- 1Department of Dermatology, Wayne State University, Detroit, MI, United States
- 2Department of Dermatology, Johns Hopkins School of Medicine, Baltimore, MD, United States
- 3Department of Medical Oncology, Rutgers Cancer Institute of New Jersey, New Brunswick, NJ, United States
- 4Department of Pediatrics, Child Health Institute of New Jersey, Rutgers Medical School, New Brunswick, NJ, United States
- 5Department of Surgical Oncology, Rutgers Cancer Institute of New Jersey, New Brunswick, NJ, United States
Background: The success of immunotherapy has made it a lifesaving treatment, but not without side effects. Currently, the risk factors for developing immune-related adverse events (irAEs) in patients who receive immunotherapy are poorly understood, and there is no risk-stratifying mechanism for potentially fatal irAEs. It is postulated that oncology patients with preexisting autoimmune diseases are likely to have flares on immunotherapy. However, some patients develop de novo autoimmune conditions on immunotherapy without a prior history. Literature reports have postulated that human leukocyte antigen (HLA) inherence may play a role in irAEs. However, this potential remains underexplored.
Methods: The oncology patients who developed autoimmune adverse events on immunotherapy for whom the continuation of treatment was prudent or lifesaving were selected. Of note, all nine patients received checkpoint inhibitors (CIs). Of the nine selected patients, only one had a prior history of an autoimmune condition. None of the nine selected patients had an active autoimmune condition at the time of CI initiation. Their HLA was typed, and the results were cross-referenced with the literature reports in PubMed and Google search with the corresponding autoimmune condition of each patient.
Results: Herein, we report nine patients with irAEs for whom retrospective HLA typing revealed the inherence of multiple related HLA alleles that may correspond to the autoimmune condition that they had developed on immunotherapy. It is to be mentioned that the inherence of enriched disease-related HLA alleles was shared among patients with the same irAEs. These patients developed a range of irAEs including bullous pemphigoid, pemphigus foliaceus/vulgaris, thyroiditis, vitiligo, and hepatitis on immunotherapy. Although some combinations of disease-related HLA were well reported in otherwise idiopathic autoimmune diseases, a frequently repeated HLA allele combination in our patient population was found to be rarely seen in the general population.
Conclusion: The authors suggest that an enriched inherence of disease-related HLA alleles may play a role in the genetic propensity for the development of irAEs in oncology patients, who receive immunotherapy, including CIs. Inherence of more than one or a cluster of particular autoimmune disease-related HLA alleles in patients who receive immunotherapy may unmask the corresponding autoimmune disease as the genotype inherence presents with the phenotype of the corresponding condition. It is suggested that enriched linked HLA genotypes, which are otherwise rare in the general population, may present as the corresponding phenotype of the autoimmune condition. Such clinical presentation, enhanced by immunotherapy, such as CIs, can play a role in risk stratifying patients for precision medicine and improve the outcome.
What is already known on this topic
Immunotherapies are known to trigger immune-relatedadverse events (irAEs), though currently there is no way to predict who will and will not develop these serious reactions. Certain HLA types have been associated with autoimmune diseases. This study proposes that HLA typing may be a way to predict who and who will not develop these adverse events.
What this study adds
This pilot study is a proof-of-concept study for the possible use of HLA biomarkers as a predictive tool for adverse events related to immunotherapy. We have found that indeed HLA types do correlate with patients' propensity for developing potentially fatal irAEs.
How this study might affect research, practice, or policy
We hope this study is the beginning of a collective effort to study HLA biomarkers on a population-wide basis. Population-based studies may allow us to narrow down a few HLA subtypes that predispose to the most dangerous irAEs. HLA subtyping prior to starting immunotherapy may allow quicker diagnosis and treatment of any irAEs that arise.
Background
Immunotherapy has significantly improved the prognosis of oncology patients. It saves and extends life, but not without side effects. Almost 60% of patients on immunotherapy experience immune-related adverse events (irAEs) (1). The risk factors for developing irAEs are poorly understood, although it is postulated that patients with preexisting autoimmune diseases are more likely to have flares on immunotherapy rather than developing a de novo autoimmune condition (2). However, some patients on immunotherapy develop an autoimmune condition without a prior history. The incidence of irAEs is increasing as the success of immunotherapy has made it one of the most frequently used and a pillar of oncologic treatment. Although studies have suggested that HLA inherence may play a role in irAEs, this area remains under investigation with the latest data showing that certain types of HLA alleles may be associated with organ or tissue-specific irAEs (3–7). Herein, we report nine patients with irAEs for whom retrospective HLA typing revealed inherence of enriched disease-related-HLA alleles, and only one of the nine patients had a prior history of a related autoimmune condition.
Methods
Nine oncology patients who developed autoimmune adverse events on CI, for whom the continuation of treatment was lifesaving, were HLA typed with high resolution by blood test as part of the diagnosis and assessment workup for the corresponding presented autoimmune AE. Simultaneously, their blood samples were also tested for the presence of serologic autoantibodies related to the corresponding irAE. In cases of cutaneous irAE, skin biopsies were also done for histologic as well as direct immunofluorescent (DIF) evaluation. Of the nine patients, only one had a prior history of an autoimmune condition. None of the nine patients had an active autoimmune condition at the time of CI initiation. The results of their high-resolution HLA type and serologic auto-antibody were cross-referenced with the literature reports in PubMed and Google search with the corresponding autoimmune condition of each patient for diagnostic assessment. Additionally, for those who had skin biopsy, their histopathologic and DIF reports were concluded in the diagnostic assessment and evaluation process.
Results
Patient A is a middle-aged woman with lymphatic metastatic melanoma who received monthly nivolumab for a year with a favorable response. After nine infusions, the patient developed an itchy rash with tense blisters on her upper and mid chest (Figure 1A). The patient's skin biopsy showed bullous pemphigoid (BP) and her blood workup was positive for BP-180 antibodies (Tables 1, 2). She was treated with a course of rapidly tapering prednisone, followed by daily topical triamcinolone ointment. Since the patient had a favorable response to Nivolumab and the irAE was well controlled and limited to her chest, Nivolumab was continued until there was no evidence of detectable melanoma on her restaging workup, when nivolumab was stopped. Thereafter, the BP-180 antibody became undetectable, and the blisters on her chest resolved. The high-resolution HLA typing revealed inherence of well-reported BP-associated HLA allele; DQA1 01:03 (Table 1) (8, 9). Additionally, a further enriched cluster of other BP-associated HLA alleles was also present (Table 2).
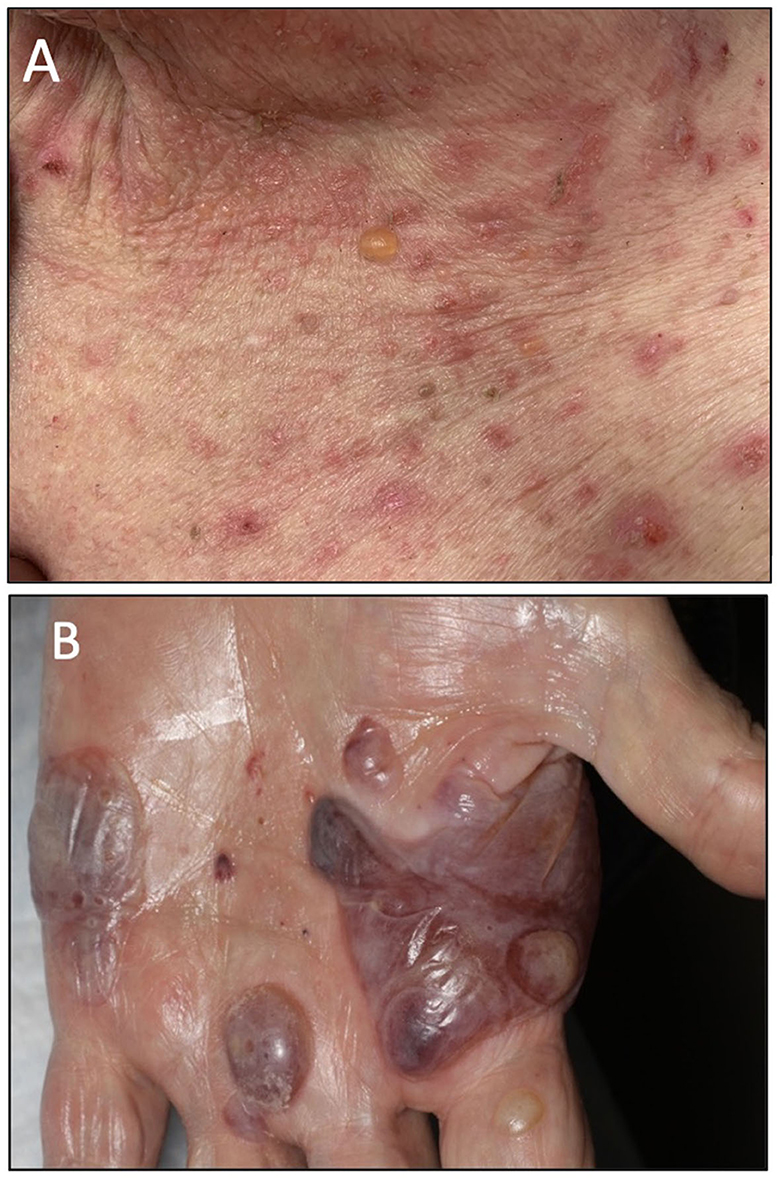
Figure 1. (A) Clinical presentation of patient A showing an intense BP blister on her chest. (B) Clinical presentation of patient B showing several tense hemorrhagic BP bullae on the left palm.

Table 1. The outline of patients' demographics, and their corresponding therapeutic agents, full high-resolution HLA report, and the primary tumor type.
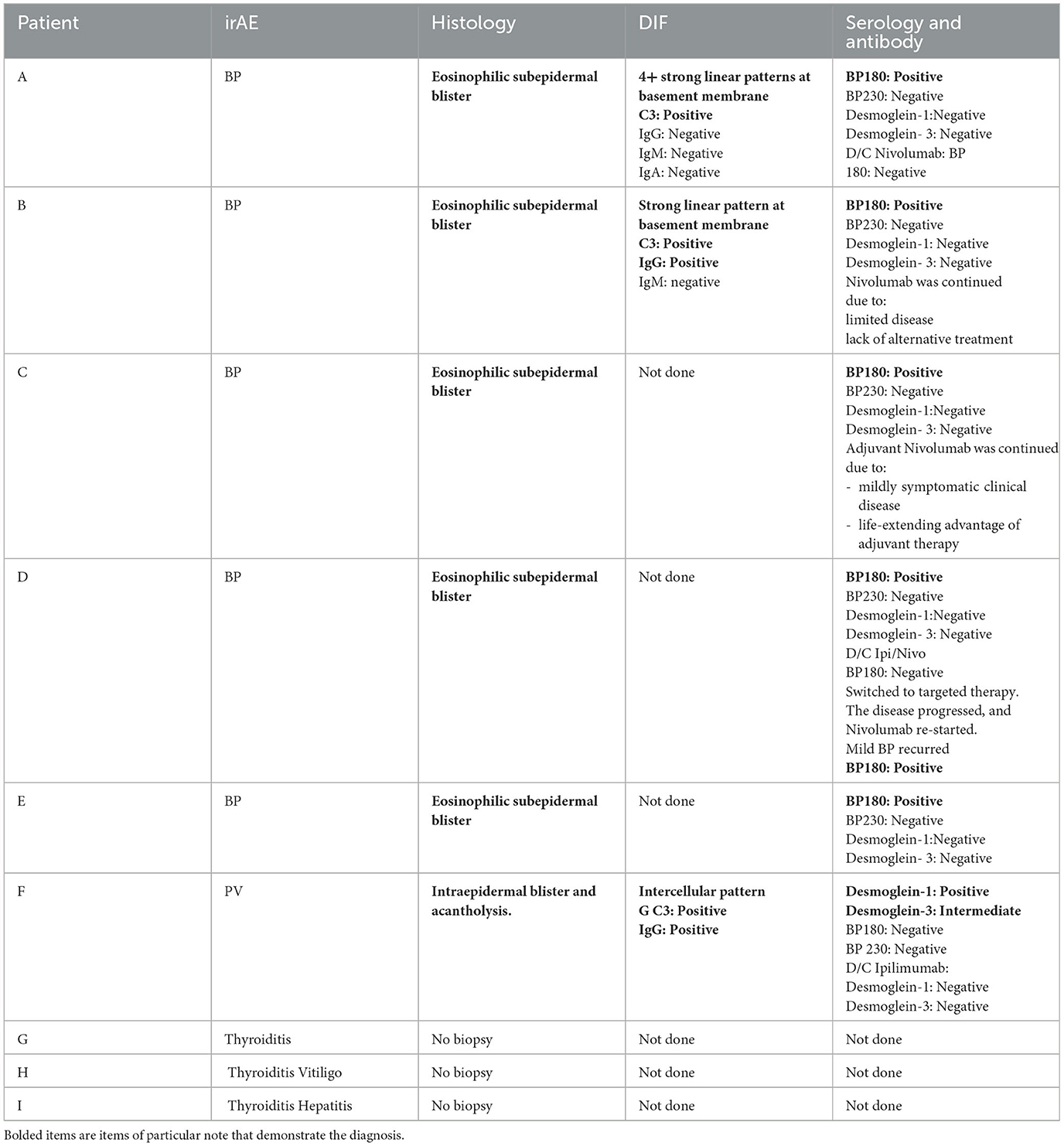
Table 2. The outline shows the patients' serologic test results, as well as the reports of the skin histopathology and DIF if relevant.
Patient B is an elderly patient with distant metastatic melanoma on nivolumab. After the third infusion, the patient developed painful, pruritic, blood-filled bullae confined to the patient's palms and feet (Figure 1B). The fourth infusion was withheld, a skin biopsy and serologic antibodies showed BP, and the patient was treated with clobetasol under occlusion given the confined distribution of the disease involving only palms and soles (Figures 2A–C). Given patient B's significant metastatic tumor burden and prior failure of other treatment options, nivolumab was continued as a lifesaving measure in light of localized non-progressing BP. Patient B eventually passed away after a year due to tumor burden. HLA typing revealed DRB1 11:04, DQA1 05:05, and DQB1*03:01 (Table 1) all of which are well reported to be associated with BP (10). Once again, an enriched cluster of other BP-associated HLA alleles was also present.
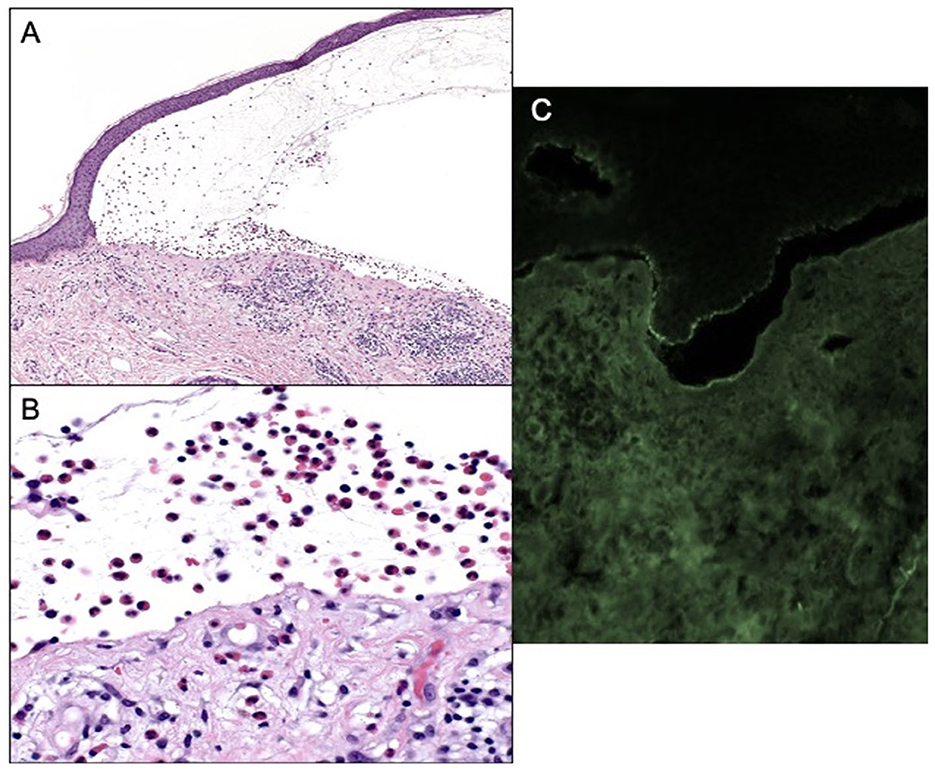
Figure 2. (A) Subepidermal blistering with an abundance of eosinophils 100× magnification and hematoxylin and eosin. (B) Eosinophils within blister cavity, 400× magnification. (C) Positive DIF, linear staining for IgG and C3 along the epidermal-dermal basement membrane.
Patients C and D with a diagnosis of lymphatic and distant metastatic melanoma, respectively, developed BP on immunotherapy. Patient C presented with mild urticarial BP involving < 30% body surface area which was well controlled with topical Triamcinolone. Nivolumab continued until restaging showed no detectable disease, at which point BP resolved once the immunotherapy stopped (Tables 1, 2). Patient D with distant metastatic melanoma was on combination Ipilimumab/Nivolumab (Ipi/Nivo) therapy and developed a blistering BP involving more than 30% of body surface area after the first infusion. The Ipi/Nivo was discontinued in favor of systemic targeted therapy based on the patient's tumor mutation, and the blisters resolved. Both patients C and D had skin biopsy and serology autoantibody levels consistent with the diagnosis of BP. Additionally, both patients had more than one inherent HLA allele reported to be associated with BP. It is worth mentioning that of all four aforementioned patients with pemphigoid irAE, patient D had the least number of enriched clusters of inherent BP-associated HLA alleles of only two, and just one was strongly associated with the disease, HLA DQB1*03:01 (10). This information became useful when patient D was considered for restarting immunotherapy when systemic targeted therapy failed and mono-immunotherapy with Nivolumab was cautiously reintroduced. New small blisters re-appeared on his chest but remained limited, and itching was well tolerated with topical Clobetasol. Of note is that all four BP patients exhibited HLA DQB1*03.
Patient E is an elderly patient with non-operable advanced ulcerating squamous cell carcinomas involving bilateral lower legs on pembrolizumab. After the three infusions, a pruritic bullous eruption appeared on her trunk and extremities and got worse after every infusion. A skin biopsy confirmed bullous pemphigoid and was consistent with positive serology autoantibody results. Pembrolizumab was stopped, the blisters resolved, and the patient was switched to cetuximab with favorable response. HLA typing was done, which revealed HLA A 11:01, B 37:01, DQB1 05:01, and DQB *03 all reported in association with BP (8) (Table 1).
Patient F is a middle-aged adult in remission from stage III melanoma on adjuvant ipilimumab, who presented with pruritic chest lesions, which worsened with each infusion. The patient's serology and skin biopsy results (Table 2) showed pemphigus vulgaris. The lesions resolved with discontinuation of adjuvant ipilimumab followed by a decline of desmoglein antibodies to an undetectable level (Table 2). HLA typing revealed DQB1*0302, DQA1 0301, and DRB1 04 (Table 1), all of which have been reported in association with pemphigus (11).
Patient G is a middle-aged patient with a history of breast ductal adenocarcinoma, who developed a thyroid storm requiring hospitalization 10 days after the first pembrolizumab infusion. Patient D's TSH level dropped to 0.02 and free T4 rose to 7.72 from a normal baseline. After the pembrolizumab was discontinued, the patient's TSH and T4 eventually normalized to 3.57 and 1.67, respectively. Patient D was not investigated for thyroid autoimmunity at the time of thyroiditis. Thyroid autoantibodies (anti-TPO, anti-TSH receptor, and anti-thyroglobulin) were investigated 3.5 years later, rendering negative results (Table 1). However, HLA typing was done to risk stratify the patient for other potentially life-threatening irAEs in order to prepare for re-challenging the patient with immunotherapy due to disease recurrence. The high-resolution HLA revealed inherence of DRB 109:01, DQB1*03:01, DQB1*03:02, and DRB4, all reported to be associated with thyroid autoimmunity and diabetes (12–14). Notably, the patient has a strong family history of type I diabetes and became pre-diabetic during the treatment course.
Patient H with metastatic acral melanoma was started on nivolumab and thyroiditis presented after the patient's fourth infusion. Serology showed a TSH of 126, a free T4 of 0.3, and a T3 of 36. Nivolumab was discontinued and the patient's laboratory values improved to a TSH of 15.2 and a free T4 of 1.7. The patient's thyroid autoantibodies were tested positive at the time of thyroiditis with a TPO antibody of >900 and a thyroglobulin antibody of 1:20. HLA typing revealed DRB1 08:04, DRB4, and A 02:179 alleles, all of which have been linked to autoimmune thyroid disease (AITD) (15, 16). The patient also developed vitiligo at the same time; the high-resolution HLA typing also revealed vitiligo-associated alleles, such as HLA Bw 4, DRB1 07:01, and HLA A 02:179 among others (Table 1) (17–21).
Patient I with stage IIIB melanoma of the nose was on adjuvant nivolumab. After the first nivolumab infusion, the patient showed serologic thyroid abnormalities with a low TSH (0.006), elevated free T4 (3.09), and elevated free T3 (6.6) while remaining clinically asymptomatic. After cycle 9, thyroid serology became more abnormal with elevated TSH (16.4), low free T3 (1.4), and low free T4 (0.38) in addition to elevated ALT (166 IU/L), AST (94 IU/L), and alkaline phosphatase (291 IU/L) accompanied with abdominal pain, vomiting, and mild diarrhea, which resulted in discontinuation of Nivolumab followed by tapering of oral prednisone. Hepatitis resolved, and thyroid hormone therapy was initiated. Restating PET/CT showed no evidence of detectable melanoma. Similar to patients F and G, patient H also showed HLA DRB4, which is a well-reported allele linked with autoimmune thyroiditis. This patient also had HLA type DRB1 04:01, which is reported in association with autoimmune hepatitis (Table 1) (22, 23).
Discussion
Checkpoint inhibitors enhance CD8 T-cell cytotoxic function by downregulating the inhibitory brakes, which can cause irAEs (24). The mechanism of irAEs is complex and occasionally life-threatening. These adverse events can occur in any organ; most commonly the skin (46–62%) and colon (22–48%) (6, 25). It is prudent to better understand such irAEs, and when possible, risk stratify patients accordingly to achieve a more precise decision at the point of care when continuation of immunotherapy is considered lifesaving (26). Though there is no current standard to predict toxicities, HLA typing has been proposed as a potential risk-stratifying parameter for irAE (26, 27). We propose that the presented patients who developed bullous pemphigoid, pemphigus vulgaris, thyroiditis, vitiligo, and hepatitis may be associated with their genetic propensity toward such autoimmune conditions, which became unmasked by immunotherapy (Table 1).
Bullous pemphigoid has been reported with almost all immunotherapies (1, 27). There are HLA associations with BP among certain populations including DQB1*03:01 in Caucasians and Iranians, and DQB1*03:02, DRB1*11:01, and DRB1 04:03 in the Japanese (12). In Brazilian populations, DQB1* 03:01, DQA1 01:03, and DQA1 05:05 alleles have been associated with BP (9). In the northern Chinese, HLA-DQA1 05:05, DQB1 05:01, and DRB1 11:04 were found in association with BP (10). In the Han Chinese population HLA-A 11:01 and HLA-B 37:01 and in the Iranian population DQB1 05:01 have been shown to be associated with BP (8, 28). All five patients who presented with immunotherapy-associated BP had an enriched cluster of multiple HLA alleles, which at times formed a haplotype linked with BP in various population study reports. Of these alleles, the HLA DQB1*03 is widely reported in association with BP, in addition to other autoimmune conditions, such as alopecia areata, thyroiditis, celiac disease, colitis, and type 2 autoimmune hepatitis (28–34). It is of note that all nine patients presented here with irAEs exhibited HLA DQB1 *03 allele in their HLA inherence. This is in light of the HLA DQB1*03 frequency of < 20% in the US with the HLA DQB1*03:01 frequency of 17.7% specifically. The enriched presence of alleles, such as HLA DQB1 *03:01 in presented patients, in light of the low prevalence in the general population, suggests that HLA typing had the potential to be considered a biomarker to stratify irAEs in high-risk patients (35, 36).
A clinically meaningful HLA association with the development of melanoma is well reported in the literature. Additionally, some of these melanoma-linked HLA alleles are reported to overlap with those associated with immunotherapy-triggered irAEs in oncology patients. Of such linked HLAs, overlap between melanoma and CI-associated BP adverse events only includes HLA-DPB1* 01 and HLADPB1*10 which were found in patients A and I who had CI-associated BP adverse events and melanoma. Of note, neither of these alleles have been found to be associated with poor outcomes in melanoma (37). It may warrant further investigation and larger data to determine if melanoma patients with the aforementioned HLA may have a clinically meaningful risk of developing BP adverse events on immunotherapy, including CI.
Notably, all nine presented patients had HLA DQB1*03 allele. This allele is not only well reported with an enriched presence in BP patients, but also has been found to be linked to many other autoimmune conditions, including alopecia areata, celiac disease, and type 2 autoimmune hepatitis (29–34). Additionally, the HLA DRB4 01:03 allele, well known to be linked with autoimmune thyroiditis, was present in all 3 thyroiditis patients presented here.
Immunotherapy-associated PV is well reported in the literature with recent studies reporting the first case of PV triggered by ipilimumab (11, 25, 38). The HLA typing of patient F revealed multiple PV-associated HLA alleles, including but not limited to the DQB1*03:02 and DQA1 03:01 (12–14, 39–41). Such an enriched inheritance of a multiple disease-associated genotype lead to the suggestion that the enhancement of the immune system by immunotherapy may unmask an otherwise underlying dormant disease to clinical presentation via HLA (42–44).
Thyroid toxicity has also been well reported with immunotherapy (45). The incidence of thyroid irAE due to pembrolizumab is reported in ~17% and usually presents within a few weeks after the first dose. Progression from thyrotoxicosis to hypothyroidism post-immunotherapy is almost universal (46).
Patients G and H had multiple thyroiditis-associated HLA alleles including DRB1 09:01, reported in the Japanese with Hashimoto's thyroiditis, DQB1*03:01 reported in the Caucasians, and DQB1*03:02 in the Greek population with autoimmune thyroiditis (13, 14). Interestingly, DQB1*03:01 and *03:02, detected in patient G, are known to present a shared genetic predisposition for type 1 diabetes and autoimmune thyroid disease (41), shedding light in part on the strong family history of diabetes in patient G (41, 47). Patient H possessed multiple predisposing autoimmune thyroiditis HLA alleles including HLA A2 and DRB1 08:04, which have been linked with Hashimoto's thyroiditis and Graves' disease, with the latter being reported with early onset of the disease. The HLA DRB4 seen in patient I is also well reported with Hashimoto's thyroiditis and other autoimmune thyroid conditions (48). In oncology patients, it is reported that time to thyrotoxicosis occurs within 6 weeks of the first immunotherapy infusion (15, 16).
Vitiligo is also well reported as an irAE of immunotherapy, especially in melanoma patients treated with PD-1 inhibitors with an incidence ranging from 7.5 to 11% (1). The mechanism of vitiligo seen in patients treated with CIs is thought to be a cross-reaction between the shared antigens in melanoma and normal melanocytes, such as MART1, GP100, or tyrosinase (1). Usually, vitiligo presents progressively, bilaterally, and symmetrically after a few months of immunotherapy and persists beyond the duration of treatment. The development of vitiligo has been proposed to be associated with favorable survival and prognosis (1). Expression of some HLA genes, such as DRB and DQB, is reported to contribute to 30% of vitiligo patients (19). Patient H with vitiligo irAE has predisposing HLA DRB1 07:01, DQB1 02:02, and DQB1 03:19, the transcription of which to cytoplasmic mRNA has been shown to increase the expression of HLA-DQ protein on the surface of antigen-presenting cells known to promote the autoreactive T cell activation with a known role in vitiligo pathophysiology (18). In otherwise healthy individuals, the onset of vitiligo is presumably due to an environmental insult and in our patient, we suggest that immunotherapy is the possible culprit. Moreover, our patient has additional vitiligo-linked HLAs, including DRB1 07:01 and A 02:179, according to the reported meta-analysis (20).
The overall incidence of hepatitis induced by immunotherapies has been reportedly low ~2–15%, and CTLA4 inhibitors have been reported to be associated with more cases of immune-mediated hepatitis (IH) than PD1/PDL1 inhibitors (49). Generally, IH leads to a hepatocellular injury with abnormal findings in liver function test (LFT). Patient I with hepatitis had HLA DRB1*04:01 with a well-reported association with IH, specifically in the Caucasian population, in the literature (22, 23).
Understanding HLA inherence in immunotherapy-associated irAEs is an important first step in the risk stratification of patients who may indeed benefit from HLA-specific modulating immune response treatments to provide the full benefit of a completed course of treatment in the relevant patient population. Although such intervention may be on the horizon and is promising, the current level of understanding warrants further research beyond the presented pilot study to further explore the role of HLA-specific TCR targeting molecules to modulate immune response. The HLA-specific TCR targeting molecules have entered clinical use for the treatments of solid malignancies, including uveal melanoma. The consideration to extend the application of such treatment strategies to the irAEs in patients with HLA-specific solid tumors would warrant risk stratification and biomarker selection. An example is tebentafusp, which is an approved treatment for uveal melanoma by targeting HLA A*02:01 to trigger T cell immune response. In the future, it may be possible to consider a similar approach in patients with HLA-specific solid tumors presenting with CI-associated irAE to treat the underlying disease while ameliorating or evading irAE. However, targeting HLA-specific TCR molecules has only been used clinically in solid tumors and chronic viral infections thus far (50–52).
Although the presented work is limited by the number of patients, it is to be mentioned that all the patients had DQB1 03. This enriched presence of the DQB1 03 allele, in light of its strong link with autoimmune diseases including BP, can present an opportunity for follow-up studies with a larger number of patients to further investigate HLA as a biomarker to stratify the risk of irAEs in patients on immunotherapy. Additionally, the co-inherence of HLA DRB3 and DPB1, which data show to be frequently found in patients with autoimmune conditions, is seen in seven of nine presented patients. It is noted that the limited number of patients in the presented work would not allow such interpretation of our patients. However, the seven aforementioned patients also exhibited an otherwise rare HLA allele; HLA DRB, which is reported in 17% of the general population. Finally, all of our nine patients were found to share the same HLA haplotype region; HLA DRB1, DQA1, and DQB, which encode proteins that are found to play a key role in presenting antigens to CD4+ T cells in the autoimmune disease processes (53). Once again, the pattern of an enriched presence of HLA allele or haplotype in our patients, although limited in number, can be observed as a potential role that HLA typing can play in irAR risk stratification in patients in whom immunotherapy is lifesaving. That being said, further studies with a larger number of patients are warranted to investigate the significance and analyze the benefit of HLA typing as a risk-stratifying tool at the point of care (Table 1).
The authors acknowledge that the presented work is one step along the collective endeavor toward what precision medicine may 1 day provide.
Conclusion
These nine patients highlight a spectrum of irAEs with various severities in oncology patients on immunotherapy, for whom such treatment was considered lifesaving. Therefore, risk stratifying these patients to continue immunotherapy in the face of such irAEs became a challenge in clinical decision-making at the point of care. While clinical information and related serologic tests were used in this process, we also applied the HLA typing to assess risk. In the process, we learned that our patients had (a) more than one HLA allele linked to their related irAEs and (b) an enriched presence of certain HLA alleles or haplotype regions competed with the general population. Although we present a limited number of patients here, the data suggest in favor of HLA typing as a risk-stratifying tool in addition to the clinical information and related serologic test to assess irAEs of immunotherapy in oncology patients, for whom this treatment is considered to be lifesaving. Currently, there is no standard method to risk stratify patients for potentially fatal toxicities or early detection of irAEs. The authors acknowledge that follow-up studies with larger data are warranted to fulfill the criteria; however, the presented data are a step toward what precision medicine may 1 day offer.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants' legal guardians/next of kin in accordance with the national legislation and the institutional requirements. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
BD: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Writing – original draft, Writing – review & editing. SG: Writing – original draft, Writing – review & editing. EN: Writing – original draft, Writing – review & editing. DT: Formal analysis, Investigation, Writing – review & editing. RS: Formal Analysis, Investigation, Writing – review & editing. LD: Formal analysis, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
HLA, human leukocyte antigen; irAEs, immune-related adverse events; BP, bullous pemphigoid; PV, pemphigus vulgaris; TSH, thyroid-stimulating hormone; T3, triiodothyronine; T4, thyroxine; TPO, thyroid peroxidase; PET/CT, positron emission tomography/computed tomography; AITD, autoimmune thyroid disease; CTLA4, cytotoxic t-lymphocyte associated protein; PD1/PDL1, programmed cell death-1, programmed cell death ligand-1; IMH, immune-mediated hepatitis; LFTs, liver function tests.
References
1. Sibaud V. Dermatologic reactions to immune checkpoint inhibitors: skin toxicities and immunotherapy. Am J Clin Dermatol. (2018) 19:345–61. doi: 10.1007/s40257-017-0336-3
2. Myers G. Immune-related adverse events of immune checkpoint inhibitors: a brief review. Curr Oncol. (2018) 25:342–7. doi: 10.3747/co.25.4235
3. Chang H, Shin YW, Keam B, Kim M, Im SA, Lee ST. HLA-B27 association of autoimmune encephalitis induced by PD-L1 inhibitor. Ann Clin Transl Neurol. (2020) 7:2243–50. doi: 10.1002/acn3.51213
4. Schoenberg E, Mehregan D, Colombe B, Hazan E, Dasgeb B. The potential role of HLA typing to risk stratify melanoma patients on immunotherapy with associated SJS: pitfalls and opportunities. Int J Dermatol. (2021) 61:e335–7. doi: 10.1111/ijd.15794
5. Ali OH, Berner F, Bomze D, Fässler M, Diem S, Cozzio A, et al. Human leukocyte antigen variation is associated with adverse events of checkpoint inhibitors. Eur J Cancer. (2019) 107:8–14. doi: 10.1016/j.ejca.2018.11.009
6. Jiang N, Yu Y, Zhang M, Tang Y, Wu D, Wang S, et al. Association between germ-line HLA and immune-related adverse events. Front Immunol. (2022) 13:952099. doi: 10.3389/fimmu.2022.952099
7. Conroy M, Naidoo J. Immune-related adverse events and the balancing act of immunotherapy. Nat Commun. (2022) 13:392. doi: 10.1038/s41467-022-27960-2
8. Fang H, Shen S, Zheng X, Dang E, Zhang J, Shao S, et al. Association of HLA class I and class II alleles with bullous pemphigoid in Chinese Hans. J Dermatol Sci. (2018) 89:258–62. doi: 10.1016/j.jdermsci.2017.11.014
9. Chagury AA, Sennes LU, Gil JM, Kalil J, Rodrigues H, Rosales CB, et al. HLA-C*17, DQB1*03:01, DQA1*01:03 and DQA1*05:05 alleles associated to bullous pemphigoid in Brazilian population. Ann Dermatol. (2018) 30:8–12. doi: 10.5021/ad.2018.30.1.8
10. Fang H, Zheng X, Sun L, Wang G. The associations of human leukocyte antigen class I and class II alleles in bullous pemphigoid. EMJ Dermatol. (2017) 5:53–4.
11. Schoenberg E, Colombe B, Cha J, Orloff M, Shalabi D, Ross NA, et al. Pemphigus associated with ipilimumab therapy. Int J Dermatol. (2021) 60, e331–3. doi: 10.1111/ijd.15405
12. Sun Y, Liu H, Wang Z, Fu X, Wang C, Mi Z, et al. The HLA-DQB1*03:01 Is associated with bullous pemphigoid in the Han Chinese population. J Invest Dermatol. (2018) 138:1874–7. doi: 10.1016/j.jid.2018.02.021
13. Katahira M, Ogata H, Takashima H, Ito T, Hodai Y, Miwata T, et al. Critical amino acid variants in HLA-DRB1 allotypes in the development of Graves' disease and Hashimoto's thyroiditis in the Japanese population. Hum Immunol. (2021) 82:226–31. doi: 10.1016/j.humimm.2020.12.007
14. Kokaraki G, Daniilidis M, Yiangou M, Arsenakis M, Karyotis N, Tsilipakou M, et al. Major histocompatibility complex class II (DRB1*, DQA1*, and DQB1*) and DRB1*04 subtypes' associations of Hashimoto's thyroiditis in a Greek population. Tissue Antigens. (2009) 73:199–205. doi: 10.1111/j.1399-0039.2008.01182.x
15. Shin D-H, Baek I-C, Kim HJ, Choi E-J, Ahn M, Jung MH, et al. HLA alleles, especially amino-acid signatures of HLA-DPB1, might contribute to the molecular pathogenesis of early-onset autoimmune thyroid disease. PLoS ONE. (2019) 14:e0216941. doi: 10.1371/journal.pone.0216941
16. Zeitlin AA, Heward JM, Newby PR, Carr-Smith JD, Franklyn JA, Gough SCL, et al. Analysis of HLA class II genes in Hashimoto's thyroiditis reveals differences compared to Graves' disease. Genes Immun. (2008) 9:358–63. doi: 10.1038/gene.2008.26
17. Li Z, Ren J, Niu X, Xu Q, Wang X, Liu Y, et al. Meta-analysis of the association between vitiligo and human leukocyte antigen-A. Biomed Res Int. (2016) 2016:5412806. doi: 10.1155/2016/5412806
18. Jin Y, Roberts GHL, Ferrara TM, Ben S, van Geel N, Wolkerstorfer A, et al. Early-onset autoimmune vitiligo associated with an enhancer variant haplotype that upregulates class II HLA expression. Nat Commun. (2019) 10:391. doi: 10.1038/s41467-019-08337-4
19. Ghaffarnia R, Saffarian Z, Shahbazi M, Zamani M. Contribution of HLA class II genes, DRB4*01:01, DRB1*07:01, and DQB1*03:03:2 to clinical features of Vitiligo disease in Iranian population. Mol Biol Rep. (2022) 49:171–8. doi: 10.1007/s11033-021-06855-3
20. Singh A, Sharma P, Kar HK, Sharma VK, Tembhre MK, Gupta S, et al. HLA alleles and amino-acid signatures of the peptide-binding pockets of HLA molecules in vitiligo. J Invest Dermatol. (2012) 132:124–34. doi: 10.1038/jid.2011.240
21. Kader Toama MA, Khattab F, Marei A. Association of human leukocyte antigen-DRB1 with the response in patients with vitiligo. Egyp J Dermatol Venerol. (2019) 39:71–7. doi: 10.4103/ejdv.ejdv_3_18
22. Oliveira LC, Porta G, Marin ML, Bittencourt PL, Kalil J, Goldberg AC. Autoimmune hepatitis, HLA and extended haplotypes. Autoimmun Rev. (2011) 10:189–93. doi: 10.1016/j.autrev.2010.09.024
23. Higuchi T, Oka S, Furukawa H, Tohma S, Yatsuhashi H, Migita K. Genetic risk factors for autoimmune hepatitis: implications for phenotypic heterogeneity and biomarkers for drug response. Hum Genomics. (2021) 15:6. doi: 10.1186/s40246-020-00301-4
24. Liudahl SM, Coussens LM. B cells as biomarkers: predicting immune checkpoint therapy adverse events. J Clin Invest. (2018) 128:577–9. doi: 10.1172/JCI99036
25. Krammer S, Krammer C, Salzer S, Bagci IS, French LE, Hartmann D. Recurrence of pemphigus vulgaris under nivolumab therapy. Front Med. (2019) 6:262. doi: 10.3389/fmed.2019.00262
26. Akturk HK, Couts KL, Baschal EE, Karakus KE, Van Gulick RJ, Turner JA, et al. Analysis of human leukocyte antigen DR alleles, immune-related adverse events, and survival associated with immune checkpoint inhibitor use among patients with advanced malignant melanoma. JAMA Network Open. (2022) 5:e2246400. doi: 10.1001/jamanetworkopen.2022.46400
27. Lopez AT, Khanna T, Antonov N, Audrey-Bayan C, Geskin L. A review of bullous pemphigoid associated with PD-1 and PD-L1 inhibitors. Int J Dermatol. (2018) 57:664–9. doi: 10.1111/ijd.13984
28. Zhang J, Wang G. Genetic predisposition to bullous pemphigoid. J Dermatol Sci. (2020) 100:86–91. doi: 10.1016/j.jdermsci.2020.05.010
29. Megiorni F, Pizzuti A. HLA-DQA1 and HLA-DQB1 in Celiac disease predisposition: practical implications of the HLA molecular typing. J Biomed Sci. (2012) 19:88. doi: 10.1186/1423-0127-19-88
30. Hiraga N, Imamura M, Kawakami Y, Aikata H, Takahashi S, Akuta N, et al. HLA-DQB1*03 confers susceptibility to chronic hepatitis C in Japanese: a genome-wide association study. PLoS ONE. (2013) 8:e84226. doi: 10.1371/journal.pone.0084226
31. Stahl E, Roda G, Dobbyn A, Hu J, Zhang Z, Westerlind H, et al. Collagenous colitis is associated with HLA signature and shares genetic risks with other immune-mediated diseases. Gastroenterology. (2020) 159:549–61.e8. doi: 10.1053/j.gastro.2020.04.063
32. Schubert MS, Hutcheson PS, Graff RJ, Santiago L, Slavin RG. HLA-DQB1 *03 in allergic fungal sinusitis and other chronic hypertrophic rhinosinusitis disorders. J Aller Clin Immunol. (2004) 114:1376–83. doi: 10.1016/j.jaci.2004.08.029
33. Ujiie H, Muramatsu K, Mushiroda T, Ozeki T, Miyoshi H, Iwata H, et al. HLA-DQB1*03:01 as a biomarker for genetic susceptibility to bullous pemphigoid induced by DPP-4 inhibitors. J Investig Dermatol. (2018) 138:1201–4. doi: 10.1016/j.jid.2017.11.023
34. Lapierre P, Alvarez F. Type 2 autoimmune hepatitis: genetic susceptibility. Front Immunol. (2022) 13:1025343. doi: 10.3389/fimmu.2022.1025343
35. Chen N, Wang W, Wang F, Dong L, Zhao S, Zhang W, et al. The distributions of HLA-A, HLA-B, HLA-C, HLA-DRB1 and HLA-DQB1 allele and haplotype at high-resolution level in Zhejiang Han population of China. Int J Immunogen. (2019) 46:7–16. doi: 10.1111/iji.12411
36. Hernández-Mejía DG, Páez-Gutiérrez IA, Dorsant Ardón V, Camacho Ramírez N, Mosquera M, Cendales PA, et al. Distributions of the HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 alleles and haplotype frequencies of 1763 stem cell donors in the Colombian Bone Marrow Registry typed by next-generation sequencing. Front Immunol. (2023) 13:1057657. doi: 10.3389/fimmu.2022.1057657
37. Dhall A, Patiyal S, Kaur H, Bhalla S, Arora C, Raghava GPS. Computing skin cutaneous melanoma outcome from the HLA-alleles and clinical characteristics. Front Genet. (2020) 11:e00221. doi: 10.3389/fgene.2020.00221
38. Ito M, Hoashi T, Endo Y, Kimura G, Kondo Y, Ishii N, et al. Atypical pemphigus developed in a patient with urothelial carcinoma treated with nivolumab. J Dermatol. (2019) 46:e90–2. doi: 10.1111/1346-8138.14601
39. Vodo D, Sarig O, Sprecher E. The genetics of pemphigus vulgaris. Front Med. (2018) 5:226. doi: 10.3389/fmed.2018.00226
40. Olbrich M, Kunstner A, Witte M, Busch H, Fahnrich A. Genetics and omics analysis of autoimmune skin blistering diseases. Front Immunol. (2019) 10:2327. doi: 10.3389/fimmu.2019.02327
41. Kahles H, Fain PR, Baker P, Eisenbarth G, Badenhoop K. Genetics of autoimmune thyroiditis in type 1 diabetes reveals a novel association with DPB1*0201: data from the type 1 diabetes genetics consortium. Diabetes Care. (2015) 38(Suppl. 2):S21–28. doi: 10.2337/dcs15-2005
42. Petzl-Erler ML. Beyond the HLA polymorphism: a complex pattern of genetic susceptibility to pemphigus. Genet Mol Biol. (2020) 43:e20190369. doi: 10.1590/1678-4685-gmb-2019-0369
43. Wolf R, Tamir A, Brenner S. Drug-induced versus drug-triggered pemphigus. Dermatologica. (1991) 182:207–10. doi: 10.1159/000247795
44. Tarhini A, Lo E, Minor DR. Releasing the brake on the immune system: ipilimumab in melanoma and other tumors. Cancer Biother Radiopharm. (2010) 25:601–13. doi: 10.1089/cbr.2010.0865
45. Imblum BA, Baloch ZW, Fraker D, LiVolsi VA. Pembrolizumab-Induced Thyroiditis. Endocr Pathol. (2019) 30:163–7. doi: 10.1007/s12022-019-9579-2
46. de Filette J, Jansen Y, Schreuer M, Everaert H, Velkeniers B, Neyns B, et al. Incidence of thyroid-related adverse events in melanoma patients treated with pembrolizumab. J Clin Endocrinol Metab. (2016) 101:4431–9. doi: 10.1210/jc.2016-2300
47. Frommer L, Kahaly GJ. Type 1 diabetes and autoimmune thyroid disease-the genetic link. Front Endocrinol. (2021) 12:618213. doi: 10.3389/fendo.2021.618213
48. Cho WK, Jung MH, Choi EJ, Choi HB, Kim TG, Suh BK. Association of HLA alleles with autoimmune thyroid disease in Korean children. Horm Res Paediatr. (2011) 76:328–34. doi: 10.1159/000331134
49. Jennings JJ, Mandaliya R, Nakshabandi A, Lewis JH. Hepatotoxicity induced by immune checkpoint inhibitors: a comprehensive review including current and alternative management strategies. Expert Opin Drug Metab Toxicol. (2019) 15:231–44. doi: 10.1080/17425255.2019.1574744
50. Dolgin E. First soluble TCR therapy opens “new universe” of cancer targets. Nat Biotechnol. (2022) 40:441–4. doi: 10.1038/s41587-022-01282-6
51. Chen LN, Carvajal RD. Tebentafusp for the treatment of HLA-A*02:01-positive adult patients with unresectable or metastatic uveal melanoma. Exp Rev Antican Ther. (2022) 22:1017–27. doi: 10.1080/14737140.2022.2124971
52. Hazini A, Fisher K, Seymour L. Deregulation of HLA-I in cancer and its central importance for immunotherapy. J Immunother Cancer. (2021) 9:e002899. doi: 10.1136/jitc-2021-002899
Keywords: irAE, oncology, checkpoint inhibitors, HLA, HLA inherence
Citation: Gandarillas S, Newland ES, Toppmeyer D, Stephenson R, Denzin L and Dasgeb B (2024) HLA inherence as a potential parameter in checkpoint inhibitor-associated autoimmune adverse event assessment. Front. Med. 10:1288844. doi: 10.3389/fmed.2023.1288844
Received: 05 September 2023; Accepted: 11 December 2023;
Published: 08 January 2024.
Edited by:
Andreas Recke, University of Lübeck, GermanyReviewed by:
Xuming Mao, University of Pennsylvania, United StatesChristoffer Gebhardt, University Medical Center Hamburg-Eppendorf, Germany
Copyright © 2024 Gandarillas, Newland, Toppmeyer, Stephenson, Denzin and Dasgeb. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bahar Dasgeb, YmFoYXJkYXNnZWJAZ21haWwuY29t
 Sophia Gandarillas
Sophia Gandarillas Elizabeth Schoenberg Newland2
Elizabeth Schoenberg Newland2 Ryan Stephenson
Ryan Stephenson Lisa Denzin
Lisa Denzin Bahar Dasgeb
Bahar Dasgeb