- 1Immunology Research Center, Mashhad University of Medical Sciences, Mashhad, Iran
- 2Department of Immunology and Allergy, School of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran
- 3Immunoregulation Research Center, Shahed University, Tehran, Iran
- 4Non-Communicable Diseases Research Center, Fasa University of Medical Science, Fasa, Iran
- 5Department of Immunology, Shahed University, Tehran, Iran
Introduction: Host genetic variations have been identified as potential influencers of COVID-19 infection. This study aimed to examine the association between transmembrane serine protease type 2 (TMPRSS2) rs2070788 single nucleotide polymorphism (SNP) and the prognosis of COVID-19 in Iranian populations.
Method: This case-control study was performed on 756 COVID-19 patients and 59 healthy individuals across Iran. Clinical data, blood samples, and the presence of the TMPRSS2 rs2070788: G>A SNP were determined using T-ARMS-PCR. Additionally, serum levels of tumor necrosis factor α (TNF-α), C-reactive protein (CRP), interleukin-6 (IL-6), and IL-1β were evaluated in the collected blood samples.
Results: No significant association was found between the genotypes and allele frequencies of TMPRSS2 rs2070788 SNP and susceptibility to or mortality from COVID-19 infection. However, we observed a substantial increase in IL-6 and CRP levels associated with the severity of COVID-19, while no such trend was observed for IL-1β and TNF-α. This study showed a considerable rise in TNF-α and IL-1β serum levels exclusively in COVID-19 patients with TT rs2070788 TMPRSS2 SNP genotype compared to healthy controls.
Conclusion: In this study conducted across multiple cities in Iran, no significant association was found between the TMPRSS2 rs2070788 SNP genotypes and COVID-19 severity or mortality.
1 Introduction
The global impact of coronavirus disease 2019 (COVID-19), triggered by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in a considerable societal burden, encompassing elevated mortality and morbidity rates along with substantial economic expenditures (1). The World Health Organization estimates that this virus has infected 771 million individuals worldwide and that it has caused 6.9 million deaths.1
The clinical manifestations in individuals with COVID-19 span from being asymptomatic or experiencing mild flu-like symptoms to developing pneumonia, experiencing multi-organ failure, or succumbing to mortality (2, 3). Age, sex, underlying diseases, viral variants, and host genetic variations are recognized elements that may play a role in the prognosis of COVID-19 (4, 5). Hence, there is supportive evidence indicating that genetic diversity could potentially affect both the susceptibility to and the clinical consequences of SARS-CoV-2 infection (6–8).
The transmembrane serine protease type 2 (TMPRSS2) protein plays a vital role in the priming of viral spike proteins in coronavirus infections (9, 10). TMPRSS2 is expressed in different organs, containing the gastrointestinal tract, heart, kidney, lung, and respiratory system, suggesting that their extensive distribution may enhance susceptibility to SARS-CoV-2 infection (11, 12).
According to the HUGO Gene Nomenclature Committee (HGNC), the TMPRSS2 rs2070788: G˃A single nucleotide polymorphism (SNP) is situated on chromosome 21q22.3 within an intronic region. Studies have shown that this polymorphism influences TMPRSS2 gene (PRSS10) expression (13–15). Several in silico studies investigated the potential role of this SNP in the prognosis of COVID-19 using genomic databases (16). A German case-control study demonstrated no association between TMPRSS2 rs2070788 and the severity of COVID-19 (17).
By examining the importance of the TMPRSS2 gene in the SARS-CoV-2 infection process, the incidence, and severity of COVID-19 may be directly related to the increased expression of the TMPRSS2 gene, potentially leading to diverse consequences for disease susceptibility in different communities. Pandey et al. identified a significant positive correlation between the rs2070788 SNP (G allele) and the case fatality rate (CFR) in the Indian population. Also observed was the relationship between rs2070788(G) allele and TMPRSS2 expression in the lungs (18). Martínez-Gómez et al. demonstrated an association between rs2070788 and the severity of COVID-19 in various Mexican populations (19). Mesquita et al. reported no significant association between the genotype distribution of rs2070788 and patient outcomes in a Brazilian population (20).
Novel approaches targeting the proteolytic action of TMPRSS2 in viral pathogenesis and its potential blockade have been suggested as promising avenues to decrease mortality associated with SARS-CoV-2 infection (21, 22).
Consequently, this information may prove valuable in elucidating the significance of the TMPRSS2 rs2070788 polymorphism concerning susceptibility and severity of COVID-19. Utilizing this polymorphism as a promising biomarker holds the potential to predict populations at risk. The extent to which host-specific genetic variations contribute to the severity of the cytokine storm and thereby influence the outcomes of COVID-19 is poorly understood and requires further investigation (23). In response to these findings, we initiated the first Iranian study to investigate the possible correlation between the TMPRSS2 rs2070788 polymorphism and COVID-19 outcomes, with a particular focus on inflammatory markers.
2 Method
2.1 Study participants
This study involved 756 individuals diagnosed with COVID-19 and 59 healthy control subjects. These participants were recruited from hospitals in different major cities across Iran between 2020 and 2021. All participants in the study had not received the vaccine for SARS-CoV-2. Confirmed cases of COVID-19 were determined through either real-time reverse transcription-polymerase chain reaction (RT-PCR) testing or chest CT scan results. The COVID-19 cases severity was classified into four groups—mild, moderate, severe, and critical—according to the classification established by the World Health Organization (WHO) (24). The research protocol received approval from the National Institute for Medical Research Development under the reference number IR.NIMAD.REC.1399.041.
2.2 Data collection
We gathered all medical histories and relevant personal information of the participants, including age, gender, underlying health conditions, and smoking habits, by administering a patient checklist. Before collecting blood samples, informed consent was obtained from all participants themselves or their respective family members. At the onset of hospitalization, 5 mL blood samples were taken from both patients and healthy control individuals. These samples were collected using tubes containing clot activator and tubes containing ethylenediaminetetraacetic acid (EDTA).
2.3 C-reactive protein and pro-inflammatory cytokines assessment
Serum samples were subjected to an automated immunoassay (IMMULITE 2000; Siemens Healthcare Diagnostics, United Kingdom) to quantify the levels of interleukin-6 (IL-6). IL-1β, tumor necrosis factor (TNF)-α, and C-reactive protein (CRP) levels were determined in serum samples using the 7,180 clinical analyzers (Hitachi, Japan). The analysis of TNF-α, IL-1β, and CRP was conducted using the R&D Systems biotech brand kit and ACE BIOLIS (Genbio, Ireland).
2.4 Genotyping of TMPRSS2 rs2070788 polymorphism
The DNA from the buffy coat samples of all participants was extracted utilizing a spin column kit (GenAll Exgene Cell SV mini kit, GenAll Biotechnology, South Korea).
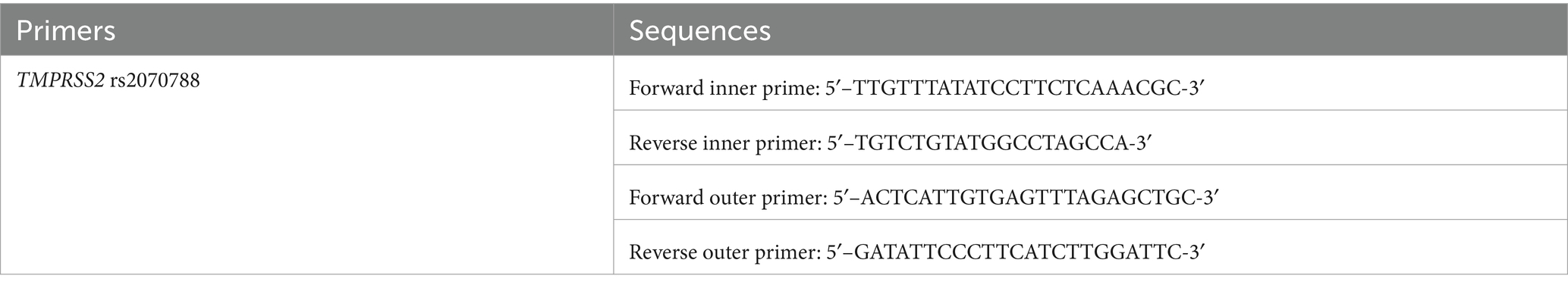
The TMPRSS2 rs2070788 polymorphism was assessed using Tetra-primer amplification refractory mutation system PCR (T-ARMS-PCR). The PRIMER1 software, available at http://primer1.soton.ac.uk/primer1.html, was employed to design T-ARMS-PCR primer pairs. The specific primers used are shown in Table 1.
The T-ARMS-PCR procedure was conducted using a total volume of 20 μL. This volume comprised 10 μL of TEMPase Hot Start 2X Master Mix (Ampliqon, Denmark), 1 μL of outer forward primer at a concentration of 10 pmol, 1 μL of outer reverse primer at a concentration of 10 pmol, 2 μL of inner forward primer at a concentration of 10 pmol, 2 μL of inner reverse primer at a concentration of 10 pmol, 2 μL of genomic DNA (ranging in concentration from 10 to 100 ng/μL), and 2 μL of Distillation Water. The PCR protocol commenced with an initial denaturation step at 95°C, lasting for 15 min. Subsequently, a total of 38 cycles were performed, consisting of denaturation at 95°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 60 s. The procedure was finalized with a concluding extension step at 72°C for 5 min. To analyze the PCR outcomes, electrophoresis was conducted on a 2% agarose gel (Figure 1).
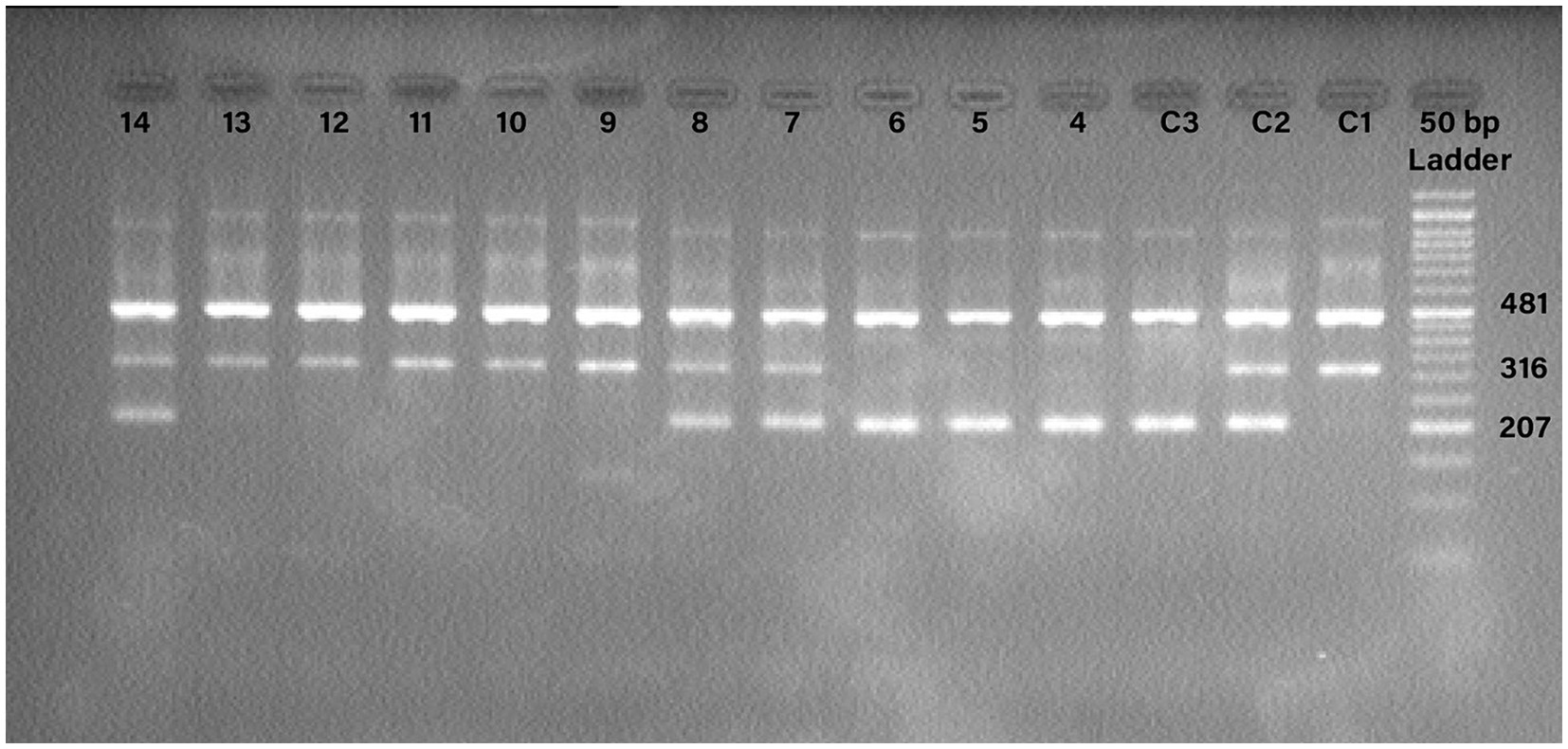
Figure 1. Detection of the PCR products for TMPRSS2 rs2070788 polymorphism. Internal control band 481 base pair (bp) is found in all wells, with band 207 bp representing allele C and band 316 bp representing allele T. C1: control TT; C2: control CT; C3: control CC; 4, 5, 6: CC; 7, 8, 14: CT; 9, 10, 11, 12, 13: TT.
To validate the T-ARMS-PCR for SNP rs2070788, approximately 10% of the samples underwent direct Sanger sequencing. Sanger sequencing was carried out on a subset of the samples using PCR primers. The alignment of the sequencing results for the rs2070788 SNP is presented in Figure 2.
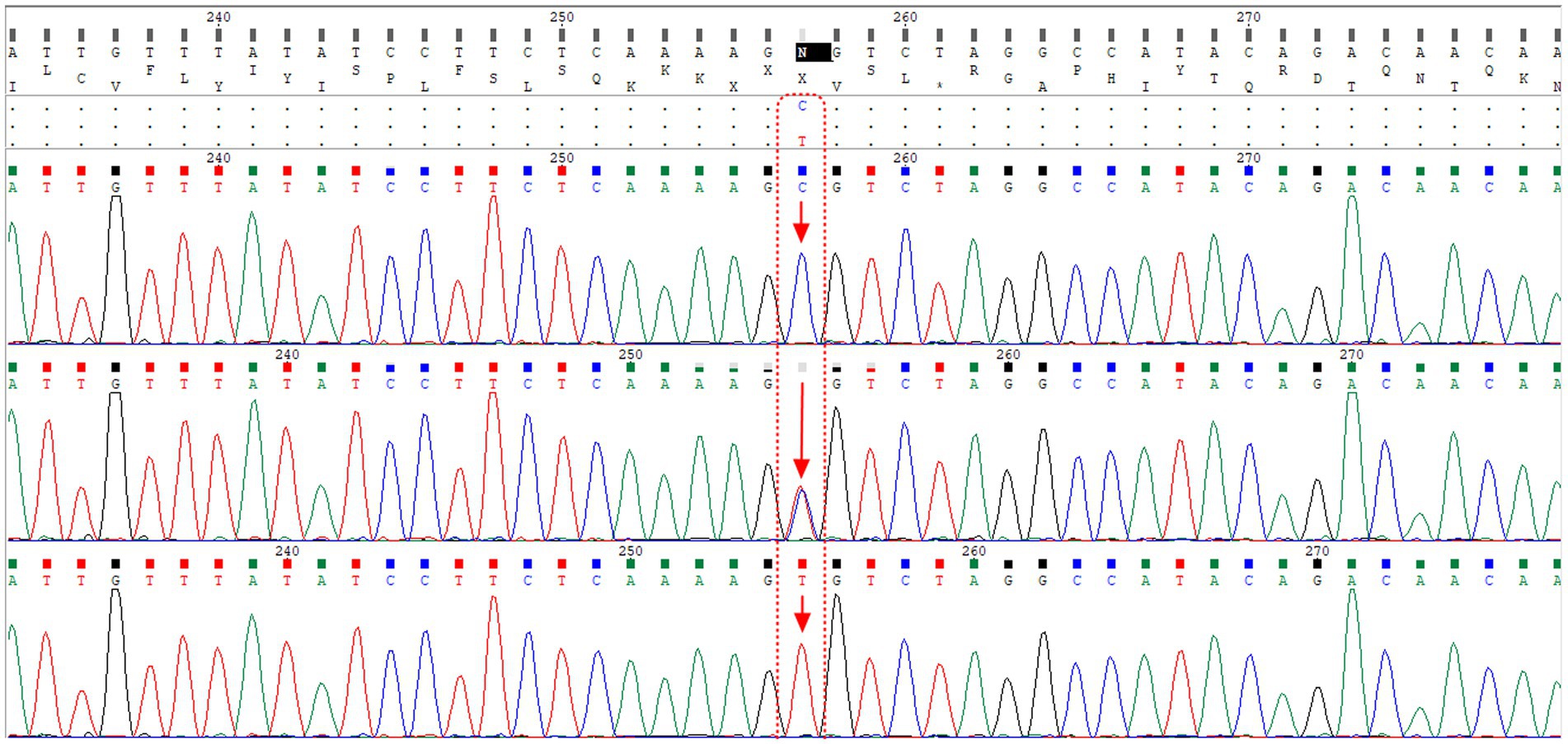
Figure 2. The alignment of sequencing results for the rs2070788 SNP, located in the TMPRSS2 gene, reveals that the first, second, and third rows correspond to samples with CC genotype, CT genotype, and TT genotype, respectively. Notably, the second row shows the presence of both peaks for both alleles.
2.5 Statistical analysis
The mean ± standard deviation was used to present the numerical variables in each group, and a Mann–Whitney test was conducted to compare the continuous data. The genotype frequencies were presented for each group as both the number and percentages (n %) and were subjected to evaluation through the Chi-square test. The Hardy–Weinberg equilibrium (HWE) was assessed using the Chi-square test. To examine the relationship between the TMPRSS2 rs2070788 polymorphism and susceptibility and severity of SARS-CoV-2 infection, multinomial or binary logistic regressions were performed. Odds ratios (ORs), both adjusted and unadjusted, along with their corresponding 95% confidence intervals (CIs), were computed. The analysis included adjustments for age, sex, diabetes mellitus (DM), cardiovascular disease (CVD), hypertension (HTN), renal disease (RD), and cigarette smoking. Statistical significance was ascertained based on a p-value below 0.05.
3 Results
3.1 Demographic characteristics
This study involved 756 patients with COVID-19 and 59 healthy controls. Patients were stratified into groups based on the severity of the disease (25). The frequencies of comorbidities such as DM, CVD, HTN, RD, and cigarette smoking can be found in Supplementary Table S1 and Table 2. Also, DM (p = 0.026), HTN (p = 0.019), RD (p = 0.005), and CVD (p = 0.021) were significantly correlated with COVID-19 mortality. No significant difference in gender was observed among the groups. The mean age showed a correlation with higher disease severity and mortality rates in COVID-19. Specifically, the mean age of various groups including healthy controls, outpatients, inpatients, ICU-admitted patients, intubated patients, and expired patients was as follows: 39.4 ± 12.1, 42.7 ± 13.5, 57.4 ± 16.7, 61.9 ± 16.6, 61.3 ± 14.6, and 65.5 ± 13.7 years, respectively. A CONSORT diagram was incorporated into the study analysis to present an overview of the entire population count process (see Supplementary Figure S1).
3.2 TMPRSS2 rs2070788 genotypes
The distribution of genotype frequencies for the TMPRSS2 rs2070788 polymorphism in both the patient and control groups demonstrated conformity to the Hardy–Weinberg equilibrium, as confirmed through Chi-square analysis (see Supplementary Table S2).
Statistical comparisons of TMPRSS2 genotypes/alleles between controls and patients (p-value).
No significant association was observed between the frequencies of different TMPRSS2 rs2070788 genotypes/alleles and the presence of comorbidities. The corresponding statistical information can be found in Supplementary Table S3.
3.3 Susceptibility to COVID-19 infection
No significant correlation was observed between the genotypes/allele frequencies of the TMPRSS2 rs2070788 SNP and the susceptibility to COVID-19 infection. Following adjustment for covariates including sex, age, DM, CVD, HTN, RD, and cigarette smoking, logistic regression analysis was conducted to compare 756 COVID-19 patients with 59 healthy controls. The findings indicated that there was no notable correlation between TMPRSS2 rs2070788 SNP genotypes and susceptibility to COVID-19 infection (Table 3).
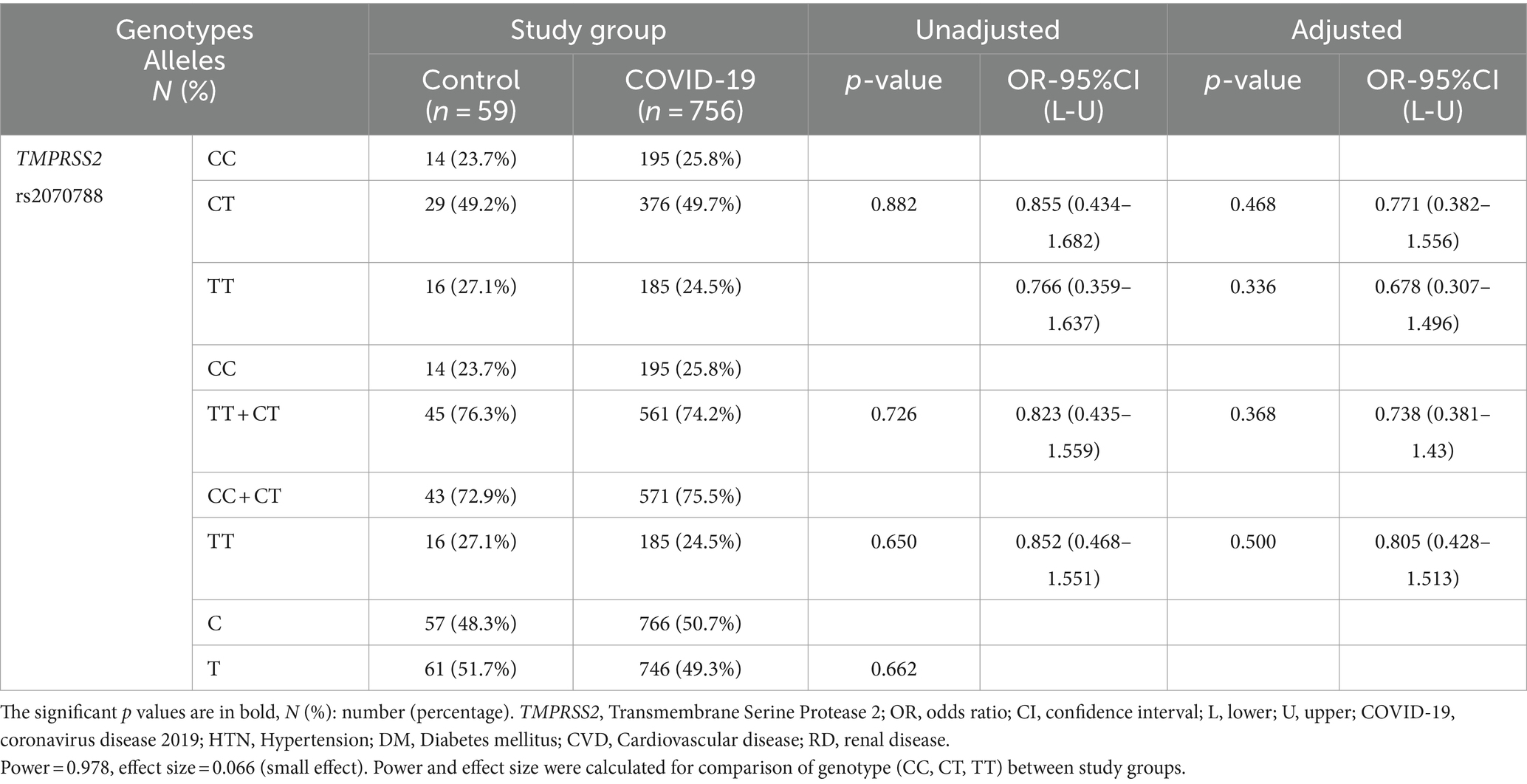
Table 3. Association of TMPRSS2 genotypes/alleles distribution with susceptibility to COVID-19, adjusted by age, sex, cigarette smoking, diabetes mellitus, HTN, CVD, and RD.
3.4 Severity and mortality of COVID-19
The frequencies of different genotypes/alleles of TMPRSS2 rs2070788 were not correlated with the severity and mortality of COVID-19 compared to different groups. After adjustment, binary logistic regression again showed no association (Tables 4, 5; Supplementary Tables S4–S6).
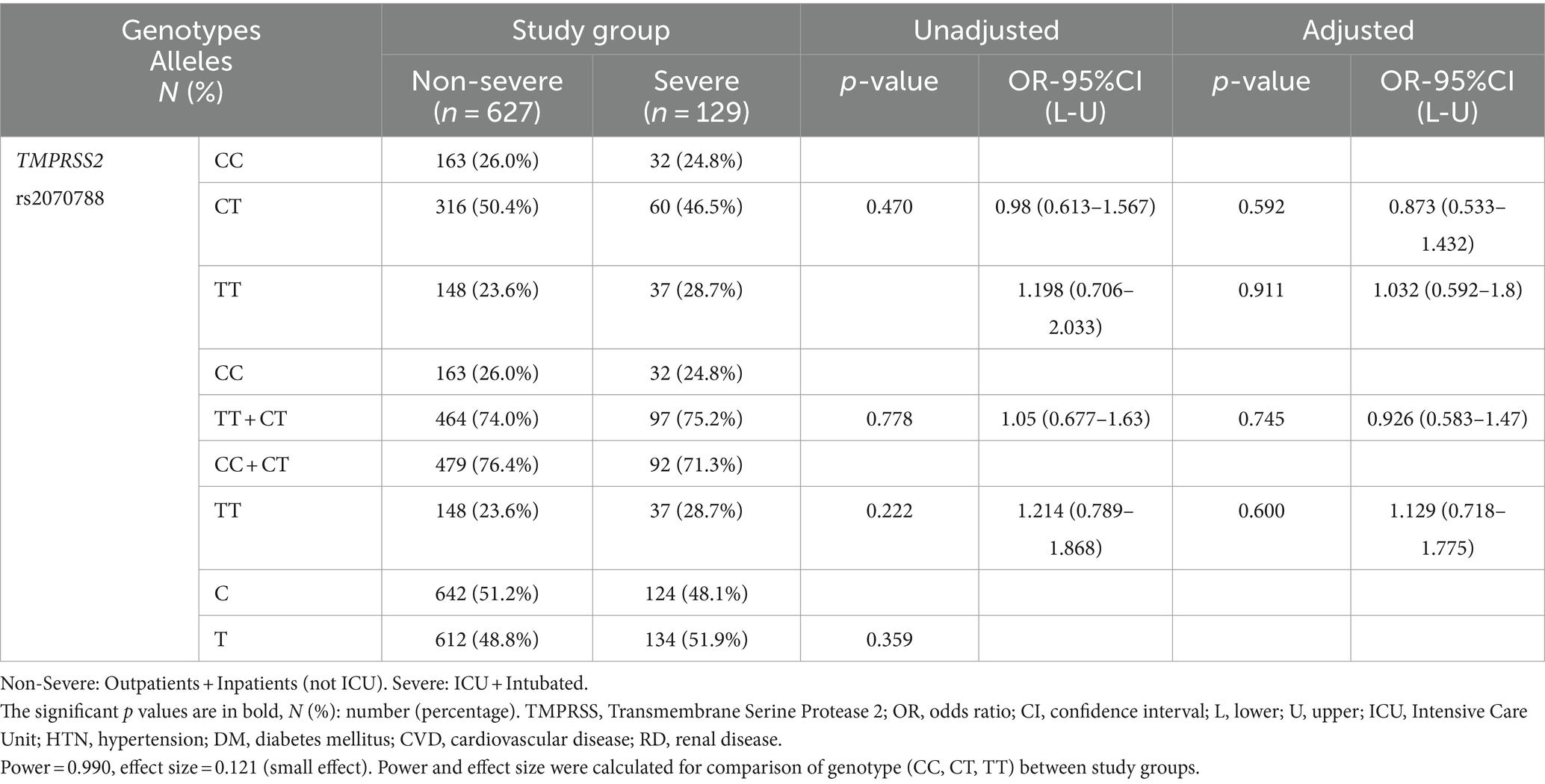
Table 4. Association TMPRSS2 genotypes/alleles distribution with susceptibility to COVID-19, adjusted by age, sex, cigarette smoking, DM, HTN, CVD, and RD.
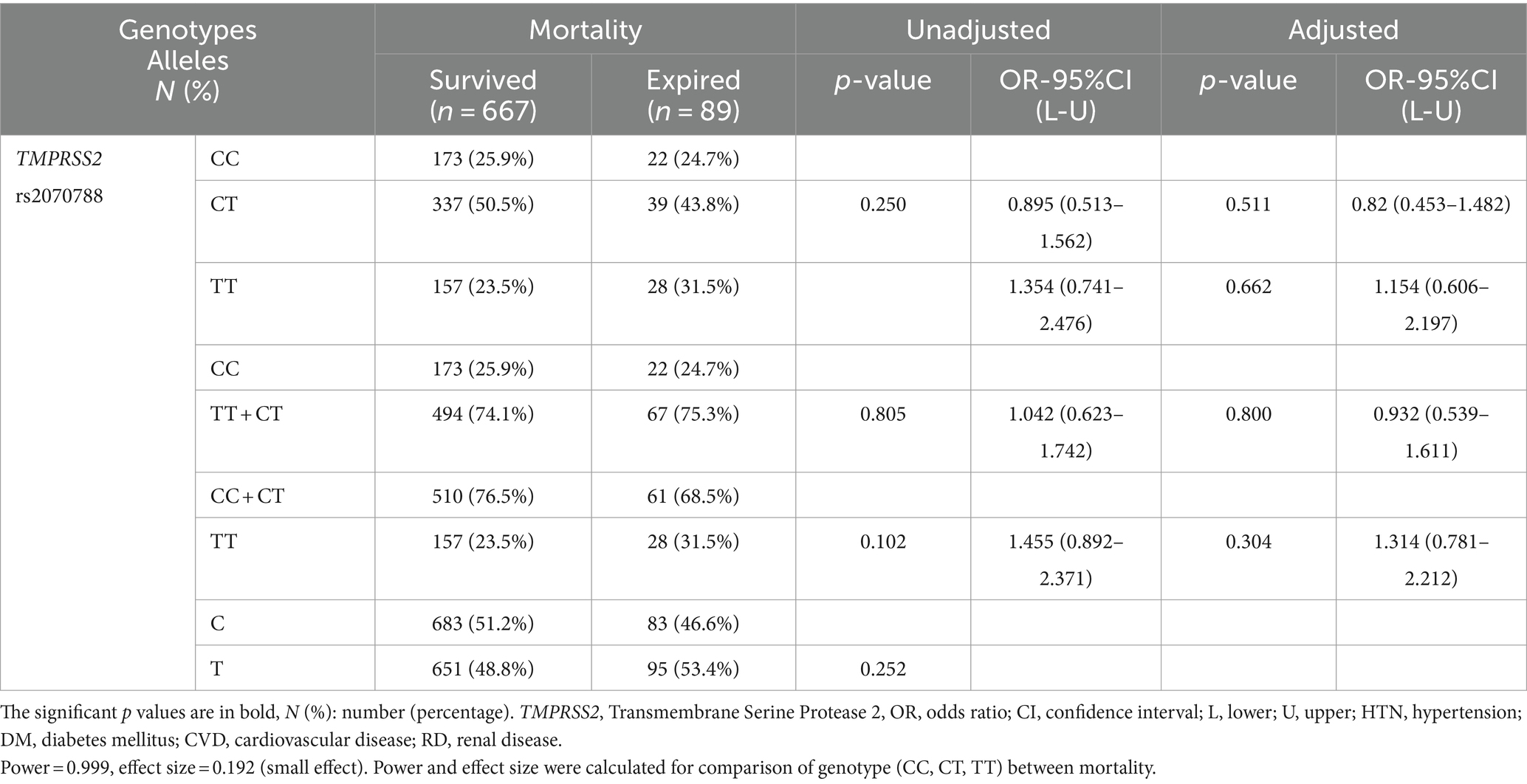
Table 5. Association of TMPRSS2 genotypes distribution with COVID-19 mortality, adjusted by age, sex, cigarette smoking, DM, HTN, CVD, and RD.
3.5 Clinical laboratory data
3.5.1 C-reactive protein
C-reactive protein (CRP) elevated along with the severity of COVID-19 in all genotypes., as shown in Supplementary Tables S7–S12. However, the mean ± SD serum levels of CRP in ICU-admitted and intubated patients only with TT + CT genotypes were shown to be significantly higher than those with TT + CT genotypes who were not admitted to the ICU or intubated (24.87 ± 20.88 vs. 19.44 ± 10.71, p = 0.001, 23.46 ± 8.05 vs. 20.26 ± 14.87, p = 0.005, respectively).
3.5.2 Interleukin-6
Among all individuals carrying TMPRSS2 rs2070788 genotypes, the IL-6 levels exhibited a significant elevation within the COVID-19 group when contrasted with the control group. Additionally, it was noticeably higher in inpatients compared to outpatients, in intubated patients compared to those not intubated, and in those who expired compared to those who survived. However, in the group that was ICU admitted, it was notably higher only in the carriers of the heterozygous genotype compared to the group that was hospitalized in the department. However, among the group that was admitted to the ICU, it was remarkably higher only in carriers of the heterozygous genotype compared to the group that was hospitalized in the regular department (Supplementary Tables S13–18; Figure 3).
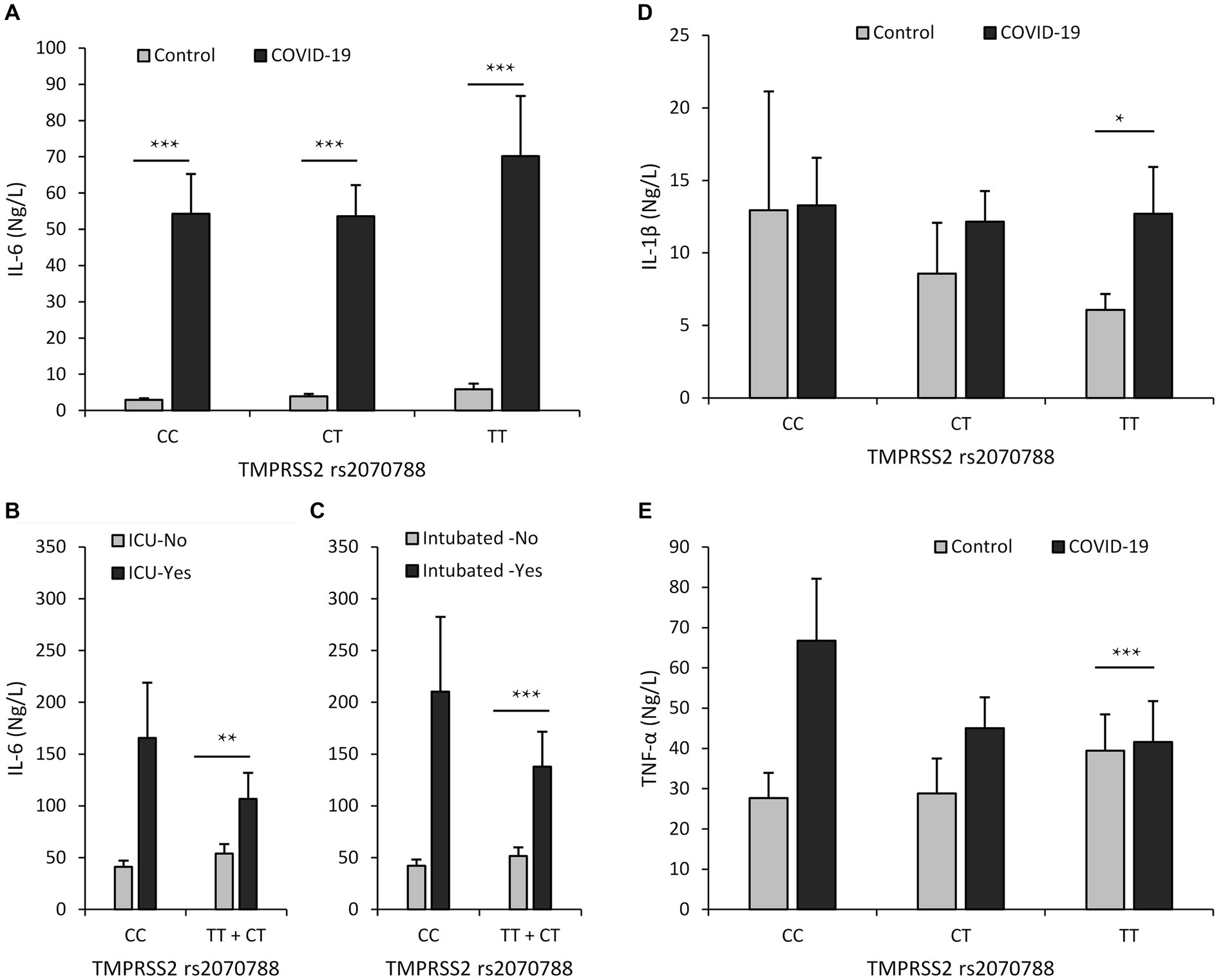
Figure 3. Relationship between serum levels of inflammatory cytokines and rs2070788 polymorphism genotypes. (A) IL-6 serum level in the COVID-19 group vs. the control group, (B) IL-6 serum level in the ICU admission group vs. ward admission, (C) IL-6 serum level in the intubated group vs. the not intubated group, (D) IL-1β serum level in the COVID-19 group vs. the control group, (E) TNF-α serum level in the COVID-19 group vs. the control group. *p < 0.05, **p < 0.01, ***p < 0.001, indicating statistically significant differences between groups.
3.5.3 Interleukin-1β
The level of Interleukin-1β (IL-1β) was elevated in COVID-19 patients compared to controls; however, this increase was determined to be significant only in carriers of the TT genotype. However, we observed that the level of IL-1β does not increase with the severity of COVID-19 disease (Supplementary Tables S19–S24; Figure 3).
3.5.4 Tumor necrosis factor-α
In COVID-19 patients, the level of tumor necrosis factor-α (TNF-α) was higher compared to controls. However, this increase was found to be significant only in individuals with the TT genotype. Nevertheless, our observations indicate that the level of TNF-α does not rise in correlation with the severity of COVID-19 disease (Supplementary Tables S25–S30; Figure 3).
4 Discussion
In this case-control study, the distribution of TMPRSS2 rs2070788 genotypes was found to be consistent with the Hardy–Weinberg equilibrium, suggesting that the chosen samples reflected the broader population. This study is the first to examine the potential correlation between the genetic factor TMPRSS2 rs2070788 SNP and COVID-19 severity in multiple predominant cities across Iran.
Following the initial evidence highlighting the significance of TMPRSS2 in the entry of SARS-CoV-2 (11), numerous studies have emerged aiming to explore the link between genetic variations in TMPRSS2 and the susceptibility to COVID-19 (26). These investigations have made use of genomic databases (13, 14, 27–30). Among the numerous single nucleotide polymorphisms (SNPs) present in the TMPRSS2 gene, certain variants, such as rs2070788 and rs12329760, have displayed potential associations as they have been linked to alterations in TMPRSS2 expression levels (13, 29, 31).
The first study to emphasize the significance of the TMPRSS2 rs2070788 SNP in the physiological mechanisms underlying viral respiratory infections was conducted on a cohort of Asian patients infected with H1N1 influenza. This study revealed that individuals carrying the rs2070788 CC genotype had a risk of severe H1N1 influenza more than twofold higher than individuals with other genotypes (31). Additionally, a recent study conducted in the Netherlands, which included 188 adult patients admitted to the hospital, exhibited a protective effect associated with the rs2070788 AA genotype against the severity of COVID-19 (32).
Conversely, a case-control study conducted in Germany, which examined 239 patients diagnosed with COVID-19, did not identify any association between the TMPRSS2 rs2070788 polymorphism and the severity of the disease (17). A cross-sectional study conducted in Spain did not find any association between TMPRSS2 rs2070788 polymorphism and long-term COVID-19 symptoms (33).
In line with these findings, our results also showed no considerable correlation between the frequency of genotypes of the TMPRSS2 rs2070788 polymorphism and the severity or mortality of COVID-19. Furthermore, across all TMPRSS2 rs2070788 genotypes, there was an observed increase in IL-6 and CRP levels in conjunction with disease severity. Conversely, no significant associations were found between disease severity and TNF-α or IL-1β levels. Previous studies have also noted a significant increase in IL-6 and CRP levels in association with disease severity (6, 34). In our study, a significant increase in IL-1β and TNF-α serum levels was exclusively observed in carriers of the TT genotype of the rs2070788 TMPRSS2 SNP in COVID-19 patients compared to healthy controls.
The precise functional mechanism through which the rs2070788 SNP, located in the noncoding region of the TMPRSS2 gene, influences the outcome of COVID-19 remains uncertain and necessitates additional research. One potential explanation is that this polymorphism could impact the stability of TMPRSS2 mRNA, including splicing processes, post-transcriptional regulation mediated by microRNAs, and the effectiveness of mRNA splicing. Factors such as the presence of silencing elements or enhancers within introns could potentially contribute to these effects.
5 Limitation
One of the study’s limitations pertained to insufficient blood sample volumes obtained from some patients, which hindered laboratory tests, including measurements of CRP, IL-1β, TNF-α, and IL-6 serum levels. Additionally, although the assessment of TMPRSS2 expression could have offered additional insights into forecasting the outcome of COVID-19, there were no remaining blood samples available to conduct these specific tests. Moreover, the control group had a relatively small sample size.
6 Conclusion
In conclusion, this case-control study examined the association between the TMPRSS2 rs2070788 SNP and COVID-19 severity in multiple cities across Iran. The findings revealed no significant correlation between the frequency of genotypes of this polymorphism and the severity or mortality of COVID-19. However, an increase in IL-6 and CRP levels was observed with disease severity across all genotypes, while no significant associations were found with TNF-α or IL-1β levels. The functional mechanism by which the rs2070788 SNP affects COVID-19 outcomes remains unclear and requires further investigation. The study focused on the need for continued research to fully understand the role of TMPRSS2 in COVID-19 pathophysiology and its potential implications for disease management and treatment strategies. Further research involving diverse ethnicities and larger sample sizes is essential to corroborate our findings. Genetic studies in the future can be influential in personalized medicine.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The studies involving humans were approved by the National Institute for Medical Research Development (IR.NIMAD.REC.1399.041), underwent a review process for studies involving human participants. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
AF: Writing – original draft, Visualization, Validation, Supervision, Project administration, Methodology, Investigation, Data curation. MM: Writing – review & editing, Visualization, Supervision, Project administration, Investigation, Conceptualization. BR: Writing – original draft, Visualization, Validation, Methodology, Investigation. MN: Writing – original draft, Visualization, Software, Resources, Methodology, Formal analysis. TG: Writing – review & editing, Validation, Supervision, Project administration, Investigation, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study received support from both the Immunoregulation Research Center at Shahed University and the Ministry of Health and Medical Education in Iran.
Acknowledgments
The authors extend their sincere gratitude to all individuals who participated in this project. They also acknowledge and appreciate the facilities and support generously provided by the universities of Mashhad, Tehran, Esfahan, Babol, and Zahedan.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2024.1425916/full#supplementary-material
Footnotes
1. ^https://covid19.who.int/ accessed on 4 October 2023.
References
1. Guo, G, Ye, L, Pan, K, Chen, Y, Xing, D, Yan, K, et al. New insights of emerging SARS-CoV-2: epidemiology, etiology, clinical features, clinical treatment, and prevention. Front Cell Dev Biol. (2020) 8:410. doi: 10.3389/fcell.2020.00410
2. Baj, J, Karakuła-Juchnowicz, H, Teresiński, G, Buszewicz, G, Ciesielka, M, Sitarz, R, et al. COVID-19: specific and non-specific clinical manifestations and symptoms: the current state of knowledge. J Clin Med. (2020) 9:1753. doi: 10.3390/jcm9061753
3. Hosseini, P, Fallahi, MS, Erabi, G, Pakdin, M, Zarezadeh, SM, Faridzadeh, A, et al. Multisystem inflammatory syndrome and autoimmune diseases following COVID-19: molecular mechanisms and therapeutic opportunities [review]. Front Mol Biosci. (2022) 9:804109. doi: 10.3389/fmolb.2022.804109
4. Dessie, ZG, and Zewotir, T. Mortality-related risk factors of COVID-19: a systematic review and meta-analysis of 42 studies and 423,117 patients. BMC Infect Dis. (2021) 21:855. doi: 10.1186/s12879-021-06536-3
5. Fricke-Galindo, I, and Falfán-Valencia, R. Genetics insight for COVID-19 susceptibility and severity: a review. Front Immunol. (2021) 12:622176. doi: 10.3389/fimmu.2021.622176
6. Faridzadeh, A, Mahmoudi, M, Ghaffarpour, S, Zamani, MS, Hoseinzadeh, A, Naghizadeh, MM, et al. The role of ACE1 I/D and ACE2 polymorphism in the outcome of Iranian COVID-19 patients: a case-control study. Front Genet. (2022) 13:955965. doi: 10.3389/fgene.2022.955965
7. Niemi, MEK, Karjalainen, J, Liao, RG, Neale, BM, Daly, M, Ganna, A, et al. Mapping the human genetic architecture of COVID-19. Nature. (2021) 600:472–7. doi: 10.1038/s41586-021-03767-x
8. Pairo-Castineira, E, Clohisey, S, Klaric, L, Bretherick, AD, Rawlik, K, Pasko, D, et al. Genetic mechanisms of critical illness in COVID-19. Nature. (2021) 591:92–8. doi: 10.1038/s41586-020-03065-y
9. Matsuyama, S, Nagata, N, Shirato, K, Kawase, M, Takeda, M, and Taguchi, F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. (2010) 84:12658–64. doi: 10.1128/jvi.01542-10
10. Shulla, A, Heald-Sargent, T, Subramanya, G, Zhao, J, Perlman, S, and Gallagher, T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. (2011) 85:873–82. doi: 10.1128/jvi.02062-10
11. Hoffmann, M, Kleine-Weber, H, Schroeder, S, Krüger, N, Herrler, T, Erichsen, S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. (2020) 181:271–280.e8. doi: 10.1016/j.cell.2020.02.052
12. Vaarala, MH, Porvari, KS, Kellokumpu, S, Kyllönen, AP, and Vihko, PT. Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues. J Pathol. (2001) 193:134–40. doi: 10.1002/1096-9896(2000)9999:9999<::Aid-path743>3.0.Co;2-t
13. Irham, LM, Chou, WH, Calkins, MJ, Adikusuma, W, Hsieh, SL, and Chang, WC. Genetic variants that influence SARS-CoV-2 receptor TMPRSS2 expression among population cohorts from multiple continents. Biochem Biophys Res Commun. (2020) 529:263–9. doi: 10.1016/j.bbrc.2020.05.179
14. Paniri, A, Hosseini, MM, and Akhavan-Niaki, H. First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations. J Biomol Struct Dyn. (2021) 39:3576–93. doi: 10.1080/07391102.2020.1767690
15. Paoloni-Giacobino, A, Chen, H, Peitsch, MC, Rossier, C, and Antonarakis, SE. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22.3. Genomics. (1997) 44:309–20. doi: 10.1006/geno.1997.4845
16. de Andrade, CC, Silva, ATP, Vasconcelos, LRS, Oliveira, PRS, de Souza, CDF, da Costa Armstrong, A, et al. A polymorphism in the TMPRSS2 gene increases the risk of death in older patients hospitalized with COVID-19. Viruses. (2022) 14:2557. doi: 10.3390/v14112557
17. Schönfelder, K, Breuckmann, K, Elsner, C, Dittmer, U, Fistera, D, Herbstreit, F, et al. Transmembrane serine protease 2 polymorphisms and susceptibility to severe acute respiratory syndrome coronavirus type 2 infection: a German case-control study. Front Genet. (2021) 12:667231. doi: 10.3389/fgene.2021.667231
18. Pandey, RK, Srivastava, A, Singh, PP, and Chaubey, G. Genetic association of TMPRSS2 rs2070788 polymorphism with COVID-19 case fatality rate among Indian populations. Infect Genet Evol. (2022) 98:105206. doi: 10.1016/j.meegid.2022.105206
19. Martínez-Gómez, LE, Martinez-Armenta, C, Tusie-Luna, T, Vázquez-Cárdenas, P, Vidal-Vázquez, RP, Ramírez-Hinojosa, JP, et al. The fatal contribution of serine protease-related genetic variants to COVID-19 outcomes [original research]. Front Immunol. (2024) 15:1335963. doi: 10.3389/fimmu.2024.1335963
20. Mesquita, FP, da Silva, JBS, de Oliveira, LLB, Lima, LB, Souza, PFN, Silva, EL, et al. Human TMPRSS2 and ACE2 genetic variability on COVID-19 outcomes in patients from Brazil. Hum Gene Ther. (2024) 41:201310. doi: 10.1016/j.humgen.2024.201310
21. Mahoney, M, Damalanka, VC, Tartell, MA, Chung, DH, Lourenço, AL, Pwee, D, et al. A novel class of TMPRSS2 inhibitors potently block SARS-CoV-2 and MERS-CoV viral entry and protect human epithelial lung cells. Proc Natl Acad Sci U S A. (2021) 118:e2108728118. doi: 10.1073/pnas.2108728118
22. Shapira, T, Monreal, IA, Dion, SP, Buchholz, DW, Imbiakha, B, Olmstead, AD, et al. A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature. (2022) 605:340–8. doi: 10.1038/s41586-022-04661-w
23. Alagbe, AE, Tonassé, WV, Pedroso, GA, Just-Junior, AS, Costa, E, Oliveira, BB, et al. Association of some host genetic polymorphisms with plasma HYPERCYTOKINEMA in COVID-19. Hematol Transfus Cell Ther. (2023) 45:S86. doi: 10.1016/j.htct.2023.09.231
24. World Health Organization . COVID-19 clinical management: living guidance, 25 January 2021. (World Health Organization) (2021).
25. COVID-19 treatment guidelines. Available at: https://www.covid19treatmentguidelines.nih.gov/ (Accessed April 21, 2020).
26. Rokni, M, Heidari Nia, M, Sarhadi, M, Mirinejad, S, Sargazi, S, Moudi, M, et al. Association of TMPRSS2 gene polymorphisms with COVID-19 severity and mortality: a case-control study with computational analyses. Appl Biochem Biotechnol. (2022) 194:3507–26. doi: 10.1007/s12010-022-03885-w
27. Asselta, R, Paraboschi, EM, Mantovani, A, and Duga, S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging (Albany NY). (2020) 12:10087–98. doi: 10.18632/aging.103415
28. Barash, A, Machluf, Y, Ariel, I, and Dekel, Y. The pursuit of COVID-19 biomarkers: putting the spotlight on ACE2 and TMPRSS2 regulatory sequences [brief research report]. Front Med. (2020) 7:582793. doi: 10.3389/fmed.2020.582793
29. David, A, Parkinson, N, Peacock, TP, Pairo-Castineira, E, Khanna, T, Cobat, A, et al. A common TMPRSS2 variant has a protective effect against severe COVID-19. Curr Res Transl Med. (2022) 70:103333. doi: 10.1016/j.retram.2022.103333
30. Hou, Y, Zhao, J, Martin, W, Kallianpur, A, Chung, MK, Jehi, L, et al. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. (2020) 18:216. doi: 10.1186/s12916-020-01673-z
31. Cheng, Z, Zhou, J, To, K. K.-WChu, H, Li, C, Wang, D, et al. Identification of TMPRSS2 as a susceptibility gene for severe 2009 pandemic a(H1N1) influenza and a(H7N9) influenza. J Infect Dis. (2015) 212:1214–21. doi: 10.1093/infdis/jiv246
32. Akin, S, Schriek, P, van Nieuwkoop, C, Neuman, RI, Meynaar, I, van Helden, EJ, et al. A low aldosterone/renin ratio and high soluble ACE2 associate with COVID-19 severity. J Hypertens. (2022) 40:606–14. doi: 10.1097/hjh.0000000000003054
33. Fernández-de-Las-Peñas, C, Arendt-Nielsen, L, Díaz-Gil, G, Gómez-Esquer, F, Gil-Crujera, A, Gómez-Sánchez, SM, et al. Genetic association between ACE2 (rs2285666 and rs2074192) and TMPRSS2 (rs12329760 and rs2070788) polymorphisms with post-COVID symptoms in previously hospitalized COVID-19 survivors. Genes (Basel). (2022) 13:1935. doi: 10.3390/genes13111935
34. Rostamian, A, Ghazanfari, T, Arabkheradmand, J, Edalatifard, M, Ghaffarpour, S, Salehi, MR, et al. Interleukin-6 as a potential predictor of COVID-19 disease severity in hospitalized patients and its association with clinical laboratory routine tests. Immunoregulation. (2020) 3:29–36. doi: 10.32598/Immunoregulation.3.1.4
Keywords: coronavirus disease 2019, TMPRSS2, SNP, polymorphism, rs2070788
Citation: Faridzadeh A, Mahmoudi M, Rahimlou B, Naghizadeh MM and Ghazanfari T (2024) Association between TMPRSS2 rs2070788 polymorphism and COVID-19 severity: a case-control study in multiple cities of Iran. Front. Med. 11:1425916. doi: 10.3389/fmed.2024.1425916
Edited by:
Gyaneshwer Chaubey, Banaras Hindu University, IndiaReviewed by:
Olga Scudiero, University of Naples Federico II, ItalyApt Lalu Muhammad Irham M. Farm, Ahmad Dahlan University, Indonesia
Copyright © 2024 Faridzadeh, Mahmoudi, Rahimlou, Naghizadeh and Ghazanfari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mahmoud Mahmoudi, bWFobW91ZGltQG11bXMuYWMuaXI=; Tooba Ghazanfari, VGdoYXphbmZhcmlAeWFob28uY29t
 Arezoo Faridzadeh
Arezoo Faridzadeh Mahmoud Mahmoudi
Mahmoud Mahmoudi Bahman Rahimlou
Bahman Rahimlou Mohammad Mehdi Naghizadeh
Mohammad Mehdi Naghizadeh Tooba Ghazanfari
Tooba Ghazanfari