
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 20 September 2022
Sec. Gastroenterology
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.995943
This article is part of the Research Topic The Role of Non-Coding RNA In the Diagnosis and Treatment of Infection Induced Gastrointestinal Cancers View all 6 articles
Objectives A bibliometric analysis for non-coding RNA and hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) was performed to describe international research status and visualize the research scope and emerging trends over the last two decades on this topic.
Materials and methods: Research data of non-coding RNA and HBV-related HCC were retrieved and extracted from the Web of Science Core Collection (WoSCC) database from 1 January 2003 to 13 June 2022 and then analyzed by means of bibliometric methods. A total of 1,036 articles published in this field were assessed for specific characteristics, including the year of publication, journal, author, institution, country/region, references, and keywords. VOSviewer was employed to perform co-authorship, co-occurrence, and co-citation analyses accompanied by constructing a visual network.
Results: Overall, 1,036 reports on non-coding RNA and HBV-related HCC from 2003 to 2022 were retrieved from WoSCC. The publication has gradually increased during the last two decades with 324 journals involved. Most research records (748 publications and 23,184 citations) were concentrated in China. A co-occurrence cluster analysis for the top 100 keywords was performed and four clusters were generated: (1) non-coding RNA as a molecular marker for the diagnosis and prognosis of HBV-related HCC; (2) dysregulation of non-coding RNA by hepatitis B virus X protein (HBx); (3) non-coding RNA affecting the biological behaviors of HBV-related HCC; and (4) epidemiological study for the effects of non-coding RNA on the risk of HBV-related HCC.
Conclusion: The publications and citations involved in non-coding RNA and HBV-related HCC have increased over the last two decades associated with many countries, institutions, and authors. Our study revealed current development trends, global cooperation models, basic knowledge, research hotspots, and emerging frontiers in this field.
According to the International Agency of Research on Cancer of the World Health Organization (WHO), primary liver cancer poses a serious threat to human health and survival. It was estimated that 906,000 new cases and 830,000 deaths occurred globally due to primary liver cancer in 2020 (1). Chronic hepatitis B virus infection is a major risk factor for the development of hepatocellular carcinoma (HCC), accounting for 43–80% of its total incidence etiologically (2, 3). In addition, prolonged HBV replication promotes immune-mediated liver inflammation to gradually progress to cirrhosis, ultimately leading to HCC (4).
Hepatitis B virus (HBV) is very adept at combining its own and host mechanisms to manage viral load and enhance persistence (5, 6). A key tool during the process is regulating the expression of non-coding RNAs by manipulating the epigenetic environment including microRNAs, lncRNAs, and circRNAs. Non-coding RNAs are aberrantly expressed from the beginning of HBV infection to liver fibrosis/cirrhosis, and finally to HBV-related HCC. They could participate in the proliferation, apoptosis, epithelial–mesenchymal transition (EMT), invasion, and migration of HCC cells via different mechanisms, such as making impacts on transcriptional regulation in nucleus, protein interactions, alternative splicing, and competing endogenous RNAs, thus affecting tumor progression (7–9).
Bibliometric analysis is a scientific and quantitative research method for publications including co-word analysis, social network analysis, and cluster analysis. Quantitatively statistical analyses are utilized to summarize the progress of research topics, figure out the contribution of authors, journals, and institutions, and explore international hotspots with emerging trends (10, 11). VOSviewer1 is a software displaying the visual map of the co-occurrence relationship between keywords and researchers (12). In recent years, the bibliometric study has been applied to the summary of research progress on clinical medicine and biomedicine (13, 14).
To our knowledge, however, no report has referred to the analysis of research progress and trends with respect to non-coding RNA and HBV-related HCC. The present study began with downloading publications information from the Web of Science Core Collection (WoSCC) from 2003 to 2022, consisting of annual distribution, country, institution, author, source journal, co-occurrence, co-citation of keywords, etc. Bibliometric methods were adopted to analyze the research status and development trends about non-coding RNA and HBV-related HCC. Our study aimed to explore the current hotspots in this field, provide novel ideas and perspectives for future investigations, and also assist researchers and experts in determining research subjects and directions on this topic.
The literature on non-coding RNA and HBV-related HCC is available for download as “plain text” in WoSCC until 13 June 2022. Systematic literature retrieval was independently conducted by two investigators (Li-rong Yan and Ao-ran Liu). Ben-gang Wang made judgments on the divergence between two independent reviewers. Data collection and retrieval strategies are shown in Figure 1. Eligible publications met the following criteria: (1) search terms TS = [(“Hepatitis B Virus” OR “HBV” OR “hepatitis b viral” OR “hepatitis b” OR “hbv hepatitis” OR “chronic hepatitis b” OR “type b hepatitis”) AND (“Hepatocellular Carcinoma” OR “liver cancer” OR “HCC” OR “hepatic carcinoma” OR “hepatoma” OR “hepatocarcinoma” OR “hepatic cancer” OR “liver carcinoma”)] AND TS = (“Non-coding RNA” OR “microRNA” OR “microRNAs” OR “Circular RNA” OR “Circular RNAs” OR “long non-coding RNA” OR “long non-coding RNA” OR “lncRNA” OR “lncRNAs” OR “Small interfering RNA” OR “SiRNA” OR “Piwi-interacting RNA” OR “small nucleolar RNA” OR “ribosomal RNA” OR “transfer RNA” OR “small nuclear RNA” OR “tRNA-Derived Fragments” OR “tRNA halves” OR “miRNA” OR “circRNA” OR “tiRNA” OR “tRF” OR “miRNAs” OR “circRNAs” OR “miR*” OR “lnc*” OR “circ_*”); (2) article document types; (3) year of publication ranging from 2003 to 2022; and (4) information collected for each article containing publication, author, country, institution, journal, keywords, and citations.
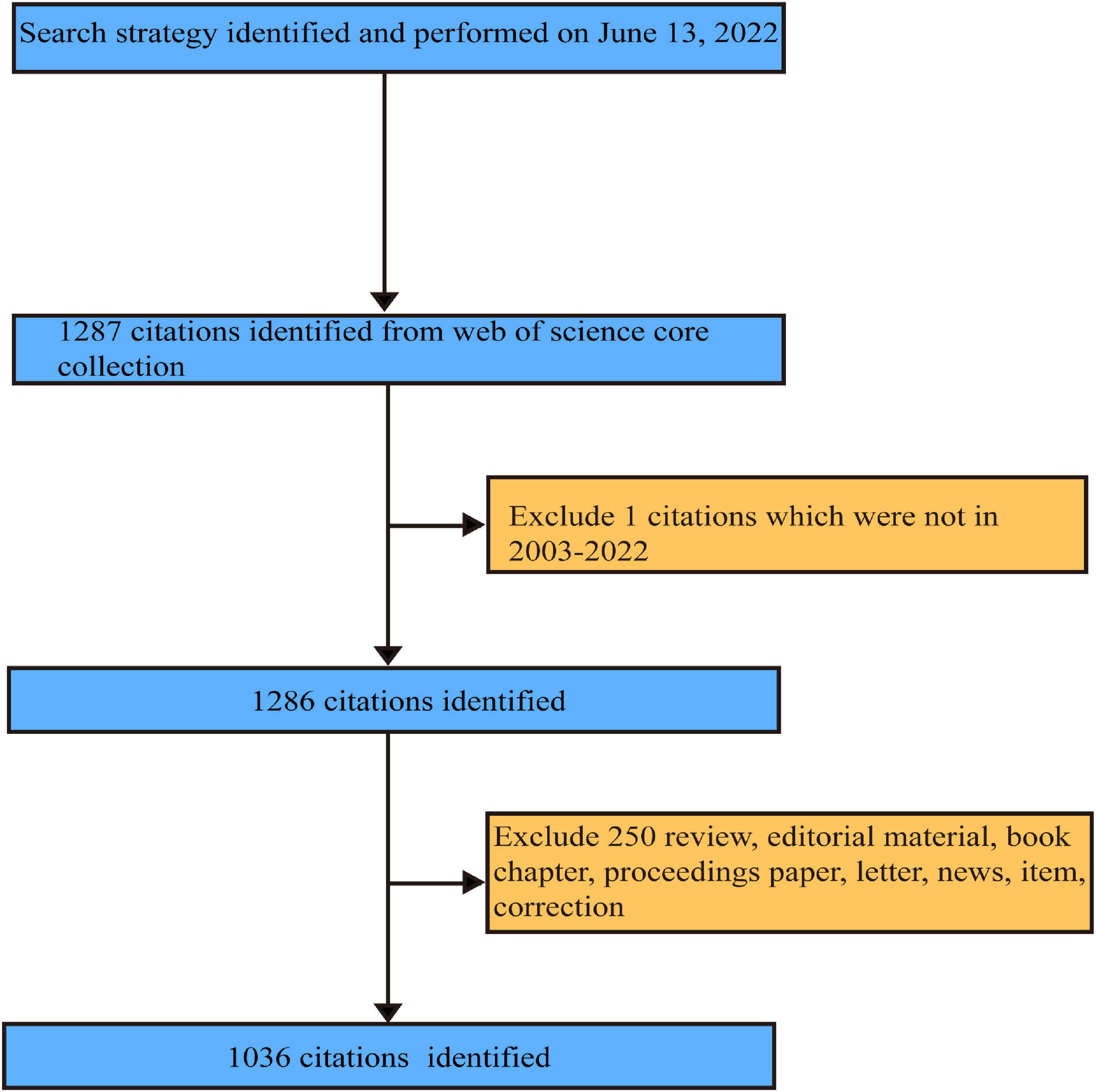
Figure 1. The flow diagram of literature selection according to the inclusion and exclusion criteria.
Bibliometrics is conducive to monitoring the development and patterns of available publications (11). Here, we extracted the literature information mentioned above from TXT files downloaded from WoSCC and then created a visual network map by means of VOSviewer Version 1.6.18. Datawrapper website was also employed for data visualization2. The most common bibliometric techniques are mainly composed of co-authorship analysis, co-occurrence analysis, and co-citation analysis. Co-authorship analysis may reveal the collaboration models among authors, institutions, and countries (15). Co-occurrence analysis could identify the relevance of multiple words by their frequency in the same article to manifest the hot topics and trends in a certain discipline. Co-citation analysis may help to establish the knowledge base of a discipline (16). These visualization maps make investigators identify bibliometric information such as active authors, institutions, countries, basic knowledge, research hotspots, and frontiers on a specific topic. The present study explored research hotspots about non-coding RNA and HBV-related HCC by co-word analysis for 100 keywords with the top frequency in retrieved publications. In the visual map of VOSviewer, each node was displayed by a circle with a label, and a larger circle represented higher frequency in co-occurrence analysis. The color of each circle was determined by the cluster it belonged to. The thickness and length of connection between nodes indicated their corresponding strength and relevance. A maximum of 1,000 connections in the network represented the 1,000 strongest relationships between nodes.
All literature downloaded from WoSCC are part of public resource. The extraction of these data takes no account of humans or animals. Therefore, no ethical issue is involved in our data utilization, and the project has no need to be approved by an ethics committee.
A total of 1,036 articles about non-coding RNA and HBV-related HCC were retrieved from WoSCC. Annual global publications are shown in Figure 2. The publication on this topic has gradually increased during the past two decades. In 2003, only two related reports suggested that the subject had just begun to be focused. The published articles exceeded 50 for the first time in 2013 and exceeded 100 in 2015, maintaining around 100 in the following years. Hence, the research topic on non-coding RNA and HBV-related HCC remains novel with many problems to be solved and needs to be further investigated.
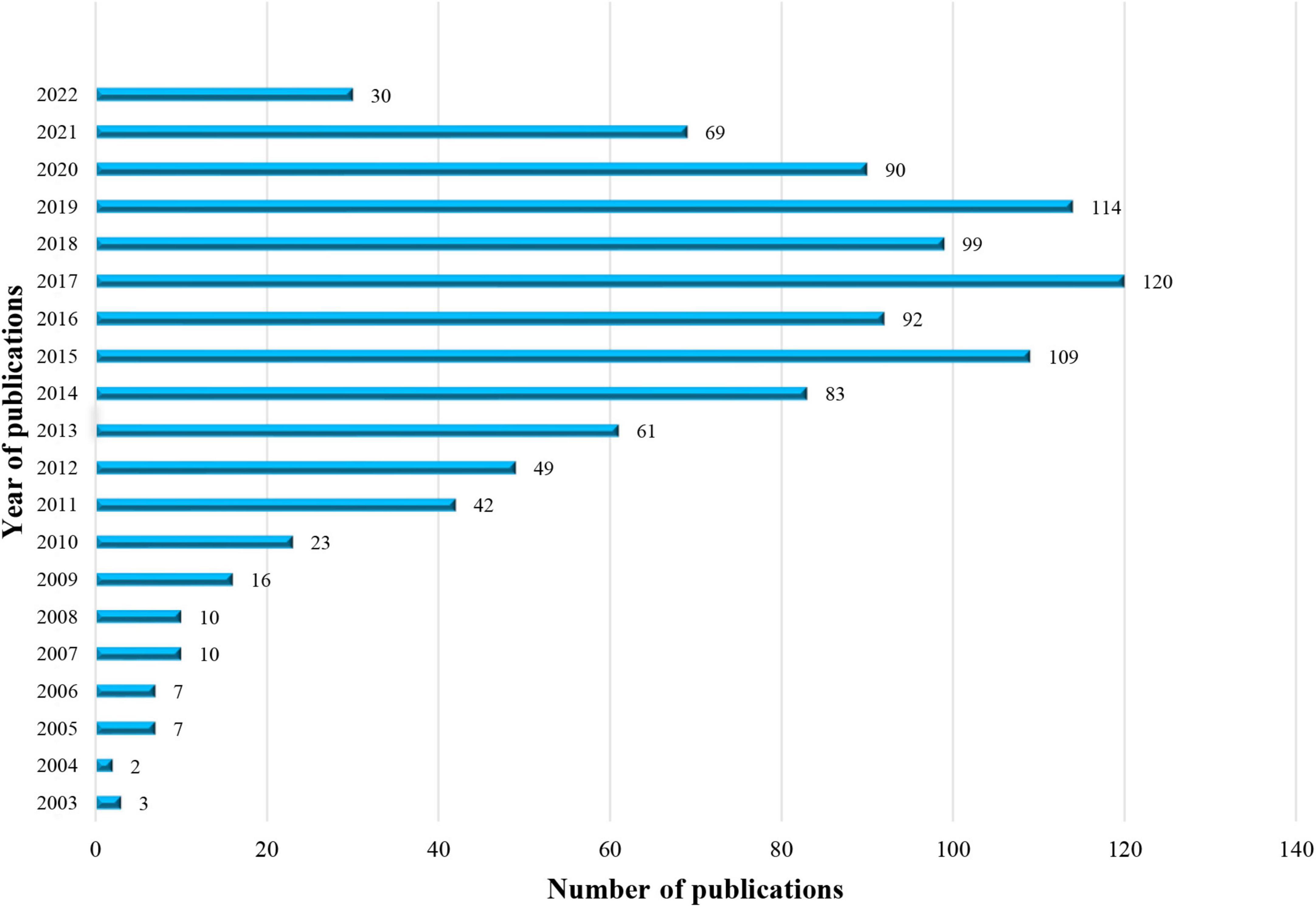
Figure 2. The number of articles about non-coding RNA and hepatitis B virus (HBV)-related hepatocellular carcinoma per year from 2003 to 2022.
Publications about non-coding RNA and HBV-related HCC were involved in 324 journals. The top 10 journals with the most articles are shown in Table 1, accounting for 24.4% (253/1036) of the whole. PLoS ONE had the most articles (n = 45) and 1,985 citations, followed by the World Journal of Gastroenterology (n = 33) and Scientific Reports (n = 32).
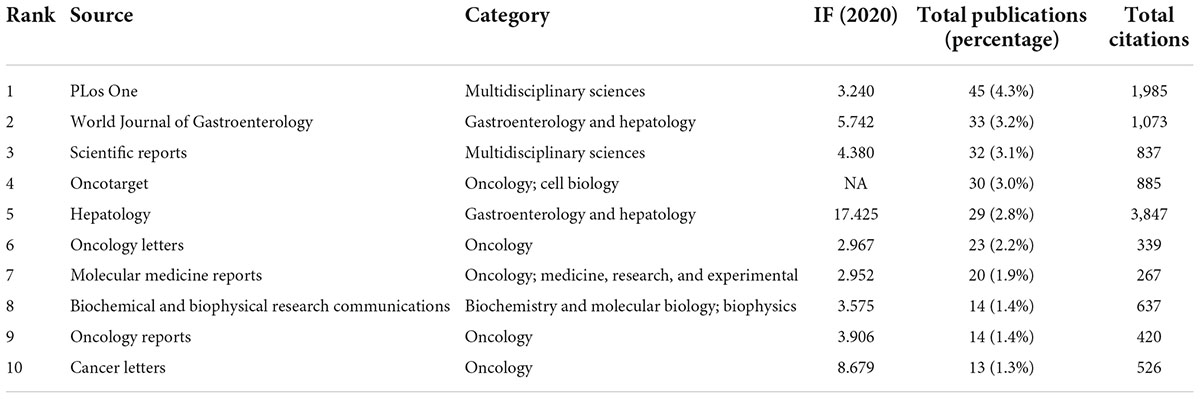
Table 1. The top 10 journals of publications on non-coding RNA and hepatitis B virus (HBV)-related hepatocellular carcinoma.
Total citations of the 1,036 articles were 33,352 with a median of 22. The top 10 articles with the most citations are shown in Table 2, with a frequency ranging from 359 to 935. The article titled “Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous carcinoma,” published on Oncogene in 2006 had the most citations of 935.
The subject of non-coding RNA and HBV-related HCC has been studied in 45 countries/regions worldwide (Figure 3A). The top 10 countries with the highest production are shown in Table 3. Among them, China had the most publications (748 publications and 23,184 citations) followed by the United States (133 publications and 8,380 citations) and India (49 publications and 3,164 citations). Co-authorship analysis of the country was performed by VOSviewer to demonstrate international collaboration in this field. The co-authorship network of countries is shown in Figure 3B, including 35 countries/regions of the whole. They were divided into five clusters represented by different colors. The largest cluster (red color) consisted of 10 countries centered on South Korea, Iran, and Australia. The United States had the most partners (n = 22) followed by Germany (n = 16), China (n = 14), Italy (n = 12), and France (n = 12).
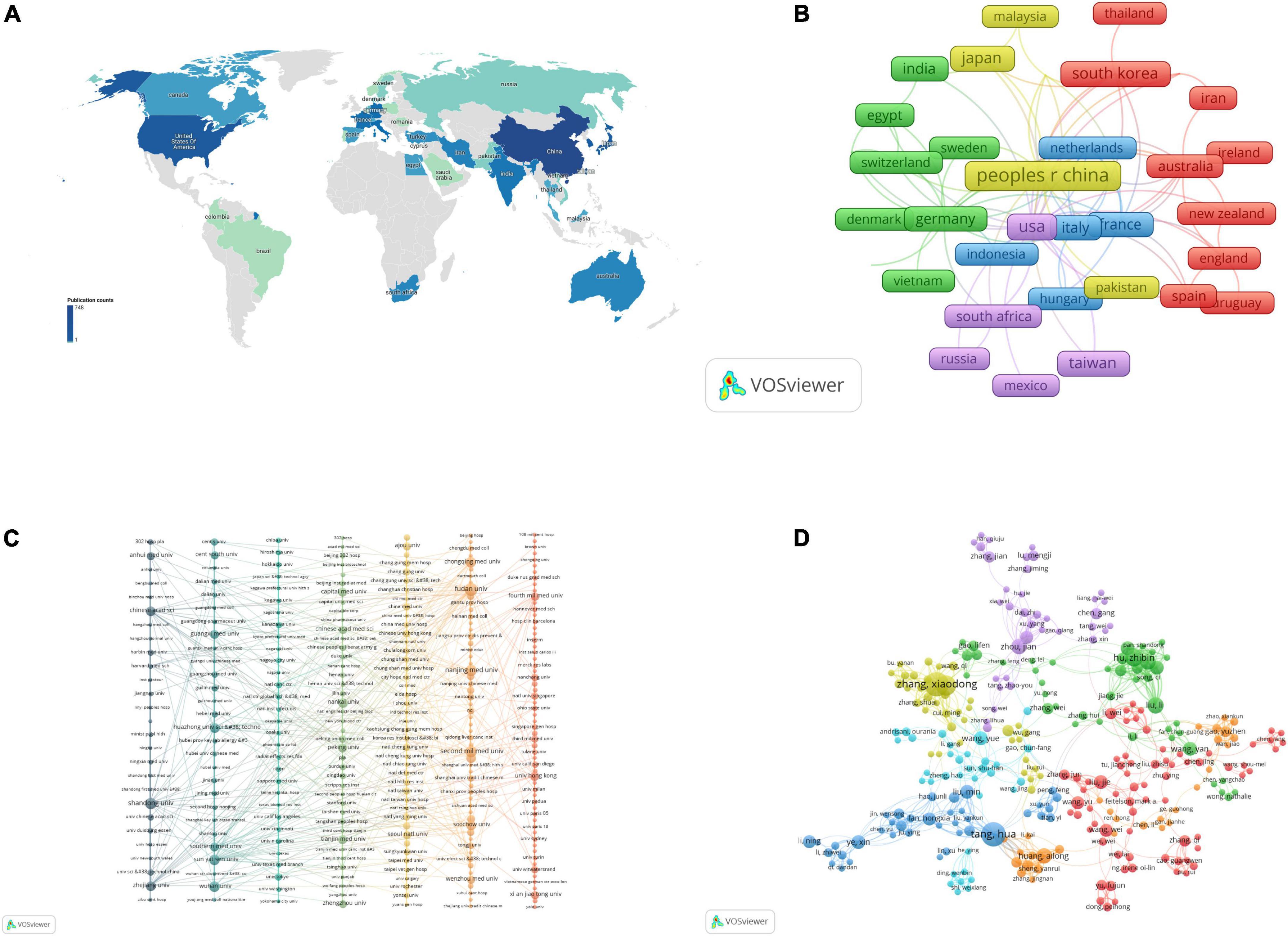
Figure 3. The co-authorship analysis of non-coding RNA and hepatitis B virus (HBV)-related hepatocellular carcinoma. (A) Distribution of country/region. (B) The co-authorship network of country/region. (C) The co-authorship network of the institution. (D) The co-authorship network of authors. Each node represents a country/region (A,B), institution (C), and author (D), respectively. The size of nodes represents the publication number. The connection between nodes represents collaboration. The distance and thickness of the connection represent the relative strength of the relationship.
A total of 1,078 institutions were involved in the study for non-coding RNA and HBV-related HCC. The top 10 institutions with the most publications are shown in Table 4. Fudan University had the most publications (60 publications and 2,649 citations) followed by The Second Military Medical University (39 publications and 3,131 citations) and Nanjing Medical University (36 publications and 1,517 citations). We selected 327 institutions with at least two published articles. The co-authoring network was constructed for them by VOSviewer (Figure 3C), including 287 institutions. They were divided into seven clusters represented by different colors. The red cluster composed of 61 institutions was the largest one centered on the University of Hong Kong, The Fourth Military Medical University, and Inserm. The University of Hong Kong had the most partners (n = 17) followed by The Fourth Military Medical University (n = 14), Xi’an Jiaotong University (n = 12), and the National University of Singapore (n = 11).
A total of 6,154 authors participated in the retrieved 1,036 articles with an average of six authors in each article. The top 20 authors with the most publications are shown in Table 5. Zhang Xiao-dong (19 publications and 1,096 citations) and Tang Hua (19 publications and 547 citations) had the highest production followed by Ye Li-hong (15 publications and 977 citations). We selected 408 authors with at least three published articles and constructed the co-authorship network with 290 authors involved. They were grouped into seven clusters represented by different colors (Figure 3D). The 70 authors centered on Liu Jie, Li Wei, and Wang Wei formed the largest red cluster. Zhang Xiao-dong had the most partners (n = 25) followed by Tang Hua (n = 23) and Fan Hong-xia (n = 19).
The retrieved 1,036 publications cited 25,975 articles in total. The top 10 articles with the highest citation frequency ranging from 50 to 137 are shown in Table 6. The article titled “MicroRNAs: genomics, biogenesis, mechanism, and function,” published on Cell in 2004 was cited the most. All the 25,975 cited articles originated from 2,555 cited sources including journals or books.
The theme of publications is covered by their keywords; thus, high-frequency keywords are well suited for co-occurrence analysis. The top 100 keywords in this study were extracted and clustered by VOSviewer (Table 7). The co-occurrence network consisted of four clusters. Keywords were displayed in node labels and the size of each node represented their frequency. The connection between two nodes indicated the co-occurrence relationship between two keywords.
The keywords of hepatocellular carcinoma (731), expression (420), hepatitis B virus (348), and microRNA (284) took up the center of the visual network. All keywords were automatically grouped into four clusters based on similarity by VOSviewer (Figure 4A). The red cluster 1 represented non-coding RNA as a diagnostic and prognostic molecular marker for HBV-related HCC; the green cluster 2 represented abnormal regulation of non-coding RNA expression by hepatitis B virus X protein (HBx); the yellow cluster 3 represented the influence of non-coding RNA on malignant biological behaviors of HCC; and the blue cluster 4 represented the influence and epidemiological study of non-coding RNA on the risk of HBV-related HCC.
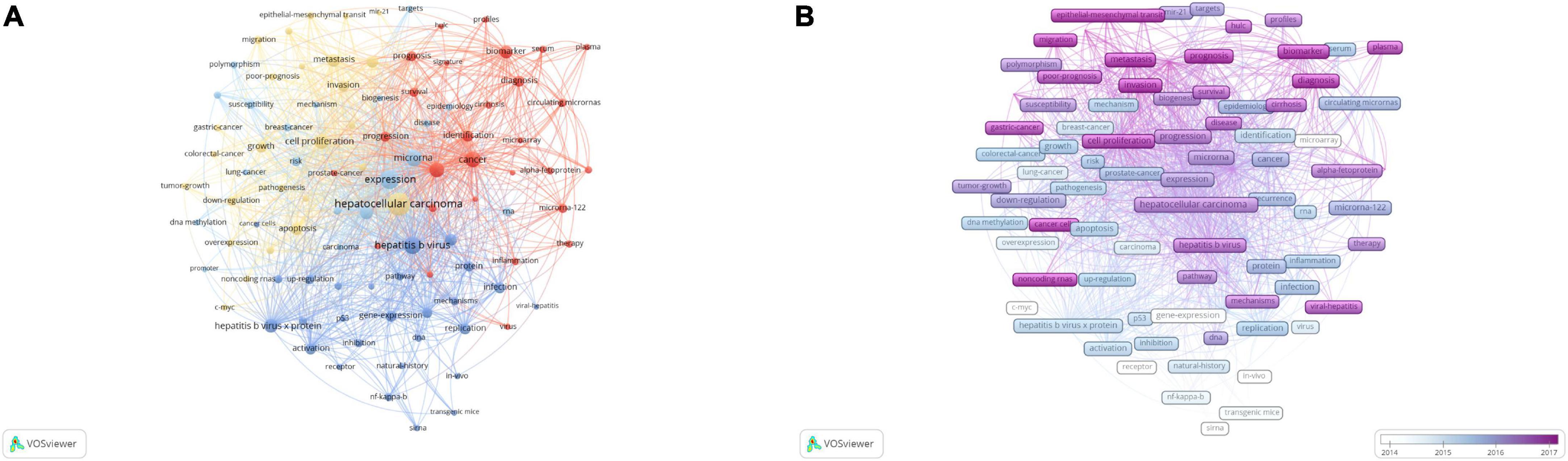
Figure 4. The co-occurrence analysis of non-coding RNA and hepatitis B virus (HBV)-related hepatocellular carcinoma. (A) The co-occurrence cluster analysis of the top 100 keywords. (B) The overlay map of the top 100 keywords. Each node represents a keyword. The size of nodes represents the publication number. The connection between nodes represents collaboration. The distance and thickness of the connection represent the relative strength of the relationship.
The evolvement trend of keywords was further explored and presented in a heatmap colored by VOSviewer based on their average appearing year (AAY) (Figure 4B). Cool colors represented keywords published earlier, while warm colors represented keywords published later. The most recent keywords included long non-coding RNA (AAY: 2018.1), epithelial–mesenchymal transition (AAY: 2017.9), migration (2017.8), invasion (AAY: 2017.6), prognosis (AAY: 2017.5), metastasis [(AAY: 2017.6) 2017.2], and biomarkers (AAY: 2017.2).
In the present study, we focused on the field of non-coding RNA and HBV-related HCC. Meanwhile, a bibliometric analysis was performed for the publications in WoSCC from 2003 to 2022, including the distribution of annual publication, country, institution, authors, source journals, co-occurrence of keywords, and co-citations. Furthermore, the development trends and current hotspots were also explored. Our study would provide novel ideas and perspectives for future investigations and also assist researchers and experts in determining the research subject and direction on this topic.
The research on non-coding RNA and HBV-related HCC has been gradually strengthened in recent years. The number of published articles showed a steady increase over the last two decades and was mainly concentrated after 2015, accounting for about 70% of the whole. They were involved in multiple disciplines. The top 10 journals sorted by publication volume consisted of five journals of oncology, two journals of gastroenterology and hepatology, two journals of multidisciplinary science, and one journal of molecular biology and biochemistry.
A total of 6,154 authors from 1,078 institutions in 45 countries/regions have published articles on non-coding RNAs and HBV-related HCC, suggesting that the topic has attracted widespread attention worldwide. Extensive cooperation has also been developed between countries/regions. China and the United States had the most published articles and formed the core of international cooperation. Therefore, China and the United States may currently grasp the frontiers of academic research in this field.
The top 10 institutions with the most published articles are all in China, suggesting that institutions in China are leading the way in terms of quantity. Moreover, 78% of research institutions in the collaborative network demonstrated extensive cooperation with each other.
Zhang Xiao-dong and Tang Hua were found to be the most productive authors in the study. Besides, Zhang Xiao-dong had the most collaborators. Only 290 of the 408 identified authors were included in the co-authorship network. We speculated that it might be due to some authors collaborating with authors outside the network.
The knowledge base and background of non-coding RNA and HBV-related HCC could be effectively revealed by co-citation analysis for cited references. The top 10 articles cited the most covered the studies in epigenetics, epidemiology, genetics, and molecular biology. Co-occurrence analysis of keywords could help to classify the main knowledge structure and hotspots. Here, co-occurrence cluster analysis was performed for the top 100 keywords, generating the four main clusters as follows.
Cluster 1 contained 28 high-frequency keywords such as hepatitis B, biomarker, diagnosis, prognosis, survival, and microRNA-122, suggesting that this cluster mainly focused on the biomarker study of HBV-related HCC. Early diagnosis of HCC is vital for the treatment outcome, and yet, effective markers remain lacking. It has attracted increasing attention for non-coding RNA to be a potential biomarker for the screening and early diagnosis of HBV-related HCC as well as the prediction of HCC prognosis.
A classification system based on seven miRNAs (miR-29a, miR-29c, miR-133a, miR-143, miR-145, miR-192, and miR-505) was suggested to have more significant sensitivity than alpha-fetoprotein (AFP) in distinguishing HCC cases from healthy controls, HBsAg inactive carriers, patients with chronic hepatitis B, and patients with HBV-cirrhosis. This miRNA-based classification was also the first biomarker established for the diagnosis of preclinical HCC and is expected to improve the clinical outcome of HCC through early diagnosis and precise treatment (17). MiR-122 is the most replicated miRNA biomarker in HCC with a sensitivity of 71–81%, a specificity of 59–83%, and an AUC of 0.63–0.87 in distinguishing HBV-related HCC from controls (18, 19). Recent studies showed that exosomal miRNAs might be better than whole-serum or plasma miRNAs for the early diagnosis of HBV-related HCC. In addition, the detection of miR-21 in exosomes was more sensitive than that in serum (20). Similarly, the exosomal miR-125b level of patients with HBV-related HCC was significantly lower than the serum level compared with patients with chronic hepatitis B or cirrhosis, which at least partially explained why the miR-125b level in exosomes, but not in serum, independently predicted HCC progression (21). In addition, exosomal miRNA levels of patients with HBV-related HCC had more significant differences than whole-serum compared with chronic hepatitis B or cirrhosis cases (22). The combination of miRNA with other classical serum markers can improve the sensitivity and specificity of early blood-based detection of HBV-related HCC (17, 23), especially for atypical HCC patients with low serum AFP levels. The expression levels of several miRNAs in liver tissue or circulation were shown to correlate with the severity and survival of patients with HBV-related HCC. For example, miR-223-3p expression in HBV-related HCC tissue was different from matched normal tissue. Circulating miR-223-3p might be a novel diagnostic and prognostic marker for patients with HBV-related HCC (24). Therefore, miRNAs are greatly potential markers for the diagnosis and prediction of HBV-related HCC.
In 2007, the expression of lncRNA HULC was first found to be significantly elevated in peripheral blood cells of 75% of patients with liver cancer (25). Subsequent research showed that the positive rate of HULC in the plasma of patients with HBV-positive HCC was higher than that of patients with HBV-negative, and the HULC level was correlated with the Edmondson score (26). These results suggested that plasma HULC was a potential biomarker for HBV-related HCC. The serum levels of lncRNA UC003WBD (27), AF085935 (27), UC00LNCR (28), and AX800134 (28) were also found to be elevated in patients with HBV-related HCC. In light of the above findings, lncRNAs may be useful biomarkers for predicting the risk and prognosis of HBV-related HCC.
Hsa_circ_0027089, a kind of plasma circRNA, was proven to distinguish HBV-related HCC from HBV-associated cirrhosis and healthy subjects, with AUC values of 0.765 and 0.794, respectively (29). It may be considered a biomarker for clinical diagnosis and evaluation of HBV-related HCC. In a microarray based on three pairs of HBV-associated HCC and adjacent tissue, aberrant expression of circRNAs was observed including 24 upregulated and 23 downregulated circRNAs (30). Hence, circRNAs may play key roles in distinguishing different types of HCC. For example, plasmatic circRNA can be used as a valuable diagnostic marker to distinguish HBV-related HCC from HBV-associated cirrhosis. Three circulating circRNAs (circ_0009582, circ_0037120, and circ_0140117) combined with AFP were reported to have higher sensitivity and specificity as potential diagnostic markers for predicting the development of HBV-related HCC (31). Zhu et al. found that plasma hsa_circ_0027089 could distinguish patients with HBV-related HCC from patients with non-HCC. Its combination with AFP had higher sensitivity to diagnose HBV-related HCC compared with patients with cirrhosis, healthy patients, and patients with no HCC but accompanied by poor specificity (29). Except for these, circRNAs can also be applied to clinical intervention targets.
In conclusion, non-coding RNA has great potential to become a marker for the diagnosis and prognosis of HBV-related HCC, which remains at the frontier of this field at present.
Cluster 2 contained 27 high-frequency keywords such as hepatitis B virus, hepatitis B virus X protein (HBX), replication, activation, hepatocarcinogenesis, and gene expression. Keywords in this cluster mainly reflected HBX, dysregulating the expression of non-coding RNA.
Hepatitis B virus X protein can affect miRNA biosynthesis, transcription, and translation in HBV-associated HCC (32), thus influencing tumor progression by altering the activity of tumor-related signaling pathways. Aberrantly expressed miRNA induced by HBx can change tumor signaling pathway activity affecting tumor progression. For instance, dysregulated HBx upregulates miR-21 (activated by IL-6) (33–35), miR-29, miR-155-5p (36), and miR-221/222 (37, 38) to inhibit the regulation of AKT/mTOR expression by PTEN. HBx also induces the upregulation of miR-21 to reduce the inhibition of WNT signaling by DCC6 (39) and enhances WNT signaling by decreasing PDCD4 expression and E-cadherin (33). Moreover, HBx downregulates let-7 to reduce its regulation on the activation of IL-6-induced JAK/STAT and mTOR-mediated transcription of oncogenic proteins such as C-MYC/MCL-1 (40–42).
In HBV-associated HCC, increased HBx level is associated with upregulated Zinc finger E-box binding homeobox 2 antisense RNA1 (ZEB2-AS1) level and the transition to mesenchymal phenotype. As an antisense lncRNA, ZEB2-AS1 forms RNA–RNA double strands by pairing with complementary bases at the 5′ splicing site of the precursor mRNA encoding ZEB2 (43). It prevents the binding of spliceosomes and keeps ZEB2 retained in mature transcripts by translating longer IRES-containing 5′ UTR, thus increased ZEB2 level inhibits E-cadherin transcription and promotes EMT. The upregulation of lncRNA UCA1 caused by HBX promotes cell growth and tumorigenesis by recruiting EZH2 and inhibiting the p27Kip1/CDK2 signaling pathway (44). HBX-associated lncRNA-ATB activated by TGF-β promotes the invasion and migration of HCC cells by inducing autophagy (45). The upregulation of MFG-AS1 caused by HBX promotes HCC cell proliferation and migration by enhancing MAFG expression and stabilizing non-myosin IIA (46).
In conclusion, dysregulated expression of non-coding RNA caused by HBX can significantly affect HCC progression. To date, research on this topic is mainly focused on miRNA and lncRNA, while circRNA and ribosomal RNA have not been involved yet, which provides us with some hotspots and blind spots, which is an important clue for selecting future research directions.
Cluster 3 contained 26 high-frequency keywords such as hepatocellular carcinoma, cell proliferation, metastasis, apoptosis, growth, invasion, migration, epithelial–mesenchymal transition, hepatitis, and non-coding RNA. This cluster mainly reflected the influence of non-coding RNA on the biological behaviors of HBV-related HCC.
Non-coding RNAs can affect the biological behaviors of HCC via multiple mechanisms. First, lncRNAs participate in the transcriptional regulation in the nucleus. For example, urothelial cancer-associated 1 (UCA1) is a lincRNA with a length of 1,400 nt (47). HBx was found to upregulate UCA1 expression in HBV-related HCC samples and HBx protein-expressing cell lines (44), associated with increased cell proliferation and inhibition of apoptosis. Similar to many other lincRNAs, UCA1 can recruit EZH2 to mediate epigenetic silencing. The inhibition of its target, tumor suppressor p27, enables cyclin-dependent kinase 2 (CDK2) to accelerate G1/S cell cycle progression and cell proliferation. Some lncRNAs can also post-translationally regulate oncogenic or tumor suppressor proteins discovered in the nucleus and the cytoplasm. They enhance protein stability by directly binding to their target proteins or preventing ubiquitination and proteasomal degradation (48). The bound lncRNAs can also inhibit function or promote the degradation of proteins. The downregulated lncRNA-6195 in HBV-related HCC is a tumor suppressor that binds to the substrate-binding site (aa 237–405) of ENO1 to inhibit tumor growth (49). lncRNAs act as competing endogenous RNAs (ceRNAs) to sequester or sponge miRNAs to inhibit miRNA-mediated posttranscriptional gene silencing. The ceRNA network mediated by lncRNAs is closely associated with the initiation and development of HBV-related HCC. LncRNA PCNAP1 is a pseudogene of PCNA (50) highly expressed in HBV-positive HCC cells and tumor tissue (51, 52). PCNAP1 acts as a ceRNA to sponge the tumor suppressor miR-154, preventing it from repressing PCNA expression. Then, PCNA binds covalently closed circular DNA (cccDNA) by interacting with HBV core protein and promoting HBV replication and cccDNA accumulation. PCNAP1 was also reported to sponge the tumor suppressor miR-340-5p and prevent it from directly inhibiting ATF7 expression in HCC cells, promoting HCC cell proliferation (52). CircRNAs may play critical roles in the progression of HBV-related HCC through the circRNA/miRNA axis. For instance, the expression of circRNA_100338 was suggested to be associated with the metastasis of tumor cells. MiR-141-3p is a direct target of circRNA_100338 that regulates gene expression required for HCC development (53). Metastatic suppressor 1 (MTSS1) is a potential target of miR-141-3p that promotes HBV-related hepatoma cell migration associated with poor prognosis. Therefore, the circRNA_100338–miR141–MTSS1 network might exert oncogenic and pro-migratory roles in HBV-related HCC (54). In conclusion, the effects of non-coding RNA on the biological behaviors of liver cancer cells are mainly concentrated on proliferation, migration, invasion, and apoptosis, while other effects are less involved needing further exploration.
Cluster 4 contained 19 high-frequency keywords such as risk, susceptibility, epidemiology, polymorphism, biogenesis, and expression. This cluster mainly reflected the impacts of non-coding RNA on the risk of HBV-related HCC and associated epidemiological research. In a cohort of independent Han Chinese patients, the variant genotypes of rs7763881 in lncRNA HULC were proven to reduce the risk of HBV-related liver cancer (55). A genome-wide association study was conducted to investigate the loci associated with the risk of familial HBV-related HCC. It demonstrated that a group of SNPs overlapping with LINCO0272 was associated with an increased risk of HBV-related HCC, although the detailed molecular mechanisms were unclear (56). MiR-196a2 rs11614913 was also significantly associated with HCC susceptibility, especially in HBV-related HCC, and individuals carrying the TC/CC genotype were more susceptible (57). The findings showed that non-coding RNA variants could affect the susceptibility to HBV-related HCC.
The overlay map analysis for the top 100 keywords suggested that the research hotspots about non-coding RNA and HBV-related HCC in the recent 5 years were mainly focused on long non-coding RNA, EMT, migration, invasion, and other cellular biological behaviors. With the depth of research into the level of mechanism, lncRNA has been paid widespread attention as a hotspot molecule. However, circRNA and other biological effects of tumor cells remain less studied, which are valuable research directions.
In our study, a bibliometric analysis was performed to analyze the evolvement trends of research on non-coding RNA and HBV-related HCC comprehensively. Some limitations should be acknowledged. First, the literature enrolled in the study was only extracted from the WoSCC database, thus some relevant publications might be omitted. Second, no quality assessment was performed for included publications, all of which were considered to have the same validity. Third, some authors might use different spellings of their names or multiple names; thus, errors could be made during the automatic extraction of author names by VOSviewer.
Information from other databases such as PubMed and China National Knowledge Internet were less than WoSCC. It will be a concern to supplement our analysis with updated information from these databases timely, and data from other sources will also be addressed in future. Some new methods should be adopted to perform a weighted analysis for publications based on quality assessment.
The study first comprehensively analyzed all publications on non-coding RNAs and HBV-related HCC with the aid of bibliometric analysis, including publication trends, global collaboration models, and research hotspots. The findings would help to identify new research subjects and frontiers as a guide to future directions.
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
B-GW conceived and designed the study. L-RY and A-RL were responsible for the data analysis and performed data interpretation. L-RY, L-YJ, and B-GW wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the Natural Science Foundation of Liaoning Province (2022-YGJC-59).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
WoSCC, Web of Science Core Collection; HBV, hepatitis B virus; HBx, hepatitis B virus X protein; HCC, hepatocellular carcinoma; AAY, average appearing year; AFP, alpha-fetoprotein.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Sartorius K, An P, Winkler C, Chuturgoon A, Li X, Makarova J, et al. The epigenetic modulation of cancer and immune pathways in hepatitis b virus-associated hepatocellular carcinoma: the influence of HBx and miRNA Dysregulation. Front Immunol. (2021) 12:661204. doi: 10.3389/fimmu.2021.661204
3. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. (2019) 16:589–604. doi: 10.1038/s41575-019-0186-y
4. Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Ann Rev Immunol. (1995) 13:29–60. doi: 10.1146/annurev.iy.13.040195.000333
5. Dandri M. Epigenetic modulation in chronic hepatitis B virus infection. Semin Immunopathol. (2020) 42:173–85. doi: 10.1007/s00281-020-00780-6
6. Koumbi L, Karayiannis P. The epigenetic control of hepatitis B Virus modulates the outcome of infection. Front Microbiol. (2015) 6:1491. doi: 10.3389/fmicb.2015.01491
7. Sartorius K, Makarova J, Sartorius B, An P, Winkler C, Chuturgoon A, et al. The regulatory role of microrna in hepatitis-b virus-associated hepatocellular carcinoma (HBV-HCC) pathogenesis. Cells. (2019) 8:1504. doi: 10.3390/cells8121504
8. Samudh N, Shrilall C, Arbuthnot P, Bloom K, Ely A. Diversity of Dysregulated Long Non-Coding RNAs in HBV-Related Hepatocellular Carcinoma. Front Immunol. (2022) 13:834650. doi: 10.3389/fimmu.2022.834650
9. Liao R, Liu L, Zhou J, Wei X, Huang P. Current molecular biology and therapeutic strategy status and prospects for circRNAs in HBV-associated hepatocellular carcinoma. Front Oncol. (2021) 11:697747. doi: 10.3389/fonc.2021.697747
10. Agarwal A, Durairajanayagam D, Tatagari S, Esteves SC, Harlev A, Henkel R, et al. Bibliometrics: tracking research impact by selecting the appropriate metrics. Asian J Androl. (2016) 18:296–309. doi: 10.4103/1008-682X.171582
11. Peng C, Kuang L, Zhao J, Ross AE, Wang Z, Ciolino JB. Bibliometric and visualized analysis of ocular drug delivery from 2001 to 2020. J Control Release. (2022) 345:625–45. doi: 10.1016/j.jconrel.2022.03.031
12. van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. (2010) 84:523–38. doi: 10.1007/s11192-009-0146-3
13. Karydakis P, Giakoumettis D, Themistocleous M. The 100 most cited papers about pediatric traumatic brain injury: a bibliometric analysis. Irish J Med Sci. (2020) 189:315–25. doi: 10.1007/s11845-019-02085-6
14. Perazzo MF, Otoni ALC, Costa MS, Granville-Granville AF, Paiva SM, Martins-Júnior PA. The top 100 most-cited papers in Paediatric Dentistry journals: A bibliometric analysis. Int J Paediatr Dent. (2019) 29:692–711. doi: 10.1111/ipd.12563
15. Newman ME. Coauthorship networks and patterns of scientific collaboration. Proc Natl Acad Sci USA. (2004) 101(Suppl 1):5200–5. doi: 10.1073/pnas.0307545100
16. Romero L, Portillo-Salido E. Trends in Sigma-1 Receptor Research: A 25-Year Bibliometric Analysis. Front Pharmacol. (2019) 10:564. doi: 10.3389/fphar.2019.00564
17. Lin XJ, Chong Y, Guo ZW, Xie C, Yang XJ, Zhang Q, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. (2015) 16:804–15. doi: 10.1016/S1470-2045(15)00048-0
18. Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. (2011) 50:136–42. doi: 10.1002/mc.20712
19. Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One. (2011) 6:e28486. doi: 10.1371/journal.pone.0028486
20. Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. BioMed Res Int. (2014) 2014:864894. doi: 10.1155/2014/864894
21. Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao B, et al. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. OncoTargets Thera. (2017) 10:3843–51. doi: 10.2147/OTT.S140062
22. Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. (2015) 47:e184. doi: 10.1038/emm.2015.68
23. Wen Y, Han J, Chen J, Dong J, Xia Y, Liu J, et al. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer. (2015) 137:1679–90. doi: 10.1002/ijc.29544
24. Pratedrat P, Chuaypen N, Nimsamer P, Payungporn S, Pinjaroen N, Sirichindakul B, et al. Diagnostic and prognostic roles of circulating miRNA-223-3p in hepatitis B virus-related hepatocellular carcinoma. PLoS One. (2020) 15:e0232211. doi: 10.1371/journal.pone.0232211
25. Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. (2007) 132:330–42. doi: 10.1053/j.gastro.2006.08.026
26. Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. BioMed Res Int. (2013) 2013:136106. doi: 10.1155/2013/136106
27. Lu J, Xie F, Geng L, Shen W, Sui C, Yang J. Investigation of serum lncRNA-uc003wbd and lncRNA-AF085935 expression profile in patients with hepatocellular carcinoma and HBV. Tumour Biol. (2015) 36:3231–6. doi: 10.1007/s13277-014-2951-4
28. Wang K, Guo WX, Li N, Gao CF, Shi J, Tang YF, et al. Serum LncRNAs Profiles Serve as Novel Potential Biomarkers for the Diagnosis of HBV-Positive Hepatocellular Carcinoma. PLoS One. (2015) 10:e0144934. doi: 10.1371/journal.pone.0144934
29. Zhu K, Zhan H, Peng Y, Yang L, Gao Q, Jia H, et al. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis. (2020) 41:296–302. doi: 10.1093/carcin/bgz154
30. Cui S, Qian Z, Chen Y, Li L, Li P, Ding H. Screening of up- and downregulation of circRNAs in HBV-related hepatocellular carcinoma by microarray. Oncol Lett. (2018) 15:423–32. doi: 10.3892/ol.2017.7265
31. Wu C, Deng L, Zhuo H, Chen X, Tan Z, Han S, et al. Circulating circRNA predicting the occurrence of hepatocellular carcinoma in patients with HBV infection. J Cell Mol Med. (2020) 24:10216–22. doi: 10.1111/jcmm.15635
32. Wang Z, Wu Z, Huang P. The function of miRNAs in hepatocarcinogenesis induced by hepatitis B virus X protein (Review). Oncol Rep. (2017) 38:652–64. doi: 10.3892/or.2017.5716
33. Qiu X, Dong S, Qiao F, Lu S, Song Y, Lao Y, et al. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene. (2013) 32:3296–305. doi: 10.1038/onc.2013.150
34. Liu C, Yu J, Yu S, Lavker RM, Cai L, Liu W, et al. MicroRNA-21 acts as an oncomir through multiple targets in human hepatocellular carcinoma. J Hepatol. (2010) 53:98–107. doi: 10.1016/j.jhep.2010.02.021
35. Murakami Y, Kawada N. MicroRNAs in hepatic pathophysiology. Hepatol Res. (2017) 47:60–9. doi: 10.1111/hepr.12730
36. Fu X, Wen H, Jing L, Yang Y, Wang W, Liang X, et al. MicroRNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI3K/Akt pathway. Cancer Sci. (2017) 108:620–31. doi: 10.1111/cas.13177
37. Wong QW, Ching AK, Chan AW, Choy KW, To KF, Lai PB, et al. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res. (2010) 16:867–75. doi: 10.1158/1078-0432.CCR-09-1840
38. Bandopadhyay M, Banerjee A, Sarkar N, Panigrahi R, Datta S, Pal A, et al. Tumor suppressor micro RNA miR-145 and onco micro RNAs miR-21 and miR-222 expressions are differentially modulated by hepatitis B virus X protein in malignant hepatocytes. BMC Cancer. (2014) 14:721. doi: 10.1186/1471-2407-14-721
39. Callegari E, Gramantieri L, Domenicali M, D’Abundo L, Sabbioni S, Negrini M. MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. (2015) 22:46–57. doi: 10.1038/cdd.2014.136
40. Wu G, Huang P, Ju X, Li Z, Wang Y. Lin28B over-expression mediates the repression of let-7 by hepatitis B virus X protein in hepatoma cells. Int J Clin Exp Med. (2015) 8:15108–16.
41. Wang Y, Wang Q, Song J. Inhibition of autophagy potentiates the proliferation inhibition activity of microRNA-7 in human hepatocellular carcinoma cells. Oncol Lett. (2017) 14:3566–72. doi: 10.3892/ol.2017.6573
42. Yu LX, Ling Y, Wang HY. Role of nonresolving inflammation in hepatocellular carcinoma development and progression. NPJ Precis Oncol. (2018) 2:6. doi: 10.1038/s41698-018-0048-z
43. Beltran M, Puig I, Peña C, García JM, Alvarez AB, Peña R, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. (2008) 22:756–69. doi: 10.1101/gad.455708
44. Hu JJ, Song W, Zhang SD, Shen XH, Qiu XM, Wu HZ, et al. HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci Rep. (2016) 6:23521. doi: 10.1038/srep23521
45. Zhang Y, Li J, Wang S, Yang F, Zhou Y, Liu Y, et al. HBx-associated long non-coding RNA activated by TGF-β promotes cell invasion and migration by inducing autophagy in primary liver cancer. Int J Oncol. (2020) 56:337–47. doi: 10.3892/ijo.2019.4908
46. Zhang F, Li Y, Gan L, Tong X, Qi D, Wang Q, et al. HBx-upregulated MAFG-AS1 promotes cell proliferation and migration of hepatoma cells by enhancing MAFG expression and stabilizing nonmuscle myosin IIA. FASEB J. (2021) 35:e21529. doi: 10.1096/fj.202002374R
47. Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. (2006) 12:4851–8. doi: 10.1158/1078-0432.CCR-06-0134
48. Zhang H, Chen X, Zhang J, Wang X, Chen H, Liu L, et al. Long non-coding RNAs in HBV-related hepatocellular carcinoma (Review). Int J Oncol. (2020) 56:18–32. doi: 10.3892/ijo.2019.4909
49. Yu S, Li N, Huang Z, Chen R, Yi P, Kang R, et al. A novel lncRNA, TCONS_00006195, represses hepatocellular carcinoma progression by inhibiting enzymatic activity of ENO1. Cell Death Dis. (2018) 9:1184. doi: 10.1038/s41419-018-1231-4
50. Stoimenov I, Lagerqvist A. The PCNA pseudogenes in the human genome. BMC Res Notes. (2012) 5:87. doi: 10.1186/1756-0500-5-87
51. Feng J, Yang G, Liu Y, Gao Y, Zhao M, Bu Y, et al. LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics. (2019) 9:5227–45. doi: 10.7150/thno.34273
52. He M, Hu L, Bai P, Guo T, Liu N, Feng F, et al. LncRNA PCNAP1 Promotes Hepatoma Cell Proliferation through Targeting miR-340-5p and is Associated with Patient Survival. J Oncol. (2021) 2021:6627173. doi: 10.1155/2021/6627173
53. Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. (2017) 7:5428. doi: 10.1038/s41598-017-05432-8
54. Huang XY, Huang ZL, Xu B, Chen Z, Re TJ, Zheng Q, et al. Elevated MTSS1 expression associated with metastasis and poor prognosis of residual hepatitis B-related hepatocellular carcinoma. J Exp Clin Cancer Res. (2016) 35:85. doi: 10.1186/s13046-016-0361-8
55. Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J, et al. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One. (2012) 7:e35145. doi: 10.1371/journal.pone.0035145
56. Lin YY, Yu MW, Lin SM, Lee SD, Chen CL, Chen DS, et al. Genome-wide association analysis identifies a GLUL haplotype for familial hepatitis B virus-related hepatocellular carcinoma. Cancer. (2017) 123:3966–76. doi: 10.1002/cncr.30851
57. Zhang Q, Xu X, Wu M, Qin T, Wu S, Liu H. MiRNA polymorphisms and hepatocellular carcinoma susceptibility: a systematic review and network meta-analysis. Front Oncol. (2020) 10:562019. doi: 10.3389/fonc.2020.562019
58. Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. (2006) 25:2537–45. doi: 10.1038/sj.onc.1209283
59. Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. (2015) 149:1226–39. doi: 10.1053/j.gastro.2015.05.061
60. Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology (Baltimore, Md). (2008) 47:1955–63. doi: 10.1002/hep.22256
61. Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology (Baltimore, Md). (2011) 54:1679–89. doi: 10.1002/hep.24563
62. McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. (2003) 21:639–44. doi: 10.1038/nbt824
63. Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. (2011) 29:4781–8. doi: 10.1200/JCO.2011.38.2697
64. Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. (2012) 22:291–303. doi: 10.1016/j.ccr.2012.07.023
65. Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. (2008) 135:257–69. doi: 10.1053/j.gastro.2008.04.003
Keywords: non-coding RNA, HBV-related hepatocellular carcinoma, bibliometric analysis, long noncoding RNA (LncRNA), microRNA (miRNA)
Citation: Yan L-r, Liu A-r, Jiang L-y and Wang B-g (2022) Non-coding RNA and hepatitis B virus-related hepatocellular carcinoma: A bibliometric analysis and systematic review. Front. Med. 9:995943. doi: 10.3389/fmed.2022.995943
Received: 16 July 2022; Accepted: 23 August 2022;
Published: 20 September 2022.
Edited by:
Jianping Liu, Karolinska Institutet (KI), SwedenReviewed by:
Xiaodan Wang, Shanghai Jiao Tong University, ChinaCopyright © 2022 Yan, Liu, Jiang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ben-gang Wang, Ymd3YW5nQGNtdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.