- 1College of Pharmacy, Cihan University-Erbil, Kurdistan Region, Erbil, Iraq
- 2Department of Biology, College of Science, Salahaddin University-Erbil, Erbil, Iraq
- 3Department of Medicine, Hawler Medical University, Erbil, Iraq
- 4College of Medicine, University of Sulaimani, Sulaymaniyah, Iraq
- 5Department of Medical Sciences: Respiratory, Allergy and Sleep Research, Uppsala University, Uppsala, Sweden
The pandemic of coronavirus disease-19 (COVID-19), provoked by the appearance of a novel coronavirus named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), required a worldwide healthcare emergency. This has elicited an immediate need for accelerated research into its mechanisms of disease, criteria for diagnosis, methods for forecasting outcomes, and treatment approaches. microRNAs (miRNAs), are diminutive RNA molecules, that are non-coding and participate in gene expression regulation post-transcriptionally, having an important participation in regulating immune processes. miRNAs have granted substantial interest in their impact on viral replication, cell proliferation, and modulation of how the host’s immune system responds. This narrative review delves into host miRNAs’ multifaceted roles within the COVID-19 context, highlighting their involvement in disease progression, diagnostics, and prognostics aspects, given their stability in biological fluids and varied expression profiles when responding to an infection. Additionally, we discuss complicated interactions between SARS-CoV-2 and host cellular machinery facilitated by host miRNAs revealing how dysregulation of host miRNA expression profiles advances viral replication, immune evasion, and inflammatory responses. Furthermore, it investigates the potential of host miRNAs as therapeutic agents, whether synthetic or naturally occurring, which could be harnessed to either mitigate harmful inflammation or enhance antiviral responses. However, searching more deeply is needed to clarify how host’s miRNAs are involved in pathogenesis of COVID-19, its diagnosis processes, prognostic assessments, and treatment approaches for patients.
Graphical Abstract. Created with BioRender.com.
1 Introduction
Since its initial appearance in the final quarter of 2019, coronavirus disease-19 which is widely known as COVID-19, having the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) as the causative virus, escalating into a health-related global emergency (1). Upon infection, accumulation of viral particles leads to the emergence of variable symptoms, including endothelial lining damage and impaired oxygen diffusion capacity. In the later severe stages, an excessive release of cytokines can contribute to organ failure and potentially result in increased mortality (2). MicroRNAs (miRNAs), which are single-stranded RNAs of 20 to 24 nucleotides, play a role in gene expression regulation by selectively binding to the 3′ untranslated region (UTRs) of mRNAs, resulting in mRNA degradation or translation inhibition, effectively stopping protein synthesis (3). Alterations in the levels of host miRNAs have been noted in individuals who have contracted the SARS-CoV-2 virus (4). Moreover, their close association with various diseases, such as cancer, cardiovascular disorders, and viral diseases, highlights their significance as biomarkers to assess disease severity and monitor treatment responses (5, 6). Human miRNAs play a significant role in regulating cellular activities and their potential interaction with viruses, specifically in the context of SARS-CoV-2 (7). Research has identified certain miRNAs that show significance in binding to essential genes of SARS-CoV-2, making them potential biomarkers for COVID-19 diagnosis (8). Specifically, miR-155-5p, miR-29a-3p, and miR-146a-3p have shown high specificity and sensitivity, indicating their potential as biomarkers for COVID-19 (9).
Genetic and epigenetic factors contribute to the determining the disease severity. Response coming from miRNAs encoded by the host is substantial for prognosticating the SARS-CoV-2 infection’s outcomes (10). In both the acute and post-acute stages of the disease, there were changes in the expression of cellular host miRNAs that make it a good candidate as a marker for disease prognosis (9). The study indicated a notable miR-155-5p elevation in COVID-19 patients that were critically ill, as evidenced by its significant upregulation among 11 host miRNAs assessed for prognostic implications. Additionally, miR-146a-3p and miR-29a-3p show a potential to serve as innovative prognostic indicators for differentiating the acute stages from the post-acute during COVID-19 infection (11).
The clinical significance of host miRNAs is of utmost importance due to their potential therapeutic impact through targeted inhibition, particularly in terms of antiviral effects (12). Host miRNAs have the capacity to augment the body’s protective responses against infections or, at the very least, it could induce pathogen latency (13). The replication/synthesis of viral proteins takes place within the host cell, whereas miRNAs function in preventing the conversion of targeted mRNA into a protein through the translation process. Consequently, miRNAs present themselves as potentially therapeutic approaches to viral diseases (14, 15). This approach can serve as an alternative strategy for managing COVID-19 disease and other related coronavirus illnesses, including utilization of fully complementary miRNAs (cc miRNA) specifically targeting viral genes, suppressing their expression post-transcriptionally. The modified cc miRNAs with a length of 25 to 27 nucleotides, including hsa-miR-2510-3p, miR-3154, hsa-miR-448-3p, miR-5197-3p, miR-7114-5p, hsa-miR-2750-3p, and hsa-miR-1851-5p, demonstrated notable attraction to SARS-CoV-2 genome, suggesting the possibility of an interaction (16). Furthermore, certain viruses possess their own miRNAs, which have the potential to manipulate the signaling pathways of the host, ultimately promoting the virus’s survival and replication. Therefore, the development of drugs that target viral miRNA through inhibition could serve as an additional approach for COVID-19 therapy. Furthermore, research has demonstrated that plant miRNAs that possess identical sequences to human miRNAs could potentially be utilized for COVID-19 treatment (16, 17). The review aims to provide a comprehensive overview of host miRNAs and their involvement in the pathogenesis of SARS-CoV-2 infection. It explores their potential as diagnostic markers to identify different phases of the disease, such as asymptomatic, mild, moderate, and severe cases. Additionally, the review investigates the prognostic value of host miRNAs in predicting unfavorable outcomes, including disease progression, development of complications, and mortality. Furthermore, it delves into the therapeutic relevance of these miRNAs in COVID-19 management, discussing their potential as targets for therapeutic interventions and the development of miRNA-based therapies.
2 The emergence and impact of coronaviruses: from SARS-CoV-1 to SARS-CoV-2
Coronaviruses are a heterogeneous group of pathogens responsible for causing illnesses in a wide variety of animal species. Moreover, they are able to cause a spectrum of human respiratory complications, from light to severe (18). Coronaviruses remained relatively obscure in terms of pathologic impact until the early 21st century, when the SARS-CoV-1 emerged in 2002. A subsequent outbreak occurred approximately a decade later with the discovery of the Middle-East respiratory syndrome coronavirus (MERS-CoV) in 2012. Both viruses are believed to have a zoonotic origin, with SARS-CoV-1 being linked to bats and palm civets and MERS-CoV to dromedary camels. These coronaviruses are well known for their ability to cause severe and frequently lethal respiratory diseases (19). An outbreak of pneumonia caused by an unknown agent was announced in Wuhan city of China in the final quarter of 2019. Subsequent investigations involving the isolation of the virus from patients with acute pneumonia, accompanied by RNA metagenomic sequencing, uncovered a novel beta coronavirus as the responsible pathogen (18). The SARS-CoV-2 emerged as a new coronavirus, and within a period of 6 months, it was reported that 215 nations were affected with a mortality rate of approximately 750 thousand humans (20). SARS-CoV-2 seemed to have a quite similar genetic sequence and structural resemblance to SARS-CoV-1 of 2002 (21). SARS-CoV-2 is highly infectious and transmissible SARS-CoV-2 virus that belongs to Orthocoronavirinae subfamily, the virus led to a global spread of COVID-19, an illness causing severe respiratory complications, which put the health and safety of the public at risk (22).
2.1 The SARS-CoV-2’s biology and its replication cycle
Coronaviruses contain the largest RNA genome compared to all the other RNA viruses, reaching a length of 27–32 kb. The positive-sense single-stranded RNA (+ssRNA) acts like a messenger RNA that can be translated directly upon host cells’ invasion (23). In SARS-CoV-2 viruses, the nucleocapsid (N) protein encases the viral RNA to form the N structure and is integral to virus replication and mRNA transcription. This N protein also disrupts cell structures and dampens immune responses by shielding viral RNA from the host’s RNA interference pathway, which typically destructs such RNA. It is triggering of antibody and cell-mediated immune reactions post-infection highlights its significance in both diagnostic and vaccine research (24). An envelope with the three associated structural proteins on its surface further surrounds the capsid. The envelope is a lipid bilayer that can be broken down using soap. This viral envelope is derived from the membrane of the host cells (23). Looking at coronaviruses, it is obvious that they have a series of glycoprotein spikes on their envelope. The spiky projections give the appearance of a crown under an electron microscope examination, hence, the given name (corona) from the Latin word coronam for crown (25). Four essential structural proteins are present in SARS-CoV-2 virus, spike (S) proteins, membrane (M) proteins, envelope (E) proteins, and nucleocapsid (N) proteins. Non-structural proteins (nsps) are present as well, which are 16 in number (nsp 1–16) (23). The S protein is a glycoprotein that the SARS-CoV-2 virus utilizes as a unique surface antigen. The significance of this protein in attaching to and invading host cells, and in disease progression, renders it a pivotal focus for devising vaccines and therapeutics against COVID-19 (26). SARS-CoV-2’s S proteins encourage the binding and following penetration of the virus into host cells subsequent to the identification of angiotensin-converting enzyme 2 (ACE2) receptors, which are ubiquitously found in many organs like the heart, lungs, and kidneys. The S glycoprotein is bifunctional, having two subunits (S1 and S2). The S1 facilitates the attachment to the ACE2 receptor, while the S2 mediates viral fusion with the host’s cell membrane, thus provoking infection (26). The M glycoprotein represents the most abundant structural protein discovered in coronaviruses. The rest of the structural proteins are bound to the M proteins. This adhering aids in the finalization of viral assemblage process through stabilizing and complex forming between RNA and N protein (27). The E protein, despite its modest dimensions, is pivotal in maintaining the structural integrity of viral particles by its interaction with the M protein (Figure 1).
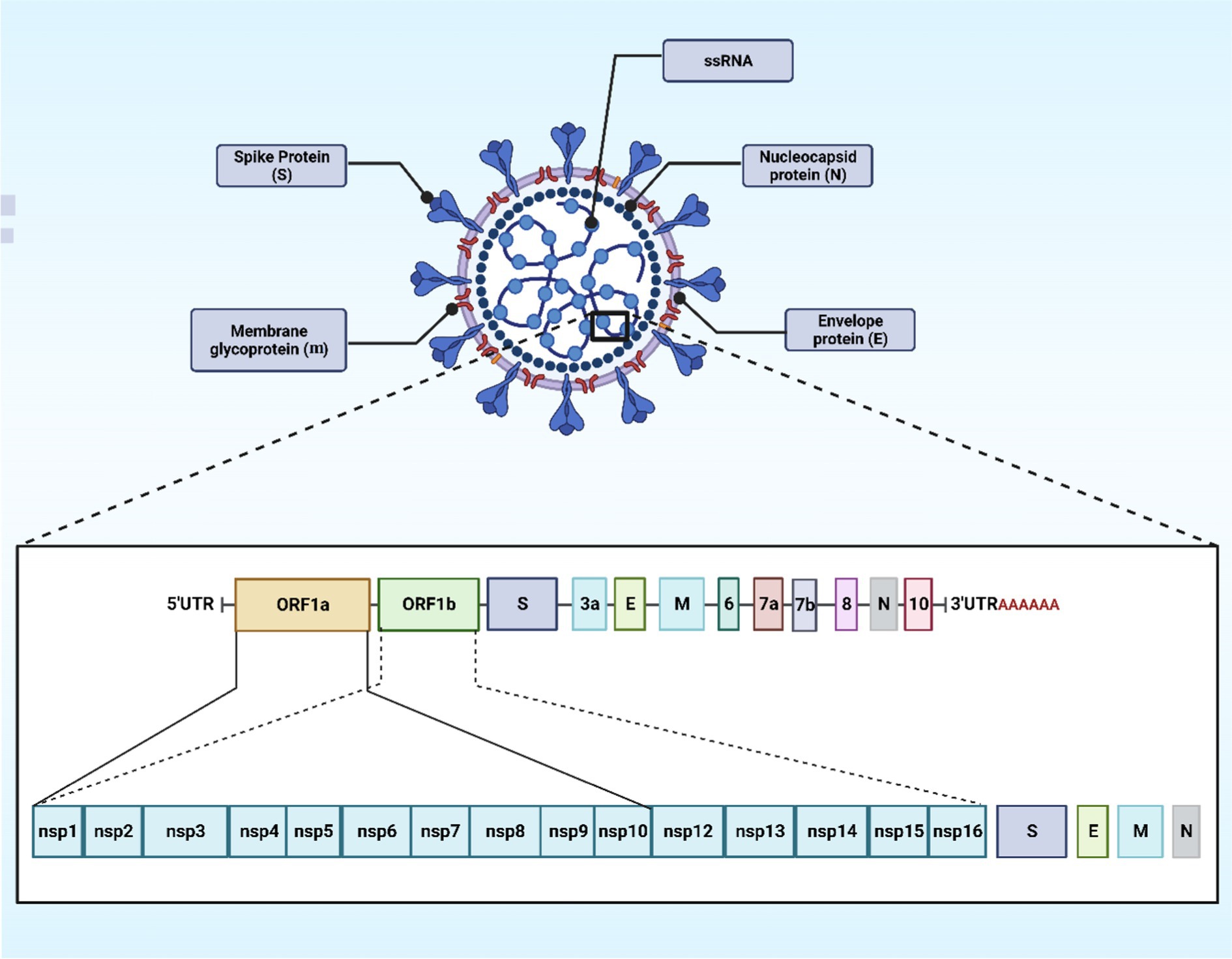
Figure 1. The schematic structure and organization of the SARS-CoV-2 genome. The image was created using the BioRender program, available at https://www.biorender.com.
E protein was found to have high expression in cells infected with the virus, having an important role in the assemblage and egress of the new viruses. Removing E protein could notably reduce or completely stop the virus’s disease induction potentiality, due to its involvement in the life cycle of the virus (28, 29). SARS-CoV-2 begins infecting cells by its S proteins latching onto ACE2 receptors, followed by the action of priming by transmembrane serine protease 2 (TMPRSS2) and cathepsin L (Figure 2). Within the cellular environment, the virus releases its RNA, which is utilized for the synthesis of replication proteins such as RNA-dependent RNA polymerase in vesicles derived from the endoplasmic reticulum (ER). The replicated RNA generates more viral components and genomes. New viruses exit cells and the infection start spreading after the assemblage of the viral structural proteins (N, S, M, and E) within the ER-Golgi intermediate compartment (ERGIC) (30).
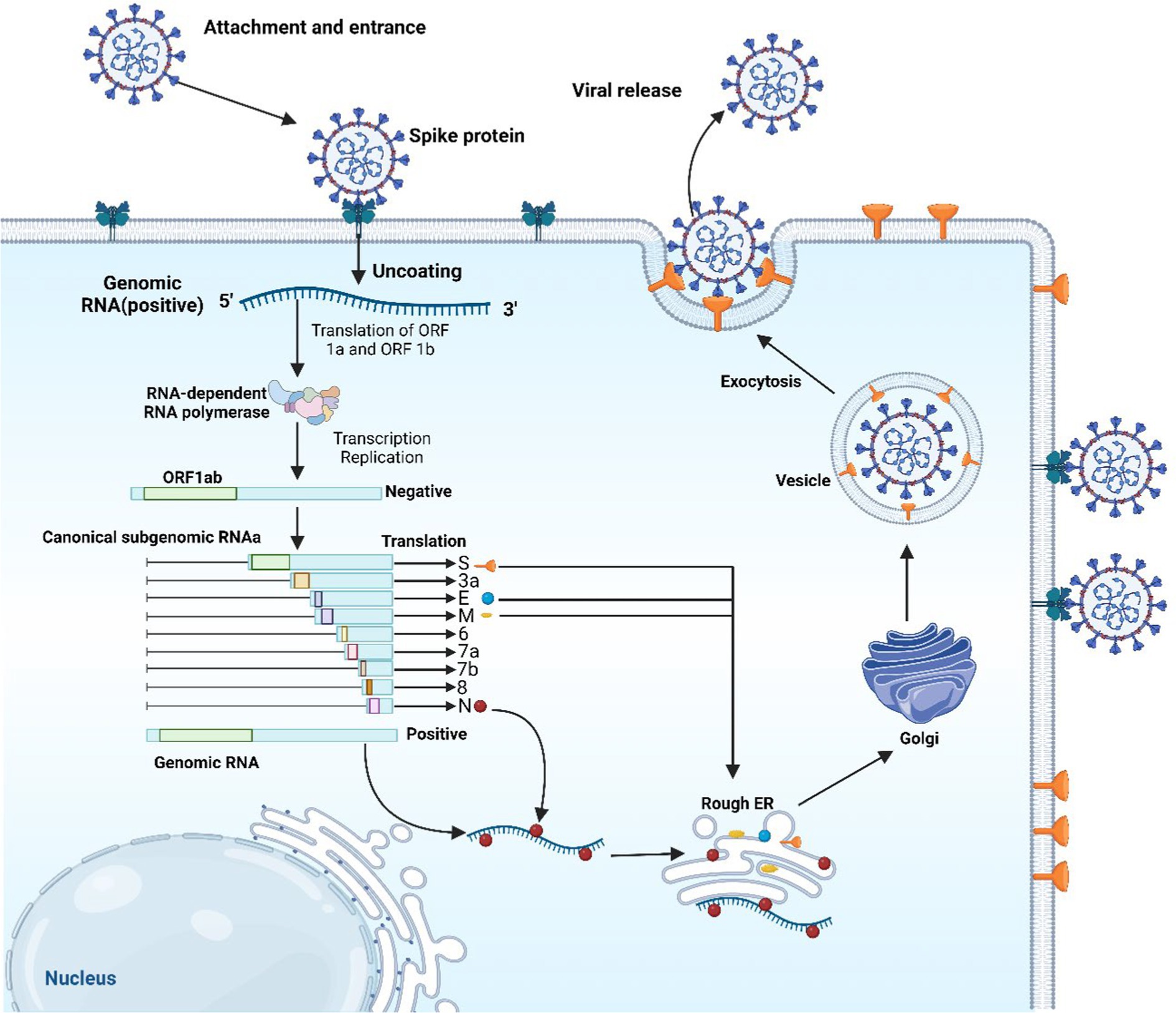
Figure 2. The replication cycle of SARS-CoV-2. The image was created using the BioRender program, available at https://www.biorender.com.
2.2 Immunological response to SARS-CoV-2 infection
Antigen-presenting cells (APCs) like macrophages and dendritic cells (DCs) having an integral role in the activation of innate and adaptive immune responses, are able to detect the virus’s viral antigen as it enters into host cells, by their diversified pattern recognition receptors (PRRs), that encompasses NOD-like receptors, RIG-I-Like receptors (RLRs), and Toll-like receptors (TLRs), the latter is the most significant type of PRR, mostly occurring on macrophage cells. TLRs is a group of transmembrane receptor proteins which recognize pathogen-associated molecular patterns (PAMPs). TLRs group with its 10 members occupy different places, TLR (1, 2, 4, 5, 6, and 10) are found on cell surface, while TLR (3, 7, 8, and 9) are found located in intracellular compartments such as endosomes (31, 32).
Once the virus is identified by TLR4 of the APC, the viral antigenic peptide is presented through class II of major histocompatibility complex (MHC) to T-helper (Th) cell, while class I of MHC represent the antigenic peptide to cytotoxic T-lymphocyte (CTL). APC releases interleukin (IL)-12, a co-stimulatory molecule, which additionally assists in the activation of Th1. For the successful eradication of a virus, the stimulation of Th1 by IL-12 and MHC class I antigen presentation is required (33, 34). When SARS-CoV-2 interacts with PRRs, interferon regulatory factor (IRF) and nuclear factor-kappa B (NF-κB) are generated and transported to the nucleus. The transcription of inflammatory cytokines [IL-1, IL-6, and tumor necrosis factor-α (TNF-α)] and interferons (IFNs) like (IFN-α, IFN-β, and IFN-γ) is facilitated by NF-κB and IRF, respectively. The anti-viral response is attributed to IFNs, which increase the cell’s immunity to viral infection (Figure 3) (35).
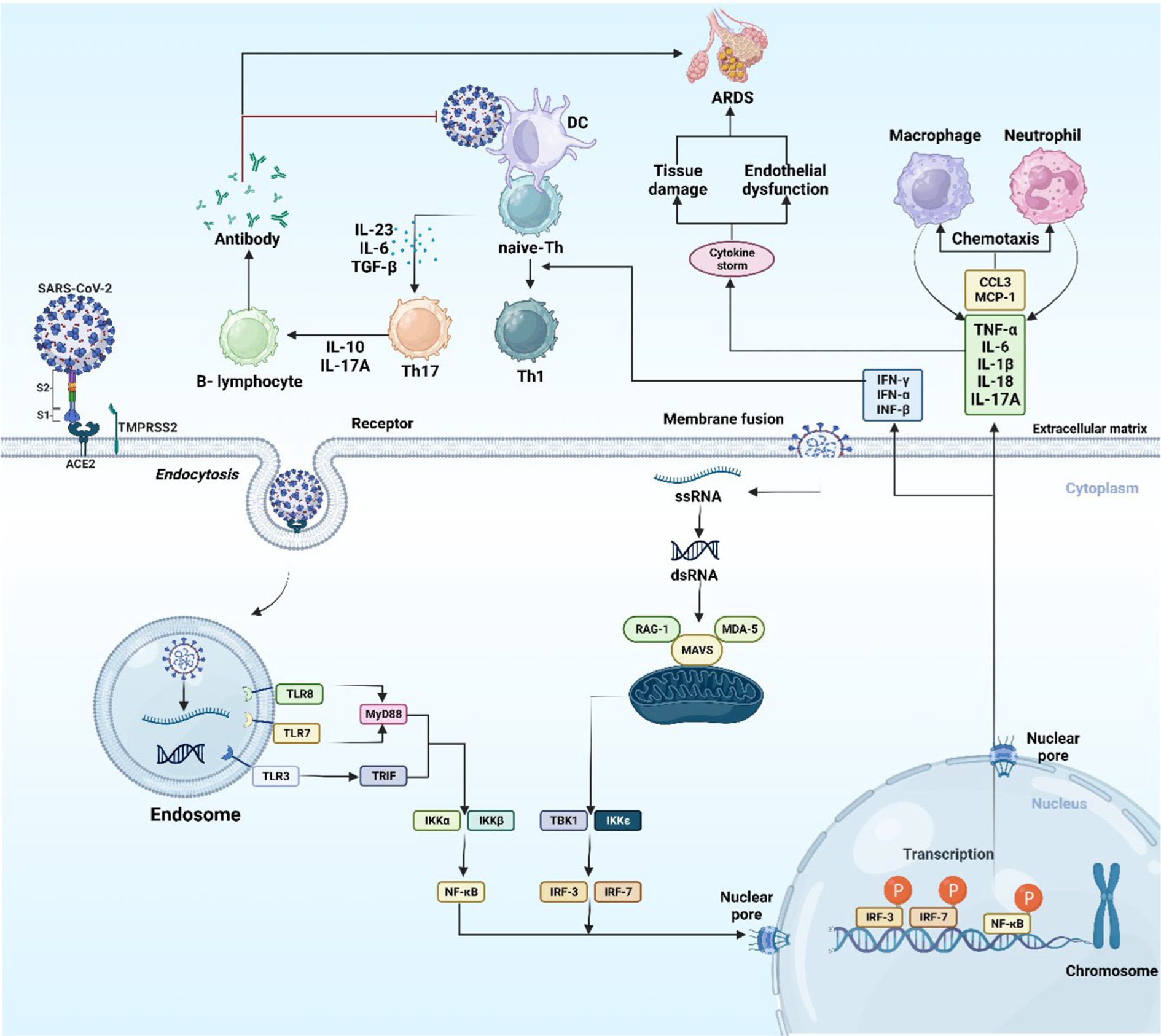
Figure 3. The host immune response to SARS-CoV-2. The image was created using the BioRender program, available at https://www.biorender.com.
The immune system relies on neutrophils to ward off infections. During the primary phases of COVID-19 infection, neutrophils dispatch to the infected area helping in controlling viral replication and impeding the virus’s spread. During advanced severe cases of COVID-19, it is evident that the body’s immune defenses might become disrupted, leading to the cytokine storm (CS). As a result of this, high level of cytokines and other inflammatory molecules are released, which can cause harm to lungs and other organs. It is proposed that neutrophils contribute to the emergence of CS in COVID-19, as they can release pro-inflammatory molecules, causing tissue damage. Additionally, it is indicated that persons with COVID-19 in their severe stages show elevated levels of circulating neutrophils in comparison with less severe cases (36).
3 The significance of microRNAs and their biogenesis
microRNAs (miRNA), typically ranging from 20 to 24 nucleotides, represent a subset of non-coding RNAs that are instrumental in controlling gene expression post-transcriptionally. These small non-coding RNAs are critical for the modulation of gene activity (37). Current research has documented approximately 2,300 mature miRNAs in human beings, and an estimated half of these miRNAs are listed in the database named miRbase (38). While the majority of these miRNAs remain within cells, a category, referred to as circulating miRNAs, are measurable extracellularly in variable bodily fluids including plasma, cerebrospinal fluid (CSF), tears, and saliva. Circulating miRNAs, carried via extracellular vesicles in association with RNA-binding protein complexes, are featured by their outstanding resilience in extreme conditions, in addition to their robustness against degradation by native RNase. Therefore, they are proposed as promising biomarkers (39). miRNAs’ biogenesis is a sophisticated, multi-sites and steps pathway, beginning within the nucleus, and finalizing structures out in the cytoplasm. The pathway relies on diverse enzymes and protein structures sequentially operating together to yield an operational mature miRNA. Among several synthesis routes, Figure 4 illustrates the canonical pathway, which is being thoroughly investigated. The pathway begins with the transcription of DNA sequences. It is worth mentioning that approximately half of the miRNAs associated with humans, are encoded by introns of genes encoding proteins (40, 41). RNA polymerase (RNA pol) II or III carries out the transcription of DNA regions, supplying the primary miRNA (pri-miRNAs), shaped as long, hairpin, double stranded RNAs, equipped with a 5′ cap and a 3′ poly-A tail having a length of 70 nucleotides, pri-miRNAs have the capacity to provide up to six miRNA precursors. Initial processing of pri-miRNAs initiates in the nucleus, via an enzymatic complex encompassing of RNase III drosha and DiGeorge syndrome critical region (DGCR8) as the assisting protein. The latter complex adheres to pri-miRNA molecule, cleaving the cap and polyadenylated tail, shaping it as shorter precursor miRNA (pre-miRNA). Exporting the pre-miRNA is done in association with exportin-5 (XPO5) (39, 41, 42). Exported to the cytoplasmic area, dicer enzyme finalizes shaping the pre-miRNAs into their mature form by removing the unpaired bases from the pre-miRNA molecule, leaving it as a duplex with a 21 to 25 nucleotides in length. Phosphorylated 5′ end 3′ ends that each overhang by two nucleotides are featuring this mature structure. Mature miRNAs serve as integral components of the RNA-induced silencing complex (RISC), comprising the incorporation of argonaute protein, including AGO2. During the process of mi-RISC assembly, the double-stranded miRNA gets separated, passenger strand gets discarded, retaining the guider strand to guide the complex. Guider strand navigates the mi-RISC complex to the targeted mRNA, base pairing results in the expression repression of the gene, either by mRNA degradation, or restraining the translation stage (39, 41, 43, 44). The binding flexibility of miRNA molecules with various mRNA sequences is due to the small number of nucleotide bases involved in the miRNA-mRNA interaction. A lower extent of base pairing can diminish protein production capacity from the mRNA, whereas a more substantial base pairing can instigate the breakdown of the mRNA (39). Recent research reveals that host miRNAs can influence the infectious cycle of RNA viruses by binding to their genomes, thus affecting viral replication and pathogenesis. Additionally, viral infection may prompt shifts in host miRNA levels, altering gene expression in ways that could favor the virus or enhance the host’s antiviral response, potentially restraining viral spread (45).
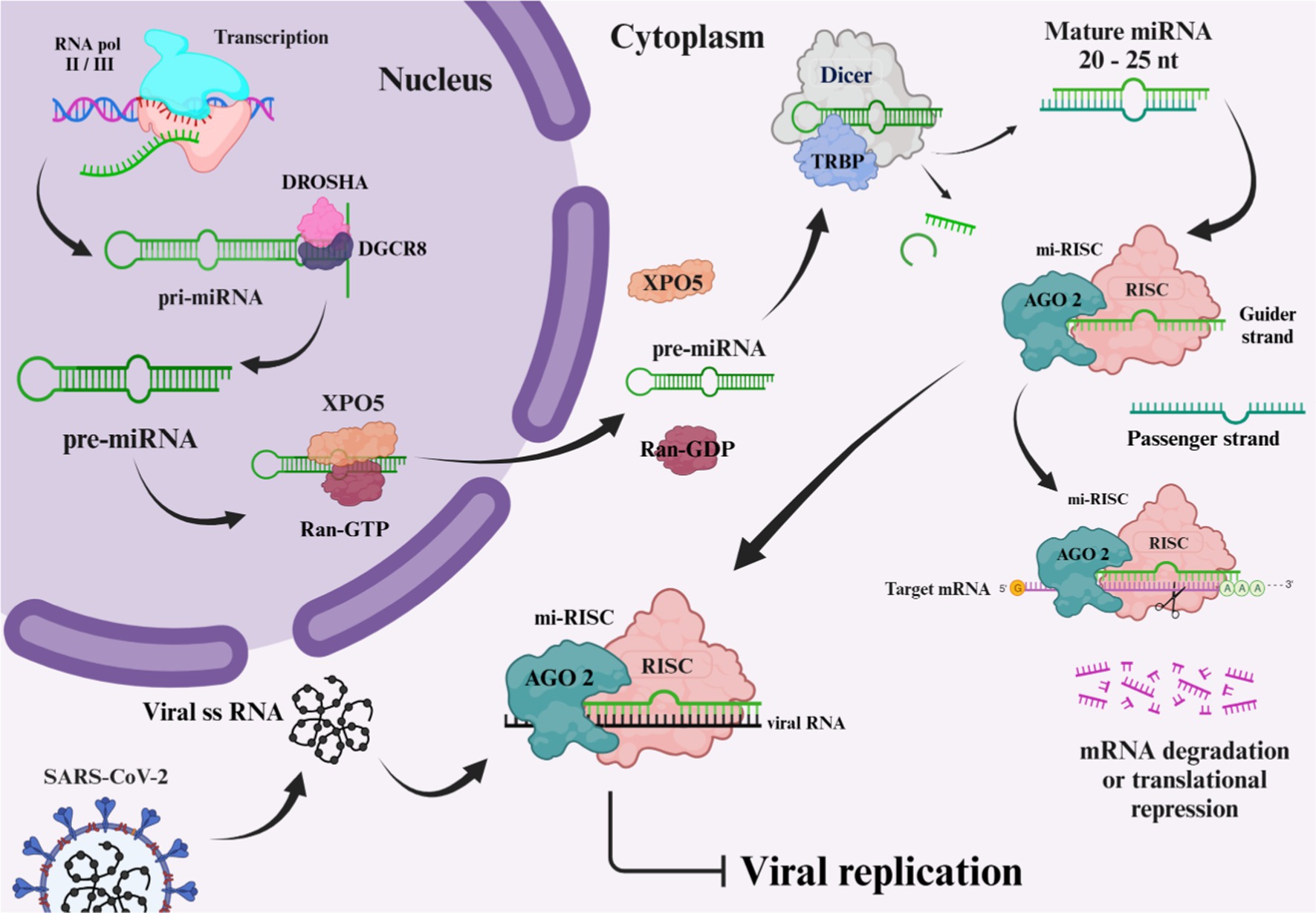
Figure 4. Biogenesis of microRNA. The image was created using the BioRender program, available at https://www.biorender.com.
3.1 Role of microRNAs in COVID-19
Cellular injury and the potential disruption of vital organs’ functionality, these include liver, lungs, brain, gut tissues, and kidneys, can be induced as the SARS-CoV-2 virus elicits an immunological and inflammatory responses (3). CS, a deep inflammatory response that follows up the primary infective stage, is initiated by the activation of inflammation-related genes including IL-6, IL-8, granulocyte colony-stimulating factor (G-CSF), and NF-κB proteins. CS initiates as the S proteins of SARS-CoV-2 bind to ACE2 receptors, provoking ACE/angiotensin II (Ang II)/angiotensin II type I receptor (AT1R) pathway giving rise to a heightened activation of NF-κB by IL-6/signal transducer along with activator of transcription (STATs) pathway (Figure 5) (46). CS can lead to multiple organ failures and may cause fatality (47). Host miRNAs are pivotal in the immune response during viral infections, as they fine-tune cytokines production. By enhancing or suppressing cytokine signaling, miRNAs can either boost beneficial immune functions or restrain detrimental inflammatory responses, thereby preserving the immune system’s equilibrium (48).
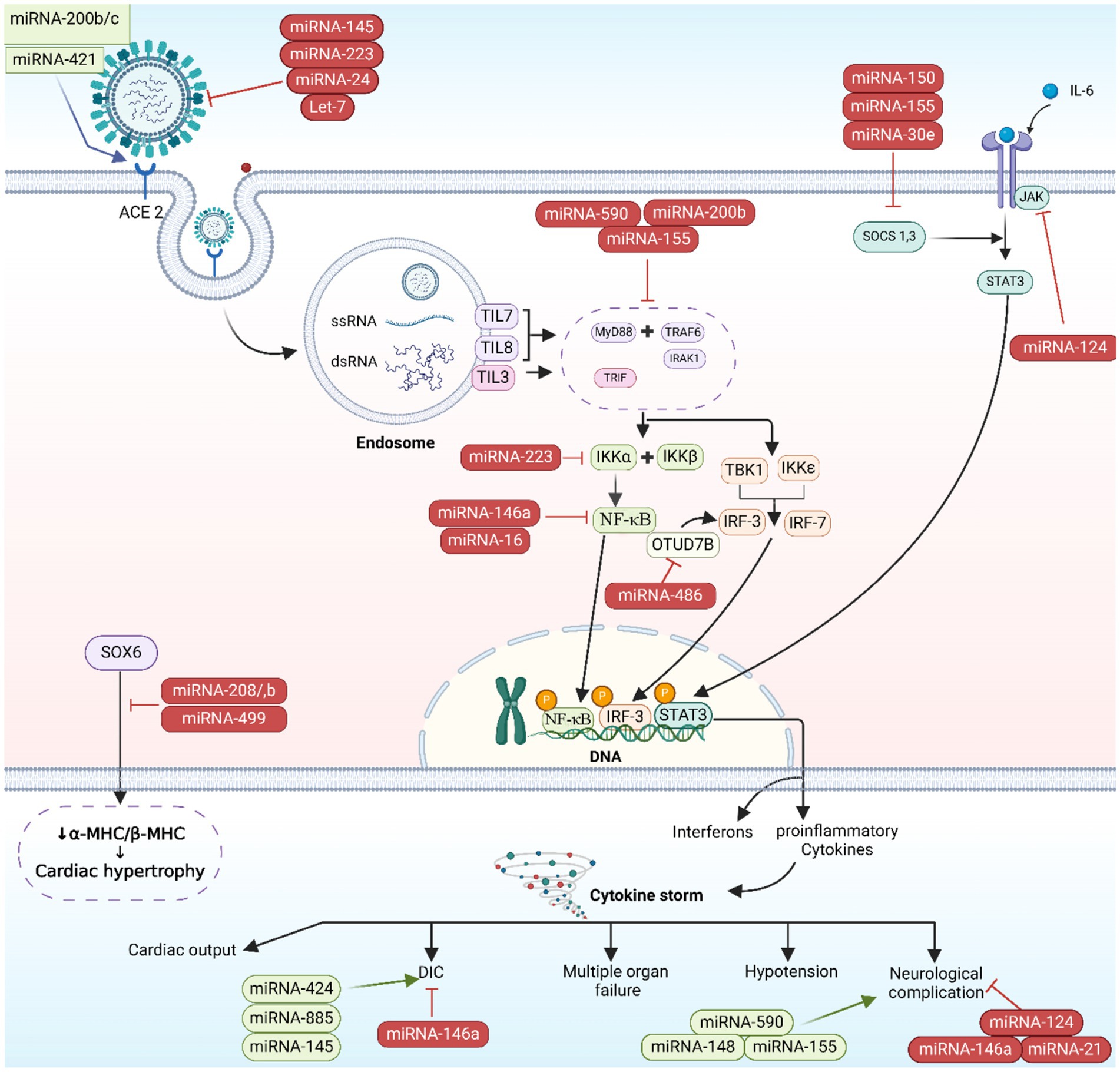
Figure 5. microRNA-induced immunopathogenesis. The image was generated utilizing the BioRender software, accessible at the website https://www.biorender.com.
3.1.1 The involvement of microRNAs in the immune response to COVID-19
In post-transcriptional gene expression, miRNAs play a vital role and affect several biological processes, such as inflammation and immune responses. Two types of miRNAs exist: pro-inflammatory and anti-inflammatory miRNAs. These miRNAs, miR-155, and miR-92a, have pro-inflammatory effects. TLR activation leads to the secretion of multiple transcription factors such as activator protein (AP-1), STATs, and (purine-rich box) PU.1, which lead to upregulate miR-155. Inhibition of SH2 containing inositol 5′ phosphatases 1 (SHIP1) and suppressor of cytokine signaling 1 (SOCS1) by miR-155 results in the release of IL-6 and IFNs, respectively (49). The suppression of SOCS1 by miR-155 leads to the activation of Janus kinase 2 (JAK2)/STAT1 and TLR/NF-κB signaling pathways of IFN-γ, resulting in macrophage 1 polarization (50). Upregulation of hypoxia-inducible factor-1 alpha (HIF-1α) and TNF-α, as well as downregulation of heme oxygenase-1 (HO-1), contribute to miR-155’s enhancement of lipopolysaccharide (LPS)-induced lung injury (51). Additionally, miR-92a triggers inflammation and lung damage by activating the phosphatase tensin homolog (PTEN)/serine/threonine protein kinase (Akt)/NF-κB signaling pathway. Therefore, inhibiting the PTEN/Akt/NF-κB signaling pathway through miR-155 antagonism helps alleviate lung injury (52). On the other hand, miR-146a and miR-124 exhibit anti-inflammatory properties. miR-146a blocks TLR, IL-1 receptor associated protein kinase 1 (IRAK1), and TNF receptor-associated factor 6 (TRAF6) to reduce inflammation, while miR-124 inhibits IL-6 and TNF-α release by targeting TNF-α converting enzyme (TACE) and STAT3 pathway (53).
The severity of COVID-19 is increased by the activation of NLPR3 inflammasome and the inhibition of vascular endothelial growth factor (VEGF), both of which are linked to miR-205-5p (54). The expression of NLPR3 inflammasome and inflammatory cytokine (IL-1β) can be decreased by miR-205-5p through the inhibition of B-cell lymphoma 6 (BCL6) (55). The study found that miR-205-5p blocks VEGF, a growth factor induced by the hypoxia-causing virus. Hypoxia leads to an increase in HIF-1 levels, causing higher VEGF levels and pulmonary edema. It is believed that miR-205-5p reduces lung edema by suppressing the HIF-1/VEGF pathway (56).
The expression of the miR-143-3p gene influences the number of neutrophils by affecting the expression of the BCL2 gene (57). miR-143-3p boosts neutrophil count through multiple mechanisms, including increasing the expression of cell adhesion molecules that facilitate neutrophil transmigration (58). The second mechanism involves miR-143-3p inhibiting BLC2, an anti-apoptotic protein. This particular miRNA hinders neutrophil apoptosis through this specific mechanism. By enhancing myeliod cell leukemia-1 (Mcl-1) levels and activating the phosphoinositide 3 kinase (PI3K)/Akt/STAT pathway, miR-143-3p promotes neutrophil survival and prolongs their lifespan (59).
The miR-19b-3p molecule attaches to specific regions of the SARS-CoV-2 virus, boosting the activity of CTLs (38). The reason behind increase activity of CTL by miR-19b-3p is due to upregulate of PTEN. In addition, PTEN also plays a significant part in CD4+ and CTL cell development. PTEN deficiency can lead to defects in the development of these T cell subsets, affecting their maturation and functionality. PTEN within the T cell lineage has diverse roles, including negative regulation of TCR and CD28 signaling, involvement in T cell development, influence on Treg development, and participation in immune checkpoint inhibitory signaling pathways. Understanding the multifaceted functions of PTEN in T cells is vital for unraveling the complexities of T cell biology and may have implications for therapeutic interventions in various immune-related disorders (60).
Decreasing the level of miR-155 in the serum severely impacts the ability of CTL (61) to respond to the virus, resulting in a reduced level of CTL and their ability to eliminate the virus (62). miR-155 plays a crucial role in the generation and function of CTL during viral infections. Further studies have shown that miR-155 regulates key pathways involved in CTL activation and effector functions. For example, it has been demonstrated that miR-155 targets SHIP-1, a negative regulator of T cell receptor signaling, leading to enhanced T cell activation and proliferation. Additionally, miR-155 has been shown to target SOCS1, a suppressor of cytokine signaling, resulting in increased production of pro-inflammatory cytokines by CTL (63).
3.1.2 Endogenous players confronting SARS-CoV-2 viruses
Human related antiviral cellular responses are enhanced by miRNAs, bringing about the disruption of vital pathways related to viral replication, these include PI3K/Akt and p38 mitogen-activated protein kinases (MAPK). miRNAs like hsa-miR-17-5p, hsa-miR-20b-5p, and hsa-miR-323a-5p have participated in SARS-CoV-2 proliferation inhibition in host cells (64). miR-146a, as an early-response miRNA to viral infections, it has the potentiality to both target genome of SARS-CoV-2 and regulate TLRs signaling in order to moderate inflammation, as it is known that TLRs are important in viral detection and subsequent innate immunity stimulation, enhancing the release of inflammation related cytokines such as IL-1, IL-6, TNF-α, and type I IFNs. The over-stimulation of immune responses is prevented through TLRs regulation (65, 66). miR-146a influencing cardiac regions, miR-4262 located in pulmonary cells, and miR-18 beside miR-125b found in renal tissues, are certain miRNAs identified as fundamental regulators of ACE2 genes expression (67). In a study, bioinformatics analysis results using RNA hybrid suggested that 125 miRNAs out of 240 correlated to aging have targeting ability of SARS-CoV-2’s genome, providing potential antiviral influence. Particularly, it is believed that miR-24-3p, miR-7-5p, miR-145-5p, and miR-223-3p have interaction with S proteins of SARS-CoV-2, as it is known that S proteins facilitate the invasion of host cells by the virus, also, these four miRNAs were low in level in old individuals, besides miR-223-3p, miR-24-3p, and miR-7-5p mostly lowered in diabetic persons. Furthermore, both circulatory exosomal miRNAs and synthetic ones were indicated to suppress the S proteins, thus potentially hindering SARS-CoV-2 replication (68). Table 1 further depicts native miRNAs’ influence during COVID-19 infection.
3.1.2.1 Influence of native microRNAs on COVID-19 severity
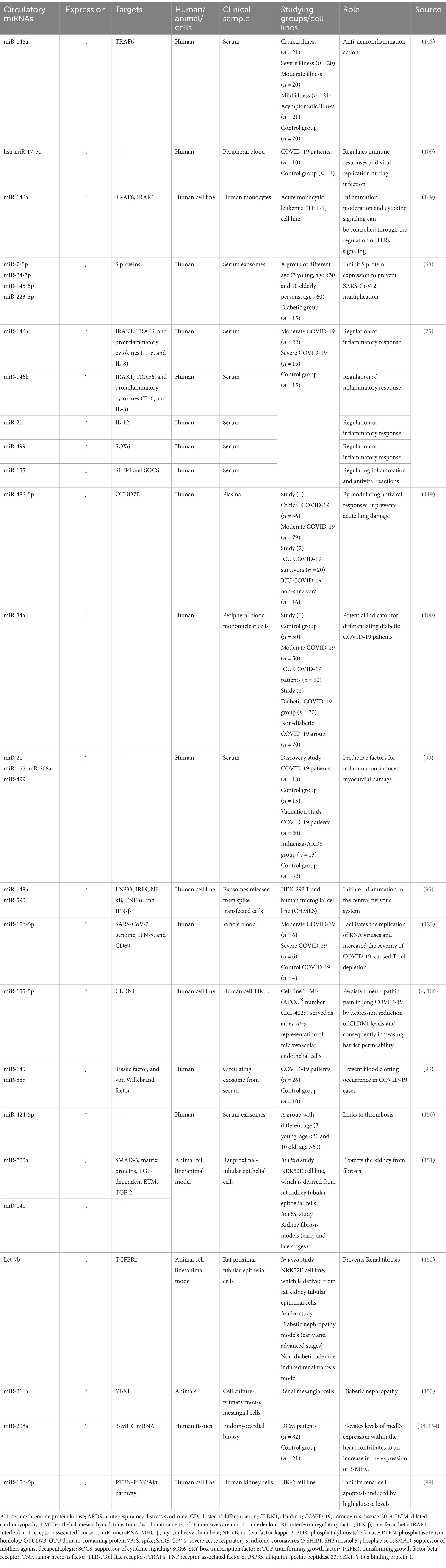
Circulatory miRNAs, ubiquitously circulate within nearly all bodily fluids, possessing the ability to serve as highly sensitive biomarkers to precisely reflect the physiological and pathological conditions of the body. Encased within extracellular vesicles, circulatory miRNAs are implicated in facilitating communication between immune cells and may offer valuable insight into disease mechanisms as biomarkers (69, 70). The extracellular vesicles are lipid bilayer-bound particles released by various cell types, including immune cells. These extracellular vesicles are taken up by recipient cells through endocytosis or fusion with the plasma membrane, leading to the delivery of miRNAs into the cytoplasm of the recipient cells. For example, miR-21 and miR-155 are known to be carried by exosomes derived from dendritic cells and can modulate the activity of T cells, influencing their differentiation and function (71, 72). By influencing adhesion molecule levels, miRNAs affect the efficiency of immune cell migration to the inflammatory sites. miRNAs can regulate the expression of adhesion molecules, which allow immune cells to attach to blood vessel walls and migrate into tissues (73). miR-146a has been shown to target TRAF6, which may further down-regulate the IL-17/intercellular adhesion molecule 1 (ICAM-1) pathway. Thus, miR-146a inhibit the expression of ICAM-1, an adhesion molecule, thereby reducing the migration of monocytes to inflammatory sites (74).
Host’s miRNAs play a significant role in managing cytokines activities in the inflammatory phase of COVID-19. In a study, miRNAs in hematologic specimens taken from COVID-19 hospitalized cases were analyzed and compared to non-infected individuals to evaluate their potential as a mortality prediction biomarker. Inflammatory gene regulation was shown to be influenced by specific miRNAs: Cytokine gene expression is managed when miR-146 targets TRAF6 and IRAK1, and modulates the pro-inflammatory cytokines (IL-6 and IL-8) levels. miR-21, miR-499, and miR-155 were shown to regulate IL-12, target SRY box 6 (SOX6) gene to influence sepsis-related lung deterioration, and down-regulate SHIP1 and SOCS1, respectively. SHIP1 and SOCS1 are known to serve as inhibitors of inflammatory reactions in macrophages (Figure 5). The same study concluded that miR-155 can be used to forecast survival and prognosis in patients after contracting SARS-CoV-2 (75). The relationship between miR-21, miR-499, miR-155, and IL-12 targeting SOX6 is complex and not completely understood. It is possible that the influence of these miRNAs on IL-12 production is indirect, occurring via their targeting of SOX6 (76). The transcription factor SOX6 can impact the expression of inflammation-related genes like IL-12 (77–79). In addition, miR-155 indirectly regulate IL-12 production by downregulating SHIP-1 as miR-155 has recently been shown to suppress SHIP-1 expression by binding to its 3′ untranslated region (80, 81). When SHIP-1 is not present, macrophages produce higher levels of IL-12 (82). Moreover, the miRNAs regulate IL-12 by targeting SOCS1 (75). Infections efficiently stimulate SOCS1 expression in macrophages. The reported inhibition of STAT mediated responses by SOCS1 is responsible for the regulation of IL-12 expression in macrophages. The biological function of numerous cytokines is mediated by STAT, and SOCS1 plays a regulatory role in IL-12 (50, 83) and eventually the miRNAs exert their effect on IL-12 by SOCS (38, 84). SHIP-1 has been found to act as a suppressor of hematopoietic transformation by blocking the PI3K/Akt signaling pathway. SHIP1 is a target of miR-155 in which miR-155 decrease the expression of SHIP-1 and it is known that SHIP-1 is a suppressor of hematopoietic transformation as its activity can inhibit the PI3K/Akt signaling pathway in acute myeloid leukemia and myelodysplastic syndromes. Therefore, these findings indicate that miR-155/SHIP-1/Akt pathways may be valuable clinical biomarkers for disease progression, even in COVID-19 (85, 86).
3.1.2.1.1 The involvement of microRNA in pulmonary injury
Oxidative stress arising when an excess of reactive oxygen species (ROS) overwhelm the body’s antioxidative mechanisms, resulting in cellular damage and potential tissue injury (87), acute lung injury and other pulmonary illnesses like chronic obstructive pulmonary disease are also caused by oxidative stress, beside its negative impact on cell communication. Research on human lung cell exosomes has shown that low oxygen levels cause a significant reduction in miR-3691-3p. This miRNA is vital for pathways associated with fibroblast growth factor 2 (FGF2), transforming growth factor-beta (TGF-β), and vascular cell adhesion molecule 1 (VCAM1) protein, that have importance in lung damage and repair (88). miR-146a and miR-486-5p level up IL-1β, IL-6, and TNF-α concentrations via the reduction of OTU domain-containing protein 7B (OTUD7B) during lung damage, beside stimulating NF-κB pathways, in which the latter leads to an elevated pulmonary tissue inflammation. Conversely, the overall inflammatory response is dampened, and the severity of acute lung injury is mitigated when PI3K, TNF-α, and IL-6 are downregulated by miR-16-5p (Figures 5, 6) (38).
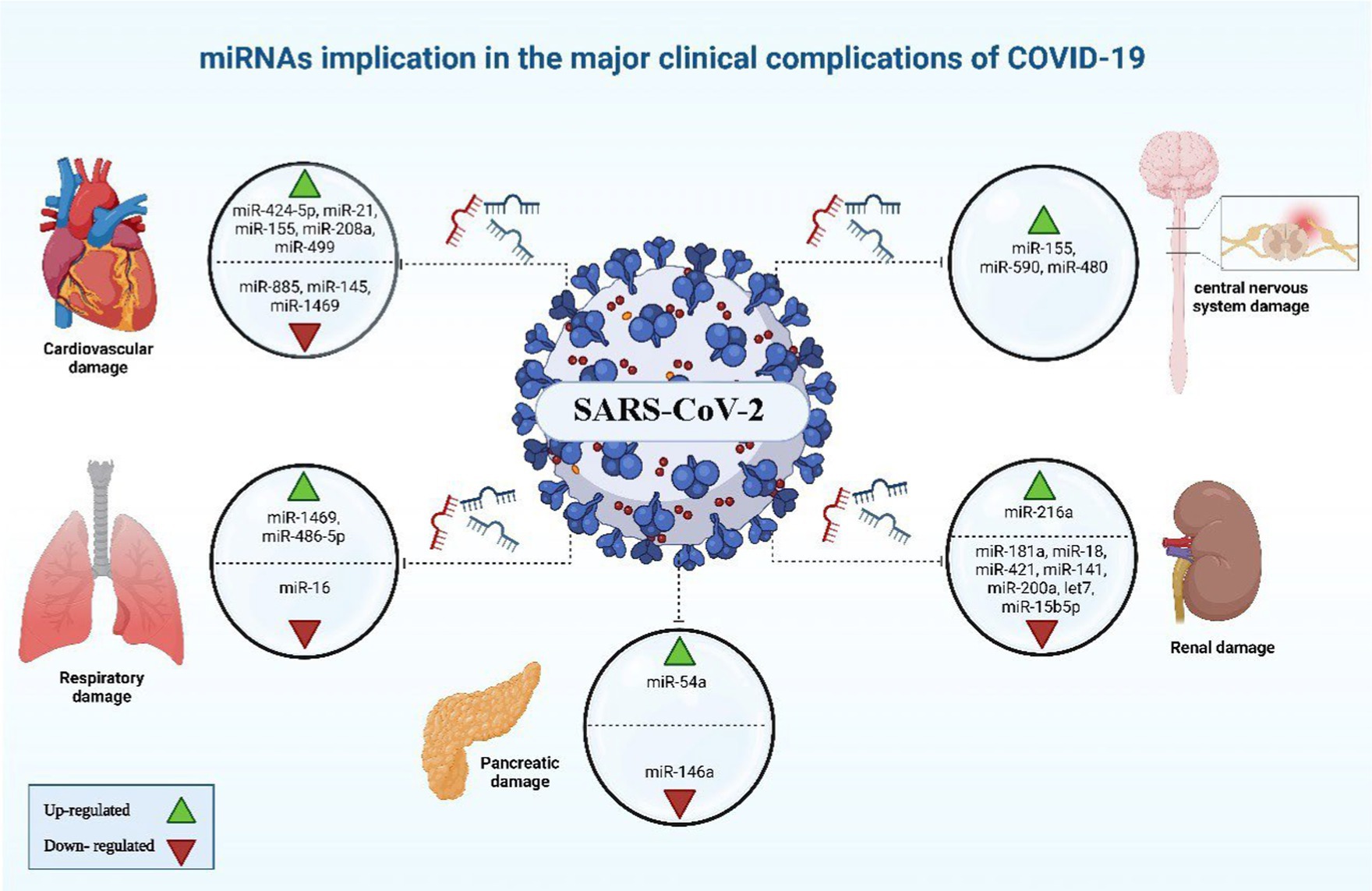
Figure 6. miRNAs implicated in the major clinical complications of COVID-19. The image was generated utilizing the BioRender software, accessible at the website https://www.biorender.com.
3.1.2.1.2 microRNA and its implications in cardiovascular problems and thrombosis
Certain patients infected with COVID-19 have been observed to develop cardiovascular complications (89). It was noted that COVID-19 cases with cardiovascular comorbidity have had elevated expression concentrations of miR-155, miR-499, miR-21, and miR-208a, acting like predictive factors for inflammation-induced myocardial damage (90). Furthermore, miR-208a demonstrated associations with arrhythmia, cardiac hypertrophy, and dilated cardiomyopathy. The mechanism underlying miR-208a-induced cardiac hypertrophy involves two pathways. Firstly, it induces the overexpression of medI3, leading to an increase in beta-myosin heavy chain (β-MHC) expression, a known hallmark of cardiac hypertrophy. Secondly, it enhances the expression of miR-499, which also contributes to the increase in β-MHC (Figures 5, 6) (91).
Alterations were observed in blood clotting indicators during the initial phase of COVID-19 pandemic, after measuring D-dimer, fibrinogen, and activated partial thromboplastin time (aPTT) of hospitalized patients by healthcare workers (92). Endothelial cells are pivotal in initiation and regulation of the thrombotic process, as they produce and release Von Willebrand factor and tissue factor. Impairment to the endothelium’s regular functionality as SARS-CoV-2 infects the body was hypothesized by researches, leading to system’s dis-balance. As a consequence, the synthesis and release of miR-885 and miR-145 by the endothelial cells is reduced, potentially leading to unregulated blood clots formation. Conclusions showed that there was a connection between the increased probability of thrombosis and mortality of COVID-19 cases with the low levels of exosomal miR-145, and miR-885 (93). Furthermore, the insufficiency of miR-146a and the elevation of miR-424-5p have also been linked to thrombosis in COVID-19 and could be considered as prognostic indicators for thrombotic conditions in COVID-19 (38).
3.1.2.1.3 microRNA’s influence on COVID-19 neurological outcomes
The COVID-19 infection is notorious as a multi-organs illness, notably the central nervous system, persisting over a prolonged period, this situation is especially troubling for the elderly, who are more vulnerable to intense illness and are at higher risk of neurological conditions, regardless of the viral infection (67). miRNAs are prevalent throughout the central nervous system and exhibit unique patterns of expression based on location and time. Therefore, alternations to their typical expression levels may contribute to the development of neurological conditions (75). In neurological complications involving Alzheimer’s disease, down syndrome-related dementia, and multiple sclerosis, miR-155’s high concentrations contribute to inflammation. This type of miRNA mentioned earlier is found in association with high COVID-19 severity when found in elevated levels. It disrupts the blood-brain barrier, activates T cells, and fosters beta-amyloid plaque accumulation (67). Conversely, research indicated a decrease in many blood miRNAs during COVID-19 and different neurological issues. miR-21 as a common factor which plays its role in inflammatory reactions within the nervous system, regulation of normal cell death, in addition to its participation in glutamate-induced toxicity, activation of astrocytes, disruption in synapse function, increased activity of microglia, and restoration of myelin sheaths (94). Thus, miR-21 is critical in the progression and onset of diseases affecting the central nervous system. Hence, low levels of miR-21 lead to an amplification in the inflammatory response in COVID-19 pathogenesis due to its role as regulator of inflammation-related genes, this causes worsening of the disease’s state as well as influences neuro-inflammatory mechanisms (67). miR-148a specifically targets ubiquitin specific protein 33 (USP33), while miR-590 targets interferon regulatory factor 9 (IRF9). This targeting activates NF-κB, resulting in the overexpression of TNF-α and IFN-α, leading to severe neuro-inflammation (38). There was an observed elevation in miR-590 and miR-148a levels in cases with COVID-19 associated with neuro-inflammation (Figures 5, 6) (95).
3.1.2.1.4 The impact of microRNA on renal diseases in COVID-19 context
Nephrons are found to express ACE2 receptors, thus, as SARS-CoV-2 cause renal impairment, it could be attributed to sepsis stemmed from cytokine storm in ill persons with severe stages of COVID-19, bringing about dehydration and could also reduce glomerular filtration rate (GFR). A correlation has been identified between SARS-CoV-2 infection, podocyte damage, and proteinuria (96). An increase in miR-216a, as well as missing let-7, miR-141, and miR-200a, serve as indication factors of kidney fibrosis during COVID-19, due to their potential in TGF-β expression activation (97). miR-181a, miR-18, and miR-421 have been linked to renal injury and the regulation of ACE2 expression (98). Diabetic nephropathy patients have a deficiency in miR-15b-5p, which is important for inhibiting apoptosis in renal cells which is provoked by the elevated levels of glucose via control of PTEN-PI3K/Akt pathway (Figures 5, 6) (99).
3.1.2.1.5 The involvement of microRNA in metabolic syndromes linked to COVID-19
Individuals with conditions such as obesity, and diabetes commonly display diminished levels of a crucial regulator of inflammatory process which is miR-146a. miR-146a expression is induced in response to inflammation instigated by NF-κB, inhibiting TNF-α and IL-1 receptor activity to dampen the inflammatory response. Deficiency in miR-146 linked to diabetes leads to elevated inflammation from improper IRAK1/TRAF6 regulation (Figures 5, 6). SARS-CoV-2 can worsen COVID-19 severity by impairing miR-146a, leading to poor antiviral defenses, excess cytokine release, and uncontrolled inflammation-related tissue damage (66). On the other hand, the study by Khatami et al. (100) revealed an upregulation of miR-34a in COVID-19 patients with diabetes. Additionally, there was an association observed between the level of miR-34a and HbA1c (101).
3.1.2.2 microRNAs in long COVID
Through systematic analysis of research data, it has been concluded that the predominant symptoms manifested by COVID-19 patients are as follows: fever, cough, fatigue, dyspnea, and sputum production (102). Headache, migraines, joint discomfort, muscle pain, and neuropathic pain sensation (4). When the pandemic began, prolonged COVID-19 symptoms were often mistaken for post-traumatic stress resulting from the disease’s novelty, lack of treatment, and associated social isolation, delaying the identification and naming of long COVID (103). Post-acute sequelae of SARS-CoV-2 or Long-haul COVID-19 as other terminologies for long COVID (104), a condition in which individuals face ongoing health complications persisting well after recovery from initial COVID-19 infection, far exceeding the typical recovery timeframe (105). Current research has identified long COVID as a separate condition with diverse symptoms, including respiratory problems like breathlessness and chest pain, neurological challenges like headaches and fatigue, and other issues such as hair loss, taste loss, and hearing impairment (4). Research explains that reduced concentrations of the tight junction protein claudin-1 (CLDN1), which are critical for preserving the integrity of the blood-brain and blood-nerve barriers, have been correlated with the manifestation of neuropathic pain. Animal studies and preliminary human data indicate a correlation between decreased claudin-1 and neuropathic discomfort. In case of long COVID, the elevation of miR-155-5p, a known miRNA for its action in claudin-1 suppression, could lead eventually to persistent neuropathic pain by expression reduction of claudin-1 levels and consequently increasing barrier permeability (4, 106).
3.1.3 Viral microRNAs as exogenous participants
Mirroring the structural and functional features of human miRNAs, SARS-CoV-2 genome has been found to encode virus’s own miRNAs which exhibit capability to change the host’s immune mechanism and manage inflammatory responses as the infection progresses (47). Host miRNAs that manage crucial processes such as kidney and heart development, metabolism, neuronal functions, insulin resistance are compromised by viral infections. These miRNAs’ reduction in cases with COVID-19 disrupts such essential bodily processes, with the impact strengthened in individuals with underlying conditions (3). Research shows that miRNAs related to SARS-CoV-2 can hinder protein synthesis by attaching to human mRNA, affecting key proteins such as hemoglobin subunits (beta and gamma-globin2), type 1 interferons, and olfaction proteins. This may lead to the oxygen transport problems, immune changes, and olfactory dysfunction observed in COVID-19 patients, due to viral miRNA-like effects (107). The insulin signaling pathway is hypothesized to be susceptible to modulation by virally encoded miRNAs, leading to potential vulnerability exacerbation to SARS-CoV-2 infection of persons with comorbidities, especially diabetes. This suggests that viral miRNAs could play a role in epigenetic control and may interact with the host organism’s native miRNAs (3).
4 The role of microRNAs: implications for diagnosis
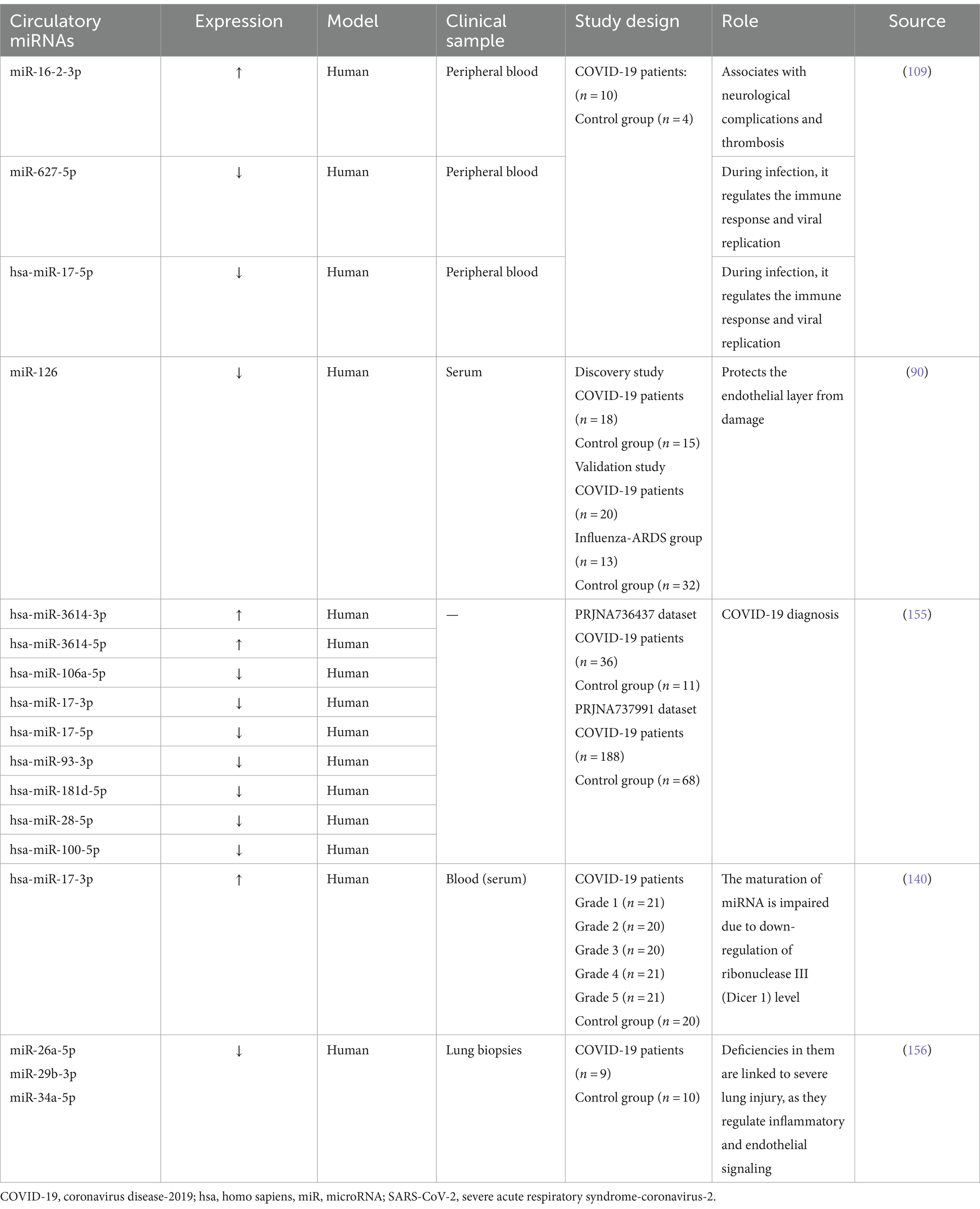
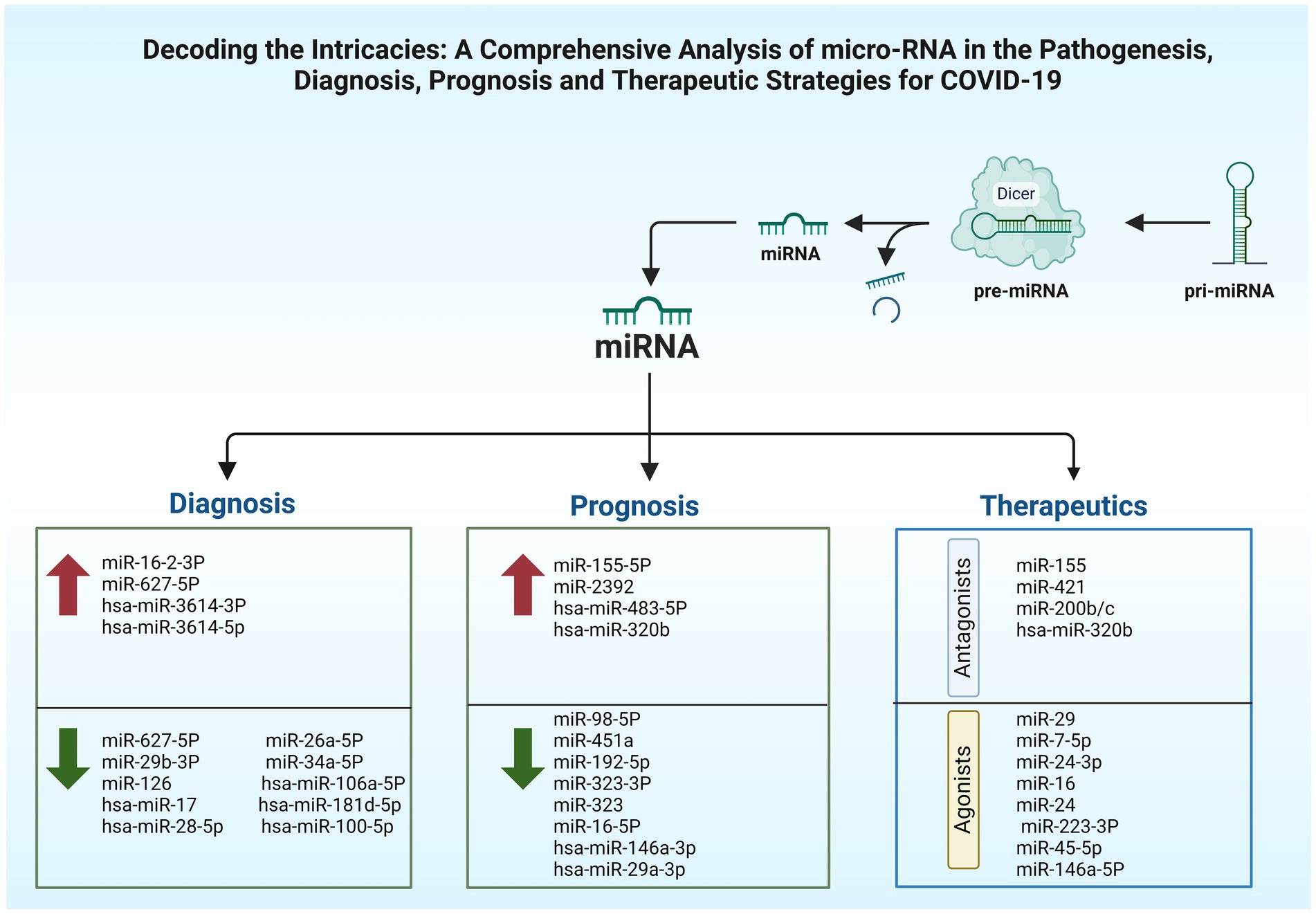
The early identification, immediate patients’ isolation, quick response to worsening health, and proper oxygen supplements along with good nutrition can notably reduce the risks of COVID-19 mortality. A standard test used to detect the virus is the nasopharyngeal swab polymerase chain reaction (PCR), posing risks to those dealing with specimens due to potential viral exposure. Thus, blood analysis for the detection of circulating miRNAs offers a safer diagnostic alternative (64). Certain circulatory miRNAs, like those found circulating in urine, tears, serum, and plasma have shown resistance against enzymatic degradation, which exhibit unique patterns associated with specific diseases. They are offered as potential non-invasive biomarkers for conditions like COVID-19 and can be detected by liquid biopsy (38, 64). Studies suggest that miRNAs can be affected by this viral infection, offering promise for their use in many aspects as shown in Table 2, such as the detection of the disease (108). In a study, the peripheral blood of cases with COVID-19 beside healthy individuals underwent high-throughput sequencing. Results demonstrated upregulation of 35 miRNAs while the rest 38 were showing downregulation. miR-16-2-3p stood out with an upregulation showing a 1.6 times elevation in comparison to healthy persons, whereas miR-627-5p showed most reduction (109). In the cases of COVID-19, research depicted alterations in nine miRNA types including upregulated hsa-miR-3614-5p and hsa-miR-3614-3p, while the downregulated ones involved hsa-miR-17-5p, hsa-miR-93-3p, hsa-miR-28-5p, hsa-miR-106a-5p, hsa-miR-17-3p, hsa-miR-100-5p, and hsa-miR-181d-5p, these changes in expression levels are proposed as reliable biomarkers to separate COVID-19 cases from healthy members (110). In comparison to healthy individuals, research findings informed a reduction in miR-126 levels in COVID-19 patients (90).
miRNAs play a crucial role in the diagnosis of COVID-19. These small non-coding RNA molecules are involved in the regulation of gene expression and have been found to be dysregulated in various diseases, including viral infections. In the case of COVID-19, studies have shown that specific miRNAs are differentially expressed in infected individuals compared to healthy controls. These miRNAs can be detected and quantified using techniques such as reverse transcription polymerase chain reaction (RT-PCR) or next-generation sequencing (NGS). By analyzing the expression levels of specific miRNAs, healthcare professionals can potentially identify individuals who are infected with COVID-19, even in the absence of symptoms. Furthermore, miRNA-based diagnostic tests have the potential to provide rapid and accurate results, making them valuable tools in the fight against the pandemic.
5 The role of microRNAs: implications for prognosis
Diversified biomarkers were tested in order to ascertain their prognostic value in prediction of COVID-19 severity, among those, laboratory ones included ILs, D-dimer, and CRP, while genetic biomarkers involved TMPRSS2, SNPs in ACE2, TNF-α, and IFN-γ (111–114). The utilization of molecular techniques relying on miRNAs has the potential to enhance risk evaluation and offer a direct strategy for informing clinical judgments regarding patient care, monitoring, and treatment.
In the light of the pandemic, it is of utmost importance to define biomarkers capable of forecasting the severity of the illness, thereby mitigating SARS-CoV-2-induced fatalities. Taking into account the fact that circulating miRNAs can act as non-invasive markers for viral diseases. Establishing a predictor of disease severity is of utmost importance, allowing for proper stratification of patients based on disease severity and progression. Therefore, employing miRNAs as prognostic markers for early mortality in severe COVID-19 patients may greatly influence therapeutic strategies and patient care. This enables early stratification and personalized intervention (Table 3).
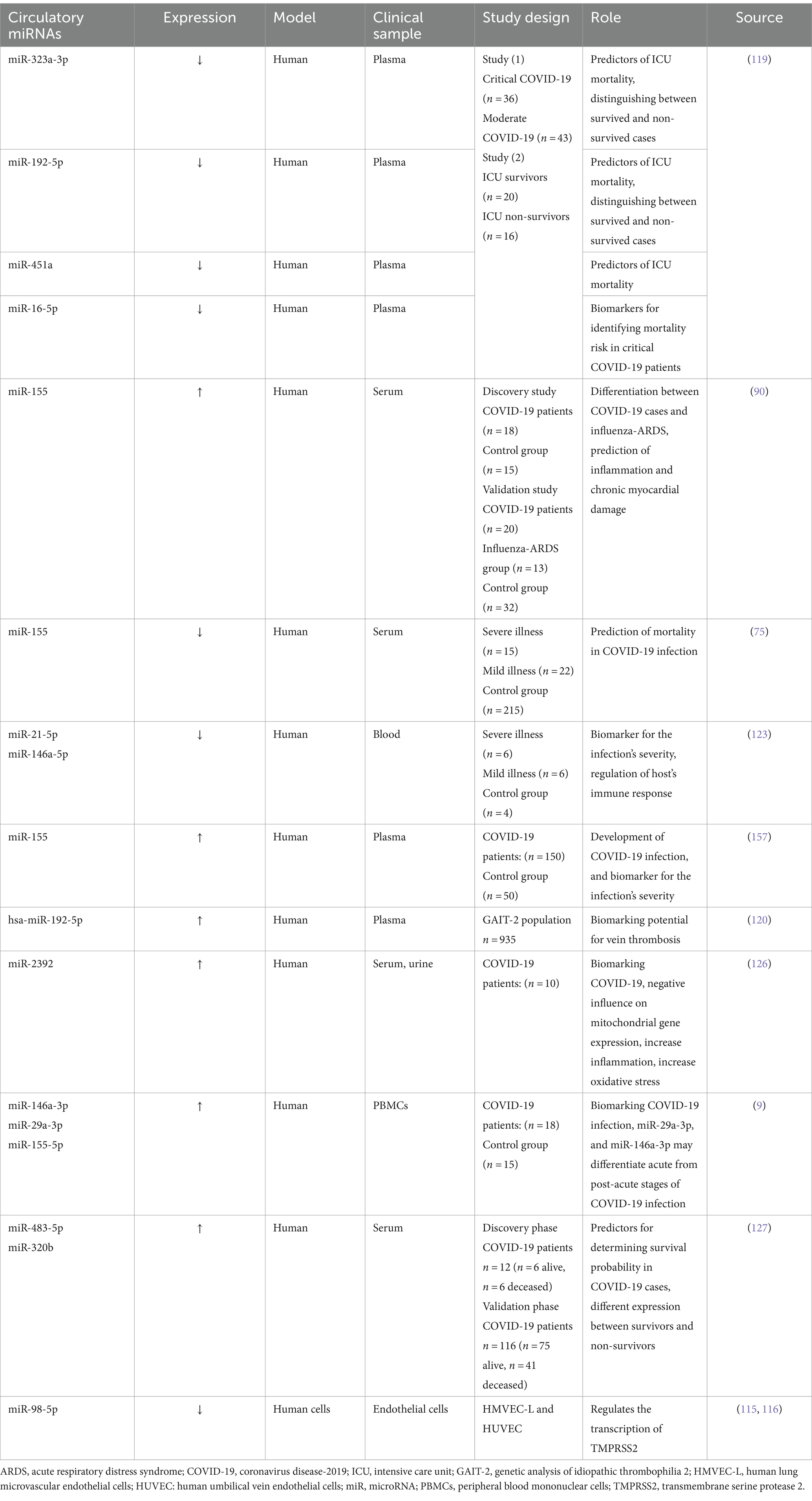
Intensity of COVID-19 infection is negatively correlated to miR-98-5p (115). miR-98-5p exhibits antiviral properties by binding to the viral genome, thereby inhibiting its replication (116). Moreover, intensely affected cases of SARS-CoV-2 infection depicted reduced patterns of miR-451a, and miR-323-3p (117). The probability of COVID-19 related CS occurrence is positively associated with the downregulation of miR-451a hypothesis suggested the interaction between miR-323-3p and the SARS-CoV-2 viral genome, as it was previously proposed to have binding ability to influenza gene exhibiting antiviral effects (118).
Based on the study performed by de Gonzalo-Calvo et al. (119), two miRNAs namely miR-192-5p and miR-323-3p were proposed to serve as prognostic biomarkers for the prediction of mortality caused by COVID-19 infection in intensive care unit (ICU), distinguishing non-survivors from those who survived. ROC analysis was taken as a support for this conclusion. Moreover, the study observed a negative relationship between period of ICU stay and expression levels of miR-323-3p and miR-192-5p. A significant connection has been established between a diminished level of miR-192-5p and the development of deep vein thrombosis (120).
The research entailed cooperation with 19 ICUs situated in diverse regions of Spain that are associated with the CIBERESUCICOVID initiative. A total of 503 plasma samples were procured from the patient population. After quantifying circulating miRNAs in plasma, the researchers established a signature composed of four miRNAs. The implementation of this signature facilitated the classification of patients according to their risk of mortality within 48 h of their admission to the ICU. The predictors of ICU mortality among COVID-19 have been confirmed to be miR-192-5p, miR-323-3p, miR-16-5p, and miR-451a, as determined through Kaplan–Meier survival analysis and LASSO regression methods (121). A study indicated that miRNAs found in macrophages had decreased levels subsequent to the exposure to SARS-CoV-2’s S proteins (122).
In a study, two potential biomarkers for determining COVID-19 intensity have been recognized, namely miR-21-5p and miR-146a-5p (123). Further research on severe cases revealed variations in cardiovascular and inflammatory miRNAs levels. Following the infection with influenza A virus, the exacerbated acute respiratory distress syndrome (ARDS) related lung damage was found to be due to an increased concentrations of miR-155-5p. It is suggested that miR-155 could be used as a potential therapeutic target given that its removal can mitigate pulmonary damage, particularly in COVID-19 illness (124). An additional beneficial attribute of miRNAs is their ability to serve as predictive biomarkers for tracking patient’s disease progression and to categorize patients, enabling health practitioners to offer more accurate and customized therapies (64). As for differentiating between severe from mild COVID-19 cases, research demonstrated that key clinical markers like C-reactive protein (CRP) and D-dimer have been outperformed by SARS-CoV-2-encoded miRNA called miR-nsp3-3p, additionally, its ability in distinguishing cases at higher risk even before the appearance of severe signs, rendered it as a prognostic indicator. This biomarker can be easily incorporated in COVID-19 diagnostic strategies due to the simplified measuring techniques (125). A study attempted to show how miR-2392 is involved in making it easier to become infected with SARS-CoV-2, it was concluded that this miRNA type negatively impacts the mitochondrial gene expression, beside its influence on inflammatory responses and glycolytic activity, relating to the poor outcomes and COVID-19’s symptoms, miR-2392 is found circulating in fluids like blood and urine of infected persons, it is shown to have an antiviral effect when reduced, as it influences severity of COVID-19 and how the disease progress (126). Distinguishing acute stage from the subsequent post-acute one as the COVID-19 progress was found to be possible by the utilization of hsa-miR-29a-3p and hsa-miR-146a-3p (9). Heightened concentrations of miR-320b and miR-483-5p are positively linked to higher risk of mortality, these two miRNAs can serve in determination of survivability in COVID-19 cases, acting like crucial predictors. The research investigating their role, indicated that patients with the highest concentrations of these miRNAs were three times more likely to die during hospitalization (127). These findings offer potential for improving patient care and creating specialized treatments.
The role of miRNAs in prognosis is of utmost importance as they have been implicated in various diseases, including cancer, cardiovascular diseases, and viral infection. By modulating gene expression, miRNAs can have profound effects on cellular processes such as cell proliferation, differentiation, and apoptosis. Furthermore, dysregulation of miRNA expression has been associated with disease progression and poor prognosis in many cases. Therefore, understanding the prognostic implications of miRNAs can provide valuable insights into disease progression, patient outcomes, and potentially guide treatment decisions for improved personalized medicine approaches.
6 Implications of microRNAs for therapeutics
In addition to the utilization of vaccines and repurposed drugs for COVID-19 prevention and treatment, scientists are also exploring novel therapeutics like miRNA (128). The growth of miRNA-derived drugs is rapidly occurring and has the potential to revolutionize standard treatments for various diseases, including COVID-19. They offer distinct benefits when compared to traditional small molecule and biologic medicines. These include cost-effectiveness, ease of manufacture, and the ability to target previously unresponsive sites. Through the implementation of chemical modifications and RNA nanocarriers, it is possible to alleviate or minimize the typical challenges associated with stability, delivery, and undesired effects.
Variations in miRNAs profiles are indicative of COVID-19 severity, with notable implications in patients with comorbidities, suggesting that targeting such altered non-coding RNAs can attenuate the complications (98). Manipulation of miRNAs, like those mentioned in Table 4, might attenuate COVID-19’s adverse effects by the inhibition of chemokines and cytokines release, potentially leading to more favorable outcomes. miRNAs could be an effective treatment strategy among the other nucleic acid approaches for addressing COVID-19 (129). Hindering viral attachment offers an effective early strategy to combat COVID-19, by targeting miRNAs regulating proteins such as ACE2 and TMPRSS2 important for viral entry into host cells (98). A therapeutic strategy that harnesses antiviral beside anti-inflammatory miRNAs could be more efficacious than a single-agent therapies, targeting miRNAs, essential proteins, or the viral genome offering a hopeful strategy in the development of disease treatments (98).
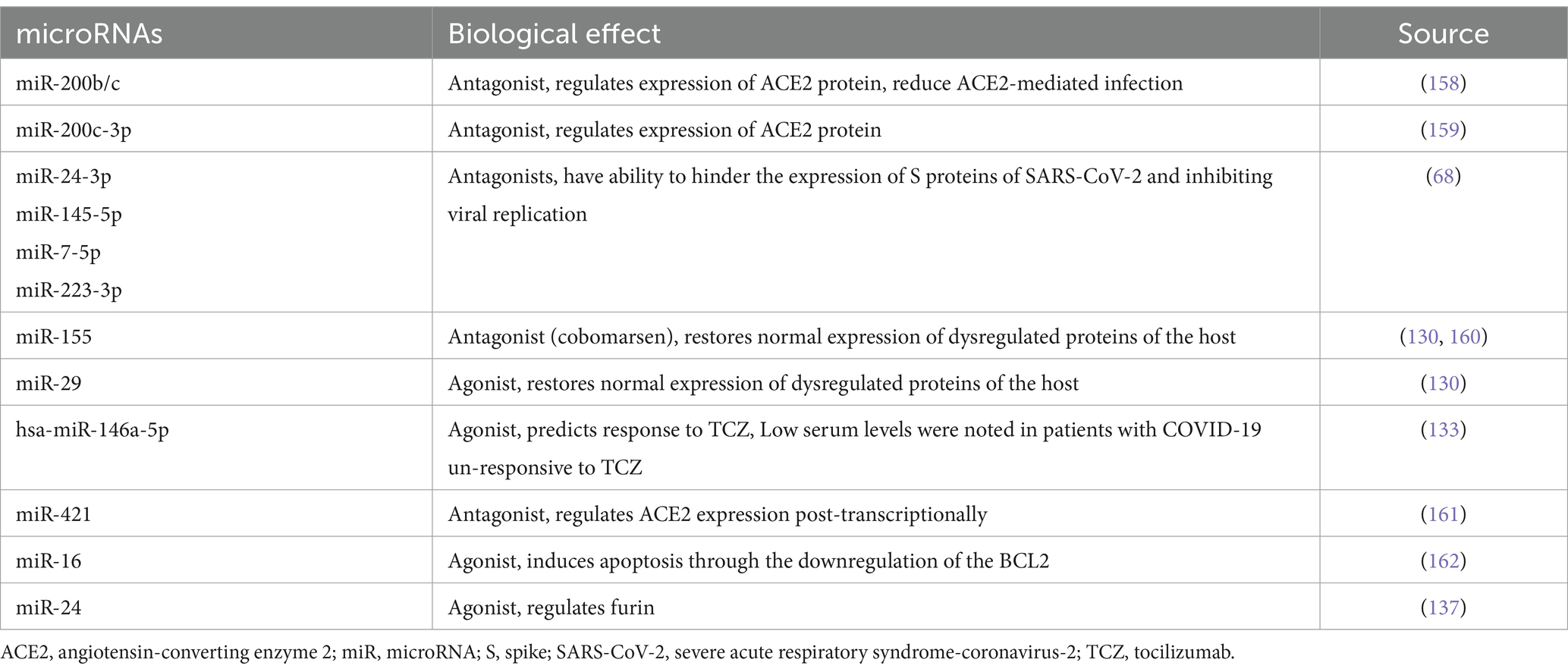
Therapeutic strategies have been emerging lately, in which miRNAs (antagonists or inhibitors) are used to target host’s miRNAs, reducing the negative impact of such native miRNAs. miR-155 inhibitor called cobomarsen and miR-29 mimic with the name remlarsen have the potentiality to restore normal expression of dysregulated proteins of the host. Thus, anti-miRs and miR mimics may serve as therapeutic options for managing COVID-19 disease (130). Extracellular vesicles (EVs) are investigated as a promising therapeutic approach due to their cell-free nature, being immunologically safe, and lack exogenous substances (131). Cells release membrane-bound structures called EVs, which encapsulate genetic components such as miRNAs, DNA, and long non-coding RNA (lncRNA), as well as other biologically active molecules derived from the parental cells (132). Two analytical methods, quantitative real-time PCR along with high-throughput sequencing indicated four miRNA types miR-24-3p, miR-223-3p, miR-145-5p, and miR-7-5p that demonstrated their ability in impeding SARS-CoV-2’s S proteins and the inhibition of viral replication either in the free or exosomal form as observed in serum of young persons, these miRNAs showed a significant reduced pattern in the elderly and those with diabetes (68). While further investigating the potentiality of miRNAs as biomarkers, it was found that specific miRNAs found within the circulatory system can serve as predictive indicators for a patient’s response to medicines. COVID-19 patients who did not respond to tocilizumab (TCZ) exhibited diminished serum levels of hsa-miR-146a-5p (133).
It is of interest to mention that miR-421 has been implicated in the regulation of ACE2, highlighting its relevance in therapeutic investigations for SARS-CoV-2 (134). miR-16 possesses additional therapeutic potential due to its role in inducing apoptosis through the downregulation of the BCL2 (135). Apoptosis plays an important role in the initial antiviral defense by prompting the clearance of virally infected cells. For instance, in the context of influenza infection in mouse cells, the induced apoptotic response facilitates the removal of infected cells by phagocytic action of phagocytes as macrophages and neutrophils. However, in the case of SARS-CoV-2, the virus can evade this innate immune pathway by downregulating miR-16 leading to the inhibition of the process of apoptosis in host cells. Moreover, it was reported that inhibition of miR-16 leads to elevated production of mitochondrial ROS as well as upregulation of TLR4 expression (136). Thus, the possibility of upregulating miR-16, potentially through the use of a miRNA mimic, emerges as a viable strategy to counteract the deleterious manifestations of COVID-19 (137).
Severe COVID-19 cases exhibit an upregulation of miR-200b/c. The reduction of ACE2 is the mechanism by which this miRNA exerts its detrimental effects. Therapeutic targeting can be achieved by blocking miR-200b/c. The decline in ACE2 levels is deleterious for the control of pulmonary fibrosis because ACE2 is key in metabolizing Ang II, a peptide hormone which is recognized for instigating lung fibrosis by stimulating TGF-β, and without sufficient ACE2, this harmful process is exacerbated. Elevated miR-200c-3p is associated with diminished ACE2 expression in H5N1 viral infection, potentially leading to lung injury. A similar reduction in ACE2 levels has been noted in infections by SARS-CoV-2 and the earlier SARS-CoV-1, resembling the H5N1 viral impact. Additional practical studies, both in vivo and in vitro are necessary to define the expression patterns of miR-200b/c throughout infections of SARS-CoV-2 and SARS-CoV-1. Research has elucidated the pivotal role of ACE2 in regulating heart function, indicating a potential correlation between the accumulation of Ang II and the initiation of cardiovascular diseases. Further studies elucidating the regulatory effect of miR-200b/c on ACE2 could potentially illuminate the cardiac complications observed in COVID-19, including acute coronary syndrome, heart failure, arrhythmias, and heart inflammation.
The serine protease furin plays a crucial role in facilitating the entry of enveloped respiratory viruses like MERS-CoV, H5N1, and SARS-CoV-2, into host cells by mediating the proteolysis of surface glycoproteins necessary for virus membrane attachment and fusion. It is implicated that miR-24 can regulate this process. Furin exhibits widespread expression across various tissues, including those within the respiratory system, making it a key factor for these viruses to exploit for successful infection and pathogenesis. Enzymatic adjustments are key to prompting membrane adherence and integration, impacting the virus’s cell tropism and pathogenic potential. The presence of a furin-like cleavage site (FCS) on the S protein of SARS-CoV-2, which is not identified in SARS-CoV-1, is crucial for the virus’s ability to bind and fuse with host cell membranes. This site might be integral to the enhanced membrane fusion capabilities that are a hallmark of SARS-CoV-2’s viral pathogenicity and infectivity. During SARS-CoV-2 infection, a decrease in miR-24 levels may lead to increased expression of furin, facilitating viral entry and replication. Additionally, the consequent activation of transforming growth factor beta 1 (TGF-β1) by furin could worsen the injury of the lung by prompting the progression of pulmonary fibrosis, which is prevalent in intense COVID-19 cases.
Mesenchymal stem cells (MSCs) giving rise to EVs express a protective service against SARS-2 coronavirus, mitigating cellular damage by lessening inflammatory response through viral spreading hindrance. Proposed as a possible treatment for COVID-19, mesenchymal stem cells are currently under examination, owing to their capacity improving the lung damage repair, as observed in pneumonia cases, in addition to their ability to stimulate macrophages to devour pathogens, successfully limiting viral spread (138). Efficient and safe delivery of miRNAs to a specific desired location within the body is slowed down by challenges such as direct targeting, stability, possibility of side effects. This highlights the importance of employing suitable delivery mechanisms to facilitate the reliable transfer of miRNAs for therapeutic purposes (98). The usage of MSCs or their derivatives is considered as a possible therapeutic approach and suitable carriers for delivering miRNAs owing to their tendency to be drawn to damaged or inflamed areas, a phenomenon known as tropism (98, 139).
The implications of miRNAs for therapeutics are highly significant. Due to their ability to target multiple genes simultaneously, miRNAs have emerged as potential therapeutic targets for various diseases, include COVID-19. By modulating the expression of specific miRNAs, it is possible to manipulate the expression of target genes and potentially correct aberrant gene expression patterns associated with disease. This opens up new avenues for the development of novel therapeutic strategies that can specifically target disease-associated pathways and potentially offer more effective and precise treatments. Furthermore, the stability and accessibility of miRNAs in body fluids, such as blood and urine, make them attractive as potential biomarkers for monitoring treatment response. Therefore, understanding the implications of miRNAs in therapeutics holds great promise for the development of personalized medicine and improved patient outcomes.
6.1 miRNA can be used as a marker to monitor treatment outcomes and drug response
miRNAs have the advantage of being potential multi-marker models for guiding treatment and assessing treatment response. The process of running multiple protein markers during research can be both expensive and time-consuming. However, using different sets of miRNA panels could provide a non-invasive method for tracking the effectiveness of drugs. There has been significant research conducted on the relationship between miRNAs and drug resistance. Tocilizumab has not been effective in treating COVID-19 cases with low levels of miR-146a (133). COVID-19 patients who were hospitalized and showed increased levels of miR-29a, miR-31, and miR-126 did not exhibit any response to treatment. On the other hand, the patients who showed a positive response to the treatment exhibited those miRNA levels within the normal range (140).
6.2 Enhancing therapeutic approaches: nutraceutical targeting of microRNAs modulating inflammation in the context of COVID-19
An alternative strategy to combat COVID-19 infection involves the employment of natural compounds in the treatment process. Inadequate nutritional status can lead to oxidative stress and inflammation, affecting the immune system’s regular functions. This review focuses on the use of polyphenol as a nutraceutical for therapeutic purposes. Polyphenol is an ideal example for several reasons, primarily because it is found abundantly in commonly consumed foods such as fruits, vegetables, and tea (141). Additionally, the mechanism of action is clearly identified against numerous viruses (142). Lastly, it’s important to note that this review does not specifically focus on the role of secondary metabolite modulation of miRNA in SARS-CoV-2 infection; polyphenol is simply used as an example. Owing to their capabilities, polyphenols can modify signals within cells and the expression of genes that are linked to oxidative stress. This capability indicates a potential impact on the severity of infections caused by SARS-CoV-2 (143). Research has demonstrated that polyphenols can combat viruses directly by damaging the pathogens or enhancing the adaptive immune response indirectly, according to both laboratory and living organism studies. A comparison between two human miRNA sets was done. Analysis demonstrated interaction of 17 miRNAs with the SARS-CoV-2 genome and are also modulated by polyphenols, including hsa-miR-148a-5p, hsa-miR-25-5p, hsa-miR-320c, and hsa-let-7a-3p (144). Quercetin, epigallocatechin-3-gallate (EGCG), resveratrol, and curcumin, are four polyphenols often utilized as nutritional therapeutic agents, showing their efficiency as prophylaxis of SARS-CoV-2 infection (64), having the following effects summarized in Table 5.
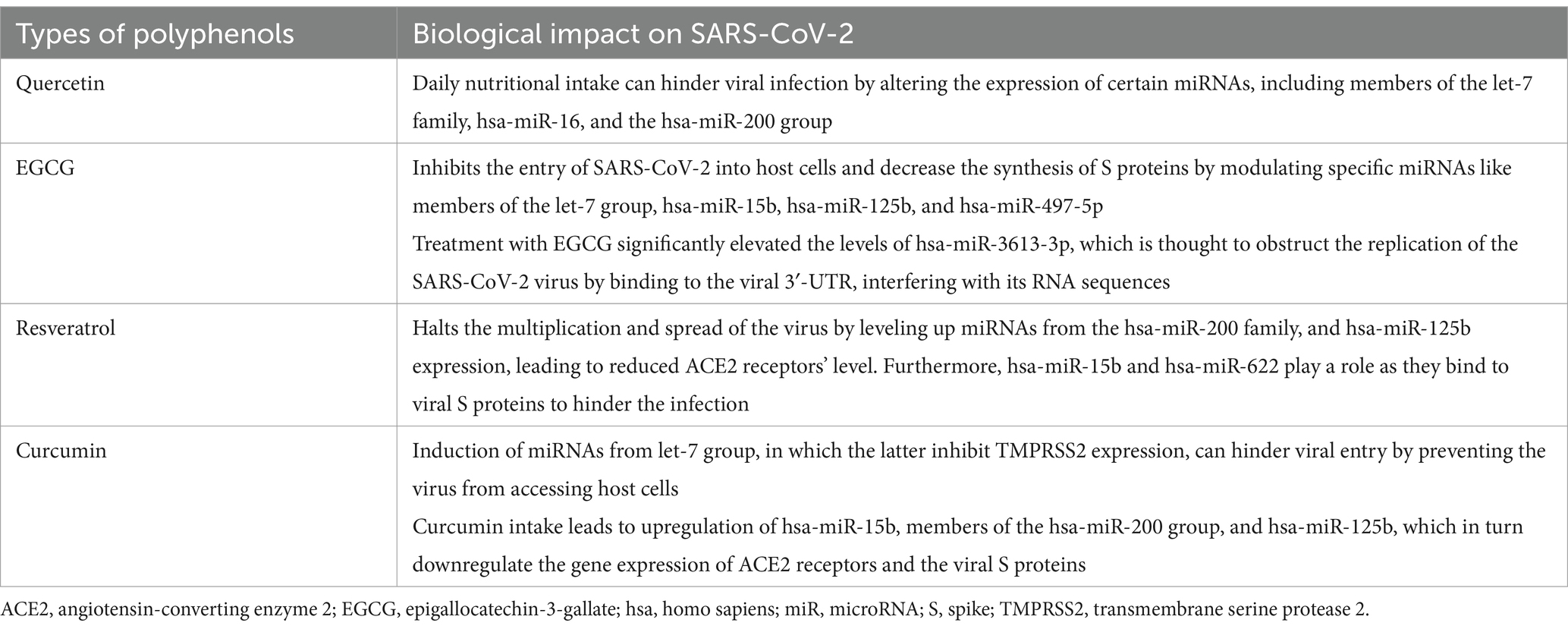
Table 5. A glance into the effect of polyphenols on SARS-CoV-2 infection by modulating microRNAs (64).
6.3 The interaction between vaccines, variants of concern, and miRNA
Vaccines, variants of concerns (VOCs), and miRNA have an intricate relationship. Vaccines aim to enhance the immune system’s response against the SARS-CoV-2 virus, while VOCs suppress the immune system in order to ensure their own survival. mRNA vaccines like Mrna-1273 and BNT162b utilize the host machinery of miRNA to combat viruses. miRNA micro-arrays are employed to determine the effectiveness of COVID-19 vaccines (145). Adverse reactions to COVID-19 vaccines and antibody levels can be determined by examining extracellular vesicle (EV) miRNAs like miR-92a and miR-148a. These miRNAs are utilized as biomarkers to evaluate the effectiveness and safety of vaccines (146). The presence of miRNAs like has-miR-4784 and has-miR-3150b-3p decreases the binding of Omicron strains to their receptors and diminishes the virulence of the virus (147).
7 Recommendations and limitations for microRNA research in COVID-19
7.1 Recommendations
7.1.1 Utilize miRNA for therapeutic purposes
Additional research is needed to test miRNAs in cell culture, animal models, and humans to inhibit SARS-CoV-2 binding, replication, or apoptosis in infected cells. Clinical trials should test miRNA mimics or anti-miRNA for treating viral infections. Testing miRNAs in combination with antiviral or immunomodulatory drugs could provide a synergistic approach to the treatment of COVID-19. By targeting both the viral replication and the dysregulated immune response, this combination therapy could potentially enhance the efficacy of treatment and reduce the severity of the disease. Further research and clinical trials are needed to evaluate the safety and effectiveness of miRNA-based combination therapies for COVID-19.
7.1.2 Analysis of additional miRNA as prognostic and diagnostic markers
More miRNA should be examined for high area under curve (AUC), specificity, and sensitivity. Additionally, miRNA levels should be assessed at various stages of COVID-19 to predict disease severity.
The identification of reliable biomarkers for early prediction and diagnosis of long COVID-19 (post-acute sequelae of COVID-19) is crucial for effective management and intervention. In recent years, miRNAs have gained attention as potential biomarkers due to their stability and role in regulating gene expression. Therefore, assessing the use of miRNAs as potential predictors of long COVID-19 could provide valuable insights into the pathophysiology of the disease and aid in the development of personalized treatment strategies.
7.2 Limitations
Using miRNA is limited by the dynamic nature of miRNA expression. The levels of miRNA can fluctuate over time, making it challenging to establish a reliable baseline for diagnostic or prognostic purposes. Additionally, the expression patterns of miRNA can be affected by various factors such as age, sex, comorbidities, and even medication use. Moreover, the genetic background of individuals plays a significant role in miRNA expression, as certain genetic variations can impact the processing and stability of miRNAs. All these factors contribute to the variability in miRNA expression, making it difficult to establish a universal miRNA signature for COVID-19 diagnosis or prognosis. However, despite these limitations, miRNA profiling still holds promise as a potential biomarker tool in COVID-19 research, especially when integrated with other clinical and molecular data. Ongoing studies are focusing on developing standardized protocols and analytical methods to overcome the challenges posed by miRNA variability and harness their potential as reliable diagnostic and prognostic markers in COVID-19.
The complexity of miRNA-target interactions poses a limitation to miRNA utilization. miRNAs often exhibit pleiotropic effects, meaning that a single miRNA can target multiple genes simultaneously. While this ability allows miRNAs to regulate various biological processes, it also increases the risk of potential side effects. When a miRNA targets multiple genes, the regulation of one gene may inadvertently affect the expression or function of other genes. This can lead to unintended consequences and disrupt normal cellular processes. Therefore, the pleiotropic effects of miRNAs must be carefully considered when studying their functions or developing therapeutic interventions involving miRNA-based therapies. Understanding the complex network of miRNA-gene interactions is crucial for predicting and minimizing potential side effects associated with miRNA-based treatments.
The method of delivery of miRNA to the target site is another significant limitation that researchers face in the field of miRNA therapeutics. Currently, there are various approaches for delivering miRNA molecules to specific tissues or cells, including viral vectors, liposomes, nanoparticles, and exosomes. However, each of these delivery methods has its own challenges and drawbacks. For instance, viral vectors can induce immune responses and have limited cargo capacity. Liposomes and nanoparticles may suffer from poor stability, low efficiency, and potential toxicity. Exosomes, on the other hand, have limited loading capacity and lack precise control over delivery. Therefore, finding an efficient and safe delivery system for miRNA remains a critical hurdle in realizing the full therapeutic potential of miRNA-based therapies. Efficient and targeted delivery of miRNA mimics or inhibitors remains a significant hurdle. Current delivery methods may not ensure the stability and bioavailability of these molecules, which is essential for their therapeutic efficacy.
One limitation of the few ongoing clinical trials for investigating the safety and efficacy of miRNA as therapeutics is the small sample size. Due to the novelty and relatively recent discovery of miRNA as potential therapeutic targets, there are currently limited clinical trials available. This small sample size can limit the generalizability of the results and make it difficult to draw definitive conclusions about the safety and efficacy of miRNA therapeutics. Additionally, the lack of diversity in the study populations can further restrict the applicability of the findings to a broader range of patients. Furthermore, the limited number of clinical trials may also hinder the identification of potential adverse effects or long-term consequences associated with miRNA therapeutics. Therefore, while the ongoing trials provide valuable insights into the potential benefits of miRNA therapeutics, the limited number of studies is a notable limitation that needs to be addressed in future research. Additionally, the role of certain miRNA in relation to COVID-19 remains unknown. Ultimately, to effectively study miRNA, one must have access to advanced bioinformatics tools and a large dataset.
8 Conclusion
To recap, this comprehensive analysis clarifies the intricate role of miRNAs in the pathogenesis, diagnostic frameworks, prognostic assessments, and therapeutic interventions in relation to COVID-19, underscoring their significance as regulators in the interplay between host and virus, influencing immune responses, inflammatory processes, along with viral replication. Moreover, miRNAs act as potent regulatory molecules of gene expression, exerting control by attaching to compatible interaction sites. Many clinical targets for COVID-19 are miRNAs, proposing that the virus might manage its entry into cells and disrupt bodily functions by altering these regulatory miRNAs. Both host-and virus-derived miRNAs play a critical role in helping the virus to evade immune system’s components, triggering inflammatory responses, leading to overproduction of cytokines, and tissue damage. Shifts in miRNAs’ concentration may potentially mark variable stages of COVID-19 illness. Furthermore, it was discovered that miRNAs performed better in terms of diagnosis than frequently used assays. Despite showing the impact that the host-and viral-encoded miRNAs have on the development of COVID-19 in several investigations, but the actual practical usage of such findings regarding the treatment and prevention are still in the early developmental stages.
Author contributions
SS: Conceptualization, Writing – original draft. SH: Writing – review & editing. AA: Resources, Writing – original draft. NA: Resources, Writing – original draft. HA: Resources, Writing – original draft. KA: Visualization, Writing – original draft. CJ: Conceptualization, Supervision, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors would like to extend their sincere appreciation to Mrs. Snur Rasool Abdullah for her invaluable assistance in the design of the figures for this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Roustai Geraylow, K, Hemmati, R, Kadkhoda, S, and Ghafouri-Fard, S. miRNA expression in COVID-19. Gene Rep. (2022) 28:101641. doi: 10.1016/j.genrep.2022.101641
2. Zhang, S, Amahong, K, Sun, X, Lian, X, Liu, J, Sun, H, et al. The miRNA: a small but powerful RNA for COVID-19. Brief Bioinform. (2021) 22:1137–49. doi: 10.1093/bib/bbab062
3. Abedi, F, Rezaee, R, Hayes, AW, Nasiripour, S, and Karimi, G. MicroRNAs and SARS-CoV-2 life cycle, pathogenesis, and mutations: biomarkers or therapeutic agents? Cell Cycle. (2021) 20:143–53. doi: 10.1080/15384101.2020.1867792
4. Reyes-Long, S, Cortes-Altamirano, JL, Bandala, C, Avendano-Ortiz, K, Bonilla-Jaime, H, Bueno-Nava, A, et al. Role of the microRNAs in the pathogenic mechanism of painful symptoms in long COVID: systematic review. Int J Mol Sci. (2023) 24:3574. doi: 10.3390/ijms24043574
5. Andrew, W, Meizi, Y, and DaLiao, X. Therapeutic implication of miRNA in human disease In: S Shashwat and K Suman, editors. Antisense therapy. Rijeka: IntechOpen (2018). 6.
6. Notarte, KI, Senanayake, S, Macaranas, I, Albano, PM, Mundo, L, Fennell, E, et al. MicroRNA and other non-coding RNAs in Epstein–Barr virus-associated cancers. Cancers. (2021) 13:3909. doi: 10.3390/cancers13153909
7. Bivona, G, Agnello, L, and Ciaccio, M. Biomarkers for prognosis and treatment response in COVID-19 patients. Ann Lab Med. (2021) 41:540–8. doi: 10.3343/alm.2021.41.6.540
8. Guterres, A, de Azeredo Lima, CH, Miranda, RL, and Gadelha, MR. What is the potential function of microRNAs as biomarkers and therapeutic targets in COVID-19? Infect Genet Evol. (2020) 85:104417. doi: 10.1016/j.meegid.2020.104417
9. Donyavi, T, Bokharaei-Salim, F, Baghi, HB, Khanaliha, K, Alaei Janat-Makan, M, Karimi, B, et al. Acute and post-acute phase of COVID-19: analyzing expression patterns of miRNA-29a-3p, 146a-3p, 155-5p, and let-7b-3p in PBMC. Int Immunopharmacol. (2021) 97:107641. doi: 10.1016/j.intimp.2021.107641
10. Farr, RJ, Rootes, CL, Rowntree, LC, Nguyen, THO, Hensen, L, Kedzierski, L, et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. (2021) 17:e1009759. doi: 10.1371/journal.ppat.1009759
11. Gedikbasi, A, Adas, G, Isiksacan, N, Kart Yasar, K, Canbolat Unlu, E, Yilmaz, R, et al. The effect of host miRNAs on prognosis in COVID-19: miRNA-155 may promote severity via targeting suppressor of cytokine signaling 1 (SOCS1) gene. Genes. (2022) 13:1146. doi: 10.3390/genes13071146
12. Janssen, HL, Reesink, HW, Lawitz, EJ, Zeuzem, S, Rodriguez-Torres, M, Patel, K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. (2013) 368:1685–94. doi: 10.1056/NEJMoa1209026
13. Girardi, E, López, P, and Pfeffer, S. On the importance of host microRNAs during viral infection. Front Genet. (2018) 9:439. doi: 10.3389/fgene.2018.00439
15. Zhdanov, VP . Intracellular miRNA or siRNA delivery and function. Biosystems. (2018) 171:20–5. doi: 10.1016/j.biosystems.2018.05.007
16. Chauhan, N, Jaggi, M, Chauhan, SC, and Yallapu, MM. COVID-19: fighting the invisible enemy with microRNAs. Expert Rev Anti-Infect Ther. (2021) 19:137–45. doi: 10.1080/14787210.2020.1812385
17. Rebolledo-Mendez, JD, Vaishnav, RA, Cooper, NG, and Friedland, RP. Cross-kingdom sequence similarities between human micro-RNAs and plant viruses. Commun Integr Biol. (2013) 6:e24951. doi: 10.4161/cib.24951
18. Hu, B, Guo, H, Zhou, P, and Shi, ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. (2021) 19:141–54. doi: 10.1038/s41579-020-00459-7
19. Cui, J, Li, F, and Shi, ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. (2019) 17:181–92. doi: 10.1038/s41579-018-0118-9
20. Dhama, K, Patel, SK, Sharun, K, Pathak, M, Tiwari, R, Yatoo, MI, et al. SARS-CoV-2 jumping the species barrier: zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus. Travel Med Infect Dis. (2020) 37:101830. doi: 10.1016/j.tmaid.2020.101830
21. Hui, DS, IA, E, Madani, TA, Ntoumi, F, Kock, R, Dar, O, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. (2020) 91:264–6. doi: 10.1016/j.ijid.2020.01.009
22. Yang, H, and Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat Rev Microbiol. (2021) 19:685–700. doi: 10.1038/s41579-021-00630-8
23. Wang, MY, Zhao, R, Gao, LJ, Gao, XF, Wang, DP, and Cao, JM. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. (2020) 10:587269. doi: 10.3389/fcimb.2020.587269
24. Wu, W, Cheng, Y, Zhou, H, Sun, C, and Zhang, S. The SARS-CoV-2 nucleocapsid protein: its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostics. Virol J. (2023) 20:6. doi: 10.1186/s12985-023-01968-6
25. Cascella, M, Rajnik, M, Aleem, A, Dulebohn, SC, and Di Napoli, R. Features, evaluation, and treatment of coronavirus (COVID-19). Treasure Island, FL: StatPearls (2023).
26. Banerjee, S, Wang, X, Du, S, Zhu, C, Jia, Y, Wang, Y, et al. Comprehensive role of SARS-CoV-2 spike glycoprotein in regulating host signaling pathway. J Med Virol. (2022) 94:4071–87. doi: 10.1002/jmv.27820
27. Thomas, S . The structure of the membrane protein of SARS-CoV-2 resembles the sugar transporter SemiSWEET. Pathog Immun. (2020) 5:342–63. doi: 10.20411/pai.v5i1.377
28. Cao, Y, Yang, R, Lee, I, Zhang, W, Sun, J, Wang, W, et al. Characterization of the SARS-CoV-2 E protein: sequence, structure, viroporin, and inhibitors. Protein Sci. (2021) 30:1114–30. doi: 10.1002/pro.4075
29. Santos-Mendoza, T . The envelope (E) protein of SARS-CoV-2 as a pharmacological target. Viruses. (2023) 15:1000. doi: 10.3390/v15041000
30. Harrison, AG, Lin, T, and Wang, P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. (2020) 41:1100–15. doi: 10.1016/j.it.2020.10.004
31. Hamming, I, Timens, W, Bulthuis, ML, Lely, AT, Navis, G, and van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. (2004) 203:631–7. doi: 10.1002/path.1570
32. Chu, KH, Tsang, WK, Tang, CS, Lam, MF, Lai, FM, To, KF, et al. Acute renal impairment in coronavirus-associated severe acute respiratory syndrome. Kidney Int. (2005) 67:698–705. doi: 10.1111/j.1523-1755.2005.67130.x
33. Chen, YZ, Liu, G, Senju, S, Wang, Q, Irie, A, Haruta, M, et al. Identification of SARS-COV spike protein-derived and HLA-A2-restricted human CTL epitopes by using a new muramyl dipeptidederivative adjuvant. Int J Immunopathol Pharmacol. (2010) 23:165–77. doi: 10.1177/039463201002300115
34. Fast, E, Altman, RB, and Chen, B. (2020). Potential T-cell and B-cell epitopes of 2019-nCoV. bioRxiv. Available at: https://doi.org/10.1101/2020.02.19.955484. [Epub ahead of preprint]
35. Prompetchara, E, Ketloy, C, and Palaga, T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. (2020) 38:1–9. doi: 10.12932/AP-200220-0772
36. Chan, L, Karimi, N, Morovati, S, Alizadeh, K, Kakish, JE, Vanderkamp, S, et al. The roles of neutrophils in cytokine storms. Viruses. (2021) 13:2318. doi: 10.3390/v13112318
37. Isenmann, M, Stoddart, MJ, Schmelzeisen, R, Gross, C, Della Bella, E, and Rothweiler, RM. Basic principles of RNA interference: nucleic acid types and in vitro intracellular delivery methods. Micromachines. (2023) 14:1321. doi: 10.3390/mi14071321
38. Liang, Y, Fang, D, Gao, X, Deng, X, Chen, N, Wu, J, et al. Circulating microRNAs as emerging regulators of COVID-19. Theranostics. (2023) 13:125–47. doi: 10.7150/thno.78164
39. Rocchi, A, Chiti, E, Maiese, A, Turillazzi, E, and Spinetti, I. MicroRNAs: an update of applications in forensic science. Diagnostics. (2020) 11:32. doi: 10.3390/diagnostics11010032
40. Steiman-Shimony, A, Shtrikman, O, and Margalit, H. Assessing the functional association of intronic miRNAs with their host genes. RNA. (2018) 24:991–1004. doi: 10.1261/rna.064386.117
41. Jorge, AL, Pereira, ER, Oliveira, CS, Ferreira, EDS, Menon, ETN, Diniz, SN, et al. MicroRNAs: understanding their role in gene expression and cancer. Einstein. (2021) 19:eRB5996. doi: 10.31744/einstein_journal/2021RB5996
42. Huang, HY, Lin, YC, Li, J, Huang, KY, Shrestha, S, Hong, HC, et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. (2020) 48:D148–54. doi: 10.1093/nar/gkz896
43. Dharap, A, Pokrzywa, C, Murali, S, Pandi, G, and Vemuganti, R. MicroRNA miR-324-3p induces promoter-mediated expression of RelA gene. PLoS One. (2013) 8:e79467. doi: 10.1371/journal.pone.0079467
44. Daka, A, and Peer, D. RNAi-based nanomedicines for targeted personalized therapy. Adv Drug Deliv Rev. (2012) 64:1508–21. doi: 10.1016/j.addr.2012.08.014
45. Trobaugh, DW, and Klimstra, WB. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol Med. (2017) 23:80–93. doi: 10.1016/j.molmed.2016.11.003
46. Gasparello, J, Finotti, A, and Gambari, R. Tackling the COVID-19 “cytokine storm” with microRNA mimics directly targeting the 3’UTR of pro-inflammatory mRNAs. Med Hypotheses. (2021) 146:110415. doi: 10.1016/j.mehy.2020.110415
47. Ergun, S, Sankaranarayanan, R, and Petrovic, N. Clinically informative microRNAs for SARS-CoV-2 infection. Epigenomics. (2023) 15:705–16. doi: 10.2217/epi-2023-0179
48. Maranini, B, Ciancio, G, Ferracin, M, Cultrera, R, Negrini, M, Sabbioni, S, et al. microRNAs and inflammatory immune response in SARS-CoV-2 infection: a narrative review. Life. (2022) 12:288. doi: 10.3390/life12020288
49. Jiang, K, Yang, J, Guo, S, Zhao, G, Wu, H, and Deng, G. Peripheral circulating exosome-mediated delivery of miR-155 as a novel mechanism for acute lung inflammation. Mol Ther. (2019) 27:1758–71. doi: 10.1016/j.ymthe.2019.07.003
50. Yoshimura, A, Naka, T, and Kubo, M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. (2007) 7:454–65. doi: 10.1038/nri2093
51. Hu, R, Zhang, Y, Yang, X, Yan, J, Sun, Y, Chen, Z, et al. Isoflurane attenuates LPS-induced acute lung injury by targeting miR-155-HIF1-alpha. Front Biosci. (2015) 20:139–56. doi: 10.2741/4302
52. Fu, L, Zhu, P, Qi, S, Li, C, and Zhao, K. MicroRNA-92a antagonism attenuates lipopolysaccharide (LPS)-induced pulmonary inflammation and injury in mice through suppressing the PTEN/AKT/NF-κB signaling pathway. Biomed Pharmacother. (2018) 107:703–11. doi: 10.1016/j.biopha.2018.08.040
53. Sun, C-M, Wu, J, Zhang, H, Shi, G, and Chen, Z-T. Circulating miR-125a but not miR-125b is decreased in active disease status and negatively correlates with disease severity as well as inflammatory cytokines in patients with Crohn’s disease. World J Gastroenterol. (2017) 23:7888–98. doi: 10.3748/wjg.v23.i44.7888
54. Vaz, CO, Hounkpe, BW, Oliveira, JD, Mazetto, B, Cardoso Jacintho, B, Aparecida Locachevic, G, et al. MicroRNA 205-5p and COVID-19 adverse outcomes: potential molecular biomarker and regulator of the immune response. Exp Biol Med. (2023) 248:1024–33. doi: 10.1177/15353702231175412
55. Zhang, S, Lin, S, Tang, Q, and Yan, Z. Knockdown of miR-205-5p alleviates the inflammatory response in allergic rhinitis by targeting B-cell lymphoma 6. Mol Med Rep. (2021) 24:818. doi: 10.3892/mmr.2021.12458
56. Sánchez, MY, Santos, MM, Oltra, M, dos Santos Pires, ME, Sancho-Pelluz, J, and Barcia, JM. Regulation of HIF-1α/VEGFA mRNA transcription by miR-205-5p in diabetic mice. Invest Ophthalmol Vis Sci. (2024) 65:842.
57. Gjorgjieva, T, Chaloemtoem, A, Shahin, T, Bayaraa, O, Dieng, MM, Alshaikh, M, et al. Systems genetics identifies miRNA-mediated regulation of host response in COVID-19. Hum Genomics. (2023) 17:49. doi: 10.1186/s40246-023-00494-4
58. Wu, X, Liu, H, Hu, Q, Wang, J, Zhang, S, Cui, W, et al. Astrocyte-derived extracellular vesicular miR-143-3p dampens autophagic degradation of endothelial adhesion molecules and promotes neutrophil transendothelial migration after acute brain injury. Adv Sci. (2024) 11:2305339. doi: 10.1002/advs.202305339
59. Vier, J, Groth, M, Sochalska, M, and Kirschnek, S. The anti-apoptotic Bcl-2 family protein A1/Bfl-1 regulates neutrophil survival and homeostasis and is controlled via PI3K and JAK/STAT signaling. Cell Death Dis. (2016) 7:e2103. doi: 10.1038/cddis.2016.23
60. Locke, FL, Zha, Y-y, Zheng, Y, Driessens, G, and Gajewski, TF. Conditional deletion of PTEN in peripheral T cells augments TCR-mediated activation but does not abrogate CD28 dependency or prevent anergy induction. J Immunol. (2013) 191:1677–85. doi: 10.4049/jimmunol.1202018
61. Gracias, DT, Stelekati, E, Hope, JL, Boesteanu, AC, Doering, TA, Norton, J, et al. The microRNA miR-155 controls CD8+ T cell responses by regulating interferon signaling. Nat Immunol. (2013) 14:593–602. doi: 10.1038/ni.2576
62. Lu, C-H, Chen, D-X, Dong, K, Wu, Y-J, Na, N, Wen, H, et al. Inhibition of miR-143-3p alleviates myocardial ischemia reperfusion injury via limiting mitochondria-mediated apoptosis. Biol Chem. (2023) 404:619–31. doi: 10.1515/hsz-2022-0334
63. Seddiki, N, Brezar, V, Ruffin, N, Lévy, Y, and Swaminathan, S. Role of miR-155 in the regulation of lymphocyte immune function and disease. Immunology. (2014) 142:32–8. doi: 10.1111/imm.12227
64. Yang, CY, Chen, YH, Liu, PJ, Hu, WC, Lu, KC, and Tsai, KW. The emerging role of miRNAs in the pathogenesis of COVID-19: protective effects of nutraceutical polyphenolic compounds against SARS-CoV-2 infection. Int J Med Sci. (2022) 19:1340–56. doi: 10.7150/ijms.76168
65. Khanmohammadi, S, and Rezaei, N. Role of toll-like receptors in the pathogenesis of COVID-19. J Med Virol. (2021) 93:2735–9. doi: 10.1002/jmv.26826
66. Roganovic, J . Downregulation of microRNA-146a in diabetes, obesity and hypertension may contribute to severe COVID-19. Med Hypotheses. (2021) 146:110448. doi: 10.1016/j.mehy.2020.110448
67. Jankovic, M, Nikolic, D, Novakovic, I, Petrovic, B, Lackovic, M, and Santric-Milicevic, M. miRNAs as a potential biomarker in the COVID-19 infection and complications course, severity, and outcome. Diagnostics. (2023) 13:1091. doi: 10.3390/diagnostics13061091
68. Wang, Y, Zhu, X, Jiang, XM, Guo, J, Fu, Z, Zhou, Z, et al. Decreased inhibition of exosomal miRNAs on SARS-CoV-2 replication underlies poor outcomes in elderly people and diabetic patients. Signal Transduct Target Ther. (2021) 6:300. doi: 10.1038/s41392-021-00716-y
69. Shukuya, T, Ghai, V, Amann, JM, Okimoto, T, Shilo, K, Kim, TK, et al. Circulating microRNAs and extracellular vesicle-containing microRNAs as response biomarkers of anti-programmed cell death protein 1 or programmed death-ligand 1 therapy in NSCLC. J Thorac Oncol. (2020) 15:1773–81. doi: 10.1016/j.jtho.2020.05.022
70. Pozniak, T, Shcharbin, D, and Bryszewska, M. Circulating microRNAs in medicine. Int J Mol Sci. (2022) 23:3996. doi: 10.3390/ijms23073996
71. O'Brien, J, Hayder, H, Zayed, Y, and Peng, C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. (2018) 9:402. doi: 10.3389/fendo.2018.00402
72. Monnot, GC, Martinez-Usatorre, A, Lanitis, E, Lopes, SF, Cheng, WC, Ho, PC, et al. miR-155 overexpression in OT-1 CD8+ T cells improves anti-tumor activity against low-affinity tumor antigen. Mol Ther Oncolytics. (2020) 16:111–23. doi: 10.1016/j.omto.2019.12.008
73. Valastyan, S, and Weinberg, RA. Roles for microRNAs in the regulation of cell adhesion molecules. J Cell Sci. (2011) 124:999–1006. doi: 10.1242/jcs.081513
74. Yin, Y, Li, F, Shi, J, Li, S, Cai, J, and Jiang, Y. MiR-146a regulates inflammatory infiltration by macrophages in polymyositis/dermatomyositis by targeting TRAF6 and affecting IL-17/ICAM-1 pathway. Cell Physiol Biochem. (2016) 40:486–98. doi: 10.1159/000452563
75. Kassif-Lerner, R, Zloto, K, Rubin, N, Asraf, K, Doolman, R, Paret, G, et al. miR-155: a potential biomarker for predicting mortality in COVID-19 patients. J Pers Med. (2022) 12:324. doi: 10.3390/jpm12020324
76. Nachtigall, PG, Dias, MC, Carvalho, RF, Martins, C, and Pinhal, D. MicroRNA-499 expression distinctively correlates to target genes sox6 and rod1 profiles to resolve the skeletal muscle phenotype in Nile tilapia. PLoS One. (2015) 10:e0119804. doi: 10.1371/journal.pone.0119804
77. Zhou, C, Hu, C, Wang, B, Fan, S, and Jin, W. Curcumin suppresses cell proliferation, migration, and invasion through modulating miR-21-5p/SOX6 axis in hepatocellular carcinoma. Cancer Biother Radiopharm. (2020) 35:267–274. doi: 10.1089/cbr.2020.3734
78. Hu, J, Huang, S, Liu, X, Zhang, Y, Wei, S, and Hu, X. miR-155: an important role in inflammation response. J Immunol Res. (2022) 2022:1–13. doi: 10.1155/2022/7437281
79. Han, X, Wang, S, Yong, Z, Zhang, X, Wang, X, and You, P. Effect of miR-499-5p/SOX6 axis on atrial fibrosis in rats with atrial fibrillation. Open Med. (2023) 18:20230654. doi: 10.1515/med-2023-0654
80. Costinean, S, Sandhu, SK, Pedersen, IM, Tili, E, Trotta, R, Perrotti, D, et al. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. (2009) 114:1374–82. doi: 10.1182/blood-2009-05-220814
81. O’Connell, RM, Chaudhuri, AA, Rao, DS, and Baltimore, D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. (2009) 106:7113–8. doi: 10.1073/pnas.0902636106
82. Hadidi, S, Antignano, F, Hughes, MR, Wang, SK, Snyder, K, Sammis, GM, et al. Myeloid cell-specific expression of Ship1 regulates IL-12 production and immunity to helminth infection. Mucosal Immunol. (2012) 5:535–43. doi: 10.1038/mi.2012.29
83. Carow, B, Ye, X, Gavier-Widén, D, Bhuju, S, Oehlmann, W, Singh, M, et al. Silencing suppressor of cytokine signaling-1 (SOCS1) in macrophages improves Mycobacterium tuberculosis control in an interferon-gamma (IFN-gamma)-dependent manner. J Biol Chem. (2011) 286:26873–87. doi: 10.1074/jbc.M111.238287
84. Ghafouri-Fard, S, Shoorei, H, and Taheri, M. Non-coding RNAs participate in the ischemia-reperfusion injury. Biomed Pharmacother. (2020) 129:110419. doi: 10.1016/j.biopha.2020.110419
85. Lee, DW, Futami, M, Carroll, M, Feng, Y, Wang, Z, Fernandez, M, et al. Loss of SHIP-1 protein expression in high-risk myelodysplastic syndromes is associated with miR-210 and miR-155. Oncogene. (2012) 31:4085–94. doi: 10.1038/onc.2011.579
86. Xue, H, Hua, LM, Guo, M, and Luo, JM. SHIP1 is targeted by miR-155 in acute myeloid leukemia. Oncol Rep. (2014) 32:2253–9. doi: 10.3892/or.2014.3435
87. Preiser, JC . Oxidative stress. JPEN J Parenter Enteral Nutr. (2012) 36:147–54. doi: 10.1177/0148607111434963
88. Arisan, ED, Dart, A, Grant, GH, Arisan, S, Cuhadaroglu, S, Lange, S, et al. The prediction of miRNAs in SARS-CoV-2 genomes: hsa-miR databases identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses. (2020) 12:614. doi: 10.3390/v12060614
89. Bansal, M . Cardiovascular disease and COVID-19. Diabetes Metab Syndr Clin Res Rev. (2020) 14:247–50. doi: 10.1016/j.dsx.2020.03.013
90. Garg, A, Seeliger, B, Derda, AA, Xiao, K, Gietz, A, Scherf, K, et al. Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur J Heart Fail. (2021) 23:468–75. doi: 10.1002/ejhf.2096
91. Huang, X-H, Li, J-L, Li, X-Y, Wang, S-X, Jiao, Z-H, Li, S-Q, et al. miR-208a in cardiac hypertrophy and remodeling. Front Cardiovasc Med. (2021) 8:773314. doi: 10.3389/fcvm.2021.773314
92. Eyileten, C, Wicik, Z, Simoes, SN, Martins-Jr, DC, Klos, K, Wlodarczyk, W, et al. Thrombosis-related circulating miR-16-5p is associated with disease severity in patients hospitalised for COVID-19. RNA Biol. (2022) 19:963–79. doi: 10.1080/15476286.2022.2100629
93. Gambardella, J, Kansakar, U, Sardu, C, Messina, V, Jankauskas, SS, Marfella, R, et al. Exosomal miR-145 and miR-885 regulate thrombosis in COVID-19. J Pharmacol Exp Ther. (2023) 384:109–15. doi: 10.1124/jpet.122.001209
94. Bai, X, and Bian, Z. MicroRNA-21 is a versatile regulator and potential treatment target in central nervous system disorders. Front Mol Neurosci. (2022) 15:842288. doi: 10.3389/fnmol.2022.842288
95. Mishra, R, and Banerjea, AC. SARS-CoV-2 spike targets USP33-IRF9 axis via exosomal miR-148a to activate human microglia. Front Immunol. (2021) 12:656700. doi: 10.3389/fimmu.2021.656700
96. Peleg, Y, Kudose, S, D’Agati, V, Siddall, E, Ahmad, S, Nickolas, T, et al. Acute kidney injury due to collapsing glomerulopathy following COVID-19 infection. Kidney Int Rep. (2020) 5:940–5. doi: 10.1016/j.ekir.2020.04.017
97. Wu, H, Kong, L, Zhou, S, Cui, W, Xu, F, Luo, M, et al. The role of microRNAs in diabetic nephropathy. J Diabetes Res. (2014) 2014:1–12. doi: 10.1155/2014/920134
98. Arghiani, N, Nissan, T, and Matin, MM. Role of microRNAs in COVID-19 with implications for therapeutics. Biomed Pharmacother. (2021) 144:112247. doi: 10.1016/j.biopha.2021.112247
99. Shen, H, Fang, K, Guo, H, and Wang, G. High glucose-induced apoptosis in human kidney cells was alleviated by miR-15b-5p mimics. Biol Pharm Bull. (2019) 42:758–63. doi: 10.1248/bpb.b18-00951
100. Khatami, A, Taghizadieh, M, Sadri Nahand, J, Karimzadeh, M, Kiani, SJ, Khanaliha, K, et al. Evaluation of microRNA expression pattern (miR-28, miR-181a, miR-34a, and miR-31) in patients with COVID-19 admitted to ICU and diabetic COVID-19 patients. Intervirology. (2023) 66:63–76. doi: 10.1159/000529985
101. Fomison-Nurse, I, Saw, EEL, Gandhi, S, Munasinghe, PE, Van Hout, I, Williams, MJA, et al. Diabetes induces the activation of pro-ageing miR-34a in the heart, but has differential effects on cardiomyocytes and cardiac progenitor cells. Cell Death Differ. (2018) 25:1336–49. doi: 10.1038/s41418-017-0047-6
102. Alimohamadi, Y, Sepandi, M, Taghdir, M, and Hosamirudsari, H. Determine the most common clinical symptoms in COVID-19 patients: a systematic review and meta-analysis. J Prev Med Hyg. (2020) 61:E304–12. doi: 10.15167/2421-4248/jpmh2020.61.3.1530
103. Baig, AM . Chronic COVID syndrome: need for an appropriate medical terminology for long-COVID and COVID long-haulers. J Med Virol. (2021) 93:2555–6. doi: 10.1002/jmv.26624
104. Pinto, MD, Downs, CA, Huang, Y, El-Azab, SA, Ramrakhiani, NS, Barisano, A, et al. A distinct symptom pattern emerges for COVID-19 long-haul: a nationwide study. Sci Rep. (2022) 12:15905. doi: 10.1038/s41598-022-20214-7
105. Mahase, E . COVID-19: what do we know about “long COVID”? BMJ. (2020) 370:m2815. doi: 10.1136/bmj.m2815
106. Maucher, D, Schmidt, B, and Schumann, J. Loss of endothelial barrier function in the inflammatory setting: indication for a cytokine-mediated post-transcriptional mechanism by virtue of upregulation of miRNAs miR-29a-3p, miR-29b-3p, and miR-155-5p. Cells. (2021) 10:2843. doi: 10.3390/cells10112843
107. Demongeot, J, and Seligmann, H. SARS-CoV-2 and miRNA-like inhibition power. Med Hypotheses. (2020) 144:110245. doi: 10.1016/j.mehy.2020.110245
108. Visacri, MB, Nicoletti, AS, Pincinato, EC, Loren, P, Saavedra, N, Saavedra, K, et al. Role of miRNAs as biomarkers of COVID-19: a scoping review of the status and future directions for research in this field. Biomark Med. (2021) 15:1785–95. doi: 10.2217/bmm-2021-0348
109. Li, C, Hu, X, Li, L, and Li, JH. Differential microRNA expression in the peripheral blood from human patients with COVID-19. J Clin Lab Anal. (2020) 34:e23590. doi: 10.1002/jcla.23590
110. Das, R, Sinnarasan, VSP, Paul, D, and Venkatesan, A. Correction: a machine learning approach to identify potential miRNA-gene regulatory network contributing to the pathogenesis of SARS-CoV-2 infection. Biochem Genet. (2023) 62:1007. doi: 10.1007/s10528-023-10511-9
111. Smail, SW, Babaei, E, and Amin, K. Hematological, inflammatory, coagulation, and oxidative/antioxidant biomarkers as predictors for severity and mortality in COVID-19: a prospective cohort-study. Int J Gen Med. (2023) 16:565–80. doi: 10.2147/IJGM.S402206
112. Smail, SW, Babaei, E, and Amin, K. Demographic, clinical and genetic factors associated with COVID-19 disease susceptibility and mortality in a Kurdish population. Ann Saudi Med. (2023) 43:125–42. doi: 10.5144/0256-4947.2023.125
113. Smail, SW, Babaei, E, Amin, K, and Abdulahad, WH. Serum IL-23, IL-10, and TNF-α predict in-hospital mortality in COVID-19 patients. Front Immunol. (2023) 14:1145840. doi: 10.3389/fimmu.2023.1145840
114. Smail, SW, Babaei, E, and Amin, K. Ct, IL-18 polymorphism, and laboratory biomarkers for predicting chemosensory dysfunctions and mortality in COVID-19. Future Sci OA. (2023) 9:FSO838. doi: 10.2144/fsoa-2022-0082
115. Fernández-Pato, A, Virseda-Berdices, A, Resino, S, Ryan, P, Martínez-González, O, Pérez-García, F, et al. Plasma miRNA profile at COVID-19 onset predicts severity status and mortality. Emerg Microbes Infect. (2022) 11:676–88. doi: 10.1080/22221751.2022.2038021
116. Matarese, A, Gambardella, J, Sardu, C, and Santulli, G. miR-98 regulates TMPRSS2 expression in human endothelial cells: key implications for COVID-19. Biomedicines. (2020) 8:462. doi: 10.3390/biomedicines8110462
117. Wilson, JC, Kealy, D, James, SR, Plowman, T, Newling, K, Jagger, C, et al. Integrated miRNA/cytokine/chemokine profiling reveals severity-associated step changes and principal correlates of fatality in COVID-19. iScience. (2022) 25:103672. doi: 10.1016/j.isci.2021.103672
118. Song, L, Liu, H, Gao, S, Jiang, W, and Huang, W. Cellular microRNAs inhibit replication of the H1N1 influenza a virus in infected cells. J Virol. (2010) 84:8849–60. doi: 10.1128/JVI.00456-10
119. de Gonzalo-Calvo, D, Benítez, ID, Pinilla, L, Carratalá, A, Moncusí-Moix, A, Gort-Paniello, C, et al. Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl Res. (2021) 236:147–59. doi: 10.1016/j.trsl.2021.05.004
120. Rodriguez-Rius, A, Lopez, S, Martinez-Perez, A, Souto, JC, and Soria, JM. Identification of a plasma MicroRNA profile associated with venous thrombosis. Arterioscler Thromb Vasc Biol. (2020) 40:1392–9. doi: 10.1161/ATVBAHA.120.314092
121. de Gonzalo-Calvo, D, Molinero, M, Benítez, ID, Perez-Pons, M, García-Mateo, N, Ortega, A, et al. A blood microRNA classifier for the prediction of ICU mortality in COVID-19 patients: a multicenter validation study. Respir Res. (2023) 24:159. doi: 10.1186/s12931-023-02462-x
122. Recchiuti, A, Patruno, S, Mattoscio, D, Isopi, E, Pomilio, A, Lamolinara, A, et al. Resolvin D1 and D2 reduce SARS-CoV-2-induced inflammatory responses in cystic fibrosis macrophages. FASEB J. (2021) 35:e21441. doi: 10.1096/fj.202001952R
123. Tang, H, Gao, Y, Li, Z, Miao, Y, Huang, Z, Liu, X, et al. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin Transl Med. (2020) 10:e200. doi: 10.1002/ctm2.200
124. Wyler, E, Mosbauer, K, Franke, V, Diag, A, Gottula, LT, Arsie, R, et al. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience. (2021) 24:102151. doi: 10.1016/j.isci.2021.102151
125. Fu, Z, Wang, J, Wang, Z, Sun, Y, Wu, J, Zhang, Y, et al. A virus-derived microRNA-like small RNA serves as a serum biomarker to prioritize the COVID-19 patients at high risk of developing severe disease. Cell Discov. (2021) 7:48. doi: 10.1038/s41421-021-00289-8
126. McDonald, JT, Enguita, FJ, Taylor, D, Griffin, RJ, Priebe, W, Emmett, MR, et al. Role of miR-2392 in driving SARS-CoV-2 infection. Cell Rep. (2021) 37:109839. doi: 10.1016/j.celrep.2021.109839
127. Giuliani, A, Matacchione, G, Ramini, D, Di Rosa, M, Bonfigli, AR, Sabbatinelli, J, et al. Circulating miR-320b and miR-483-5p levels are associated with COVID-19 in-hospital mortality. Mech Ageing Dev. (2022) 202:111636. doi: 10.1016/j.mad.2022.111636
128. Smail, SW, Saeed, M, Alkasalias, T, Khudhur, ZO, Younus, DA, Rajab, MF, et al. Inflammation, immunity and potential target therapy of SARS-COV-2: a total scale analysis review. Food Chem Toxicol. (2021) 150:112087. doi: 10.1016/j.fct.2021.112087
129. Ahmed, JQ, Maulud, SQ, Dhawan, M, Priyanka,, Choudhary, OP, Jalal, PJ, et al. MicroRNAs in the development of potential therapeutic targets against COVID-19: a narrative review. J Infect Public Health. (2022) 15:788–99. doi: 10.1016/j.jiph.2022.06.012
130. Ying, H, Ebrahimi, M, Keivan, M, Khoshnam, SE, Salahi, S, and Farzaneh, M. miRNAs; a novel strategy for the treatment of COVID-19. Cell Biol Int. (2021) 45:2045–53. doi: 10.1002/cbin.11653
131. Lener, T, Gimona, M, Aigner, L, Borger, V, Buzas, E, Camussi, G, et al. Applying extracellular vesicles based therapeutics in clinical trials - an ISEV position paper. J Extracell Vesicles. (2015) 4:30087. doi: 10.3402/jev.v4.30087
132. Casado-Diaz, A, Quesada-Gomez, JM, and Dorado, G. Extracellular vesicles derived from mesenchymal stem cells (MSC) in regenerative medicine: applications in skin wound healing. Front Bioeng Biotechnol. (2020) 8:146. doi: 10.3389/fbioe.2020.00146
133. Sabbatinelli, J, Giuliani, A, Matacchione, G, Latini, S, Laprovitera, N, Pomponio, G, et al. Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients. Mech Ageing Dev. (2021) 193:111413. doi: 10.1016/j.mad.2020.111413
134. Lambert, DW, Lambert, LA, Clarke, NE, Hooper, NM, Porter, KE, and Turner, AJ. Angiotensin-converting enzyme 2 is subject to post-transcriptional regulation by miR-421. Clin Sci. (2014) 127:243–9. doi: 10.1042/CS20130420
135. Cimmino, A, Calin, GA, Fabbri, M, Iorio, MV, Ferracin, M, Shimizu, M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. (2005) 102:13944–9. doi: 10.1073/pnas.0506654102
136. Li, X, Chu, Q, and Wang, H. MicroRNA-16 regulates lipopolysaccharide-induced inflammatory factor expression by targeting TLR4 in normal human bronchial epithelial cells. Exp Ther Med. (2021) 22:982. doi: 10.3892/etm.2021.10414
137. Hum, C, Loiselle, J, Ahmed, N, Shaw, TA, Toudic, C, and Pezacki, JP. MicroRNA mimics or inhibitors as antiviral therapeutic approaches against COVID-19. Drugs. (2021) 81:517–31. doi: 10.1007/s40265-021-01474-5
138. Park, JH, Choi, Y, Lim, CW, Park, JM, Yu, SH, Kim, Y, et al. Potential therapeutic effect of microRNAs in extracellular vesicles from mesenchymal stem cells against SARS-CoV-2. Cells. (2021) 10:2393. doi: 10.3390/cells10092393
139. Chen, Y, He, Y, Wang, X, Lu, F, and Gao, J. Adipose-derived mesenchymal stem cells exhibit tumor tropism and promote tumorsphere formation of breast cancer cells. Oncol Rep. (2019) 41:2126–36. doi: 10.3892/or.2019.7018
140. Keikha, R, Hashemi-Shahri, SM, and Jebali, A. The relative expression of miR-31, miR-29, miR-126, and miR-17 and their mRNA targets in the serum of COVID-19 patients with different grades during hospitalization. Eur J Med Res. (2021) 26:75. doi: 10.1186/s40001-021-00544-4
141. Cory, H, Passarelli, S, Szeto, J, Tamez, M, and Mattei, J. The role of polyphenols in human health and food systems: a mini-review. Front Nutr. (2018) 5:87. doi: 10.3389/fnut.2018.00087
142. Montenegro-Landívar, MF, Tapia-Quirós, P, Vecino, X, Reig, M, Valderrama, C, Granados, M, et al. Polyphenols and their potential role to fight viral diseases: an overview. Sci Total Environ. (2021) 801:149719. doi: 10.1016/j.scitotenv.2021.149719
143. Iddir, M, Brito, A, Dingeo, G, Fernandez Del Campo, SS, Samouda, H, La Frano, MR, et al. Strengthening the immune system and reducing inflammation and oxidative stress through diet and nutrition: considerations during the COVID-19 crisis. Nutrients. (2020) 12:1562. doi: 10.3390/nu12061562
144. Milenkovic, D, Ruskovska, T, Rodriguez-Mateos, A, and Heiss, C. Polyphenols could prevent SARS-CoV-2 infection by modulating the expression of miRNAs in the host cells. Aging Dis. (2021) 12:1169–82. doi: 10.14336/AD.2021.0223
145. Lin, Y-P, Hsieh, Y-S, Cheng, M-H, Shen, C-F, Shen, C-J, and Cheng, C-M. Using microRNA arrays as a tool to evaluate COVID-19 vaccine efficacy. Vaccine. (2022) 10:1681. doi: 10.3390/vaccines10101681
146. Miyashita, Y, Yoshida, T, Takagi, Y, Tsukamoto, H, Takashima, K, Kouwaki, T, et al. Circulating extracellular vesicle microRNAs associated with adverse reactions, proinflammatory cytokine, and antibody production after COVID-19 vaccination. npj Vaccines. (2022) 7:16. doi: 10.1038/s41541-022-00439-3
147. Capistrano, KJ, Richner, J, Schwartz, J, Mukherjee, SK, Shukla, D, and Naqvi, AR. Host microRNAs exhibit differential propensity to interact with SARS-CoV-2 and variants of concern. Biochim Biophys Acta Mol basis Dis. (2023) 1869:166612. doi: 10.1016/j.bbadis.2022.166612
148. Keikha, R, Hashemi-Shahri, SM, and Jebali, A. The miRNA neuroinflammatory biomarkers in COVID-19 patients with different severity of illness. Neurologia. (2023) 38:e41–51. doi: 10.1016/j.nrl.2021.06.005
149. Taganov, KD, Boldin, MP, Chang, KJ, and Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. (2006) 103:12481–6. doi: 10.1073/pnas.0605298103
150. Starikova, I, Jamaly, S, Sorrentino, A, Blondal, T, Latysheva, N, Sovershaev, M, et al. Differential expression of plasma miRNAs in patients with unprovoked venous thromboembolism and healthy control individuals. Thromb Res. (2015) 136:566–72. doi: 10.1016/j.thromres.2015.07.005
151. Wang, B, Koh, P, Winbanks, C, Coughlan, MT, McClelland, A, Watson, A, et al. miR-200a prevents renal fibrogenesis through repression of TGF-β2 expression. Diabetes. (2011) 60:280–7. doi: 10.2337/db10-0892
152. Wang, B, Jha, JC, Hagiwara, S, McClelland, AD, Jandeleit-Dahm, K, Thomas, MC, et al. Transforming growth factor-beta1-mediated renal fibrosis is dependent on the regulation of transforming growth factor receptor 1 expression by let-7b. Kidney Int. (2014) 85:352–61. doi: 10.1038/ki.2013.372
153. Kato, M, Wang, L, Putta, S, Wang, M, Yuan, H, Sun, G, et al. Post-transcriptional up-regulation of Tsc-22 by Ybx1, a target of miR-216a, mediates TGF-beta-induced collagen expression in kidney cells. J Biol Chem. (2010) 285:34004–15. doi: 10.1074/jbc.M110.165027
154. Satoh, M, Minami, Y, Takahashi, Y, Tabuchi, T, and Nakamura, M. Expression of microRNA-208 is associated with adverse clinical outcomes in human dilated cardiomyopathy. J Card Fail. (2010) 16:404–10. doi: 10.1016/j.cardfail.2010.01.002
155. Das, R, Sinnarasan, VSP, Paul, D, and Venkatesan, A. A machine learning approach to identify potential miRNA-gene regulatory network contributing to the pathogenesis of SARS-CoV-2 infection. Biochem Genet. (2023) 62:987–1006. doi: 10.1007/s10528-023-10458-x
156. Centa, A, Fonseca, AS, da Silva Ferreira, SG, Azevedo, MLV, de Paula, CBV, Nagashima, S, et al. Deregulated miRNA expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients. Am J Physiol Lung Cell Mol Physiol. (2021) 320:L405–12. doi: 10.1152/ajplung.00457.2020
157. Haroun, RA, Osman, WH, Amin, RE, Hassan, AK, Abo-Shanab, WS, and Eessa, AM. Circulating plasma miR-155 is a potential biomarker for the detection of SARS-CoV-2 infection. Pathology. (2022) 54:104–10. doi: 10.1016/j.pathol.2021.09.006
158. Lu, D, Chatterjee, S, Xiao, K, Riedel, I, Wang, Y, Foo, R, et al. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J Mol Cell Cardiol. (2020) 148:46–9. doi: 10.1016/j.yjmcc.2020.08.017
159. Liu, Q, Du, J, Yu, X, Xu, J, Huang, F, Li, X, et al. miRNA-200c-3p is crucial in acute respiratory distress syndrome. Cell Discov. (2017) 3:17021. doi: 10.1038/celldisc.2017.21
160. Seto, AG, Beatty, X, Lynch, JM, Hermreck, M, Tetzlaff, M, Duvic, M, et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br J Haematol. (2018) 183:428–44. doi: 10.1111/bjh.15547
161. Kianmehr, A, Faraoni, I, Kucuk, O, and Mahrooz, A. Epigenetic alterations and genetic variations of angiotensin-converting enzyme 2 (ACE2) as a functional receptor for SARS-CoV-2: potential clinical implications. Eur J Clin Microbiol Infect Dis. (2021) 40:1587–98. doi: 10.1007/s10096-021-04264-9
Keywords: microRNAs, COVID-19, biomarkers, SARS-CoV-2, therapeutic strategies
Citation: Smail SW, Hirmiz SM, Ahmed AA, Albarzinji N, Awla HK, Amin K and Janson C (2024) Decoding the intricacies: a comprehensive analysis of microRNAs in the pathogenesis, diagnosis, prognosis and therapeutic strategies for COVID-19. Front. Med. 11:1430974. doi: 10.3389/fmed.2024.1430974
Edited by:
Pier Paolo Piccaluga, University of Bologna, ItalyReviewed by:
Abdou Kamal Allayeh, National Research Centre (Egypt), EgyptErica Diani, University of Verona, Italy
Copyright © 2024 Smail, Hirmiz, Ahmed, Albarzinji, Awla, Amin and Janson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christer Janson, Q2hyaXN0ZXIuSmFuc29uQG1lZHNjaS51dS5zZQ==
 Shukur Wasman Smail
Shukur Wasman Smail Sarah Mousa Hirmiz
Sarah Mousa Hirmiz Akhter Ahmed Ahmed
Akhter Ahmed Ahmed Niaz Albarzinji3
Niaz Albarzinji3 Harem Khdir Awla
Harem Khdir Awla Kawa Amin
Kawa Amin Christer Janson
Christer Janson