- 1Department of Clinical Laboratory, Wuhan Asia General Hospital, Wuhan Asia General Hospital Affiliated to Wuhan University of Science and Technology, Wuhan, China
- 2Respiratory and Critical Care Medicine, Wuhan Asia General Hospital, Wuhan Asia General Hospital Affiliated to Wuhan University of Science and Technology, Wuhan, China
- 3Department of Radiology, Wuhan Asia General Hospital, Wuhan Asia General Hospital Affiliated to Wuhan University of Science and Technology, Wuhan, China
Background: Psittacosis is a zoonotic infectious disease caused by Chlamydia psittaci (C. psittaci) infection, which can be transmitted by birds, poultry and wild animals. The symptoms and imaging findings of C. psittaci pneumonia are atypical and primarily rely on etiological diagnosis. The incidence of C. psittaci infection has been significantly underestimated because of the low sensitivity and poor timeliness of traditional diagnostic methods. Therefore, early and accurate diagnosis of psittacosis remains a challenge.
Case presentation: A case series with six pneumonia patients who were admitted to our hospital in the period from January 2023 to June 2023 is presented. These patients exhibited acute onset and symptoms, including fever, cough, poor appetite, dry mouth, dizziness, chills, and chest tightness. Despite comprehensive laboratory and radiological examinations, the cause of the pneumonia remained unidentified. Therefore, a sample of bronchoalveolar lavage fluid (BALF) was tested via target next-generation sequencing (tNGS), which revealed a positive result for C. psittaci. Prompt adjustment of the treatment regimens upon identification of the pathogen led to favorable outcomes in all patients.
Conclusion: tNGS is a novel diagnostic technology that enables rapid, accurate and cost-effective detection of C. psittaci pneumonia. Early detection of C. psittaci can improve patient outcomes through timely adjustment of therapies.
1 Introduction
According to the Global Burden of Disease Collaboration, it is estimated that nearly 600 million individuals were estimated to be affected by pneumonia and other lower respiratory tract infections globally in 2019, resulting in 2.5 million deaths (1). The epidemiologic distribution of pneumonia pathogens has shifted in recent years, with an increase in some rare pathogens. C. psittaci, an intracellular gram-negative pathogen, is commonly found in birds (especially parrots and pigeons) and mammals (2). Inhalation of aerosols from infected bird feathers or avian excreta can lead to C. psittaci pneumonia in humans. This pathogen is frequently misdiagnosed, leading to the potential misuse of antimicrobials and the development of drug resistance (3, 4). Patients with C. psittaci pneumonia typically exhibit sepsis and a rapidly deteriorating condition. Early diagnosis and treatment are crucial for improving patient prognosis because of the rapid progression of the disease (5).
The diagnosis of C. psittaci pneumonia is challenging because of its nonspecific clinical presentation and often relies on laboratory testing for the pathogen. Currently, culture, serology and molecular testing are the main methods of detection. Cell cultures for C. psittaci are time-consuming and require a high level of laboratory biosafety, making them unsuitable for routine use, and serologic tests have limited early diagnostic value, as they are more appropriate for retrospective diagnosis (6). PCR reagents for C. psittaci are not readily available and are limited by clinician judgment, although PCR is considered the gold standard for detection (7). Metagenomic next-generation sequencing (mNGS) employs high-throughput sequencing technology to capture the comprehensive range of microbial nucleic acid sequences present in samples. These sequences are then compared and analyzed with existing microbial nucleic acid sequences stored in a database. This approach facilitates the efficient and accurate identification of suspected pathogenic microorganisms in samples. In recent years, many reported cases of C. psittaci pneumonia have been definitively diagnosed by mNGS (8, 9). More recently, target next-generation sequencing (tNGS), a new technology based on the combination of ultra-multiplex PCR amplification and high-throughput sequencing, has gradually entered clinic practice as a faster and less expensive molecular detection method than mNGS. However, reports on the application of tNGS for C. psittaci detection are scarce. Therefore, this study was conducted to analyze the clinical data of six patients with C. psittaci pneumonia at our hospital to evaluate the effectiveness of tNGS for early and rapid diagnosis and to better understand the clinical characteristics of these patients.
2 Case presentation
Between January 2023 and June 2023, a total of 297 inpatients underwent tNGS testing, and six of these patients were diagnosed with C. psittaci pneumonia. Data regarding the demographic characteristics of the six patients, including age, gender, occupational history, and avian exposure, were collected. Clinical manifestations, signs, as well as laboratory and imaging findings, were documented. The aetiological diagnosis was based on the sequence of the C. psittaci obtained from BALF samples via tNGS. Furthermore, the use of antibiotics prior to and following the diagnosis of C. psittaci pneumonia, along with the time intervals from symptom onset to admission and from admission to diagnosis, were recorded.
2.1 Baseline characteristics and clinical manifestations
Table 1 presents general information on the six patients, comprising three men and three women aged between 18 and 66 years, who were included in this study. Among them, two patients had a history of hypertension, three had a history of diabetes, one had a history of atrial fibrillation, one had a history of ulcers, and one had abnormal liver function. Additionally, three patients had a documented history of exposure to poultry, whereas two had a history of exposure to parrots. All patients experienced fever, with the highest body temperature recorded ranging from 38.0 to 40.0°C. Common accompanying symptoms included cough (3/6), headache (2/6), dizziness (2/6), cold symptoms (2/6), stiffness (3/6), chest tightness (2/6), pharyngeal pain (1/6), fatigue (4/6), muscle pain (2/6), and frequent urination urgency (2/6).
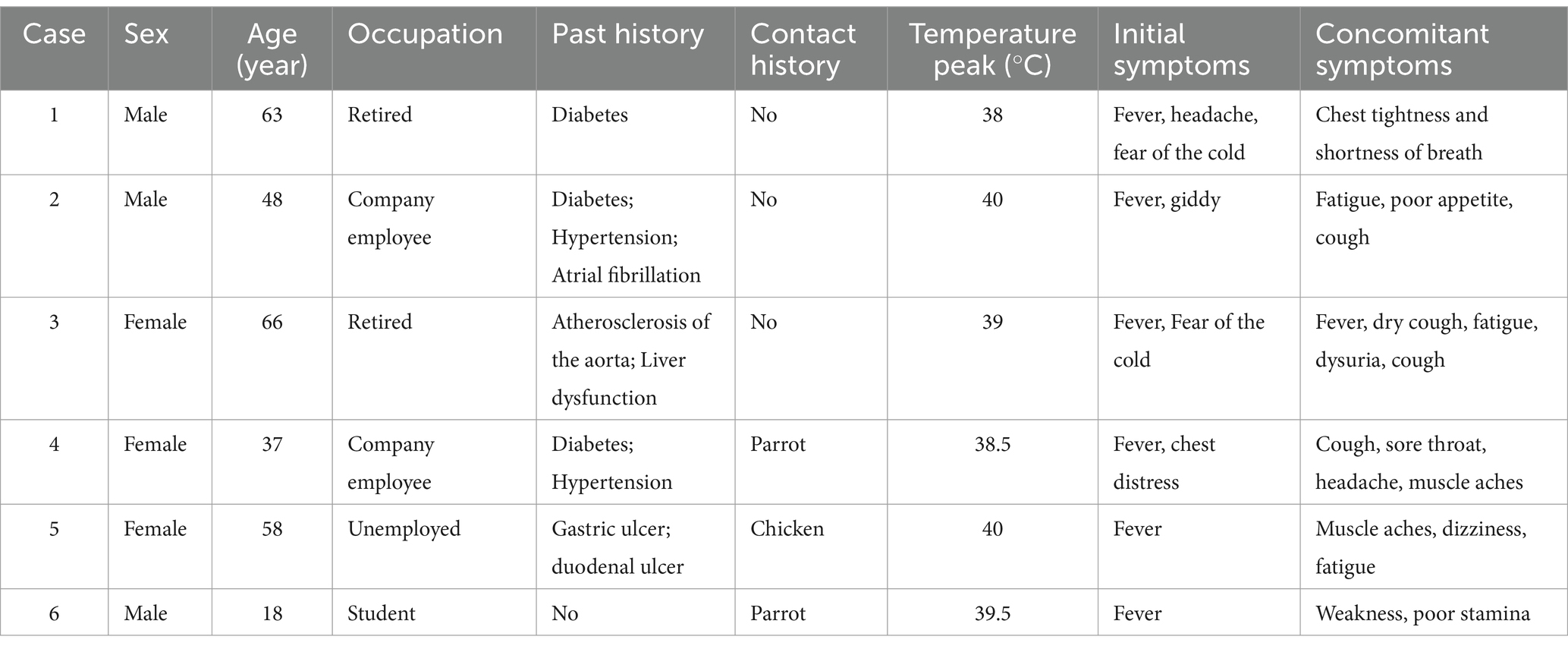
Table 1. Baseline characteristics and clinical manifestations of patients with Chlamydia psittaci pneumonia.
2.2 Clinical laboratory testing
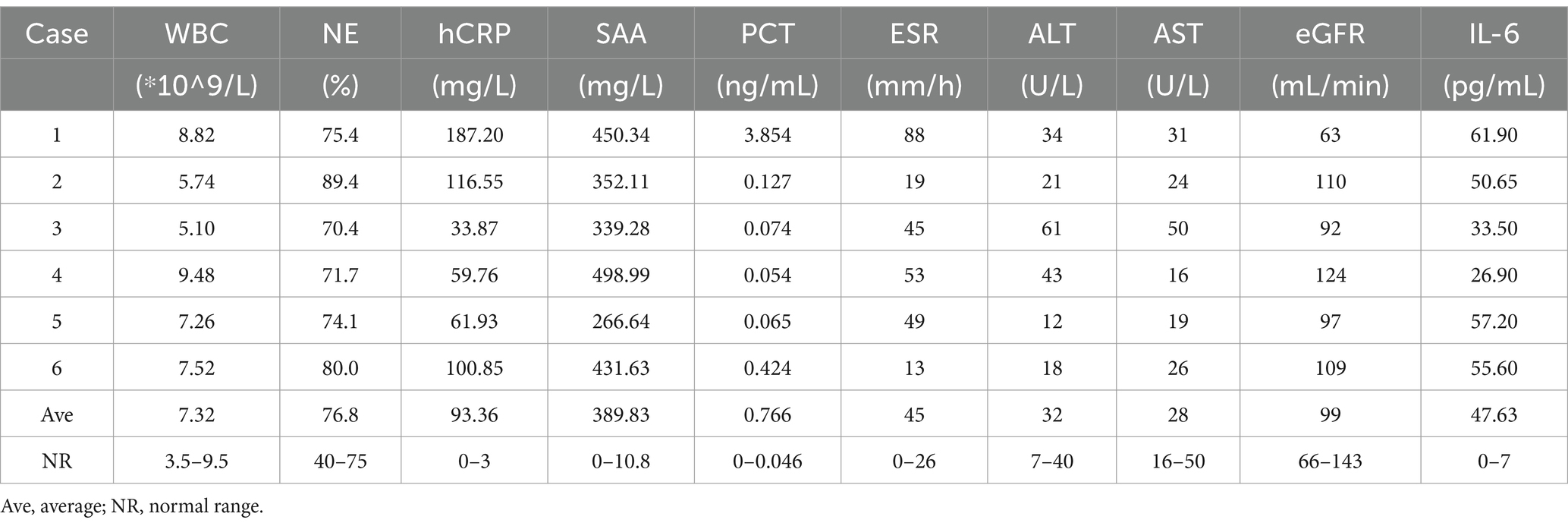
Various parameters, including the white blood cell (WBC) count, neutrophil (NE) percentage, ultrasensitive c-reactive protein (hCRP) level, serum amyloid A (SAA) level, procalcitonin (PCT) level, erythrocyte sedimentation rate (ESR), alanine aminotransferase (ALT) level, aspartate aminotransferase (AST) level, glomerular filtration rate (eGFR) and interleukin 6 (IL-6) level, were assessed upon admission (Table 2). All six patients presented normal WBC counts, with three patients showing elevated NE percentages. Furthermore, all patients presented increased levels of hCRP, SAA, PCT, and IL-6. During the initial visit, the recorded levels ranged from 33.87–187.20 mg/L, 266.64–498.99 mg/L, 0.054–3.854 ng/mL and 26.90–61.90 pg./L, respectively. The ESR ranged from 13 to 88 mm/h, with increases noted in 4 of 6 patients. ALT levels ranged from 18 to 61 U/L, with elevated levels observed in 2 of the 6 patients. AST levels fell within the range of 16–50 U/L, and elevated levels were observed in 1 of the 6 patients. Additionally, the eGFRs ranged from 63 to 124 mL/min, with elevated levels noted in 5 of the 6 patients.
2.3 Imaging findings
Chest CT was conducted for all patients upon admission (Figure 1). Among the six patients, five had lesions affecting one lung, whereas one patient had bilateral lung involvement. The lesions were diffusely distributed ground-glass shadows with localized solid lesions, and two patients presented bronchial air signs.
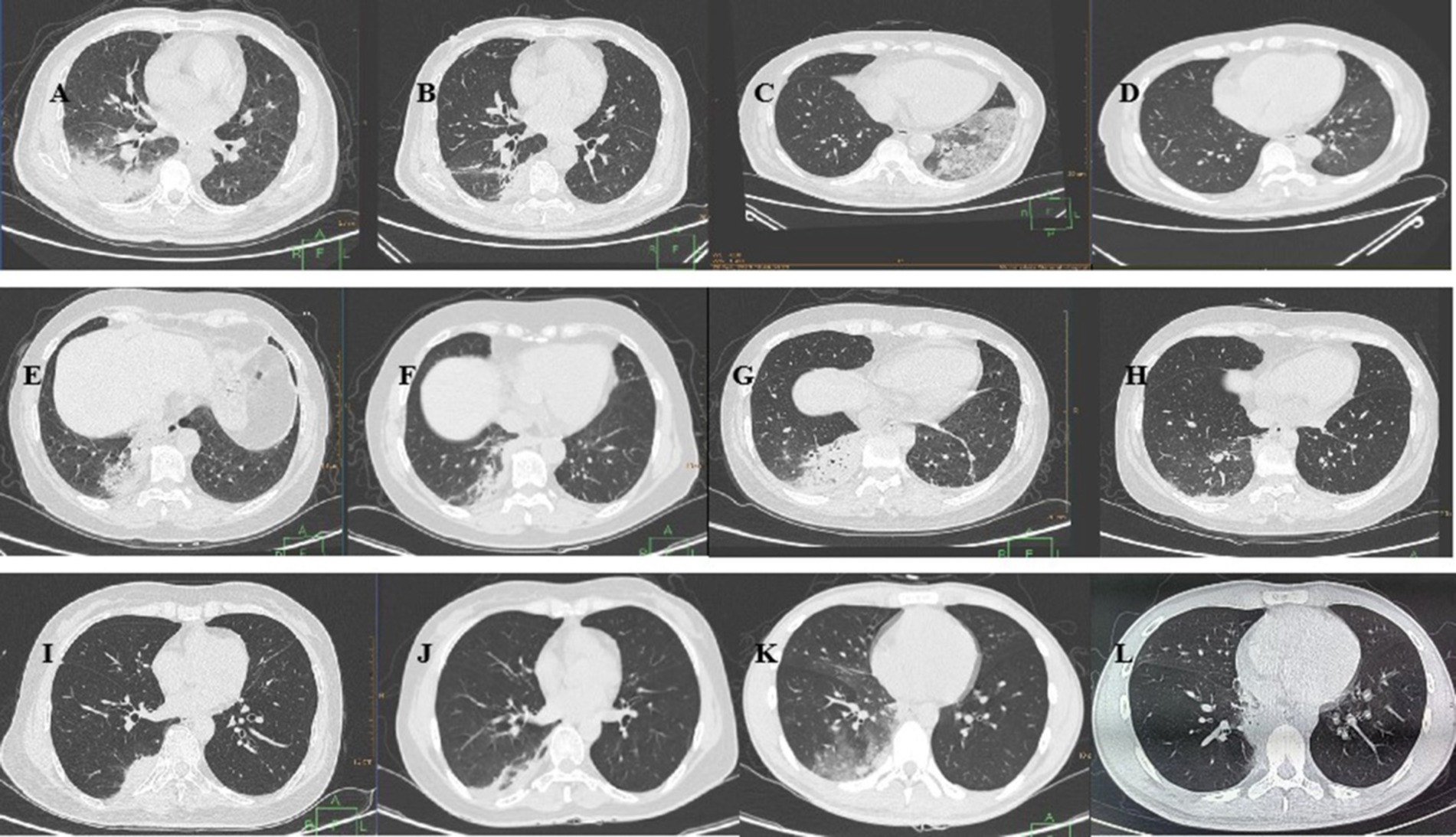
Figure 1. Chest CT of the six patients with Chlamydia psittaci pneumonia. Case 1: (A) On January 30, the lower lobe of the right lung had a large area of consolidation, and the ventilation bronchus was visible. (B) On February 10, shadow absorption in the lower lobe of the right lung was significantly reduced. Case 2: (C) On February 9, a chest CT revealed diffusely distributed ground–glass shadows with patchy solid shadows in the lower lobe of the left lung. (D) On February 28, a small number of pale shadows were observed in the lower lobe of the left lung, and the foci of intrapulmonary infection were mostly resorbed. Case 3: (E) On March 11, a solid lesion was detected in the posterior basal segment of the lower lobe of the right lung. (F) On March 18, there was a decreased lesional area and partial thinning of the posterior basal segment of the lower lobe of the right lung. Case 4: (G) On April 10, a chest CT revealed a large solid lesion in the lower lobe of the right lung and a striated shadow in the lower lobe of the left lung. (H) On April 17, chest CT revealed that the solid lesion in the original lung was largely absorbed, as was the striated shadow in the left lower lobe. Case 5: (I) On April 12, a chest CT revealed a solid shadow in the posterior basal segment of the lower lobe of the right lung. (J) On April 21, a chest CT revealed inflammatory thinning of the lower lobe of the right lung. Case 6: (K) On June 26, there were multiple ground glass shadows in the middle lobe and lower lobe of the right lung, and patchy consolidation was observed in the lower lobe of the right lung. (L) On July 8, a chest CT revealed that the inflammation in the lower lobe of the right lung had resolved.
2.4 Targeted next-generation sequencing
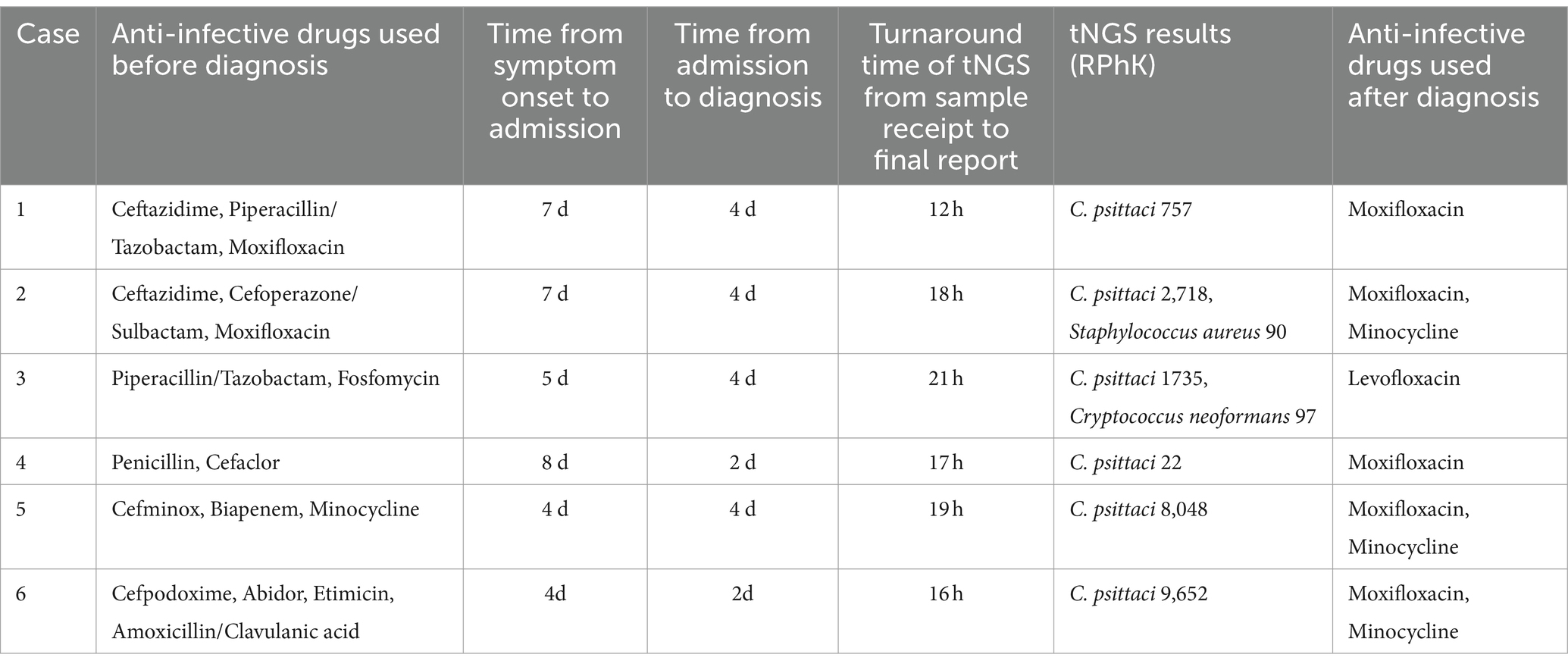
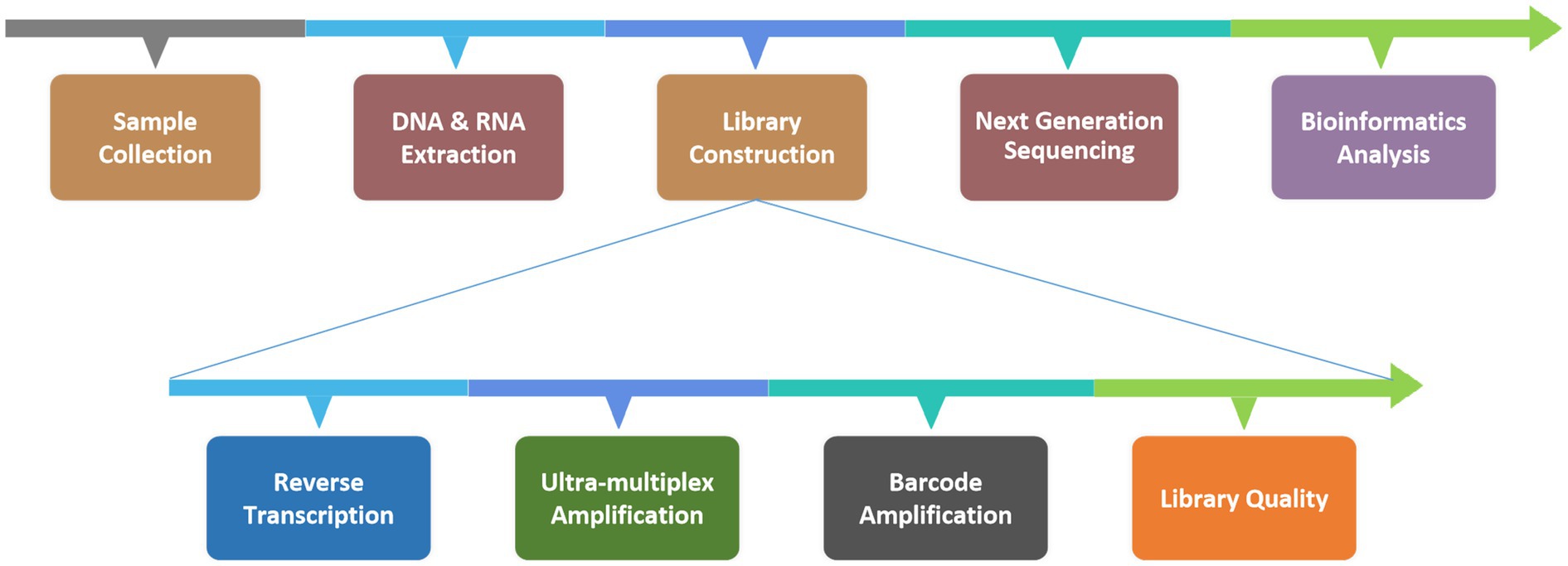
Six patients with suspected C. psittaci pneumonia underwent bronchoscopy to obtain BALF samples which were then sent to the KingMed Diagnostics Group Co., Ltd., Guangzhou, China for tNGS. C. psittaci was detected in all six cases, other pathogens were identified in two of the cases (Table 3). The protocol for the tNGS assay is showed in Figure 2. After the samples were collected in the clinical laboratory, nucleic acids (DNA and RNA) were extracted, after which the tNGS target library was constructed and sequenced. The target 198 pathogens (Supplementary Table S1) of the tNGS assay include 80 bacteria, 79 viruses, 32 fungi and 7 atypical pathogens. Three amplicons (Supplementary Table S2) were designed for the detection of C. psittaci.
2.4.1 Material processing and nucleic acid extraction
BALF samples were collected according to the established standard procedures. For viscous BALF samples, an equal volume of 0.1 mol/L dithiothreitol (DTT) liquefaction agent was added to the collection tube; the mixture was then vortexed thoroughly and incubated at room temperature for 3–5 min to ensure complete liquefaction. Non-viscous BALF samples did not require a 0.1 mol/L DTT treatment step. A total of 13 μL of exogenous endogenous reference was added to 1.3 mL of the liquefied mix; the mixture was vortexed to ensure thorough mixing and then centrifuged at 12,000 rpm for 5 min. The supernatant was discarded, and the residual sample volume was adjusted to 500 μL by pipetting. This aliquot was transferred into the bead mill tube provided in the extraction kit, to which 50 μL of SDS was added, and the mixture was subjected to a wall-breaking apparatus for mechanical lysis. Following mechanical lysis, the samples were centrifuged at 12,000 rpm for 5 min, and 250 μL of the supernatant was used for nucleic acid extraction. The extraction was performed via the MagPure Pathogen RNA/DNA Extraction Kit (Magen Biotechnology, Guangzhou, China), following the manufacturer’s protocol. The extracted nucleic acids were quantified via an Equalbit DNA HS Assay Kit (Vazyme Biotech, Nanjing, China) with an Invitrogen™ Qubit™ 4.0 (Thermo Fisher Scientific, Massachusetts, United States), and the input nucleic acids did not exceed 100 ng for library construction.
2.4.2 Library preparation and sequencing
Library preparation was performed via the RP100TM Respiratory Pathogen Microorganisms Multiplex Testing Kit (KingCreate Biotechnology, Guangzhou, China). cDNA was synthesized via reverse transcription of the extracted nucleic acids, followed by steps such as ultra-multiplex PCR amplification, PCR product purification, adapter ligation and library purification to complete library construction. The constructed libraries were pooled into homogeneous masses. The size of the library fragments was determined via the Qsep100 Bio-Fragment Analyzer (BiOptic, Taiwan, China). The size of the library fragments should be of from 250 to 350 bp. The qualified pooled library was diluted and denatured, 500 μL of which was subjected to the Illumina MiniSeq Platform (Illumina, California, United States) for sequencing.
2.4.3 Bioinformatics analysis
The raw sequencing read data were subjected to a quality control procedure. Fastp v0.20.1 was employed for adapter trimming and quality trimming using default parameters, followed by mapping to the reference via Bowtie2 v2.4.1 in ‘very-sensitive’ mode. The reference sequence used for read mapping was a database curated from various sources including the GenBank, RefSeq, and Nucleotide databases from NCBI.1 To identify positive signals for specific pathogens, the number of mapped reads was counted and normalized to the number of reads per 100,000 (RPhK). Cases with specific RPhKs were considered positive for each sample.
2.4.4 Interpretation of tNGS results
For bacteria (excluding the Mycobacterium tuberculosis complex), fungi, viruses, and atypical pathogens, the criteria for positivity are as follows: (1) RPhK ≥10 when amplicon coverage is 100% and the number of amplicons is greater than 1; (2) RPhK ≥30 when 50% ≤ amplicon coverage <100% and the number of amplicons is equal to 1; (3) RPhK ≥50 when amplicon coverage is less than 50%. For Mycobacterium tuberculosis complex: a positive result is considered when RPhK ≥1, no other samples in the same batch detect Mycobacterium tuberculosis, and the retest result is ≥1. Additionally, the detected pathogens are classified according to their pathogenicity: (A) specifically pathogenic in respiratory specimens or clinically common pathogens; (B) opportunistic (conditional) pathogens in respiratory specimens that may cause infection in patients with systemic or local immunocompromise/compromise/deficiency, respiratory barrier disruption, or lower respiratory microecological imbalance; (C) the normal microecological flora of the respiratory tract, usually does not lead to infection. Finally, we need to perform a thorough assessment of the patient’s clinical profile to determine the presence of a lung infection and the clinical relevance of the potential pathogen. This evaluation includes the patient’s medical history, symptoms, imaging findings, tNGS results and other laboratory findings.
2.5 Treatment and outcome
The diagnosis and treatment histories of the six patients with C. psittaci pneumonia are shown in Table 3. All patients received empiric anti-infective therapy with two to four antibiotics sequentially prior to diagnosis, with one patient also receiving antiviral therapy using abidor. The six patients experienced a mean time of 5.8 days from symptom onset to hospital admission (range 4–8 days) and a mean time of 3.3 days from hospital admission to diagnosis (range 2–4 days). For all samples, the tNGS turnaround time from sample receipt to the final report was less than 24 h. Additional pathogens were found in the tNGS results of two patients, but were subsequently excluded by other laboratory findings and clinical manifestations. Six patients were eventually diagnosed with C. psittaci pneumonia. Following diagnosis, the treatment regimen was promptly adjusted to quinolone antibiotic therapy for anti-infection purposes. After 1 week of anti-infection treatment, all six patients were discharged once their body temperature normalized and their clinical symptoms significantly improved. Prior to discharge, all patients underwent chest CT examinations, which revealed the disappearance of inflammatory lesions.
3 Discussion
Studies have indicated that the global prevalence of C. psittaci infection in birds is as high as to 20%, with all birds posing a potential risk for C. psittaci infection in humans (10). In the present study, three patients had confirmed contact with birds in their prehistory. This case report suggests that since close contact with birds is one of the major risk factors for infection with the pathogen, it is important to elicit the patient’s history in detail to provide direction for pathogen detection. All six patients in this case series experienced a fever between 38.0 and 40.0°C and symptoms such as cough, headache, sore throat, malaise and other flu-like symptoms. These signs and symptoms lack specificity and are similar to those of community-acquired pneumonia caused by various pathogens including viruses, fungi and bacteria. Owing to the diverse and atypical nature of the symptoms of psittacosis, its incidence has been significantly underestimated compared with that of other atypical pathogens (11). Our data further support previous reports on this matter.
Similarly, inflammatory biomarkers are nonspecific for C. psittaci pneumonia. The WBC count, PCT level, and ESR demonstrated limited diagnostic value for C. psittaci pneumonia in this study. hCRP, SAA, and IL-6 showed some diagnostic potential, but their elevation is not exclusive to this pathogen and lacks specificity. In summary, the laboratory findings in C. psittaci pneumonia are nonspecific, which aligns with the findings from previous studies (12). Chest CT scans of all patients revealed extensive lung lesions with varying degrees of pulmonary infiltrates. Unilateral lung involvement was common, but bilateral lesions were also observed, both of which were nonspecific. While C. psittaci infection is often linked to bird contact, it is not a prerequisite for diagnosis. Therefore, pathogen detection plays a crucial role in confirming the diagnosis of C. psittaci pneumonia (13).
Nonetheless, conventional methods for detecting pathogens such as culture or serological tests, are not suitable for early detection of the disease (14). Compared with traditional methods, mNGS, known for its high sensitivity and broad detection range, plays a crucial role in guiding the diagnosis and treatment of pneumonia (15). Previous studies have investigated the use of mNGS for the detection of C. psittaci infection, and this method has demonstrated high detection efficacy (8, 16–19). Moreover, mNGS has gained popularity for its ability to detect multiple pathogens simultaneously (20, 21). The rise in the diagnosis and reporting of psittacosis in recent years may be linked to the increased clinical utilization of mNGS (22). However, owing to its cost and long reporting time, mNGS may not be feasible in all cases and is typically used as a complementary option for challenging clinical scenarios. In China, the current price of mNGS in 2024 is US$ 500–700 per sample, whereas the price of tNGS is US$ 100–200. To control the cost of mNGS, batch processing of samples is often necessary, resulting in longer turnaround times. The tNGS test offers advantages such as fast detection speed, wide coverage and high accuracy, similar to those of the mNGS test, but incurs only a quarter of the cost (23). The time span from admission to diagnosis for the six patients in this study ranged from 2 to 4 days, which was notably shorter than the 5 to 11 days reported by Dai et al. for six patients diagnosed with C. psittaci pneumonia via mNGS (24). tNGS focuses on dozens to hundreds of clinically common and subrare pathogens, dramatically reducing the amount of sequencing data to 0.1 M through targeted enrichment technology, which in turn dramatically reduces the cost of testing (25). Furthermore, tNGS allows for dual-process DNA and RNA assays in a single experiment, is less impacted by prior antibiotic exposure, and excels in diagnosing rare, atypical pathogens while simultaneously identifying coinfecting pathogens. In this study, C. psittaci was consistently detected in all the BALF samples tested. Following prompt and targeted anti-infective treatment, the patients experienced a significant improvement in their symptoms, and imaging results revealed resolution of most of the infected foci. The use of tNGS technology in this study proved to be crucial in identifying the source of infection.
BALF is regarded as the most reliable specimen for the detection of pathogens in cases of lower respiratory tract infection (26, 27). However, BALF samples are challenging to obtain in some hospitals where bronchoscopy is unfeasible. Compared with BALF, sputum specimens are relatively easy to collect and have wider application and operability in practice. Zhenfeng Deng et al. (28) compared the detection of 209 sputum samples with tNGS and conventional microbiological testing methods, and found that the overall microbial detection rate of tNGS was significantly higher than that of conventional microbiological testing (96.7% vs. 36.8%). Their study suggests that tNGS may also be a valuable pathogenic diagnostic method for patients for whom alveolar lavage fluid collection is not appropriate.
In summary, the clinical manifestations of C. psittaci pneumonia can be complex, diverse and nonspecific. Clinicians should have a high index of suspicion for C. psittaci infection in patients with a history of live poultry exposure, who present with high fever and cough, along with significantly elevated levels of hCRP, SAA, and IL-6, slightly elevated PCT levels, normal WBC counts, and negative results of routine aetiological tests. Under the above circumstances, tNGS, as an emerging diagnostic technique for pathogenic microorganisms, is expected to be an economical, rapid and accurate method for diagnosing C. psittaci pneumonia.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by the ethical review committee of the Wuhan Asia General Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
XY: Data curation, Writing – original draft. HF: Data curation, Writing – review & editing. WD: Data curation, Writing – review & editing. ZZ: Conceptualization, Writing – review & editing. DW: Data curation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Fund of Wuhan Municipal Health Commission (No. WX21Q29) and the Research and Innovation Fund of Wuhan Asia General Hospital (No. 2022KYCX1-B09).
Acknowledgments
We thank the Wuhan KingMed Diagnostics for their technical assistance. We also thank the colleagues in Wuhan Asia General Hospital for their contributions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2024.1491838/full#supplementary-material
Abbreviations
mNGS, metagenomics next-generation sequencing; tNGS, target next-generation sequencing; CT, computed tomography; hCRP, hypersensitive C-reactive protein; WBC, white blood cell; SAA, serum amyloid A; ALT, alanine aminotransferase; AST, aspartate aminotransferase; ESR, erythrocyte sedimentation rate; PCT, procalcitonin; IL-6, interleukin 6; eGFR, estimated glomerular filtration rate; C. psittaci, Chlamydia psittaci; BALF, bronchoalveolar lavage fluid; PCR, polymerase chain reaction; RPhK, reads per 100,000.
Footnotes
References
1. GBD. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the global burden of disease study 2019. Lancet (North American ed). (2022) 400:2221–48. doi: 10.1016/s0140-6736(22)02185-7
2. Stewardson, AJ, and Grayson, ML. Psittacosis. Infect Dis Clin N Am. (2010) 24:7–25. doi: 10.1016/j.idc.2009.10.003
3. Han, X, Liu, X, Chen, L, Wang, Y, Li, H, Zhou, F, et al. Disease burden and prognostic factors for clinical failure in elderly community acquired pneumonia patients. BMC Infect Dis. (2020) 20:668. doi: 10.1186/s12879-020-05362-3
4. Torres, A, Chalmers, JD, Dela Cruz, CS, Dominedò, C, Kollef, M, Martin-Loeches, I, et al. Challenges in severe community-acquired pneumonia: a point-of-view review. Intensive Care Med. (2019) 45:159–71. doi: 10.1007/s00134-019-05519-y
5. Hogerwerf, L, de Gier, B, Baan, B, and van der Hoek, W. Chlamydia Psittaci (psittacosis) as a cause of community-acquired pneumonia: a systematic review and Meta-analysis. Epidemiol Infect. (2017) 145:3096–105. doi: 10.1017/s0950268817002060
6. Missault, S, de Meyst, A, van Elslande, J, van den Abeele, AM, Steen, E, van Acker, J, et al. Three cases of atypical pneumonia with Chlamydia Psittaci: the role of laboratory vigilance in the diagnosis of psittacosis. Pathogens. (2023) 12:65. doi: 10.3390/pathogens12010065
7. Nieuwenhuizen, AA, Dijkstra, F, Notermans, DW, and van der Hoek, W. Laboratory methods for case finding in human psittacosis outbreaks: a systematic review. BMC Infect Dis. (2018) 18:442. doi: 10.1186/s12879-018-3317-0
8. Tang, J, Tan, W, Luo, L, Xu, H, Li, N, and Li, P. Application of metagenomic next-generation sequencing in the diagnosis of pneumonia caused by Chlamydia Psittaci. Microbiol Spectr. (2022) 10:e0238421. doi: 10.1128/spectrum.02384-21
9. Gu, L, Liu, W, Ru, M, Lin, J, Yu, G, Ye, J, et al. The application of metagenomic next-generation sequencing in diagnosing Chlamydia Psittaci pneumonia: a report of five cases. BMC Pulm Med. (2020) 20:65. doi: 10.1186/s12890-020-1098-x
10. Sukon, P, Nam, NH, Kittipreeya, P, Sara-in, A, Wawilai, P, Inchuai, R, et al. Global prevalence of chlamydial infections in birds: a systematic review and Meta-analysis. Prev Vet Med. (2021) 192:105370. doi: 10.1016/j.prevetmed.2021.105370
11. Rybarczyk, J, Versteele, C, Lernout, T, and Vanrompay, D. Human psittacosis: a review with emphasis on surveillance in Belgium. Acta Clin Belg. (2020) 75:42–8. doi: 10.1080/17843286.2019.1590889
12. Shi, Y, Chen, J, Shi, X, Hu, J, Li, H, Li, X, et al. A case of Chlamydia Psittaci caused severe pneumonia and meningitis diagnosed by metagenome next-generation sequencing and clinical analysis: a case report and literature review. BMC Infect Dis. (2021) 21:621. doi: 10.1186/s12879-021-06205-5
13. Balsamo, G, Maxted, AM, Midla, JW, Murphy, JM, Wohrle, R, Edling, TM, et al. Compendium of measures to control Chlamydia Psittaci infection among humans (psittacosis) and pet birds (avian Chlamydiosis), 2017. J Avian Med Surg. (2017) 31:262–82. doi: 10.1647/217-265
14. Liu, J, and Gao, Y. Tigecycline in the treatment of severe pneumonia caused by Chlamydia Psittaci: a case report and literature review. Front Med (Lausanne). (2022) 9:1040441. doi: 10.3389/fmed.2022.1040441
15. Gu, W, Miller, S, and Chiu, CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol. (2019) 14:319–38. doi: 10.1146/annurev-pathmechdis-012418-012751
16. Chen, X, Cao, K, Wei, Y, Qian, Y, Liang, J, Dong, D, et al. Metagenomic next-generation sequencing in the diagnosis of severe pneumonias caused by Chlamydia Psittaci. Infection. (2020) 48:535–42. doi: 10.1007/s15010-020-01429-0
17. Wu, HH, Feng, LF, and Fang, SY. Application of metagenomic next-generation sequencing in the diagnosis of severe pneumonia caused by Chlamydia Psittaci. BMC Pulm Med. (2021) 21:300. doi: 10.1186/s12890-021-01673-6
18. Liu, K, Wu, L, Chen, G, Zeng, D, Zhong, Q, Luo, L, et al. Clinical characteristics of Chlamydia psittaci infection diagnosed by metagenomic next-generation sequencing: a retrospective multi-center study in Fujian, China. Infect Drug Resist. (2024) 17:697–708. doi: 10.2147/idr.S443953
19. Teng, XQ, Gong, WC, Qi, TT, Li, GH, Qu, Q, Lu, Q, et al. Clinical analysis of metagenomic next-generation sequencing confirmed Chlamydia Psittaci pneumonia: a case series and literature review. Infect Drug Resist. (2021) 14:1481–92. doi: 10.2147/idr.S305790
20. Yin, XW, Mao, ZD, Zhang, Q, Ou, QX, Liu, J, Shao, Y, et al. Clinical metagenomic sequencing for rapid diagnosis of pneumonia and meningitis caused by Chlamydia Psittaci. World J Clin Cases. (2021) 9:7693–703. doi: 10.12998/wjcc.v9.i26.7693
21. Zhang, Q, Li, S, Zhou, W, Zheng, L, Ren, Y, Dong, L, et al. Application of metagenomic next-generation sequencing (Mngs) combined with rapid on-site cytological evaluation (Rosce) for the diagnosis of Chlamydia Psittaci pneumonia. Int J Clin Exp Pathol. (2021) 14:389–98. Available at: https://pubmed.ncbi.nlm.nih.gov/33936360
22. Xu, W, Wang, Q, Li, L, Zhu, B, Cai, Q, Yi, X, et al. Case report: metagenomic next-generation sequencing applied in diagnosing psittacosis caused by Chlamydia Psittaci infection. FCIMB. (2023) 13:1249225. doi: 10.3389/fcimb.2023.1249225
23. Li, S, Tong, J, Li, H, Mao, C, Shen, W, Lei, Y, et al. L. pneumophila infection diagnosed by tNGS in a lady with lymphadenopathy. Infect Drug Resist. (2023) 16:4435–42. doi: 10.2147/idr.S417495
24. Dai, J, Lian, X, Mo, J, Li, X, Mo, W, Wang, H, et al. Case report: a clinical case study of six patients with Chlamydia Psittaci pneumonia. Front Cell Infect Microbiol. (2023) 13:1084882. doi: 10.3389/fcimb.2023.1084882
25. Yin, Y, Zhu, P, Guo, Y, Li, Y, Chen, H, Liu, J, et al. Enhancing lower respiratory tract infection diagnosis: implementation and clinical assessment of multiplex Pcr-based and hybrid capture-based targeted next-generation sequencing. EBioMedicine. (2024) 107:105307. doi: 10.1016/j.ebiom.2024.105307
26. Liu, J, Zhao, F, Lu, J, Xu, H, Liu, H, Tang, X, et al. High Mycoplasma Pneumoniae loads and persistent long-term Mycoplasma Pneumoniae DNA in lower airway associated with severity of pediatric Mycoplasma Pneumoniae pneumonia. BMC Infect Dis. (2019) 19:1045. doi: 10.1186/s12879-019-4667-y
27. Li, Z, Zhang, Y, Zhang, W, Zhang, Y, Zhou, S, Chen, W, et al. Study on the detection and infection distribution of multidrug-resistant organisms in different specimens. Infect Drug Resist. (2022) 15:5945–52. doi: 10.2147/idr.S375682
Keywords: psittacosis, Chlamydia psittaci , target next-generation sequencing, tNGS, pathogen detection
Citation: Yan X, Fu H, Deng W, Zhang Z and Wang D (2024) Early and rapid diagnosis of Chlamydia psittaci pneumonia by tNGS in six patients: a case series. Front. Med. 11:1491838. doi: 10.3389/fmed.2024.1491838
Edited by:
Uday Kishore, United Arab Emirates University, United Arab EmiratesReviewed by:
Yutaka Yoshii, The Jikei University School of Medicine, JapanZheng Jin Tu, Cleveland Clinic, United States
Copyright © 2024 Yan, Fu, Deng, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong Wang, d2FuZ2Rvbmd3dWhhbjAwMUAxNjMuY29t
 Xinsheng Yan
Xinsheng Yan Huali Fu2
Huali Fu2 Dong Wang
Dong Wang