- 1Department of Pathobiology, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran
- 2Department of Microbiology, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
- 3Department of Microbiology, Karaj Branch, Islamic Azad University, Karaj, Iran
- 4Research Center for Antibiotic Stewardship and Antimicrobial Resistance, Tehran University of Medical Sciences, Tehran, Iran
Introduction: Stenotrophomonas maltophilia is a little-known environmental opportunistic bacterium that can cause broad-spectrum infections. Despite the importance of this bacterium as an emerging drug-resistant opportunistic pathogen, a comprehensive analysis of its prevalence and resistance to antibiotics has not yet been conducted.
Methods: A systematic search was performed using four electronic databases (MEDLINE via PubMed, Embase, Scopus, and Web of Science) up to October 2019. Out of 6,770 records, 179 were documented in the current meta-analysis according to our inclusion and exclusion criteria, and 95 studies were enrolled in the meta-analysis.
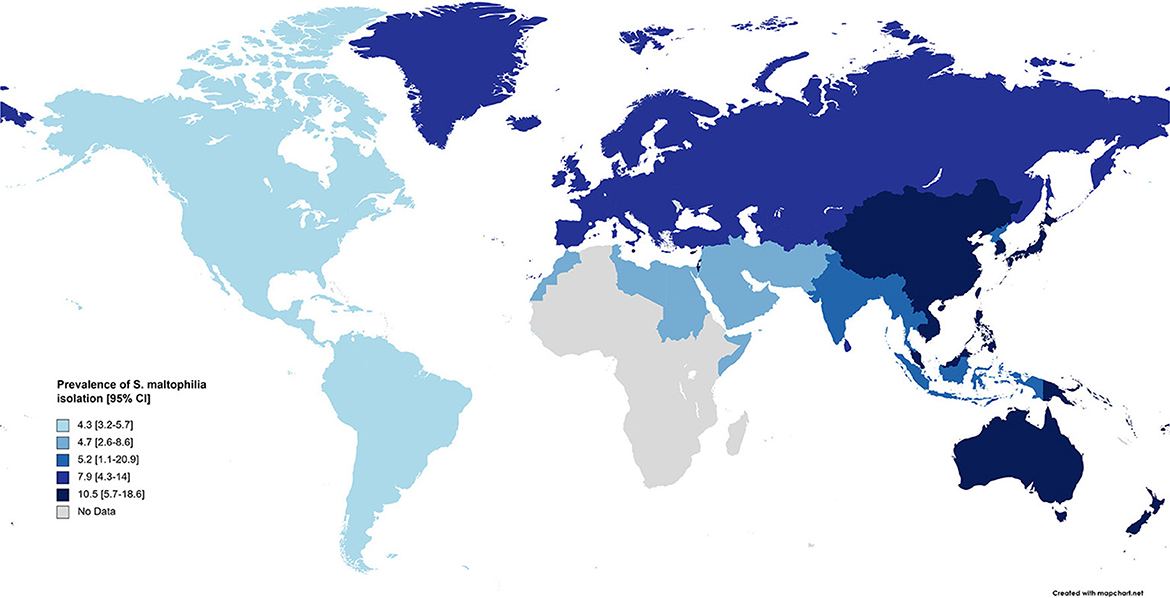
Results: Present analysis revealed that the global pooled prevalence of S. maltophilia was 5.3 % [95% CI, 4.1–6.7%], with a higher prevalence in the Western Pacific Region [10.5%; 95% CI, 5.7–18.6%] and a lower prevalence in the American regions [4.3%; 95% CI, 3.2–5.7%]. Based on our meta-analysis, the highest antibiotic resistance rate was against cefuroxime [99.1%; 95% CI, 97.3–99.7%], while the lowest resistance was correlated with minocycline [4·8%; 95% CI, 2.6–8.8%].
Discussion: The results of this study indicated that the prevalence of S. maltophilia infections has been increasing over time. A comparison of the antibiotic resistance of S. maltophilia before and after 2010 suggested there was an increasing trend in the resistance to some antibiotics, such as tigecycline and ticarcillin-clavulanic acid. However, trimethoprim-sulfamethoxazole is still considered an effective antibiotic for treating S. maltophilia infections.
Introduction
Stenotrophomonas maltophilia is an environmental Gram-negative bacillus that has been the subject of extensive research over the last two decades due to its status as the only known species of Stenotrophomonas to cause opportunistic infections in humans (1). Before the 1970s, this bacterium was underestimated and was considered a rare opportunistic pathogen with low invasiveness. However, advances in medical interventions and pharmacological treatments have led to an increase in the population of immunocompromised patients, such as those undergoing chemotherapy, organ transplantations, or complex surgeries, who are prone to infection with this bacterium. In addition, the development of diagnostic methods in clinical microbiology resulted in more precise identification of this pathogen. Therefore, the number of reported S. maltophilia infections has increased, and it is recognized as an emerging nosocomial pathogen (2). S. maltophilia causes infections of the soft tissue, urinary tract, eye, and wound. In addition, it causes pneumonia, bacteremia, sepsis, endocarditis, osteochondritis, mastoiditis, and meningitis (3). Predisposing factors associated with S. maltophilia infections include underlying malignancy, indwelling devices, chronic respiratory disease, particularly cystic fibrosis, immune compromisation, prolonged antibiotic use, and long-term hospitalization or admission to an intensive care unit (ICU) (3, 4). The treatment of infections caused by this bacterium presents several challenges. Distinguishing colonization from invasive infections is problematic, and physicians often fail to recognize their associated risk factors and clinical characteristics, which leads to delayed antibiotic prescription and high mortality (5).
Because of the high-level intrinsic resistance of S. maltophilia to several classes of antibiotics, there are restricted therapeutic choices for its infections. This bacterium can resist the β-lactam antibiotics (most notably carbapenems) by producing ß-lactamase enzymes, including L1 and L2. It also disrupts the action of aminoglycosides by hydrolyzing enzymes such as acetyl-transferases or modifying the structure of lipopolysaccharide. In addition, low membrane permeability and the overproduction of efflux pumps are other mechanisms that render S. maltophilia resistant to a broad range of antibiotics (2, 6). Additionally, they can acquire resistance genes and genetic mutations (7, 8), further limiting the choice of effective antimicrobials. This increasing prevalence of drug-resistant S. maltophilia has presented one of the biggest challenges in treating patients in recent years (3, 9).
The Infectious Diseases Society of America (IDSA) has approved a guideline document with recommendations for treating S. maltophilia infections (10). Trimethoprim-sulfamethoxazole (TMP/SMX) is the antibiotic of choice for treating these infections, but its use is limited by allergy, intolerance, and increased resistance (11). Other drugs with good susceptibility impact include ticarcillin-clavulanate, ceftazidime, and fluoroquinolones, although resistance to these drugs has been reported. Tetracyclines such as minocycline, tigecycline, and doxycycline are also efficacious in treating S. maltophilia infections, and their efficacy has been reported in different geographic areas (3, 12).
The main objective of this study was to assess the global prevalence of S. maltophilia and its resistance to commonly used antibiotics. We conducted this systematic review of global human infections due to S. maltophilia over the last 31 years.
Methods
Search strategy and selection criteria
Four electronic databases, including MEDLINE (via PubMed), Embase, Web of Science, and Scopus, were systematically searched using different combinations of the following keywords: “Stenotrophomonas maltophilia” OR “Xanthomonas maltophilia” AND “antibiotic resistance” AND “minimum inhibitory concentration” AND “disk agar diffusion” AND “multilocus sequence typing” AND “E-test” AND “antimicrobial resistance gene”. The databases were searched up to 20 October 2019 without any start time limitation.
The study was carried out based on the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) (13). Two distinct reviewers applied the inclusion and exclusion criteria for article selection and screened the titles and abstracts of all studies; then, two autonomous researchers qualified the screened papers. Any disagreements between the reviewers were resolved by consensus.
Inclusion criteria
Articles were included if they reported the prevalence of S. maltophilia isolation among diverse patients in combination with the antibiotic resistance rates of the isolates to various antibiotics, or reported only the antibiotic resistance rates of the isolates. Only articles about the clinical isolates of S. maltophilia were enrolled, and studies on the environmental isolates were not considered.
Exclusion criteria
Conference papers were not evaluated as they did not provide sufficient information for quality assessment. Dissertations and theses were excluded. Articles with unrelated topics, duplicates or overlapping studies, reviews, meta-analyses or systematic reviews, case reports, brief reports, notes, editorials, correspondence, short communications, and letters to the editors were not included. Studies with languages other than English or with unavailable full text were dismissed. Studies that evaluated species other than S. maltophilia or tested a total isolate <10 were not assessed. Articles that reported antibiotic resistance as MIC 90 or those that evaluated the combinatorial effects of antibiotics were not enrolled. Studies that considered S. maltophilia a Gram-negative bacterium and reported a total antibiotic resistance rate in Gram-negative bacteria were excluded. Articles were removed if they tested only the resistant isolates or reported only the prevalence of S. maltophilia infection.
Study selection and data extraction
Two independent researchers read the included articles in full text and extracted the following details: first author's name, year of study, year of publication, location of the study (country and region), sample size (N/total), type of samples, antibiotic susceptibility testing methods used (agar dilution, broth microdilution, broth macrodilution, E-test, disk agar diffusion [DAD], MIC test strip, Vitek, Phoenix, and Microscan), the antibiotic resistance rate of isolates against various antibiotics, frequency of resistance genes, and frequency of different sequence types. Any discrepancy between the two reviewers was settled by consensus.
Quality assessment
Two reviewers separately evaluated the quality of the included studies using the Joanna Briggs Institute (JBI) critical appraisal checklist for studies reporting prevalence data (14). This scale rates each criterion out of 1, with a total score ranging from 0 to 10. Studies with a score of ≥5 were classified as high quality.
Meta-analysis
The meta-analysis was carried out using Comprehensive Meta-Analysis (CMA) software version 2.0 (Biostat, Englewood, NJ). A random-effect model was used for meta-analysis and to pool the estimations. The prevalence of the investigated phenomenon was presented as a forest plot diagram, which shows the estimated prevalence and its relevant 95% confidence interval (CI). Heterogeneity between studies was reported by I2 statistics. An I2 between 0 and 25% suggests low heterogeneity, 25–50% indicates moderate heterogeneity, 50–75% represents substantial heterogeneity, and 75–100% shows considerable heterogeneity. Subgroup meta-analysis was employed to compare the prevalence of S. maltophilia based on WHO-defined regions and 5-year time intervals. In addition, the antibiotic resistance rates of isolates were compared based on world regions and whether they were reported before or after 2010. To assess the potential risk of publication bias, Begg's rank correlation and Egger's weighted regression methods in combination with a funnel plot were used (P < 0.05 was regarded as indicative of a statistically notable publication bias) (15).
Results
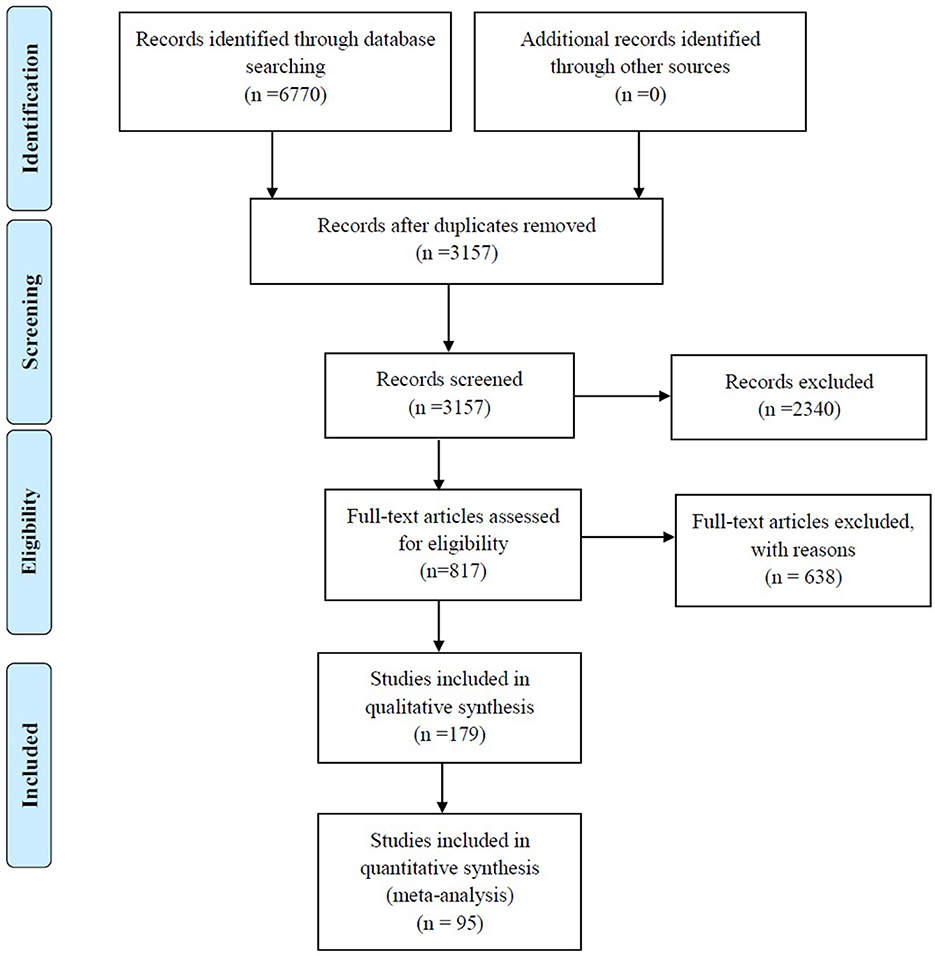
A total of 6,770 records were identified through searches of the four aforementioned electronic databases (Figure 1). After removing the 3,613 duplicates, 3,157 unique records were screened based on titles and abstracts, and 2,340 articles were excluded, such as studies with non-relevant topics (n = 1,245), repetitive articles (n = 470), reviews (n = 234), systematic reviews (n = 3), case reports (n = 64), letters to the editors (n = 60), conference abstracts (n = 111), editorials (n = 9), short surveys (n = 10), correspondence (n = 2), notes (n = 12), reports (n = 3), a book (n = 1), articles with a total sample of <10 strains (n = 17), non-English studies (n = 47), and articles that studied environmental samples (n = 21). In addition, 30 articles were removed because their full texts were not available. The eligibility of 817 full-text articles was assessed and, ultimately, 179 studies met the inclusion criteria and were enrolled in the qualitative analysis. Of these, 95 studies reporting the prevalence of S. maltophilia infection were selected for quantitative analysis (meta-analysis). The characteristics of the 179 included studies are summarized in Table 1.
Table 1. Characteristics of the studies that reported Stenotrophomonas maltophilia isolation in different parts of the world.
Overall, 179 studies conducted during the 31-year period between 1986 and 2017 were included. The articles had a wide geographical distribution, and the studies featured in them were carried out in different parts of the world. According to the World Health Organization's (WHO) regions, most studies were from the European Region (n = 57, 32%), followed by the West-Pacific Region (n = 39, 22%), the Region of the Americas (n = 37, 21%), the Eastern Mediterranean Region (n = 14, 8%), and the South-East Asian Region (n = 8, 4%). There was no independent study from the African Region. Twenty-four studies (13%) were conducted across different continents and were, therefore, classified as multiple region studies and did not conform to the WHO categories (Table 1).
The studies had very different sample sizes, ranging from 10 to 130,033. A total of 580,963 samples were examined, of which 25,596 were positive for S. maltophilia. Of the 179 studies, only 58 reported the types and details of examined specimens (5,106 samples). The most frequent sources of S. maltophilia isolation were respiratory samples (n = 3,434, 67%) and blood (n = 1,223, 24%) (Table 1). The qualities of all the reviewed studies were evaluated using the JBI critical appraisal checklist. Of the 95 studies included in the meta-analysis, 78 (82%) scored seven, 16 (17%) scored six, and one (1%) scored five. Therefore, all the studies enrolled in the meta-analysis had a high-quality score (a score of five or more) (Table 1).
Prevalence of Stenotrophomonas maltophilia by WHO regional offices
Based on the meta-analysis, the pooled prevalence rate of global S. maltophilia infection was estimated to be 5.3 % [95% CI, 4.1–6.7%] (Table 2 and Figure 2). Egger's test did not demonstrate publication bias (P > 0.05). However, Begg's test showed evidence of publication bias in the 95 analyzed studies (P = 0.017). Additionally, the corresponding funnel plot indicated publication bias (Supplementary File 1). Results demonstrated high heterogeneity (I2= 99.428%; P = 0.000) among the selected studies (Table 2).
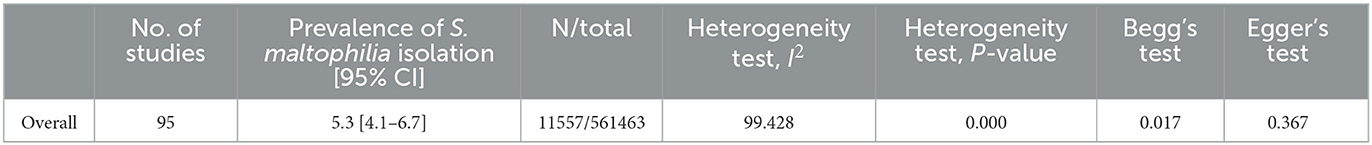
Table 2. Meta-analysis of the global prevalence rate of Stenotrophomonas maltophilia isolation from clinical samples.
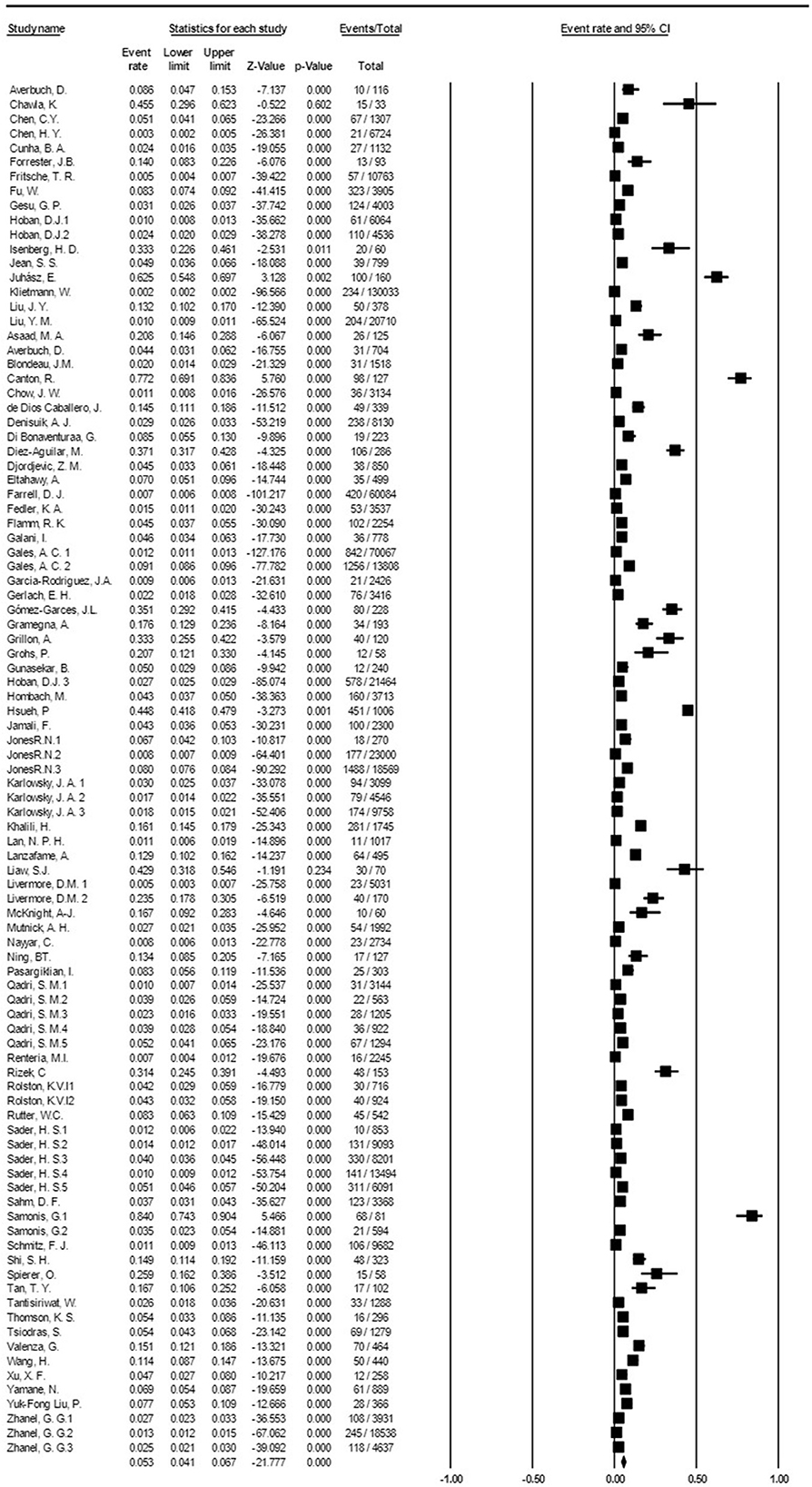
Figure 2. Forest plot diagram of the global prevalence rate of S. maltophilia isolation from clinical samples. The middle point of each line indicates the prevalence rate, and the length of the line indicates the 95% confidence interval of each study.
Subgroup meta-analysis based on the publication period of the studies (from 1991 to 2019) revealed that the prevalence rate of S. maltophilia isolation had an increasing trend over time, from 1.7% [95% CI, 0.7–4%] between 1991 and 1995 to 6.5% [95% CI, 4.1–10.1%] between 2016 and 2019. The highest prevalence rate [7.7%; 95% CI, 4.3–13.4 %] was observed between 2011 and 2015 (See Figure 3 and Table 3) (Supplementary File 1).
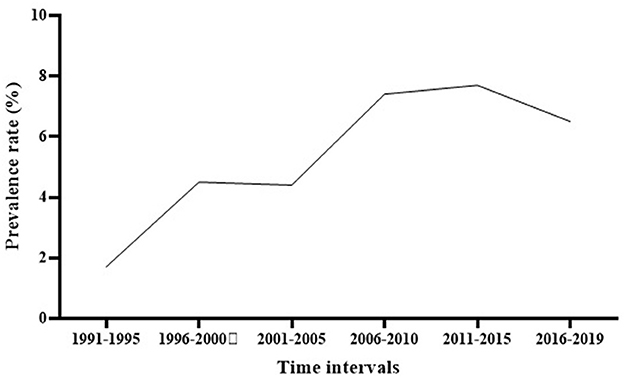
Figure 3. The global prevalence of S. maltophilia isolation based on the publication time of studies.
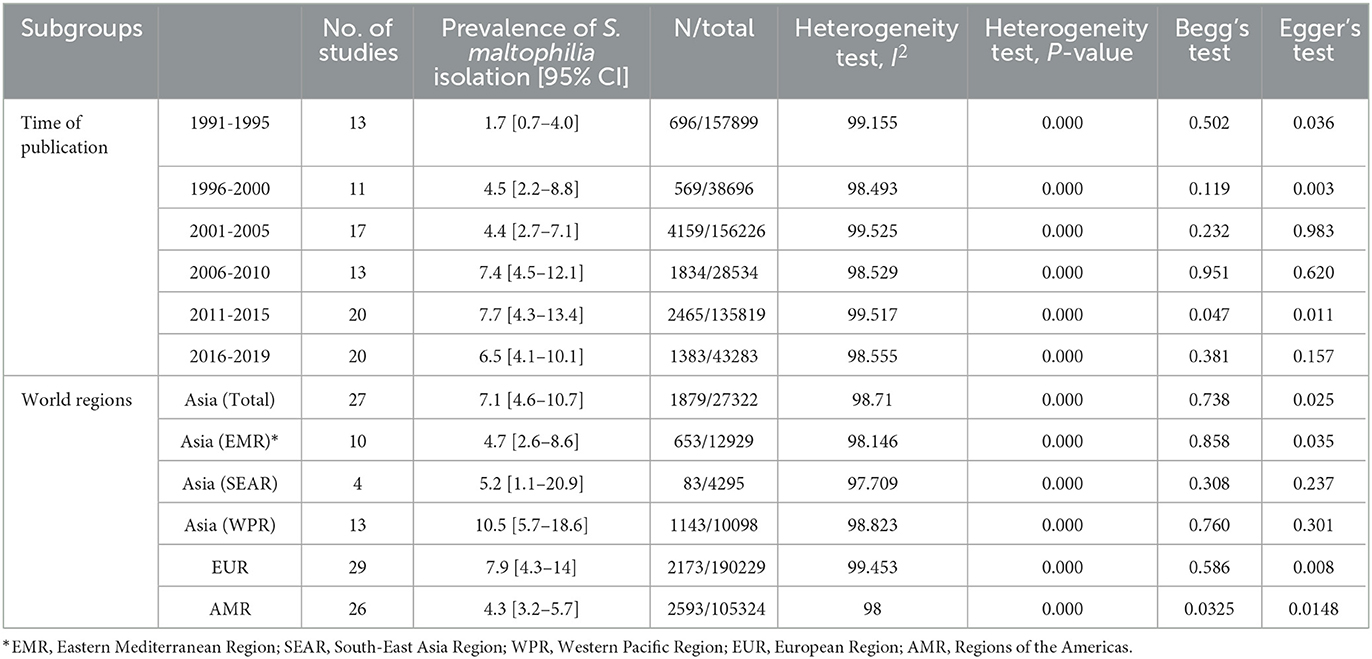
Table 3. Subgroup meta-analysis of the global prevalence rate of Stenotrophomonas maltophilia isolation from clinical samples.
Subgroup meta-analysis based on the world regions defined by WHO revealed that the highest prevalence of S. maltophilia infections occurred in the Western Pacific Region [10.5%; 95% CI, 5.7–18.6%] and the European Region [7.9%; 95% CI, 4.3–14%]. The lowest prevalence occurred in the Region of the Americas [4.3%; 95% CI, 3.2–5.7%] (see Table 3 and Figure 4).
Evaluation of the regional prevalence of S. maltophilia isolation based on the publication time of studies (from 1991 to 2019) showed an overall increasing trend. In the Western Pacific Region, the prevalence rate of S. maltophilia decreased from 2006 to 2010; however, the prevalence rates in the European Region and the Regions of America increased after this time interval (Figure 5 and Supplementary File 1).
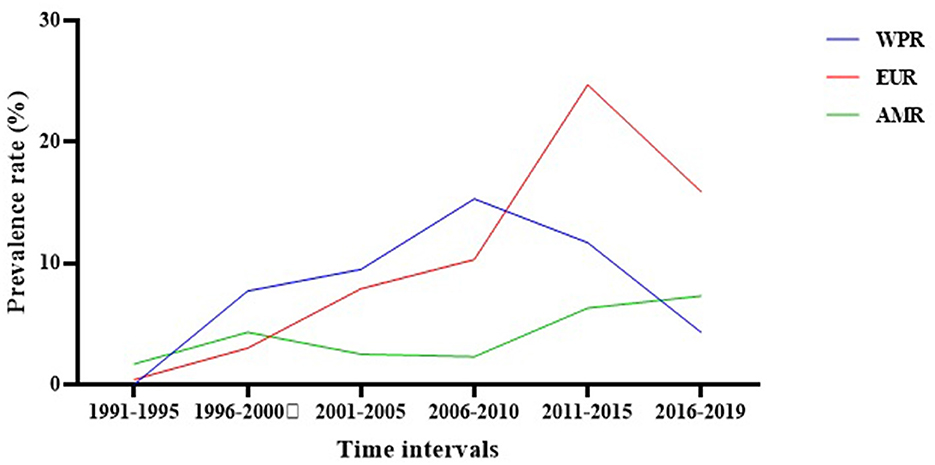
Figure 5. The regional prevalence of S. maltophilia isolation based on the publication time of studies.
The antibiotic resistance rate of Stenotrophomonas maltophilia
The susceptibility of S. maltophilia isolates to various antibiotics was determined using various methods, including broth micro-dilution, broth macro-dilution, agar dilution, disk agar diffusion (DAD), E-test, and automated methods (e.g., VITEK, Phoenix, and micro-scan systems). Broth micro-dilution was the most frequently used assay. The standards used for interpreting the results of susceptibility assays varied, with different breakpoints used, such as those of the Clinical and Laboratory Standards Institute (CLSI), National Committee for Clinical Laboratory Standards (NCCLS), European Committee on Antimicrobial Susceptibility Testing (EUCAST), U.S. Food and Drug Administration (FDA), British Society for Antimicrobial Chemotherapy (BSAC), TRUST, and Comité de l'Antibiogramme de la Société Française de Microbiologie (CA-SFM) (Supplementary File 2).
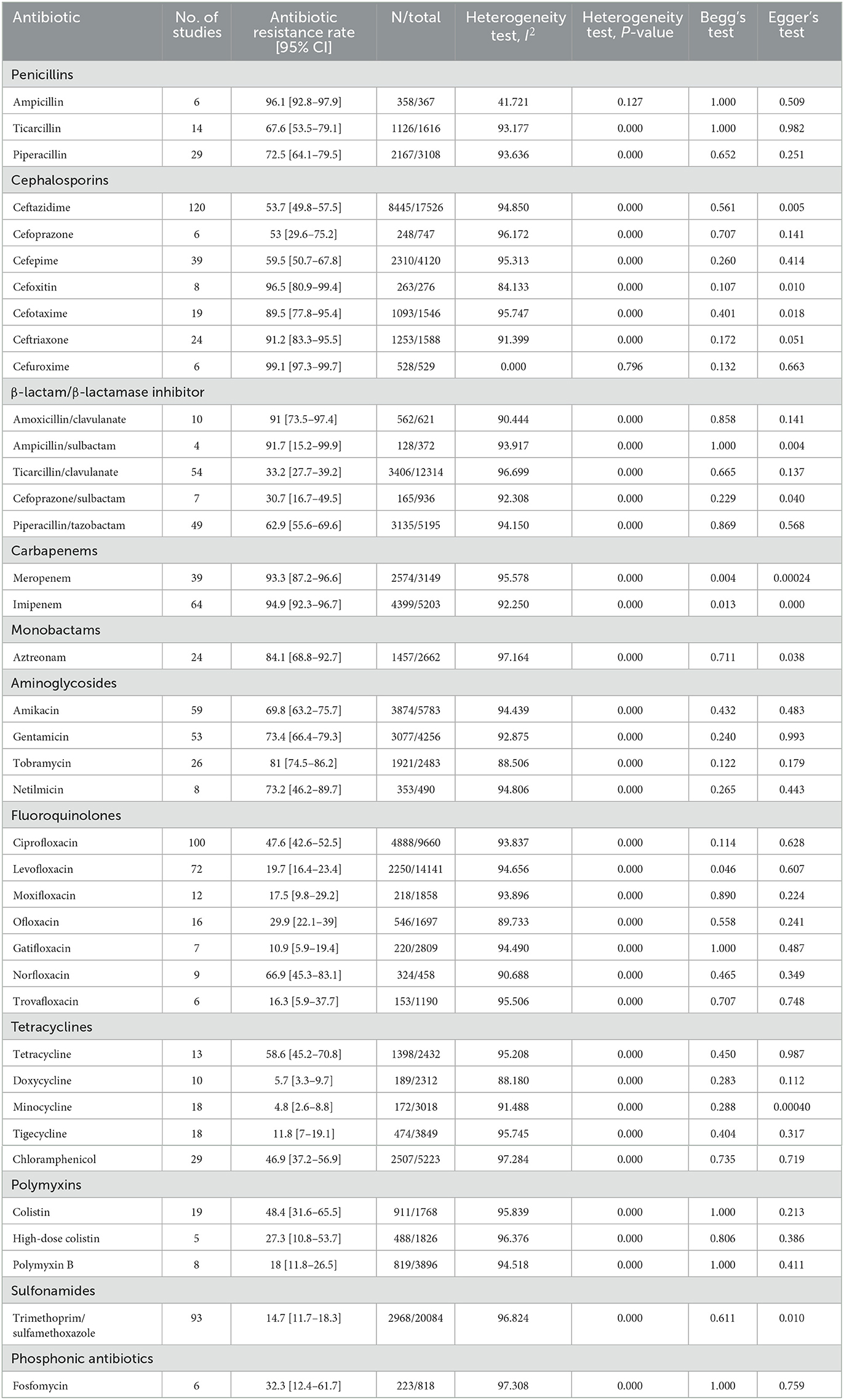
As shown in Table 4, the highest resistance rates of S. maltophilia isolates were to cefuroxime [99.1%; 95% CI, 97.3–99.7%], cefoxitin [96.5%; 95% CI, 80.9–99.4%], ampicillin [96.1%; 95% CI, 92.8–97.9%], imipenem [94.9%; 95% CI, 92.3–96.7%], and meropenem [93.3%; 95% CI, 87.2–96.6%], while the lowest resistance rates were to doxycycline [5.7%; 95% CI, 3.3–9.7%] and minocycline [4.8%; 95% CI, 2.6–8.8%].
A comparison of antibiotic resistance rates of S. maltophilia before and after 2010 (Figure 6) revealed an increasing trend for some antibiotics, such as chloramphenicol (12.3%), TMP/SMX (11.6%), ceftazidime (8.6%), and levofloxacin (1.8%). Conversely, the resistance rate against minocycline (2.2%) decreased.
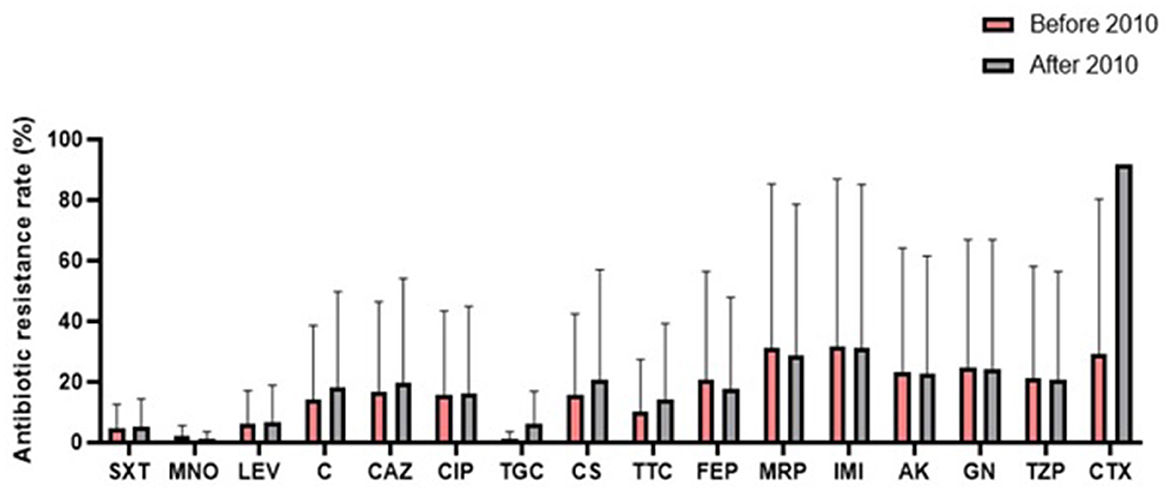
Figure 6. Comparison of the global antibiotic resistance rates of S. maltophilia before and after 2010 (SXT, trimethoprim-sulfamethoxazole; MNO, minocycline; LEV, levofloxacin; C, chloramphenicol; CAZ, ceftazidime; CIP, ciprofloxacin; TGC, tigecycline; CS, colistin; TTC, ticarcillin-clavulanic acid; FEP, cefepime; MRP, meropenem; IMI, imipenem; AK, amikacin; GN, gentamicin; TZP, piperacillin-tazobactam; CTX, cefotaxime).
The results of the subgroup meta-analysis based on the world regions and antibiotic resistance rates, presented in Figures 7–9, as well as in Supplementary File 1, showed that the highest resistance rate across all regions was to ceftazidime, while the lowest rate was to minocycline.
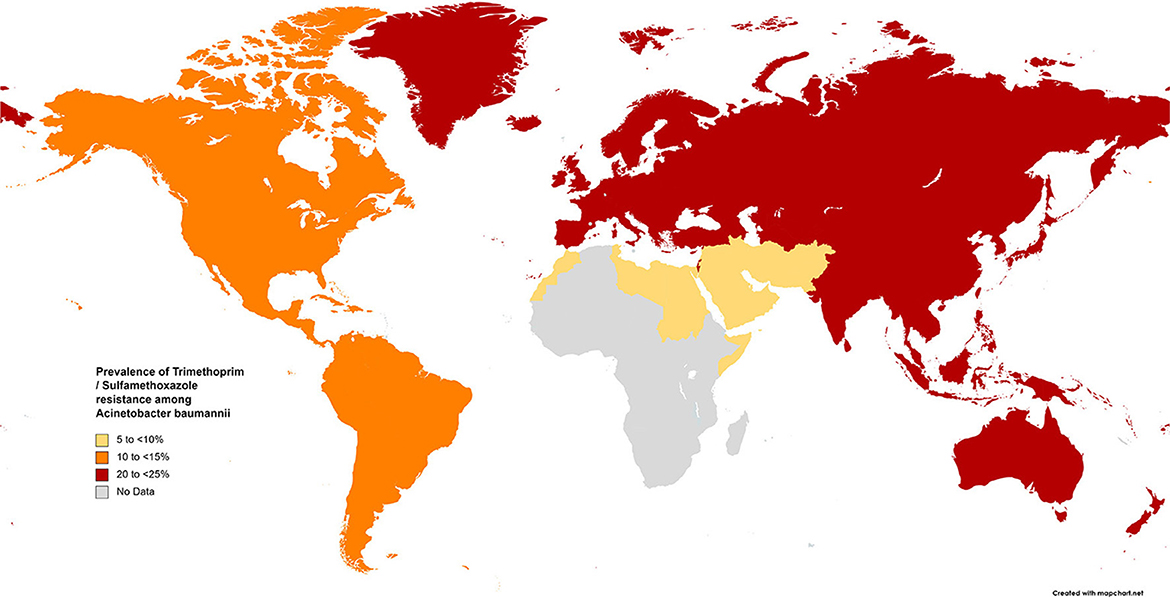
Figure 7. Prevalence of trimethoprim/sulfamethoxazole resistance in S. maltophilia isolated from clinical samples, by WHO regions.
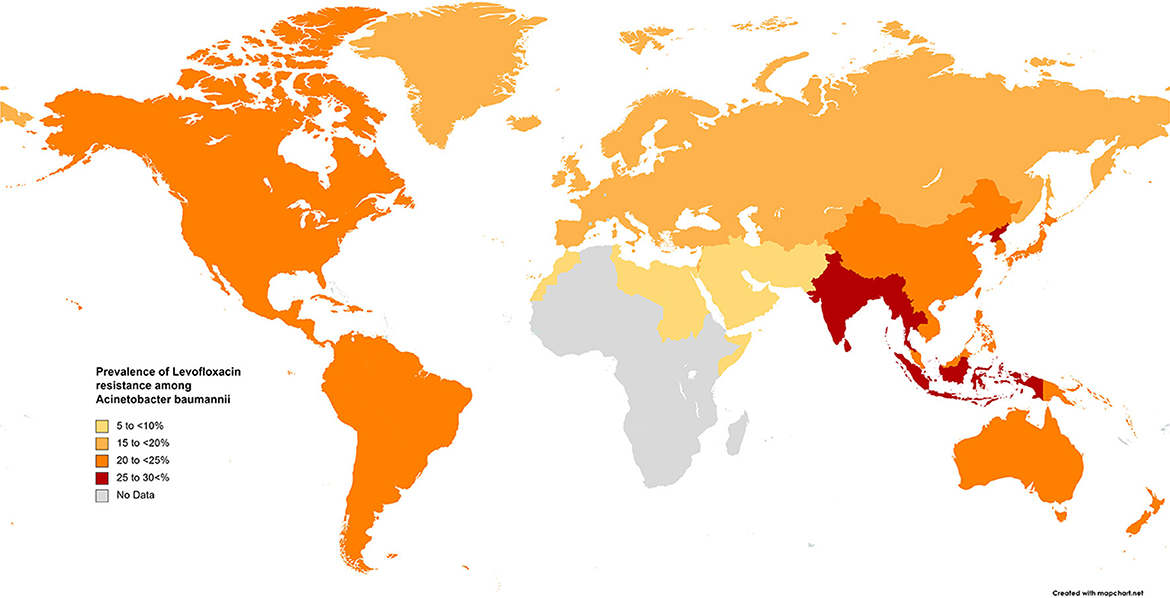
Figure 8. Prevalence of levofloxacin resistance in S. maltophilia isolated from clinical samples, by WHO regions.
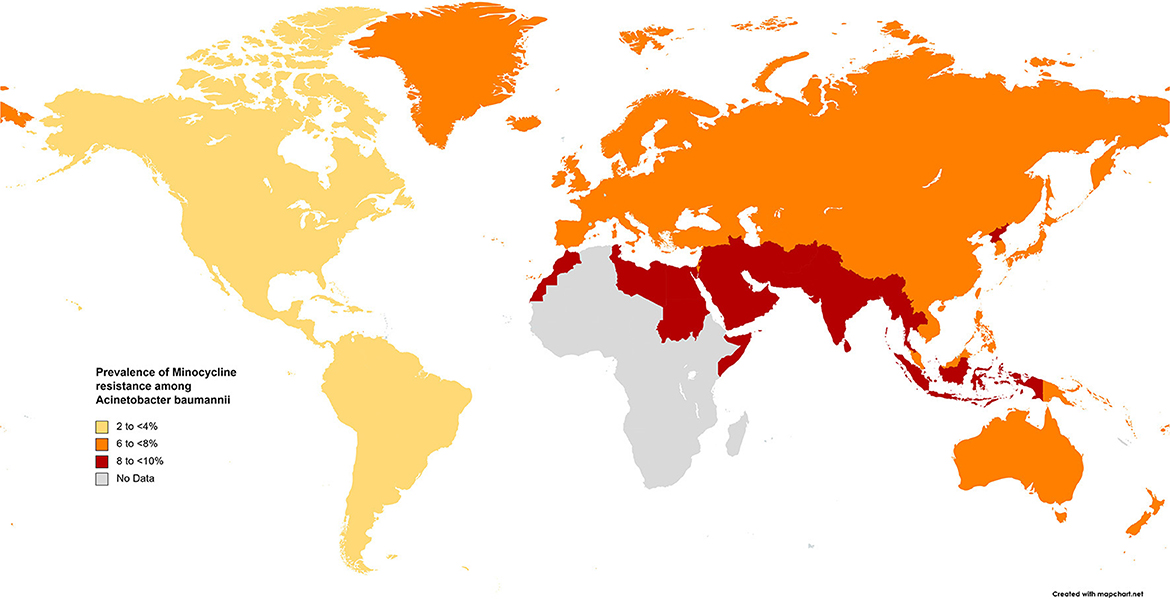
Figure 9. Prevalence of minocycline resistance in S. maltophilia isolated from clinical samples, by WHO regions.
Discussion
Although S. maltophilia shows limited invasiveness in immunocompetent individuals, it can lead to severe infections in immunocompromised patients. Moreover, its high intrinsic resistance to a large number of antimicrobial agents results in treatment failure and mortality in patients infected by this microorganism (191–194). Thus, the undertaking of a first systematic review and meta-analysis addressing the prevalence rate of isolation and antibiotic resistance rates of S. maltophilia in different regions of the world may be of great value in managing infections caused by this bacterium.
Based on the present meta-analysis, most studies were reported from the European Region (n = 57, 32%), while in a similar investigation (12), the majority of cases were reported and managed in the United States of America (n = 72, 27.7%). The differences between the inclusion and exclusion criteria applied in these two studies may explain the differing results. In the current study, the global prevalence rate of S. maltophilia isolation from clinical samples was 5.3%, and according to the WHO classification, the highest prevalence rate of S. maltophilia isolation was observed in the Western Pacific Region (10.5%), followed by the European Region (7.9%), which may be due to their long-shared land border. Among the reasons for the discrepancies in the prevalence of Stenotrophomonas maltophilia infection in different world regions, we can mention the following: disparate health policies in each country affect the importance of pathogens, so, in some countries, Stenotrophomonas maltophilia is still considered an unimportant opportunistic pathogen, so few studies have been reported. For example, most of the cases were documented in European (195), Asian (86), and American (196) countries, while there was no relevant study performed in the African continent. This difference can cause publication bias and affect the overall results. Additionally, the differences in health levels of various countries and the numbers and types of examined patients all influence the reported prevalence of Stenotrophomonas maltophilia.
In this meta-analysis, among different clinical samples, respiratory samples were the most frequent source (67%), followed by blood samples (24%). This finding is consistent with other studies, in which S. maltophilia was most commonly associated with respiratory tract infections, followed by bloodstream infections (74, 197). However, in another systematic review, blood was the most prevalent site of S. maltophilia isolation (12). In a large study performed in the USA and fifteen centers in European countries in 2012, 6.3% of the isolates obtained from respiratory tract infections were identified as S. maltophilia. These data suggest that the rate of respiratory tract infections caused by S. maltophilia is increasing (3, 198). The bacterium's capability for adherence to plastic surfaces and biofilm formation on hospital devices, such as those inserted into the respiratory tract, may explain its high rate in the aforementioned samples (199, 200). For example, among patients with ventilator-associated pneumonia (VAP), the most common nosocomial infection in mechanically ventilated patients, S. maltophilia is the probable causative pathogen (196, 201). Moreover, its adaptation to the airways of individuals with cystic fibrosis (CF) has led it to being recognized as an emerging multi-drug resistant opportunistic pathogen (86).
The prevalence rate of infections caused by this bacterium increased from 1.7% to 6.5% during the 31 investigated years, suggesting that it is emerging as an opportunistic pathogen, particularly among immunocompromised hosts. This rapid rise may be due to its resistance to a wide range of antimicrobial agents, as well as the increased focus on this bacterium as a cause of infection. The treatment of S. maltophilia infections is challenging due to the difficulty of differentiating colonization from infection and the intrinsic resistance of this bacterium to multiple classes of antibiotics. The WHO has classified S. maltophilia as one of the leading multidrug-resistant organisms in hospital settings (202). Additionally, recent antibiotic treatment and other known factors associated with acquiring S. maltophilia infections demonstrate specific features of this bacterium (195).
Based on our data, the highest and the lowest global resistant rates were to cefuroxime and minocycline, respectively (Figure 3). The lowest resistance to TMP-SMX was observed in the EMR (4.5%) and AMR (13.1%), while in other geographical regions, resistance was higher than 20%. Consequently, TMP-SMX may be the first choice for treatment based on antibiotic susceptibility and therapeutic success (3, 60, 203). Fortunately, in the present study, a comparison of global antibiotic resistance rates of S. maltophilia before and after 2010 (Figure 4) confirmed the effectiveness of this medication for treating infections of this opportunistic organism. However, there is not always a logical correlation between laboratory sensitivity and clinical results. Other antibiotics for treating Stenotrophomonas infections include fluoroquinolones, tetracyclines, and selected β-lactams, such as ceftazidime and ticarcillin/clavulanate. However, the development of resistance to some of these antibiotics renders them unreliable.
Fluoroquinolones are prescribed for treating infections caused by TMP-SMX-resistant S. maltophilia and for patients for whom this drug has adverse effects. Studies comparing treatments with fluoroquinolones and TMP-SMX have proposed that levofloxacin has similar effectiveness with fewer adverse effects than TMP-SMX (204, 205). Our study indicates that resistance rates to levofloxacin vary geographically, ranging from 6.4% in EMR to 15%−22% in EUR, AMR, and WPR, and up to 26% in SEAR. However, the rapid emergence of resistance against quinolones in vitro and in vivo is of concern when levofloxacin is used to treat S. maltophilia infections.
In surveillance studies of the efficacy of tigecycline and related tetracycline antibiotics, minocycline was found to be effective against S. maltophilia (206). In this study, resistance to minocycline was <10% in all geographical areas and global resistance to tigecycline was 11.8%. A comparison of the antibiotic resistance rates of S. maltophilia before and after 2010 revealed an increase in resistance to tigecycline from 4.1% to 18.6%. Several studies have revealed that minocycline is not inferior to TMP-SMX and may even be more suitable than TMP-SMX in terms of susceptibility. These results suggest that minocycline and TMP-SMX may be the first-line therapy in S. maltophilia infections, even in TMP-SMX-resistant strains (59).
Ceftazidime and ticarcillin/clavulanate have previously been reported as the most effective β-lactam drugs against S. maltophilia. However, reduced sensitivity to ceftazidime has been documented in recent studies. Owing to β-lactamase production, a high resistance rate to β-lactams such as cefuroxime, cefoxitin, imipenem, and meropenem (> 90%, Table 4) has been observed, thus reducing their role in the treatment of S. maltophilia infections (207). According to this analysis, ceftazidime has a high resistance rate in all regions classified by the WHO (AMR, 56.4%; EMR, 42.9%; SEAR, 65.1%; WPR, 52.6%). Our study suggests that the rate of resistance to ticarcillin/clavulanate globally is 33.2%. Therefore, these current resistance rates to ceftazidime and ticarcillin/clavulanate render them unreliable. However, the use of ceftazidime in combination with other antibiotics (typically vancomycin, amikacin, TMP-SMX, or fluoroquinolones) is an effective treatment for infections caused by S. maltophilia (13). A systemic literature review by Gibb and Wong (208) offers recommendations for a treatment strategy for Stenotrophomonas infection based on current evidence. The first-line drugs suggested are TMP-SMX, fluoroquinolones, and tetracyclines.
Our study presents several limitations. First, a large number of the included studies (84 articles) evaluated a specific number of S. maltophilia isolates but did not report the prevalence rate of isolation; thus, these studies were not included in the meta-analysis, which could affect the pooled prevalence rate of S. maltophilia isolation and the antibiotic resistance rates. Second, the number of published studies reporting the resistance mechanism of strains isolated from clinical samples (see Supplementary File 2) is relatively small, and the specific genes conferring antibiotic resistance in these isolates remain unclear. Third, a few studies used typing methods to evaluate S. maltophilia isolates (see Supplementary File 2), so we could not report the most prevalent types of this bacterium at the global and regional levels.
Conclusion
In conclusion, despite the undeniable clinical impact of S. maltophilia, compared with other Gram-negative species, this bacterium is remarkably understudied. Thus, collecting and analyzing data related to different aspects of S. maltophilia may assist in improving the clinical management of challenges caused by this bacterium. This meta-analysis presents the global antibiotic resistance of S. maltophilia over the last 31 years and demonstrates different rates of resistance in world geographical regions, as well as the growing trend of resistance to most antibiotics. The variations in antibiotic resistance of S. maltophilia isolates in different regions may be the result of the use of different protocols for patient treatment. Additionally, the improper and experimental use of antibiotics plays an important role in increasing resistance, leading to an increased risk of treatment failure. To address this issue, it is necessary to carry out antibiotic sensitivity tests before prescribing antibiotics and implementing an antimicrobial stewardship program for every hospital, as well as provide continuous training for clinicians about their performance in the hospital environment. Finally, collecting and preparing local sensitivity patterns will be effective in allowing the selection of the optimal empiric treatment for S. maltophilia infections.
Author contributions
MB contributed to the study design, data extraction, data analysis, design and production of figures, and wrote and revised the final manuscript. AS-M contributed to the study design, data extraction, data analysis, and writing of the manuscript. GB contributed to the data analysis and statistical analysis, designed and produced figures, and writing of the manuscript. EE contributed to the study design, data extraction, and writing of the manuscript. LJ contributed to the study design and the writing and revision of the final manuscript. RB contributed to the study design, data analysis and interpretation, and the writing of the manuscript. ME designed the study, oversaw the analysis, and wrote and revised the final manuscript. FJ designed the study, was the arbiter for the study searches and data extraction, and wrote and revised the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Tehran University of Medical Sciences and Health Services (97-01-30-38043).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2023.1163439/full#supplementary-material
References
1. Cerezer VG, Bando SY, Pasternak J, Franzolin MR, Moreira-Filho CA. Phylogenetic analysis of Stenotrophomonas spp isolates contributes to the identification of nosocomial and community-acquired infections. BioMed Research Int. (2014) 2014. doi: 10.1155/2014/151405
2. Gajdacs M, Urban E. Prevalence and antibiotic resistance of Stenotrophomonas maltophilia in respiratory tract samples: a 10-year epidemiological snapshot. Health Serv Res Manag Epidemiol. (2019) 6:2333392819870774. doi: 10.1177/2333392819870774
3. Chang YT, Lin CY, Chen YH, Hsueh PR. Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front Microbiol. (2015) 6:893. doi: 10.3389/fmicb.2015.00893
4. Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev. (2012) 25:2–41. doi: 10.1128/CMR.00019-11
5. Mojica MF, Humphries R, Lipuma JJ, Mathers AJ, Rao GG, Shelburne SA, et al. Clinical challenges treating Stenotrophomonas maltophilia infections: an update. JAC-Antimicrob Resis. (2022) 4:dlac040. doi: 10.1093/jacamr/dlac040
6. Cruz-Cordova A, Mancilla-Rojano J, Luna-Pineda VM, Escalona-Venegas G, Cazares-Dominguez V, Ormsby C, et al. Molecular epidemiology, antibiotic resistance, and virulence traits of Stenotrophomonas maltophilia strains associated with an outbreak in a Mexican tertiary care hospital. Front Cell Infect Microbiol. (2020) 10:50. doi: 10.3389/fcimb.2020.00050
7. Sanchez MB, Hernandez A, Martinez JL. Stenotrophomonas maltophilia drug resistance. Future Microbiol. (2009) 4:655–60. doi: 10.2217/fmb.09.45
8. Insuwanno W, Kiratisin P, Jitmuang A. Stenotrophomonas maltophilia Infections: Clinical characteristics and factors associated with mortality of hospitalized patients. Infect Drug Resist. (2020) 13:1559. doi: 10.2147/IDR.S253949
9. Chung HS, Kim K, Hong SS, Hong SG, Lee K, Chong Y, et al. The sul1 gene in Stenotrophomonas maltophilia with high-level resistance to trimethoprim/sulfamethoxazole. Annal Lab Med. (2015) 35:246. doi: 10.3343/alm.2015.35.2.246
10. Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, Van Duin D, Clancy CJ, et al. Infectious diseases society of america guidance on the treatment of ampc β-lactamase–producing enterobacterales, carbapenem-resistant acinetobacter baumannii, and Stenotrophomonas maltophilia infections. Clin Infect Dis. (2022) 74:2089–114. doi: 10.1093/cid/ciab1013
11. Chong SY, Lee K, Chung HS, Hong SG, Suh Y, Chong Y, et al. Levofloxacin efflux and smeD in clinical isolates of Stenotrophomonas maltophilia. Microb. Drug Resis. (2017) 23:163–8. doi: 10.1089/mdr.2015.0228
12. Andelković MV, Janković SM, Kostić MJ, Živković Zarić RS, Opančina VD, Živić MŽ, et al. Antimicrobial treatment of Stenotrophomonas maltophilia invasive infections: systematic review. J Chemother. (2019) 31:297–306. doi: 10.1080/1120009X.2018.1542551
13. Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol. (2009) 62:e1–e34. doi: 10.1016/j.jclinepi.2009.06.006
14. Joanna Briggs Institute. Joanna Briggs Institute Reviewers' Manual: 2017 edition. Australia: The Joanna Briggs Institute (2017).
15. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. (2002) 21:1539–58. doi: 10.1002/sim.1186
16. Al-Lawati AM, Crouch ND, Elhag KM. Antibiotic consumption and development of resistance among gram-negative bacilli in intensive care units in Oman. Ann Saudi Med. (2000) 20:324–7. doi: 10.5144/0256-4947.2000.324
17. Asaad AM, Al-Ayed MS, Qureshi MA. Emergence of unusual nonfermenting Gram-negative nosocomial pathogens in a Saudi hospital. Jpn J Infect Dis. (2013) 66:507–11. doi: 10.7883/yoken.66.507
18. Bostanghadiri N, Ghalavand Z, Fallah F, Yadegar A, Ardebili A, Tarashi S, et al. Characterization of phenotypic and genotypic diversity of Stenotrophomonas maltophilia strains isolated from selected hospitals in Iran. Front Microbiol. (2019) 10:1191. doi: 10.3389/fmicb.2019.01191
19. Cunha BA, Qadri SM, Ueno Y, Walters EA, Domenico P. Antibacterial activity of trovafloxacin against nosocomial Gram-positive and Gram-negative isolates. J Antimicrob Chemother. (1997) 39:29–34. doi: 10.1093/jac/39.suppl_2.29
20. Ebrahim-Saraie HS, Heidari H, Soltani B, Mardaneh J, Motamedifar M. Prevalence of antibiotic resistance and integrons, sul and Smqnr genes in clinical isolates of Stenotrophomonas maltophilia from a tertiary care hospital in Southwest Iran. Iran J Basic Med Sci. (2019) 22:872–7. doi: 10.22038/ijbms.2019.31291.7540
21. El Tahawy ATAE, Khalaf RMF. Antibiotic resistance among gram-negative non-fermentative bacteria at a teaching hospital in Saudi Arabia. J Chemother. (2001) 13:260–4. doi: 10.1179/joc.2001.13.3.260
22. Jamali F, Boroumand MA, Yazdani F, Anvari MS, Pourgholi L, Mahfouzi S, et al. Minimal inhibitory concentration of ceftazidime and Co-trimoxazole for Stenotrophomonas maltophilia using E-test. J Glob Infect Dis. (2011) 3:254–8. doi: 10.4103/0974-777X.83531
23. Khalili H, Dashti-Khavidaki S, Shahidi MR, Abdollahi A, Jafari S, Jahangard-Rafsanjani Z, et al. Changes in gram negative microorganisms resistance pattern during 4years period in a referral teaching hospital; a surveillance study. DARU J Pharm Sci. (2012) 20. doi: 10.1186/2008-2231-20-28
24. Morsi SS, Sharaf HE, Gerges MA. Association of sul genes and class 1 integron with trimethoprim-sulfamethoxazole Resistance in Stenotrophomonas maltophilia clinical isolates in Zagazig University, Egypt. Afr J Clin Exp Microbiol. (2016) 17:158–65. doi: 10.4314/ajcem.v17i3.1
25. Qadri SM, Lee GC, Ellis ME. In vitro activity of lomefloxacin, a difluorinated quinolone, compared with other antimicrobials. Chemother. (1991) 37:166–74. doi: 10.1159/000238850
26. Qadri SM, Ueno Y, Burns JJ, Almodovar E, Rabea N. In vitro activity of sparfloxacin (CI-978), a new broad-spectrum fluoroquinolone. Chemother. (1992) 38:99–106. doi: 10.1159/000238948
27. Qadri SM, Ueno Y, Saldin H, Cunha BA. In vitro activity of Ro 23-9424, a dual-acting cephalosporin-quinolone antimicrobial agent. J Clin Pharmacol. (1993) 33:923–8. doi: 10.1002/j.1552-4604.1993.tb01923.x
28. Qadri SMH, Ueno Y, Almodovar E, Tullo D, Alahdal MN. Comparative invitro evaluation of cefepime, an aminothiazolyl methoxyamino cephem. Drug Investigation. (1993) 5:127–34. doi: 10.1007/BF03259584
29. Qadri SMH, Ueno Y, Saldin H, Burdette JM, Lee GC. Comparative antibacterial activity of the new fluoroquinolone pd-131628. Drug Investigation. (1992) 4:409–15. doi: 10.1007/BF03258419
30. Cha MK, Kang CI, Kim SH, Cho SY, Ha YE, Chung DR, et al. Emergence of fluoroquinolone-resistant Stenotrophomonas maltophilia in blood isolates causing bacteremia: Molecular epidemiology and microbiologic characteristics. Diagn Microbiol Infect Dis. (2016) 85:210–2. doi: 10.1016/j.diagmicrobio.2016.02.020
31. Chang LL, Chen HF, Chang CY, Lee TM, Wu WJ. Contribution of integrons, and SmeABC and SmeDEF efflux pumps to multidrug resistance in clinical isolates of Stenotrophomonas maltophilia. J Antimicrob Chemother. (2004) 53:518–21. doi: 10.1093/jac/dkh094
32. Chen CY, Tsay W, Tang JL, Tien HF, Chen YC, Chang SC, et al. Epidemiology of bloodstream infections in patients with haematological malignancies with and without neutropenia. Epidemiol Infect. (2010) 138:1044–51. doi: 10.1017/S0950268809991208
33. Cho SY, Lee DG, Choi SM, Park C, Chun HS, Park YJ, et al. Stenotrophomonas maltophilia bloodstream infection in patients with hematologic malignancies: a retrospective study and in vitro activities of antimicrobial combinations. BMC Infect Dis. (2015) 15. doi: 10.1186/s12879-015-0801-7
34. Cho HH, Sung JY, Kwon KC, Koo SH. Expression of Sme efflux pumps and multilocus sequence typing in clinical isolates of Stenotrophomonas maltophilia. Ann Lab Med. (2012) 32:38–43. doi: 10.3343/alm.2012.32.1.38
35. Chung HS, Hong SG, Kim YR, Shin KS, Whang DH, Ahn JY, et al. Antimicrobial susceptibility of Stenotrophomonas maltophilia isolates from Korea, and the activity of antimicrobial combinations against the isolates. J Korean Med Sci. (2013) 28:62–6. doi: 10.3346/jkms.2013.28.1.62
36. Fu W, Demei Z, Shi W, Fupin H, Yingyuan Z. The susceptibility of non-fermentative Gram-negative bacilli to cefperazone and sulbactam compared with other antibacterial agents. Int J Antimicrob Agents. (2003) 22:444–8. doi: 10.1016/S0924-8579(03)00109-2
37. Fujita J, Yamadori I, Xu G, Hojo S, Negayama K, Miyawaki H, et al. Clinical features of Stenotrophomonas maltophilia pneumonia in immunocompromised patients. Respir Med. (1996) 90:35–8. doi: 10.1016/S0954-6111(96)90242-5
38. Friedman ND, Korman TM, Fairley CK, Franklin JC, Spelman DW. Bacteraemia due to Stenotrophomonas maltophilia: an analysis of 45 episodes. J Infect. (2002) 45:47–53. doi: 10.1053/jinf.2002.0978
39. Hsueh PR, Chen WH, Luh KT. Relationships between antimicrobial use and antimicrobial resistance in Gram-negative bacteria causing nosocomial infections from 1991-2003 at a university hospital in Taiwan. Int J Antimicrob Agents. (2005) 26:463–72. doi: 10.1016/j.ijantimicag.2005.08.016
40. Hu LF, Chang X, Ye Y, Wang ZX, Shao YB, Shi W, et al. Stenotrophomonas maltophilia resistance to trimethoprim/sulfamethoxazole mediated by acquisition of sul and dfrA genes in a plasmid-mediated class 1 integron. Int J Antimicrob Agents. (2011) 37:230–4. doi: 10.1016/j.ijantimicag.2010.10.025
41. Hu LF, Chen GS, Kong QX, Gao LP, Chen X, Ye Y, et al. Increase In The prevalence of resistance determinants to trimethoprim/ Sulfamethoxazole in clinical Stenotrophomonas maltophilia isolates in China. PLoS ONE. (2016) 11:693. doi: 10.1371/journal.pone.0157693
42. Hu LF, Gao LP, Ye Y, Chen X, Zhou XT, Yang HF, et al. Susceptibility of Stenotrophomonas maltophilia clinical strains in China to antimicrobial combinations. J Chemother. (2014) 26:276–81. doi: 10.1179/1973947814Y.0000000168
43. Hu LF, Xu XH Li HR, Gao LP, Chen X, Sun N, et al. Surveillance of antimicrobial susceptibility patterns among Stenotrophomonas maltophilia isolated in China during the 10-year period of 2005–2014. J Chemother. (2018) 30:25–30. doi: 10.1080/1120009X.2017.1378834
44. Ismail N, Zam Z, Hassan SA, Rahman ZA, A. Combination of trimethoprim-sulfamethoxazole and ceftazidime showed good in vitro activity against Stenotrophomonas maltophilia. Malays J Med Sci. (2017) 24:21–7. doi: 10.21315/mjms2016.24.2.3
45. Jean SS, Liao CH, Sheng WH, Lee WS, Hsueh PR. Comparison of commonly used antimicrobial susceptibility testing methods for evaluating susceptibilities of clinical isolates of Enterobacteriaceae and nonfermentative Gram-negative bacilli to cefoperazone–sulbactam. J Microbiology Immunol Inf. (2017) 50:454–63. doi: 10.1016/j.jmii.2015.08.024
46. Kanamori H, Yano H, Tanouchi A, Kakuta R, Endo S, Ichimura S, et al. Prevalence of Smqnr and plasmid-mediated quinolone resistance determinants in clinical isolates of Stenotrophomonas maltophilia from Japan: novel variants of Smqnr. New Microbes New Inf. (2015) 7:8–14. doi: 10.1016/j.nmni.2015.04.009
47. Liaw SJ, Lee YL, Hsueh PR. Multidrug resistance in clinical isolates of Stenotrophomonas maltophilia: roles of integrons, efflux pumps, phosphoglucomutase (SpgM), and melanin and biofilm formation. Int J Antimicrob Agents. (2010) 35:126–30. doi: 10.1016/j.ijantimicag.2009.09.015
48. Liu JY, Wang FD, Ho MW, Lee CH, Liu JW, Wang JT, et al. In vitro activity of aminoglycosides against clinical isolates of Acinetobacter baumannii complex and other nonfermentative Gram-negative bacilli causing healthcare-associated bloodstream infections in Taiwan. J Microbiol Immunol Inf. (2016) 49:918–23. doi: 10.1016/j.jmii.2015.07.010
49. Lan NPH, Hien NH, Le Thi Phuong T, Thanh DP, Thieu NTV, Ngoc DTT, et al. Phenotypic and genotypic characteristics of ESBL and AmpC producing organisms associated with bacteraemia in Ho Chi Minh City, Vietnam. Antimicrob Resist Infect Control. (2017) 6:265. doi: 10.1186/s13756-017-0265-1
50. Neela V, Rankouhi SZ, Van Belkum A, Goering RV, Awang R. Stenotrophomonas maltophilia in Malaysia: molecular epidemiology and trimethoprim-sulfamethoxazole resistance. Int J Infect Dis. (2012) 16:e603–7. doi: 10.1016/j.ijid.2012.04.004
51. Ning BT, Zhang CM, Liu T, Ye S, Yang ZH, Chen ZJ, et al. Pathogenic analysis of sputum from ventilator-associated pneumonia in a pediatric intensive care unit. Exp Ther Med. (2013) 5:367–71. doi: 10.3892/etm.2012.757
52. Rhee JY, Choi JY, Choi MJ, Song JH, Peck KR, Ko KS, et al. Distinct groups and antimicrobial resistance of clinical Stenotrophomonas maltophilia complex isolates from Korea. J Med Microbiol. (2013) 62:748–53. doi: 10.1099/jmm.0.053355-0
53. Shi SH, Kong HS, Xu J, Zhang WJ, Jia CK, Wang WL, et al. Multidrug resistant gram-negative bacilli as predominant bacteremic pathogens in liver transplant recipients. Transpl Infect Dis. (2009) 11:405–12. doi: 10.1111/j.1399-3062.2009.00421.x
54. Sun EL, Liang GH, Wang LN, Wei WJ, Lei MD, Song SD, et al. Antimicrobial susceptibility of hospital acquired Stenotrophomonas maltophilia isolate biofilms. Brazilian J Infectious Diseases. (2016) 20:365–73. doi: 10.1016/j.bjid.2016.04.002
55. Tan TY, Ng SY. The in-vitro activity of colistin in gram-negative bacteria. Singapore Med J. (2006) 47:621–4.
56. Tanimoto K. Stenotrophomonas maltophilia strains isolated from a university hospital in Japan: genomic variability and antibiotic resistance. J Med Microbiol. (2013) 62:565–70. doi: 10.1099/jmm.0.051151-0
57. Wang H, Yu Y, Xie X, Wang C, Zhang Y, Yuan Y, et al. In-vitro antibacterial activities of cefpiramide and other broad- spectrum antibiotics against 440 clinical isolates in China. J Inf Chemother. (2000) 6:81–5. doi: 10.1007/PL00012156
58. Wang WS, Liu CP, Lee CM, Huang FY. Stenotrophomonas maltophilia bacteremia in adults: four years' experience in a medical center in northern Taiwan. J Microbiol Immunol Infect. (2004) 37:359–65.
59. Wei C, Ni W, Cai X, Zhao J, Cui J. Evaluation of trimethoprim/sulfamethoxazole (SXT), minocycline, tigecycline, moxifloxacin, and ceftazidime alone and in combinations for sxt-susceptible and sxt-resistant Stenotrophomonas maltophilia by in vitro time-kill experiments. PLoS ONE. (2016) 11:132. doi: 10.1371/journal.pone.0152132
60. Wu H, Wang JT, Shiau YR, Wang HY, Yang Lauderdale TL, Chang SC, et al. A multicenter surveillance of antimicrobial resistance on Stenotrophomonas maltophilia in Taiwan. J Microbiol Immunol Inf. (2012) 45:120–6. doi: 10.1016/j.jmii.2011.09.028
61. Watanabe K, Zhu H, Willcox M. Susceptibility of Stenotrophomonas maltophilia clinical isolates to antibiotics and contact lens multipurpose disinfecting solutions. Inves Ophthalmol Visual Sci. (2014) 55:8475–9. doi: 10.1167/iovs.14-15667
62. Xu XF, Ma XL, Chen Z, Shi LP, Du LZ. Clinical characteristics of nosocomial infections in neonatal intensive care unit in eastern China. J Perinat Med. (2010) 38:431–7. doi: 10.1515/jpm.2010.063
63. Liu PY, Lau YJ, Hu BS, Shyr JM, Shi ZY, Tsai WS, et al. Comparison of susceptibility to extended-spectrum beta-lactam antibiotics and ciprofloxacin among gram-negative bacilli isolated from intensive care units. Diagn Microbiol Infect Dis. (1995) 22:285–91. doi: 10.1016/0732-8893(95)00096-S
64. Zhao S, Yang L, Liu H, Gao F. Stenotrophomonas maltophilia in a university hospital of traditional Chinese medicine: molecular epidemiology and antimicrobial resistance. J Hospital Infection. (2017) 96:286–9. doi: 10.1016/j.jhin.2017.04.001
65. Zhao J, Xing Y, Liu W, Ni W, Wei C, Wang R, et al. Surveillance of dihydropteroate synthase genes in Stenotrophomonas maltophilia by LAMP: implications for infection control and initial therapy. Front Microbiol. (2016) 7:1723. doi: 10.3389/fmicb.2016.01723
66. Zhao J, Liu YX, Liu Y, Wang D, Ni WT, Wang R, et al. frequency and genetic determinants of tigecycline resistance in clinically isolated Stenotrophomonas maltophilia in Beijing, China. Front Microbiol. (2018) 9:549. doi: 10.3389/fmicb.2018.00549
67. Zhang R, Sun Q, Hu YJ Yu H, Li Y, Shen Q, et al. Detection of the Smqnr quinolone protection gene and its prevalence in clinical isolates of Stenotrophomonas maltophilia in China. J Med Microbiol. (2012) 61:535–9. doi: 10.1099/jmm.0.037309-0
68. Chawla K, Vishwanath S, Munim FC. Nonfermenting Gram-negative Bacilli other than Pseudomonas aeruginosa and Acinetobacter Spp. causing resp tract infections in a tertiary care center. J Glob Infect Dis. (2013) 5:144–8. doi: 10.4103/0974-777X.121996
69. Chawla K, Vishwanath S, Gupta A. Stenotrophomonas maltophilia in lower respiratory tract infections. J Clin Diag Res. (2014) 8:DC20–2. doi: 10.7860/JCDR/2014/10780.5320
70. Garg A, Garg J, Kumar S, Bhattacharya A, Agarwal S, Upadhyay GC, et al. Molecular epidemiology and therapeutic options of carbapenem-resistant Gram-negative bacteria. Indian J Med Res. (2019) 149:285–9. doi: 10.4103/ijmr.IJMR_36_18
71. Gunasekar B, Shameembanu AS, Kalyani M. Non-fermenting gram-negative bacilli: Phenotypic identification and a correlation between biofilm formation and antibiotic susceptibility testing. Int J Res Pharm Sci. (2018) 9:1229–34.
72. Kaur P, Gautam V, Tewari R. Distribution of class 1 integrons, sul1 and sul2 genes among clinical isolates of Stenotrophomonas maltophilia from a tertiary care hospital in North India. Microb Drug Resistance. (2015) 21:380–5. doi: 10.1089/mdr.2014.0176
73. Nayyar C, Thakur P, Tak V, Saigal K. Stenotrophomonas maltophilia: An Emerging Pathogen in Paediatric Population. J Clin Diagn Res. (2017) 11:Dc08–dc11. doi: 10.7860/JCDR/2017/24304.9318
74. Paopradit P, Srinitiwarawong K, Ingviya N, Singkhamanan K, Vuddhakul V. Distribution and characterization of Stenotrophomonas maltophilia isolates from environmental and clinical samples in Thailand. J Hospital Infection. (2017) 97:185–91. doi: 10.1016/j.jhin.2017.06.006
75. Tantisiriwat W, Linasmita P. In vitro activity of sitafloxacin and other antibiotics against bacterial isolates from HRH princess maha chakri sirindhorn medical center, srinakharinwirot university and Samitivej Sukhumvit hospital. J Med Assoc Thailand. (2017) 100:469–78.
76. Averbuch D, Avaky C, Harit M, Stepensky P, Fried I, Ben-Ami T, et al. Non-fermentative Gram-negative rods bacteremia in children with cancer: a 14-year single-center experience. Infection. (2017) 45:327–34. doi: 10.1007/s15010-017-0988-1
77. Averbuch D, Tridello G, Hoek J, Mikulska M, Akan H, Yanez San Segundo L. Antimicrobial resistance in gram-negative rods causing bacteremia in hematopoietic stem cell transplant recipients: intercontinental prospective study of the infectious diseases working party of the european bone marrow transplantation group. Clin Infect Dis. (2017) 65:1819–28. doi: 10.1093/cid/cix646
78. Bousquet A, Malfuson JV, Sanmartin N, Konopacki J, Macnab C, Souleau B, et al. An 8-year survey of strains identified in blood cultures in a clinical haematology unit. Clin Microbiol Inf. (2014) 20:O7–O12. doi: 10.1111/1469-0691.12294
79. Canton R, Valdezate S, Vindel A, Del Saz BS, Maiz L, Baquero F, et al. Antimicrobial susceptibility profile of molecular typed cystic fibrosis Stenotrophomonas maltophilia isolates and differences with noncystic fibrosis isolates. Pediatr Pulmonol. (2003) 35:99–107. doi: 10.1002/ppul.10216
80. Chen HY, Bonfiglio G, Allen M, Piper D, Edwardson T, Mcvey D, et al. Multicentre survey of the comparative in-vitro activity of piperacillin/tazobactam against bacteria from hospitalized patients in the British Isles. J Antimicrob Chemother. (1993) 32:247–66. doi: 10.1093/jac/32.2.247
81. De Dios Caballero J, Del Campo R, Royuela A, Solé A, Máiz L, Olveira C, et al. Bronchopulmonary infection-colonization patterns in Spanish cystic fibrosis patients: Results from a national multicenter study. J Cystic Fibrosis. (2016) 15:357–65. doi: 10.1016/j.jcf.2015.09.004
82. Cikman A, Parlak M, Bayram Y, Guducuoglu H, Berktas M. Antibiotics resistance of Stenotrophomonas maltophilia strains isolated from various clinical specimens. Afr Health Sci. (2016) 16:149–52. doi: 10.4314/ahs.v16i1.20
83. Bonaventura G, D'antonio D, Catamo G, D'ercole S, Piccolomini R. Comparative in vitro activity of levofloxacin and ciprofloxacin against bacterial isolates from neutropenic patients. Chemother. (2002) 48:134–7. doi: 10.1159/000064918
84. Bonaventura G, D'antonio D, Catamo G, D'ercole S, Piccolomini R. Biofilm Formation by Stenotrophomonas maltophilia: Modulation by Quinolones, Trimethoprim-Sulfamethoxazole, and Ceftazidime. Antimicrob Agents Chemother. (2004) 48:151–60. doi: 10.1128/AAC.48.1.151-160.2004
85. Djordjevic ZM, Folic MM, Jankovic SM. Distribution and antibiotic susceptibility of pathogens isolated from adults with hospital-acquired and ventilator-associated pneumonia in intensive care unit. J Infect Public Health. (2017) 10:740–4. doi: 10.1016/j.jiph.2016.11.016
86. Esposito A, Pompilio A, Bettua C, Crocetta V, Giacobazzi E, Fiscarelli E. Evolution of Stenotrophomonas maltophilia in cystic fibrosis lung over chronic infection: a genomic and phenotypic population study. Front Microbiol. (2017) 8:15. doi: 10.3389/fmicb.2017.01590
87. Frank U, Jonas D, Lüpke T, Ribeiro-Ayeh B, Schmidt-Eisenlohr E, Rüden H, et al. Antimicrobial susceptibility among nosocomial pathogens isolated in intensive care units in Germany. European J Clin Microbiol Inf Diseases. (2000) 19:888–91. doi: 10.1007/s100960000389
88. Fadda G, Spanu T, Ardito F, Taddei C, Santangelo R, Siddu A, et al. Antimicrobial resistance among non-fermentative Gram-negative bacilli isolated from the respiratory tracts of Italian inpatients: a 3-year surveillance study by the Italian Epidemiological Survey. Int J Antimicrob Agents. (2004) 23:254–61. doi: 10.1016/j.ijantimicag.2003.07.017
89. Galani I, Kontopidou F, Souli M, Rekatsina PD, Koratzanis E, Deliolanis J, et al. Colistin susceptibility testing by Etest and disk diffusion methods. Int J Antimicrob Agents. (2008) 31:434–9. doi: 10.1016/j.ijantimicag.2008.01.011
90. Garcia-Rodriguez JA, Fresnadillo MJ, Garcia Garcia MI, Garcia-Sanchez E, Garcia-Sanchez JE, et al. Multicenter Spanish study of ciprofloxacin susceptibility in gram-negative bacteria. European J Clin Microbiol Inf Dis. (1995) 14:456–9. doi: 10.1007/BF02114906
91. Garcia-Rodriguez JA, Garcia Sanchez JE, Garcia Garcia MI, Garcia Sanchez E, Munoz Bellido JL. Antibiotic susceptibility profile of Xanthomonas maltophilia. In vitro activity of β-lactam/β-lactamase inhibitor combinations. Diag Microbiol Inf Disease. (1991) 14:239–43. doi: 10.1016/0732-8893(91)90038-H
92. Garcia-Rodriguez JA, Garcia Sanchez JE, Munoz Bellido JL, Garcia Sanchez E, Garcia Garcia MI. In-vitro activity of meropenem, a new carbapenem, against imipenem-resistant Pseudomonas aeruginosa and Xanthomonas maltophilia. J Chemother. (1991) 3:143–6. doi: 10.1080/1120009X.1991.11739081
93. Gesu GP, Marchetti F, Piccoli L, Cavallero A. Levofloxacin and ciprofloxacin in vitro activities against 4,003 clinical bacterial isolates collected in 24 Italian laboratories. Antimicrob Agents Chemother. (2003) 47:816–9. doi: 10.1128/AAC.47.2.816-819.2003
94. Glupczynski Y, Delmee M, Goossens H, Struelens M, Belgian Multicenter ICUSG. Distribution and prevalence of antimicrobial resistance among Gram-negative isolates in intensive care units (ICU) in Belgian hospitals between 1996 and 1999. Acta Clin Belg. (2001) 56:297–306. doi: 10.1179/acb.2001.044
95. Gomez-Garces JL, Aracil B, Gil Y, Burillo A. Susceptibility of 228 non-fermenting gram-negative rods to tigecycline and six other antimicrobial drugs. J Chemother. (2009) 21:267–71. doi: 10.1179/joc.2009.21.3.267
96. Goncalves-Vidigal P, Grosse-Onnebrink J, Mellies U, Buer J, Rath PM, Steinmann J, et al. Stenotrophomonas maltophilia in cystic fibrosis: Improved detection by the use of selective agar and evaluation of antimicrobial resistance. J Cystic Fibrosis. (2011) 10:422–7. doi: 10.1016/j.jcf.2011.06.010
97. Gordon NC, Wareham DW. Novel variants of the Smqnr family of quinolone resistance genes in clinical isolates of Stenotrophomonas maltophilia. J Antimicrob Chemother. (2010) 65:483–9. doi: 10.1093/jac/dkp476
98. Gospodarek E, Kania I, Bialek M. Sensitivity to antibiotics of Burkholderia (Pseudomonas) cepacia and Stenotrophomonas (Xanthomonas) maltophilia strains isolated from hospitalised patients. Medical Science Monitor. (1997) 3:807–12.
99. Gramegna A, Millar BC, Blasi F, Elborn JS, Downey DG, Moore JE, et al. In vitro antimicrobial activity of ceftolozane/tazobactam against Pseudomonas aeruginosa and other non-fermenting Gram-negative bacteria in adults with cystic fibrosis. J Global Antimicro Resistance. (2018) 14:224–7. doi: 10.1016/j.jgar.2018.03.002
100. Grillon A, Schramm F, Kleinberg M, Jehl F. Comparative activity of ciprofloxacin, levofloxacin and moxifloxacin against Klebsiella pneumoniae, Pseudomonas aeruginosa and Stenotrophomonas maltophilia assessed by minimum inhibitory concentrations and time-kill studies. PLoS ONE. (2016) 11:690. doi: 10.1371/journal.pone.0156690
101. Grohs P, Taieb G, Morand P, Kaibi I, Podglajen I, Lavollay M, et al. In vitro activity of ceftolozane-tazobactam against multidrug-resistant nonfermenting Gram-negative bacilli isolated from patients with cystic fibrosis. Antimicrob Agents Chemother. (2017) 61. doi: 10.1128/AAC.02688-16
102. Guembe M, Cercenado E, Alcala L, Marin M, Insa R, Bouza E, et al. Evolution of antimicrobial susceptibility patterns of aerobic and facultative gram-negative bacilli causing intra-abdominal infections: results from the SMART studies 2003-2007. Rev ESP Quimioter. (2008) 21:166–73.
103. Gulmez D, Cakar A, Sener B, Karakaya J, Hascelik G. Comparison of different antimicrobial susceptibility testing methods for Stenotrophomonas maltophilia and results of synergy testing. J Infect Chemother. (2010) 16:322–8. doi: 10.1007/s10156-010-0068-2
104. Gülmez D, Hasçelik G. Stenotrophomonas maltophilia: Antimicrobial resistance and molecular typing of an emerging pathogen in a Turkish university hospital. Clin Microbiol Inf. (2005) 11:880–6. doi: 10.1111/j.1469-0691.2005.01257.x
105. Güriz H, Çiftçi E, Ayberkin E, Aysev D, Ince E, Arsan S, et al. Stenotrophomonas maltophilia bacteraemia in Turkish children. Ann Trop Paediatr. (2008) 28:129–36. doi: 10.1179/146532808X302152
106. Hohl P, Frei R, Aubry P. In vitro susceptibility of 33 clinical case isolates of Xanthomonas maltophilia. Inconsistent correlation of agar dilution and of disk diffusion test results. Diagnostic Microbiol Inf Dis. (1991) 14:447–50. doi: 10.1016/0732-8893(91)90072-N
107. Hombach M, Bloemberg GV, Böttger EC. Effects of clinical breakpoint changes in CLSI guidelines 2010/2011 and EUCAST guidelines 2011 on antibiotic susceptibility test reporting of Gram-negative bacilli. J Antimicrob Chemother. (2012) 67:622–32. doi: 10.1093/jac/dkr524
108. Hoban D, Bouchillon S, Johnson J, Zhanel G, Butler D, Miller L, et al. Comparative in vitro potency of gemifloxacin and fluoroquinolones against recent European clinical isolates from a global surveillance study. European J Clin Microbiol Inf Dis. (2001) 20:814–9. doi: 10.1007/s100960100604
109. Juhász E, Pongrácz J, Iván M, Kristóf K. Antibiotic susceptibility of sulfamethoxazole-trimethoprim resistant Stenotrophomonas maltophilia strains isolated at a tertiary care centre in Hungary. Acta Microbiol Immunol Hung. (2015) 62:295–305. doi: 10.1556/030.62.2015.3.7
110. Klietmann W, Focht J, Nosner K. Retrospective resistance pattern of clinical isolates in vitro against imipenem and other antimicrobial agents between 1986 and 1989. Drug Inv. (1991) 3:270–7. doi: 10.1007/BF03259577
111. Koseoglu O, Sener B, Gulmez D, Altun B, Gur D. Stenotrophomonas maltophilia as a nosocomial pathogen. New Microbiol. (2004) 27:273–9.
112. Kucukates E. Antimicrobial resistance among Gram-negative bacteria isolated from intensive care units in a cardiology institute in Istanbul, Turkey. JPN J Infect Dis. (2005) 58:228–31.
113. Lakatos B, Jakopp B, Widmer A, Frei R, Pargger H, Elzi L, et al. Evaluation of treatment outcomes for Stenotrophomonas maltophilia bacteraemia. Infection. (2014) 42:553–8. doi: 10.1007/s15010-014-0607-3
114. Lanzafame A, Bonfiglio G, Santini L, Mattina R. In vitro activity of levofloxacin against recent gram-negative nosocomial pathogens. Chemother. (2005) 51:44–50. doi: 10.1159/000084418
115. Livermore DM, Mushtaq S, James D, Potz N, Walker RA, Charlett A, et al. In vitro activity of piperacillin/tazobactam and other broad-spectrum antibiotics against bacteria from hospitalised patients in the British Isles. Int J Antimicrob Agents. (2003) 22:14–27. doi: 10.1016/S0924-8579(03)00108-0
116. Livermore DM, Mushtaq S, Warner M, Woodford N. Comparative in vitro activity of sulfametrole/trimethoprim and sulfamethoxazole/trimethoprim and other agents against multiresistant Gram-negative bacteria. J Antimicrob Chemother. (2014) 69:1050–6. doi: 10.1093/jac/dkt455
117. Madi H, Lukic J, Vasiljevic Z, Biocanin M, Kojic M, Jovcic B, et al. Genotypic and Phenotypic Characterization of Stenotrophomonas maltophilia Strains from a Pediatric Tertiary Care Hospital in Serbia. PLoS ONE. (2016) 11:e0165660. doi: 10.1371/journal.pone.0165660
118. McKnight AJ, Shaw A, Goldsmith CE, Clarke L, Millar BC, Mccaughan J, et al. Comparison of in vitro susceptibilities to levofloxacin and ciprofloxacin with Pseudomonas aeruginosa and Stenotrophomonas maltophilia isolated from cystic fibrosis patients in Northern Ireland. Br J Biomed Sci. (2005) 62:30–2. doi: 10.1080/09674845.2005.11978067
119. Micozzi A, Venditti M, Monaco M, Friedrich A, Taglietti F, Santilli S, et al. Bacteremia due to Stenotrophomonas maltophilia in patients with hematologic malignancies. Clin Infect Dis. (2000) 31:705–11. doi: 10.1086/314043
120. Milne KE, Gould IM. Combination antimicrobial susceptibility testing of multidrug-resistant Stenotrophomonas maltophilia from cystic fibrosis patients. Antimicrob Agents Chemother. (2012) 56:4071–7. doi: 10.1128/AAC.00072-12
121. Pasargiklian I, Lusco G, Paizis G, Mascheroni E. Ticarcillin/clavulanic acid: determination of minimal inhibitory concentrations against bacterial strains isolated from patients in intensive care units. Comparison with other agents. J Chemother. (1996) 8:113–21. doi: 10.1179/joc.1996.8.2.113
122. Samonis G, Karageorgopoulos DE, Maraki S, Levis P, Dimopoulou D, Spernovasilis NA, et al. Stenotrophomonas maltophilia infections in a general hospital: patient characteristics, antimicrobial susceptibility, and treatment outcome. PLoS ONE. (2012) 7:e37375. doi: 10.1371/journal.pone.0037375
123. Samonis G, Maraki S, Rafailidis PI, Kapaskelis A, Kastoris AC, Falagas ME, et al. Antimicrobial susceptibility of Gram-negative nonurinary bacteria to fosfomycin and other antimicrobials. Future Microbiol. (2010) 5:961–70. doi: 10.2217/fmb.10.47
124. Schmitz FJ, Verhoef J, Fluit AC. Comparative activities of six different fluoroquinolones against 9,682 clinical bacterial isolates from 20 European university hospitals participating in the European SENTRY surveillance programme. The SENTRY participants group. Int J Antimicrob Agents. (1999) 12:311–7. doi: 10.1016/S0924-8579(99)00091-6
125. Traub WH, Spohr M. Comparative disk and broth dilution susceptibility test results with ticarcillin and timentin against Pseudomonas aeruginosa and Pseudomonas maltophilia. Chemother. (1987) 33:340–6. doi: 10.1159/000238519
126. Traub WH, Leonhard B, Bauer D. Antibiotic susceptibility of Stenotrophomonas (Xanthomonas) maltophilia: chemotherapy 44 164–173. Comparative (NCCLS criteria) evaluation of antimicrobial drugs with the agar dilution and the agar disk diffusion (Bauer-Kirby) tests. Chemother. (1998) 44:164–73. doi: 10.1159/000007111
127. Tripodi MF, Andreana A, Sarnataro G, Ragone E, Adinolfi LE, Utili R, et al. Comparative activities of isepamicin, amikacin, cefepime, and ciprofloxacin alone or in combination with other antibiotics against Stenotrophomonas maltophilia. Eur J Clin Microbiol Inf Diseases. (2001) 20:73–5. doi: 10.1007/PL00011239
128. Tunger O, Vural S, Cetin CB, Keles G, Borand H, Gazi H, et al. Clinical aspects and risk factors of nosocomial Stenotrophomonas maltophilia bacteremia episodes in a Turkish intensive care unit. J Chemother. (2007) 19:658–64. doi: 10.1179/joc.2007.19.6.658
129. Usarek P, Dobrzaniecka K, Szymanek-Majchrzak K, Sawicka-Grzelak A, Mlynarczyk A, Durlik M, et al. Drug susceptibility assessment in Stenotrophomonas maltophilia strains isolated from the blood of organ transplantation recipients in a warsaw teaching hospital during 2011 to 2014. Transplant Proc. (2016) 48:1411–3. doi: 10.1016/j.transproceed.2016.01.072
130. Valenza G, Tappe D, Turnwald D, Frosch M, Konig C, Hebestreit H, et al. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cyst Fibros. (2008) 7:123–7. doi: 10.1016/j.jcf.2007.06.006
131. Adams-Sapper S, Sergeevna-Selezneva J, Tartof S, Raphael E, An Diep B, Perdreau-Remington F, et al. Globally dispersed mobile drug-resistance genes in Gram-negative bacterial isolates from patients with bloodstream infections in a US urban general hospital. J Med Microbiol. (2012) 61:968–74. doi: 10.1099/jmm.0.041970-0
132. Alcaraz E, Garcia C, Papalia M, Vay C, Friedman L, Rossi D. Stenotrophomonas maltophilia isolated from patients exposed to invasive devices in a university hospital in Argentina: Molecular typing, susceptibility and detection of potential virulence factors. J Med Microbiol. (2018) 67:992–1002. doi: 10.1099/jmm.0.000764
133. Blondeau JM, Laskowski R, Borsos S. In-vitro activity of cefepime and seven other antimicrobial agents against 1518 non-fermentative Gram-negative bacilli collected from 48 Canadian health care facilities. J Antimicrobial Chemother. (1999) 44:545–8. doi: 10.1093/jac/44.4.545
134. Church D, Lloyd T, Peirano G, Pitout J. Antimicrobial susceptibility and combination testing of invasive Stenotrophomonas maltophilia isolates. Scand J Infect Dis. (2013) 45:265–70. doi: 10.3109/00365548.2012.732240
135. Denisuik AJ, Garbutt LA, Golden AR, Adam HJ, Baxter M, Nichol KA, et al. Antimicrobial-resistant pathogens in Canadian ICUs: Results of the CANWARD 2007 to 2016 study. J Antimicrobial Chemother. (2019) 74:645–53. doi: 10.1093/jac/dky477
136. Flamm RK, Rhomberg PR, Watters AA, Sweeney K, Ellis-Grosse EJ, Shortridge D, et al. Activity of fosfomycin when tested against US contemporary bacterial isolates. Diagn Microbiol Infect Dis. (2019) 93:143–6. doi: 10.1016/j.diagmicrobio.2018.08.010
137. Flores-Treviño S, Gutiérrez-Ferman JL, Morfín-Otero R, Rodríguez-Noriega E, Estrada-Rivadeneyra D, Rivas-Morales C, et al. Stenotrophomonas maltophilia in Mexico: Antimicrobial resistance, Biofilm formation and clonal diversity. J Med Microbiol. (2014) 63:1524–30. doi: 10.1099/jmm.0.074385-0
138. Forrester JB, Steed LL, Santevecchi BA, Flume P, Palmer-Long GE, Bosso JA, et al. In vitro activity of ceftolozane/tazobactam vs nonfermenting, gram- negative cystic fibrosis isolates. Open Forum Inf Diseases. (2018) 5:158. doi: 10.1093/ofid/ofy158
139. Fuchs PC, Barry AL, Brown SD. Survey of antimicrobial activity of four commonly used third generation cephalosporins tested against recent bacterial isolates from ten American medical centers, and assessment of disk diffusion test performance. AST Surveillance Group Diagn Microbiol Infect Dis. (1996) 24:213–9. doi: 10.1016/0732-8893(96)00028-4
140. Gerlach EH, Jones RN, Allen SD, Koontz FP, Murray PR, Pfaller MA, et al. Cefdinir (FK482), an orally administered cephalosporin in vitro activity comparison against recent clinical isolates from five medical centers and determination of MIC quality control guidelines. Diagn Microbiol Infect Dis. (1992) 15:537–43. doi: 10.1016/0732-8893(92)90105-3
141. Herrera-Heredia SA, Pezina-Cantú C, Garza-González E, Bocanegra-Ibarias P, Mendoza-Olazarán S, Morfín-Otero R, et al. Risk factors and molecular mechanisms associated with trimethoprim–sulfamethoxazole resistance in Stenotrophomonas maltophilia in Mexico. J Med Microbiol. (2017) 66:1102–9. doi: 10.1099/jmm.0.000550
142. Hoban DJ, Bouchillon SK, Johnson JL, Zhanel GG, Butler DL, Saunders KA, et al. Comparative in vitro potency of amoxycillin-clavulanic acid and four oral agents against recent North American clinical isolates from a global surveillance study. Int J Antimicrob Agents. (2003) 21:425–33. doi: 10.1016/S0924-8579(03)00038-4
143. Isenberg HD, Alperstein P, France K. In vitro activity of ciprofloxacin, levofloxacin, and trovafloxacin, alone and in combination with β-lactams, against clinical isolates of Pseudomonas aeruginosa, Stenotrophomonas maltophilia, and Burkholderia cepacia. Diagn Microbiol Infect Dis. (1999) 33:81–6. doi: 10.1016/S0732-8893(98)00126-6
144. Jones RN, Pfaller MA, Marshall SA, Hollis RJ, Wilke WW. Antimicrobial activity of 12 broad-spectrum agents tested against 270 nosocomial blood. Stream infection isolates caused by non-enteric gram-negative bacilli: occurrence of resistance molecular epidemiology and screening for metallo-enzymes. Diag Microbiol Inf Dis. (1997) 29:187–92. doi: 10.1016/S0732-8893(97)81808-1
145. Jones RN, Croco MAT, Pfaller MA, Beach ML, Kugler KC. Antimicrobial activity evaluations of gatifloxacin, a new fluoroquinolone: contemporary pathogen results from a global antimicrobial resistance surveillance program (SENTRY, 1997). Clin Microbiol Inf. (1999) 5:540–6. doi: 10.1111/j.1469-0691.1999.tb00432.x
146. Karlowsky JA, Adam HJ, Baxter MR, Lagacé-Wiens PRS, Walkty AJ, Hoban DJ, et al. In Vitro activity of ceftaroline-avibactam against gram-negative and gram-positive pathogens isolated from patients in canadian hospitals from 2010 to 2012: results from the CANWARD surveillance study. Antimicrob Agents Chemother. (2013) 57:5600–11. doi: 10.1128/AAC.01485-13
147. Karlowsky JA, Adam HJ, Decorby MR, Lagacé-Wiens PRS, Hoban DJ, Zhanel GG, et al. In vitro activity of ceftaroline against gram-positive and gram-negative pathogens isolated from patients in Canadian hospitals in 2009. Antimicrob Agents Chemother. (2011) 55:2837–46. doi: 10.1128/AAC.01787-10
148. Karlowsky JA, Kelly LJ, Thornsberry C, Jones ME, Evangelista AT, Critchley IA, et al. Susceptibility to fluoroquinolones among commonly isolated Gram-negative bacilli in 2000: TRUST and TSN data for the United States. Int J Antimicrob Agents. (2002) 19:21–31. doi: 10.1016/S0924-8579(01)00466-6
149. Krueger TS, Clark EA, Nix DE. In vitro susceptibility of Stenotrophomonas maltophilia to various antimicrobial combinations. Diagn Microbiol Infect Dis. (2001) 41:71–8. doi: 10.1016/S0732-8893(01)00281-4
150. Mutnick AH, Kirby JT, Jones RN, CANCER. resistance surveillance program: initial results from hematology-oncology centers in North America. Annal Pharmacother. (2003) 37:47–56. doi: 10.1345/aph.1C292
151. Nicodemo AC, Araujo MR, Ruiz AS, Gales AC. In vitro susceptibility of Stenotrophomonas maltophilia isolates: comparison of disc diffusion, Etest and agar dilution methods. J Antimicrob Chemother. (2004) 53:604–8. doi: 10.1093/jac/dkh128
152. Passerini De Rossi B, García C, Calenda M, Vay C, Franco M. Activity of levofloxacin and ciprofloxacin on biofilms and planktonic cells of Stenotrophomonas maltophilia isolates from patients with device-associated infections. Int J Antimicrob Agents. (2009) 34:260–4. doi: 10.1016/j.ijantimicag.2009.02.022
153. Poulos CD, Matsumura SO, Willey BM, Low DE, Mcgeer A. In vitro activities of antimicrobial combinations against Stenotrophomonas (Xanthomonas) maltophilia. Antimicrobial Agents Chemother. (1995) 39:2220–3. doi: 10.1128/AAC.39.10.2220
154. Rizek C, Ferraz JR, Van Der Heijden IM, Giudice M, Mostachio AK, Paez J, et al. In vitro activity of potential old and new drugs against multidrug-resistant gram-negatives. J Inf Chemother. (2015) 21:114–7. doi: 10.1016/j.jiac.2014.10.009
155. Rolston KV, Frisbee-Hume S, Leblanc B, Streeter H, Ho DH. In vitro antimicrobial activity of moxifloxacin compared to other quinolones against recent clinical bacterial isolates from hospitalized and community-based cancer patients. Diagn Microbiol Infect Dis. (2003) 47:441–9. doi: 10.1016/S0732-8893(03)00115-9
156. Rolston KV, Ho DH, Leblanc B, Streeter H, Dvorak T. In-vitro activity of trovafloxacin against clinical bacterial isolates from patients with cancer. J Antimicrob Chemother. (1997) 39:15–22. doi: 10.1093/jac/39.suppl_2.15
157. Rutter WC, Burgess DR, Burgess DS. Increasing incidence of multidrug resistance among cystic fibrosis respiratory bacterial isolates. Microbial Drug Resistance. (2017) 23:51–5. doi: 10.1089/mdr.2016.0048
158. Sader HS, Castanheira M, Mendes RE, Flamm RK. Frequency and antimicrobial susceptibility of Gram-negative bacteria isolated from patients with pneumonia hospitalized in ICUs of US medical centres (2015–17). J Antimicrobial Chemother. (2018) 73:3053–9. doi: 10.1093/jac/dky279
159. Sader HS, Jones RN. Antimicrobial activity of the new carbapenem biapenem compared to imipenem, meropenem and other broad-spectrum beta-lactam drugs. Eur J Clin Microbiol Infect Dis. (1993) 12:384–91. doi: 10.1007/BF01964439
160. Sahm DF, Critchley IA, Kelly LJ, Karlowsky JA, Mayfield DC, Thornsberry C, et al. Evaluation of current activities of fluoroquinolones against gram-negative bacilli using centralized in vitro testing and electronic surveillance. Antimicrob Agents Chemother. (2001) 45:267–74. doi: 10.1128/AAC.45.1.267-274.2001
161. San Gabriel P, Zhou JY, Tabibi S, Chen YH, Trauzzi M, Saiman L, et al. Antimicrobial susceptibility and synergy studies of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis. Antimicrob Agents Chemother. (2004) 48:168–71. doi: 10.1128/AAC.48.1.168-171.2004
162. Sattler CA, Mason EO, Kaplan SL. Nonrespiratory Stenotrophomonas maltophilia infection at a children's hospital. Clin Infect Dis. (2000) 31:1321–30. doi: 10.1086/317473
163. Travassos LH, Pinheiro MN, Coelho FS, Sampaio JL, Merquior VL, Marques EA, et al. Phenotypic properties, drug susceptibility and genetic relatedness of Stenotrophomonas maltophilia clinical strains from seven hospitals in Rio de Janeiro, Brazil. J Appl Microbiol. (2004) 96:1143–50. doi: 10.1111/j.1365-2672.2004.02248.x
164. Spierer O, Miller D, O'Brien TP. Comparative activity of antimicrobials against Pseudomonas aeruginosa, Achromobacter xylosoxidans and Stenotrophomonas maltophilia keratitis isolates. British J Ophthalmol. (2018) 102:708–12. doi: 10.1136/bjophthalmol-2017-311751
165. Zhanel GG, Adam HJ, Low DE, Blondeau J, Decorby M, Karlowsky JA, et al. Antimicrobial susceptibility of 15,644 pathogens from Canadian hospitals: results of the CANWARD 2007-2009 study. Diagn Microbiol Infect Dis. (2011) 69:291–306. doi: 10.1016/j.diagmicrobio.2010.10.025
166. Zhanel GG, Baxter MR, Adam HJ, Sutcliffe J, Karlowsky JA. In vitro activity of eravacycline against 2213 Gram-negative and 2424 Gram-positive bacterial pathogens isolated in Canadian hospital laboratories: CANWARD surveillance study 2014–2015. Diagn Microbiol Infect Dis. (2018) 91:55–62. doi: 10.1016/j.diagmicrobio.2017.12.013
167. Zhanel GG, Decorby M, Nichol KA, Wierzbowski A, Baudry PJ, Karlowsky JA, et al. Antimicrobial susceptibility of 3931 organisms isolated from intensive care units in Canada: canadian National Intensive Care Unit Study, 2005/2006. Diagn Microbiol Infect Dis. (2008) 62:67–80. doi: 10.1016/j.diagmicrobio.2008.04.012
168. Chow JW, Satishchandran V, Snyder TA, Harvey CM, Friedland IR, Dinubile MJ, et al. In vitro susceptibilities of aerobic and facultative gram-negative bacilli isolated from patients with intra-abdominal infections worldwide: the 2002 Study for Monitoring Antimicrobial Resistance Trends (SMART). Surg Infect. (2005) 6:439–47. doi: 10.1089/sur.2005.6.439
169. Corlouer C, Lamy B, Desroches M, Ramos-Vivas J, Mehiri-Zghal E, Lemenand O. Stenotrophomonas maltophilia healthcare-associated infections: identification of two main pathogenic genetic backgrounds. J Hospital Inf. (2017) 96:183–8. doi: 10.1016/j.jhin.2017.02.003
170. Diez-Aguilar M, Ekkelenkamp M, Morosini MI, Merino I, Dios Caballero D. Antimicrobial susceptibility of non-fermenting Gram-negative pathogens isolated from cystic fibrosis patients. Int J Antimicrob Agents. (2019) 53:84–8. doi: 10.1016/j.ijantimicag.2018.09.001
171. Farrell DJ, Flamm RK, Sader HS, Jones RN. Ceftobiprole activity against over 60,000 clinical bacterial pathogens isolated in Europe, Turkey, and Israel from 2005 to 2010. Antimicrobial Agents Chemother. (2014) 58:3882–8. doi: 10.1128/AAC.02465-14
172. Farrell DJ, Sader HS, Jones RN. Antimicrobial susceptibilities of a worldwide collection of Stenotrophomonas maltophilia isolates tested against tigecycline and agents commonly used for S. maltophilia infections. Antimicrobial Agents Chemother. (2010) 54:2735–7. doi: 10.1128/AAC.01774-09
173. Fedler KA, Biedenbach DJ, Jones RN. Assessment of pathogen frequency and resistance patterns among pediatric patient isolates: report from the 2004 SENTRY Antimicrobial Surveillance Program on 3 continents. Diagn Microbiol Infect Dis. (2006) 56:427–36. doi: 10.1016/j.diagmicrobio.2006.07.003
174. Flamm RK, Castanheira M, Streit JM, Jones RN. Minocycline activity tested against Acinetobacter baumannii complex, Stenotrophomonas maltophilia, and Burkholderia cepacia species complex isolates from a global surveillance program (2013). Diagn Microbiol Infect Dis. (2016) 85:352–5. doi: 10.1016/j.diagmicrobio.2016.03.019
175. Frei R, Jones RN, Pignatari AC, Yamane N, Marco F, Hoban DJ, et al. Antimicrobial activity of FK-037, a new broad-spectrum cephalosporin. International in vitro comparison with cefepime and ceftazidime. Diagn Microbiol Infect Dis. (1994) 18:167–73. doi: 10.1016/0732-8893(94)90087-6
176. Fritsche TR, Sader HS, Stilwell MG, Dowzicky MJ, Jones RN. Antimicrobial activity of tigecycline tested against organisms causing community-acquired respiratory tract infection and nosocomial pneumonia. Diagn Microbiol Infect Dis. (2005) 52:187–93. doi: 10.1016/j.diagmicrobio.2005.05.004
177. Gales AC, Jones RN, Sader HS. Global assessment of the antimicrobial activity of polymyxin B against 54 731 clinical isolates of Gram-negative bacilli: report from the SENTRY antimicrobial surveillance programme (2001-2004). Clin Microbiol Inf. (2006) 12:315–21. doi: 10.1111/j.1469-0691.2005.01351.x
178. Gales AC, Jones RN, Sader HS. Antimicrobial susceptibility profile of contemporary clinical strains of Stenotrophomonas maltophilia isolates: can moxifloxacin activity be predicted by levofloxacin MIC results? J Chemother. (2008) 20:38–42. doi: 10.1179/joc.2008.20.1.38
179. Hoban DJ, Jones RN, Yamane N, Frei R, Trilla A, Pignatari AC, et al. In vitro activity of three carbapenem antibiotics. Comparative studies with biapenem (L-627) imipenem and meropenem against aerobic pathogens isolated worldwide. Diagn Microbiol Infect Dis. (1993) 17:299–305. doi: 10.1016/0732-8893(93)90039-A
180. Jones RN, Sader HS, Beach ML. Contemporary in vitro spectrum of activity summary for antimicrobial agents tested against 18 569 strains non-fermentative Gram-negative bacilli isolated in the SENTRY Antimicrobial Surveillance Program (1997-2001). Int J Antimicrob Agents. (2003) 22:551–6. doi: 10.1016/S0924-8579(03)00245-0
181. Liu YM, Chen YS, Toh HS, Huang CC, Lee YL, Ho CM, et al. In vitro susceptibilities of non-Enterobacteriaceae isolates from patients with intra-abdominal infections in the Asia-Pacific region from 2003 to 2010: Results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). Int J Antimicrob Agents. (2012) 40:S11–7. doi: 10.1016/S0924-8579(12)70004-3
182. Renteria MI, Biedenbach DJ, Bouchillon SK, Hoban DJ, Raghubir N, Sajben P, et al. In vitro activity of tigecycline against isolates collected from complicated skin and skin structure infections and intra-abdominal infections in Africa and Middle East countries: TEST 2007-2012. Diagn Microbiol Infect Dis. (2014) 79:54–9. doi: 10.1016/j.diagmicrobio.2014.01.017
183. Sader HS, Castanheira M, Farrell DJ, Flamm RK, Mendes RE, Jones RN, et al. Tigecycline antimicrobial activity tested against clinical bacteria from Latin American medical centres: results from SENTRY Antimicrobial Surveillance Program (2011–2014). Int J Antimicrob Agents. (2016) 48:144–50. doi: 10.1016/j.ijantimicag.2016.04.021
184. Sader HS, Farrell DJ, Flamm RK, Jones RN. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalised with pneumonia in US and European hospitals: results from the SENTRY Antimicrobial Surveillance Program, 2009-2012. Int J Antimicrob Agents. (2014) 43:328–34. doi: 10.1016/j.ijantimicag.2014.01.007
185. Sader HS, Flamm RK, Jones RN. Tigecycline activity tested against antimicrobial resistant surveillance subsets of clinical bacteria collected worldwide (2011). Diagn Microbiol Infect Dis. (2013) 76:217–21. doi: 10.1016/j.diagmicrobio.2013.02.009
186. Sader HS, Jones RN, Dowzicky MJ, Fritsche TR. Antimicrobial activity of tigecycline tested against nosocomial bacterial pathogens from patients hospitalized in the intensive care unit. Diagn Microbiol Infect Dis. (2005) 52:203–8. doi: 10.1016/j.diagmicrobio.2005.05.002
187. Thomson KS, Moland ES, Sanders CC. Activity of trovafloxacin against antibiotic-resistant bacterial pathogens. Inf Dis Clin Prac. (1999) 8:S7–S16. doi: 10.1097/00019048-199905001-00003
188. Toleman MA, Bennett PM, Bennett DMC, Jones RN, Walsh TR. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg Infect Dis. (2007) 13:559–65. doi: 10.3201/eid1304.061378
189. Tsiodras S, Pittet D, Carmeli Y, Eliopoulos G, Boucher H, Harbarth S, et al. Clinical implications of Stenotrophomonas maltophilia resistant to trimethoprim-sulfamethoxazole: a study of 69 patients at 2 University Hospitals. Scand J Infect Dis. (2000) 32:651–6. doi: 10.1080/003655400459577
190. Yamane N, Jones RN, Frei R, Hoban DJ, Pignatari AC, Marco F, et al. Levofloxacin in vitro activity: results from an international comparative study with ofloxacin and ciprofloxacin. J Chemother. (1994) 6:83–91. doi: 10.1080/1120009X.1994.11741134
191. Abdel-Aziz N, Morsy MMF, Amin SS, Mohammed KI, Alharbi AE, Alshami I, et al. Threatening problem of Stenotrophomonas maltophilia producing extended-spectrum beta-lactamases: prevalence and automated antibiotic susceptibility pattern. Clin Microbiol. (2013) 12:108. doi: 10.4172/2327-5073.1000108
192. Jia W, Wang J, Xu H, Li G. Resistance of Stenotrophomonas maltophilia to fluoroquinolones: prevalence in a university hospital and possible mechanisms. Int J Environ Res Public Health. (2015) 12:5177–95. doi: 10.3390/ijerph120505177
193. Nicodemo A, Paez J. Antimicrobial therapy for Stenotrophomonas maltophilia infections. Eur J Clin Microbiol Inf Dis. (2007) 26:229–37. doi: 10.1007/s10096-007-0279-3
194. Waters V. New treatments for emerging cystic fibrosis pathogens other than Pseudomonas. Curr Pharm Des. (2012) 18:696–725. doi: 10.2174/138161212799315939
195. Al-Anazi KA, Al-Jasser AM. Infections caused by Stenotrophomonas maltophilia in recipients of hematopoietic stem cell transplantation. Front Oncol. (2014) 4:232. doi: 10.3389/fonc.2014.00232
196. Scholte JB, Zhou TL, Bergmans DC, Rohde GG, Winkens B, Van Dessel HA, et al. Stenotrophomonas maltophilia ventilator-associated pneumonia. A Retrospective Matched case-control study. Inf Dis. (2016) 48:738–43. doi: 10.1080/23744235.2016.1185534
197. Gales AC, Jones R, Forward K, Linares J, Sader HS, Verhoef J, et al. Emerging importance of multidrug-resistant Acinetobacter species and Stenotrophomonas maltophilia as pathogens in seriously ill patients: geographic patterns, epidemiological features, and trends in the SENTRY Antimicrobial Surveillance Program (1997–1999). Clin Infect Dis. (2001) 32:S104–13. doi: 10.1086/320183
198. Farrell DJ, Sader HS, Flamm RK, Jones RN. Ceftolozane/tazobactam activity tested against Gram-negative bacterial isolates from hospitalised patients with pneumonia in US and European medical centres (2012). Int J Antimicrob Agents. (2014) 43:533–9. doi: 10.1016/j.ijantimicag.2014.01.032
199. Rogues A, Maugein J, Allery A, Fleureau C, Boulestreau H, Surcin S, et al. Electronic ventilator temperature sensors as a potential source of respiratory tract colonization with Stenotrophomonas maltophilia. J Hospital Infection. (2001) 49:289–92. doi: 10.1053/jhin.2001.1099
200. Abreu Vidipó D, De Andrade Marques L, Puchelle EE, Plotkowski MC. Stenotrophomonas maltophilia interaction with human epithelial respiratory cells in vitro. Microbiol Immunol. (2001) 45:563–9. doi: 10.1111/j.1348-0421.2001.tb01287.x
201. Kalanuria AA, Mirski M, Ziai W. Ventilator-associated pneumonia in the ICU. Ann Update Int Care Emerg Med. (2014) 2014:65–77. doi: 10.1007/978-3-319-03746-2_6
202. Brooke JS. New Strategies Against Stenotrophomonas maltophilia: A Serious Worldwide Intrinsically Drug-Resistant Opportunistic Pathogen. New York, NY: Taylor and Francis. (2014).
203. Ratjen A, Yau Y, Wettlaufer J, Matukas L, Zlosnik JE, Speert DP, et al. In vitro efficacy of high-dose tobramycin against Burkholderia cepacia complex and Stenotrophomonas maltophilia isolates from cystic fibrosis patients. Antimicrob Agents Chemother. (2015) 59:711–3. doi: 10.1128/AAC.04123-14
204. Cho SY, Kang CI, Kim J, Ha YE, Chung DR, Lee NY, et al. Can levofloxacin be a useful alternative to trimethoprim-Sulfamethoxazole for treating Stenotrophomonas maltophilia bacteremia? Antimicrob Agents Chemother. (2014) 58:581–3. doi: 10.1128/AAC.01682-13
205. Sarzynski SH, Warner S, Sun J, Matsouaka R, Dekker JP, Babiker A, et al. Trimethoprim-sulfamethoxazole versus levofloxacin for Stenotrophomonas maltophilia infections: a retrospective comparative effectiveness study of electronic health records from 154 US hospitals. Open Forum Infectious Diseases. (2022) 17:ofab644. doi: 10.1093/ofid/ofab644
206. Biagi M, Tan X, Wu T, Jurkovic M, Vialichka A, Meyer K, et al. Activity of potential alternative treatment agents for Stenotrophomonas maltophilia isolates nonsusceptible to levofloxacin and/or trimethoprim-sulfamethoxazole. J Clin Microbiol. (2020) 58:e01603–19. doi: 10.1128/JCM.01603-19
207. Mojica M, Rutter J, Taracila M, Abriata L, Fouts D, Papp-Wallace K, et al. Population structure, molecular epidemiology, and beta-lactamase diversity among Stenotrophomonas maltophilia isolates in the United States. MBio. (2019) 10:e00405–19. doi: 10.1128/mBio.00405-19
Keywords: Stenotrophomonas maltophilia, prevalence, antibiotic resistance, global, meta-analysis
Citation: Banar M, Sattari-Maraji A, Bayatinejad G, Ebrahimi E, Jabalameli L, Beigverdi R, Emaneini M and Jabalameli F (2023) Global prevalence and antibiotic resistance in clinical isolates of Stenotrophomonas maltophilia: a systematic review and meta-analysis. Front. Med. 10:1163439. doi: 10.3389/fmed.2023.1163439
Received: 10 February 2023; Accepted: 14 April 2023;
Published: 05 May 2023.
Edited by:
Asad U. Khan, Aligarh Muslim University, IndiaReviewed by:
Nayeem Ahmad, Arabian Gulf University, BahrainShraddha Karve, Ashoka University, India
Copyright © 2023 Banar, Sattari-Maraji, Bayatinejad, Ebrahimi, Jabalameli, Beigverdi, Emaneini and Jabalameli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fereshteh Jabalameli, amFiYWxhbWZAdHVtcy5hYy5pcg==
 Maryam Banar
Maryam Banar Azin Sattari-Maraji1
Azin Sattari-Maraji1 Mohammad Emaneini
Mohammad Emaneini Fereshteh Jabalameli
Fereshteh Jabalameli