- 1Department of Molecular Biology and Cell Pathology, Third Faculty of Medicine, Charles University, Prague, Czechia
- 2Institute for the Care of the Mother and Child, Third Faculty of Medicine, Charles University, Prague, Czechia
Background: Placenta previa is the abnormal implantation of the placenta into the lower segment of the uterus, is associated with adverse maternal and fetal outcomes such as placenta accreta spectrum disorders, antepartum and postpartum hemorrhage, fetal growth restriction, prematurity, stillbirth and neonatal death, thrombophlebitis, and septicemia. The aim of the study was to assess retrospectively how the later onset of placenta previa affects the microRNA expression profile in the whole peripheral blood during the first trimester of gestation.
Methods: Regarding the occurrence of the association between aberrant microRNA expression profiles at early stages of gestation and later onset of various pregnancy-related complications, we selected for the study pregnancies developing placenta previa as the only pregnancy-related disorder. In total, 24 singleton pregnancies diagnosed with placenta previa that underwent first-trimester prenatal screening and delivered on-site within the period November 2012–May 2018 were included in the study. Overall, 80 normal pregnancies that delivered appropriate-for-gestational age newborns after completing 37 weeks of gestation were selected as the control group based on the equality of the length of biological sample storage.
Results: Downregulation of multiple microRNAs (miR-20b-5p, miR-24-3p, miR-26a-5p, miR-92a-3p, miR-103a-3p, miR-130b-3p, miR-133a-3p, miR-145-5p, miR-146a-5p, miR-155-5p, miR-181a-5p, miR-195-5p, miR-210-3p, miR-342-3p, and miR-574-3p) was observed in pregnancies destined to develop placenta previa. The combination of seven microRNAs (miR-130b-3p, miR-145-5p, miR-155-5p, miR-181a-5p, miR-210-3p, miR-342-3p, and miR-574-3p) showed the highest accuracy (AUC 0.937, p < 0.001, 100.0% sensitivity, 83.75% specificity) to differentiate, at early stages of gestation, between pregnancies with a normal course of gestation and those with placenta previa diagnosed in the second half of pregnancy. Overall, 75% of pregnancies destined to develop placenta previa were correctly identified at 10.0% FPR.
Conclusion: Consecutive large-scale analyses must be performed to verify the reliability of the proposed novel early predictive model for placenta previa occurring as the only pregnancy-related disorder.
1 Introduction
Placenta previa is diagnosed when the placenta obstructs the internal cervical os. There are three classification grades of placenta previa: marginal (placenta reaches the margin of the internal cervical os), partial (placenta partially covers the internal cervical os), and complete (placenta completely covers the internal cervical os) (1). The prevalence of placenta previa differs regionally but, overall, it reaches 5.2 cases per 1,000 pregnancies (2, 3). The pathogenesis of this placental disorder remains to be resolved. Placenta previa is associated with adverse maternal and fetal outcomes such as placenta accreta spectrum (PAS) disorders, antepartum and postpartum hemorrhage, fetal growth restriction (FGR), prematurity, stillbirth and neonatal death, thrombophlebitis, and septicemia (2, 4–7). Prior spontaneous or induced abortion, male fetus, smoking, advanced maternal age, C-section, and assisted reproductive techniques (singleton pregnancy) represent the main risk factors associated with placenta previa (8–10). The recommendations for diagnosis and classification of placenta previa and for managing the care of women have been summarized in the SOGC Clinical Practice Guideline (11). Screening for placenta previa is a part of the routine antenatal care performed at 18–22 gestational weeks and 32–34 gestational weeks (12). Currently, there is no screening protocol for placenta previa in the first trimester of gestation. However, recently, first-trimester screening for PAS disorders based on the early and late first-trimester sonographic markers suitable for individuals with a history of cesarean delivery has been introduced. A finding of the placenta under or within the scar niche should be referred to specialized centers (13).
MicroRNAs are small non-coding RNAs (18–25 nucleotides) that regulate gene expression at the post-transcriptional level (14, 15). MicroRNA upregulation results in the blockage of translation or degradation of mRNAs. On the other hand, microRNA downregulation results in overexpression of potential target genes. Usually, an altered microRNA expression profile accompanies certain diseases and may be used for the diagnosis and/or the assessment of prognosis (16–18).
The aim of the study was to assess if there are any changes in the microRNA expression profile in the whole peripheral blood in the first trimester of gestation in pregnancies developing placenta previa only in the second half of pregnancy. Recently, we observed an altered expression profile of microRNAs that play a role in the homeostasis and maintenance of the cardiovascular system and the pathophysiology of cardiovascular and cerebrovascular diseases in women at risk of adverse pregnancy outcomes (19–25). We have demonstrated the association between aberrant microRNA expression profiles at early stages of gestation and the presence of chronic hypertension (19), later onset of gestational hypertension (GH) (19), preeclampsia (PE) (19), FGR (20), small for gestational age (SGA) (20), preterm delivery (21), gestational diabetes mellitus (GDM) (22), HELLP syndrome (23), and stillbirth (24). Therefore, we intentionally excluded from the study pregnancies with placenta previa simultaneously affected with chronic hypertension and other pregnancy-related complications. We selected for the study pregnancies with placenta previa only (without the presence of abnormally invasive placenta such as placenta accreta, increta, or percreta).
2 Materials and methods
2.1 Patients cohort
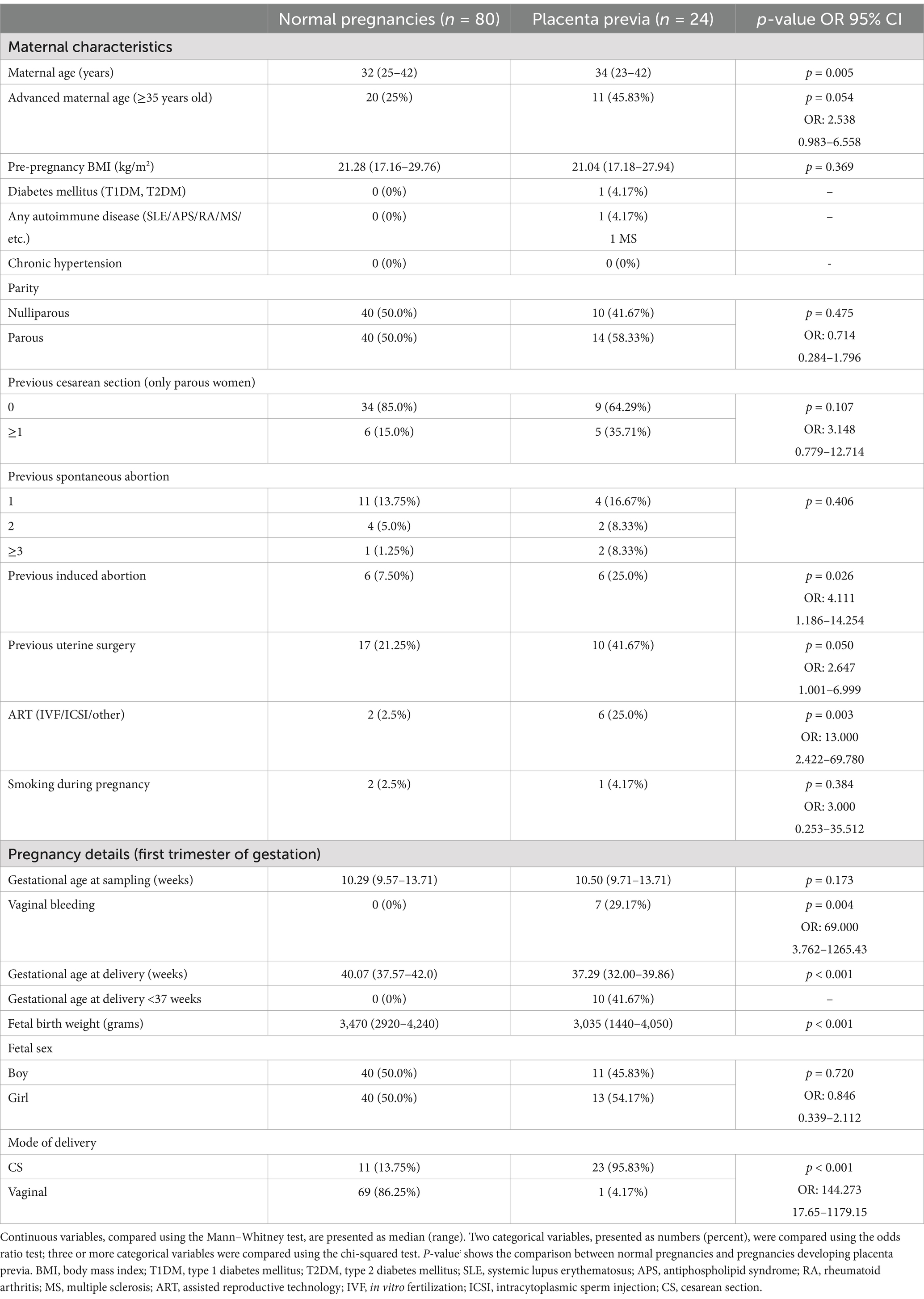
The retrospective study in pregnancies of Caucasian descent was performed within the period 11/2012–5/2018. In total, 24 singleton pregnancies diagnosed with placenta previa in the second half of gestation, but with an otherwise normal course of gestation, were identified from a cohort of 3,028 pregnancies that underwent first-trimester prenatal screening and were delivered on-site. Only pregnancies without abnormally invasive placenta (placenta accreta, increta, or percreta) were included in the study. The reference group consisted of 80 normal pregnancies that delivered appropriate-for-gestational age newborns after completing 37 weeks of gestation. The selection of gestational-age-matched normal-term pregnancies at sampling (weeks) with equal biological sample-storage time ensured the homogeneity and comparability between the studied groups. A peripheral venous blood sampling was performed between 10 and 13 gestational weeks. Relevant clinical characteristics of patients are summarized in Table 1.
All the included patients provided informed written consent for participation in the study. The Ethics Committee of the Third Faculty of Medicine, Charles University, granted initial approval for this study (implication of placental-specific microRNAs in maternal circulation for diagnosis and prediction of pregnancy-related complications, date of approval: 7 April 2011). Ongoing approval for the study was obtained from the Ethics Committee of the Third Faculty of Medicine, Charles University (long-term monitoring of complex cardiovascular profiles in mother, fetus, and offspring descending from pregnancy-related complications, date of approval: 27 March 2014) and the Ethics Committee of the Institute for the Care of the Mother and Child, Charles University (long-term monitoring of complex cardiovascular profiles in mother, fetus, and offspring descending from pregnancy-related complications, date of approval: 28 May 2015, number of approval: 1/4/2015). Informed consent is a complex process as it involves attaining consent for collecting peripheral blood samples at the beginning of pregnancy. In addition, it also includes gaining consent for collecting peripheral blood samples at the onset of pregnancy-related complications and collecting placental samples during childbirth in case of the onset of pregnancy-related complications.
2.2 Processing of samples and real-time RT-PCR analyses
Sample processing and real-time RT-PCR analysis were performed as previously described (19–25).
2.3 Statistical analysis
MicroRNA gene expression was compared between pregnancies with a normal course of gestation with and without the presence of placenta previa diagnosed in the second half of pregnancy using the Mann–Whitney test. The adjustment for covariates was performed using the Quade Non-parametric Analysis of Covariance (ANCOVA) (IBM SPSS Statistics 29.0.2.0). Benjamini–Hochberg-adjusted p-values were used to evaluate the statistical significance at α = 0.05 (p < 0.025*), α = 0.01 (p < 0.005**), and α = 0.001 (p < 0.001***) (26). Box plots were produced using the Statistica software (version 9.0; StatSoft, Inc., Tulsa, OK, United States).
Receiver operating characteristic (ROC) curves were produced using MedCalc software (MedCalc Software bvba, Ostend, Belgium). ROC curves displayed the areas under the curves (AUC) and the cutoff points associated with sensitivities, specificities, positive and negative likelihood ratios (LR+, LR−), and sensitivities at a 10.0% false-positive rate (FPR). To estimate the area under the curve (AUC) in the case of a combination of microRNA biomarkers, logistic regression was initially performed (MedCalc Software bvba, Ostend, Belgium). MicroRNAs entered into the initial logistic regression model as independent variables and the diagnosis as a dependent variable. Subsequently, a ROC curve analysis was performed (MedCalc Software bvba, Ostend, Belgium). The predictive probabilities gained from the logistic regression analysis were used as the new variable and the diagnosis as the classification variable.
2.4 Information on microRNA-gene-biological pathways interactions
The DIANA miRPath v.3 database (DIANA TOOLS-mirPath v.31) and the genes union mode were used as an a priori analysis method to perform KEGG pathway enrichment analysis. This approach aimed to investigate the regulatory mechanisms of microRNAs dysregulated at the early stages of gestation in the whole peripheral blood of mothers destined to develop placenta previa. The TarBase v7.0 database, which contains experimentally verified microRNA targets, was preferentially used for this analysis. In case the TarBase v7.0 database did not provide a sufficient list of experimentally verified microRNA targets, the target prediction algorithm (microT-CDS v5.0) was used as an alternative. In addition, the search for interactions between microRNAs and genes associated with pathways (KEGG) was performed using miRWalk database v.3 and TargetScan, miRDB, and miRTarBase filters (miRWalk2).
3 Results
3.1 Altered expression profile of microRNAs during the first trimester of gestation in pregnancies with normal course of gestation that developed placenta previa in the second half of pregnancy
The expression profile of microRNAs was compared during the first trimester of gestation in whole peripheral blood samples between pregnancies with a normal course of gestation with and without the presence of placenta previa diagnosed in the second half of pregnancy. Downregulation of miR-20b-5p (p = 0.016*), miR-24-3p (p = 0.001**), miR-26a-5p (p = 0.023*), miR-92a-3p (p = 0.009*), miR-103a-3p (p = 0.024*), miR-130b-3p (p < 0.001***), miR-133a-3p (p = 0.001**), miR-145-5p (p < 0.001***), miR-146a-5p (p = 0.001**), miR-155-5p (p < 0.001***), miR-181a-5p (p < 0.001***), miR-195-5p (p = 0.008*), miR-210-3p (p < 0.001***), miR-342-3p (p < 0.001***), and miR-574-3p (p < 0.001***) was detected in pregnancies destined to develop placenta previa (Figure 1).

Figure 1. MicroRNA expression profile at early stages of gestation in pregnancies developing placenta previa. In total, 15 microRNAs differentiate between pregnancies with a normal course of gestation with and without the presence of placenta previa diagnosed in the second half of pregnancy. (A) Statistically significant microRNAs that were not used in the final microRNA combination. (B) Statistically significant microRNAs that were used in the final microRNA combination since they display excellent levels of differentiation using ROC analyses.
After adjustment for covariates (maternal age, pre-pregnancy BMI, previous cesarean section, previous uterine surgery, and ART), seven microRNAs remained statistically significant [miR-130b-3p (p = 0.009*), miR-145-5p (p < 0.001***), miR-155-5p (p = 0.006*), miR-181a-5p (p = 0.012*), miR-210-3p (p = 0.008*), miR-342-3p (p = 0.002**), and miR-574-3p (p = 0.005**)]. These seven microRNAs with excellent and acceptable levels of area under the curve (AUC) were able to differentiate between normal pregnancies with and without the presence of placenta previa with the following sensitivities at 10.0% false-positive rate (FPR) [miR-130b-3p (25.0%), miR-145-5p (41.67%), miR-155-5p (25.0%), miR-181a-5p (12.5%), miR-210-3p (41.67%), miR-342-3p (12.5%), and miR-574-3p (20.83%)] (Figure 1).
3.2 Combination of seven microRNA biomarkers differentiates at early stages of gestation between pregnancies with normal course of gestation with and without the presence of placenta previa diagnosed in the second half of pregnancy
The combination of seven microRNA biomarkers was able to differentiate, during the first trimester of gestation, between pregnancies with a normal course and those with placenta previa diagnosed in the second half of pregnancy, with very high accuracy (AUC 0.937, p < 0.001, 100.0% sensitivity, 83.75% specificity, and cutoff >0.17329). Overall, 75% of pregnancies destined to develop placenta previa were identified at 10.0% FPR (Figure 2).
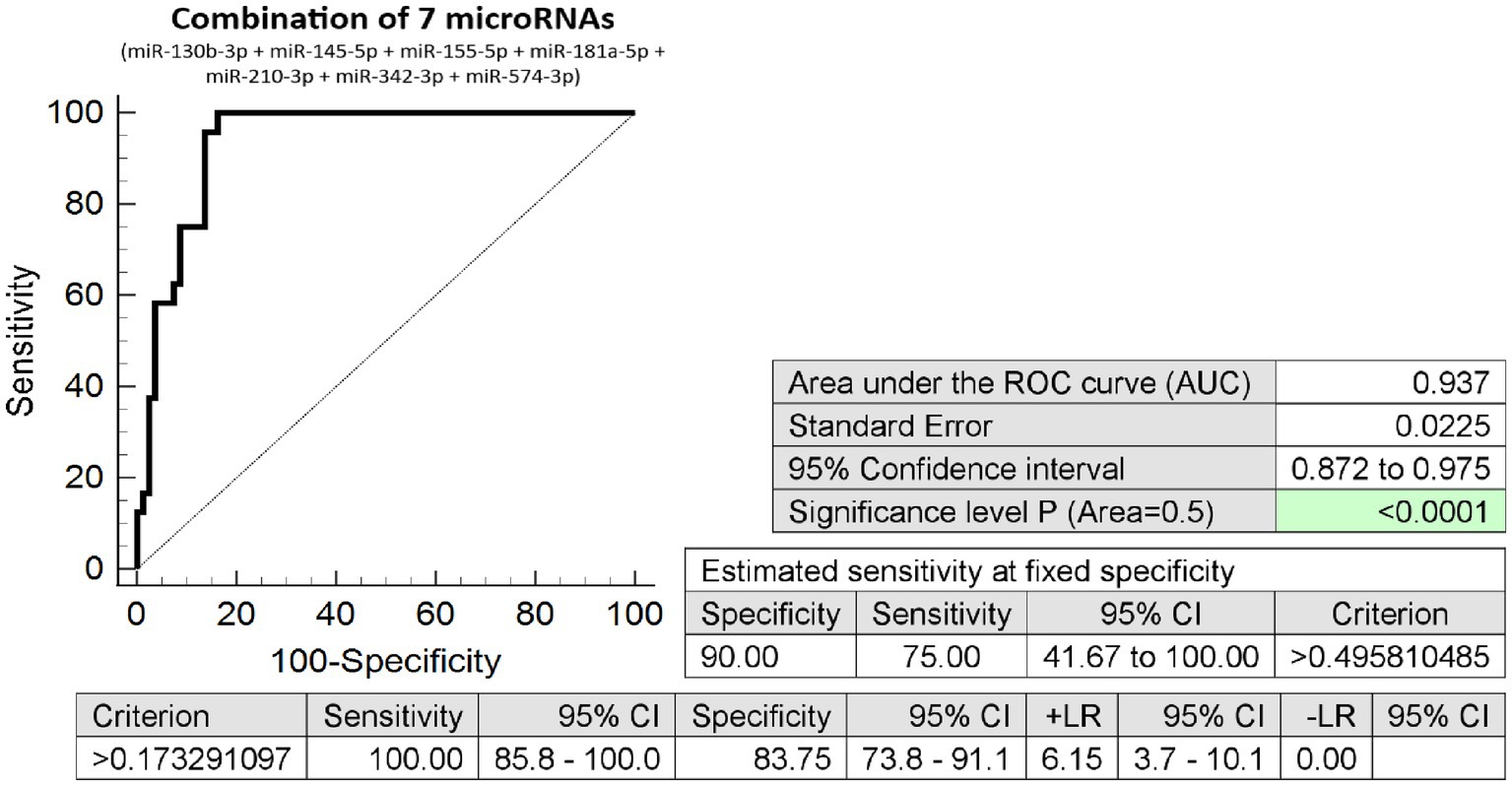
Figure 2. Combination of seven microRNA biomarkers – differentiation between pregnancies with and without the presence of placenta previa. The combination of 7 microRNA biomarkers (miR-130b-3p, miR-145-5p, miR-155-5p, miR-181a-5p, miR-210-3p, miR-342-3p, and miR-574-3p). Overall, 75% of pregnancies destined to develop placenta previa were revealed at early stages of gestation at 10.0% FPR.
3.3 Information on microRNA-gene-biological pathways interactions
The microRNA/gene/KEGG pathway analyses revealed the involvement of particular microRNAs with altered expression in various biological pathways and processes involved in placental development and maintenance of placental homeostasis (Table 2).
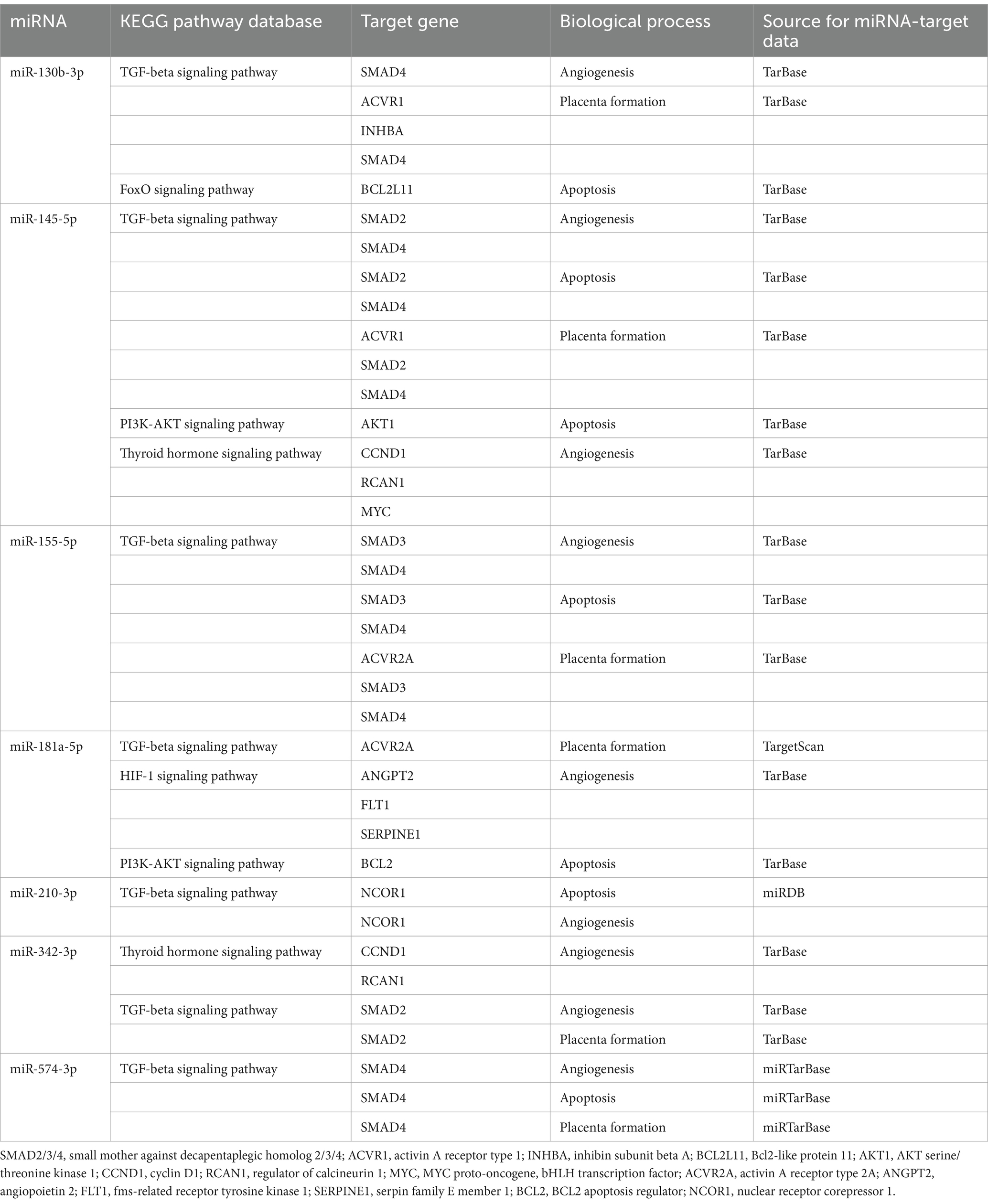
Table 2. Involvement of microRNAs in biological pathways and processes involved in placental development and maintenance of placental homeostasis.
4 Discussion
At early stages of gestation, we detected decreased expression of multiple microRNAs (miR-20b-5p, miR-24-3p, miR-26a-5p, miR-92a-3p, miR-103a-3p, miR-130b-3p, miR-133a-3p, miR-145-5p, miR-146a-5p, miR-155-5p, miR-181a-5p, miR-195-5p, miR-210-3p, miR-342-3p, and miR-574-3p) in pregnancies that developed placenta previa in the second half of pregnancy as the only pregnancy-related disorder. MicroRNA expression profiles associated with placenta previa resembled microRNA expression profiles associated with preterm birth [spontaneous preterm birth (PTB) or preterm prelabor rupture of membranes (PPROM)] in the absence of other pregnancy-related complications (21), where some of these microRNAs (miR-20b-5p, miR-24-3p, miR-26a-5p, miR-92a-3p, miR-133a-3p, miR-145-5p, miR-146a-5p, miR-155-5p, miR-210-3p, and miR-342-3p) were also downregulated at the early stages of gestation. The microRNA expression profile associated with placenta previa was identical to the microRNA expression profile associated with late miscarriage or stillbirth (24), where microRNAs such as miR-130b-3p, miR-145-5p, miR-210-3p, miR-342-3p, and miR-574-3p were also observed to have decreased expression at the early stages of gestation. The combination of seven microRNAs (miR-130b-3p, miR-145-5p, miR-155-5p, miR-181a-5p, miR-210-3p, miR-342-3p, and miR-574-3p) showed the highest accuracy (AUC 0.937, p < 0.001, 100.0% sensitivity, 83.75% specificity) to differentiate, at early stages of gestation, between pregnancies with normal course of gestation and those with placenta previa diagnosed in the second half of pregnancy. Overall, 75% of pregnancies destined to develop placenta previa were correctly identified at 10.0% FPR.
To the best of our knowledge, no study on the microRNA expression profile in the whole peripheral blood at the first trimester of gestation in pregnancies destined to develop placenta previa as the only pregnancy-related complication is available. Only a few studies examined microRNA expression profiles in pregnancies with placenta previa or abnormally invasive placenta (placenta accreta, increta, and percreta) in various biological samples (mainly in plasma or serum) at various stages of gestation (mainly at the third trimester of gestation) with the aim to predict these placental disorders. Hasegawa et al. reported that plasma miR-517a may serve at 32 weeks of gestation in pregnancies with placenta previa as a predictive marker for the risk of alert bleeding and massive hemorrhage at the delivery (27). Timofeeva et al. (28) observed abnormal plasma levels of miR-17-5p, miR-21-5p, miR-25-3p, miR-92a-3p, and miR-320a-3p at 30–34 gestational weeks in pregnancies with placenta accreta, increta, or percreta. The study by Munoz et al. (29) showed that plasma exosomal microRNAs (miR-92, −103, and −192) may represent additional potential biomarkers for detecting PAS within 25–36 gestational weeks. Similarly, Chen et al. (30) validated four other serum microRNAs (miR-139-3p, miR-196a-5p, miR-518a-3p, and miR-671-3p) that could be potentially used for non-invasive prenatal PAS screening during the third trimester and before the delivery. None of these stated microRNA biomarkers are identical to those we demonstrated to be dysregulated at early stages of gestation in pregnancies affected with placenta previa with otherwise normally ongoing gestation.
Consecutive large-scale analyses must be performed to verify the reliability of the proposed novel early predictive model for placenta previa occurring as the only pregnancy-related disorder based on the combination of microRNA biomarkers (miR-130b-3p, miR-145-5p, miR-155-5p, miR-181a-5p, miR-210-3p, miR-342-3p, and miR-574-3p). Multicenter studies will be needed to acquire a sufficient number of novel cases to validate the data resulting from the current pilot study. In addition, future studies tracking microRNAs with abnormal expression at early stages of gestation throughout the pregnancy are needed as well. If satisfactory discrimination power is achieved, gynecologists and obstetricians could have at their disposal a feasible, cost-effective way of identifying pregnancies at risk of placenta previa at early gestational stages when it occurs as the only pregnancy-related disorder.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the Ethics Committee of the Third Faculty of Medicine, Charles University and Ethics Committee of the Institute for the Care of the Mother and Child, Charles University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
IH: Writing – original draft, Supervision, Resources, Project administration, Funding acquisition, Conceptualization, Writing – review & editing, Validation, Software, Methodology, Formal analysis, Data curation. KK: Visualization, Investigation, Writing – review & editing, Validation, Software, Methodology, Formal analysis, Data curation. LK: Writing – review & editing, Validation, Supervision, Software, Resources, Project administration, Funding acquisition, Conceptualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Charles University Research Program Cooperation—Mother and Childhood Care (no. 207035) and a research grant SVV (no. 260645).
Acknowledgments
We thank the staff of the Institute for the Care of Mother and Child for their assistance with the collection of the patient biological samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Bi, S, Zhang, L, Wang, Z, Chen, J, Tang, J, Gong, J, et al. Effect of types of placenta previa on maternal and neonatal outcomes: a 10-year retrospective cohort study. Arch Gynecol Obstet. (2021) 304:65–72. doi: 10.1007/s00404-020-05912-9
2. Sahu, SA, and Shrivastava, D. Maternal and perinatal outcomes in placenta Previa: a comprehensive review of evidence. Cureus. (2024) 16:e59737. doi: 10.7759/cureus.59737
3. Jansen, CHJR, Kastelein, AW, Kleinrouweler, CE, Van Leeuwen, E, De Jong, KH, Pajkrt, E, et al. Development of placental abnormalities in location and anatomy. Acta Obstet Gynecol Scand. (2020) 99:983–93. doi: 10.1111/aogs.13834
4. Karami, M, Jenabi, E, and Fereidooni, B. The association of placenta previa and assisted reproductive techniques: a meta-analysis. J Matern Fetal Neonatal Med. (2018) 31:1940–7. doi: 10.1080/14767058.2017.1332035
5. Hessami, K, Mitts, M, Zargarzadeh, N, Jamali, M, Berghella, V, and Shamshirsaz, AA. Ultrasonographic cervical length assessment in pregnancies with placenta previa and risk of perinatal adverse outcomes: a systematic review and meta-analysis. Am J Obstet Gynecol MFM. (2024) 6:101172. doi: 10.1016/j.ajogmf.2023.101172
6. Hu, H, Wang, L, Gao, J, Chen, Z, Chen, X, Tang, P, et al. Risk factors of severe postpartum hemorrhage in pregnant women with placenta previa or low-lying placenta: a retrospective cohort study. BMC Pregnancy Childbirth. (2024) 24:674. doi: 10.1186/s12884-024-06876-3
7. Kumari, U, Naniwal, A, Rani, V, Chandat, R, Yadav, S, and Pipal, DK. A study of clinical characteristics, demographic characteristics, and fetomaternal outcomes in cases of placenta Previa: an experience of a tertiary care center. Cureus. (2022) 14:e32125. doi: 10.7759/cureus.32125
8. Jenabi, E, Salimi, Z, Bashirian, S, Khazaei, S, and Ayubi, E. The risk factors associated with placenta previa: an umbrella review. Placenta. (2022) 117:21–7. doi: 10.1016/j.placenta.2021.10.009
9. Pun, I, and Singh, A. Feto-maternal outcomes in placenta Previa with and without previous cesarean section. J Nepal Health Res Counc. (2022) 20:142–6. doi: 10.33314/jnhrc.v20i01.3640
10. Post, RJ, Chang, J, Ziogas, A, Crosland, BA, Silver, RM, Haas, DM, et al. Risk factors and perinatal outcomes for persistent placenta previa in nulliparas. Am J Obstet Gynecol MFM. (2023) 5:101136. doi: 10.1016/j.ajogmf.2023.101136
11. Jain, V, Bos, H, and Bujold, E. Guideline No. 402: diagnosis and Management of Placenta Previa. J Obstet Gynaecol Can. (2020) 42:906–917.e1. doi: 10.1016/j.jogc.2019.07.019
12. Bhide, A. Routine screening for placenta accreta spectrum. Best Pract Res Clin Obstet Gynaecol. (2023) 90:102392. doi: 10.1016/j.bpobgyn.2023.102392
13. Dar, P, and Doulaveris, G. First-trimester screening for placenta accreta spectrum. Am J Obstet Gynecol MFM. (2024) 6:101329. doi: 10.1016/j.ajogmf.2024.101329
14. Lai, EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. (2002) 30:363–4. doi: 10.1038/ng865
15. Bartel, DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/s0092-8674(04)00045-5
16. Piletič, K, and Kunej, T. MicroRNA epigenetic signatures in human disease. Arch Toxicol. (2016) 90:2405–19. doi: 10.1007/s00204-016-1815-7
17. Wang, J, Chen, J, and Sen, S. MicroRNA as biomarkers and diagnostics. J Cell Physiol. (2016) 231:25–30. doi: 10.1002/jcp.25056
18. Condrat, CE, Thompson, DC, Barbu, MG, Bugnar, OL, Boboc, A, Cretoiu, D, et al. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells. (2020) 9:276. doi: 10.3390/cells9020276
19. Hromadnikova, I, Kotlabova, K, and Krofta, L. Cardiovascular disease-associated MicroRNA dysregulation during the first trimester of gestation in women with chronic hypertension and normotensive women subsequently developing gestational hypertension or preeclampsia with or without fetal growth restriction. Biomedicines. (2022) 10:256. doi: 10.3390/biomedicines10020256
20. Hromadnikova, I, Kotlabova, K, and Krofta, L. First-trimester screening for fetal growth restriction and small-for-gestational-age pregnancies without preeclampsia using cardiovascular disease-associated MicroRNA biomarkers. Biomedicines. (2022) 10:718. doi: 10.3390/biomedicines10030718
21. Hromadnikova, I, Kotlabova, K, and Krofta, L. First trimester prediction of preterm delivery in the absence of other pregnancy-related complications using cardiovascular-disease associated MicroRNA biomarkers. Int J Mol Sci. (2022) 23:3951. doi: 10.3390/ijms23073951
22. Hromadnikova, I, Kotlabova, K, and Krofta, L. Cardiovascular disease-associated MicroRNAs as novel biomarkers of first-trimester screening for gestational diabetes mellitus in the absence of other pregnancy-related complications. Int J Mol Sci. (2022) 23:10635. doi: 10.3390/ijms231810635
23. Hromadnikova, I, Kotlabova, K, and Krofta, L. First-trimester screening for HELLP syndrome-prediction model based on MicroRNA biomarkers and maternal clinical characteristics. Int J Mol Sci. (2023) 24:5177. doi: 10.3390/ijms24065177
24. Hromadnikova, I, Kotlabova, K, and Krofta, L. First-trimester screening for miscarriage or stillbirth-prediction model based on MicroRNA biomarkers. Int J Mol Sci. (2023) 24:10137. doi: 10.3390/ijms241210137
25. Hromadnikova, I, Kotlabova, K, and Krofta, L. First-trimester predictive models for adverse pregnancy outcomes—a base for implementation of strategies to prevent cardiovascular disease development. Front Cell Dev Biol. (2024) 12:1461547. doi: 10.3389/fcell.2024.1461547
26. Haynes, W. Benjamini–Hochberg Method In: W Dubitzky, O Wolkenhauer, KH Cho, and H Yokota, editors. Encyclopedia of systems biology. New York, NY: Springer (2013)
27. Hasegawa, Y, Miura, K, Higashijima, A, Abe, S, Miura, S, Yoshiura, K, et al. Increased levels of cell-free miR-517a and decreased levels of cell-free miR-518b in maternal plasma samples from placenta Previa pregnancies at 32 weeks of gestation. Reprod Sci. (2015) 22:1569–76. doi: 10.1177/1933719115589407
28. Timofeeva, AV, Fedorov, IS, Pirogova, MM, Vasilchenko, ON, Chagovets, VV, Ezhova, LS, et al. Clusterin and its potential regulatory microRNAs as a part of Secretome for the diagnosis of abnormally invasive placenta: Accreta, increta, and Percreta cases. Life. (2021) 11:270. doi: 10.3390/life11040270
29. Munoz, JL, Einerson, BD, Silver, RM, Mulampurath, S, Sherman, LS, Rameshwar, P, et al. Serum exosomal microRNA pathway activation in placenta accreta spectrum: pathophysiology and detection. AJOG Glob Rep. (2024) 4:100319. doi: 10.1016/j.xagr.2024.100319
Keywords: first trimester screening, gene expression, microRNAs, prediction, placenta previa, whole peripheral venous blood
Citation: Hromadnikova I, Kotlabova K and Krofta L (2024) Abnormal microRNA expression profile at early stages of gestation in pregnancies destined to develop placenta previa. Front. Med. 11:1469855. doi: 10.3389/fmed.2024.1469855
Edited by:
Ali Çetin, University of Health Sciences, TürkiyeReviewed by:
Bilge Yaylak Gediksiz, University of Virginia, United StatesSeyda Berk, Cumhuriyet University, Türkiye
Duygu Sari Ak, University of Health Sciences, Türkiye
Elisa Spataro, Ospedale Santo Stefano, Italy
Copyright © 2024 Hromadnikova, Kotlabova and Krofta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ilona Hromadnikova, aWxvbmEuaHJvbWFkbmlrb3ZhQGxmMy5jdW5pLmN6
 Ilona Hromadnikova
Ilona Hromadnikova Katerina Kotlabova
Katerina Kotlabova Ladislav Krofta
Ladislav Krofta