- 1Department of Pathology, School of Basic Medical Sciences, Southern Medical University, Guangzhou, China
- 2Department of Pathology, Guangdong Provincial People's Hospital, Guangdong Academy of Medical Sciences, Guangzhou, China
- 3Department of Ophthalmology, Shenzhen Nanshan People's Hospital and the 6th Affiliated Hospital of Shenzhen University Health Science Center, Shenzhen, China
- 4Department of Hepatopancreatobiliary Surgery, Institute of Translational Medicine, Shenzhen University Health Science Center, Shenzhen University School of Medicine, First Affiliated Hospital of Shenzhen University, Shenzhen Second People's Hospital, Shenzhen, China
- 5Department of Pathology and Pathophysiology School of Medicine, Tongji University, China
- 6Department of Biochemistry, College of Science, Northeastern University, Boston, MA, United States
- 7Imaging Department, Institute of Translational Medicine, Shenzhen University Health Science Center, Shenzhen University School of Medicine, First Affiliated Hospital of Shenzhen University, Shenzhen Second People's Hospital, Shenzhen, China
Age-related macular degeneration (AMD) causes central vision impairment with increased incidence. In the pathogenesis of AMD, reactive oxygen species (ROS) are associated with RPE cell apoptosis. H2O2 is an oxidative toxicant and is used to establish the AMD in vitro model. However, the mechanisms of ROS in H2O2-induced AMD are still unclear. Fullerenol, a promising antioxidant of nanomaterials, protects RPE cells from ROS attack. In addition to working as a scavenger, little is known about the antioxidant mechanism of fullerenol in RPE cells. In this study, transcriptome sequencing was performed to examine the global changes in mRNA transcripts induced by H2O2 in human ARPE-19 cells. Moreover, we comprehensively investigated the protective effects of fullerenol against H2O2-induced oxidative injury by RNA sequencing. Gene Ontology enrichment analysis showed that those pathways related to the release of positive regulation of DNA-templated transcription and negative regulation of apoptotic process were affected. Finally, we found that 12 hub genes were related to the oxidative-protection function of fullerenol. In summary, H2O2 affected these hub genes and signaling pathways to regulate the senescence of RPE cells. Moreover, fullerenol is a potent nanomaterial that protects the RPE and would be a promising approach for AMD prevention.
Introduction
Aged-related macular degeneration (AMD) causes severe vision damage and loss by affecting the macular region of the retina (1). In Caucasians, for people aged over 70 (including 70), the overall early AMD prevalence is 13.2% (2). AMD is usually associated with the destruction of photoreceptors, abnormalities in the retinal pigment epithelium (RPE), and degeneration of the choriocapillaris (3, 4). The key contributor to AMD pathogenesis is oxidative damage-induced RPE senescence (5, 6). Oxidative stress can result in permanent cell senescence and overproduction of reactive oxygen species (ROS) (7). Previous studies have shown that reactive oxygen species contribute significantly to AMD. Accumulated ROS drives DNA damage, and the permanent DNA damage response induces the state of senescence of RPE cells (8, 9). Therefore, AMD can be prevented and delayed by avoiding oxidative damage to RPE cells.
Hydrogen peroxide (H2O2)-induced RPE damage is a method for establishing the AMD in vitro model (10–12). A previous study showed that a low concentration of H2O2 caused RPE senescence (13), and a high concentration caused cell death in a dose-dependent manner. Antioxidants, including melatonin (14), quercetin (15), farrerol (10), and kaempferol, can protect RPE cells from H2O2-induced apoptosis (16). In addition, H2O2 was reported to trigger necrosis in the RPE (17). Further investigation is needed to study H2O2-induced RPE cell senescence.
Fullerenol (C60[OH]n), derived from fullerene C60, shows great antioxidative potential in pharmaceutics and medical treatment of oxidative stress-related diseases (18). The good water solubility of fullerenol makes it useful in pharmaceutics and medical treatment of oxidative stress-related diseases (19, 20). It can remove free radicals such as superoxide anion radicals, hydroxyl radicals, lipid peroxyl radicals, and nitrous oxide radicals, making it effective in anti-aging, antioxidant stress, anti-inflammation, and anti-apoptosis (21, 22). In addition, ROS can bind to the electron-deficient position of fullerenol. As a result, fullerenol has the ability to reduce oxidative stress in cells (23). The cytotoxicity of fullerenol was proven to be low (24). Therefore, these advantages make fullerenol nanoparticles significant and promising in oxidative damage-induced disease research and treatment. Nanoparticle fullerenol was also proven to protect the RPE from oxidatively induced senescence by activating the SIRT1 pathway (25). However, the comprehensive transcriptional profile of fullerenol nanoparticles on senescent RPE is still unclear.
We comprehensively profiled the gene expression of ARPE-19 cells treated with H2O2 and/or fullerenol for the first time. We also identified 12 hub genes that were rescued by fullerenol treatment in H2O2-induced ARPE-19 senescent cells. These genes showed promise as therapeutic targets for AMD. In summary, our results provide evidence for a deeper investigation into the function of fullerenol nanoantioxidants in AMD treatment.
Materials and methods
Reagents
Fullerenol was synthesized by Jing-Ying Xu with previous published methods (26). The Senescence-Associated β-Galactosidase (SA-β-galactosidase) Staining Kit was from Cell Signaling Technology, Beverly, MA, USA. The PrimeScript TM RT Reagent Kit was from Takara, Dalian, China. TRIzolTM Reagent was purchased from Thermo Fisher Scientific (Waltham, MA, USA). DMEM/F12 1:1 (1X), FBS, P/S, and trypsin–EDTA were from GIBCO (Carlsbad, CA, USA), and the CCK-8 assay was performed with a cell counting kit by Yeasen Biotech (Jiangsu, China). 3% (w/w) H2O2 was purchased from Sigma Aldrich (St. Louis, MO, USA).
Cell viability and in situ staining for SA-β-galactosidase activity
ARPE-19 cells were grown in 1:1 (1X) DMEM/F12 with 10% (v/v) FBS and 1% (v/v) P/S. Cells were passaged with 0.25% (v/v) trypsin/0.2% EDTA every 3–4 days. ARPE-19 cells were plated in a 96-well plate and treated at 80% confluence. Then, 3% (w/w) H2O2 was used to make a medium with the intended H2O2 concentration. H2O2 solution was freshly diluted each time. For the H2O2 exposure, the medium used for the cells was changed to DMEM/F12 with the desired concentration of H2O2. Before establishing the cell senescence model, the concentration of H2O2 best used for stimulating ARPE-19 cells was explored. H2O2 (0 μM, 50 μM, 100 μM, 150 μM, 200 μM, and 400 μM) diluted with the cell culture medium was tested to find the most suitable concentrations. For the fullerenol exposure, after 2 h of exposure to H2O2, 5ug/mL fullerenol was added and further incubated for 22 h before analysis. Cell viability was analyzed by CCK-8 assay following manufacturers instructions. Experiments were repeated at least 3 times. In the SA-β-galactosidase staining assay and RNA sequencing, the treatment of the H2O2 exposure group and fullerenol treatment group followed previous studies (25).
RNA sequencing
The RNA-Seq Samples Consisted of Three Groups, Represented by the Control Group, the H2O2 Group, and the Fullerenol Group. For Each Group, Three Biological Replicates Were Considered, for a Total of 9 Samples. CDNA Libraries From These Samples Were Sequenced and Analyzed According to the Protocols for RNA-Seq (Novogene Company, Beijing, China). The Gene Expression Distribution of Each Sample That Passed Quality Control Was Used for Further Analysis (Supplementary Figures S1A–D).
Identification of differentially expressed genes (DEGs)
DESeq2 R Pakage Is Used for Differential Analysis of Comparative High-Throughput Sequencing Assays, Based on Gene Count Data. Shrinkage Estimation for Dispersions and Fold Changes Are Used to Improve Stability and Interpretability of Estimates. DEGs of the Three Groups in Our Study Were Analyzed by the DESeq2 R Package (1.16.1) With Fold Change > 1.5 and P < 0.05. The Overlay of DEGs Was Performed by Evenn (Http://www.Ehbio.com/Test/Venn/#/).
Gene ontology (GO) enrichment analysis
DAVID is a popular bioinformatics resource system including a web server and web service for functional annotation and enrichment analyses of gene lists. It consists of a comprehensive knowledge base and a set of functional analysis tools. We examined the DEGs by GO enrichment analysis of DAVID (https://david.ncifcrf.gov/) with a significance threshold of P < 0.05. Biological processes enriched by DEGs were obtained for further analysis.
Functional network analysis of the hub genes
The potential roles and related genes of the genes were analyzed by GeneMANIA (27, 28). GeneMANIA identifies other genes that are associated with the input genes. GeneMANIA used very large datasets of functional association data including protein and genetic interactions, pathways, co-expression, co-localization, and similarity of the protein domain (27, 28).
Ingenuity Pathway Analysis software (IPA, QIAGEN) was used to analyze the potential regulatory networks and related diseases of the hub genes. Ingenuity Pathway Analysis (IPA) can interpret the biological changes, altered canonical pathways, upstream transcriptional regulators, and gene networks, which contains large knowledge of gene functions and interaction networks based on published literature collected by the Ingenuity Pathway Analysis software.
Statistical analysis
The statistical analyses were conducted in R 4.0.5. The student's t-test was used for statistical analysis. P < 0.05 was considered statistically significant. The heatmap plots were computed and visualized using the R package pheatmap.
Results
H2O2-induced senescence in ARPE-19 cells
A cell senescence model and fullerenol treatment model were first established by exposing ARPE-19 cells to H2O2, as illustrated in Figure 1A. The cell cultures were exposed to H2O2 at various concentrations. As detected by the CCK-8 assay, statistical significance was found between the control and the H2O2-treated groups at 200 μM and 400 μM, respectively (Figure 1B). Thus, H2O2 at a 200 μM concentration was used in the following experiments. The results suggested that ARPE-19 cells treated with 200 μM H2O2 for 2 hours for five consecutive days were able to establish a senescence model, as confirmed by senescence-associated β-galactosidase staining (SA-β-galactosidase) (Figure 1C). In the control group, few cells were positive with SA-β-galactosidase staining (2%) (Figure 1C). In the H2O2 group, the ratio of cells that were positive with SA-β-galactosidase staining increased to 16% (Figure 1C).
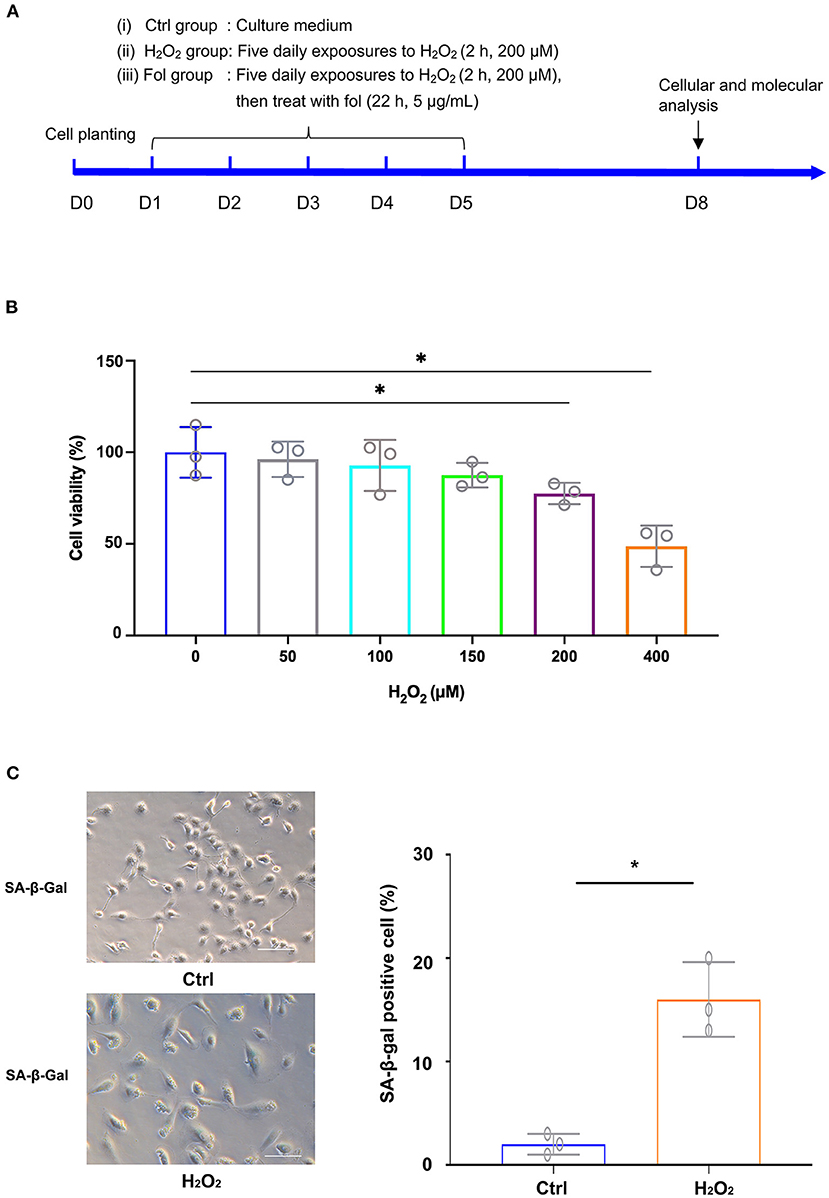
Figure 1. H2O2-Induced Senescence in ARPE-19 Cells. (A) Experimental design and reference time frame. (B) ARPE-19 cells were treated with H2O2 (50 μM, 100 μM, 150 μM, 200 μM, 400 μM) for 2 h daily. Cell viability was analyzed with the CCK8 method. (C) ARPE-19 cells in the control group and H2O2 group of ARPE-19 cells were stained with senescence-associated β-galactosidase (SA-β-Gal). Fol, fullerenol; *p < 0.05.
Analysis of differentially expressed mRNAs by RNA sequencing in H2O2-treated ARPE-19 cells
To determine the transcriptome profile of H2O2-treated ARPE-19 cells, we performed RNA sequencing of the H2O2-treated group and the control group of ARPE-19 cells, with each group containing three biological replicates. We performed differentially expressed gene analysis by DESeq2. In total 2,926 and 825 were considered as significantly up-or down- regulated genes after H2O2 treatment, respectively, compared with the control sample, respectively (Figures 2A,B). Among them, CXCL8, SOD2, PLAT, CLSTN2, TXNIP, BIRC3, and CLDN1 were the top up-regulated genes after H2O2 treatment, while DIO2 was the top down-regulated gene (Figure 2A).
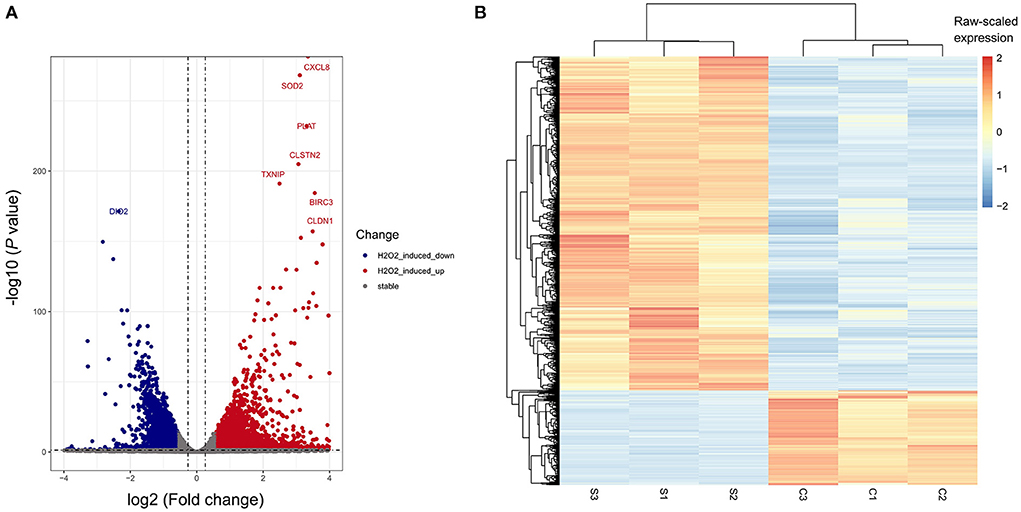
Figure 2. Differentially-expressed genes (DEGs) of H2O2-treated ARPE-19 cells. (A) Volcano plot of DEGs of RPE cells induced by H2O2. (B) Heatmap plot of DEGs of RPE cells induced by H2O2. S, H2O2 group; C, Ctrl group.
Fullerenol-induced dynamic gene expression changes in H2O2-treated ARPE-19 cells
Fullerenol was shown to protect the RPE from oxidatively induced senescence in a previous study (25), which was also confirmed in our study (Figure 3A). Therefore, we performed RNA-seq to analyze the cytoprotective effect of fullerenol. DESeq2 was used to analyze differentially expressed genes with fold change ≥ 1.5 and P < 0.05 (Figure 3B). As a result, 2,926 and 825 genes were up- or downregulated after H2O2 induction in ARPE-19 cells, respectively (Figures 3B,C). At the same time, 2,274 and 3,070 genes were up- or downregulated after H2O2 and fullerenol treatment, respectively, indicating the dynamic gene expression regulation induced by fullerenol. Among them, we found that 95 and 44 genes were up- or downregulated between fullerenol and H2O2 treatment, respectively (Figure 3B). These genes might play crucial roles in the biological processes induced by fullerenol.
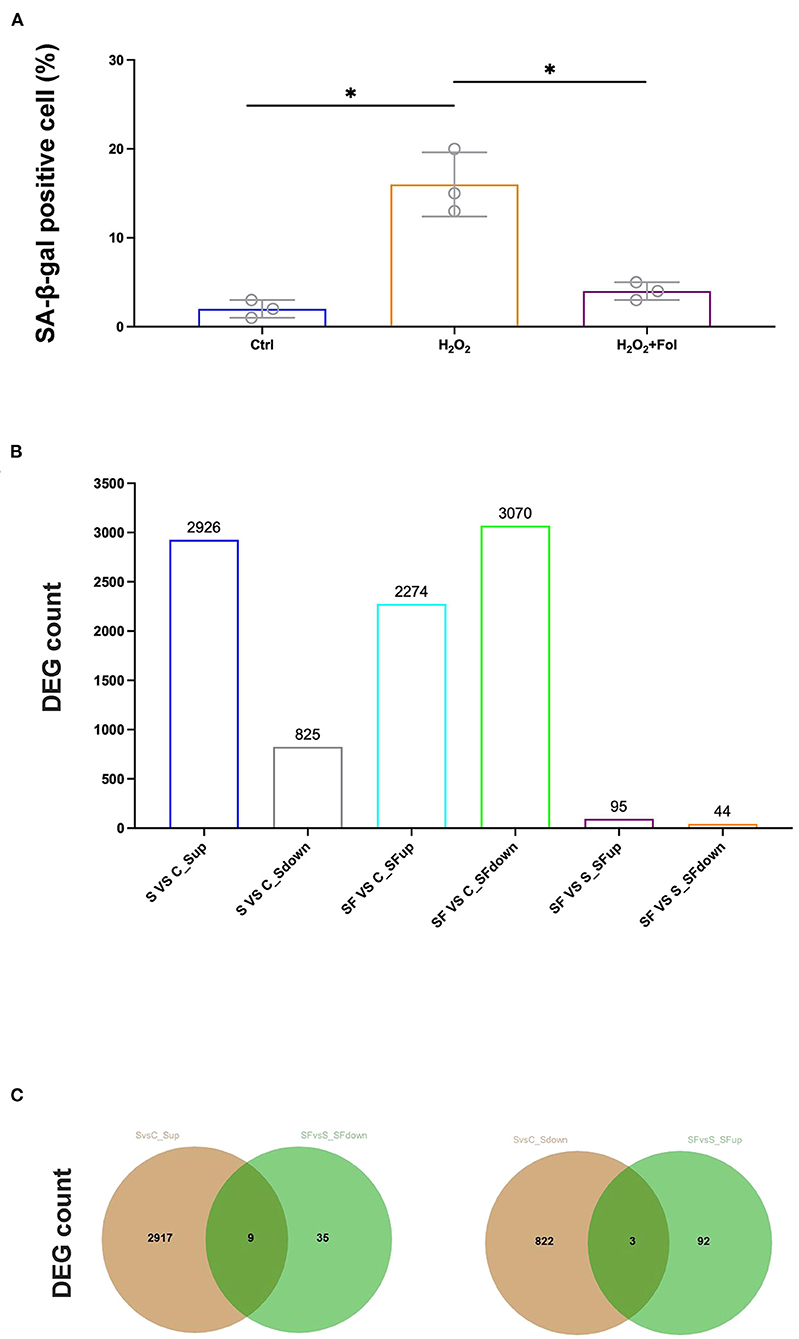
Figure 3. Hub genes and pathways affected by H2O2 and fullerenol. (A) ARPE-19 cells in the control group, H2O2 group, and fullerenol group were stained with SA-β-Gal. (B) Counts of DEGs among all the groups. (C) The overlay of genes up-regulated after H2O2 treatment and down-regulated after fullerenol treatment is showed in the left panel, while genes down-regulated after H2O2 treatment and up-regulated after fullerenol treatment is showed in the right panel. Fol, fullerenol; S, H2O2 group; C, Ctrl group; SF, Fullerenol group. *p < 0.05.
Furthermore, after Gene Ontology enrichment by DEGs among the three groups, we found that 11 GO terms were enriched by the DEGs up-regulated after H2O2 but down-regulated after fullerenol treatment, or down-regulated after H2O2 but up-regulated after fullerenol treatment. Positive regulation of DNA-templated transcription, positive regulation of endothelial cell migration, and negative regulation of apoptotic process pathways were both enriched by DEGs down-regulated after H2O2 treatment and up-regulated after fullerenol treatment, indicating the potential roles of fullerenol (Figure 4A). At the same time, the steroid hormone-mediated signaling pathway, gene expression, cell proliferation, cell migration, and negative regulation of gene expression pathways were both enriched by DEGs up-regulated after H2O2 treatment and down-regulated after fullerenol treatment (Figure 4A). We further identified 9 genes (ARHGEF37, KRT38, AC005307.1, FLG, HRNR, AC79466.1, PPFIA4, PFKFB4, and SPP1) that were up-regulated after H2O2 treatment and down-regulated after fullerenol treatment (Figure 4B), and 3 genes (EGR1, AC010343.1, and PFN1P1) were down-regulated after H2O2 treatment and up-regulated after fullerenol treatment (Figure 4B). These pathways and genes were reversed by fullerenol in ARPE-19 cells.
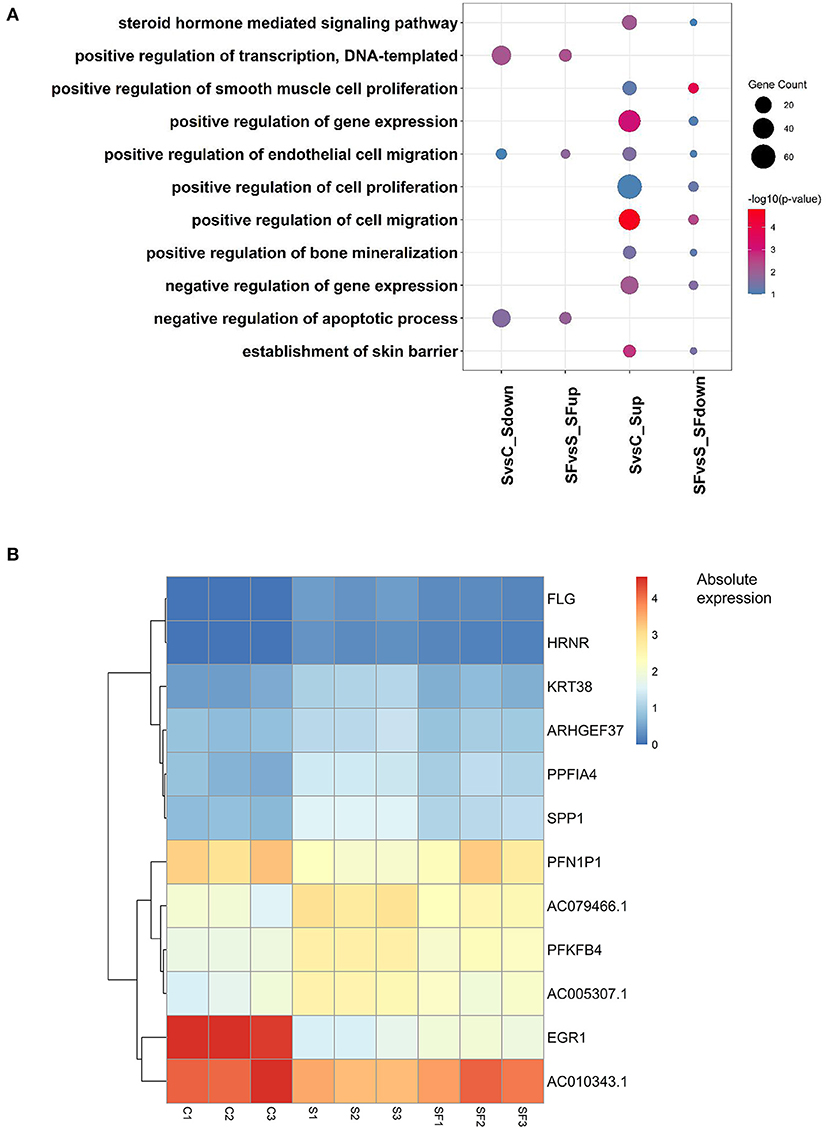
Figure 4. Functions and genes of genes rescued by fullerenol treatment. (A) Common GO terms of DEGs induced by H2O2 and fullerenol. (B) Heatmap plot of the expression of hub genes affected by H2O2 and fullerenol. S, H2O2 group; C, Ctrl group; SF, Fullerenol group.
Interaction networks of the 12 hub genes
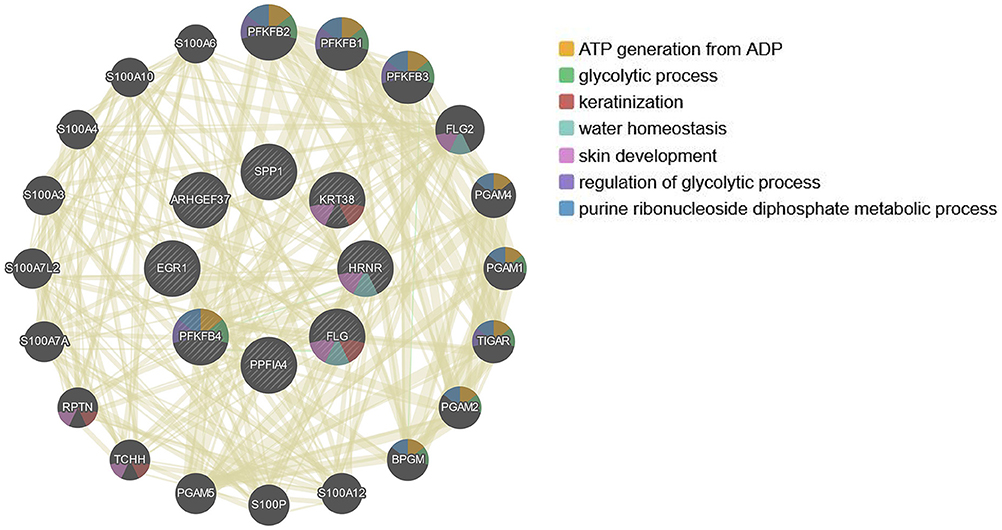
Since the 12 hub genes serve as potential targets of fullerenol treatment for aged-related macular degeneration (AMD) to investigate the potential roles and related genes of those genes, we performed GeneMANIA analysis and constructed a gene interaction network (Figure 5). In the outer circle, GeneMANIA identified functionally associated genes with eight of 12 hub genes (in the inner circle: ARHGEF37, KRT38, FLG, HRNR, PPFIA4, PFKFB4, and SPP1). GeneMANIA pathway analysis using the eight hub genes (the inner circle) connected to 20 associated genes (the outer circle) by genetic, physical, or pathway analysis identifies seven significant pathways (Figure 5). The top two pathways are ATP generation and glycolytic processes (Figure 5). Furthermore, ingenuity pathway analysis (IPA) showed detailed related biological pathways of the 12 crucial genes which include three major networks (Figure 6A). The first described a network characterizing signaling pathways including CXCR4 which is reported with the AMD pathogenesis (29, 30); the second included 3-phosphoinositide biosynthesis which is important for the initiation of early pathological events in retinal degenerative diseases under the presence of oxidative stress (31–33); the third included AMPK signaling which prevents degeneration of photoreceptors and the RPE cells (34, 35). In addition to a number of pathways, genes including CREB, AR, ERK1/2, CREB1, HIF1A, and HNRNPL genes, were also related to the 12 hub genes (Figure 6B). Among these genes, activation of ERK1/2 and CREB showed protection function for human RPEs from H2O2-induced oxidative damage (36). AR was proved to be a hub gene during the pathogenesis of AMD (37). These results provide further insights into the biological processes and potential roles regulated by the 12 hub genes. These targets and pathways may serve as potential targets for fullerenol treatment for AMD.
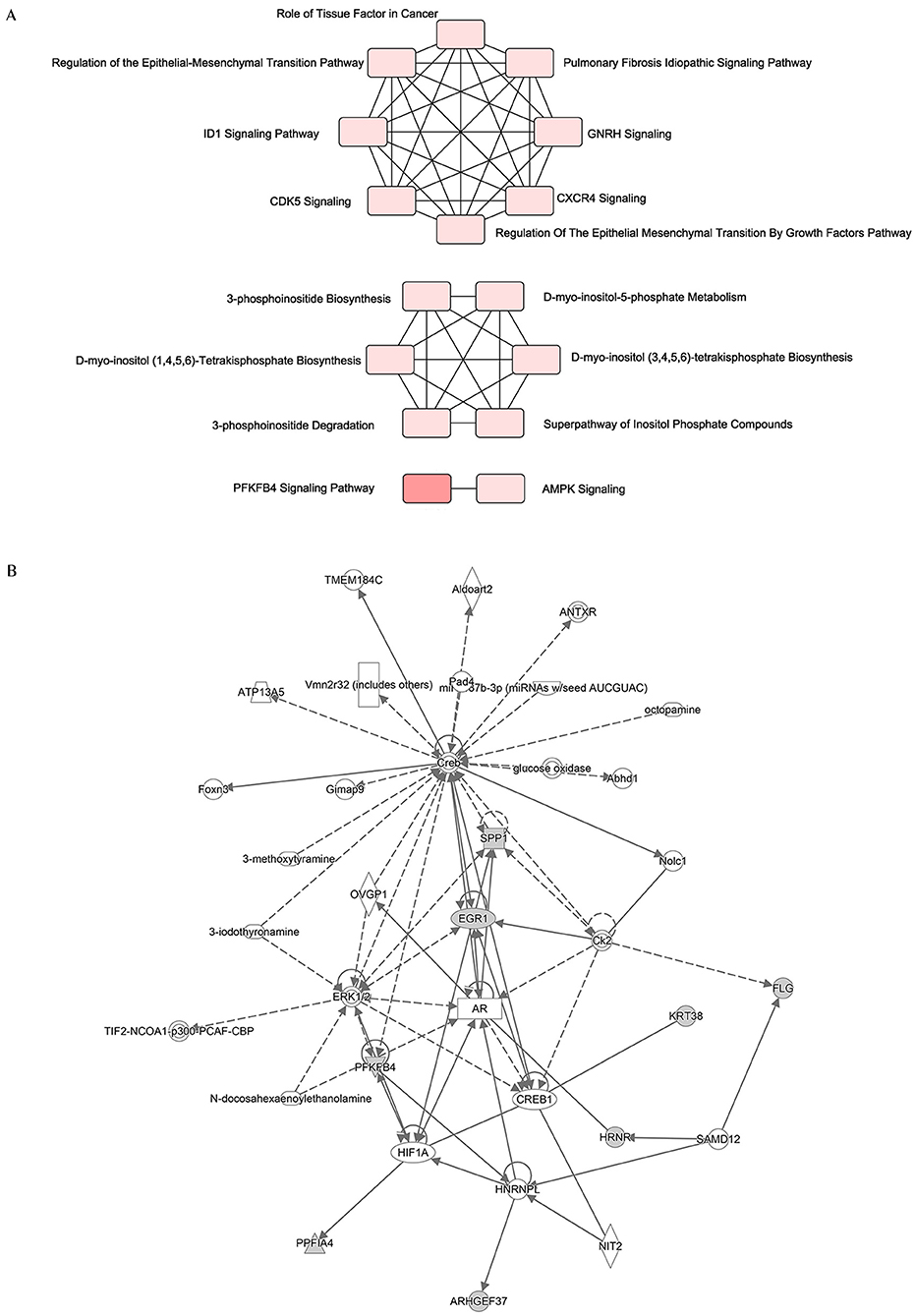
Figure 6. Functional network of the twelve hub genes. (A) An analysis of the core functions of twelve hub genes by QIAGEN Ingenuity Pathway Analysis (IPA). (B) An analysis of the core interaction network of twelve hub genes by IPA.
Discussion
In this study, we first profiled the comprehensive gene expression levels of H2O2-induced senescent ARPE-19 cells by RNA sequencing. ROS were found to affect the expression of multiple genes, such as CXCL8, SOD2, PLAT, CLSTN2, TXNIP, BIRC3, CLDN1, and DIO2. We further demonstrated that in the presence of oxidants, fullerenol inhibited the steroid hormone-mediated signaling pathway, cell proliferation, cell migration, and negative regulation of gene expression pathways. On the other hand, genes influenced by fullerenol were involved in the positive regulation of DNA-templated transcription and the negative regulation of apoptotic process pathways to protect RPE senescence.
To explore the biological role of these affected genes between the H2O2-treated and fullerenol-treated groups, we annotated the GO functions of these genes. In our study, we found that genes involved in the negative regulation of apoptotic processes were down-regulated after H2O2 treatment and up-regulated after fullerenol treatment, indicating that the apoptotic process is activated by H2O2 damage but inhibited by fullerenol. A previous study supported that the apoptotic process of RPEs was changed after various damages, such as light irradiation and the progression of AMD. As described in our study, genes involved in the positive regulation of the DNA-templated transcription pathway were also down-regulated after H2O2 treatment and up-regulated after fullerenol treatment. This pathway may also play crucial roles in RPE cells after H2O2 and fullerenol treatment. For the first time, our results showed that genes in the steroid hormone mediated signaling pathway were up-regulated after H2O2 treatment and down-regulated after fullerenol treatment, indicating the effects of fullerenol on the steroid hormone signaling pathway in RPE cells. A previous study showed that the level of serum cortisol (a steroid hormone) was positively correlated with RPE alterations in diabetic retinopathy. In addition, there were three more pathways, whose genes were up-regulated after H2O2 treatment and down-regulated after fullerenol treatment: negative regulation of gene expression, cell proliferation, and cell migration pathways. These pathways play crucial roles in RPE cells after H2O2 and fullerenol treatment. Our study provides global insight into the transcriptome changes in RPE cells, and hub genes in those pathways have the potential to serve as targets of fullerenol treatment.
Notably, we identified nine genes (ARHGEF37, KRT38, AC005307.1, FLG, HRNR, AC79466.1, PPFIA4, PFKFB4, and SPP1) that were upregulated after H2O2 treatment and downregulated after fullerenol treatment, and three genes (EGR1, AC010343.1, PFN1P1) were downregulated after H2O2 treatment and upregulated after fullerenol treatment. The 12 genes play crucial roles in metabolism-related pathways and biological signaling pathways. For example, ARHGEF37 was reported to serve as a regulatory protein involved in endocytosis (38). SPP1 is related to the activation of the PI3K/AKT and ERK1/2 pathways (39). Our study revealed their new roles in the regulation of H2O2 damage and fullerenol treatment of RPE cells. They may serve as useful biomarkers to illustrate the potential functions of fullerenol.
This study has several limitations. Firstly, when compared with the control group, the fullerenol treatment group had more DEGs than the H2O2-induced group. We speculate that this is due to the function of fullerenol itself. A previous study used RNA sequencing analysis to confirm that fullerenol itself can alleviate corneal oxidative injury by downregulation of oxidative stress-associated genes and upregulation of proliferation-related genes (19). However, the function of fullerenol in RPE cells needs further investigation in the future. Secondly, we only did the relation network analysis of the 12 hub genes by GeneMANIA and IPA analysis. Therefore, further molecular and cellular studies are needed to confirm the mechanistic basis for our conclusions.
In this study, we comprehensively profiled the gene expression of an H2O2-induced ARPE-19 senescence model and a nanoantioxidant fullerenol rescue model for the first time. Through dynamic transcriptome analysis, we identified positive regulation of the DNA-templated transcription and the negative regulation of apoptotic processes were down-regulated after H2O2 treatment and up-regulated after fullerenol treatment. In addition, there were four more pathways, whose genes were up-regulated after H2O2 treatment and down-regulated after fullerenol treatment: the steroid hormone mediated, negative regulation of gene expression, cell proliferation, and cell migration pathways. We also identified 12 hub genes that were rescued by fullerenol treatment. The 12 hub genes showed promise as therapeutic targets for AMD, which is worth further investigation. In summary, our results provide evidence for the nanomaterial fullerenol as an antioxidant in AMD treatment (Figure 7).
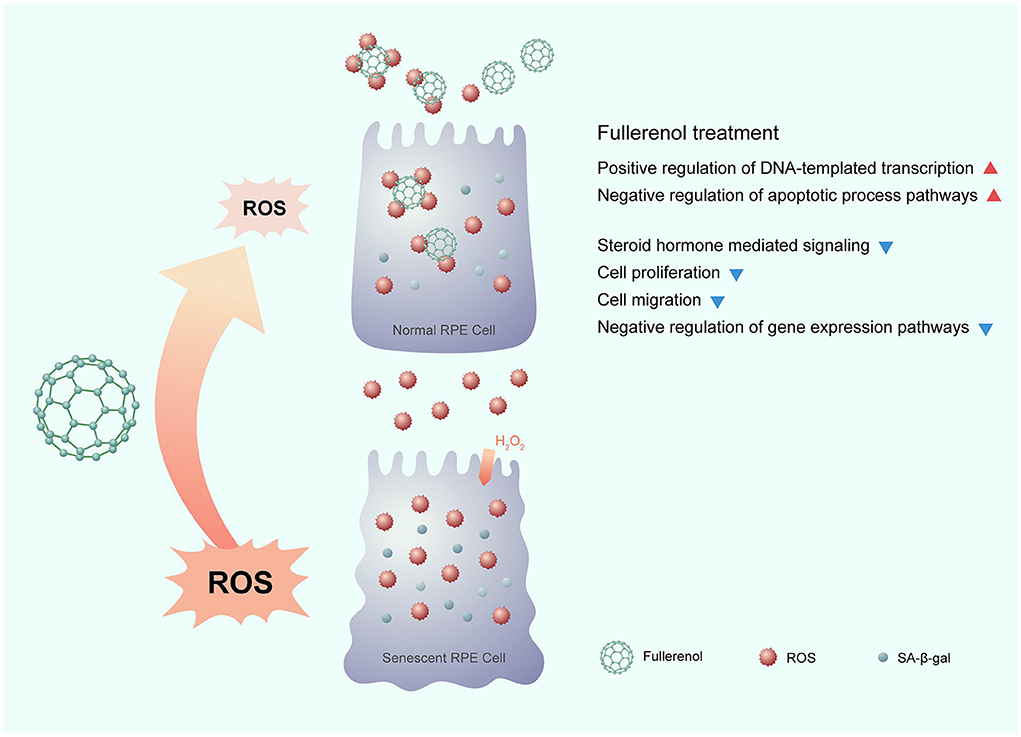
Figure 7. Potential mechanism of the nanomaterial fullerenol as an antioxidant in senescent RPE cell treatment.
Data availability statement
The raw data were submitted to the NCBI-SRA database with the Bioproject ID: PRJNA722601 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA722601).
Author contributions
LM and QZ initiated the study. XW, FY, J-YX, and YL performed the data analysis. JC and WL performed cell culture. XW and FY wrote and revised the manuscript. JD discussed and optimized the pictures in this manuscript. ZP, LM, and QZ designed the study and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Shenzhen High-level Hospital Construction Fund (2019).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.996280/full#supplementary-material
Supplementary Figure S1. Gene expression distribution across all the samples. (A) Boxplot of the distribution of FPKMs of all the genes in each sample. (B) Density plot of FPKMs of all the genes in each group. (C) PCA plot for each sample based on FPKMs of all the genes. (D) Correlation heatmap plot of each sample based on FPKMs of all the genes. S, H2O2 group; C, Ctrl group; SF, Fullerenol group.
Abbreviations
AMD, aged-related macular degeneration; RPE, retinal pigment epithelium; DEGs, differentially expressed genes; GSEA, gene set enrichment analysis; GO, Gene Ontology.
References
1. Macchioni L, Chiasserini D, Mezzasoma L, Davidescu M, Orvietani PL, Fettucciari K, et al. Crosstalk between long-term sublethal oxidative stress and detrimental inflammation as potential drivers for age-related retinal degeneration. Antioxidants (Basel). (2020) 10:25. doi: 10.3390/antiox10010025
2. Colijn JM, Buitendijk GHS, Prokofyeva E, Alves D, Cachulo ML, Khawaja AP, et al. Prevalence of age-related macular degeneration in Europe. Ophthalmology. (2017) 124:1753–63.
3. Blasiak J. Senescence in the pathogenesis of age-related macular degeneration. Cell Mol Life Sci. (2020) 77:789–805. doi: 10.1007/s00018-019-03420-x
4. Mitchell P, Liew G, Gopinath B, Wong TY. Age-Related macular degeneration. Lancet. (2018) 392:1147–59. doi: 10.1016/S0140-6736(18)31550-2
5. Sun Y, Zheng Y, Wang C, Liu Y. Glutathione depletion induces ferroptosis, autophagy, and premature cell senescence in retinal pigment epithelial cells. Cell Death Dis. (2018) 9:753. doi: 10.1038/s41419-018-0794-4
6. Datta S, Cano M, Ebrahimi K, Wang L, Handa JT. The impact of oxidative stress and inflammation on RPE degeneration in non-neovascular AMD. Prog Retin Eye Res. (2017) 60:201–18. doi: 10.1016/j.preteyeres.2017.03.002
7. Nita M, Grzybowski A. The role of the reactive oxygen species and oxidative stress in the pathomechanism of the age-related ocular diseases and other pathologies of the anterior and posterior eye segments in adults. Oxid Med Cell Longev. (2016) 2016:734. doi: 10.1155/2016/3164734
8. Mantha AK, Sarkar B, Tell G. A short review on the implications of base excision repair pathway for neurons: relevance to neurodegenerative diseases. Mitochondrion. (2014) 16:38–49. doi: 10.1016/j.mito.2013.10.007
9. Blasiak J, Piechota M, Pawlowska E, Szatkowska M, Sikora E, Kaarniranta K. Cellular senescence in age-related macular degeneration: can autophagy and DNA damage response play a role? Oxid Med Cell Longev. (2017) 2017:5293258. doi: 10.1155/2017/5293258
10. Ma N, Yang X, Qi C, Yu Q, Zhu C, Ren H. Farrerol enhances Nrf2-mediated defense mechanisms against hydrogen peroxide-induced oxidative damage in human retinal pigment epithelial cells by activating Akt and MAPK. Oxid Med Cell Longev. (2021) 2021:8847844. doi: 10.1155/2021/8847844
11. Du Y, You L, Ni B, Sai N, Wang W, Sun M, et al. Phillyrin mitigates apoptosis and oxidative stress in hydrogen peroxide-treated RPE cells through activation of the Nrf2 signaling pathway. Oxid Med Cell Longev. (2020) 2020:2684672. doi: 10.1155/2020/2684672
12. Mitter SK, Song C, Qi X, Mao H, Rao H, Akin D, et al. Dysregulated autophagy in the RPE is associated with increased susceptibility to oxidative stress and AMD. Autophagy. (2014) 10:1989. doi: 10.4161/auto.36184
13. Marazita MC, Dugour A, Marquioni-Ramella MD, Figueroa JM, Suburo AM. Oxidative stress-induced premature senescence dysregulates VEGF and CFH expression in retinal pigment epithelial cells: implications for age-related macular degeneration. Redox Biol. (2016) 7:78–87. doi: 10.1016/j.redox.2015.11.011
14. Chang C-C, Huang T-Y, Chen H-Y, Huang T-C, Lin L-C, Chang Y-J, et al. Protective effect of melatonin against oxidative stress-induced apoptosis and enhanced autophagy in human retinal pigment epithelium cells. Oxid Med Cell Longev. (2018) 2018:9015765. doi: 10.1155/2018/9015765
15. Kook D, Wolf AH Yu AL, Neubauer AS, Priglinger SG, Kampik A, Welge-Lüssen UC. The protective effect of quercetin against oxidative stress in the human RPE in vitro. Invest Ophthalmol Vis Sci. (2008) 49:1712–20. doi: 10.1167/iovs.07-0477
16. Du W, An Y, He X, Zhang D, He W. Protection of kaempferol on oxidative stress-induced retinal pigment epithelial cell damage. Oxid Med Cell Longev. (2018) 2018:1610751. doi: 10.1155/2018/1610751
17. Hanus J, Zhang H, Wang Z, Liu Q, Zhou Q, Wang S. Induction of necrotic cell death by oxidative stress in retinal pigment epithelial cells. Cell Death Dis. (2013) 4:e965. doi: 10.1038/cddis.2013.478
18. Ye S, Chen M, Jiang Y, Chen M, Zhou T, Wang Y, et al. Polyhydroxylated fullerene attenuates oxidative stress-induced apoptosis via a fortifying Nrf2-regulated cellular antioxidant defence system. Int J Nanomedicine. (2014) 9:2073–87. doi: 10.2147/IJN.S56973
19. Chen X, Yang J, Li M, Zhu S, Zhao M, Yang C, et al. Fullerenol protects cornea from ultraviolet B exposure. Redox Biol. (2022) 54:102360. doi: 10.1016/j.redox.2022.102360
20. Grebowski J, Kazmierska-Grebowska P, Cichon N, Piotrowski P, Litwinienko G. The Effect of Fullerenol C60(OH)36 on the antioxidant defense system in erythrocytes. Int J Mol Sci. (2021) 23:119. doi: 10.3390/ijms23010119
21. Grebowski J, Konopko A, Krokosz A, DiLabio GA, Litwinienko G. Antioxidant activity of highly hydroxylated fullerene C60 and its interactions with the analogue of α-tocopherol. Free Radic Biol Med. (2020) 160:734–44. doi: 10.1016/j.freeradbiomed.2020.08.017
22. Cong W, Wang P, Qu Y, Tang J, Bai R, Zhao Y. Chunying Chen null, Bi X. Evaluation of the influence of fullerenol on aging and stress resistance using Caenorhabditis elegans. Biomaterials. (2015) 42:78–86. doi: 10.1016/j.biomaterials.2014.11.048
23. Hao T, Li J, Yao F, Dong D, Wang Y, Yang B, et al. Injectable Fullerenol/Alginate hydrogel for suppression of oxidative stress damage in brown adipose-derived stem cells and cardiac repair. ACS Nano. (2017) 11:5474–88. doi: 10.1021/acsnano.7b00221
24. Nahle S, Safar R, Grandemange S, Foliguet B, Lovera-Leroux M, Doumandji Z, et al. Single wall and multiwall carbon nanotubes induce different toxicological responses in rat alveolar macrophages. J Appl Toxicol. (2019) 39:764–72. doi: 10.1002/jat.3765
25. Zhuge C-C, Xu J-Y, Zhang J, Li W, Li P, Li Z, et al. Fullerenol protects retinal pigment epithelial cells from oxidative stress-induced premature senescence via activating SIRT1. Invest Ophthalmol Vis Sci. (2014) 55:4628–38. doi: 10.1167/iovs.13-13732
26. Qingnuan L. yan X, Xiaodong Z, Ruili L, qieqie D, Xiaoguang S, Shaoliang C, Wenxin L. Preparation of (99m)Tc-C(60)(OH)(x) and its biodistribution studies. Nucl Med Biol. (2002) 29:707–10. doi: 10.1016/S0969-8051(02)00313-X
27. Franz M, Rodriguez H, Lopes C, Zuberi K, Montojo J, Bader GD, et al. GeneMANIA update 2018. Nucleic Acids Res. (2018) 46:W60–4. doi: 10.1093/nar/gky311
28. Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. (2010) 38:W214–220. doi: 10.1093/nar/gkq537
29. Tanaka M, Kakihara S, Hirabayashi K, Imai A, Toriyama Y, Iesato Y, et al. Adrenomedullin-Receptor activity-modifying protein 2 system ameliorates subretinal fibrosis by suppressing epithelial-mesenchymal transition in age-related macular degeneration. Am J Pathol. (2021) 191:652–68. doi: 10.1016/j.ajpath.2020.12.012
30. Wang J, Feng Y, Han P, Wang F, Luo X, Liang J, et al. Photosensitization of A2E triggers telomere dysfunction and accelerates retinal pigment epithelium senescence. Cell Death Dis. (2018) 9:178. doi: 10.1038/s41419-017-0200-7
31. Kang KH, Lemke G, Kim JW. The PI3K-PTEN tug-of-war, oxidative stress and retinal degeneration. Trends Mol Med. (2009) 15:191–8. doi: 10.1016/j.molmed.2009.03.005
32. Hwang N, Kwon M-Y, Woo JM, Chung SW. Oxidative stress-induced pentraxin 3 expression human retinal pigment epithelial cells is involved in the pathogenesis of age-related macular degeneration. Int J Mol Sci. (2019) 20:E6028. doi: 10.3390/ijms20236028
33. Datta S, Cano M, Satyanarayana G, Liu T, Wang L, Wang J, et al. Mitophagy initiates retrograde mitochondrial-nuclear signaling to guide retinal pigment cell heterogeneity. Autophagy. (2022) doi: 10.1080/15548627.2022.2109286 [Epub ahead of print].
34. Xu L, Kong L, Wang J, Ash JD. Stimulation of AMPK prevents degeneration of photoreceptors and the retinal pigment epithelium. Proc Natl Acad Sci U S A. (2018) 115:10475–80. doi: 10.1073/pnas.1802724115
35. Blasiak J, Pawlowska E, Sobczuk A, Szczepanska J, Kaarniranta K. The aging stress response and its implication for AMD pathogenesis. Int J Mol Sci. (2020) 21:E8840. doi: 10.3390/ijms21228840
36. Chong C-M, Zheng W. Artemisinin protects human retinal pigment epithelial cells from hydrogen peroxide-induced oxidative damage through activation of ERK/CREB signaling. Redox Biol. (2016) 9:50–6. doi: 10.1016/j.redox.2016.06.002
37. Liang G, Ma W, Luo Y, Yin J, Hao L, Zhong J. Identification of differentially expressed and methylated genes and construction of a co-expression network in age-related macular degeneration. Ann Transl Med. (2022) 10:223. doi: 10.21037/atm-21-7043
38. Viplav A, Saha T, Huertas J, Selenschik P, Ebrahimkutty MP, Grill D, et al. ArhGEF37 assists dynamin 2 during clathrin-mediated endocytosis. J Cell Sci. (2019) 132:jcs226530. doi: 10.1242/jcs.226530
Keywords: fullerenol, nanomaterial, RNA sequencing, oxidative stress, senescence, RPE, AMD
Citation: Wu X, Yao F, Xu J-Y, Chen J, Lu Y, Li W, Deng J, Mou L, Zhang Q and Pu Z (2022) The transcriptome profile of RPE cells by the fullerenol against hydrogen peroxide stress. Front. Med. 9:996280. doi: 10.3389/fmed.2022.996280
Received: 17 July 2022; Accepted: 22 August 2022;
Published: 14 September 2022.
Edited by:
Shaochong Zhang, Shenzhen Eye Hospital, ChinaReviewed by:
Zepeng Qu, National Institutes of Health (NIH), United StatesYin Shen, Renmin Hospital of Wuhan University, China
Yang Cheng, Huazhong University of Science and Technology, China
Copyright © 2022 Wu, Yao, Xu, Chen, Lu, Li, Deng, Mou, Zhang and Pu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisha Mou, bGlzaGFtb3VAZ21haWwuY29t; Qingling Zhang, enFsbGM4QDEyNi5jb20=; Zuihui Pu, cHVwZXRlcjE5MEAxNjMuY29t
†These authors have contributed equally to this work
 Xiaojun Wu1,2,3†
Xiaojun Wu1,2,3† Fuwen Yao
Fuwen Yao Ying Lu
Ying Lu Wei Li
Wei Li Lisha Mou
Lisha Mou Qingling Zhang
Qingling Zhang Zuihui Pu
Zuihui Pu