- Department of Microbiology, University of Chicago, Chicago, IL, USA
The pathogenic lifecycle of obligate intracellular bacteria presents a superb opportunity to develop understanding of the interaction between the bacteria and host under the pretext that disruption of these processes will likely lead to death of the pathogen and prevention of associated disease. Species of the genus Rickettsia contain some of the most hazardous of the obligate intracellular bacteria, including Rickettsia rickettsii and R. conorii the causative agents of Rocky Mountain and Mediterranean spotted fevers, respectively. Spotted fever group Rickettsia species commonly invade and thrive within cells of the host circulatory system whereby the endothelial cells are severely perturbed. The subsequent disruption of circulatory continuity results in much of the severe morbidity and mortality associated with these diseases, including macropapular dermal rash, interstitial pneumonia, acute renal failure, pulmonary edema, and other multisystem manifestations. This review describes current knowledge of the essential pathogenic processes of adherence to and invasion of host cells, efforts to disrupt these processes, and potential for disease prevention through vaccination with recently identified bacterial adherence and invasion proteins. A more complete understanding of these bacterial proteins will provide an opportunity for prevention and treatment of spotted fever group Rickettsia infections.
Introduction
The Gram-negative α-proteobacteria of the genus Rickettsia are small (0.3–0.5 × 0.8–1.0 μm), obligate intracellular organisms. They are categorized into two major groups, the spotted fever group (SFG) and typhus group (TG), which can be distinguished by antigenicity and intracellular actin-based motility. Members of this genus are responsible for severe human diseases and several species including Rickettsia rickettsii, R. prowazekii, and R. typhi, have been classified as Category B and C Priority Pathogens by the National Institute of Allergy and Infectious Diseases (NIAID) and as Select Agents (R. rickettsii and R. prowazekii) by the Centers for Disease Control and Prevention (CDC) for their potential use as tools for biological terrorism.
Spotted fever group rickettsiae including R. rickettsii (Rocky Mountain spotted fever, RMSF) and R. conorii (Mediterranean spotted fever, MSF) are pathogenic organisms transmitted to humans through tick salivary contents during the blood meal. RMSF is one of the most severe SFG rickettsioses in the western hemisphere, causing severe morbidity and up to 20% mortality in the absence of timely and appropriate antibiotic treatment (Walker, 1989). MSF, endemic to southern Europe, North Africa and India, has previously been characterized as a milder rickettsiosis in humans, with 2–3% mortality; however, in light of improved molecular diagnostic tools, recent accumulating evidence has unveiled that MSF exhibits an expansive geographic distribution, now including central Europe and central and southern Africa, and increased disease severity commensurate to RMSF, with mortality rates reported as high as 32% in Portugal in 1997 (de Sousa et al., 2003). This raises concern over the challenges posed by R. conorii infection.
Symptoms from rickettsial disease manifest 2–14 days following inoculation by an infected ixodid tick. Early indications of infection are unremarkable and include headache, fever, and malaise. Soon after the tick bite, localized replication of rickettsiae at the inoculation site and ensuing tissue damage may give rise to a necrotic lesion, or eschar. Damage to the vascular endothelium and infiltration of perivascular mononuclear cells leads to fluid leakage into the interstitial space resulting in a dermal rash in 90% of cases. Endothelial cells are the main targets during rickettsial infection. Bacterial replication within the endothelial tissues and subsequent damage to the integrity of the vasculature leads to complications such as encephalitis, non-cardiogenic pulmonary edema, interstitial pneumonia, hypovolemia, hypotensive shock, and acute renal failure (Walker et al., 1994).
The TG rickettsiae include R. prowazekii, the etiologic agent of epidemic typhus, and R. typhi, the causative agent of murine typhus. Epidemic typhus outbreaks have been documented in disparate global communities (Bise and Coninx, 1997; Raoult et al., 1997, 1998), thriving in areas of poor sanitation and hygiene. TG rickettsiae are normally and stably transmitted through the excrement of human body lice and are inoculated into abraded skin by scratching. Early signs of infection are characterized by headaches, fever, delirium, and rash. Delayed or inappropriate treatment can result in mortality rates ranging from 10 to 60%. Unlike SFG rickettsial diseases, R. prowazekii can cause latent infections, where recurrence results in Brill–Zinsser disease, a less severe but chronic R. prowazekii infection that can be transmitted to feeding lice, thus fueling epidemics.
Rickettsia species are obligate intracellular bacteria and as such have evolved to take full advantage of the nutrient and energy-rich environment of the cytosol of host cells. In doing so, they have undergone reductive evolution, discarding many of their own genes necessary for metabolite synthesis. Their succinct genome has made them completely dependent on the intracellular environment of the mammalian host cell for growth and survival. During an infection, rickettsial pathogenesis depends initially on the bacteria’s ability to attach to and invade the host’s cells. This requires successful recognition and interaction with specific cellular receptors, and is thought to be dependent on the presence of heat-labile proteins on the rickettsial surface (Li and Walker, 1992). While Rickettsia primarily infect the host endothelium, in vitro they are seen to adhere to and invade diverse types of mammalian cells (Cohn et al., 1959; Ramm and Winkler, 1973; Winkler, 1974, 1977; Winkler and Ramm, 1975; Stork and Wisseman, 1976; Wisseman et al., 1976; Walker and Winkler, 1978; Rikihisa and Ito, 1979; Turco and Winkler, 1982; Walker, 1984; Teysseire et al., 1995), in a mechanism requiring host membrane cholesterol (Ramm and Winkler, 1976; Martinez and Cossart, 2004). This review addresses current knowledge of SFG Rickettsia adherence to and invasion of host cells, with particular emphasis on the host signaling mechanisms induced by the bacteria, the bacterial proteins that mediate these processes, and efforts to utilize these rickettsial proteins in vaccination.
Rickettsia Invasion
Intracellular bacterial pathogens have been shown to facilitate their entry into non-phagocytic host cells by either of two mechanistically and morphologically distinct means: the “zipper” or “trigger” mechanisms (reviewed in Alonso and Garcia-del Portillo, 2004). The zipper invasion mechanism is a receptor-mediated invasion strategy, whereby a bacterial protein induces host intracellular signaling through extracellular stimulation of a membrane receptor. These signals modulate local host cytoskeletal rearrangements and recruitment of endocytic machinery at the site of interaction, culminating in membrane “zippering” around the pathogen (Cossart, 2004). Hallmark examples of this are seen in Listeria monocytogenes with InlA and InlB, and Yersinia pseudotuberculosis YadA and invasin (Mengaud et al., 1996; Wong and Isberg, 2003; Alonso and Garcia-del Portillo, 2004; Veiga and Cossart, 2005, 2007). In contrast, the “trigger” mechanism relies on Type III secretion system (TTSS)-mediated delivery of bacterial effectors into the host cell to modulate cellular GTPases and give rise to dramatic actin and membrane rearrangements (Alonso and Garcia-del Portillo, 2004). Genome analyses of sequenced strains of the genus Rickettsia show the absence of genes encoding for components of the TTSS, suggesting that rickettsial species likely induce their internalization into non-phagocytic mammalian cells by a mechanism independent of this secretion machinery. Interestingly, transmission electron micrographs of R. conorii entry illustrate that, like L. monocytogenes, rickettsial invasion is characterized by intimate localized rearrangement of cellular plasma membrane around the bacteria morphologically resembling a zipper-induced entry mechanism (Teysseire et al., 1995; Gouin et al., 1999).
Numerous detailed analyses of invasion into non-phagocytic mammalian cells have demonstrated that host actin polymerization plays a critical role during the induced entry of rickettsial species (Walker and Winkler, 1978; Walker, 1984; Gouin et al., 1999; Martinez and Cossart, 2004). An initial examination of proteins directly involved in modulating actin dynamics during R. conorii invasion illustrated the involvement of the small GTP-binding protein, Cdc42, and the actin nucleating complex, Arp2/3, in the invasion process (Martinez and Cossart, 2004). These proteins are required for rickettsial entry and localize to the sites of R. conorii invasion as revealed by immunofluorescence microscopy. Specific perturbation of these players, for example, inhibition of Arp2/3 function by transfection of the WASP family member, Scar, or Cdc42 through transfection of a dominant-negative variant, diminishes rickettsial invasion. The GTPase activity governing the actin cytoskeletal rearrangements was seen to be specific for Cdc42, as similar inhibition of Rac1 had no effect on the efficiency of R. conorii invasion. Pharmacological inhibition of pathways that both directly and indirectly modulate the activities of Cdc42 and Arp2/3 showed that actin polymerization, PI 3-kinases, phosphotyrosine kinase (PTK) activities and the activity of a specific PTK, c-Src, contribute to R. conorii invasion of non-phagocytic mammalian cells (Martinez and Cossart, 2004). Examination of c-Src and one of its downstream effectors, cortactin, by immunofluorescence microscopy showed that both are also recruited to sites of rickettsial internalization, highlighting the importance of these proteins to the entry process (Martinez and Cossart, 2004). In addition, R. conorii invasion coincides with the tyrosine phosphorylation of several host proteins including focal adhesion kinase (FAK) (Martinez and Cossart, 2004), a protein involved in cytoskeletal reorganization and in the invasion of other “zippering” pathogens (Persson et al., 1997; Martinez et al., 2000). Together, these findings indicate that signaling events from these various components are likely coordinated to ultimately activate Arp2/3, thus instigating localized actin recruitment around the invading bacteria. While these studies provide insight into the signaling events involved in stimulating R. conorii entry of non-phagocytic mammalian cells, until recently, little was known about the identity of the rickettsial proteins and cellular receptors and their interplay at the extracellular interfaces that govern the induction of these pathways.
Identification of A R. Conorii Host Cell Receptor
In an effort to identify putative host cell receptors involved in rickettsial entry, whole, intact R. conorii were used as an affinity matrix to identify interacting mammalian host cell proteins (Martinez et al., 2005). This investigation led to the identification of host Ku70 as one of several cellular proteins that specifically interact with purified R. conorii and not other invasive pathogens. Ku70’s role as a receptor for rickettsial entry was initially highlighted by immunofluorescence microscopy, which demonstrated co-localization of plasma membrane Ku70 with invading bacteria (Martinez et al., 2005). Further, pre-incubation of cells with antibody directed against a surface-exposed, extracellular epitope of Ku70 reduces R. conorii entry into mammalian cells, suggesting interaction of rickettsiae with this protein is important for stimulating bacterial internalization. An analysis of R. conorii invasion in cells either deficient (Ku70−/− murine embryonic fibroblasts) or siRNA-depleted of Ku70 reveal that invasion into these cells is perturbed at least 50% compared to controls (Martinez et al., 2005), indicating that the presence of cellular Ku70 is important for the rickettsial entry process. Interestingly, cells lacking Ku70 are not completely impervious to infection by R. conorii, implying that additional host proteins are likely utilized in rickettsial invasion (Martinez et al., 2005).
Ku70 is ubiquitously expressed in mammalian cells and is generally localized to the nucleus and cytoplasm (Muller et al., 2005). Expression of Ku70 at the plasma membrane is restricted to specific cell types including endothelial cells, monocytes and macrophages, which are the main target cells during rickettsial infection in vivo, and cultured tumor cell lines such as HeLa and Vero (Muller et al., 2005), both commonly utilized to culture Rickettsia in vitro (Martinez and Cossart, 2004; Ammerman et al., 2008). Because Ku70 lacks the canonical signal sequence that typically directs plasma membrane-destined polypeptides through the endoplasmic reticulum/golgi secretory pathway, its presentation in the extracellular environment was baffling. Recent studies examining induced Ku70 plasma membrane display in monocytes have shown that Ku70 is transported via a non-classical vesicle secretion mechanism (Paupert et al., 2007). Here, cytoplasmic Ku70 is gathered into membrane-bound vesicles and trafficked in the cytoplasm along the actin cytoskeleton for secretion and display at the plasma membrane. Biochemical fractionations of mammalian cells have shown that plasma membrane-associated Ku70 is present in cholesterol-rich, detergent-resistant microdomains or “lipid-rafts” (Martinez et al., 2005), and that depletion of cholesterol from HeLa or Vero cells by methyl-β-cyclodextrin disrupts the integrity of these microdomains and prevents rickettsial entry (Martinez et al., 2005). While these studies illustrate the presence of Ku70 at the plasma membrane, and its integral role in rickettsial invasion, the exact topology of the protein is unknown and is currently under investigation.
Attachment of invasive pathogens to host cell receptors commonly leads to receptor modification and subsequent endocytic signaling that enable bacterial entry (Su et al., 1999; Eto et al., 2007; Veiga and Cossart, 2007). Examination of the modification status of Ku70 consequent to rickettsial infection reveal that upon bacterial attachment, Ku70 is ubiquitinated by c-Cbl, an E3 ubiquitin ligase, which by fluorescence microscopy is seen to localize to R. conorii entry foci (Martinez et al., 2005). Inhibition of endogenous c-Cbl expression by siRNA depletion effectively reduces rickettsial invasion and also prevents R. conorii-induced ubiquitination of Ku70, suggesting that c-Cbl modification of Ku70 may contribute to R. conorii entry (Martinez et al., 2005). Mechanistically this process resembles L. monocytogenes InlB induction of c-Met receptor mono-ubiquitination, which is involved in subsequent clathrin-dependent endocytosis (Veiga and Cossart, 2005). The morphological and mechanistic similarities shared amongst “zippering” pathogens would suggest that ubiquitin-modification of bacterial receptors may be a generalized strategy to recruit components of the endocytic machinery to entry foci (see below).
The Rickettsial Cell Surface
Since rickettsial adherence and invasion are most likely mediated by components present on the surface of these bacteria, much research has focused on the identification and characterization of outer membrane-associated rickettsial proteins. Bioinformatic analyses of sequenced rickettsial genomes have identified families of proteins that likely localize to the outer membrane and are therefore predicted to be involved in mediating interactions with target host cells. Of interest was the identification of a family of at least 17 different genes termed sca for surface cell antigens whose products resemble autotransporter proteins, many of which are known virulence factors in Gram-negative bacteria (Henderson and Lam, 2001; Jacob-Dubuisson et al., 2004; Blanc et al., 2005). These proteins have modular structures, including an N-terminal signal peptide, a central passenger domain, and a C-terminal translocation module (β-peptide). Following translation, these proteins are secreted across the inner membrane using information encoded in the N-terminal signal sequence. The C-terminal domain then inserts into the outer membrane to form a β-barrel-rich transmembrane pore believed to serve in the secretion of the passenger domain into the extracellular milieu (Jacob-Dubuisson et al., 2004; Figure 1). While a significant amount of degradation of genes in the sca family has occurred in many phylogenetically related and divergent rickettsial species, five genes, namely sca0 (rompA), sca1, sca2, sca4, and sca5 (rompB) are highly conserved among the majority of SFG rickettsiae (Roux and Raoult, 2000; Blanc et al., 2005); the evolutionary maintenance of these genes suggests they may exhibit a functional role in rickettsial pathogenesis. Thus, a great deal of emphasis has been put forth to determine the contribution of these gene products in rickettsial host cell interactions.
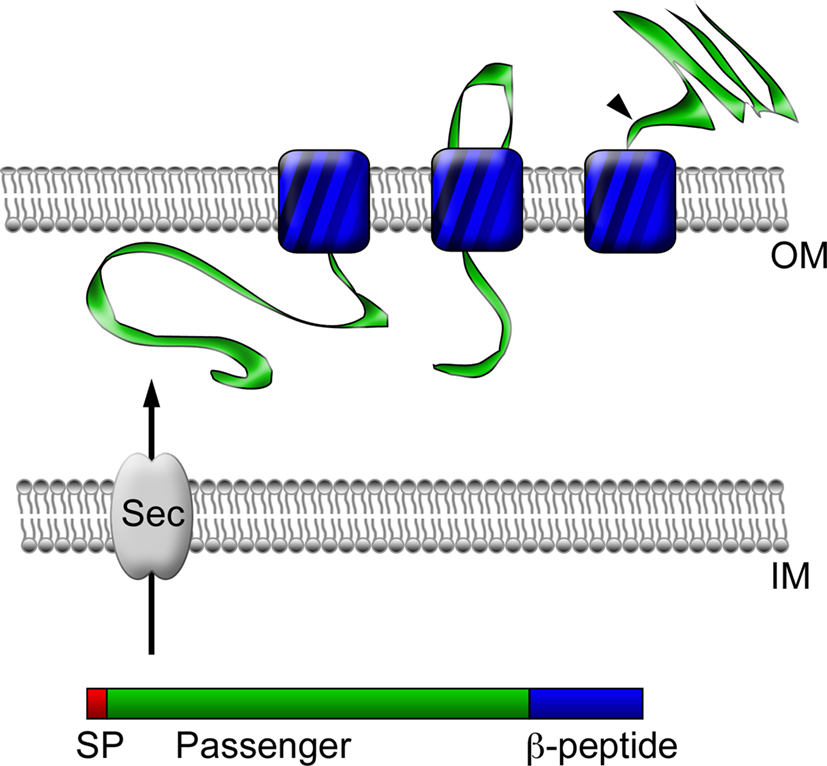
Figure 1. Autotransporter protein structure and secretion. Autotransporter proteins have modular structures, including an N-terminal signal peptide (SP, red), a central passenger domain (green), and a C-terminal translocation module called a β-peptide (blue). Following translation, the polypeptide is secreted across the inner membrane (IM) through the Sec translocon into the periplasmic space using information encoded in the N-terminal signal sequence. Through an undefined mechanism, the β-peptide next inserts into the bacterial outer membrane (OM) to form a β-barrel-rich transmembrane pore for the secretion of the central passenger domain into the extracellular space. In the case of some proteins, such as rOmpB, the passenger domain is subsequently proteolyzed from the β-peptide portion of the protein, but may remain associated with the outer leaflet of the bacterial outer membrane.
Rickettsia Surface Cell Antigens
rOmpB (Sca5)
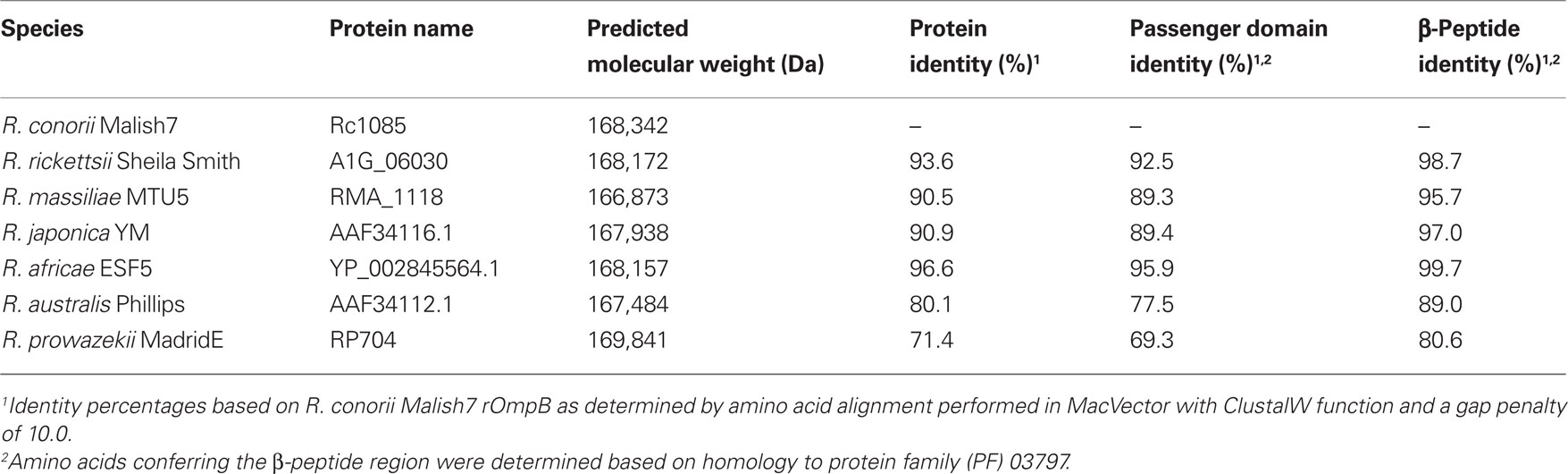
rOmpB is evolutionarily conserved amongst all known rickettsial species, and is present in both SFG and TG rickettsia. Originally referred to as SpaP, SPA, or p120, its persistent and current nomenclature is rOmpB protein encoded by the rompB gene. Electron microscopic analyses of rickettsiae have revealed the presence of a 7–16 nm monolayer of protein arranged in a tetragonal array attached externally to the outer membrane (Palmer et al., 1974a,b; Popov and Ignatovich, 1976; Silverman and Wisseman, 1978; Silverman et al., 1978). This proteinaceous layer consists of 10–15% of total cellular protein and is composed of a diverse set of proteins, most predominantly rOmpB (Smith and Winkler, 1979; Dasch, 1981; Dasch et al., 1981; McDonald et al., 1988; Hechemy et al., 1989). Sequence analyses of rOmpB reveals a high level of conservation amongst diverse groups of rickettsiae (Blanc et al., 2005), exhibiting 70–95% identity on the amino acid level (Table 1). This conservation is evidence of positive selection which is apparent not only in the β-barrel domain which is restricted by its known function, but also proceeds into the predicted passenger domain (Table 1). These areas of conservation suggest rOmpB serves a unified function in rickettsial pathogenesis. rOmpB is expressed as a pre-protein (168 kDa) and cleaved to release the passenger domain (120 kDa) from the β-barrel translocation domain (32 kDa), leaving the mature 120-kDa domain associated with the outer leaflet of the outer membrane (Hackstadt et al., 1992). Interestingly, through affinity chromatography methods, Martinez et al. (2005) identified rOmpB as a bacterial ligand of the host receptor Ku70 suggesting that rOmpB–Ku70 serves as a bona fide adhesin-receptor pair in rickettsia-host cell interactions.
Recently, the roles of rOmpB in rickettsial pathogenesis, with regard to its contribution to bacterial adherence to and invasion of host cells, have been examined. Investigation of rOmpB function from related SFG rickettsiae, R. conorii and R. japonica, through heterologous expression of these proteins in Escherichia coli have shown that rOmpB is an integral player in initiating bacterial infection of mammalian host cells. When expressed on normally inert E. coli, rOmpB is sufficient to facilitate bacterial adherence to and invasion of non-phagocytic cells (Uchiyama et al., 2006; Chan et al., 2009; Figure 2). A previous study examining R. conorii entry showed that host protein, Ku70, served as a receptor for rickettsial invasion and interacted with rOmpB (Martinez et al., 2005). Bacterial invasion mediated by rOmpB alleles from both R. conorii and R. japonica were demonstrated to be Ku70-dependent (Chan et al., 2009), suggesting not only that rOmpB is a rickettsial adhesion and invasion molecule, but that it is the cognate bacterial ligand for the host receptor Ku70. Mammalian cells depleted of Ku70 no longer support rOmpB-mediated E. coli invasion, indicating that interaction of rOmpB and Ku70 are requisite to stimulate signals for bacterial uptake (Chan et al., 2009).
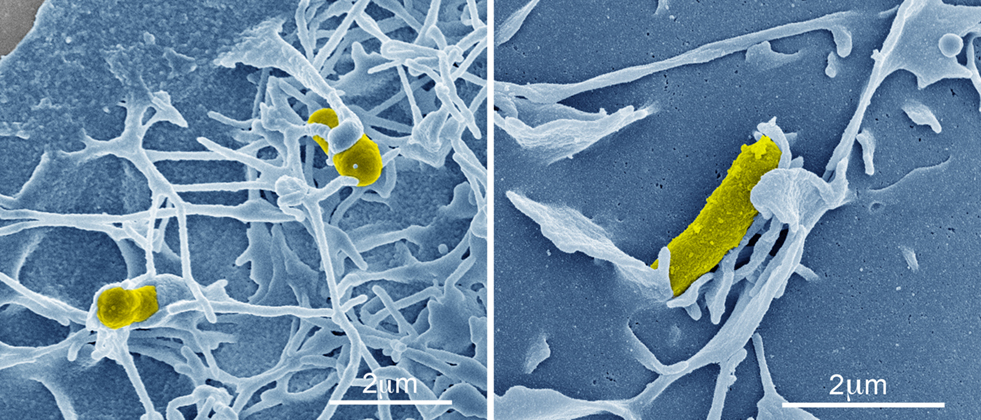
Figure 2. rOmpB-mediated bacterial invasion. Pseudo-colored scanning electron micrographs of a rOmpB-expressing Escherichia coli (yellow) inducing host membrane rearrangements during infection of a mammalian HeLa cell (blue).
Mechanistic interrogation of the rOmpB–Ku70-mediated invasion process revealed that the interaction of these two proteins is sufficient to recruit the actin cytoskeleton to the bacterial entry foci on the cell (Chan et al., 2009). This rOmpB-mediated process, resembling that observed during R. conorii invasion, is manifested through the activities of PTKs and PI 3-kinases, and dependent on the cellular microtubule infrastructure (Martinez and Cossart, 2004; Chan et al., 2009). Similar to findings by Martinez et al., and paralleling the entry mechanisms observed in R. conorii infection, rOmpB-induced invasion via Ku70 is dependent on c-Cbl ubiquitin ligase activity, suggesting c-Cbl may be a signaling molecule involved in commandeering host endocytic pathways (Martinez and Cossart, 2004; Veiga and Cossart, 2005; Chan et al., 2009). Whether rOmpB is sufficient to mediate c-Cbl-dependent Ku70 ubiquitination during the entry process is not known. However, siRNA depletion of cellular players of endocytosis revealed that invasion through rOmpB is in part dependent on clathrin and caveolin-2 expression (Chan et al., 2009).
The specificity and physiological significance of rOmpB in its role in bacterial entry is underscored by the ability for recombinant rOmpB to competitively impede not only rOmpB-mediated E. coli association (Chan et al., 2009), but also R. conorii association with host cells (Y. G. Chan, unpublished data). The perturbation of this infection process can be elicited by the purified passenger domain (36–1334 aa), which additionally interacts directly with Ku70 (Chan et al., 2009). Although a previous study has proposed a putative role of the rOmpB β-peptide in adhesion (Renesto et al., 2006), these findings show that an effector region of this protein is in part contained within the passenger domain (Martinez et al., 2005; Chan et al., 2009).
rOmpA (Sca0)
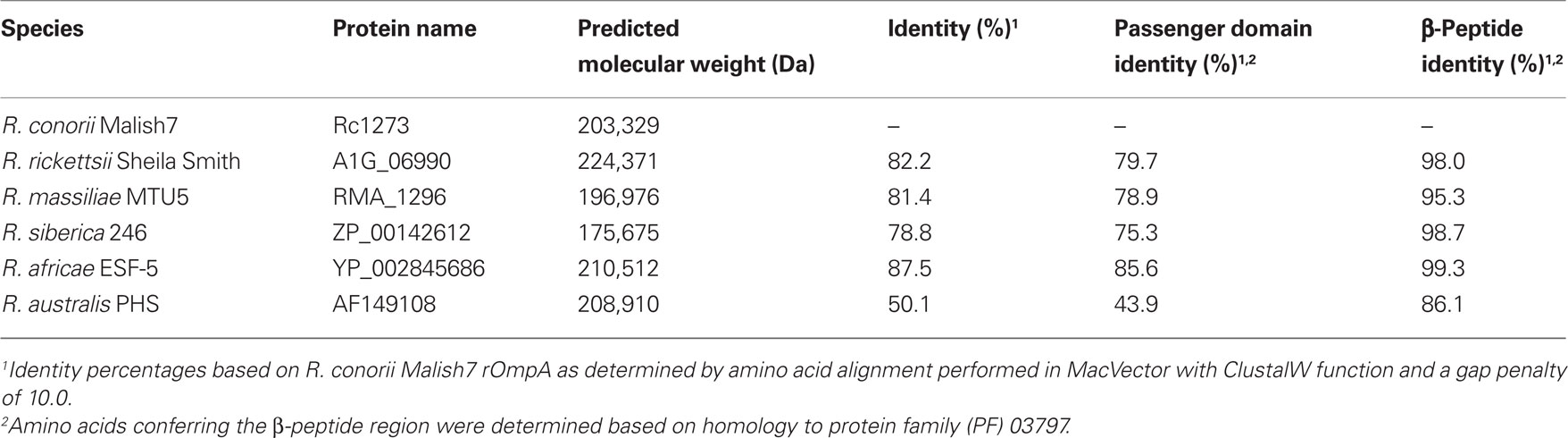
The rompA gene is found in several SFG rickettsial species but is absent in TG Rickettsia (Blanc et al., 2005). In R. rickettsii strain Sheila Smith, rompA encodes for a protein of 247 kDa, while in R. conorii Malish 7 rompA encodes for a protein of 224 kDa. The final predicted lengths of rOmpA alleles vary from species to species, as is apparent in the disparate molecular weights of this protein between rickettsial strains. These differences are largely the result of variances in the number of tandem repeat sequences present within the passenger domain region of the protein (Eremeeva et al., 2003; Ellison et al., 2008). While nothing is known about the function of these repeat regions, their presence and conservation in a diverse set of rickettsial species suggests a conserved function in this protein (Table 2).
In comparison to rOmpB, very little is known about the putative processing of rOmpA in rickettsial species. The predicted rOmpA β-peptides of various Rickettsia species are nearly identical (Table 2). While bioinformatic models predict that the rOmpA signal sequence from R. conorii is demarcated by amino acids 1–38 (Emanuelsson et al., 2007), the identity of putative cleavage sites to release the passenger domain from the β-peptide is not known. Western immunoblot analyses of R. conorii detergent soluble lysates with rOmpA antisera have illuminated a reactive species of approximately 190 kDa, approximately 34 kDa less than the predicted mass of the full-length protein (Hillman and Martinez, unpublished observations) and (Vishwanath, 1991); similar results were observed in the mobility of the R. rickettsii rOmpA protein (Anderson et al., 1990). The molecular weight differences between the predicted and observed rOmpA might be due to aberrant mobility of the protein on SDS–PAGE; alternatively, these data suggest the intriguing possibility that rOmpA is proteolytically cleaved to release the passenger domain from an approximately 32–34 kDa beta-peptide translocon domain in the pre-protein, as has been observed for other autotransporters.
rOmpA has been shown to mediate rickettsial adherence (Li and Walker, 1998). Previously, monoclonal antibodies to R. rickettsii rOmpA and F(ab′)2 fragments derived from these antibodies have been demonstrated to inhibit the adhesion of rickettsiae to L-929 cells (Li and Walker, 1998). rOmpA extracted from intact R. rickettsii was further shown to be sufficient in competitively inhibiting attachment of R. rickettsii to these cells (Li and Walker, 1998), implicating a role for rOmpA in rickettsial association to mammalian cells. Another analysis of rOmpA function demonstrated that like rOmpB, rOmpA from R. conorii, when expressed at the outer membrane of E. coli, is sufficient not only to mediate bacterial adherence to but also invasion of cultured mammalian cells (Hillman and Martinez, unpublished observations). Together, these results suggest a critical role for rOmpA in mediating SFG Rickettsia-host cell interactions.
Sca1
sca1 is present in nearly all sequenced Rickettsia genomes, with the exceptions of R. prowazekii and R. canadensis, where it is present as a pseudo-gene (Ngwamidiba et al., 2006). Like rOmpA and rOmpB, Sca1 possesses all of the hallmarks of a surface-exposed autotransporter (Blanc et al., 2005). While selective pressure has limited the diversity of particular regions of the N-terminal and the β-peptide portions, the overall size of Sca1 varies significantly, ranging from 594 to 1976 aa (Ngwamidiba et al., 2006; Riley et al., 2010). However, the conservation of specific domains of Sca1 has led to the hypothesis that it serves universal functions amongst SFG Rickettsia.
Riley et al. (2010) demonstrated that Sca1 is expressed and present on the surface of R. conorii grown in mammalian cell culture. Flow cytometric analysis and immunofluorescence microscopy using a polyclonal antibody generated against the N- terminal portion (29–327 aa) of Sca1 provided evidence for surface localization of this protein in R. conorii, demonstrating that indeed Sca1 is exposed to the extracellular milieu (Riley et al., 2010). Interestingly, immunoblot analysis of R. conorii lysates with specific anti-Sca1 antisera revealed that Sca1 in R. conorii appeared to be processed to an approximately 120 kDa mature protein from the predicted greater than 200 kDa polypeptide (Riley et al., 2010). This finding complicates the historical assumption that rOmpB is the only antigenic protein at 120 kDa, since both the mature forms of Sca1 and rOmpB migrate similarly by SDS–PAGE analyses.
The sca1 gene from R. conorii was expressed in E. coli and was demonstrated to be present in the outer membrane of these bacteria. The presence of Sca1 in E. coli was sufficient to mediate attachment to but not invasion of a panel of host cells (Riley et al., 2010). Finally, pre-incubation with a recombinant fragment of the Sca1 protein blocked some rickettsial association with host cells (Riley et al., 2010). Together, these findings indicate that adherence and invasion are two distinct events that can be mediated through multiple rickettsial proteins. Examination of the relative contribution of the rickettsial Sca proteins to adherence, invasion, and other potential cellular processes will need to be examined further.
Sca2
Sca2 is conserved and present in most SFG Rickettsia and either split or absent in TG species (Blanc et al., 2005; Haglund et al., 2010). The gene encodes for an approximately 200–220 kDa protein that ranges from 80 to 95% identity among SFG species, exhibiting the greatest conservation in the β-peptide and signal sequence (Ngwamidiba et al., 2005; Cardwell and Martinez, 2009). To date nothing is known about its potential processing and gene regulation, although numerous studies have demonstrated the gene is expressed during infection of mammalian cells and the protein is presented at the rickettsial surface (Cardwell and Martinez, 2009; Haglund et al., 2010).
The contribution of Sca2 to bacterial entry was examined by heterologous expression in E. coli (Cardwell and Martinez, 2009). In this system, the R. conorii Sca2 protein sufficiently mediated bacterial association to and invasion of numerous non-phagocytic mammalian cell types, most notably primary human endothelial cell lines, which are major targets during rickettsial infection. Furthermore, competitive inhibition of both E. coli as well as R. conorii invasion mediated through Sca2 was achieved using the N-terminal half of the Sca2 passenger domain (34–794 aa), suggesting this portion of the protein may be involved in mediating interactions with the host cell surface.
While Cardwell et al. were able to demonstrate a role for Sca2 in R. conorii entry (Cardwell and Martinez, 2009), new evidence has emerged indicating this may not be its sole function during infection of mammalian cell culture. In a recent transposon mutagenesis study, a R. rickettsii sca2 mutant was discovered and seen to exhibit a defect in actin-based motility (Kleba et al., 2010). Proteomic analyses of the N-terminal portion of the Sca2 passenger domain sequence elucidated motifs resembling those present in eukaryotic actin modulating proteins, including three actin-binding WASP homology 2 domains and a predicted secondary structure resembling the formin homology 2 domain, suggesting a role for Sca2 in producing the observed parallel actin bundles involved in SFG rickettsial intra- and inter-cellular motility (Gouin et al., 1999; Haglund et al., 2010; Kleba et al., 2010). Comprehensive analyses of R. parkeri Sca2 by Haglund et al. (2010) further endorse the function of this protein in actin-based motility, showing Sca2 localization to actin-polymerizing surfaces of the bacteria, and demonstrating its in vitro capacity to polymerize actin monomers, and elongate filaments in a profilin-dependent manner. Together, these findings suggest that in the face of the limited genome size, rickettsial proteins may serve multiple pathogenic roles. Whether the invasion and actin-polymerizing functions of R. conorii Sca2 are composed of two distinct domains or are contained within the same region of the polypeptide is under investigation.
Putative Rickettsial Adhesins
A biochemical approach whereby rickettsial proteins resolved by SDS–PAGE were examined for interaction with surface-biotinylated mammalian cell extracts has unveiled several potential rickettsial adhesins. In addition to the rOmpB β-peptide, rickettsial gene products of RC1281 from R. conorii and RP828, its homologue in R. prowazekii, were identified as putative interactors of cellular membrane proteins (Renesto et al., 2006). RC1281/RP828 is conserved and ubiquitously present in the genomes of Rickettsia species and is paralogous to the adjacent gene RC1282 in R. conorii, which is also similarly conserved and prevalent but was not identified in that screen. BLAST analyses of these genes show a low level of homology to other bacterial surface proteins, some which have adhesin and toxin functions. Balraj et al. (2009) have reported that antibodies generated against the recombinant proteins encoded by RC1281 and RC1282 reduce R. conorii infectivity of mammalian cell culture, suggesting they may be surface-exposed antigens. Further functional examination is required to validate the role of these proteins as rickettsial adhesins.
Sca Proteins as Vaccine Candidates
The immune responses generated from a prior rickettsial infection are seen to be protective and detectible for years following an infection (Bengtson and Topping, 1942; Fox et al., 1957). This has led to a hypothesis that the Sca proteins will provide attractive targets for recombinant vaccine production. There is currently no preventative therapy available for protection against rickettsial pathogens and antibiotic treatment with broad-spectrum antibiotics such as doxycycline, tetracycline, or chloramphenicol is effective only when administered within the first week of symptom manifestation, thus relying on prompt and proper diagnosis. Rickettsiae also pose a great biodefense threat and have historically been researched and employed as biological weapons (Harris, 1992; Kelly et al., 2002; Croddy et al., 2005).
Throughout the last 60 years, researchers have been exploring the potential use of whole bacterial immunization procedures through various treatments of the pathogens to prevent disease while maintaining the protective effects of exposure to the bacteria. Whole cell vaccines utilized in the early to mid-twentieth century against RMSF or typhus were comprised of formalin-fixed R. rickettsii or R. prowazekii isolated from ticks, embyronated chicken eggs, chick embryo fibroblasts, lice, or rabbit lungs. These vaccines failed to completely protect individuals from disease onset, but did reduce the severity of the symptoms and facilitated an expedited response to antibiotic therapy (DuPont et al., 1973; Kenyon et al., 1975; Oster et al., 1977; Clements et al., 1983). Live attenuated strains of R. prowazekii (Madrid E and Δpld) and R. rickettsii (Iowa) exist and have been demonstrated to be protective in vivo in animal models (Cox, 1941; Fox et al., 1957; Ellison et al., 2008; Driskell et al., 2009), and in the case of the R. prowazekii E strain, in human subjects as well (Fox et al., 1954, 1957).
Since CD8+ T lymphocytes play a crucial role in the anti-rickettsial adaptive immune response and are critical effectors against intracellular infections, anti-Rickettsia vaccine development has focused heavily on understanding how to optimally stimulate cell-mediated immunity. Numerous studies have elucidated regions of rompA and rompB in DNA immunizations that exhibit the capacity to prime T cells for INF-γ production and cellular proliferation in response to stimulation with formalin-fixed rickettsial antigen preparations (Crocquet-Valdes et al., 2001; Diaz-Montero et al., 2001; Li et al., 2003). Although these regions are able to activate these requisite cell-mediated mechanisms in vitro, their protective effects in vivo have been limited. In these studies, complete protection of mice was achieved only with a combination of DNA/protein vaccinations (Crocquet-Valdes et al., 2001; Diaz-Montero et al., 2001). Interestingly, adoptive transfer of CD8+ T cells from mice sub-lethally infected with R. conorii or R. typhi into naïve mice, fully protects mice against lethal homologous and heterologous rickettsial challenge (Valbuena et al., 2004), showing that effective priming of T lymphocytes can independently afford complete protection. Notably, the protection afforded by T lymphocytes was shown to be cross protective against divergent SFG and TG rickettsiae.
Humoral responses are believed to play a minor role in immune clearance of primary rickettsial infections, but instead contribute as a protective mechanism against re-infection. Several studies in mice and guinea pig animal models have examined the time-course of anti-SFG rickettsial antibody production in relation to disease resolution in sub-lethal rickettsial challenges, and determined that the antibody response matures after the primary infection has resolved, suggesting that the contribution of humoral immunity is in the prevention of secondary rickettsial infections (Lange and Walker, 1984; Feng et al., 2004a). Despite the minor role antibodies might play in resolving a primary rickettsial infection, anti-rickettsial antibodies have been demonstrated to perturb disease progression (Topping Norman, 1940; Anigstein et al., 1943; Feng et al., 2004a). Evidence from numerous studies indicate that antibodies against TG and SFG rickettsia species can facilitate bacterial clearance both in vivo and in vitro (Topping Norman, 1940; Anigstein et al., 1943; Gambrill and Wisseman, 1973; Beaman and Wisseman, 1976a,b; Lange and Walker, 1984; Anacker et al., 1985, 1987; Li et al., 1988; Keysary et al., 1989; Feng and Walker, 2003; Feng et al., 2004a,b). Humoral responses to live SFG rickettsial immunizations generate antibodies that specifically recognize high molecular weight rickettsial antigens (115–200 kDa), believed to be rOmpA and rOmpB based on SDS–PAGE migration, as well as protease-resistant species thought to be lipopolysaccharide (LPS) (Anacker et al., 1985; Li et al., 1988; Xu et al., 1997; Feng et al., 2004a). Characterization of monoclonal antibodies produced from these live rickettsial immunizations in various passive transfer studies have revealed that only a subset of antibodies recognizing epitopes belonging to the high molecular proteins afford protection, while those against LPS do not (Feng et al., 2004a). These findings led to the hypothesis that rOmpA and rOmpB could elicit protective immune responses in the host (Anacker et al., 1985, 1987; Li et al., 1988; Xu et al., 1997; Feng et al., 2004a); however, their roles as definitive protective antigens remain to be defined.
What may importantly be gleaned from these studies is that antibody recognition of single epitopes of single rickettsial antigens is potentially sufficient to neutralize rickettsial infection. In light of subsequent studies revealing the humoral immune-recognition of other high molecular weight Scas, including Sca1, Sca2, and Sca4 (Schuenke and Walker, 1994; Uchiyama et al., 1996; Uchiyama, 1997, and our unpublished data), it cannot be ruled out that the protective antibody responses attributed to rOmpA and rOmpB might also be associated with other less characterized rickettsial antigens. Notably, the humoral responses generated from a prior rickettsial infection have been seen to be long-lived and can be detected years following an infection (Bengtson and Topping, 1942; Fox et al., 1957). Though the robustness and longevity of anti-rickettsial humoral immunity must be examined in greater depth, these observations reasonably support the case that a vaccine comprised of one or more rickettsial antigens could be potentially efficacious. Further interrogation into the underlying features of rickettsial antigens necessary to elicit protective immunity is warranted.
Perspective
Exploration of the contribution of various conserved Sca outer membrane proteins have led to the current model of rickettsial invasion of host cells (Figure 3). An overlapping but distinct set of proteins mediate adherence and/or invasion of these pathogens. These multiple interactions contribute to activation of an array of signaling pathways that ultimately recruit actin to the entry focus and result in uptake of the invasive Rickettsia. Undoubtedly, this model of invasion does not represent a complete picture of the host and bacterial factors involved in the entry process. However, the currently defined interactions serve as primary targets for therapeutics. Through detailed biochemical and genetic analyses these host–pathogen interactions may be inhibited, thus resulting in a loss of bacterial invasion and death of the pathogen.
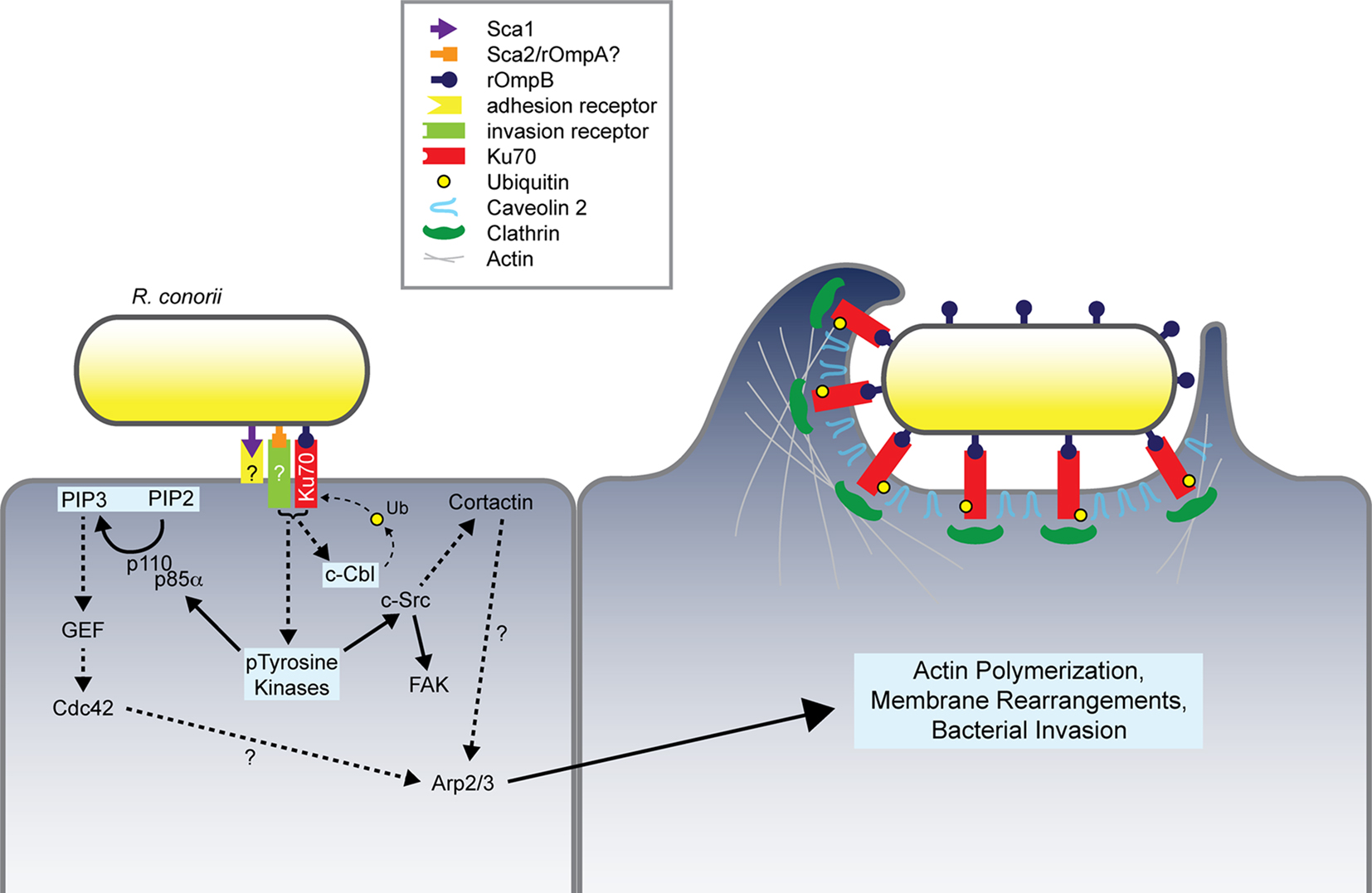
Figure 3. Model of Rickettsia conorii invasion of non-phagocytic mammalian cells. R. conorii interaction with host receptors, including Ku70, activates numerous signaling pathways whose coordinated activities lead to modulation of the actin cytoskeleton and predicted localized recruitment of endocytic players including clathrin and caveolin 2. R. conorii internalization depends on the stimulation of multiple pathways involving PTKs, Cdc42, PI 3-kinase, Src, FAK, c-Cbl, and cortactin, which ultimately result in Arp2/3-dependent actin polymerization at the bacterial entry foci. Pathways and proteins involved in rOmpB–Ku70-mediated bacterial invasion have been highlighted in light blue boxes. Dashed arrows and question marks indicate putative protein interplay during the invasion process.
A fact to be noted is the extraordinary amount of effort and creativity that was put into the above described research. The development of methods for selective deletion of rickettsial genes (Driskell et al., 2009) allows for direct examination of the contribution of adherence and invasion genes on pathogenesis of these bacteria. As it appears that many different rickettsial proteins contribute to invasion of host cells (Li and Walker, 1998; Renesto et al., 2006; Uchiyama et al., 2006; Cardwell and Martinez, 2009; Chan et al., 2009; Riley et al., 2010), it seems plausible that any single gene encoding these functions may be successfully deleted. These analyses will provide great insight into rickettsial diseases including the relative contribution of each protein to pathogenesis, their roles in disease progression or resolution, potential organ tropism, stimulation of host cellular signaling, and so forth. Determination of rickettsial factors and host proteins involved in Rickettsia interactions with target cells will provide valuable information both into the pathogenic process and also serve as prime targets for interventional therapy.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank members of the Martinez laboratory for their useful discussions with regard to the content of this review. Research in my laboratory was in part sponsored by NIH award RO1-AI072606-01 and NIH/NIAID Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (RCE) Program (NIH award 1-U54-AI-057153). The authors wish to acknowledge membership within and support from the Region V ‘Great Lakes’ RCE (GLRCE).
References
Alonso, A., and Garcia-del Portillo, F. (2004). Hijacking of eukaryotic functions by intracellular bacterial pathogens. Int. Microbiol. 7, 181–191.
Ammerman, N. C., Beier-Sexton, M., and Azad, A. F. (2008). Laboratory maintenance of Rickettsia rickettsii. Curr. Protoc. Microbiol. Chapter 3, Unit 3A 5.
Anacker, R. L., List, R. H., Mann, R. E., Hayes, S. F., and Thomas, L. A. (1985). Characterization of monoclonal antibodies protecting mice against Rickettsia rickettsii. J. Infect. Dis. 151, 1052–1060.
Anacker, R. L., McDonald, G. A., List, R. H., and Mann, R. E. (1987). Neutralizing activity of monoclonal antibodies to heat-sensitive and heat-resistant epitopes of Rickettsia rickettsii surface proteins. Infect. Immun. 55, 825–827.
Anderson, B. E., McDonald, G. A., Jones, D. C., and Regnery, R. L. (1990). A protective protein antigen of Rickettsia rickettsii has tandemly repeated, near-identical sequences. Infect. Immun. 58, 2760–2769.
Anigstein, L., Bader, M. N., and Young, G. (1943). Protective effect of separate inoculation of spotted fever virus and immune serum by intradermal route. Science 98, 285–286.
Balraj, P., Renesto, P., and Raoult, D. (2009). Advances in rickettsia pathogenicity. Ann. N.Y. Acad. Sci. 1166, 94–105.
Beaman, L., and Wisseman, C. L. Jr. (1976a). Mechanisms of immunity in typhus infections. V. Demonstration of Rickettsia mooseri-specific antibodies in convalescent mouse and human serum cytophilic for mouse peritoneal macrophages. Infect. Immun. 14, 1065–1070.
Beaman, L., and Wisseman, C. L. Jr. (1976b). Mechanisms of immunity in typhus infections. VI. Differential opsonizing and neutralizing action of human typhus rickettsia-specific cytophilic antibodies in cultures of human macrophages. Infect. Immun. 14, 1071–1076.
Bengtson, I. A., and Topping, N. H. (1942). Complement-fixation in rickettsial diseases. Am. J. Public Health Nations Health 32, 48–58.
Bise, G., and Coninx, R. (1997). Epidemic typhus in a prison in Burundi. Trans. R. Soc. Trop. Med. Hyg. 91, 133–134.
Blanc, G., Ngwamidiba, M., Ogata, H., Fournier, P. E., Claverie, J. M., and Raoult, D. (2005). Molecular evolution of rickettsia surface antigens: evidence of positive selection. Mol. Biol. Evol. 22, 2073–2083.
Cardwell, M. M., and Martinez, J. J. (2009). The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infect. Immun. 77, 5272–5280.
Chan, Y. G., Cardwell, M. M., Hermanas, T. M., Uchiyama, T., and Martinez, J. J. (2009). Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell Microbiol. 11, 629–644.
Clements, M. L., Wisseman, C. L. Jr., Woodward, T. E., Fiset, P., Dumler, J. S., McNamee, W., Black, R. E., Rooney, J., Hughes, T. P., and Levine, M. M. (1983). Reactogenicity, immunogenicity, and efficacy of a chick embryo cell-derived vaccine for Rocky Mountain spotted fever. J. Infect. Dis. 148, 922–930.
Cohn, Z. A., Bozeman, F. M., Campbell, J. M., Humphries, J. W., and Sawyer, T. K. (1959). Study on growth of Rickettsia. V. Penetration of Rickettsia tsutsugamushi into mammalian cells in vitro. J. Exp. Med. 109, 271–292.
Cossart, P. (2004). Bacterial invasion: a new strategy to dominate cytoskeleton plasticity. Dev. Cell 6, 314–315.
Cox, H. R. (1941). Cultivation of Rickettsiae of the Rocky Mountain spotted fever, typhus and Q fever groups in the embryonic tissues of developing chicks. Science 94, 399–403.
Crocquet-Valdes, P. A., Diaz-Montero, C. M., Feng, H. M., Li, H., Barrett, A. D., and Walker, D. H. (2001). Immunization with a portion of rickettsial outer membrane protein A stimulates protective immunity against spotted fever rickettsiosis. Vaccine 20, 979–988.
Croddy, E. A., Wirtz, J.J., and Larsen, J. A. (2005). “Typhus (Rickettsia prowazekii),” in Weapons of Mass Destruction: An Encyclopedia of Worldwide Policy, Technology, and History, eds E. A. Croddy and J. J. Wirtz (Santa Barbara: ABC-CLIO Inc.), 293.
Dasch, G. A. (1981). Isolation of species-specific protein antigens of Rickettsia typhi and Rickettsia prowazekii for immunodiagnosis and immunoprophylaxis. J. Clin. Microbiol. 14, 333–341.
Dasch, G. A., Samms, J. R., and Williams, J. C. (1981). Partial purification and characterization of the major species-specific protein antigens of Rickettsia typhi and Rickettsia prowazekii identified by rocket immunoelectrophoresis. Infect. Immun. 31, 276–288.
de Sousa, R., Nobrega, S. D., Bacellar, F., and Torgal, J. (2003). Mediterranean spotted fever in Portugal: risk factors for fatal outcome in 105 hospitalized patients. Ann. N.Y. Acad. Sci. 990, 285–294.
Diaz-Montero, C. M., Feng, H. M., Crocquet-Valdes, P. A., and Walker, D. H. (2001). Identification of protective components of two major outer membrane proteins of spotted fever group Rickettsiae. Am. J. Trop. Med. Hyg. 65, 371–378.
Driskell, L. O., Yu, X. J., Zhang, L., Liu, Y., Popov, V. L., Walker, D. H., Tucker, A. M., and Wood, D. O. (2009). Directed mutagenesis of the Rickettsia prowazekii pld gene encoding phospholipase D. Infect. Immun. 77, 3244–3248.
DuPont, H. L., Hornick, R. B., Dawkins, A. T., Heiner, G. G., Fabrikant, I. B., Wisseman, C. L. Jr., and Woodward, T. E. (1973). Rocky Mountain spotted fever: a comparative study of the active immunity induced by inactivated and viable pathogenic Rickettsia rickettsii. J. Infect. Dis. 128, 340–344.
Ellison, D. W., Clark, T. R., Sturdevant, D. E., Virtaneva, K., Porcella, S. F., and Hackstadt, T. (2008). Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect. Immun. 76, 542–550.
Emanuelsson, O., Brunak, S., von Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971.
Eremeeva, M. E., Klemt, R. M., Santucci-Domotor, L. A., Silverman, D. J., and Dasch, G. A. (2003). Genetic analysis of isolates of Rickettsia rickettsii that differ in virulence. Ann. N.Y. Acad. Sci. 990, 717–722.
Eto, D. S., Jones, T. A., Sundsbak, J. L., and Mulvey, M. A. (2007). Integrin-mediated host cell invasion by type 1-piliated uropathogenic Escherichia coli. PLoS Pathog. 3, e100. doi: 10.1371/journal.ppat.0030100.
Feng, H. M., and Walker, D. H. (2003). Cross-protection between distantly related spotted fever group rickettsiae. Vaccine 21, 3901–3905.
Feng, H. M., Whitworth, T., Olano, J. P., Popov, V. L., and Walker, D. H. (2004a). Fc-dependent polyclonal antibodies and antibodies to outer membrane proteins A and B, but not to lipopolysaccharide, protect SCID mice against fatal Rickettsia conorii infection. Infect. Immun. 72, 2222–2228.
Feng, H. M., Whitworth, T., Popov, V., and Walker, D. H. (2004b). Effect of antibody on the rickettsia-host cell interaction. Infect. Immun. 72, 3524–3530.
Fox, J. P., Everritt, M. G., Robinson, T. A., and Conwell, D. P. (1954). Immunization of man against epidemic typhus by infection with avirulent Rickettsia prowazekii (strain E); observations as to post-vaccination reactions, the relation of serologic response to size and route of infecting dose, and the resistance to challenge with virulent typhus strains. Am. J. Hyg. 59, 74–88.
Fox, J. P., Jordan, M. E., and Gelfand, H. M. (1957). Immunization of man against epidemic typhus by infection with avirulent Rickettsia prowazekii strain E. IV. Persistence of immunity and a note as to differing complement-fixation antigen requirements in post-infection and post-vaccination sera. J. Immunol. 79, 348–354.
Gambrill, M. R., and Wisseman, C. L. Jr. (1973). Mechanisms of immunity in typhus infections. 3. Influence of human immune serum and complement on the fate of Rickettsia mooseri within the human macrophages. Infect. Immun. 8, 631–640.
Gouin, E., Gantelet, H., Egile, C., Lasa, I., Ohayon, H., Villiers, V., Gounon, P., Sansonetti, P. J., and Cossart, P. (1999). A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J. Cell Sci. 112(Pt 11), 1697–1708.
Hackstadt, T., Messer, R., Cieplak, W., and Peacock, M. G. (1992). Evidence for proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect. Immun. 60, 159–165.
Haglund, C. M., Choe, J. E., Skau, C. T., Kovar, D. R., and Welch, M. D. (2010). Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat. Cell Biol. 12, 1057–1063.
Harris, S. (1992). Japanese biological warfare research on humans: a case study of microbiology and ethics. Ann. N.Y. Acad. Sci. 666, 21–52.
Hechemy, K. E., Raoult, D., Fox, J., Han, Y., Elliott, L. B., and Rawlings, J. (1989). Cross-reaction of immune sera from patients with rickettsial diseases. J. Med. Microbiol. 29, 199–202.
Henderson, I. R., and Lam, A. C. (2001). Polymorphic proteins of Chlamydia spp.– autotransporters beyond the Proteobacteria. Trends Microbiol. 9, 573–578.
Jacob-Dubuisson, F., Fernandez, R., and Coutte, L. (2004). Protein secretion through autotransporter and two-partner pathways. Biochim. Biophys. Acta. 1694, 235–257.
Kelly, D. J., Richards, A. L., Temenak, J., Strickman, D., and Dasch, G. A. (2002). The past and present threat of rickettsial diseases to military medicine and international public health. Clin. Infect. Dis. 34, S145–S169.
Kenyon, R. H., Sammons, L. S., and Pedersen, C. E. Jr. (1975). Comparison of three rocky mountain spotted fever vaccines. J. Clin. Microbiol. 2, 300–304.
Keysary, A., McCaul, T. F., and Winkler, H. H. (1989). Roles of the Fc receptor and respiratory burst in killing of Rickettsia prowazekii by macrophagelike cell lines. Infect. Immun. 57, 2390–2396.
Kleba, B., Clark, T. R., Lutter, E. I., Ellison, D. W., and Hackstadt, T. (2010). Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect. Immun. 78, 2240–2247.
Lange, J. V., and Walker, D. H. (1984). Production and characterization of monoclonal antibodies to Rickettsia rickettsii. Infect. Immun. 46, 289–294.
Li, H., Lenz, B., and Walker, D. H. (1988). Protective monoclonal antibodies recognize heat-labile epitopes on surface proteins of spotted fever group rickettsiae. Infect. Immun. 56, 2587–2593.
Li, H., and Walker, D. H. (1992). Characterization of rickettsial attachment to host cells by flow cytometry. Infect. Immun. 60, 2030–2035.
Li, H., and Walker, D. H. (1998). rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb. Pathog. 24, 289–298.
Li, Z., Diaz-Montero, C. M., Valbuena, G., Yu, X. J., Olano, J. P., Feng, H. M., and Walker, D. H. (2003). Identification of CD8 T-lymphocyte epitopes in OmpB of Rickettsia conorii. Infect. Immun. 71, 3920–3926.
Martinez, J. J., and Cossart, P. (2004). Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. J. Cell Sci. 117, 5097–5106.
Martinez, J. J., Mulvey, M. A., Schilling, J. D., Pinkner, J. S., and Hultgren, S. J. (2000). Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 19, 2803–2812.
Martinez, J. J., Seveau, S., Veiga, E., Matsuyama, S., and Cossart, P. (2005). Ku70, a component of DNA-dependent protein kinase, is a mammalian receptor for Rickettsia conorii. Cell 123, 1013–1023.
McDonald, G. A., Anacker, R. L., Mann, R. E., and Milch, L. J. (1988). Protection of guinea pigs from experimental Rocky Mountain spotted fever with a cloned antigen of Rickettsia rickettsii. J. Infect. Dis. 158, 228–231.
Mengaud, J., Ohayon, H., Gounon, P., Mege, R. M., and Cossart, P. (1996). E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84, 923–932.
Muller, C., Paupert, J., Monferran, S., and Salles, B. (2005). The double life of the Ku protein: facing the DNA breaks and the extracellular environment. Cell Cycle 4, 438–441.
Ngwamidiba, M., Blanc, G., Ogata, H., Raoult, D., and Fournier, P. E. (2005). Phylogenetic study of Rickettsia species using sequences of the autotransporter protein-encoding gene sca2. Ann. N.Y. Acad. Sci. 1063, 94–99.
Ngwamidiba, M., Blanc, G., Raoult, D., and Fournier, P. E. (2006). Sca1, a previously undescribed paralog from autotransporter protein-encoding genes in Rickettsia species. BMC Microbiol. 6, 12. doi:10.1186/1471-2180-6-12.
Oster, C. N., Burke, D. S., Kenyon, R. H., Ascher, M. S., Harber, P., and Pedersen, C. E. Jr. (1977). Laboratory-acquired Rocky Mountain spotted fever. The hazard of aerosol transmission. N. Engl. J. Med. 297, 859–863.
Palmer, E. L., Mallavia, L. P., Tzianabos, T., and Obijeski, J. F. (1974a). Electron microscopy of the cell wall of Rickettsia prowazekii. J. Bacteriol. 118, 1158–1166.
Palmer, E. L., Martin, M. L., and Mallavia, L. (1974b). Ultrastucture of the surface of Rickettsia prowazekii and Rickettsia akari. Appl. Microbiol. 28, 713–716.
Paupert, J., Dauvillier, S., Salles, B., and Muller, C. (2007). Transport of the leaderless protein Ku on the cell surface of activated monocytes regulates their migratory abilities. EMBO Rep. 8, 583–588.
Persson, C., Carballeira, N., Wolf-Watz, H., and Fallman, M. (1997). The PTPase YopH inhibits uptake of Yersinia, tyrosine phosphorylation of p130Cas and FAK, and the associated accumulation of these proteins in peripheral focal adhesions. EMBO J. 16, 2307–2318.
Popov, V. L., and Ignatovich, V. F. (1976). Electron microscopy of surface structures of Rickettsia prowazekii stained with ruthenium red. Acta Virol. 20, 424–428.
Ramm, L. E., and Winkler, H. H. (1973). Rickettsial hemolysis: adsorption of rickettsiae to erythrocytes. Infect. Immun. 7, 93–99.
Ramm, L. E., and Winkler, H. H. (1976). Identification of cholesterol in the receptor site for rickettsiae on sheep erythrocyte membranes. Infect. Immun. 13, 120–126.
Raoult, D., Ndihokubwayo, J. B., Tissot-Dupont, H., Roux, V., Faugere, B., Abegbinni, R., and Birtles, R. J. (1998). Outbreak of epidemic typhus associated with trench fever in Burundi. Lancet 352, 353–358.
Raoult, D., Roux, V., Ndihokubwayo, J. B., Bise, G., Baudon, D., Marte, G., and Birtles, R. (1997). Jail fever (epidemic typhus) outbreak in Burundi. Emerg. Infect. Dis. 3, 357–360.
Renesto, P., Samson, L., Ogata, H., Azza, S., Fourquet, P., Gorvel, J. P., Heinzen, R. A., and Raoult, D. (2006). Identification of two putative rickettsial adhesins by proteomic analysis. Res. Microbiol. 157, 605–612.
Rikihisa, Y., and Ito, S. (1979). Intracellular localization of Rickettsia tsutsugamushi in polymorphonuclear leukocytes. J. Exp. Med. 150, 703–708.
Riley, S. P., Goh, K. C., Hermanas, T. M., Cardwell, M. M., Chan, Y. G., and Martinez, J. J. (2010). The Rickettsia conorii autotransporter protein Sca1 promotes adherence to nonphagocytic mammalian cells. Infect. Immun. 78, 1895–1904.
Roux, V., and Raoult, D. (2000). Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB). Int. J. Syst. Evol. Microbiol. 50(Pt 4), 1449–1455.
Schuenke, K. W., and Walker, D. H. (1994). Cloning, sequencing, and expression of the gene coding for an antigenic 120-kilodalton protein of Rickettsia conorii. Infect. Immun. 62, 904–909.
Silverman, D. J., and Wisseman, C. L. Jr. (1978). Comparative ultrastructural study on the cell envelopes of Rickettsia prowazekii, Rickettsia rickettsii, and Rickettsia tsutsugamushi. Infect. Immun. 21, 1020–1023.
Silverman, D. J., Wisseman, C. L. Jr., Waddell, A. D., and Jones, M. (1978). External layers of Rickettsia prowazekii and Rickettsia rickettsii: occurrence of a slime layer. Infect. Immun. 22, 233–246.
Smith, D. K., and Winkler, H. H. (1979). Separation of inner and outer membranes of Rickettsia prowazekii and characterization of their polypeptide compositions. J. Bacteriol. 137, 963–971.
Stork, E., and Wisseman, C. L. Jr. (1976). Growth of Rickettsia prowazekii in enucleated cells. Infect. Immun. 13, 1743–1748.
Su, B., Johansson, S., Fallman, M., Patarroyo, M., Granstrom, M., and Normark, S. (1999). Signal transduction-mediated adherence and entry of Helicobacter pylori into cultured cells. Gastroenterology 117, 595–604.
Teysseire, N., Boudier, J. A., and Raoult, D. (1995). Rickettsia conorii entry into Vero cells. Infect. Immun. 63, 366–374.
Topping Norman, H. (1940). Rocky Mountain Spotted Fever, Treatment of Infected Laboratory Animals With Immune Rabbit Serum, Vol. 1. (Washington, DC: U.S. Govt. Print).
Turco, J., and Winkler, H. H. (1982). Differentiation between virulent and avirulent strains of Rickettsia prowazekii by macrophage-like cell lines. Infect. Immun. 35, 783–791.
Uchiyama, T. (1997). Intracytoplasmic localization of antigenic heat-stable 120- to 130-kilodalton proteins (PS120) common to spotted fever group rickettsiae demonstrated by immunoelectron microscopy. Microbiol. Immunol. 41, 815–818.
Uchiyama, T., Kawano, H., and Kusuhara, Y. (2006). The major outer membrane protein rOmpB of spotted fever group rickettsiae functions in the rickettsial adherence to and invasion of Vero cells. Microbes Infect. 8, 801–809.
Uchiyama, T., Zhao, L., and Uchida, T. (1996). Demonstration of a heat-stable 120-kilodalton protein of Rickettsia japonica as a spotted fever group-common antigen. Microbiol. Immunol. 40, 133–139.
Valbuena, G., Jordan, J. M., and Walker, D. H. (2004). T cells mediate cross-protective immunity between spotted fever group rickettsiae and typhus group rickettsiae. J. Infect. Dis. 190, 1221–1227.
Veiga, E., and Cossart, P. (2005). Listeria hijacks the clathrin-dependent endocytic machinery to invade mammalian cells. Nat. Cell Biol. 7, 894–900.
Vishwanath, S. (1991). Antigenic relationships among the rickettsiae of the spotted fever and typhus groups. FEMS. Microbiol. Lett. 65, 341–344.
Walker, D. H. (1989). Rocky Mountain spotted fever: a disease in need of microbiological concern. Clin. Microbiol. Rev. 2, 227–240.
Walker, D. H., Popov, V. L., Wen, J., and Feng, H. M. (1994). Rickettsia conorii infection of C3H/HeN mice. A model of endothelial-target rickettsiosis. Lab. Invest. 70, 358–368.
Walker, T. S. (1984). Rickettsial interactions with human endothelial cells in vitro: adherence and entry. Infect. Immun. 44, 205–210.
Walker, T. S., and Winkler, H. H. (1978). Penetration of cultured mouse fibroblasts (L cells) by Rickettsia prowazekii. Infect. Immun. 22, 200–208.
Winkler, H. H. (1974). Inhibitory and restorative effects of adenine nucleotides on rickettsial adsorption and hemolysis. Infect. Immun. 9, 119–126.
Winkler, H. H. (1977). Rickettsial hemolysis: adsorption, desorption, readsorption, and hemagglutination. Infect. Immun. 17, 607–612.
Winkler, H. H., and Ramm, L. E. (1975). Adsorption of typhus rickettsiae to ghosts of sheep erythrocytes. Infect. Immun. 11, 1244–1251.
Wisseman, C. L. Jr., Edlinger, E. A., Waddell, A. D., and Jones, M. R. (1976). Infection cycle of Rickettsia rickettsii in chicken embryo and L-929 cells in culture. Infect. Immun. 14, 1052–1064.
Wong, K. W., and Isberg, R. R. (2003). Arf6 and phosphoinositol-4-phosphate-5-kinase activities permit bypass of the Rac1 requirement for beta1 integrin-mediated bacterial uptake. J. Exp. Med. 198, 603–614.
Keywords: Rickettsia, adherence, invasion
Citation: Chan YGY, Riley SP and Martinez JJ (2010) Adherence to and invasion of host cells by spotted fever group Rickettsia species. Front. Microbio. 1:139. doi: 10.3389/fmicb.2010.00139
Received: 03 November 2010;
Paper pending published: 16 November 2010;
Accepted: 09 December 2010;
Published online: 20 December 2010
Edited by:
Robert Heinzen, National Institute of Health, National Institute of Allergy and Infectious Diseases-Rocky Mountain Laboratories, USAReviewed by:
Jon Audia, University of South Alabama School of Medicine, USAHélène Marquis, Cornell University, USA
Copyright: © 2010 Chan, Riley and Martinez. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Juan Jose Martinez, Department of Microbiology, Cummings Life Sciences Center, University of Chicago, 920 E. 58th Street, CLSC701A, Chicago, IL 60637, USA. e-mail:am1hcnRpbmVAYnNkLnVjaGljYWdvLmVkdQ==