
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 28 August 2023
Sec. Dermatology
Volume 10 - 2023 | https://doi.org/10.3389/fmed.2023.1207993
 Priyanka Madaan1
Priyanka Madaan1 Uttam Sharma2
Uttam Sharma2 Nipanshi Tyagi3
Nipanshi Tyagi3 Balvinder Kaur Brar4
Balvinder Kaur Brar4 Shivani Bansal5
Shivani Bansal5 Hemant Rituraj Kushwaha3
Hemant Rituraj Kushwaha3 Harmanpreet Singh Kapoor6
Harmanpreet Singh Kapoor6 Aklank Jain2
Aklank Jain2 Manju Jain1*
Manju Jain1*Psoriasis is a chronic inflammatory skin disease with keratinocyte hyperproliferation and T cells as key mediators of lesional and systemic inflammatory changes. To date, no suitable differential biomarkers are available for the disease diagnosis. More recently, microRNAs have been identified as critical regulators of lesional and systemic immune changes in psoriasis with diagnostic potential. We have performed expression profiling of T cell-specific miRNAs in 38 plasma samples from psoriasis vulgaris patients and an equal number of age- and gender-matched healthy subjects. Our findings have identified a panel of five blood-based circulatory miRNAs with a significant change in their expression levels, comprising miR-215, miR-148a, miR-125b-5p, miR-223, and miR-142-3p, which can differentiate psoriasis vulgaris patients from healthy individuals. The receiver operating characteristic (ROC) curves for all five miRNAs individually and in combination exhibited a significant disease discriminatory area under the curve with an AUC of 0.762 and a p < 0.0001 for all the miRNAs together. Statistically, all five miRNAs in combination depicted the best-fit model in relation to disease severity (PASI) compared with individual miRNAs, with the highest R2 value of 0.94 and the lowest AIC score of 131.8. Each of the miRNAs also exhibited a significant association with at least one of the other miRNAs in the panel. Importantly, the five miRNAs in the panel regulate one or more immune-inflammation pathways based on target prediction, pathway network analysis, and validated roles in the literature. The miRNA panel provides a rationalized combination of biomarkers that can be tested further on an expanded cohort of patients for their diagnostic value.
Psoriasis is a chronic inflammatory skin disease with itchy and painful cutaneous manifestations that affects nearly 2% of the world's population (1, 2). Psoriatic lesions are characterized by abnormal differentiation and hyperproliferation of keratinocytes with immune cell infiltration. Disease etiology involves a complex interplay between genetic and environmental factors that disturbs the homeostatic cross-talk between the skin keratinocytes and different immune cells normally observed in healthy skin. Despite substantial studies on the initiation and progression of the disease, the exact molecular mechanisms that dysregulate the complex interactions among the lesional cells to create a chronic inflammatory environment are not fully known. Also, the significance of systemic immune changes associated with the disease is not well understood (1). Immunologically, new insights into the pathogenesis of psoriasis imply a major role of T cells in the initiation and maintenance of the inflammatory state, which can potentially lead to keratinocyte-specific changes observed in the disease lesions (3, 4). Generally, the disease is diagnosed by clinical evaluation of skin lesions by expert dermatologists, with occasional histopathological examination (5). With no reliable molecular biomarkers as criteria for clinical diagnostics to date, there is an urgent requirement for differential biomarkers with diagnostic potential and predictive value.
Psoriasis is the result of several genetic–epigenetic, environmental, and immunological factors, of which miRNAs have recently emerged as critical regulators of disease pathogenesis (6–11). miRNAs are small (~20–25 bp) non-coding RNAs that control gene expression in a cell-specific manner with wide functional implications in processes such as development, growth, apoptosis, plasticity, activation, survival, proliferation, and differentiation (12, 13). In this context, miRNAs that regulate keratinocyte and heterogeneous T cell populations become significant in the pathophysiology of psoriasis (12–16). Although produced within cells, miRNAs with altered cellular expression can potentially mirror dysregulated stable cell-free molecules in peripheral circulation under disease conditions (17, 18). Thus, a disease-specific circulatory miRNA pool makes it amenable to developing minimally invasive blood-based diagnostic biomarkers in relation to disease prediction and severity. Several studies on miRNA expression analysis have been carried out across various sample types, viz., blood, PBMCs, hair, and lesional skin, for the potential diagnosis of psoriasis (19–22). Explorations of differentially expressed systemic miRNAs in sera/plasma samples with select miRNA candidates have been carried out in a few studies based on human miRNA arrays and small RNA sequencing analysis (19, 20, 22–27). Heterogeneous findings on miRNA candidates with respect to disease-specific differential expression, low abundance, a non-significant correlation with disease severity, a lack of knowledge on the role of altered miRNAs in disease progression, a small sample size used, and variation in blood components used as samples in these studies are limiting the transition to their clinical use.
In the present study, a set of immunologically relevant, T cell-specific miRNA candidates has been selected to access their differential expression in the plasma of psoriatic patients (28–35). With the potential to drive inflammatory disease processes, we have deciphered a combination of five significant miRNAs that can provide a robust diagnostic panel in light of their functional implication in disease etiology and progression.
A total of 40 psoriasis vulgaris patients clinically confirmed by dermatologists along with 40 gender-matched healthy subjects were enrolled in the study. The patients with psoriasis were recruited from the outpatient dermatology clinic at the Guru Gobind Singh Medical College and Hospital, Faridkot, Punjab, India, and the All India Institute of Medical Sciences, Bathinda, Punjab, India. Patients aged 18 years or older, those with a clinical diagnosis of mild-to-severe psoriasis vulgaris (PV), those who have not received topical therapy for at least 2 weeks, and those who have been receiving systemic immunosuppressive treatment or phototherapy for at least 1 month were included, while psoriasis patients with other infections or co-morbidities were not enrolled in the study. Disease severity was graded by the psoriasis area and severity index (PASI) score and body surface area (BSA) into mild (PASI score ≤ 5/BSA < 3%), moderate (PASI score 5–10/BSA 3–10%), and severe (PASI score >10/BSA> 10%) (36); 5-mm punch biopsies were taken from the lesional skin of 16 patients. Non-lesional biopsies from the same 16 patients from the uninvolved skin away from the lesion were also collected for comparative analysis. Healthy age- and gender-matched controls were recruited from the staff and graduate students at the Central University of Punjab, Bathinda, Punjab, India. Written informed consent, along with a subject information form, was obtained from all the participants. The present study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committees of Guru Gobind Singh Medical College and Hospital, Faridkot, Punjab, India (GGS/IEC/19/57), AIIMS, Bathinda, India (IEC/AIIMS/BTI/150), and Central University of Punjab, Bathinda, Punjab, India (CUPB/IEC/2016/045).
Of the 40 patients enrolled in the study, lesional and non-lesional tissues from 16 psoriasis patients were processed for disease-specific histopathological examination, and 5-mm lesional and non-lesional punch biopsies were collected and fixed in 10% neutral buffered formaldehyde (NBF). Paraffin-embedded tissue was processed into 5 μm sections and stained with hematoxylin and eosin (H&E) for histopathological analysis. Whole blood samples from 40 psoriasis patients and 40 healthy controls were collected in EDTA-coated vials. The blood samples were centrifuged at 1,800 rpm for 10 min at RT to collect the straw-colored platelet-rich plasma. The plasma was centrifuged again at 2,000 rpm for 10 min at 4°C to remove the remaining cellular components. The purified plasma samples were stored in aliquots at −80°C until further assays and 200 μl of each plasma sample was used for miRNA extraction.
miRNA extraction from plasma samples was performed using a miRNeasy serum/plasma kit (Catalog No. 217184: Qiagen, Inc., USA) according to the manufacturer's instructions. The concentration and purity of miRNA were determined with a NanoDrop UV-Vis Spectrophotometer 2000cc (Thermo Fisher Scientific, CA, USA), and 100 ng of total isolated miRNA was used for cDNA synthesis, using the miScript II RT kit following the manufacturer's protocol (Catalog no. 218161: Qiagen, Inc., USA). A set of 15 immunologically relevant miRNAs with respective sequences enlisted in Supplementary Table S1 were quantified in plasma samples of psoriasis patients compared with a healthy cohort by real-time PCR using the Qiagen miScript SYBR Green PCR Kit and miR-specific primers (Catalog no. 218073: Qiagen, Inc., USA, Cat. no. MS00004900; Qiagen, Inc., QuantStudio 3 Applied Biosystems real-time PCR). Cel-miR-39 was used as an internal control. qRT-PCR comprised 6.25 μl of SYBR Green Master Mix, 1.25 μl each of miR-specific primer and universal primer, and 3.5 μl of nuclease-free water with the required amount of cDNA. The reaction conditions were as follows: initial activation was performed at 95°C for 15 min, denaturation at 94°C for 15 s, annealing at 55°C for 30 s, and extension at 70°C for 30 s for 40 cycles with melting curve analysis for quality check. A non-template control reaction for each primer was also carried out to check for contamination by including all components except the cDNA template. The threshold cycle (Ct) value for each sample with primers was recorded. All samples were tested at least three times. The relative expression between psoriasis patients and healthy controls was estimated by the 2−ΔCt method (37, 38). Briefly, the miRNA-specific Ct values for each of the psoriasis and control samples were normalized using the respective Ct values for Cel-miR-39 as a spike-in control. The average of normalized Ct values for the psoriasis group and control group was further used for the calculation of fold changes for each miRNA tested.
Target genes for five miRNAs of interest, viz., miR-215, miR-142-3p, miR-223, miR-148a, and miR-125b-5p, were predicted using four different miRNA-target prediction tools: miRDB (http://www.mirdb.org/), TargetScan (https://www.targetscan.org/vert_80/), DIANA-microT (https://dianalab.e-ce.uth.gr/html/dianauniverse/index.php?r=microT_CDS), and Mirabel (http://bioinfo.univ-rouen.fr/mirabel/). For each miRNA, common targets were deciphered based on the four databases. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were enriched for each miRNA using Starbase (https://starbase.sysu.edu.cn/). The enriched pathways were analyzed for the signaling components that overlapped with the common set of targets for each miRNA retrieved from the four databases. Significant pathways involving miRNA-specific targets were reviewed for potential roles in T cells and/or keratinocyte-associated biological functions and were mapped for interaction networks using Cytoscape.
The non-parametric Mann–Whitney U-test was performed to compare the qRT-PCR-based miRNA expression differences in the psoriasis patient group and the age- and gender-matched healthy control group. Furthermore, 2−ΔCt values were used to generate receiver operating characteristic (ROC) curves with a calculation of the area under the curve (AUC) as an index for evaluating the diagnostic potential of miRNAs of interest to differentiate psoriasis patients from healthy controls (39). A p ≤ 0.05 was considered statistically significant for qRT-PCR data and ROC analysis. All statistical calculations were performed using GraphPad Prism 8.0.2 and R software 4.2.2. Pearson correlation coefficients were derived to study the pairwise relationship between the expression levels of each of the miRNAs among study subjects (psoriasis patients and control subjects). A p ≤ 0.05 was considered statistically significant. A non-linear model b-spline was used to study the individual and the joint impact of miRNA levels in relation to the PASI score.
A total of 40 patients clinically confirmed for psoriasis vulgaris and 40 healthy subjects enrolled in the study were grouped into age- and gender-matched study cohorts. Patients including men and women comprised clinically identifiable individuals with typical dry, raised, erythematous psoriatic plaques exhibiting silvery scales and were graded as mild, moderate, and severe based on disease severity, with the PASI score ranging from 3.5 to 15.6 and the BSA from 2% to 58% (Figure 1A, Table 1); 5 mild, 18 moderate, and 17 severe psoriasis patients with no ongoing treatment at the time of diagnosis and sample collection were enrolled in experimental studies. Histopathological analysis of lesional vs. non-lesional tissue was performed on biopsies from 16 patients. Hematoxylin and eosin-stained skin lesional sections exhibited psoriasis-specific dermal and epidermal changes to variable extents (Figure 1B, Table 1). All 16 patients exhibited thickening of the epidermis designated as acanthosis, with severe manifestations leading to elongation of rete ridges in 12 of the 16 patients. Hyperkeratosis with thickened stratum corneum, parakeratosis with abnormal retention of nuclei in the stratum corneum layer, and dermal leukocyte infiltrates were apparent in 13 specimens with hypogranulosis in 12 patients. An exploration of the systemic expression profile of miRNAs of interest was further carried out in the study cohorts.

Figure 1. (A) Representative images of psoriasis patients showing well-demarcated scaly, erythematous lesions. (B) Representation of histopathologic features as observed in hematoxylin- and eosin-stained sections of lesional and non-lesional tissue from psoriasis vulgaris patients. a. Hyperkeratosis and acanthosis (black arrow), original magnification x10. b. Extension of rete ridges (black arrow) and leukocytic dermal infiltration (dashed circle with white arrow) in lesional tissue as compared to the non-lesional tissue, original magnification x10 c. Reduced granular layer (hypogranulosis) in lesional tissue, original magnification ×20.
A set of 15 immunologically relevant miRNA candidates were explored for qRT-based differential expression in the plasma samples of 12 psoriasis patients and 12 age- and gender-matched healthy controls (Supplementary Table 1). In total, five miRNAs, viz. miR-215, miR-148a, miR-125b, miR-223, and miR-142-3p, exhibited an expression pattern with consistent and significant changes. miR-215 was significantly decreased (fold change = −2.24, p = 0.043), while four miRNAs miR-148a, miR-125b, miR-223, and miR-142-3p were significantly upregulated (fold change = 1.82, 1.84, 2.42, and 2.56 and p = 0.032, 0.034, 0.028, 0.045, respectively) in psoriasis patients compared with healthy controls. miR-146a and miR-21 showed a trend toward upregulation with no significant change, while no change was observed in miR-155 expression levels. The remaining miRNA candidates, miR-590-5p, miR-15b, miR-568, miR-150, miR-23b, miR-27b, and miR-184, were associated with high sample-to-sample variation in terms of threshold detection or low abundance. The five miRNAs with significant dysregulation in expression levels were further validated in an extended cohort of psoriasis patients and healthy study subjects. In line with initial results, cumulative study cohorts with 38 psoriasis patients versus 38 healthy subjects exhibited significant downregulation in miR-215 expression (fold change = −3.34, p < 0.001) and significant upregulation in miR-148a (fold change = 2.43, p < 0.05), miR-125b (fold change = 1.75, p < 0.05), miR-223 (fold change = 2.79, p < 0.001), and miR-142-3p (fold change = 2.27, p < 0.01), as shown in Figure 2A and Table 2. The five miRNAs of significance were tested for their diagnostic potential to differentiate psoriasis vulgaris patients from healthy individuals.
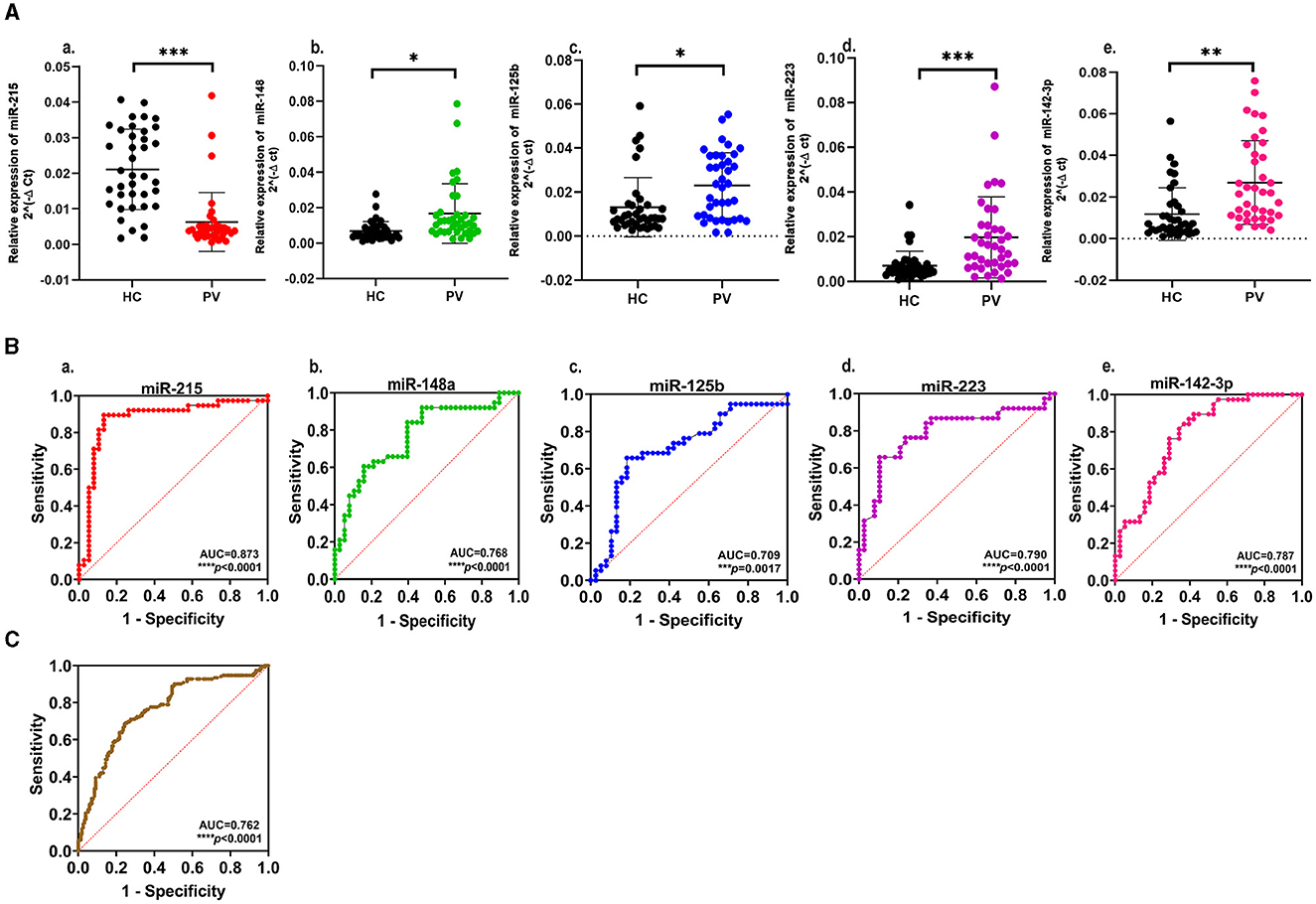
Figure 2. (A) Expression profiling of select miRNAs in plasma samples from psoriasis vulgaris patients (PV) in comparison to healthy controls (HC): a. miR-215; b. miR-148a; c. miR-125b; d. miR-223; e. miR-142-3p. The scatter plot represents normalized 2−ΔCt values of individual samples. Black dots represent healthy controls, and PV patients are represented by red dots for miR-215, green dots for miR-148a, blue dots for miR-125b, purple dots for miR-223, and pink dots for miR-142-3p. Data are expressed as mean ± SD. ***p < 0.001, **p < 0.01, and *p < 0.05 calculated using the Mann–Whitney U-test. Data represent three independent qRT-PCR experiments. (B) Receiver operating characteristic (ROC) curve with area under the curve showing the diagnostic value of the select miRNAs a. miR-215; b. miR-148a; c. miR-125b; d. miR-223; e. miR-142-3p; ****p < 0.0001 and ***p <0.001. (C) Receiver operating characteristic (ROC) curve showing the significant diagnostic value of five miRNAs in combination, ****p < 0.0001.
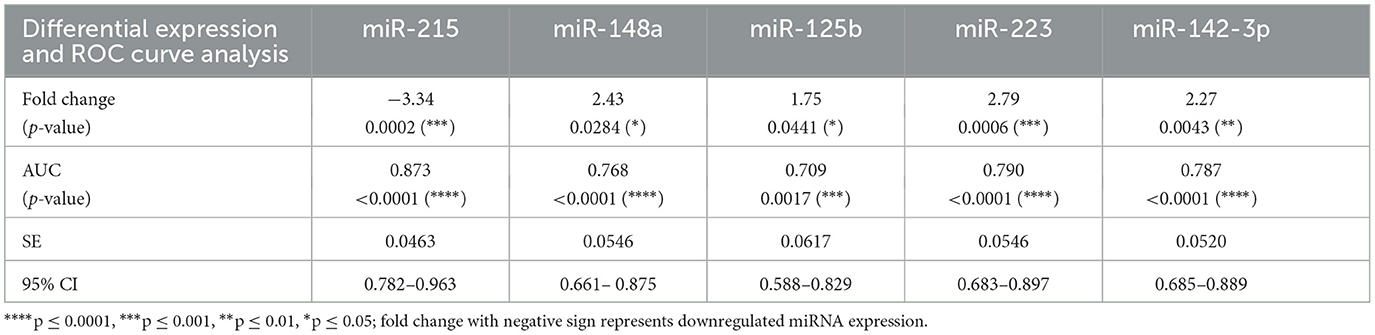
Table 2. Differential gene expression and receiver operating curve (ROC) analysis with the area under the curve (AUC) for the expression of miRNAs in plasma samples from psoriasis vulgaris patients and healthy controls.
The receiver operating characteristic (ROC) curves were generated for each of the significantly altered miRNAs, and the area under the receiver operating characteristic curve (AUC) was employed as an accuracy index to evaluate the diagnostic performance of the five miRNAs with significant expression changes. The ROC analysis of miR-215, miR-148a, miR-125b, miR-223, and miR-142-3p corresponded to AUCs of 0.873, 0.768, 0.709, 0.790, and 0.787, respectively, indicating a robust discriminatory potential of each of the miRNAs (Figure 2B, Table 2). The combination of all five miRNAs together also showed significant disease discrimination readout with an AUC of 0.762 and a p < 0.0001 (Figure 2C).
A correlation analysis between each of the five miRNAs for the control and psoriasis subjects was performed to explore the association between their expression patterns (Figure 3A, Table 3). Based on the Pearson correlation coefficient as a readout, each of the five miRNAs exhibited a significant correlation with at least one of the miRNAs in the panel. miR-215 showed a negative correlation with all other 4 miRNAs and a significant association only with miR-223 (p < 0.05). miR-148a, miR-142-3p, miR-223, and miR-125b showed a trend toward a positive correlation between each other with significant positive correlations between miR-148a and miR-142-3p (p < 0.05), miR-142-3p and miR-223 (p < 0.05), and miR-223 and miR-125b (p < 0.05). The AUC values along with the correlation matrix imply that the miRNA panel with the combination of a downregulated miR-215 and upregulated miRNAs miR-148a, miR-125b, miR-223, and miR-142-3p is a promising set of diagnostic miRNA biomarkers for psoriasis vulgaris patients.
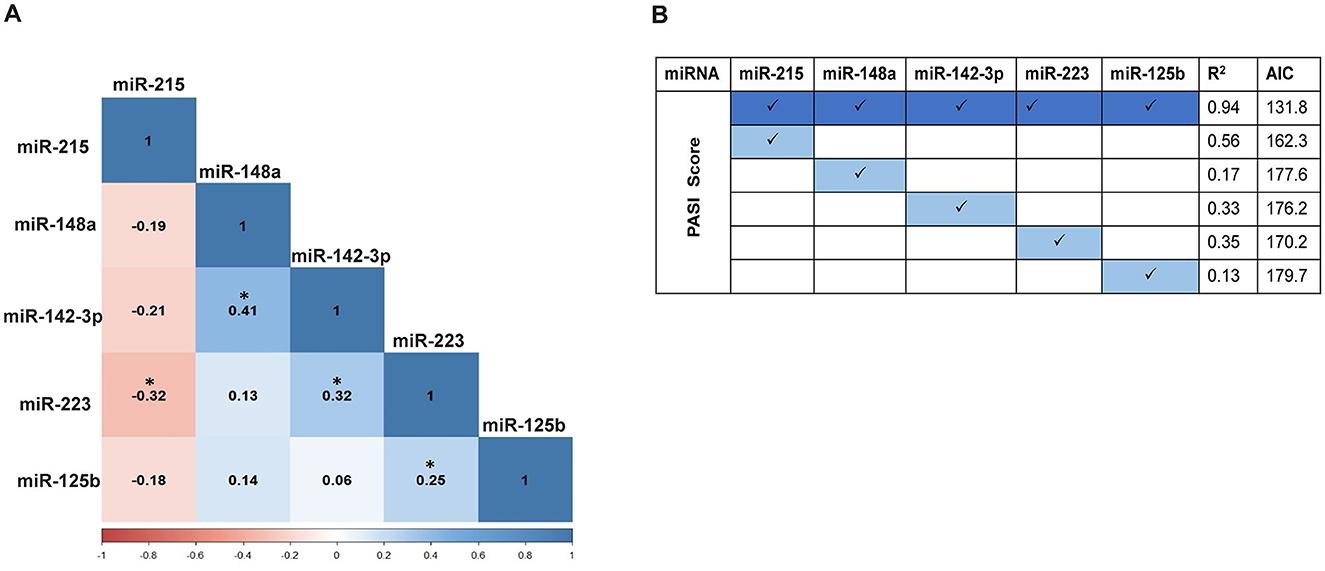
Figure 3. (A) Pairwise correlation between the expression levels of the five miRNAs using Pearson correlation. The values shown depict the correlation coefficient (r) with a significant correlation indicated by *p ≤ 0.05. (B) Correlation analysis of miRNA expression levels with PASI score using the b-spline model. All five miRNAs together exhibit the best-fit model with a coefficient of determination, R2 = 0.94, Akaike information criterion (AIC) = 131.8. The b-spline model for individual miRNAs with PASI scores does not exhibit a good-fit model.
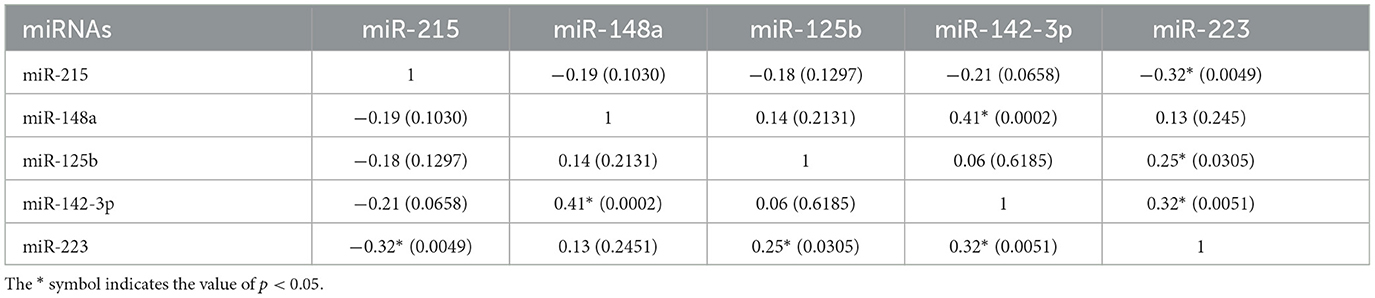
Table 3. Pairwise correlation between miRNAs represented as Pearson correlation coefficient (r) with p-values given in parentheses.
All five miRNAs were further analyzed for their correlation with disease severity in terms of patient PASI score, alone or in combination. With a non-linear association pattern derived between miRNA expression levels and PASI scores, the b-spline model was used to decipher their relationship. Based on the best-fit b-spline model, all five miRNAs together exhibited the best-fit model with an R2 value = 0.94 and the lowest AIC score of 131.8 (Figure 3B). None of the individual miRNAs showed a good fit with the PASI score, with lower R2 values and higher AIC readouts. Individually, miR-215 alone showed the highest correlation (R2 value = 0.56 and AIC = 162.3) compared with each of the other miRNAs. The additive effect derived from the b-spline model in terms of R2 and AIC values with all five miRNAs taken together implies a robust correlation with disease severity, unlike individual miRNAs.
Enrichment of significant pathways involving direct targets of the dysregulated panel of five miRNAs, i.e.,miR-215, miR-142-3p, miR-125b, miR-148a, and miR-223, was performed using a combination of the miRNA-specific targets and KEGG pathways as detailed in the Materials and Methods section. A set of signaling pathways downstream of one or more than one miRNA were delineated in light of the literature as shown in Table 4. All the relevant targets involved in the enriched pathways were mapped as a miRNA–mRNA regulatory network. The panel of five dysregulated miRNAs exhibited a network of interconnected targets as part of one or more enriched pathways of interest, which is color-coded in Figure 4 with supporting information in Table 4. Each of the pathways was found to be relevant to the pathophysiology of psoriasis based on their validated or predicted role in T cell and/or keratinocyte biology. miR-142-3p, miR-125b, miR-148a, and miR-223 are upstream of multiple components of Wnt signaling implicated in keratinocyte proliferation and apoptosis in psoriatic lesions, along with a key role in T cell development and differentiation of peripheral T cell subsets (40, 41, 71–74). With a role in vast cellular functions, MAPK signaling enriched with targets specific for miR125b and miR-223, miR-142-3p, and miR-148a is known to be involved in keratinocyte proliferation and differentiation with a multidimensional role in T cell activation, differentiation, and effector function (42–44, 71–74).
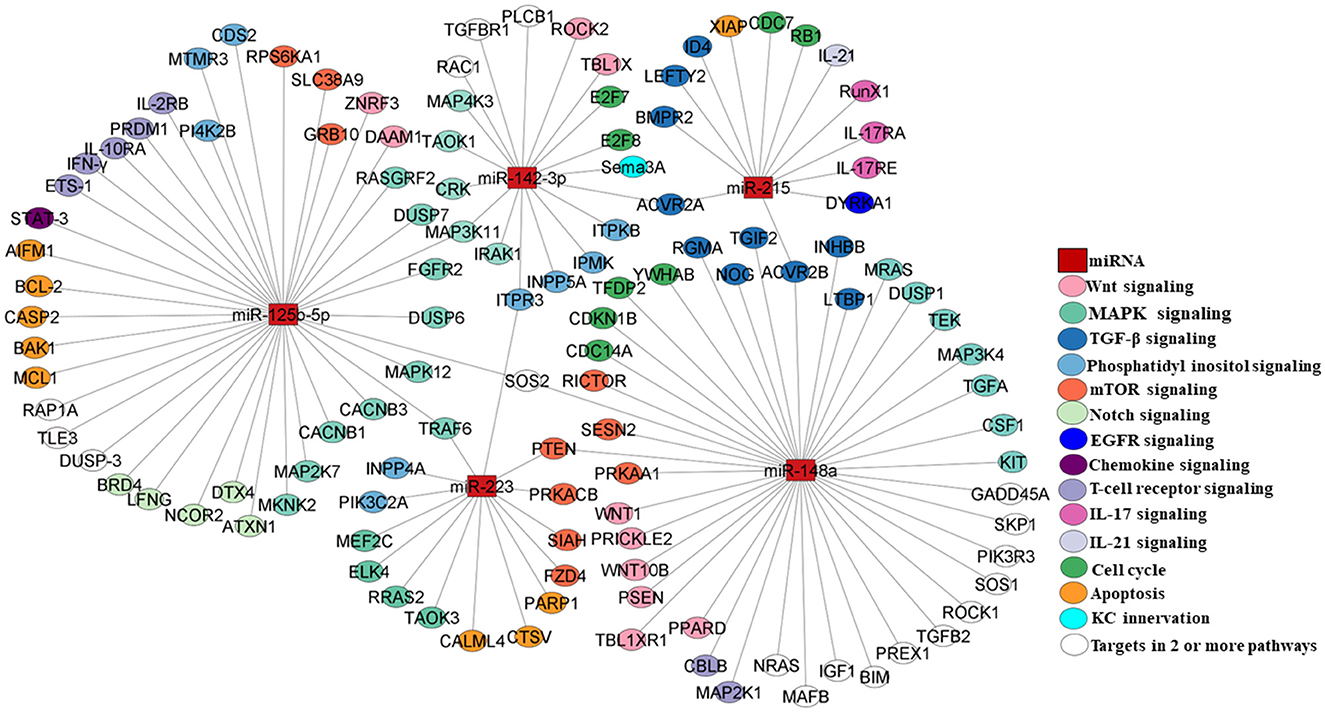
Figure 4. miRNA–mRNA network mapping for miR-215, miR-148a, miR-223, miR-125b, and miR-142-3p as select miRNAs of interest with their direct targets (please see Table 4). miRNAs are depicted as rectangular boxes, and their targets are represented as circles. The color coding of the targets represents their functionality in the corresponding pathways as highlighted in different colors. Targets in empty circles represent those candidates involved in two or more pathways.
TGF-β signaling involving targets of miR-142-3p, miR-148a, and miR-215 constitutes a pathway with relevance in psoriasis, affecting keratinocyte proliferation and T cell homeostasis and differentiation in response to the altered cytokine milieu (45, 46, 71, 74, 75). A phosphatidylinositol signaling network with pleiotropic upstream and downstream mediators was enriched and involves targets of miR-223, miR-125b-5p, and miR-142-3p that can mediate altered keratinocyte proliferation and T cell activation/differentiation in response to varied input stimuli (47–49, 71–73). One of the important downstream extensions of the phosphatidylinositol pathway works through mTOR signaling, which is known to regulate cellular growth, proliferation, and metabolism. In our analysis, multiple mTOR signaling complex molecules are direct targets of miR125b and miR-148 in light of the key role of the axis in abnormal keratinocyte proliferation and differentiation and modulating T cell metabolism, cell proliferation, activation, and differentiation (50, 51, 72, 74, 76). EGFR signaling, modulable via miR-215-targeted dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRKA1), is another molecular module that leads to keratinocyte hyperproliferation (52). The KC innervation pathway comprises Semaphorin 3A (Sema 3A) as a direct target of miR-142-3p, with implications for keratinocyte proliferation, apoptosis, and the production of inflammatory mediators (53). BRD4/Notch signaling works via miR-125b, resulting in altered keratinocyte proliferation (54). Notch signaling is known to play a pivotal role in T cell development, activation, proliferation, and effector function (55).
Multiple chemokines produced by keratinocytes and immunocytes may contribute to the inflammatory psoriatic lesions and systemic changes involving direct targets of miR-125b and miR-148 (56). Stat3, as a direct target of miR125b and downstream of multiple chemokine/cytokine inflammatory pathways, has been shown to dysregulate keratinocyte proliferation and apoptosis (57). T cell receptor signaling constitutes the primary event for a homeostatic effector T cell function, which is modulated in the autoimmune condition of psoriasis with a pathogenic T cell response (58, 59). The perpetual T cell activation drives the keratinocyte inflammatory loop with the manifestation of psoriatic hyperplasia (77). miR-125b-5p and miR-148 regulate multiple targets directly involved in T cell receptor signaling (72, 74). IL-21, a T cell-derived cytokine, leads to epidermal hyperplasia in psoriasis by inducing keratinocyte proliferation (60). IL-21 signaling regulates T cell activation, proliferation, and differentiation (61). Among our miRNAs of interest, miR-215 targets the IL-21 transcript (62). IL-17 signaling plays a major role in setting the inflammatory pathophysiological changes associated with psoriasis with enhanced IL-17-producing T cells and inflamed hyperproliferative keratinocytes in response to amplified IL-17 expression in psoriatic lesions and systemic circulation (63, 78). IL-17 signaling components and effector molecules, viz. IL-17RA, IL-17RE, and RUNX1, show up as direct targets of miR-215 (64, 65). Altered cell cycle progression and apoptosis in an inflammatory skin microenvironment are hallmarks of psoriatic lesions with epidermal hyperplasia (66, 68, 73–75). Specific molecules involved in these cellular processes are direct targets of miRNAs of interest. The miR-215-XIAP axis alters cellular apoptosis, while miR-142-3p targets E2F7 and E2F8, the transcription factors that regulate T cell cycling (67, 69, 70). All five miRNAs with significant expression changes in peripheral circulation showed downstream target pathways with functional roles in keratinocytes and/or T cells implying their potential role in the etiology of psoriasis.
Psoriasis is a cutaneous manifestation with T cells as crucial mediators of lesional and systemic inflammatory changes. The diagnosis of the disease is largely dependent on the clinical identification of typical psoriatic lesions in conjunction with histopathological findings. To date, there is a lack of reliable molecularly defined diagnostic markers for the disease, despite substantial knowledge of key cellular and molecular processes that define psoriasis. In this context, miRNAs, representing specific and stable small non-coding RNAs involved in the pathophysiology of psoriasis, comprise a promising set of molecules with diagnostic potential. A large number of such studies on the differential expression of miRNAs in varied sample types from psoriasis patients have been performed to elucidate their diagnostic and prognostic value. Limitations with respect to significant differential expression, heterogeneous abundance and correlation with disease severity, lack of knowledge on the role of altered miRNAs in disease progression, small sample test size, and variable sample types used are in the way of translating these findings to clinical use.
In the present study, 15 immune miRNAs were tested with the rationale of their functional involvement in T cell-mediated immune inflammation associated with lesional and systemic psoriatic changes. Of all the miRNAs, five candidates showed significant differential expression, with the downregulation of miR-215 and the upregulation of miR-148a, miR-223, miR-142-3p, and miR-125b-5p. All five miRNAs, individually and combinedly, exhibited promising diagnostic potential to differentiate psoriasis patients from healthy individuals as per significant and acceptable AUC values (39).
The expression pattern data of the five significant miRNAs and their correlation trend among each other and with the PASI score as a panel signify a promising combination of miRNAs with diagnostic value. The panel of five miRNAs deciphered in the present study with two newly profiled miRNAs (miR-215 and miR-148a) and three previously studied miRNAs (miR-223, miR-142-3p, and miR-125b-5p) can be tested in a larger cohort of psoriasis vulgaris patients along with a control group of patients with other skin diseases to further validate its differential diagnostic value in a clinical setting. Among the various exploratory searches for circulating diagnostic miRNAs that include global miRNA expression profiling and/or miRNA candidate-specific expression studies, this is the pilot report on a set of promising miRNAs that can be taken up as a panel for the diagnosis of psoriasis vulgaris (19, 20, 22–27).
Interestingly, all five miRNAs exhibited a validated and/or predicted role in dysregulating T cell and/or keratinocyte function, thus implying their involvement in inflammatory psoriatic manifestations. The altered expression of the five miRNAs in the peripheral circulation found in our study may reflect their dysregulation in keratinocytes and/or T cells, which constitute the key cell types involved in the pathophysiology of the disease, as depicted in Figure 5. Much of the literature on these miRNAs has demonstrated specific targets and associated pathways involved in the dysregulation of psoriatic keratinocytes, albeit with limited knowledge of their T cell-centric role in disease development.
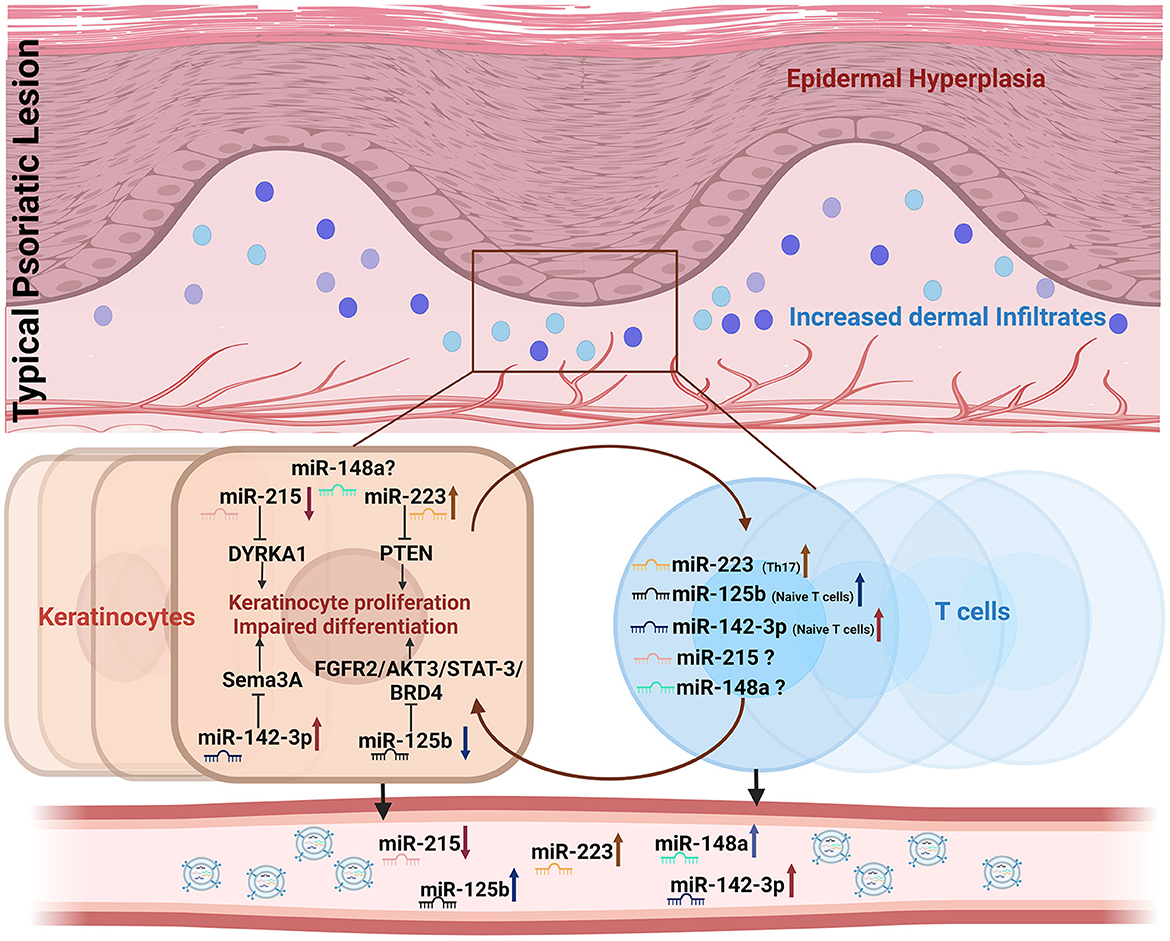
Figure 5. Representation of candidate miRNAs involved in keratinocyte and T cell crosstalk dysregulated in psoriasis. Dysregulated expression of miR-215, miR-148a, miR-125b, miR-223, and miR-142-3p in keratinocytes and/or T cells is shown to be altered in cell-free peripheral circulation.
miR-215 is associated with multiple cellular processes, viz., proliferation, apoptosis, migration, invasion, and epithelial–mesenchymal transition, in various cancers (79, 80). In psoriasis, miR-215 is reported to be downregulated in the skin lesion tissues in humans and mouse models, with the functional role validated in a keratinocyte-specific study, wherein miR-215 is shown to target DYRK1A and dysregulate keratinocyte proliferation via EGFR signaling (6, 52, 81). In the context of T cells, miR-215 has been reported to be differentially expressed in the Th2 subset in healthy human subjects, with no T cell-associated report in psoriasis (30, 82). IL-17 receptors, RUNX1, and IL-21 with miR-215-binding UTRs constitute Th17 subset-specific signature molecules, implying the potential role of miR-215 in psoriasis-associated Th17 dysregulation (62, 64, 65). Also, not a single report on expression patterns in peripheral circulation in psoriasis patients is available on miR-215, possibly due to its low abundance and the downregulated expression detected in our study. miR-148a has been demonstrated to contribute toward the maintenance of a chronic inflammatory immune-environment by regulating the persistence of activated Th1 cells in the murine helper T cell model (83). With upregulation in its expression in PBMCs from psoriasis patients, miRNA-148a has been shown to facilitate differentiation of inflammatory monocyte-derived dendritic cells via PU.1–miR-148a–MAFB axis, albeit with no report in the context of psoriatic lesions, Keratinocytes, T cells, or peripheral circulation (84). miR-223 is another miRNA associated with pathogenic T cells in autoimmune diseases like rheumatoid arthritis and multiple sclerosis (85–87). In the context of psoriasis, miR-223 is reported as one of the important miRNAs altered in skin lesions with changes in epidermal and dermal infiltrate along with peripheral Th17 subsets involved in the inflammatory disease outcome (7, 81, 88). Mechanistically, miR-223 has been shown to mediate keratinocyte hyperproliferation and apoptosis via the PTEN/Akt pathway in a HaCaT cell line model (48). Additionally, miR-223 is shown to be differentially upregulated in PBMCs, with variable reports on extracellular systemic circulation in psoriasis patients. Lovendorf et al. and Pivarcsi et al. reported no change in peripheral circulation in two independent reports, while Garcia et al. demonstrated an increase similar to our finding in psoriatic plasma samples (20, 26, 89). miR-142-3p is known to be a prominent hematopoietic miRNA with a role in T cell cycling and involvement in controlling T cell subset cAMP levels, with functional implications for Treg suppressor function (67, 90). The miRNA is shown to be differentially detected in the psoriatic skin miRNAome with differential expression in the psoriatic epidermis, dermal infiltrate, and peripheral T cells in the same patient cohort (7, 81, 88). Mechanistically, miR-142-3p has been shown to dysregulate keratinocyte proliferation and apoptosis via targeting Sema3A based on HaCAT cell line studies (53). Blood-based studies on miR-142 exhibit variable findings with no change to a downregulated expression pattern, unlike the significant increase found in plasma samples in our study (25, 26). miR-125b constitutes a signature miRNA responsible for the maintenance of T cell naivety and thus regulates effector T cell function based on its high expression in naive T cells in healthy individuals and naive T cells from psoriatic patients (30, 88). Lesional miRNA profiling is also reported with miR-125b as one of the most downregulated miRNAs in the epidermal layer and in dermal infiltrates (6, 88). The major cell type in psoriatic lesions with decreased expression is reported to be keratinocytes that exhibit miR-125b-mediated hyperproliferation and abnormal differentiation via multiple signaling pathways validated in in vitro studies (43, 54, 57, 91). Studies on serum expression patterns demonstrate different findings, with downregulation reported by Koga et al. and Pan M et al. and no change reported by Hernandez et al., unlike the significant upregulation detected in our study (19, 25, 54).
All five miRNAs, with their predicted or validated targets, constituted regulators of one or more immune inflammation-associated pathways, such that changes in their levels in keratinocytes and/or T cells can potentially lead to the development of autoimmune-psoriatic manifestations. The miR–mRNA network analysis with predicted/validated targets highlights the dysregulation of multiple autoimmune disease-related pathways that may contribute to keratinocyte hyperproliferation and abnormal differentiation, along with altered T cell activation, differentiation, and effector function. All the pathways, namely Wnt, MAPK, TGF-β, PI3K, mTOR, Notch, IL-21, IL-17, chemokine signaling, and TCR signaling, have been reported to directly impinge on activation and proliferative capabilities of keratinocytes and T cells with the elaboration of an inflammatory milieu as the result in disease development (41, 42, 45, 47, 51, 55, 56, 58, 61, 63). Based on our findings, much exploration of the downstream miRNA mechanisms involving multiple predicted targets still needs to be explored for their role in psoriasis.
Importantly, the five miRNAs as a promising disease diagnostic panel, along with their robust correlation with disease severity, corroborate the involvement of multiple miR-target pathways that may drive the disease. In the context of specific limitations in our study, each of the five miRNAs exhibited a significant correlation with only one or two of the other miRNAs in the panel. This could be because of the heterogeneous expression of different miRNAs in both healthy and psoriasis patients. An extended cohort of control and diseased subjects with the inclusion of more severe psoriasis patients may reveal a better relationship among the five miRNAs. Importantly, experimental validation of the downstream miRNA targets can further provide proof of concept for our in silico-derived miRNA–mRNA regulatory network. Nonetheless, in light of the existing literature, the plasma miRNAs, viz., miR-215, miR-142-3p miR-223, miR-125b-5p, and miR-148a, constitute a promising panel of miRNA-based biomarkers potentially involved in a pathogenic inflammatory response as a result of a dysregulated keratinocyte–T cell crosstalk.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by the Ethics Committee of Guru Gobind Singh Medical College and Hospital, Faridkot, Punjab, India (GGS/IEC/19/57), AIIMS, Bathinda, India (IEC/AIIMS/BTI/150), and Central University of Punjab, Bathinda, Punjab, India (CUPB/IEC/2016/045). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
PM and MJ conceived the original idea, planned the experiments, and wrote the manuscript. PM carried out the experiments. BB and SB provided the clinical samples and patient information. HSK provided support for the statistical analysis. NT and HRK helped with the miRNA-target network image. US and AJ critically revised the manuscript. MJ supervised and supported the research. All authors contributed to the article and approved the submitted version.
We would like to acknowledge financial support from the University Grants Commission (UGC BSR Grant No. F.30-432/2018), New Delhi, India and the Department of Biotechnology, India, for supporting PM with a Ph.D. fellowship (DBT/2018/CUP/975). We also acknowledge the use of equipment setup under the DST-FIST grant at the Department of Biochemistry, Central University of Punjab, Bathinda, Punjab, India.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2023.1207993/full#supplementary-material
1. Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. (2009) 361:496–509. doi: 10.1056/NEJMra0804595
2. Griffiths CEM, van der Walt JM, Ashcroft DM, Flohr C, Naldi L, Nijsten T, et al. The global state of psoriasis disease epidemiology: a workshop report. Br J Dermatol. (2017) 177:e4–7. doi: 10.1111/bjd.15610
3. Cai Y, Fleming C, Yan J. New insights of T cells in the pathogenesis of psoriasis. Cell Mol Immunol. (2012) 9:302–9. doi: 10.1038/cmi.2012.15
4. Diani M, Altomare G, Reali ET. Helper cell subsets in clinical manifestations of psoriasis. J Immunol Res. (2016) 2016:7692024. doi: 10.1155/2016/7692024
5. Raychaudhuri SK, Maverakis E, Raychaudhuri SP. Diagnosis and classification of psoriasis. Autoimmun Rev. (2014) 13:490–5. doi: 10.1016/j.autrev.2014.01.008
6. Sonkoly E, Wei TL, Janson PCJ, Saaf A, Lundeberg L, Tengvall-Linder M, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS ONE. (2007) 2:8. doi: 10.1371/journal.pone.0000610
7. Joyce CE, Zhou X, Xia J, Ryan C, Thrash B, Menter A, et al. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet. (2011) 20:4025–40. doi: 10.1093/hmg/ddr331
8. Di Meglio P, Villanova F, Nestle FO. Psoriasis. Cold Spring Harb Perspect Med. (2014) 4:a015354. doi: 10.1101/cshperspect.a015354
9. Xia J, Zhang W. MicroRNAs in normal and psoriatic skin. Physiol Genomics. (2014) 46:113–22. doi: 10.1152/physiolgenomics.00157.2013
10. Huang RY Li L, Wang MJ, Chen XM, Huang QC, Lu CJ. An exploration of the role of microRNAs in psoriasis: a systematic review of the literature. Medicine (Baltimore). (2015) 94:10. doi: 10.1097/MD.0000000000002030
11. Masalha M, Sidi Y, Avni D. The contribution of feedback loops between miRNAs, cytokines and growth factors to the pathogenesis of psoriasis. Exp Dermatol. (2018) 27:603–10. doi: 10.1111/exd.13520
12. Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol. (2013) 13:666–78. doi: 10.1038/nri3494
13. Sethi A, Kulkarni N, Sonar S, Lal G. Role of miRNAs in CD4 T cell plasticity during inflammation and tolerance. Front Genet. (2013) 4:8. doi: 10.3389/fgene.2013.00008
14. Joly-Tonetti N, Viñuelas J, Gandrillon O, Lamartine J. Differential miRNA expression profiles in proliferating or differentiated keratinocytes in response to gamma irradiation. BMC Genomics. (2013) 14:184. doi: 10.1186/1471-2164-14-184
15. Sommers CL, Rouquette-Jazdanian AK, Robles AI, Kortum RL, Merrill RK Li W, et al. miRNA signature of mouse helper T cell hyper-proliferation. PLoS ONE. (2013) 8:e66709. doi: 10.1371/journal.pone.0066709
16. Lee AY. The role of microRNAs in epidermal barrier. Int J Mol Sci. (2020) 21:16. doi: 10.3390/ijms21165781
17. Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. (2010) 285:17442–52. doi: 10.1074/jbc.M110.107821
18. Murillo OD, Thistlethwaite W, Rozowsky J, Subramanian SL, Lucero R, Shah N, et al. exRNA atlas analysis reveals distinct extracellular RNA cargo types and their carriers present across human biofluids. Cell. (2019) 177:463–77.e15. doi: 10.1016/j.cell.2019.02.018
19. Koga Y, Jinnin M, Ichihara A, Fujisawa A, Moriya C, Sakai K, et al. Analysis of expression pattern of serum microRNA levels in patients with psoriasis. J Dermatol Sci. (2014) 74:170–1. doi: 10.1016/j.jdermsci.2014.01.005
20. Lovendorf MB, Zibert JR, Gyldenlove M, Ropke MA, Skov L. MicroRNA-223 and miR-143 are important systemic biomarkers for disease activity in psoriasis. J Dermatol Sci. (2014) 75:133–9. doi: 10.1016/j.jdermsci.2014.05.005
21. Tsuru Y, Jinnin M, Ichihara A, Fujisawa A, Moriya C, Sakai K, et al. miR-424 levels in hair shaft are increased in psoriatic patients. J Dermatol. (2014) 41:382–5. doi: 10.1111/1346-8138.12460
22. Xiao S, Liu X, Wang X, Lv H, Zhao J, Guo X, et al. Plasma microRNA expression profiles in psoriasis. J Immunol Res. (2020) 2020:1561278. doi: 10.1155/2020/1561278
23. Guo S, Zhang W, Wei C, Wang L, Zhu G, Shi Q, et al. Serum and skin levels of miR-369-3p in patients with psoriasis and their correlation with disease severity. Eur J Dermatol: EJD. (2013) 23:608–13. doi: 10.1684/ejd.2013.2148
24. Ichihara A, Jinnin M, Oyama R, Yamane K, Fujisawa A, Sakai K, et al. Increased serum levels of miR-1266 in patients with psoriasis vulgaris. Eur J Dermatol. (2012) 22:68–71. doi: 10.1684/ejd.2011.1600
25. Martínez-Hernández R, Fuente H, Lamana A, Sampedro-Núñez M, Ramos-Levi A, Serrano-Somavilla A, et al. Utility of circulating serum miRNA profiles to evaluate the potential risk and severity of immune-mediated inflammatory disorders. J Autoimmun. (2020) 111:102472. doi: 10.1016/j.jaut.2020.102472
26. Pivarcsi A, Meisgen F, Xu N, Stahle M, Sonkoly E. Changes in the level of serum microRNAs in patients with psoriasis after antitumour necrosis factor-alpha therapy. Br J Dermatol. (2013) 169:563–70. doi: 10.1111/bjd.12381
27. Garcia-Rodriguez S, Arias-Santiago S, Orgaz-Molina J, Magro-Checa C, Valenzuela I, Navarro P, et al. Abnormal levels of expression of plasma microRNA-33 in patients with psoriasis. Actas Dermosifiliogr. (2014) 105:497–503. doi: 10.1016/j.adengl.2014.04.003
28. Podshivalova K, Salomon DR. MicroRNA regulation of T-lymphocyte immunity: modulation of molecular networks responsible for T cell activation, differentiation, and development. Crit Rev Immunol. (2013) 33:435–76. doi: 10.1615/CritRevImmunol.2013006858
29. Rodriguez-Galan A, Fernandez-Messina L, Sanchez-Madrid F. Control of immunoregulatory molecules by miRNAs in T cell activation. Front Immunol. (2018) 9:2148. doi: 10.3389/fimmu.2018.02148
30. Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, Bonnal RJP, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol. (2011) 12:796. doi: 10.1038/ni.2057
31. Li W, Kong LB Li JT, Guo ZY, Xue Q, Yang T, et al. MiR-568 inhibits the activation and function of CD4(+) T cells and Treg cells by targeting NFAT5. Int Immunol. (2014) 26:269–81. doi: 10.1093/intimm/dxt065
32. Liu Q, Gao Q, Zhang Y, Li ZB, Mei XX. MicroRNA-590 promotes pathogenic Th17 cell differentiation through targeting Tob1 and is associated with multiple sclerosis. Biochem Biophys Res Commun. (2017) 493:901–8. doi: 10.1016/j.bbrc.2017.09.123
33. Weitzel RP, Lesniewski ML, Haviernik P, Kadereit S, Leahy P, Greco NJ, et al. microRNA 184 regulates expression of NFAT1 in umbilical cord blood CD4+ T cells. Blood. (2009) 113:6648–57. doi: 10.1182/blood-2008-09-181156
34. Chen Z, Stelekati E, Kurachi M, Yu S, Cai Z, Manne S, et al. miR-150 regulates memory CD8 T cell differentiation via c-Myb. Cell Rep. (2017) 20:2584–97. doi: 10.1016/j.celrep.2017.08.060
35. Ghisi M, Corradin A, Basso K, Frasson C, Serafin V, Mukherjee S, et al. Modulation of microRNA expression in human T cell development: targeting of NOTCH3 by miR-150. Blood. (2011) 117:7053–62. doi: 10.1182/blood-2010-12-326629
36. Imafuku S, Zheng M, Tada Y, Zhang X, Theng C, Thevarajah S, et al. Asian consensus on assessment and management of mild to moderate plaque psoriasis with topical therapy. J Dermatol. (2018) 45:805–11. doi: 10.1111/1346-8138.14338
37. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. (2008) 3:1101–8. doi: 10.1038/nprot.2008.73
38. Khandelwal A, Seam RK, Gupta M, Rana MK, Prakash H, Vasquez KM, et al. Circulating microRNA-590-5p functions as a liquid biopsy marker in non-small cell lung cancer. Cancer Sci. (2020) 111:826–39. doi: 10.1111/cas.14199
39. Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol. (2010) 5:1315–6. doi: 10.1097/JTO.0b013e3181ec173d
40. Zhang Y, Tu C, Zhang D, Zheng Y, Peng Z, Feng Y, et al. Wnt/β-catenin and Wnt5a/Ca pathways regulate proliferation and apoptosis of keratinocytes in psoriasis lesions. Cell Physiol Biochem. (2015) 36:1890–902. doi: 10.1159/000430158
41. van Loosdregt J, Coffer PJ. The role of WNT signaling in mature T cells: T cell factor is coming home. J Immunol. (2018) 201:2193–200. doi: 10.4049/jimmunol.1800633
42. Meng X, Qiu L, Song H, Dang N, MAPK. Pathway involved in epidermal terminal differentiation of normal human epidermal keratinocytes. Open Med. (Warsaw, Poland). (2018) 13:189–95. doi: 10.1515/med-2018-0029
43. Xu N, Brodin P, Wei TL, Meisgen F, Eidsmo L, Nagy N, et al. MiR-125b, a microRNA downregulated in psoriasis, modulates keratinocyte proliferation by targeting FGFR2. J Invest Dermatol. (2011) 131:1521–9. doi: 10.1038/jid.2011.55
44. Rincón M. MAP-kinase signaling pathways in T cells. Curr Opin Immunol. (2001) 13:339–45. doi: 10.1016/S0952-7915(00)00224-7
45. Han G, Williams CA, Salter K, Garl PJ Li AG, Wang XJ, A. role for TGFbeta signaling in the pathogenesis of psoriasis. J Invest Dermatol. (2010) 130:371–7. doi: 10.1038/jid.2009.252
46. Oh SA, Li MO. TGF-β: guardian of T cell function. J Immunol. (2013) 191:3973–9. doi: 10.4049/jimmunol.1301843
47. Huang T, Lin X, Meng X, Lin M. Phosphoinositide-3 kinase/protein kinase-B/mammalian target of rapamycin pathway in psoriasis pathogenesis. A potential therapeutic target? Acta Dermato-Venereologica. (2014) 94:371–9. doi: 10.2340/00015555-1737
48. Wang R, Wang FF, Cao HW, Yang JY. MiR-223 regulates proliferation and apoptosis of IL-22-stimulated HaCat human keratinocyte cell lines via the PTEN/Akt pathway. Life Sci. (2019) 230:28–34. doi: 10.1016/j.lfs.2019.05.045
49. Han JM, Patterson SJ, Levings MK. The role of the PI3K signaling pathway in CD4(+) T cell differentiation and function. Front Immunol. (2012) 3:245. doi: 10.3389/fimmu.2012.00245
50. Buerger C. Epidermal mTORC1 signaling contributes to the pathogenesis of psoriasis and could serve as a therapeutic target. Front Immunol. (2018) 9:2786. doi: 10.3389/fimmu.2018.02786
51. Pollizzi KN, Powell JD. Regulation of T cells by mTOR: the known knowns and the known unknowns. Trends Immunol. (2015) 36:13–20. doi: 10.1016/j.it.2014.11.005
52. Liu A, Zhang B, Zhao W, Tu Y, Wang Q, Li J. MicroRNA-215-5p inhibits the proliferation of keratinocytes and alleviates psoriasis-like inflammation by negatively regulating DYRK1A and its downstream signalling pathways. Exp Dermatol. (2021) 30:932–42. doi: 10.1111/exd.14188
53. Zhang D, Wang Y, Xia Y, Huo J, Zhang Y, Yang P, et al. Repression of miR-142-3p alleviates psoriasis-like inflammation by repressing proliferation and promoting apoptosis of keratinocytes via targeting Sema3A. Mol Cell Probes. (2020) 52:101573. doi: 10.1016/j.mcp.2020.101573
54. Pan M, Huang Y, Zhu X, Lin X, Luo D. miR125bmediated regulation of cell proliferation through the Jagged1/Notch signaling pathway by inhibiting BRD4 expression in psoriasis. Mol Med Rep. (2019) 19:5227–36. doi: 10.3892/mmr.2019.10187
55. Brandstadter JD, Maillard I. Notch signalling in T cell homeostasis and differentiation. Open Biol. (2019) 9:190187. doi: 10.1098/rsob.190187
56. Zdanowska N, Kasprowicz-Furmańczyk M, Placek W, Owczarczyk-Saczonek A. The role of chemokines in psoriasis-an overview. Medicina. (2021) 57:8. doi: 10.3390/medicina57080754
57. Yang Z, Chen Z, Wang C, Huang P, Luo M, Zhou R. STAT3/SH3PXD2A-AS1/miR-125b/STAT3 positive feedback loop affects psoriasis pathogenesis via regulating human keratinocyte proliferation. Cytokine. (2021) 144:155535. doi: 10.1016/j.cyto.2021.155535
58. Gaud G, Lesourne R, Love PE. Regulatory mechanisms in T cell receptor signalling. Nat Rev Immunol. (2018) 18:485–97. doi: 10.1038/s41577-018-0020-8
59. Prinz JC. The role of T cells in psoriasis. JEADV. (2003) 17:257–70. doi: 10.1046/j.1468-3083.2003.00720.x
60. Caruso R, Botti E, Sarra M, Esposito M, Stolfi C, Diluvio L, et al. Involvement of interleukin-21 in the epidermal hyperplasia of psoriasis. Nat Med. (2009) 15:1013–5. doi: 10.1038/nm.1995
61. Liu SM, King C. IL-21-producing Th cells in immunity and autoimmunity. J Immunol. (2013) 191:3501–6. doi: 10.4049/jimmunol.1301454
62. Enomoto Y, Takagi R, Naito Y, Kiniwa T, Tanaka Y, Hamada-Tsutsumi S, et al. Identification of the novel 3′ UTR sequences of human IL-21 mRNA as potential targets of miRNAs. Sci Rep. (2017) 7:7780. doi: 10.1038/s41598-017-07853-x
63. Mosca M, Hong J, Hadeler E, Hakimi M, Liao W, Bhutani T. The role of IL-17 cytokines in psoriasis. Immuno Targets Ther. (2021) 10:409–18. doi: 10.2147/ITT.S240891
64. Sun Y, Pan J, Mao S, Jin J. IL-17/miR-192/IL-17Rs regulatory feedback loop facilitates multiple myeloma progression. PLoS ONE. (2014) 9:e114647. doi: 10.1371/journal.pone.0114647
65. Li N, Zhang QY, Zou JL Li ZW, Tian TT, Dong B, et al. miR-215 promotes malignant progression of gastric cancer by targeting RUNX1. Oncotarget. (2016) 7:4817–28. doi: 10.18632/oncotarget.6736
66. Henri P, Prevel C, Pellerano M, Lacotte J, Stoebner PE, Morris MC, et al. Psoriatic epidermis is associated with upregulation of CDK2 and inhibition of CDK4 activity. Br J Dermatol. (2020) 182:678–89. doi: 10.1111/bjd.18178
67. Sun Y, Oravecz-Wilson K, Mathewson N, Wang Y, McEachin R, Liu C, et al. Mature T cell responses are controlled by microRNA-142. J Clin Invest. (2015) 125:2825–40. doi: 10.1172/JCI78753
68. Laporte M, Galand P, Fokan D, de Graef C, Heenen M. Apoptosis in established and healing psoriasis. Dermatology. (2000) 200:314–6. doi: 10.1159/000018394
69. Ye M, Zhang J, Zhang J, Miao Q, Yao L, Zhang J. Curcumin promotes apoptosis by activating the p53-miR-192-5p/215-XIAP pathway in non-small cell lung cancer. Cancer Lett. (2015) 357:196–205. doi: 10.1016/j.canlet.2014.11.028
70. Lu C, Hong M, Chen B, Liu K, Lv Y, Zhou X, et al. MicroRNA-215 regulates the apoptosis of HCT116 colon cancer cells by inhibiting x-linked inhibitor of apoptosis protein. Cancer Biother Radiopharm. (2021) 36:728–36. doi: 10.1089/cbr.2019.3011
71. miR-142-3p. Available online at: https://starbase.sysu.edu.cn/mirTarPathways.php?source=mRNA&flag=miRNA&clade=mammal&genome=human&assembly=hg19&miRNA=hsa-miR-142-3p&clipNum=&deNum=&proNum=&program=&panNum=&pathway (accessed February 26, 2023).
72. miR-125b-5p. Available online at: https://starbase.sysu.edu.cn/mirTarPathways.php?source=mRNA&flag=miRNA&clade=mammal&genome=human&assembly=hg19&miRNA=hsa-miR-125b-5p&clipNum=&deNum=&proNum=&program=&panNum=&pathway= (accessed February 26, 2023).
73. miR-223-3p. Available online at: https://starbase.sysu.edu.cn/mirTarPathways.php?source=mRNA&flag=miRNA&clade=mammal&genome=human&assembly=hg19&miRNA=hsa-miR-223-3p&clipNum=&deNum=&proNum=&program=&panNum=&pathway (accessed February 26, 2023).
74. miR-148a. Available online at: https://starbase.sysu.edu.cn/mirTarPathways.php?source=mRNA&flag=miRNA&clade=mammal&genome=human&assembly=hg19&miRNA=hsa-miR-148a-3p&clipNum=&deNum=&proNum=&program=&panNum=&pathway= (accessed February 26, 2023).
75. miR-215. Available online at: https://starbase.sysu.edu.cn/mirTarPathways.php?source=mRNA&flag=miRNA&clade=mammal&genome=human&assembly=hg19&miRNA=hsa-miR-215-5p&clipNum=&deNum=&proNum=&program=&panNum=&pathway (accessed February 26, 2023).
76. Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. (2012) 12:325–38. doi: 10.1038/nri3198
77. Kim J, Krueger JG. The immunopathogenesis of psoriasis. Dermatol Clin. (2015) 33:13–23. doi: 10.1016/j.det.2014.09.002
78. Kagami S, Rizzo HL, Lee JJ, Koguchi Y, Blauvelt A. Circulating Th17, Th22, and Th1 cells are increased in psoriasis. J Invest Dermatol. (2010) 130:1373–83. doi: 10.1038/jid.2009.399
79. Zhang P, Tao X, Li P, Ma L. miR-215 controls proliferation, invasion, and apoptosis of human retinoblastoma cells by regulating RB1 expression. Int J Clin Exp Med. (2018) 11:8978–87.
80. Hou Y, Zhen J, Xu X, Zhen K, Zhu B, Pan R, et al. miR-215 functions as a tumor suppressor and directly targets ZEB2 in human non-small cell lung cancer retraction in/10.3892/ol.2021.12861. Oncol Lett. (2015) 10:1985–92. doi: 10.3892/ol.2015.3587
81. Ichihara A, Jinnin M, Yamane K, Fujisawa A, Sakai K, Masuguchi S, et al. microRNA-mediated keratinocyte hyperproliferation in psoriasis vulgaris. Br J Dermatol. (2011) 165:1003–10. doi: 10.1111/j.1365-2133.2011.10497.x
82. Pagani M, Rossetti G, Panzeri I, de Candia P, Bonnal RJP, Rossi RL, et al. Role of microRNAs and long-non-coding RNAs in CD4+T cell differentiation. Immunol Rev. (2013) 253:82–96. doi: 10.1111/imr.12055
83. Haftmann C, Stittrich AB, Zimmermann J, Fang Z, Hradilkova K, Bardua M, et al. miR-148a is upregulated by Twist1 and T-bet and promotes Th1-cell survival by regulating the proapoptotic gene Bim. Eur J Immunol. (2015) 45:1192–205. doi: 10.1002/eji.201444633
84. Meng Y, Li J, Ye Z, Yin Z, Sun Q, Liao Z, et al. MicroRNA-148a facilitates inflammatory dendritic cell differentiation and autoimmunity by targeting MAFB. JCI Insight. (2020) 5:8. doi: 10.1172/jci.insight.133721
85. Hosseini A, Ghaedi K, Tanhaei S, Ganjalikhani-Hakemi M, Teimuri S, Etemadifar M, et al. Upregulation of CD4(+) T cell derived MiR-223 in the relapsing phase of multiple sclerosis patients. Cell J. (2016) 18:371–80. doi: 10.22074/cellj.2016.4565
86. Lu MC Yu CL, Chen HC Yu HC, Huang HB, Lai NS. Increased miR-223 expression in T cells from patients with rheumatoid arthritis leads to decreased insulin-like growth factor-1-mediated interleukin-10 production. Clin Exp Immunol. (2014) 177:641–51. doi: 10.1111/cei.12374
87. Satoorian T, Li B, Tang X, Xiao J, Xing W, Shi W, et al. MicroRNA223 promotes pathogenic T cell development and autoimmune inflammation in central nervous system in mice. Immunology. (2016) 148:326–38. doi: 10.1111/imm.12611
88. Lovendorf MB, Mitsui H, Zibert JR, Ropke MA, Hafner M, Dyring-Andersen B, et al. Laser capture microdissection followed by next-generation sequencing identifies disease-related microRNAs in psoriatic skin that reflect systemic microRNA changes in psoriasis. Exp Dermatol. (2015) 24:187–93. doi: 10.1111/exd.12604
89. Garcia-Rodriguez S, Arias-Santiago S, Blasco-Morente G, Orgaz-Molina J, Rosal-Vela A, Navarro P, et al. Increased expression of microRNA-155 in peripheral blood mononuclear cells from psoriasis patients is related to disease activity. J Eur Acad Dermatol Venereol. (2017) 31:312–22. doi: 10.1111/jdv.13861
90. Huang B, Zhao J, Lei Z, Shen S, Li D, Shen GX, et al. miR-142-3p restricts cAMP production in CD4+CD25- T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep. (2009) 10:180–5. doi: 10.1038/embor.2008.224
Keywords: psoriasis, diagnostics, circulatory miRNAs, miR-215, miR-148a, miR-223, miR-125b, miR-142-3p
Citation: Madaan P, Sharma U, Tyagi N, Brar BK, Bansal S, Kushwaha HR, Kapoor HS, Jain A and Jain M (2023) A panel of blood-based circulatory miRNAs with diagnostic potential in patients with psoriasis. Front. Med. 10:1207993. doi: 10.3389/fmed.2023.1207993
Received: 18 April 2023; Accepted: 28 July 2023;
Published: 28 August 2023.
Edited by:
Emi Dika, University of Bologna, ItalyReviewed by:
Vincenzo Dattilo, Magna Græcia University of Catanzaro, ItalyCopyright © 2023 Madaan, Sharma, Tyagi, Brar, Bansal, Kushwaha, Kapoor, Jain and Jain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manju Jain, bWFuanVqYWlubWRhQGdtYWlsLmNvbQ==; bWFuanUuamFpbkBjdXAuZWR1Lmlu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.