- 1 Institute of Microbiology and Genetics, Technische Universität Darmstadt, Darmstadt, Germany
- 2 Max-Planck Institute of Biochemistry, Martinsried, Germany
Background: The thermoacidophilic and chemolithotrophic archaeon Acidianus ambivalens is routinely grown with sulfur and CO2-enriched air. We had described a membrane-bound, tetrathionate (TT) forming thiosulfate:quinone oxidoreductase. Here we describe the first TT hydrolase (TTH) from Archaea. Results: A. ambivalens cells grown aerobically with TT as sole sulfur source showed doubling times of 9 h and final cell densities of up to 8 × 108/ml. TTH activity (≈0.28 U/mg protein) was found in cell-free extracts of TT-grown but not of sulfur-grown cells. Differential fractionation of freshly harvested cells involving a pH shock showed that about 92% of the TTH activity was located in the pseudo-periplasmic fraction associated with the surface layer, while 7.3% and 0.3% were present in the soluble and membrane fractions, respectively. The enzyme was enriched 54-fold from the cytoplasmic fraction and 2.1-fold from the pseudo-periplasmic fraction. The molecular mass of the single subunit was 54 kDa. The optimal activity was at or above 95°C at pH 1. Neither PQQ nor divalent cations had a significant effect on activity. The gene (tth1) was identified following N-terminal sequencing of the protein. Northern hybridization showed that tth1 was transcribed in TT-grown cells in contrast to a second paralogous tth2 gene. The deduced amino acid sequences showed similarity to the TTH from Acidithiobacillus and other proteins from the PQQ dehydrogenase superfamily. It displayed a β-propeller structure when being modeled, however, important residues from the PQQ-binding site were absent. Conclusion: The soluble, extracellular, and acidophilic TTH identified in TT-grown A. ambivalens cells is essential for TT metabolism during growth but not for the downstream processing of the TQO reaction products in S°-grown cells. The liberation of TTH by pH shock from otherwise intact cells strongly supports the pseudo-periplasm hypothesis of the S-layer of Archaea.
Introduction
The dissimilatory oxidation of inorganic sulfur compounds (ISC) is one of the most important sources of metabolic energy for CO2 fixation in light-independent ecosystems like solfataras and other volcanic hot springs (reviewed for example by Kelly et al., 1997; Friedrich et al., 2005; Stetter, 2006; Kletzin, 2007; Ghosh and Dam, 2009). In addition, ISC oxidation is essential for bioleaching of precious and/or toxic metals from low-grade ores and slag heaps, regardless whether this effect is desired (metal extraction) or undesired (acid mine drainage; Sand et al., 2001; Gadd, 2010). Sulfur oxidation pathways and especially the SOX complex from neutrophilic chemo- or phototrophic bacteria are fairly well understood (for review, see Friedrich et al., 2005, 2008; Frigaard and Dahl, 2009; Sakurai et al., 2010). In contrast, there are many gaps in our knowledge concerning the mechanisms of ISC oxidation in acidophilic Bacteria and Archaea, the dominant microorganisms in volcanic hot springs and bioleaching environments (Kletzin, 2007, 2008; Ghosh and Dam, 2009).
Our model organism Acidianus ambivalens belongs to the Sulfolobales order within the archaeal domain (Zillig et al., 1986; Fuchs et al., 1996). A. ambivalens is a chemolithoautotrophic acidophile growing optimally at 80°C and pH 2.5. It oxidizes S° to sulfuric acid under aerobic conditions and uses hydrogen as the electron donor for S° reduction under anaerobic conditions. A soluble sulfur oxygenase reductase (SOR) mediates the initial step in the S° oxidation pathway of aerobically grown cells, a unique enzyme catalyzing the simultaneous S° oxygenation and disproportionation to sulfite, thiosulfate, and sulfide (Kletzin, 1989; Urich et al., 2004; Veith et al., 2011; this volume). It is not known whether thiosulfate is a primary product of the enzyme or whether it originates from a rapid non-enzymatic reaction between S° and sulfite under the assay conditions (Kletzin, 1989; Müller et al., 2004). Regardless of that, S°-grown A. ambivalens cells contain significant amounts of a membrane-bound thiosulfate:quinone oxidoreductase (TQO) oxidizing thiosulfate to tetrathionate (TT) and reducing quinones (Müller et al., 2004). This type of thiosulfate oxidation is known as the S4 intermediate pathway (Ghosh and Dam, 2009). Two observations suggest that the active site of the TQO is facing the cytoplasm. First, the SOR, which delivers thiosulfate, is a soluble and cytoplasmic enzyme. Secondly, thiosulfate is not stable at the growth conditions of A. ambivalens (acidic pH and high temperature; Johnston and McAmish, 1973) so that it rapidly decomposes, when being added to the medium forming sulfur and sulfite. In contrast, TT is not very stable at the near-neutral pH (6.5; Johnston and McAmish, 1973; Moll and Schäfer, 1988) and the reducing conditions in the cytoplasm. We had hypothesized that the TQO and a non-enzymatic TT reduction to thiosulfate mediated by hydrogen sulfide and sulfite in the cytoplasm ultimately feeds electrons into the respiratory chain (Müller et al., 2004; Kletzin, 2008).
As the fate of the TT produced during the TQO reaction is not clear, we initiated a search for TT-metabolizing enzymes in A. ambivalens. Three ways of TT metabolism are known: (1) the reduction by either multiheme or molybdenum-containing reductases enabling mesophilic proteobacteria like Salmonella enterica to grow anaerobically with TT as a terminal electron acceptor (Hensel et al., 1999; Price-Carter et al., 2001; Mowat et al., 2004); (2) the oxidation by still unknown subunits of SOX multienzyme complexes (Mukhopadhyaya et al., 2000; Lahiri et al., 2006); or (3) a disproportionation by tetrathionate hydrolases (TTH) forming sulfate, thiosulfate or sulfite, and sulfur or pentathionate (De Jong et al., 1997a; Bugaytsova and Lindström, 2004; Kanao et al., 2007; Rzhepishevska et al., 2007). TTHs isolated from various acidophilic Acidithiobacillus species and Acidiphilium acidophilum (formerly Thiobacillus acidophilus) are usually monomeric and/or homodimeric, membrane-associated, or soluble enzymes with pH optima around 2.5–4 (De Jong et al., 1997a; Bugaytsova and Lindström, 2004; Egorova et al., 2004; Kanao et al., 2007; Mangold et al., 2011; this volume). Sequence comparisons predicted that the TTHs belong to a pyrroloquinoline quinone (PQQ) containing protein family. Kanao et al. (2010) however showed that recombinant Acidithiobacillus ferrooxidans TTH purified from E. coli inclusion bodies could be refolded into the active state without PQQ and under acidic conditions.
In this contribution, we demonstrate that A. ambivalens grows well with TT as the sole sulfur source thereby producing a highly active TTH with an optimum at pH 1. We also show that the protein is extracellular and associated with the S-layer of the archaeon.
Materials and Methods
Organism and Growth Conditions
Acidianus ambivalens DSM 3772 was grown as described (Zillig et al., 1986) with slight modifications. The medium contained per liter: KH2PO4, 2.8 g; (NH4)SO4, 1.5 g; MgSO4 × 7 H2O, 0.25 g; CaCl2 × 2 H2O, 70 mg; FeSO4 × 7 H2O, 28 mg; Na2B2O7 × 10 H2O, 9 mg; MnCl2 × 4 H2O, 3.6 mg; ZnSO4 × 7 H2O, 0.44 mg; CuCl2 × 2 H2O, 0.1 mg; VaSO4 × 5 H2O, 0.07 mg; Na2MoO4 × 2 H2O, 0.06 mg; CoSO4 × 7 H2O, 0.02 mg. The pH was adjusted to 2.5 with sterile 50% H2SO4. About 10 ml/l of freshly prepared and filter-sterilized 1 M K2S4O6 solution (Merck, Darmstadt, Germany) was added as sulfur source. Additionally, the same amounts were added 24 and 48 h post-inoculation. Alternatively, elemental sulfur was used (sulfur flower, 10 g/l).
Cultures of 13–14 l were grown at 80°C in a 15-l glass bottle heated in a silicon oil bath and stirred with a magnetic stirrer at a speed of 500/min. The culture was continuously bubbled with CO2-enriched air. The fermenter was inoculated with 500 ml of a late-exponential culture and harvested by centrifugation (10,000 × g) 72 h after inoculation. Cell densities were estimated by measuring both the optical density at 600 nm and the cell counts with a Hemocytometer of 10 μm depth.
Enzyme Assay
Tetrathionate hydrolase (TTH) activity was routinely measured following the decrease of TT concentration in the assay mixture by cyanolysis (Kelly et al., 1969). The mixture (1 ml, pH 1) contained 100 mM maleic acid, 1 mM MgCl2, 1.5 or 1.8 mM K2S4O6, and 25–190 μl of protein extracts. About 4 μM Pyrroloquinoline quinone (PQQ, methoxatin; Sigma, München, Germany), 2 mM Mg2+, or 2 mM Ca2+ were added if appropriate. The assay mixture including the protein extracts was preheated to 80°C for 5 min prior to TT addition. Aliquots (50 μl) were taken at appropriate time points, chilled on ice water, and kept at 4°C pending cyanolysis. One unit is defined as the conversion of 1 μmol TT in 1 min. Pyrophosphatase activity was measured according to Richter and Schäfer (1992).
Protein Purification from Whole Cells
Washed cells were resuspended in 40 mM KPi pH 7.0 (10 ml/g of cells) and homogenized in a potter after addition of 1 mM MgCl2, 25 μg/ml RNase A, and 10 μg/ml DNase I. Cells were disrupted by two passages through a high-pressure homogenizer at 175 MPa (Constant Cell Disruption Systems, Daventry, UK). Undisrupted cells and cell debris were removed by centrifugation at 10,000 × g for 30 min at 4°C. Particle fractions were removed by a second centrifugation step at 135,000 × g and 4°C for 60 min. The supernatant (i.e., soluble extract) was dialyzed against 40 mM KPi, 1 mM MgCl2, 1 nM PQQ, pH 7.0 for 16 h.
The soluble extract was applied onto a DEAE Sepharose fast-flow column [36 ml column volume (CV); GE Healthcare Europe, Munich, Germany] equilibrated with 40 mM KPi, 1 mM MgCl2, pH 7.0. Proteins were eluted with a linear gradient from 0 to 0.15 M NaCl in 6 CV followed by a linear gradient from 0.15 to 0.6 M NaCl in 6 CV. Fractions showing TTH activity were pooled and dialyzed against 50 mM Tris/HCl, pH 8.5 for 16 h at 4°C prior to addition of 2 M ammonium sulfate followed by centrifugation for 1 h at 40,000 × g and 4°C. The supernatant was applied onto a Phenyl-Sepharose Fast-Flow column (bed volume 24 ml; GE Healthcare). Proteins were eluted with a linear gradient from 2 to 0 M ammonium sulfate. Fractions showing TTH activity were pooled, concentrated with Centriprep YM-30 Devices (Millipore, Bedford, MA, USA), and applied on a Superdex 200 size exclusion column (30 mm × 600 mm; GE Healthcare). Elution was performed with 50 mM Tris/HCl, 150 mM NaCl, pH 8.5, and a constant flow rate of 1 ml/min. The fraction with the highest specific activity of the TTH was dialyzed against 50 mM Tris/HCl, 5 mM NaCl, pH 7.5 for 16 h at 4°C, and applied on a fast-flow Q-Sepharose column (GE Healthcare; bed volume 3.5 ml). Proteins were eluted with a linear gradient from 187.5 to 675 mM NaCl.
Protein Purification from Spheroblasts
Freshly harvested cells (wet mass 8.6 g) of a late-exponential and TT-grown A. ambivalens culture were split into 10 equal parts of 0.86 g and each resuspended in 10 ml spheroblast buffer of various pH values or sterile culture supernatant, followed by overnight incubation (12 h) at 4°C and centrifugation at 1000 × g for 15 min. The buffers were composed each of 500 mM sucrose, 100 mM NaCl, and the buffering substance, i.e., 50 mM KH2PO4/K2HPO4 (pH 5–6.5), 50 mM formic acid/NaOH (pH 3–4.5), or 50 mM maleic acid (pH 2). The supernatants containing the pseudo-periplasmic fraction were used for TTH activity assays and gel electrophoresis (Figure 1C).
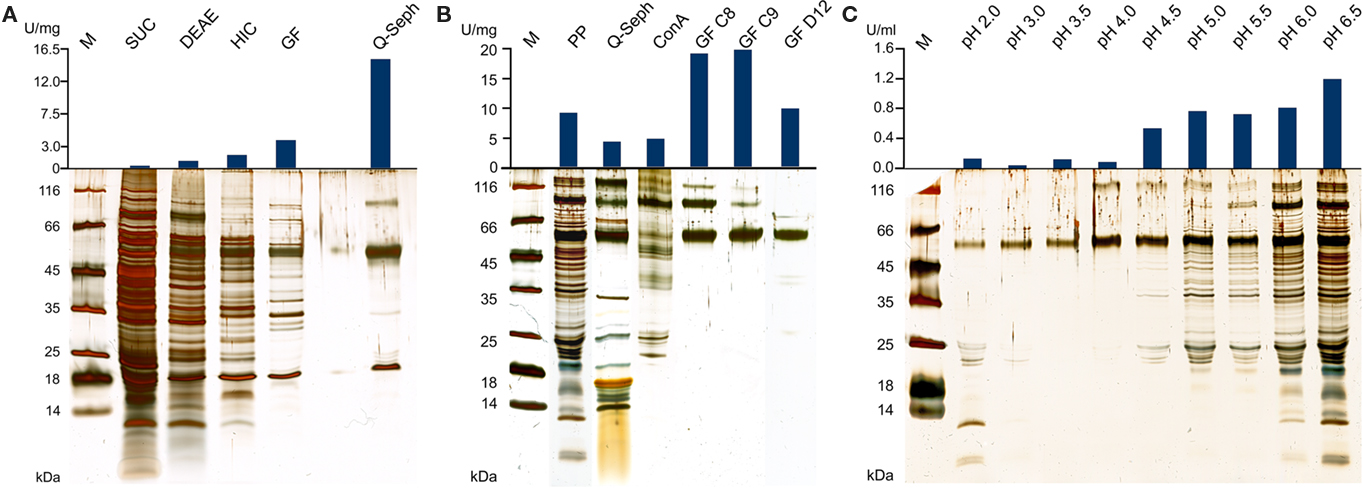
Figure 1. Silver-stained SDS gels and activities of TTH fractions from A. ambivalens. (A) Purification of the TTH from soluble cell extracts and specific activities; SUC, supernatant ultracentrifugation; DEAE and HIC, active fractions of DEAE anion exchange and hydrophobic interaction chromatographies, respectively; GF, size exclusion chromatography; Q-seph, Q-sepharose chromatography. (B) Purification of the TTH from the pseudo-periplasmic fraction [obtained by incubation in buffer pH 6.5; see (C)] and specific activities; PP, pseudo-periplasmic fraction; Q-seph, Q-sepharose chromatography; Con A, concanavalin A chromatography; GF C8 and GF C9, two consecutive fractions of the dimer peak of the size exclusion chromatography; GF D12, main fraction of the monomer peak (lane from a different SDS gel). (C) Silver-stained SDS gel of supernatants from spheroblast formation after incubation of whole cells in buffers of the pH values given above as described in the Section “Materials and Methods” (pseudo-periplasmic fractions), 10 μl of the supernatants were separated in the gel without prior concentration; top, resulting total TTH activities per milliliter of supernatant (total: 10 ml from of 0.86 g of cells each).
For protein purification, the pseudo-periplasmic fraction of 6 g of A. ambivalens cells obtained by incubation in 500 mM sucrose, 100 mM NaCl, and 50 mM KH2PO4/K2HPO4 pH 6.5 was dialyzed against 20 mM Tris/HCl, 5 mM NaCl, pH 7.5, and applied to a fast-flow Q-Sepharose column (bed volume 3.5 ml; GE Healthcare). Proteins were eluted with a linear gradient from 187 to 675 mM NaCl. Fractions showing TTH activity were pooled and the salt concentration was adjusted to 500 mM NaCl, 1 mM MnCl2, and 1 mM CaCl2. This protein solution was applied to a concanavalin A column (Hi-Trap Con A 4B, GE Healthcare; bed volume 1 ml) at a flow rate of 0.1 ml/min. Proteins were eluted with 20 mM Tris/HCl, 500 mM NaCl, 500 mM methyl-D-glucoside (Acros, Fisher Scientific, Bonn, Germany) pH 7.5 with a linear gradient of 0–500 mM methyl-D-glucoside in 2.5 CV. Fractions showing TTH activity were pooled, concentrated with Centriprep YM-30 Devices (Millipore, Bedford, MA, USA), and applied to a Superdex 200 size exclusion column (30 mm × 600 mm; Amersham GE Healthcare). The elution was performed with 50 mM Tris/HCl, 150 mM NaCl, pH 8.5, and a constant flow rate of 1 ml/min.
Sequence Analysis
The amino acid sequence of A. ferrooxidans TTH was used as probe in searches of a partial genomic database of A. ambivalens, which resulted in the identification of two homologs (tth1 and tth2). The deduced amino acid sequences were used in standard BLASTP searches conducted against the non-redundant protein database1. The A. ambivalens TTH1 and the A. ferrooxidans TTH were used as probes in a BLAST search against the Sulfobacillus acidophilus DSM 10332 genome sequence. A multiple alignment and a phylogenetic dendrogram were generated using MAFFT2 with the L–INS–i algorithm, the BLOSUM30 similarity matrix, a gap penalty of 1.56, and an offset value of 0.1 (Katoh et al., 2005). All homologs of TTH1 and TTH2 with e-values lower than 1 × e−10 and 1 × e−20, respectively, were selected from the BLAST results and included in the alignment. In addition, a group of sequences from methanogenic Archaea was added for outgroup alignment. For better visualization the tree (see below; Figure 5) was edited with NjPlot (Perriere and Gouy, 1996).
Secondary structure predictions were done using PSIPRED3. Predictions of topology and signal sequences were done with TMHMM and SignalP4 and MEMSAT (at the Psipred server). Three-dimensional models of TTH1 and TTH2 were predicted using the Phyre server5 (Kelley and Sternberg, 2009). Energy minimization of the PHYRE models was performed using UCSF Chimera6 (Pettersen et al., 2004). The figures were generated with Pymol (DeLano, 2002).
The nucleotide sequences of the tth1 and tth2 genes were submitted to EMBL database; they were assigned the accession numbers FR734215 and FR734216, respectively.
Isolation of RNA and Northern Analysis
Acidianus ambivalens cells were grown on tetrathionate or elemental sulfur. Samples (2 l each) were taken at 24, 48, and 72 h after inoculation and harvested by centrifugation (10 min, 6000 × g). RNA was prepared from these samples by the guanidinium isothiocyanide extraction method (Chomczynski and Sacchi, 1987) and quantified by UV absorption measurements. Five micrograms of each RNA preparation were separated on a denaturing agarose gel and subsequently transferred to a nylon membrane (Biodyne A; Pall Filtron; Seldon, 1988). RNA ladder mix (Invitrogen, Darmstadt, Germany) was used as a size marker.
After heat-fixation for 2 h at 80°C, the membrane was stained with methylene blue (Herrin and Schmidt, 1988). The DNA fragments used for probe synthesis were amplified with the primers TTH1_sig_fwd (ggaat catga accta aaaat cattg ttgg) and TTH1_1018_rev (attcc tcgag ataat agatg tatgc agga) for the tth1 gene and TTH2_sig2_fwd (tctac catgg caatg aagag agag) and TTH2_442_rev (tggct cgagt gttaa acagt aaaga ggtaa) for the tth2 gene. The primers sorn_fwd and sorc_rev (Urich et al., 2005) were used for the amplification of the sor gene. All primer sets amplified the entire open reading frames (ORF). The PCR products were Digoxigenin-labeled using the random hexamer labeling kit (No. 11175033910; Roche, Mannheim, Germany). The hybridization solution contained 50% formamide, 10% (w/v) dextran sulfate (Sigma, Germany), 1% (w/v) SDS, and 0.5% (w/v) skim milk powder. The detection reaction was performed using the DIG luminescence detection kit (11363514910; Roche) and Amersham hyperfilm ECL (GE Healthcare, Munich, Germany) as described by the manufacturers.
Analytical Procedures
Protein concentration was determined by the bicinchoninic acid method using the BCA Protein Assay Kit (Thermo Scientific Pierce, Rockford, IL, USA). The molecular masses and subunit composition were determined by size exclusion chromatography in the course of the purification (see Protein Purification from Spheroblasts). The monomer, dimer, and trimer peaks of bovine serum albumin were used for calibration. SDS/PAGE was performed according to the method of Schägger and von Jagow (1987) using 12% (w/v) resolving and 4% (w/v) stacking gels. Samples were mixed with SDS-loading buffer (60 mM Tris/HCl, 2% SDS (v/v), 10% glycerol (v/v), 0.01% bromophenol blue (w/v), 0.015 g/ml mercaptoethanol) and incubated at 95°C for 5 min before loading onto the gel. Protein Molecular Weight Marker (SM0431, Fermentas) was used as marker. Gels were stained with colloidal Coomassie blue (Roti Blue, Roth, Karlsruhe, Germany) or with silver nitrate (Blum et al., 1987). For N-terminal sequence analysis, the concentrated protein was run on 12% SDS gel and transferred to a PVDF (polyvinylidene difluoride) membrane using a Multiphor NovaBlot system (GE Healthcare) according to the method of Kyhse-Andersen (1984). The protein bands were visualized with conventional Coomassie brilliant blue R-250 [0.1% (w/v), 1% (v/v) acetic acid, 40% (v/v) methanol] followed by N-terminal sequence analysis by Edman degradation using a 492-cLC protein sequencer (Lifetech-Applied Biosystems, Darmstadt, Germany) according to the manufacturer’s instructions.
Results
Acidianus ambivalens is Growing with Tetrathionate as Sole Sulfur Source
Acidianus ambivalens grown aerobically with 10 mM tetrathionate (TT) as sole sulfur and energy source reached cell densities of 5.9–8.0 × 108/ml after 72 h. Doubling times were 9 h and the final pH of the culture was 0.8–1.0. The cell yields with TT were 0.26–0.36 g/l of culture volume (wet mass). Cells grown with elemental sulfur as previously reported (Zillig et al., 1986; Teixeira et al., 1995) reached cell densities of 9.9 × 108/ml at doubling times of 8.4 h. Cell yields were 0.35–0.47 g/l, however, cell pellets still contained elemental sulfur. When cells were grown at TT concentrations of 20 mM or more, the cultures showed a whitish turbidity after 12 h from transiently formed hydrophilic and finely dispersed sulfur droplets. The turbidity decreased and vanished after 36–48 h due to elemental sulfur oxidation. TT concentrations of more than 40 mM were inhibitory (data not shown).
Purification of the Tetrathionate Hydrolase from Whole Cell Extracts and Identification of the Gene
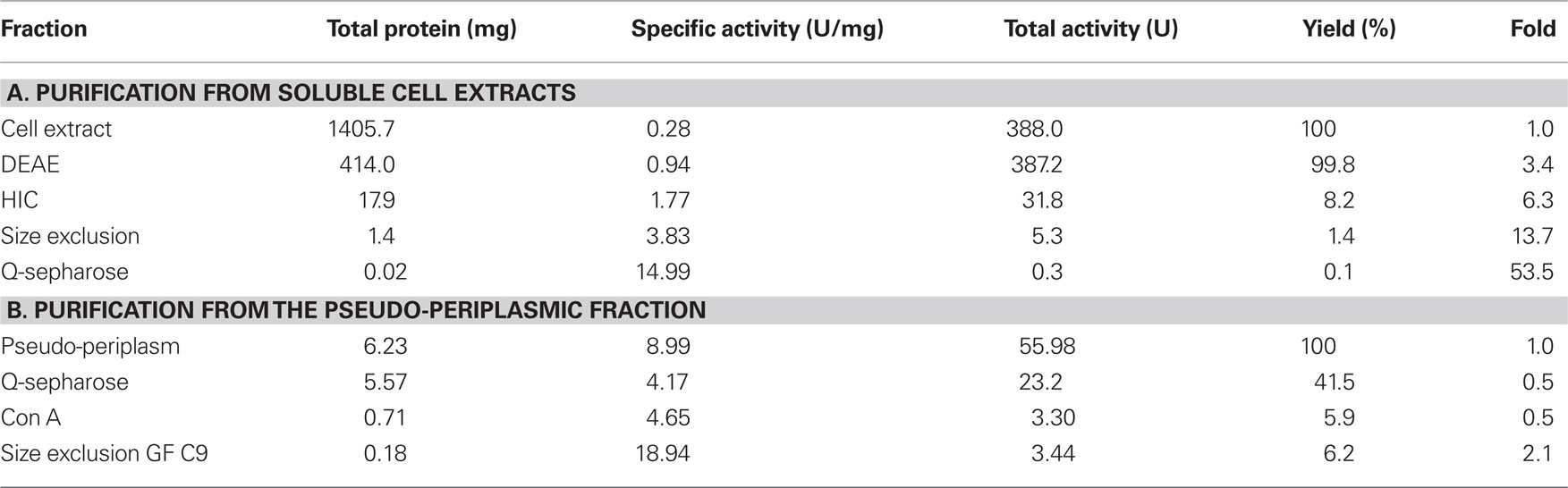
A specific TTH activity of 40 mU/mg protein was found in total cell extracts of A. ambivalens grown on TT. The particle-free extract showed a specific activity of 280 mU/mg protein (Table 1). After four chromatographic purification steps, the specific TTH activity was increased to 15 U/mg protein equaling a 54-fold purification and 0.1% recovery.
In the SDS/PAGE (Figure 1A) of different purification steps, a clear relation is visible between the increase of the specific TTH activity and a protein band at 54 kDa. N-terminal sequencing of this band gave the amino acids Gly-Pro-Ile-Val-Tyr-Thr-Tyr-Thr-Glu-Tyr-Asn*-Gly-Thr-Tyr with Asn* being glycosylated. These residues correspond to the amino acid sequence encoded by the tth1 gene (Gly40-Tyr53) previously identified by similarity searches in a database of partial A. ambivalens genome sequences (5 MBp) using the amino acid sequence of the A. ferrooxidans TTH as a query. Residues 1–39 obviously form a signal peptide that was also predicted by the SignalP web server7. Another paralogous gene termed tth2 was identified in the genome sequences as well. The smaller band with a molecular mass of ≈20 kDa seen in the SDS gel was identified as a pyrophosphatase (not shown), the larger weak protein band (approximate molecular mass of 90–100 kDa) did not give an N-terminal sequence. Specific pyrophosphatase activities (Richter and Schäfer, 1992) were up to 107 mU/mg in enrichment fractions.
Based on the TTH liberation by pH shock (see below), an alternative purification protocol was developed. The pseudo-periplasmic fraction of 6 g A. ambivalens cells (after incubation in pH 6.5 buffer) displayed a specific activity of 9 U/mg and a total activity of 56 U. When being fractionated with a fast-flow Q-Sepharose column (GE Healthcare Europe, Freiburg, Germany) the TTH activity eluted at about 320 mM NaCl. A pool of the active fractions showed a specific activity of 4 U/mg and a total activity of 23 U. Lectin affinity chromatography was carried out for the removal of non-glycosylated proteins. The TTH activity eluted at a concentration of 325 mM methyl-D-glucoside. Active fractions were pooled and concentrated prior to size exclusion chromatography with a Superdex 200 column. The TTH eluted in two major 280 nm absorption peaks (not shown) with specific TTH activities of 19 U/mg (Vr = 165 ml; corresponding to a molecular mass of 105 kDa) and 10 U/mg (Vr = 190 ml; 54 kDa), respectively (Figure 1B). The elution pattern points to equilibrium formation between monomer and homodimer. The alternative three-step purification scheme from the pseudo-periplasmic fraction resulted in a 2.1-fold higher specific TTH activity – despite an intermediate drop – and a 62-fold higher yield of enzyme recovery compared to the purification from total soluble extracts (Table 1). The fractions did not have pyrophosphatase activity and the 20-kDa band was not seen in SDS gels. The 54-kDa TTH band was predominant in the dimer peak and almost homogeneous in the monomer peak (Figure 1B).
TTH has an Extracellular Location
TTH activity was not detectable in cell-free culture supernatants (pH 1) immediately after harvesting. This changed when cells were resuspended in a 20-fold volume of spent supernatant after harvesting and incubated overnight at 4°C followed by another round of centrifugation: the TTH activity was 7.5 mU/ml (0.3 U/mg protein) in the secondary supernatant suggesting a slow release of the protein (Table 2). After overnight incubation of cells in pH 6.5 phosphate buffer, 92% of the TTH activity was found in the supernatant. 7.3% and 0.3% were found in the cytoplasmic and membrane fractions, respectively. In order to determine the effect of pH on the solubilization of the TTH from the cells, buffers of different pH values were used for overnight incubation after harvesting. The TTH activities in the supernatants increased significantly between pH 4 and 4.5, while the highest amount of TTH activity and the highest protein concentration were found in the pH 6.5 supernatant (Table 2; Figure 1C).
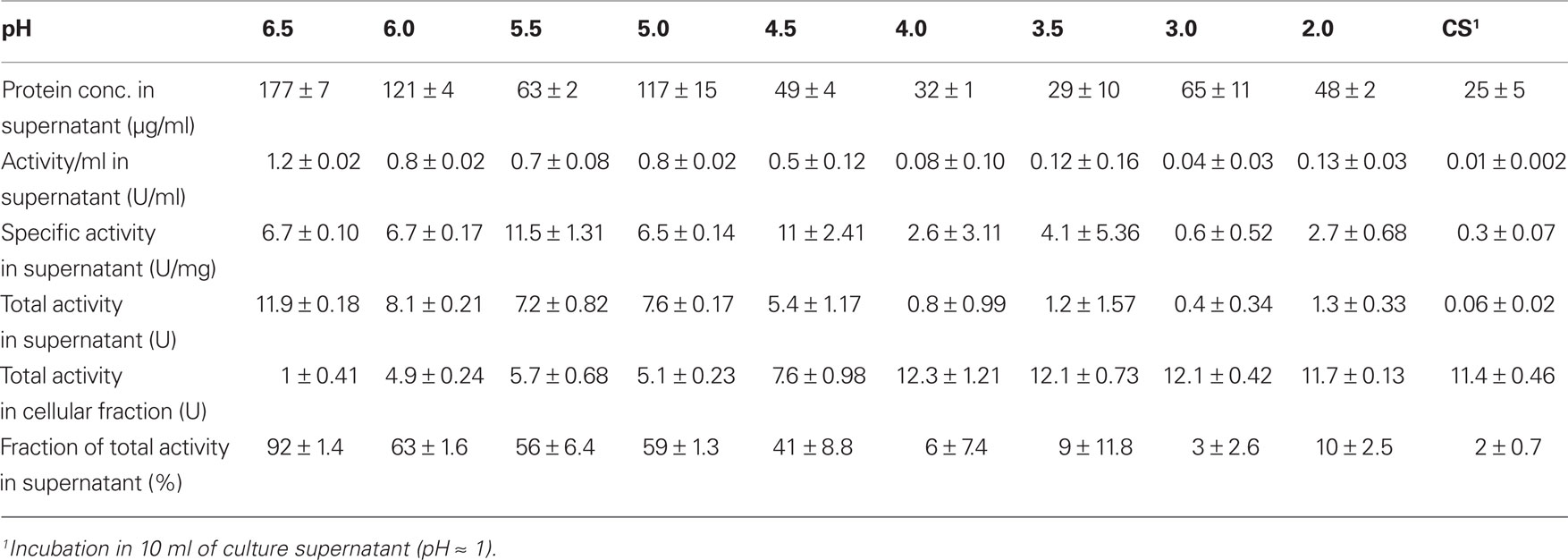
Table 2. TTH activities in the cell-free supernatants following incubation of 0.86 g each of A. ambivalens cells in 10 ml of buffer of different pH values; average values from two to three enzyme assays.
TTH is an Extremely Thermophilic and Acidophilic Enzyme
Decrease of TT catalyzed by the TTH showed a sigmoidal behavior and an activation period whose duration is depended on the amount of enzyme in the assay mixture. The slope of the regression line was directly dependent on the amount of protein used for the assay (Figure 2). The specific activity increased in an almost linear way with the temperature with no activity at 40°C, modest activity at 50°C, and maximal activity at 95°C (Figure 3). The optimal and maximal temperatures were not determined for lack of suitable assay equipment. The highest enzymatic activity was recorded at pH 1. From pH 2–5, the TTH showed a specific activity of 1–0.75 U/mg (Figure 3). No enzyme activity was detected at pH 6. Addition of PQQ and/or Mg2+ or Ca2+ did not have a significant effect on the TTH activity (not shown). The KM was recorded using a different TTH preparation of higher purity. At 80°C and pH 1, the KM of the TTH was 0.8 mM TT when assayed using 2.7 μg of protein. The observed Vmax was 43.8 nmol/min corresponding to a specific activity of 16 U/mg.
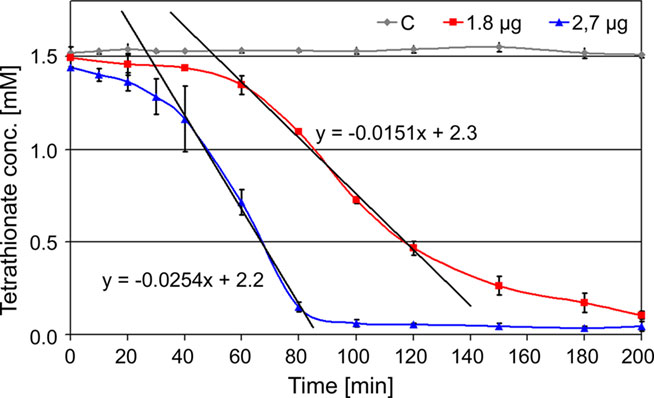
Figure 2. Kinetics of TTH activity. 1.8 μg (red line) and 2.7 μg of protein (blue line) assayed at 80°C and pH 1; C: assay mixture without enzyme (control; gray line); formula, gradients of the trend lines used for activity calculation.
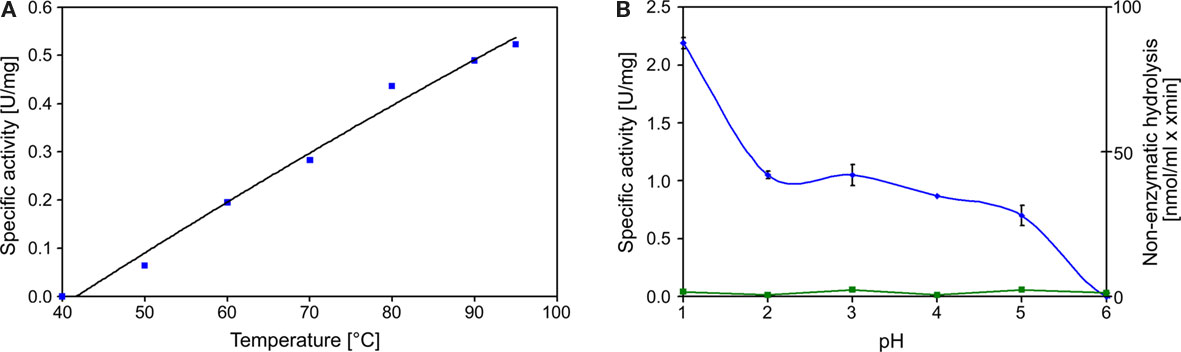
Figure 3. Temperature and pH profiles of TTH activity. (A) Temperature profile of TTH activity from 40 to 95°C; (B) pH profile of TTH activity assayed at 80°C (blue line, left axis) and non-enzymatic TT hydrolysis (green line, right axis).
Only the tth1 Gene is Expressed in TT-Grown A. ambivalens Cells
In order to analyze expression of the two tth genes, total RNA was isolated from A. ambivalens grown aerobically with sulfur or TT at different stages of the growth curve. Northern hybridization was carried out with probes derived from the tth1 and tth2 genes and, as a control, with a sor probe from the gene encoding the sulfur oxygenase reductase (Kletzin, 1992). Hybridization signals appeared at a size of about 1500 nt with the tth1 gene pointing toward a monocistronic transcription (Figure 4). Signals developed in hybridization reactions with RNA from cells grown under aerobic but not under anaerobic conditions (not shown). Signals were stronger in RNA from TT-grown cells as compared to sulfur-grown cells at late-exponential phase. The tth2 gene did not give hybridization signals under any condition tested. The sor probe resulted in signals of approximately 900 nt in length with RNA from aerobically grown cells both with sulfur and TT as substrates, however signals were stronger in S°-grown cells (Figure 4). These results showed that the sor gene is expressed in TT-grown cells presumably due to transient formation of elemental sulfur.
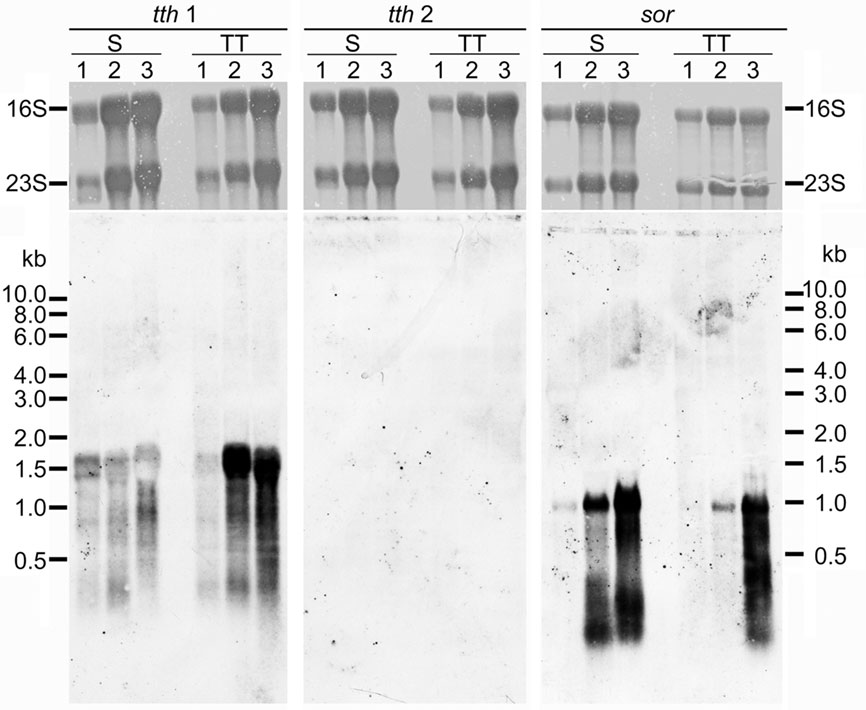
Figure 4. Northern hybridization of tth1, tth2, and sor genes. All lanes with total A. ambivalens RNA (5 μg each) isolated from cells grown with sulfur (S) or tetrathionate (TT) after 24 h (1); 48 h (2), and 72 h (3); upper panels, methylene blue staining for 16S and 23S rRNA; lower panels, hybridization results.
Tetrathionate Hydrolase is Similar to PQQ-Containing Dehydrogenases
The tth1 gene encodes an ORF of 538 codons. The deduced amino acid sequence includes a signal sequence of 39 residues, so that the mature protein has a length of 499 aa. The calculated molecular mass of 53,642 without glycosylation corresponds well with the apparent molecular masses in SDS gels and gel filtration (54 kDa). The tth2 gene encoded an ORF of 449 aa including a predicted signal sequence of 23–32 aa in length. Both proteins are 29–32% identical (depending on alignment parameters), which is less than the pairwise identity of TTH1 with the orthologous enzyme from A. ferrooxidans (40–42%).
The top hits in BLASTP searches of TTH1 at NCBI (pairwise identity >33%; e-value <1 × 10−54) were restricted to close homologs from acidophilic Bacteria and Archaea (TTH cluster; Figure 5), which seem to be true tetrathionate hydrolases. Comparable results were obtained with the TTH2 protein. Both proteins and their homologs gave distinct clades in the dendrogram. These proteins belong to a large superfamily of β-propeller proteins, whose best-known members are pyrroloquinoline quinone (PQQ) containing dehydrogenases. Secondary structure predictions confirmed that TTH1 and TTH2 are all-beta proteins except for the transmembrane helices within the signal sequences (not shown). Six copies of a PQQ enzyme β-propeller repeat are present in each protein (PFAM family PF01011)8.
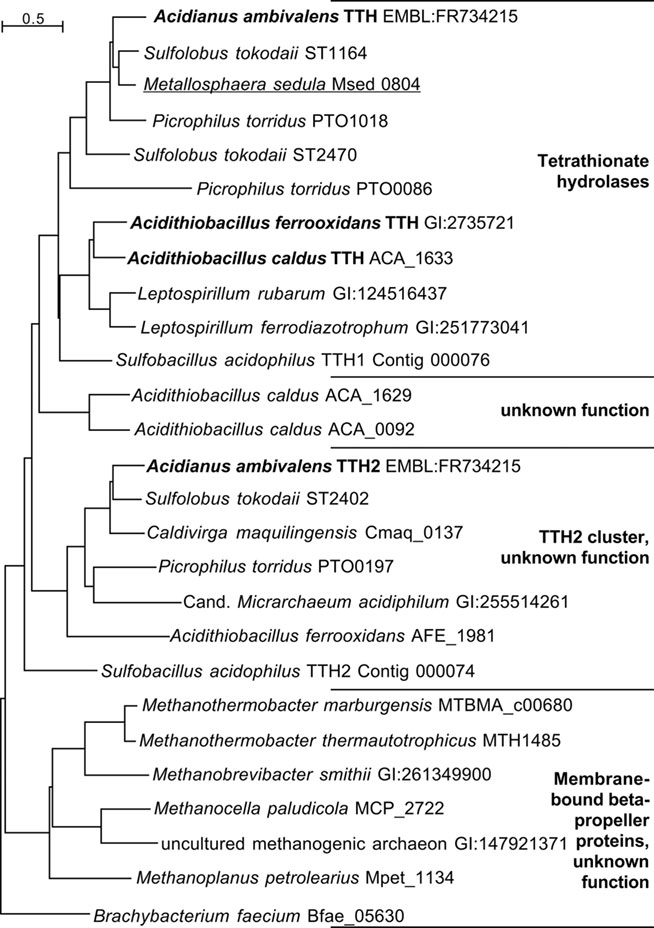
Figure 5. Phylogenetic dendrogram of the TT hydrolases with 26 of the closest homologs from Archaea and Bacteria. Bold face, biochemically characterized TTH enzymes (Bugaytsova and Lindström, 2004; Kanao et al., 2007; Rzhepishevska et al., 2007); underlined, gene described in transcriptome study (Auernik and Kelly, 2008). The Sulfobacillus TTH homologs were identified in the draft genome sequence available at http://www.jgi.doe.gov (as of January, 2011).
Nine out of 10 top hits in homology modeling of TTH1 (and of TTH2) were PQQ-containing dehydrogenases with an eight-bladed β-propeller structure, a calcium site, and a cytochrome c domain (e.g., alcohol dehydrogenase from Pseudomonas putida HK5; PDB accession 1kv9; e-value 1 × e−25; Figure 6). Homologs of the cytochrome c domain were missing in both TTHs. Closer examination showed that the Ca2+-coordinating residues were absent as well. Likewise, a nearby disulfide from two adjacent cysteine residues was missing (Figure 7). The models predict that the glycosylated Asn of TTH1 is located in the center of the β-propeller.
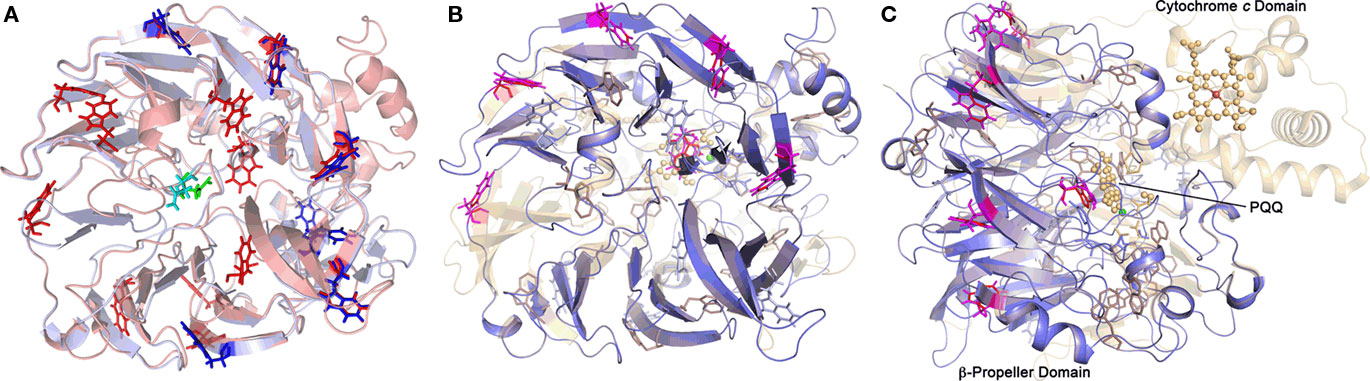
Figure 6. Predictions of the 3D structures of A. ambivalens TTH1 and TTH2 modeled with the quinoprotein alcohol dehydrogenase from Pseudomonas putida as template (PDB accession number 1kv9). (A) TTH1 model (red) overlaid with TTH2 (blue); tryptophane residues characteristic of the β-propeller wings (see text) are highlighted in sticks representation; the glycosylated Asn resdidue of TTH1 is highlighted in green, the corresponding Asn of TTH2 in cyan. (B,C) Overlay of the TTH1 model (blue) with the 1kv9 template (red), top and side views; spheres, PQQ and cytochrome c molecules; green sphere, Ca2+ ion; brown sphere, iron ion; the tryptophane residues are highlighted as above.
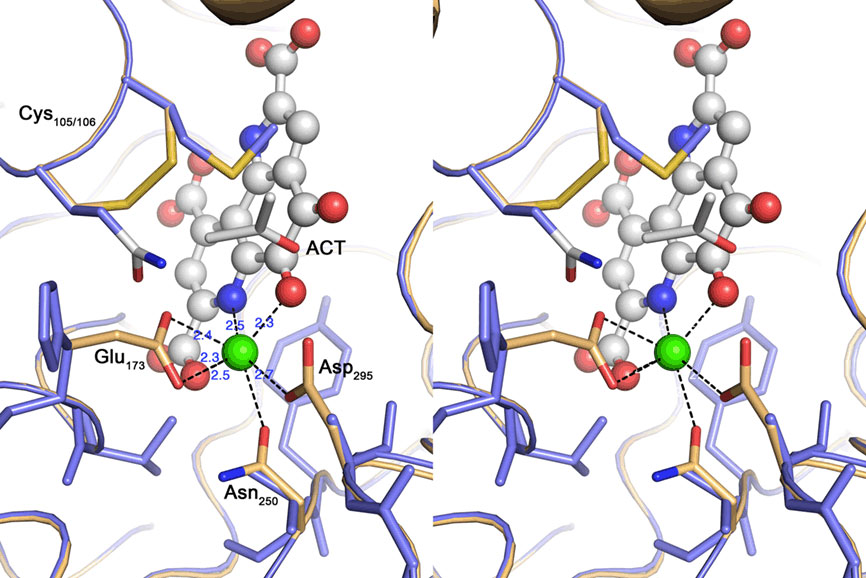
Figure 7. Cross-eyed stereo view of the active site of the quinoprotein alcohol dehydrogenase from Pseudomonas putida (beige) overlayed with the TTH1 model (blue). Spheres represent the Calcium–PQQ moiety (carbon, gray; oxygen, red; nitrogen, blue; calcium, green); sticks acetone (ACT) and amino acid side chains. The calcium-coordinating side chains and the disulfide bridge characteristic for this type of proteins are highlighted; note the absence of the equivalent residues in the TTH model.
Discussion
Growth of (Acidi-) Thiobacillus species and other bacteria by TT oxidation is known for a long time. Likewise, TT-oxidizing Archaea were described previously, most of them acidophiles (Wood et al., 1987; Kurosawa et al., 2003). In contrast, most of the known TT-metabolizing enzymes come from neutrophilic proteobacteria and are molybdenum or heme-containing reductases allowing these organisms to grow anaerobically by TT respiration (Hensel et al., 1999; Price-Carter et al., 2001; Mowat et al., 2004). Although TT hydrolysis had been described already in the 1960s (Trudinger, 1964) and TT hydrolases in the 1990s (De Jong et al., 1997a), the link between purification and identification of the gene was established only recently by Kanao et al. (2007) and Rzhepishevska et al. (2007) for the TTHs from A. ferrooxidans and Acidithiobacillus caldus, respectively. Enzymes oxidizing TT to sulfate without intermediate(s) are not known, with the exception of SOX complexes from some but not all neutrophilic ISC-oxidizing bacteria (Mukhopadhyaya et al., 2000; Lahiri et al., 2006). We report here on the identification, enzyme characterization, and expression data of the first archaeal TTH described so far.
Growth of A. ambivalens on TT, TTH Gene Expression Data, and Enzyme Properties
Acidianus ambivalens growth rates and yields were comparable with TT and elemental sulfur. An indication for the presence of a TTH came from the turbidity observed in cultures growing at high TT concentrations, which showed that elemental sulfur is transiently formed. TTH activity was measured in TT-grown but not in sulfur-grown cells suggesting that the protein is formed only in the presence of the substrate. This observation is supported by the Northern data, which showed a stronger signal of the tth gene in TT-grown cells (Figure 4).
The TTH activity was initially localized in the soluble fraction. The enzyme was highly enriched during various chromatographic steps but the resulting preparation was not electrophoretically homogeneous. A band of approximately 54 kDa was co-enriched with the TTH activity, whose N-terminus was identical to a protein that was previously identified by sequence similarity search with the A. ferrooxidans TTH. A pyrophosphatase and a protein of unknown identity were still present in the preparation (Figure 1A). The same 54 kDa band was enriched during purification from the pseudo-periplasmic fraction (Figure 1B). Again, two larger protein bands were visible in the gel but their stoichiometries were not constant. Therefore, we concluded that the 54-kDa protein represents the sole subunit of the TTH. This conclusion is supported by the results from the gel permeation chromatography, which showed two peaks with TTH activity, corresponding to molecular masses of 54 and 105 kDa and representing the monomer and the homodimer.
As expected, the optimal reaction temperature of the A. ambivalens TTH is higher (≥95°C; Figure 3) than for the enzymes from mesophilic species (optima 40–65°C; Sugio et al., 1996; Tano et al., 1996; De Jong et al., 1997a,b; Bugaytsova and Lindström, 2004; Kanao et al., 2007). In addition, the optimal pH was lower, being around pH 1. The protein was glycosylated at Asn50 and probably other residues as well. Glycosylation seems to be a common protection mechanism of membrane-bound and extracellular proteins from A. ambivalens and other Sulfolobales species: the TQO, the terminal oxidase, and the large S-layer protein could be stained with glycosylation-specific dyes (Müller et al., 2004; Veith et al., 2009; Peyfoon et al., 2010).
TTH is Found in the Pseudo-Periplasmic Space of TT-Grown Cells
Cell pellets had been washed with pH 6.5 buffer prior to disruption in order to remove excess acid (not shown). It turned out that this procedure releases the enzyme from the cells (Figure 1C). Treatment of freshly harvested cells with buffers of increasing pH values resulted in the liberation of almost all of the TTH activity in buffers of pH 4.5 and above. A 54-kDa protein band is constantly present in all of the supernatant, that showed TTH activity. Although the amount and the number of proteins seen in the SDS gel are increasing with the pH (Figure 1B) the cells apparently remained intact under the microscope (not shown). Therefore, we think that the protein is liberated by pH shock presumably due to changes in S-layer topology but not by (partial) cell lysis. The TTH has a canonical signal sequence but no membrane anchor according to the analysis of the deduced amino acid sequence of the protein. Therefore, an extracytoplasmic location was expected.
As most Archaea including Acidianus have proteinaceous S-layers (Baumeister et al., 1988; Baumeister and Lembcke, 1992; König et al., 2007; Veith et al., 2009) as their sole cell wall component, the question arises whether the TTH is secreted into the medium or whether it is somehow attached to the S-layer. As we were unable to detect activity in spent growth medium, the TTH must be attached to the outside of the cytoplasmic membrane or to the S-layer. S-layers consist of a proteinaceous canopy supported by stalks at a constant and species-specific distance above the archaeal cytoplasmic membrane. Baumeister et al. (1988) and Baumeister and Lembcke (1992) proposed from electron microscopic images that S-layers form a space equivalent to the periplasm of Gram-negative Bacteria. Narrow pores in the S-layer connect the medium with the so-called quasi- or pseudo-periplasmic space as porins do in the outer membrane of bacteria. It had been speculated that the electron-light pseudo-periplasmic space in between the electron-dense stalks might accommodate secreted proteins, which are prevented from diffusion into the medium by the porous protein canopy (Baumeister and Lembcke, 1992). We showed here that the TTH is liberated from the cells by pH shift alone. We deduced from these results that the shift in pH causes a rearrangement of the S-layer thereby liberating the TTH. Alternatively, the TTH might be attached to the S-layer by ionic and/or hydrophobic interactions, however, the total lack of TTH activity in the medium argues against this hypothesis. Therefore, we conclude that our results support the pseudo-periplasm hypothesis and that the TTH is one of few enzymes so far shown to be located in this compartment. This conclusion is consistent with the acidic pH optimum of the A. ambivalens TTH (around pH 1) corresponding to the media conditions.
Acidithiobacillus and Acidianus TTHs are Similar Enzymes
The extracellular location of the A. ambivalens enzyme is consistent with bacterial TTHs, which were localized in the periplasm or attached to the membrane. The localization was verified experimentally in the case of the A. caldus TTH (Bugaytsova and Lindström, 2004). In most other cases the localization was inferred by the acidic pH optima of the enzymes (pH 2.5–4; Sugio et al., 1996; Tano et al., 1996; De Jong et al., 1997a,b). Comparison of the amino acid sequences showed that almost all of the TTHs including TTH2 contain a canonical signal sequence (not shown) thereby confirming the presumed or verified periplasmic or extracellular location. The molecular masses of the TTHs purified so far are fairly similar and differ from the only known trithionate hydrolase, which is a homotrimeric enzyme with a subunit mass of 34 kDa (Meulenberg et al., 1992). Meulenberg’s results suggest that trithionate hydrolases represent separate and unrelated entities, however, this cannot be confirmed because the genes are not known.
The sequence analysis and comparisons of the TTH genes showed that they are rare and present only in obligatory or facultative chemolithoautotrophs. In addition, most of the species represented in the dendrogram (Figure 5) harbor at least two tth genes. The gene products fall into two separate phylogenetic clades. The clade around the TTH1 encompasses true tetrathionate hydrolases from their similarity and from the known enzyme activities. The TTH2 proteins are paralogous proteins with a different and unknown function, however, they also contain a signal sequence and should therefore be secreted (Figure 5).
TTH in Lithoautotrophic and Organoheterotrophic Sulfolobales Species
The absence of tth genes in many organoheterotrophic Sulfolobales species is of particular interest as for example Sulfolobus acidocaldarius had originally been described as a facultatively chemolithoautotrophic, sulfur-dependent species (Brock et al., 1972). Neither S. acidocaldarius nor Sulfolobus solfataricus/Sulfolobus islandicus species have tth genes suggesting that they are unable to grow at the expense of TT oxidation. An analogous observation was made with the sulfur oxygenase reductase (SOR): (facultatively) chemolithoautotrophs like Sulfolobus metallicus, Metallosphaera sedula, and Sulfolobus tokodaii possess sor genes, while S. acidocaldarius, S. solfataricus, and S. islandicus lack the gene and corresponding enzyme activities (Kletzin, 1989; Veith et al., 2011; this volume). Both observations suggest that the colony-isolated, heterotrophic Sulfolobales species are not sulfur-dependent at all and unable to grow oxidatively with sulfur or tetrathionate.
TTH: β-Propeller Structure without PQQ
It can be concluded from sequence analysis and structure prediction that the TTHs are all-beta and also β-propeller-forming proteins with a conserved 3D structure similar but not identical to PQQ-containing dehydrogenases. These proteins, e.g., type II alcohol dehydrogenases and glucose dehydrogenases, consist of a PQQ domain, composed of seven to eight conserved, propeller-forming PQQ repeats (PFAM family PF01011; Figure 6; Oubrie et al., 1999; Chen et al., 2002). The alcohol dehydrogenases contain an additional heme c domain (Chen et al., 2002). The PQQ repeats contain a conserved motif of amino acids (Ala-x5-Gly-x3-Trp) thought to stabilize the structure (Chen et al., 2002).
The model of the A. ambivalens TTH1 and the sequence alignments used for dendrogram calculation (Figure 5; alignment not shown) contain five to six of these tryptophane motifs, whereas TTH2 contains four (Figure 6). In contrast, the heme c domain and conserved PQQ-binding site residues are missing including the disulfide-forming cysteines and the calcium-coordinating amino acids (Figure 7). The glycosylated Asn of TTH1 sits in the middle of the β-propeller and far from the region where the dehydrogenases bind PQQ. The alignments did not show a conserved cysteine residue – the usual suspect for sulfur binding – so that the reaction mechanism remains enigmatic at present, especially in the absence of PQQ and metal ions.
Conclusion
Some final conclusions for the A. ambivalens sulfur metabolism can be drawn: One of the initial questions was, how TT produced in the TQO reaction is metabolized (Müller et al., 2004). The TTH found here does not give an answer to this question because the protein was found only in TT-grown cells and the gene is poorly expressed in sulfur-grown cells. The localization and pH optimum of the TTH argue against a role in the oxidative metabolism of elemental sulfur. In contrast, the SOR does seem to play an important role in TT-grown cells, presumably by disproportionation of the sulfur generated by the TTH.
The TTH itself seems to be a monomeric or homodimeric enzyme with an overall β-propeller structure. The enzyme has a high temperature optimum and very low pH optimum, both of which are in accordance with the extracellular location shown by the pH shock-mediated enzyme liberation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Felicitas Pfeifer (Darmstadt, Germany) for her generosity and encouragement. This work was supported by grants of the Deutsche Forschungsgemeinschaft (Az Kl885-3/3 and Kl885-5/1).
Footnotes
References
Auernik, K. S., and Kelly, R. M. (2008). Identification of components of electron transport chains in the extremely thermoacidophilic crenarchaeon Metallosphaera sedula through iron and sulfur compound oxidation transcriptomes. Appl. Environ. Microbiol. 74, 7723–7732.
Baumeister, W., and Lembcke, G. (1992). Structural features of archaebacterial cell envelopes. J. Bioenerg. Biomembr. 24, 567–575.
Baumeister, W., Wildhaber, I., and Engelhardt, H. (1988). Bacterial surface proteins. Some structural, functional and evolutionary aspects. Biophys. Chem. 29, 39–49.
Blum, H., Beier, H., and Gross, H. J. (1987). Improved silver staining of plant proteins, RNA and DNA in polyacylamide gels. Electrophoresis 8, 93–99.
Brock, T. D., Brock, K. M., Belly, R. T., and Weiss, R. L. (1972). Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Microbiol. 84, 54–68.
Bugaytsova, Z., and Lindström, E. B. (2004). Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus. Eur. J. Biochem. 271, 272–280.
Chen, Z. W., Matsushita, K., Yamashita, T., Fujii, T. A., Toyama, H., Adachi, O., Bellamy, H. D., and Mathews, F. S. (2002). Structure at 1.9 Å resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5. Structure 10, 837–849.
Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159.
De Jong, G. A., Hazeu, W., Bos, P., and Kuenen, J. G. (1997a). Isolation of the tetrathionate hydrolase from Thiobacillus acidophilus. Eur. J. Biochem. 243, 678–683.
De Jong, G. A. H., Hazeu, W., Bos, P., and Kuenen, J. G. (1997b). Polythionate degradation by tetrathionate hydrolase of Thiobacillus ferrooxidans. Microbiology 143, 499–504.
DeLano, W. L. (2002). The PyMOL Molecular Graphics System. 0.97 ed. San Carlos, CA: DeLano Scientific.
Egorova, M. A., Tsaplina, I. A., Zakharchuk, L. M., Bogdanova, T. I., and Krasil’nikova, E. N. (2004). Effect of cultivation conditions on the growth and activities of sulfur metabolism enzymes and carboxylases of Sulfobacillus thermosulfidooxidans subsp. asporogenes strain 41. Prikl. Biokhim. Mikrobiol. 40, 448–454.
Friedrich, C. G., Bardischewsky, F., Rother, D., Quentmeier, A., and Fischer, J. (2005). Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8, 253–259.
Friedrich, C. G., Quentmeier, A., Bardischewsky, F., Rother, D., Orawski, G., Hellwig, P., and Fischer, J. (2008). “Redox control of chemotrophic sulfur oxidation of Paracoccus pantotrophus,” in Microbial Sulfur Metabolism, eds C. Dahl and C. G. Friedrich (Berlin: Springer Verlag), 139–150.
Frigaard, N. U., and Dahl, C. (2009). Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 54, 103–200.
Fuchs, T., Huber, H., Burggraf, S., and Stetter, K. O. (1996). 16S rDNA-based phylogeny of the archaeal order Sulfolobales and reclassification of Desulfurolobus ambivalens as Acidianus ambivalens comb. nov. Syst. Appl. Microbiol. 19, 56–60.
Gadd, G. M. (2010). Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology 156, 609–643.
Ghosh, W., and Dam, B. (2009). Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 33, 999–1043.
Hensel, M., Hinsley, A. P., Nikolaus, T., Sawers, G., and Berks, B. C. (1999). The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol. Microbiol. 32, 275–287.
Herrin, D. L., and Schmidt, G. W. (1988). Rapid, reversible staining of northern blots prior to hybridization. BioTechniques 6, 196–197, 199–200.
Johnston, F., and McAmish, L. (1973). Study of rates of sulfur production in acid thiosulfate solutions using S-35. J. Colloid Interface Sci. 42, 112–119.
Kanao, T., Kamimura, K., and Sugio, T. (2007). Identification of a gene encoding a tetrathionate hydrolase in Acidithiobacillus ferrooxidans. J. Biotechnol. 132, 16–22.
Kanao, T., Matsumoto, C., Shiraga, K., Yoshida, K., Takada, J., and Kamimura, K. (2010). Recombinant tetrathionate hydrolase from Acidithiobacillus ferrooxidans requires exposure to acidic conditions for proper folding. FEMS Microbiol. Lett. 309, 43–47.
Katoh, K., Kuma, K., Miyata, T., and Toh, H. (2005). Improvement in the accuracy of multiple sequence alignment program MAFFT. Genome Inform. 16, 22–33.
Kelley, L. A., and Sternberg, M. J. (2009). Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 4, 363–371.
Kelly, D. P., Chambers, L. A., and Trudinger, P. A. (1969). Cyanolysis and spectrophotometric determination of trithionate in mixtures with thiosulfate and tetrathionate. Anal. Chem. 41, 898–901.
Kelly, D. P., Shergill, J. K., Lu, W. P., and Wood, A. P. (1997). Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Van Leeuwenhoek 71, 95–107.
Kletzin, A. (1989). Coupled enzymatic production of sulfite, thiosulfate, and hydrogen sulfide from sulfur: purification and properties of a sulfur oxygenase reductase from the facultatively anaerobic archaebacterium Desulfurolobus ambivalens. J. Bacteriol. 171, 1638–1643.
Kletzin, A. (1992). Molecular characterization of the sor gene, which encodes the sulfur oxygenase reductase of the thermoacidophilic archaeon Desulfurolobus ambivalens. J. Bacteriol. 174, 5854–5859.
Kletzin, A. (2007). “Metabolism of inorganic sulfur compunds in Archaea,” in Archaea. Evolution, Physiology, and Molecular Biology, eds R. A. Garrett and H.-P. Klenk (Oxford: Blackwell Publishing), 261–274.
Kletzin, A. (2008). “Oxidation of sulfur and inorganic sulfur compounds in Acidianus ambivalens,” in Microbial Sulfur Metabolism, eds C. Dahl and C. G. Friedrich (Berlin: Springer), 184–201.
König, H., Rachel, R., and Claus, H. (2007). “Proteinaceous surface layers of Archaea: ultrastructure and biochemistry,” in Archaea. Molecular and Cellular Biology, ed. R. Cavicchioli (Washington, DC: ASM Press), 315–340.
Kurosawa, N., Itoh, Y. H., and Itoh, T. (2003). Reclassification of Sulfolobus hakonensis Takayanagi et al. 1996 as Metallosphaera hakonensis comb. nov. based on phylogenetic evidence and DNA G + C content. Int. J. Syst. Evol. Microbiol. 53, 1607–1608.
Kyhse-Andersen, J. (1984). Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10, 203–209.
Lahiri, C., Mandal, S., Ghosh, W., Dam, B., and Roy, P. (2006). A novel gene cluster soxSRT is essential for the chemolithotrophic oxidation of thiosulfate and tetrathionate by Pseudaminobacter salicylatoxidans KCT001. Curr. Microbiol. 52, 267–273.
Mangold, S., Valdés, J., Holmes, D. S., and Dopson, M. (2011). Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front. Microbiol. 2:17. doi: 10.3389/fmicb.2011.00017
Meulenberg, R., Pronk, J. T., Frank, J., Hazeu, W., Bos, P., and Kuenen, J. G. (1992). Purification and partial characterization of a thermostable trithionate hydrolase from the acidophilic sulphur oxidizer Thiobacillus acidophilus. Eur. J. Biochem. 209, 367–374.
Moll, R., and Schäfer, G. (1988). Chemiosmotic H+ cycling across the plasma membrane of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. FEBS Lett. 232, 359–363.
Mowat, C. G., Rothery, E., Miles, C. S., McIver, L., Doherty, M. K., Drewette, K., Taylor, P., Walkinshaw, M. D., Chapman, S. K., and Reid, G. A. (2004). Octaheme tetrathionate reductase is a respiratory enzyme with novel heme ligation. Nat. Struct. Mol. Biol. 11, 1023–1024.
Mukhopadhyaya, P. N., Deb, C., Lahiri, C., and Roy, P. (2000). A soxA gene, encoding a diheme cytochrome c, and a sox locus, essential for sulfur oxidation in a new sulfur lithotrophic bacterium. J. Bacteriol. 182, 4278–4287.
Müller, F. H., Bandeiras, T. M., Urich, T., Teixeira, M., Gomes, C. M., and Kletzin, A. (2004). Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate:quinone oxidoreductase. Mol. Microbiol. 53, 1147–1160.
Oubrie, A., Rozeboom, H. J., Kalk, K. H., Olsthoorn, A. J., Duine, J. A., and Dijkstra, B. W. (1999). Structure and mechanism of soluble quinoprotein glucose dehydrogenase. EMBO J. 18, 5187–5194.
Perriere, G., and Gouy, M. (1996). WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78, 364–369.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., and Ferrin, T. E. (2004). UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612.
Peyfoon, E., Meyer, B., Hitchen, P. G., Panico, M., Morris, H. R., Haslam, S. M., Albers, S. V., and Dell, A. (2010). The S-layer glycoprotein of the crenarchaeote Sulfolobus acidocaldarius is glycosylated at multiple sites with chitobiose-linked N-glycans. Archaea 2010, Article ID 754101. doi: 10.1155/2010/754101
Price-Carter, M., Tingey, J., Bobik, T. A., and Roth, J. R. (2001). The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183, 2463–2475.
Richter, O.-M., and Schäfer, G. (1992). Purification and enzymic characterization of the cytoplasmic pyrophosphatase from the thermoacidophilic archaebacterium Thermoplasma acidophilum. Eur. J. Biochem. 209, 343–349.
Rzhepishevska, O. I., Valdes, J., Marcinkeviciene, L., Gallardo, C. A., Meskys, R., Bonnefoy, V., Holmes, D. S., and Dopson, M. (2007). Regulation of a novel Acidithiobacillus caldus gene cluster involved in metabolism of reduced inorganic sulfur compounds. Appl. Environ. Microbiol. 73, 7367–7372.
Sakurai, H., Ogawa, T., Shiga, M., and Inoue, K. (2010). Inorganic sulfur oxidizing system in green sulfur bacteria. Photosyn. Res. 104, 163–176.
Sand, W., Gehrke, T., Jozsa, P. G., and Schippers, A. (2001). (Bio) chemistry of bacterial leaching – direct vs. indirect bioleaching. Hydrometallurgy 59, 159–175.
Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379.
Seldon, R. F. (1988). “Preparation and analysis of RNA: 4.9, analysis of RNA by northern hybridization,” in Current Protocols in Molecular Biology, eds F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (New York: Greene Publishing Associates and Wiley-Interscience), 4.9.1–4.9.8.
Stetter, K. O. (2006). Hyperthermophiles in the history of life. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1837–1842; discussion 1842–1833.
Sugio, T., Kanao, T., Furukawa, H., Nagasawa, T., and Blake, R. C. (1996). Isolation and identification of an iron-oxidizing bacterium which can grow on tetrathionate medium and the properties of a tetrathionate-decomposing enzyme isolated from the bacterium. J. Ferment. Bioeng. 82, 233–238.
Tano, T., Kitaguchi, H., Harada, M., Nagasawa, T., and Sugio, T. (1996). Purification and some properties of a tetrathionate decomposing enzyme from Thiobacillus thiooxidans. Biosci. Biotech. Biochem. 60, 224–227.
Teixeira, M., Batista, R., Campos, A. P., Gomes, C., Mendes, J., Pacheco, I., Anemüller, S., and Hagen, W. R. (1995). A seven-iron ferredoxin from the thermoacidophilic Desulfurolobus ambivalens. Eur. J. Biochem. 227, 322–327.
Trudinger, P. A. (1964). Effects of thiosulphate and oxygen concentration on tetrathionate oxidation by Thiobacillus X and Thiobacillus thioparus. Biochem. J. 90, 640–646.
Urich, T., Bandeiras, T. M., Leal, S. S., Rachel, R., Albrecht, T., Zimmermann, P., Scholz, C., Teixeira, M., Gomes, C. M., and Kletzin, A. (2004). The sulphur oxygenase reductase from Acidianus ambivalens is a multimeric protein containing a low-potential mononuclear non-haem iron centre. Biochem. J. 381, 137–146.
Urich, T., Kroke, A., Bauer, C., Seyfarth, K., Reuff, M., and Kletzin, A. (2005). Identification of core active site residues of the sulfur oxygenase reductase from Acidianus ambivalens by site-directed mutagenesis. FEMS Microbiol. Lett. 248, 171–176.
Veith, A., Klingl, A., Zolghadr, B., Lauber, K., Mentele, R., Lottspeich, F., Rachel, R., Albers, S.-V., and Kletzin, A. (2009). Acidianus, Sulfolobus and Metallosphaera surface layers: structure, composition and gene expression. Mol. Microbiol. 73, 58–72.
Veith, A., Urich, T., Seyfarth, K., Protze, J., Frazão, C., and Kletzin, A. (2011). Substrate pathways and mechanisms of inhibition in the sulfur oxygenase reductase of Acidianus ambivalens. Front. Microbiol. 2:37. doi: 10.3389/fmicb.2011.00037
Keywords: Archaea, sulfur metabolism, tetrathionate hydrolase, S4 intermediate pathway, surface layer, disproportionation
Citation: Protze J, Müller F, Lauber K, Naß B, Mentele R, Lottspeich F and Kletzin A (2011) An extracellular tetrathionate hydrolase from the thermoacidophilic archaeon Acidianus ambivalens with an activity optimum at pH 1. Front. Microbio. 2:68. doi: 10.3389/fmicb.2011.00068
Received: 03 January 2011;
Paper pending published: 21 January 2011;
Accepted: 25 March 2011;
Published online: 25 April 2011.
Edited by:
Martin G. Klotz, University of Louisville, USAReviewed by:
Biswarup Mukhpadhyay, Virginia Bioinformatics Institute, USAUwe Deppenmeier, University of Bonn, Germany
Copyright: © 2011 Protze, Müller, Lauber, Naß, Mentele, Lottspeich and Kletzin. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Arnulf Kletzin, Institute of Microbiology and Genetics, Technische Universität Darmstadt, Schnittspahnstraße 10, 64287 Darmstadt, Germany. e-mail:a2xldHppbkBiaW8udHUtZGFybXN0YWR0LmRl

 Fabian Müller1
Fabian Müller1