- 1Dipartimento di Scienze di Laboratorio e Infettivologiche, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Rome, Italy
- 2Dipartimento di Scienze biotecnologiche di base, cliniche infettivologiche e peri-operatorie – Sezione di Microbiologia, Università Cattolica del Sacro Cuore, Rome, Italy
- 3Dipartimento di Scienze dell'Emergenza, Anestesiologiche e della Rianimazione, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Rome, Italy
- 4Defence Institute for Biomedical Sciences, Rome, Italy
- 5Dipartimento di Malattie Infettive, Istituto Superiore di Sanità, Rome, Italy
- 6National Reference Centre for Legionella, Hospices Civils de Lyon, Lyon, France
- 7ESCMID Study Group for Legionella Infections (ESGLI), Basel, Switzerland
Legionnaires' disease (LD) is a serious type of pneumonia, typically contracted by susceptible people through the inhalation of aerosols contaminated with Legionella pneumophila (Lp). In this report, the first case of coinfection with Lp–Bordetella bronchiseptica (Bb) is described. A possible source of the Lp infection may be the hotel in Paris (France) where the patient had stayed before developing the symptoms. The Bb infection may have been transmitted by the dog with which he had constant contact, although this has not been proven.
Introduction
Legionnaires' disease (LD) is an infection caused by Legionella pneumophila (Lp), a Gram-negative waterborne pathogen noted by the World Health Organization as posing the highest health burden in the European Union (1). Lp infection is acquired through the inhalation of infectious aerosols originating from water systems in buildings, such as showers, fountains, spa pools, and cooling towers. LD cases can be acquired in the community, through travel, or in nosocomial settings. Male individuals aged 65 years and older, especially those with underlying diseases, alcohol abuse, smoking habits, or immunosuppression, are mainly susceptible (2). In Europe, Italy is one of the four countries, along with France, Germany, and Spain, responsible for 72% of all notified cases; the majority (82%) of the cases are travel-associated legionnaires' disease (TALD) cases (3). Few cases both in Europe (10%) and in Italy (0.8%) are diagnosed through the culture method. Consequently, since the strain is not available, it is difficult to trace the origin of the infection (3, 4).
Bordetella is a strictly aerobic, non-fermentative, catalase-positive, and oxidase-positive Gram-negative coccobacillus, consisting of 16 species. Bordetella pertussis (Bp), Bordetella parapertussis (Bpp), and Bordetella bronchiseptica (Bb) are defined as “Classical Bordetellae,” all of which cause respiratory infections ranging from severe to mild or asymptomatic (5, 6). However, while Bp and Bpp can infect only humans and cannot survive in the environment, Bb is the major causative agent of the canine infectious respiratory disease complex (7). Although Bb is seldom considered infectious for humans, transmission from the dog to the owner is possible (8). Sporadic cases of Bb infection have primarily been linked to debilitated or immunosuppressed patients (9, 10).
In this article, we present for the first time a coinfection with Lp–Bb in a 69-year-old immunocompromised man with chronic renal failure, hypertension, and IgG Lambda multiple myeloma (MM) diagnosed in 2014.
Methods
Case description
On 13 January 2023, a patient was admitted to the emergency room (ER) with fever and worsening dyspnea, which had started 5 days earlier. He reported having traveled to Paris for New Year's Eve and had previously undergone autologous hematopoietic cell stem transplantation with a positive outcome in 2015. Due to multiple recrudescences of the MM, he was actively receiving fourth-line chemotherapy with isatuximab plus pomalidomide and dexamethasone (IsaPd), the last dose of which was administrated 10 days before his travel to Paris (France).
During the first 2 days in the ER, he rapidly developed severe hypoxemic respiratory failure and hemodynamic instability. He was initially treated with non-invasive mechanical ventilation, followed by invasive mechanical ventilation, which required sedation, paralysis, and orotracheal intubation. While he was in the ER, vasopressors, such as norepinephrine, were administered to the patient, and empiric antibiotic therapy was initiated with three doses of intravenous (IV) clarithromycin (500 mg, twice a day) and one dose of Meropenem (500 mg).
From the beginning of the antibiotic therapy, the patient clinically improved, and we were able to extubate him 48 h after his admission to the intensive care unit (ICU). When the patient was experiencing spontaneous breathing, we initially supported oxygenation using high-flow oxygen therapy delivered by a nasal cannula but soon switched to conventional oxygen therapy. Norepinephrine was reduced and stopped during the first 24 h in the ICU. On 15 January, the patient was moved to the general intensive care unit (ICU); however, he remained deeply hemodynamically unstable and required a high-dose noradrenaline infusion and continuous renal replacement therapy. Hematologic parameters showed severe leukopenia (0.74 × 109/L), thrombocytopenia (72 × 109/L), and elevated serum levels of procalcitonin and C-reactive protein (271.57 ng/ml and 43.41 mg/dl, respectively). Urine, blood cultures, and bronchoalveolar lavage (BAL) samples were collected immediately and forwarded to the hospital's microbiology service for urine antigen testing (UAT) and BAL sample culture to accurately diagnose Legionella infection.
The patient remained in the ICU for 7 days and was discharged to the ward, awake, without any kind of organ support, and maintaining only therapy against Lp and Bb.
FilmArray analysis, culture examination, MALDI Biotyper identification, and serological assay
The BioFire FilmArray Pneumonia plus Panel (FAPP, Biomerieux), a multiplex PCR technique based on a syndromic approach, was used (11). This test can simultaneously identify 27 of the most common pathogens involved in lower respiratory tract infections (semi-quantitative results for 11 Gram-negative and 4 Gram-positive bacteria and qualitative results for 3 atypical bacteria and 9 viruses) and 7 antibiotic resistance genes. The BAL culture was performed on buffered charcoal yeast extract (BCYE, ThermoFisher, United Kingdom) and MacConkey agar plates media (ThermoFisher, United Kingdom) for Lp and Bb, respectively, and both were checked every 2 days. Suspected colonies were tested using a latex agglutination test.
Water samples from the water system of the hotel where the patient had stayed and from his home were collected and analyzed by culture according to ISO 11731.
Quantification of IgG and IgM against Lp was performed with anti-Legionella pneumophila (IgG–IgM) ELISA (Euroimmun, Germany). The results were interpreted and calculated according to manufacturers' instructions.
Furthermore, the isolated colonies of Bb were pre-treated using the ethanol/formic acid extraction procedure, as previously described (12), while MALDI Biotyper software, version 3.1 (Bruker Daltonics, Bremen, Germany), was used to process the raw spectra and to compare them for strain classification.
Whole genome sequencing and typing
Lp serogroup 1 (Lp1), isolated from the patient's BAL, was analyzed by whole genome sequencing using Illumina technology. To this end, sequencing libraries were prepared using the Nextera XT DNA Library Preparation Kit (Illumina) and then run on two different Illumina platforms: a 150-bp paired-end sequencing run was performed on the NextSeq 500 (Illumina) using a Mid Output Kit v2 and a 250-bp paired-end sequencing run was performed on the MiSeq (Illumina) (BioProject: PRJNA1126987).
Core genome multi-locus sequence typing (cgMLST) was carried out on 46 ST1, including the patient's and other Lp1 strains isolated in France and Italy. The cgMLST scheme, based on 1,521 core genes (13), was converted using chewBBACA software (Galaxy version 2; 32), and a minimum spanning tree was visualized using GrapeTree software (14). The Lp1 clinical isolate was further typed using sequence-based typing and monoclonal antibody typing (15–17).
Antimicrobial susceptibility test
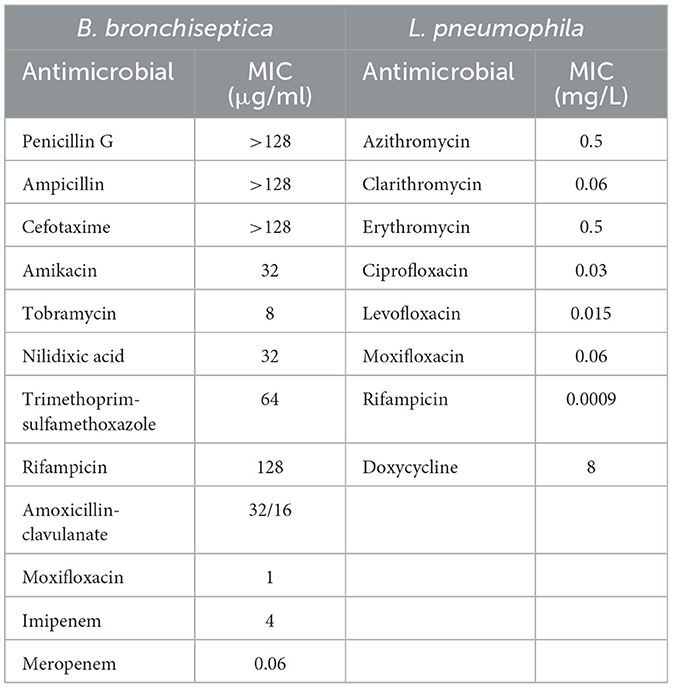
The Bp colonies were analyzed for antimicrobial susceptibility according to the Clinical and Laboratory Standards Institute (CLSI). ETEST (bioMérieux, France) for Bb was performed on Mueller–Hinton agar with a 0.5 McFarland standard, using the following antimicrobials: penicillin G, ampicillin, cefotaxime, amikacin, tobramycin, nalidixic acid, trimethoprim-sulfamethoxazole, rifampicin, amoxicillin-clavulanate, moxifloxacin, imipenem, and meropenem. Minimum inhibitory concentration (MIC) values were determined after incubation at 37°C for 18–24 h.
The following eight antibiotics were tested against the Lp clinical strain using broth microdilution: azithromycin (0.015–8 mg/ml), clarithromycin, ciprofloxacin, levofloxacin, moxifloxacin (0.0009–0.5 μg/ml), erythromycin and doxycycline (0.03–16 μg/ml), and rifampicin (0.00005–0.03 μg/ml), as reported in the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidance document. For Bp susceptibility, tests were performed according to CLSI standards (18, 19).
Results
FilmArray and culture examination
Lp DNA was detected using the FilmArray method shortly after admission to the ICU. This enabled the ICU physicians to initiate targeted therapy with IV levofloxacin (750 mg) and empirical IV piperacillin/tazobactam (a 9 g loading dose followed by a continuous infusion for a total of 18 g per day was stopped on the 5th day). After 4 days of incubation, the BAL culture provided the isolation of Lp serogroup 1 (Lp1). The UAT was positive and remained positive even after 7 months. In addition, the IgG and IgM titers (IgG = 0.3 U/ml; IgM = 2.7 U/ml) were positive.
The culture of the water samples collected from the water system of the hotel where the patient stayed did not provide any isolate, while the culture of the water samples from the patient's home provided Lp non-serogroup 1 at 3.5 × 103-6.1 × 102 CFU/L.
Interestingly, the microbiological culture of the BAL on selective MacConkey agar revealed Bb-positive colonies, identified using MALDI-TOF MS.
Typing and WGS analysis
The Lp1 clinical isolate was typed as the Philadelphia monoclonal subgroup, ST1. cgMLST was produced and the gene profiles were visualized using a minimum spanning tree (Figure 1A). When the patient's ST1 was compared with other unrelated ST1 strains isolated in France and Italy, very similar gene profiles, ranging from 1 to 126 different loci, were found. In particular, an environmental ST1 strain isolated in a French hospital in 2019 showed a single locus of difference compared to the ST1 isolated from the patient (Figure 1A, red circle). As shown in Figure 1B, when the comparison was extended to a large number of ST1 genomes isolated in France and Italy, the phylogenetic analysis revealed that all these genomes are quite similar.
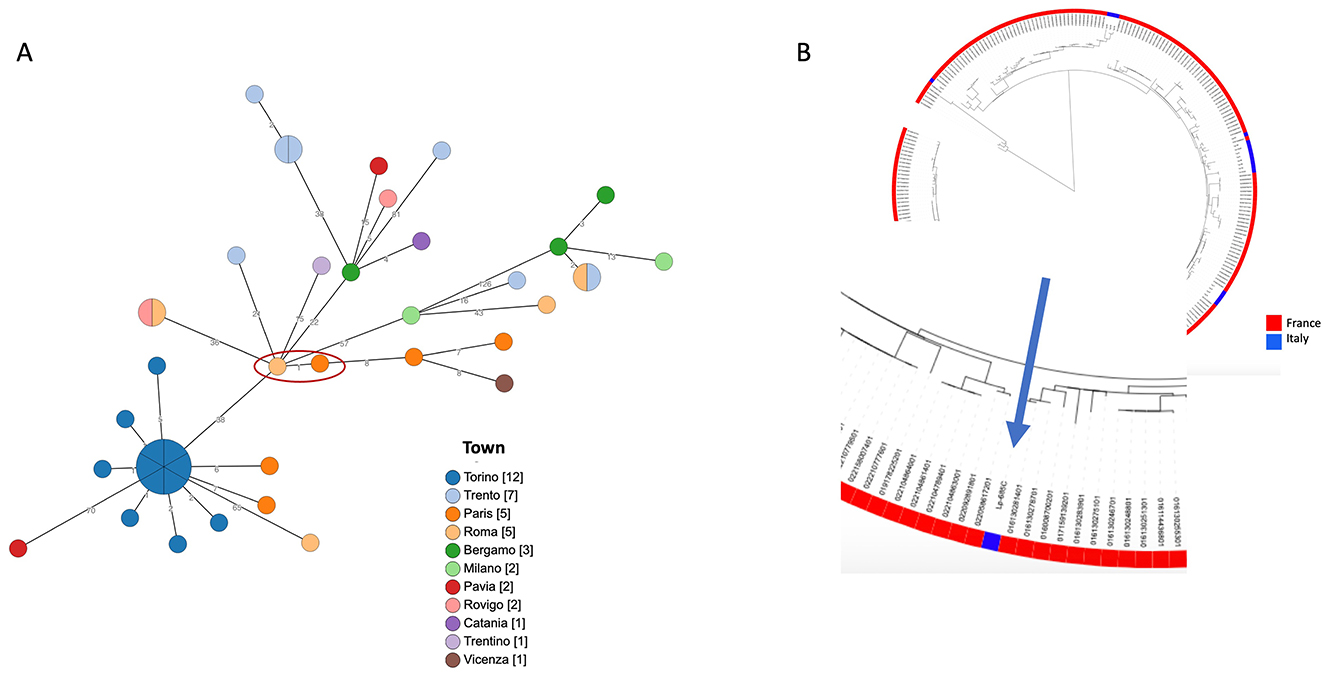
Figure 1. Phylogenetic analysis of ST1. (A) cgMLST based on the subset 1,471 cgMLST targets of the 1,521 core gene scheme is shown. Patient's ST1 and the most phylogenetically related strain are highlighted by a red circle. Node colors correspond to the town where strains were isolated, and on the branches the number of different loci is reported. In (B), a maximum likelihood tree of a large number of ST1 genomes, available in GenBank, (in red those isolated in France, in blue those from Italy) is shown. The zoom on the patient's ST1 is highlighted by the arrow.
Antimicrobial susceptibility test
MIC values determined for Bb and Lp are shown in Table 1.
Discussion
To the best of our knowledge, we report the first case of Lp-Bb coinfection in an immunocompromised patient. Concomitants or sequential coinfections of Lp with other pathogens have rarely been reported, primarily involving viral and few bacterial coinfections, but never with Bb (20–24). Bb is primarily a zoonotic organism that is rarely encountered as a pathogen in cases of acute respiratory infections, which mainly involve immunocompromised people (10, 25). Although it was not possible to isolate Bb from the patient's dog, the regular contact between the patient and his dog (presumably with its upper respiratory tract colonized by Bb) likely allowed the transmission of the infection, exacerbated by the patient's immunosuppressive conditions. Although vaccination against kennel cough is a common practice in domestic pets, there is no official surveillance for Bb that could alert to the emergence of resistant strains (26). In addition, in the absence of reference methods and breakpoints from the CLSI or EUCAST for this organism, the results of susceptibility testing are difficult to interpret from a clinical point of view. No specific guidelines for the treatment of Bb infection are available. Furthermore, only a few specific studies have been published on the mechanisms of resistance in Bb clinical isolates (26). Bb has evolved a species-specific β-lactamase of the BOR-1 class, which could explain the highest level of penicillin MICs. Efflux mechanisms and/or reduced membrane permeability may also be related to the highest level of MICs for other β-lactams and cephalosporins (27).
Similarly, for Lp, reference protocols, breakpoints, and Epidemiological cut-off (ECOFF) values have not yet been established. However, based on the data reported in the literature, the isolated strain appears sensitive to all the antibiotics tested; even for azithromycin, it shows an MIC value considered on average higher than the value observed for most strains (28). Lp1 ST1 is globally spread in clinical cases and has been reported as a leading cause of community- and hospital-acquired LD in many countries (29). It is frequently found in the water systems of various buildings. New insights into the ST1 population genome structure, prone to recombination events that are at the basis of genetic diversity, revealed a wide divergence within this ST (30). For all these reasons, molecular epidemiological investigation requires not only genome matching between clinical and environmental strains but also an accurate analysis of ST1 genomes. Unfortunately, in this case, no environmental ST1 strain was isolated from the hotel where the patient stayed during the 10 days before the onset of the symptoms and where he presumably contracted the infection. The phylogenetic analysis highlighted a high similarity between the ST1 strains isolated in France and the patient's ST1, as well as with other unrelated ST1 strains isolated in Italy. Therefore, no correlation with the source of infection could be established. In this LD case, only the availability of both clinical and environmental strains, possibly in comparison with other unrelated ST1 genomes, can clarify the true source of infection.
Prompt molecular tests allowed for the initiation of the specific antibiotic therapy, which was fundamental in improving the patient's condition. Fortunately, no increased resistance to macrolides and fluoroquinolones was found for Lp1, although a possible increase has been reported recently. In addition, ascertaining sensitivity to all the antibiotics suggested by the EUCAST for Bb allowed the administration of the correct therapy for an immunocompromised patient (31, 32).
This case report focused on the following: (a) Bb as an opportunistic pathogen, particularly in patients with a previous history of respiratory disease and in those who are immunocompromised. In these patients, susceptibility testing should be routinely performed. Interpretative criteria for the clinical interpretation of MICs should be developed. (b) Lp is responsible for serious pneumonia, and the risk of contracting additional infections can further aggravate the patient's condition, making the outcome of the disease much more serious or even fatal. Based on the literature, no relationship can be established between this type of coinfection and the severity of the patient's disease. However, we can empirically assume that the patient likely experiences some degree of immunosuppression due to his hematological disorder (multiple myeloma with multiple relapses, treated with multiple cycles of chemotherapy), making him susceptible to both infections. For this reason, it is important to maintain a high suspicion of other possible infections, especially in elderly people and immunocompromised patients, by conducting as broad a screening as possible for other pathogens using rapid and sensitive nucleic acid amplification tests. This allows for targeted therapy and promotes better control against the infection caused by Lp, with a consequent reduction in severe sequela, hospital stays, and costs for public spending.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
Ethical approval was not required for the studies involving humans because ethical approval was not necessary. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by- product of routine care or industry. Written informed consent to participate in this study was not required from the participants or the participants' legal guardians/next of kin in accordance with the national legislation and the institutional requirements. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
ML: Conceptualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. IP: Conceptualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. DN: Methodology, Writing – review & editing. SF: Methodology, Writing – review & editing. FG: Methodology, Writing – review & editing. FP: Methodology, Writing – review & editing. AM: Methodology, Writing – review & editing. FL: Supervision, Writing – review & editing. FM: Methodology, Writing – review & editing. AG: Methodology, Writing – review & editing. MR: Supervision, Writing – review & editing. MC: Methodology, Writing – review & editing. RR: Methodology, Writing – review & editing. CG: Formal analysis, Writing – review & editing. SJ: Supervision, Writing – review & editing. MSa: Supervision, Writing – review & editing. MSc: Conceptualization, Formal analysis, Writing – review & editing, Writing – original draft. MR: Conceptualization, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by One Helth Basic and Translational Reserch Actions Addressing Unmet Needs on Emerging infectious Diseases (INF-ACT), (project no. PE00000007).
Acknowledgments
We would like to thank our colleague Cristine Campese of the National Institute for Public Health Surveillance in Saint Maurice, France, for providing us with the information on the sampling carried out at the hotel in Paris where the patient had stayed. We would also like to thank the operators of health authority in Paris who conducted the investigation and contributed to this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Directive (EU) 2020/2184. Directive (EU) 2020/2184 of the European Parliament and of the Council of 16 December 2020 on the quality of water intended for human consumption. (2020). Available at: https://eur-lex.europa.eu/eli/dir/2020/2184/oj (accessed October 1, 2024).
2. Phin N, Parry-Ford F, Harrison T, Stagg HR, Zhang N, Kumar K, et al. Epidemiology and clinical management of Legionnaires' disease. Lancet Infect Dis. (2014) 14:1011–21. doi: 10.1016/S1473-3099(14)70713-3
3. European Centre for Disease Prevention and Control. Legionnaires' disease. In: ECDC Annual Epidemiological Report for 2021. Stockholm: ECDC (2023).
4. Rota MC, Caporali MG, Giannitelli S, Urciuoli R, Scaturro M, Ricci ML. I risultati del sistema di sorveglianza della legionellosi nel 2021. Bollettino Epidemiologico Nazion. (2023) 4:25–32. doi: 10.53225/BEN_045
5. Miguelena Chamorro B, De Luca K, Swaminathan G, Longet S, Mundt E, Paul S. Bordetella bronchiseptica and Bordetella pertussis: similarities and differences in infection, immuno-modulation, and vaccine considerations. Clin Microbiol Rev. (2023) 36:e00164–22. doi: 10.1128/cmr.00164-22
6. Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. (2003) 35:32–40. doi: 10.1038/ng1227
7. Mattoo S, Cherry JD. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev. (2005) 18:326–82. doi: 10.1128/CMR.18.2.326-382.2005
8. Woolfrey BF, Moody JA. Human infections associated with Bordetella bronchiseptica. Clin Microbiol Rev. (1991) 4:243–55. doi: 10.1128/CMR.4.3.243
9. Brady C, Ackerman P, Johnson M, McNamara J. Bordetella bronchiseptica in a pediatric Cystic Fibrosis center. J Cystic Fibr. (2014) 13:43–8. doi: 10.1016/j.jcf.2013.08.002
10. Wernli D, Emonet S, Schrenzel J, Harbarth S. Evaluation of eight cases of confirmed Bordetella bronchiseptica infection and colonization over a 15-year period. Clin Microbiol Infect. (2011) 17:201–3. doi: 10.1111/j.1469-0691.2010.03258.x
11. Ginocchio CC, Garcia-Mondragon C, Mauerhofer B, Rindlisbacher C. Multinational evaluation of the BioFire® FilmArray® Pneumonia plus Panel as compared to standard of care testing. Eur J Clin Microbiol Infect Dis. (2021) 40:1609–22. doi: 10.1007/s10096-021-04195-5
12. Sanguinetti M, Posteraro B. Identification of molds by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. (2017) 55:369–79. doi: 10.1128/JCM.01640-16
13. Moran-Gilad J, Prior K, Yakunin E, Harrison TG, Underwood A, Lazarovitch T, et al. Design and application of a core genome multilocus sequence typing scheme for investigation of Legionnaires' disease incidents. Eurosurveillance. (2015) 20:21186. doi: 10.2807/1560-7917.ES2015.20.28.21186
14. Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. (2018) 28:1395–404. doi: 10.1101/gr.232397.117
15. Gaia V, Fry NK, Afshar B, Lück PC, Meugnier H, Etienne J, et al. Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J Clin Microbiol. (2005) 43:2047–52. doi: 10.1128/JCM.43.5.2047-2052.2005
16. Helbig J, Bernander S, Castellani Pastoris M, Etienne J, Gaia V, Lauwers S, et al. Pan-European study on culture-proven Legionnaires' disease: distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur J Clin Microbiol Infect Dis. (2002) 21:710–6. doi: 10.1007/s10096-002-0820-3
17. Ratzow S, Gaia V, Helbig JH, Fry NK, Lück PC. Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J Clin Microbiol. (2007) 45:1965–8. doi: 10.1128/JCM.00261-07
18. ECDC. Guidance Document on Antimicrobial Susceptibility Testing of Legionella pneumophila, 2021. Basel: ESCMID; EUCAST (2023).
20. Rota MC, Caporali MG, Scaturro M, Girolamo A, Andrianou X, Ricci ML. Legionella pneumophila and SARS-COV-2 co-infection: the importance of laboratory diagnosis. Annali dell'Istituto Super Sanità. (2021) 57:199–200. doi: 10.4415/ANN_21_03_01
21. Tan MJ, Tan JS, File TM. Legionnaires disease with bacteremic coinfection. Clin Infect Dis. (2002) 35:533–9. doi: 10.1086/341771
22. Oggioni C, Za A, Auxilia F, Faccini M, Senatore S, Vismara C, et al. Legionnaires' disease contracted from patient workplace: First report of a severe case of coinfection with varicella-zoster virus. Am J Infect Control. (2016) 44:1164–5. doi: 10.1016/j.ajic.2016.03.057
23. Rizzo C, Caporali MG, Rota MC. Pandemic influenza and pneumonia due to Legionella pneumophila: a frequently underestimated coinfection. Clin Infect Dis. (2010) 51:115. doi: 10.1086/653444
24. Scaturro M, Girolamini L, Pascale MR, Mazzotta M, Marino F, Errico G, et al. Case report: First report of fatal Legionella pneumophila and Klebsiella pneumoniae coinfection in a kidney transplant recipient. Front Med. (2022) 9:912649. doi: 10.3389/fmed.2022.912649
25. Brady TM, Redwine KM, Flynn JT. American Society of Pediatric Nephrology. Screening blood pressure measurement in children: are we saving lives? Pediatr Nephrol. (2014) 29:947–50. doi: 10.1007/s00467-013-2715-1
26. Kadlec K, Schwarz S. Antimicrobial resistance in Bordetella bronchiseptica. Microbiol Spectr. (2018) 6:10–128. doi: 10.1128/microbiolspec.ARBA-0024-2017
27. Lartigue MF, Poirel L, Fortineau N, Nordmann P. Chromosome-borne class A BOR-1 β-lactamase of Bordetella bronchiseptica and Bordetella parapertussis. Antimicr Agents Chemother. (2005) 49:2565–7. doi: 10.1128/AAC.49.6.2565-2567.2005
28. Vandewalle-Capo M, Massip C, Descours G, Charavit J, Chastang J, Billy PA, et al. Minimum inhibitory concentration (MIC) distribution among wild-type strains of Legionella pneumophila identifies a subpopulation with reduced susceptibility to macrolides owing to efflux pump genes. Int J Antimicrob Agents. (2017) 50:684–9. doi: 10.1016/j.ijantimicag.2017.08.001
29. Fontana S, Scaturro M, Rota MC, Caporali MG, Ricci ML. Molecular typing of Legionella pneumophila serogroup 1 clinical strains isolated in Italy. Int J Med Microbiol. (2014) 304:597–602. doi: 10.1016/j.ijmm.2014.04.004
30. Mercante JW, Caravas JA, Ishaq MK, Kozak-Muiznieks NA, Raphael BH, Winchell JM. Genomic heterogeneity differentiates clinical and environmental subgroups of Legionella pneumophila sequence type 1. PLoS ONE. (2018) 13:e0206110. doi: 10.1371/journal.pone.0206110
31. Cruz C, Rodrigues L, Fernandes F, Santos R, Paixão P, Chasqueira MJ. Antibiotic susceptibility pattern of Portuguese environmental Legionella isolates. Front Cell Infect Microbiol. (2023) 13:1141115. doi: 10.3389/fcimb.2023.1141115
Keywords: legionnaires' disease, Legionella pneumophila, Bordetella bronchiseptica, coinfection, cgMLST
Citation: La Sorda M, Palucci I, Natalini D, Fillo S, Giordani F, Paglione F, Monte A, Lista F, Mancini F, Girolamo A, Rota MC, Caporali MG, Ricci R, Ginevra C, Jarraud S, Sanguinetti M, Scaturro M and Ricci ML (2024) Case report: First report of Legionella pneumophila and Bordetella bronchiseptica coinfection in an immunocompromised patient. Front. Med. 11:1470567. doi: 10.3389/fmed.2024.1470567
Received: 25 July 2024; Accepted: 20 September 2024;
Published: 22 October 2024.
Edited by:
Daniele Roberto Giacobbe, University of Genoa, ItalyReviewed by:
Giovanni Gherardi, Campus Bio-Medico University, ItalyDimosthenis Chochlakis, University of Crete, Greece
Copyright © 2024 La Sorda, Palucci, Natalini, Fillo, Giordani, Paglione, Monte, Lista, Mancini, Girolamo, Rota, Caporali, Ricci, Ginevra, Jarraud, Sanguinetti, Scaturro and Ricci. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Scaturro, bWFyaWEuc2NhdHVycm8mI3gwMDA0MDtpc3MuaXQ=
†These authors have contributed equally to this work
 Marilena La Sorda1†
Marilena La Sorda1† Ivana Palucci
Ivana Palucci Daniele Natalini
Daniele Natalini Silvia Fillo
Silvia Fillo Francesco Paglione
Francesco Paglione Florigio Lista
Florigio Lista Fabiola Mancini
Fabiola Mancini Christophe Ginevra
Christophe Ginevra Sophie Jarraud
Sophie Jarraud Maurizio Sanguinetti
Maurizio Sanguinetti Maria Scaturro
Maria Scaturro Maria Luisa Ricci
Maria Luisa Ricci