- 1School of Medical Information Engineering, Anhui University of Chinese Medicine, Hefei, China
- 2Center for Xin’an Medicine and Modernization of Traditional Chinese Medicine of IHM, Anhui University of Chinese Medicine, Hefei, China
Introduction: Large Language Models (LLMs) play a crucial role in clinical information processing, showcasing robust generalization across diverse language tasks. However, existing LLMs, despite their significance, lack optimization for clinical applications, presenting challenges in terms of illusions and interpretability. The Retrieval-Augmented Generation (RAG) model addresses these issues by providing sources for answer generation, thereby reducing errors. This study explores the application of RAG technology in clinical gastroenterology to enhance knowledge generation on gastrointestinal diseases.
Methods: We fine-tuned the embedding model using a corpus consisting of 25 guidelines on gastrointestinal diseases. The fine-tuned model exhibited an 18% improvement in hit rate compared to its base model, gte-base-zh. Moreover, it outperformed OpenAI’s Embedding model by 20%. Employing the RAG framework with the llama-index, we developed a Chinese gastroenterology chatbot named “GastroBot,” which significantly improves answer accuracy and contextual relevance, minimizing errors and the risk of disseminating misleading information.
Results: When evaluating GastroBot using the RAGAS framework, we observed a context recall rate of 95%. The faithfulness to the source, stands at 93.73%. The relevance of answers exhibits a strong correlation, reaching 92.28%. These findings highlight the effectiveness of GastroBot in providing accurate and contextually relevant information about gastrointestinal diseases. During manual assessment of GastroBot, in comparison with other models, our GastroBot model delivers a substantial amount of valuable knowledge while ensuring the completeness and consistency of the results.
Discussion: Research findings suggest that incorporating the RAG method into clinical gastroenterology can enhance the accuracy and reliability of large language models. Serving as a practical implementation of this method, GastroBot has demonstrated significant enhancements in contextual comprehension and response quality. Continued exploration and refinement of the model are poised to drive forward clinical information processing and decision support in the gastroenterology field.
1 Introduction
In recent years, there has been significant advancement in large language models (LLMs), with notable models like ChatGPT demonstrating remarkable performance in question answering, summarization, and content generation (1–3). These models exhibit robust generalization not only within natural language processing (NLP) tasks but also across various interdisciplinary domains (4). However, models akin to ChatGPT, trained on general datasets, lack specific optimizations for clinical applications and encounter issues in question generation characterized by “perceptual illusions” and “unrealistic” features (5–7), potentially resulting in the provision of incomplete or inaccurate information and posing inherent risks (8, 9). To specialize LLMs, three methods have been proposed: optimizing the original LLM model (10), employing prompt engineering (11–13), and Retrieval-Augmented Generation (RAG) (14).
RAG, introduced in 2020, is a retrieval-augmented technique capable of fetching information from external knowledge sources, thus significantly enhancing answer accuracy and relevance (15). In recent years, RAG technology has proven effective in the biomedical field (16). Wang et al. (17) developed Almanac, which improved medical guideline retrieval. Ge et al. (18) created Li Versa for liver disease queries, while Ranjit et al. (19) applied RAG to radiology reports. Yu et al. (20) utilized RAG for diagnosing heart disease and sleep apnea, whereas Lozano et al. (21) and Manathunga et al. (22) applied it to medical literature and education.
This study focuses on the application of RAG in the field of clinical gastroenterology in China, aiming to address the issue associated with the continuous increase in the infection rate of Helicobacter pylori and the rising incidence of gastric cancer (23, 24). Given the substantial patient population with gastrointestinal diseases and the complexity of diagnosis and treatment, the “illusions” brought about by LLMs may pose additional challenges to the diagnosis and treatment of gastrointestinal diseases (5, 7). Integrating RAG is crucial for enhancing the accuracy of clinical practitioners in managing these diseases and can effectively mitigate this issue.
The aim of this study is to leverage RAG and large-scale language models, utilizing 25 guidelines on gastrointestinal diseases and 40 recent gastrointestinal literature sources as external knowledge bases, to develop a dedicated chatbot for gastrointestinal diseases named GastroBot. Furthermore, to enhance the relevance between retrieved content and user queries, this study conducted domain-specific fine-tuning of the embedding model tailored to gastrointestinal diseases, directly enhancing the performance of RAG. GastroBot is capable of providing precise diagnosis and treatment recommendations for gastrointestinal patients, thereby improving treatment efficacy. Figure 1 illustrates the comprehensive workflow of GastroBot.
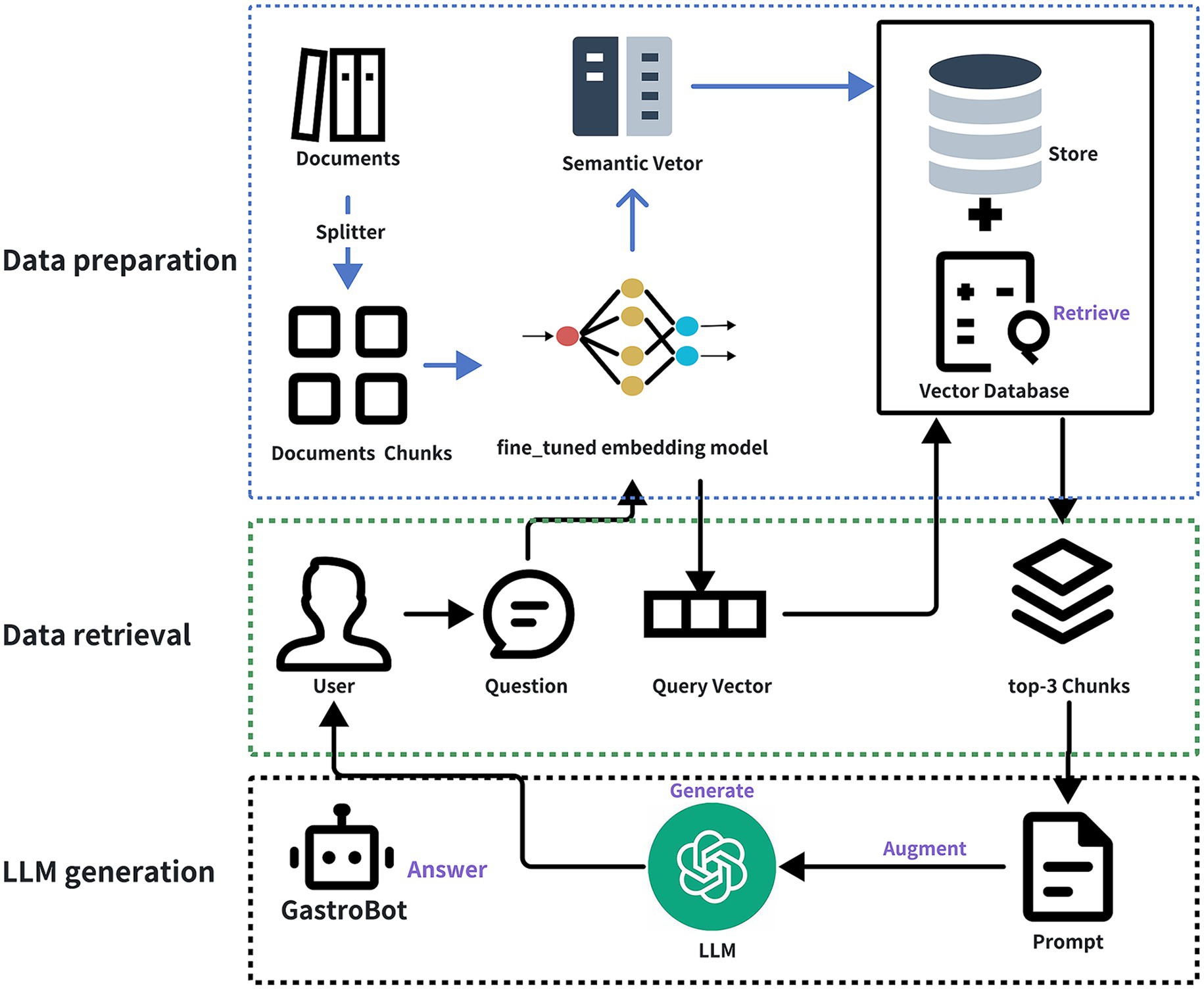
Figure 1. The overview of GastroBot. The process begins with the data preparation stage, where documents are initially split using a Splitter, dividing them into multiple document chunks. Subsequently, each chunk is encoded using the fine-tuned embedding model, producing semantic vectors stored in a Vector Database. Moving on to the data retrieval stage, the user inputs a question (User Question), and based on the question vector (Query Vector), the most relevant chunks (top-3 Chunks) are retrieved from the vector database. Finally, in the LLM generation phase, the large model generates answers by combining the top-3 chunks and the prompt, submitting them together to the LLM to obtain the final answer (Answer).
In summary, the main contributions can be summarized as follows:
• We have created a specialized dataset named the “EGD Database” specifically for Chinese gastrointestinal diseases.
• We performed domain-specific fine-tuning of the embedding model to enhance retrieval performance for gastrointestinal diseases.
• We utilized 25 gastrointestinal disease guidelines and 40 related literature articles as the knowledge base to develop a gastrointestinal disease chatbot named GastroBot using RAG and LLM.
2 Materials and methods
2.1 Dataset and data preprocessing
In order to develop a gastrointestinal chatbot tailored for the Chinese context, we initially sourced 25 clinical guideline documents related to gastrointestinal diseases from the Chinese Medical Journal Full-text Database.1 These guidelines were selected based on their alignment with the most current official guidelines in the field, ensuring comprehensive coverage across various dimensions. Additionally, we integrated the latest literature on gastroenterology from the China National Knowledge Infrastructure (CNKI) database,2 categorized under the discipline of digestive system diseases. These articles covered a range of topics including gastroesophageal reflux disease, Helicobacter pylori, clinical observations, H. pylori infection, and peptic ulcer diseases, all published in 2024, totaling 40 articles.
Subsequently, we conducted data preprocessing on the collected dataset, removing elements such as English abstracts and references that were irrelevant to our research objectives. Given RAG’s inability to process images, all image data were excluded during this preprocessing stage. The resulting refined dataset was named the “EGD Database,” with EGD standing for Expert Guidelines for Gastrointestinal Diseases.
2.2 Experiment
The experimental section comprises two crucial steps aimed at developing a dedicated Chinese gastrointestinal disease chatbot, named GastroBot, for knowledge-based question answering on gastrointestinal diseases. The initial steps involve fine-tuning the embedding model specifically for gastrointestinal diseases. Subsequently, the LlamaIndex3 is employed to construct the RAG pipeline.
2.2.1 Fine-tuning embedding model
The objective of fine-tuning is to strengthen the correlation between retrieved content and queries. Fine-tuning the embedding model aims to optimize the influence of retrieved content on generating outputs. Particularly in the medical domain, characterized by evolving or rare terminology, these tailored embedding techniques can enhance retrieval relevance. The GTE (25) embedding model is renowned for its high performance. In this study, the gte-base-zh model from Alibaba DAMO Academy served as the foundational embedding model and underwent domain-specific fine-tuning.
For fine-tuning the gte-base-zh model, we employed GPT-3.5 Turbo to aid in generating question-answer pairs. During the fine-tuning process, the LLM generated questions based on document blocks, forming pairs with their respective answers. Then, the SentenceTransformersFinetuneEngine in LlamaIndex was utilized for fine-tuning. The fine-tuning of the customized Chinese gastrointestinal domain embedding model was accomplished through the steps illustrated in Figure 2.
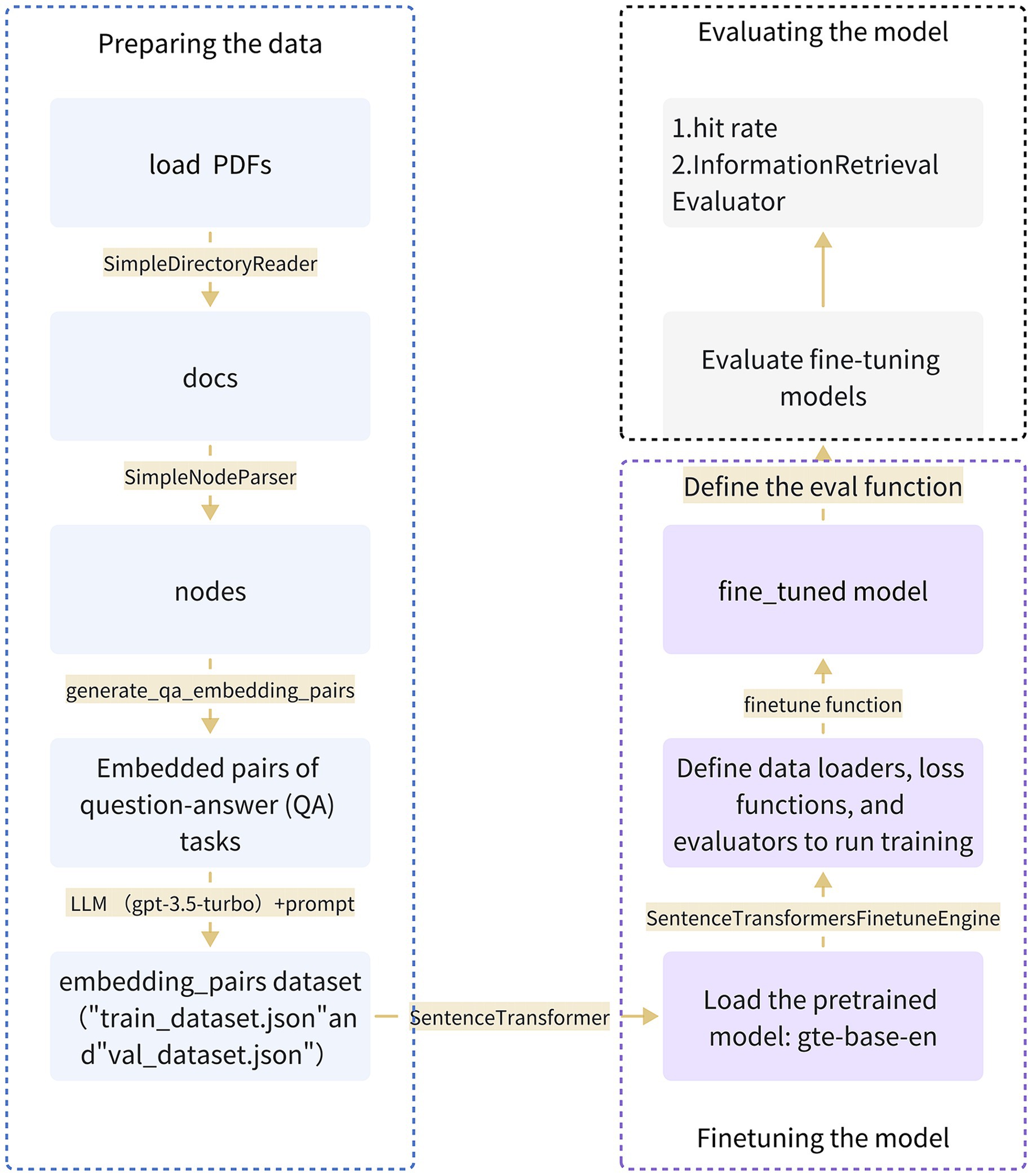
Figure 2. Provides a comprehensive overview of the entire process involved in fine-tuning the embedding model specific to the Chinese gastroenterology domain. The process encompasses three key steps: data preparation, model fine-tuning, and model evaluation.
2.2.1.1 Data preprocessing in the fine-tuning stage
In this study, we leveraged the EGD database as the source of fine-tuning data and underwent a multi-step preprocessing procedure to adapt it into a corpus suitable for training and evaluation. Initially, we loaded cleaned and processed PDF files containing essential text information. Subsequently, the SimpleDirectoryReader tool was employed to extract data from designated files, generating a list containing all documents. Then, utilizing the SimpleNodeParser, we extracted meaningful node information from the documents. This parser was encapsulated within a callable function named load_corpus. After obtaining textual nodes, the data underwent transformation and processing through the generate_qa_embedding_pairs function in LlamaIndex to produce QA pairs suitable for fine-tuning embedding models. Furthermore, we utilized gpt-3.5-turbo to define context information and questions, establishing prompt generation templates. The resulting output is an embedded pair dataset, saved as “train_dataset,” which includes “train_dataset.json” and “val_dataset.json.” This process laid the groundwork for subsequent training and evaluation of the fine-tuned Chinese gastrointestinal embedding model.
2.2.1.2 Training the fine-tuned model
We chose the gte-base-zh model from the GTE series models trained by Alibaba DAMO Academy as the embedding model for the fine-tuning phase. Throughout the fine-tuning process, the SentenceTransformersFinetuneEngine was employed to carry out various subtasks. This involved constructing a pre-training model using SentenceTransformer, defining a data loader responsible for loading the training dataset and parsing it into queries, corpus, and relevant_docs. Leveraging the gte-base-zh model, the engine mapped node_ids from relevant_docs to text nodes in the corpus and compiled a list of Input Examples. Training employed the multiple negatives ranking (MNR) loss from sentence_transformers, with an evaluator monitoring the model’s performance using the eval dataset throughout training. The entire process was seamlessly integrated into the training pipeline, encapsulated within the SentenceTransformersFinetuneEngine in LlamaIndex, and executed by invoking its fine-tuning function.
2.2.2 Implementing retrieval-augmented generation with Llamaindex
The LlamaIndex framework is employed to construct the pipeline for RAG implementation. Initially, data loading is conducted to facilitate subsequent experiments. The entire document is segmented into smaller text units termed nodes, facilitating processing within the LlamaIndex framework. Using the SimpleNodeParser, the loaded documents are parsed and transformed into these nodes. Following this, within the global configuration object, the embedding model and LLM are explicitly specified. The chosen embedding model is fine-tuned for gastrointestinal diseases, streamlining the conversion of text into vector representations crucial for subsequent computations. The gpt-3.5 turbo is selected as the generative LLM and is utilized throughout the process for answer generation. Finally, three core components—index, retriever, and query engine—are instantiated via the LlamaIndex framework, collectively supporting question-answering functionality based on user data or documents. The index serves as a data structure for swiftly retrieving information pertinent to user queries from external documents. This is accomplished through the vector storage index, which generates vector embeddings for the text of each node. The retriever is responsible for acquiring and retrieving information relevant to user queries, while the query engine, built upon the index and retriever, furnishes a universal interface for posing questions to the data.
2.2.2.1 Data preparation
2.2.2.1.1 Load data
We are populating GastroBot’s knowledge base with a curated selection of 25 guidelines on gastrointestinal diseases and 40 research papers on gastroenterology. Leveraging Llamaindex’s SimpleDirectoryReader and PDFReader classes facilitates the loading and extraction of data from PDF documents, ensuring a streamlined dataset that prioritizes information pertinent to clinical gastroenterology.
2.2.2.1.2 Chunking
Subsequently, in the text segmentation step, once the data extraction is complete, the document is partitioned into multiple text blocks referred to as chunks within LlamaIndex. Each chunk’s size is defined as 512 characters. Although the default ID for each node is a randomly generated text string, we have the flexibility to format it into a specific pattern as required.
2.2.2.1.3 Embedding
Each chunk undergoes encoding using the fine-tuned embedding model to generate semantic vectors that encapsulate nuanced information within the captured segments. This fine-tuned embedding model excels particularly well in capturing specialized vocabulary associated with gastrointestinal diseases. Vectorization is pivotal as it transforms text data into a matrix of vectors, directly influencing the effectiveness of subsequent retrieval operations. While existing generic embedding models may serve adequately in many scenarios, in the medical domain, where rare specialized vocabulary or terminology is prevalent, we opted to fine-tune GTE to suit our specific application needs and enhance retrieval efficiency.
2.2.2.1.4 Vector database
The semantic vectors generated are stored within the Vector Database, establishing an indexed repository optimized for swift and semantically aligned searches. This Vector Database forms the cornerstone for efficient retrieval in the subsequent phases of the RAG model. The intricacies of these steps are elaborated upon in the data preparation section of Figure 1.
2.2.2.2 Building RAG
Having completed the data preparation phase, the second step involves the selection of the embedding model and LLM. The embedding model is tasked with generating vector embeddings for each text chunk, for which we employ a fine-tuned embedding model. Meanwhile, the LLM handles user queries and related text chunks, producing contextually relevant answers. To achieve this, we opt to utilize the gpt-3.5-turbo model via API calls. Both models collaborate synergistically within the service framework, playing indispensable roles in the indexing and querying processes. Subsequently, in the third step, we call upon LlamaIndex to construct the index, retriever, and query engine—these three pivotal components collectively facilitate question-answering based on user data or documents.
The index facilitates swift retrieval of information relevant to user queries directly from the external knowledge base. This is achieved through the creation of vector embeddings for the text of each node within the vector storage index. The retriever’s role is to acquire and retrieve information pertinent to user queries, while the query engine, positioned atop the index and retriever, offers a universal interface for posing inquiries to the data. The fundamental implementation of RAG, based on LlamaIndex, streamlines this process.
When a user poses a question, it undergoes conversion into a vector representation. Using this query vector, the most relevant segments (top-3 chunks) are retrieved from the vector database, constituting the Data retrieval phase depicted in Figure 1. Following this, the top-3 chunks, along with the prompt, are fed into the gpt-3.5-turbo model for answer generation, culminating in the final answer, as illustrated in the LLM generation phase of Figure 1. Throughout the process delineated in Figure 1, the user query is embedded into the same vector space as the additional context retrieved from the vector database, facilitating similarity-based search and returning the most proximate data objects, denoted as retrieval (labeled as Retrieve in the figure). The amalgamation of the user query and the supplementary context obtained from the prompt template is referred to as augmentation (labeled as Augment in the figure). Ultimately, the augmented prompt is input into the LLM for answer generation, a step termed generation (labeled as Generate in the figure).
2.2.3 Comparative experiment
To demonstrate the superior performance of GastroBot, we conducted a comparative analysis between GastroBot and three baseline models utilizing RAG. When selecting these comparative models, we considered their scale, performance metrics, and diversity. Firstly, we selected Llama2 (26), an open-source language model that consistently outperforms other models across various external benchmark tests, including inference, encoding, proficiency, and knowledge evaluation. Secondly, we included ChatGLM-6B (27) and Qwen-7B (28), representing the latest advancements in Chinese artificial intelligence, as additional comparative models for this study, both demonstrating robust capabilities across multiple natural language processing tasks. We randomly selected 20 questions related to gastrointestinal diseases and compared the answers generated by GastroBot with those generated by the other three models. The performance of GastroBot relative to the other three models will be evaluated using human assessment methods.
2.3 Evaluation
2.3.1 Embedding model evaluation
In this section, we will evaluate three different embedding models: OpenAI text-embedding-ada-002 (29), gte-base-zh, and our fine-tuned embedding model. We will employ two distinct evaluation methods:
Hit rate (30): Conducting a straightforward top-k retrieval for each query/relevant_doc pair. A retrieval is considered successful if the search result includes the relevant_doc, defining it as a “hit.” The hit ratio is shown in Equation (1):
Information Retrieval Evaluator: A comprehensive metric suite provided by the LlamaIdex for the evaluation of open-source embeddings. This class evaluates an Information Retrieval (IR) (31) setting. Given a set of queries and a large corpus set. It will retrieve for each query the top-k most similar document. It measures Mean Reciprocal Rank (MRR) (32), Recall@k, and Normalized Discounted Cumulative Gain (NDCG) (33, 34).
2.3.2 Using RAGAs to evaluate RAG
Ragas (35) is a large-scale model evaluation framework designed to assess the effectiveness of Retrieval-Augmented Generation (RAG). It aids in analyzing the output of models, providing insights into their performance on a given task.
To assess the RAG system, Ragas requires the following information:
Questions: Queries provided by users.
Answers: Responses generated by the RAG system (elicited from a large language model, LLM).
Contexts: Documents relevant to the queries retrieved from external knowledge sources.
Ground Truths: Authentic answers provided by humans, serving as the correct references based on the questions. This is the sole required input.
Once Ragas obtains this information, it will utilize LLMs to evaluate the RAG system.
Ragas’s evaluation metrics comprise Faithfulness, Answer Relevance, Context Precision, Context Relevancy, Context Recall, Answer Semantic Similarity, Answer Correctness, and Aspect Critique. For this study, our chosen evaluation metrics are Faithfulness, Answer Relevance, and Context Recall.
2.3.2.1 Faithfulness
Faithfulness is evaluated by assessing the consistency of generated answers with the provided context, derived from both the answer itself and the context of retrieval. Scores are scaled from 0 to 1, with higher scores indicating greater faithfulness.
An answer is deemed reliable if all assertions within it can be inferred from the given context. To compute this value; a set of assertions needs identification from the generated answer, followed by cross-checking each assertion against the provided context. The Equation (2) for computing faithfulness is as follows:
Where represents the number of statements supported by LLM, and denotes the total number of statements.
2.3.2.2 Answer relevance
To evaluate the relevance of answers: utilize LLM to generate potential questions and compute their similarity to the original question. The relevance score of an answer is determined by averaging the similarity between all generated questions and the original question.
Let the original question be q, and the answer to the question be . The context segment relevant to question q is denoted as If the claims presented in the answer can be inferred from the context, we assert that the answer is faithful to the context . To gauge credibility, we initially employ LLM to extract a set of statements . If the answer directly and appropriately addresses the question, we consider it relevant. Particularly, our evaluation of answer relevance does not account for factual accuracy but penalizes incomplete or redundant information in the answers. To estimate answer relevance, given an answer , we prompt LLM to generate n potential questions based on . Subsequently, we use the text-embedding-ada-002 model from the OpenAI API to obtain embeddings for all questions. For each , we calculate the similarity with the original question . The specific formula for Answer relevance is Equation (3):
2.3.2.3 Context recall
Context recall assesses how well the retrieved context aligns with the authentic answers provided by humans. It is calculated by comparing the ground truth with the retrieved context, with scores ranging from 0 to 1, where higher scores indicate better performance.
To estimate context recall based on the authentic answers, each sentence in the authentic answers is examined to determine its relevance to the retrieved context. Ideally, all sentences in the authentic answers should be relevant to the retrieved context. The context relevance score is calculated using the following Equation (4):
2.3.3 Human evaluation
Although RAGAS has contributed to assessing the performance of RAG to some extent, human evaluation remains essential from the perspectives of safety, validation of professional knowledge, flexibility and adaptability, as well as ethical considerations. Therefore, this study incorporated human assessment. Our investigation presents the SUS (Safety, Usability, and Smoothness) evaluation method for human assessors SUS encompasses three dimensions: safety, usability, and smoothness (36). The “safety” dimension evaluates whether the model-generated content could potentially mislead users, posing health risks. Assessments from the “usability” dimension reflect the depth of professional expertise, while the “smoothness” dimension gauges the proficiency of the generated model functioning as an LLM. The skill enhancement program employs a three-tier scoring system, ranging from 1 (unsatisfactory) to 3 (good), with 2 indicating acceptable performance.
3 Results
3.1 Illustrative Q&A examples from GastroBot
To demonstrate the capabilities of GastroBot, we developed a web application using Streamlit. This application enables users to input questions related to gastrointestinal diseases. Additionally, a question dialogue box is located below the main interface, allowing users to input their queries. The interface of the web application is depicted in Figure 3.
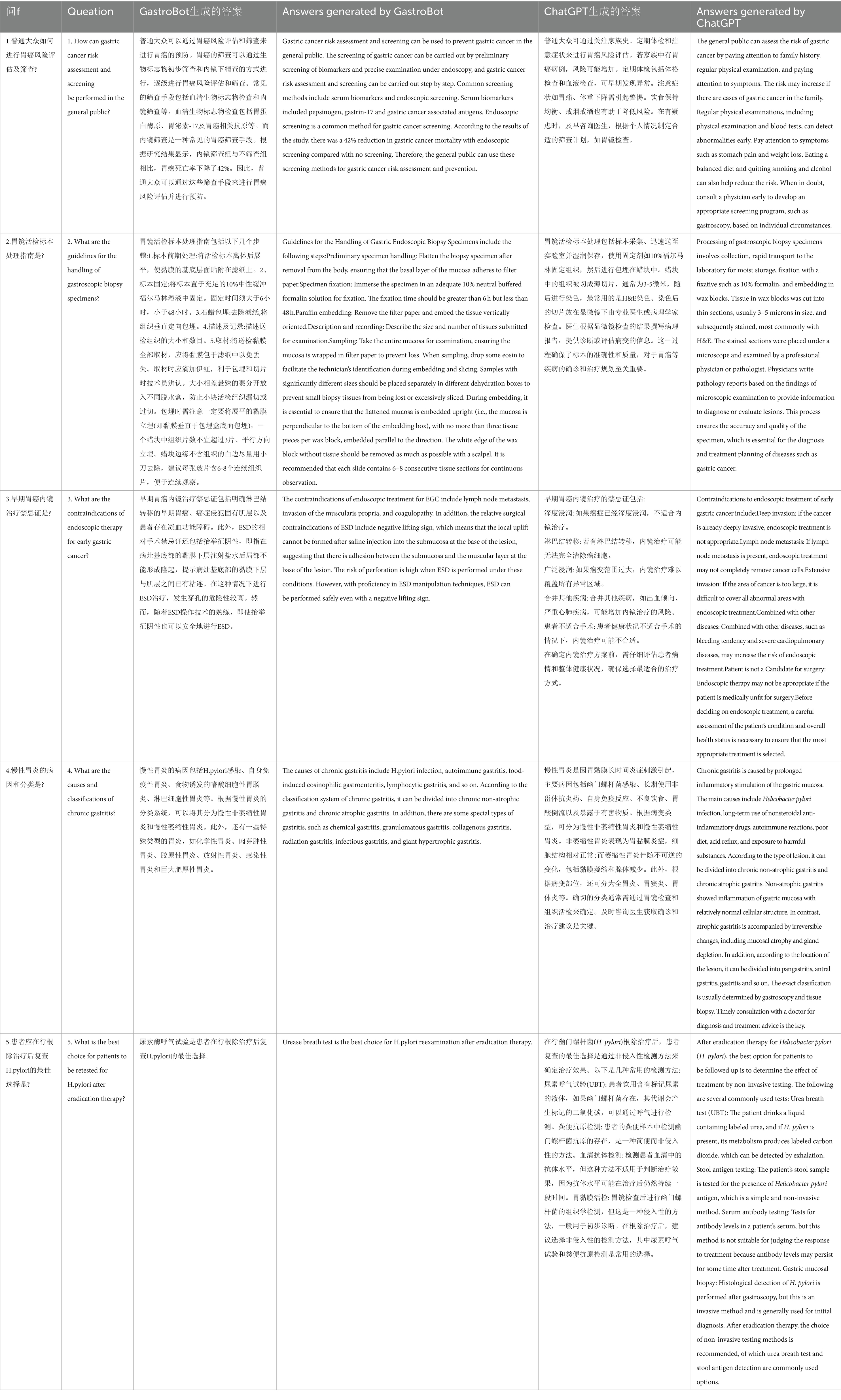
We randomly present five questions related to gastric diseases and generate answers using both GastroBot and ChatGPT. Table 1 compares the answers generated by GastroBot and ChatGPT for the five sample questions. Through this comparison, we observe that GastroBot’s answers are more precise, contextually relevant, and effectively mitigate the production of misleading information. For instance, in question 2, where the user in quires about the guidelines for handling gastric biopsy specimens, GastroBot provides a detailed response comprising five steps, each accompanied by precise explanations and requirements, and devoid of errors. In contrast, ChatGPT’s response offers only a series of steps without clear time references and lacks sufficient clarity in explanations.
3.2 Fine-tuned embedding model improved performance
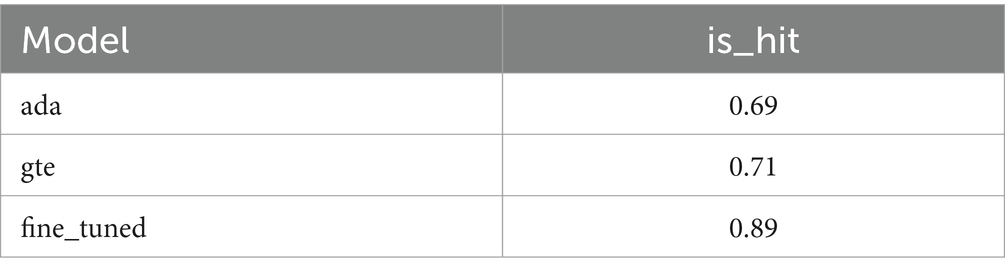
The results of the hit rate are illustrated in Figure 4 and presented in Table 2. Our fine-tuned model exhibits an 18% improvement in performance compared to its base model, gte-base-zh. When contrasted with OpenAI’s embedding model, text-embedding-ada-002, our fine-tuned model demonstrates a 20% enhancement in performance. The results of the Information Retrieval Evaluator evaluation are presented in Figure 5, showing a 21% improvement in performance for the fine-tuned model compared to the base model. Additionally, the fine-tuned model exhibits improvements in each of the 30 evaluation metrics columns.
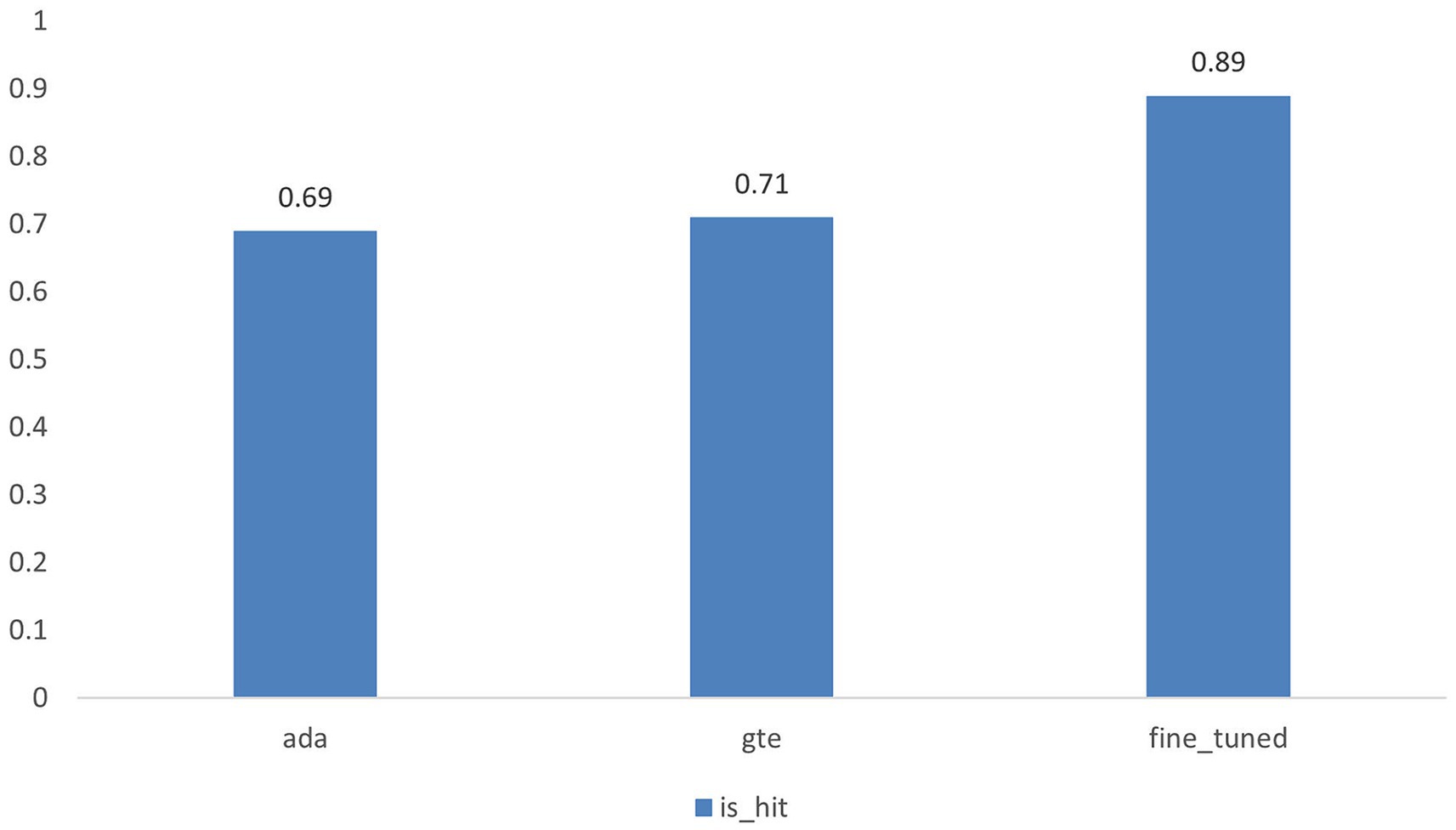
Figure 4. Illustrates the hit rates of the text-embedding-ada-002 model, the gte-base-zh model, and the fine-tuned model.

Figure 5. Information retrieval evaluator is a comprehensive metric suite provided by LlamaIndex, showcasing the outcomes of 30 evaluation metrics.
3.3 RAG evaluation
3.3.1 RAGAs scores
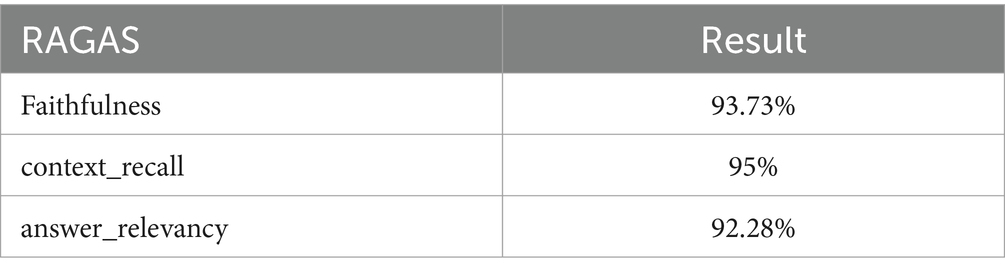
Our Ragas evaluation relies on GPT-3.5-Turbo. To ensure diversity and representativeness in the test set, we meticulously designed the distribution of questions across categories such as “simple,” “inference,” “multi-context,” and “conditional.” Adhering to these guidelines, we curated a test set comprising 20 questions for assessment. The evaluation outcomes of Ragas are consolidated in Table 3. When employing the RAGAS framework to evaluate GastroBot, we attained a context recall rate of 95%, with faithfulness reaching 93.73%. Moreover, the answer relevancy score achieved 92.28%.
3.3.2 SUS scores
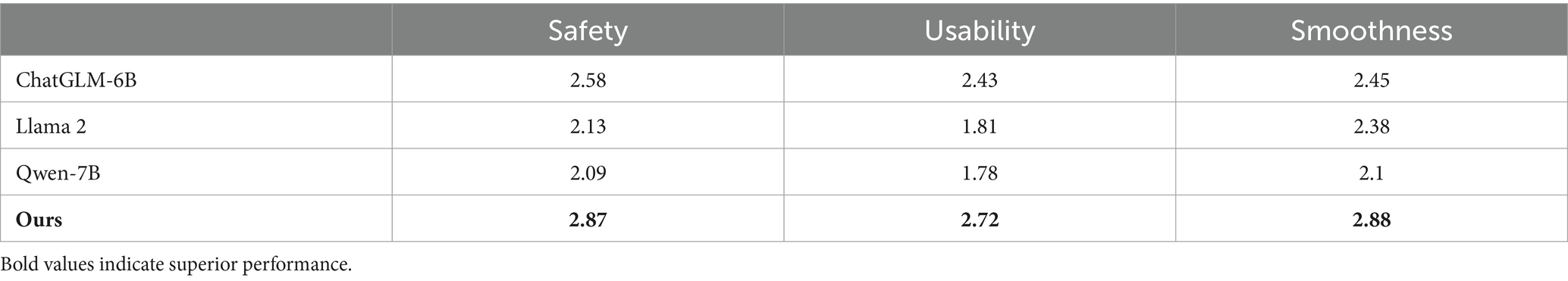
To assess the model’s performance, we enlisted 5 professionals with medical expertise and randomly selected 20 questions recommended by GastroBot for evaluation. The experimental results of the SUS scores are detailed in Table 4. In comparison with the other three models, GastroBot scored remarkably high in terms of safety, usability, and smoothness, achieving scores of 2.87, 2.72, and 2.88, respectively. These scores signify that GastroBot’s responses are exceptionally smooth and notably enhance the accessibility of knowledge while maintaining safety.
4 Discussion
4.1 Previous research background
The extensive utilization of LLM in natural language processing showcases remarkable generalization capabilities. However, challenges such as hallucinations and interpretability issues persist in clinical applications. Our research tackles these challenges by introducing the RAG method, which improves the accuracy and relevance of answers by retrieving information from external knowledge sources. RAG has previously proven successful in biomedical fields (16), liver disease research (18), clinical test diagnostics (19), and electrocardiogram data diagnostics (20).
In Wang et al.’s study, the application of the Almanac framework improved the retrieval function of medical guidelines and treatment recommendations, showcasing the potential effectiveness of LLM in clinical decision-making (17). Moreover, Ge et al. (18) utilized RAG technology to develop Li Versa, a specialized model for liver diseases, which offered more targeted answers compared to ChatGPT. These studies robustly support our work, affirming the practical application potential of RAG technology in professional domains. The high prevalence of Helicobacter pylori infection in China correlates with the increasing incidence of gastric cancer, ranking third in both incidence and mortality among all cancers (23, 24). Given the substantial patient population and the complexity of managing gastrointestinal diseases, implementing RAG is paramount for enhancing the accuracy of diagnosis and treatment for these diseases.
4.2 Novelty discovered
We employed a corpus consisting of 25 guideline documents and 40 relevant literature articles to apply RAG technology in clinical practice within the field of Chinese gastroenterology. By fine-tuning the embedding models, we achieved a significant enhancement in performance. Post fine-tuning, our model exhibited an 18% increase in hit rate compared to the base model gte-base-zh. Furthermore, our fine-tuned model demonstrated a 20% performance improvement compared to OpenAI’s embedding model. Concurrently, leveraging the RAG framework, we developed GastroBot, a Chinese gastroenterology chatbot. Evaluation using the RAGAS framework showcased a context recall rate of 95%, faithfulness of 93.73%, and a high answer relevancy of 92.28%. Human assessments indicated GastroBot’s excellent performance in safety, usability, and smoothness, with scores of 2.87, 2.72, and 2.88, respectively. These findings underscore the significant advantages and innovative potential of RAG technology in addressing clinical challenges.
4.3 Explanation of potential drawbacks and limitations
Although RAG technology has shown significant improvements in accuracy and relevance, it still faces inherent limitations. The quality and accuracy of external knowledge sources directly impact the quality of generated responses. In our study, using 25 guideline documents and 40 literature articles, we encountered challenges related to the comprehensiveness and timeliness of knowledge. Responses may lack necessary details or specificity, requiring subsequent clarification. Additionally, responses may sometimes be overly vague or generic, failing to effectively meet user needs. In future work, we intend to explore complex retrieval strategies to address these challenges. It is important to note that our current research primarily focuses on the Chinese region, thus exhibiting geographical limitations. Future research endeavors will strive to enhance the model’s applicability, encompassing broader regions and cultural backgrounds. Furthermore, RAG technology’s reliance on large datasets may hinder its performance in scenarios with limited sample sizes. Subsequent research plans should aim to alleviate these challenges to bolster the model’s resilience and adaptability.
4.4 Integration with current problem understanding and advancement
Our research provides a fresh perspective and solution to the information processing challenges faced in today’s clinical environment. Currently, AI chatbots are making significant strides in healthcare, particularly in pain management (37, 38). Inspired by these advancements, we successfully integrated RAG technology, which combines LLM’s reasoning capabilities with domain-specific knowledge retrieval, to develop an AI chatbot tailored to addressing the interpretation and understanding challenges in clinical gastroenterology. This advancement not only deepens our understanding of clinical problem-solving but also holds promise for extending this technology to other clinical domains, especially in underserved regions with limited medical resources, thereby aiding in early diagnosis.
4.5 Theoretical hypotheses for future directions and testing
Future research endeavors may concentrate on refining the selection and updating mechanisms of external knowledge sources to ensure the model’s access to the latest and most comprehensive clinical data. To enhance GastroBot’s performance, we intend to explore the utilization of advanced RAG technology. Moreover, we will seek collaboration with experts in gastroenterology to enrich GastroBot’s domain-specific knowledge. Integrating electronic medical record systems into GastroBot is also part of our future enhancement agenda. Additionally, we anticipate the emergence of high-performance LLMs deployable locally, facilitating the deployment of relevant chatbots by researchers in various fields. Fundamentally, our research introduces an innovative solution to the clinical information processing domain and provides valuable insights for future studies. Ongoing improvements and deeper exploration suggest that RAG technology may play a crucial role in the foreseeable future, particularly in fields such as clinical decision support systems.
Data availability statement
The original data for this research is sourced from the Chinese Medical Journal Full-text Database (https://www.yiigle.com) and China National Knowledge Infrastructure (https://www.cnki.net). Other experimental-related code, exported datasets, and model weights have been released on GitHub (https://github.com/hujili007/ragbot).
Author contributions
QZ: Writing – original draft. CL: Validation, Writing – review & editing. YD: Data curation, Writing – review & editing. KS: Writing – review & editing. YL: Writing – review & editing. HK: Writing – review & editing. ZG: Writing – review & editing. JS: Writing – review & editing, Investigation. JH: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Industry-University Cooperation Collaborative Education Project of the Ministry of Education of the People’s Republic of China (Grant no. 202101123001), the University Synergy Innovation Program of Anhui Province (Grant no. GXXT-2023-071), and Major Natural Science Research Projects of Anhui Provincial Department of Education (Grant no. 2023AH040102). Central Finance ‘Hua Tuo of the North, Xin’an of the South’ Traditional Chinese Medicine (TCM) Heritage and Innovation Project (Grant no. 2022BHTNXA01).
Acknowledgments
We extend heartfelt appreciation to all participants whose invaluable contributions have significantly bolstered the success of this study. A special acknowledgment is reserved for mentor, JH, whose sagacious guidance and unwavering assistance have been instrumental throughout the entire research endeavor. We would also like to express profound gratitude to the funding organization for their generous support, playing an indispensable role in bringing this research to fruition. The collective efforts of these esteemed individuals and institutions have undeniably enriched the outcomes of this scholarly work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Tan, Y, Min, D, Li, Y, Li, W, Hu, N, Chen, Y, et al. Can ChatGPT replace traditional KBQA models? An in-depth analysis of the question answering performance of the GPT LLM family. Cham: Springer Nature Switzerland (2023).
2. Luo, Z, Xie, Q, and Ananiadou, S. ChatGPT as a factual inconsistency evaluator for text summarization. arXiv (2023).
4. Zhao, WX, Zhou, K, Li, J, Tang, T, Wang, X, Hou, Y, et al. A survey of large language models. ArXiv. (2023)
5. Raunak, V, Menezes, A, and Junczys-Dowmunt, M. The Curious Case of Hallucinations in Neural Machine Translation. In: Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Online. Association for Computational Linguistics (2021). p. 1172–1183.
6. Lee, P, Bubeck, S, and Petro, J. Benefits, limits, and risks of GPT-4 as an AI Chatbot for medicine. N Engl J Med. (2023) 388:1233–9. doi: 10.1056/NEJMsr2214184
7. Ji, Z, Lee, N, Frieske, R, Yu, T, Su, D, Xu, Y, et al. Survey of hallucination in natural language generation. ACM Comput Surv. (2023) 55:1–38. doi: 10.1145/3571730
8. Dave, T, Athaluri, SA, and Singh, S. ChatGPT in medicine: an overview of its applications, advantages, limitations, future prospects, and ethical considerations. Front Artif Intell. (2023):1169595:6. doi: 10.3389/frai.2023.1169595
9. Meskó, B, and Topol, EJ. The imperative for regulatory oversight of large language models (or generative AI) in healthcare. NPJ Digital Medicine. (2023):120:6. doi: 10.1038/s41746-023-00873-0
10. Vanian, J, and Leswing, K. ChatGPT and generative AI are booming, but the costs can be extraordinary. CNBC News. (2023)
11. Kojima, T, Gu, SS, Reid, M, Matsuo, Y, and Iwasawa, Y. Large language models are zero-shot reasoners. ArXiv. (2022) abs/2205.11916. Available at: https://api.semanticscholar.org/CorpusID:249017743
12. Zheng, C, Liu, Z, Xie, E, Li, Z, and Li, Y. Progressive-hint prompting improves reasoning in large language models. ArXiv. (2023)
13. Parnami, A, and Lee, M. Learning from few examples: a summary of approaches to few-shot learning. arXiv (2022).
14. Lewis, P, Perez, E, Piktus, A, Petroni, F, Karpukhin, V, Goyal, N, et al. Retrieval-augmented generation for knowledge-intensive NLP tasks. In: Larochelle H, Ranzato M, Hadsell R, Balcan MF, Lin H, editors. Advances in Neural Information Processing Systems. Curran Associates, Inc. (2021).
15. Shuster, K, Poff, S, Chen, M, Kiela, D, and Weston, J. Retrieval augmentation reduces hallucination in conversation. In: Findings of the Association for Computational Linguistics: EMNLP 2021. Punta Cana: Dominican Republic. Association for Computational Linguistics. (2021). p. 3784–3803.
16. Soong, D, Sridhar, S, Si, H, Wagner, JS, S’a, ACC, Yu, CY, et al. Improving accuracy of GPT-3/4 results on biomedical data using a retrieval-augmented language model. ArXiv. (2023)
17. Zakka, C, Chaurasia, A, Shad, R, Dalal, AR, Kim, JL, Moor, M, et al. Almanac: retrieval-augmented language models for clinical medicine. Res. Square. (2023)
18. Ge, J, Sun, S, Owens, J, Galvez, V, Gologorskaya, O, Lai, JC, et al. Development of a liver disease-specific large language model chat Interface using retrieval augmented generation. medRxiv. (2023)
19. Ranjit, M, Ganapathy, G, Manuel, R, and Ganu, T. Retrieval augmented chest x-ray report generation using openai gpt models. arXiv preprint arXiv:230503660 (2023)
20. Yu, H, Guo, P, and Sano, A. Zero-shot ECG diagnosis with large language models and retrieval-augmented generation. Mach Learn Health. (2023) 225:650–63.
21. Lozano, A, Fleming, SL, Chiang, C-C, and Shah, N. Clinfo.Ai: an open-source retrieval-augmented large language model system for answering medical questions using scientific literature. Biocomputing. 2024. (2023) 29:8–23. doi: 10.1142/9789811286421_0002
22. Manathunga, SS, and Illangasekara, YA. Retrieval augmented generation and representative vector summarization for large unstructured textual data in medical education. (2023). Available at: http://arxiv.org/abs/2308.00479
23. Bai, Y, Li, ZS, Zou, DW, Wu, RP, Yao, YZ, Jin, ZD, et al. Alarm features and age for predicting upper gastrointestinal malignancy in Chinese patients with dyspepsia with high background prevalence of Helicobacter pylori infection and upper gastrointestinal malignancy: an endoscopic database review of 102 665 patients from 1996 to 2006. Gut. (2010) 59:722–8. doi: 10.1136/gut.2009.192401
24. Yang, W-J, Zhao, H-P, Yu, Y, Wang, J-H, Guo, L, Liu, J-Y, et al. Updates on global epidemiology, risk and prognostic factors of gastric cancer. World J Gastroenterol. (2023) 29:2452–68. doi: 10.3748/wjg.v29.i16.2452
25. Li, Z, Zhang, X, Zhang, Y, Long, D, Xie, P, and Zhang, M. Towards general text Embeddings with multi-stage contrastive learning. ArXiv. (2023) abs/2308.03281. Available at: https://api.semanticscholar.org/CorpusID:260682258
26. Touvron, H, Martin, L, Stone, K, Albert, P, Almahairi, A, Babaei, Y, et al. Llama 2: open foundation and fine-tuned chat models. arXiv preprint arXiv:230709288 (2023).
27. Du, Z, Qian, Y, Liu, X, Ding, M, Qiu, J, Yang, Z, et al.. GLM: general language model pretraining with autoregressive blank infilling. In: Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics. Dublin, Ireland: Association for Computational Linguistics (2022). p. 320–335.
28. Bai, J, Bai, S, Chu, Y, Cui, Z, Dang, K, Deng, X, et al. Qwen technical report. arXiv preprint arXiv:230916609 (2023).
29. Neelakantan, A, Xu, T, Puri, R, Radford, A, Han, JM, Tworek, J, et al. Text and code Embeddings by contrastive pre-training. ArXiv. (2022) abs/2201.10005. Available at: https://api.semanticscholar.org/CorpusID:246275593
30. Alsini, A, Huynh, DQ, and Datta, A. Hit ratio: an evaluation metric for hashtag recommendation. ArXiv. (2020) abs/2010.01258 Available at: https://api.semanticscholar.org/CorpusID:222134056
31. Levkowitz, H. Introduction to information retrieval (IR). (2008). Available at: https://api.semanticscholar.org/CorpusID:61517903
33. Järvelin, K, and Kekäläinen, J. IR evaluation methods for retrieving highly relevant documents. In: Proceedings of the 23rd Annual International ACM SIGIR Conference on Research and Development in Information Retrieval. Athens: ACM (2000). 41–48.
34. Järvelin, K, and Kekäläinen, J. Cumulated gain-based evaluation of IR techniques. ACM Trans Inform Syst. (2002) 20:422–46. doi: 10.1145/582415.582418
35. Shahul, ES, James, J, Anke, LE, and Schockaert, S. RAGAS: automated evaluation of retrieval augmented generation. ArXiv. (2023) abs/2309.15217. Available at: https://api.semanticscholar.org/CorpusID:263152733
36. Wang, H, Liu, C, Xi, N, Qiang, Z, Zhao, S, Qin, B, et al.. Huatuo: tuning llama model with chinese medical knowledge. arXiv preprint arXiv:230406975 (2023).
37. Rossettini, G, Cook, C, Palese, A, Pillastrini, P, and Turolla, A. Pros and cons of using artificial intelligence chatbots for musculoskeletal rehabilitation management. J Orthop Sports Phys Ther. (2023) 53:728–34. doi: 10.2519/jospt.2023.12000
38. Gianola, S, Bargeri, S, Castellini, G, Cook, C, Palese, A, Pillastrini, P, et al. Performance of ChatGPT compared to clinical practice guidelines in making informed decisions for lumbosacral radicular pain: a cross-sectional study. J Orthop Sports Phys Ther. (2024) 54:222–8. doi: 10.2519/jospt.2024.12151
Keywords: retrieval-augmented generation, gastrointestinal disease, natural language process, large language models, embedding model, chatbot, question-answering, GPT
Citation: Zhou Q, Liu C, Duan Y, Sun K, Li Y, Kan H, Gu Z, Shu J and Hu J (2024) GastroBot: a Chinese gastrointestinal disease chatbot based on the retrieval-augmented generation. Front. Med. 11:1392555. doi: 10.3389/fmed.2024.1392555
Edited by:
Md. Mohaimenul Islam, The Ohio State University, United StatesReviewed by:
Ibrahim M. Mehedi, King Abdulaziz University, Saudi ArabiaGiacomo Rossettini, University of Verona, Italy
Copyright © 2024 Zhou, Liu, Duan, Sun, Li, Kan, Gu, Shu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jili Hu, aHVqaWxpQGFodGNtLmVkdS5jbg==
 Qingqing Zhou
Qingqing Zhou Can Liu
Can Liu Yuchen Duan
Yuchen Duan Kaijie Sun
Kaijie Sun Yu Li
Yu Li Hongxing Kan1,2
Hongxing Kan1,2 Zongyun Gu
Zongyun Gu Jianhua Shu
Jianhua Shu Jili Hu
Jili Hu