
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 06 December 2010
Sec. Cellular and Infection Microbiology - closed section
Volume 1 - 2010 | https://doi.org/10.3389/fmicb.2010.00130
Circadian clock genes are vital features of eukaryotes that have evolved such that organisms can adapt to our planet’s rotation in order to anticipate the coming day or night as well as unfavorable seasons. This circadian clock uses oscillation as a timekeeping element. However, circadian clock mechanisms exist also in prokaryotes. The circadian clock of Cyanobacteria is well studied. It is regulated by a cluster of three genes: kaiA, kaiB, and kaiC. In this review, we will discuss the circadian system in cyanobacteria, and provide an overview and updated phylogenetic analysis of prokaryotic organisms that contain the main circadian genes. It is evident that the evolution of the kai genes has been influenced by lateral transfers but further and deeper studies are needed to get an in depth understanding of the exact evolutionary history of these genes. Interestingly, Legionella pneumophila an environmental bacterium and opportunistic human pathogen that parasitizes protozoa in fresh water environments also contains kaiB and kaiC, but their functions are not known. All of the residues described for the biochemical functions of the main pacemaker KaiC in Synechococcus elongatus are also conserved in the L. pneumophila KaiC protein.
Many living organisms from bacteria to eukaryotes have evolved internal mechanisms to synchronize their physiological functions with the Earth’s rotation cycle. These internal timing mechanisms of about 24 h are named circadian cycles. In the molecular context, circadian cycles are self-sustained biochemical oscillations that are temperature compensated and persist in absence of external environmental cues. This endogenous mechanism is able to respond to changes in the environment (like intensities of light and dark) and to entrain its phase to the local environmental cues. The distribution of circadian clocks in eukaryotic organisms is wide (Jolma et al., 2010). In contrast in prokaryotes the circadian clock genes are found only in some groups of bacteria (e.g., Cyanobacteria, Proteobacteria, Bacteroidetes) and archea (see Table 1 and Dvornyk et al., 2003). Among prokaryotes the circadian mechanism was first reported in the cyanobacterium Synechococcus elongatus PCC 7942 (Ishiura et al., 1998), which since then became a model to study this complex system. The circadian clock core genes in cyanobacteria are named kaiABC (kai after the Japanese word kaiten for a cycle or “turning of the heavens”).
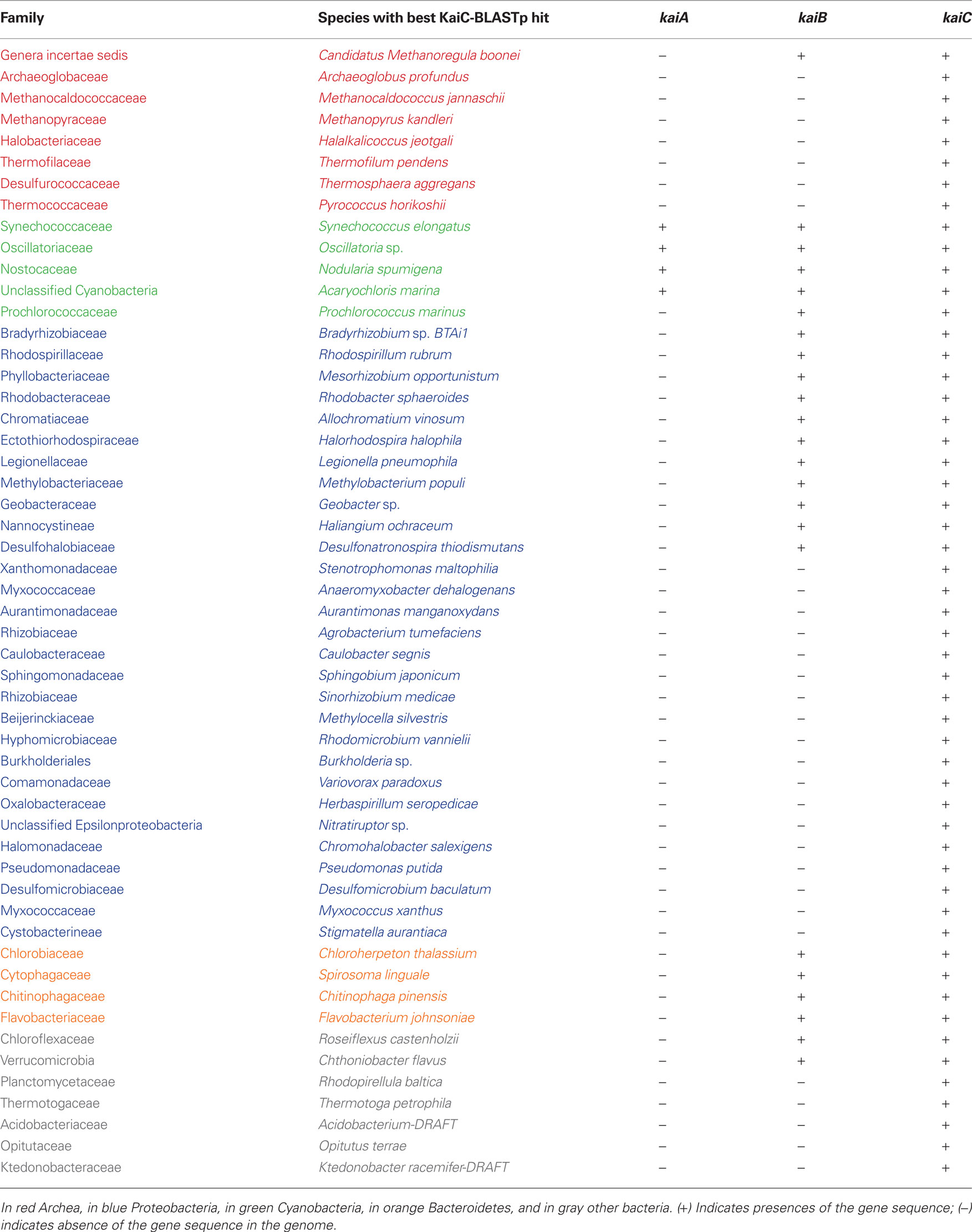
Table 1. Distribution of KaiA, KaiB, and KaiC homologues in completely sequenced prokaryotic genomes.
Cyanobacteria play an important role in the history of the Earth as the primary producers of oxygen (Tomitani et al., 2006). Since their appearance on Earth, they have evolved different strategies to survive the harsh environmental conditions they have to face like UV radiation, changes in temperature or changes in the redox state of the atmosphere (Garcia-Pichel, 1998). Furthermore, cyanobacteria are organisms that depend on the light/dark cycle, allowing oxygenic photosynthesis to occur during the day, in the presence of sunlight, and oxygen-sensitive nitrogen fixation during the dark period. Circadian clock systems have evolved as important tools that have been pivotal for their adaptation to a wide range of environmental conditions where cellular functions are linked to photochemical processes.
Apart from the circadian clock rhythm described and extensively studied in S. elongatus (Ishiura et al., 1998) there is only little evidence of circadian rhythms in other cyanobacterial species (Aoki and Onai, 2009) and even less in other prokaryotes that contain kai genes. The only other bacterium where the presence of a circadian-like timing mechanism was described is the purple bacterium Rhodobacter sphaeroides (Min et al., 2005). However, other prokaryotes apart from cyanobacteria and purple bacteria might be influenced by the daily Earth’s life cycle leading to enhanced fitness by improving their ability to adapt to extrinsic influences, specifically the daily changes in environmental factors such as light, temperature, and humidity (Woelfle et al., 2004). This might explain, why several other prokaryotes acquired the kai genes and components of the circadian system in their genomes. Insight into the roles of the circadian clock proteins in other prokaryotes would also provide a better understanding of their evolutionary history. These Kai proteins might be involved in metabolic processes, chromosome compaction (Woelfle et al., 2007), cell division (Mori et al., 1996; Mori, 2009), and UV stress resistance (Garcia-Pichel, 1998; Dvornyk and Nevo, 2003) as described for cyanobacteria or may have evolved divergent functions.
Surprisingly, L. pneumophila, an environmental bacterium parasitizing aquatic protozoa, but also an important human pathogen causing a severe pneumonia called Legionnaires’ disease, contains in its genome genes encoding homologues of KaiB and KaiC (Cazalet et al., 2004; Albert-Weissenberger et al., 2007). These genes are conserved in the sequenced L. pneumophila genomes, but KaiC of L. pneumophila strain Lens contains a frameshift at its N-terminus, that truncates the first walker motif. In contrast, these genes do not belong to the core genome of the genus Legionella, as Legionella longbeachae, does not encode putative circadian clock proteins (Cazalet et al., 2010) neither Legionella micdadei (unpublished) nor Legionella drancourtii. Intrigued by the identification of these proteins in L. pneumophila, we investigated the circadian clock genes in prokaryotes further. Here we provide an updated overview of prokaryotic organisms whose genome contains kai genes and analyze their evolutionary history. We then discuss what is known about the biochemistry of the Kai proteins and their roles reported for cyanobacteria and conclude with hypothesis of the role they could play in L. pneumophila.
The core element of the circadian system in the model organism S. elongatus PCC 7942 (hereafter S. elongatus) is a cluster of three tandemly located genes called: kaiA, kaiB, and kaiC that are essential for circadian rhythm in Synechococcus. Among these genes, kaiB and kaiC operate with the same promoter (Ishiura et al., 1998) and both genes are conserved among many other prokaryotes including several archea (Dvornyk et al., 2003). In some cyanobacterial genomes there are also paralogs of kaiB and kaiC at other loci (Dvornyk et al., 2003; Aoki and Onai, 2009). For example, Synechocystis sp. strain PCC6803 contains one copy of kaiA but kaiB and kaiC are present in triplicates in its genome, located in four different loci: kaiABC, kaiC2B2, kaiB3, and kaiC3 (Aoki and Onai, 2009). In contrast, kaiA is missing in many prokaryotic genomes that contain kaiBC. However, it has been shown that the marine cyanobacterium Prochlorococcus displays circadian cycles, despite the fact that it contains only the kaiB and kaiC genes but not kaiA, which has suffered a stepwise deletion, together with significant genome streamlining (Holtzendorff et al., 2008). Natural Prochlorococcus populations and laboratory cultures are strongly synchronized by the alternation of day and night, displaying 24-h rhythms in DNA replication. It was shown that the KaiB and KaiC proteins fulfill their known biochemical functions and that a mechanism that is less robust than the well-characterized KaiABC protein clock of Synechococcus may be sufficient for biological timing suited to the very stable environment where Prochlorococcus is found (Axmann et al., 2009). This finding opens the possibility that other prokaryotes like proteobacteria or archea that contain the kaiBC genes could also display an oscillator machinery similar to the one in Prochlorococcus. It was believed that the Kai proteins were ubiquitous among cyanobacteria, but an “exception to the rule” was reported when the genome of Gloeobacter violaceus PPC7421 was sequenced (Nakamura et al., 2003). This cyanobacterium lacks the kaiABC genes as well as the sasA, cikA, and pex genes, which are other major components of the clock system (see below). So far it is not known if G. violaceus displays circadian rhythmicity, possibly through another, not yet identified system, or not. However, this organism also shows ancestral characteristics for oxygenic photosynthesis and lacks thylakoids (Nakamura et al., 2003) characteristics reflecting its phylogenetic distance to other cyanobacteria.
Although cyanobacteria have become a major model system for analyzing clock phenomena in prokaryotes, efforts have been undertaken to get insight into the possible circadian rhythms of other prokaryotes. Indeed, recently a circadian oscillator has been reported for the purple bacterium Rhodobacter sphaeroides (Min et al., 2005). This proteobacterium displays circadian oscillations under aerobic conditions. Rh. Sphaeroides contains the kaiB and kaiC cyanobacterial homologous genes, but lacks the kaiA gene. One possible explanation of the absence of KaiA in Rh. sphaeroides could be that KaiB and KaiC have evolved functions different from those in cyanobacteria, and thus KaiA is not necessary anymore (Min et al., 2005). So far, the work of Min et al. (2005) has been the first report with evidence of a circadian rhythm in a proteobacterium. Rhythmic oscillations have also been reported in an other purple bacteria, Rhodospirillum rubrum (Van Praag et al., 2000), however it remains unknown if the gene expression in this organism is rhythmically controlled or not and whether the cyanobacterial homologs of kaiB and kaiC are involved.
Elucidation of the circadian clock behavior in purple bacteria as well as efforts to understand a possible circadian system in other non-photosynthetic proteobacteria may also help to get a better understanding on its evolution and origin(s). In a first approach to better understand the distribution and evolution of the circadian clock core genes, we have undertaken a search for the presence of kaiC genes in all completely sequenced prokaryotic and archeal genomes available in the NCBI database (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). To search for KaiC proteins we used the S. elongatus KaiC amino acid sequence as seed sequence and Blastp similarity searches. All hits covering at least 80% of the length of the S. elongatus KaiC protein sequence with a minimum e-value of e−5 were taken into account. Finally, the sequence showing the best hit with S. elongatus KaiC of each family of bacteria or archea, containing KaiC was chosen as representative for further analysis. In total 53 sequences belonging each to different species were retained (Table 1). To date (September 2010) the number of complete microbial genome sequences available in the NCBI database has increased to 1408 (1332 bacteria and 76 archea) allowing to identify additional organisms containing kai genes in their genome. Dvornyk et al. (2003) reported the presence of kai genes in four groups of organisms: Cyanobacteria, Proteobacteria, Archea, and Chloroflexi. The list of organisms has now increased in each of these groups (Table 1) and in addition we identified kai genes in the phylum Bacteroidetes. Furthermore, we have found independent cases of occurrence of the kai genes in different families of organisms that we named “other bacteria,” which comprise Chlamydiae, Planctomycetes, and Acidobacteria (Table 1). Interestingly, only Cyanobacteria (except Prochlorococcus sp.) encode KaiA in their genomes, but none of the other prokaryotes sequenced to date contain KaiA, even when they encode KaiB and KaiC or KaiC alone. KaiB sequences are present in all cyanobacteria sequenced and Bacteroidetes, but only in some proteobacteria. KaiB is present in some taxa belonging to the α-proteobacteria, γ-proteobacteria, and δ-proteobacteria, but not in β- or ε- proteobacteria listed in Table 1. This irregularity in the occurrence of circadian clock proteins in proteobacteria has also been described by Dvornyk et al. (2003). Here we report an interesting, new example, the family Legionellaceae. As stated above only L. pneumophila strains contain both, genes encoding homologues of KaiC and KaiB but none of the other Legionella species of which the genome sequence is known. Thus, L. pneumophila is the first bacterial human pathogen reported to contain the circadian genes (kaiBC). It will be an interesting question to answer whether they are involved in virulence and intracellular survival and replication of L. pneumophila or if they enhance its fitness in the environment.
Many different groups have been investigating the biological clock machinery of the cyanobacterial model S. elongatus PCC 7942 to get major insight into the mechanisms and the role of this system. In general, this circadian system shares three major components: (1) a central oscillator generating the fundamental rhythm of approximately 24 h; in S. elongatus the oscillator core consists of the KaiA, KaiB, and KaiC proteins (Ishiura et al., 1998); (2) an input pathway conveying environmental signals to the oscillator and modifying the oscillation to synchronize it precisely with the daily cycle. In S. elongatus the proteins CikA, LdpA, and Pex are key components for this input pathway (Kutsuna et al., 1998; Schmitz et al., 2000; Katayama et al., 2003; Ivleva et al., 2006); (3) an output pathway that relays temporal information from the oscillator to a variety of downstream biochemical processes. In the output pathway, the key players in S. elongatus are SasA (Iwasaki et al., 2000), RpaA (Takai et al., 2006), and LabA (Taniguchi et al., 2007) (Figure 1).
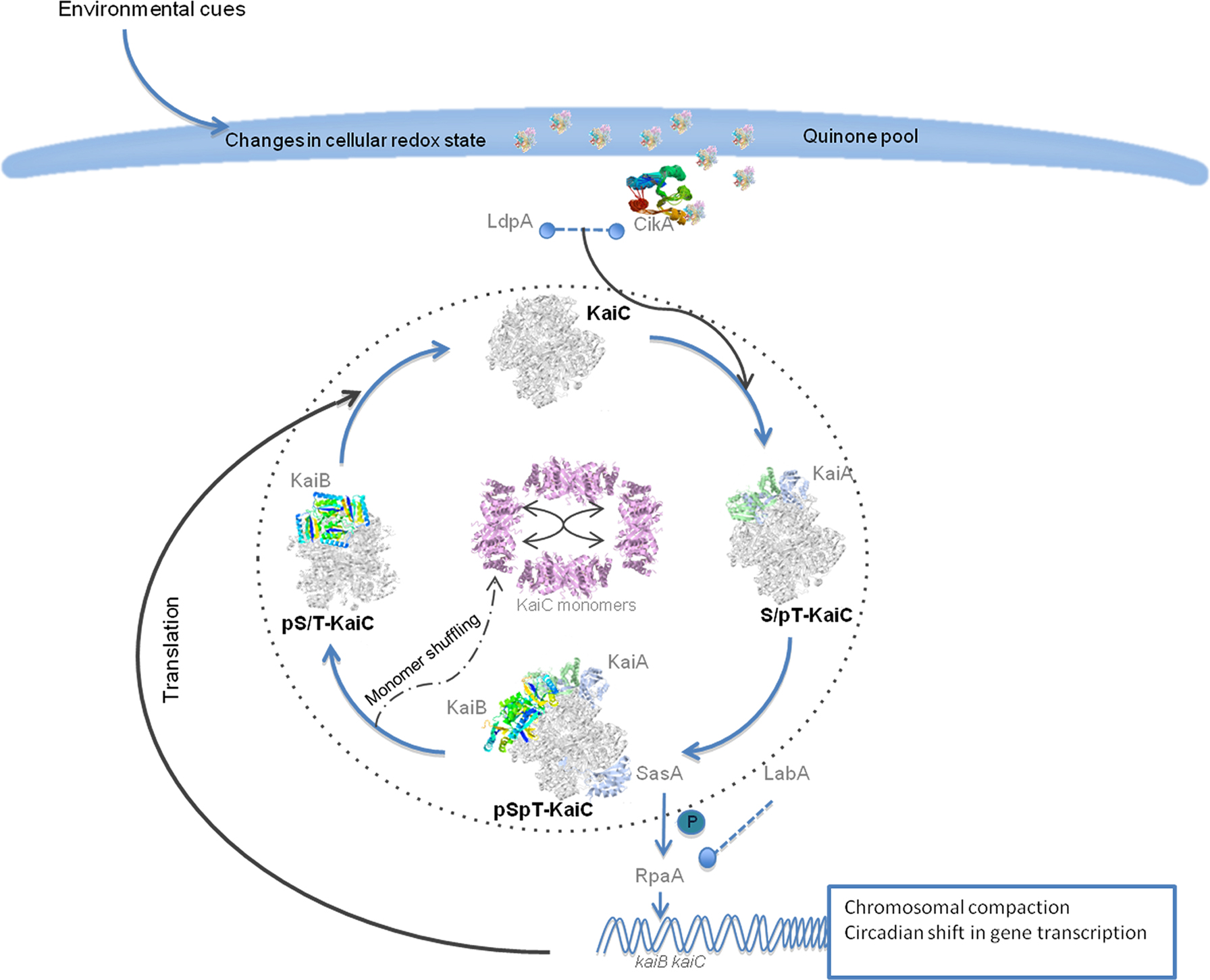
Figure 1. Schema of the molecular mechanisms of the circadian clock in S. elongatus. The input pathway responds to the environmental signals like light and temperature to entrain the circadian oscillator. Both CikA and LdpA can sense changes in the redox state of the cell. CikA interacts with quinones through its pseudo receiver domain. The components of the input pathway influence the phosphorylation state of the KaiC hexameric complex. The KaiA homodimer complex binds to KaiC and stimulates its phosphorylation. KaiC is first autophosphorylated at the T432 conserved residue and reaches the highest phosphorylation state when on each KaiC monomer the T321 and S431 residues are phosphorylated. Then KaiB tetramers bind to the KaiC complex and also SasA binds to the complex to transmit the signals downstream and to shift the global gene expression, regulate metabolic functions and regulate chromosomal compaction. The circadian cycle will have transcriptional–translational feedback from the global gene expression increasing its robustness. Finally the dephosphorylation state starts and in the early phase KaiC monomer shuffling occurs that might help to synchronize the clock with the environmental cues. The KaiC hexameric complex first loses the phosphorylation on the T432 and then in the S431 before the cycle starts again.
In bacteria, the Kai proteins constitute the core for the circadian system, but they do not have homologues in the genomes of mammals, insects or fungi. However, both prokaryotic and eukaryotic circadian systems show autoregulatory features that involve a negative feedback loop in which the clock genes encode proteins that repress their own transcription (Ishiura et al., 1998; Glossop et al., 1999; Cheng et al., 2001).
The input pathway of the circadian clock transmits signals from the environment to entrain the oscillator. The signals that can be sensed are usually light and temperature. For example, the circadian clock of S. elongatus can be reset by signals such as light and dark pulses (Schmitz et al., 2000) or pulses of high kaiC expression levels (Ishiura et al., 1998). The main components of the input pathway in S. elongatus are CikA (circadian input kinase) that sense the cellular redox state and environmental light intensity (Ivleva et al., 2006) and LdpA (light-dependent period) that is also sensitive to the redox state of the cell. CikA senses light and dark from the environment through the direct interaction with quinone molecules whose redox state depends on light and the metabolic redox state of the bacteria (Ivleva et al., 2006). CikA has a GAF domain, histidine protein kinase (HPK) and a pseudo receiver domain (PsR) necessary for quinone sensitivity (Mutsuda et al., 2003; Ivleva et al., 2006).
Co-purification assays showed that in addition to the Kai proteins CikA and LdpA form part of the periodosome (the heteromultimeric complex necessary to sustain circadian rhythms; Golden, 2004) together with a protein of the output pathway named SasA (Ivleva et al., 2005). LdpA entrains the clock with the intensity of the light through sensing the changes in electron transport and molecules like O2 or reactive oxygen species (Ivleva et al., 2005). The LdpA protein contains an HcyB domain and two specific, conserved terminal domains and belongs to the ferredoxin family of proteins (Katayama et al., 2003). cikA mutants are defective in the ability to reset the phase of the rhythm after a dark pulse and affect phosphorylation of KaiC (Ivleva et al., 2006). In contrast ldpA mutants show a conditional alteration in the circadian period as compared to the wild type due to an insensitivity to light gradient that normally modulates the circadian period resulting in a lengthening of the period at low light intensities. Thus the LdpA protein seems to modulate the circadian clock as an indirect function of light intensity by sensing changes in cellular physiology (Katayama et al., 2003). Another protein involved in the input pathway is Pex (period-extender) a transcriptional repressor of KaiA (Kutsuna et al., 1998). Constitutive expression of pex leads to a prolongation of the circadian period to 28 h. In contrast cells lacking pex, show a 1-h shorter circadian period due to the increase of KaiA that leads to a faster phosphorylation of KaiC (Kutsuna et al., 1998).
KaiC is the central oscillatory protein. Its primary sequence contains a double KaiC domain (CI and CII) and each domain contains a Walker motif that can bind ATP (Nishiwaki et al., 2000). KaiC belongs to the RecA/DnaB protein superfamily. Although the members of this family have the ability to bind DNA, KaiC lacks a DNA binding motif and does not show affinity for single or double stranded DNA. However, KaiC can bind forked DNA (Mori et al., 2002). KaiC is the only of the three proteins with enzymatic activity. It was reported to function as kinase, autophosphorylase, and ATPase (Nishiwaki et al., 2007; Terauchi et al., 2007) and that its activity is modulated through the interaction with KaiA and KaiB (Iwasaki et al., 2002; Kitayama et al., 2003). Indeed, self-sustainable oscillation of KaiC phosphorylation could be reconstituted in vitro by incubating KaiC with KaiA, KaiB, and adenosine triphosphate. The period of the in vitro oscillation was stable despite temperature change, and the circadian periods observed in vivo in KaiC mutant strains were consistent with those measured in vitro (Nakajima et al., 2005). The KaiC phosphorylation state persists in absence of transcriptional feedback and protein synthesis (Tomita et al., 2005), however the transcription and translation feedback increases the robustness of the circadian clock system (see Johnson et al., 2008).
KaiA forms a homodimer that interacts directly with hexamers of KaiC by binding at the C-terminal tail (Kim et al., 2008). This interaction stimulates its phosphorylation and inhibits dephosphorylation steps whereas tetramers of KaiB antagonize the effects of KaiA (Xu et al., 2003). In the in vitro system, Kai protein complexes assemble and disassemble dynamically over the KaiC phosphorylation cycle (Kageyama et al., 2006; Mori et al., 2007). In the initial step hexamers of KaiC are in unphosphorylated form. It associates with KaiA and subsequently KaiC autophosphorylates at residue T432 and then the double phosphorylated forms of KaiC appear (T432-S431-KaiC). This is the highest level of phosphorylation, KaiB binds to KaiC and when the KaiABC complex predominates the dephosphorylation phase starts. In the early phase of dephosphorylation “monomer shuffling” occurs among the KaiC hexamers probably helping to synchronize KaiC with the environmental cues (Ito et al., 2007). Finally, KaiC appears monophosphorylated at residue S431, dissociates from KaiB and the cycle starts a new (Nishiwaki et al., 2004, 2007; Rust et al., 2007) (Figure 1). Recently a third phosphorylation site, a threonine residue at position 42, has been reported to also be important to maintain rhythmicity (Xu et al., 2009). The phosphorylation state of the oscillator has been proposed as the pacemaker of the circadian clock, but there is evidence that circadian gene expression persists also when the phosphorylation cycle is disrupted (Kitayama et al., 2008).
Phosphorylation phase and ATPase activity levels of the oscillator complex determine the information transmitted to the downstream transcriptional regulatory system (Nishiwaki et al., 2004; Dong et al., 2010). This regulatory system is constituted of the SasA (Synechococcus adaptive sensor A)-RpaA (regulator of phycobilisome-associated) two-component system (Takai et al., 2006). The HPK SasA forms a complex with KaiC by binding KaiC through its KaiB-like sensory domain (Iwasaki et al., 2000) leading to autophosphorylation and the transfer of a phosphate to the DNA binding response regulator RpaA (Takai et al., 2006). Both, SasA autophosphorylation and the SasA-to-RpaA phosphotransfer are modulated according to the KaiC phosphorylation state. Consequently the target genes of RpaA can be activated or repressed in a circadian manner. Thus the KaiC-SasA-RpaA interaction is so far the major positive pathway known to regulate the circadian shift in gene expression (Takai et al., 2006). Mutants in sasA grow slowly in comparison to the wild type bacteria in a light/dark cycle (Iwasaki et al., 2000) and produce unstable short periods of transcription rhythms (Takai et al., 2006). Null mutants of rpaA show arrhythmia under continues light conditions (Takai et al., 2006).
An alternative output pathway by which the circadian clock modulates circadian gene expression is LabA (low-amplitude and bright). LabA is needed for negative feedback regulation of KaiC (Taniguchi et al., 2007). Mutants in labA increase the levels of circadian gene expression due to the high levels of non-regulated-KaiC resulting in a low amplitude phenotype. In contrast, labA overexpression results in low circadian gene expression (Taniguchi et al., 2007). Furthermore, it has been suggested that the LabA pathway is implicated in chromosomal compaction (Woelfle et al., 2007). Both pathways LabA and SasA work in parallel to regulate the expression of kaiBC in S. elongates. RpaA is situated downstream in this cascade to generate the robust modulation of circadian gene expression (Kondo, 2007) (Figure 1).
Many efforts have been undertaken to decipher the biochemical basis of the oscillatory machinery and the pathways it controls. However, most approaches have been based on genetics, as it is not easy to predict and model the biochemical processes for such a complex mechanism. Thus, mainly mutants that target a component gene of the clock machinery have been analyzed to infer their possible roles and their involvement in the circadian clock. Most of the knowledge about the cellular processes that the circadian clock gates comes from alterations in such cellular functional pathways as result of the mutation of one clock component or by determining the rhythmicity of cellular processes.
One of the first physiological evidence that suggested the presence of circadian clock mechanisms in cyanobacteria was the observation that nitrogen fixation onset occurs in a rhythmic way (Grobbelaar et al., 1986). Apart from photosynthesis and nitrogen fixation, circadian clocks regulate amino acid uptake (Chen et al., 1991), cell division (Sweeney and Borgese, 1989; Mori et al., 1996), global patterns of gene expression (Liu et al., 1995), and topological chromosomal compaction (Smith and Williams, 2006). Circadian expression in S. elongatus was shown to be pervasive using bioluminescence to monitor the promoter activity of many different genes. Liu et al. (1995) reported that no special set of genes controlled by the circadian clock exists, but that many of these genes probably are regulated by cis- and trans-acting regulatory factors that can incorporate non-specific circadian control and circadian regulation from components in the downstream circadian output pathway.
Many mechanisms that impact the functioning of the cell and how they make the circadian clock work, remain to be discovered and deciphered, but new parts in the puzzle are added constantly. For example it was known for many years that the circadian clock gates the cell cycle process but the mechanisms that allow this phenomenon were unknown (Mori et al., 1996; Mori and Johnson, 2000). However, very recently Dong et al. (2010) reported that the circadian clock has a checkpoint on cell division that can inhibit cytokinesis at specific points of the circadian cycle. Normally, the “gate” is open so cell division can happen independently of the circadian clock (Mori et al., 1996; Mori, 2009), but the gate closes when high levels of KaiC ATPase activity are present. Then, the signal downstream is transmitted through the SasA-RpaA pathway and it results in the inhibition of the midcell FtsZ ring assembly, blocking cell division until the levels of ATPase activity decrease (Dong et al., 2010). The biological implication of the circadian checkpoint in cell division may impact in different ways by protecting other cellular functions that can be damaged as result of cell division in vulnerable conditions (Dong et al., 2010). It is important to notice that in S. elongatus the gate occurs with the onset of darkness when oxidative stress and UV damage would be lower than during the day, but cell division is not restricted to the dark period. In addition, DNA replication also remains constant during the circadian cycle suggesting that the most evident target is cytokinesis (Mori et al., 1996; Mori, 2009; Dong et al., 2010). Much knowledge to better understand the biological significance of the circadian clock gating different cell functions has been gained in the last years, but further studies are needed to complete our understanding of the impact of the prokaryotic circadian clock on the global physiology of the cell and research should be expanded on putative circadian clocks of other prokaryotes.
Considering that the biochemistry of the KaiC protein depends on conserved motifs (Walker motifs) for its activity as ATPase and some conserved residues for its autokinase activity, we searched for the presence of these motifs in other prokaryotic organisms, and specifically in L. pneumophila. We selected those prokaryotes that contained both, kaiB and kaiC sequences including L. pneumophila KaiC (Table 1) and aligned the protein sequence using the program ClustalX v.2.0.12 (Larkin et al., 2007) (Figure 2). Interestingly, all prokaryotic KaiC sequences aligned contained a well conserved Walker motif (GxxxxGK(T/S)) in the first KaiC domain (Figure 2). It has been reported that in bacteria the KaiC proteins usually contain a double KaiC domain but in some archea shorter KaiC sequences containing only a single domain can occur (Dvornyk et al., 2003), as also shown in Figure 2. Those organisms that have both KaiC domains show also a well conserved second Walker motif, but L. pneumophila and Flavobacterium johnsoniae exhibit a variant in the motif (GxxxxAK(T/S)). Furthermore, in most of the KaiC protein sequences, the potential N-terminal phosphorylation sites are well conserved (T426, S431, and T432 in S. elongatus). However, the T426 residue is also missing in F. johnsoniae and three δ-proteobacteria (Figure 2). The S431 phosphorylation residue is not conserved in Allochromatium vinosum and the T432 residue is replaced by an S residue in L. pneumophila, and others like Geobacter sp., Rhodobacter sphaeroides, and Spirosoma linguale but that it may still be a phosphorylation target residue (Figure 2).
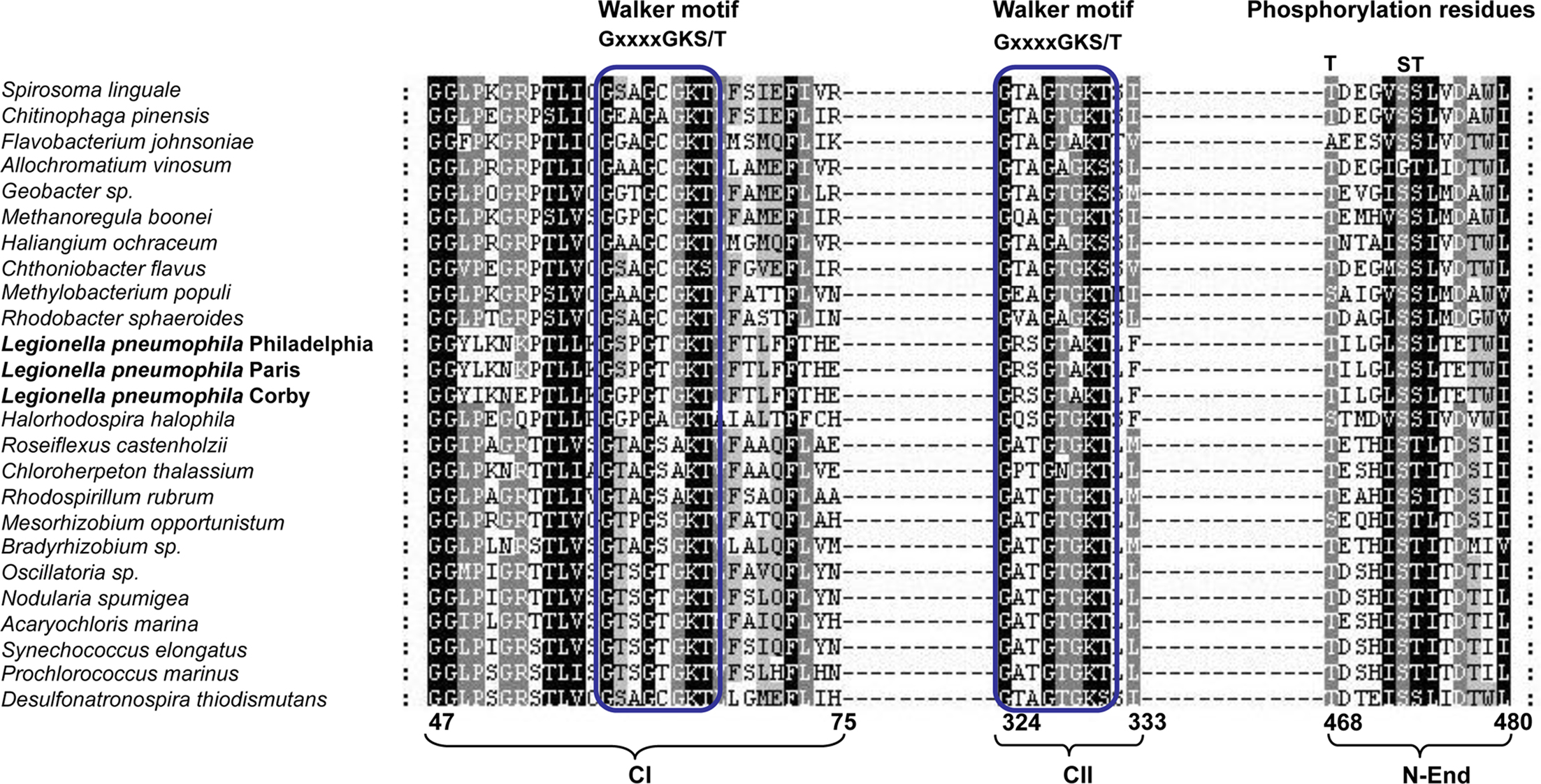
Figure 2. Alignment of the KaiC sequences from prokaryotes containing both kaiB and kaiC sequences in their genome. Walker motifs and phosphorylation residues are indicated. The numbers below indicate the amino acid position with respect to the full protein sequence. Parentheses below the alignment indicate the location of each motif or residue: CI (first KaiC domain), CII (second KaiC domain), and N-End (N-terminus of the protein).
In S. elongates, mutations in any of the kai genes lead to arrhythmic patterns of the luciferase gene driven by different promoters (Ishiura et al., 1998). However, circadian clock systems exist also in organisms that do not have the kaiA gene like P. marinus and Rh. sphaeroides. With respect to the evolutionary analysis of the circadian components in cyanobacteria it was logical to infer that in cyanobacteria more than one circadian system might exist. First evidence came from the finding that some cyanobacteria lack the kaiA gene (Dvornyk et al., 2003), one of the three circadian core proteins in S. elongatus PCC7942 (Ishiura et al., 1998) but still have circadian rhythms. Based on these observations, Dvornyk and colleagues proposed that different circadian systems might exist in bacteria. The kaiABC system that has been extensively studied in S. elongatus PCC7942, and the kaiBC system that evolved probably from the kaiABC through the loss of kaiA and other components of the input or output circadian pathways like cikA (Baca et al., 2010). An example of the kaiBC system is the circadian-like rhythm present in the purple bacterium Rh. sphaeroides (Min et al., 2005).
Although, circadian systems might also be present in other prokaryotes, the kai sequences in these prokaryotes might also have evolved functional divergence/convergence at least in those prokaryotes that do not show signs of lateral transfer. To get further insight into the evolution of the KaiC proteins we have undertaken an updated phylogenetic analysis of the KaiC sequence of all organisms listed in Table 1. In order to have a phylogenetic marker allowing to infer possible horizontal gene transfer, we also retrieved the RpoB sequence for the selected organisms from the NCBI database. As expected the phylogeny based on RpoB allowed good phylogenetic resolution comparable to the one obtained with the 16S rRNA. However, the advantage is, that rpoB is a single copy gene and thus no intra-genomic heterogeneity among copies as seen for the 16S rRNA is present. Both groups of sequences were aligned using the program ClustalX v.2.0.9 (Larkin et al., 2007). The best evolutionary model was selected using Prottest (Abascal et al., 2005). Using the best model according to AIC criterion, phylogenetic trees were constructed using both a distance method (Neighbor-joining) and likelihood with 300 replicates of bootstrap. Distance methods were run using MEGA4 (Tamura et al., 2007) with the option “Pairwise deletion” in the case of RpoB and the likelihood tree was obtained by using Phyml (Guindon et al., 2005). As shown in Figure 3, the RpoB tree allowed to clearly separate the different taxonomic groups and was supported by high bootstrap values in both, distance and likelihood trees. Both methods gave very similar topologies and the few differences involved only the positions of bacteria not belonging to any of the main taxonomic groups (classified as “Others” in Figure 3).
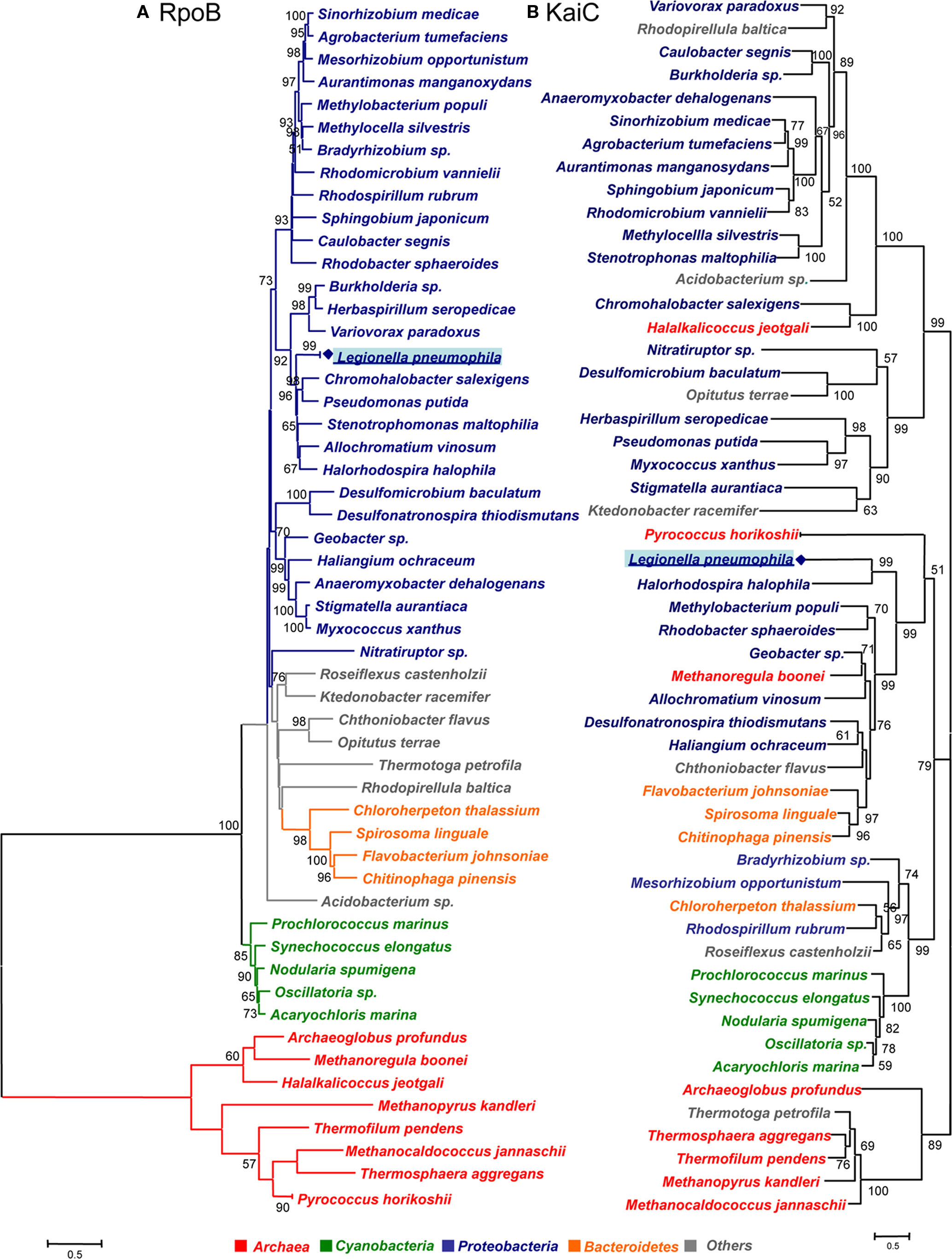
Figure 3. Phylogenetic tree of rpoB and kaiC sequences of prokaryotes containing kaiB and kaiC in their genome. (A) RpoB amino acid sequence tree obtained by the neighbor-joining method. (B) KaiC amino acid sequences obtained by the likelihood method. The bootstrap values presented at corresponding nodes were obtained from 300 replicates. Values under 50 are not indicated.
Phylogenetic analysis of the KaiC sequences, gave also very similar topologies with both methods although not identical. The main difference was that in the likelihood tree (Figure 3), the clade containing Legionella, some Proteobacteria and Bacteroidetes grouped together with the clade containing among others Cyanobacteria, however in the distance tree the mentioned clade grouped with the rest of proteobacteria. Whereas in the likelihood tree the grouping node was supported by a bootstrap value of 79, there was no bootstrap value supporting this node in the distance tree. Thus we refer in our analysis to the likelihood tree shown in Figure 3.
Interestingly, the organisms containing both the kaiB and kaiC genes grouped in two main clades: one containing cyanobacteria and some α-Proteobacteria and a second clade containing two subclades of Proteobacteria and Bacteroidetes. In contrast, most of the organisms that do not contain the kaiB gene sequence form another clade. Thus our phylogenetic analysis grouped organisms containing the putative KaiABC or KaiBC systems in prokaryotes in a coherent way (Figure 4). Although the KaiC tree groups the main taxonomical groups (Cyanobacteria, and Proteobacteria) in separated clades, numerous non-correspondences with respect to the RpoB tree topology are indicating possible multiple horizontal transfer events in the evolution of this gene as previously suggested (Dvornyk et al., 2003). Some clear evidence of horizontal transfer events well supported are the ones from proteobacteria to the archea Halalkalicoccus jeotgali or from proteobacteria to the Ktedonobacteria Ktedonobacter racemifer. In contrast for L. pneumophila we cannot infer clear evidence of horizontal gene transfer since Legionella is grouping with other Proteobacteria in the same clade. However the fact that this clade groups with Cyanobacteria and less with other proteobacteria may indicate either a lack of phylogenetic resolution of this gene or a different origin for KaiC in different Proteobacteria resulting from horizontal transfer or convergent evolution.
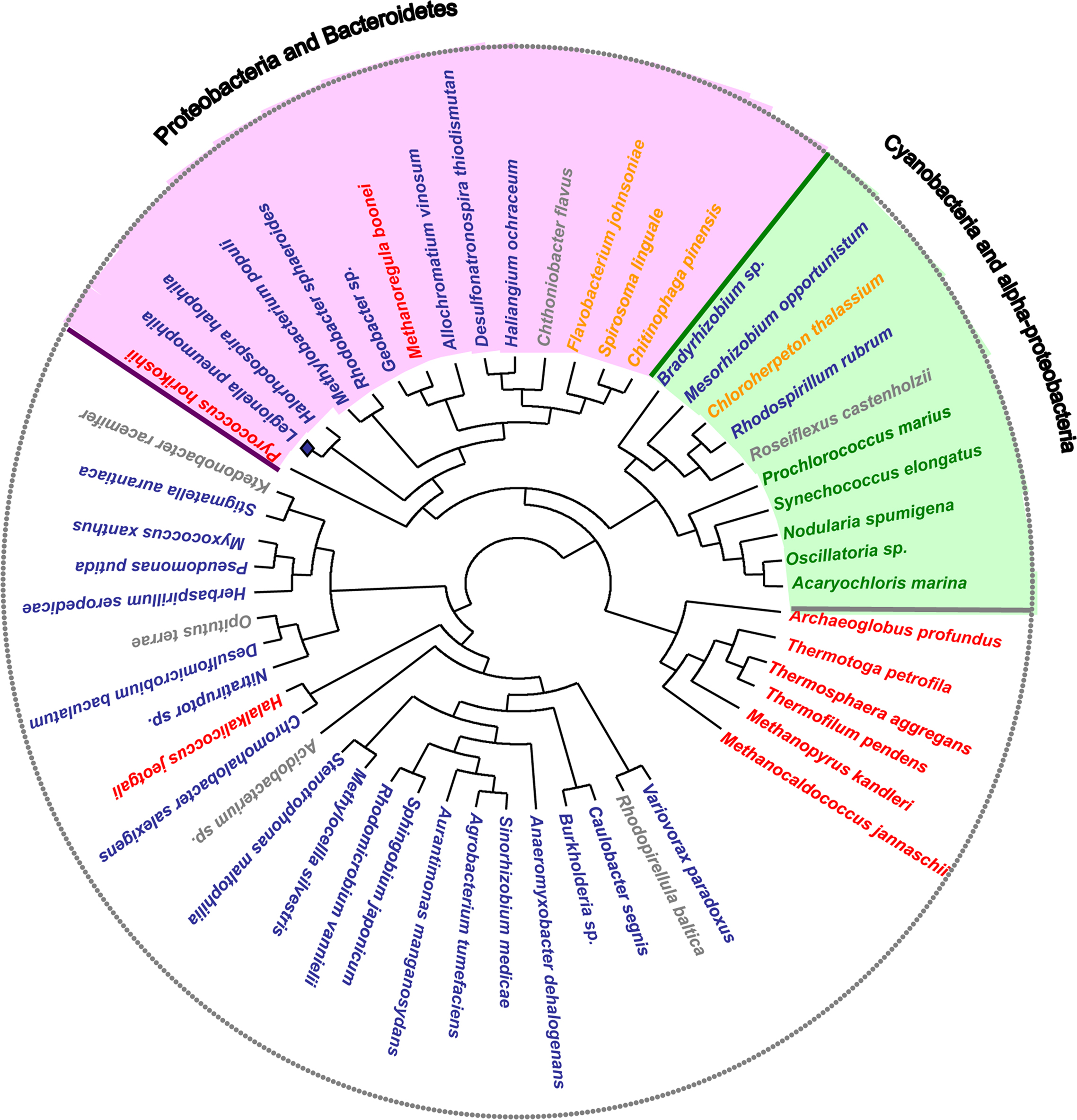
Figure 4. Maximum likelihood unrooted KaiC amino acid sequence tree. Organisms containing both kaiB and kaiC systems are grouped in two main clades: Cyanobacteria and α-Proteobacteria (green), and Proteobacteria and Bacteroidetes (pink). In these groups some organisms that do not belong to the main phylogenetic group are present. This grouping indicates potential events of lateral gene transfer.
Circadian rhythms are daily cycles of metabolic activity, gene expression, and other biological processes that are regulated by self-sustained intracellular oscillators. Cyanobacteria are among the oldest organisms on earth and they are very successful in ecological plasticity and adaptability. Cyanobacteria were the first prokaryotes reported to have the circadian clock regulated by three genes, kaiA, kaiB, and kaiC allowing adaptation and enhancing fitness in response to changing environments. The massive acquisition of genome sequence of the most diverse organisms in the last years has now identified many additional prokaryotic and archeal genomes that contain homologues of kai genes. Although the model circadian clock is composed of the kaiABC genes, only some cyanobacteria encode KaiA, which is also not present in the rest of the prokaryotes containing the kaiB and/or kaiC genes. However, recently it was shown that the cyanobacterium Prochlorococcus sp. strain MED4 lacking kaiA displays the same biochemical functions than those from S. elongatus, having a KaiABC system. It might be expected that an evolutive adjustment of mechanistical features of the cyclic process occurred due to the absence of kaiA. Indeed Axmann et al. (2009) have shown that there is a difference in the effect that KaiB exerts on KaiC dephosphorylation in Prochlorococcus sp. strain MED4. Thus, also in other bacteria timing mechanism might exist that are regulated by the kaiBC system. It will be challenging to investigate the function of the diverse Kai homologues present in prokaryotes and archea. We are particularly interested in the KaiBC proteins encoded by L. pneumophila an environmental organism that also inhabits natural fresh waters like cyanobacteria. Thus L. pneumophila is also exposed to a series of photochemical reactions in water that could lead to the production of reactive oxygen species. Furthermore, L. pneumophila is an intracellular pathogen that parasitizes fresh water protozoa and that can also infect human alveolar macrophages causing a sever pneumonia. L. pneumophila has thus also evolved mechanisms to evade the host defense and the kaiBC proteins might influence the adaptation capabilities and enhancing fitness of L. pneumophila in these hostile conditions. Further studies will provide new knowledge and a better understanding of the function of the circadian clock proteins and their evolution in bacteria different from cyanobacteria.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work received financial support from the Institut Pasteur and the Centre National de la Recherche (CNRS). Maria Loza-Correa is holder of a fellowship from the Ministère délégué à l’enseignement supérieur et à la recherche (France) and Laura Gomez-Valero is holder of a postdoctoral fellowship from the Fondation pour la Recherche Médicale (FRM).
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105.
Albert-Weissenberger, C., Cazalet, C., and Buchrieser, C. (2007). Legionella pneumophila – a human pathogen that co-evolved with fresh water protozoa. Cell. Mol. Life Sci. 64, 432–448.
Aoki, S., and Onai, K. (2009). “Circadian clocks of Synechocystis sp. strain PCC 6803, Thermosynechococcus elongatus, Prochlorococcus spp., Trichodesmium spp. and other species,” in Bacterial Circadian Programs, eds J. L. Ditty, S. Mackey and C. H. Johnson, (Berlin, Heidelberg: Springer), 241–258.
Axmann, I. M., Duhring, U., Seeliger, L., Arnold, A., Vanselow, J. T., Kramer, A., and Wilde, A. (2009). Biochemical evidence for a timing mechanism in prochlorococcus. J. Bacteriol. 191, 5342–5347.
Baca, I., Sprockett, D., and Dvornyk, V. (2010). Circadian input kinases and their homologs in Cyanobacteria: evolutionary constraints versus architectural diversification. J. Mol. Evol. 70, 453–465.
Cazalet, C., Gomez-Valero, L., Rusniok, C., Lomma, M., Dervins-Ravault, D., Newton, H. J., Sansom, F. M., Jarraud, S., Zidane, N., Ma, L., Bouchier, C., Etienne, J., Hartland, E. L., and Buchrieser, C. (2010). Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires’ disease. PLoS Genet. 6, e1000851. doi: 10.1371/journal.pgen.1000851.
Cazalet, C., Rusniok, C., Bruggemann, H., Zidane, N., Magnier, A., Ma, L., Tichit, M., Jarraud, S., Bouchier, C., Vandenesch, F., Kunst, F., Etienne, J., Glaser, P., and Buchrieser, C. (2004). Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36, 1165–1173.
Chen, T. H., Chen, T. L., Hung, L. M., and Huang, T. C. (1991). Circadian rhythm in amino acid uptake by Synechococcus RF-1. Plant Physiol. 97, 55–59.
Cheng, P., Yang, Y., and Liu, Y. (2001). Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc. Natl. Acad. Sci. U.S.A. 98, 7408–7413.
Dong, G., Yang, Q., Wang, Q., Kim, Y. I., Wood, T. L., Osteryoung, K. W., van Oudenaarden, A., and Golden, S. S. (2010). Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell 140, 529–539.
Dvornyk, V., and Nevo, E. (2003). Genetic polymorphism of cyanobacteria under permanent natural stress: a lesson from the “Evolution Canyons”. Res. Microbiol. 154, 79–84.
Dvornyk, V., Vinogradova, O., and Nevo, E. (2003). Origin and evolution of circadian clock genes in prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 100, 2495–2500.
Garcia-Pichel, F. (1998). Solar ultraviolet and the evolutionary history of cyanobacteria. Orig. Life Evol. Biosph. 28, 321–347.
Glossop, N. R., Lyons, L. C., and Hardin, P. E. (1999). Interlocked feedback loops within the Drosophila circadian oscillator. Science 286, 766–768.
Golden, S. S. (2004). Meshing the gears of the cyanobacterial circadian clock. Proc. Natl. Acad. Sci. U.S.A. 101, 13697–13698.
Grobbelaar, N., Huang, T. C., Lin, H. Y., Chow, T. J. u., and T. C. (1986). Dinitrogen-fixing endogenous rhythm in Synechococcus RF-1. FEMS Microbiol. Lett. 37, 173–177.
Guindon, S., Lethiec, F., Duroux, P., and Gascuel, O. (2005). PHYML Online – a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33, W557– W559.
Holtzendorff, J., Partensky, F., Mella, D., Lennon, J. F., Hess, W. R., and Garczarek, L. (2008). Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J. Biol. Rhythms 23, 187–199.
Ishiura, M., Kutsuna, S., Aoki, S., Iwasaki, H., Andersson, C. R., Tanabe, A., Golden, S. S., Johnson, C. H., and Kondo, T. (1998). Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523.
Ito, H., Kageyama, H., Mutsuda, M., Nakajima, M., Oyama, T., and Kondo, T. (2007). Autonomous synchronization of the circadian KaiC phosphorylation rhythm. Nat. Struct. Mol. Biol. 14, 1084–1088.
Ivleva, N. B., Bramlett, M. R., Lindahl, P. A., and Golden, S. S. (2005). LdpA: a component of the circadian clock senses redox state of the cell. EMBO J. 24, 1202–1210.
Ivleva, N. B., Gao, T., LiWang, A. C., and Golden, S. S. (2006). Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc. Natl. Acad. Sci. U.S.A. 103, 17468–17473.
Iwasaki, H., Nishiwaki, T., Kitayama, Y., Nakajima, M., and Kondo, T. (2002). KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 99, 15788–15793.
Iwasaki, H., Williams, S. B., Kitayama, Y., Ishiura, M., Golden, S. S., and Kondo, T. (2000). A kaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell 101, 223–233.
Johnson, C. H., Egli, M., and Stewart, P. L. (2008). Structural insights into a circadian oscillator. Science 322, 697–701.
Jolma, I. W., Laerum, O. D., Lillo, C., and Ruoff, P. (2010). Circadian oscillators in eukaryotes. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 533–549.
Kageyama, H., Nishiwaki, T., Nakajima, M., Iwasaki, H., Oyama, T., and Kondo, T. (2006). Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol. Cell 23, 161–171.
Katayama, M., Kondo, T., Xiong, J., and Golden, S. S. (2003). ldpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC 7942. J. Bacteriol. 185, 1415–1422.
Kim, Y. I., Dong, G., Carruthers, C. W. Jr., Golden, S. S., and LiWang, A. (2008). The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 12825–12830.
Kitayama, Y., Iwasaki, H., Nishiwaki, T., and Kondo, T. (2003). KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 22, 2127–2134.
Kitayama, Y., Nishiwaki, T., Terauchi, K., and Kondo, T. (2008). Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes Dev. 22, 1513–1521.
Kondo, T. (2007). A cyanobacterial circadian clock based on the Kai oscillator. Cold Spring Harb. Symp. Quant. Biol. 72, 47–55.
Kutsuna, S., Kondo, T., Aoki, S., and Ishiura, M. (1998). A period-extender gene, pex, that extends the period of the circadian clock in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 180, 2167–2174.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948.
Liu, Y., Tsinoremas, N. F., Johnson, C. H., Lebedeva, N. V., Golden, S. S., Ishiura, M., and Kondo, T. (1995). Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 9, 1469–1478.
Min, H., Guo, H., and Xiong, J. (2005). Rhythmic gene expression in a purple photosynthetic bacterium, Rhodobacter sphaeroides. FEBS Lett. 579, 808–812.
Mori, T. (2009). “Cell division and circadian clock,” in Bacterial Circadian Programs, eds J. L. Ditty, S. Mackey, and C. H. Johnson (Berlin/Heidelberg: Springer-Verlag), 241–258.
Mori, T., Binder, B., and Johnson, C. H. (1996). Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc. Natl. Acad. Sci. U.S.A. 93, 10183–10188.
Mori, T., and Johnson, C. H. (2000). Circadian control of cell division in unicellular organisms. Prog. Cell Cycle Res. 4, 185–192.
Mori, T., Saveliev, S. V., Xu, Y., Stafford, W. F., Cox, M. M., Inman, R. B., and Johnson, C. H. (2002). Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc. Natl. Acad. Sci. U.S.A. 99, 17203–17208.
Mori, T., Williams, D. R., Byrne, M. O., Qin, X., Egli, M., McHaourab, H. S., Stewart, P. L., and Johnson, C. H. (2007). Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol. 5, e93. doi: 10.1371/journal.pbio.0050093.
Mutsuda, M., Michel, K. P., Zhang, X., Montgomery, B. L., and Golden, S. S. (2003). Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongatus PCC 7942. J. Biol. Chem. 278, 19102–19110.
Nakajima, M., Imai, K., Ito, H., Nishiwaki, T., Murayama, Y., Iwasaki, H., Oyama, T., and Kondo, T. (2005). Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308, 414–415.
Nakamura, Y., Kaneko, T., Sato, S., Mimuro, M., Miyashita, H., Tsuchita, T., Sasamoto, S., Watanabe, A., Kawashima, K., Kishida, Y., Kiyokawa, C., Kohara, M., Matsumoto, M., Nakazaki, N., Shimpo, S., Takeuchi, C., Yamada, M., and Tabata, S. (2003). Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res. 10, 137–145.
Nishiwaki, T., Iwasaki, H., Ishiura, M., and Kondo, T. (2000). Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 97, 495–499.
Nishiwaki, T., Satomi, Y., Kitayama, Y., Terauchi, K., Kiyohara, R., Takao, T., and Kondo, T. (2007). A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 26, 4029–4037.
Nishiwaki, T., Satomi, Y., Nakajima, M., Lee, C., Kiyohara, R., Kageyama, H., Kitayama, Y., Temamoto, M., Yamaguchi, A., Hijikata, A., Go, M., Iwasaki, H., Takao, T., and Kondo, T. (2004). Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc. Natl. Acad. Sci. U.S.A. 101, 13927–13932.
Rust, M. J., Markson, J. S., Lane, W. S., Fisher, D. S., and O’Shea, E. K. (2007). Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science 318, 809–812.
Schmitz, O., Katayama, M., Williams, S. B., Kondo, T., and Golden, S. S. (2000). CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science 289, 765–768.
Smith, R. M., and Williams, S. B. (2006). Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. U.S.A. 103, 8564–8569.
Sweeney, B. M., and Borgese, M. B. (1989). A circadian rhythm in cell division in a prokaryote, the cyanobacterium Synechococcus WH7830. J. Phycol. 25, 183–186.
Takai, N., Nakajima, M., Oyama, T., Kito, R., Sugita, C., Sugita, M., Kondo, T., and Iwasaki, H. (2006). A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 12109–12114.
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599.
Taniguchi, Y., Katayama, M., Ito, R., Takai, N., Kondo, T., and Oyama, T. (2007). labA: a novel gene required for negative feedback regulation of the cyanobacterial circadian clock protein KaiC. Genes Dev. 21, 60–70.
Terauchi, K., Kitayama, Y., Nishiwaki, T., Miwa, K., Murayama, Y., Oyama, T., and Kondo, T. (2007). ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 104, 16377–16381.
Tomita, J., Nakajima, M., Kondo, T., and Iwasaki, H. (2005). No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307, 251–254.
Tomitani, A., Knoll, A. H., Cavanaugh, C. M., and Ohno, T. (2006). The evolutionary diversification of cyanobacteria: molecular-phylogenetic and paleontological perspectives. Proc. Natl. Acad. Sci. U.S.A. 103, 5442–5447.
Van Praag, E., Degli Agosti, R., and Bachofen, R. (2000). Rhythmic activity of uptake hydrogenase in the prokaryote Rhodospirillum rubrum. J. Biol. Rhythms 15, 218–224.
Woelfle, M. A., Ouyang, Y., Phanvijhitsiri, K., and Johnson, C. H. (2004). The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr. Biol. 14, 1481–1486.
Woelfle, M. A., Xu, Y., Qin, X., and Johnson, C. H. (2007). Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 104, 18819–18824.
Xu, Y., Mori, T., and Johnson, C. H. (2003). Cyanobacterial circadian clockwork: roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J. 22, 2117–2126.
Keywords: circadian clock genes, Legionella, archea, proteobacteria, cyanobacteria, evolution
Citation: Loza-Correa M, Gomez-Valero L and Buchrieser C (2010) Circadian clock proteins in prokaryotes: hidden rhythms? Front. Microbio. 1:130. doi: 10.3389/fmicb.2010.00130
Received: 19 September 2010;
Paper pending published: 12 October 2010;
Accepted: 30 October 2010;
Published online: 06 December 2010.
Edited by:
Elizabeth L. Hartland, University of Melbourne, AustraliaReviewed by:
Elizabeth L. Hartland, University of Melbourne, AustraliaCopyright: © 2010 Loza-Correa, Gomez-Valero and Buchrieser. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Carmen Buchrieser, Biology of Intracellular Bacteria, Institut Pasteur, 25, rue du Dr. Roux, 75724 Paris Cedex 15, France. e-mail:Y2FybWVuLmJ1Y2hyaWVzZXJAcGFzdGV1ci5mcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.