- 1Department of Traditional Chinese Medicine, Jiangxi Maternal and Child Health Hospital, Nanchang, China
- 2Key Laboratory of Women's Reproductive Health of Jiangxi Province, Jiangxi Maternal and Child Health Hospital, Nanchang, China
- 3Key Research Unit of Female Reproduction with Integrated Chinese and Western Medicine of Jiangxi Province, Jiangxi Maternal and Child Health Hospital, Nanchang, China
- 4Central Laboratory, Jiangxi Maternal and Child Health Hospital, Nanchang, China
- 5Quality Control Office, Ganzhou People's Hospital, Ganzhou, China
- 6Graduate School of Clinical Medicine, Jiangxi University of Traditional Chinese Medicine, Nanchang, China
Background: Recurrent spontaneous abortion (RSA) is a multifactorial disease, the exact causes of which are still unknown. Environmental, maternal, and genetic factors have been shown to contribute to this condition. The aim of this study was to investigate the presence of mutations in the ANXA4 gene in patients with RSA.
Methods: Genomic DNA was extracted from 325 patients with RSA and 941 control women with a normal reproductive history for whole-exome sequencing (WES). The detected variants were annotated and filtered, and the pathogenicity of the variants was predicted through the SIFT online tool, functional enrichment analyses, Sanger sequencing validation, prediction of changes in protein structure, and evolutionary conservation analysis. Furthermore, plasmid construction, Western blotting, RT–qPCR, and cell migration, invasion and adhesion assays were used to detect the effects of ANXA4 mutations on protein function.
Results: An ANXA4 mutation (p.G8D) in 1 of the 325 samples from patients with RSA (RSA-219) was identified through WES. This mutation was not detected in 941 controls or included in public databases. Evolutionary conservation analysis revealed that the amino acid residue affected by the mutation (p.G8D) was highly conserved among 13 vertebrate species, and the SIFT program and structural modeling analysis predicted that this mutation was harmful. Furthermore, functional assays revealed that this mutation could inhibit cell migration, invasion and adhesion.
Conclusion: Our study suggests that an unreported novel ANXA4 mutation (p.G8D) plays an important role in the pathogenesis of RSA and may contribute to the genetic diagnosis of RSA.
1 Introduction
Recurrent spontaneous abortion (RSA) is a common complication in women of childbearing age during pregnancy, with an incidence rate ranging from 1 to 5% (1).
The number of miscarriages required for a diagnosis of RSA is controversial because the risk of another miscarriage after two consecutive miscarriages is similar to that after three consecutive miscarriages, and recent studies have suggested that predicting pregnancy loss after two consecutive miscarriages is more reasonable (2, 3). The American Reproductive Association defines recurrent spontaneous abortion as two or more consecutive miscarriages (4). Obviously, the pathogenesis of RSA is complex. To date, the potential risk factors for RSA reported include genetic, immunologic, anatomic, and infectious anomalies (5–10). However, approximately 50% of RSAs are unexplained or poorly understood (11).
For almost all human diseases, individual susceptibility is influenced by genetic variations to some degree (12). In recent years, whole-genome sequencing (WGS) and whole-exome sequencing (WES) have proven to be powerful new approaches for identifying disease-associated variants across the full minor allele frequency (MAF) spectrum in animals and humans (13–15). Genome-wide screening has shown that siblings of patients with RSA have a greater risk of miscarriage (16), Variants associated with RSA have been detected in genes involved in the immune response (IFNG, IL10, KIR2DS2, KIR2DS3, KIR2DS4, MBL, and TNF), coagulation (F2, F5, PAI-1, and PROZ), metabolism (GSTT1 and MTHFR), hormonal regulation (ESR1 and ADRB2), angiogenesis (NOS3 and VEGFA), and endometrial and placental function (ENOS and ACE) (17–19). Moreover, several studies have shown that rare variants play crucial roles in multiple human diseases, including RSA (20–22). However, few studies have confirmed that rare variants may play certain roles in the pathogenesis of RSA at the whole-exome/genome scale.
Annexin A4 (ANXA4) is a cytosolic calcium-binding protein with four repeat domains, each containing one calcium-binding site (CBS), and belongs to a ubiquitous family of Ca2+-dependent membrane-binding proteins thought to be involved in membrane trafficking and membrane organization within cells (23). Previous studies have shown that ANXA4 modulates membrane permeability and membrane trafficking, participates in cellular growth and apoptosis, and enhances tumor invasion (24). ANXA4 has been shown to be upregulated in various clinical epithelial tumors, including colorectal, pancreatic, ovarian, breast, and prostate cancers (24). Furthermore, a previous study revealed that the ANXA4 protein is localized to the glandular and luminal epithelium of the human endometrium and is present at high levels throughout the menstrual cycle, except during the early secretory (ES) phase (25).
In this study, we used WES to explore the potential involvement of rare variants in RSA risk in 325 patients with RSA. Among these rare variants, a novel missense mutation in the ANXA4 gene (c.23G > A, p.G8D) was detected in 1 patient with RSA (RSA-219). This variant was not found in either 941 controls or public databases. Functional assays revealed that this novel mutation could influence cell migration, invasion and adhesion, indicating that the ANXA4 mutation p.G8D contributes to the pathogenesis of RSA in women of Chinese ethnicity.
2 Materials and methods
2.1 Clinical samples
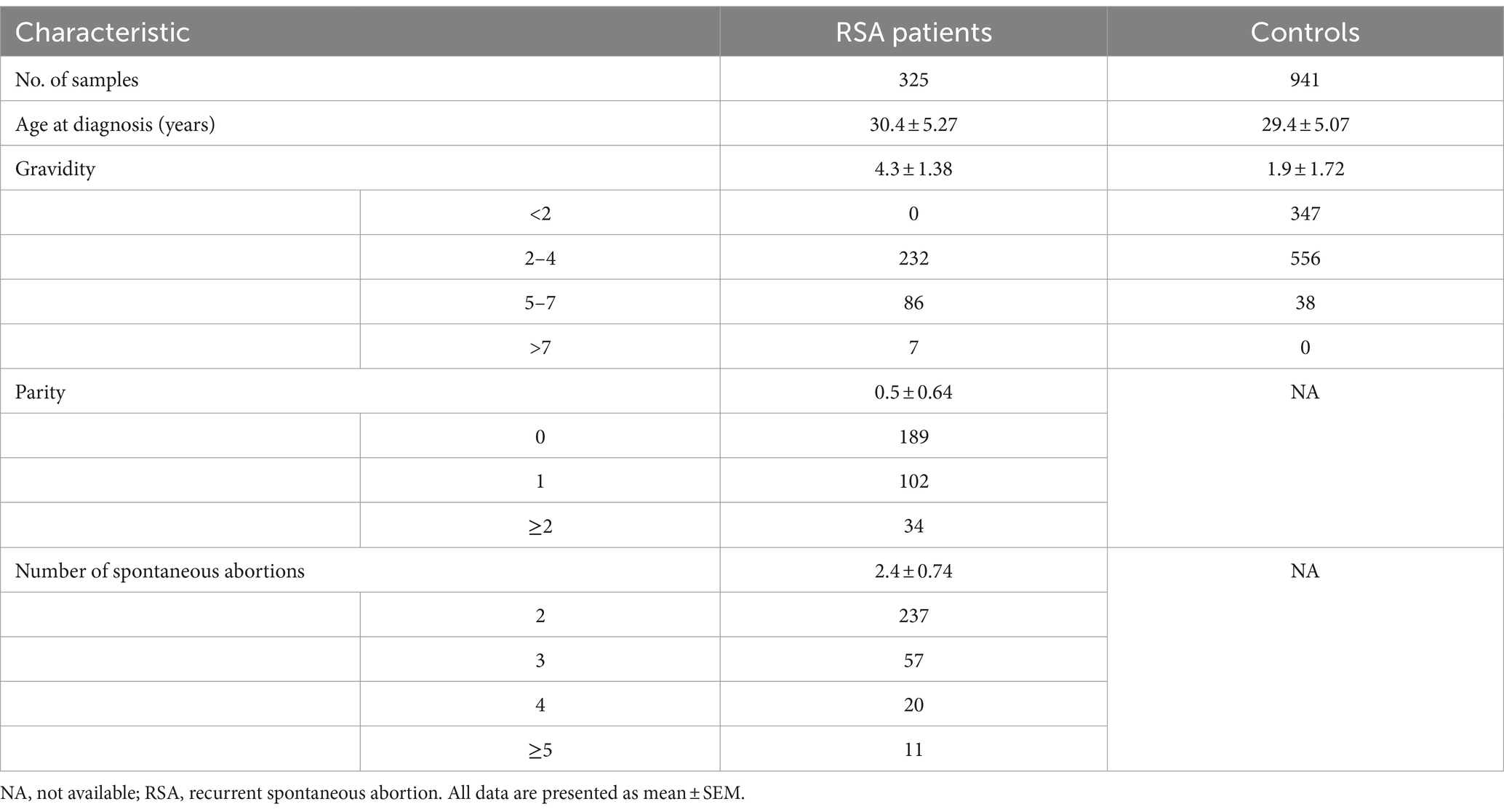
A total of 325 Han Chinese RSA patients and 941 control individuals at the Maternal and Child Health Hospital of Jiangxi Province between August 2018 and October 2019 were enrolled in this study. The RSA patients had experienced more than 2 unexplained and consecutive spontaneous abortions at less than 10 weeks of gestation. Patients who had genital abnormalities; chronic hypertension; diabetes; liver, kidney, cardiovascular, and thyroid diseases; autoimmune diseases; or infectious diseases were excluded. The control group was composed of women who underwent induced abortions of a normal pregnancy at a gestational age matched to that of the RSA group. The individuals in the control group were not treated with any drugs and had no pregnancy risk. The basic information of our study population is summarized in Table 1.
All patients signed an informed consent form before being enrolled in the study, and this study was approved by the Ethics Committee of Jiangxi Provincial Maternal and Child Health Hospital (EC-KY-202149).
2.2 Exome sequencing and data analysis
Genomic DNA was extracted from the peripheral blood of each sample with the AxyPrep Blood Genomic DNA Miniprep Kit (Axygen Scientific, Inc., 33,210 Central Avenue, Union City, CA 94587, United States) according to the instructions provided by the manufacturer. DNA quality and concentration were determined with a NanoDrop-1000 spectrophotometer (Thermo Fisher, United States) and gel electrophoresis, respectively. Exome capture was performed with a BGI Exome V4 (59 Mb) Kit (BGI, China) according to the manufacturer’s protocols. DNA sequencing was performed on BGISEQ-500 sequencers (BGI, China) in high-output mode with 100 bp paired-end reads. Exome data were mapped to the human reference genome GRCh37 using the Burrows–Wheeler Aligner (BWA-MEM, version 0.6.2) (26). The credibility and quality of single-nucleotide polymorphisms (SNPs) and small insertions/deletions (InDels) were detected with the Genome Analysis Tool Kit (GATK, version 3.7) (27). ANNOVAR was used for the annotation and classification of SNPs and InDels, respectively (28). Variants with a read depth > 20 were extracted for further analysis. Finally, the variants identified through the above pipeline were filtered to eliminate benign variants with minor allele frequency (MAF) ≥1% in the dbSNP database,1 1,000 Genomes database,2 Exome Aggregation Consortium (ExAC, http://exac.broadinstitute.org/) and BGI’s in-house databases containing exome data from >100,000 subjects (29, 30). We ranked the genes and their potential for damage using the online prediction program SIFT to analyze whether these variants were potentially pathogenic. Variants that received lower SIFT scores (typically ≤0.05), indicating a greater likelihood of affecting protein function, were predicted to be damaging.
2.3 Validation by Sanger sequencing
Sanger sequencing was used to verify the mutation in the gene of interest in RSA patients. The polymerase chain reaction (PCR) primer was designed according to the mutation site in the ANXA4 gene with Primer 3 (F: 5’-GGCCTCGAAGAACTTCTGCT-3′, R: 5’-TGGGCATCTTCCATGGCATT-3′). PCR amplification was performed using the rTaq enzyme (Takara Biotechnology, Dalian, China) with 50 ng of genomic DNA as a template. PCR was performed with an ABI 2720 thermocycler (Applied Biosystems, Waltham, MA), and the reaction conditions were as follows: the first step was 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and extension at 72°C for 30 s and a final extension at 72°C for 10 min. The amplified PCR products were subjected to 1% agarose gel electrophoresis and then purified from the gel with a DNA purification kit (Tiangen, Beijing, China). The purified PCR products were sequenced with an ABI 3730 Automatic Capillary DNA Sequencer (Applied Biosystems, Waltham, MA). The obtained DNA sequences were analyzed using DNASTAR Lasergene version 7.2 software (DNASTAR, Inc., Madison, WI, United States) with the reference sequence from the NCBI database.
2.4 Evolutionary conservation analysis of the rare ANXA4 variant (p.G8D)
To predict the potential pathogenicity of the mutation, evolutionary conservation analysis was performed by aligning the amino acid sequences of ANXA4 proteins from 13 species obtained from the GenBank database,3 including Human (NP_001144.1), Chimpanzee (XP_016804171.1), Pongo abelii (XP_002811982.2), Green monkey (XP_007968539.1), Nomascus leucogenys (XP_030683909.1), Mouse (NP_001318049.1), Rat (NP_077069.3), Cattle (NP_001001440.2), Pig (NP_001161111.1), Dog (NP_001003039.2), Horse (XP_023474726.1), Rabbit (XP_017196184.1) and Goat (XP_017910949.1). Evolutionary conservation analysis was performed using MEGA4 software.
2.5 Protein structural modeling
The modeling of the protein template between the reference and the modified (p.G8D) mutation of the ANXA4 gene was conducted using the SWISS-MODEL repository database.4 Then, we employed the Chimera 1.14rc package to concurrently compare the protein models.
2.6 Plasmid construction
Wild-type and mutant ANXA4 (NM_001153.5) were obtained from GeneCreate Biological Engineering Co., Ltd. (Wuhan, China). Human ANXA4 cDNA was inserted into the pcDNA3.1 vector to generate the wild-type plasmid. The mutant plasmid was generated with the wild-type plasmid as the template using a KOD-Plus-Mutagenesis Kit (Toyobo, Osaka, Japan). All the plasmids were confirmed by Sanger sequencing.
2.7 Cell culture and transfection
The THESCs cell line (human endometrial stromal cell line) was obtained from ATCC (Manassas, VA, http://www.lgcstandards-atcc.org). The cells were cultured in DMEM/F12 medium (HyClone, GE, United States) supplemented with 10% fetal bovine serum (FBS) (SA211.02, Minhai Bioengineering Co., Ltd., Lanzhou, China) and 1% penicillin–streptomycin (PS) (P1400, Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) in an incubator at 37°C with a humidified atmosphere of 5% CO2.
The wild-type or mutant ANXA4 plasmids were transfected into THESCs with FuGENE HD transfection reagent (Promega, E2311) according to the manufacturer’s instructions. Briefly, 1 × 105 THESCs were seeded in 6-well plates and cultured for 24 h. The medium in the 6-well plate was subsequently discarded, and 800 μL of DMEM (without FBS or antibiotics) was added to each well. A total of 100 μL of DMEM (without FBS or antibiotics), 2 μg of plasmid and 5 μL of FuGENE HD transfection reagent were mixed together, and the mixture was incubated for 10 min at room temperature. Finally, the transfection mixture was added to the cells gently and incubated in an incubator with 5% CO2 at 37°C for 2 h. Then, 1 mL of DMEM with 10% FBS was added to each well, and the cells were cultured in an incubator with 5% CO2 at 37°C.
2.8 Western blotting
Protein expression was detected by Western blotting analysis. Forty-eight hours after transfection, the cells were treated with cell lysis buffer (Applygen Technologies Co., Ltd., Beijing). The protein concentration was determined by using a bicinchoninic acid (BCA) protein analysis kit (Thermo Fisher Biochemical, Beijing). The protein samples (20–30 μg) were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE), transferred to polyvinylidene difluoride (PVDF) membranes (Merck KGaA, United States), and blocked in 5% skim milk for 1 h at room temperature. The membranes were subsequently incubated with the following primary antibodies at 4°C overnight: anti-ANXA4 (1:1000, TA803051; Origene, China) and anti-β-actin (1,2000, sc-47778; Santa Cruz). After being washed with 1× TBST three times, the membranes were incubated with the corresponding secondary antibodies for 2 h at room temperature. Protein bands were visualized using luminol reagent (sc-2048; Santa Cruz Biotechnology Company, United States) or enhanced chemiluminescence reagent (Santa Cruz Biotechnology Company, United States) with a ChemiDoc XRS instrument (Bio-Rad Laboratories, CA, United States). Relative protein expression levels were measured using ImageJ software (NIH).
2.9 RNA isolation and real-time quantitative PCR
RT–qPCR was used to assess the mRNA expression of ANXA4. The quality and concentration of total RNA were confirmed with a Nanodrop 2000 instrument (Thermo Scientific, United States), followed by extraction from treated cells using TRIzol reagent (Invitrogen, United States). Five hundred nanograms of RNA was reverse transcribed to synthesize cDNA (20 μL) using a PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa, Japan) according to the manufacturer’s protocol. Then, qPCR was performed with SYBR Green PCR Master Mix (TaKaRa, Japan) on an ABI 7500 thermocycler (Applied Biosystems, Waltham, MA). ANXA4 was used as the target gene, and GAPDH was used as an internal reference. The primer sequences for both genes were designed and validated for specificity. The primer sequences used for RT–qPCR were as follows: ANXA4-F (5’-TGCCCTGCTGAGCTGGACTT-3′) and ANXA4-R (5’-AAAGCTGCTCAGGACCATGT-3′); GAPDH-F (5’-GGAGCGAGATCCCTCCAAAAT-3′) and GAPDH-R (5′- GGCTGTTGTCATACTTCTCATGG-3′). The following qPCR conditions were used: initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 10 s, and extension at 72°C for 15 s and a final extension at 72°C for 10 min. The 2−ΔΔCt method was used to calculate and normalize the relative expression of ANXA4 to that of GAPDH.
2.10 Cell migration and invasion assays
The migration and invasion capabilities of THESCs transfected with wild-type or mutant ANXA4 plasmids were assessed with Transwell assays (Corning, Toledo, OH, United States).
For the migration analysis, after 48 h of transfection, the cells (3 × 105/ml) were collected, resuspended in DMEM/F12 without FBS, and then inoculated into the upper Transwell chamber, while the lower chamber was filled with 500 μL of DMEM/F12 supplemented with 20% FBS. After incubation for 24 h, the upper chambers were washed, fixed with methanol for 20 min, stained with 1% crystal violet dye solution for 30 min at room temperature, and washed with phosphate-buffered saline (PBS) several times. The chamber membranes were then photographed via light microscopy (IX71, OLYMPUS, Japan), and the number of migratory cells on the lower surface was counted in five random fields to assess the migration capability.
For invasion analysis, a total of 4 × 105 cells were seeded into the upper Transwell chamber, after which the upper chamber was precoated with Matrigel matrix (3.5 mg/mL) (BD Biosciences, United States). After 48 h, the same protocol as in the migration analysis was performed.
2.11 Cell adhesion assay
Briefly, after 48 h of transfection, a total of 4 × 103 cells/well were seeded in a 96-well plate, which was pretreated with 50 μL/well of Matrigel matrix (BD Biosciences, United States) and incubated at 37°C for 30 min to allow polymerization. Then, the 96-well plates were incubated in an incubator with 5% CO2 at 37°C for 20 min, 40 min or 60 min. The plates were washed with PBS three times to remove nonadherent cells, and the adherent cells were fixed with methanol for 15 min and stained with 1% crystal violet dye solution for 30 min at room temperature. The numbers of adherent cells were counted using light microscopy (IX71, OLYMPUS, Japan).
2.12 Statistical analyses
All the statistical analyses were performed using the statistical analysis software SPSS 21.0. Data are expressed as the mean ± standard error of the mean (S.E.M.). An unpaired Student’s t test was used to evaluate the significant differences among the groups. Statistical significance was established at a p value of less than 0.05.
3 Results
3.1 Identification of one novel rare variant in ANXA4 in one patient with RSA using WES
In total, 325 patients with RSA and 941 local women without RSA as controls underwent WES. WES performed on BGISEQ-500 sequencers achieved, on average, 21,266.42 Mb of raw bases. After removing low-quality reads, we obtained an average of 212,622,806 clean reads (21,255.71 Mb). The clean reads of each sample had high Q20 and Q30 values, indicating high sequencing quality. The average GC content was 52.14%. The chromosomal positions of the SNPs were based on the UCSC GRCh37/hg19 genome.
We obtained a total of 79,534 variants, including synonymous, missense, stop-loss, stop-gain, start-loss and splicing variants. The SnpEff tool5 was used to perform variant annotation and prediction. After excluding variants with MAFs greater than or equal to 1% in publicly available databases, e.g., dbSNP, 1,000 Genomes Project, ExAC and BGI’s in-house databases, 262 variants were retained. Using the in silico prediction algorithm SIFT, we selected candidate variants with potentially damaging impacts. The results showed that the ANXA4 gene was prominent on the basis of its functional annotation related to cell migration and invasion and the pathogenicity predictions of its mutations.
A novel mutation (c.23G > A, p.G8D) in the ANXA4 gene (NM_001153.5) was detected in 1 of the 325 patients with RSA (RSA-219) but was not detected in either the 941 controls or public databases (dbSNP, 1,000 Genomes Project, ExAC and BGI in-house databases). Primers were designed for PCR to amplify the ANXA4 fragment containing the rare variant, and Sanger sequencing was performed to validate this mutation (Figure 1A). The patient (RSA-219) was confirmed to harbor this mutation by Sanger sequencing. The ANXA4 gene is located on chromosome 2p13.3 and consists of 22 exons. The mutation is positioned in the first exon.
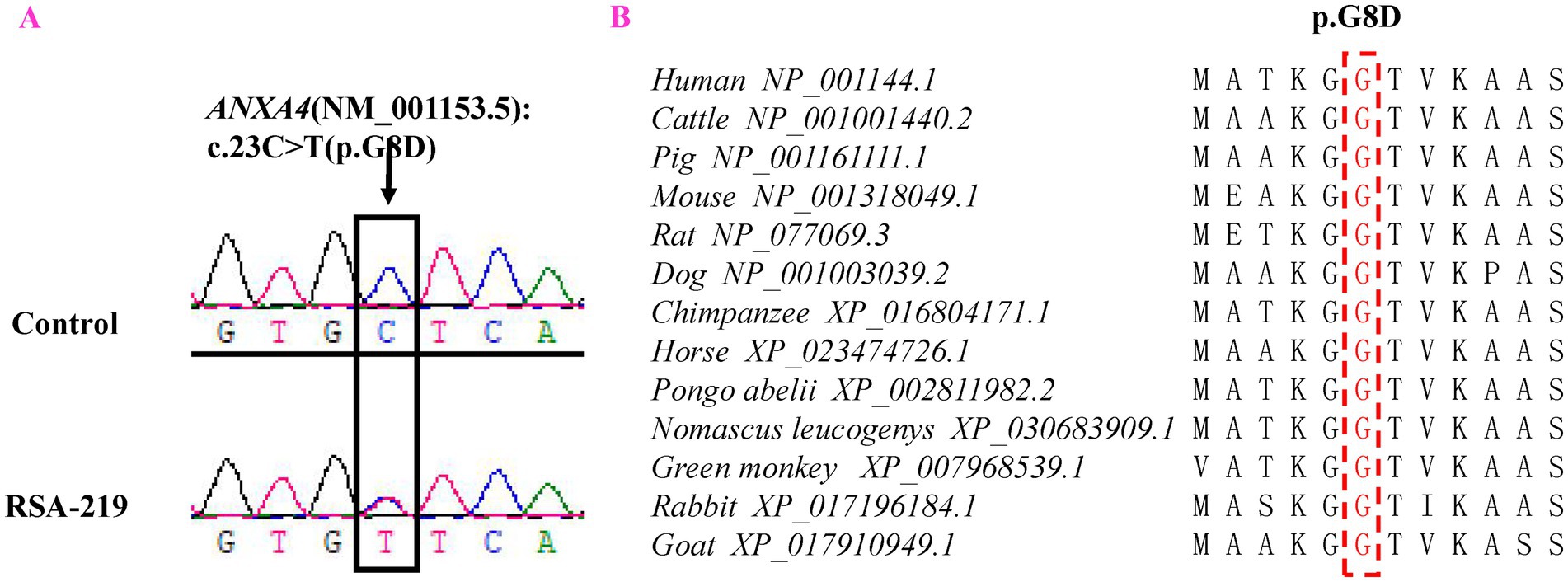
Figure 1. Representative sequencing electropherogram of the ANXA4 variant (p.G8D). The arrow indicates the location of the mutation, and patient “RSA-219” harbored the ANXA4 p.G8D variant (A). Evolutionary conservation analysis of ANXA4 p.G8D in vertebrate species (B).
3.2 Evolutionary conservation analysis and protein structural modeling
Evolutionary conservation analysis revealed that the amino acid residue affected by the p.G8D mutation was highly conserved among 13 vertebrate species (Figure 1B). Furthermore, we analyzed the changes in the 3D structure of ANXA4 between the wild-type and mutant proteins using SWISS-MODEL. Protein structural modeling revealed that the p.G8D ANXA4 mutation caused structural changes in the ANXA4 protein (Figure 2).
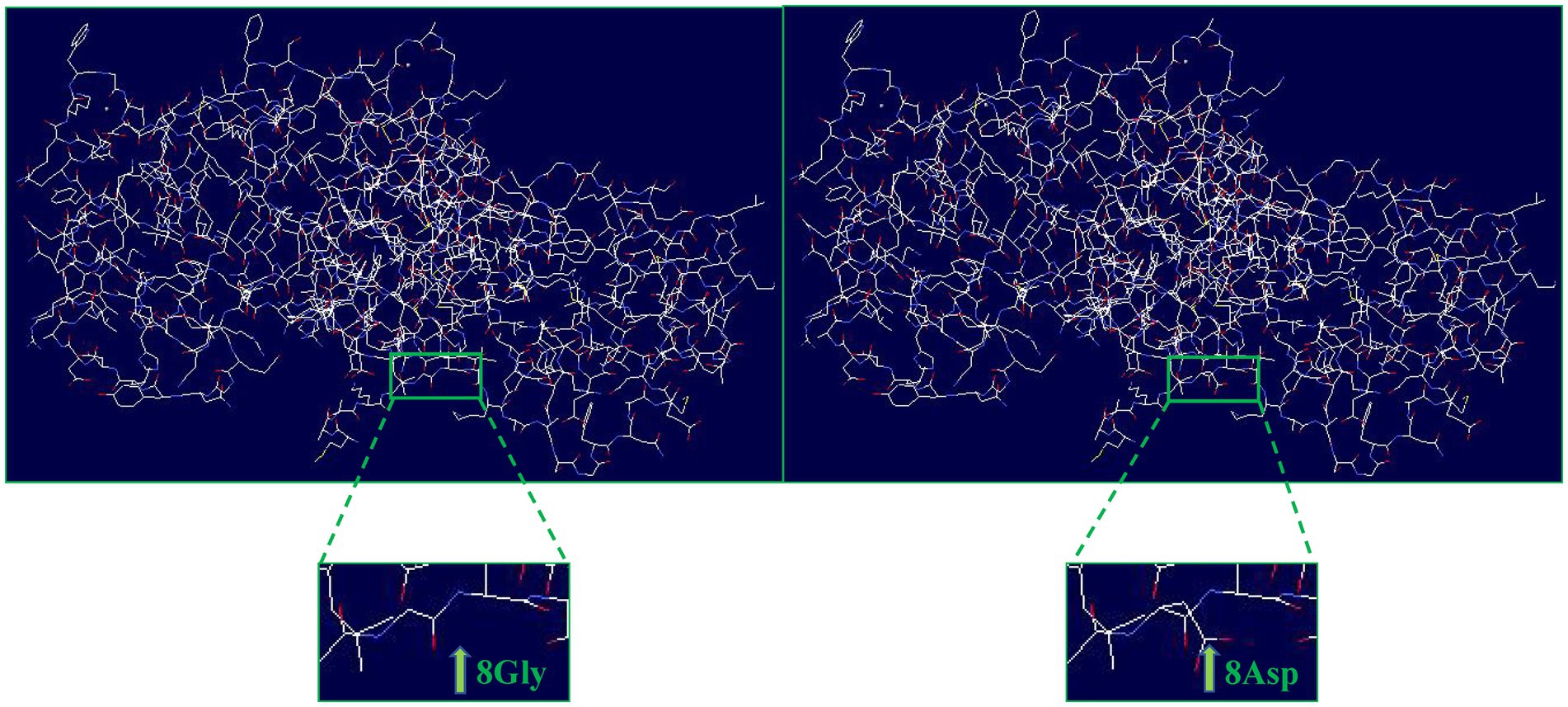
Figure 2. Structural differences between wild-type ANXA4 (p.G8) and mutated ANXA4 (p.D8) proteins. The protein structures of the wild-type and mutated ANXA4 proteins were modeled based on the crystal model of the human ANXA4 protein.
3.3 A novel mutation in ANXA4 (p.G8D) inhibits the migration, invasion and adhesion of THESCs
Annexins have been suggested to promote cellular migration, invasion and adhesion, and adhesion is an essential step required for blastocysts to adhere to the endometrium (31). Next, we examined whether the p.G8D mutation in ANXA4 affects protein function.
THESCs were cultured and transfected with ANXA4 wild-type (WT) or mutant-type (MT) plasmids, and pcDNA3.1 was used as a control plasmid (Con). Twenty-four hours after transfection, G418 was used to screen stably transfected cells, and one individual clone was separated by dilution. The expression of ANXA4 in stably transfected cells was confirmed by RT–qPCR and WB (Figures 3A–C).
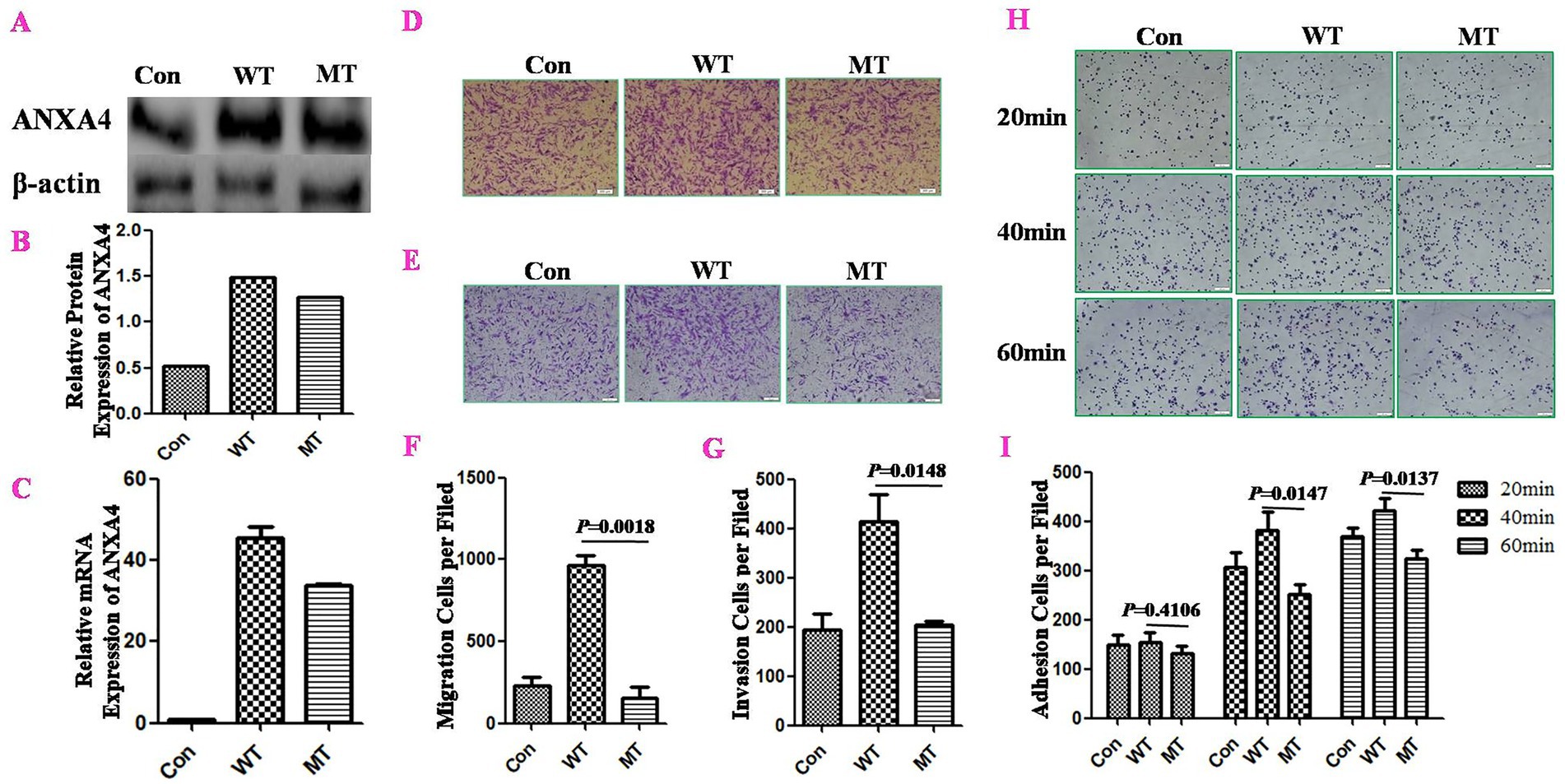
Figure 3. The ANXA4 p.G8D mutation inhibited cell migration, invasion and adhesion in stably transfected THESCs. THESCs were cultured and transfected with ANXA4 wild-type (WT) or mutant-type (MT) plasmids, while pcDNA3.1 was used as a control plasmid (Con). Twenty-four hours after transfection, G418 was used to screen stably transfected cells, and one individual clone was separated by dilution. ANXA4 expression in stably transfected cells was confirmed by WB (A,B) and RT–qPCR (C). The effects of ANXA4 mutation on cell migration (D,F) and invasion (E,G) were determined by Transwell assays, and cell adhesion (H,I) was determined by Matrigel adhesion assays. These assays revealed that ANXA4 p.G8D mutation inhibited cell migration, invasion and adhesion in stably transfected cells compared with ANXA4 WT. Scale bars, 100 μm. The data are shown as the means ± SDs. p < 0.05 was considered to indicate a significant difference.
Transwell assays were used to detect the effects of ANXA4 WT/MT on migration and invasion. The results showed that MT ANXA4 could inhibit both cell migration (p = 0.0018) (Figures 3D,F) and invasion (p = 0.0148) (Figures 3E,G) of stable transfected cells, compared to WT ANXA4.
We examined the effect of the ANXA4 mutation on cell adhesion with a Matrigel adhesion assay. Compared with WT ANXA4, MT ANXA4 inhibited cell adhesion, especially at 40 min (p = 0.0147) and 60 min (p = 0.0137) (Figures 3H,I).
These results indicated that the novel ANXA4 mutation inhibited cell migration, invasion and adhesion.
4 Discussion
The main cause of abortion is implantation failure. Embryo implantation is a dynamic development process that requires a series of interactions between the blastocyst and the endometrium. The three prerequisites for successful implantation are an embryo with implantation ability, a receptive endometrium and simultaneous development of the embryo-endometrium. Many existing animal studies have confirmed that the uterus can keep the embryo in a viable but dormant state until the uterus is in an acceptable state for implantation, indicating that the proper regulation of the uterus by ovarian hormones mainly determines the success of implantation (32). Decidualization of the endometrium leads to corresponding vascular changes preparing for the invasion of human endometrial trophoblasts and the formation of a functional blood placenta; in other words, decidualization contributes to the functional transition of the endometrium from a nonreceptive state to a receptive state (33). An increasing amount of experimental and clinical data indicates that impaired or disrupted decidualization plays a role in the development of an unsuitable maternal–fetal interface. This situation has important clinical implications, including repeated implantation failures and recurrent pregnancy losses in the early stages of pregnancy, as well as several serious complications during late gestation (34). Studies have shown that in the presence of trophoblasts, the motility and invasive ability of decidualized endometrial stromal cells are enhanced (35), and trophoblast cell-derived CXCL12 has been shown to promote the expression and invasiveness of CXCR4 (the receptor of CXCL12) in decidual stromal cells isolated from early pregnancy (36). Many studies have shown that the coordinated migration and invasion of decidualized endometrial stromal cells in response to signals from embryo and trophoblast cells is the key to successful implantation. The ability of mature endometrial stromal cells to migrate and invade is increasingly recognized as the basis of intense tissue remodeling associated with endometrial regeneration, decidualization, embryo implantation, and trophoblast invasion (37, 38).
The annexin family is a class of proteins that are widely present in cell membranes and intracellular organs and are known mainly for their ability to bind phospholipids and their dependence on calcium ions. Given that the annexin family is highly expressed at the maternal–fetal interface and that both the endometrium and blastocyst undergo swift growth and differentiation throughout pregnancy, annexin is believed to perform various functions in both the placental tissues originating from the fetus and the decidual cells derived from the mother (39). Defective expression of endometrial ANXA2 may impair decidualization of endometrial stromal cells in vitro and in vivo, leading to the development of preeclampsia (40). Previous studies have demonstrated that the binding of antibodies and the apoptosis of syncytiotrophoblasts, can hinder the secretion of trophoblast gonadotropin. These factors may be crucial mechanisms through which anti-annexin V antibodies influence embryo implantation upon binding and the results of pregnancy (41). Annexin A4 (ANXA4) is a member of the annexin family that belongs to the multigene family of calcium ion (Ca2+) and phospholipid-binding proteins. The gene encoding human ANXA4 is located on chromosome 2q13.3. The C-terminal conserved domain contains four annexin repeats, each of which has a Ca2+-binding site and five α-helical structures, which can bind phospholipids in a calcium-dependent manner. The N-terminal domain is 12 amino acids long and determines the function of ANXA4 in vivo. ANXA4 can regulate membrane permeability and membrane transport, participate in cell growth and apoptosis, increase tumor invasion and promote antitumor drug resistance (24). An increasing number of studies have shown that ANXA4 is highly expressed in various clinical tumors and is an indicator of tumor development, invasion, chemoresistance, and poor outcomes in cancer patients (42–45). For example, ANXA4 is upregulated and translocated to the nucleus in ovarian clear cell carcinoma and colorectal cancer (44, 45), and the knockdown of ANXA4 weakens the migration and invasion of ovarian cancer and breast cancer cells (43, 46). In addition, several studies have shown that ANXA4 expression is significantly increased in the oviductal fluid of pregnant mares and bovines, and in the uterine endometrium during early pregnancy in pigs, ANXA4 has been identified as an embryo-interacting protein originating from oviductal fluid (47–49). Additionally, the ANXA4 transcript in the human endometrium is significantly upregulated during the menstrual secretory phase compared with the proliferative phase (50). Previous studies reported that elevated ANXA4 mRNA levels are correlated with an increasing level of progesterone (51), and might play a crucial role in the receptive process (52). Proportionately, proteomic PLCD4 (p.L696P) analyses of human placental tissues have shown that ANXA4 expression is downregulated in preeclampsia (PE) placentas and PE placenta-derived extravillous cytotrophoblasts compared with the expression in normal placentas (53). Additionally, ANXA4 overexpression alleviated rat PE progression, accompanied by increases in the expression of PI3K, p-Akt, and p-eNOS in rat placentas, which indicated that ANXA4 may promote trophoblast invasion through the PI3K/Akt/eNOS pathway (53). Furthermore, the ANXA4 gene may play an important role in pregnancy, but there are currently no reports on whether it is involved in the occurrence of miscarriage.
RSA is a multifactorial disease, the exact causes of which are still unknown. Environmental, maternal, and genetic factors have been shown to contribute to this condition (54). WES is widely used to detect genetic variations associated with human diseases, including RSA. In recent years, using WES, studies in families with inherited recurrent pregnancy loss (RPL) confirmed the associations of a number of gene variations with RPL, including IFT122 (p.V553G, p.S373F, p.W7C, p.G546R), ASIC5 (p.R227I), DYNC2H1 (p.Y2016C, p.D2184V), ALOX15 (p.Y139C, p.T560M), FKBP4 (p.A16E, p.N125S, p.Q381L, p.R399Q), PLCD4 (p.L696P) and OSBPL5 (p.G385R) (55–59). The compromised expression of FKBP4, resulting from the increased expression of miR-29c, leads to impaired progesterone signaling and defects in decidualization (60). This may contribute to the development of endometriosis and infertility. In addition, studies in unrelated RSA patients and controls revealed potentially harmful gene variants, including APP (p.K510N), FN1 (p.M1874T), KDR (p.D814N), POLR2B (p.G136C), ITGB1 (p.Y219H), PLK1 (p.A404S), COL4A2 (p.H1603P), LAMA4 (p.D1053G), FOXA2 (p.Y420X), FGA (p.A762V), F13A1 (p.Q401X, p.R612C), KHDC3L (p.146_156del), ANXA5 (p.G317R), DNMT1 (p.G876R), THBS1 (p.N700S), and MSH2 (p.L390F) (11, 61). Earlier research has offered significant understanding regarding the function of human KHDC3L in the modulation of homologous recombination (HR) repair, the activation of PARP1, and the maintenance of genome stability, while also recognizing human KHDC3L as an emerging risk gene associated with RPL (62). Previous studies have demonstrated that reduced expression of ANXA5, associated with the M2 haplotype, increases the risk of vascular thrombosis during pregnancy, thereby increasing the occurrence of RPL, primary maternal peripheral complications (PMPC), and recurrent implantation failure (RIF) (63). In the present study, a total of 325 patients with RSA and 941 local women without RSA as controls were included, and we identified specific gene variants that are pathogenic or possibly pathogenic through WES analysis. Among those variants, a novel ANXA4 mutation (p.G8D) in 1 of the 325 patients with RSA (RSA-219) was detected. This mutation was not detected in either the 941 controls or public databases (dbSNP, 1,000 Genomes Project, ExAC and BGI in-house databases). The SIFT program prediction and structural modeling analysis show that this mutation is harmful. Furthermore, functional analyses revealed that this mutation could inhibit cell migration, invasion and adhesion. Moreover, despite appropriate fertility preservation treatment, the patient experienced another recurrent miscarriage in 2020 during the follow-up period. These results indicate that the ANXA4 mutation p.G8D plays an important role in the pathogenesis of RSA. To the best of our knowledge, this is the first report revealing a novel ANXA4 mutation that may be associated with RSA. Insights from these studies can help us better identify recurrent miscarriages. By discovering new mutations in the ANXA4 gene that are associated with recurrent miscarriage, geneticists can work with clinicians to identify patients with deleterious variants in orthologous genes, thereby playing an active role in personalized medicine. However, the underlying mechanisms by which this mutation affects RSA need further investigation.
5 Conclusion
In conclusion, for the first time, we detected a novel ANXA4 mutation (p.G8D) in 325 samples from women with RSA using WES analysis, this mutation was not detected in either 941 local control women or public databases. Furthermore, functional analyses revealed that this mutation could inhibit cell migration, invasion and adhesion. Taken together, our findings suggest that the ANXA4 mutation p.G8D plays an important role in the pathogenesis of RSA and may contribute to its genetic diagnosis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The studies involving humans were approved by the Ethics Committee of Jiangxi Maternal and Child Health Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
QY: Data curation, Writing – original draft, Validation. F-YL: Data curation, Validation, Writing – review & editing. X-JX: Data curation, Investigation, Resources, Writing – original draft. X-YC: Data curation, Investigation, Writing – original draft. LZ: Investigation, Writing – original draft. H-MW: Validation, Writing – original draft. D-DL: Investigation, Writing – original draft. C-NX: Validation, Writing – original draft. TH: Investigation, Writing – original draft. YC: Data curation, Methodology, Writing – review & editing, Writing – original draft. YZ: Data curation, Methodology, Resources, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Key Research and Development Program of Jiangxi Province (20203BBGL73135) and the Natural Science Foundation of Jiangxi Province (20181BBG70018).
Acknowledgments
We thank all participants involved in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Practice Committee of the American Society for Reproductive Medicine . Electronic address aao. Definitions of infertility and recurrent pregnancy loss: a committee opinion. Fertil Steril. (2020) 113:533–5. doi: 10.1016/j.fertnstert.2019.11.025
2. Zhang, LM, Yang, YN, Zhang, RX, Luo, L, Tan, JF, Zhou, L, et al. Comparison of the etiological constitution of two and three or more recurrent miscarriage. Zhonghua Fu Chan Ke Za Zhi. (2018) 53:855–9. doi: 10.3760/cma.j.issn.0529-567x.2018.12.010
3. Youssef, A, Vermeulen, N, Lashley, E, Goddijn, M, and van der Hoorn, MLP. Comparison and appraisal of (inter) national recurrent pregnancy loss guidelines. Reprod Biomed Online. (2019) 39:497–503. doi: 10.1016/j.rbmo.2019.04.008
4. Practice Committee of the American Society for Reproductive Medicine . Evaluation and treatment of recurrent pregnancy loss: a committee opinion. Fertil Steril. (2012) 98:1103–11. doi: 10.1016/j.fertnstert.2012.06.048
5. Wu, M, Zhu, Y, Zhao, J, Ai, H, Gong, Q, Zhang, J, et al. Soluble costimulatory molecule sTim3 regulates the differentiation of Th1 and Th2 in patients with unexplained recurrent spontaneous abortion. Int J Clin Exp Med. (2015) 8:8812–9.
6. Popescu, F, Jaslow, CR, and Kutteh, WH. Recurrent pregnancy loss evaluation combined with 24-chromosome microarray of miscarriage tissue provides a probable or definite cause of pregnancy loss in over 90% of patients. Hum Reprod. (2018) 33:579–87. doi: 10.1093/humrep/dey021
7. Rodrigues, VO, Soligo, A, and Pannain, GD. Antiphospholipid antibody syndrome and infertility. Rev Bras Ginecol Obstet. (2019) 41:621–7. doi: 10.1055/s-0039-1697982
8. Ticconi, C, Pietropolli, A, Di Simone, N, Piccione, E, and Fazleabas, A. Endometrial immune dysfunction in recurrent pregnancy loss. Int J Mol Sci. (2019) 20:5332. doi: 10.3390/ijms20215332
9. di Simone, N, Castellani, R, Raschi, E, Borghi, MO, Meroni, PL, and Caruso, A. Anti-beta-2 glycoprotein I antibodies affect Bcl-2 and Bax trophoblast expression without evidence of apoptosis. Ann N Y Acad Sci. (2006) 1069:364–76. doi: 10.1196/annals.1351.034
10. D'Ippolito, S, Ticconi, C, Tersigni, C, Garofalo, S, Martino, C, Lanzone, A, et al. The pathogenic role of autoantibodies in recurrent pregnancy loss. Am J Reprod Immunol. (2020) 83:e13200. doi: 10.1111/aji.13200
11. Mou, JT, Huang, SX, Yu, LL, Xu, J, Deng, QL, Xie, YS, et al. Identification of genetic polymorphisms in unexplained recurrent spontaneous abortion based on whole exome sequencing. Ann Transl Med. (2022) 10:603. doi: 10.21037/atm-22-2179
12. Claussnitzer, M, Cho, JH, Collins, R, Cox, NJ, Dermitzakis, ET, Hurles, ME, et al. A brief history of human disease genetics. Nature. (2020) 577:179–89. doi: 10.1038/s41586-019-1879-7
13. Erdenee, S, Akhatayeva, Z, Pan, C, Cai, Y, Xu, H, Chen, H, et al. An insertion/deletion within the CREB1 gene identified using the RNA-sequencing is associated with sheep body morphometric traits. Gene. (2021) 775:145444. doi: 10.1016/j.gene.2021.145444
14. Savoldi, IR, Ibelli, AMG, Cantao, ME, Peixoto, JO, Pires, MP, Mores, MAZ, et al. A joint analysis using exome and transcriptome data identifiescandidate polymorphisms and genes involved with umbilical hernia in pigs. BMC Genomics. (2021) 22:818. doi: 10.1186/s12864-021-08138-4
15. Li, C, Ou, R, Hou, Y, Chen, Y, Wei, Q, Zhang, L, et al. Rare variant analysis of PTRHD1 in Parkinson's disease in the Chinese population. J Parkinsons Dis. (2022) 12:1917–20. doi: 10.3233/JPD-223337
16. Kolte, AM, Nielsen, HS, Moltke, I, Degn, B, Pedersen, B, Sunde, L, et al. A genome-wide scan in affected sibling pairs with idiopathic recurrent miscarriage suggests genetic linkage. Mol Hum Reprod. (2011) 17:379–85. doi: 10.1093/molehr/gar003
17. Karim, S, Jamal, HS, Rouzi, A, Ardawi, MSM, Schulten, HJ, Mirza, Z, et al. Genomic answers for recurrent spontaneous abortion in Saudi Arabia: an array comparative genomic hybridization approach. Reprod Biol. (2017) 17:133–43. doi: 10.1016/j.repbio.2017.03.003
18. Pereza, N, Ostojic, S, Kapovic, M, and Peterlin, B. Systematic review and meta-analysis of genetic association studies in idiopathic recurrent spontaneous abortion. Fertil Steril. (2017) 107:150–159.e2. doi: 10.1016/j.fertnstert.2016.10.007
19. Loizidou, EM, Kucherenko, A, Tatarskyy, P, Chernushyn, S, Livshyts, G, Gulkovskyi, R, et al. Risk of recurrent pregnancy loss in the Ukrainian population using a combined effect of genetic variants: a case-control study. Genes (Basel). (2021) 12:64. doi: 10.3390/genes12010064
20. Povysil, G, Petrovski, S, Hostyk, J, Aggarwal, V, Allen, AS, and Goldstein, DB. Rare-variant collapsing analyses for complex traits: guidelines and applications. Nat Rev Genet. (2019) 20:747–59. doi: 10.1038/s41576-019-0177-4
21. Gourhant, L, Bocher, O, De Saint, ML, Ludwig, TE, Boland, A, Deleuze, JF, et al. Whole exome sequencing, a hypothesis-free approach to investigate recurrent early miscarriage. Reprod Biomed Online. (2021) 42:789–98. doi: 10.1016/j.rbmo.2021.01.008
22. Smail, C, Ferraro, NM, Hui, Q, Durrant, MG, Aguirre, M, Tanigawa, Y, et al. Integration of rare expression outlier-associated variants improves polygenic risk prediction. Am J Hum Genet. (2022) 109:1055–64. doi: 10.1016/j.ajhg.2022.04.015
23. Arii, Y, Butsusihta, K, and Fukuoka, S. Role of calcium-binding sites in calcium-dependent membrane association of annexin A4. Biosci Biotechnol Biochem. (2015) 79:978–85. doi: 10.1080/09168451.2014.1003131
24. Yao, H, Sun, C, Hu, Z, and Wang, W. The role of annexin A4 in cancer. Front Biosci (Landmark Ed). (2016) 21:949–57. doi: 10.2741/4432
25. Ponnampalam, AP, and Rogers, PA. Cyclic changes and hormonal regulation of annexin IV mRNA and protein in human endometrium. Mol Hum Reprod. (2006) 12:661–9. doi: 10.1093/molehr/gal075
26. Li, H, and Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. (2009) 25:1754–60. doi: 10.1093/bioinformatics/btp324
27. Van der Auwera, GA, Carneiro, MO, Hartl, C, Poplin, R, Del Angel, G, Levy-Moonshine, A, et al. From fast Q data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. (2013) 43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43
28. Yang, H, and Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. (2015) 10:1556–66. doi: 10.1038/nprot.2015.105
29. Sherry, ST, Ward, MH, Kholodov, M, Baker, J, Phan, L, Smigielski, EM, et al. Db SNP: the NCBI database of genetic variation. Nucleic Acids Res. (2001) 29:308–11. doi: 10.1093/nar/29.1.308
30. Lek, M, Karczewski, KJ, Minikel, EV, Samocha, KE, Banks, E, Fennell, T, et al. Analysis of protein-coding genetic variation in 60, 706 humans. Nature. (2016) 536:285–91. doi: 10.1038/nature19057
31. Greening, DW, Nguyen, HP, Evans, J, Simpson, RJ, and Salamonsen, LA. Modulating the endometrial epithelial proteome and secretome in preparation for pregnancy: the role of ovarian steroid and pregnancy hormones. J Proteome. (2016) 144:99–112. doi: 10.1016/j.jprot.2016.05.026
32. Zhang, S, Lin, H, Kong, S, Wang, S, Wang, H, Wang, H, et al. Physiological and molecular determinants of embryo implantation. Mol Asp Med. (2013) 34:939–80. doi: 10.1016/j.mam.2012.12.011
33. Brosens, JJ, Pijnenborg, R, and Brosens, IA. The myometrial junctional zone spiral arteries in normal and abnormal pregnancies: a review of the literature. Am J Obstet Gynecol. (2002) 187:1416–23. doi: 10.1067/mob.2002.127305
34. Ticconi, C, Di Simone, N, Campagnolo, L, and Fazleabas, A. Clinical consequences of defective decidualization. Tissue Cell. (2021) 72:101586. doi: 10.1016/j.tice.2021.101586
35. Gellersen, B, Reimann, K, Samalecos, A, Aupers, S, and Bamberger, AM. Invasiveness of human endometrial stromal cells is promoted by decidualization and by trophoblast-derived signals. Hum Reprod. (2010) 25:862–73. doi: 10.1093/humrep/dep468
36. Ren, L, Liu, YQ, Zhou, WH, and Zhang, YZ. Trophoblast-derived chemokine CXCL12 promotes CXCR4 expression and invasion of human first-trimester decidual stromal cells. Hum Reprod. (2012) 27:366–74. doi: 10.1093/humrep/der395
37. Weimar, CH, Macklon, NS, Post Uiterweer, ED, Brosens, JJ, and Gellersen, B. The motile and invasive capacity of human endometrial stromal cells: implications for normal and impaired reproductive function. Hum Reprod Update. (2013) 19:542–57. doi: 10.1093/humupd/dmt025
38. Murata, H, Tanaka, S, and Okada, H. The regulators of human endometrial stromal cell Decidualization. Biomol Ther. (2022) 12:1275. doi: 10.3390/biom12091275
39. Hu, J, Chen, L, Ruan, J, and Chen, X. The role of the annexin a protein family at the maternal-fetal interface. Front Endocrinol (Lausanne). (2024) 15:1314214. doi: 10.3389/fendo.2024.1314214
40. Garrido-Gomez, T, Quiñonero, A, Dominguez, F, Rubert, L, Perales, A, Hajjar, KA, et al. Preeclampsia: a defect in decidualization is associated with deficiency of Annexin A2. Am J Obstet Gynecol. (2020) 222:376.e1–e17. doi: 10.1016/j.ajog.2019.11.1250
41. Di Simone, N, Castellani, R, Caliandro, D, and Caruso, A. Monoclonal anti-annexin V antibody inhibits trophoblast gonadotropin secretion and induces syncytiotrophoblast apoptosis. Biol Reprod. (2001) 65:1766–70. doi: 10.1095/biolreprod65.6.1766
42. Konopka-Postupolska, D, Clark, G, and Hofmann, A. Structure, function and membrane interactions of plant annexins: an update. Plant Sci. (2011) 181:230–41. doi: 10.1016/j.plantsci.2011.05.013
43. Mogami, T, Yokota, N, Asai-Sato, M, Yamada, R, Koizume, S, Sakuma, Y, et al. Annexin A4 is involved in proliferation, chemo-resistance and migration and invasion in ovarian clear cell adenocarcinoma cells. PLoS One. (2013) 8:e80359. doi: 10.1371/journal.pone.0080359
44. Liu, J, Wang, H, Zheng, M, Deng, L, Zhang, X, and Lin, B. P 53 and ANXA4/NF-kappaB p50 complexes regulate cell proliferation, apoptosis and tumor progression in ovarian clear cell carcinoma. Int J Mol Med. (2020) 46:2102–14. doi: 10.3892/ijmm.2020.4757
45. Peng, Y, Zhang, Z, Zhang, A, Liu, C, Sun, Y, Peng, Z, et al. Membrane-cytoplasm translocation of annexin A4 is involved in the metastasis of colorectal carcinoma. Aging (Albany NY). (2021) 13:10312–25. doi: 10.18632/aging.202793
46. Li, L, Zhang, R, Liu, Y, and Zhang, G. ANXA4 activates JAK-STAT3 signaling by interacting with ANXA1 in basal-like breast Cancer. DNA Cell Biol. (2020) 39:1649–56. doi: 10.1089/dna.2020.5570
47. Smits, K, Willems, S, Van Steendam, K, Van De Velde, M, De Lange, V, Ververs, C, et al. Proteins involved in embryo-maternal interaction around the signalling of maternal recognition of pregnancy in the horse. Sci Rep. (2018) 8:5249. doi: 10.1038/s41598-018-23537-6
48. Banliat, C, Tsikis, G, Labas, V, Teixeira-Gomes, AP, Com, E, Lavigne, R, et al. Identification of 56 proteins involved in embryo-maternal interactions in the bovine oviduct. Int J Mol Sci. (2020) 21:466. doi: 10.3390/ijms21020466
49. Pierzchala, D, Liput, K, Korwin-Kossakowska, A, Ogluszka, M, Polawska, E, Nawrocka, A, et al. Molecular characterisation of uterine endometrial proteins during early stages of pregnancy in pigs by MALDI TOF/TOF. Int J Mol Sci. (2021) 22:6720. doi: 10.3390/ijms22136720
50. Ponnampalam, AP, Weston, GC, Trajstman, AC, Susil, B, and Rogers, PA. Molecular classification of human endometrial cycle stages by transcriptional profiling. Mol Hum Reprod. (2004) 10:879–93. doi: 10.1093/molehr/gah121
51. Talbi, S, Hamilton, AE, Vo, KC, Tulac, S, Overgaard, MT, Dosiou, C, et al. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology. (2006) 147:1097–121. doi: 10.1210/en.2005-1076
52. Mirkin, S, Arslan, M, Churikov, D, Corica, A, Diaz, JI, Williams, S, et al. In search of candidate genes critically expressed in the human endometrium during the window of implantation. Hum Reprod. (2005) 20:2104–17. doi: 10.1093/humrep/dei051
53. Xu, Y, Sui, L, Qiu, B, Yin, X, Liu, J, and Zhang, X. ANXA4 promotes trophoblast invasion via the PI3K/Akt/eNOS pathway in preeclampsia. Am J Physiol Cell Physiol. (2019) 316:C481–91. doi: 10.1152/ajpcell.00404.2018
54. Guo, W, Zhu, X, Yan, L, and Qiao, J. The present and future of whole-exome sequencing in studying and treating human reproductive disorders. J Genet Genomics. (2018) 45:517–25. doi: 10.1016/j.jgg.2018.08.004
55. Tsurusaki, Y, Yonezawa, R, Furuya, M, Nishimura, G, Pooh, RK, Nakashima, M, et al. Whole exome sequencing revealed biallelic IFT122 mutations in a family with CED1 and recurrent pregnancy loss. Clin Genet. (2014) 85:592–4. doi: 10.1111/cge.12215
56. Filges, I, Manokhina, I, Penaherrera, MS, McFadden, DE, Louie, K, Nosova, E, et al. Recurrent triploidy due to a failure to complete maternal meiosis II: whole-exome sequencing reveals candidate variants. Mol Hum Reprod. (2015) 21:339–46. doi: 10.1093/molehr/gau112
57. Qiao, Y, Wen, J, Tang, F, Martell, S, Shomer, N, Leung, PC, et al. Whole exome sequencing in recurrent early pregnancy loss. Mol Hum Reprod. (2016) 22:364–72. doi: 10.1093/molehr/gaw008
58. Demetriou, C, Chanudet, E, GOSgeneJoseph, A, Topf, M, Thomas, AC, et al. Exome sequencing identifies variants in FKBP4 that are associated with recurrent fetal loss in humans. Hum Mol Genet. (2019) 28:3466–74. doi: 10.1093/hmg/ddz203
59. Al Qahtani, NH, AbdulAzeez, S, Almandil, NB, Fahad Alhur, N, Alsuwat, HS, Al Taifi, HA, et al. Whole-genome sequencing reveals Exonic variation of ASIC5 gene results in recurrent pregnancy loss. Front Med (Lausanne). (2021) 8:699672. doi: 10.3389/fmed.2021.699672
60. Joshi, NR, Miyadahira, EH, Afshar, Y, Jeong, JW, Young, SL, Lessey, BA, et al. Progesterone resistance in endometriosis is modulated by the altered expression of Micro RNA-29c and FKBP4. J Clin Endocrinol Metab. (2017) 102:141–9. doi: 10.1210/jc.2016-2076
61. Xiang, H, Wang, C, Pan, H, Hu, Q, Wang, R, Xu, Z, et al. Exome-sequencing identifies novel genes associated with recurrent pregnancy loss in a Chinese cohort. Front Genet. (2021) 12:746082. doi: 10.3389/fgene.2021.746082
62. Zhang, W, Chen, Z, Zhang, D, Zhao, B, Liu, L, Xie, Z, et al. KHDC3L mutation causes recurrent pregnancy loss by inducing genomic instability of human early embryonic cells. PLoS Biol. (2019) 17:e3000468. doi: 10.1371/journal.pbio.3000468
Keywords: ANXA4 mutation, recurrent spontaneous abortion, cell invasion, migration, adhesion
Citation: Ye Q, Liu F-Y, Xia X-J, Chen X-Y, Zou L, Wu H-M, Li D-D, Xia C-N, Huang T, Cui Y and Zou Y (2024) Whole exome sequencing identifies a novel mutation in Annexin A4 that is associated with recurrent spontaneous abortion. Front. Med. 11:1462649. doi: 10.3389/fmed.2024.1462649
Edited by:
J. Francis Borgio, Imam Abdulrahman Bin Faisal University, Saudi ArabiaReviewed by:
Wen-Jing Wang, Beijing Genomics Institute (BGI), ChinaAndrey Glotov, D. O. Ott Research Institute of Obstetrics, Gynecology and Reproductology, Russia
Nicoletta Di Simone, Humanitas University, Italy
Copyright © 2024 Ye, Liu, Xia, Chen, Zou, Wu, Li, Xia, Huang, Cui and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Cui, Y3VpeWluZzM4OEAxMjYuY29t; Yang Zou, em91eWFuZzgxQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Qian Ye
Qian Ye Fa-Ying Liu2,3,4†
Fa-Ying Liu2,3,4† Yang Zou
Yang Zou