- 1Changsha KingMed Diagnostics Group Co., Ltd., Changsha, Hunan, China
- 2Department of Infection and Hepatology, Taizhou People's Hospital Affiliated to Nanjing Medical University, Taizhou, China
- 3Nanjing KingMed Diagnostics Group Co., Ltd., Nanjing, Jiangsu, China
Introduction/background: Vibrio cholerae is the causative agent of the human intestinal infectious disease cholera, which includes a variety of serogroups. However, there have been very few cases of hepatic space-occupying lesions associated with this infection. Currently, there are various methods for detecting this pathogen, including metagenomic sequencing, which enables quicker and more accurate identification. In this study, metagenomic sequencing is employed to accurately identify non-O1/O139 Vibrio cholerae infections by analyzing the genetic material present in clinical samples.
Presentation of case: A 75-year-old man presented with diarrhea and fever after consuming crabs. The initial treatment improved the diarrhea, but a liver abscess developed later. Magnetic resonance imaging (MRI) of the liver revealed a hepatic space-occupying lesion. Upon further investigation, a Gram-negative, rod-shaped bacterium was cultured from the patient’s liver puncture fluid, and Vibrio cholerae was detected in the same fluid using metagenomic next-generation sequencing (mNGS). The pathogen was confirmed to be non-O1/non-O139 Vibrio cholerae (NOVC) using polymerase chain reaction (PCR). Following treatment with piperacillin/tazobactam sodium and moxifloxacin, the patient’s body temperature returned to normal, the liver abscess improved significantly, and he was subsequently discharged from the hospital.
Discussion: This case study describes an elderly male patient with a hepatic space-occupying lesion. Multiple cultures of specimens failed to identify the underlying cause; however, advanced techniques such as mNGS and PCR confirmed an NOVC infection. This indicates that mNGS can serve as a valuable tool in diagnosing cases of unexplained liver infections.
Conclusion: The use of mNGS is significant for detecting and clinically diagnosing infectious pathogens in patients with unexplained space-occupying lesions.
1 Introduction
Vibrio cholerae is a Gram-negative bacteria, belonging to the genus Vibrio. It is an aerobic, water-borne pathogen. Currently, there are more than 200 Vibrio cholerae serogroups (1). Among these, O1 and O139 isolates typically produce the cholera toxin (2). Infection after consuming water or food contaminated with Vibrio cholerae can lead to cholera, a severe intestinal infectious disease characterized by severe diarrhea and vomiting. If not treated in time, the mortality rate can reach up to 70%.
The non-O1/non-O139 Vibrio cholerae (NOVC) isolates cannot produce toxins and, therefore, cannot cause cholera. However, they can cause bacteremia and invasive extraintestinal diseases (3, 4). The most common clinical manifestations of NOVC infection include gastroenteritis, but they can also cause septicemia (5), chronic otitis media (6), oral infections (7), necrotizing fasciitis (8), endophthalmitis (9), meningitis (10), and other invasive infections. There have been only a few reports of this bacterium causing hepatic space-occupying lesions.
Metagenomic next-generation sequencing (mNGS) is a culture-free and bias-free pathogen detection technology based on next-generation sequencing technology. It can simultaneously detect a variety of pathogens including bacteria, fungi, viruses, and parasites using high-throughput sequencing of DNA and/or RNA directly extracted from clinical samples, followed by data comparison and bioinformatics analysis (11, 12). Currently, it is gradually being transferred from research to clinical laboratories and used to identify pathogens in different infected parts of the body, such as respiratory infections, central nervous system infections, bloodstream infections, gastrointestinal infections, ocular infections, urinary tract infections, and hepatobiliary infections (11–13). Studies have shown that mNGS detection can be used to diagnose unknown infections in body fluids. This study tested 182 body fluids from 160 patients with acute illness using two sequencing platforms for mNGS. The results were compared to other diagnostic methods, such as culture, 16S bacterial PCR, and/or 28S internal donated ribosomal gene spacer (28S ITS) functional PCR. The sensitivity and specificity of the test for bacteria and fungi were over 75%, as determined by Illumina sequencing (14).
2 Case report
A 75-year-old man was admitted with fever and diarrhea. Routine examination and inquiry revealed that the patient had a clear history of seafood consumption before the onset of the disease. The patient consumed crabs that had been stored in the refrigerator for 8 days before developing symptoms of diarrhea and fever. Levofloxacin was used for anti-infection treatment in a local hospital, and the symptoms of diarrhea were relieved, but liver abscesses appeared. Later, the patient was transferred to our hospital for treatment. After undergoing an MRI examination of the liver at our hospital, a hepatic space-occupying lesion was identified.
On admission, his vital signs were as follows: white blood cell (WBC) count was 10.03 × 10^9/L, with 69.3% neutrophils, 21.5% lymphocytes, and 7.0% monocytes. The concentration of procalcitonin (PCT) was 0.120 ng/mL, and the serum C-reactive protein concentration was 81.76 mg/L. Additionally, the patient had a mild fever upon admission (Figure 1a). Furthermore, the liver MRI scan revealed a hepatic space-occupying lesion (Figure 1c). No white blood cells were found in the smear of the liver puncture fluid.
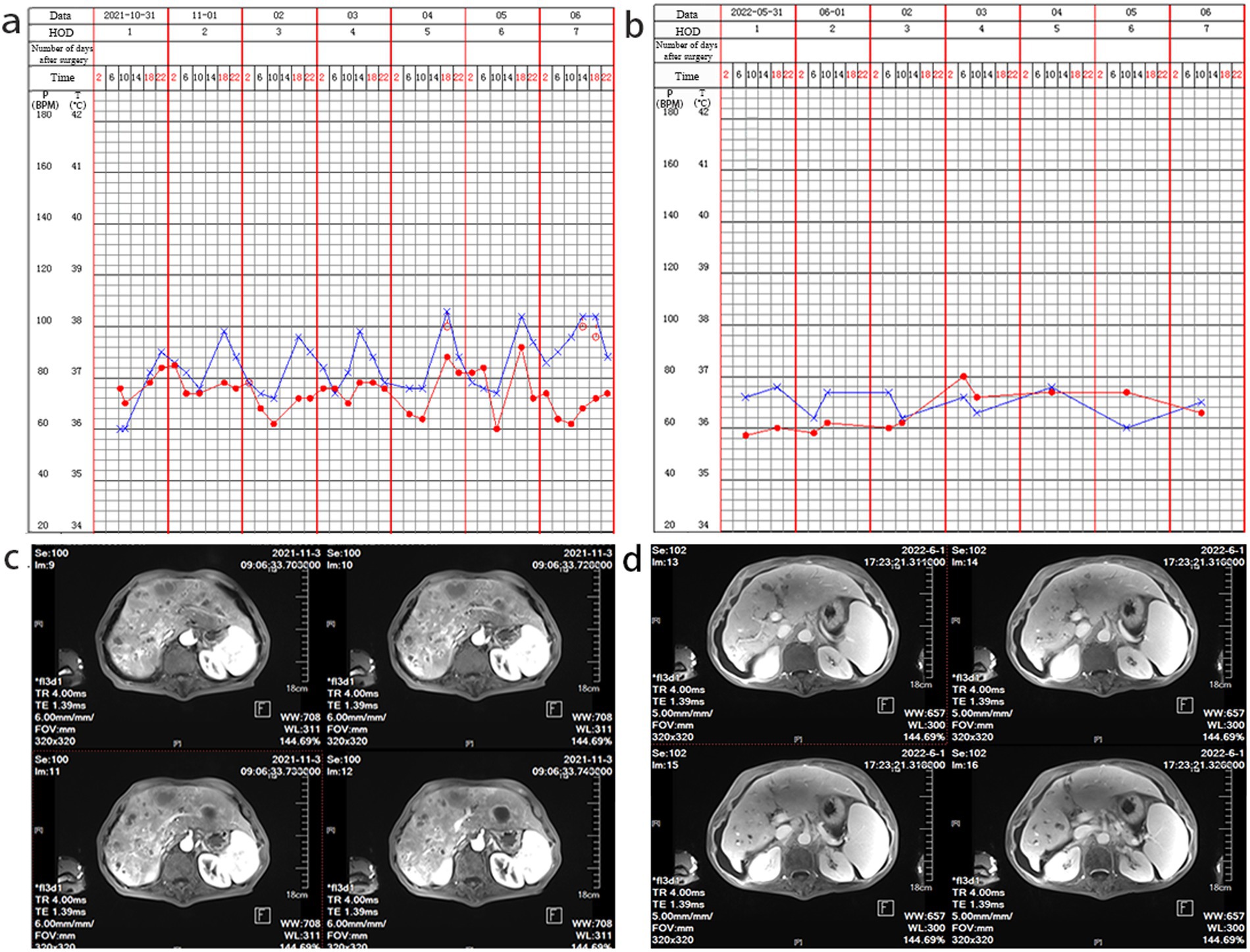
Figure 1. Nursing record sheet and liver MRI examination. (a) At the time of admission, a low fever was observed. (b) A follow-up after treatment indicated that the admission examination reveals a normal body temperature. (c) The initial liver MRI examination shows hepatic space-occupying lesions; (d) After treatment, a follow-up liver MRI examination shows improvement in liver abscess.
Based on the MRI results, a liver abscess was suspected 1 week after admission, and a 21-day regimen of piperacillin/tazobactam sodium (4.5 g every 8 h) and moxifloxacin hydrochloride (0.4 g once daily) was started to treat the infection. Although there was a reduction in the size of the lesion, it did not completely disappear. Then, the patient’s condition was reassessed. Blood, feces, and liver puncture fluid samples were collected. After disinfecting the skin, approximately 5 mL of blood was collected through venipuncture. For the stool sample, a swab was used to collect an amount equivalent to the size of a peanut (approximately 5 g).
In addition, under the guidance of CT or ultrasound, an 8-ml sample of the liver abscess fluid was aspirated through a puncture. All of the specimens were preserved and cultured at Nanjing KingMed Diagnostics (a third-party laboratory). Blood culture results were negative. Escherichia coli and Enterococcus faecium were identified in stool culture. The liver puncture fluid was inoculated onto a Columbia blood plate. After 24 h of incubation, a small amount of curved, Gram-negative, rod-shaped bacterium was observed growing on the blood agar under the microscope (Figure 2a).
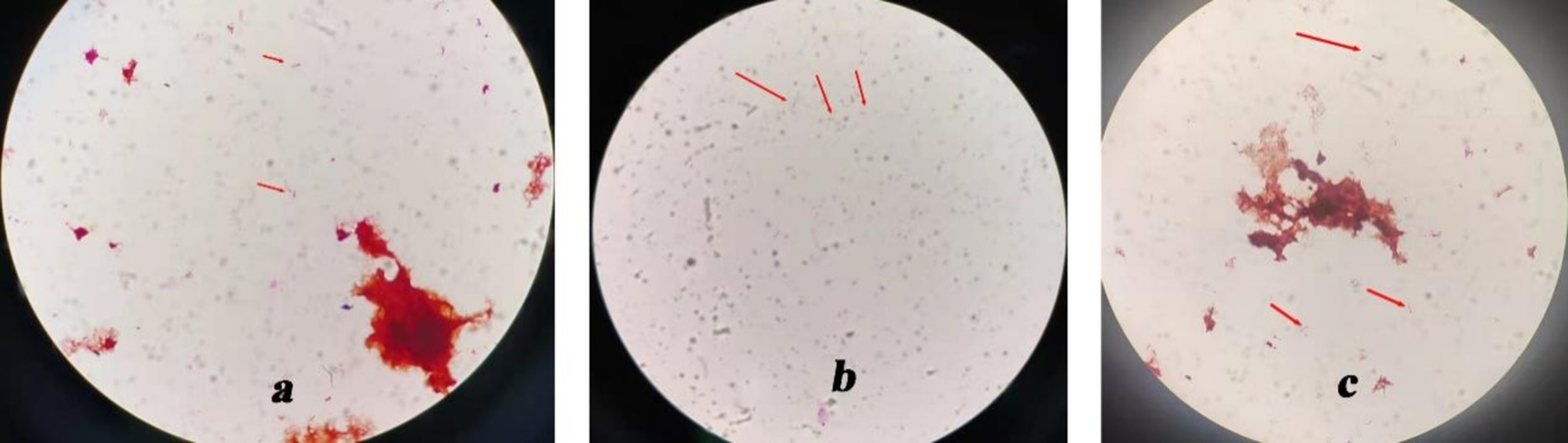
Figure 2. Culture results show a Gram-negative curved rod-shaped bacterium. (×1,000). (a) Gram-negative curved rod-shaped bacterium in blood agar. (b) Gram-negative curved rod-shaped bacterium in BacT/ALERT® FA culture bottles. (c) Gram-negative curved rod-shaped bacterium in a blood plate, isolated from BacT/ALERT® FA culture bottles.
The liver puncture fluid was also inoculated into a BacT/ALERT® FA culture bottle. After 120 h of incubation, the culture medium was taken from the BacT/ALERT® FA culture bottle, and Gram-negative rod-shaped bacteria with slight swelling and curvature were observed under a microscope (Figure 2b). The colony morphology suggested the presence of a rod-shaped bacterium. Subsequently, the liquid in the bottle was transferred to Columbia blood plates for further cultivation. After 24 h, curved Gram-negative rod-shaped bacteria were observed under the microscope (Figure 2c). Unfortunately, pathogen identification was not possible.
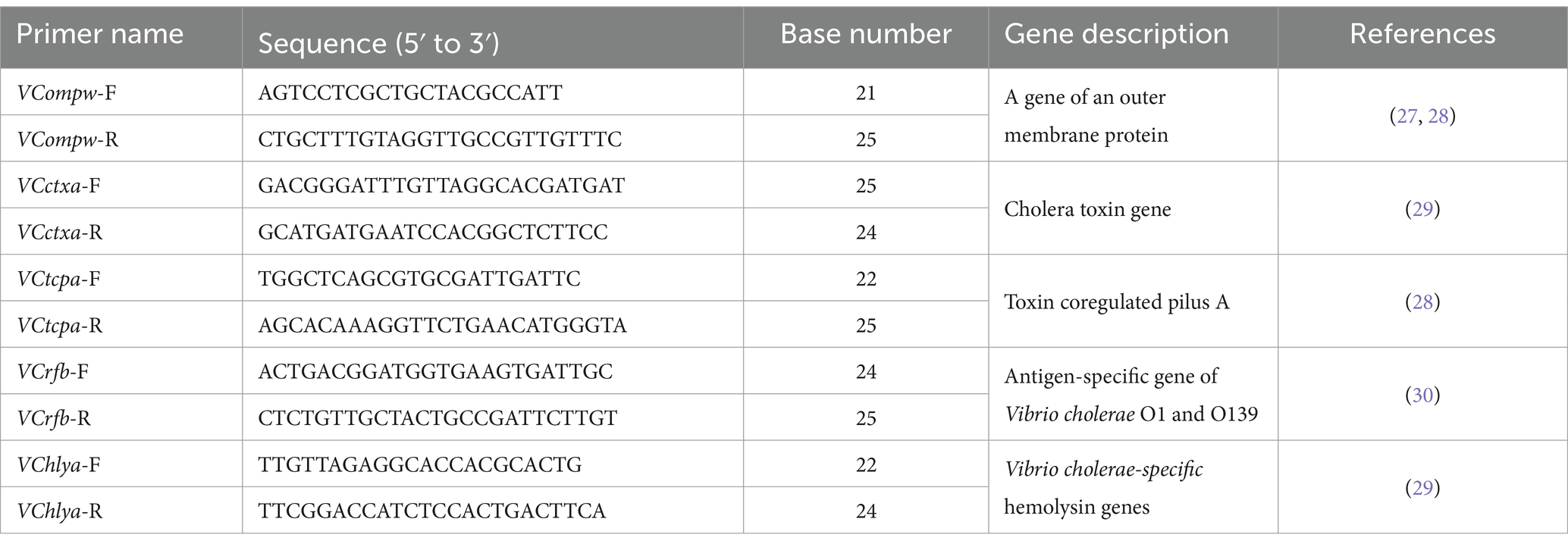
To further confirm the pathogen of infection, mNGS detection was conducted. Subsequently, the liver puncture fluid was sent for mNGS testing (KingMed Diagnostics, Changsha, China). The experimental process of mNGS includes the extraction of nucleic acids (DNA and/or RNA), fragmentation, library preparation, and sequencing. By analyzing and processing the sequencing data, the microorganisms found in the specimen can be identified and studied. The mNGS results reported a V. cholerae infection. DNA mNGS detected 108 sequences that can be mapped to V. cholerae, with a coverage of 0.19% (Figure 3a). RNA mNGS detected 14 sequences that can be mapped to V. cholerae, with a coverage of 0.02% (Figure 3b). To verify the serotype of V. cholerae, five genes were amplified by PCR in the sliver puncture fluid, including ompw, ctxa, tcpa, rfb, and hlya (Table 1). However, ctxa, tcpa, rfb, and hlya did not amplify any specific fragments (Figure 4). Finally, the confirmation of the NOVC strain was achieved by mNGS and PCR at Changsha KingMed Diagnostics (a third-party laboratory).
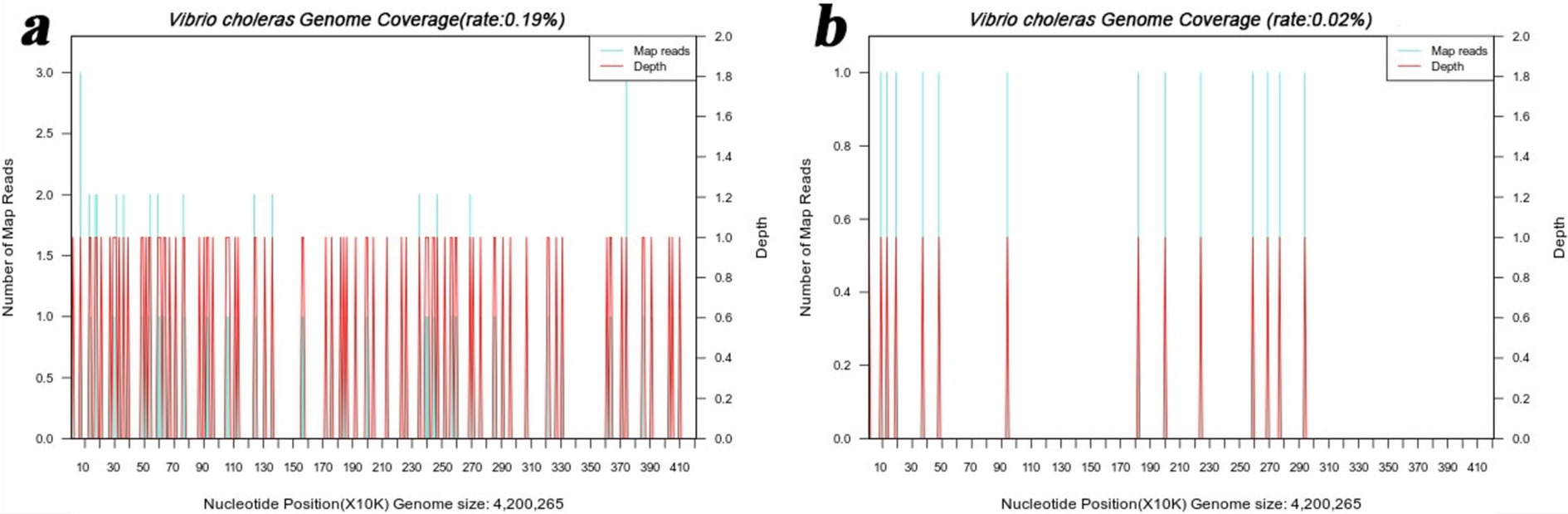
Figure 3. mNGS results of the liver puncture fluid. (a) The DNA mNGS results show that the coverage of Vibrio cholerae was 0.19%. (b) The RNA mNGS results show that the coverage of Vibrio cholerae was 0.02%.
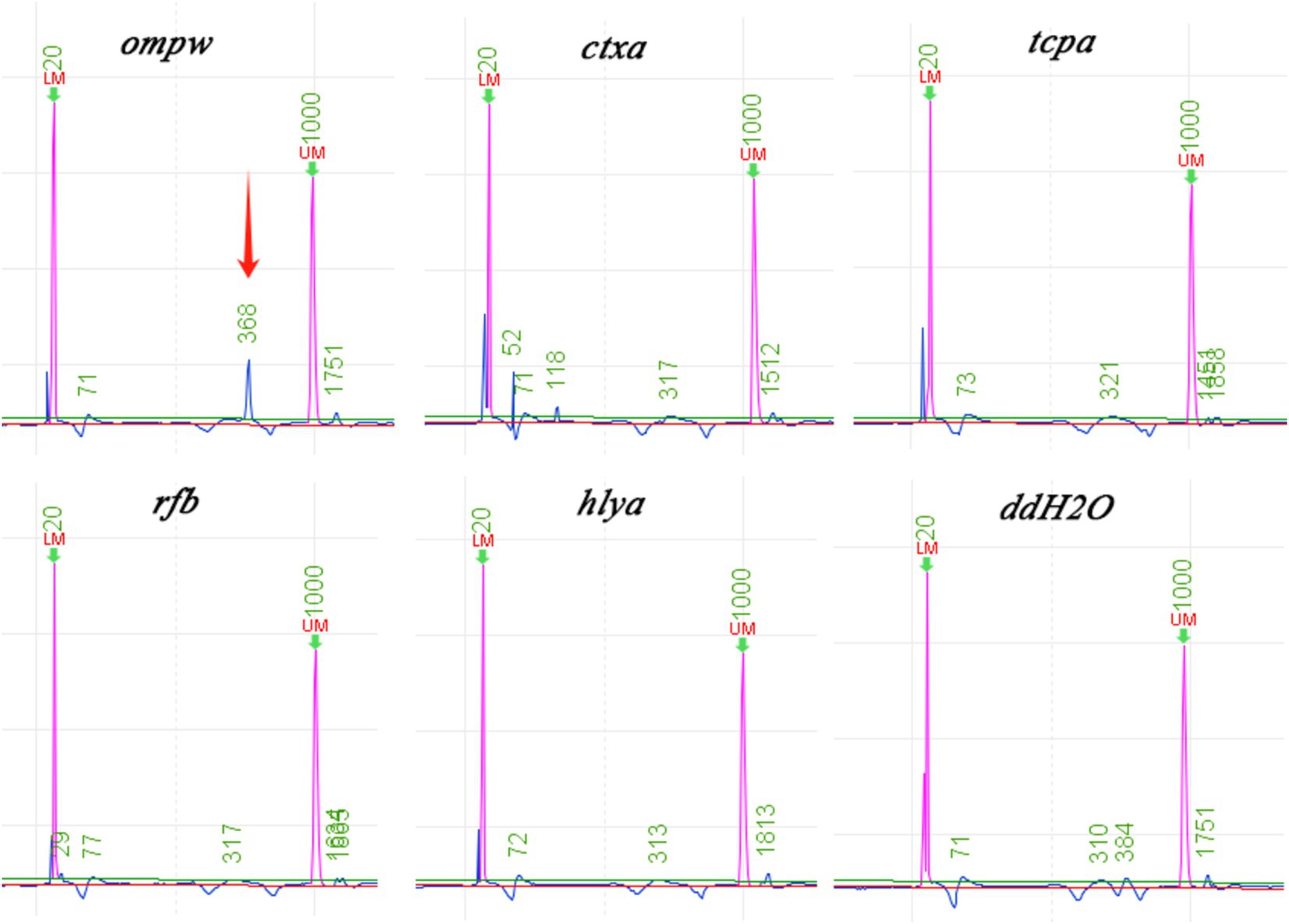
Figure 4. Results of the gene PCR. Except for ompw, other O1/O139 Vibrio cholerae toxicity-related genes are not amplified by PCR. (ompw-positive, ctxa-negative, tcpa-negative, rfb-negative, and hlya-negative).
This patient had a clear history of seafood consumption before the onset of the disease. Subsequently, it was confirmed to be NOVC through mNGS and PCR testing. Based on experience and recommendations from the ninth edition of “Infectious Diseases” by the People’s Health Publishing House, quinolone drugs are preferred for treating cholera infection. Therefore, the use of a moxifloxacin antibiotic was continued for treatment. After 40 days of treatment, the patient’s temperature tended to be stable, and he was discharged without fever. After 7 months of follow-up, the body temperature was normal (Figure 1b). After anti-infection treatment, the hepatic space-occupying lesions improved (Figure 1d).
3 Discussion
In this report, a 75-year-old man with NOVC infection was described. He suffered from hepatic space-occupying lesions and had consumed crabs that had been refrigerated for a long time. To the best of our knowledge, this is the first case of NOVC infection identified using mNGS in a patient with hepatic space-occupying lesions.
Vibrio cholerae is widely distributed in aquatic environments. Humans can contract an infection by consuming contaminated food, especially seafood and fish (15). Over the last 20 years, diseases caused by NOVC have increased steadily worldwide (15).
Studies have shown that infections caused by NOVC are more common in patients with underlying diseases or low immunity. Cirrhosis is the most common risk factor in mainland China, followed by malignancy, hematological malignancies, and diabetes (5, 16). NOVC bacteremia (17) and septicemia (18) have been found in patients with liver cirrhosis. However, NOVC sepsis was also reported in a patient who had no underlying disease (19). As stated in the case, the patient had liver space-occupying symptoms, but the early symptoms were insufficient to support a diagnosis of NOVC infection.
The patient’s WBC count was slightly higher, whereas the cell proportion and PCT were normal. In addition, C-reactive protein (CRP) levels increased significantly, and the patient’s body temperature increased, indicating an infection. After culturing samples from the blood, feces, and liver puncture fluid, no bacteria were detected. Escherichia coli and Enterococcus faecium were cultured from feces. Curved Gram-negative, rod-shaped bacteria were found in the liver puncture fluid, and the bacterial morphology was consistent with that of V. cholerae. However, the culture method is time-consuming and has limitations (20). Due to limited conditions, it was not possible to cultivate a single colony and conduct drug sensitivity tests. Afterward, Vibrio cholerae was detected using mNGS, and the toxicity-related genes of O1/O139 Vibrio cholerae were not amplified by PCR, confirming that the bacterium is NOVC. Following treatment based on clinical experience and mNGS results, the patient’s physical signs improved. NOVC was not found in blood and fecal cultures, demonstrating the importance of selecting the appropriate sample type. Due to the rarity of the NOVC infection, its diagnosis may be challenging. Agglutination with O1 antiserum can be used to exclude other types of Vibrio cholerae infection, which is typically performed in clinical laboratories and can be difficult for initial diagnosis (21).
However, mNGS is increasingly used to detect pathogens directly from clinical specimens (20). At present, mNGS is used to detect and diagnose secondary infection in patients with severe Legionella pneumonia after treatment (22). In addition, research has shown that mNGS has advantages in terms of speed and sensitivity for detecting pathogens in mixed lung infections (23). Now, mNGS can be used to identify pathogens in infectious CNS diseases, providing certain advantages over traditional detection methods (24). Rare pathogens can also be identified using mNGS, such as Chlamydia psittaci, Orientia tsutsugamushi, and others (25, 26). In this case, the liver puncture fluid was detected using mNGS technology, which confirmed the presence of V. cholerae. In general, this shows that the detection of unidentified infected body fluids through mNGS is of significant importance for clinical diagnosis.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
WZ: Writing – original draft, Writing – review & editing. LX: Writing – original draft, Writing – review & editing. XS: Writing – review & editing. BD: Writing – review & editing. CT: Writing – review & editing. JX: Writing – review & editing. YY: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We thank Hu Ge, Yuan Cao, Changlin Liu, Qiuran Lu, and Han Zhou from Changsha KingMed Diagnostics for the technical support of this study.
Conflict of interest
WZ, BD, CT, and YY were employed by Changsha KingMed Diagnostics Group Co., Ltd. XS was employed by Nanjing KingMed Diagnostics Group Co., Ltd.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Clemens, JD, Nair, GB, Ahmed, T, Qadri, F, and Holmgren, J. Cholera Lancet. (2017) 390:1539–49. doi: 10.1016/S0140-6736(17)30559-7
2. Bhandari, M, Jennison, AV, Rathnayake, IU, and Huygens, F. Evolution, distribution and genetics of atypical Vibrio cholerae - a review. Infect Genet Evol. (2021) 89:104726. doi: 10.1016/j.meegid.2021.104726
3. Ko, WC, Chuang, YC, Huang, GC, and Hsu, SY. Infections due to non-O1 Vibrio cholerae in southern Taiwan: predominance in cirrhotic patients. Clin Infect Dis. (1998) 27:774–80. doi: 10.1086/514947
4. Zhang, X, Lu, Y, Qian, H, Liu, G, Mei, Y, Jin, F, et al. Non-O1, non-O139 Vibrio cholerae (NOVC) bacteremia: case report and literature review, 2015-2019. Infect Drug Resist. (2020) 13:1009–16. doi: 10.2147/IDR.S245806
5. Zmeter, C, Tabaja, H, Sharara, AI, and Kanj, SS. Non-O1, non-O139 Vibrio cholerae septicemia at a tertiary care center in Beirut, Lebanon; a case report and review. J Infect Public Health. (2018) 11:601–4. doi: 10.1016/j.jiph.2018.01.001
6. Van Bonn, SM, Schraven, SP, Schuldt, T, Heimesaat, MM, Mlynski, R, and Warnke, PC. Chronic otitis media following infection by non-O1/non-O139 Vibrio cholerae: a case report and review of the literature. Eur J Microbiol Immunol. (2020) 10:186–91. doi: 10.1556/1886.2020.00013
7. Xie, H, Wu, Y, Liu, C, Guo, J, Ma, J, Li, X, et al. Oral infection caused by non-O1/non-O139 Vibrio cholerae in a patient with esophageal Cancer undergoing Esophagectomy and Chemoradiotherapy: a case report. Infect Drug Resist. (2020) 13:3923–7. doi: 10.2147/IDR.S274077
8. Tsuruta, K, Ueyama, T, Watanabe, T, Nakano, K, Uno, K, and Fukushima, H. Intensive care management of a patient with necrotizing fasciitis due to non-O1/O139 Vibrio cholerae after traveling to Taiwan: a case report. BMC Infect Dis. (2020) 20:618. doi: 10.1186/s12879-020-05343-6
9. Yang, CC, Lee, BJ, Yang, SS, Lin, YH, and Lee, YL. A case of non-O1 and non-O139 Vibrio cholerae septicemia with endophthalmitis in a cirrhotic patient. Jpn J Infect Dis. (2008) 61:475–6. doi: 10.7883/yoken.JJID.2008.475
10. Hao, Y, Wang, Y, Bi, Z, Sun, B, Jin, Y, Bai, Y, et al. A case of non-O1/non-O139 Vibrio cholerae septicemia and meningitis in a neonate. Int J Infect Dis. (2015) 35:117–9. doi: 10.1016/j.ijid.2015.05.004
11. Chiu, CY, and Miller, SA. Clinical metagenomics. Nat Rev Genet. (2019) 20:341–55. doi: 10.1038/s41576-019-0113-7
12. Gu, W, Miller, S, and Chiu, CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol. (2019) 14:319–38. doi: 10.1146/annurev-pathmechdis-012418-012751
13. Han, D, Li, Z, Li, R, Tan, P, Zhang, R, and Li, J. mNGS in clinical microbiology laboratories: on the road to maturity. Crit Rev Microbiol. (2019) 45:668–85. doi: 10.1080/1040841X.2019.1681933
14. Gu, W, Deng, X, Lee, M, Sucu, YD, Arevalo, S, Stryke, D, et al. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat Med. (2021) 27:115–24. doi: 10.1038/s41591-020-1105-z
15. Špačková, M, Košťálová, J, and Fabiánová, K. Non-O1/non-O139 vibrios - occurrence not only in Europe in recent years. Epidemiol Mikrobiol Imunol. (2021) 70:131–8.
16. Li, X, Wu, Y, Sun, X, Ma, J, Li, X, Liu, C, et al. Non-O1/non-O139 Vibrio cholerae bacteraemia in mainland China from 2005 to 2019: clinical, epidemiological and genetic characteristics. Epidemiol Infect. (2020) 148:e186. Published 2020 Jul 8. doi: 10.1017/S0950268820001545
17. Gallardo-Cartagena, JA, Chiappe-Gonzalez, AJ, Astocondor-Salazar, LM, Salazar-Mesones, BN, Narcizo Susanibar, JA, Cucho-Espinoza, C, et al. Bacteremia por Vibrio cholerae NO-O1/NO-O139 en un paciente cirrótico. Primer reporte de caso en el Perú y revisión de la literatura [Vibrio cholerae NO-O1/NO-O139 bacteremia in a cirrhotic patient. First case report in Peru and literatura review]. Revista Gastroenterologia Peru. (2018) 38:301–5.
18. Inoue, T, Kitai, S, Hayaishi, S, and Kudo, M. Septicemia due to Vibrio cholerae serogroup non-O1/non-O139 strain in a cirrhotic patient. Clin J Gastroenterol. (2012) 5:383–7. doi: 10.1007/s12328-012-0332-3
19. Hwang, S, Kim, Y, Jung, H, Chang, HH, Kim, SJ, Park, HK, et al. A fatal case of bacteremia caused by Vibrio cholerae non-O1/O139. Infect Chemother. (2021) 53:384–90. doi: 10.3947/ic.2020.0301
20. Filkins, LM, Bryson, AL, Miller, SA, and Mitchell, SL. Navigating clinical utilization of direct-from-specimen metagenomic pathogen detection: clinical applications, limitations, and testing recommendations. Clin Chem. (2020) 66:1381–95. doi: 10.1093/clinchem/hvaa183
21. Khan, S, Kumar, A, Meparambu, D, Thomas, S, Harichandran, D, and Karim, S. Fatal non-O1, non-O139 Vibrio cholerae septicaemia in a patient with chronic liver disease. J Med Microbiol. (2013) 62:917–21. doi: 10.1099/jmm.0.049296-0
22. Yue, R, Wu, X, Li, T, Chang, L, Huang, X, and Pan, L. Early detection of Legionella pneumophila and aspergillus by mNGS in a critically ill patient with Legionella pneumonia after extracorporeal membrane oxygenation treatment: case report and literature review. Front Med. (2021) 8:686512. doi: 10.3389/fmed.2021.686512
23. Wang, J, Han, Y, and Feng, J. Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm Med. (2019) 19:252. doi: 10.1186/s12890-019-1022-4
24. Xing, XW, Zhang, JT, Ma, YB, He, MW, Yao, GE, Wang, W, et al. Metagenomic next-generation sequencing for diagnosis of infectious encephalitis and meningitis: a large, prospective case series of 213 patients. Front Cell Infect Microbiol. (2020) 10:88. doi: 10.3389/fcimb.2020.00088
25. Gu, L, Liu, W, Ru, M, Lin, J, Yu, G, Ye, J, et al. The application of metagenomic next-generation sequencing in diagnosing Chlamydia psittaci pneumonia: a report of five cases. BMC Pulm Med. (2020) 20. Published 2020 Mar 17:65. doi: 10.1186/s12890-020-1098-x
26. Li, J, Chen, C, Zou, F, Liu, L, Wang, B, Kang, H, et al. Diagnosing scrub typhus without eschar: a case report using metagenomic next-generation sequencing (mNGS). Annal Transl Med. (2021) 9:1190. doi: 10.21037/atm-21-3015
27. Fu, X, Zhang, J, Li, T, Zhang, M, Li, J, and Kan, B. The outer membrane protein OmpW enhanced V. cholerae growth in hypersaline conditions by transporting carnitine. Front Microbiol. (2018) 8:2703. Published 2018 Jan 22. doi: 10.3389/fmicb.2017.02703
28. Zareitaher, T, Sadat Ahmadi, T, and Gargari, LM. Immunogenic efficacy of DNA and protein-based vaccine from a chimeric gene consisting OmpW, TcpA and CtxB, of Vibrio cholerae. Immunobiology. (2022) 227:152190. doi: 10.1016/j.imbio.2022.152190
29. Meena, B, Anburajan, L, Sathish, T, Das, AK, Vinithkumar, NV, Kirubagaran, R, et al. Studies on diversity of Vibrio sp. and the prevalence of hapA, tcpI, st, rtxA&C, acfB, hlyA, ctxA, ompU and toxR genes in environmental strains of Vibrio cholerae from Port Blair bays of south Andaman, India. Mar Pollut Bull. (2019) 144:105–16. doi: 10.1016/j.marpolbul.2019.05.011
Keywords: non-O1/non-O139 Vibrio cholerae, hepatic space-occupying lesions, culture, metagenomic next-generation sequencing, polymerase chain reaction
Citation: Zhang W, Xiao L, Shan X, Dai B, Tang C, Xian J and Yu Y (2024) Case report: Detection of non-O1/non-O139 Vibrio cholerae in a patient with hepatic space-occupying lesions using metagenomic next-generation sequencing. Front. Med. 11:1483027. doi: 10.3389/fmed.2024.1483027
Edited by:
Shisan (Bob) Bao, The University of Sydney, AustraliaReviewed by:
Susanne Fleischmann, Free University of Berlin, GermanySilas Onyango Awuor, Jaramogi Oginga Odinga Teaching and Referral Hospital, Kenya
Copyright © 2024 Zhang, Xiao, Shan, Dai, Tang, Xian and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Yu, aG4teXV5YW5Aa2luZ21lZC5jb20uY24=
†These authors have contributed equally to this work
 Wei Zhang
Wei Zhang Li Xiao
Li Xiao Xingxing Shan3
Xingxing Shan3 Jianchun Xian
Jianchun Xian