
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Med. , 10 March 2025
Sec. Hepatobiliary Diseases
Volume 12 - 2025 | https://doi.org/10.3389/fmed.2025.1482089
Many biomolecules and signaling pathways are involved in the development of alcoholic liver disease (ALD). The molecular mechanisms of ALD are not fully understood and there is no effective treatment. Numerous studies have demonstrated the critical role of non-coding RNAs, including long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), in ALD. miRNAs play an important regulatory role in the pathogenesis of ALD by controlling critical biological processes such as inflammation, oxidative stress, lipid metabolism, apoptosis and fibrosis. Among them, miR-155, miR-223 and miR-34a play a central role in these processes and influence the pathological process of ALD. In addition, lncRNAs are involved in regulating liver injury and repair by interacting with miRNAs to form a complex regulatory network. These findings help to elucidate the molecular mechanisms of ALD and provide a scientific basis for the development of new diagnostic markers and therapeutic targets. In this article, we review the roles and mechanisms of LncRNAs and miRNAs in ALD and their potential use as diagnostic markers and therapeutic targets.
Alcoholic liver disease (ALD) is a group of liver injuries and diseases caused by chronic excessive alcohol consumption (1). With the improvement in living standards and lifestyle changes in society, alcohol consumption has increased, leading to a yearly increase in the incidence of alcoholic liver disease, which has become a major global public health challenge (2). The pathophysiological process of ALD is very complex and includes different stages such as fatty liver, alcoholic hepatitis (AH) and alcoholic cirrhosis (3). In the early stages, alcohol causes fat to accumulate in the liver, resulting in fatty liver. Over time, secondary inflammation and fibrosis can lead to AH, which can eventually lead to serious consequences such as cirrhosis and hepatocellular carcinoma. This process involves the complex regulation of multiple cellular and molecular mechanisms such as apoptosis, oxidative stress and the release of inflammatory mediators (4). In addition to direct damage to the liver, ALD is often associated with a number of systemic complications, including malnutrition (e.g., vitamin deficiency), neurological damage (e.g., hepatic encephalopathy, peripheral neuropathy), cardiovascular disease (e.g., alcoholic cardiomyopathy), and immune dysfunction (5). For example, miR-155 was found to exacerbate cardiovascular injury by upregulating systemic inflammatory factors (e. g., TNF- α, IL-6) release (6). The miR-34a promotes hepatocyte apoptosis through the SIRT 1 / p53 pathway, and its released exosomes may affect the CNS (7, 8). lncRNA CRNDE By activating inflammatory factors such as IL-6, it may mediate multiorgan inflammatory (9, 10) through circulating exosomes.Therefore, in-depth research and effective intervention in ALD are of great importance.
Most (76–97%) of the human genome encodes RNAs that are not translated into proteins, called non-coding RNAs (ncRNAs). NcRNAs are functional RNA molecules that are not directly involved in translation and play an important role in fine-tuning cellular functions. Based on the difference in length, ncRNAs can be divided into short and long ncRNAs. Among them, miRNAs are a specific class of short ncRNAs, approximately 22 nucleotides in length. miRNAs achieve degradation or inhibition of mRNA translation by complementary pairing with the 3' untranslated region (3'UTR) of the target mRNA, and thus play an important role in regulating gene expression at the level of gene expression. Specifically, miRNAs inhibit the conversion of mRNAs into proteins by two main mechanisms: degradation of mRNAs and inhibition of translation initiation (11). lncRNAs are more diverse and, like miRNAs, have specific functions depending on their cellular localization (12). Nuclear-localized lncRNAs act as transcription factors or chromatin-modifying complexes (13), and cytoplasmic LncRNAs can directly regulate mRNA stabilization or act as endogenous competing RNAs that regulate functional proteins (14). Most lncRNAs are transcribed by RNA polymerase II or other RNA polymerases and undergo a splicing process similar to that of mRNAs, with 5-co-terminal capping [7-methyl ouabain (m7G)] and 3 m-terminal polyadenylation. lncRNAs have a wide range of biological functions. They are involved in the regulation of gene expression, including DNA replication and transcription, protein translation, and epigenetic lncRNAs are involved in a wide range of biological roles in the regulation of gene expression, including DNA replication and transcription, protein translation, and epigenetic regulation (15). lncRNAs mediate epigenetic modification of DNA by recruiting chromatin remodeling complexes to specific sites (16, 17).
In exploring the interactions between lncRNAs and miRNAs, numerous studies have found that they are jointly involved in post-transcriptional regulatory processes and significantly impact critical aspects of disease onset, metastatic progression and drug resistance (18). By revealing the potential association between lncRNAs and miRNAs, we can gain a deeper understanding of the biological functions of these two RNA molecules and their roles in the pathogenesis of complex diseases (Table 1). This study not only helps us to deepen our understanding of RNA regulatory networks but may also provide new strategies and directions for disease treatment.
In this paper, we review the research progress of lncRNAs and miRNAs in ALD and describe the biological functions and therapeutic potential of lncRNAs, miRNAs and their interactions in the disease.
In recent years, an increasing number of studies have shown that miRNA play a crucial regulatory role in the pathological process of many liver diseases. During the onset and development of ALD, miRNA are not only involved in the regulation of critical biological processes such as hepatocyte steatosis, inflammatory response, fibrosis and apoptosis, but also interact with other ncRNAs such as lncRNAs through a complex molecular network that further influences the disease process. Alcohol-induced oxidative stress and inflammatory response are key aspects of the pathological mechanisms of ALD, and miRNA play an important role in this process by regulating the expression of relevant signaling pathways and target genes. For example, certain miRNA can directly target and regulate genes involved in adipogenesis, inflammatory response, and fibrosis, thereby influencing pathological changes in the liver (19). In addition, alcohol consumption can further exacerbate the condition by affecting the expression profile of miRNA and altering the physiological state of liver cells. This section will review the biological functions of miRNA in ALD and their specific mechanisms, including their roles in steatosis, inflammatory response, fibrosis, and hepatocyte apoptosis (Figure 1).
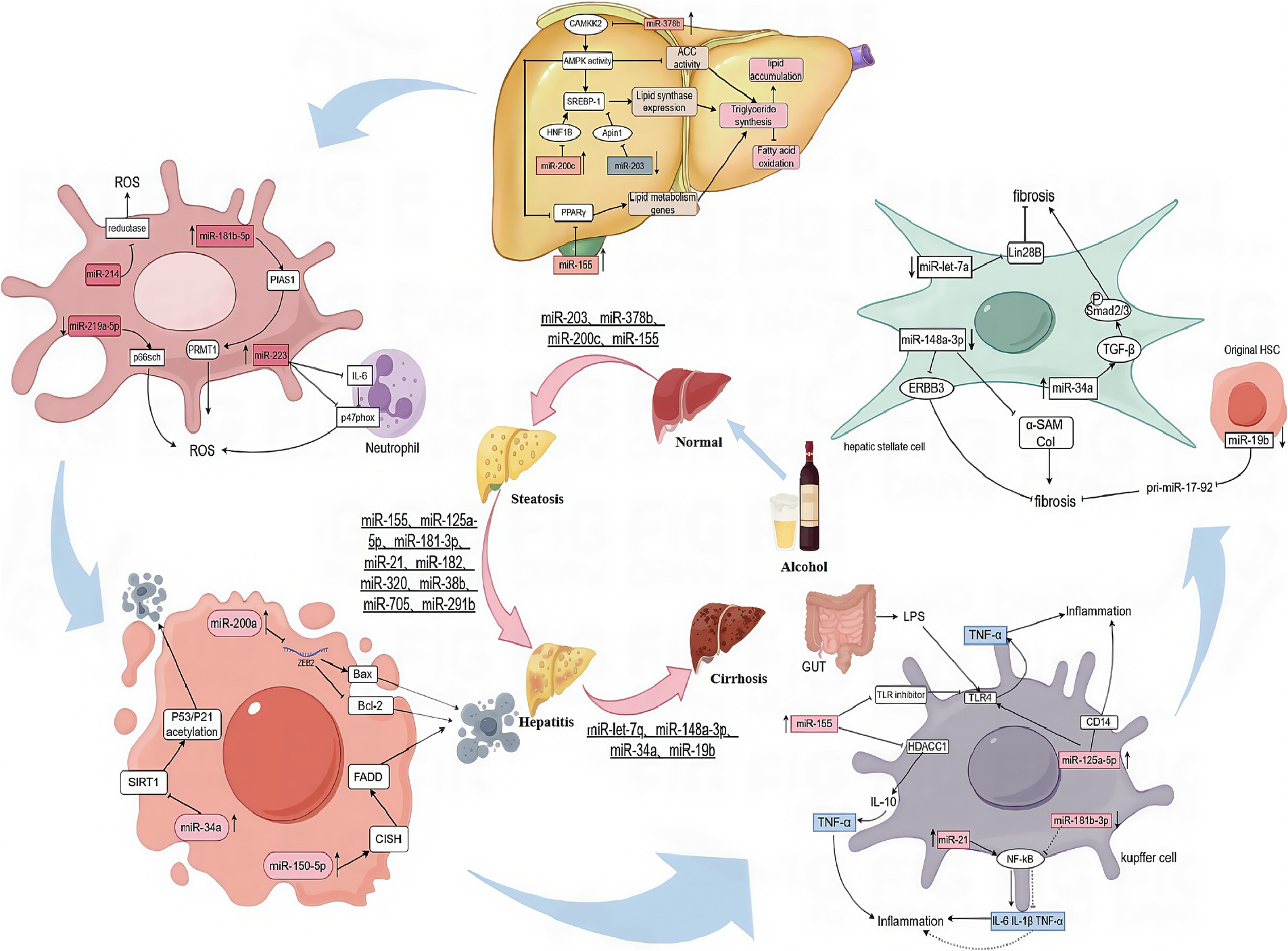
Figure 1. The mechanisms of miRNAs action in steatosis, inflammatory, fibrosis, apoptosis, and oxidative stress.
miRNA play a key role in lipid metabolism in ALD. Sterol Regulatory Element Binding Protein 1 (SREBP1), a major transcriptional regulator of lipid synthesis, is involved in lipid synthesis. miR-203 was able to directly inhibit the downstream target Lipin1, which in turn reduced SREBP1 expression, thereby inhibiting the dual functions of SREBP1 in promoting lipid synthesis and inhibiting fatty acid oxidation. However, in ALD, miR-203 was significantly down-regulated, leading to the over-expression of Lipin1, resulting in lipid over-accumulation and promoting disease progression. miR-203 reduced intracellular lipid synthesis in hepatocytes by targeting and downregulating Lipin1, suggesting that miR-203 is a specific target for the treatment of alcoholic fatty liver disease (19). miR-378b inhibits CaMKK2, which in turn inhibits the activation of AMPK, a key enzyme in the regulation of lipid metabolism that controls the process of lipid metabolism by phosphorylating downstream targets such as acetyl coenzyme A carboxylase ACC. AMPK can phosphorylate SREBP1, inhibiting its activity and repressing the expression of genes involved in its biosynthesis. Another study found that activation of AMPK can affect the protein stability of the peroxisome proliferator-activated receptor γ (PPARγ), which is particularly important in lipid metabolism, regulating fatty acid uptake and storage, lipoprotein synthesis and cholesterol metabolism (20). Reduced expression of AMPK leads to an imbalance in the regulation of lipid metabolism, with increased fatty acid synthesis and decreased fatty acid oxidation, exacerbating lipid accumulation in the liver. Inhibition of miR-378b can effectively alleviate ethanol-induced lipid metabolism dysfunction (21). miR-200c directly targets HNF1B (HNF1 homology box B) and ApoO (apolipoprotein O) to regulate lipid homeostasis. These two genes play important roles in lipid metabolism. The expression of miR-200c is significantly upregulated under chronic ethanol exposure. miR-200c contributes to the development of AFL by inhibiting HNF1B and ApoO, leading to reduced TG secretion and decreased fatty acid oxidation, which exacerbates lipid accumulation in the liver (22). Bala S et al. analyzed the critical role of miR-155 in the progression of alcohol-induced liver disease. miR-155 can directly target PPARγ to promote the formation and function of mature adipocytes by activating the expression of lipid metabolism genes such as ACC1 (acetyl CoA carboxylase), FABP4 (fatty acid binding protein), LDLR (low-density lipoprotein receptor-associated protein) and LXR (hepatic X receptor), which increase fatty acid uptake and storage in adipose tissue. By knocking down miR-155, mice were able to significantly resist alcohol-induced steatosis and inflammation, while reducing the degree of alcohol-induced fibrosis. This finding provides strong evidence that miR-155 plays a driving role in promoting alcohol-induced steatohepatitis and fibrosis (6).
miRNA also play an important role in alcohol-associated oxidative stress. miR-219a-5p specifically binds to and represses the expression of p66shc, which causes oxidative cellular damage by regulating ROS (reactive oxygen species) formation in alcoholic liver injury. By inhibiting the expression of p66shc, miR-219a-5p in turn reduces ROS production, thereby alleviating oxidative stress and ROS-induced cellular damage (23). miR-181b-5p indirectly promotes PRMT1 activity by targeting and inhibiting PIAS1 expression, thereby exacerbating oxidative stress and inflammatory responses in AFLD (24). p47phox is a key molecule in the response to oxidative stress and its main function is to stimulate the cell when it receives a stimulus, leading to the activation of NADPH oxidase, the assembly of the enzyme complex and the production of ROS. miR-223 reduces the activation of NADPH oxidase and thus ROS production in neutrophils by directly inhibiting the expression of IL-6 in neutrophils, which in turn inhibits the downstream expression of p47phox. miR-223 is a key molecule in the oxidative stress response (25). Alcohol also induces the expression of miR-214 in hepatocytes. miR-214 reduces ROS scavenging by targeting and inhibiting the expression of genes encoding certain antioxidant enzymes, such as superoxide dismutase (SOD) and glutathione peroxidase (GPx). During alcohol-induced oxidative stress, miR-214 was found to further inhibit the expression and activity of GSR and POR by specifically binding to the 3' untranslated region (3'UTR) of glutathione reductase (GSR) and cytochrome P450 oxidoreductase (POR). By down-regulating the expression of these enzymes, it exacerbates oxidative stress (26).
Alcohol consumption can significantly stimulate the expression of specific miRNA, which influence the cell life cycle by regulating their target genes. miR-200a, whose expression is upregulated in ALD, effectively inhibits the expression of ZEB2 mRNA by specifically binding to the 3'UTR of ZEB2 mRNA, which further leads to a decrease in the antiapoptotic protein Bcl-2 and an increase in the pro-apoptotic protein Bax increases, ultimately triggering the apoptotic process (27). miR-150-5p indirectly affects the ubiquitinated degradation of death domain proteins (FADDs) by inhibiting the expression of cytokine signaling inhibitor (CISH), induces exogenous apoptosis through the FADD/procaspase-8/caspase-dependent signaling cascade and further affects the apoptotic and survival status of hepatocytes (28). miR-34a maintains cellular homeostatic balance by finely regulating cell survival, proliferation, and cycling. miR-34a plays a key role in the response to cellular oxidative stress and DNA damage (29). However, overexpression of miR-34a inhibits the expression of its target gene SIRT1, which in turn triggers the acetylation process of P53 and P21, ultimately leading to apoptosis. Alcohol induces increased miR-34a expression, which further promotes apoptosis in hepatocytes and hepatic stellate cells through the SIRT1-p53 acetylation pathway (7, 30). These findings not only improve our understanding of the pathogenesis of ALD, but also provide new ideas and targets for the future diagnosis and treatment of ALD.
The inflammatory response plays a central role in the development and progression of ALD. Several miRNA have been shown to influence this process by regulating key signaling pathways and gene expression. Chronic alcohol consumption led to the upregulation of miR-155 in hepatic Kupffer cells, which further led to the downregulation of negative regulators of the TLR4 pathway, such as IRAK-M and SOCS1, thereby attenuating the suppression of inflammatory responses. In addition, miR-155 exacerbated the inflammatory response by down- regulating the expression of histone deacetylase HDAC11 in Kupffer cells, which in turn promoted the expression of IL-10, enhanced the responsiveness of Kupffer cells to LPS, and increased the release of inflammatory factors such as TNF-α (31). Therefore, miR-155 knockdown not only attenuates alcohol-induced lipid accumulation by targeting PPAR-α, but also prevents alcohol- induced inflammation. One study found that miR-125a-5p reduced Kupffer cell tolerance to LPS by directly targeting TLR4, CD14 and effector cytokines (e.g., TNF-α, IL-6, etc.) (32). Significant down- regulation of miR-181b-3p in chronically alcohol-fed rats, which led to up-regulation of NFκB, a key transcription factor regulating inflammatory responses, exacerbated alcohol-induced inflammation. On the other hand, miR-21 promotes hepatic stellate cell activation and inflammatory responses through activation of the VHL/NF-κB pathway, which further releases inflammatory cytokines such as IL-6 and IL-1 β, further exacerbating the inflammatory state of hepatocytes (33). In addition, miR-182 is elevated in the liver ducts and hepatocytes of ALD patients, which directly affects the expression of inflammatory mediators (34).
In the pathological process of alcoholic liver injury, downregulation of miR-let-7a expression leads to an increase in Lin28B expression, which further promotes HSC activation and accelerates the progression of liver fibrosis (35). miR-148a-3p in hepatocytes inhibits ERBB3 expression, reduces cell viability, and promotes stellate cell apoptosis by regulating the signaling of its target gene ERBB3 (36, 37). On the other hand, in alcohol-treated rat livers, increasing the level of miR-148a-3p inhibited the expression of α-smooth muscle actin (α-SAM) and type I collagen, thereby ameliorating alcohol-induced liver fibrosis (38). In addition, miR-34a expression is upregulated in ALD, which contributes to the progression of liver fibrosis. In animal studies, inhibition of miR-34a suppressed TGF-β1/Smad2/3 signaling and attenuated liver fibrosis (39). The effect of alcohol on miRNA expression is also reflected in miR-19b and pri-miR-17-92. In primary hsc and LX2 cells, alcohol was able to down-regulate miR-19b expression and significantly up-regulate pri-miR-17-92, a change that further contributed to liver fibrosis (40). In conclusion, a variety of miRNA, including miR-let-7a, miR-let-7b, miR-148a-3p, miR-34a, and miR-19b, regulate the onset and progression of liver fibrosis through different molecular mechanisms, providing new potential targets for the treatment of liver fibrosis.
lncRNAs are closely associated with the development of liver disease and are not only involved in the regulation of various processes associated with liver injury, but also offer new possibilities for the diagnosis and treatment of ALD (Figure 2).
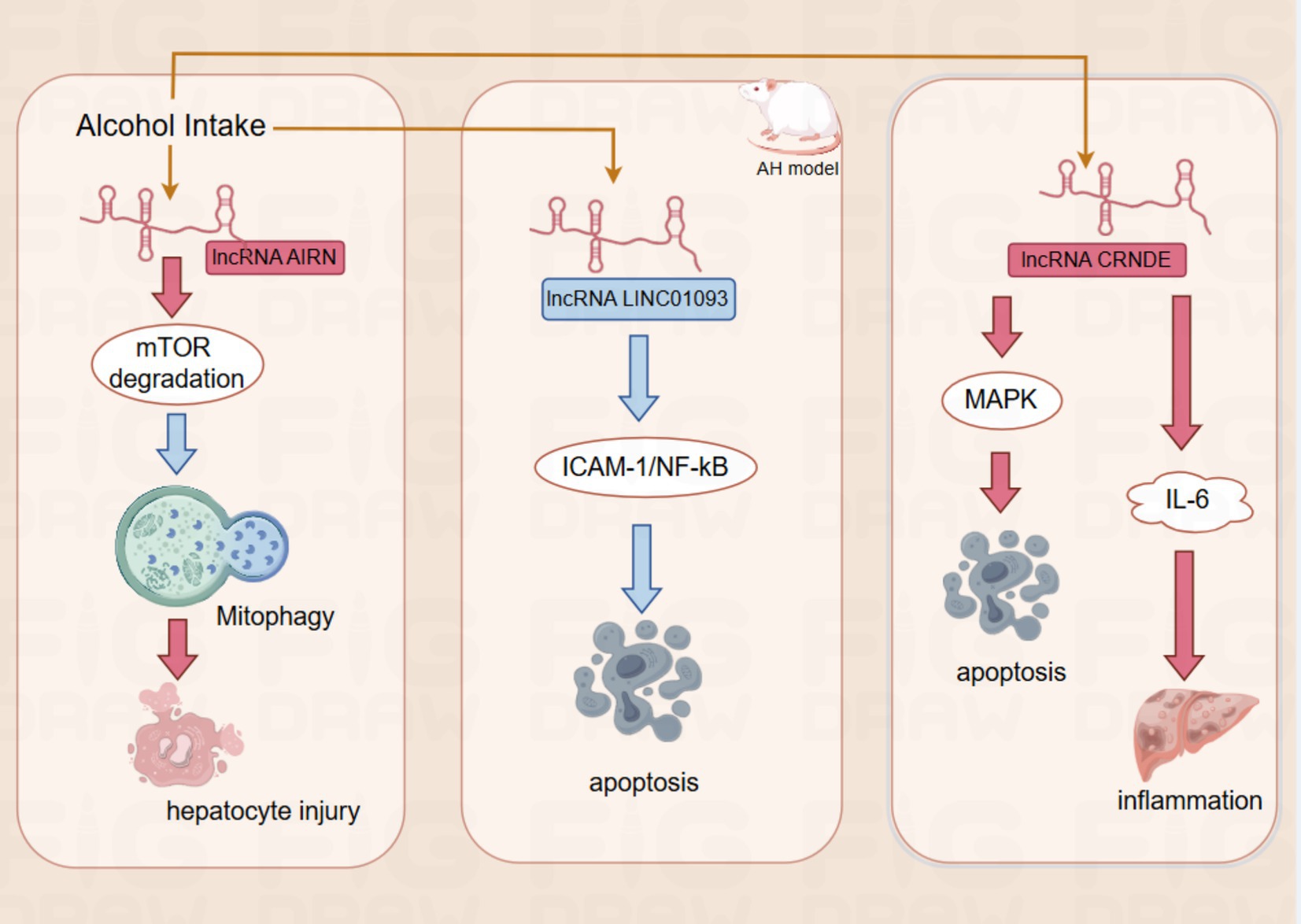
Figure 2. The red rectangles for upregulation, the blue rectangle for downregulation.The red arrows for promotion, the blue arrows for inhibition.AIRN by promoting mTOR degradation, it further suppresses mitophagy and eventually leads to hepatocyte injury; LINC01093 Inhibition of hepatocyte apoptosis by inhibiting the ICAM-1-mediated NF- κ B signaling pathway;CRNDE promotes apoptosis of hepatocytes by altering MAPK activity. Moreover, CRNDE also activates cytokines such as IL-6, which participate in the inflammatory response.
In ALD, the expression of the lncRNA AIRN is upregulated, which regulates the protein expression level of mTOR (mammalian target of rapamycin) by promoting its ubiquitination and degradation. mTOR is a key regulator of cell growth and metabolism, and reducing its activity contributes to promoting mitochondrial autophagy. Interfering with the expression of lncRNA AIRN can reduce the activity of mTOR, which in turn promotes mitochondrial autophagy and helps hepatocytes resist alcohol-induced injury (41). In addition, the expression of lncRNA LINC01093 was downregulated in an AH mouse model, resulting in its reduced inhibitory effect on the ICAM-1-mediated NF-κB signaling pathway, which further led to an increase in hepatocellular apoptosis (42). The lncRNA CRNDE was up-regulated in patients with ALD. CRNDE was directly involved in the development of ALD by altering the activity of specific signaling pathways such as MAPK (mitogen-activated protein kinase) activity, CRNDE directly regulates intracellular signaling in hepatocytes, thereby affecting the apoptotic process of hepatocytes. In addition, CRNDE activates cytokines such as IL-6, which are involved in the inflammatory response. These findings further demonstrate the important role of lncRNAs in the pathogenesis of ALD and provide a basis for their use as potential therapeutic targets and diagnostic markers (9). Validation in animal experiments and cellular models showed that the expression levels of lncRNAs AK054921 and AK128652 in the serum of ALD patients were closely associated with disease progression. Their expression levels were inversely correlated with the survival of patients with alcoholic cirrhosis, suggesting that these lncRNAs may serve as serum biomarkers to differentiate the progression of ALD and may also have predictive value for patient prognosis and survival (43). These findings not only improve our understanding of the pathogenesis of ALD but also provide new ideas and tools for clinical treatment and diagnosis. Although many studies have investigated the expression of lncRNAs in alcohol abuse-related diseases, little is known about their mechanism of action in ALD.
In ALD, the expression patterns of AIRN, LINC01093, and CRNDE in liver tissues do not show similar changes in other organs or diseases, suggesting that these lncRNAs may be liver-specific regulatory factors (9, 41, 42). This tissue-specific expression characteristic makes them potential biomarkers for liver diseases, particularly for early diagnosis of ALD. Monitoring the expression changes of AIRN, LINC01093, and CRNDE in liver tissue or serum could aid in the early detection of ALD and in tracking disease progression. Furthermore, the expression levels of AK054921 and AK128652 in the serum of ALD patients are closely associated with disease progression, and their changes are negatively correlated with the survival rate of patients with alcoholic liver cirrhosis (43). Therefore, they could serve as serum biomarkers to differentiate between various stages of ALD and provide valuable information for prognostic evaluation.
Thus, the tissue specificity of these lncRNAs not only makes them diagnostically relevant in ALD but also potentially offers new directions for targeted therapies for liver diseases. By modulating the expression of these lncRNAs, it may be possible to alleviate liver damage caused by ALD and improve clinical outcomes for patients.
The synergistic effect of lncRNAs with microRNAs is more convincing in the study of ALD than the single effect of lncRNAs. With the development of high-throughput sequencing and other technologies, multiple interactions of lncRNAs with microRNAs are gradually being discovered, forming interconnected complex networks (Figure 3; Table 2).
In mouse liver tissue from AH model and alcohol-treated AML12 hepatocytes, lncRNA 1700020I14Rik indirectly increased the expression of AKR1B10 by inhibiting its activity through interaction with miR-137, which in turn activated the Erk pathway and enhanced the damage response of hepatocytes (44). In addition, alcohol-induced downregulation of the expression of lncRNA Gm5091 in hepatic stellate cells (HSCs). Gm5091 contains sites that bind to miR-27b, miR- 23b, and miR-24 and reduce the availability of miRNA by forming a complex with these miRNA, which reduces the expression of HSC activation markers α-SMA, desmin, and type I collagen expression. Overexpression of Gm5091 also significantly reduced secretion of the inflammatory factor IL-1β and intracellular ROS production, thereby attenuating hepatocyte inflammation, oxidative stress, and fibrosis (45). In the ASH mouse model, lncRNA NEAT1 slowed the liver fibrosis process in ASH by binding to miR-129-5p, reducing its availability in cells and further inhibiting its targeted inhibitory effect on the translation of SOCS2 mRNA. lncRNA NEAT1 is involved in the onset and progression of ALD by mediating the targeting of miR-129-5p to SOCS2 (46). The transcription factor KLF5 regulates the expression of pro-inflammatory cytokines (e.g., TNF-α, IL-1β, and IL-6) and affects hepatocyte survival and death by modulating the expression of apoptosis-related genes (e.g., Bcl-2 family proteins) (47). It was found that alcohol- induced upregulation of lncRNA UCA1. lncRNA UCA1 binds to miR-214 through its sequence, reduces the inhibitory effect of miR-214 on its target gene KLF5, promotes the expression of KLF5, and affects cellular inflammatory response and apoptosis. Knockdown of UCA1 expression can increase the level of miR-214, which in turn regulates the expression of KLF5, suggesting that the UCA1/miR-214/KLF5 axis plays a key role in alcohol-induced hepatocyte injury (48). In addition, it was found that the lncRNA HOTAIR can specifically bind to miR-148a-3p, regulate the expression of S1PR1 (sphingosine 1-phosphate receptor), and affect the proliferation and apoptosis of hepatic stellate cells. HOTAIR reduces the inhibitory effect of miR-148a-3p on its target S1PR1 by binding to miR-148a-3p, thereby upregulating S1PR1 expression, which promotes hepatic stellate cell activity, including proliferation and migration, and exacerbates liver injury and fibrosis. The important role of this axis in alcoholic liver injury has been demonstrated in the AH rat model by altering the phenotype of hepatic stellate cells through regulation of the HOTAIR/miR-148a-3p/S1PR1 axis (49). It was also found that the expression levels of lncRNA MEG3 and NLRC5 were significantly increased in an alcohol-fed model, whereas that of miR-let-7c-5p was significantly decreased. lncRNA MEG3, as a competing endogenous RNA (ceRNA) for miR-let-7c-5p, inhibited the interaction of miR-let-7c-5p with the binding of NLRC5’s 3' untranslated region (3'UTR), which promoted NLRC5 expression and further aggravated alcohol-induced steatosis and apoptosis (50).
In the future, the relationship between specific lncRNAs-miRNA needs to be studied in depth, and a deeper understanding of the synergistic effects of lncRNAs and microRNAs may provide new clues and significant discoveries for the treatment of ALD and other diseases.
Autophagy is a process by which cells maintain intracellular homeostasis through the degradation of damaged organelles and proteins (51). It plays a crucial role in the pathogenesis of ALD. Alcohol disrupts hepatic lipid homeostasis through multiple pathways, including promoting lipid synthesis, reducing lipoprotein secretion, and inhibiting fatty acid oxidation. Lipophagy, a specialized form of autophagy, is responsible for the degradation of lipid droplets in hepatocytes. Studies have shown that alcohol exposure can suppress lipophagy, leading to hepatic steatosis (52). Dynamin2 (DYN2) and GTPase RAB7 are critical for lipid droplet degradation and endosome-lysosome transport. Chronic alcohol consumption reduces their activity, thereby inhibiting lipophagy and promoting lipid accumulation (8, 10, 53, 54). Furthermore, alcohol decreases the number and functionality of hepatic lysosomes, impairing autophagic flux. TFEB, a master regulator of lysosomal biogenesis, plays a key role in this process. Acute alcohol intake increases nuclear TFEB levels in hepatocytes, whereas chronic alcohol exposure reduces its nuclear localization. TFEB knockdown exacerbates alcohol-induced liver damage, while its overexpression alleviates ALD-related pathological changes (55, 56). Chronic alcohol consumption activates the mTOR pathway, inhibiting TFEB nuclear translocation and thereby reducing autophagic activity. However, treatment with mTOR inhibitors such as Torin-1 can restore TFEB activity and mitigate liver damage (57, 58). Finally, chaperone-mediated autophagy (CMA) also plays a significant role in ALD. Studies have demonstrated that SNX10-deficient mice exhibit resistance to alcohol-induced liver damage. The absence of SNX10 upregulates LAMP2A, enhances CMA activity, and alleviates liver injury via the NRF2 and AMPK signaling pathways (59). Additionally, LCN2 deficiency sustains CMA activity and reduces liver damage caused by chronic alcohol consumption (60). These findings indicate that autophagy, particularly lipophagy, TFEB regulation, and CMA, plays a vital role in the progression of ALD and provides potential therapeutic targets for future interventions.
Alcoholic fatty liver disease (AFLD) is the primary manifestation of ALD, and its pathogenesis involves liver toxicity induced by ethanol and its metabolites, leading to oxidative stress, endoplasmic reticulum (ER) stress, and mitochondrial dysfunction. These processes trigger hepatocellular damage, inflammation, and liver fibrosis. ER stress occurs due to the accumulation of misfolded or unfolded proteins in the ER, resulting in ER dysfunction. The cell responds to this stress by activating the unfolded protein response (UPR) (61). ER stress is the trigger for UPR, which is a protective response by the cell to cope with ER stress. Studies have found that alcohol impairs the activity of methionine synthase, leading to hyperhomocysteinemia and ER stress, which further disrupts lipid metabolism and increases triglyceride and cholesterol production (62). Alcohol-induced UPR is typically transient, but chronic ALD leads to severe oxidative damage, inflammation, and cell apoptosis (63, 64), especially through the PERK-ATF6 signaling pathway, which upregulates NNMT and promotes lipid generation (65). In liver specimens from ALD patients, the ER protein Nogo-B is inhibited by CHOP signaling, which promotes M1 polarization of Kupffer cells and exacerbates liver damage (66). ER stress is influenced by factors such as acetaldehyde, hyperhomocysteinemia, and oxidative stress. The activation of UPR signaling in the later stages of ALD worsens lipid generation, inflammation, and apoptosis. Inhibiting UPR may offer potential therapeutic benefits for ALD treatment.
In addition to their roles in ALD, these ncRNAs are also widely involved in the regulation of various alcohol-independent diseases. Studies have shown that miR-155 promotes inflammatory signaling pathways in immune system diseases, such as rheumatoid arthritis (67), and in various cancers, such as breast cancer and lymphoma, by inhibiting tumor suppressor factors, thereby promoting cancer cell proliferation and metastasis (68). miR-223 plays a protective role in atherosclerosis and cardiovascular diseases by regulating inflammation and lipid metabolism (69), while in hematological diseases like leukemia, it exhibits dual roles, either promoting or suppressing cancer progression (70). As a downstream regulator of p53, miR-34a inhibits cell proliferation in various cancers, including ovarian and prostate cancers (71, 72), and impacts neuronal survival and synaptic plasticity in neurodegenerative diseases like Alzheimer’s disease (73). Furthermore, lncRNA HOTAIR has been widely demonstrated to promote cancer cell invasion and metastasis through chromatin remodeling in various malignancies, including lung, breast, and colorectal cancers (74), while also regulating inflammation and vascular remodeling in cardiovascular diseases (75, 76). These studies suggest that these ncRNAs not only play roles in ALD but are also involved in the key regulatory processes of various alcohol-independent diseases. Therefore, further investigation into their functions in different pathological contexts will help to more clearly distinguish their specific roles in ALD and provide new insights for the precision treatment of these diseases.
The study of ncRNAs in ALD has provided insights into the pathogenesis of this complex disease at the molecular level and led to new directions for innovative diagnostic and therapeutic tools. However, specific, non-invasive, and more sensitive biomarkers for the diagnosis of ALD are still lacking.
Extracellular vesicles (EVs), including exosomes, transport a variety of bioactive molecules. As mediators of cell to cell communication of functional RNA (77, 78), these exosome-derived miRNA play important roles in disease initiation, development and resistance mechanism (79). The membrane structure of the exosome provides a natural protective barrier for miRNA, effectively preventing the degradation of enzymes and other chemicals, thus significantly improving miRNA stability (80, 81). In the field of hepatology, EVs has recently become a new player in the onset and progression of ALD (82–84). It has been shown that stressed hepatocytes promote exosome release and increase their content in specific proteins, lipids, and miRNA. These proteins, lipids, and miRNA modulate the transcriptional programs of adjacent hepatocytes and non-parenchymal cells (e. g., hepatic stellate cells, endothelial cells, cholangiocytes, Kupffer cells, and hepatic endothelial cells) (85). Thus regulating inflammation and fibrosis. EVs can also be secreted from other cells and affect liver inflammation. It has been shown that, On ethanol induction, the increased release of EVs from monocytes, and by delivering miR-27a with M2 polarization, stimulation of adjacent naive monocytes to polarization into M2 macrophages with increased IL-10 and TGF β secretion, thus contributing to the resolution of inflammation (86). Another study using an alcohol gastric infusion model developing ALD found that the let7f, miR-29a, and miR-340 were enriched in Evs from mouse blood, possibly as barcodes identifying ASH (87). EVs have been shown to be associated with the diagnosis and prognosis of AH. A study in trauma patients showed that only in the circulating EVs obtained by drinkers with evidence of liver injury, its specific miR barcode (miR-122 and let7f), and systemic inflammation-related markers IL-6 and IL-33 (88). This suggests that these miRNA can be used to detect “high-risk” populations for ALD. However, in ALD and other fields, its specificity and practical effect still need further study and verification.
Methylation modification is not only an important regulator of ncRNA expression and function, but ncRNAs can also influence methylation status (89), and RNA-based therapeutic strategies show great potential. For example, significant hepatic lipid accumulation is observed in a mouse model in which the hepatocyte histone methyltransferase Setdb1 is knocked out. In this process, upregulated miR-216b-5p binds to the 3'UTR of Setdb1 mRNA and interferes with its mRNA stability, which in turn exacerbates hepatic steatosis (90). Therefore, targeting strategies against hepatic Setdb1 may provide new ideas for the diagnosis and treatment of ALD. However, given the broad role of miRNA in many biological processes, their potential off-target effects need to be assessed comprehensively and cautiously.
In recent years, ncRNA-targeted therapeutic studies for ALD have gradually increased, but are still in the exploratory stage. And there are many studies using ncRNAs to regulate related liver diseases (e.g., viral hepatitis, non-alcoholic fatty liver disease (NAFLD), and hepatocellular carcinoma) at present. For example, miR-34a is a miRNA that is upregulated in ALD and other liver diseases and is involved in apoptosis and inflammatory responses. One study evaluated the safety and efficacy of a miR-34a mimic (MRX34) in patients with advanced solid-state tumors. Although this study does not directly target ALD, it provides preliminary data on miR-34a in clinical care.The findings show that MRX34 regulates apoptosis-related pathways while treating tumors, which provides important information on miR-34a as a therapeutic target for ALD (91). Another study found that in hepatitis C (HCV), miR-122 inhibitor (Miravirsen) was effective in reducing HCV viral load and improving liver function with good tolerability and safety. This is a miRNA therapy for HCV. However, this treatment also provides a theoretical basis for miR-122 therapeutic regimens in other liver diseases (e.g., ALD) (92). MALAT1 inhibition effectively slows down the course of NAFLD and beneficially affects the inflammatory response in the liver (93). Although this study focused on NAFLD, it demonstrates the potential of targeting the lncRNA MALAT1 in the treatment of liver disease, which also provides a potential therapeutic strategy for future applications in ALD.
The results of these clinical trials provide practical clinical data for the utilization of ncRNAs to modulate liver disease, and although most of the studies were not directly targeted at ALD, they provide valuable clinical experience and data support for the application of miRNA and lncRNAs in the treatment of ALD.
The study of lncRNAs and miRNA has deeply revealed the molecular biological mechanism of ALD, covering key biological pathways such as lipid metabolism, oxidative stress, inflammatory response, apoptosis and fibrosis, which not only helps to fully understand the development of ALD, but also helps in the early diagnosis and treatment of diseases.
However, there are still more limitations. First, the expression of many lncRNAs and miRNA is significantly affected by the stage of the disease and external environmental factors (e.g., amount of alcohol consumed, duration of alcohol consumption, etc.).This dynamic change may lead to instability of the biomarker, limiting its widespread clinical use. Second, because many lncRNAs and miRNA have similar expression patterns in other liver diseases (e.g., nonalcoholic steatohepatopathy, hepatocellular carcinoma), it reduces their specificity in the diagnosis of ALD and increases the risk of misdiagnosis. Finally, studies on lncRNAs and miRNA in ALD are relatively few and mostly focused on laboratory and animal models, with limited clinical sample size and lack of support from large-scale multicenter studies, which affects the generalizability and reproducibility of the findings.
Although lncRNAs and miRNA studies in ALD provide promising insights into disease mechanisms, diagnostic potential and therapeutic strategies, deeper studies are needed to overcome challenges such as specificity, delivery systems and large-scale validation to be used for future clinical applications.
LZ: Writing – original draft. RW: Writing – review & editing. YN: Writing – review & editing. LK: Methodology, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by 2023 Government-funded Clinical Medicine Excellence Talent Project, ZF2023077; Medical Science Research Project of Hebei, 20240207; and Hebei Medical Applicable Technology Tracking Project, GZ2024049.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Singal, AK, Bataller, R, Ahn, J, Kamath, PS, and Shah, VH. ACG clinical guideline: alcoholic liver disease. Am J Gastroenterol. (2018) 113:175–94. doi: 10.1038/ajg.2017.469
2. Ren, J, Fu, L, Nile, SH, Zhang, J, and Kai, G. Salvia miltiorrhiza in treating cardiovascular diseases: a review on its pharmacological and clinical applications. Front Pharmacol. (2019) 10:753. doi: 10.3389/fphar.2019.00753
3. Seitz, HK, Bataller, R, Cortez-Pinto, H, Gao, B, Gual, A, and Lackner, C. Alcoholic liver disease. Nat Rev Dis Primers. (2018) 4:16. doi: 10.1038/s41572-018-0014-7
4. Dukić, M, Radonjić, T, Jovanović, I, Zdravković, M, Gual, A, Lackner, C, et al. Alcohol, inflammation, and microbiota in alcoholic liver disease. Int J Mol Sci. (2023) 24:3735. doi: 10.3390/ijms24043735
5. Day, CP. Treatment of alcoholic liver disease. Liver Transpl. (2007) 13:S69–75. doi: 10.1002/lt.21336
6. Bala, S, Csak, T, Saha, B, Zatsiorsky, J, Kodys, K, Catalano, D, et al. The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J Hepatol. (2016) 64:1378–87. doi: 10.1016/j.jhep.2016.01.035
7. Lee, J, Padhye, A, Sharma, A, Song, G, Miao, J, Mo, YY, et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J Biol Chem. (2010) 285:12604–11. doi: 10.1074/jbc.M109.094524
8. Schulze, RJ, Rasineni, K, Weller, SG, Schott, MB, Schroeder, B, Casey, CA, et al. Ethanol exposure inhibits hepatocyte lipophagy by inactivating the small guanosine triphosphatase Rab7. Hepatol Commun. (2017) 1:140–52. doi: 10.1002/hep4.1021
9. Yan, Y, Ren, L, Liu, Y, and Liu, L. Long non-coding RNA CRNDE as potential biomarkers facilitate inflammation and apoptosis in alcoholic liver disease. Aging. (2021) 13:23233–44. doi: 10.18632/aging.203614
10. Schulze, RJ, Weller, SG, Schroeder, B, Krueger, EW, Chi, S, Casey, CA, et al. Lipid droplet breakdown requires dynamin 2 for vesiculation of autolysosomal tubules in hepatocytes. J Cell Biol. (2013) 203:315–26. doi: 10.1083/jcb.201306140
11. Shi, T, Morishita, A, Kobara, H, and Masaki, T. The role of long non-coding RNA and microRNA networks in hepatocellular carcinoma and its tumor microenvironment. Int J Mol Sci. (2021) 22:10630. doi: 10.3390/ijms221910630
12. Jiang, M-C, Ni, J-J, Cui, W-Y, Wang, B-Y, and Zhuo, W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. (2019) 9:1354–66.
13. Dragomir, MP, Kopetz, S, Ajani, JA, and Calin, GA. Non-coding RNAs in GI cancers: from cancer hallmarks to clinical utility. Gut. (2020) 69:748–63. doi: 10.1136/gutjnl-2019-318279
14. Rajtmajerov’a,, Trailin, A, Liˇska, V, Hemminki, K, and Ambrozkiewicz, F. Long noncoding RNA and microRNA interplay in colorectal cancer and their effect on the tumor microenvironment. Cancers. (2022) 14:5450. doi: 10.3390/cancers14215450
15. Akhade, VS, Pal, D, and Kanduri, C. Long noncoding RNA: genome organization and mechanism of action. Adv Exp Med Biol. (2017) 1008:47–74. doi: 10.1007/978-981-10-5203-3_2
16. Navarro, P, Page, DR, Avner, P, and Rougeulle, C. Tsix-mediated epigenetic switch of a CTCF-flanked region of the Xist promoter determines the Xist transcription program. Genes Dev. (2006) 20:2787–92. doi: 10.1101/gad.389006
17. Herman, AB, Tsitsipatis, D, and Gorospe, M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol Cell. (2022) 82:2252–66. doi: 10.1016/j.molcel.2022.05.027
18. Sheng, N, Huang, L, Gao, L, Cao, Y, Xie, X, and Wang, Y. A survey of computational methods and databases for lncRNA-MiRNA interaction prediction. IEEE/ACM Trans Comput Biol Bioinform. (2023) 20:2810–26. doi: 10.1109/TCBB.2023.3264254
19. Cheng, XY, Liu, JD, Lu, XY, Yan, X, Huang, C, Meng, XM, et al. miR-203 inhibits alcohol-induced hepatic steatosis by targeting Lipin1. Front Pharmacol. (2018) 9:275. doi: 10.3389/fphar.2018.00275
20. Yuan, W, Fang, W, Zhang, R, Lyu, H, Xiao, S, Guo, D, et al. Therapeutic strategies targeting AMPK-dependent autophagy in cancer cells. Biochim Biophys Acta, Mol Cell Res. (2023) 1870:119537. doi: 10.1016/j.bbamcr.2023.119537
21. Wang, YZ, Lu, J, Li, YY, Zhong, YJ, Yang, CF, Zhang, Y, et al. microRNA-378b regulates ethanol-induced hepatic steatosis by targeting CaMKK2 to mediate lipid metabolism. Bioengineered. (2021) 12:12659–76. doi: 10.1080/21655979.2021.2003677
22. Mostofa, MG, Tran, M, Gilling, S, Lee, G, Fraher, O, Jin, L, et al. MicroRNA-200c coordinates HNF1 homeobox B and apolipoprotein O functions to modulate lipid homeostasis in alcoholic fatty liver disease. J Biol Chem. (2022) 298:101966. doi: 10.1016/j.jbc.2022.101966
23. Fu, R, Zhou, J, Wang, R, Sun, R, Feng, D, Wang, Z, et al. Protocatechuic acid-mediated miR-219a-5p activation inhibits the p66shc oxidant pathway to alleviate alcoholic liver injury. Oxidative Med Cell Longev. (2019) 2019:1–15. doi: 10.1155/2019/3527809
24. Wang, W, Zhong, GZ, Long, KB, Liu, Y, Liu, YQ, and Xu, AL. Silencing miR-181b-5p upregulates PIAS1 to repress oxidative stress and inflammatory response in rats with alcoholic fatty liver disease through inhibiting PRMT1. Int Immunopharmacol. (2021) 101:108151. doi: 10.1016/j.intimp.2021.108151
25. Li, M, He, Y, Zhou, Z, Ramirez, T, Gao, Y, Gao, Y, et al. MicroRNA-223 ameliorates alcoholic liver injury by inhibiting the IL-6-p47-oxidative stress pathway in neutrophils. Gut. (2017) 66:705–15. doi: 10.1136/gutjnl-2016-311861
26. Dong, X, Liu, H, Chen, F, Li, D, and Zhao, Y. MiR-214 promotes the alcohol-induced oxidative stress via down- regulation of glutathione reductase and cytochrome P450 oxidoreductase in liver cells. Alcohol Clin Exp Res. (2014) 38:68–77. doi: 10.1111/acer.12209
27. Zhao, YX, Sun, YY, Huang, AL, Li, XF, Huang, C, Ma, TT, et al. MicroRNA-200a induces apoptosis by targeting ZEB2 in alcoholic liver disease. Cell Cycle. (2018) 17:250–62. doi: 10.1080/15384101.2017.1417708
28. Fan, X, Wu, J, Poulsen, KL, Kim, A, Wu, X, Huang, E, et al. Identification of a MicroRNA-E3 ubiquitin ligase regulatory network for hepatocyte death in alcohol-associated hepatitis. Hepatol Commun. (2021) 5:830–45. doi: 10.1002/hep4.1677
29. He, W, Wang, Y, Zhang, MZ, You, L, Davis, LS, Fan, H, et al. Sirt1 activation protects the mouse renal medulla from oxidative injury. J Clin Invest. (2010) 120:1056–68. doi: 10.1172/JCI41563
30. Shen, Z, Liang, X, Rogers, CQ, Rideout, D, and You, M. Involvement of adiponectin-SIRT1-AMPK signaling in the protective action of rosiglitazone against alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol. (2010) 298:G364–74. doi: 10.1152/ajpgi.00456.2009
31. Bala, S, Csak, T, Kodys, K, Catalano, D, Ambade, A, Furi, I, et al. Alcohol-induced miR-155 and HDAC11 inhibit negative regulators of the TLR4 pathway and lead to increased LPS responsiveness of Kupffer cells in alcoholic liver disease. J Leukoc Biol. (2017) 102:487–98. doi: 10.1189/jlb.3A0716-310R
32. Curtale, G, Renzi, TA, Mirolo, M, Drufuca, L, Albanese, M, De Luca, M, et al. Multi-step regulation of the TLR4 pathway by the miR-125a~99b~let-7e cluster. Front Immunol. (2018) 9:2037. doi: 10.3389/fimmu.2018.02037
33. Wu, N, McDaniel, K, Zhou, T, Ramos-Lorenzo, S, Wu, C, Huang, L, et al. Knockout of microRNA-21 attenuates alcoholic hepatitis through the VHL/NF-κB signaling pathway in hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol. (2018) 315:G385–98. doi: 10.1152/ajpgi.00111.2018
34. Blaya, D, Coll, M, Rodrigo-Torres, D, Vila-Casadesús, M, Altamirano, J, Llopis, M, et al. Integrative microRNA profiling in alcoholic hepatitis reveals a role for microRNA-182 in liver injury and inflammation. Gut. (2016) 65:1535–45. doi: 10.1136/gutjnl-2015-311314
35. McDaniel, K, Huang, L, Sato, K, Wu, N, Annable, T, Zhou, T, et al. The let-7/Lin28 axis regulates activation of hepatic stellate cells in alcoholic liver injury. J Biol Chem. (2017) 292:11336–47. doi: 10.1074/jbc.M116.773291
36. Xiong, J, Ni, J, Chen, C, and Wang, K. miR-148a-3p regulates alcoholic liver fibrosis through targeting ERBB3. Int J Mol Med. (2020) 46:1003–12. doi: 10.3892/ijmm.2020.4655
37. Scheving, LA, Zhang, X, Threadgill, DW, and Russell, WE. Hepatocyte ERBB3 and EGFR are required for maximal CCl4-induced liver fibrosis. Am J Physiol Gastrointest Liver Physiol. (2016) 311:G807–16. doi: 10.1152/ajpgi.00423.2015
38. Mimura, S, Iwama, H, Kato, K, Nomura, K, Kobayashi, M, Yoneyama, H, et al. Profile of microRNAs associated with aging in rat liver. Int J Mol Med. (2014) 34:1065–72. doi: 10.3892/ijmm.2014.1892
39. Wan, Y, McDaniel, K, Wu, N, Ramos-Lorenzo, S, Glaser, T, Venter, J, et al. Regulation of cellular senescence by miR-34a in alcoholic liver injury. Am J Pathol. (2017) 187:2788–98. doi: 10.1016/j.ajpath.2017.08.027
40. Brandon-Warner, E, Feilen, NA, Culberson, CR, Field, CO, deLemos, AS, Russo, MW, et al. Processing of miR17-92 cluster in hepatic stellate cells promotes hepatic Fibrogenesis during alcohol-induced injury. Alcohol Clin Exp Res. (2016) 40:1430–42. doi: 10.1111/acer.13116
41. Shen, S, Wang, J, and Lin, LM. Downregulation of long non-coding RNA AIRN promotes mitophagy in alcoholic fatty hepatocytes by promoting ubiquitination of mTOR. Physiol Res. (2021) 70:245–53. doi: 10.33549/physiolres.934549
42. Shi, X, Jiang, X, Yuan, B, Liu, T, Tang, Y, Che, Y, et al. LINC01093 upregulation protects against alcoholic hepatitis through inhibition of NF-κB signaling pathway. Mol Ther Nucleic Acids. (2019) 17:791–803. doi: 10.1016/j.omtn.2019.06.018
43. Yang, Z, Ross, RA, Zhao, S, Tu, W, Liangpunsakul, S, and Wang, L. LncRNA AK054921 and AK128652 are potential serum biomarkers and predictors of patient survival with alcoholic cirrhosis. Hepatol Commun. (2017) 1:513–23. doi: 10.1002/hep4.1061
44. Wu, Y, Qi, Y, Bai, Y, Zhang, H, Zhu, W, Zhou, S, et al. LncRNA 1700020I14Rik promotes AKR1B10 expression and activates Erk pathway to induce hepatocyte damage in alcoholic hepatitis. Cell Death Discov. (2022) 8:374. doi: 10.1038/s41420-022-01135-w
45. Zhou, B, Yuan, W, and Li, X. LncRNA Gm5091 alleviates alcoholic hepatic fibrosis by sponging miR-27b/23b/24 in mice. Cell Biol Int. (2018) 42:1330–9. doi: 10.1002/cbin.11021
46. Ye, J, Lin, Y, Yu, Y, and Sun, D. LncRNA NEAT1/microRNA-129-5p/SOCS2 axis regulates liver fibrosis in alcoholic steatohepatitis. J Transl Med. (2020) 18:445. doi: 10.1186/s12967-020-02577-5
47. Luo, Y, and Chen, C. The roles and regulation of the KLF5 transcription factor in cancers. Cancer Sci. (2021) 112:2097–117. doi: 10.1111/cas.14910
48. Xiang, H, Tu, B, Luo, M, Hou, P, Wang, J, Zhang, R, et al. Knockdown of UCA1 attenuated the progression of alcoholic fatty disease by sponging miR-214. Mamm Genome. (2022) 33:534–42. doi: 10.1007/s00335-022-09953-0
49. Chen, D, Lu, P, Sun, T, and Ding, A. Long non-coding RNA HOX transcript antisense intergenic RNA depletion protects against alcoholic hepatitis through the microRNA-148a-3p/sphingosine 1-phosphate receptor 1 axis. Cell Tissue Res. (2023) 394:471–85. doi: 10.1007/s00441-023-03835-w
50. Wang, Q, Li, M, Shen, Z, Bu, F, Yu, H, Pan, X, et al. The Long non-coding RNA MEG3/miR-let-7c-5p Axis regulates ethanol-induced hepatic steatosis and apoptosis by targeting NLRC5. Front Pharmacol. (2018) 9:302. doi: 10.3389/fphar.2018.00302
51. Liu, S, Yao, S, Yang, H, Liu, S, and Wang, Y. Autophagy: regulator of cell death. Cell Death Dis. (2023) 14:648. doi: 10.1038/s41419-023-06154-8
52. Wang, L, Zhou, J, Yan, S, Lei, G, Lee, CH, and Yin, XM. Ethanol-triggered Lipophagy requires SQSTM1 in AML12 hepatic cells. Sci Rep. (2017) 7:12307. doi: 10.1038/s41598-017-12485-2
53. Rasineni, K, Donohue, TM Jr, Thomes, PG, Yang, L, Tuma, DJ, McNiven, MA, et al. Ethanol-induced steatosis involves impairment of lipophagy, associated with reduced Dynamin2 activity. Hepatol Commun. (2017) 1:501–12. doi: 10.1002/hep4.1063
54. Schroeder, B, Schulze, RJ, Weller, SG, Sletten, AC, Casey, CA, and McNiven, MA. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology. (2015) 61:1896–907. doi: 10.1002/hep.27667
55. Chao, X, Wang, S, Zhao, K, Li, Y, Williams, JA, Li, T, et al. Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology. (2018) 155:865–879.e12. doi: 10.1053/j.gastro.2018.05.027
56. Thomes, PG, Trambly, CS, Fox, HS, Tuma, DJ, and Donohue, TM Jr. Acute and chronic ethanol administration differentially modulate hepatic autophagy and transcription factor EB. Alcohol Clin Exp Res. (2015) 39:2354–63. doi: 10.1111/acer.12904
57. Laplante, M, and Sabatini, DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. (2013) 126:1713–9. doi: 10.1242/jcs.125773IF:3.3Q3
58. Settembre, C, Zoncu, R, Medina, DL, Vetrini, F, Erdin, S, Erdin, S, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. (2012) 31:1095–108. doi: 10.1038/emboj.2012.32
59. You, Y, Li, WZ, Zhang, S, Hu, B, Li, YX, Li, HD, et al. SNX10 mediates alcohol-induced liver injury and steatosis by regulating the activation of chaperone-mediated autophagy. J Hepatol. (2018) 69:129–41. doi: 10.1016/j.jhep.2018.01.038
60. Cai, Y, Jogasuria, A, Yin, H, Xu, MJ, Hu, X, Wang, J, et al. The detrimental role played by Lipocalin-2 in alcoholic fatty liver in mice. Am J Pathol. (2016) 186:2417–28. doi: 10.1016/j.ajpath.2016.05.006
61. Xia, SW, Wang, ZM, Sun, SM, Su, Y, Li, ZH, Shao, JJ, et al. Endoplasmic reticulum stress and protein degradation in chronic liver disease. Pharmacol Res. (2020) 161:105218. doi: 10.1016/j.phrs.2020.105218
62. Ji, C, and Kaplowitz, N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology. (2003) 124:1488–99. doi: 10.1016/S0016-5085(03)00276-2
63. Galligan, JJ, Smathers, RL, Shearn, CT, Fritz, KS, Backos, DS, Jiang, H, et al. Oxidative stress and the ER stress response in a murine model for early-stage alcoholic liver disease. J Toxicol. (2012) 2012:207594:1–12. doi: 10.1155/2012/207594
64. Galligan, JJ, Fritz, KS, Backos, DS, Shearn, CT, Smathers, RL, Jiang, H, et al. Oxidative stress-mediated aldehyde adduction of GRP78 in a mouse model of alcoholic liver disease: functional independence of ATPase activity and chaperone function. Free Radic Biol Med. (2014) 73:411–20. doi: 10.1016/j.freeradbiomed.2014.06.002
65. Song, Q, Chen, Y, Wang, J, Hao, L, Huang, C, Griffiths, A, et al. ER stress-induced upregulation of NNMT contributes to alcohol-related fatty liver development. J Hepatol. (2020) 73:783–93. doi: 10.1016/j.jhep.2020.04.038
66. Yang, S, and Rao, J. Comment on "an endoplasmic reticulum protein, Nogo-B, facilitates alcoholic liver disease through regulation of Kupffer cell polarization". Hepatology. (2017) 66:1701–2. doi: 10.1002/hep.29402
67. Xu, WD, Feng, SY, and Huang, AF. Role of miR-155 in inflammatory autoimmune diseases: a comprehensive review. Inflamm Res. (2022) 71:1501–17. doi: 10.1007/s00011-022-01643-6
68. Bayraktar, R, and Van Roosbroeck, K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. (2018) 37:33–44. doi: 10.1007/s10555-017-9724-7
69. Nguyen, MA, Hoang, HD, Rasheed, A, Duchez, AC, Wyatt, H, Cottee, ML, et al. miR-223 exerts translational control of Proatherogenic genes in macrophages. Circ Res. (2022) 131:42–58. doi: 10.1161/CIRCRESAHA.121.319120
70. Rainer, J, Ploner, C, Jesacher, S, Ploner, A, Eduardoff, M, Mansha, M, et al. Glucocorticoid-regulated microRNAs and mirtrons in acute lymphoblastic leukemia. Leukemia. (2009) 23:746–52. doi: 10.1038/leu.2008.370
71. Dirimtekin, E, Mortoglou, M, Alavanda, C, Benomar Yemlahi, A, Arslan Ates, E, Guney, I, et al. miR-34a-FOXP1 loop in ovarian Cancer. ACS Omega. (2023) 8:27743–50. doi: 10.1021/acsomega.3c03867
72. Li, WJ, Liu, X, Dougherty, EM, and Tang, DG. MicroRNA-34a, prostate Cancer stem cells, and therapeutic development. Cancers (Basel). (2022) 14:4538. doi: 10.3390/cancers14184538
73. Santos-Bezerra, DP, Cavaleiro, AM, Santos, AS, Suemoto, CK, Pasqualucci, CA, Jacob-Filho, W, et al. Alcohol use disorder is associated with upregulation of MicroRNA-34a and MicroRNA-34c in hippocampal postmortem tissue. Alcohol Clin Exp Res. (2021) 45:64–8. doi: 10.1111/acer.14505
74. Bhan, A, and Mandal, SS. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. (2015) 1856:151–64. doi: 10.1016/j.bbcan.2015.07.001
75. Gao, L, Liu, Y, Guo, S, Yao, R, Wu, L, Xiao, L, et al. Circulating Long noncoding RNA HOTAIR is an essential mediator of acute myocardial infarction. Cell Physiol Biochem. (2017) 44:1497–508. doi: 10.1159/000485588
76. Woo, CJ, and Kingston, RE. HOTAIR lifts noncoding RNAs to new levels. Cell. (2007) 129:1257–9. doi: 10.1016/j.cell.2007.06.014
77. Raab-Traub, N, and Dittmer, DP. Viral effects on the content and function of extracellular vesicles. Nat Rev Microbiol. (2017) 15:559–72. doi: 10.1038/nrmicro.2017.60
78. Kogure, A, Kosaka, N, and Ochiya, T. Cross-talk between cancer cells and their neighbors via miRNA in extracellular vesicles: an emerging player in cancer metastasis. J Biomed Sci. (2019) 26:7. doi: 10.1186/s12929-019-0500-6
79. Anastasiadou, E, Jacob, LS, and Slack, FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. (2018) 18:5–18. doi: 10.1038/nrc.2017.99
80. Xu, YX, Pu, SD, Li, X, Yu, ZW, Zhang, YT, Tong, XW, et al. Exosomal ncRNAs: novel therapeutic target and biomarker for diabetic complications. Pharmacol Res. (2022) 178:106135. doi: 10.1016/j.phrs.2022.106135
81. Hamdy, NM, Basalious, EB, El-Sisi, MG, Nasr, M, Kabel, AM, Nossier, ES, et al. Advancements in current one-size-fits-all therapies compared to future treatment innovations for better improved chemotherapeutic outcomes: a step-toward personalized medicine. Curr Med Res Opin. (2024) 40:1943–61. doi: 10.1080/03007995.2024.2416985
82. Urban, SK, Mocan, T, Sänger, H, Lukacs-Kornek, V, and Kornek, M. Extracellular vesicles in liver diseases: diagnostic, prognostic, and therapeutic application. Semin Liver Dis. (2019) 39:070–7. doi: 10.1055/s-0038-1676122
83. Lemoinne, S, Thabut, D, Housset, C, Moreau, R, Valla, D, Boulanger, CM, et al. The emerging roles of microvesicles in liver diseases. Nat Rev Gastroenterol Hepatol. (2014) 11:350–61. doi: 10.1038/nrgastro.2014.7
84. Szabo, G, and Momen-Heravi, F. Extracellular vesicles in liver disease and potential as biomarkers and therapeutic targets. Nat Rev Gastroenterol Hepatol. (2017) 14:455–66. doi: 10.1038/nrgastro.2017.71
85. Chen, L, Chen, R, Kemper, S, and Brigstock, DR. Pathways of production and delivery of hepatocyte exosomes. J Cell Commun Signal. (2018) 12:343–57. doi: 10.1007/s12079-017-0421-7
86. Saha, B, Momen-Heravi, F, Kodys, K, and Szabo, G. MicroRNA cargo of extracellular vesicles from alcohol-exposed monocytes signals naive monocytes to differentiate into M2 macrophages. J Biol Chem. (2016) 291:149–59. doi: 10.1074/jbc.M115.694133
87. Eguchi, A, Lazaro, RG, Wang, J, Kim, J, Povero, D, Willliams, B, et al. Extracellular vesicles released by hepatocytes from gastric infusion model of alcoholic liver disease contain a MicroRNA barcode that can be detected in blood. Hepatology. (2017) 65:475–90. doi: 10.1002/hep.28838
88. Eguchi, A, Franz, N, Kobayashi, Y, Iwasa, M, Wagner, N, Hildebrand, F, et al. Circulating extracellular vesicles and their miR "barcode" differentiate alcohol drinkers with liver injury and those without liver injury in severe trauma patients. Front Med. (2019) 6:30. doi: 10.3389/fmed.2019.00030
89. Liu, Y, Leng, P, Liu, Y, Guo, J, and Zhou, H. Crosstalk between methylation and ncRNAs in breast Cancer: therapeutic and diagnostic implications. Int J Mol Sci. (2022) 23:15759. doi: 10.3390/ijms232415759
90. Zhang, Y, Li, Y, Liu, Y, Wang, H, Chen, Y, Zhang, B, et al. Alcoholic Setdb1 suppression promotes hepatosteatosis in mice by strengthening Plin2. Metabolism. (2023) 146:155656. doi: 10.1016/j.metabol.2023.155656
91. Ling, H, Fabbri, M, and Calin, GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. (2013) 12:847–65. doi: 10.1038/nrd4140
92. Panigrahi, M, Palmer, MA, and Wilson, JA. MicroRNA-122 regulation of HCV infections: insights from studies of miR-122-independent replication. Pathogens. (2022) 11:1005. doi: 10.3390/pathogens11091005
93. Xiang, J, Deng, YY, Liu, HX, and Pu, Y. LncRNA MALAT1 promotes PPARα/CD36-mediated hepatic lipogenesis in nonalcoholic fatty liver disease by modulating miR-206/ARNT Axis. Front Bioeng Biotechnol. (2022) 10:858558. doi: 10.3389/fbioe.2022.858558
ALD - Alcoholic liver disease
AH - Alcoholic hepatitis
AFL - Alcoholic fatty liver
lncRNAs - Long non-coding RNAs
miRNA - MicroRNAs
mTOR - Mammalian target of rapamycin
MAPK - Mitogen-activated protein kinase
HSCs - Hepatic stellate cells
GSR - Glutathione reductase
S1PR1 - Sphingosine 1-phosphate receptor
CAMKK2 - Calcium/calmodulin-dependent protein kinase kinase 2
AMPK - AMP-activated protein kinase
AKR1B10 - Aldo-keto reductase family 1 member B10
SREBP-1 - Sterol regulatory element-binding protein 1
S1PR1 - Sphingosine-1-phosphate receptor 1
HNF-1B - Hepatocyte nuclear factor 1 beta
ROS - Reactive oxygen species
PIAS1 - Protein inhibitor of activated STAT 1
PRMT1 - Protein arginine methyltransferase 1
TLR - Toll-like receptor
TGF - Transforming growth factor
ERBB3 - Epidermal growth factor receptor 3
ErK - Extracellular signal-regulated kinase
NLRC5 - NOD-like receptor family CARD domain containing 5
KLF5 - Kruppel-like factor 5
SOCS2 - Suppressor of cytokine signaling 2
α-SMA - Alpha-smooth muscle actin
EVs - Extracellular vesicles
EMT - Epithelial-mesenchymal transition
ACC - Coenzyme A carboxylase
PPARγ - Peroxisome proliferator-activated receptor γ
HNF1B - HNF1 homology box B
Apo - Oapolipoprotein O
ACC1 - Acetyl CoA carboxylase
FABP4 - Fatty acid binding protein
LDLR - Low-density lipoprotein receptor-associated protein
LXR - Liver X-activated receptor
3'UTR - 3' untranslated region
GSR - Glutathione reductase
CISH - Cytokine signaling inhibitor
Keywords: alcoholic liver disease, MicroRNAs, long non-coding RNAs, gene regulation, therapeutic targets
Citation: Zhang L, Wang R, Nan Y and Kong L (2025) Molecular regulators of alcoholic liver disease: a comprehensive analysis of microRNAs and long non-coding RNAs. Front. Med. 12:1482089. doi: 10.3389/fmed.2025.1482089
Received: 17 August 2024; Accepted: 20 February 2025;
Published: 10 March 2025.
Edited by:
Pradeep Kumar Shukla, University of Tennessee Health Science Center (UTHSC), United StatesReviewed by:
Manikandan Murugesan, University of California, San Diego, United StatesCopyright © 2025 Zhang, Wang, Nan and Kong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingbo Kong, a2xiaGR6aEAxNjMuY29t
†ORCID: Lingbo Kong, orcid.org/0000-0002-6899-6559
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.