
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 19 April 2024
Sec. Infectious Diseases: Pathogenesis and Therapy
Volume 11 - 2024 | https://doi.org/10.3389/fmed.2024.1378207
Objective: To outline the epidemiology of puerperal mastitis caused by methicillin-resistant Staphylococcus aureus (MRSA) and evaluate the effect of an infection control bundle on its incidence.
Methods: A surge in MRSA puerperal mastitis was noted in a community hospital in September 2009. MRSA samples from mastitis cases and the environment underwent typing using multilocus sequence typing (MLST), staphylococcal cassette chromosome mec (SCCmec), gene encoding surface protein A (spa), accessory gene regulator (agr), and pulsed-field gel electrophoresis (PFGE). The phenotypic characteristics, including superantigen toxin profiles, gene encoding Panton-Valentine leucocidin (pvl), and minimal inhibitory concentration (MIC) against vancomycin, were ascertained. Subsequently, an infection control bundle emphasizing contact precautions was introduced, and mastitis incidence rates pre- and post-intervention were compared.
Results: The majority of cases occurred within 6 weeks post-delivery in first-time mothers. Of the 42 S. aureus isolates (27 from mastitis and 15 from colonized staff and environmental sources), 25 (92.6%) clinical and 3 (20%) colonized MRSA were identified as ST59-SCCmecVT-spa t437-agr group I with a vancomycin MIC of 1 mg/L, pvl-positive, and predominantly with a consistent toxin profile (seb-selk-selr). PFGE revealed 13 patterns; pulsotype B exhibited clonal relatedness between two clinical and three colonized MRSA samples. Post-intervention, the incidence of both mastitis and MRSA mastitis notably decreased from 13.01 to 1.78 and from 3.70 to 0.99 episodes per 100 deliveries, respectively.
Conclusion: Distinct community-associated MRSA (CA-MRSA) clones were detected among puerperal mastitis patients and colonized staff. The outbreak was effectively controlled following the implementation of a targeted infection control bundle.
Staphylococcus aureus, particularly methicillin-resistant S. aureus (MRSA), is a primary pathogen causing skin and soft tissue infections, including mastitis (1, 2). Mastitis predominantly occurs within 3 months post-delivery (2, 3), with an estimated prevalence of 10–33% in breastfeeding women (4). This condition is a leading cause for discontinuing breastfeeding (2, 5), prompting the World Health Organization (WHO) to highlight the significance of managing and preventing puerperal mastitis to sustain breastfeeding (6). Thus, insights into the epidemiology, characteristics, and risk factors of mastitis are imperative.
While numerous studies have detailed the epidemiology and clinical characteristics of human mastitis (1, 7), few have addressed the molecular types and phenotypes of the causative organisms (1, 8). Furthermore, prevention measures, including hand hygiene, antibiotic prophylaxis, and breast emptying to avert milk stasis, have been documented (5, 6, 9). In this study, we analyze the demographic and clinical profiles of mastitis patients, ascertain the molecular types and phenotypes of S. aureus isolates from these patients and colonized staff, and assess the influence of a specific infection control bundle on mastitis prevalence.
From January 1st, 2008, to December 31st, 2010, we retrieved demographic data of patients diagnosed with mastitis based on International Classification of Diseases, 9th Revision - ICD-9 codes 610.1, 675.1, and 675.2. This data included age, type of visit (outpatient or hospitalization) when seeking medical aid, number and type of childbirth (normal spontaneous delivery [NSD] or cesarean section [C/S]), episodes with or without breastfeeding, and site of mastitis (unilateral or bilateral involvement). This information was sourced from the medical records of a 505-bed community hospital in central Taiwan.
The 36-month study was segmented into three distinct periods based on the incidence of mastitis and the implementation timing of an infection control bundle: the baseline period from January 1st, 2008, to August 31st, 2009 (20 months); the outbreak period from September 1st, 2009, to June 30th, 2010 (10 months); and the control bundle implementation period from July 1st to December 31st, 2010 (6 months).
For mastitis patients from whom S. aureus was isolated, data were also collected for molecular and phenotypic studies. These patients were further assessed for underlying diseases, birth rank, the interval between symptom onset and childbirth, lesion size, fever presence, suitability of empiric and target antibiotic therapies, and overall clinical outcomes.
Staphylococcus aureus isolates were cultivated and identified from mastitis foci through aspiration or surgical debridement using standard techniques. Each S. aureus isolate was confirmed with colonies exhibiting β-hemolysis on 5% sheep blood agar (BD Difco & BBL, NJ, USA), Gram-staining positive cocci, and giving positive results for catalase, coagulase, DNase, and mannitol fermentation tests. Staff from both the Department of Gynecology and Obstetrics and the Department of Pediatrics were subjected to three swabbing cultures: two from the surfaces of both hands and a third taken 1.5 cm deep into the nasal vestibule using sterile swabs (CultureSwab Transport System, Difco, Detroit, USA). Cultivated and identified colonized S. aureus isolates were obtained from these swabs. Environmental screening was conducted on surfaces like tables, desktops, computer mice, and work carts within the mentioned departments using sterile swabs rinsed with normal saline. The study protocol, encompassing isolate collection, staff and environmental screening, and the implementation of the infection control bundle, received approval from the hospital’s Institutional Review Board (CSMUH no. CS10218).
After screening with a 30 μg cefoxitin disc on the Műeller-Hinton agar (MHA) (BD Difco & BBL, NJ, USA) according to the Clinical and Laboratory Standards Institutes (CLSI) guidelines (10), the existence of the mecA gene, responsible for MRSA genotype, was performed using the polymerase chain reaction (PCR) (11). The antimicrobial susceptibility of S. aureus was assessed using disc diffusion against a range of antibiotics (erythromycin 15 μg, clindamycin 2 μg, tetracycline 30 μg, oxacillin 1 μg, vancomycin 30 μg, teicoplanin 30 μg, and tigecycline 15 μg) (BD Difco & BBL, NJ, USA) and fusidic acid (Thermo Fisher Scientific, MA, USA) following CLSI protocol (12) and manufacturer’s instructions. Resistance to three or more classes of antimicrobials was categorized as multidrug resistance. The minimum inhibitory concentrations (MICs) of vancomycin using reference standard compound (Sigma Aldrich Fine Chemicals Biosciences, MA, USA) against S. aureus were determined using agar dilution from vancomycin concentration of 0.0625 mg/L with 2-fold increase up to 64 mg/L in line with CLSI guidelines (12).
Genomic DNA was extracted from each isolate using the Genomic DNA Mini Kit (Geneaid, Taiwan) and was digested with the SmaI restriction enzyme (Promega Corp., Madison, Wisconsin, USA). PFGE was conducted using a contour-clamped homogeneous electric field apparatus (DR-III, Bio-Rad, Hercules, California, USA) as described previously (13). The dendrograms were constructed employing the Dice coefficient and the unweighted pair group method with arithmetic average (14). Isolates with PFGE banding patterns displaying a similarity of 70% or greater were classified under one pulsotype.
Genomic DNA from each MRSA strain served as the template. The gene types encoding the cassette chromosome recombinase (ccr) complex and the mec gene complex were determined using multiplex PCR (M-PCR) (Supplementary Table) (11). SCCmec types I through V in MRSA strains were identified by comparing the M-PCR banding patterns of the isolates with those of the reference strains: ATCC 10442 (SCCmec type I), N315 (SCCmec type II), 85/2082 (SCCmec type III), MW2 (SCCmec type IVa), WIS (SCCmec type V), and TSGH-17 (SCCmec type VT).
Each S. aureus isolate’s seven housekeeping genes encoding the carbamate kinase, (arc), shikimate dehydrogenase (aroE), glycerol kinase (glp); guanylate kinase (gmk), phosphate acetyltransferase (pta), triosephosphate isomerase (tpi), and acetyl coenzyme A acetyltransferase (yqiL) were amplified and sequenced (Supplementary Table) (15). By comparing sequences to those in the S. aureus MLST database,1 the allelic number of each gene and the allelic profile, which determined the sequence type (ST) of each isolate, were identified.
The X region of the spa gene in each MRSA was amplified using PCR (Supplementary Table) (16). The amplified products were then sequenced and analyzed with the Ridom StaphType software (version 1.4; Ridom, GmbH, Wurzburg, Germany).2 This software automatically identified the repeat profile and the spa type for each isolate.
The agr gene of each S. aureus was amplified using PCR, employing specialized primers designed for agr group I (441-bp), agr group II (575-bp), agr group III (323-bp), and agr group IV (659-bp) (Supplementary Table) (17).
Amplification of the pvl gene was conducted using PCR with the primers luk-PV-1 and luk-PV-2, as previously described (Supplementary Table) (18). Reference strains TSGH-17 and 85/2082 served as the positive and negative control strains, respectively.
Each S. aureus isolate was tested for the presence of 18 genes encoding both classical and recently described superantigenic toxins, ranging from sea to selr and tsst-1, using four M-PCRs. The genes femA and femB served as positive controls (Supplementary Table) (19). The toxin gene profile for each isolate was established based on toxin gene assembly.
From July 1st, 2010 (the start of the third study period), all staff, patients, and their caregivers were instructed to rigorously follow the “My five moments for hand hygiene” approach recommended by the WHO (20). Measures to enhance breastfeeding management were given to patients and their caregivers, which included ensuring complete breast emptying, proper cleaning of the breast, milkers, and feeding bottles, mastering the breastfeeding technique, and being vigilant for signs of milk stasis or infection, such as continuous breast swelling and discomfort post-breastfeeding, with or without fever (6). Staff found to have S. aureus colonization during screening were treated with mupirocin applied to their anterior nares daily for 5 days (21). Surveillance and monitoring of patients with potential and confirmed mastitis continued until December 31st, 2010.
Differences between consecutive samples within the same population were assessed using the Mann–Whitney-U test for non-normally distributed populations (nonparametric). A p value of <0.05 was deemed indicative of a statistically significant difference between the samples.
During the three-year study period at the community hospital, 3,311 births were recorded, comprising 2,300 NSD and 1,011C/S. Concurrently, 249 patients received a mastitis diagnosis, with the majority (226, 90.8%) presenting during the puerperal period, equating to a prevalence of 6.8%. The patients’ average age was 30.4 years, ranging from 15 to 51 years, and all were female. The affected sites were the right side in 105 (42.2%) cases, the left side in 85 (34.1%) cases, and bilateral in 19 (7.6%) cases. The location was not recorded in 40 instances (16.1%). A majority (198, 79.5%) underwent outpatient treatment. Over the 3 years, the mastitis prevalence rates were 0.95 episodes per 1,000 patient-days of hospitalization and 7.52 episodes per 100 deliveries. The peak rates were observed in the second period, with 1.42 episodes per 1,000 patient-days and 13.01 episodes per 100 deliveries (p < 0.001 and 0.03 when compared to the first and third periods, respectively) (Table 1). The prevalence rates of mastitis during the second period were significantly different from the other two periods for both NSD and C/S patients (Table 1).
Of the 249 mastitis patients, only 61 (24.5%) underwent microbiologic surveys. From these, S. aureus was the sole microorganism isolated in 57 cases, which included 44 MRSA and 13 MSSA patients. During the second period, the prevalence rates of S. aureus-induced mastitis were 0.50 episodes per 1,000 patient-days and 4.67 episodes per 100 deliveries. The rates for MRSA were 0.40 episodes per 1,000 patient-days and 3.70 episodes per 100 deliveries. These rates were notably higher than those observed in the first and third periods (p = 0.02 and p < 0.001, respectively) (Table 1). Importantly, a marked decline in mastitis prevalence was observed in the third period following the adoption of the infection control bundle (Table 1 and Figure 1).
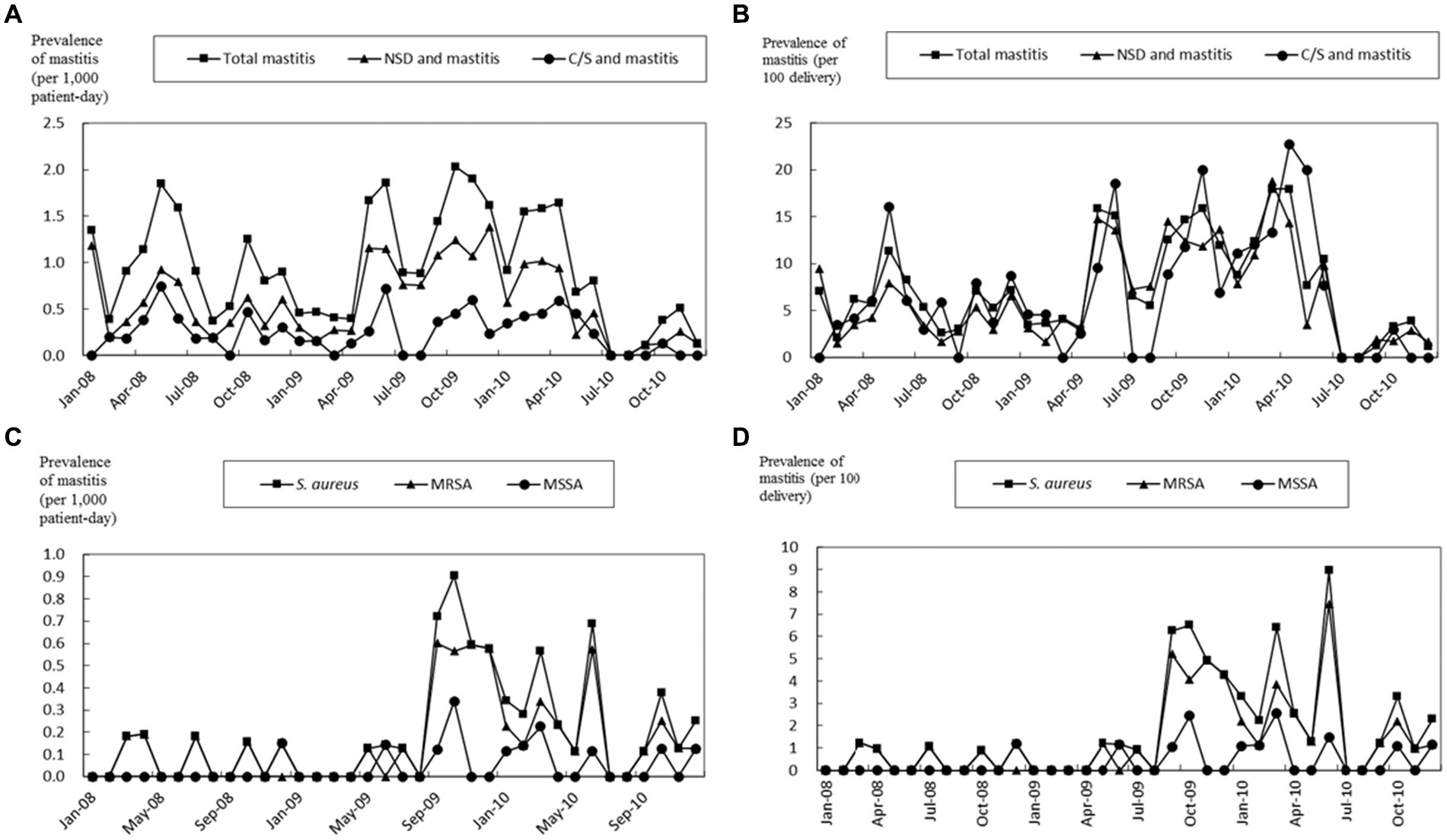
Figure 1. Prevalence of all patients with mastitis, normal spontaneous delivery (NSD) and mastitis, and cesarean section (C/S) and mastitis. The prevalence of mastitis is shown as episodes divided by 1,000 patient-days (A) and 100 deliveries (B). The prevalence of mastitis by S. aureus, MRSA, and MSSA is shown as episodes divided by 1,000 patient-days (C) and 100 deliveries (D). The study period was from January 1st, 2008 to December 31st, 2010.
During the second period, 26 S. aureus isolates were collected from 26 mastitis patients, including 25 MRSA (M001 to M027, excluding M003 and M024) and one MSSA (M024). Additionally, one MRSA (M003) isolated from a patient with a buttock abscess served as a control. The demographic and clinical details of these mastitis patients are presented in Table 2. Among the 24 patients diagnosed with puerperal mastitis, the majority (16, 66.7%) experienced it during their first childbirth. Most patients (23, 88.5%) had no underlying medical conditions, with the average time from delivery to the onset of mastitis symptoms being 24.3 days (range, 5–78 days). The right breast was most frequently affected (14, 53.8%), followed by the left (8, 30.8%) and both breasts (4, 15.4%). Fever was observed in 14 patients (53.8%). The majority did not receive suitable initial antibiotic treatment (21, 80.8%), yet once susceptibility data were accessible, most (22, 84.6%) were given appropriate targeted antibiotics. All affected individuals underwent fine needle aspiration or surgical debridement. All but two patients, who were lost to follow-up, achieved clinical recovery (Table 2).
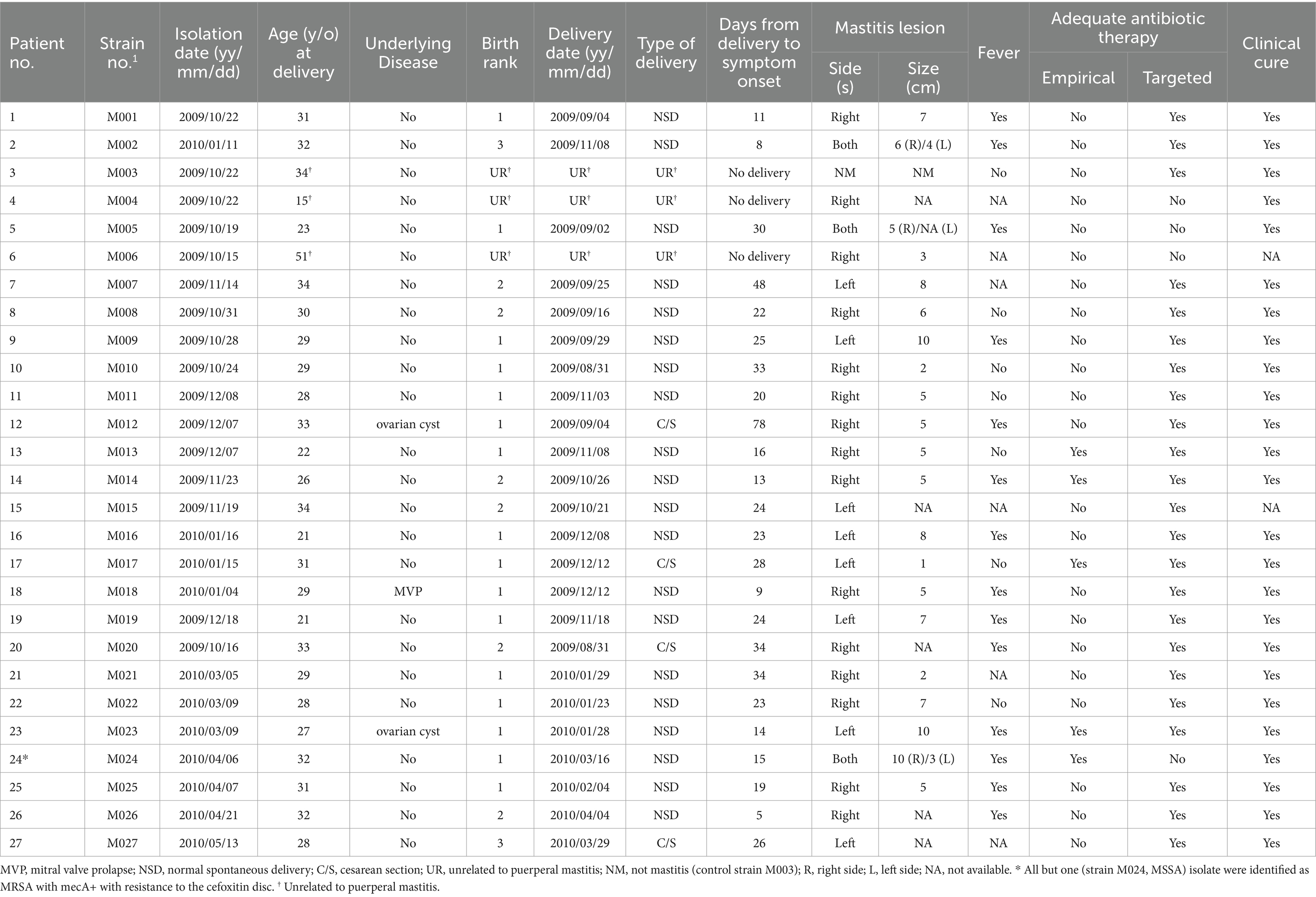
Table 2. Demographic and clinical data of 26 mastitis patients and one patient with a buttock abscess (patient no. 3 as control) caused by S. aureus.
Thirty-one staff members (93 samples) from the Departments of Gynecology and Obstetrics and Pediatrics, along with 15 associated environmental sites, underwent swabbing culture screenings. From the mastitis patients, 26 S. aureus isolates were obtained, comprising 25 MRSA and 1 MSSA (M024). Additionally, 15 S. aureus isolates (10 MRSA and 5 MSSA – strains 1–3, 7–3, 20–1, 39–2, and 43–3) were procured from the hands and nostrils of nine staff members. Environmental screenings did not yield any S. aureus. All 35 MRSA isolates, 25 from mastitis patients and 10 from colonized staff, demonstrated resistance to a 30-μg cefoxitin disc in MHA and the presence of the mecA gene. The antibiogram revealed that a vast majority (23/25, 92%) of the clinical MRSA mastitis isolates exhibited multidrug resistance (Table 3). Two isolates (strains 24–3 and 42–3), which initially displayed satisfactory inhibitory zones against oxacillin discs, were subsequently verified to be MRSA due to cefoxitin resistance and the mecA gene presence. Conversely, the 6 MSSA isolates demonstrated lower resistance levels compared to MRSA. Vancomycin MICs, ascertained by agar dilution, indicated that the MICs for the 42 S. aureus isolates ranged from 1 to 2 mg/L, with a median of 1 mg/L (Table 3).
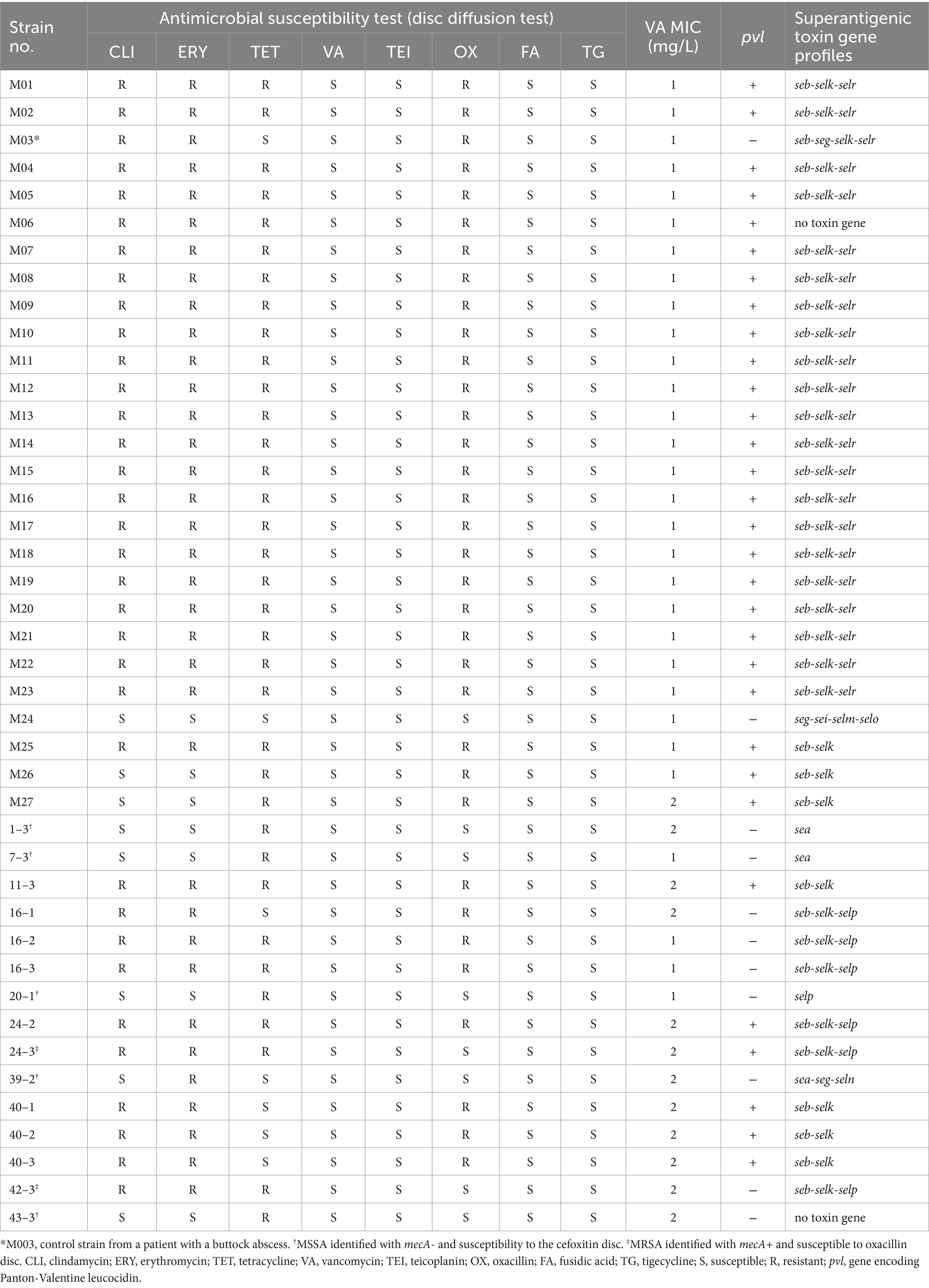
Table 3. Phenotypes of 42 S. aureus isolates from 26 mastitis patients (25 MRSA and 1 MSSA), one patient with a buttock abscess (1 MRSA), and 15 environmental samples (10 MRSA and 5 MSSA).
Figure 2 depicts the molecular profiles of 42 S. aureus isolates, as characterized by typing methods such as PFGE, SCCmec, agr, MLST, and spa typing. SCCmec was not able to type one clinical MSSA (M024) and 5 colonized MSSA isolates. All 25 clinical MRSA and 6 of the 10 colonized MRSA isolates were identified as SCCmecVT-ST59. Four colonized MRSA isolates belonged to SCCmecIV-ST59. The spa types of all 25 clinical MRSA and 7 of the 10 colonized MRSA isolates were t437, while the remaining 3 colonized MRSA isolates were categorized as spa t529. All S. aureus isolates, with the exception of one (MSSA strain 39–2), fell under the agr group I; this exception was categorized as agr group III. The 6 MSSA isolates, which included 1 clinical isolate M024 and 5 colonized isolates, exhibited varied MLST and spa types (Figure 2). Thirteen distinct pulsotypes (ranging from A to M) were discerned among the 42 S. aureus isolates through PFGE, with their correlations illustrated in Figure 2. Pulsotype A encompassed 15 clinical MRSA isolates. Notably, two clinical (M020 and M027) and three colonized (11-3, 24-2, and 24-3) isolates exhibited identical pulsotypes, all classified as pulsotype B. The control strain, M003, sourced from a buttock abscess, was designated as SCCmecIV-ST59-spa t437-agr group I-pulsotype F.
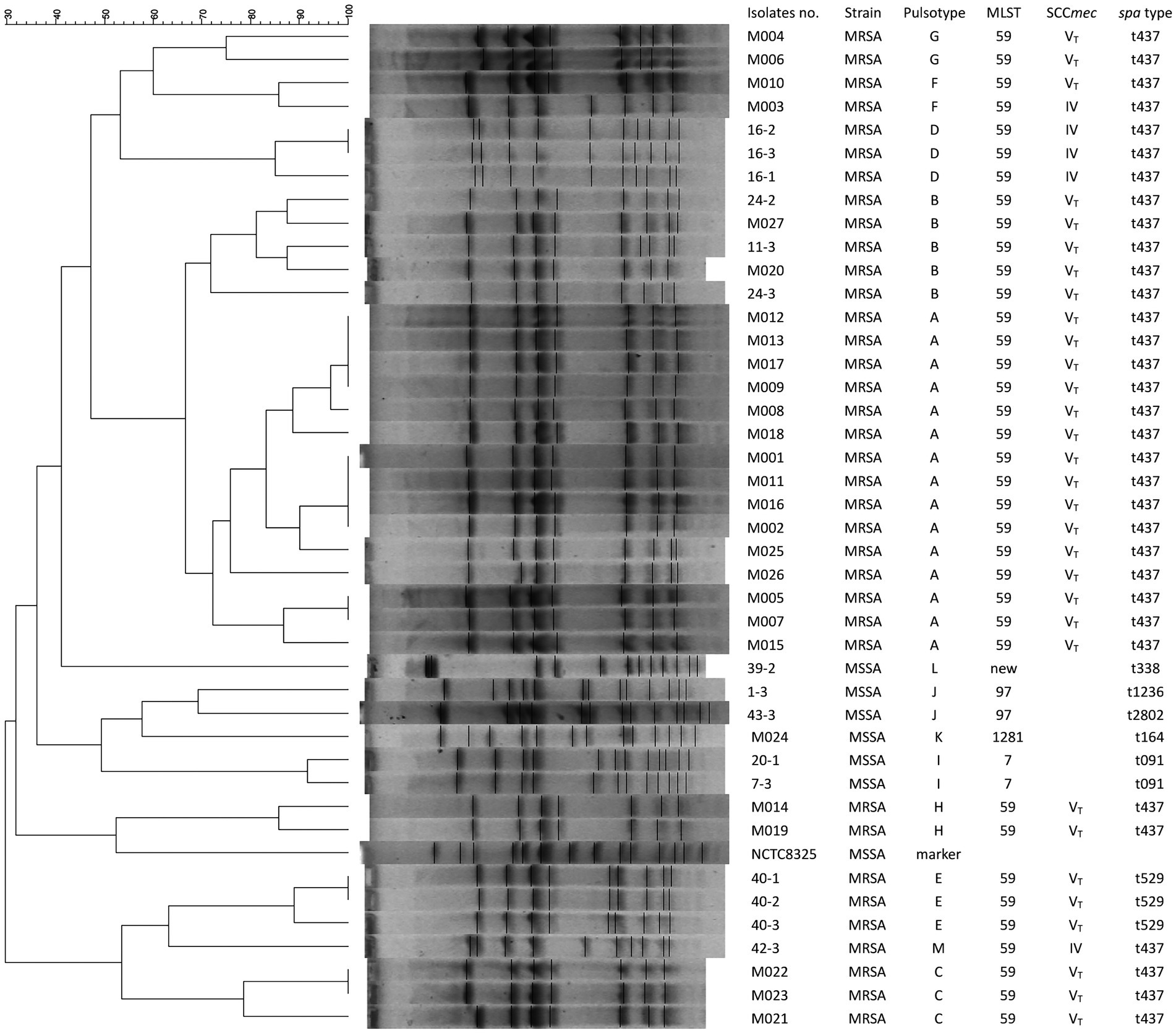
Figure 2. Pulsotypes of S. aureus digested by SmaI and their corresponding molecular typing results including MLST, SCCmec, and spa types. Fifteen pulsotypes (from A to M) were determined in 42 S. aureus isolates, including 26 from patients with mastitis, 15 from the environmental screening, and one control strain (M003) from a patient with a buttock abscess. NCTC 8325, S. aureus reference strain used as a marker.
Figure 2 and Table 3 reveal that 31 pvL + MRSA isolates (comprising 25 clinical MRSA and 6 colonized MRSA) were categorized as SCCmecVT. The remaining 5 pvL- MRSA, inclusive of the control strain M003, were identified as SCCmecIV. None of the six MSSA isolates, which include the clinical strain M024 and 5 colonized strains, carried the pvl gene (as seen in Figure 2 and Table 3). The superantigenic toxin gene profiles indicated that the seb-selk-selr profile was predominant in clinical MRSA isolates (21, 84%), succeeded by seb-selk (3, 12%). A diverse range of toxin gene profiles was observed in the 6 MSSA and 10 colonized MRSA isolates (Table 3).
Staphylococcus aureus has been recognized as the predominant etiology of human mastitis (1, 6, 7). In our study, only a quarter of the cases underwent microbiologic examinations, and S. aureus was the sole organism isolated. This is consistent with prior findings where S. aureus was the only identified organism, and a significant majority (63.6%) were MRSA (22).
The majority of mastitis cases are linked to breastfeeding (4, 6), and manifest within 6 months post-delivery, with a prevalence reaching 33% (4, 23). In our 3-year investigation, 90.8% of the mastitis cases were related to breastfeeding, presenting a prevalence of 6.8%. Risk factors for puerperal mastitis encompass a prior history of the condition, younger age, blocked ducts, and cracked nipples (5, 7). The average age at the commencement of mastitis in our study was 30.4 years, notably higher than the previously reported 23.4 years (7). Of the 24 patients with puerperal mastitis caused by S. aureus (Table 2), two-thirds were in their first pregnancy, with an average duration of 24.3 days between delivery and the appearance of symptoms. These results align with earlier research (6, 7). We postulate that younger and first-time mothers might lack the familiarity with optimal breastfeeding techniques compared to those who have had multiple pregnancies.
Common symptoms of mastitis encompass fever, malaise, and breast tenderness accompanied by swelling (1, 4). The recommended treatment approaches are needle aspiration, incision and drainage, breast emptying, and suitable antibiotics (1, 4, 6). As detailed in Table 2, half of the 26 patients with puerperal mastitis in our study exhibited fever, and each underwent needle aspiration or incision. A significant majority (80.8%) did not receive the appropriate antibiotics specific to MRSA. All patients, except for two, were diligently monitored with positive results. These observations align with earlier reports (1, 7). It is our contention that managing the source is of greater significance than antibiotic use for the effective treatment of mastitis.
The WHO advocates for routine handwashing by both staff and mothers as a preventive measure against puerperal mastitis (6, 20). Peters et al. noted a decline in the rate of puerperal mastitis from 2.9 to 0.66% after introducing bedside disinfectants (24). A sudden surge in mastitis cases was observed at our institution in September 2009. However, following the introduction of an infection control bundle, which included strict hand hygiene and bacterial decolonization via topical mupirocin, there was a marked reduction in the cases of both mastitis and MRSA-associated mastitis. Harbarth et al. found that nasal mupirocin was effective in averting recurrent soft tissue infections when in close proximity to MRSA carriers (21). Yet, the prophylactic use of mupirocin for puerperal mastitis remains a topic of debate (25). We consider that the comprehensive adoption of the infection control bundle, emphasizing contact precautions and decolonization, will yield better results compared to any other singular intervention.
PFGE is recognized as the gold standard for genotyping due to its exceptional discriminatory power and capacity for outbreak identification (13). While the 25 MRSA isolates possessed SCCmecVT-ST59-spa t437, PFGE enabled their differentiation and classification into specific clones based on chromosomal restriction patterns. Notably, both the control strain (M003) and the mastitis strain (M010) shared the pulsotype (F), suggesting that F pulsotype strains are prevalent in the community. Strains with pulsotype B comprised two from mastitis patients and three from colonized staff. These observations align with the findings of Manoharan et al. (26), suggesting that certain MRSA clones are horizontally transmitted between the asymptomatic staff and mastitis patients.
Panton-Valentine leucocidin (PVL) has been linked to soft tissue infections (27) and the clinical severity of mastitis (28). Saiman et al. reported four postpartum MRSA mastitis cases in New York characterized as SCCmecIV- spa t131-pvl+ (29). An outbreak of postpartum MRSA breast infections identified as ST22-SCCmecIV-spa t852 or t005-pvl + was documented in Mumbai (27). In our study, all 25 clinical MRSA isolates were identified as ST59-SCCmecVT-spa t437-pvl+. These findings emphasize the significant role of PVL in mastitis pathogenesis, with molecular variations potentially attributable to regional differences in CA-MRSA clones (1, 8).
Borderline oxacillin-resistant S. aureus (BORSA) has been characterized by MIC to oxacillin close to or just above the resistance breakpoint owing to production of large amounts of staphylococcal ß-lactamase (30). In addition, BORSA could not be identified by traditional latex agglutination test for MecA (31). Stańkowska et al. reported BORSA with SCCmecIV-spa t437 were isolated from patients with skin and soft tissue infections (32). The authors have recently identified 14 (1.7%) mecA- and 70 (8.3%) mecA+ BORSA among 842 invasive S. aureus isolates, which SCCmecIV and V comprised most of them (33). The authors have not determined oxacillin MIC using agar dilution among these 42 S. aureus isolates included in this study. We believe there may be BORSA, especially with SCCmecIV and V genotypes, from mastitis cases and environment sampling.
The current study has several limitations. Primarily, both epidemiologic and molecular data were sourced from a singular institution, which might not fully reflect national trends. Additionally, the follow-up periods differed among mastitis patients, leaving the long-term outcomes of these patients still to be ascertained.
To conclude, our research illustrates that specific CA-MRSA clones led to clusters of puerperal mastitis, with some clones transmitted between patients and staff. The outbreak of mastitis was effectively managed through the application of an infection control bundle focused on contact precautions.
The authors acknowledge that the data presented in this study must be deposited and made publicly available in an acceptable repository, prior to publication. Frontiers cannot accept a manuscript that does not adhere to our open data policies.
The studies involving humans were approved by Chung Shan Medical University Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Y-CL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Y-LL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Y-HC: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing, Data curation, Software. S-MT: Data curation, Formal analysis, Investigation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing, Conceptualization, Funding acquisition, Resources, Supervision. W-YW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was supported by Chung Shan Medical University (grant number: CSMU-TSMH-104-02). This funding source played no role in study design or conduction, data collection, analysis or interpretation, writing of the manuscript, or decision to submit it for publication.
We thank Aaron Coughman for his assistance in editing English grammar of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2024.1378207/full#supplementary-material
1. Reddy, P, Qi, C, Zembower, T, Noskin, GA, and Bolon, M. Postpartum mastitis and community-acquired methicillin-resistant Staphylococcus aureus. Emerg Infect Dis. (2007) 13:298–301. doi: 10.3201/eid1302.060989
2. Bates, K. Breast mastitis or abscess. JAAPA. (2021) 34:51–2. doi: 10.1097/01.JAA.0000795040.66115.51
3. Wheaton, N, Al-Abdullah, A, and Haertlein, T. Postdelivery emergencies. Emerg Med Clin North Am. (2019) 37:287–300. doi: 10.1016/j.emc.2019.01.014
4. Angelopoulou, A, Field, D, Ryan, CA, Stanton, C, Hill, C, and Ross, RP. The microbiology and treatment of human mastitis. Med Microbiol Immunol. (2018) 207:83–94. doi: 10.1007/s00430-017-0532-z
6. World Health Organization (2000). Mastitis: causes and management. WHO/FCH/CAH/00.13. Available at: http://whqlibdoc.who.int/hq/2000/WHO_FCH_CAH_00.13.pdf.
7. Stafford, I, Hernandez, J, Laibl, V, Sheffield, J, and Roberts, SGW. Community-acquired Staphylococcus aureus among patients with puerperal mastitis requiring hospitalization. Obstet Gynecol. (2008) 112:533–7. doi: 10.1097/AOG.0b013e31818187b0
8. Holmes, MA, and Zadoks, RN. Methicillin resistant S. aureus in human and bovine mastitis. J Mammary Gland Biol Neoplasia. (2011) 16:373–82. doi: 10.1007/s10911-011-9237-x
9. Mitchell, KB, Johnson, HM, Rodríguez, JM, Eglash, A, Scherzinger, C, Zakarija-Grkovic, I, et al. Academy of breastfeeding medicine clinical protocol #36: the mastitis Spectrum, revised 2022. Breastfeed Med. (2022) 17:360–76. doi: 10.1089/bfm.2022.29207.kbm
10. CLSI. Performance standards for antimicrobial disk susceptibility tests. Approved standard (M100-Ed32). Wayne, PA: CLSI (2022).
11. Kondo, Y, Ito, T, Ma, XX, Watanabe, S, Kreiswirth, BN, Etienne, J, et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. (2007) 51:264–74. doi: 10.1128/AAC.00165-06
12. CLSI. Performance standards for antimicrobial susceptibility testing; CLSI document M100. 30th ed. Wayne, PA: CLSI (2022).
13. Tenover, FC, Arbeit, RD, Goering, RV, Mickelsen, PA, Murray, BE, Persing, DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. (1995) 33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995
14. Li, WH. Simple method for constructing phylogenetic trees from distance matrices. Proc Natl Acad Sci USA. (1981) 78:1085–9. doi: 10.1073/pnas.78.2.1085
15. Enright, MC, Day, NP, Davies, CE, Peacock, SJ, and Spratt, BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. (2000) 38:1008–15. doi: 10.1128/JCM.38.3.1008-1015.2000
16. Shopsin, B, Gomez, M, Montgomery, SO, Smith, DH, Waddington, M, Dodge, DE, et al. Evaluation of protein a gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. (1999) 37:3556–63. doi: 10.1128/JCM.37.11.3556-3563.1999
17. Gilot, P, Lina, G, Cochard, T, and Poutrel, B. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J Clin Microbiol. (2002) 40:4060–7. doi: 10.1128/JCM.40.11.4060-4067.2002
18. Lina, G, Piemont, Y, Godail-Gamot, F, Bes, M, Peter, MO, Gauduchon, V, et al. Involvement of Panton-valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. (1999) 29:1128–32. doi: 10.1086/313461
19. Omoe, K, Hu, DL, Takahashi-Omoe, H, Nakane, A, and Shinagawa, K. Comprehensive analysis of classical and newly described staphylococcal superantigenic toxin genes in Staphylococcus aureus isolates. FEMS Microbiol Lett. (2005) 246:191–8. doi: 10.1016/j.femsle.2005.04.007
20. World Health Organization. WHO guidelines on hand hygiene in health care. (2009). Available at: http://whqlibdoc.who.int/publications/2009/9789241597906_eng.pdf.
21. Harbarth, S, Dharan, S, Liassine, N, Herrault, P, Auckenthaler, R, and Pittet, D. Randomized, placebo-controlled, double-blind trial to evaluate the efficacy of mupirocin for eradicating carriage of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. (1999) 43:1412–6. doi: 10.1128/AAC.43.6.1412
22. Berens, P, Swaim, L, and Peterson, B. Incidence of methicillin-resistant Staphylococcus aureus in postpartum breast abscesses. Breastfeed Med. (2010) 5:113–5. doi: 10.1089/bfm.2009.0030
23. Foxman, B, D'Arcy, H, Gillespie, B, Bobo, JK, and Schwartz, K. Lactation mastitis: occurrence and medical management among 946 breastfeeding women in the United States. Am J Epidemiol. (2002) 155:103–14. doi: 10.1093/aje/155.2.103
24. Peters, F, Flick-Fillies, D, and Ebel, S. Hand disinfection as the central factor in prevention of puerperal mastitis. Clinical study and results of a survey. Geburtshilfe Frauenheilkd. (1992) 52:117–20. doi: 10.1055/s-2007-1022965
25. Crepinsek, MA, Crowe, L, Michener, K, and Smart, NA. Interventions for preventing mastitis after childbirth. Cochrane Database Syst Rev. (2010):CD007239. doi: 10.1002/14651858.CD007239.pub2
26. Manoharan, A, Zhang, L, Poojary, A, Bhandarkar, L, Koppikar, G, and Robinson, DA. An outbreak of post-partum breast abscesses in Mumbai, India caused by ST22-MRSA-IV: genetic characteristics and epidemiological implications. Epidemiol Infect. (2012) 140:1809–12. doi: 10.1017/S0950268812000593
27. Tristan, A, Ferry, T, Durand, G, Dauwalder, O, Bes, M, Lina, G, et al. Virulence determinants in community and hospital meticillin-resistant Staphylococcus aureus. J Hosp Infect. (2007) 65:105–9. doi: 10.1016/S0195-6701(07)60025-5
28. Rimoldi, SG, Pileri, P, Mazzocco, MI, Romeri, F, Bestetti, G, Calvagna, N, et al. The role of Staphylococcus aureus in mastitis: a multidisciplinary working group experience. J Hum Lact. (2020) 36:503–9. doi: 10.1177/0890334419876272
29. Saiman, L, O'Keefe, M, Graham, PL 3rd, Wu, F, Said-Salim, B, Kreiswirth, B, et al. Hospital transmission of community-acquired methicillin-resistant Staphylococcus aureus among postpartum women. Clin Infect Dis. (2003) 37:1313–9. doi: 10.1086/379022
30. McDougal, LK, and Thornsberry, C. The role of beta-lactamase in staphylococcal resistance to penicillinase-resistant penicillins and cephalosporins. J Clin Microbiol. (1986) 23:832–9. doi: 10.1128/jcm.23.5.832-839.1986
31. Buchan, BW, and Ledeboer, NA. Identification of two borderline oxacillin-resistant strains of Staphylococcus aureus from routine nares swab specimens by one of three chromogenic agars evaluated for the detection of MRSA. Am J Clin Pathol. (2010) 134:921–7. doi: 10.1309/AJCPO9TOID1EPUIM
32. Stankowska, M, Garbacz, K, Piechowicz, L, and Bronk, M. Dissemination of t437-SCCmecIV and coagulase-negative t037-SCCmecIII types among borderline oxacillin-resistant Staphylococcus aureus isolated from skin infections and diabetic foot ulcers. Infect Drug Resist. (2019) 12:3197–203. doi: 10.2147/IDR.S219557
33. Wang, WY, Chen, YH, Lee, YL, Chiu, CF, and Tsao, SM. Comparative analysis of two commercial automated systems with agar dilution for oxacillin susceptibility and their association with genotypes of invasive Staphylococcus aureus isolates (2011-2021). Infect Drug Resist. (2024) 17:1121–9. doi: 10.2147/IDR.S445211
Keywords: puerperal mastitis, MLST, SCCmec, PFGE, CA-MRSA, infection control bundle
Citation: Lin Y-C, Lee Y-L, Chen Y-H, Tsao S-M and Wang W-Y (2024) Puerperal mastitis caused by limited community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) clones. Front. Med. 11:1378207. doi: 10.3389/fmed.2024.1378207
Received: 29 January 2024; Accepted: 02 April 2024;
Published: 19 April 2024.
Edited by:
Daniele Roberto Giacobbe, University of Genoa, ItalyReviewed by:
Katarzyna Garbacz, Medical University of Gdańsk, PolandCopyright © 2024 Lin, Lee, Chen, Tsao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Yao Wang, d3l3YW5nMTk2NkBuZG1jdHNnaC5lZHUudHc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.