- 1Institute of Disaster and Emergency Medicine, Tianjin University, Tianjin, China
- 2Wenzhou Safety (Emergency) Institute of Tianjin University, Wenzhou, China
Small noncoding RNAs, known as microRNAs (miRNAs), are vital for the regulation of diverse biological processes. miR-223, an evolutionarily conserved anti-inflammatory miRNA expressed in cells of the myeloid lineage, has been implicated in the regulation of monocyte–macrophage differentiation, proinflammatory responses, and the recruitment of neutrophils. The biological functions of this gene are regulated by its expression levels in cells or tissues. In this review, we first outline the regulatory role of miR-223 in granulocytes, macrophages, endothelial cells, epithelial cells and dendritic cells (DCs). Then, we summarize the possible role of miR-223 in chronic obstructive pulmonary disease (COPD), acute lung injury (ALI), coronavirus disease 2019 (COVID-19) and other pulmonary inflammatory diseases to better understand the molecular regulatory networks in pulmonary inflammatory diseases.
1. Introduction
The body’s non-specific immune response to injury or infection is called inflammation. It is a delicate matter of homeostasis and is highly controlled. Excessive inflammation can lead to organ dysfunction, tissue destruction or even malignancies. When injury or infection occurs, Toll-like receptors (TLRs), a family of pattern recognition receptors in sentinel cells can sense risk factors and then activate the signaling pathways associated with inflammatory responses. This results in the generation of chemokines and cytokines that possess inflammatory properties, thereby facilitating the attraction of other types of immune cells (1).
In mammals, the exchange of gases is facilitated by the lung, which serves as a crucial organ. The lung is in direct contact with the external environment and is the certain target of pathogens, allergens and poisons, which easily cause infection or inflammation, such as ALI, COPD and tuberculosis. In particular, the emergence of COVID-19 in December 2019, caused an unparalleled disturbance and drew a wide range of attention in human society (2). Thus, it is imperative to create secure and efficient therapies that can restrict inflammatory reactions in damaged lungs.
MicroRNAs (miRNAs or miRs) are a type of noncoding RNA molecules with a length of approximately 18–22 nt (3). They are highly conserved and single-stranded. miRNAs have the ability to control the expression of target genes at the posttranscriptional level (4). The targeting of a specific gene can be carried out by various miRNAs, and a particular miRNA can target several genes within a single cell type, which forms a complex regulatory network (5). Numerous studies have shown that miRNAs participate in several biological processes (6). In recent years, many studies have shown that miRNAs serve as biomarkers and treatments for different kinds of diseases (7), such as allergic diseases (8–11), cardiovascular disease (12–15), cancer (16–19) and diabetes (20–22).
The development and function of the myeloid lineage are dependent on miR-223, a microRNA that is conserved in vertebrates (23, 24). miR-223 is highly conserved in various organisms, its expression in myeloid lineage cells is characterized in humans and mice (25). miR-223 can be expressed in hematopoietic cells particularly in granulocytes and their precursors (26). During lineage commitment, miR-223 has been shown to inhibit the differentiation of progenitor cells into erythrocytes and facilitate their differentiation into granulocytes (27). It has been discovered by researchers that miR-223 is crucial in the regulation of inflammation in the lungs (28).
This review provides a comprehensive summary of the involvement of miR-223 in the inflammatory response across different cell types and its significance in pulmonary inflammatory diseases, aiming to enhance the understanding of clinical manifestations related to pulmonary inflammation.
2. The biogenesis, expression regulation and function of miR-223
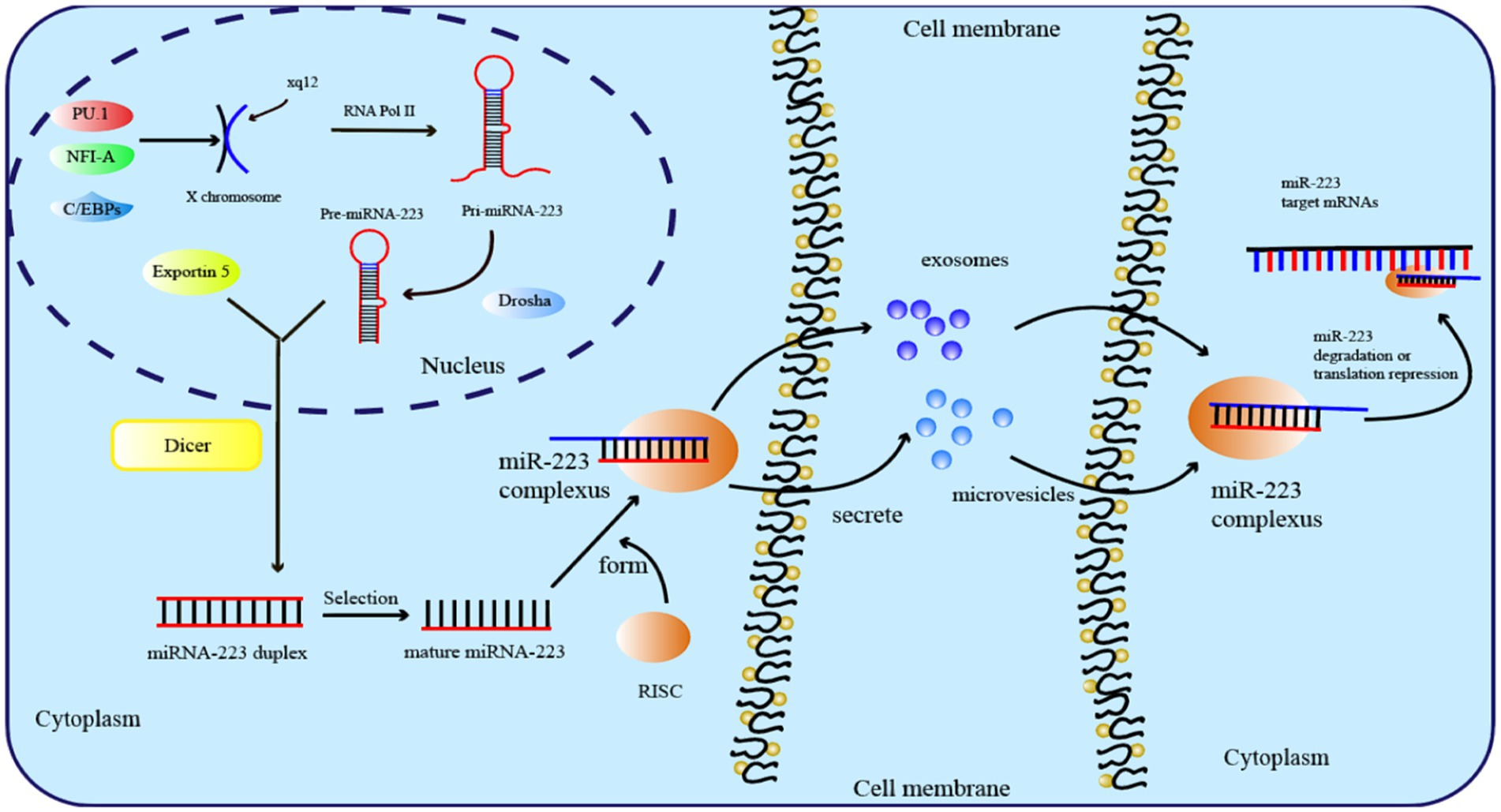
During evolution, miR-223 has been highly conserved and is located at the q12 site of the X chromosome, with a length of 22 nt (29). Before mature miR-223 can bind to the complementary sequences in the 3′ or 5′ untranslated region, a complex biosynthesis process is necessary, as is the case with all miRNAs (30). The transcription of miRNA genes into primary transcripts (pri-miRNA) occurs through RNA polymerase II in the nucleus (31). The primary transcript of miR-223 is known as pri-miRNA-223, which contains a hairpin structure located in the third exon and primarily generates the miR-223-3p strand. Then, RNA endonucleases (RNase) catalyze pri-miRNA-223 via two successive cleavage steps (32). In the first step, the RNase III-type endonuclease DROSHA cleaves pri-miRNA-223 transcripts and releases a short oligo nucleotide known as pre-miRNA (33). After being recognized by Exportin-5, pre-miRNA-223 is exported to the cytoplasm for further processing by the RNase III enzyme Dicer (34). Finally, the RNA-induced silencing complex (RISC) is formally formed as mature miRNA-223 binds to the Argonaute protein (Figure 1) (35).
Several transcription factors control the expression of miR-223. Early in 2007, PU.1 and C/EBPs known as myeloid transcription factors, were found to drive the transcription of miR-223 (36). Furthermore, upstream of pre-miR-223, miR-223 expression can be upregulated directly by PPAR-γ regulatory elements in macrophages derived from bone marrow (37). In contrast, Krüppel-like Factor 6 (KLF6) is a negative regulator of miR-223, and the overexpression of KLF6 has been proven to downregulate the expression level of miR-223 in macrophages and promote the polarization of the M1 phenotype (38). The amount of miR-223 produced is also regulated by epigenetics. For example, Fazi et al. reported that in patients with acute myeloid leukemias, the recruitment of chromatin remodeling enzymes by AML1/ETO to the AML1 binding site on the pre-miR-223 gene results in the induction of heterochromatic silencing of miR-223 (39).
Functionally, miRNAs silence genes by causing mRNA destabilization, degradation, and translational repression through binding to the complementary 3′-untranslated regions (3′ UTRs) of mRNA (40). Long et al. demonstrated that miR-223 targets NLRP3 by directly binding to its 3′UTR, resulting in the inhibition of NLRP3 expression and subsequently suppressing inflammasome activation and pyroptosis in endothelial cells infected with T. pallidum (41). In neuroblastoma (NB), miR-223 can bind to the 3′UTR of FOXO1, resulting in decreased expression of FOXO1, thus increasing the malignant ability of NB cells (42). In breast cancer, miR-223 can directly bind to the 3′UTR of the tumor suppressor gene FBXW7 and consequently facilitate the infiltration and spread of breast cancer cells (43).
3. The role of miR-223 in the inflammatory response
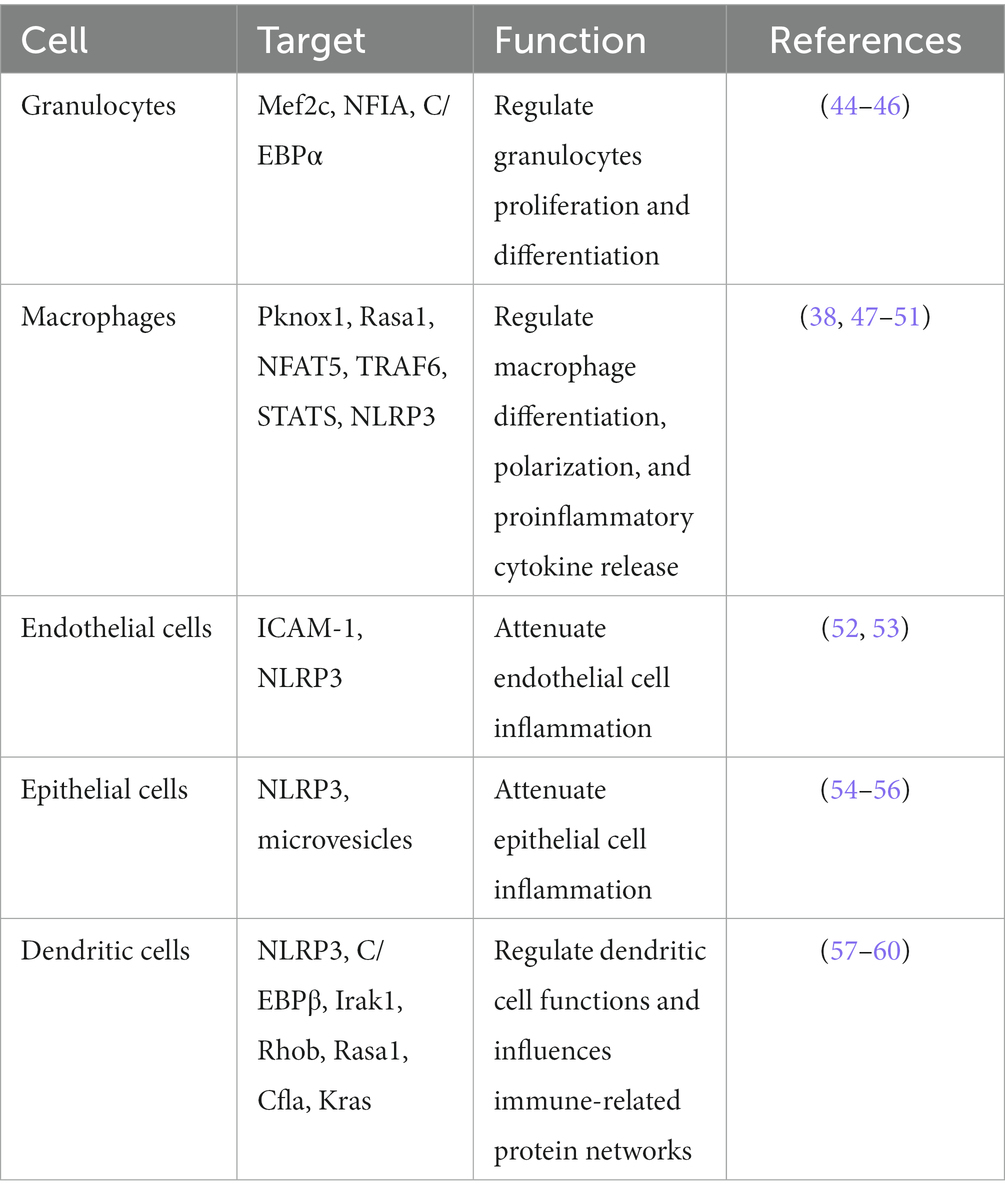
During the inflammatory response, miR-223 expression undergoes changes in various cell types. Modulations the level of miR-223 expression can modulate the diverse functions of various cells, thereby ameliorating or aggravating the resulting tissue inflammation. During inflammatory responses, by binding to specific targets, miR-223 can regulate the differentiation and proliferation of granulocytes, macrophages, and DCs, as well as the polarization of macrophages. In addition, through binding to specific genes, miR-223 can suppress proinflammatory cytokines or inflammatory signals within these cells. This section aims to give a summary of how miR-223 contributes to various biological processes in granulocytes, endothelial cells, epithelial cells, macrophages and dendritic cells (Table 1).
3.1. MiR-223’s involvement in the differentiation and activation of granulocytes
Granulocytes have three main granulocyte subsets: eosinophils, basophils and neutrophils. Within eosinophils, there exist sizable granules containing distinct eosinophilic proteins, such as eosinophil-derived neurotoxin, eosinophil peroxidase, major basic protein, and eosinophil cationic protein (61). The characteristic morphology of basophils is distinguished by the presence of large granules that are heavily stained and contain heparin, histamine sulfate, chondroitin, as well as other mediators like platelet-activating factor (PAF), slow-reacting kallikrein, neutrophil chemotactic factor (NCF). Under steady-state conditions, billions of neutrophils undergo apoptosis and are quickly replenished, as these cells have a short lifespan (62). During instances of inflammation or infections, emergency granulopoiesis is initiated, leading to a rise in the daily output of neutrophils (63). MiR-223 is highly expressed in the hematopoietic system and is a powerful regulator of granulopoiesis, with the highest expression in granulocytes. During the retinoic acid (RA)-induced granulocytic differentiation, inhibiting miR-223 expression reduces the efficiency of RA-induced differentiation of granulocyte-monocyte progenitors into monocytes (64, 65).
In 2008, Jonathan and his colleagues found that miR-223 can regulate granulocyte function and neutrophil progenitor cell proliferation by targeting myocyte enhancer Factor 2C (Mef2c) in mice. Hypermature and hypersensitive granulocytes, which display increased fungicidal activity upon activating stimuli, have been observed in mice lacking miR-223 (44). In the mouse model of allergic rhinitis (AR) inflammation, upregulation of miR-223 increased eosinophils granule protein expression, aggravated mucosal destruction and enhanced AR inflammation (66). When lung tissue injury occurs, miR-223 is rapidly upregulated in lung infiltrating granulocytes to attenuate injury during lung inflammation (67).
Studies have demonstrated that in the process of progenitor cell differentiation into granulocytes and monocytopoiesis, miR-223 is significantly upregulated by PU.1 and C/EBPβ known as myeloid transcription factors (45). Yang et al. utilized microarray profiling to assess the expression of miRNAs in neutrophils that were collected from patients suffering from severe traumatic injuries, they found that miR-223 participates in the traumatic pathogenesis of these patients (68). According to Vian’s research findings, the modulation of miR-223 activity has a significant influence on the differentiation and maturation of myeloid cell lines toward erythroid, granulocytic, monocytic, macrophagic lineages. Furthermore, overexpression of miR-223 promotes granulopoiesis and disrupts the differentiation of erythroid and monocytic macrophages (45). Other studies have also indicated that cytokines such as nuclear Factor I A (NFIA) can bind to the miR-223 gene promoter to repress its expression during the differentiation of granulocytes (46).
3.2. The function of miR-223 in regulating macrophage polarization
Macrophages are vital coordinators of immune activity and homeostasis. Based on temporal and environmental factors, macrophages can change their polarization direction to promote host immune defense mechanisms (69). In 2000, the nomenclature for macrophage polarization into M1 and M2 phenotypes was introduced (70). In vitro, cytokines have the ability to polarize macrophages. The classical activation of macrophages is initiated by LPS and IFN-γ, resulting in the development of a proinflammatory phenotype with pathogen-killing capabilities. These macrophages are commonly referred to M1 macrophages. Macrophages alternatively activated by IL-4 in vitro were named M2, which can promote cell proliferation and tissue repair (71). In the presence of pathological conditions, along with resident macrophages, additional macrophages are recruited to affected tissues and polarize into either M1 (proinflammatory) or M2 (anti-inflammatory) phenotypes.
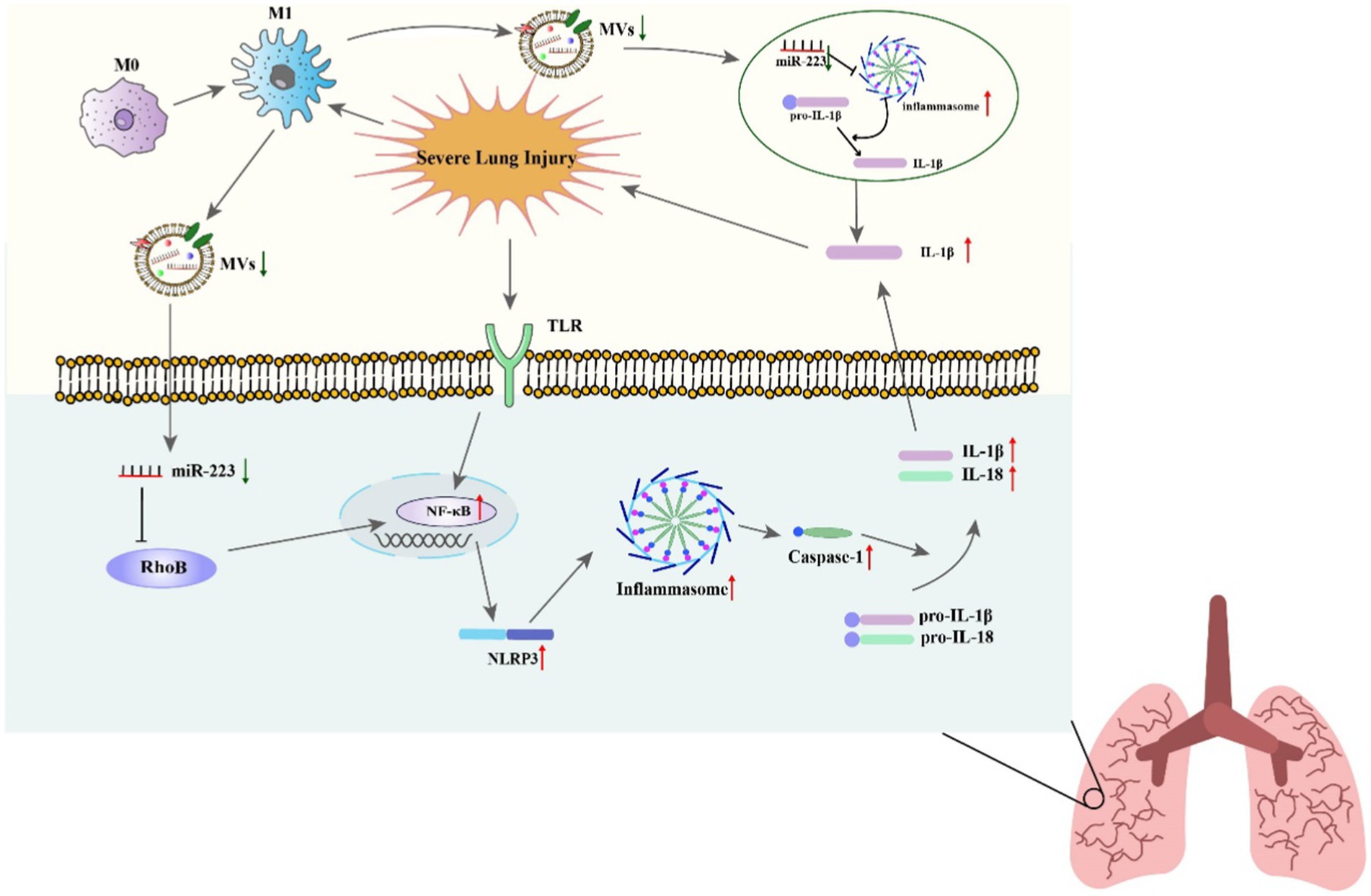
Studies have shown that miR-223 can promote the polarization of macrophages toward the M1 and M2 phenotypes under different conditions. On the one hand, the downregulation of miR-223 can promote classic inflammatory M1-type macrophage activation. Chen’s study found that downregulation of miR-223, through its binding to signal transducer and activator of transcription 3 (STAT3), increases the production of proinflammatory factors like IL-6, and IL-1β (72). Further research revealed that in macrophages, by targeting Ras homolog gene family member B (Rhob), the downregulation of miR-223 can enhance NF-κB signaling pathways and the mitogen-activated protein kinase (MAPK). This, in turn, induces the polarization of M1 proinflammatory macrophages and increases production of proinflammatory cytokines (73).
On the other hand, the upregulation of miR-223 can promote M2 (anti-inflammatory) phenotypes. Zhuang et al. discovered that in diet-induced adipose tissue inflammation, miR-223 can trigger macrophages to differentiate into an anti-inflammatory M2 phenotype to alleviate adipose tissue inflammation caused by a high-fat diet (74). Also, miR-223 expression was found to be significantly downregulated in heart tissues and heart-infiltrating macrophages in a mouse model of coxsackievirus B3 (CVB-3)-induced viral myocarditis. miR-223 directly targeting and inhibiting the expression of Pknox 1, thus to suppressed the expression of the M1 marker level and promoted the M2 marker level (47). Similarly, He and his colleagues found that miR-223 carried by exosomes can be taken in by macrophages and induce them to differentiate into M2 phenotype to accelerate wound healing by controlling Pknox1 gene expression. These studies revealed the role of the miR-223-Pknox1 axis in macrophage polarization (48). Wang et al. showed that miR-223 can mitigate sepsis by binding to the mRNA of NFAT5 and Rasa1, leading to IL-4-mediated differentiation of M2 macrophages (49). Moreover, a study on viral myocarditis (VMC) in mice discovered that long noncoding RNA maternally expressed gene 3 can facilitate miR-223 expression in macrophages, inhibiting M1 and promoting M2 macrophage polarization. Thus, upregulated miR-223 can inhibit TNF receptor-associated Factor 6 (TRAF 6) and suppress myocarditis and inflammation via NF-κB pathway inactivation in VMC mice (50). Additionally, Wang and his colleagues found that in macrophages, overexpression of miR-223 can reduce NF-κB activation by inhibiting IL-1 receptor-associated kinase-1 (IRAK-1), which leads to the polarization of M2 macrophages (75). In summary, at the molecular level, by targeting Pknox1, Rasa1, NFAT5, and TRAF6, upregulated miR-223 can induce polarization of macrophages toward the M2 anti-inflammatory phenotype during inflammation.
To summarize, miR-223 plays a crucial role in regulating the balance between M1 or M2 macrophage polarization and the development of inflammatory diseases.
3.3. The function of miR-223 in regulating inflammation in endothelial cells
The endothelium plays a critical role in maintaining multiorgan health and homeostasis. The healthy endothelium has a variety of biological functions, such as acting as a semi-permeable barrier, regulating the exchange and transport of substances, maintaining innate immunity, and balancing the production of vasodilators to regulate vascular tone, accelerating re-endothelialization to repair vascular injury and secreting antiplatelet and anticoagulant molecules to regulate hemostasis and regulating angiogenesis by producing factors (76). Lung endothelium undergoes continuous stretching during respiration and remains consistently exposed to external environmental substances. Impairment of the lung endothelium has been observed in various clinical conditions, such as acute respiratory distress syndrome (ARDS), COPD, pulmonary fibrosis, pneumonia, autoimmune disorders, pulmonary hypertension, and additional ailments (77). miR-223 plays an important role in endothelial cell inflammation and vascular endothelial injury.
On the one hand, miR-223 regulates endothelial cell inflammation. Upregulated miR-223 can be observed in peripheral microvesicles (MVs) in the plasma samples of nephritis, enteritis, hepatitis and atherosclerosis patients. miR-223 derived from platelets is conveyed to human umbilical vein endothelial cells (HUVECs) through peripheral MVs (78). Li and colleagues demonstrated that the transfection of miR-223 into HUVECs under TNF-α stimulation resulted in the inhibition of intercellular adhesion molecule-1 (ICAM-1) level. These findings suggest that miR-223 is a crucial factor in platelet-derived exosomes and is involved in inflammation response by impeding the phosphorylation of p38, JNK, and ERK, and hindering the nuclear translocation of NF-κB p65 (52). Another investigation demonstrated that platelets released microparticles (PMPs) containing functional miR-223 during sepsis can reduce the expression of ICAM-1 in endothelial cells, thereby potentially providing protection against excessive vascular inflammation induced by sepsis (79). In addition to the NF-κB and MAPK signaling pathways, the NLRP3 inflammasome pathway can also be regarded as a potential therapeutic target in vascular inflammation. The inhibition of NLRP3 inflammasome in endothelial cells through tree peony bark (Pae) leads to an increase in the expression of miR-223 in exosomes derived from the plasma of hyperlipidemic rats (53). What is more, Pae-exo increased the expression of miR-223 and decreased the expression of NLRP3, apoptosis-related spot-like protein (ASC), caspase-1 and ICAM-1 to relieve the endothelial dysfunction in atherosclerosis (AS) (80). Moreover, Zhang and his colleagues’ study on aortic damage caused by Se deficiency revealed that miR-223 is significantly downregulated, which leads to an increase in the expression of NLRP3 and the downstream targets, such as ASC, caspase-1, IL-18 and IL-1β (81).
On the other hand, miR-223 is a crucial regulator of vascular endothelial injury. Wang noted that miR-223 can reduce vascular endothelial injury by inhibiting the production of IL-6 and TNF-α during Kawasaki disease (KD) (82). Moreover, differential miR-223 expression in vascular endothelial cell (VEC) extracellular vesicles (EVs) can influence VEC generation and apoptosis. MiR-223-3p overexpression can reduce injury to mouse cardiac microvascular endothelial cells (MCMECs) and inhibit endothelial cell apoptosis in mice by regulating the expression of NLRP3 (83). In addition, Alexandre demonstrated for the first time that activation of neutrophils with various inflammatory stimuli induces the release of EVs that are internalized by endothelial cells, thus leading to the transfer of miR-223 and subsequent endothelial damage (84). These studies show that miR-223 can attenuate endothelial cell injuries.
3.4. Role of miR-223 in epithelial inflammation
Epithelial tissues constitute the physical barrier between our internal tissues, organs, etc. and the external environment exposes individuals to a wide range of inflammatory stimuli. The barrier is established through the action of stem cells that maintain a balance between self-renewal and differentiation to replenish lost cells and facilitate tissue repair in response to injury (85). Epithelial cells possess pattern recognition receptors that allow them to detect and react to various signals. These signals can guide the response of epithelial stem cells through signaling pathways that strengthen barrier function, signal nearby epithelial cells, attract immune cells, and promote tissue repair (86). The lung is a highly intricate organ consisting of different sections, which are covered by numerous specialized epithelial cells that have evolved to facilitate the exchange of gases necessary for the survival of air-breathing organisms on land. Under normal conditions, the epithelial cells in the lungs of adult mammals exhibit remarkable quiescence. Acute respiratory distress syndrome, coronavirus-induced acute lung injury, childhood interstitial lung diseases, cystic fibrosis, chronic obstructive pulmonary disease, and pulmonary fibrosis syndromes all involve, to some extent, dysfunction of the lung epithelium as a contributing factor (87).
miR-223 plays crucial functions in epithelial cells. Besides controlling the process of growth and morphology (88), the analysis of bioinformatic has shown that NLRP3 is a vital target of miR-223 in regulating epithelial cells. In renal tissue injury of mouse model induced by LPS, upregulated miR-223 can inhibit TXNIP and the NLRP3 inflammasome and attenuate LPS-induced injury in proximal tubule epithelial cells (54). In lung epithelial cells, upregulation of miR-223 by STIM1 can alleviate influenza A virus induced inflammatory injury in lung epithelial cells by deactivating NLRP3 and inflammasomes (55). Moreover, suppression of miR-223 in A549 cells induced by B(a)P resulted in elevated levels of TNF-α and IL-6 in the supernatant, as well as increased protein level of NLRP3, IL-1β, IL-18, caspase-1 (56). A recent study from Ren revealed that in animal models of dry eye (DE), miR-223 can suppress inflammation induced by hyperosmolarity in corneal epithelial cells by reducing the activation of NLRP3. The fact that miR-223 expression levels are inversely correlated with NLRP3 expression levels indicates that selectively increasing miR-223 expression could be a potential approach to alleviate chronic inflammation in DE (89). In addition to targeting NLRP3, in a model of pulmonary arterial hypertension (PAH), miR-223 can suppress the expression of the Integrin-β 3 subunit gene (ITGB3) to alleviate the progression of PAH and avoid the dysfunction of pulmonary arterial endothelial cells (90). What is more, by targeting to Rhob and deactivation of NF-κB gene activity in dairy cows, miR-223 attenuated LPS-induced inflammatory responses in mammary epithelial cells (91). Thus, through diverse signaling pathways, miR-223 reduces tissue damage by regulating the epithelial inflammation process.
3.5. The function of miR-223 in the differentiation of dendritic cells
DCs, which are antigen-presenting cells, have vital role in maintaining and inducing immunity as well as tolerance (92). DCs are bone marrow-derived cells arising from lympho-myeloid hematopoiesis that coordinate innate and adaptive immune responses (93). DCs exist in two functional states: immature and mature. Recognition of tissue homeostasis disturbances through damage-associated molecular patterns or pathogen-associated molecular patterns leads to DC maturation (94). DCs are critical innate immune cells at barrier sites, including the lung, playing a decisive role in initiation of adaptive immune responses against foreign material, infection, commensals or tissue damage (95). DCs are present in various regions of the lung tissue, positioned beneath the epithelial layer, ready to come into contact with foreign substances, infections, or tissue damage. In this capacity, they benefit from their capability to actively collect samples from the airways (96).
In process of hematopoietic stem cell (HSC) differentiation into DCs, the miR-223 expression level is altered in myeloid stem cells, HSCs and DCs, which proves that miR-223 takes part in DC differentiation (97). Bros et al. discovered that miR-223 has a negative regulatory effect on DCs activation in response to inflammatory. Upon DCs being stimulated with LPS, a significant decrease in miR-223 expression was observed. In response to glucocorticoids and anti-inflammatory cytokines, miR-223 expression is increased in DCs during the differentiation of bone marrow cells into BMDCs. Subsequently, the high level of miR-223 contributes to decreased Cfla, Kras and Rasa1 mRNA expression and influences immune protein regulatory networks (57). Tang’s research revealed that miR-223 binding to Rhob can also regulate DC differentiation and inhibit antigen uptake and presentation (98). In the autoimmune myocarditis (EAM) mouse model, Chen observed that miR-223 expression is markedly reduced in comparison to normal mice, whereas overexpressing miR-223 in DCs suppresses NLRP3 inflammasome production and facilitates the polarization of DCs into a tolerogenic phenotype (58). In mouse model of allogeneic heterotopic heart transplantation, overexpression of miR-223 in immature dendritic cells was found to result in longer graft survival and reduced infiltration of immune cells. The regulation of DC function by miR-223 through Irak1, Treg differentiation, and IL-10 secretion suppressed allogeneic heart graft rejection (59). In addition to EAM, miR-223 has been shown to regulate function and differentiation of DCs by targeting C/EBP-β in a mouse model of colitis. When miR-223 was deficient, monocytes produced more proinflammatory cytokines and gave rise to more monocyte-derived DCs upon stimulation (60). In conclusion, the aforementioned research demonstrates that miR-223 operates as a repressor of DC activation and sustains a state of maturation resistance that promotes a tolerogenic response during instances of inflammation.
4. The role of miR-223 in pulmonary inflammation
The alveoli constitute a delicate tissue structure, which makes the regulation of pulmonary inflammation as important as that of the respiratory tract. The extensive surface area of the lung is crucial for its vital role in gas exchange, and it also act as the primary barrier against the external environment. Due to its constant exposure to an airflow containing various types of proinflammatory stimuli, such as radioactive, infective, pro-allergenic or toxic agents, the lung plays a significant role in protecting the body from harmful environmental factors (99). Although the inflammatory response in the lung is thought to be a straightforward series of reactions, inflammation is a delicate matter of homeostasis and is tightly controlled. Excessive inflammation will lead to acute organ dysfunction and multiorgan failure or chronic tissue remodeling and destruction.
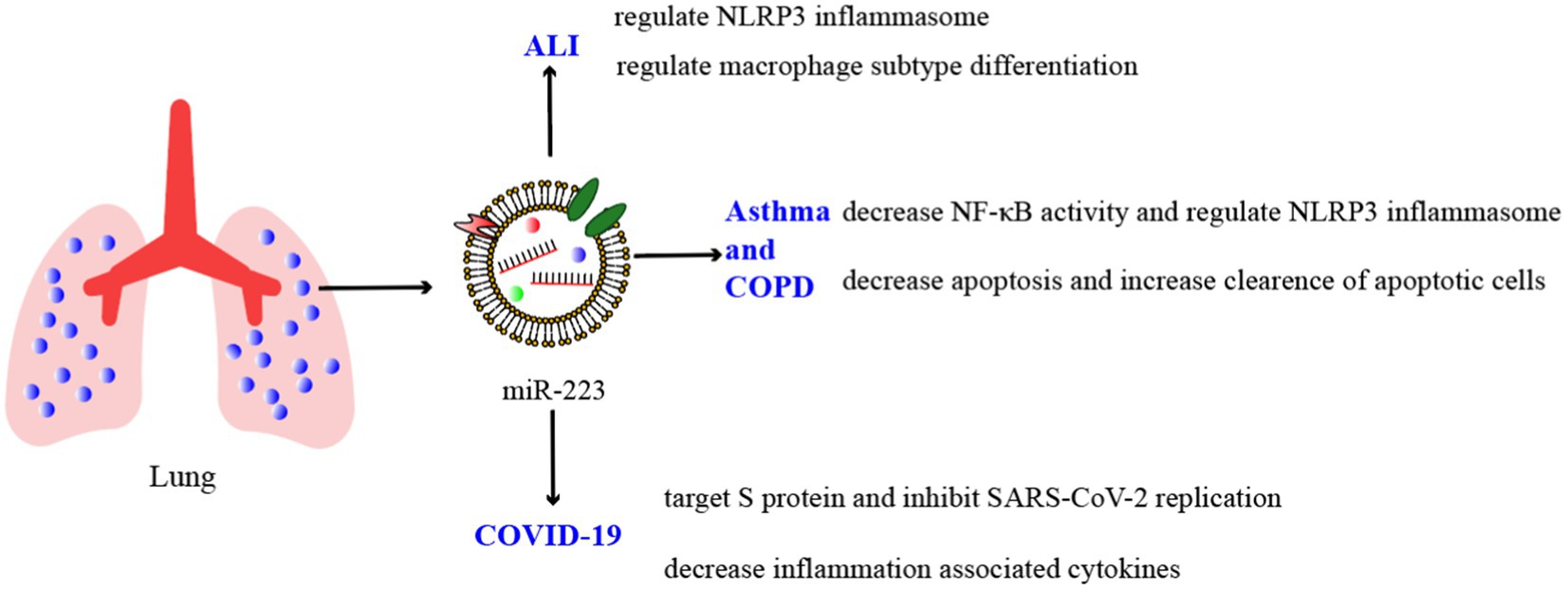
As we mentioned before, miRNAs are crucial regulators of various developmental and cellular processes. In certain pulmonary inflammatory conditions like pneumonia, COPD, and tuberculosis, miR-223 can target specific genes and suppress the synthesis of inflammatory mediators or hinder inflammation signaling pathways, thereby safeguarding the body from inflammatory damages (Figure 2; Table 2).
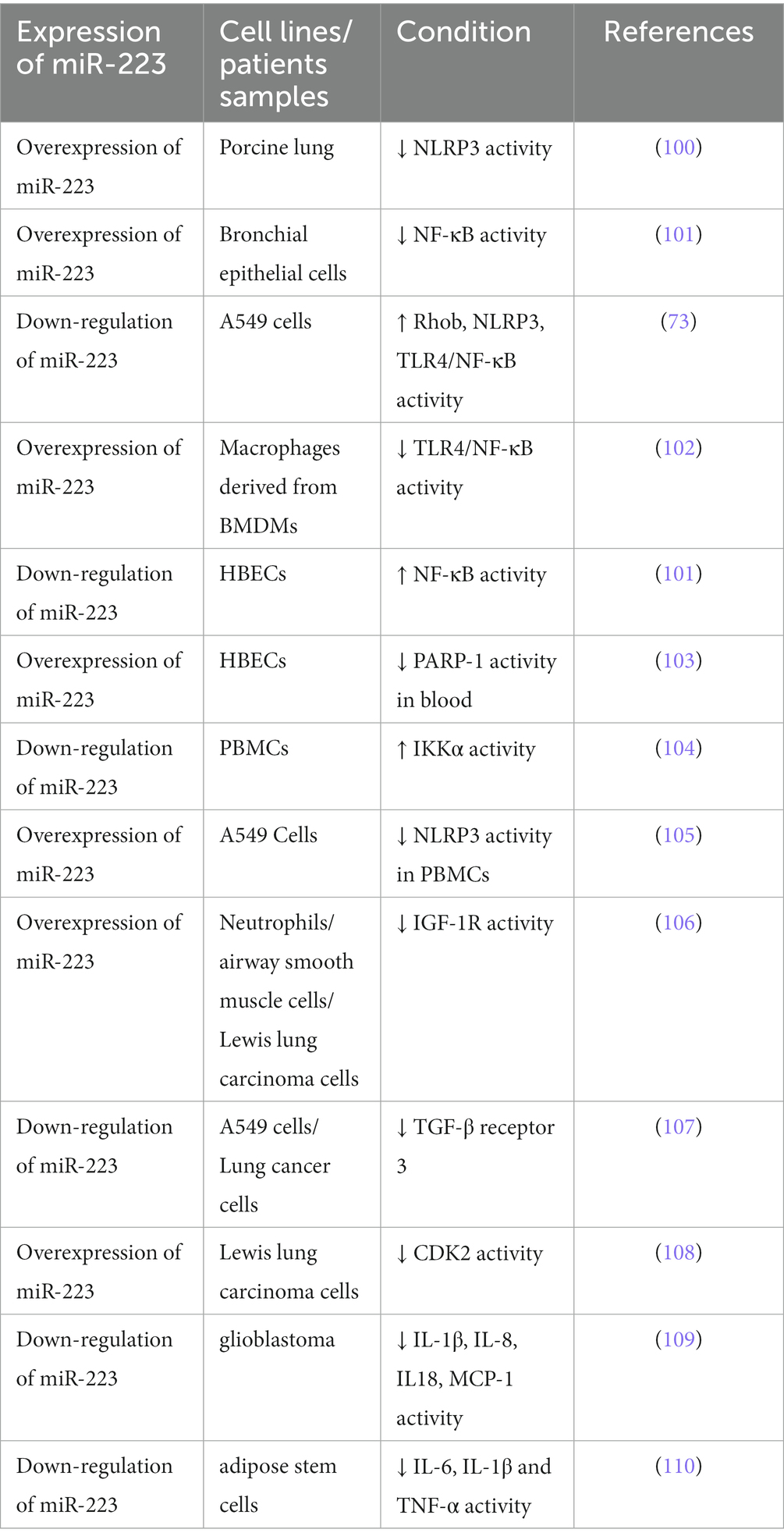
Table 2. miR-223 expression varies across different cell types and is influenced by various conditions.
4.1. Acute lung injury
ALI along with its severe form named ARDS, are both acute inflammatory lung conditions that can result in significant morbidity and mortality rates every year (111). ALI/ARDS can result from a range of lung injuries, including direct injuries like embolism, inhalation of toxic gases, lung contusion, and pneumonia, as well as indirect injuries such as major trauma, hemorrhagic shock, sepsis, drug overdose, or reperfusion injury (112, 113). Direct and indirect lung injuries exhibit comparable final pathophysiological features, such as compromised alveolar-capillary membrane function, reduced alveolar fluid clearance, and increased inflammation, which can lead to gas exchange difficulties and hypoxemia (114, 115). Although there have been extensive studies on ALI/ARDS, there are still no specific medical treatments. Therefore, the study of specific treatment strategies is urgently needed.
Studies have indicated that miR-223 can regulate the inflammatory response during ALI/ARDS (116). Reduced miR-223 expression has been demonstrated in in vitro experiments to diminish the NLRP3 inflammasome, the inhibition of RHOB and TLR4/NF-κB signaling pathway, leading to the exacerbation of lung injury (73). A further study found that miR-223 overexpression can alleviate LPS-induced ALI/ARDS in vivo by directly targeting NLRP3 (117, 118). Moreover, in macrophages, miR-223 can regulate subtype differentiation (73). miR-223 has been found to regulate macrophage differentiation by targeting the NLRP3 inflammasome, as shown in studies using a mouse model where lung inflammation is mediated by macrophages (28). In septic mice, miR-223 is an important regulator of the M2-type polarized target genes Nfat5 and Rasa1 mediated by IL-4 (49). What is more, treat with miR-223 from epithelium and endothelium derived exosomes can regulate the immune balance of alveolar macrophages (AMs) by targeting RGS1 mediated calcium signaling-dependent immune response (119). Furthermore, He’s study showed that miR-223 can also alleviates ALI by targeting STK39 in AMs (120). These results indicate that miR-223 can regulate the immune balance of AMs and can be used as potential therapeutic drugs for ALI/ARDS (Figure 3).
miR-223 is also an inhibitor of neutrophil development and function, and it can alleviate neutrophilic airway inflammation by inhibiting the release of IL-1β and the NLRP3 inflammasome. miR-223 can restrict the differentiation of Ly6G+ neutrophils derived from bone marrow and suppress the activity of the NLRP3 inflammasome and the production of IL-1β in ALI caused by mitochondrial damage-associated molecular patterns. However, the lack of miR-223 leads to continuous activation of NLRP3 and IL-1β (100). Moreover, neutrophil-derived microRNAs can modify mucosal gene expression during acute lung injury by transferring to pulmonary epithelial cells. In mouse neutrophils, miR-223 can be transferred to lung epithelial cells through MVs, where it inhibits poly (ADP-ribose) polymerase 1 (PARP-1) to impede acute lung injury. Therefore, overexpression of miR-223 in mice provides protection during acute lung injury (121). Additionally, Tan proved that miR-223 directly targeted high-mobility group box 2 (HMGB2) gene and the downregulation of miR-223 increased HMGB2 protein level, which activated the JNK signaling pathway and thus induced oxidative stress and autophagy in LPS-treated alveolar epithelial cells. Knockdown of HMGB2 protein deactivated the JNK signaling pathway and inhibited autophagy and oxidative stress in alveolar epithelial cells (122). In summary, miR-223 is regarded as a potential therapeutic target in ALI as it can regulate the process of inflammation in ALI.
4.2. Asthma and COPD
COPD and asthma represent the most frequently occurring chronic inflammatory respiratory conditions (123, 124). Asthma is a frequent inflammatory condition affecting the lower respiratory tract and is characterized by airway obstruction and hyperresponsiveness (124, 125) with symptoms such as wheezing, difficulty breathing, chest tightness, and coughing (126). COPD is a diverse disease marked by lung inflammation, rapid decline in lung function, chronic bronchitis, and damage to the alveolar tissue caused by pulmonary inflammation (124, 127). This persistent inflammation results in symptoms such as breathlessness, chronic cough, and wheezing (128). Asthma and COPD are among the diseases with the highest incidence and socioeconomic burden worldwide. Therefore, research on asthma and COPD is extremely urgent.
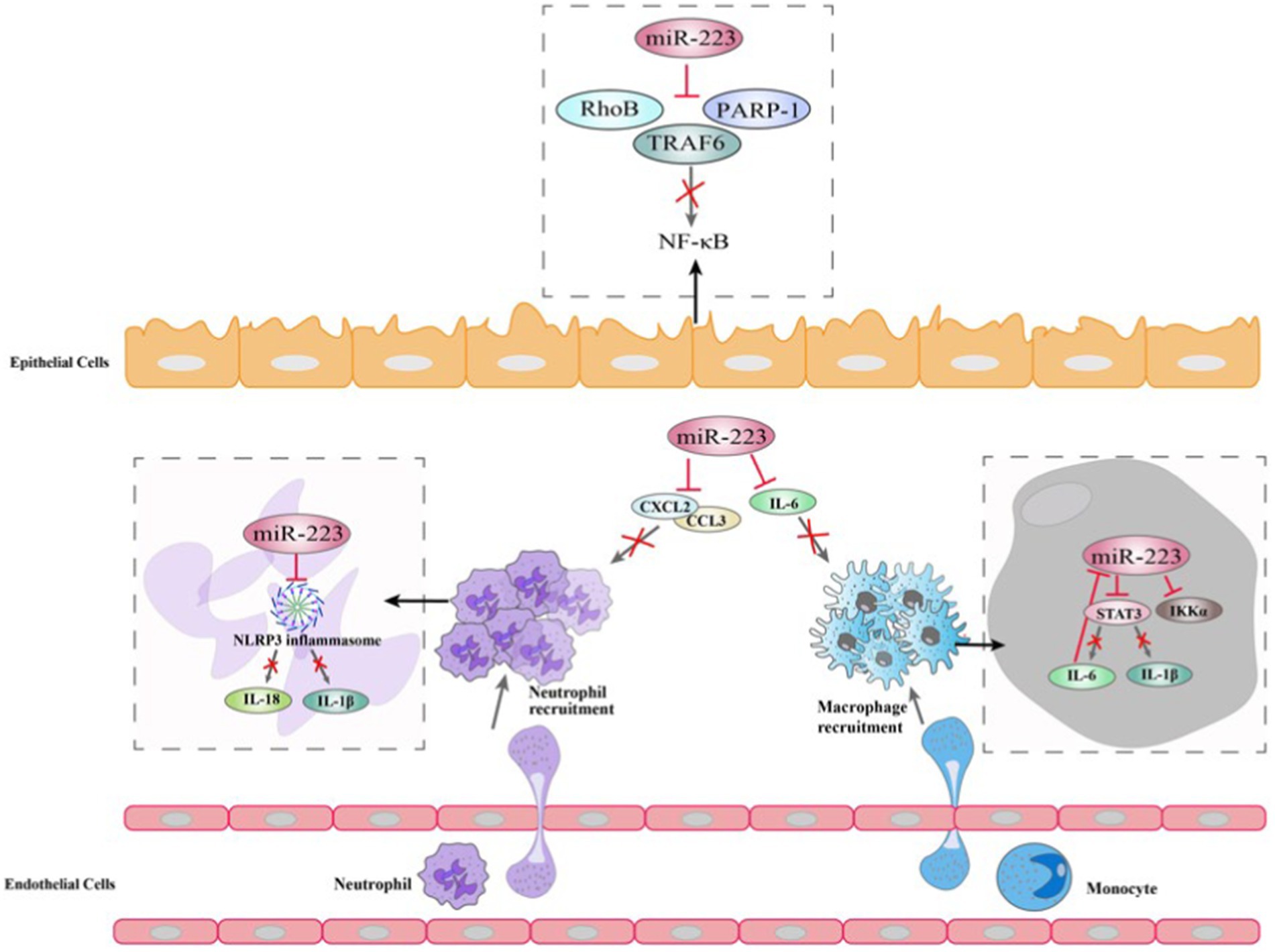
Studies have shown that miR-223 is involved in the processes of COPD and asthma. miRNA profiling shows that compared to healthy groups, miR-223 is more highly expressed in bronchial brushings (129) and induced sputum supernatant, the stimulated secretion of fluid from the respiratory tract containing various inflammatory cells, which quantitative cell count is the reference standard to reflect the airway inflammation of asthma patients (130, 131). Likewise, in neutrophilic asthma patients, the expression level of miR-223 is increased compared to that in healthy controls and eosinophilic asthmatic patients (131). Ezzie et al. analyzed the differential expression of miRNAs in lung tissues of smokers with or without COPD and found that miR-223 expression levels were higher in patients with COPD. A recent study showed that miR-223 had good combinatory predictive ability in differentiating between health and mild COPD. Furthermore, miR-223 was correlated with airway eosinophilia and were able to distinguish pure eosinophilic COPD from other airway inflammatory subtypes (132). In addition, Hirai found that miR-223 had lower expression levels in asthma-COPD overlap (ACO) patients and could discriminate between ACO patients and patients with either asthma or COPD (133). These findings highlight that miR-223 expression is distinctively regulated in obstructive lung diseases (134).
The NF-κB signaling pathway can be triggered by environmental stimuli in the bronchial biopsies of patients with COPD and asthma, resulting in proinflammatory reactions (101). Several studies suggest that elevated levels of miR-223 in human bronchial epithelial cells due to overexpression can decrease NF-κB activity by influencing the activation of NF-κB targets such as PARP-1 (103) and IκB kinase α (IKKα) (135). On the one hand, PARP-1 activation is observed in vitro in response to cigarette smoke and oxidative stress (136). miR-223 can repress PARP-1 to reverse the excessive inflammatory response (121). On the other hand, during human monocytes differentiate to macrophages, the increase in IKKα levels is correlated with a decrease in miR-223 expression (135). The levels of IKKα in peripheral blood mononuclear cells were found to be similar between asthma and COPD patients and healthy groups, according to some researchers. However, COPD patients and control smokers have higher p-IKKα levels than nonsmoking controls (104). A further study revealed that miR-223 can regulate the expression of mucin 5 AC (MUC5AC), eotaxin-2 (CCL24) and thymic stromal lymphopoietin (TSLP) by targeting NF-kB signaling pathway, this suggests that miR-223 is a regulator of allergic inflammation and could potentially consider as novel and targeted therapy for asthma (117) (Figure 4).
In addition to the inflammatory response, important characteristics of asthma and COPD also include cell dysfunction and death (137). Insulin-like growth factor-1 receptor (IGF-1R), known as the controller of cell proliferation, has been confirmed as a target of miR-223. Liu’s research confirmed that miR-223 targets the 3′UTR of IGF-1R, which subsequently led to the suppression of IGF-1R expression (106). IGF-1 can activate PI3K/Akt and mTOR to control apoptosis, survival, growth, and cell proliferation by binding with IGF-1R (138). Abnormal IGF-1 signaling has been found in asthma and COPD (139). Moreover, a reduction in serum IGF-1 levels has been observed in COPD patients experiencing acute exacerbation in comparison to healthy individuals and stable COPD patients. Conversely, IGF-1 mRNA levels in lung tissue are elevated in COPD patients when compared to healthy people (140). In addition to IGF-1R, miR-223 also targets Mef2c, which is a transcription factor that promotes the proliferation of myeloid progenitors to inhibit the proliferation of myeloid progenitor cells. Within the bronchial epithelial cells of individuals with severe asthma, the expression of Mef2c is decreased compared to that in healthy controls (141).
In addition to cell differentiation and proliferation, several vitro studies have explored the impact of miR-223 on cell survival, invasion, and apoptosis. The contribution of TGF-β and its receptors to airway remodeling in COPD and asthma is well-established, and miR-223 can target TGF-β receptor 3 to promote cell invasion and viability and reduce apoptosis (142). In the serum of COPD patients and in bronchial biopsies of asthma patients, the expression of TGF-β1 is increased (143), while the expression levels of TGF-β1 observed in both bronchial epithelial cells and alveolar macrophages of COPD patients are decreased compared to control groups (107). This may potentially be attributed to elevated expression levels of miR-223 in individuals suffering from asthma and COPD. Other research has also revealed that miR-223 participates in cell death by targeting p53 and cyclin-dependent kinase 2 (CDK2) to inhibit migration and proliferation (108). While p53 expression is typically low during homeostasis, it can be induced by oxidative stress in airway epithelial cells, such as those exposed to smoke, leading to cell apoptosis (144). Although miR-223 levels are increased in COPD patients, its validated target p53 expression levels are found to be higher in the lung tissue of COPD patients as compared to non-COPD controls, particularly in smokers with COPD (145). This indicates that miR-223 may not effectively reduce apoptosis by regulating p53 in COPD patients compared to non-COPD people. Moreover, MO’s study showed that the expression levels of miR-223 was reduced and the expression level of NLRP3 was increased by lncRNA GAS5, which subsequently promoted pyroptosis in COPD (146). In addition, Xu and his colleges found that overexpression of miR-223 via treatment with miR-223 agomirs attenuated airway inflammation, NLRP3 levels and IL-1β release. This revealed a crucial role for miR-223 in regulating the immune inflammatory responses by depressing the NLRP3/IL-1β axis in neutrophilic asthma (147).
In summary, miR-223 can target several genes, such as CDK2, TGF-β, IGF-1R, Mef2c and p53, to regulate cell viability, cell invasion and cell apoptosis during asthma and COPD.
4.3. COVID-19
An uncontrolled or excessive innate immune response in individuals with COVID-19 can result in the development of ARDS and a cytokine storm, which manifests as severe alveolar inflammation. Usually, COVID-19 causes mild respiratory symptoms, but it can also lead to severe complications. When COVID-19 infects macrophages, they present viral antigens to T cells, which produce cytokines associated with different kind of T-cell subtypes. The subsequent release of large quantities of cytokines amplifies the immune response, resulting in a cytokine storm. Massive secretion of chemokines and cytokines like TNF-β, MCP-1, IL-8, IL-21, IL-6, and IL-1 is promoted in cells in response to COVID-19 infection. The cytokines and chemokines then recruit leukocytes and lymphocytes to infection site. CD8 T cells are able to generate potent mediators that can effectively eliminate COVID-19, but the ongoing release of these mediators may result in viral persistence and have a detrimental impact on the activation of CD8 T cells (148).
When COVID-19 infects a host cell, its S protein binds to the dipeptidyl peptidase-4 receptor (DPP4R) and causes genomic RNA to appear in the host cell cytoplasm. TLR3 is then sensitized by dsRNA, activating the signaling cascades of nuclear factor-κB (NF-κB) and interferon-regulatory factors (IRFs), which produce interferons (IFNs) and proinflammatory cytokines. IFN-III, IFN-II, and IFN-I are induced by engagement of IFN receptors, which also activate different members of the signal transducer and activator of transcription families and Janus kinase (JAK), forming specific transcription factor complexes. At the same time, JAK1 and JAK2 are activated by IFN-II, resulting in the formation of a phosphorylated STAT1 homodimer (149).
Active peptide angiotensin II (Ang II), converted from the inactive decapeptide Ang I from the renin-angiotensin-system (RAS), can lead to chronic tissue injury, hypertension and vasoconstriction through the JAK–STAT signaling pathway. Furthermore, the RAS mediates the production of proinflammatory cytokines. Ang II increases the infiltration of immune cells, which leads to the local production of proinflammatory cytokines such as IL-1, IL-6, IFN-γ and TNF-α in target tissues (150). Moreover, some studies have shown that macrophage infiltration along with the apoptosis of epithelial cells and pneumocytes occurs in lung tissue (151). This may be mediated by MCP-1 through TGF-β and TNF-α, which leads to cytokine production.
Some miRNAs have been reported to be vital for virus entry into host cells and ACE2 level regulation (152). Furthermore, diverse studies have demonstrated that miRNAs suppress the replication and expression of the SARS-CoV-2 spike protein (153). Considering this, miRNAs can play a positive or negative role during virus-related processes in different ways: binding to host transcripts; binding to viral transcripts and direct binding to the viral genome (154). In humans, miRNAs have the ability to serve two functions, which include either enhancing the durability and propagation of viral RNA or strengthening the host’s anti-viral response. Due to their potential to regulate these responses, miRNAs are regarded as potential instruments for exploring the networks that regulate the immune response to both COVID-19 infection and vaccination (155).
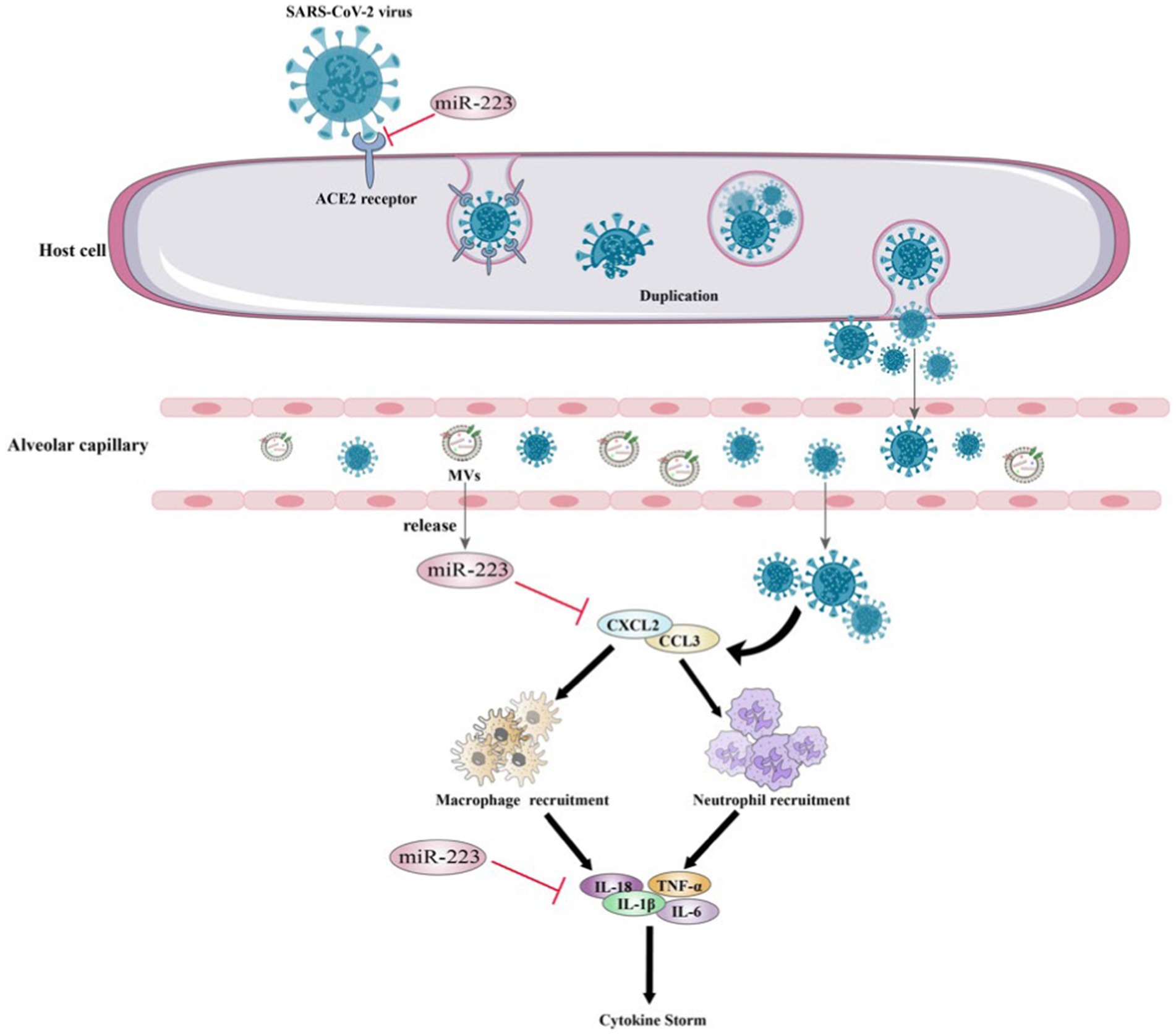
Wang et al. conducted a study and showed that miR-223-3p could directly target the S protein and inhibit the replication of SARS-CoV-2 (153). Which indicate that there is a correlation between low levels of circulating miR-223 and increased SARS-CoV-2 replication in elderly individuals and those with diabetes. Patients who are older and have comorbidities are at an increased risk of developing severe complications and experiencing higher mortality rates from COVID-19 as a result of infection with SARS-CoV-2.
In addition to targeting the S protein to inhibit SARS-CoV-2 replication, miR-223 may play a crucial role in cytokine storm regulation. During the COVID-19 pandemic, several studies have suggested a correlation between the cytokine storm in patients’ bodies and severe deterioration of their health (156). Ding et al. conducted an in vitro study on glioblastoma and found that treated with miR-223-3p mimic reduced the expression of several inflammation-associated cytokines, including IL-8, IL-18, IL-1β and MCP-1 resulting in inhibited cell proliferation and migration (109). Moreover, it has been demonstrated that miR-223-3p directly impacts the expression of inflammatory cytokines in adipose stem cells, which are known to be associated with cytokine storms (110) (Figure 5). These researches suggest that miR-223 can regulate cytokine levels in COVID-19 and thereby regulate the inflammatory process and disrupt the immune response.
4.4. Other lung inflammatory diseases
In addition to the diseases mentioned above, miR-223 also participates in the inflammatory response of other lung inflammatory diseases, including pulmonary tuberculosis (TB), sarcoidosis and pulmonary fibrosis (Table 3).
TB, as a typical infectious disease, is a chronic illness distinguished by continuous inflammation (164, 165). TB is caused by Mycobacterium tuberculosis (Mtb), which is accountable for high rates of mortality and morbidity across the globe (166). Dorhoi found that in the blood of patients with active pulmonary TB, miR-223 is abundantly expressed. TB-resistant mice that lack miR-223 expression exhibit high susceptibility to acute lung infection. miR-223 can reverse the mAb depletion of neutrophils by targeting the chemoattractants C-X-C motif chemokine ligand-2 (CXCL2), chemokine C-C motif chemokine ligand 3 (CCL3), and IL-6 to delete chemokine C-X-C motif receptor 2 (CXCR2) (157). Further research revealed that in plasma from patients with pulmonary TB, miR-223 is significantly downregulated. The variance in miR-223 expression levels may be linked to the diversity among the TB strains causing infection (158). As STAT1 has a significant impact in the immune defense against TB infection, the STAT1-related molecule miR-223 has been identified as a potential biomarker in the development of active TB (159). The above studies indicate the important role of miR-223 in the treatment of TB.
Sarcoidosis is a medical condition that is distinguished by the development of granulomas and an exaggerated immune response to unknown agents (167). To identify sarcoidosis biomarkers, Zhou et al. studied microRNA and protein-coding gene expression data of peripheral blood mononuclear cells (PBMCs) in healthy people and patients with noncomplex or complex sarcoidosis. They found that miR-223 may be associated with the severity of sarcoidosis (168). Neli et al. isolated the peripheral blood (PB) and bronchoalveolar (BAL) fluid of sarcoidosis patients for miRNA analysis. Through the analysis of PB’s and BAL’s global transcriptome, they recognize the alteration of miR-223 in TLR-2 and NF-κB apoptosis and proliferation signals. The expression of miR-223 is increased in regulatory T cells (Tregs) from patients with pulmonary sarcoidosis (160). Huppertz et al. recruited 19 healthy volunteers and 19 sarcoid patients, and demonstrated that the mRNA levels of miR-223 are decreased in BALF. And miR-223 KO mice can increase granuloma formation compared to wild-type (161). These data provide strong evidence that miR-223 is an essential part of the network in pulmonary sarcoidosis.
Pulmonary fibrosis is a chronic condition that is defined by the gradual loss of lung function (169). Most cases of fibrosis can be classified as idiopathic (170). Idiopathic pulmonary fibrosis is a lethal form of interstitial pneumonia that is characterized by the gradual scarring of lung tissue (171). Stachowiak et al. included 30 pediatric patients diagnosed with cystic fibrosis and collected their biologic material during pulmonary exacerbation. miRNA profiling showed that miR-223 is significantly altered during pulmonary exacerbation in sputum (162). In the pulmonary fibrosis rat model, Qu et al. found that the model group exhibited a significantly higher pulmonary inflammation score compared to the control group. Furthermore, the expression level of miR-223 decreased with increasing fibrosis (163). However, further research is needed to explore the specific role of miR-223 in pulmonary fibrosis.
5. The potential role of miR-223 in clinical treatment
Ever since miRNAs were first discovered in 1993, understanding the function of miRNAs in development and disease has rendered them appealing resources and objectives for innovative diagnosis and prognosis biomarkers, as well as therapeutic strategies.
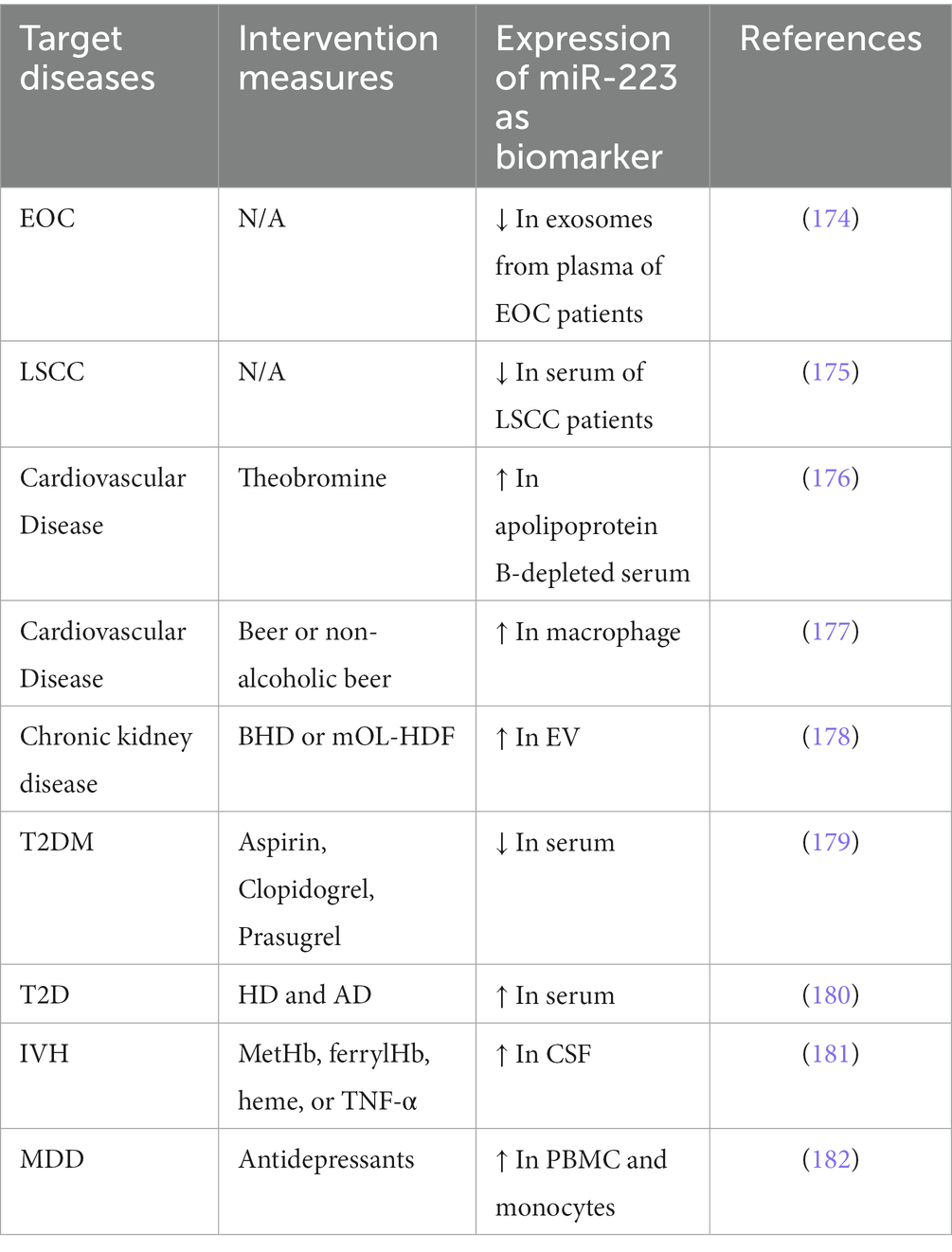
Due to the multiple functions of miRNAs in disease development, the disorder of one or a group of specific miRNAs may be closely related to human disease progression. Therefore, miRNAs are considered as potential diagnosis and prognosis biomarkers in disease. For example, miR-21, miR-20a, miR-103a, miR-106b, miR-143 and miR-215 et al. are considered as the biomarker in cancer (17), miR-155, miR-27a, miR-21, miR-146a, and miR-223 et al. are proved to play an essential role in regulating ALI/ARDS (172), miR-143, miR133, miR-145, miR-15, miR-126 et al. have been demonstrated to play a part in cardiovascular diseases (173). In recent years, the potential of miR-223 as diagnosis and prognosis biomarkers has also explored (Table 4). Pan et al. found that compared with healthy women, the levels of miR-223 are underrepresented in exosomes from plasma of epithelial ovarian cancer (EOC) patients, this indicates that miR-223 decrease its oncogenic potential in exosomes (174). Another clinical trial reveals that the expression level of miR-223 from the serum of laryngeal squamous cell carcinoma (LSCC) patients is down-regulated (175). In addition to cancer, the application of miR-223 has also been studied in cardiovascular disease. Charlotte et al. first investigates the postprandial responses of miR-223 levels. Their study shows that high-fat meal intake increases miR-223 levels (176). Lidia et al. show that miR-223 macrophage levels in non-smoker men with high cardiovascular risk are increased after beer and decreased after non-alcoholic beer consumption (177). In other diseases, Claudia et al. reveals that bicarbonate hemodialysis (BHD)-derived EV of chronic kidney disease patients had an increased expression of the proatherogenic miR-223 with respect to healthy subjects or mixed online hemodiafiltration (mOL-HDF). The switch from BHD to mOL-HDF significantly reduced systemic inflammation and miR-223 expression in plasma EV, thus improving HUVEC angiogenesis (178). In a double-blind randomized controlled trial of the effects on thrombotic markers and miRNA levels, the author found that compared with aspirin, during prasugrel-therapy of type 2 diabetes mellitus (T2DM), the expression of miR-223 decreased (179). A further study shows that miR-223 from plasma of high dairy and adequate dairy are upregulated. This indicates that miR-223 is related to T2D (180). In addition, an research analyzed miRNAs in cerebrospinal fluid (CSF) from intraventricular hemorrhage (IVH) infants showed that the levels of miR-223 were elevated in CSF after the onset of IVH (181). Hung and his colleges measured the levels of intracellular regulatory miRNAs in PBMCs and monocytes isolated from patients with major depressive disorder (MDD) before and after treatment with antidepressants. They found that the level of miR-223 was increased significantly in those who achieved remission. This suggested that miR-223 may act as biomarkers for remission during the treatment of MDD (182).
At present, clinical trials using miRNAs as therapeutic targets mainly focus on tumors and cardiovascular diseases. For cancer diseases, considering the complex interplay between miRNAs and target genes, the targets chosen for ongoing clinical trials typically exhibit confirmed interactions with multiple oncogenes or tumor suppressor genes. Moreover, these miRNAs function as Onco-miRs or Suppressor-miRs across various malignant tumors (6, 183), and several miRNA-targeted therapies are already in clinical development (184–187). For cardiovascular diseases, miRNAs can be used to treat dyslipidemia that increases the risk of cardiovascular disease, such as atherosclerosis (188). It is worth mentioning that, CDR132L, the first miR-132 inhibitor for heart failure has entered clinical trials which marks the entry of miRNA in the treatment of heart disease (189).
At present, strategies that target miR-223 in diseases treatment are mainly in animal models. For example, in the mouse model of KD, the excessive arterial damage could be rescued by administration of miR-223 mimics (190). In addition, in vivo and in vitro, miR-223 mimics were proved to protect against atherosclerosis by reducing the inflammation and the MEK1/ERK1/2 signaling pathway (191). A recent study also showed that miR-223 mimics could reduce the NLRP3 protein expression level and suppress chronic inflammation in DE (89). For radiation-induced heart disease (RIHD), miR-223 mimics could improve myocardial injury by activating adenosine monophosphate activated protein kinase (AMPK) (192). On the other hand, miR-223 inhibitors could cause hyper-activation of NLRP3 in the J774A.1 murine macrophages infected by subsp. zooepidemicus (SEZ) (193). What is more, miR-223 inhibitors can upregulate the expression of NLRP3 and increase the pyroptosis of fibroblast-like synoviocytes (FLSs) in gout arthritis (GA) rats (194). These examples indicate that in recent years, a considerable number of animal experiments have employed miR-223 mimics and inhibitors. This suggests that the development of miR-223 drugs is not impossible, and the future development and utilization of miR-223 clinical drugs will have significant implications.
As miR-223 plays an important role in pulmonary inflammations, it has great prospects in the clinical treatment of lung inflammations. Therefore, clinical trials of miR-223 as therapeutic targets in pulmonary inflammations are urgently needed.
6. Conclusions and future perspectives
In this review, we summarized the role of miR-223 during the inflammatory response in various cell types, as well as the role of miR-223 in some representative pulmonary inflammatory diseases, especially ALI, asthma, COPD and COVID-19. As we mentioned before, during tissue or cellular inflammation, the expression of miR-223, a multifunctional miRNA, is particularly elevated and regulated by several transcription factors. This miRNA is involved in regulating immune cell differentiation, polarization and proliferation, and plays a critical role in mediating cell-tissue, cell–cell, cell-tissue-inflammatory disease interactions. The molecular mechanisms of immune modulation by miRNAs can provide insights into the early diagnosis or treatment of inflammation.
miR-223, which has immunomodulatory effects in some tissues, can regulate immune cell polarization, proliferation and differentiation during pulmonary inflammation. It can also serve as a messenger of inflammation in the immune system. We can provide some insights into the early diagnosis or treatment of pulmonary inflammatory diseases by elucidating the molecular mechanisms of miR-223 in inflammation modulation. By establishing the regulatory network of miR-223, drugs targeted by miR-223 have better efficacy in treating pulmonary inflammatory diseases.
Since December 2019, COVID-19 has emerged as a global pandemic, posing serious public health threat. The symptoms of COVID-19 include fever, cough, myalgia, fatigue, or dyspnea, and elderly people and those with comorbidities are at higher risk of infection, which can lead to severe complications and high mortality rates. In this context, miR-223 has been recognized as a crucial controller of the lung inflammatory response, as it targets a variety of factors such as TLR4, CXCL2, PI3K/AKT, TRAF6, PARP-1, IFN-I, CCL3, STMN1, IKKα, IL-6, IL-1β, IL-18, NLRP3, Caspase-1 and NF-κB. These suggest that miR-223 may play a crucial role in COVID-19 pathogenesis. Since there is no authorized medication available to eliminate the virus, examining miR-223 as a possible target for therapy could provide novel approaches to manage COVID-19.
To sum up, the involvement of miR-223 in lung inflammation has been established, and the information about its targets provides valuable insights that may lead to the development of innovative therapeutic approaches for pulmonary inflammation pathogenesis.
Author contributions
HF and YZ initiated the project, made suggestions, and revised the article. MS searched the database and wrote the first draft of the manuscript. QL, ZD, SY, JL, MJ, and SH revised and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Scientific Research Translational Foundation of Wenzhou Safety (Emergency) Institute of Tianjin University (TJUWYY2022002 and TJUWYY2022011),Open Project of the Key Laboratory of Trauma and Orthopedics Research Medicine in Henan Province (HZKFKT20220504), National Natural Science Foundation of China (grant number 32000877), and Open Scientific Research Program of Military Logistics (BLB19J006 and BLB20J009).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Medzhitov, R. Origin and physiological roles of inflammation. Nature. (2008) 454:428–35. doi: 10.1038/nature07201
2. Asgharzade, S, Alizadeh, A, and Arab, S. Regenerative medicine approaches in Covid-19 pneumonia. Curr Stem Cell Res Ther. (2021) 16:647–55. doi: 10.2174/1574888X16999210112205826
3. Saliminejad, K, Khorram Khorshid, HR, Soleymani Fard, S, and Ghaffari, SH. An overview of Micrornas: biology, functions, therapeutics, and analysis methods. J Cell Physiol. (2019) 234:5451–65. doi: 10.1002/jcp.27486
4. Gebert, LFR, and MacRae, IJ. Regulation of Microrna function in animals. Nat Rev Mol Cell Biol. (2019) 20:21–37. doi: 10.1038/s41580-018-0045-7
5. Bartel, DP. Micrornas: target recognition and regulatory functions. Cells. (2009) 136:215–33. Epub 2009/01/27. doi: 10.1016/j.cell.2009.01.002
7. Paul, P, Chakraborty, A, Sarkar, D, Langthasa, M, Rahman, M, Bari, M, et al. Interplay between Mirnas and human diseases. J Cell Physiol. (2018) 233:2007–18. doi: 10.1002/jcp.25854
8. Ogulur, I, Pat, Y, Ardicli, O, Barletta, E, Cevhertas, L, Fernandez-Santamaria, R, et al. Advances and highlights in biomarkers of allergic diseases. Allergy. (2021) 76:3659–86. doi: 10.1111/all.15089
9. Weidner, J, Bartel, S, Kılıç, A, Zissler, UM, Renz, H, Schwarze, J, et al. Spotlight on Micrornas in allergy and asthma. Allergy. (2021) 76:1661–78. doi: 10.1111/all.14646
10. Snowhite, IV, Allende, G, Sosenko, J, Pastori, RL, Messinger Cayetano, S, and Pugliese, A. Association of Serum Micrornas with islet autoimmunity, disease progression and metabolic impairment in relatives at risk of type 1 diabetes. Diabetologia. (2017) 60:1409–22. doi: 10.1007/s00125-017-4294-3
11. Specjalski, K, and Jassem, E. Micrornas: potential biomarkers and targets of therapy in allergic diseases? Arch Immunol Ther Exp. (2019) 67:213–23. doi: 10.1007/s00005-019-00547-4
12. Pattarayan, D, Thimmulappa, RK, Ravikumar, V, and Rajasekaran, S. Diagnostic potential of extracellular Microrna in respiratory diseases. Clin Rev Allergy Immunol. (2018) 54:480–92. doi: 10.1007/s12016-016-8589-9
13. Barwari, T, Joshi, A, and Mayr, M. Micrornas in cardiovascular disease. J Am Coll Cardiol. (2016) 68:2577–84. doi: 10.1016/j.jacc.2016.09.945
14. Su, Q, and Lv, X. Revealing new landscape of cardiovascular disease through circular Rna-Mirna-Mrna Axis. Genomics. (2020) 112:1680–5. doi: 10.1016/j.ygeno.2019.10.006
15. Song, R, Hu, XQ, and Zhang, L. Mitochondrial Mirna in cardiovascular function and disease. Cells. (2019) 8:1475. doi: 10.3390/cells8121475
16. Hill, M, and Tran, N. Mirna interplay: mechanisms and consequences in Cancer. Dis Model Mech. (2021) 14:dmm047662. doi: 10.1242/dmm.047662
17. He, B, Zhao, Z, Cai, Q, Zhang, Y, Zhang, P, Shi, S, et al. Mirna-based biomarkers, therapies, and resistance in Cancer. Int J Biol Sci. (2020) 16:2628–47. doi: 10.7150/ijbs.47203
18. Ali Syeda, Z, Langden, SSS, Munkhzul, C, Lee, M, and Song, SJ. Regulatory mechanism of Microrna expression in Cancer. Int J Mol Sci. (2020) 21:1723. doi: 10.3390/ijms21051723
19. Hussen, BM, Hidayat, HJ, Salihi, A, Sabir, DK, Taheri, M, and Ghafouri-Fard, S. Microrna: a signature for Cancer progression. Biomed Pharmacother. (2021) 138:111528. doi: 10.1016/j.biopha.2021.111528
20. He, X, Kuang, G, Wu, Y, and Ou, C. Emerging roles of Exosomal Mirnas in diabetes mellitus. Clin Transl Med. (2021) 11:e468. doi: 10.1002/ctm2.468
21. Shi, Y, and Vanhoutte, PM. Macro- and microvascular endothelial dysfunction in diabetes. J Diabetes. (2017) 9:434–49. doi: 10.1111/1753-0407.12521
22. Vasu, S, Kumano, K, Darden, CM, Rahman, I, Lawrence, MC, and Naziruddin, B. Microrna signatures as future biomarkers for diagnosis of diabetes states. Cells. (2019) 8:1533. doi: 10.3390/cells8121533
23. Ye, D, Zhang, T, Lou, G, and Liu, Y. Role of Mir-223 in the pathophysiology of liver diseases. Exp Mol Med. (2018) 50:1–12. doi: 10.1038/s12276-018-0153-7
24. Roberto, VP, Tiago, DM, Gautvik, K, and Cancela, ML. Evidence for the conservation of Mir-223 in Zebrafish (Danio Rerio): implications for function. Gene. (2015) 566:54–62. doi: 10.1016/j.gene.2015.04.022
25. Baek, D, Villén, J, Shin, C, Camargo, FD, Gygi, SP, and Bartel, DP. The impact of Micrornas on protein output. Nature. (2008) 455:64–71. doi: 10.1038/nature07242
26. Chen, CZ, Li, L, Lodish, HF, and Bartel, DP. Micrornas modulate hematopoietic lineage differentiation. Science. (2004) 303:83–6. doi: 10.1126/science.1091903
27. Haneklaus, M, Gerlic, M, O'Neill, LA, and Masters, SL. Mir-223: infection, inflammation and Cancer. J Intern Med. (2013) 274:215–26. doi: 10.1111/joim.12099
28. Zhang, D, Lee, H, Wang, X, Groot, M, Sharma, L, Dela Cruz, CS, et al. A potential role of microvesicle-containing Mir-223/142 in lung inflammation. Thorax. (2019) 74:865–74. doi: 10.1136/thoraxjnl-2018-212994
29. Gu, J, Xu, H, Chen, Y, Li, N, and Hou, X. Mir-223 as a regulator and therapeutic target in liver diseases. Front Immunol. (2022) 13:860661. doi: 10.3389/fimmu.2022.860661
30. Chipman, LB, and Pasquinelli, AE. Mirna targeting: growing beyond the seed. Trends Genet. (2019) 35:215–22. doi: 10.1016/j.tig.2018.12.005
31. Michlewski, G, and Cáceres, JF. Post-transcriptional control of Mirna biogenesis. RNA. (2019) 25:1–16. doi: 10.1261/rna.068692.118
32. Pillai, RS. Microrna function: multiple mechanisms for a tiny Rna? RNA. (2005) 11:1753–61. doi: 10.1261/rna.2248605
33. Cho, SJ, Lee, M, Stout-Delgado, HW, and Moon, JS. Drosha-dependent Mirna and Aim2 Inflammasome activation in idiopathic pulmonary fibrosis. Int J Mol Sci. (2020) 21:1668. doi: 10.3390/ijms21051668
34. Yang, JS, and Lai, EC. Alternative Mirna biogenesis pathways and the interpretation of Core Mirna pathway mutants. Mol Cell. (2011) 43:892–903. doi: 10.1016/j.molcel.2011.07.024
35. Lui, PY, Jin, DY, and Stevenson, NJ. Microrna: master controllers of intracellular signaling pathways. Cell Mol Life Sci. (2015) 72:3531–42. doi: 10.1007/s00018-015-1940-0
36. Fukao, T, Fukuda, Y, Kiga, K, Sharif, J, Hino, K, Enomoto, Y, et al. An evolutionarily conserved mechanism for Microrna-223 expression revealed by Microrna gene profiling. Cells. (2007) 129:617–31. doi: 10.1016/j.cell.2007.02.048
37. Engin, AB. Microrna and Adipogenesis. Adv Exp Med Biol. (2017) 960:489–509. doi: 10.1007/978-3-319-48382-5_21
38. Date, D, Das, R, Narla, G, Simon, DI, Jain, MK, and Mahabeleshwar, GH. Kruppel-like transcription factor 6 regulates inflammatory macrophage polarization. J Biol Chem. (2014) 289:10318–29. doi: 10.1074/jbc.M113.526749
39. Fazi, F, Racanicchi, S, Zardo, G, Starnes, LM, Mancini, M, Travaglini, L, et al. Epigenetic silencing of the Myelopoiesis regulator Microrna-223 by the Aml1/Eto Oncoprotein. Cancer Cell. (2007) 12:457–66. doi: 10.1016/j.ccr.2007.09.020
40. Jonas, S, and Izaurralde, E. Towards a molecular understanding of Microrna-mediated gene silencing. Nat Rev Genet. (2015) 16:421–33. doi: 10.1038/nrg3965
41. Long, FQ, Kou, CX, Li, K, Wu, J, and Wang, QQ. Mir-223-3p inhibits Rtp17-induced Inflammasome activation and Pyroptosis by targeting Nlrp3. J Cell Mol Med. (2020) 24:14405–14. doi: 10.1111/jcmm.16061
42. Han, LL, Zhou, XJ, Li, FJ, Hao, XW, Jiang, Z, Dong, Q, et al. Mir-223-3p promotes the growth and invasion of Neuroblastoma cell via targeting Foxo1. Eur Rev Med Pharmacol Sci. (2019) 23:8984–90. doi: 10.26355/eurrev_201910_19298
43. Wang, Y, Shi, S, Wang, Y, Zhang, X, Liu, X, Li, J, et al. Mir-223-3p targets Fbxw7 to promote epithelial-Mesenchymal transition and metastasis in breast Cancer. Thorac Cancer. (2022) 13:474–82. doi: 10.1111/1759-7714.14284
44. Johnnidis, JB, Harris, MH, Wheeler, RT, Stehling-Sun, S, Lam, MH, Kirak, O, et al. Regulation of progenitor cell proliferation and granulocyte function by Microrna-223. Nature. (2008) 451:1125–9. doi: 10.1038/nature06607
45. Vian, L, Di Carlo, M, Pelosi, E, Fazi, F, Santoro, S, Cerio, AM, et al. Transcriptional fine-tuning of Microrna-223 levels directs lineage choice of human hematopoietic progenitors. Cell Death Differ. (2014) 21:290–301. doi: 10.1038/cdd.2013.145
46. O'Connell, RM. Micrornas function on a new level. Blood. (2012) 119:3875–6. doi: 10.1182/blood-2012-02-410977
47. Gou, W, Zhang, Z, Yang, C, and Li, Y. Mir-223/Pknox1 Axis protects mice from Cvb3-induced viral myocarditis by modulating macrophage polarization. Exp Cell Res. (2018) 366:41–8. doi: 10.1016/j.yexcr.2018.03.004
48. He, X, Dong, Z, Cao, Y, Wang, H, Liu, S, Liao, L, et al. Msc-derived exosome promotes M2 polarization and enhances cutaneous wound healing. Stem Cells Int. (2019) 2019:7132708. doi: 10.1155/2019/7132708
49. Wang, X, Zhang, H, Guo, R, Li, X, Liu, H, Wang, Z, et al. Microrna-223 modulates the Il-4-medicated macrophage M2-type polarization to control the Progress of Sepsis. Int Immunopharmacol. (2021) 96:107783. doi: 10.1016/j.intimp.2021.107783
50. Xue, YL, Zhang, SX, Zheng, CF, Li, YF, Zhang, LH, Su, QY, et al. Long non-coding Rna Meg3 inhibits M2 macrophage polarization by activating Traf6 via Microrna-223 Down-regulation in viral myocarditis. J Cell Mol Med. (2020) 24:12341–54. doi: 10.1111/jcmm.15720
51. Ying, W, Tseng, A, Chang, RC, Morin, A, Brehm, T, Triff, K, et al. Microrna-223 is a crucial mediator of Pparγ-regulated alternative macrophage activation. J Clin Invest. (2015) 125:4149–59. doi: 10.1172/JCI81656
52. Li, J, Tan, M, Xiang, Q, Zhou, Z, and Yan, H. Thrombin-activated platelet-derived Exosomes regulate endothelial cell expression of Icam-1 via Microrna-223 during the thrombosis-inflammation response. Thromb Res. (2017) 154:96–105. doi: 10.1016/j.thromres.2017.04.016
53. Zegeye, MM, Lindkvist, M, Fälker, K, Kumawat, AK, Paramel, G, Grenegård, M, et al. Activation of the Jak/Stat3 and Pi3k/Akt pathways are crucial for Il-6 trans-signaling-mediated pro-inflammatory response in human vascular endothelial cells. Cell Commun Signal. (2018) 16:55. doi: 10.1186/s12964-018-0268-4
54. Sun, Y, Liu, MW, Zhao, YH, Lu, YX, Wang, YA, and Tong, CW. Baicalin attenuates lipopolysaccharide-induced renal tubular epithelial cell injury by inhibiting the Txnip/Nlrp3 Signalling pathway via increasing Mir-223-3p expression. J Biol Regul Homeost Agents. (2020) 34:69–82. doi: 10.23812/19-502-A
55. Liu, CC, Miao, Y, Chen, RL, Zhang, YQ, Wu, H, Yang, SM, et al. Stim1 mediates Iav-induced inflammation of lung epithelial cells by regulating Nlrp3 and Inflammasome activation via targeting Mir-223. Life Sci. (2021) 266:118845. doi: 10.1016/j.lfs.2020.118845
56. Kong, X, Gao, M, Liu, Y, Zhang, P, Li, M, Ma, P, et al. Gsdmd-Mir-223-Nlrp3 Axis involved in B(a)P-induced inflammatory injury of alveolar epithelial cells. Ecotoxicol Environ Saf. (2022) 232:113286. doi: 10.1016/j.ecoenv.2022.113286
57. Bros, M, Youns, M, Kollek, V, Buchmüller, D, Bollmann, F, Seo, EJ, et al. Differentially Tolerized mouse antigen presenting cells share a common Mirna signature including enhanced Mmu-Mir-223-3p expression which is sufficient to imprint a Protolerogenic state. Front Pharmacol. (2018) 9:915. doi: 10.3389/fphar.2018.00915
58. Chen, L, Hou, X, Zhang, M, Zheng, Y, Zheng, X, Yang, Q, et al. Microrna-223-3p modulates dendritic cell function and ameliorates experimental autoimmune myocarditis by targeting the Nlrp3 Inflammasome. Mol Immunol. (2020) 117:73–83. doi: 10.1016/j.molimm.2019.10.027
59. Yuan, S, Chen, Y, Zhang, M, Wang, Z, Hu, Z, Ruan, Y, et al. Overexpression of Mir-223 promotes Tolerogenic properties of dendritic cells involved in heart transplantation tolerance by targeting Irak1. Front Immunol. (2021) 12:676337. doi: 10.3389/fimmu.2021.676337
60. Zhou, H, Xiao, J, Wu, N, Liu, C, Xu, J, Liu, F, et al. Microrna-223 regulates the differentiation and function of intestinal dendritic cells and macrophages by targeting C/Ebpβ. Cell Rep. (2015) 13:1149–60. doi: 10.1016/j.celrep.2015.09.073
61. Rosenberg, HF, Dyer, KD, and Foster, PS. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol. (2013) 13:9–22. doi: 10.1038/nri3341
62. Yvan-Charvet, L, and Ng, LG. Granulopoiesis and neutrophil homeostasis: a metabolic, Daily Balancing Act. Trends Immunol. (2019) 40:598–612. doi: 10.1016/j.it.2019.05.004
63. Manz, MG, and Boettcher, S. Emergency Granulopoiesis. Nat Rev Immunol. (2014) 14:302–14. doi: 10.1038/nri3660
64. Ge, Q, Brichard, S, Yi, X, and Li, Q. Micrornas as a new mechanism regulating adipose tissue inflammation in obesity and as a novel therapeutic strategy in the metabolic syndrome. J Immunol Res. (2014) 2014:987285. doi: 10.1155/2014/987285
65. Ramkissoon, SH, Mainwaring, LA, Ogasawara, Y, Keyvanfar, K, McCoy, JP Jr, Sloand, EM, et al. Hematopoietic-specific Microrna expression in human cells. Leuk Res. (2006) 30:643–7. doi: 10.1016/j.leukres.2005.09.001
66. Wu, S, Wang, Z, Zhu, Y, Zhu, X, Guo, L, Fu, Y, et al. Mir-223-3p regulates the eosinophil degranulation and enhances the inflammation in allergic rhinitis by targeting Fbxw7. Int Immunopharmacol. (2023) 118:110007. doi: 10.1016/j.intimp.2023.110007
67. Deny, M, Romano, M, Denis, O, Casimir, G, and Chamekh, M. Progressive control of Streptococcus Agalactiae-induced innate inflammatory response is associated with time course expression of Microrna-223 by neutrophils. Infect Immun. (2020) 88:20. doi: 10.1128/IAI.00563-20
68. Yang, J, Liang, Y, Han, H, and Qin, H. Identification of a Mirna signature in neutrophils after traumatic injury. Acta Biochim Biophys Sin. (2013) 45:938–45. doi: 10.1093/abbs/gmt100
69. Li, C, Xu, MM, Wang, K, Adler, AJ, Vella, AT, and Zhou, B. Macrophage polarization and Meta-inflammation. Transl Res. (2018) 191:29–44. doi: 10.1016/j.trsl.2017.10.004
70. Mills, CD, Kincaid, K, Alt, JM, Heilman, MJ, and Hill, AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. (2000) 164:6166–73. doi: 10.4049/jimmunol.164.12.6166
71. Italiani, P, and Boraschi, D. From monocytes to M1/M2 macrophages: Phenotypical Vs. Functional Differentiation. Front Immunol. (2014) 5:514. doi: 10.3389/fimmu.2014.00514
72. Chen, Q, Wang, H, Liu, Y, Song, Y, Lai, L, Han, Q, et al. Inducible Microrna-223 Down-regulation promotes Tlr-triggered Il-6 and Il-1β production in macrophages by targeting Stat3. PLoS One. (2012) 7:e42971. doi: 10.1371/journal.pone.0042971
73. Yan, Y, Lu, K, Ye, T, and Zhang, Z. Microrna-223 attenuates Lps-induced inflammation in an acute lung injury model via the Nlrp3 Inflammasome and Tlr4/Nf-Κb signaling pathway via Rhob. Int J Mol Med. (2019) 43:1467–77. doi: 10.3892/ijmm.2019.4075
74. Zhuang, G, Meng, C, Guo, X, Cheruku, PS, Shi, L, Xu, H, et al. A novel regulator of macrophage activation: Mir-223 in obesity-associated adipose tissue inflammation. Circulation. (2012) 125:2892–903. doi: 10.1161/CIRCULATIONAHA.111.087817
75. Wang, J, Wu, J, Cheng, Y, Jiang, Y, and Li, G. Over-expression of Microrna-223 inhibited the Proinflammatory responses in Helicobacter Pylori-infection macrophages by Down-regulating Irak-1. Am J Transl Res. (2016) 8:615–22.
76. Xu, S, Ilyas, I, Little, PJ, Li, H, Kamato, D, Zheng, X, et al. Endothelial dysfunction in atherosclerotic cardiovascular diseases and beyond: from mechanism to pharmacotherapies. Pharmacol Rev. (2021) 73:924–67. doi: 10.1124/pharmrev.120.000096
77. Goncharova, EA, Chan, SY, Ventetuolo, CE, Weissmann, N, Schermuly, RT, Mullin, CJ, et al. Update in pulmonary vascular diseases and right ventricular dysfunction 2019. Am J Respir Crit Care Med. (2020) 202:22–8. doi: 10.1164/rccm.202003-0576UP
78. Pan, Y, Liang, H, Liu, H, Li, D, Chen, X, Li, L, et al. Platelet-secreted Microrna-223 promotes endothelial cell apoptosis induced by advanced Glycation end products via targeting the insulin-like growth factor 1 receptor. J Immunol. (2014) 192:437–46. doi: 10.4049/jimmunol.1301790
79. Szilágyi, B, Fejes, Z, Rusznyák, Á, Fenyvesi, F, Pócsi, M, Halmi, S, et al. Platelet microparticles enriched in Mir-223 reduce Icam-1-dependent vascular inflammation in septic conditions. Front Physiol. (2021) 12:658524. doi: 10.3389/fphys.2021.658524
80. Shi, X, Xie, X, Sun, Y, He, H, Huang, H, Liu, Y, et al. Paeonol inhibits Nlrp3 mediated inflammation in rat endothelial cells by elevating Hyperlipidemic rats plasma Exosomal Mirna-223. Eur J Pharmacol. (2020) 885:173473. doi: 10.1016/j.ejphar.2020.173473
81. Qing, Z, Kaixin, Z, Yanfei, H, Yiming, Z, Hua, X, Ling, Z, et al. Microrna-223 triggers inflammation in porcine aorta by activating Nlrp3 Inflammasome under selenium deficiency. J Cell Physiol. (2021) 236:4555–64. doi: 10.1002/jcp.30178
82. Wang, X, Ding, YY, Chen, Y, Xu, QQ, Qian, GH, Qian, WG, et al. Mir-223-3p alleviates vascular endothelial injury by targeting Il6st in Kawasaki disease. Front Pediatr. (2019) 7:288. doi: 10.3389/fped.2019.00288
83. Deng, B, Hu, Y, Sheng, X, Zeng, H, and Huo, Y. Mir-223-3p reduces high glucose and high fat-induced endothelial cell injury in diabetic mice by regulating Nlrp3 expression. Exp Ther Med. (2020) 20:1514–20. doi: 10.3892/etm.2020.8864
84. Glémain, A, Néel, M, Néel, A, André-Grégoire, G, Gavard, J, Martinet, B, et al. Neutrophil-derived extracellular vesicles induce endothelial inflammation and damage through the transfer of Mirnas. J Autoimmun. (2022) 129:102826. doi: 10.1016/j.jaut.2022.102826
85. Blanpain, C, Horsley, V, and Fuchs, E. Epithelial stem cells: turning over new leaves. Cells. (2007) 128:445–58. Epub 2007/02/10. doi: 10.1016/j.cell.2007.01.014
86. Larsen, SB, Cowley, CJ, and Fuchs, E. Epithelial cells: liaisons of immunity. Curr Opin Immunol. (2020) 62:45–53. doi: 10.1016/j.coi.2019.11.004
87. Alysandratos, KD, Herriges, MJ, and Kotton, DN. Epithelial stem and progenitor cells in lung repair and regeneration. Annu Rev Physiol. (2021) 83:529–50. doi: 10.1146/annurev-physiol-041520-092904
88. Citron, F, Segatto, I, Vinciguerra, GLR, Musco, L, Russo, F, Mungo, G, et al. Downregulation of Mir-223 expression is an early event during mammary transformation and confers resistance to Cdk4/6 inhibitors in luminal breast Cancer. Cancer Res. (2020) 80:1064–77. doi: 10.1158/0008-5472.CAN-19-1793
89. Ren, Y, Feng, J, Lin, Y, Reinach, PS, Liu, Y, Xia, X, et al. Mir-223 inhibits Hyperosmolarity-induced inflammation through Downregulating Nlrp3 activation in human corneal epithelial cells and dry eye patients. Exp Eye Res. (2022) 220:109096. doi: 10.1016/j.exer.2022.109096
90. Liu, A, Liu, Y, Li, B, Yang, M, Liu, Y, and Su, J. Role of Mir-223-3p in pulmonary arterial hypertension via targeting Itgb3 in the Ecm pathway. Cell Prolif. (2019) 52:e12550. doi: 10.1111/cpr.12550
91. Jiao, P, Wang, J, Yang, J, Wang, X, and Luoreng, Z. Bta-Mir-223 targeting the Rhob gene in dairy cows attenuates Lps-induced inflammatory responses in mammary epithelial cells. Cells. (2022) 11:3144. doi: 10.3390/cells11193144
92. Cora, D, Re, A, Caselle, M, and Bussolino, F. Microrna-mediated regulatory circuits: outlook and perspectives. Phys Biol. (2017) 14:045001. doi: 10.1088/1478-3975/aa6f21
93. Collin, M, and Ginhoux, F. Human dendritic cells. Semin Cell Dev Biol. (2019) 86:1–2. doi: 10.1016/j.semcdb.2018.04.015
94. Patente, TA, Pinho, MP, Oliveira, AA, Evangelista, GCM, Bergami-Santos, PC, and Barbuto, JAM. Human dendritic cells: their heterogeneity and clinical application potential in Cancer immunotherapy. Front Immunol. (2018) 9:3176. doi: 10.3389/fimmu.2018.03176
95. Merad, M, Sathe, P, Helft, J, Miller, J, and Mortha, A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. (2013) 31:563–604. doi: 10.1146/annurev-immunol-020711-074950
96. Cook, PC, and MacDonald, AS. Dendritic cells in lung immunopathology. Semin Immunopathol. (2016) 38:449–60. doi: 10.1007/s00281-016-0571-3
97. Johanson, TM, Cmero, M, Wettenhall, J, Lew, AM, Zhan, Y, and Chong, MM. A Microrna expression atlas of mouse dendritic cell development. Immunol Cell Biol. (2015) 93:480–5. doi: 10.1038/icb.2014.109
98. Tang, HC, Lai, YY, Zheng, J, Jiang, HY, and Xu, G. Mir-223-3p inhibits antigen endocytosis and presentation and promotes the Tolerogenic potential of dendritic cells through targeting mannose receptor signaling and Rhob. J Immunol Res. (2020) 2020:1379458. doi: 10.1155/2020/1379458
99. Sittka, A, and Schmeck, B. Micrornas in the lung. Adv Exp Med Biol. (2013) 774:121–34. doi: 10.1007/978-94-007-5590-1_7
100. Feng, Z, Qi, S, Zhang, Y, Qi, Z, Yan, L, Zhou, J, et al. Ly6g+ neutrophil-derived Mir-223 inhibits the Nlrp3 Inflammasome in mitochondrial damp-induced acute lung injury. Cell Death Dis. (2017) 8:e3170. doi: 10.1038/cddis.2017.549
101. Zhou, W, Pal, AS, Hsu, AY, Gurol, T, Zhu, X, Wirbisky-Hershberger, SE, et al. Microrna-223 suppresses the canonical Nf-Κb pathway in basal keratinocytes to dampen Neutrophilic inflammation. Cell Rep. (2018) 22:1810–23. doi: 10.1016/j.celrep.2018.01.058
102. Qu, H, Liu, R, Chen, J, Zheng, L, and Chen, R. Aerobic exercise inhibits Cums-depressed mice hippocampal inflammatory response via activating hippocampal Mir-223/Tlr4/Myd88-Nf-Κb pathway. Int J Environ Res Public Health. (2020) 17:676. doi: 10.3390/ijerph17082676
103. Sethi, GS, Dharwal, V, and Naura, AS. Poly(Adp-ribose)Polymerase-1 in lung inflammatory disorders: a review. Front Immunol. (2017) 8:1172. doi: 10.3389/fimmu.2017.01172
104. Gagliardo, R, Chanez, P, Profita, M, Bonanno, A, Albano, GD, Montalbano, AM, et al. Iκb kinase-driven nuclear factor-Κb activation in patients with asthma and chronic obstructive pulmonary disease. J Allergy Clin Immunol. (2011) 128:635–45.e1-2. doi: 10.1016/j.jaci.2011.03.045
105. Wang, H, Lv, C, Wang, S, Ying, H, Weng, Y, and Yu, W. Nlrp3 Inflammasome involves in the acute exacerbation of patients with chronic obstructive pulmonary disease. Inflammation. (2018) 41:1321–33. doi: 10.1007/s10753-018-0780-0
106. Liu, D, Pan, J, Zhao, D, and Liu, F. Microrna-223 inhibits deposition of the extracellular matrix by airway smooth muscle cells through targeting Igf-1r in the Pi3k/Akt pathway. Am J Transl Res. (2018) 10:744–52.
107. Mak, JC, Chan-Yeung, MM, Ho, SP, Chan, KS, Choo, K, Yee, KS, et al. Elevated plasma Tgf-Beta1 levels in patients with chronic obstructive pulmonary disease. Respir Med. (2009) 103:1083–9. doi: 10.1016/j.rmed.2009.01.005
108. Gouda, MM, Shaikh, SB, Chengappa, D, Kandhal, I, Shetty, A, and Bhandary, Y. Changes in the expression level of Il-17a and P53-Fibrinolytic system in smokers with or without Copd. Mol Biol Rep. (2018) 45:2835–41. doi: 10.1007/s11033-018-4398-y
109. Zhuang, W, Niu, T, and Li, Z. Microrna Mir-145-5p regulates cell proliferation and cell migration in Colon Cancer by inhibiting chemokine (C-X-C motif) ligand 1 and integrin Α2. Bioengineered. (2021) 12:9909–17. doi: 10.1080/21655979.2021.2000243
110. Wu, J, Niu, P, Zhao, Y, Cheng, Y, Chen, W, Lin, L, et al. Impact of Mir-223-3p and Mir-2909 on inflammatory factors Il-6, Il-1ß, and Tnf-Α, and the Tlr4/Tlr2/Nf-Κb/Stat3 signaling pathway induced by lipopolysaccharide in human adipose stem cells. PLoS One. (2019) 14:e0212063. doi: 10.1371/journal.pone.0212063
111. Mart, MF, and Ware, LB. The Long-lasting effects of the acute respiratory distress syndrome. Expert Rev Respir Med. (2020) 14:577–86. doi: 10.1080/17476348.2020.1743182
112. Matthay, MA, Zemans, RL, Zimmerman, GA, Arabi, YM, Beitler, JR, Mercat, A, et al. Acute respiratory distress syndrome. Nat Rev Dis Primers. (2019) 5:18. doi: 10.1038/s41572-019-0069-0
113. Malarkkan, N, Snook, NJ, and Lumb, AB. New aspects of ventilation in acute lung injury. Anaesthesia. (2003) 58:647–67. doi: 10.1046/j.1365-2044.2003.03201.x
114. Kaku, S, Nguyen, CD, Htet, NN, Tutera, D, Barr, J, Paintal, HS, et al. Acute respiratory distress syndrome: etiology, pathogenesis, and summary on management. J Intensive Care Med. (2020) 35:723–37. doi: 10.1177/0885066619855021
115. Xu, B, Chen, SS, Liu, MZ, Gan, CX, Li, JQ, and Guo, GH. Stem cell derived Exosomes-based therapy for acute lung injury and acute respiratory distress syndrome: a novel therapeutic strategy. Life Sci. (2020) 254:117766. doi: 10.1016/j.lfs.2020.117766
116. Haneklaus, M, O'Neill, LA, and Coll, RC. Modulatory mechanisms controlling the Nlrp3 Inflammasome in inflammation: recent developments. Curr Opin Immunol. (2013) 25:40–5. doi: 10.1016/j.coi.2012.12.004
117. Langwiński, W, Szczepankiewicz, D, Narożna, B, Stegmayr, J, Wagner, D, Alsafadi, H, et al. Allergic inflammation in lungs and nasal epithelium of rat model is regulated by tissue-specific Mirna expression. Mol Immunol. (2022) 147:115–25. doi: 10.1016/j.molimm.2022.04.017
118. Fu, Z, Wu, X, Zheng, F, and Zhang, Y. Sevoflurane anesthesia ameliorates Lps-induced acute lung injury (Ali) by modulating a novel Lncrna Linc00839/Mir-223/Nlrp3 Axis. BMC Pulm Med. (2022) 22:159. doi: 10.1186/s12890-022-01957-5
119. Feng, Z, Zhou, J, Liu, Y, Xia, R, Li, Q, Yan, L, et al. Epithelium- and endothelium-derived Exosomes regulate the alveolar macrophages by targeting Rgs1 mediated calcium signaling-dependent immune response. Cell Death Differ. (2021) 28:2238–56. doi: 10.1038/s41418-021-00750-x
120. He, N, Tan, H, Deng, X, Shu, L, Qing, B, and Liang, H. Mir-223-3p-loaded Exosomes from Bronchoalveolar lavage fluid promote alveolar macrophage autophagy and reduce acute lung injury by inhibiting the expression of Stk39. Hum Cell. (2022) 35:1736–51. doi: 10.1007/s13577-022-00762-w
121. Neudecker, V, Brodsky, KS, Clambey, ET, Schmidt, EP, Packard, TA, Davenport, B, et al. Neutrophil transfer of Mir-223 to lung epithelial cells dampens acute lung injury in mice. Sci Transl Med. (2017) 9:5360. doi: 10.1126/scitranslmed.aah5360
122. Tan, HY, Qing, B, Luo, XM, and Liang, HX. Downregulation of Mir-223 promotes Hmgb2 expression and induces oxidative stress to activate Jnk and promote autophagy in an in vitro model of acute lung injury. J Inflamm. (2021) 18:29. doi: 10.1186/s12950-021-00295-3
123. Lozano, R, Naghavi, M, Foreman, K, Lim, S, Shibuya, K, Aboyans, V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study 2010. Lancet. (2012) 380:2095–128. doi: 10.1016/s0140-6736(12)61728-0
124. Roffel, MP, Bracke, KR, Heijink, IH, and Maes, T. Mir-223: a key regulator in the innate immune response in asthma and Copd. Front Med. (2020) 7:196. doi: 10.3389/fmed.2020.00196
125. Cevhertas, L, Ogulur, I, Maurer, DJ, Burla, D, Ding, M, Jansen, K, et al. Advances and recent developments in asthma in 2020. Allergy. (2020) 75:3124–46. doi: 10.1111/all.14607
126. Heaney, LG, and McGarvey, LP. Personalised medicine for asthma and chronic obstructive pulmonary disease. Respir Int Rev Thorac Dis. (2017) 93:153–61. doi: 10.1159/000455395
127. Osei, ET, Florez-Sampedro, L, Timens, W, Postma, DS, Heijink, IH, and Brandsma, CA. Unravelling the complexity of Copd by Micrornas: It's a small world after all. Eur Respir J. (2015) 46:807–18. doi: 10.1183/13993003.02139-2014
128. Postma, DS, and Rabe, KF. The asthma-Copd overlap syndrome. N Engl J Med. (2015) 373:1241–9. doi: 10.1056/NEJMra1411863
129. Solberg, OD, Ostrin, EJ, Love, MI, Peng, JC, Bhakta, NR, Hou, L, et al. Airway epithelial Mirna expression is altered in asthma. Am J Respir Crit Care Med. (2012) 186:965–74. doi: 10.1164/rccm.201201-0027OC
130. Eguiluz-Gracia, I, Tay, TR, Hew, M, Escribese, MM, Barber, D, O'Hehir, RE, et al. Recent developments and highlights in biomarkers in allergic diseases and asthma. Allergy. (2018) 73:2290–305. doi: 10.1111/all.13628
131. Maes, T, Cobos, FA, Schleich, F, Sorbello, V, Henket, M, De Preter, K, et al. Asthma inflammatory phenotypes show differential Microrna expression in sputum. J Allergy Clin Immunol. (2016) 137:1433–46. doi: 10.1016/j.jaci.2016.02.018
132. Burke, H, Cellura, D, Freeman, A, Hicks, A, Ostridge, K, Watson, A, et al. Pulmonary Ev Mirna profiles identify disease and distinct inflammatory Endotypes in Copd. Front Med. (2022) 9:1039702. doi: 10.3389/fmed.2022.1039702
133. Hirai, K, Shirai, T, Shimoshikiryo, T, Ueda, M, Gon, Y, Maruoka, S, et al. Circulating Microrna-15b-5p as a biomarker for asthma-Copd overlap. Allergy. (2021) 76:766–74. doi: 10.1111/all.14520
134. Ezzie, ME, Crawford, M, Cho, JH, Orellana, R, Zhang, S, Gelinas, R, et al. Gene expression networks in Copd: Microrna and Mrna regulation. Thorax. (2012) 67:122–31. doi: 10.1136/thoraxjnl-2011-200089
135. Li, T, Morgan, MJ, Choksi, S, Zhang, Y, Kim, YS, and Liu, ZG. Micrornas modulate the noncanonical transcription factor Nf-Kappab pathway by regulating expression of the kinase Ikkalpha during macrophage differentiation. Nat Immunol. (2010) 11:799–805. doi: 10.1038/ni.1918
136. Hwang, JW, Chung, S, Sundar, IK, Yao, H, Arunachalam, G, McBurney, MW, et al. Cigarette smoke-induced autophagy is regulated by Sirt1-Parp-1-dependent mechanism: implication in pathogenesis of Copd. Arch Biochem Biophys. (2010) 500:203–9. doi: 10.1016/j.abb.2010.05.013
137. Sauler, M, Bazan, IS, and Lee, PJ. Cell death in the lung: the apoptosis-Necroptosis Axis. Annu Rev Physiol. (2019) 81:375–402. doi: 10.1146/annurev-physiol-020518-114320
138. Lu, TX, Lim, EJ, Besse, JA, Itskovich, S, Plassard, AJ, Fulkerson, PC, et al. Mir-223 deficiency increases eosinophil progenitor proliferation. J Immunol. (2013) 190:1576–82. doi: 10.4049/jimmunol.1202897
139. Wang, Z, Li, W, Guo, Q, Wang, Y, Ma, L, and Zhang, X. Insulin-like growth Factor-1 signaling in lung development and inflammatory lung diseases. Biomed Res Int. (2018) 2018:6057589. doi: 10.1155/2018/6057589
140. Ye, M, Yu, H, Yu, W, Zhang, G, Xiao, L, Zheng, X, et al. Evaluation of the significance of circulating insulin-like growth Factor-1 and C-reactive protein in patients with chronic obstructive pulmonary disease. J Int Med Res. (2012) 40:1025–35. doi: 10.1177/147323001204000321
141. Sheu, CC, Tsai, MJ, Chen, FW, Chang, KF, Chang, WA, Chong, IW, et al. Identification of novel genetic regulations associated with airway epithelial homeostasis using next-generation sequencing data and bioinformatics approaches. Oncotarget. (2017) 8:82674–88. doi: 10.18632/oncotarget.19752
142. Huang, L, Li, F, Deng, P, and Hu, C. [article withdrawn] Microrna-223 promotes tumor progression in lung Cancer A549 cells via activation of the Nf-Κb signaling pathway. Oncol Res. (2016) 24:405–13. doi: 10.3727/096504016X14685034103437
143. Saito, A, Horie, M, and Nagase, T. Tgf-Β signaling in lung health and disease. Int J Mol Sci. (2018) 19:2460. doi: 10.3390/ijms19082460
144. van der Toorn, M, Rezayat, D, Kauffman, HF, Bakker, SJ, Gans, RO, Koëter, GH, et al. Lipid-soluble components in cigarette smoke induce mitochondrial production of reactive oxygen species in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. (2009) 297:L109–14. doi: 10.1152/ajplung.90461.2008
145. Mizuno, S, Bogaard, HJ, Gomez-Arroyo, J, Alhussaini, A, Kraskauskas, D, Cool, CD, et al. Microrna-199a-5p is associated with hypoxia-inducible factor-1α expression in lungs from patients with Copd. Chest. (2012) 142:663–72. doi: 10.1378/chest.11-2746
146. Mo, R, Li, J, Chen, Y, and Ding, Y. Lncrna Gas5 promotes Pyroptosis in Copd by functioning as a Cerna to regulate the Mir-223-3p/Nlrp3 Axis. Mol Med Rep. (2022) 26:12735. doi: 10.3892/mmr.2022.12735
147. Xu, W, Wang, Y, Ma, Y, and Yang, J. Mir-223 plays a protecting role in Neutrophilic asthmatic mice through the inhibition of Nlrp3 Inflammasome. Respir Res. (2020) 21:116. doi: 10.1186/s12931-020-01374-4
148. Li, G, Fan, Y, Lai, Y, Han, T, Li, Z, Zhou, P, et al. Coronavirus infections and immune responses. J Med Virol. (2020) 92:424–32. doi: 10.1002/jmv.25685
149. Widiasta, A, Sribudiani, Y, Nugrahapraja, H, Hilmanto, D, Sekarwana, N, and Rachmadi, D. Potential role of Ace2-related Micrornas in Covid-19-associated nephropathy. Non-coding RNA Res. (2020) 5:153–66. doi: 10.1016/j.ncrna.2020.09.001
150. Seif, F, Khoshmirsafa, M, Aazami, H, Mohsenzadegan, M, Sedighi, G, and Bahar, M. The role of Jak-Stat signaling pathway and its regulators in the fate of T helper cells. Cell Commun Signal. (2017) 15:23. doi: 10.1186/s12964-017-0177-y
151. Qin, H, and Zhao, A. Mesenchymal stem cell therapy for acute respiratory distress syndrome: from basic to clinics. Protein Cell. (2020) 11:707–22. doi: 10.1007/s13238-020-00738-2
152. Lu, D, Chatterjee, S, Xiao, K, Riedel, I, Wang, Y, Foo, R, et al. Micrornas targeting the Sars-Cov-2 entry receptor Ace2 in Cardiomyocytes. J Mol Cell Cardiol. (2020) 148:46–9. doi: 10.1016/j.yjmcc.2020.08.017
153. Wang, Y, Zhu, X, Jiang, XM, Guo, J, Fu, Z, Zhou, Z, et al. Decreased inhibition of Exosomal Mirnas on Sars-Cov-2 replication underlies poor outcomes in elderly people and diabetic patients. Signal Transduct Target Ther. (2021) 6:300. doi: 10.1038/s41392-021-00716-y
154. Trobaugh, DW, and Klimstra, WB. Microrna regulation of Rna virus replication and pathogenesis. Trends Mol Med. (2017) 23:80–93. doi: 10.1016/j.molmed.2016.11.003
155. Oshiumi, H. Circulating extracellular vesicles carry immune regulatory Mirnas and regulate vaccine efficacy and local inflammatory response after vaccination. Front Immunol. (2021) 12:685344. doi: 10.3389/fimmu.2021.685344
156. Hu, B, Huang, S, and Yin, L. The cytokine storm and Covid-19. J Med Virol. (2021) 93:250–6. doi: 10.1002/jmv.26232
157. Dorhoi, A, Iannaccone, M, Maertzdorf, J, Nouailles, G, Weiner, J 3rd, and Kaufmann, SH. Reverse translation in tuberculosis: neutrophils provide clues for understanding development of active disease. Front Immunol. (2014) 5:36. doi: 10.3389/fimmu.2014.00036
158. Elizabeth, MC, Hernández de la Cruz, ON, and Mauricio, CA. Infection of J774a.1 with different Mycobacterium species induces differential immune and Mirna-related responses. Microbiol Immunol. (2016) 60:356–63. doi: 10.1111/1348-0421.12380
159. Yi, XH, Zhang, B, Fu, YR, and Yi, ZJ. Stat1 and its related molecules as potential biomarkers in Mycobacterium tuberculosis infection. J Cell Mol Med. (2020) 24:2866–78. doi: 10.1111/jcmm.14856
160. Kachamakova-Trojanowska, N, Jazwa-Kusior, A, Szade, K, Kasper, L, Soja, J, Andrychiewicz, A, et al. Molecular profiling of regulatory T cells in pulmonary Sarcoidosis. J Autoimmun. (2018) 94:56–69. doi: 10.1016/j.jaut.2018.07.012
161. Huppertz, C, Jäger, B, Wieczorek, G, Engelhard, P, Oliver, SJ, Bauernfeind, FG, et al. The Nlrp3 Inflammasome pathway is activated in Sarcoidosis and involved in granuloma formation. Eur Respir J. (2020) 55:1900119. doi: 10.1183/13993003.00119-2019
162. Stachowiak, Z, Wojsyk-Banaszak, I, Jończyk-Potoczna, K, Narożna, B, Langwiński, W, Kycler, Z, et al. Mirna expression profile in the airways is altered during pulmonary exacerbation in children with cystic fibrosis-a preliminary report. J Clin Med. (2020) 9:1887. doi: 10.3390/jcm9061887
163. Qu, SJ, Zhao, L, Song, ZZ, Shen, WP, Ju, P, and Li, YM. Expression and significance of Mir-223 in rats with pulmonary fibrosis. Eur Rev Med Pharmacol Sci. (2020) 24:3951–8. doi: 10.26355/eurrev_202004_20864
164. Lyon, SM, and Rossman, MD. Pulmonary tuberculosis. Microbiol Spectr. (2017) 5:1. doi: 10.1128/microbiolspec.TNMI7-0032-2016
165. Yuan, S, Wu, Q, Wang, Z, Che, Y, Zheng, S, Chen, Y, et al. Mir-223: an immune regulator in infectious disorders. Front Immunol. (2021) 12:781815. doi: 10.3389/fimmu.2021.781815
166. Rajaram, MV, Ni, B, Dodd, CE, and Schlesinger, LS. Macrophage Immunoregulatory pathways in tuberculosis. Semin Immunol. (2014) 26:471–85. doi: 10.1016/j.smim.2014.09.010
167. Llanos, O, and Hamzeh, N. Sarcoidosis. Med Clin North Am. (2019) 103:527–34. doi: 10.1016/j.mcna.2018.12.011
168. Zhou, T, Casanova, N, Pouladi, N, Wang, T, Lussier, Y, Knox, KS, et al. Identification of Jak-Stat signaling involvement in Sarcoidosis severity via a novel Microrna-regulated peripheral blood mononuclear cell gene signature. Sci Rep. (2017) 7:4237. doi: 10.1038/s41598-017-04109-6
169. Ni, K, Liu, M, Zheng, J, Wen, L, Chen, Q, Xiang, Z, et al. Pd-1/Pd-L1 pathway mediates the alleviation of pulmonary fibrosis by human Mesenchymal stem cells in humanized mice. Am J Respir Cell Mol Biol. (2018) 58:684–95. doi: 10.1165/rcmb.2017-0326OC
170. Cahill, EF, Kennelly, H, Carty, F, Mahon, BP, and English, K. Hepatocyte growth factor is required for Mesenchymal stromal cell protection against Bleomycin-induced pulmonary fibrosis. Stem Cells Transl Med. (2016) 5:1307–18. doi: 10.5966/sctm.2015-0337
171. Raghu, G, Wells, AU, Nicholson, AG, Richeldi, L, Flaherty, KR, Le Maulf, F, et al. Effect of Nintedanib in subgroups of idiopathic pulmonary fibrosis by diagnostic criteria. Am J Respir Crit Care Med. (2017) 195:78–85. doi: 10.1164/rccm.201602-0402OC
172. Lu, Q, Yu, S, Meng, X, Shi, M, Huang, S, Li, J, et al. Micrornas: important regulatory molecules in acute lung injury/acute respiratory distress syndrome. Int J Mol Sci. (2022) 23:5545. doi: 10.3390/ijms23105545
173. Kalayinia, S, Arjmand, F, Maleki, M, Malakootian, M, and Singh, CP. Micrornas: roles in cardiovascular development and disease. Cardiovasc Pathol. (2021) 50:107296. doi: 10.1016/j.carpath.2020.107296
174. Pan, C, Stevic, I, Müller, V, Ni, Q, Oliveira-Ferrer, L, Pantel, K, et al. Exosomal Micrornas as tumor markers in epithelial ovarian Cancer. Mol Oncol. (2018) 12:1935–48. doi: 10.1002/1878-0261.12371
175. Lucas Grzelczyk, W, Szemraj, J, Kwiatkowska, S, and Józefowicz-Korczyńska, M. Serum expression of selected Mirnas in patients with laryngeal squamous cell carcinoma (Lscc). Diagn Pathol. (2019) 14:49. doi: 10.1186/s13000-019-0823-3
176. Talbot, CPJ, Mensink, RP, Smolders, L, Bakeroot, V, and Plat, J. Theobromine does not affect fasting and postprandial Hdl cholesterol efflux capacity, while it decreases fasting Mir-92a levels in humans. Mol Nutr Food Res. (2018) 62:e1800027. doi: 10.1002/mnfr.201800027
177. Daimiel, L, Micó, V, Díez-Ricote, L, Ruiz-Valderrey, P, Istas, G, Rodríguez-Mateos, A, et al. Alcoholic and non-alcoholic beer modulate plasma and macrophage Micrornas differently in a pilot intervention in humans with cardiovascular risk. Nutrients. (2020) 13:10069. doi: 10.3390/nu13010069
178. Cavallari, C, Dellepiane, S, Fonsato, V, Medica, D, Marengo, M, Migliori, M, et al. Online Hemodiafiltration inhibits inflammation-related endothelial dysfunction and vascular calcification of uremic patients modulating Mir-223 expression in plasma extracellular vesicles. J Immunol. (2019) 202:2372–83. doi: 10.4049/jimmunol.1800747
179. Parker, WAE, Schulte, C, Barwari, T, Phoenix, F, Pearson, SM, Mayr, M, et al. Aspirin, Clopidogrel and Prasugrel Monotherapy in patients with type 2 diabetes mellitus: a double-blind randomised controlled trial of the effects on thrombotic markers and Microrna levels. Cardiovasc Diabetol. (2020) 19:3. doi: 10.1186/s12933-019-0981-3
180. Khorraminezhad, L, and Rudkowska, I. Dairy product intake modifies Microrna expression among individuals with Hyperinsulinemia: a post-intervention cross-sectional study. Lifestyle Genomics. (2022) 15:77–86. doi: 10.1159/000523809
181. Fejes, Z, Erdei, J, Pócsi, M, Takai, J, Jeney, V, Nagy, A, et al. Elevated pro-inflammatory cell-free Microrna levels in cerebrospinal fluid of premature infants after Intraventricular hemorrhage. Int J Mol Sci. (2020) 21:6870. doi: 10.3390/ijms21186870
182. Hung, YY, Wu, MK, Tsai, MC, Huang, YL, and Kang, HY. Aberrant expression of intracellular let-7e, Mir-146a, and Mir-155 correlates with severity of depression in patients with major depressive disorder and is ameliorated after antidepressant treatment. Cells. (2019) 8:647. doi: 10.3390/cells8070647
183. Rupaimoole, R, and Slack, FJ. Microrna therapeutics: towards a new era for the management of Cancer and other diseases. Nat Rev Drug Discov. (2017) 16:203–22. doi: 10.1038/nrd.2016.246
184. Zhang, JX, Song, W, Chen, ZH, Wei, JH, Liao, YJ, Lei, J, et al. Prognostic and predictive value of a Microrna signature in stage ii Colon Cancer: a Microrna expression analysis. Lancet Oncol. (2013) 14:1295–306. doi: 10.1016/S1470-2045(13)70491-1
185. Wiemer, EAC, Wozniak, A, Burger, H, Smid, M, Floris, G, Nzokirantevye, A, et al. Identification of Microrna biomarkers for response of advanced soft tissue sarcomas to Eribulin: translational results of the Eortc 62052 trial. Eur J Cancer. (2017) 75:33–40. doi: 10.1016/j.ejca.2016.12.018
186. Beg, MS, Brenner, AJ, Sachdev, J, Borad, M, Kang, YK, Stoudemire, J, et al. Phase I study of Mrx34, a liposomal Mir-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig New Drugs. (2017) 35:180–8. doi: 10.1007/s10637-016-0407-y
187. Cortez, MA, Ivan, C, Valdecanas, D, Wang, X, Peltier, HJ, Ye, Y, et al. Pdl1 regulation by P53 via Mir-34. J Natl Cancer Inst. (2016) 108:djv303. doi: 10.1093/jnci/djv303
188. Rayner, KJ, Esau, CC, Hussain, FN, McDaniel, AL, Marshall, SM, van Gils, JM, et al. Inhibition of Mir-33a/B in non-human Primates raises plasma Hdl and lowers Vldl triglycerides. Nature. (2011) 478:404–7. doi: 10.1038/nature10486
189. Täubel, J, Hauke, W, Rump, S, Viereck, J, Batkai, S, Poetzsch, J, et al. Novel antisense therapy targeting Microrna-132 in patients with heart failure: results of a first-in-human phase 1b randomized, double-blind, Placebo Controlled Study. Eur Heart J. (2021) 42:178–88. doi: 10.1093/eurheartj/ehaa898
190. Zhang, Y, Wang, Y, Zhang, L, Xia, L, Zheng, M, Zeng, Z, et al. Reduced platelet Mir-223 induction in Kawasaki disease leads to severe coronary artery pathology through a Mir-223/Pdgfrβ vascular smooth muscle cell Axis. Circ Res. (2020) 127:855–73. doi: 10.1161/CIRCRESAHA.120.316951
191. You, D, Qiao, Q, Ono, K, Wei, M, Tan, W, Wang, C, et al. Mir-223-3p inhibits the progression of atherosclerosis via Down-regulating the activation of Mek1/Erk1/2 in macrophages. Aging. (2022) 14:1865–78. doi: 10.18632/aging.203908
192. Zhang, DM, Deng, JJ, Wu, YG, Tang, T, Xiong, L, Zheng, YF, et al. Microrna-223-3p protect against radiation-induced cardiac toxicity by alleviating myocardial oxidative stress and programmed cell death via targeting the Ampk pathway. Front Cell Dev Biol. (2021) 9:801661. doi: 10.3389/fcell.2021.801661
193. Li, G, Zong, X, Cheng, Y, Xu, J, Deng, J, Huang, Y, et al. Mir-223-3p contributes to suppressing Nlrp3 Inflammasome activation in Streptococcus Equi ssp. Zooepidemicus. Infect Vet Microbiol. (2022) 269:109430. doi: 10.1016/j.vetmic.2022.109430
Keywords: miR-223, inflammatory disease, acute lung injury (ALI), asthma, chronic obstructive pulmonary disease (COPD), COVID-19
Citation: Shi M, Lu Q, Zhao Y, Ding Z, Yu S, Li J, Ji M, Fan H and Hou S (2023) miR-223: a key regulator of pulmonary inflammation. Front. Med. 10:1187557. doi: 10.3389/fmed.2023.1187557
Edited by:
Laurent Metzinger, University of Picardie Jules Verne, FranceReviewed by:
Prabhash Kumar Jha, Brigham and Women's Hospital and Harvard Medical School, United StatesGan Zhao, University of Pennsylvania, United States
Copyright © 2023 Shi, Lu, Zhao, Ding, Yu, Li, Ji, Fan and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanmei Zhao, emhhb3lhbm1laUAxMjYuY29t; Haojun Fan, aGFvanVuZmFuODZAMTYzLmNvbQ==; Shike Hou, aG91c2hpa2VAdGp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Mingyu Shi
Mingyu Shi Qianying Lu
Qianying Lu Yanmei Zhao
Yanmei Zhao Ziling Ding1
Ziling Ding1 Haojun Fan
Haojun Fan