
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Med., 12 October 2023
Sec. Dermatology
Volume 10 - 2023 | https://doi.org/10.3389/fmed.2023.1278232
This article is part of the Research TopicArtificial Intelligence in Cutaneous Lesions: Where do we Stand and What is Next?View all 11 articles
This paper provides an overview of artificial-intelligence (AI), as applied to dermatology. We focus our discussion on methodology, AI applications for various skin diseases, limitations, and future opportunities. We review how the current image-based models are being implemented in dermatology across disease subsets, and highlight the challenges facing widespread adoption. Additionally, we discuss how the future of AI in dermatology might evolve and the emerging paradigm of large language, and multi-modal models to emphasize the importance of developing responsible, fair, and equitable models in dermatology.
Recent advancements in artificial intelligence (AI) have fueled an interest in the utility of AI models in medicine (1). These models range from computer vision models that can interpret medical images (2) to large language models (LLM) that have capabilities for analyzing text data (3, 4) to multi-modal models that take both images and text as input (5). These AI models now have the capacity to analyze unstructured data such as clinical notes (3, 6), identify novel correlations in large datasets (7), and generate synthetic image data for improving model training (8, 9).
One medical specialty poised to benefit from these emerging AI technologies is dermatology. Its inherent visual diagnostic process, combined with an increasing volume of clinical photographs, dermoscopy images, and electronic health records (EHR) data (10) underscores its suitability for AI-augmented patient care. Moreover, the shortage of specialists-3.65 dermatologists per 100,000 people in the US (11, 12) and limited access to dermatological services in many regions (13, 14) provides a compelling case for augmented intelligent systems to help bridge this access gap (15). However, clinical integration of AI in dermatology workflow remains challenging. As novel medical applications arise, they also unveil problems that necessitate further research.
In this paper, we present a comprehensive overview of the fundamental principles of AI methodology as applied to dermatology, diving into categories and training approaches. Special emphasis is placed on the role of AI in the diagnosis and prognostication of an array of skin conditions. We also address the limitations of the AI models used in dermatology, notably issues of generalizability, bias, and explainability. Finally, we examine what the future might hold for dermatology-AI, while highlighting some research opportunities to help improve real-world utility of AI models. Our goal is to provide the readers with a panoramic view of AI’s principles and evolving role in dermatology, while equipping them with the knowledge to navigate this dynamic field.
AI is the ability of a computer system to mimic human cognitive functions and encompasses many computational subfields, including machine learning and natural language processing (Figure 1). Currently, major developments in AI are within the field of machine learning (ML), which are algorithms that make predictions about data without explicit programming. In other words, the machines are “learning” from the data and providing analyses without being explicitly told what features to prioritize. Examples from dermatology include identifying melanomas from clinical images (16), predicting efficacy of biologic therapies in psoriasis (17), and analyzing physician notes in electronic health records to determine focus of atopic dermatitis clinic visits (18).
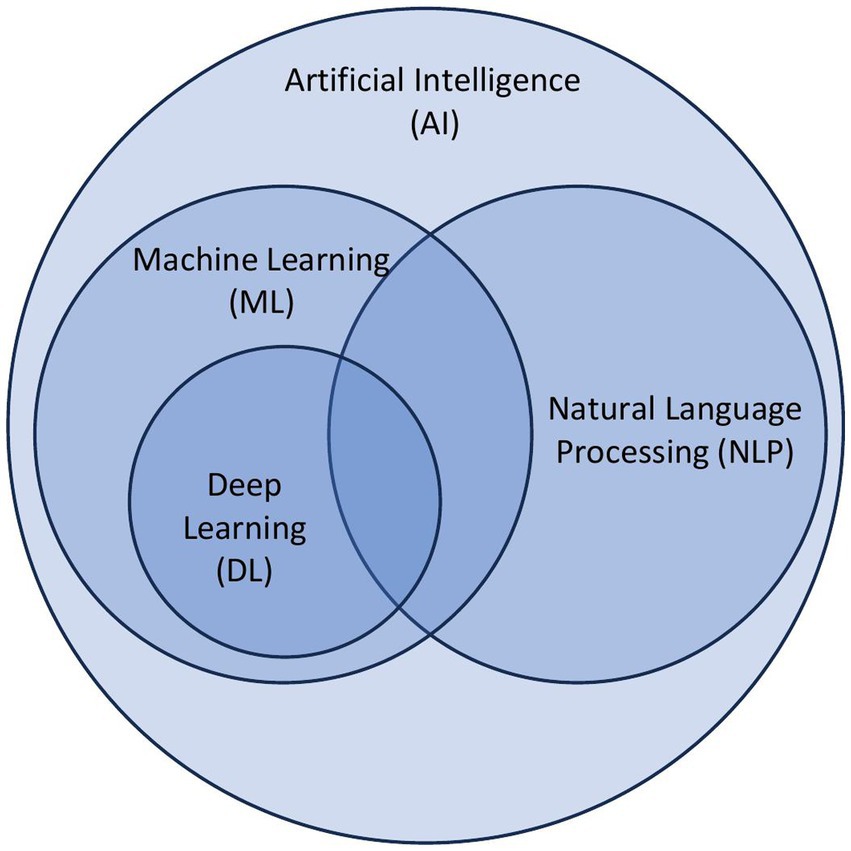
Figure 1. Overview of principles of artificial intelligence. Artificial intelligence (AI) is a broad categorization of algorithms that encompass subcategories including machine learning (ML), natural language processing (NLP), and deep learning (DL).
Deep learning (DL) (19) is a subset of ML that uses algorithms modeled off human neurons that can model complex patterns and relationships in the data. ML techniques prior to the introduction of DL required domain expertise and human engineering to convert raw data into features that the algorithm can understand and detect patterns from. On the other hand, in DL, raw data can be inputted into the algorithm, and the machine is able to create its own representation needed for pattern recognition. These representations are typically arranged in sequential layers, where each layer is inputted into the next layer, increasing the abstraction of the data, collectively known as neural networks (6) (Figure 2). Within DL, there are multiple algorithms that are implemented, including convolutional neural networks (CNN) (20), traditionally used in image processing, and transformer models (21), which are neural networks that learn context and track relationships in sequential data.
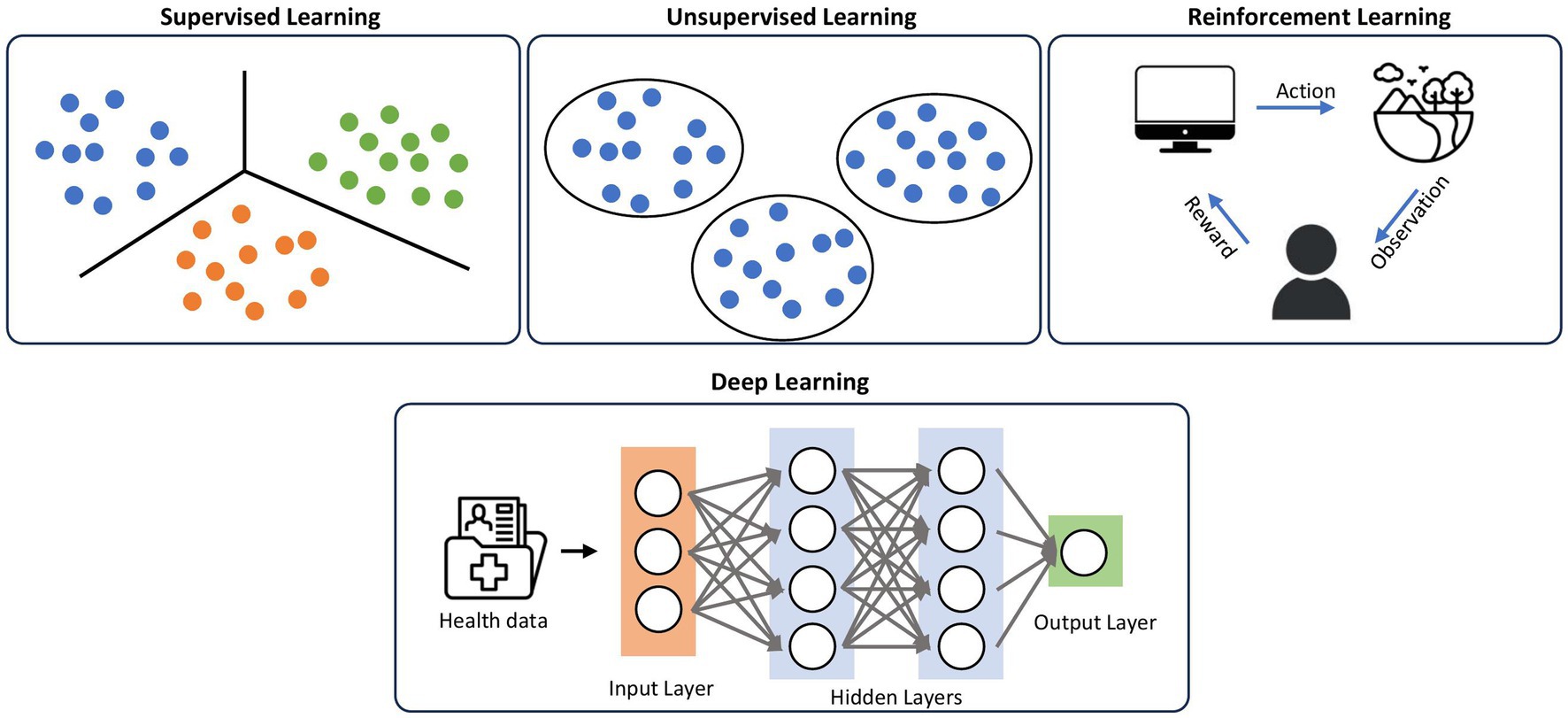
Figure 2. Classifications of machine learning. Supervised learning uses labeled datasets to categorize the data, while unsupervised learning does not have labeled datasets, using patterns and relationships in the data to create categories. Reinforcement learning uses iterative feedback loops to teach the algorithm. DL utilizes representation layers in a neural network to increase abstraction of the data, and employs techniques from supervised, unsupervised, and reinforcement learning.
Within ML, there are different ways that algorithms can learn, including supervised learning, unsupervised learning, and reinforcement learning (Figure 2). Supervised learning, the most common form of machine learning, uses a labeled dataset to predict results. The algorithm learns to map the input data to the correct output, allowing it to make predictions on unseen data. The algorithm is given the data and the correct answers (ground truths) in a training set, which the algorithm uses to set its weights. Once the algorithm has learned from the training data, its performance is measured against a held-out test set that it has never encountered previously. This category of machine learning includes what most people are familiar with, such as logistic regression, linear regression, etc. Most of the image-based deep learning models in dermatology use supervised learning. Unsupervised learning is training a model on unlabeled datasets, meaning the data input does not have the ground truth. This algorithm aims to find patterns and relationships within the data, such as clustering similar data points together. Finally, reinforcement learning is when the agent (the algorithm) interacts with an environment to achieve specific goals. The agent receives feedback from the user (the human) in the form of rewards or penalties based on its actions, and it learns to optimize its behavior to maximize rewards. Compared to supervised and unsupervised learning, reinforcement learning has no predefined data input, but rather learns from the iterative feedback loops.
Natural language processing (NLP) is a branch of artificial intelligence that focuses on interpreting, analyzing, and generating human language. It combines linguistics with statistics, machine learning, and DL to process human language (22). NLP is generally divided into two subfields-natural language understanding (NLU), and natural language generation (NLG). NLU is focused on determining the understanding of the text, while NLG is focused on generating new text. Recent advancements in large language models, including OpenAI’s (San Francisco, United States) publicly-available ChatGenerative Pre-trained Transformer (ChatGPT) (23), fall under the subfield of NLG.
There are also recent emerging concepts of multimodal approaches, where algorithms are utilizing multiple data types to train their algorithms. Medicine is inherently a multimodal discipline, with clinicians interpreting lab values, clinical notes, radiology images, genomic data, etc. New development has been focused on utilizing the rich diversity of data to build more robust models and algorithms, including Med-PaLM Multimodal (Med-PaLM M) (24), LLaVa-Med (25), Med-Flamingo (5), and MiniGPT-4 (26). These new technologies are built on foundation models (FMs), which are models that are trained on a broad range of unlabeled data that are then adapted (fine-tuned) to specific downstream applications (27). These models can learn from the large amounts of data, and then transfer their learnings to a more specific application, like medicine.
There has been an abundance of work done to explore artificial intelligence use in all aspects of dermatology (28–30), ranging from skin malignancies to inflammatory skin conditions, to dermatopathology, to text-based analyses. The visual nature of Dermatology lends itself to many advancements that are image-based, though researchers are exploring other multimodal approaches that use patient characteristics and clinical texts. Here, we will provide a broad overview of the different applications of AI in dermatology.
Applications of AI in dermatologic malignancies, which have been well described in the literature (31, 32), include identifying and distinguishing between benign nevi and melanoma. Researchers break down images of skin lesions to the pixel level for individual analysis and then utilize the techniques described above to predict and classify malignancies. There have been multiple landmark papers for AI applications in skin malignancies (16, 31, 33), resulting in high sensitivities and specificities when distinguishing malignant from benign lesions. Esteva et al. trained a CNN using a large dataset of over 100,000 biopsy-proven clinical images to determine keratinocyte carcinomas versus benign seborrheic keratoses, and malignant melanomas versus benign nevi (16). Han et al. fine-tuned a previously-built CNN model with clinical images to classify multiple malignancies, including basal cell carcinoma, squamous cell carcinoma, melanoma, etc. (34). There is also an annual international skin imaging competition, which provides publicly accessible dermatology images for researchers to build melanoma-classifying models (32, 35). Aside from identifying the primary lesion, there are also studies exploring metastases. Jansen et al. utilized histological tissue sections of sentinel lymph nodes in their convolutional neural network models to identify presence of metastases with high sensitivity and specificity (36).
Aside from classification of melanomas and other malignant skin conditions, researchers are also exploring the identification and management of inflammatory skin conditions, including psoriasis, dermatitis and acne. Similar to malignancy classification, a majority of work is focused on psoriasis identification and classification through images of skin (37–40), nails (41), and scalp (42) using CNN’s and other DL techniques. In addition to diagnosing psoriasis via image recognition, researchers have utilized machine learning techniques to identify patients with increased risk of associated psoriatic conditions, including psoriatic arthritis (43). Work has also been done to determine the efficacy of psoriasis management by predicting outcomes of biologic therapies by using parameters such as patient demographics, clinical history of psoriasis, treatment history, and presence of other comorbidities (17, 44). These preliminary models could be used to eventually optimize therapy and management for patients. Finally, aside from determining outcomes of current biological treatments, AI techniques have been applied to genomic studies to help with drug target identification and drug repurposing (45), as well as screening for psoriasis biomarkers (46) and gene expression profiling (47).
Similar to diagnostic tasks with psoriasis, many researchers have explored using machine learning algorithms in dermatitis (48), ranging from image-based algorithms (49) to electronic health record text-based algorithms (50). Aside from determining diagnoses, researchers have developed proof-of-concept algorithms using self-reported eczema flare scores, patient demographics and treatment history to predict atopic dermatitis severity, resulting in a biologically interpretable model that focuses on patient’s responsiveness to treatment (51). AI models have also been used to help prevent contact dermatitis by predicting skin sensitization potential and potency of substances (52).
In addition to psoriasis and dermatitis, researchers have developed acne lesion segmentation and evaluation tools (53–55) that can grade acne severity from easily-accessible smartphone images (56). There is also exploration in identifying lichen planus (41), and assessing the severity of hidradenitis suppurativa (57).
One of the primary methods in identifying and classifying skin lesions is segmenting the lesion from the backdrop of normal skin. Multiple studies have explored determining and measuring ill-defined wound boundaries using techniques that simplify images down to the pixel level (58–61). Recent work has been done to apply these techniques into broader hospital systems to predict pressure ulcers (62), with the ultimate goal of pressure ulcer prevention (63, 64). Groups have even explored using body heat maps from pressure mats to identify poor in-bed position posture that could cause pressure ulcers (65). These proof-of-concept works, after validation in clinical trials, may ultimately translate into clinical-assist tools to aid clinicians in the management of ulcers.
Beyond identifying diagnoses via clinical images and electronic health record notes, machine learning techniques are being applied in dermatopathology (66, 67). Groups have developed models to classify basal cell carcinoma in digitized Mohs micrographic surgery histology slides to reduce the workload of manually examining these slides (68). Likewise, Hekler et al. used CNNs to aid in histopathologic melanoma diagnoses (69). There have also been studies done to interpret indirect immunofluorescence microscopies to classify bullous dermatoses (70).
To better replicate real-world clinical scenarios of multiple differential diagnoses from a single skin lesion, technologies take a broader approach to solve multi-class classification problems. Many of the problems discussed above were binary classification, where algorithms strived to identify if a lesion was a specific disease or not; multi-class classification presents a more challenging problem with multiple possible diagnoses. Liu et al. created a DL system that provided a differential diagnosis for skin lesions, creating a ranked list of the most likely diagnoses for the skin lesion (71). Taking another multi-class approach, Sitaru et al. have worked to classify body parts from dermatology clinical images, creating body distribution maps for different diagnoses (72).
While dermatology is a visual specialty that focuses on using visual cues for diagnoses, there are aspects of written data that can be used to aid in better understanding questions posed by the research community. Frequently, this written data is unstructured and freeform, using natural human language; to understand and interpret this data, one needs to implement NLP techniques. Researchers have used NLP methods to examine dermatology discussion forums on social media to understand patient perceptions of the field (73). Others conducted analyses of clinical notes in the electronic health records to identify specific topics that providers and patients discuss during clinical visits (18). This analysis provided insights into the lack of documentation of the disease’s impact on a patient’s life, which may ultimately affect management and treatment options.
In addition to understanding natural language, there are also recent advances in technologies that generate new text, including ChatGPT. This technology could be utilized to guide patients, aid clinicians with administrative tasks, educate trainees, etc. Groups are exploring ChatGPT’s ability to generate responses to patient inquiries about melanoma (74), create patient education guides for acne (75), and even triage surgical management of cutaneous neoplasms (76).
Given all the innovation that is occurring at the intersection of AI and dermatology, the logical next step is to evaluate the performance of these AI algorithms against clinicians (77, 78). Esteva et al.’s landmark study was the first to compare a DL algorithm against dermatologists, showing that their model was able to match the performance of 21 dermatologists in melanoma classification (16). Others have even shown that, in a group of 58 international dermatologists, many were outperformed by a CNN model (79). Because of the incredible ability of the technology to perform diagnostic tasks, many researchers are exploring ways to incorporate AI in a clinical workflow to help clinicians. There have now been multiple studies creating AI-based assistive tools to aid clinicians in interpreting clinical images. Groups have designed pipelines with the ultimate goal of real-time AI analysis of skin lesions in the clinics (77). Marchetti et al. prospectively assessed the diagnostic accuracy and utility of a melanoma AI algorithm used in real-world clinical settings to help determine the necessity of biopsying a suspicious lesion (33). Han et al. conducted a randomized trial and showed that AI can augment the accuracy of non-expert physicians in the real-world setting (80). With these smaller pilot studies showing promising results, the AI research community may be looking to increase prospective studies and randomized trials to help further assess AI’s application in the real-world clinical setting.
AI research in dermatology is still in its infancy and encounters a myriad of challenges. From biases and lack of interpretability to regulatory hurdles and difficulty in integrating with existing clinical workflows, these issues are complex and need to be tackled before AI can become ubiquitous in clinical practice. Robust, transparent, and equitable AI algorithms are needed in order to truly enhance patient care without introducing new problems.
AI algorithms learn by identifying features and patterns found in their training datasets and then use this knowledge to make future predictions. However, the presence of confounders in these datasets can influence the validity of AI algorithms. Confounders are features that may be correlated with the AI algorithm’s outcome through spurious associations. An illustrative example involves the presence of surgical pen markers or rulers on clinical dermatology images. As demonstrated in research by Winkler et al., lesions marked with surgical pen markers are more likely to be classified as malignant by the models (81). This finding is due to the fact that these markings are frequently used during biopsy procedures, which are typically performed on lesions suspected of malignancy. Therefore, an algorithm may incorrectly learn an association between these markers and malignancy, when in fact the markings only indicate which lesions were biopsied, not necessarily those that are malignant. This example highlights the importance of identifying and managing confounders during the training phase of AI models to ensure their accuracy and validity.
Bias in training datasets can also inadvertently perpetuate pre-existing inequities in healthcare. In dermatology, this issue is particularly highlighted by early AI models trained on datasets that predominantly featured lighter Fitzpatrick skin types (I-IV). Daneshjou et al. has shown that some of these existing algorithms tend to underperform when assessed with images of darker Fitzpatrick skin types (V-VI) (15). Fortunately, fine-tuning these original algorithms with a dataset featuring darker Fitzpatrick skin types improved their performance, effectively closing the gap in performance between different skin types. Diverse and equitable data representation in the training dataset is primordial to ensure accurate and fair outputs in AI algorithms.
Standardizing images in Dermatology AI research is important to preserve data quality. Images can originate from diverse sources, including various devices (e.g., iPhones, Android smartphones, or professional cameras) and with or without the help of diagnostic tools such as dermatoscopes (82). Additionally, the images may be captured under various settings (e.g., at home or in a clinic) and by different individuals (e.g., patients or healthcare providers). These factors result in a highly heterogeneous dataset comprising images of differing quality. Just as human interpretation can be affected by image quality, AI algorithms are equally sensitive. Blurry images with poor lightning have been shown to negatively impact the performance of AI models (83). Simple image manipulations such as rotation can change the output of an algorithm (84). These considerations underscore the importance of establishing robust image capturing standards and Digital Imaging and Communications in Medicine (DICOM) standards similar to those in other medical fields such as cardiology and radiology (85).
The mechanisms behind traditional medical devices are often transparent and logical in nature. In contrast, AI algorithms appear more mysterious and impenetrable, like a “black box.” This phenomenon makes it difficult for humans to understand its reasoning process and to trust its outputs. Various techniques have been developed by researchers to tackle this problem including saliency maps (e.g., highlighting relevant areas on a picture) and content-based image retrieval (e.g., retrieving similar images from a database based on the query image). As AI penetrates high stake fields such as medicine, it becomes increasingly important to bring transparency and interpretability to AI models.
Implementing AI into clinical practice presents a number of challenges that extend beyond technological complexity. The rapid advancement of AI technologies has created a complex landscape of medical-legal challenges regarding its use in the healthcare sector, spanning from concerns about patient consent and data privacy to liability in the event of AI-induced medical errors (86, 87). Scholars and professionals must work collaboratively to devise sound and comprehensive guidance to navigate the ethical and legal intricacies of integrating AI into our healthcare systems (87). Medical AI devices, by their very nature, will evolve as they learn from newly acquired data, a process that may continue long after receiving approval from regulatory bodies such as the Food and Drug Administration (FDA). This continual learning and adaptation, while a strength in many respects, also presents a challenge in ensuring the devices’ sustained reliability and performance over time. Without vigilant monitoring and a robust framework for ongoing validation, there may be unforeseen shifts in the accuracy or effectiveness of these tools, which could potentially negatively impact patient care. Moreover, there is a lack of high quality prospective randomized controlled trials of AI algorithms. While AI holds immense promise in dermatology, the absence of prospective trials hinders the validation of AI models in real-world clinical situations where there will be a diverse photo quality, image capturing modalities and demographically diverse population (33). For these reasons, an AI model validated in a hospital in Asia might not perform similarly in another hospital in North America. Wu et al. have shown that 126 out of 130 FDA approved medical AI devices were trained on retrospective data at the time of their approval (88). Most of the datasets used are not publicly available, thus preventing regulatory bodies and researchers from auditing their algorithms. Future AI models should undergo multi-site validation on a diverse and representative population in order to assess the generalizability of AI models. Furthermore, establishing trust among AI and various stakeholders will be vital in realizing AI’s full potential in the field. While model accuracy is very important, research has shown that dermatologists and patients value the potential of augmented intelligence in dermatology and also put a high priority on the human physician-patient relationship (89, 90).
In recent months, advanced language models, in the form of chatbots, have gained popularity in medicine (4, 91–93). For dermatology, an extension of these models—Vision-Language Models (VLMs) and multi-modal models—offer immense potential. VLMs are large-scale models adept at associating visual inputs, such as images and videos, with text data (5). Their capabilities span generative tasks (creating new content), retrieval of information, and navigation. Recent studies underscore their impact on dermatology. For instance, Skin-GPT4, a VLM, can provide descriptions and diagnosis from clinical skin lesion photos (94). Further, research by Moor et al. and Tu et al. show the accuracy of VLMs in medical visual question-answering tasks (5, 24). In a related vein, Kim et al.’s study on FMs underscore the capacity of this new class of models to generate accurate skin images annotations (95).
The rapid advancements in this domain have the potential to usher a future of a generalist medical AI (96). These generalist models could be capable of giving approximate diagnoses from clinical photos, generate treatment options, and offer deeper insights into patient data by integrating demographics, visual inspection, and genetic data when applicable. Their potential applications can range from patient chatbots to triage tools (96). Additionally, the inclusion of genetic data could improve the diagnosis of orphan skin conditions. As dermatological datasets expand and computing power increases, FMs are on track to become more accurate and prove utility in dermatology. They could augment the practice of dermatology to provide more precise and holistic care.
Medical data, including skin images, are difficult to access largely due to privacy, legal, and the ethical risks associated with sharing health data. Currently, many dermatological images reside in data silos within healthcare institutions all over the world. Also, medical data is hard to collate, and often requires years of planning with significant costs (97). This is even more pronounced in resource-limited settings, where there is less infrastructure to support collection and sharing of data. Since the DL model’s performance significantly improves with more diverse data (98, 99), new approaches are needed to expand model access to more distributed high-quality datasets.
Federated learning (FL) is a concept that enables DL models to be trained on different datasets without the need to leave their original locations (100). In FL, multiple collaborators can train a model on separate institutional datasets. It is an approach that can enable the preservation of data privacy, and it has already demonstrated similar performance—compared to centralizing the data—in fields like radiology and oncology (100). Although in some cases, there have been drawbacks in which the model sometimes memorizes the data inputs (101). Appropriately implemented, FL has the potential to enable fairer and more generalizable dermatology models by incorporating diverse demographics, thereby capturing the nuances in skin conditions across different societies. This is crucial in dermatology where the popular models significantly perform worse on underrepresented skin types of Fitzpatrick IV–VI (15).
Beyond FL, the concept of FMs introduces the possibility of local models. FMs have the distinct capability to learn from unlabeled data and can be adapted for a variety of downstream tasks without the necessity of specific training (96). This characteristic allows FMs to be fine-tuned with local data, from which they can glean insights and achieve impressive performance across diverse tasks. Given that the fine-tuning procedure is more cost-efficient than full-scale training (27), it amplifies the appeal of FMs within institutional contexts. Consequently, dermatology institutions can harness bespoke models attuned to their unique demographics and guidelines. While promising, progress towards this will require resolving data quality, aggregation, and infrastructural challenges. However, these new techniques could be instrumental in building invaluable dermatology-AI models.
Recent years have witnessed notable advancements in the architectures of AI models, leading to enhanced performance across numerous medical tasks as previously discussed. As the industry attracts more investment and data generation surges, new architectures will likely further improve on existing tasks and expand into new areas. However, with these advancements arises a vital question: how should we holistically evaluate these models? While metrics like accuracy, area under the curve are common, comprehensive model evaluation will need to go beyond mere percentages. Clinical value needs to be demonstrated. As reported by Wornow et al., standard evaluations are lacking for evaluating emerging models (102). In addition, many models fail to be evaluated on fairness and transparency metrics, and in many cases there’s no standard for this evaluation frameworks (103). Holistic model evaluation is likely to emerge in the near future as the desire for clinical integration increases. This could include uncertainty, model interpretability, and subpopulation analysis—which is important for dermatology.
Developing these types of model benchmarks will require collaboration among dermatologists, researchers, and patients. We posit that soon, more robust consensus guidelines are likely to emerge.
The current rapid model evolution underscores the pressing need for robust regulation (104). Such regulatory measures serve a two-fold purpose: Firstly, they shield the dermatology community from prematurely adopting under-tested models by establishing stringent benchmarks. Secondly, they foster trust, ensuring that AI tools resonate with the foundational clinical values practitioners hold dear. For AI models to achieve widespread adoption, especially in dermatology, they must be both reproducible and generalizable (105). As these models seek to bridge the dermatology access gap, especially in resource-limited settings, generalizability becomes even more pivotal. Also, standardizing the data collection process is another important factor towards optimizing model training and thus, performance. As highlighted in the position statement by the American Academy of Dermatology Augmented Intelligence working group, research in the dermatology-AI space needs to be directed towards prospective and randomized clinical trials that rigorously vet models before deployment (29, 106). Also, although most DL models are in the form of “black-boxes” (107), emerging FMs could further obfuscate their inner workings, making the issue of explainability vital. Research addressing explainability will be invaluable for model advancement and deployment.
Dermatology presents both opportunities and challenges to integrate AI into its daily workings. Whilst in its infancy, with many regulatory standards that are specific to the field yet to be developed, current trajectories of innovation and advance showcase the potential of AI is likely to emerge a critical element of the dermatologists workflow, with the need for the clinician to have a global understanding of its workings. Steering this ship towards a future of a transparent, fair, safe, and responsible dermatology-AI will be an interdisciplinary effort that involves the leadership of the dermatology community.
JO: Conceptualization, Project administration, Supervision, Writing – original draft, Writing – review & editing. HG: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. RD: Resources, Supervision, Writing – review & editing. ZC: Supervision, Writing – original draft, Writing – review & editing. VM: Conceptualization, Resources, Supervision, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
RD has served as an advisor to MDAlgorithms and Revea and received consulting fees from Pfizer, L’Oreal, Frazier Healthcare Partners, and DWA, and research funding from UCB.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Dorr, DA , Adams, L , and Embí, P . Harnessing the promise of artificial intelligence responsibly. JAMA. (2023) 329:1347–8. doi: 10.1001/jama.2023.2771
2. Rajpurkar, P , and Lungren, MP . The current and future state of AI interpretation of medical images. N Engl J Med. (2023) 388:1981–90. doi: 10.1056/NEJMra2301725
3. Goh, KH , Wang, L , Yeow, AYK , Poh, H , Li, K , Yeow, JJL, et al. Artificial intelligence in sepsis early prediction and diagnosis using unstructured data in healthcare. Nat Commun. (2021) 12:711. doi: 10.1038/s41467-021-20910-4
4. Lee, P , Bubeck, S , and Petro, J . Benefits, limits, and risks of GPT-4 as an AI Chatbot for medicine. N Engl J Med. (2023) 388:1233–9. doi: 10.1056/NEJMsr2214184
5. Moor, M. , Huang, Q. , Wu, S. , Yasunaga, M. , Zakka, C. , Dalmia, Y., et al. (2023) ‘Med-flamingo: a multimodal medical few-shot learner’, arXiv [cs.CV]. Available at: http://arxiv.org/abs/2307.15189.
6. Esteva, A , Robicquet, A , Ramsundar, B , Kuleshov, V , DePristo, M , Chou, K, et al. A guide to deep learning in healthcare. Nat Med. (2019) 25:24–9. doi: 10.1038/s41591-018-0316-z
7. Bohr, A , and Memarzadeh, K . Chapter 2 – The rise of artificial intelligence in healthcare applications In: A Bohr and K Memarzadeh, editors. Artificial intelligence in healthcare. Cambridge, MA: Academic Press (2020). 25–60.
8. Castiglioni, I , Rundo, L , Codari, M , di Leo, G , Salvatore, C , Interlenghi, M, et al. AI applications to medical images: from machine learning to deep learning. Phys Med. (2021) 83:9–24. doi: 10.1016/j.ejmp.2021.02.006
9. Chen, RJ , Lu, MY , Chen, TY , Williamson, DFK , and Mahmood, F . Synthetic data in machine learning for medicine and healthcare. Nat Biomed Eng. (2021) 5:493–7. doi: 10.1038/s41551-021-00751-8
10. Raghupathi, W , and Raghupathi, V . Big data analytics in healthcare: promise and potential. Health Inf Sci Syst. (2014) 2:3. doi: 10.1186/2047-2501-2-3
11. Feng, H , Berk-Krauss, J , Feng, PW , and Stein, JA . Comparison of dermatologist density between urban and rural counties in the United States. JAMA Dermatol. (2018) 154:1265–71. doi: 10.1001/jamadermatol.2018.3022
12. Kimball, AB , and Resneck, JS Jr. The US dermatology workforce: a specialty remains in shortage. J Am Acad Dermatol. (2008) 59:741–5. doi: 10.1016/j.jaad.2008.06.037
13. Coustasse, A , Sarkar, R , Abodunde, B , Metzger, BJ , and Slater, CM . Use of Teledermatology to improve dermatological access in rural areas. Telemed J E Health. (2019) 25:1022–32. doi: 10.1089/tmj.2018.0130
14. Tsang, MW , and Resneck, JS Jr. Even patients with changing moles face long dermatology appointment wait-times: a study of simulated patient calls to dermatologists. J Am Acad Dermatol. (2006) 55:54–8. doi: 10.1016/j.jaad.2006.04.001
15. Daneshjou, R , Vodrahalli, K , Novoa, RA , Jenkins, M , Liang, W , Rotemberg, V, et al. Disparities in dermatology AI performance on a diverse, curated clinical image set. Sci Adv. (2022) 8:eabq6147. doi: 10.1126/sciadv.abq6147
16. Esteva, A , Kuprel, B , Novoa, RA , Ko, J , Swetter, SM , Blau, HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. (2017) 542:115–8. doi: 10.1038/nature21056
17. Emam, S , Du, AX , Surmanowicz, P , Thomsen, SF , Greiner, R , and Gniadecki, R . Predicting the long-term outcomes of biologics in patients with psoriasis using machine learning. Br J Dermatol. (2020) 182:1305–7. doi: 10.1111/bjd.18741
18. Pierce, EJ , Boytsov, NN , Vasey, JJ , Sudaria, TC , Liu, X , Lavelle, KW, et al. A qualitative analysis of provider notes of atopic dermatitis-related visits using natural language processing methods. Dermatol Ther. (2021) 11:1305–18. doi: 10.1007/s13555-021-00553-5
19. LeCun, Y , Bengio, Y , and Hinton, G . Deep learning. Nature. (2015) 521:436–44. doi: 10.1038/nature14539
20. Gu, J , Wang, Z , Kuen, J , Ma, L , Shahroudy, A , Shuai, B, et al. Recent advances in convolutional neural networks. Pattern Recogn. (2018) 77:354–77. doi: 10.1016/j.patcog.2017.10.013
21. Wolf, T. , Debut, L. , Sanh, V. , Chaumond, J. , Delangue, C. , Moi, A., et al. (2020) ‘Transformers: state-of-the-art natural language processing’, in Proceedings of the 2020 conference on empirical methods in natural language processing: system demonstrations. Online: Association for Computational Linguistics, pp. 38–45.
22. Nadkarni, PM , Ohno-Machado, L , and Chapman, WW . Natural language processing: an introduction. J Am Med Inform Assoc. (2011) 18:544–51. doi: 10.1136/amiajnl-2011-000464
23. Introducing ChatGPT . (no date). Available at: https://openai.com/blog/chatgpt (Accessed November 30, 2022).
24. Tu, T. , Azizi, S. , Driess, D. , Schaekermann, M. , Amin, M. , Chang, P.-C., et al. (2023) ‘Towards generalist biomedical AI’, arXiv [csCL]. Available at: http://arxiv.org/abs/2307.14334.
25. Li, C. , Wong, C. , Zhang, S. , Usuyama, N. , Liu, H. , Yang, J., et al. (2023) ‘LLaVA-med: training a large language-and-vision assistant for biomedicine in one day’, arXiv [cs.CV]. Available at: http://arxiv.org/abs/2306.00890.
26. Zhu, D. , Chen, J. , Shen, X. , Li, X. , and Elhoseiny, M. (2023) ‘MiniGPT-4: enhancing vision-language understanding with advanced large language models’, arXiv [csCV]. Available at: http://arxiv.org/abs/2304.10592.
27. Bommasani, R. , Hudson, D. A. , Adeli, E. , Altman, R. , Arora, S. , von Arx, S., et al. (2021) ‘On the opportunities and risks of foundation models’, arXiv [csLG]. Available at: http://arxiv.org/abs/2108.07258
28. du-Harpur, X , Watt, FM , Luscombe, NM , and Lynch, MD . What is AI? Applications of artificial intelligence to dermatology. Br J Dermatol. (2020) 183:423–30. doi: 10.1111/bjd.18880
29. Gomolin, A , Netchiporouk, E , Gniadecki, R , and Litvinov, IV . Artificial intelligence applications in dermatology: where do we stand? Front Med. (2020) 7:100. doi: 10.3389/fmed.2020.00100
30. Young, AT , Xiong, M , Pfau, J , Keiser, MJ , and Wei, ML . Artificial intelligence in dermatology: a primer. J Invest Dermatol. (2020) 140:1504–12. doi: 10.1016/j.jid.2020.02.026
31. Jones, OT , Matin, RN , van der Schaar, M , Prathivadi Bhayankaram, K , Ranmuthu, CKI , Islam, MS, et al. Artificial intelligence and machine learning algorithms for early detection of skin cancer in community and primary care settings: a systematic review. Lancet Digit Health. (2022) 4:e466–76. doi: 10.1016/S2589-7500(22)00023-1
32. Marchetti, MA , Codella, NCF , Dusza, SW , Gutman, DA , Helba, B , Kalloo, A, et al. Results of the 2016 international skin imaging collaboration international symposium on biomedical imaging challenge: comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J Am Acad Dermatol. (2018) 78:270–277.e1. doi: 10.1016/j.jaad.2017.08.016
33. Marchetti, MA , Cowen, EA , Kurtansky, NR , Weber, J , Dauscher, M , DeFazio, J, et al. Prospective validation of dermoscopy-based open-source artificial intelligence for melanoma diagnosis (PROVE-AI study). NPJ Digit Med. (2023) 6:127. doi: 10.1038/s41746-023-00872-1
34. Han, SS , Kim, MS , Lim, W , Park, GH , Park, I , and Chang, SE . Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J Invest Dermatol. (2018) 138:1529–38. doi: 10.1016/j.jid.2018.01.028
35. Gutman, D. , Codella, N. C. F. , Celebi, E. , Helba, B. , Marchetti, M. , Mishra, N., et al. (2016) ‘Skin lesion analysis toward melanoma detection: a challenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC)’, arXiv [cs.CV]. Available at: http://arxiv.org/abs/1605.01397.
36. Jansen, P , Baguer, DO , Duschner, N , Arrastia, JL’C , Schmidt, M , Landsberg, J, et al. Deep learning detection of melanoma metastases in lymph nodes. Eur J Cancer. (2023) 188:161–70. doi: 10.1016/j.ejca.2023.04.023
37. Lu, J , Kazmierczak, E , Manton, JH , and Sinclair, R . Automatic segmentation of scaling in 2-D psoriasis skin images. IEEE Trans Med Imaging. (2013) 32:719–30. doi: 10.1109/TMI.2012.2236349
38. Mohan, S , and Kasthuri, N . Automatic segmentation of psoriasis skin images using adaptive chimp optimization algorithm-based CNN. J Digit Imaging. (2023) 36:1123–36. doi: 10.1007/s10278-022-00765-x
39. Shrivastava, VK , Londhe, ND , Sonawane, RS , and Suri, JS . Computer-aided diagnosis of psoriasis skin images with HOS, texture and color features: a first comparative study of its kind. Comput Methods Prog Biomed. (2016) 126:98–109. doi: 10.1016/j.cmpb.2015.11.013
40. Shrivastava, VK , Londhe, ND , Sonawane, RS , and Suri, JS . A novel and robust Bayesian approach for segmentation of psoriasis lesions and its risk stratification. Comput Methods Prog Biomed. (2017) 150:9–22. doi: 10.1016/j.cmpb.2017.07.011
41. Keser, G , Bayrakdar, İŞ , Pekiner, FN , Çelik, Ö , and Orhan, K . A deep learning algorithm for classification of oral lichen planus lesions from photographic images: a retrospective study. J Stomatol Oral Maxillofac Surg. (2023) 124:101264. doi: 10.1016/j.jormas.2022.08.007
42. Yu, Z , Kaizhi, S , Jianwen, H , Guanyu, Y , and Yonggang, W . A deep learning-based approach toward differentiating scalp psoriasis and seborrheic dermatitis from dermoscopic images. Front Med. (2022) 9:965423. doi: 10.3389/fmed.2022.965423
43. Lee, LT-J , Yang, HC , Nguyen, PA , Muhtar, MS , and Li, YCJ . Machine learning approaches for predicting psoriatic arthritis risk using electronic medical records: population-based study. J Med Internet Res. (2023) 25:e39972. doi: 10.2196/39972
44. du, AX , Ali, Z , Ajgeiy, KK , Dalager, MG , Dam, TN , Egeberg, A, et al. Machine learning model for predicting outcomes of biologic therapy in psoriasis. J Am Acad Dermatol. (2023) 88:1364–7. doi: 10.1016/j.jaad.2022.12.046
45. Zhan, Y-P , and Chen, B-S . Drug target identification and drug repurposing in psoriasis through systems biology approach, DNN-based DTI model and genome-wide microarray data. Int J Mol Sci. (2023) 24:10033. doi: 10.3390/ijms241210033
46. Zhou, Y , Wang, Z , Han, L , Yu, Y , Guan, N , Fang, R, et al. Machine learning-based screening for biomarkers of psoriasis and immune cell infiltration. Eur J Dermatol. (2023) 33:147–56. doi: 10.1684/ejd.2023.4453
47. Guo, P , Luo, Y , Mai, G , Zhang, M , Wang, G , Zhao, M, et al. Gene expression profile based classification models of psoriasis. Genomics. (2014) 103:48–55. doi: 10.1016/j.ygeno.2013.11.001
48. McMullen, EP , Syed, SA , Espiritu, KD , Grewal, RS , Elder, GA , Morita, PP, et al. The therapeutic applications of machine learning in atopic dermatitis: a scoping review. J Cutan Med Surg. (2023) 27:286–7. doi: 10.1177/12034754231168846
49. Rasheed, A , Umar, AI , Shirazi, SH , Khan, Z , Nawaz, S , and Shahzad, M . Automatic eczema classification in clinical images based on hybrid deep neural network. Comput Biol Med. (2022) 147:105807. doi: 10.1016/j.compbiomed.2022.105807
50. Gustafson, E , Pacheco, J , Wehbe, F , Silverberg, J , and Thompson, W . A machine learning algorithm for identifying atopic dermatitis in adults from electronic health records. IEEE Int Conf Healthc Inform. (2017) 2017:83–90. doi: 10.1109/ICHI.2017.31
51. Hurault, G , Domínguez-Hüttinger, E , Langan, SM , Williams, HC , and Tanaka, RJ . Personalized prediction of daily eczema severity scores using a mechanistic machine learning model. Clin Exp Allergy. (2020) 50:1258–66. doi: 10.1111/cea.13717
52. Wilm, A , Kühnl, J , and Kirchmair, J . Computational approaches for skin sensitization prediction. Crit Rev Toxicol. (2018) 48:738–60. doi: 10.1080/10408444.2018.1528207
53. Liu, S , Fan, Y , Duan, M , Wang, Y , Su, G , Ren, Y, et al. AcneGrader: an ensemble pruning of the deep learning base models to grade acne. Skin Res Technol. (2022) 28:677–88. doi: 10.1111/srt.13166
54. Liu, S , Chen, R , Gu, Y , Yu, Q , Su, G , Ren, Y, et al. AcneTyper: an automatic diagnosis method of dermoscopic acne image via self-ensemble and stacking. Technol Health Care. (2023) 31:1171–87. doi: 10.3233/THC-220295
55. Zhang, H , and Ma, T . Acne detection by ensemble neural networks. Sensors. (2022) 22:6828. doi: 10.3390/s22186828
56. Seité, S , Khammari, A , Benzaquen, M , Moyal, D , and Dréno, B . Development and accuracy of an artificial intelligence algorithm for acne grading from smartphone photographs. Exp Dermatol. (2019) 28:1252–7. doi: 10.1111/exd.14022
57. Hernández Montilla, I , Medela, A , Mac Carthy, T , Aguilar, A , Gómez Tejerina, P , Vilas Sueiro, A, et al. Automatic International Hidradenitis Suppurativa Severity Score System (AIHS4): a novel tool to assess the severity of hidradenitis suppurativa using artificial intelligence. Skin Res Technol. (2023) 29:e13357. doi: 10.1111/srt.13357
58. Liu, TJ , Wang, H , Christian, M , Chang, CW , Lai, F , and Tai, HC . Automatic segmentation and measurement of pressure injuries using deep learning models and a LiDAR camera. Sci Rep. (2023) 13:680. doi: 10.1038/s41598-022-26812-9
59. Manohar Dhane, D , Maity, M , Mungle, T , Bar, C , Achar, A , Kolekar, M, et al. Fuzzy spectral clustering for automated delineation of chronic wound region using digital images. Comput Biol Med. (2017) 89:551–60. doi: 10.1016/j.compbiomed.2017.04.004
60. Mukherjee, R , Manohar, DD , Das, DK , Achar, A , Mitra, A , and Chakraborty, C . Automated tissue classification framework for reproducible chronic wound assessment. Bio Med Res Int. (2014) 2014:851582. doi: 10.1155/2014/851582
61. Wang, L , Pedersen, PC , Agu, E , Strong, DM , and Tulu, B . Area determination of diabetic foot ulcer images using a cascaded two-stage SVM-based classification. IEEE Trans Biomed Eng. (2017) 64:2098–109. doi: 10.1109/TBME.2016.2632522
62. Toffaha, KM , Simsekler, MCE , and Omar, MA . Leveraging artificial intelligence and decision support systems in hospital-acquired pressure injuries prediction: a comprehensive review. Artif Intell Med. (2023) 141:102560. doi: 10.1016/j.artmed.2023.102560
63. Dweekat, OY , Lam, SS , and McGrath, L . A hybrid system of Braden scale and machine learning to predict hospital-acquired pressure injuries (bedsores): a retrospective observational cohort study. Diagnostics (Basel, Switzerland). (2022) 13:31. doi: 10.3390/diagnostics13010031
64. Dweekat, OY , Lam, SS , and McGrath, L . An integrated system of multifaceted machine learning models to predict if and when hospital-acquired pressure injuries (bedsores) occur. Int J Environ Res Public Health. (2023) 20:828. doi: 10.3390/ijerph20010828
65. Stern, L , and Roshan Fekr, A . In-bed posture classification using deep neural network. Sensors. (2023) 23:2430. doi: 10.3390/s23052430
66. Gorman, BG , Lifson, MA , and Vidal, NY . Artificial intelligence and frozen section histopathology: a systematic review. J Cutan Pathol. (2023) 50:852–9. doi: 10.1111/cup.14481
67. Sauter, D , Lodde, G , Nensa, F , Schadendorf, D , Livingstone, E , and Kukuk, M . Deep learning in computational dermatopathology of melanoma: a technical systematic literature review. Comput Biol Med. (2023) 163:107083. doi: 10.1016/j.compbiomed.2023.107083
68. van Zon, MCM , van der Waa, JD , Veta, M , and Krekels, GAM . Whole-slide margin control through deep learning in Mohs micrographic surgery for basal cell carcinoma. Exp Dermatol. (2021) 30:733–8. doi: 10.1111/exd.14306
69. Hekler, A , Utikal, JS , Enk, AH , Berking, C , Klode, J , Schadendorf, D, et al. Pathologist-level classification of histopathological melanoma images with deep neural networks. Eur J Cancer. (2019) 115:79–83. doi: 10.1016/j.ejca.2019.04.021
70. Hocke, J , Krauth, J , Krause, C , Gerlach, S , Warnemünde, N , Affeldt, K, et al. Computer-aided classification of indirect immunofluorescence patterns on esophagus and split skin for the detection of autoimmune dermatoses. Front Immunol. (2023) 14:1111172. doi: 10.3389/fimmu.2023.1111172
71. Liu, Y , Jain, A , Eng, C , Way, DH , Lee, K , Bui, P, et al. A deep learning system for differential diagnosis of skin diseases. Nat Med. (2020) 26:900–8. doi: 10.1038/s41591-020-0842-3
72. Sitaru, S , Oueslati, T , Schielein, MC , Weis, J , Kaczmarczyk, R , Rueckert, D, et al. Automatic body part identification in real-world clinical dermatological images using machine learning. J Dtsch Dermatol Ges. (2023) 21:863–9. doi: 10.1111/ddg.15113
73. Okon, E , Rachakonda, V , Hong, HJ , Callison-Burch, C , and Lipoff, JB . Natural language processing of Reddit data to evaluate dermatology patient experiences and therapeutics. J Am Acad Dermatol. (2020) 83:803–8. doi: 10.1016/j.jaad.2019.07.014
74. Young, JN , Ross O'Hagan,, Poplausky, D , Levoska, MA , Gulati, N , Ungar, B, et al. The utility of ChatGPT in generating patient-facing and clinical responses for melanoma. J Am Acad Dermatol. (2023) 89:602–4. doi: 10.1016/j.jaad.2023.05.024
75. Mondal, H , Mondal, S , and Podder, I . Using ChatGPT for writing articles for patients’ education for dermatological diseases: a pilot study. Indian Dermatol Online J. (2023) 14:482–6. doi: 10.4103/idoj.idoj_72_23
76. O’Hern, K , Yang, E , and Vidal, NY . ChatGPT underperforms in triaging appropriate use of Mohs surgery for cutaneous neoplasms. JAAD Int. (2023) 12:168–70. doi: 10.1016/j.jdin.2023.06.002
77. Jain, A , Way, D , Gupta, V , Gao, Y , de Oliveira Marinho, G , Hartford, J, et al. Development and assessment of an artificial intelligence-based tool for skin condition diagnosis by primary care physicians and nurse practitioners in teledermatology practices. JAMA Netw Open. (2021) 4:e217249. doi: 10.1001/jamanetworkopen.2021.7249
78. Liu, X , Faes, L , Kale, AU , Wagner, SK , Fu, DJ , Bruynseels, A, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health. (2019) 1:e271–97. doi: 10.1016/S2589-7500(19)30123-2
79. Haenssle, HA , Fink, C , Schneiderbauer, R , Toberer, F , Buhl, T , Blum, A, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol. (2018) 29:1836–42. doi: 10.1093/annonc/mdy166
80. Han, SS , Kim, YJ , Moon, IJ , Jung, JM , Lee, MY , Lee, WJ, et al. Evaluation of artificial intelligence-assisted diagnosis of skin neoplasms: a single-center, paralleled, unmasked, randomized controlled trial. J Invest Dermatol. (2022) 142:2353–2362.e2. doi: 10.1016/j.jid.2022.02.003
81. Winkler, JK , Fink, C , Toberer, F , Enk, A , Deinlein, T , Hofmann-Wellenhof, R, et al. Association between surgical skin markings in dermoscopic images and diagnostic performance of a deep learning convolutional neural network for melanoma recognition. JAMA Dermatol. (2019) 155:1135–41. doi: 10.1001/jamadermatol.2019.1735
82. Vodrahalli, K , Daneshjou, R , Novoa, RA , Chiou, A , Ko, JM , and Zou, J . TrueImage: a machine learning algorithm to improve the quality of telehealth photos. Pac Symp Biocomput. (2020) 26:220–31. doi: 10.1142/9789811232701_0021
83. Maier, K , Zaniolo, L , and Marques, O . Image quality issues in teledermatology: a comparative analysis of artificial intelligence solutions. J Am Acad Dermatol. (2022) 87:240–2. doi: 10.1016/j.jaad.2021.07.073
84. Young, AT , Fernandez, K , Pfau, J , Reddy, R , Cao, NA , von Franque, MY, et al. Stress testing reveals gaps in clinic readiness of image-based diagnostic artificial intelligence models. NPJ Digit Med. (2021) 4:10. doi: 10.1038/s41746-020-00380-6
85. Caffery, LJ , Rotemberg, V , Weber, J , Soyer, HP , Malvehy, J , and Clunie, D . The role of DICOM in artificial intelligence for skin disease. Front Med. (2020) 7:619787. doi: 10.3389/fmed.2020.619787
86. Jones, C , Thornton, J , and Wyatt, JC . Artificial intelligence and clinical decision support: clinicians’ perspectives on trust, trustworthiness, and liability. Med Law Rev. (2023):fwad013. doi: 10.1093/medlaw/fwad013
87. Naik, N , Hameed, BMZ , Shetty, DK , Swain, D , Shah, M , Paul, R, et al. Legal and ethical consideration in artificial intelligence in healthcare: who takes responsibility? Front Surg. (2022) 9:862322. doi: 10.3389/fsurg.2022.862322
88. Wu, E , Wu, K , Daneshjou, R , Ouyang, D , Ho, DE , and Zou, J . How medical AI devices are evaluated: limitations and recommendations from an analysis of FDA approvals. Nat Med. (2021) 27:582–4. doi: 10.1038/s41591-021-01312-x
89. Nelson, CA , Pérez-Chada, LM , Creadore, A , Li, SJ , Lo, K , Manjaly, P, et al. Patient perspectives on the use of artificial intelligence for skin cancer screening: a qualitative study. JAMA Dermatol. (2020) 156:501–12. doi: 10.1001/jamadermatol.2019.5014
90. Nelson, CA , Pachauri, S , Balk, R , Miller, J , Theunis, R , Ko, JM, et al. Dermatologists ‘perspectives on artificial intelligence and augmented intelligence – a cross-sectional survey. JAMA Dermatol. (2021) 157:871–4. doi: 10.1001/jamadermatol.2021.1685
91. Haug, CJ , and Drazen, JM . Artificial intelligence and machine learning in clinical medicine, 2023. N Engl J Med. (2023) 388:1201–8. doi: 10.1056/NEJMra2302038
92. Kassab, J , el Dahdah, J , Chedid el Helou, M , Layoun, H , Sarraju, A , Laffin, LJ, et al. Assessing the accuracy of an online chat-based artificial intelligence model in providing recommendations on hypertension management in accordance with the 2017 American College of Cardiology/American Heart Association and 2018 European Society of Cardiology/European Society of Hypertension Guidelines. Hypertension. (2023) 80:e125–7. doi: 10.1161/HYPERTENSIONAHA.123.21183
93. Li, H , Moon, JT , Purkayastha, S , Celi, LA , Trivedi, H , and Gichoya, JW . Ethics of large language models in medicine and medical research. Lancet Digit Health. (2023) 5:e333–5. doi: 10.1016/S2589-7500(23)00083-3
94. Zhou, J. , He, X. , Sun, L. , Xu, J. , Chen, X. , Chu, Y., et al. (2023) ‘SkinGPT-4: an interactive dermatology diagnostic system with visual large language model’, arXiv [eessIV]. Available at: http://arxiv.org/abs/2304.10691.
95. Kim, C. , Gadgil, S.U. , DeGrave, A. , Cai, Z.R. , Daneshjou, R. , and Lee, S.I. (2023) ‘Fostering transparent medical image AI via an image-text foundation model grounded in medical literature’, medRxiv [Preprint]. doi: 10.1101/2023.06.07.23291119
96. Moor, M , Banerjee, O , Abad, ZSH , Krumholz, HM , Leskovec, J , Topol, EJ, et al. Foundation models for generalist medical artificial intelligence. Nature. (2023) 616:259–65. doi: 10.1038/s41586-023-05881-4
97. van Panhuis, WG , Paul, P , Emerson, C , Grefenstette, J , Wilder, R , Herbst, AJ, et al. A systematic review of barriers to data sharing in public health. BMC Public Health. (2014) 14:1144. doi: 10.1186/1471-2458-14-1144
98. Hosny, A , Parmar, C , Quackenbush, J , Schwartz, LH , and Aerts, HJWL . Artificial intelligence in radiology. Nat Rev Cancer. (2018) 18:500–10. doi: 10.1038/s41568-018-0016-5
99. Wang, F , Casalino, LP , and Khullar, D . Deep learning in medicine-promise, progress, and challenges. JAMA Intern Med. (2019) 179:293–4. doi: 10.1001/jamainternmed.2018.7117
100. Rieke, N , Hancox, J , Li, W , Milletarì, F , Roth, HR , Albarqouni, S, et al. The future of digital health with federated learning. NPJ Digit Med. (2020) 3:119. doi: 10.1038/s41746-020-00323-1
101. Carlini, N. , Liu, C. , Erlingsson, Ú. , Kos, J. , and Song, D. (2019) ‘The secret sharer: evaluating and testing unintended memorization in neural networks’, in 28th USENIX security symposium (USENIX security 19), pp. 267–284.
102. Wornow, M , Xu, Y , Thapa, R , Patel, B , Steinberg, E , Fleming, S, et al. The shaky foundations of large language models and foundation models for electronic health records. NPJ Digit Med. (2023) 6:135. doi: 10.1038/s41746-023-00879-8
104. Meskó, B , and Topol, EJ . The imperative for regulatory oversight of large language models (or generative AI) in healthcare. NPJ Digit Med. (2023) 6:120. doi: 10.1038/s41746-023-00873-0
105. Daneshjou, R , Smith, MP , Sun, MD , Rotemberg, V , and Zou, J . Lack of transparency and potential bias in artificial intelligence data sets and algorithms: a scoping review. JAMA Dermatol. (2021) 157:1362–9. doi: 10.1001/jamadermatol.2021.3129
106. Kovarik, C , Lee, I , Ko, J , Adamson, A , Otley, C , Kvedar, J, et al. Commentary: position statement on augmented intelligence (AuI). J Am Acad Dermatol. (2019) 81:998–1000. doi: 10.1016/j.jaad.2019.06.032
Keywords: dermatology, artificial intelligence (AI), large language models (LLM), machine learning, melanoma, federated learning
Citation: Omiye JA, Gui H, Daneshjou R, Cai ZR and Muralidharan V (2023) Principles, applications, and future of artificial intelligence in dermatology. Front. Med. 10:1278232. doi: 10.3389/fmed.2023.1278232
Received: 16 August 2023; Accepted: 27 September 2023;
Published: 12 October 2023.
Edited by:
Mara Giavina-Bianchi, Albert Einstein Israelite Hospital, BrazilReviewed by:
Federica Veronese, Azienda Ospedaliero Universitaria Maggiore della Carità, ItalyCopyright © 2023 Omiye, Gui, Daneshjou, Cai and Muralidharan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jesutofunmi A. Omiye, dG9taXllQHN0YW5mb3JkLmVkdQ==
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.