- 1Faculty of Computer Science, AI and Digital Science Institute, International Laboratory of Bioinformatics, Higher School of Economics University, Moscow, Russia
- 2Department of Cardiology, Surgut State University, Surgut, Russia
- 3Institute of Chemical Biology and Fundamental Medicine, Novosibirsk, Russia
- 4Federal Research Center Institute of Cytology and Genetics, Novosibirsk, Russia
- 5Department of General Pathology and Pathophysiology, Surgut State University, Surgut, Russia
- 6Ugra Center for Diagnostics and Cardiovascular Surgery, Surgut, Russia
Background: The development of prognostic models for the identification of high-risk myocardial infarction (MI) patients is a crucial step toward personalized medicine. Genetic factors are known to be associated with an increased risk of cardiovascular diseases; however, little is known about whether they can be used to predict major adverse cardiac events (MACEs) for MI patients. This study aimed to build a machine learning (ML) model to predict MACEs in MI patients based on clinical, imaging, laboratory, and genetic features and to assess the influence of genetics on the prognostic power of the model.
Methods: We analyzed the data from 218 MI patients admitted to the emergency department at the Surgut District Center for Diagnostics and Cardiovascular Surgery, Russia. Upon admission, standard clinical measurements and imaging data were collected for each patient. Additionally, patients were genotyped for VEGFR-2 variation rs2305948 (C/C, C/T, T/T genotypes with T being the minor risk allele). The study included a 9-year follow-up period during which major ischemic events were recorded. We trained and evaluated various ML models, including Gradient Boosting, Random Forest, Logistic Regression, and AutoML. For feature importance analysis, we applied the sequential feature selection (SFS) and Shapley’s scheme of additive explanation (SHAP) methods.
Results: The CatBoost algorithm, with features selected using the SFS method, showed the best performance on the test cohort, achieving a ROC AUC of 0.813. Feature importance analysis identified the dose of statins as the most important factor, with the VEGFR-2 genotype among the top 5. The other important features are coronary artery lesions (coronary artery stenoses ≥70%), left ventricular (LV) parameters such as lateral LV wall and LV mass, diabetes, type of revascularization (CABG or PCI), and age. We also showed that contributions are additive and that high risk can be determined by cumulative negative effects from different prognostic factors.
Conclusion: Our ML-based approach demonstrated that the VEGFR-2 genotype is associated with an increased risk of MACEs in MI patients. However, the risk can be significantly reduced by high-dose statins and positive factors such as the absence of coronary artery lesions, absence of diabetes, and younger age.
1 Introduction
Despite significant advances in cardiovascular surgery, interventional procedures, and pharmacotherapy, the in-hospital and long-term rates of major adverse cardiac events (MACEs)—including death, recurrent acute coronary syndrome (ACS), stroke, and myocardial revascularization—remain high worldwide among myocardial infarction (MI) patients (1, 2).
Secondary prevention programs for these individuals are based on identification of high-risk patients using models that focus on clinical features of MI (ST-segment deviation, left ventricular ejection fraction (LVEF), Killip class, and coronary artery lesions) and conventional risk factors (hypertension, diabetes, chronic kidney disease, dyslipidemia, and smoking) that worsen cardiac outcomes (3, 4).
Identifying patients at high risk of MACEs is crucial. Various clinical scores, such as GRACE, ACEF, CADILLAC, TIMI, PROCAM, PREDICT, PURSUIT, DAPT, and PRECISE-DAPT are available, These scores, primarily based on regression models using clinical parameters, estimate cardiovascular (CV) risk over a 1- to 3-year period after an index MI. Nevertheless, modern personalized approaches to the treatment of post-MI individuals already require more precise clinical scoring systems incorporating biomarkers and genetics data (5, 6).
The pathological basis of coronary artery disease (CAD) and acute coronary syndrome (ACS) is atherosclerosis. Factors such as lipid metabolism disorders, vascular endothelial cell damage, inflammation, and immune dysfunction can promote the development and progression of coronary atherosclerosis, potentially leading to CAD and ACS. The vascular endothelial growth factor (VEGF) family is involved in angiogenesis, inflammation, oxidative stress, and lipid metabolism, presenting potential therapeutic and prognostic value for ACS and MI (7, 8).
The prognostic performance of VEGF serum levels following cardiac ischemia shortly after MI was previously investigated in the Coronary Disease Cohort Study. VEGFs primarily bind to three tyrosine kinase receptors (VEGFR-1, VEGFR-2, and VEGFR-3) with different affinities. Single nucleotide polymorphisms (SNPs) from VEGFR-2 are associated with the development of cardiovascular disease (7). Moreover, three SNPs of the VEGFR-2 gene were identified and significantly associated with coronary atherosclerosis: +1192C > T (rs2305948), −604 T > C (rs2071559), and + 1719A > T (rs1870377) (9–13). Consequently, VEGFR-2 gene allelic variants have the potential to be evaluated as prognostic markers in MI (14).
Recently, machine learning (ML) approaches have proven efficient in predicting patient outcomes based on clinical, imaging, and biomarker data. The largest study, based on the clinical data including 23,000 patients in the BleeMACS and RENAMI registries, showed that ML models can predict all-cause death, recurrent MI, and major bleeding after ACS (15) using 25 clinical features. Other approaches, based on considerably smaller datasets, have also proven the efficacy of ML models for cardiovascular outcomes research (16–20). One advantage of ML modeling is its ability to perform feature importance analysis and rank features according to their contribution to model performance. This not only reveals the most important features but also helps to select a minimal set that can reduce overfitting and improve the accuracy of the model. Additionally, an ML model with a small set of features can be easily applied in practice. Thus, ML models have shown that serum creatinine and LVEF alone can predict all-cause death (16). In another study (21), an ML model trained to predict all-cause death reduced the feature space from 430 to 25. Other models provide evidence of the predictive power of ML models and present the most important features. These include models for prognosis of post-thrombotic syndrome (22), prognostication of the time to death of patients in the Coronary Care Unit (23), prediction of mortality and heart failure (HF) hospitalization in patients with preserved LVEF (17), prediction of long-term risk of MI and cardiac death (24), and in-hospital all-cause mortality in HF patients (20). All the aforementioned ML models were trained using clinical features routinely measured at hospital admission. In some studies, these features were complemented by demographic and social characteristics.
All proposed prognostic ML models do not have high-performance metrics with ROC AUC lying in the interval of 70–80%. One way to improve ML model prediction power is to include more information about patient status, with genetic markers potentially serving a key role. Adding genetic markers as features to an ML model does not mean that clinical features are not prognostic or that, in the future, they are supposed to be replaced with genetic markers only. However, genetic markers can significantly improve patient risk assessment when taken together with other clinical characteristics. This kind of model is limited due to the high-cost genetic testing, and most MI patient clinical datasets do not contain genetic information. Here, for the first time, we incorporate genetic information into an ML model to predict MACEs in MI patients.
2 Methods
2.1 Clinical data collection
The prospective observational study consecutively included 218 patients with acute MI who were admitted to the Surgut District Cardiology Clinic of the Center for Diagnostics and Cardiovascular Surgery. This study was approved by the local ethics committee at the clinic. During the initial admission to the emergency department, medical researchers (cardiologists) explained the main points of the study protocol to the patients and obtained their informed consent to participate. Then, the patients were transported to the operating unit to undergo emergent coronary angiography (CAG). Significant lesions (stenoses) in the coronary arteries were considered present when the lumen was narrowed by more than 70%. For the left main coronary artery (LMCA), significant stenosis was considered present when the lumen was narrowed by more than 50%. Based on the CAG results, balloon angioplasty and stenting or coronary artery bypass grafting (CABG) were performed. Standard transthoracic echocardiography was also performed during in-hospital treatment.
All patients received guideline-based therapy, including RAAS-blockers, beta-blockers, statins, and dual antiplatelet therapy. High-dose statin therapy was defined as atorvastatin 40–80 mg and rosuvastatin 20–40 mg daily, whereas low-dose statin therapy was defined as atorvastatin 10–20 mg and rosuvastatin 5–10 mg daily, for at least 12 months post-MI.
The inclusion criteria were as follows: acute myocardial infarction with or without ST-segment elevation and age range of 30–70 years. The exclusion criteria were as follows: contraindications to the use of statins; pregnancy and lactation; advanced tumor disease; HIV; and patient’s refusal to participate in the study.
During the laboratory stage, DNA was isolated from the leukocyte rings in the collected blood samples and then frozen at −80°C for future genetic testing. VEGFR-2 (rs2305948) genotypes (C/C, C/T, T/T) were determined by real-time polymerase chain reaction (PCR) using a Real-Time CFX96 Touch device (Bio-Rad Laboratories, United States). The T-allele of VEGFR-2 (rs2305948) is commonly regarded as a risk allele for unfavorable ischemic post-MI outcomes. Glomerular filtration rate (GFR) was assessed using a CKD-EPI calculator to classify chronic kidney disease. Major ischemic events (cardiovascular death, recurrent ACS, stroke, and myocardial revascularization) were registered based on the results of clinical observation between 9 and years (2015–2024) after the indexed MI. The information was collected from in-hospital medical records of patients who were re-admitted to the clinic after the index MI. Additionally, patients were contacted by phone, and the fact of hospital treatment for ischemic events in other medical organizations was confirmed by requesting corresponding medical records. Cases of death in the post-infarction period were recorded in the health insurance policy database, while cardiovascular (ischemic) causes of death were specified by clinical autopsy services based on the patient’s residence. Basic statistics analysis was performed in the SPSS 23.0 software package.
2.2 Data preprocessing
For logistic regression, all non-categorical features were normalized using Standard Scaler. For tree-based models that do not require normalization, data transformation was not performed. There was no additional preprocessing of categorical features as all categorical features were either binary or ordered.
For data imputation, we used the Multivariate Imputation by Chained Equations (MICE) method (25). A Linear Regression model with default parameters was used as an estimator for MICE with 100 iterations for numerical data. For categorical and binary features, we used the logistic regression model with a ‘liblinear’ solver as the default estimator for MICE with 100 iterations.
The dataset was randomly split in a 75:25 ratio into training (n = 163) and test (n = 55) sets respectively, to develop and validate the models. Data imputation and scaling, hyperparameter tuning, feature selection, and the final model training were performed on the training set. Evaluation of metrics of the developed models was performed on the test set.
2.3 Machine learning models
We built machine learning models based on Gradient Boosting (Catboost and LightGBM), Random Forest, and Logistic Regression algorithms with hyperparameter tuning using 10-fold cross-validation on the training dataset. For the AutoML approach, we used FLAML (26), a Python package for automatic machine learning that automates the process of model selection, hyperparameter tuning, and feature construction.
To optimize model performance, we used Optuna (27), a Python package that automates hyperparameter tuning using Bayesian optimization algorithms. We utilized a tree-structured Parzen Estimator with 10 startup trials as our search algorithm and used 10-fold cross-validation during the optimization process to obtain a robust estimate of the ROC AUC metric. The sequential forward selection (SFS) method was used to determine the optimal number of training features. During the feature selection cycle, we used 5-fold cross-validation and used the mean ROC AUC as the evaluation metric. The model with the best average metric was chosen as the baseline model for the feature selection procedure. We added one feature at a time until we explored all the features and then selected the subset with the best metric as the final feature subset.
Feature importance analysis was performed using Shapley’s scheme of additive explanation (SHAP) (28). SHAP provided visualization of the feature importance for the selected model and individual contributions of features to the predictive power of the model. Additionally, we constructed partial dependence plots (PDP) and individual conditional expectation (ICE) (29) plots.
To evaluate the predictive performance of the developed models, we scored data samples from the test set. Specifically, we generated 1,000 bootstrap samples of the initial test sample size to obtain the distribution of PR AUC and ROC AUC. By averaging the metrics across all iterations, we obtained the average PR AUC and ROC AUC as the final evaluation metrics.
3 Results
3.1 Characteristics of MI patients
The dataset used in this study included 218 patients admitted to the emergency department at the Surgut District Center for Diagnostics and Cardiovascular Surgery, Russia, with MI. For each patient, standard clinical measurements, including laboratory tests and imaging indices were selected for inclusion in the model (Table 1). Additionally, each patient was genotyped for VEGFR-2 variation, rs2305948 which includes the C/C, C/T, and T/T genotypes.
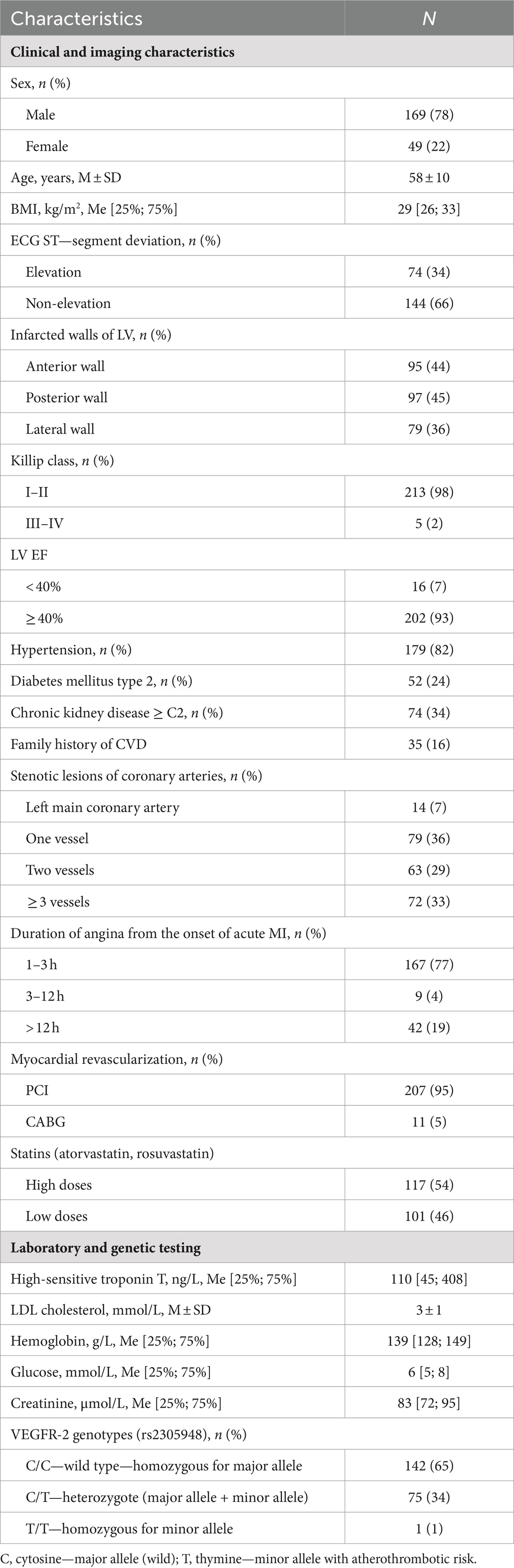
Table 1. Clinical and imaging characteristics, laboratory, and genetic testing of patients with MI (n = 218).
3.2 Machine learning models
The data underwent imputation and preprocessing and were split into training and test sets in a 75–25% ratio for training and validation. In order to choose the best model, we trained and tested the following ML algorithms: Gradient Boosting (CatBoost and LightGBM), Random Forest, Logistic Regression, and AutoML approach (see Methods section for details). For model performance metrics, we took an average ROC AUC calculated on the bootstrap samples from the test set. Model performance comparison is presented in Figure 1A, and the best metric was obtained for the CatBoost model (ROC AUC of 0.787), which was significantly better than the second-best Random Forest (p < 1e−03, t-test) (Figure 1A). The CatBoost ROC curves for training and test sets are given in Supplementary Figure S1. We chose the CatBoost model for feature selection and feature importance analysis.
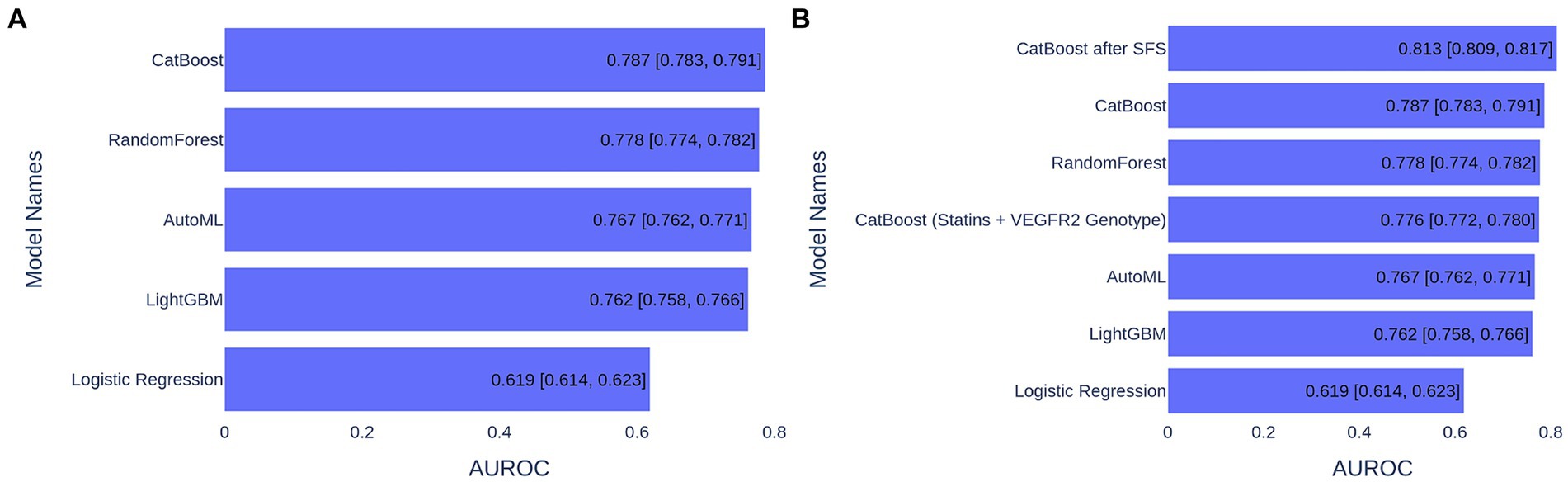
Figure 1. ML model performance comparison based on ROC AUC metrics for 1,000 bootstrapped test sets. (A) ML models trained on 39 features. (B) ML models trained on various number of features.
3.3 Feature selection to find the best model
The initial ML models were trained on 39 features (Table 2). We explored the contribution of each feature to model performance using the forward sequential feature selection (SFS) method (see Figure 2). We chose the SFS method due to its balance between computational efficiency and robustness. SFS directly evaluates the contribution of each feature to model performance using cross-validation at each step, ensuring a resilient and accurate feature selection process. This avoids potential inaccuracies associated with heuristic methods like feature importance in tree-based methods or feature imputation, which can be misleading, especially with high cardinality features. Thus, SFS is a simple and straightforward method for the selection of a minimal set of features that provide maximal precision of an ML model.
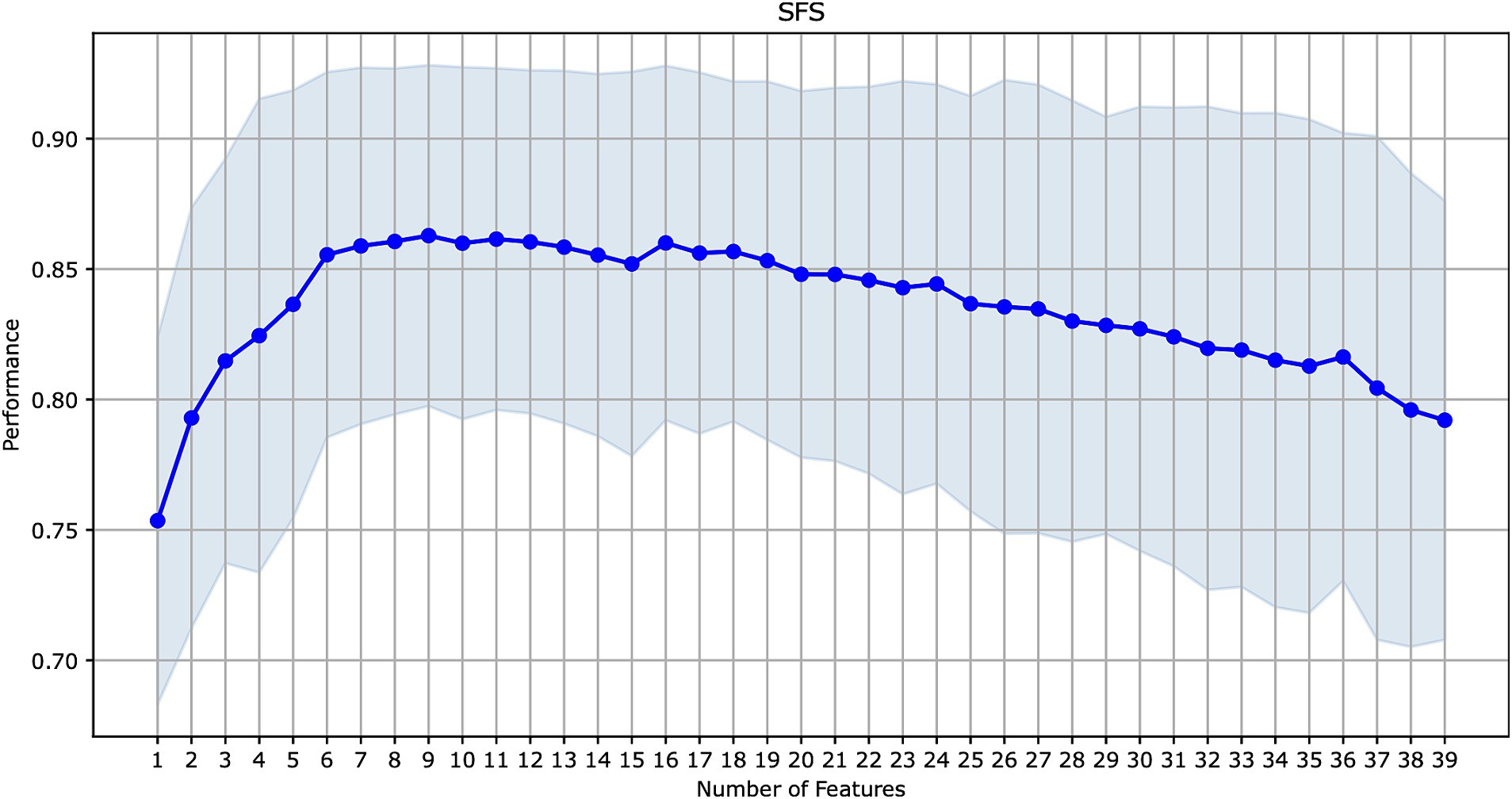
Figure 2. Sequential forward feature selection for the model with the best performance on all features (CatBoost).
Since CatBoost showed the best performance with 39 features, it was used as the baseline model for SFS. The model’s performance increased gradually with the addition of the first nine features, but then began to decline as additional features were included. These first nine features (presented in Figure 3) were selected for retraining the CatBoost model (Supplementary Table S1). The CatBoost model trained on the nine selected features showed statistically significant improvement in the model’s performance compared to the CatBoost model trained on 39 features (ROC AUC of 0.813 vs. 0.787, p = 5e−19, t-test) (Figure 1B and Supplementary Figure S2). Thus, the CatBoost model trained on nine features was chosen for further analysis of MI patients.
3.4 Feature importance analysis
For feature importance analysis, we chose SHAP because it estimates the contribution of the selected features to the model performance, and it considers the cooperative effect of other features. SHAP values can be added to show the joint contribution of several features to a final ML model prediction. This helps to understand the combined effect of multiple features on risk assessment.
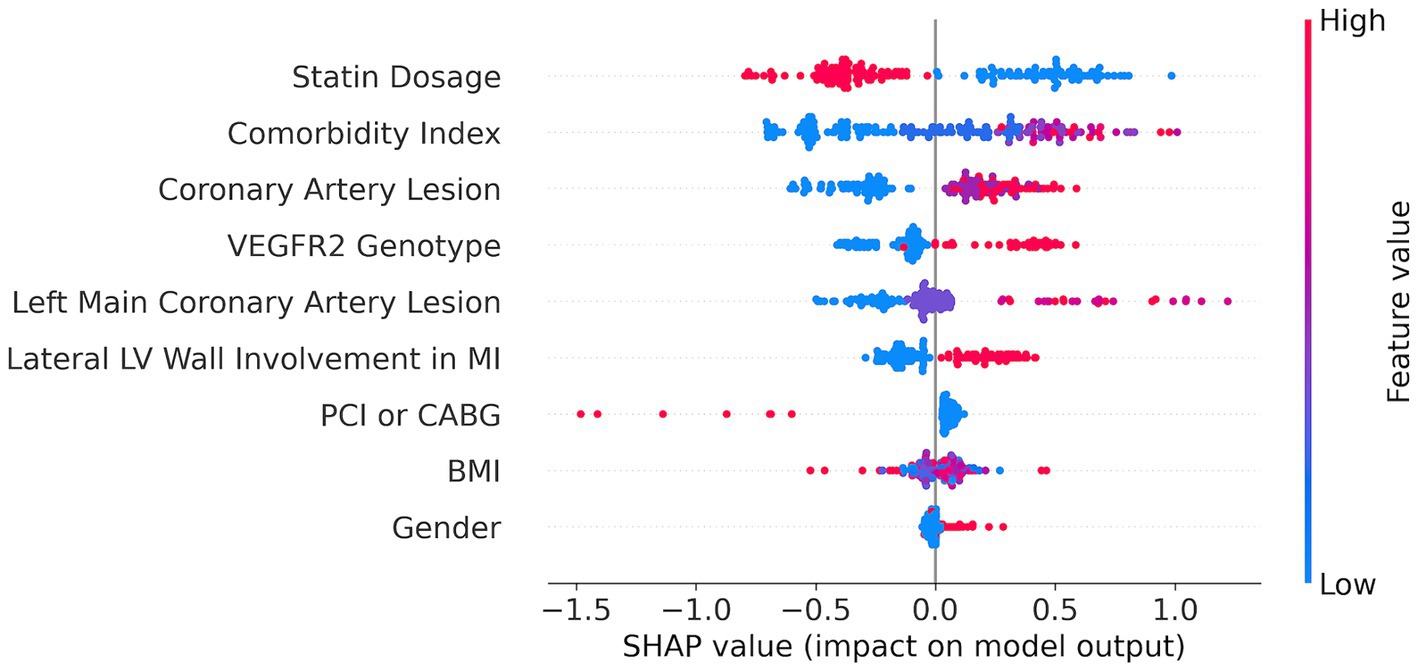
We applied the SHAP method using the best CatBoost model with the nine features selected by the SFS method (Figure 3). The nine features are ranked from the most important (top) to the least important (bottom). A higher SHAP value for a feature indicates a greater risk of MACEs. Feature values are color-coded, with red representing higher values and blue representing lower values. We can see from the SHAP plot that the most important feature is the dose of statins with low values of the dose corresponding to a high risk of MACEs. Interestingly, the VEGFR-2 genotypes appeared to be the fourth top important feature with the risk T-allele corresponding to higher risk of MACEs. The other important features are comorbidity index, coronary artery lesions, LV parameters, such as lateral LV wall, CABG or PCI, body mass index (BMI), and sex (see Figure 3).
Additionally, we constructed partial dependence plots (PDP) and individual conditional expectation (ICE) plots for key features to provide a more intuitive understanding of their impact on predictions (Supplementary Figure S3). PDP shows the average effect of a feature on the predicted outcome, whereas ICE plots illustrate the impact of a feature at the individual observation level, highlighting variability across different data points. The provided PDP and ICE curves offer a detailed visualization of how individual features influence the predicted risk of MACEs in MI patients. PDP graphs depict the marginal effect of each feature, whereas the ICE curves illustrate the variability in predictions across different patients.
For continuous variables, the plots revealed the following trends: an increased BMI and a higher comorbidity index are associated with a greater risk, while a higher statin dosage is linked to a reduced risk. Similarly, greater severity in coronary artery lesions and left main coronary artery lesions leads to an increased risk. The average effect is represented by the orange dashed line, and individual patient trajectories are shown in blue (Supplementary Figure S3).
For categorical features, the bar plots highlight distinct differences in risk levels: male sex, undergoing PCI instead of CABG, lateral LV wall involvement in MI, and the presence of the risk T-allele in the VEGFR2 genotype—all correspond to a greater risk. These visualizations underscore the importance of considering both average trends and individual variances in the assessment of ML model predictive power.
We built the ML model based on only two features: the most important feature—a dose of statins and the genetics—VEGFR2 genotype, and the model performance reached 0.80 ROC AUC, which is only 0.013 less than the best model (Figure 1B). This means that these two predictors can serve as markers in assessing the risk of MACEs in MI patients.
3.5 Model risk assessment for individual patients
With SHAP methods, one can evaluate the contributions of each parameter to the individual risk of a patient. The ML model risk assessment for high- and low-risk patients with corresponding SHAP values is presented in Figures 4, 5.
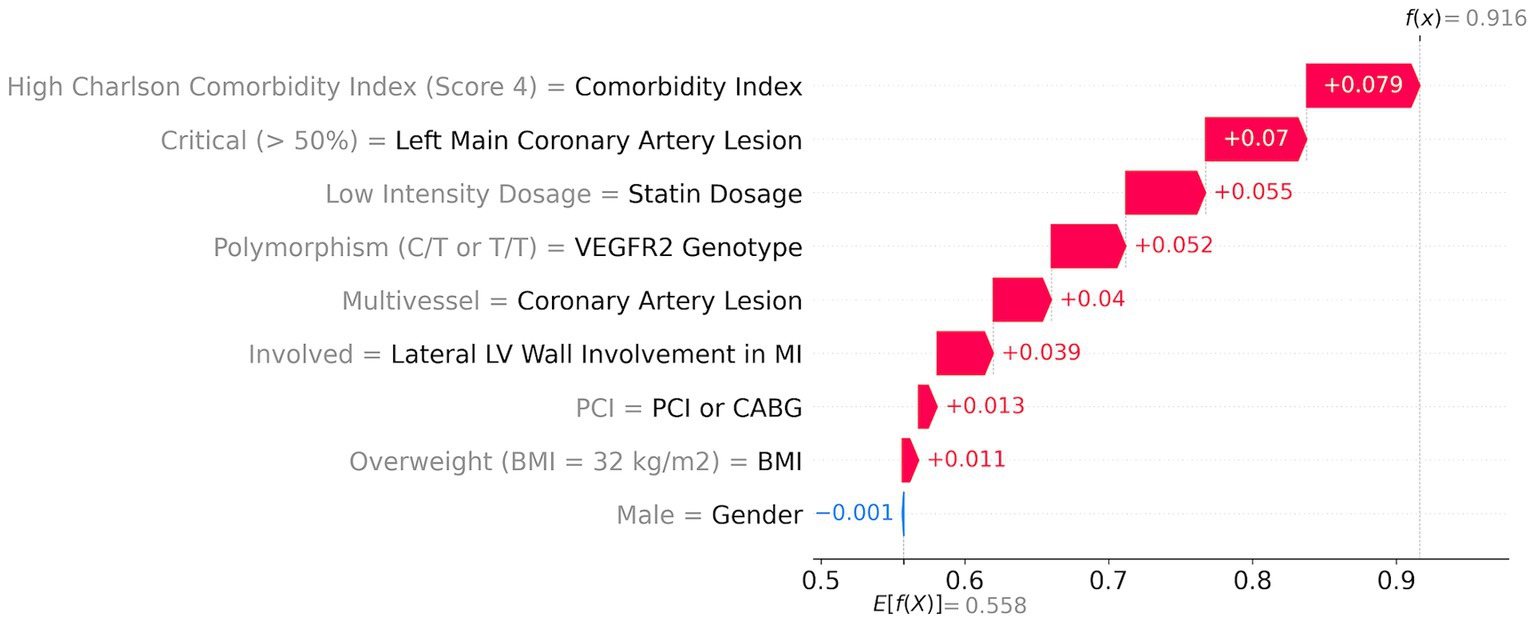
A high-risk patient with a 92% risk (Figure 4) has the following top five major contributors: high comorbidity index (score = 4; red arrow in Figure 4) with a 7.9% contribution, followed by the critical left main coronary artery lesion (>50%) with a 7% contribution, low dosage of statins with a 5.5% contribution, risk T-allele with a 5.2% contribution, and multivessel coronary artery lesion with a 4% contribution. Additionally, lateral LV wall involvement, PCI, and a BMI of 32 kg/m2 contribute to the risk by 3.9, 1.3, and 1.1%, respectively.
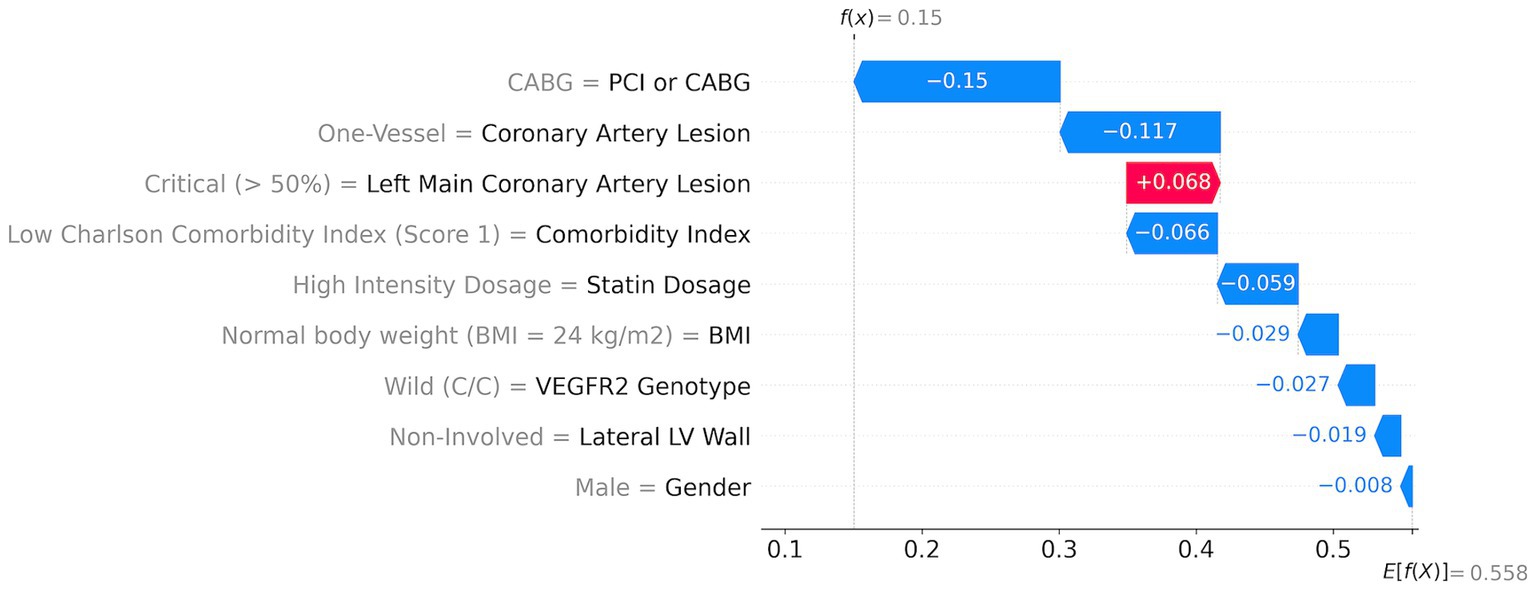
An example of a patient with a low risk of MACEs among the investigated patients (15% risk) shows that all features contribute positively (indicated by blue arrows) to reducing the total risk (Figure 5). The first risk-reducing parameter is CABG followed by the absence of coronary artery lesion. However, left coronary artery lesions increase the risk by almost 7% (indicated by the red arrow in Figure 5). A low comorbidity index and high statin dosage reduce the risk by 6.6 and 5.9%, respectively. Other factors such as normal body weight, non-risk genetics, non-involved lateral LV walk, and sex further reduce the risk cumulatively by 8%.
4 Discussion
Several studies have shown strong associations between VEGFs, VEGFRs serum levels, and other growth factors with clinical characteristics and prognosis in ACS (30). VEGFR-2 SNPs have previously been shown to be associated with the development of CVDs such as CAD, ACS, and Kawasaki disease (11) and have also been evaluated as prognostic markers in stroke and ACS. Thus, in the meta-analysis of Qui et al. (10), VEGFR-2 rs2305948 and rs1870377 (both found in exon regions of VEGF receptor-2 and lead to aminoacidic substitutions that reduce the binding affinity of VEGF to VEGF receptor-2), but not rs2071559, were associated with an increased risk of stroke. In another meta-analysis, Wang et al. (31) proved that VEGFR2 polymorphisms (rs1870377, rs2071559, and rs2305948) could be used to identify individuals with increased susceptibility to atherosclerotic cardiovascular disease. In another study, based on the Nanjing Stroke Registry, rs1870377 could predict the 3-month outcome of patients with large artery atherosclerotic stroke (32). Marks et al. showed that none of the VEGFR-2 SNPs (rs1870377, rs2071559, and rs2305948) were associated with mortality in 2067 ACS patients of the New Zealand Coronary Disease Cohort Study (14). We, for the first time, included the VEGFR-2 genotype as a feature in the prognostic model for MI patients’ outcomes and found that the genetic factor is included in the top five most important features.
We also evaluated the impact of various factors—clinical, imaging, genetic, and treatment-related—on risk prognosis. Using the developed ML approach, we demonstrated a sufficient negative impact of various factors on combined clinical endpoints, including cardiovascular death, recurrent ACS, stroke, and myocardial revascularization. The key factors are as follows: clinical factors—male sex, high body mass index, and complex comorbidity; imaging factors—multivessel coronary lesions and LV lateral wall involvement; genetic factors—minor allelic variants of VEGFR-2 rs2305948; and treatment factors—PCI only for the infarct-related coronary artery instead of CABG in cases of multivessel coronary artery disease, and low-intensity statin dosage. At the same time, high versus low doses of statins and CABG versus PCI in the study cohort of MI patients showed significant benefits in reducing MACEs during the 9-year follow-up.
Advanced age, female sex at menopause (33), and comorbidities such as diabetes, obesity, and dyslipidemia are well-known conventional risk factors for MACEs (recurrent ACS, stroke, and death) in MI patients. A dramatic shift has been observed over the last 25 years in the epidemiology of cardiovascular events. The incidence of MI and ischemic stroke has decreased 3- to 4-fold, which can be attributed to major changes in population risk factors with substantial decreases in systolic blood pressure, cholesterol concentrations, and smoking rates, although this has been offset by increases in body mass index, diabetes, and other comorbidities prevalence. Additionally, a positive impact has been made by the wider post-MI use of cardioprotective medications, such as high-dose statins, P2Y12-platelet inhibitors, beta-blockers, and newer drugs in real clinical practice (34–37). Involvement of the lateral LV wall in MI is a negative prognostic marker of maladaptive cardiac remodeling, often requiring more advanced treatment options (e.g., surgical ventricular restoration) to prevent the development of congestive heart failure (38). During the last two decades, the role of multivessel cardiac disease (including the left main coronary artery lesions) has been actively discussed in the prognosis of MI patients (39, 40). According to data from the SWEDEHEART registry, CABG in patients with LMCA disease is associated with lower mortality and fewer MACEs compared to PCI (41). Complete PCI revascularization following MI reduces all-cause mortality, cardiovascular mortality, recurrent ACS, and repeat symptom-driven revascularization. Immediate complete PCI or CABG revascularization in MI may be equally beneficial, but it requires additional head-to-head trials (42).
Comparison to other ML prognostic models revealed similarities in our findings with respect to clinical and imaging factors. In the study (15), feature importance analysis revealed LVEF, age, diabetes, estimated GFR, multivessel disease, peripheral artery disease, complete revascularization, hemoglobin level, previous bleeding, malignancy, ACE inhibitors or ARB at discharge, and statin therapy at discharge as the most important features to predict all-cause death and recurrent MI. Similar to our results, significant risk factors of MACEs included comorbidities (age, multivessel disease, diabetes, chronic kidney disease, and malignancies), complete myocardial revascularization, and in-hospital and post-discharge high-intensity statin usage.
ML models for predicting all-cause death at 3-year follow-up, based on clinical data from the nationwide perspective registry of AMI in Korea (n = 13,104) (43), identified the following top 10 predictors: statin use at discharge, sex, body mass index, use of glycoprotein IIb/IIIa inhibitors, in-hospital duration, coronary lesion classification, NT-proBNP, total cholesterol, door-to-balloon time, and peak troponin I. The majority of the predictors were also shown as significant for the Russian MI population in our prognostic model for the combined end-point. Specifically, the top important features include: coronary lesions, (high-intensity dose) statin use at discharge, sex, and body mass index. Wang et al. (44) presented the ML risk model including 21 patient characteristic variables for a 6-month prediction of MACEs for 1,004 Chinese patients who had undergone coronary revascularization. The model also found age and CABG as the top important factors.
In another large prospective study (45) based on the UK Biobank cohort, the authors revealed 10 predictors for 473,611 CVD-free participants, namely age, sex, cholesterol and blood pressure medications, cholesterol ratio (total/high-density lipoprotein), systolic blood pressure, previous angina or heart disease, number of medications taken, cystatin C, chest pain, and pack-years of smoking. Direct comparison is difficult due to different cohorts (CVD-free participants or post-MI patients); however, cholesterol medications, age, coronary artery disease, and comorbidities (such as diabetes) were also detected as top important factors.
As for the best model performance, the initial model selection was guided by an AutoML framework (FLAML) that evaluated multiple algorithms, including Random Forest, CatBoost, and LightGBM. The AutoML process identified CatBoost as the optimal base estimator, highlighting its balance between accuracy and generalization. Although Random Forest showed higher performance metrics on the test dataset, this can be attributed to overfitting as indicated by its exceptionally high results. CatBoost, on the other hand, demonstrated performance without overfitting, making it a more reliable choice for our clinical application. Boosting algorithms, in general, are well-documented for providing state-of-the-art performance for tabular data. In particular, CatBoost with hyperparameter tuning has been shown to be the best-performing model on average as detailed in (46). This consistency in performance underscores the reliability of CatBoost for our study. Ultimately, CatBoost showed superior performance on the test dataset, reinforcing its suitability as the primary algorithm.
The present study demonstrates that VEGFR-2 genotypes can be used as a predictor of MACEs. While these findings might be specific to the study population due to the lack of external validation, the results provide a solid foundation for conducting larger studies. Such research could lead to the development of a universal prognostic tool for MACEs using this genetic biomarker.
To our knowledge, genetic factors have not previously been used in ML models for predicting MACEs in MI patients, mostly due to the fact that sequencing or even single nucleotide genotyping is not routinely performed in hospitals. In this respect, this study is pioneering in showing that the risk allele of the VEGFR-2 variant is among the top 5 most important factors for predicting long-term outcomes in MI patients.
We see a large potential for integration of the model into current clinical practice. Patient MACE risk assessment can be integrated into telemedicine service and automatically assessed based on patients’ clinical and genetic data. Telemedicine service will provide feedback on patient status and facilitate corrections to the model, thereby improving the prognostic accuracy of the model.
Directions for future research include the improvement of the ML model by training on a larger dataset, performing external validation, and investigating other genetic markers known to be associated with the risk of MACEs. The latter is the subject of extensive research worldwide, and patient genetic information will help to improve personalized treatment strategies.
5 Conclusion
We developed an ML model that can predict long-term (9 years) ischemic outcomes in MI patients with ROC AUC of 0.813, based on nine features selected from an initial set of 39. For the first time, we included genetic factors in the ML predictive model for assessing the risk of MI patients from a long-term perspective. VEGFR-2 rs2305948 (C/C, C/T, T/T) genotypes showed high predictive power with risk T-allele increasing the risk of MACEs. Additionally, we demonstrated that high-dose statin therapy for 12 months post-MI, along with other factors, can minimize cardiac risk for patients carrying the risk T-allele.
Integration of VEGFR-2 genotypes in MACE prediction models requires a larger study. However, we find this model suitable for the risk assessment for patients at the Surgut District Cardiology Clinic. Immediate integration of this model into clinical practice requires an easy-to-use risk assessment application.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Surgut State University, Surgut, Russia. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
AK: Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing, Visualization. KB: Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. AV: Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Funding acquisition, Project administration, Supervision. EZb: Formal analysis, Investigation, Project administration, Writing – review & editing. GL: Formal analysis, Writing – review & editing. KN: Methodology, Resources, Writing – review & editing, Investigation, Funding acquisition, Supervision, Data curation, Formal analysis. EZe: Data curation, Formal analysis, Methodology, Writing – review & editing. MD: Formal analysis, Writing – review & editing, Investigation. LK: Formal analysis, Writing – review & editing, Funding acquisition, Resources, Supervision. IU: Funding acquisition, Resources, Supervision, Writing – review & editing, Investigation, Methodology, Project administration. MP: Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Validation, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The article was prepared within the framework of the project “Mirror Laboratories” at HSE University.
Acknowledgments
We thank the reviewers for their fruitful comments that helped considerably improve the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2024.1452239/full#supplementary-material
References
1. Hall, M, Smith, L, Wu, J, Hayward, C, Batty, JA, Lambert, PC, et al. Health outcomes after myocardial infarction: a population study of 56 million people in England. PLoS Med. (2024) 21:e1004343. doi: 10.1371/journal.pmed.1004343
2. Chi, G, Lee, JJ, Kazmi, SH, Fitzgerald, C, Memar Montazerin, S, Kalayci, A, et al. Early and late recurrent cardiovascular events among high-risk patients with an acute coronary syndrome: meta-analysis of phase iii studies and implications on trial design. Clin Cardiol. (2022) 45:299–307. doi: 10.1002/clc.23773
3. Grundy, SM, Stone, NJ, Bailey, AL, Beam, C, Birtcher, KK, Blumenthal, RS, et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APHA/ASPC/NLA/PCNA guideline on the Management of Blood Cholesterol: executive summary: a report of the American College of Cardiology/American Heart Association task force on clinical practice guidelines. Circulation. (2019) 139:e1046–81. doi: 10.1161/CIR.0000000000000624
4. Liang, S, Wang, C, Zhang, J, Liu, Z, Bai, Y, Chen, Z, et al. Triglyceride-glucose index and coronary artery disease: a systematic review and meta-analysis of risk, severity, and prognosis. Cardiovasc Diabetol. (2023) 22:170. doi: 10.1186/s12933-023-01906-4
5. Hung, J, Roos, A, Kadesjö, E, McAllister, DA, Kimenai, DM, Shah, AS, et al. Performance of the Grace 2.0 score in patients with type 1 and type 2 myocardial infarction. Eur Heart J. (2021) 42:2552–61. doi: 10.1093/eurheartj/ehaa375
6. Scirica, BM. Acute coronary syndrome: emerging tools for diagnosis and risk assessment. J Am Coll Cardiol. (2010) 55:1403–15. doi: 10.1016/j.jacc.2009.09.071
7. Zhou, Y, Zhu, X, Cui, H, Shi, J, Yuan, G, Shi, S, et al. The role of the Vegf family in coronary heart disease. Front Cardiovasc Med. (2021) 8:738325. doi: 10.3389/fcvm.2021.738325
8. Paradowska-Gorycka, A, Stypinska, B, Pawlik, A, Malinowski, D, Romanowska-Prochnicka, K, Manczak, M, et al. Kdr (Vegfr2) genetic variants and serum levels in patients with rheumatoid arthritis. Biomol Ther. (2019) 9:355. doi: 10.3390/biom9080355
9. Camaré, C, Pucelle, M, Nègre-Salvayre, A, and Salvayre, R. Angiogenesis in the atherosclerotic plaque. Redox Biol. (2017) 12:18–34. doi: 10.1016/j.redox.2017.01.007
10. Qiu, S, Wu, T, Wang, P, Li, J, Li, Q, and Du, J. The association between Vegfr gene polymorphisms and stroke: a meta-analysis. PLoS One. (2016) 11:e0151371. doi: 10.1371/journal.pone.0151371
11. Liu, D, Song, J, Ji, X, Liu, Z, Cong, M, and Hu, B. Association of genetic polymorphisms on Vegfa and Vegfr2 with risk of coronary heart disease. Medicine. (2016) 95:e3413. doi: 10.1097/MD.0000000000003413
12. Jin, F, Hagemann, N, Brockmeier, U, Schäfer, ST, Zechariah, A, and Hermann, DM. Ldl attenuates Vegf-induced angiogenesis via mechanisms involving Vegfr2 internalization and degradation following endosome-trans-Golgi network trafficking. Angiogenesis. (2013) 16:625–37. doi: 10.1007/s10456-013-9340-2
13. Holmes, K, Roberts, OL, Thomas, AM, and Cross, MJ. Vascular endothelial growth factor Receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell Signal. (2007) 19:2003–12. doi: 10.1016/j.cellsig.2007.05.013
14. Marks, EC, Wilkinson, TM, Frampton, CM, Skelton, L, Pilbrow, AP, Yandle, TG, et al. Plasma levels of soluble Vegf receptor isoforms, circulating Pterins and Vegf system Snps as prognostic biomarkers in patients with acute coronary syndromes. BMC Cardiovasc Disord. (2018) 18:1–10. doi: 10.1186/s12872-018-0894-1
15. D'Ascenzo, F, De Filippo, O, Gallone, G, Mittone, G, Deriu, MA, Iannaccone, M, et al. Machine learning-based prediction of adverse events following an acute coronary syndrome (praise): a modelling study of pooled datasets. Lancet. (2021) 397:199–207. doi: 10.1016/S0140-6736(20)32519-8
16. Chicco, D, and Jurman, G. Machine learning can predict survival of patients with heart failure from serum creatinine and ejection fraction alone. BMC Med Inform Decis Mak. (2020) 20:1–16. doi: 10.1186/s12911-020-1023-5
17. Angraal, S, Mortazavi, BJ, Gupta, A, Khera, R, Ahmad, T, Desai, NR, et al. Machine learning prediction of mortality and hospitalization in heart failure with preserved ejection fraction. JACC Heart Fail. (2020) 8:12–21. doi: 10.1016/j.jchf.2019.06.013
18. Chirinos, JA, Orlenko, A, Zhao, L, Basso, MD, Cvijic, ME, Li, Z, et al. Multiple plasma biomarkers for risk stratification in patients with heart failure and preserved ejection fraction. J Am Coll Cardiol. (2020) 75:1281–95. doi: 10.1016/j.jacc.2019.12.069
19. Zhao, H, Li, P, Zhong, G, Xie, K, Zhou, H, Ning, Y, et al. Machine learning models in heart failure with mildly reduced ejection fraction patients. Front Cardiovasc Med. (2022) 9:1042139. doi: 10.3389/fcvm.2022.1042139
20. Mpanya, D, Celik, T, Klug, E, and Ntsinjana, H. Predicting in-hospital all-cause mortality in heart failure using machine learning. Front Cardiovasc Med. (2023) 9:1032524. doi: 10.3389/fcvm.2022.1032524
21. Lin, H, Xue, Y, Chen, K, Zhong, S, and Chen, L. Acute coronary syndrome risk prediction based on gradient boosted tree feature selection and recursive feature elimination: a dataset-specific modeling study. PLoS One. (2022) 17:e0278217. doi: 10.1371/journal.pone.0278217
22. Yu, T, Shen, R, You, G, Lv, L, Kang, S, Wang, X, et al. Machine learning-based prediction of the post-thrombotic syndrome: model development and validation study. Front Cardiovasc Med. (2022) 9:990788. doi: 10.3389/fcvm.2022.990788
23. Wang, X, Zhu, T, Xia, M, Liu, Y, Wang, Y, Wang, X, et al. Predicting the prognosis of patients in the coronary care unit: a novel multi-category machine learning model using Xgboost. Front Cardiovasc Med. (2022) 9:764629. doi: 10.3389/fcvm.2022.764629
24. Commandeur, F, Slomka, PJ, Goeller, M, Chen, X, Cadet, S, Razipour, A, et al. Machine learning to predict the long-term risk of myocardial infarction and cardiac death based on clinical risk, coronary calcium, and epicardial adipose tissue: a prospective study. Cardiovasc Res. (2020) 116:2216–25. doi: 10.1093/cvr/cvz321
25. Van Buuren, S, and Groothuis-Oudshoorn, K. Mice: multivariate imputation by chained equations in R. J Stat Softw. (2011) 45:1–67. doi: 10.18637/jss.v045.i03
26. Wang, C, Wu, Q, Weimer, M, and Zhu, E. Flaml: a fast and lightweight Automl library. Proc Mach Learn Res. (2021) 3:434–47. doi: 10.48550/arXiv.1911.04706
27. Akiba, T, Sano, S, Yanase, T, Ohta, T, and Koyama, M, editors. Optuna: A next-generation Hyperparameter optimization framework. Proceedings of the 25th ACM SIGKDD international conference on knowledge discovery & data mining; (2019).
28. Lundberg, SM, and Lee, S-I. A unified approach to interpreting model predictions. Adv Neural Inf Proces Syst. (2017) 30:4768–77.
29. Goldstein, A, Kapelner, A, Bleich, J, and Pitkin, E. Peeking inside the black box: visualizing statistical learning with plots of individual conditional expectation. J Comput Graph Stat. (2015) 24:44–65. doi: 10.1080/10618600.2014.907095
30. Ferrara, N, Gerber, HP, and LeCouter, J. The biology of Vegf and its receptors. Nat Med. (2003) 9:669–76. doi: 10.1038/nm0603-669
31. Wang, L, Ge, H, Peng, L, and Wang, B. A meta-analysis of the relationship between Vegfr2 polymorphisms and atherosclerotic cardiovascular diseases. Clin Cardiol. (2019) 42:860–5. doi: 10.1002/clc.23233
32. Li, Z, Wang, M, Gu, J, Zhao, L, Guo, Y, Zhang, Z, et al. Missense variants in hypoxia-induced Vegfa/Vegfr2 signaling predict the outcome of large artery atherosclerotic stroke. Cell Mol Neurobiol. (2021) 41:1217–25. doi: 10.1007/s10571-020-00890-7
33. Tschiderer, L, Seekircher, L, Willeit, P, and Peters, SA. Assessment of cardiovascular risk in women: progress so far and progress to come. Int J Women's Health. (2023) 15:191–212. doi: 10.2147/IJWH.S364012
34. Shah, AS, Lee, KK, Pérez, JAR, Campbell, D, Astengo, F, Logue, J, et al. Clinical burden, risk factor impact and outcomes following myocardial infarction and stroke: a 25-year individual patient level linkage study. Lancet Reg Health Eur. (2021):7. doi: 10.1016/j.lanepe.2021.100141
35. Li, L, Scott, CA, Rothwell, PM, and Study, OV. Trends in stroke incidence in high-income countries in the 21st century: population-based study and systematic review. Stroke. (2020) 51:1372–80. doi: 10.1161/STROKEAHA.119.028484
36. Safi, S, Sethi, NJ, Korang, SK, Nielsen, EE, Feinberg, J, Gluud, C, et al. Beta-blockers in patients without heart failure after myocardial infarction. Cochrane Database Syst Rev. (2021) 11:CD012565. doi: 10.1002/14651858.CD012565.pub2
37. Yu, D, and Liao, JK. Emerging views of statin pleiotropy and cholesterol lowering. Cardiovasc Res. (2022) 118:413–23. doi: 10.1093/cvr/cvab032
38. Patel, ND, Williams, JA, Nwakanma, LU, Weiss, ES, and Conte, JV. Impact of lateral wall myocardial infarction on outcomes after surgical ventricular restoration. Ann Thorac Surg. (2007) 83:2017–28. doi: 10.1016/j.athoracsur.2007.02.026
39. Gripenberg, T, Jokhaji, F, Östlund-Papadogeorgos, N, Ekenbäck, C, Linder, R, Samad, B, et al. Outcome and selection of revascularization strategy in left main coronary artery stenosis. Scand Cardiovasc J. (2018) 52:100–7. doi: 10.1080/14017431.2018.1429648
40. Puymirat, E, Nakache, A, Saint Etienne, C, Marcollet, P, Fichaux, O, Decomis, MP, et al. Is coronary multivessel disease in acute myocardial infarction patients still associated with worse clinical outcomes at 1-year? Clin Cardiol. (2021) 44:429–37. doi: 10.1002/clc.23567
41. Persson, J, Yan, J, Angerås, O, Venetsanos, D, Jeppsson, A, Sjögren, I, et al. Pci or Cabg for left Main coronary artery disease: the Swedeheart registry. Eur Heart J. (2023) 44:2833–42. doi: 10.1093/eurheartj/ehad369
42. Banning, AP. Revascularization strategies in patients with MI and MVD: Have we got the answers? J Am Coll Cardiol. (2024) 84:295–7. doi: 10.1016/j.jacc.2024.05.019
43. Jeong, JH, Lee, KS, Park, SM, Kim, SR, Kim, MN, Chae, SC, et al. Prediction of longitudinal clinical outcomes after acute myocardial infarction using a dynamic machine learning algorithm. Front Cardiovasc Med. (2024) 11:1340022. doi: 10.3389/fcvm.2024.1340022
44. Wang, J, Wang, S, Zhu, MX, Yang, T, Yin, Q, and Hou, Y. Risk prediction of major adverse cardiovascular events occurrence within 6 months after coronary revascularization: machine learning Study. JMIR Med Inform. (2022) 10:e33395. doi: 10.2196/33395
45. You, J, Guo, Y, Kang, JJ, Wang, HF, Yang, M, Feng, JF, et al. Development of machine learning-based models to predict 10-year risk of cardiovascular disease: a prospective cohort study. Stroke Vasc Neurol. (2023) 8:475–85. doi: 10.1136/svn-2023-002332
Keywords: machine learning, myocardial infarction, vascular endothelial growth factor receptor 2 gene, statins, prognostic modeling, MACEs, left ventricular wall
Citation: Kirdeev A, Burkin K, Vorobev A, Zbirovskaya E, Lifshits G, Nikolaev K, Zelenskaya E, Donnikov M, Kovalenko L, Urvantseva I and Poptsova M (2024) Machine learning models for predicting risks of MACEs for myocardial infarction patients with different VEGFR2 genotypes. Front. Med. 11:1452239. doi: 10.3389/fmed.2024.1452239
Edited by:
Xianghong Ma, Aston University, United KingdomReviewed by:
Syed Nurul Hasan, New York University, United StatesNandini Nair, The Pennsylvania State University, United States
Copyright © 2024 Kirdeev, Burkin, Vorobev, Zbirovskaya, Lifshits, Nikolaev, Zelenskaya, Donnikov, Kovalenko, Urvantseva and Poptsova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Poptsova, bXBvcHRzb3ZhQGhzZS5ydQ==
†These authors have contributed equally to this work and share first authorship
 Alexander Kirdeev
Alexander Kirdeev Konstantin Burkin1†
Konstantin Burkin1† Galina Lifshits
Galina Lifshits Maria Poptsova
Maria Poptsova