
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 26 February 2025
Sec. Pulmonary Medicine
Volume 12 - 2025 | https://doi.org/10.3389/fmed.2025.1531853
This article is part of the Research Topic Unveiling Distinctions: Active Tuberculosis versus Latent Tuberculosis Infection - Immunological Insights, Biomarkers, and Innovative Approaches View all 4 articles
Background: MPT64 protein is an effective marker for detecting Mycobacterium tuberculosis (MTB) in liquid culture and clinical tissue samples. However, some MTB clinical isolates test negative for this antigen because of varied mutation types across different regions.
Methods: DNA samples of MPT64 antigen assay-negative MTB strains were collected from a tertiary hospital from January 2016 to January 2024, and mpt64 gene mutations were detected by sequencing. Clinical records of patients with negative MPT64 antigen results were collected and compared with those of patients with positive results. The global distribution of mpt64 gene mutations was analyzed using MTB genome sequences from the National Center for Biotechnology Information (NCBI) database.
Results: Among 821 mycobacterial specimens with negative MPT64 antigen assay results, 77 MTB strains were collected from 73 patients. Compared with MPT64-positive patients (n = 301), a higher percentage of MPT64-negative patients had a history of anti-tuberculosis therapy (n = 7, 11.1%; P = 0.01). Moreover, MPT64-negative patients demonstrated a lower percentage of positive Gene Xpert results than MPT64-positive patients (73.8% vs 95.1%, P < 0.001). Several gene mutations were detected in the MPT64-negative MTB strains, including 63 bp deletion, single nucleotide mutations, and IS6110 insertion. Among 7,324 MTB genomes from the NCBI database, 87 strains had mutations in the mpt64 gene sequence, with four common mutation sites causing single amino acid changes, including G34A (8.0%), A103G (27.6%), T128A (9.2%), and C477A (24.1%).
Conclusion: A negative MPT64 antigen result in MTB cultures can be attributed to mutations in the mpt64 gene, and infections caused by these strains are more likely to be misdiagnosed.
Tuberculosis remains a major public health concern worldwide, although its prevalence and incidence have gradually decreased over the years. According to the Global Tuberculosis Report of the World Health Organization, over 10.3 million individuals developed tuberculosis in 2022 (1). Disease transmission can be prevented through early molecular diagnosis and treatment using a combination of anti-tuberculous agents.
Patients with tuberculosis are screened using several markers specific to Mycobacterium tuberculosis (MTB) (2). MPT64, a soluble protein encoded by a gene located in the region of difference (RD) 2, is expressed in MTB isolates but not in the live attenuated Mycobacterium bovis (Bacillus Calmette-Guérin) vaccine strain (3). Therefore, the positive MPT64 expression in laboratory cultures and clinical samples provides direct evidence of MTB infection (4, 5). In general, MPT64 is detected via an immunochromatographic test using specific antibodies. Commonly used commercial kits include Capilia TB and SD Bioline TB Ag MPT64. While MPT64 detection in tissue specimens is superior to Gene Xpert assay and mycobacterial cultures for diagnosing extrapulmonary tuberculosis (6, 7), its diagnostic performance is heterogeneous across other specimens.
Nonetheless, inconsistencies have been observed between the MPT64 and nucleic acid-based assays for MTB infections (8, 9). The negative results are largely caused by mpt64 gene mutations, as supported by a few studies (4, 9–12). For negative MPT64 results, further species identification usingmolecular techniques is necessary to confirm whether the strainbelongs to non-tuberculous mycobacteria (NTM). Certain MTB lineages appear to have a higher prevalence of mpt64 mutations; however, mutation types vary across different regions (9, 12, 13). Furthermore, whether clinical features vary across patients with infections by MTB strains with negative MPT64 detection remains unknown.
This study was conducted per the World Medical Association Declaration of Helsinki. Verbal informed consent was obtained via telephone call upon identifying the existence of MTB strain. The study did not involve patients younger than 18 years. It was approved by the Ethics Committee and Institutional Review Board of Dongyang People’s Hospital (2023-YX-319).
First, mycobacterial culture was obtained by positive results in the BACTEC MGIT960 System (BD, United States) and confirmed by positive acid-fast test. Second, the MPT64 antigen assay was conducted in the culture supernatants using a kit (Genesis Corporation, Hangzhou, China) according to the manufacturer’s instructions. Finally, species were identified by hsp65 amplification and sequencing, as described in our previous study (14). The heated and lysed supernatants of MPT64-negative strains were collected from patients with tuberculosis admitted to our hospital between January 2016 and January 2024 and stored at −80°C until further analysis.
The clinical data of patients with positive MPT64 assay results from 2019 were retrospectively extracted from medical records. Comorbidities were identified based on physical, laboratory, and radiological examinations. Results of the acid-fast smear test and Gene Xpert assay (Cepheid, United States) using sputum and bronchoalveolar lavage samples, as well as TB T spot test using venous blood samples, were collected. The history of anti-tuberculosis therapy was obtained by consulting the medical history, regardless of whether the treatment was completed. Additionally, computer tomography images showing nodules, cavities, and pleural effusion were obtained from medical records.
The mpt64 gene was amplified using primers specific for regions outside the coding sequence (12). The RD105 sequence was detected using two pairs of primers targeting the deleted type and intact type, respectively (15, 16). For samples with an intact RD105, the pks15/1 region was amplified and sequenced. The Beijing family strain was defined based on the RD105 deletion (17), whereas the non-Beijing family type was further analyzed by detecting RD239 and RD711 and sequencing pks15/1 (18–20). Amplification condition and procedure have been described previously (14). The amplified products were analyzed by electrophoresis in 1% agarose gel. The sequence of mpt64 was obtained by Sanger sequencing (GENEWIZ, China). The primer sequences are listed in Supplementary Table 1.
All assembled genomes of MTB were accessed from the National Center for Biotechnology Information (NCBI) genome database (by June 15, 2024) and downloaded using the NCBI Datasets command-line tool “datasets.” The mpt64 gene sequences were extracted using the R “seqinir” and “Biostrings” packages and matched using two flanking regions (CTAGGCCAGCATCGAGTCGA and ATGAAGATCTTGATGCGCAC). Genomes without matched sequences or mutations, compared with the H37Rv-derived mpt64 gene, were ignored. The sequences were complementally reversed and aligned using MEGA version 11.0.13 (21). The DNA sequences were translated into amino acid sequences to confirm the impact of gene mutation on the encoded protein.
Categorical data were expressed as numbers with percentages, and differences between groups were analyzed by chi-square test. Continuous data were expressed as mean ± standard deviation, and significance was determined using independent-sample t-tests. All analyses were conducted using the Statistical Package for the Social Sciences Software version 27 (International Business Machines Corporation, United States).
A total of 821 mycobacterial strains with negative MPT64 antigen assay results were selected. After excluding 744 NTM strains, 77 MTB strains were isolated from 73 patients who were negative for the MPT64 assay. After excluding cases with missing clinical data and repeated strains from the same patients, 64 MTB strains corresponding to 64 cases were analyzed. Compared with MPT64-positive patients with tuberculosis (n = 8, 2.9%), MPT64-negative patients (n = 7, 11.1%) demonstrated a higher percentage of a history of anti-tuberculosis therapy (P = 0.01, Table 1). Only one MPT64-negative patient (n = 1, 1.6%) had chronic obstructive pulmonary disease, lower than 7.9% of MPT64-positive patients (P = 0.093). Moreover, MPT64-negative patients demonstrated a lower percentage of positive Gene Xpert results in the original specimens than MPT64-positive patients (73.8% vs 95.1%, P < 0.001).
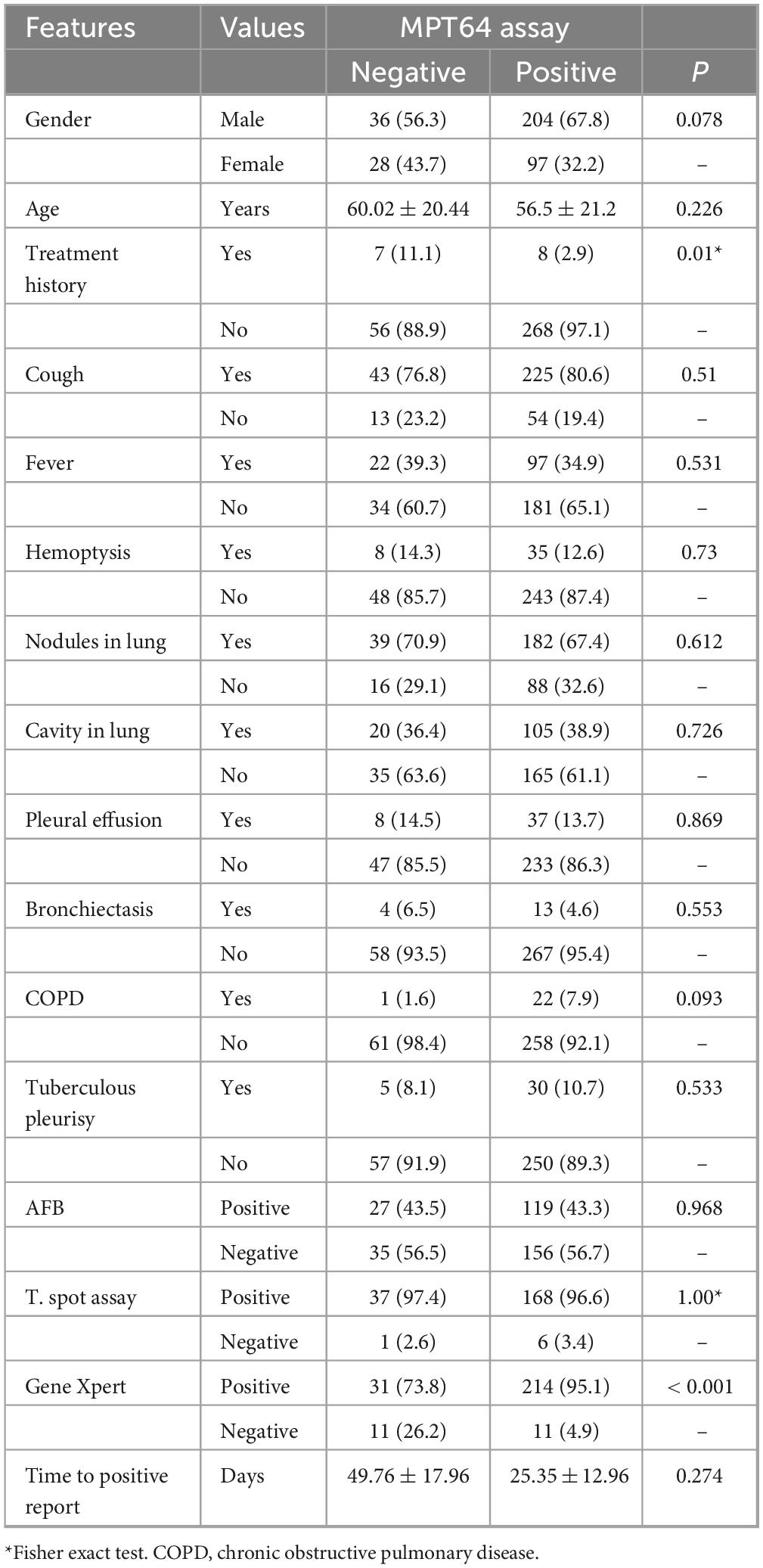
Table 1. The clinical features for tuberculosis patients with different MPT64 antigen assay results.
The mpt64 sequences of all MTB strains with negative MPT64 results were amplified and sequenced. A 63 bp region was deleted in 53 strains, resulting in a truncated protein consisting of 21 amino acids, compared with the wild-type protein (Table 2). Two strains had a copy of the IS6110 sequence in the mpt64 gene at the 313 bp position, which introduced a stop codon leading to premature termination. Finally, one strain had a 13 bp insertion, which resulted into a longer novel protein sequence.
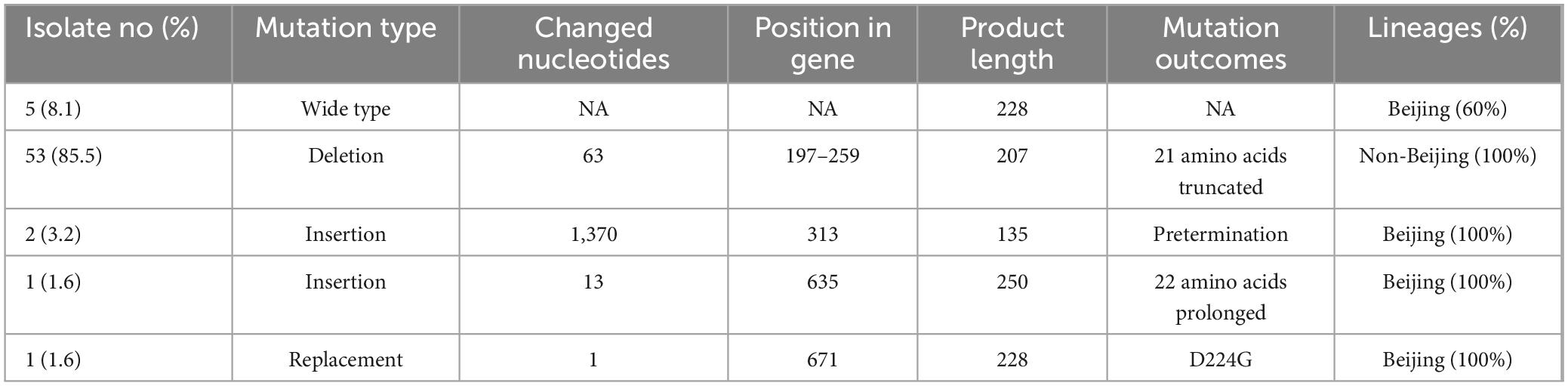
Table 2. The distribution of mutation types in the mpt64 gene in Mycobacterium tuberculosis (MTB) strains with negative MPT64 assay.
The 63 bp deletion was exclusively present in strains with the RD105 region, which corresponded to the non-Beijing family genotype. Strains with other mutation types belonged to the Beijing family genotype. Furthermore, the non-Beijing family strains had intact RD239, RD711, TbD1, and RD750 regions. The pks15/1 sequences were identical to the H37Rv-derived sequence. Based on these sequence markers, all strains with the 63 bp deletion belonged to the Euro-American lineage.
A total of 7,324 MTB genomes were obtained from the NCBI, of which 87 strains showed mpt64 mutations, compared with the reference strain (Table 3). G34A (8%), A103G (27.6%), T128A (9.2%), and C477A (24.1%) were the most common sites with single amino acid changes. All strains with G34A mutations were isolated from Peru, whereas strains with other mutations were distributed in different regions (Supplementary Table 2). Furthermore, two strains had a 21 amino acid deletion because of the 63 bp deletion. Other mutated loci were distributed sporadically in MTB isolates.
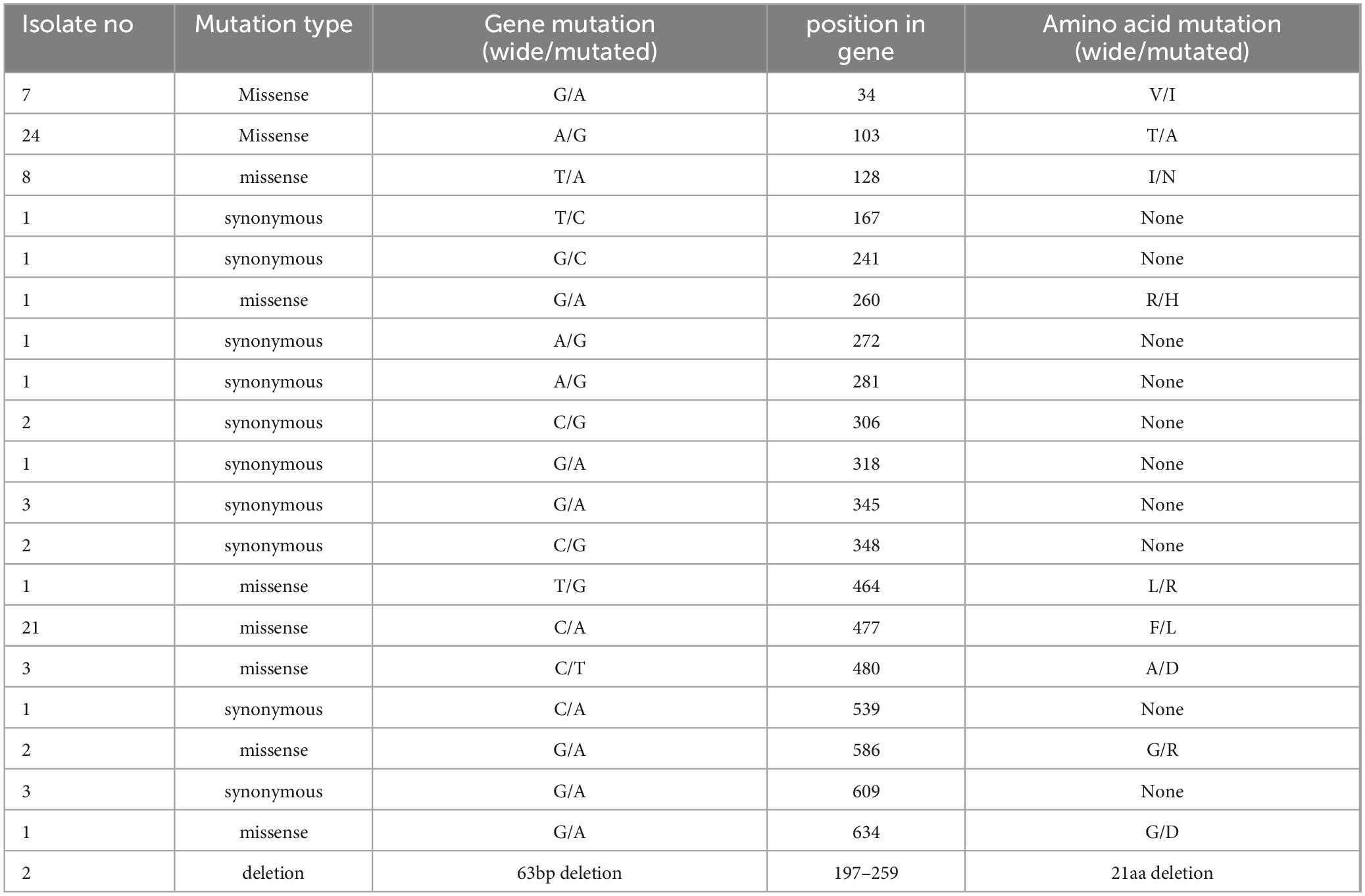
Table 3. The distribution of mutation sites across the mpt64 gene from accessible genomes of Mycobacterium tuberculosis.
The MPT64 antigen assay is routinely used to detect MTB in mycobacterial cultures and clinical samples. However, negative results caused by mpt64 mutations can lead to misdiagnosis in the absence of other molecular assays. In practice, a cultured mycobacterial strain with a negative MPT64 assay result is considered an NTM, which requires a different drug regimen, compared with MTB (8, 22). Moreover, patients with NTM infection have not been routinely monitored in most regions because NTM rarely leads to transmission in immunocompetent individuals. Moreover, multiple antibiotic agent-containing therapy is not recommended for patients with mild symptoms (23). Thus, false-negative results in the MPT64 assay possibly resulted in MTB misdiagnosis as NTM infection, causing more susceptible individuals becoming infected by MPT64-negative patients because of a lack of therapy or resistance acquirement from an inappropriate drug regimen (24, 25). For mycobacterial strains with negative MPT64 assay results, additional time and money are warranted to conduct nucleotide acid-based assays using mycobacterial culture to identify the mycobacterial species (8). However, alternative assays (next-generation sequencing, nucleotide acid amplification, and nucleic acid probe detection) for all mycobacterial cultures are not recommended because of higher costs and the need for professional instruments and trained staff.
With the application of several molecular detection techniques in clinical laboratories for mycobacterial identification, negative MPT64 antigen assay results in MTB strains have been reported globally (8, 10–13, 26). Nevertheless, the clinical relevance of negative MPT64 antigen results has not been thoroughly investigated. In this study, a higher percentage of patients who tested negative for the MPT64 antigen received anti-tuberculosis treatment previously. This phenomenon could be partially explained by the insufficient duration of routine treatment for these patients because MPT64-negative MTB strains might have lower division activity than MPT64-positive patients (8, 27). The bacilli load influences the detection of the MPT64 antigen because the cultured MTB strains negative for the MPT64 antigen are more likely to be negative for MTB-specific nucleic acids in clinical samples, such as Gene Xpert assay (8). Moreover, the time to report tended to be longer for strains with negative MPT64 assay results, despite no significance in this study, indicating the possibility of a lower bacilli load in the specimen.
Mpt64 mutations are the major reason for a negative MPT64 result (8, 10, 12, 13). Mpt64 mutation types appear to have a specific association with certain lineages (12, 26). In this study, all strains carrying the 63 bp deletion in the mpt64 gene belonged to the non-Beijing lineage, aligning with previous data suggesting that this deletion is commonly observed in L4 and L1 lineages (10, 13). The truncated protein encoded by the mpt64 gene with a 63 bp deletion may not be recognized by the antibodies in enzyme-linked immunosorbent assay or immunochromatographic commercial kits, yielding false-negative results (10). A 63 bp deletion is predicted to drastically change the protein structure because of an alpha helix, explaining the phenomenon (13).
Single-nucleotide mutations have been reported in the mpt64 gene. Missense mutations can alter protein structure and stability; however, the corresponding strains remained positive for the MPT64 protein assay (9, 11, 13). For example, a common mutation at 477 bp that changes phenylalanine to leucine does not affect MPT64 antigen detection (9, 11). Similar results have been reported for the missense mutation at 128 T > A, which appears specific to L5 lineage strains (9). Furthermore, a longer incubation period (8, 12) or the use of L-J culture instead of liquid culture (8) may reverse the decreased sensitivity of the MPT64 assay because of a single amino acid change. For MTB strains with negative MPT64 assay results from Australia, the mpt64 mutation type is the deletion of two nucleotides, resulting in a frameshift in protein expression (26). Further analysis indicated that all the strains carrying this mutation belong to Lineage 4, which has been attributed to European migration and colonization (28). IS6110 insertion is an uncommon mpt64 mutation, and previously reported insertion sites (501 bp) (29) is different from our study (313 bp). The influence of IS6110 on protein structure or function depends on the insertion site (30). In detail, insertion at 313 bp in this study introduced a termination codon, resulting in a shorter polypeptide of 135 amino acids.
Mpt64 mutations are specific to certain MTB lineages or sublineages; nonetheless, whether these strains possess unique features concerning resistance phenotype and virulence remains elusive (28, 31). In the future, multi-omic investigations might elucidate the mechanism by which these strains interact with geographic populations (32–34). Moreover, the lower sensitivity of the MPT64 antigen-based assay could be overcome by combining molecular assays for different targets or adopting more convenient and economical methods for MTB diagnosis (35, 36). Furthermore, novel antibodies detecting both the mutated and wild MPT64 proteins would improve diagnostic efficiency in liquid cultures and minimize false negatives (37).
This study has several limitations. MTB strains were collected from a single center, which may not entirely represent broader genetic diversity. Strains carrying the wild-type mpt64 gene might have yielded positive results after longer incubation in liquid culture; however, these strains were not preserved and only the nucleic acids were available. Furthermore, clinical information for some patients was missing because of retrospective data collection, particularly for outpatients, possibly introducing bias in the comparison of clinical characteristics. Finally, detailed lineage data was inaccessible for some strains because of the lower DNA quality. In the future, whole genome sequencing of mutated MTB strains would provide a more comprehensive mutational landscape.
Patients with MTB infections who are negative for the MPT64 antigen are more likely to be misdiagnosed as having NTM because of lower positive rates in the Gene Xpert assay. Mpt64 mutations, primarily the 63 bp deletion and IS6110 insertion, were associated with negative MPT64 antigen assay results. In contrast, single nucleotide mutations in the mpt64 gene partly contributed to the lower sensitivity of antigen assays, which may be improved by prolonged incubation. Other nucleic acid-based tests for MTB should be conducted for patients who tested negative for the MPT64 antigen but positive for mycobacterial culture.
The authors acknowledge that the data presented in this study must be deposited and made publicly available in an acceptable repository, prior to publication. Frontiers cannot accept a manuscript that does not adhere to our open data policies.
The studies involving humans were approved by Ethics Committee and Institutional Review Board of Dongyang People’s Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by- product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
XP: Conceptualization, Funding acquisition, Methodology, Writing – original draft, Writing – review and editing. SZ: Methodology, Software, Writing – original draft, Writing – review and editing. LJ: Conceptualization, Funding acquisition, Writing – original draft, Writing – review and editing. SJ: Data curation, Methodology, Writing – original draft, Writing – review and editing. XL: Investigation, Methodology, Resources, Supervision, Writing – original draft, Writing – review and editing. BL: Data curation, Resources, Writing – original draft, Writing – review and editing. JZ: Data curation, Funding acquisition, Investigation, Resources, Writing – original draft, Writing – review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (grant number 2023KY386) and the Science and Technology Bureau of Jinhua (2022-3-013 and 2022-3-011).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2025.1531853/full#supplementary-material
1. World Health Organization. Global Tuberculosis Report 2023. Licence: CC BY-NC-SA 3.0 IGO. Geneva: World Health Organization (2023).
2. Druszczynska M, Wawrocki S, Szewczyk R, Rudnicka W. Mycobacteria-derived biomarkers for tuberculosis diagnosis. Indian J Med Res. (2017) 146:700–7.
3. Muchwa C, Akol J, Etwom A, Morgan K, Orikiriza P, Mumbowa F, et al. Evaluation of Capilia TB assay for rapid identification of Mycobacterium tuberculosis complex in BACTEC MGIT 960 and BACTEC 9120 blood cultures. BMC Res Notes. (2012) 5:44. doi: 10.1186/1756-0500-5-44
4. Cao XJ, Li YP, Wang JY, Zhou J, Guo XG. MPT64 assays for the rapid detection of Mycobacterium tuberculosis. BMC Infect Dis. (2021) 21:336. doi: 10.1186/s12879-021-06022-w
5. Armstrong DT, Pretty L, D’Agostino K, Redhead-Harper R, Parrish N. Diagnostic accuracy of the abbott SD bioline MPT64 antigen test for identification of MTB complex in a U.S. clinical mycobacteriology laboratory. Heliyon. (2024) 10:e30501. doi: 10.1016/j.heliyon.2024.e30501
6. Helle OMB, Kanthali M, Ishtiaq S, Ambreen A, Purohit MR, Mustafa T. Diagnosing adult and pediatric extrapulmonary tuberculosis by MPT64 antigen detection with immunohistochemistry and immunocytochemistry using reproduced polyclonal antibodies. J Pathol Clin Res. (2024) 10:e12373. doi: 10.1002/2056-4538.12373
7. Hoel IM, Sviland L, Syre H, Dyrhol-Riise AM, Skarstein I, Jebsen P, et al. Diagnosis of extrapulmonary tuberculosis using the MPT64 antigen detection test in a high-income low tuberculosis prevalence setting. BMC Infect Dis. (2020) 20:130. doi: 10.1186/s12879-020-4852-z
8. Ofori-Anyinam B, Kanuteh F, Agbla SC, Adetifa I, Okoi C, Dolganov G, et al. Impact of the Mycobaterium africanum West Africa 2 lineage on TB diagnostics in West Africa: decreased sensitivity of rapid identification tests in The Gambia. PLoS Negl Trop Dis. (2016) 10:e0004801. doi: 10.1371/journal.pntd.0004801
9. Zamir H, Ahmad B, Ali S, Khan SA, Sarwar R, Khan A, et al. Molecular characterization of Mycobacterium tuberculosis through MPT64 gene polymorphism using next-generation sequencing technology. Fut Microbiol. (2022) 17:763–72. doi: 10.2217/fmb-2020-0231
10. Jiang Y, Liu H, Wang H, Dou X, Zhao X, Bai Y, et al. Polymorphism of antigen MPT64 in Mycobacterium tuberculosis strains. J Clin Microbiol. (2013) 51:1558–62. doi: 10.1128/JCM.02955-12
11. Muhammad N, Khan MT, Ali S, Khan TA, Khan AS, Ullah N, et al. Novel mutations in MPT64 secretory protein of Mycobacterium tuberculosis complex. Int J Environ Res Public Health. (2023) 20:2530. doi: 10.3390/ijerph20032530
12. Sanoussi CN, de Jong BC, Odoun M, Arekpa K, Ali Ligali M, Bodi O, et al. Low sensitivity of the MPT64 identification test to detect lineage 5 of the Mycobacterium tuberculosis complex. J Med Microbiol. (2018) 67:1718–27. doi: 10.1099/jmm.0.000846
13. Song Z, He W, Pei S, Zhao B, Cao X, Wang Y, et al. Association of lineage 4.2.2 of Mycobacterium tuberculosis with the 63-bp deletion variant of the mpt64 gene. Microbiol Spectr. (2023) 11:e0184223. doi: 10.1128/spectrum.01842-23
14. Pan X, Zhou Y, Li Z, Zhang J, Hong L, Shi Y, et al. Investigation of non-tuberculous mycobacteria in a primary hospital from southeastern China. J Infect Dev Ctries. (2019) 13:1095–100. doi: 10.3855/jidc.11772
15. Yuan L, Huang Y, Mi LG, Li YX, Liu PZ, Zhang J, et al. There is no correlation between sublineages and drug resistance of Mycobacterium tuberculosis Beijing/W lineage clinical isolates in Xinjiang, China. Epidemiol Infect. (2015) 143:141–9. doi: 10.1017/S0950268814000582
16. Luo T, Yang C, Gao Q. Mycobacterial interspersed repetitive-unit locus PCR amplification and Beijing strains of Mycobacterium tuberculosis. J Clin Microbiol. (2011) 49:4026–7; author reply 8. doi: 10.1128/JCM.05389-11
17. Rindi L, Lari N, Cuccu B, Garzelli C. Evolutionary pathway of the Beijing lineage of Mycobacterium tuberculosis based on genomic deletions and mutT genes polymorphisms. Infect Genet Evol. (2009) 9:48–53. doi: 10.1016/j.meegid.2008.09.006
18. Zenteno-Cuevas R, Silva-Hernandez FX, Mendoza-Damian F, Ramirez-Hernandez MD, Vazquez-Medina K, Widrobo-Garcia L, et al. Characterisation of pks15/1 in clinical isolates of Mycobacterium tuberculosis from Mexico. Mem Inst Oswaldo Cruz. (2013) 108:718–23. doi: 10.1590/0074-0276108062013007
19. Sales ML, Fonseca AA Jr., Sales EB, Cottorello AC, Issa MA, Hodon MA, et al. Evaluation of molecular markers for the diagnosis of Mycobacterium bovis. Folia Microbiol (Praha). (2014) 59:433–8. doi: 10.1007/s12223-014-0317-3
20. Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. (2006) 103:2869–73. doi: 10.1073/pnas.0511240103
21. Tamura KSG, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 33:6. doi: 10.1093/molbev/msab120
22. Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ, Andrejak C, et al. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Clin Infect Dis. (2020) 71:905–13.
23. Nguyen MH, Haas MK, Kasperbauer SH, Calado Nogueira de Moura V, Eddy JJ, Mitchell JD. Nontuberculous mycobacterial pulmonary disease: patients, principles, and prospects. Clin Infect Dis. (2024) 79:e27–47.
24. Datta D, Jamwal S, Jyoti N, Patnaik S, Kumar D. Actionable mechanisms of drug tolerance and resistance in Mycobacterium tuberculosis. FEBS J. (2024) 291:4433–52. doi: 10.1111/febs.17142
25. Roemhild R, Bollenbach T, Andersson DI. The physiology and genetics of bacterial responses to antibiotic combinations. Nat Rev Microbiol. (2022) 20:478–90.
26. Bainomugisa A, Pandey S, O’Connor B, Syrmis M, Whiley D, Sintchenko V, et al. Sustained transmission over two decades of a previously unrecognised MPT64 negative Mycobacterium tuberculosis strain in Queensland, Australia: a whole genome sequencing study. Lancet Reg Health West Pac. (2024) 47:101105. doi: 10.1016/j.lanwpc.2024.101105
27. Wang Z, Potter BM, Gray AM, Sacksteder KA, Geisbrecht BV, Laity JH. The solution structure of antigen MPT64 from Mycobacterium tuberculosis defines a new family of beta-grasp proteins. J Mol Biol. (2007) 366:375–81. doi: 10.1016/j.jmb.2006.11.039
28. Stucki D, Brites D, Jeljeli L, Coscolla M, Liu Q, Trauner A, et al. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat Genet. (2016) 48:1535–43. doi: 10.1038/ng.3704
29. Hirano K, Aono A, Takahashi M, Abe C. Mutations including IS6110 insertion in the gene encoding the MPB64 protein of Capilia TB-negative Mycobacterium tuberculosis isolates. J Clin Microbiol. (2004) 42:390–2. doi: 10.1128/JCM.42.1.390-392.2004
30. Gonzalo-Asensio J, Perez I, Aguilo N, Uranga S, Pico A, Lampreave C, et al. New insights into the transposition mechanisms of IS6110 and its dynamic distribution between Mycobacterium tuberculosis complex lineages. PLoS Genet. (2018) 14:e1007282. doi: 10.1371/journal.pgen.1007282
31. Alvarez-Maya I, Garcia-Ulloa M, Martinez-Guarneros A, Vazquez-Chacon CA, Martinez-Urtaza J. nationwide phylogenomic surveillance of Mycobacterium tuberculosis in Mexico reveals pathogenic and drug resistant signatures of the prevailing L4 sublineage. J Glob Antimicrob Resist. (2025) 41:224–32. doi: 10.1016/j.jgar.2025.01.013
32. Borah K, Xu Y, McFadden J. Dissecting host-pathogen interactions in TB using systems-based omic approaches. Front Immunol. (2021) 12:762315. doi: 10.3389/fimmu.2021.762315
33. Rajwani R, Galata C, Lee AWT, So PK, Leung KSS, Tam KKG, et al. A multi-omics investigation into the mechanisms of hyper-virulence in Mycobacterium tuberculosis. Virulence. (2022) 13:1088–100. doi: 10.1080/21505594.2022.2087304
34. Baysoy A, Tian X, Zhang F, Renauer P, Bai Z, Shi H, et al. Spatially Resolved in vivo CRISPR Screen Sequencing via Perturb-DBiT. bioRxiv [preprint]. (2024). doi: 10.1101/2024.11.18.624106
35. Shrestha S, Addae A, Miller C, Ismail N, Zwerling A. Cost-effectiveness of targeted next-generation sequencing (tNGS) for detection of tuberculosis drug resistance in India, South Africa and Georgia: a modeling analysis. EClinicalMedicine. (2025) 79:103003. doi: 10.1016/j.eclinm.2024.103003
36. Rickman HM, Phiri MD, Mbale H, Horton KC, Henrion MYR, Nightingale ES, et al. Low concordance between QIAreach QuantiFERON-TB, a novel interferon-gamma release assay, and QuantiFERON-TB Gold Plus, in a population-based survey in Blantyre, Malawi. J Clin Microbiol. (2025) 63:e0132324. doi: 10.1128/jcm.01323-24
37. Magalhaes CG, Moreira G, Ferreira MRA, Santos LMD, Finger PF, Ramos DF, et al. Novel phage display-derived recombinant antibodies recognizing both MPT64 native and mutant (63-bp deletion) are promising tools for tuberculosis diagnosis. Biologicals. (2021) 72:54–7. doi: 10.1016/j.biologicals.2021.07.002
Keywords: Mycobacterium tuberculosis, MPT64, gene mutation, false negative, clinical features
Citation: Pan X, Zhou S, Jin L, Ji S, Lou X, Lu B and Zhao J (2025) mpt64 mutations in Mycobacterium tuberculosis with negative MPT64 antigen assay results from a tertiary hospital in Southeastern China. Front. Med. 12:1531853. doi: 10.3389/fmed.2025.1531853
Received: 21 November 2024; Accepted: 18 February 2025;
Published: 26 February 2025.
Edited by:
Ying Luo, UT Southwestern Medical Center, United StatesReviewed by:
Fu Gao, Yale University, United StatesCopyright © 2025 Pan, Zhou, Jin, Ji, Lou, Lu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Zhao, MTU5ODg1MDQzNTZAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.