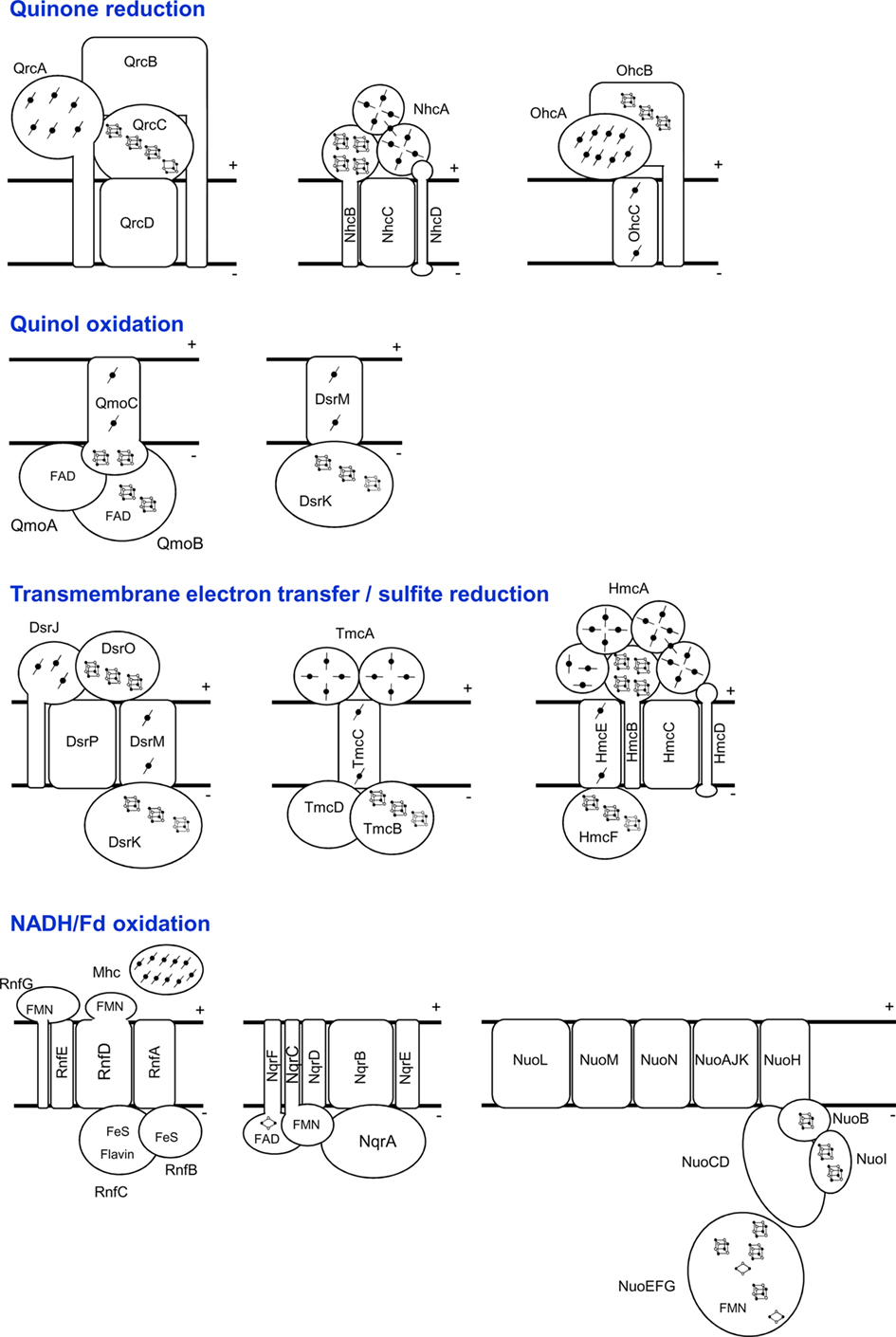
Figure 2. Schematic representation of the SRO membrane-bound electron-transfer complexes, grouped in different categories according to expected function. The NuoEFG proteins are shown as one module, which is not always present.
QmoABC (for Quinone-interacting membrane-bound oxidoreductase complex) was first described in D. desulfuricans ATCC 27774 (Pires et al., 2003). It includes three subunits binding two hemes b, two FAD groups and several iron–sulfur centers. QmoA and QmoB are both soluble proteins homologous to HdrA, a flavin-containing subunit of the soluble heterodisulfide reductases (HDRs; Hedderich et al., 2005). HDRs are key enzymes in methanogens that catalyze the reduction of the CoM-S-S-CoB heterodisulfide, formed in the last step of methanogenesis, to the corresponding thiols (Hedderich et al., 2005). The function of HdrA is still not clear, but it has been proposed to be involved in flavin-based electron bifurcation by an HdrABC/MvhADG complex, where the endergonic reduction of ferredoxin by H2 is coupled to the exergonic reduction of the CoM-S-S-CoB heterodisulfide by H2 (Thauer et al., 2008). QmoC is a fusion protein that contains a cytochrome b transmembrane domain related to HdrE and a hydrophilic iron–sulfur domain related to HdrC. QmoB includes also a domain similar to MvhD, a subunit of F420-non-reducing hydrogenase (Mvh; Thauer et al., 2010). Since the qmo genes are usually adjacent to aprAB, and both QmoC hemes are reduced by a menaquinol analog, it has been proposed that Qmo transfers electrons from the quinone pool to AprAB, in a process that may result in energy conservation (Pires et al., 2003; Venceslau et al., 2010). Although direct electron transfer has not been reported, it was recently shown that in D. vulgaris Hildenborough the Qmo complex is essential for sulfate, but not for sulfite, reduction (Zane et al., 2010). Our analysis confirmed that a gene locus containing sat, aprAB and the qmoABC genes is present in the majority of SRO analyzed. The exceptions are the archaeon C. maquilingensis for which no qmo genes are detected, and the Gram-positive bacteria where the qmoC gene is absent. In Desulfotomaculum acetoxidans and Candidatus Desulforudis audaxviator the qmoC gene is replaced by the hdrBC genes that code for soluble subunits of HDRs (Junier et al., 2010). This suggests that in Gram-positive bacteria the reduction of APS reductase may derive from soluble pathways, rather than quinones, and not be coupled to energy conservation.
The dsrMKJOP genes were first reported in the sulfur-oxidizing bacterium Allochromatium vinosum as part of a dsr locus encoding also the dsrAB and dsrC genes, among others (Pott and Dahl, 1998). The DsrMKJOP complex was isolated from Archaeoglobus fulgidus (Mander et al., 2002; where it was named Hme) and D. desulfuricans ATCC 27774 (Pires et al., 2006). It is a transmembrane complex with redox subunits in the periplasm – the triheme cytochrome c DsrJ, and the iron–sulfur protein DsrO; in the membrane – the cytochrome b DsrM (NarI family), and DsrP (NrfD family); and in the cytoplasm – the iron–sulfur protein DsrK that is homologous to HdrD, the catalytic subunit of the membrane-bound HdrED. DsrK and HdrD are both members of the CCG protein family, named after the CysCysGly residues present in the conserved cysteine-rich sequence (CXnCCGXmCXXC), which includes over 2000 archaeal and bacterial proteins (Hedderich et al., 1999; Hamann et al., 2007). This Cys sequence binds a special [4Fe4S] cluster, which in HDR is responsible for heterodisulfide reduction (Hedderich et al., 2005), and is also present in Dsr (Pires et al., 2006). Sequence analysis suggests that there may be two modules in the Dsr complex. One module, formed by DsrMK (based on its similarity to HdrED), may be involved in menaquinol oxidation and reduction of a cytoplasmic substrate, probably the DsrC disulfide (Oliveira et al., 2008); a second module formed by DsrJOP may be involved in electron transfer between the menaquinone pool and a periplasmic component, but it is not clear in which direction. The dsrMKJOP genes are present in all SRO genomes analyzed, with the exception of the Gram-positive bacteria (Junier et al., 2010) and C. maquilingensis, for which only dsrMK are present. This indicates that only these two proteins are essential for sulfite reduction. Gram-positive bacteria lack a periplasmic space, which may explain the absence of DsrJO, and in these organisms DsrMK must transfer electrons between the menaquinone pool and the cytoplasm, whereas in organisms with DsrMKJOP electron transfer likely involves also periplasmic components. Several SRO contain both dsrMKJOP and one or more copies of dsrMK. A DsrMK protein was isolated from Archaeoglobus profundus (Mander et al., 2004).
The dsrC gene is also strictly conserved in all SRO. It is one of the most highly expressed genes in D. vulgaris Hildenborough (Haveman et al., 2003; Wall et al., 2008) and also environmental samples (Canfield et al., 2010), pointing to an important role in sulfur metabolism. All organisms encoding a dsrAB sulfite reductase (sulfate/sulfite reducers or sulfur oxidizers) also contain the dsrC and dsrMK genes. DsrC is a small protein with a C-terminal swinging arm containing two strictly conserved cysteines (Cort et al., 2001; Mander et al., 2005). It belongs to a larger family of proteins, present also in organisms that do not perform dissimilatory sulfur metabolism (e.g., E. coli TusE), where they are involved in sulfur-transfer reactions (Ikeuchi et al., 2006). In these cases, a single cysteine, the penultimate residue of the C-terminal arm, is conserved. This suggests the involvement of a disulfide bond between the two DsrC cysteines as a redox-active center in the sulfite reduction pathway. DsrC was initially described as a subunit of DsrAB, with which it forms a tight complex (Pierik et al., 1992). However, DsrC is not a subunit, but rather a protein with which DsrAB interacts. The crystal structure of the DsrAB–DsrC complex from D. vulgaris revealed that the DsrC swinging arm inserts into a cleft between DsrA and DsrB, such that its penultimate cysteine comes in close proximity to the sulfite binding site at the catalytic siroheme (Oliveira et al., 2008). A mechanism for sulfite reduction involving DsrC was proposed, in which a DsrC persulfide is formed and gives rise to oxidized DsrC (DsrCox) with a disulfide bond between the two cysteines (Oliveira et al., 2008). DsrCox is then proposed to be reduced by the DsrK subunit of the Dsr complex, which contains a catalytic iron–sulfur center for putative reduction of disulfide bonds, as described in HDRs (Pires et al., 2006). The involvement of the Dsr complex provides a link between membrane quinol oxidation and sulfite reduction that may explain the fact that proton translocation is associated with this reduction (Kobayashi et al., 1982). In vitro sulfite reduction by desulfoviridin, the dissimilatory sulfite reductase of Desulfovibrio spp. does not produce sulfide as observed in the assimilatory enzymes, but a mixture of products including thiosulfate and trithionate (Rabus et al., 2007). This led to the proposal that sulfite reduction in SRO proceeds with thiosulfate and trithionate as intermediates (Akagi, 1995). In Desulfovibrio gigas, flavoredoxin was implicated in thiosulfate reduction (Broco et al., 2005). However, flavoredoxin is not conserved across the SRO analyzed and there is also no evidence for enzymes to handle trithionate. Most likely the in vitro polythionate products observed originate from the absence of other proteins required for physiological sulfite reduction, namely DsrC (Oliveira et al., 2008).
Our genomic analysis of SRO supports the interaction between DsrC, DsrAB and the DsrMKJOP complex: In A. profundus and T. yellowstonii dsrC is in the same gene cluster as dsrMKJOP, and in the three Gram-positive organisms and Ammonifex degensii, a dsrMK–dsrC gene cluster is present (Figure 3). Strikingly, this cluster is preceded by a gene encoding a ferredoxin (Fd), and a Fd gene is also present after the dsrMKJOP genes and in close proximity to dsrAB in three Deltaproteobacteria. This suggests that a Fd may also be involved in the electron transfer pathway between the Dsr complex, DsrC, and DsrAB. The involvement of Fd provides a link between the sulfite reduction step and other soluble electron transfer pathways.
One of the most discussed models for energy conservation in SRO is the hydrogen-cycling mechanism proposed by Odom and Peck (1981). In this mechanism the reducing power from lactate oxidation is transferred to a cytoplasmic hydrogenase to generate H2 that diffuses to the periplasm. There its reoxidation generates electrons that are transferred back across the membrane for the cytoplasmic reduction of sulfate, resulting in a transmembrane proton gradient to drive ATP synthesis. This intracellular redox cycling proposal has been extended to include other possible intermediates like formate and CO (Voordouw, 2002). Hydrogen and formate are also important energy sources for SRO in natural habitats. Oxidation of these substrates by periplasmic enzymes contributes to a proton gradient as electrons are transferred to the quinone pool or directly across the membrane for cytoplasmic sulfate reduction. The common bacterial uptake hydrogenases (Hases) and formate dehydrogenases (FDHs) are composed of three subunits: a large catalytic subunit, a small electron-transfer subunit and a membrane-associated protein responsible for quinone reduction. Desulfovibrio organisms are unusual in that most of their periplasmic Hases and FDHs lack the membrane subunit, and instead transfer electrons to one or several cytochromes c (Heidelberg et al., 2004; Matias et al., 2005).
Two of the SRO analyzed contain no Hases at all: the archaeon C. maquilingensis and the Deltaproteobacterium Dc. oleovorans. In addition, Desulfonatronospira thiodismutans contains no periplasmic Hases (Table 1). The total absence of Hases in two SRO was unexpected and indicates that hydrogen metabolism is not essential for sulfate reduction. The other SRO contain from one to four periplasmic enzymes, the most common of which is the soluble [NiFe] HynAB. All Deltaproteobacteria contain at least one copy of HynAB. In two archaea and three Deltaproteobacteria this protein is membrane-anchored by an additional subunit for quinone reduction (HynABC). Eight organisms also contain the [NiFeSe] HysAB Hase (Valente et al., 2005). The HynAB and HysAB enzymes use as electron acceptor the Type I cytochrome c3 (TpIc3; Matias et al., 2005). Finally, only two organisms contain a copy of a HynABC3, in which another dedicated cytochrome c3 is encoded next to the hynAB genes. A periplasmic [FeFe] Hase is present in all Desulfovibrio organisms, except D. piger, and is also found in S. fumaroxidans. This enzyme is soluble and also uses TpIc3 as electron acceptor. A membrane-anchored [FeFe] Hase is present in the four Clostridial organisms. A Tat signal peptide present in the catalytic subunit indicates that the enzyme is translocated to the extracytoplasmic side of the cellular membrane, which is somewhat unexpected for the Gram-positive organisms that lack a periplasmic compartment. The enzyme is anchored to the membrane through a NrfD-like transmembrane protein that should transfer electrons to the menaquinone pool.
Overall, the analysis indicates that a periplasmic Hase is found in most SRO, which functions in the uptake of H2. The Desulfovibrionacae organisms contain a higher number of periplasmic enzymes compared to the others. In D. vulgaris Hildenborough, which has four periplasmic Hases, it has been shown that expression of these enzymes is fine tuned to respond to metal availability (Valente et al., 2006) and hydrogen concentration (Caffrey et al., 2007). The Clostridial organisms contain a novel membrane-anchored [FeFe] Hase.
As in the case of Hases, the periplasmic FDHs can be either soluble, comprising only the catalytic and small subunits (FdhAB; Almendra et al., 1999) or additionally a dedicated cytochrome c3 (FdhABC3; Sebban et al., 1995), or they can be of the typical membrane-associated form, in which a subunit for quinone reduction is present. This can either be a NarI-like cytochrome b (FdhABC) or a larger protein of the NrfD family (FdhABD). The physiological electron acceptor for FdhAB is also likely to be the soluble TpIc3 (ElAntak et al., 2003; Venceslau et al., 2010). Of the SRO analyzed, two Archaea contain neither periplasmic or cytoplasmic FDHs (Table A1 in Appendix), again indicating that formate metabolism is not essential for sulfate reduction. All other SRO analyzed contain from one to three periplasmic FDHs, the most widespread of which is FdhAB. Six organisms contain one FdhABC3. Only three organisms contain FdhABC. Two Gram-positive bacteria contain FdhABD where FdhB has a twin-arginine signal peptide, indicating that these enzymes are translocated to outside of the cellular membrane, as observed for the [FeFe] Hase. In D. vulgaris Hildenborough the gene locus for FdhABD includes also two cytochromes c. Several of the FDHs contain selenocysteine (Sec), and in some organisms only Sec-containing FDHs are present, whereas others contain also Cys-containing enzymes.
The Desulfovibrionacae organisms are characterized by a very high level of multiheme cytochromes c, the most abundant and well studied of which is the TpIc3 (Matias et al., 2005). The genome of D. vulgaris Hildenborough first revealed that a pool of cytochromes c is present in the periplasm (Heidelberg et al., 2004), some of which belong to the cytochrome c3 family, but not all (Matias et al., 2005; Pereira et al., 2007). Several multiheme cytochromes c are associated with membrane complexes, and these will be discussed in the following section. Most SRO analyzed contain a high number of multiheme cytochromes c (Table A2 in Appendix) but several exceptions are observed: C. maquilingensis, Dm. acetoxidans, and Desulfotomaculum reducens contain no cytochromes c at all; A. profundus contains only DsrJ; A. fulgidus contains DsrJ and an octaheme cytochrome, and Dm. reducens contains only the NrfHA proteins (Rodrigues et al., 2006), both with signal peptides again indicating an extracytoplasmic location. In general terms, the Deltaproteobacteria and T. yellowstonii have multiple cytochromes c, contrary to the Archaea, Gram-positive SRO, and A. degensii. The TpIc3 is present in all the Deltaproteobacteria (except Dt. psychrophila and Dv. alkaliphilus) and in T. yellowstonii. Often there are two to four copies of monocistronic cytochromes c3, whereas others are associated with periplasmic Hases and FDHs. Tetraheme cytochromes of the c554 family (Iverson et al., 1998) are also present in several organisms, including one associated with a methyl-accepting chemotaxis sensory transducer protein, suggesting an involvement in regulation. The monoheme cytochrome c553 is only present in five Deltaproteobacteria, often in the same locus as cytochrome c oxidase, suggesting it acts as its electron donor. The nitrite reductase complex formed by the two cytochromes NrfH and NrfA (Rodrigues et al., 2006) is one of the more widespread cytochromes in SRO. Nitrite is a powerful inhibitor of SRO and NrfHA acts as a detoxifying enzyme (Greene et al., 2003).
A family of transmembrane redox complexes that include a multiheme cytochrome c subunit has been described in Desulfovibrio (Pereira, 2008). The first complex identified was the Hmc complex composed of HmcABCDEF (Rossi et al., 1993). The subunit composition of Hmc is strikingly similar to the Dsr complex in terms of the type of proteins present: a cytoplasmic CCG protein related to HdrD, two integral membrane proteins of the NarI and NrfD families, a periplasmic ferredoxin-like protein and a periplasmic cytochrome c (Figures 1 and 2). This suggests that both complexes have related functions, but the sequence identity between subunits is very low. The cytochrome c subunit is a large 16 heme cytochrome in Hmc (HmcA) and a small triheme cytochrome in Dsr (DsrJ). HmcA can accept electrons from periplasmic hydrogenases via the TpIc3 (Pereira et al., 1998; Matias et al., 2005), but this is not observed for DsrJ (Pires et al., 2006). This cytochrome has a heme with unusual histidine/cysteine ligation, but its function has not been elucidated (Pires et al., 2006; Grein et al., 2010). It is not clear if Hmc exchanges electrons with the quinone pool, or directly between the periplasm and cytoplasm. Some studies have indicated that the function of Hmc is in electron transfer to the cytoplasm during growth with hydrogen (Dolla et al., 2000; Voordouw, 2002), but the hmc genes are downregulated under these conditions (Caffrey et al., 2007; Pereira et al., 2008). More recently this complex was shown to play a role during syntrophic growth of D. vulgaris, where it was proposed to be implicated in electron transfer from the cytoplasm to the periplasm (Walker et al., 2009).
The TmcABCD complex seems to be a simplified version of Hmc. It includes a tetraheme cytochrome c3 (TmcA, first described as acidic cytochrome c3 or Type II c3, Valente et al., 2001), a CCG protein homologous to HmcF (TmcB), a cytochrome b (TmcC), and a tryptophan-rich protein (TmcD; Pereira et al., 2006). TmcA is an efficient electron acceptor of the periplasmic Hase/TpIc3 couple (Valente et al., 2001, 2005; Pieulle et al., 2005). All redox centers of the Tmc complex are reduced with H2 (Pereira et al., 2006), and the tmc genes are upregulated in growth with hydrogen versus lactate (Pereira et al., 2008), indicating that Tmc acts to transfer electrons from periplasmic H2 oxidation to the cytoplasm.
Two other complexes related to Tmc and Hmc are present in the genomes of SRO. One includes a nine-heme cytochrome and is designated as Nhc complex (for nine-heme cytochrome complex; Saraiva et al., 2001), and the other includes an eight-heme cytochrome and was designated as Ohc (for octaheme cytochrome complex; Pereira et al., 2007). The structure of the NhcA cytochrome is similar to the C-terminal domain of the HmcA, and it is also reduced by the Hase/TpIc3 couple (Matias et al., 1999), whereas OhcA belongs to a different cytochrome family. OhcC is a cytochrome b of the NarI family, whereas NhcC membrane subunit is of the NrfD family. The subunits of the Hmc, Tmc, Nhc, and Ohc complexes are homologous to each other, indicating they belong to the same family. However, the Nhc and Ohc complexes lack the cytoplasmic CCG protein, so they should transfer electrons from the periplasm to the quinone pool. In contrast, both Hmc and Tmc include the CCG protein related to DsrK and HdrD, containing a binding site for a putative catalytic [4Fe4S] center, which hints that they are implicated in similar thiol/disulfide redox chemistry as DsrK possibly involving DsrCox.
The genomic analysis indicates that the Hmc, Tmc, Nhc, and Ohc complexes (Table 2) are present in Deltaproteobacteria, with the exception of the two members of the Desulfobulbaceae family. They are not present in the Archaea organisms or members of Clostridia, and T. yellowstonii has only Hmc. This distribution correlates well with the presence of their putative electron donor, TpIc3. All organisms that have Hmc, usually also have Tmc, and some organisms have two copies of Tmc. In D. desulfuricans ATCC 27774 a three-subunit complex is found including a triheme cytochrome c7, homologous to the N-terminal part of Hmc. Although its subunits are more similar to Hmc, the subunit composition is more characteristic of a Tmc. The Nhc complex has a more limited distribution, and in some organisms the cytochrome subunit has 13 hemes. In Dt. thiodismutans the cytochrome subunit is not present.
Recently, a new membrane complex named Qrc (for quinone reductase complex) was isolated from D. vulgaris (Venceslau et al., 2010). It is composed of four subunits, QrcABCD, including a hexaheme cytochrome c (QrcA), a large protein of the molybdopterin-containing family, but which does not bind molybdenum (QrcB), a periplasmic iron–sulfur protein (QrcC) and an integral membrane protein of the NrfD family (QrcD). The Qrc complex accepts electrons from periplasmic Hases and FDHs through TpIc3 and has activity as a TpIc3:menaquinone oxidoreductase (Venceslau et al., 2010). A D. desulfuricans G20 mutant lacking the qrcB gene was selected from a library of transposon mutants by its inability to grow syntrophically with a methanogen on lactate (Li et al., 2009). This mutant is unable to grow with H2 or formate as electron donors but grows normally with lactate, confirming the role of Qrc in H2 and formate oxidation. It has been proposed that the Qrc and Qmo complexes constitute the two arms of an energy conserving redox loop (Simon et al., 2008), contributing to proton motive force generation during sulfate reduction with H2 or formate (Venceslau et al., 2010). This previous study showed that the qrc genes are present in sulfate reducers that have periplasmic Hases and/or FDHs that lack a membrane subunit for quinone reduction. Our present analysis confirms this and shows that the qrc genes are found in many Deltaproteobacteria, but not in other SRO (Table 2). D. piger and Dt. thiodismutans both have soluble periplasmic Hases and FDHs but lack a Qrc. In both cases an alternative complex for quinone reduction is present, like Nhc and Ohc. An exception is T. yellowstonii that also has soluble periplasmic Hases and FDHs and for which only the Hmc complex was identified. In this case maybe electrons go directly to the cytoplasm through Hmc or this is also capable of quinone reduction.
Although it has long been known that NADH and ferredoxin (Fd) are important cytoplasmic components of energy metabolism in SRO, it is still not clear what role they play in the electron-transfers chains of these organisms. The membrane-bound Rnf complex mediates electron transfer between NADH and Fd and is found in numerous organisms (Li et al., 2006; McInerney et al., 2007; Müller et al., 2008; Seedorf et al., 2008). It was first described in Rhodobacter capsulatus where it is proposed to catalyze the reverse electron transport from NADH to Fd driven by the transmembrane proton potential (Schmehl et al., 1993). In other organisms it is proposed to carry out electron transport from reduced Fd to NAD+, coupled to electrogenic Na+ or H+ translocation (Müller et al., 2008). The Rnf complexes are constituted by six to eight subunits (Figures 1 and 2), which show similarity to Na+-translocating NADH:quinone oxidoreductases (Nqr; Steuber, 2001). There is yet no direct biochemical confirmation that Rnf translocates ions, but recent inhibitor studies obtained with membrane vesicles of the acetogenic bacterium Acetobacterium woodii are consistent with the proposal that Rnf catalyzes reduction of NAD+ from Fd coupled to electrogenic Na+ transport (Biegel and Müller, 2010). Both Rnf and Nqr are small complexes compared to the usual 14 subunits of the Nuo NADH:quinone oxidoreductases (Complex I; Efremov et al., 2010).
Our analysis shows that most organisms analyzed contain one, or more, of the Nuo, Rnf, and Nqr complexes (except C. Dr. audaxviator and A. degensii; Table 2). A surprisingly high number of SRO contain the nuo genes for complex I. Only Nuo is detected in the four Clostridia organisms, and F420H2:quinone oxidoreductase in the case of the Archaea (Kunow et al., 1994). In most cases the NuoEFG subunits that form the NADH dehydrogenase module are absent, as observed for the complex from cyanobacteria and chloroplasts (Friedrich and Scheide, 2000), suggesting that NADH is not the actual electron donor. It is tempting to speculate that these complexes also oxidize Fd. In Desulfovibrio magneticus and Desulfovibrio sp. FW1012B two clusters of nuo genes are present, one of which includes the nuoEFG genes.
The Rnf complex is present in most organisms, with the exception of the Clostridia and Archaea, suggesting it plays a key role in the energy conservation strategies of many sulfate reducers. In most cases a multiheme cytochrome c encoding gene (with 4–10 hemes) is found next to the rnf genes as reported for Methanosarcina acetivorans (Li et al., 2006). Interestingly, Desulfobacterium autotrophicum and Dc. oleovorans have two copies of the rnf genes, and only one includes the cytochrome c gene. The presence of this cytochrome provides an electron input/output module in the periplasm, which may link the cytochrome c pool with NAD(H) and/or Fd. The Nqr complex has a more limited distribution and is detected in only 5 of the 25 genomes analyzed. Of these, four are marine organisms and the other (Desulfurivibrio alkaliphilus) is a haloalkaliphilic bacterium isolated from soda lakes, and thus all are likely to have Na+-based bioenergetics. Two of these organisms have genes for all three complexes (Nuo, Rnf, and Nqr).
The Gram-positive organisms, C. maquiligensis and a few Deltaproteobacteria contain ion-translocating pyrophosphatases, which are probably involved in energy conservation (Table 2). This is likely to compensate for the absence of other transmembrane complexes in some of these organisms. A bc1 complex is also present in C. maquiligensis, S. fumaroxidans, and T. yellowstonii. A bd quinol oxidase is present in 19 of the 25 organisms, and 7 contain a cytochrome c oxidase (Table A2 in Appendix).
The Ech Hases belong to the energy-conserving membrane-bound [NiFe] Hases that are closely related to complex I (Hedderich and Forzi, 2005; Hedderich et al., 2005). They catalyze the reduction of H+ with Fd coupled to chemiosmotic energy conservation, or reduction of Fd with H2 driven by reverse electron transport. Thus, Ech Hases and Rnf constitute the two complexes in SRO capable of performing endergonic reduction of Fd based on chemiosmotic coupling. A closely related group are the CooMKLXUH CO-induced Hases of chemolithoautotrophic bacteria that oxidize CO to CO2 with reduction of H+ to H2 (Hedderich et al., 2005; Singer et al., 2006). The presence of an Ech Hase in SRO was first reported in Desulfovibrio gigas, where it was proposed to constitute the cytoplasmic Hase required for the hydrogen-cycling hypothesis (Rodrigues et al., 2003). The genome of D. vulgaris Hildenborough encodes both an Ech and Coo Hase (Heidelberg et al., 2004), and it was shown that this organism produces CO transiently from pyruvate during growth on sulfate (Voordouw, 2002). In D. vulgaris the ech genes are very upregulated during growth with H2, and also upregulated with pyruvate as electron donors relative to lactate, whereas the coo genes are downregulated in H2 (Pereira et al., 2008). This agrees with an expected higher level of CO during growth with lactate, leading to production of the Coo Hase, and suggests that Ech may work bidirectionaly, to reduce Fd for carbon fixation during growth with H2 or to produce H2 from reduced Fd during growth with pyruvate. Recently, the coo genes were shown to be upregulated during syntrophic growth of D. vulgaris on lactate with a methanogen (Walker et al., 2009). In addition, mutation of the coo genes severely impaired syntrophic growth while not affecting sulfate respiration, suggesting that Coo is an essential Hase to produce H2 from lactate in these conditions.
Despite these interesting results the Ech and Coo Hases are restricted to Desulfovibrio organisms, with a single exception of C. Dr. audaxviator that has a set of ech genes (Table 1). In contrast, the other organisms have soluble cytoplasmic Hases that are not present in Desulfovibrio.
In recent years several studies unraveled a novel process of coupling endergonic to exergonic redox reactions in anaerobic organisms, through a flavin-based electron bifurcation mechanism involving only soluble proteins (Herrmann et al., 2008; Li et al., 2008; Thauer et al., 2008; Schut and Adams, 2009). This mechanism involves the two-step reduction/oxidation of a flavin cofactor, through a flavin-semiquinone intermediate, in which each step is associated with a different reductant/oxidant (Thauer et al., 2008), in analogy to the quinone-based electron bifurcating mechanism of the bc1 complex (Xia et al., 2007). Five examples have been described including: (i) the coupling of Fd reduction with NADH to reduction of butyryl-CoA with NADH by the butyryl-CoA dehydrogenase-EtfAB complex (Herrmann et al., 2008; Li et al., 2008), (ii) coupling of Fd reduction with H2 to the reduction of the methanogenic CoM-S-S-CoB heterodisulfide with H2 catalyzed by the MvhADG/HdrABC complex (Thauer et al., 2008, 2010), (iii) coupling of Fd reduction with formate to the reduction of the methanogenic CoM-S-S-CoB heterodisulfide with formate catalyzed by a FdhAB/HdrABC complex (Costa et al., 2010), (iv) coupling of H2 formation from NADH with H2 formation from reduced Fd catalyzed by the multimeric [FeFe] Hases (Schut and Adams, 2009), and (v) coupling of NADP+ reduction with reduced Fd with NADP+ reduction with NADH catalyzed by NfnAB (Wang et al., 2010). These cases stress the important role Fd plays in anaerobic metabolism. The reduced Fd produced through a bifurcating reaction may be oxidized by membrane-associated ion-translocating complexes (such as Rnf or Ech), resulting in energy conservation, or it may be used as electron donor in other metabolic reactions. Our genomic analysis of SRO revealed there are several examples of soluble proteins in these organisms with the potential to carry out electron bifurcation from H2, formate or other carbon-based electron donors. In particular, a very high number of proteins related to HDRs were identified (see below).
An unexpectedly high number of soluble cytoplasmic hydrogenases, of both [NiFe] and [FeFe] families, were detected in the present analysis (Table 1). Most organisms contain a cytoplasmic-facing Hase, either soluble or membrane-bound, except the two organisms that contain no Hases at all and Desulfomicrobium baculatum. In numerous cases, the gene organization indicates that the cytoplasmic Hases are likely to be involved in electron bifurcation mechanisms, either involving NADH dehydrogenases or HdrA-like proteins. A large number of the [NiFe] Hases detected are related to the MvhADG Hases of methanogens (Thauer et al., 2010). In these organisms MvhADG reduces the cytoplasmic heterodisulfide reductase HdrABC, and the two proteins have been shown to form a large complex (Stojanowic et al., 2003). The activity of this complex is increased in the presence of Fd, and MvhADG/HdrABC are proposed to couple the endergonic reduction of Fd with H2 to the exergonic reduction of the heterodisulfide with H2 by electron bifurcation, probably involving the FAD group of HdrA (Thauer et al., 2008, 2010). In the SRO analyzed the mvhADG genes are found next to an hdrA gene (six organisms) or hdrABC genes (four organisms), suggesting these act as electron acceptors in a process that may involve electron bifurcation. In five organisms no hdr genes are close by. Another type of closely related [NiFe] Hase, of the Hox type, is present only in three organisms. Hox Hases are bidirectional NAD(P)-linked Hases common in cyanobacteria, and also found in other organisms (Vignais and Billoud, 2007). In the three SRO the Hox gene cluster includes hoxHY coding for the catalytic and small subunits, and hoxEFG that are homologous to nuoEFG, and code for the diaphorase module of the Hase. It is striking that in all SRO analyzed, with a single exception (C. Dr. audaxviator), the organisms that contain the energy-conserving Hases Ech or Coo do not contain other cytoplasmic [NiFe] Hases, and organisms that contain cytoplasmic [NiFe] Hases do not contain either Ech or Coo. This suggests that in SRO energy coupling through [NiFe] Hases involves either a chemiosmotic or an electron bifurcating mechanism. In the Archaea, only MvhADG/HdrABC is detected, and in the Clostridia only two isolated MvhADG Hases are present. In two organisms, genes for another [NiFe] Hase are found next to genes encoding sensor/response-regulator proteins and histidine kinases, suggesting they are regulatory Hases.
Many cytoplasmic [FeFe] Hases are also present in the SRO analyzed, and are particularly abundant in the Clostridia class. Many of these are monomeric Fd-dependent Hases (Table 1). Another large group of [FeFe] Hases detected is formed by multimeric NAD(P)-dependent Hases similar to the tetrameric Hases from D. fructosovorans (Malki et al., 1995) and Thermoanaerobacter tengcongensis (Soboh et al., 2004). These enzymes include one flavoprotein subunit that binds NAD(P). Another member of this group is the trimeric Hase of Thermotoga maritima that was shown to use Fd and NADH synergistically as electron donors for production of H2 (Schut and Adams, 2009). This is proposed to be also an electron bifurcating mechanism in which the exergonic oxidation of Fd is coupled to the unfavorable oxidation of NADH to give H2. In D. fructosovorans cell extracts no NAD+-reducing activity was detected and it was proposed that the enzyme functions as a NADP+-reducing H2-uptake Hase (Malki et al., 1995). The enzyme from T. tengcongensis was isolated and shown to work bidirectionally with NAD(H), but not with NADP(H) (Soboh et al., 2004). In the organisms analyzed the enzyme may be tetrameric, trimeric and in two organisms (D. vulgaris and Db. autotrophicum) dimeric. At this point it is not clear if the function of these Hases in the SRO is of H2 production from Fd/NAD(P)H, the reverse, or both depending on the metabolic conditions.
A novel and interesting group of [FeFe] Hases genes is found next to a gene coding for a type I FDH (Matson et al., 2010), suggesting the two units may form a soluble formate–hydrogen lyase complex (FHLs). This gene cluster is present only in five Deltaproteobacteria, and includes minimally the gene coding for the iron-only Hase, the gene for the catalytic subunit of FDH and two four-cluster electron-transfer proteins related to HydN. All subunits are soluble in contrast to the E. coli FHL complex (Sawers, 2005). In some organisms, the iron–sulfur protein encoded next to the hydA gene has a predicted signal peptide, but this is absent in other organisms. This raises doubts about the cellular location of the Hase. It is possible that this sequence is not cleaved and acts as a membrane anchor. This putative FHLs complex is equivalent of the one recently described to be present in the termite gut acetogen Treponema primitia, where it is proposed to carry out H2-dependent CO2 reduction (Matson et al., 2010). However, the function of these proteins in SRO remains for now unknown.
Finally, in six organisms an [FeFe] Hase including a PAS sensor domain was identified, which is very similar to the HsfB protein recently reported in Thermoanaerobacterium saccharolyticum (Shaw et al., 2009). This Hase is likely to be involved in H2 sensing and regulation.
A cytoplasmic FDH is present in many, but not all SRO (Table A1 in Appendix). It is absent in the Archaea, for which a single periplasmic FDH is detected. A NAD(P)H-linked FDH is present in many organisms, but not in Desulfovibrionacae and Desulfobacteraceae. In these cases the catalytic FDH gene is found next to two nuoEF-like genes. Another noteworthy group is that of the putative soluble FHLs described above. Only in two organisms (Df. alkenivorans and Db. autotrophicum) is an isolated fdhA gene present that may encode a Fd-dependent FDH. In other cases an fdhA gene is part of a more complex gene cluster, including in some cases hdr genes (see below).
A heterodimeric transhydrogenase was recently reported from Clostridium kluyveri (Wang et al., 2010). The enzyme, named NfnAB, catalyzes the reversible NADH-dependent reduction of NADP+ by reduced Fd, or the NAD+-dependent reduction of Fd by NADPH. It is another example of a bifurcating reaction as it couples the exergonic reduction of NADP+ with reduced Fd to the endergonic reduction of NADP+ with NADH. The nfnAB genes, both encoding iron–sulfur flavoproteins, are present in several organisms (Wang et al., 2010). They are often annotated as sulfide dehydrogenase, as this enzyme was initially reported in Pyrococcus furiosus to act as sulfide dehydrogenase (Ma and Adams, 1994), but later described to act physiologically as a Fd:NADP+ oxidoreductase (Ma and Adams, 2001). We found that the nfnAB genes are also present in the great majority of SRO, with the exception of the Archaea, and three bacteria (Table A3 in Appendix), suggesting it plays an important role also in the metabolism of SRO.
In methanogens without cytochromes the HDR enzyme is soluble and composed of three subunits, HdrABC, whereas in methanogens with cytochromes it is membrane-associated and formed by two subunits, HdrDE (Hedderich et al., 2005; Thauer et al., 2008). HdrA is an iron–sulfur flavoprotein, HdrC is a small iron–sulfur protein and HdrB contains two CCG domains and harbors a special [4Fe4S] catalytic site. HdrE is a membrane-bound cytochrome b and HdrD has both HdrB- and HdrC-like domains and includes a similar catalytic cofactor to HdrB. The HdrDE protein receives electrons from methanophenazine and reduction of the heterodisulfide is coupled to energy conservation by a redox loop mechanism involving also the membrane-associated VhoACG Hase (Hedderich et al., 2005; Thauer et al., 2008). The soluble HdrABC enzyme forms a complex with the soluble MvhADG Hase that catalyzes heterodisulfide reduction with H2. This exergonic reaction is proposed to be coupled to the endergonic reduction of Fd by flavin-based electron bifurcation involving HdrA (Thauer et al., 2008). As discussed above, the membrane complexes Qmo, Dsr, Tmc, and Hmc all include subunits related to HDRs (Pereira, 2008). The abundance of HDR-like proteins in SRO has been highlighted in recent genomes of SRO (Strittmatter et al., 2009; Junier et al., 2010). Recently, Strittmatter et al. (2009) proposed two new types of HDR subunits, based on proteins encoded in the Db. autotrophicum genome. The first, HdrF includes HdrB- and HdrC-like domains fused to a third transmembrane domain. Thus, HdrF is like a fusion of HdrE and HdrD. The second, HdrL, is a large protein containing an HdrA domain and one or two NADH-binding domains (Strittmatter et al., 2009). We have analyzed genes coding for HdrA-, HdrB-, and HdrD-like proteins as these are the most relevant subunits of HDRs. In general, we found few HdrB-like proteins and they are either associated with HdrAs or they are domains of HdrDs. In contrast, we found a very high number of HdrA- and HdrD-related proteins in the genomes of SRO, so our analysis focuses on these two protein families.
The majority of HdrA-like proteins are encoded in two types of gene loci (Figure 4; Table 3). In the first type an hdrA gene or a set of hdrABC genes are found next to mvhDGA genes coding for a soluble Mvh [NiFe] Hase as discussed above. In the second type, again a single hdrA gene or a set of hdrABC genes are found next to four genes that we named floxABCD genes (for flavin oxidoreductase). The floxABCD/hdrABC gene cluster was first identified in D. vulgaris Hildenborough as encoding a putative Hase–HDR complex (Haveman et al., 2003), as the flox genes are annotated as putative Hase genes because they code for proteins related to subunits of P. furiosus NAD(P)-dependent soluble Hases (SH) I and II (Jenney and Adams, 2008). However, a gene coding for a catalytic Hase subunit is not present, so Flox is not a Hase. The floxA gene codes for a protein with both FAD and NAD(P)-binding domains and is similar to P. furiosus SH subunit γ. The floxB and floxC genes are related to rnfC and both code for iron–sulfur proteins similar to P. furiosus SH subunit β. The floxD gene codes for a protein similar to MvhD, which in methanogens is involved in electron transfer from Mvh Hase to Hdr (Stojanowic et al., 2003). In several organisms the floxCD genes are fused into a single gene. Thus, the Flox proteins are likely to oxidize NAD(P)H and transfer electrons to the HdrABC proteins. In D. vulgaris and other organisms the floxABCD/hdrABC genes are found next to a co-regulated adh gene coding for an alcohol dehydrogenase (Haveman et al., 2003). The Adh may reduce NAD+ to NADH, which will be oxidized by Flox. The floxABCD/hdrA or floxABCD/hdrABC genes are present in the majority of the SRO analyzed. This suggests they play an important physiological role, and indeed these genes have been reported in several gene expression and proteomic studies of D. vulgaris energy metabolism (Haveman et al., 2003; Zhang et al., 2006a,b; Caffrey et al., 2007; Pereira et al., 2008; Walker et al., 2009). The HdrA-associated Mvh and Flox proteins probably constitute parallel pathways for HdrA reduction from H2 or NAD(P)H. It seems likely that these proteins may be involved in electron bifurcating reactions involving HdrA as previously suggested (Thauer et al., 2008). We further propose that the electron acceptor of the HdrBC proteins may be DsrCox, also thought to be a substrate for DsrK (Oliveira et al., 2008). Thus, in SRO the HdrABC/MvhDGA and HdrABC/FloxABCD complexes may provide a soluble route of electron transfer to sulfite reduction through DsrC, where energy coupling occurs through electron bifurcation rather than chemiosmotically through DsrMK. In support of this hypothesis the dsrC gene of Db. autotrophicum is found next to a hdrA(L)/floxACBD gene cluster (Figure 3).
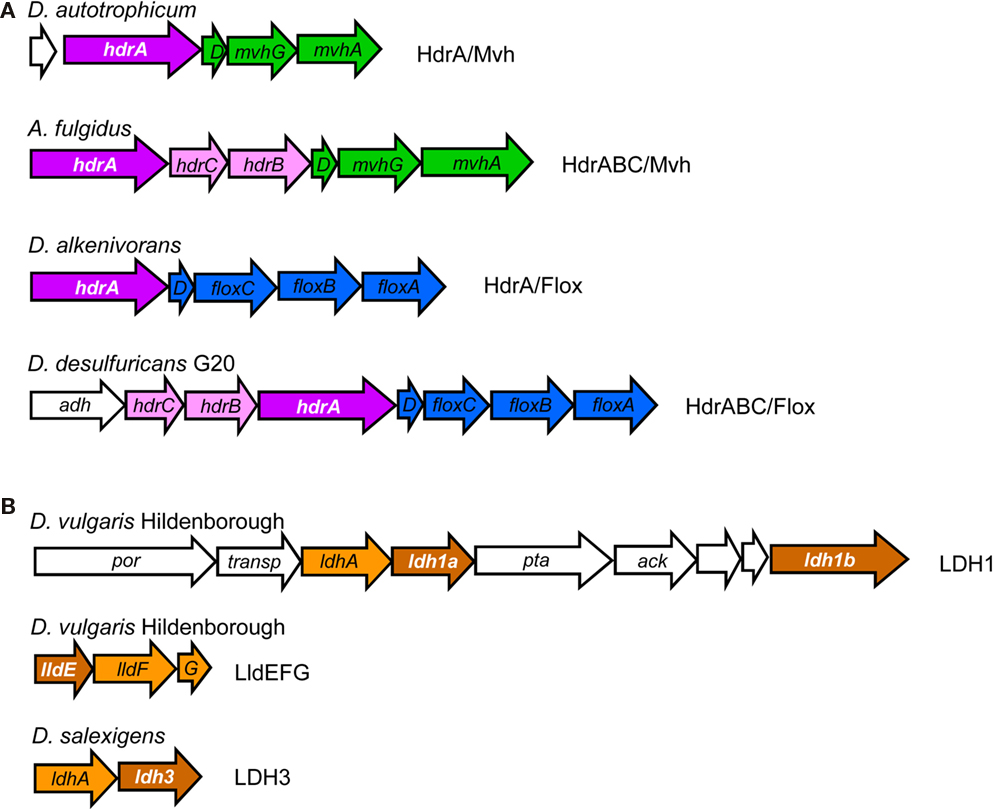
Figure 4. Examples of gene loci for (A) hdrA-related genes (in white lettering) and (B) hdrD-related genes (in white lettering).
Other gene loci in SRO containing hdrA-like genes include a fdhA gene (and an hdrL) or genes for a pyruvate:Fd oxidoreductase (Por), suggesting that formate and pyruvate may also be the source of electrons for HdrA reduction.
The analysis of hdrD-like genes also provided interesting results, one of which was the identification of the iron–sulfur subunit of three putative lactate dehydrogenases (LDH) as belonging to the CCG family (Figure 4; Table 3). One of the LDH gene clusters was previously identified as an “organic acid oxidation region” in the genome of D. vulgaris and D. desulfuricans G20 (Pereira et al., 2007; Wall et al., 2008). It includes genes for pyruvate:Fd oxidoreductase (por), putative lactate permease, the putative LDH catalytic subunit, a putative LDH iron–sulfur subunit that has two CCG domains, phosphate acetyl transferase (pta) and acetate kinase (ack). A larger HdrD-related protein is also present in this gene cluster. A novel three-subunit L-lactate dehydrogenase that was named LldEFG (or LutABC) was recently identified in Bacillus subtilis (Chai et al., 2009), Shewanella oneidensis (Pinchuk et al., 2009), and Campylobacter jejuni (Thomas et al., 2011). LldEFG is also present in several of the SRO genomes analyzed and the LldE protein is a small HdrD-related iron–sulfur protein with one CCG domain. The LldEFG enzyme is membrane-associated although no transmembrane helices are present in any of its subunits. A third putative LDH with an HdrD-like subunit was also identified. The role of the LDH HdrD-like subunits is uncertain, as the electron acceptor for LDH has not been identified.
Other proteins related to HdrD include one membrane-associated HdrF protein found next to the etfAB genes coding for electron-transfer flavoprotein, a large flavoprotein with two CCG domains and a putative FAD-binding site, and a protein encoded next to a gene for a molybdenum-containing aldehyde oxidoreductase. These HdrD-related proteins suggest the presence of different electron-transfer pathways (from lactate, β-oxidation, and others) as possible donors for reduction of the menaquinone pool or DsrCox.
The comparative genomic analysis reported in this work provides new insights into the energy metabolism of SRO. By comparing phylogenetically distinct organisms capable of sulfate reduction we identified the proteins that can be considered as comprising the minimal set required for this metabolic activity: a sulfate transporter, Sat, a pyrophosphatase, AprAB, DsrAB, DsrC, DsrMK, and Fd. The QmoAB proteins are also present in most organisms, being absent only in C. maquiligensis. In addition, we identified a higher diversity of possible energy conserving pathways than classically has been considered to be present in these organisms. The intracellular redox cycling of metabolites (like H2, formate or CO) is not a universal mechanism, but should play a role in bioenergetics of Deltaproteobacteria and T. yellowstonii, which are characterized by a large number of cytochromes c and cytochrome c-associated membrane redox complexes. A large number of cytochromes c has previously been correlated with increased respiratory versatility in anaerobes (Thomas et al., 2008), and such versatility is also suggested by the apparent redundancy of periplasmic redox proteins and membrane complexes found in many Deltaproteobacteria. Redox cycling is associated with energy conservation though charge separation or redox loop mechanisms. In contrast, the Archaea and Clostridia groups contain practically no cytochromes c or associated membrane complexes. The Gram-positive organisms analyzed present some unique traits including the absence of QmoC and DsrJOP proteins. Despite the absence of a periplasmic space, three extracytoplasmic proteins are predicted for these organisms, namely NrfHA and membrane-anchored [FeFe] Hase and FDH.
Overall, this analysis suggests that all SRO use diverse processes for energy conservation involving membrane-based chemiosmotic mechanisms, or soluble flavin-based electron bifurcation ones. Many organisms include nuo genes for an ion-translocating complex I, which in most cases uses a still unidentified electron donor. Another widespread ion-translocating complex is Rnf, which together with Ech and Coo Hases, provides coupling sites for Fd-associated processes such as electron bifurcation. Regarding soluble processes, we identified a surprisingly high number of cytoplasmic Hases and FDHs as likely candidates for electron bifurcation coupling involving NAD(P)/H, Fd, or HDRs. A large number of HDR-related proteins were also detected. We propose that these proteins are part of electron-transfer pathways involving energy coupling through electron bifurcation, from diverse electron donors such as H2, formate, pyruvate, NAD(P)H, β-oxidation, and others. These pathways may constitute alternatives to Dsr and other transmembrane complexes for reduction of DsrCox, the protein we propose is central to the sulfite reduction step.
A few novel redox proteins were identified in SRO, including a FloxABCD/HdrA(BC) complex proposed to perform electron bifurcation with NAD(P)H, Fd, and DsrCox, a new type of membrane-anchored periplasmic [FeFe] Hase, and a putative soluble FHL also comprising an [FeFe] Hase. In conclusion, this analysis indicates that energy metabolism of SRO is far more versatile than previously considered; both chemiosmotic and flavin-based electron bifurcating mechanisms provide alternative strategies for energy conservation. An interesting aspect of (at least some) SRO is their ability to grow syntrophically in the absence of sulfate. In such situation some modules of this versatile redox machinery may operate in the opposite direction to that of respiratory conditions. Finally, it should be stressed that although drawing theories based on comparative genomic analysis is an attractive and even convincing exercise, no definite conclusions can be drawn until experimental evidence is provided. Thus, much work remains to be carried out to elucidate the bioenergetic mechanisms of SRO.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by grant QUI-BIQ/10059/2008 funded by FCT, Portugal.
The locus tags for all genes can be found in http://www.frontiersin.org/Microbial_Physiology_and_Metabolism/10.3389/fmicb.2011.00069/abstract
Akagi, J. M. (1995). “Respiratory sulfate reduction,” in Sulfate-Reducing Bacteria, ed. L. L. Barton (New York: Plenum Press), 89–111.
Almendra, M. J., Brondino, C. D., Gavel, O., Pereira, A. S., Tavares, P., Bursakov, S., Duarte, R., Caldeira, J., Moura, J. J. G., and Moura, I. (1999). Purification and characterization of a tungsten-containing formate dehydrogenase from Desulfovibrio gigas. Biochemistry 38, 16366–16372.
Biegel, E., and Müller, V. (2010). Bacterial Na+-translocating ferredoxin: NAD+ oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 107, 18138–18142.
Broco, M., Rousset, M., Oliveira, S., and Rodrigues-Pousada, C. (2005). Deletion of flavoredoxin gene in Desulfovibrio gigas reveals its participation in thiosulfate reduction. FEBS Lett. 579, 4803–4807.
Caffrey, S. A., Park, H. S., Voordouw, J. K., He, Z., Zhou, J., and Voordouw, G. (2007). Function of periplasmic hydrogenases in the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 189, 6159–6167.
Canfield, D. E., Stewart, F. J., Thamdrup, B., De Brabandere, L., Dalsgaard, T., Delong, E. F., Revsbech, N. P., and Ulloa, O. (2010). A cryptic sulfur cycle in oxygen-minimum-zone waters off the chilean coast. Science 330, 1375–1378.
Chai, Y. R., Kolter, R., and Losick, R. (2009). A widely conserved gene cluster required for lactate utilization in Bacillus subtilis and its involvement in biofilm formation. J. Bacteriol. 191, 2423–2430.
Cort, J. R., Mariappan, S. V., Kim, C. Y., Park, M. S., Peat, T. S., Waldo, G. S., Terwilliger, T. C., and Kennedy, M. A. (2001). Solution structure of Pyrobaculum aerophilum DsrC, an archaeal homologue of the gamma subunit of dissimilatory sulfite reductase. Eur. J. Biochem. 268, 5842–5850.
Costa, K. C., Wong, P. M., Wang, T., Lie, T. J., Dodsworth, J. A., Swanson, I., Burn, J. A., Hackett, M., and Leigh, J. A. (2010). Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc. Natl. Acad. Sci. U.S.A. 107, 11050–11055.
Dolla, A., Pohorelic, B. K. J., Voordouw, J. K., and Voordouw, G. (2000). Deletion of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox-potential niche establishment. Arch. Microbiol. 174, 143–151.
Efremov, R. G., Baradaran, R., and Sazanov, L. A. (2010). The architecture of respiratory complex I. Nature 465, 441–445.
ElAntak, L., Morelli, X., Bornet, O., Hatchikian, C., Czjzek, M., Alain, D. A., and Guerlesquin, F. (2003). The cytochrome c3-[Fe]-hydrogenase electron-transfer complex: structural model by NMR restrained docking. FEBS Lett. 548, 1–4.
Friedrich, T., and Scheide, D. (2000). The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 479, 1–5.
Greene, E. A., Hubert, C., Nemati, M., Jenneman, G. E., and Voordouw, G. (2003). Nitrite reductase activity of sulphate-reducing bacteria prevents their inhibition by nitrate-reducing, sulphide-oxidizing bacteria. Environ. Microbiol. 5, 607–617.
Grein, F., Venceslau, S. S., Schneider, L., Hildebrandt, P., Todorovic, S., Pereira, I. A. C., and Dahl, C. (2010). DsrJ, an essential part of the DsrMKJOP transmembrane complex in the purple sulfur bacterium Allochromatium vinosum, is an unusual triheme cytochrome c. Biochemistry 49, 8290–8299.
Hamann, N., Mander, G. J., Shokes, J. E., Scott, R. A., Bennati, M., and Hedderich, R. (2007). Cysteine-rich CCG domain contains a novel [4Fe-4S] cluster binding motif as deduced from studies with subunit B of heterodisulfide reductase from Methanothermobacter marburgensis. Biochemistry 46, 12875–12885.
Haveman, S. A., Brunelle, V., Voordouw, J. K., Voordouw, G., Heidelberg, J. F., and Rabus, R. (2003). Gene expression analysis of energy metabolism mutants of Desulfovibrio vulgaris Hildenborough indicates an important role for alcohol dehydrogenase. J. Bacteriol. 185, 4345–4353.
Hedderich, R., and Forzi, L. (2005). Energy-converting [NiFe] hydrogenases: more than just H2 activation. J. Mol. Microbiol. Biotechnol. 10, 92–104.
Hedderich, R., Hamann, N., and Bennati, M. (2005). Heterodisulfide reductase from methanogenic archaea: a new catalytic role for an iron-sulfur cluster. Biol. Chem. 386, 961–970.
Hedderich, R., Klimmek, O., Kröger, A., Dirmeier, R., Keller, M., and Stetter, K. O. (1999). Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol. Rev. 22, 353–381.
Heidelberg, J. F., Seshadri, R., Haveman, S. A., Hemme, C. L., Paulsen, I. T., Kolonay, J. F., Eisen, J. A., Ward, N., Methe, B., Brinkac, L. M., Daugherty, S. C., Deboy, R. T., Dodson, R. J., Durkin, A. S., Madupu, R., Nelson, W. C., Sullivan, S. A., Fouts, D., Haft, D. H., Selengut, J., Peterson, J. D., Davidsen, T. M., Zafar, N., Zhou, L. W., Radune, D., Dimitrov, G., Hance, M., Tran, K., Khouri, H., Gill, J., Utterback, T. R., Feldblyum, T. V., Wall, J. D., Voordouw, G., and Fraser, C. M. (2004). The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22, 554–559.
Herrmann, G., Jayamani, E., Mai, G., and Buckel, W. (2008). Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 190, 784–791.
Ikeuchi, Y., Shigi, N., Kato, J., Nishimura, A., and Suzuki, T. (2006). Mechanistic insights into multiple sulfur mediators sulfur relay by involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell 21, 97–108.
Iverson, T. M., Arciero, D. M., Hsu, B. T., Logan, M. S. P., Hooper, A. B., and Rees, D. C. (1998). Heme packing motifs revealed by the crystal structure of the tetra-heme cytochrome c554 from Nitrosomonas europaea. Nat. Struct. Mol. Biol. 5, 1005–1012.
Jenney, F. E., and Adams, M. W. W. (2008). Hydrogenases of the model hyperthermophiles. Ann. N. Y. Acad. Sci. 1125, 252–266.
Jørgensen, B. B. (1982). Mineralization of organic matter in the sea-bed – the role of sulphate reduction. Nature 390, 364–370.
Junier, P., Junier, T., Podell, S., Sims, D. R., Detter, J. C., Lykidis, A., Han, C. S., Wigginton, N. S., Gaasterland, T., and Bernier-Latmani, R. (2010). The genome of the Gram-positive metal- and sulfate-reducing bacterium Desulfotomaculum reducens strain MI-1. Environ. Microbiol. 12, 2738–2754.
Kobayashi, K., Hasegawa, H., Takagi, M., and Ishimoto, M. (1982). Proton translocation associated with sulfite reduction in a sulfate-reducing bacterium, Desulfovibrio vulgaris. FEBS Lett. 142, 235–237.
Kunow, J., Linder, D., Stetter, K. O., and Thauer, R. K. (1994). F420H2: quinone oxidoreductase from Archaeoglobus fulgidus. Characterization of a membrane-bound multisubunit complex containing FAD and iron-sulfur clusters. Eur. J. Biochem. 223, 503–511.
Li, F., Hinderberger, J., Seedorf, H., Zhang, J., Buckel, W., and Thauer, R. K. (2008). Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J. Bacteriol. 190, 843–850.
Li, Q., Li, L., Rejtar, T., Lessner, D. J., Karger, B. L., and Ferry, J. G. (2006). Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J. Bacteriol. 188, 702–710.
Li, X., Luo, Q., Wofford, N. Q., Keller, K. L., McInerney, M. J., Wall, J. D., and Krumholz, L. R. (2009). A molybdopterin oxidoreductase is involved in H2 oxidation in Desulfovibrio desulfuricans G20. J. Bacteriol. 191, 2675–2682.
Ma, K., and Adams, M. W. (2001). Ferredoxin:NADP oxidoreductase from Pyrococcus furiosus. Meth. Enzymol. 334, 40–45.
Ma, K., and Adams, M. W. W. (1994). Sulfide dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus – a new multifunctional enzyme involved in the reduction of elemental sulfur. J. Bacteriol. 176, 6509–6517.
Malki, S., Saimmaime, I., De Luca, G., Rousset, M., Dermoun, Z., and Belaich, J. P. (1995). Characterization of an operon encoding an NADP-reducing hydrogenase in Desulfovibrio fructosovorans. J. Bacteriol. 177, 2628–2636.
Mander, G. J., Duin, E. C., Linder, D., Stetter, K. O., and Hedderich, R. (2002). Purification and characterization of a membrane-bound enzyme complex from the sulfate-reducing archaeon Archaeoglobus fulgidus related to heterodisulfide reductase from methanogenic archaea. Eur. J. Biochem. 269, 1895–1904.
Mander, G. J., Pierik, A. J., Huber, H., and Hedderich, R. (2004). Two distinct heterodisulfide reductase-like enzymes in the sulfate-reducing archaeon Archaeoglobus profundus. Eur. J. Biochem. 271, 1106–1116.
Mander, G. J., Weiss, M. S., Hedderich, R., Kahnt, J., Ermler, U., and Warkentin, E. (2005). X-ray structure of the gamma-subunit of a dissimilatory sulfite reductase: fixed and flexible C-terminal arms. FEBS Lett. 579, 4600–4604.
Matias, P. M., Coelho, R., Pereira, I. A., Coelho, A. V., Thompson, A. W., Sieker, L. C., Gall, J. L., and Carrondo, M. A. (1999). The primary and three-dimensional structures of a nine-haem cytochrome c from Desulfovibrio desulfuricans ATCC 27774 reveal a new member of the Hmc family. Structure 7, 119–130.
Matias, P. M., Pereira, I. A., Soares, C. M., and Carrondo, M. A. (2005). Sulphate respiration from hydrogen in Desulfovibrio bacteria: a structural biology overview. Prog. Biophys. Mol. Biol. 89, 292–329.
Matson, E. G., Zhang, X. N., and Leadbetter, J. R. (2010). Selenium controls transcription of paralogous formate dehydrogenase genes in the termite gut acetogen, Treponema primitia. Environ. Microbiol. 12, 2245–2258.
McInerney, M. J., Rohlin, L., Mouttaki, H., Kim, U., Krupp, R. S., Rios-Hernandez, L., Sieber, J., Struchtemeyer, C. G., Bhattacharyya, A., Campbell, J. W., and Gunsalus, R. P. (2007). The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc. Natl. Acad. Sci. U.S.A. 104, 7600–7605.
Meuer, J., Kuettner, H. C., Zhang, J. K., Hedderich, R., and Metcalf, W. W. (2002). Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc. Natl. Acad. Sci. U.S.A. 99, 5632–5637.
Müller, V., Imkamp, F., Biegel, E., Schmidt, S., and Dilling, S. (2008). Discovery of a ferredoxin:NAD+-oxidoreductase (Rnf) in Acetobacterium woodii: a novel potential coupling site in acetogens. Ann. N. Y. Acad. Sci. 1125, 137–146.
Muyzer, G., and Stams, A. J. (2008). The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6, 441–454.
Odom, J. M., and Peck, H. D. Jr. (1981). Hydrogen cycling as a general mechanism for energy coupling in the sulfate-reducing bacteria, Desulfovibrio sp. FEMS Microbiol. Lett. 12, 47–50.
Oliveira, T. F., Vonrhein, C., Matias, P. M., Venceslau, S. S., Pereira, I. A., and Archer, M. (2008). The crystal structure of Desulfovibrio vulgaris dissimilatory sulfite reductase bound to DsrC provides novel insights into the mechanism of sulfate respiration. J. Biol. Chem. 283, 34141–34149.
Pereira, I. A. C. (2008). “Membrane complexes in Desulfovibrio,” in Microbial Sulfur Metabolism, eds C. Friedrich and C. Dahl (Berlin: Springer-Verlag), 24–35.
Pereira, I. A. C., Haveman, S. A., and Voordouw, G. (2007). “Biochemical, genetic and genomic characterization of anaerobic electron transport pathways in sulphate-reducing delta-proteobacteria,” in Sulphate-Reducing Bacteria: Environmental and Engineered Systems, eds L. L. Barton and W. A. Allan Hamilton (Cambridge: Cambridge University Press), 215–240.
Pereira, I. A. C., Romão, C. V., Xavier, A. V., LeGall, J., and Teixeira, M. (1998). Electron transfer between hydrogenases and mono and multiheme cytochromes in Desulfovibrio spp. J. Biol. Inorg. Chem. 3, 494–498.
Pereira, P. M., He, Q., Valente, F. M. A., Xavier, A. V., Zhou, J. Z., Pereira, I. A. C., and Louro, R. O. (2008). Energy metabolism in Desulfovibrio vulgaris Hildenborough: insights from transcriptome analysis. Antonie Van Leeuwenhoek 93, 347–362.
Pereira, P. M., Teixeira, M., Xavier, A. V., Louro, R. O., and Pereira, I. A. (2006). The Tmc complex from Desulfovibrio vulgaris Hildenborough is involved in transmembrane electron transfer from periplasmic hydrogen oxidation. Biochemistry 45, 10359–10367.
Pierik, A. J., Duyvis, M. G., van Helvoort, J. M., Wolbert, R. B., and Hagen, W. R. (1992). The third subunit of desulfoviridin-type dissimilatory sulfite reductases. Eur. J. Biochem. 205, 111–115.
Pieulle, L., Morelli, X., Gallice, P., Lojou, E., Barbier, P., Czjzek, M., Bianco, P., Guerlesquin, F., and Hatchikian, E. C. (2005). The type I/type II cytochrome c3 complex: an electron transfer link in the hydrogen-sulfate reduction pathway. J. Mol. Biol. 354, 73–90.
Pinchuk, G. E., Rodionov, D. A., Yang, C., Li, X. Q., Osterman, A. L., Dervyn, E., Geydebrekht, O. V., Reed, S. B., Romine, M. F., Collart, F. R., Scott, J. H., Fredrickson, J. K., and Beliaev, A. S. (2009). Genomic reconstruction of Shewanella oneidensis MR-1 metabolism reveals a previously uncharacterized machinery for lactate utilization. Proc. Natl. Acad. Sci. U.S.A. 106, 2874–2879.
Pires, R. H., Lourenco, A. I., Morais, F., Teixeira, M., Xavier, A. V., Saraiva, L. M., and Pereira, I. A. (2003). A novel membrane-bound respiratory complex from Desulfovibrio desulfuricans ATCC 27774. Biochim. Biophys. Acta 1605, 67–82.
Pires, R. H., Venceslau, S. S., Morais, F., Teixeira, M., Xavier, A. V., and Pereira, I. A. (2006). Characterization of the Desulfovibrio desulfuricans ATCC 27774 DsrMKJOP complex-A membrane-bound redox complex involved in the sulfate respiratory pathway. Biochemistry 45, 249–262.
Pott, A. S., and Dahl, C. (1998). Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 144, 1881–1894.
Rabus, R., Hansen, T., and Widdel, F. (2007). “Dissimilatory sulfate- and sulfur-Reducing prokaryotes,” in The Prokaryotes, ed. M. Dworkin (New York: Springer-Verlag), 659–768.
Rodrigues, M. L., Oliveira, T. F., Pereira, I. A., and Archer, M. (2006). X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel haem coordination. EMBO J. 25, 5951–5960.
Rodrigues, R., Valente, F. M., Pereira, I. A., Oliveira, S., and Rodrigues-Pousada, C. (2003). A novel membrane-bound Ech [NiFe] hydrogenase in Desulfovibrio gigas. Biochem. Biophys. Res. Commun. 306, 366–375.
Rossi, M., Pollock, W. B. R., Reij, M. W., Keon, R. G., Fu, R., and Voordouw, G. (1993). The hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough encodes a potential transmembrane redox protein complex. J. Bacteriol. 175, 4699–4711.
Saraiva, L. M., da Costa, P. N., Conte, C., Xavier, A. V., and LeGall, J. (2001). In the facultative sulphate/nitrate reducer Desulfovibrio desulfuricans ATCC 27774, the nine-haem cytochrome c is part of a membrane-bound redox complex mainly expressed in sulphate-grown cells. Biochim. Biophys. Acta 1520, 63–70.
Sawers, R. G. (2005). Formate and its role in hydrogen production in Escherichia coli. Biochem. Soc. Trans. 33, 42–46.
Schmehl, M., Jahn, A., Meyer zu Vilsendorf, A., Hennecke, S., Masepohl, B., Schuppler, M., Marxer, M., Oelze, J., and Klipp, W. (1993). Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 241, 602–615.
Schut, G. J., and Adams, M. W. (2009). The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J. Bacteriol. 191, 4451–4457.
Sebban, C., Blanchard, L., Bruschi, M., and Guerlesquin, F. (1995). Purification and characterization of the formate dehydrogenase from Desulfovibrio vulgaris Hildenborough. FEMS Microbiol. Lett. 133, 143–149.
Seedorf, H., Fricke, W. F., Veith, B., Bruggemann, H., Liesegang, H., Strittmatter, A., Miethke, M., Buckel, W., Hinderberger, J., Li, F., Hagemeier, C., Thauer, R. K., and Gottschalk, G. (2008). The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc. Natl. Acad. Sci. U.S.A. 105, 2128–2133.
Serrano, A., Perez-Castineira, J. R., Baltscheffsky, M., and Baltscheffsky, H. (2007). H+-PPases: yesterday, today and tomorrow. IUBMB Life 59, 76–83.
Shaw, A. J., Hogsett, D. A., and Lynd, L. R. (2009). Identification of the [FeFe]-hydrogenase responsible for hydrogen generation in Thermoanaerobacterium saccharolyticum and demonstration of increased ethanol yield via hydrogenase knockout. J. Bacteriol. 191, 6457–6464.
Simon, J., van Spanning, R. J., and Richardson, D. J. (2008). The organisation of proton motive and non-proton motive redox loops in prokaryotic respiratory systems. Biochim. Biophys. Acta 1777, 1480–1490.
Singer, S. W., Hirst, M. B., and Ludden, P. W. (2006). CO-dependent H2 evolution by Rhodospirillum rubrum: role of CODH:CooF complex. Biochim. Biophys. Acta 1757, 1582–1591.
Soboh, B., Linder, D., and Hedderich, R. (2004). A multisubunit membrane-bound [NiFe] hydrogenase and an NADH-dependent Fe-only hydrogenase in the fermenting bacterium Thermoanaerobacter tengcongensis. Microbiology 150, 2451–2463.
Steuber, J. (2001). Na(+) translocation by bacterial NADH:quinone oxidoreductases: an extension to the complex-I family of primary redox pumps. Biochim. Biophys. Acta 1505, 45–56.
Stojanowic, A., Mander, G. J., Duin, E. C., and Hedderich, R. (2003). Physiological role of the F420-non-reducing hydrogenase (Mvh) from Methanothermobacter marburgensis. Arch. Microbiol. 180, 194–203.
Strittmatter, A. W., Liesegang, H., Rabus, R., Decker, I., Amann, J., Andres, S., Henne, A., Fricke, W. F., Martinez-Arias, R., Bartels, D., Goesmann, A., Krause, L., Pühler, A., Klenk, H. P., Richter, M., Schüler, M., Glöckner, F. O., Meyerdierks, A., Gottschalk, G., and Amann, R. (2009). Genome sequence of Desulfobacterium autotrophicum HRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environ. Microbiol. 11, 1038–1055.
Thauer, R. K., Kaster, A. K., Goenrich, M., Schick, M., Hiromoto, T., and Shima, S. (2010). Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu. Rev. Biochem. 79, 507–536.
Thauer, R. K., Kaster, A. K., Seedorf, H., Buckel, W., and Hedderich, R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591.
Thomas, M. T., Shepherd, M., Poole, R. K., van Vliet, A. H., Kelly, D. J., and Pearson, B. M. (2011). Two respiratory enzyme systems in Campylobacter jejuni NCTC 11168 contribute to growth on L-lactate. Environ. Microbiol. 13, 48–61.
Thomas, S. H., Wagner, R. D., Arakaki, A. K., Skolnick, J., Kirby, J. R., Shimkets, L. J., Sanford, R. A., and Löffler, F. E. (2008). The mosaic genome of Anaeromyxobacter dehalogenans strain 2CP-C suggests an aerobic common ancestor to the delta-proteobacteria. PLoS ONE 3, e2103. doi: 10.1371/ journal.pone.0002103
Valente, F. A. A., Almeida, C. C., Pacheco, I., Carita, J., Saraiva, L. M., and Pereira, I. A. C. (2006). Selenium is involved in regulation of periplasmic hydrogenase gene expression in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 188, 3228–3235.
Valente, F. M. A., Oliveira, A. S. F., Gnadt, N., Pacheco, I., Coelho, A. V., Xavier, A. V., Teixeira, M., Soares, C. M., and Pereira, I. A. C. (2005). Hydrogenases in Desulfovibrio vulgaris Hildenborough: structural and physiologic characterisation of the membrane-bound [NiFeSe] hydrogenase. J. Biol. Inorg. Chem. 10, 667–682.
Valente, F. M. A., Saraiva, L. M., LeGall, J., Xavier, A. V., Teixeira, M., and Pereira, I. A. C. (2001). A membrane-bound cytochrome c3: a type II cytochrome c3 from Desulfovibrio vulgaris Hildenborough. Chembiochem 2, 895–905.
Venceslau, S. S., Lino, R. R., and Pereira, I. A. (2010). The Qrc membrane complex, related to the alternative complex III, is a menaquinone reductase involved in sulfate respiration. J. Biol. Chem. 285, 22774–22783.
Vignais, P. M., and Billoud, B. (2007). Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–4272.
Voordouw, G. (2002). Carbon monoxide cycling by Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 184, 5903–5911.
Walker, C. B., He, Z. L., Yang, Z. K., Ringbauer, J. A., He, Q., Zhou, J. H., Voordouw, G., Wall, J. D., Arkin, A. P., Hazen, T. C., Stolyar, S., and Stahl, D. A. (2009). The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J. Bacteriol. 191, 5793–5801.
Wall, J. D., Arkin, A. P., Balci, N. C., and Rapp-Giles, B. (2008). “Genetics and genomics of sulfate respiration in Desulfovibrio,” in Microbial Sulfur Metabolism, eds C. Dahl and C. G. Friedrich (Heidelbeg: Springer-Verlag), 1–12.
Wang, S., Huang, H., Moll, J., and Thauer, R. K. (2010). NADP+ reduction with reduced ferredoxin and NADP+ reduction with NADH are coupled via an electron-bifurcating enzyme complex in Clostridium kluyveri. J. Bacteriol. 192, 5115–5123.
Xia, D., Esser, L., Yu, L., and Yu, C. A. (2007). Structural basis for the mechanism of electron bifurcation at the quinol oxidation site of the cytochrome bc1 complex. Photosyn. Res. 92, 17–34.
Zane, G. M., Yen, H. C., and Wall, J. D. (2010). Effect of the deletion of qmoABC and the promoter-distal gene encoding a hypothetical protein on sulfate reduction in Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 76, 5500–5509.
Zhang, W. W., Culley, D. E., Scholten, J. C. M., Hogan, M., Vitiritti, L., and Brockman, F. J. (2006a). Global transcriptomic analysis of Desulfovibrio vulgaris on different electron donors. Antonie Van Leeuwenhoek 89, 221–237.
Zhang, W. W., Gritsenko, M. A., Moore, R. J., Culley, D. E., Nie, L., Petritis, K., Strittmatter, E. F., Camp, D. G., Smith, R. D., and Brockman, F. J. (2006b). A proteomic view of Desulfovibrio vulgaris metabolism as determined by liquid chromatography coupled with tandem mass spectrometry. Proteomics 6, 4286–4299.
Keywords: energy metabolism, sulfate reducing bacteria, membrane complexes, electron bifurcation, hydrogenase, formate dehydrogenase, cytochrome, Desulfovibrio
Citation: Pereira IAC, Ramos AR, Grein F, Marques MC, Marques da Silva S and Venceslau SS (2011) A comparative genomic analysis of energy metabolism in sulfate reducing bacteria and archaea. Front. Microbio. 2:69. doi: 10.3389/fmicb.2011.00069
Received: 03 February 2011;
Paper pending published: 07 March 2011;
Accepted: 25 March 2011;
Published online: 19 April 2011.
Edited by:
Martin G. Klotz, University of Louisville, USAReviewed by:
Kathleen Scott, University of South Florida, USACopyright: © 2011 Pereira, Ramos, Grein, Marques, Marques da Silva and Venceslau. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Inês A. Cardoso Pereira, Instituto de Tecnologia Química e Biológica, Avenida da Republica – Estação Agronómica Nacional, 2780-157 Oeiras, Portugal. e-mail:aXBlcmVpcmFAaXRxYi51bmwucHQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.