- 1Department of Microbiology and Microbial Biotechnology, Faculty of Life Sciences and Biotechnology, Shahid Beheshti University, Tehran, Iran
- 2Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 4Basic and Molecular Epidemiology of Gastrointestinal Disorders Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Background: Although the etiopathogenesis of inflammatory bowel disease (IBD) is still poorly understood, Escherichia coli has been described as a potential causative microorganism in IBD pathogenesis and also disease progression, offering a potential therapeutic target for disease management. Therefore, we conducted this study to investigate the pathotypes, phylogenetic groups, and antimicrobial resistance of E. coli isolates from patients with IBD in Iran.
Methods: Fecal and biopsy colonic samples were collected from IBD patients experiencing flare-up episodes referred to Taleghani hospital in Tehran, Iran, between August 2020 and January 2021. Identification of E. coli strains was performed based on biochemical and molecular methods. Antibiotic susceptibility testing was performed as recommended by the Clinical and Laboratory Standards Institute. Phylogrouping and pathotyping of each isolate were carried out using polymerase chain reaction (PCR) and multilocus sequence typing (MLST) assays.
Results: A total of 132 non-duplicate E. coli strains were isolated from 113 IBD patients, including 96 ulcerative colitis (UC), and 17 Crohn’s disease (CD) patients. In our study, 55% of CD-related E. coli and 70.5% of UC-related isolates were non-susceptible to at least three or more unique antimicrobial classes, and were considered as multidrug-resistant (MDR) strains. E. coli strains exhibited a high level of resistance to cefazolin, ampicillin, tetracycline, ceftazidime, ciprofloxacin, and cefotaxime. Enterotoxigenic E. coli (ETEC) and diffusely adherent E. coli (DAEC) were the most prevalent pathotypes, and groups B2 and D were the predominant phylogroups.
Conclusion: In the present study, we found that E. coli strains that colonize the gut of Iranian patients with IBD most frequently belonged to phylogenetic groups B2 and D. We also conclude that E. coli isolates from IBD patients have been revealed to be resistant to commonly used antibiotics, in which most of them harbored strains that would be categorized as MDR.
Background
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), are chronic, idiopathic inflammatory disorders characterized by relapsing and remitting episodes of intestinal inflammation (1). CD and UC are distinguished by the location, clinical manifestations, and hypothesized pathogenic mechanisms. The clinical features of IBD patients depend on the site and extent of mucosal inflammation that may include diarrhea, rectal bleeding, abdominal pain, malnutrition, and weight loss (2). The etiopathogenesis of IBD is still poorly understood; however, genetic susceptibility, environmental factors, altered intestinal microbiota, and immune-mediated intestinal injury are thought to be seriously involved in this complex disorder (3).
Altered microbial communities, termed dysbiosis, are associated with changes in microbial abundance, composition and a reduction in the overall biodiversity. Previous studies have suggested a decrease in intestinal species richness, more specifically a significant drop in Firmicutes, and an increase in the prevalence of Proteobacteria phylum, particularly Enterobacteriaceae, in IBD patients (4, 5). Recent researches have also identified Escherichia coli as a potential causative microorganism in IBD pathogenesis and also disease progression (6–8).
Based on the genetic and clinical criteria, E. coli strains can be classified into three major groups, including commensal strains, intestinal pathogenic or diarrheagenic E. coli (DEC), and extraintestinal pathogenic E. coli (ExPEC) (9). There are six classic pathotypes among the DECs according to the virulence factors they possess or pathological effects they cause, including enteropathogenic E. coli (EPEC), Shiga toxin-producing E. coli (STEC), enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC) and diffusely adherent E. coli (DAEC) (10).
It has been previously described that IBD-associated E. coli isolates commonly carry ExPEC-associated genetic markers (11, 12). On the other hand, several lines of evidence indicated that some commensal E. coli strains in the gut, which are better described as pathobionts, are associated with immune-mediated disorders like IBD. Importantly, the pathogenicity of these strains are depending on environmental and host genetic factors to cause disease (13, 14). Recently, several additional studies have suggested the involvement of adherent-invasive E. coli (AIEC) in the pathogenesis of CD, with demonstrations that these heterogeneous strains can invade human intestinal epithelial cells as well as survive in macrophages, resulting in tissue damage and inflammation (15–19). Although DEC strains have not been comprehensively recognized to implicate in the pathogenesis of IBD, some authors have described that they may promote intestinal inflammation (14). They have also shown that DAEC strains possessing adherence factors reside in the large intestine and can attach to the rectal mucosa, irrespective of the presence of colitis in UC patients (14).
Among typing methods applied for characterization of E. coli strains, such as pulsed-field gel electrophoresis (PFGE), enterobacterial repetitive intergenic consensus sequence (ERIC)-PCR, randomly amplified polymorphic DNA (RAPD), ribotyping, multiplex phylogrouping PCR has been widely employed because of its simplicity and rapidity (20–22). Phylogrouping technique is also useful for genetic structure analysis of E. coli populations, classification of extraintestinal pathogenic and commensal strains, and also host-source relationships (23). Additionally, it has been reported that different phylogroups vary in the presence of virulence factors, ecological niches and their antibiotic-resistance profiles (24). E. coli isolates from IBD patients frequently belong to certain phylogroups, particularly phylogroups B2 and D, compared to healthy controls (12).
Since the 1970s, various investigators have described increased numbers of E. coli isolates with certain virulence characteristics from IBD patients compared to those from healthy controls, particularly when focusing on CD patients during disease relapses (8, 25–27). For instance, the adhesion index of active CD-associated E. coli isolates was significantly higher than those isolated from healthy subjects (8). The adhesion capabilities of E. coli allow the bacterium to colonize the intestinal mucosa and induce intestinal inflammation (18). In addition, it was found that hemolysin- and necrotoxin-producing E. coli were associated with relapse of UC, however, the authors suggested that the inflammation during a relapse of colitis tends to favor the presence of these organisms, rather than that these organisms cause the relapse (28). Moreover, most IBD-associated E. coli isolates showed higher invasion properties than those from healthy subjects, in which the invasive properties are associated with induction of bowel inflammation (18, 29).
The frequent recovery of E. coli strains possessing certain virulence markers from patients with CD and UC has increased interest in these strains over the last two decades (6, 19, 29). However, there are only very limited data reported on the association between E. coli and IBD pathogenesis in Iran (30, 31). With mounting evidence confirming the impact of dysbiotic microbiome in IBD, some antibiotics are exploited for treating bacterial overgrowth, as well as for the treatment of IBD flare-up and septic complications of the disease, such as abscesses and post-operative wound infections. In this regard, different antibiotics are used as empirical antibiotic therapy for IBD patients, most often ciprofloxacin and metronidazole, each alone or in combination (32). The aim of this study principally was to investigate the pathotypes, phylogenetic groups, and antimicrobial resistance of E. coli isolates from patients with IBD in Iran.
Materials and methods
Patients, biopsies, and fecal samples
In this cross-sectional study, 156 patients with confirmed diagnosis of IBD who referred to Taleghani hospital in Tehran, Iran were enrolled in this study between August 2020 and January 2021. A combination of clinical, radiological, endoscopic, and pathological criteria was considered for a reliable diagnosis of IBD (33). Biopsy specimens were obtained from 56 IBD patients during colonoscopy, and 100 fecal samples were collected from the rest of the patients. All the biopsies were taken from inflamed tissue from the ileum and/or colon. Demographic and disease variables such as; age, gender, disease duration, medication, and clinical features were recorded for all patients through a questionnaire. The flare-up of CD and UC patients was diagnosed according to Crohn’s disease activity index (CDAI) and Powell-Tuck index, respectively (34). This study was given ethical approval by the Institutional Ethical Review Committee of Research Institute for Gastroenterology and Liver Diseases (RIGLD) at Shahid Beheshti University of Medical Sciences (Project No. IR.SBMU.RIGLD.REC.1399.052). Informed consent was obtained from all subjects and/or their legal guardians prior to sample collection.
Bacterial isolation and identification
Biopsy samples were taken with sterile forceps, placed in brain heart infusion (BHI) broth solution, and then immediately delivered to the microbiology laboratory of Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases (35). The fresh biopsy samples were homogenized in BHI broth, then the tissue homogenates were plated on MacConkey agar and incubated for 24 h at 37°C in aerobic conditions. Subsequently, single pink (lactose fermenting) colonies on MacConkey agar were picked for further confirmation (36).
The freshly collected stool sample was immediately transported to the microbiology laboratory for the microbial examination within 1 h of collection. Briefly, fecal sample suspension was prepared in phosphate buffer solution (PBS) and plated onto MacConkey agar followed by 24 h incubation at 37°C aerobically (37). The identification of E. coli strains was done based on morphological and biochemical tests (38). Additionally, molecular confirmation was performed by PCR on E. coli 16S rRNA gene using specific primers (ECB75F: 5′-GGAAGAAGCTTGCTTCTTTGCTG-3′, ECR620R: 5′-GAGCCCGGGGATTTCACAT-3′) as previously described (39).
Antimicrobial susceptibility testing
Antibiotic susceptibility testing was performed using the Kirby–Bauer disk diffusion method as recommended by the Clinical and Laboratory Standards Institute (CLSI) guidelines (40). Commercially available antibiotic disks (Mast Co., United Kingdom) used in this study included ampicillin (10 μg), piperacillin/tazobactam (100/10 μg), cefazolin (30 μg), cefoxitin (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), cefepime (30 μg), imipenem (10 μg), aztreonam (30 μg), ciprofloxacin (5 μg), ofloxacin (5 μg), gentamicin (10 μg), amikacin (30 μg), tetracycline (30 μg). In addition, susceptibility to colistin was performed using minimum inhibitory concentration (MIC). Since there are no CLSI breakpoints for Enterobacteriaceae for MIC testing of colistin, its MICs were interpreted based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines as follows: ≤2 mg/L, susceptible; >2 mg/L, resistant (41). Multidrug-resistant (MDR) phenotype was defined as non-susceptibility to at least one agent in three or more antimicrobial classes (42).
DNA extraction
Total genomic DNA of E. coli isolates was extracted from the fresh culture of the bacterium using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The DNA concentration was measured by NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA, United States) and the integrity of DNA was evaluated using electrophoresis on 0.8% (w/v) agarose gels. DNA samples were kept at −20°C until used for PCR assays.
Virulence detection and pathotyping of E. coli isolates
Molecular characterization of E. coli pathotypes was carried out based on virulence gene detection of all 6 categories of intestinal pathogenic E. coli by PCR method as described previously (43–45). Genes used to screen for identification of different pathotypes include lt and stII for ETEC, eae for atypical (aEPEC), eae and bfp for typical EPEC, aggR and pvcD for EAEC, stx1 and stx2 for EHEC, virF and ipaH for EIEC, and daaD for DAEC. The reaction mixture contained 12.5 μl of Taq DNA Polymerase Master Mix (Ampliqon, Denmark), 1 μl (10 pM/μl) of each primer, 8.5 μl of distilled water, and 2 μl (100 ng) of DNA template in a final volume of 25 μl. PCR reactions were performed under the following conditions: 96°C for 4 min, 94°C for 20 s, 55°C for 20 s, and 72°C for 10 s for 30 cycles, with a final extension at 72°C for 7 min. PCR products were visualized following electrophoresis through 1.5% agarose gel (Gibco Life Technologies, Paisley, United Kingdom) stained with ethidium bromide.
E. coli phylogrouping
The phylogroup of each isolate was determined based on E. coli phylogrouping method described by Clermont et al. (46). Briefly, this method assigns strains to phylogroups A, B1, B2, C, D, E, F that belong to E. coli sensu stricto, whereas the eighth is the Escherichia cryptic clade I. This technique has been designed based on extended quadruplex PCR and multilocus sequence typing (MLST) scheme (47, 48). All PCR reactions were carried out in a 20 μl final volume containing 2 μl of 10X buffer, 2 μM of dNTPs, 2 U of Taq polymerase (Ampliqon, Denmark), 2 μl (100 ng) of DNA template and the appropriate primers. The amounts of primer used are 20 pmol, except for AceK.f (40 pmol), ArpA1.r (40 pmol), trpBA.f (12 pmol) and trpBA.r (12 pmol). PCR reactions were performed under the following conditions: denaturation 4 min at 94°C, 30 cycles of 5 s at 94°C and 20 s at 57°C (group E) or 59°C (quadruplex and group C), and a final extension step of 5 min at 72°C. The primers used for the allele-specific phylogroups E and C PCRs were ArpAgpE.f and ArpAgpE.r and trpAgpC.f and trpAgpC.r, respectively. In E- and C-specific PCR reactions, the primers trpBA.f and trpBA.r are added to provide an internal control.
Statistical analysis
Data analysis was performed using IBM SPSS Statistics 20.0 (IBM Corp., Armonk, NY, United States) and GraphPad Prism software version 8 (GraphPad Software, Inc., CA, United States). Categorical variables among groups were compared using the Chi-square test. Results were presented as the average ± standard error of the mean (SEM) of at least three experiments unless otherwise stated. Differences were considered statistically significant when *P < 0.05.
Results
Baseline demographics and clinical characteristics
One hundred and fifty-six IBD patients experiencing flare-ups consisting of 129 (82.7%) UC and 27 (17.3%) CD were enrolled in this study. The median ages of UC and CD patients were 36.08 ± 14.37 years and 39.32 ± 17.22 years, respectively. Proctitis was the most common extent of disease among UC patients (65, 50.4%), followed by left-sided colitis (21, 16.3%), backwash ileitis (15, 11.6%), and pancolitis (11, 8.5%). The ileocolonic region was the most predominantly affected area among CD patients, followed by the right-sided colitis. More detailed demographic and clinical characteristics of the patients are summarized in Table 1. According to the Chi-square test there were no significant differences (P > 0.05) between CD and UC patients regarding to the demographic and clinical characteristics.
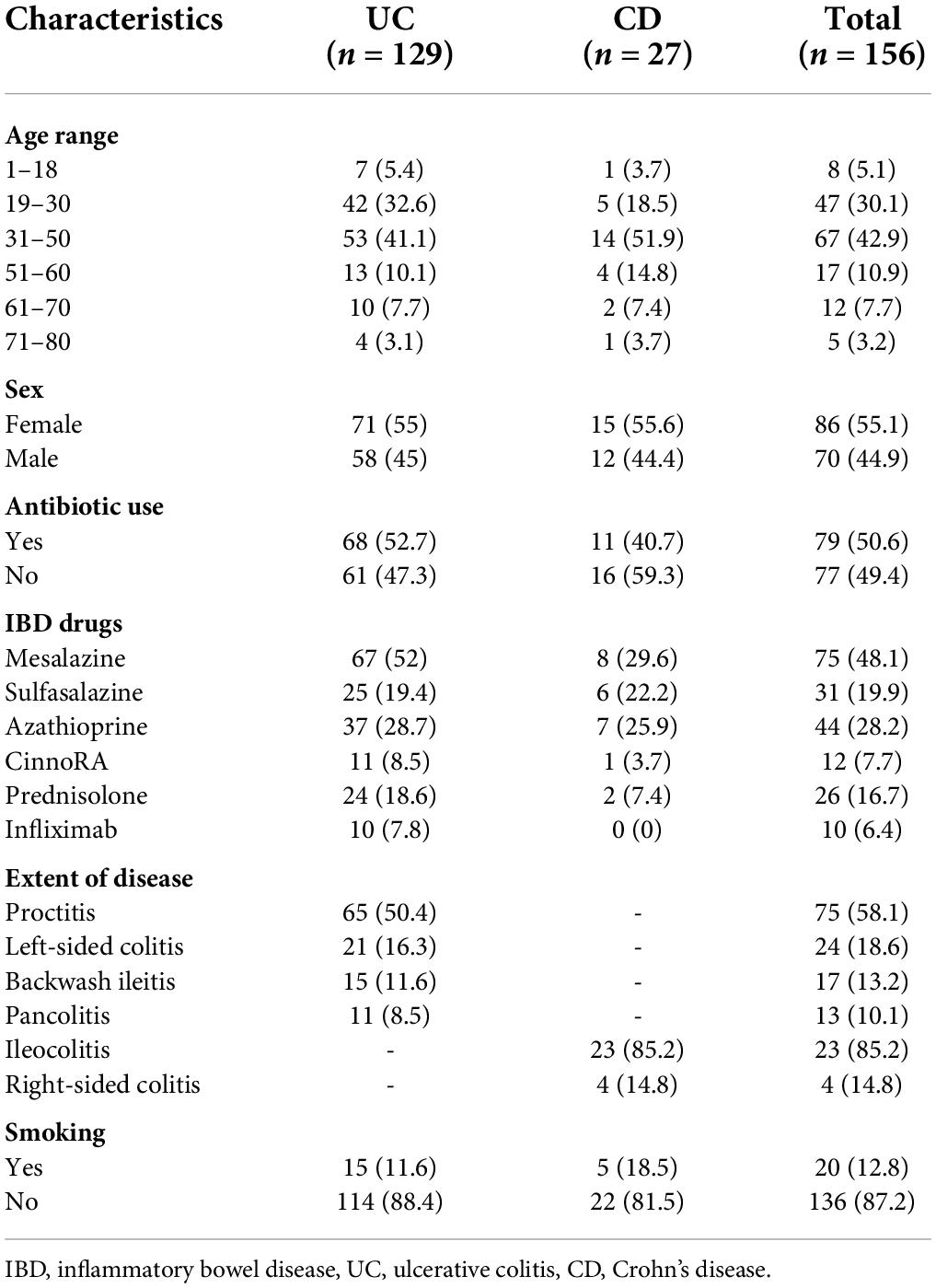
Table 1. Demographic data and clinical characteristics of the inflammatory bowel disease (IBD) patients.
E. coli isolates and antibiotic resistance patterns
A total of 132 non-duplicate E. coli strains were isolated from 113 IBD patients, including 96 UC and 17 CD patients. Of these, 47 strains were recovered from 39 biopsy samples and 85 from 74 stool samples. From 12 patients with UC and 3 with CD at least two different E. coli strains from each patient were recovered based on pathotyping, phylogenetic typing, and/or antibiotic resistance patterns. Table 2 shows the antimicrobial resistance patterns according to disease type and sample type. E. coli isolates were highly resistant to cefazolin (80.3%, n = 106), ampicillin (78.8%, n = 104), and tetracycline (54.5%, n = 72) (Figure 1). All isolates were susceptible to colistin. The concentrations that inhibited 50% (MIC50) and 90% (MIC90) of colistin for E. coli isolates were 0.125 and 0.25 μg/mL, respectively. In our study, 55% of CD- and 70.5% of UC-associated E. coli isolates were non-susceptible (intermediate or resistant) to at least three or more unique antimicrobial classes and, thus were considered as MDR strains. Resistance to ciprofloxacin, an antibiotic commonly selected for use in IBD patients, was present in 50% of CD- associated and 72% of UC- associated E. coli strains. Chi-square test revealed that there was no significant difference (P > 0.05) between CD- and UC- E. coli isolates with regarding to the antimicrobial patterns.
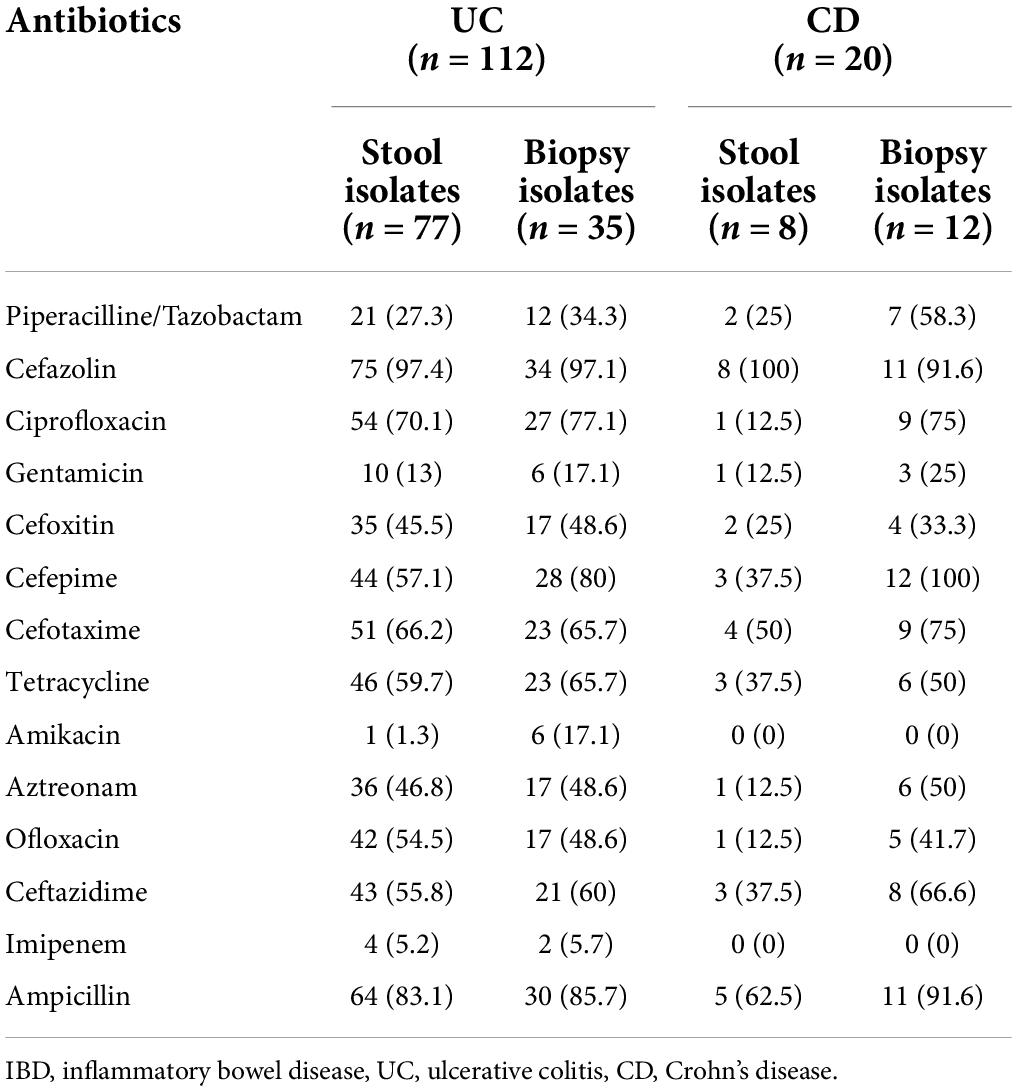
Table 2. Antimicrobial resistance profile (including resistant and intermediate) of the IBD-associated E. coli isolates according to the type of the disease.
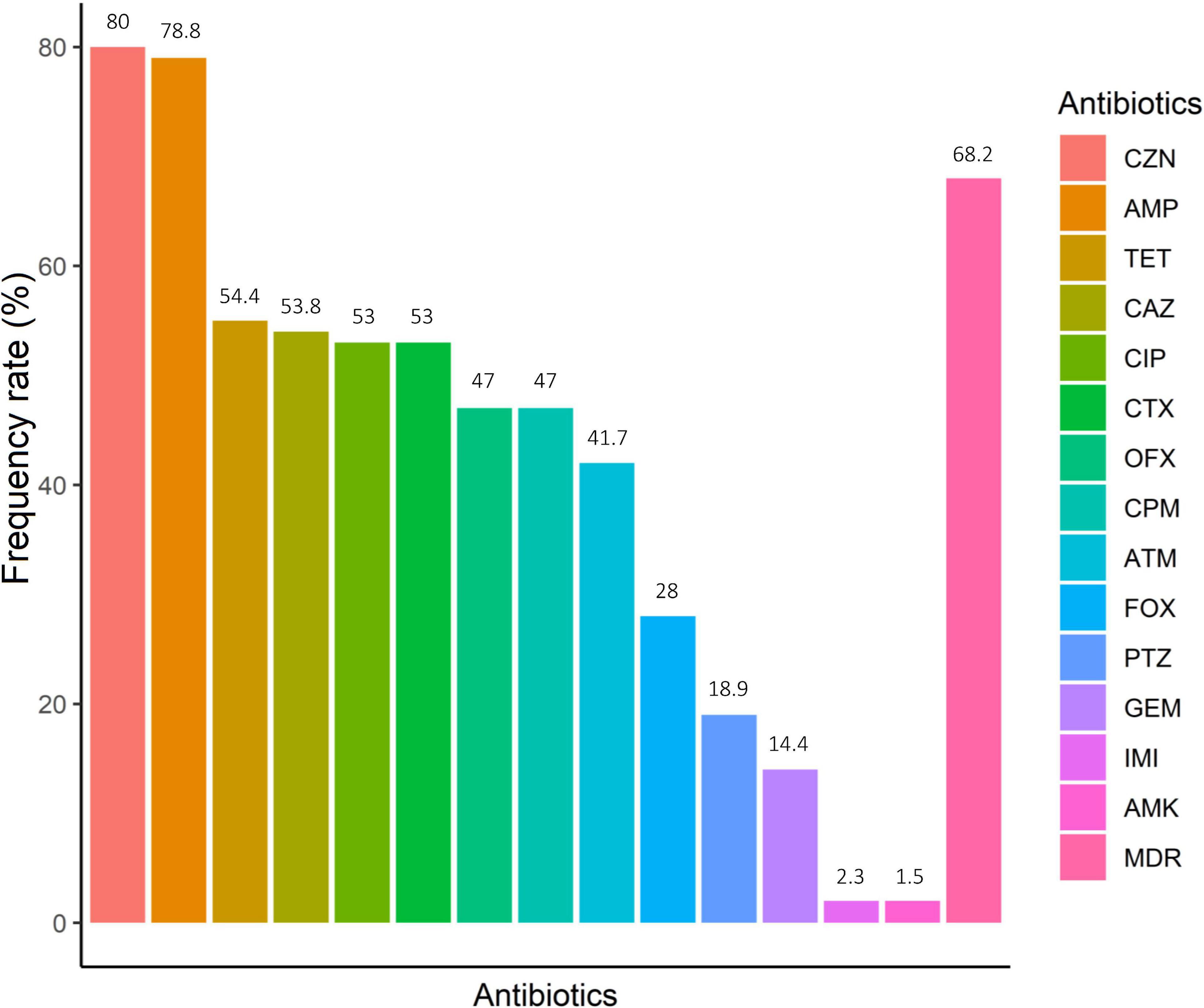
Figure 1. Antimicrobial resistance of IBD-associated E. coli. Amikacin (AMK), ampicillin (AMP), cefotaxime (CTX), piperacillin/tazobactam (PTZ), cefazolin (CZN), cefoxitin (FOX), ceftazidime (CAZ), cefepime (CPM), imipenem (IMI), aztreonam (ATM), ciprofloxacin (CIP), ofloxacin (OFX), gentamicin (GEM) and tetracycline (TET). *MDR, multidrug-resistant.
Pathotyping, phylogrouping, and multilocus sequence typing
E. coli isolates were studied concerning virulence factors and pathotypes, in which lt, daaD, pvcD, aggR, eae genes were detected in 21 (15.9%), 11 (8.3%), 3 (2.3%), 3 (2.3%), and 1 (0.8%) strains, respectively. No STEC strain was detected among E. coli isolates examined, while, one EPEC, 21 ETEC, 11 DAEC, and 3 EAEC strains were isolated. All EAEC strains were cultured from patients with UC, whereas DAEC was recovered from CD patients with a higher frequency than UC subjects, however these differences did not reach statistical significance.
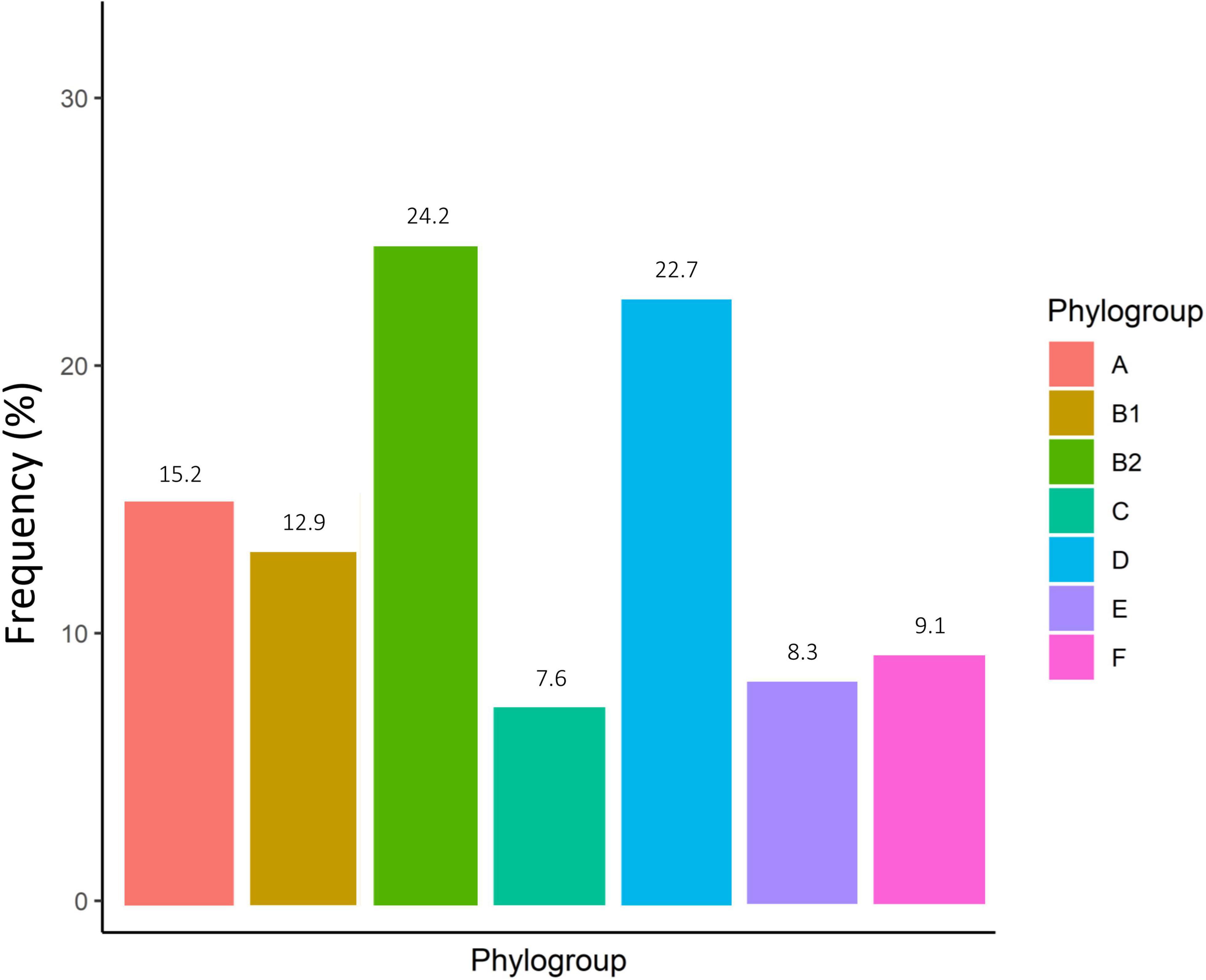
According to the genomic similarity analysis using phylogrouping, phylogroup B2 (24.2%: 20.5% in UC and 30% in CD) and phylogroup D (22.7%: 17.8% in UC and 25% in CD) were the most prevalent phylogroups among the IBD patients. The percentage of other phylogroups were determined 15.2%, 12.9%, 9.1%, 8.3%, 7.6% for A, B1, F, E, and C, respectively. In addition, 6 isolates could not be assigned to none of the phylogroups using quadruplex PCR assay, which then were characterized using MLST. We found six ST types including ST1049, ST10, ST2817, ST12, ST1279, and ST74 allocated to phylogroups B2, A, A, B2, B1, and B2 respectively. Figure 2 and Supplementary Table 1 represent the distribution of E. coli strains according to the phylogrouping and MLST analysis. Analysis of the phylogroup distribution of E. coli strains and demographics of IBD patients showed no significant correlations. Figure 3 represents the frequency of phylogenetic groups according to the gender of patients, type of samples, type of disease, and the used biologics and anti-inflammatory drugs. Figure 4 depicts resistance to commonly used antibiotics according to the phylogenetic groups.
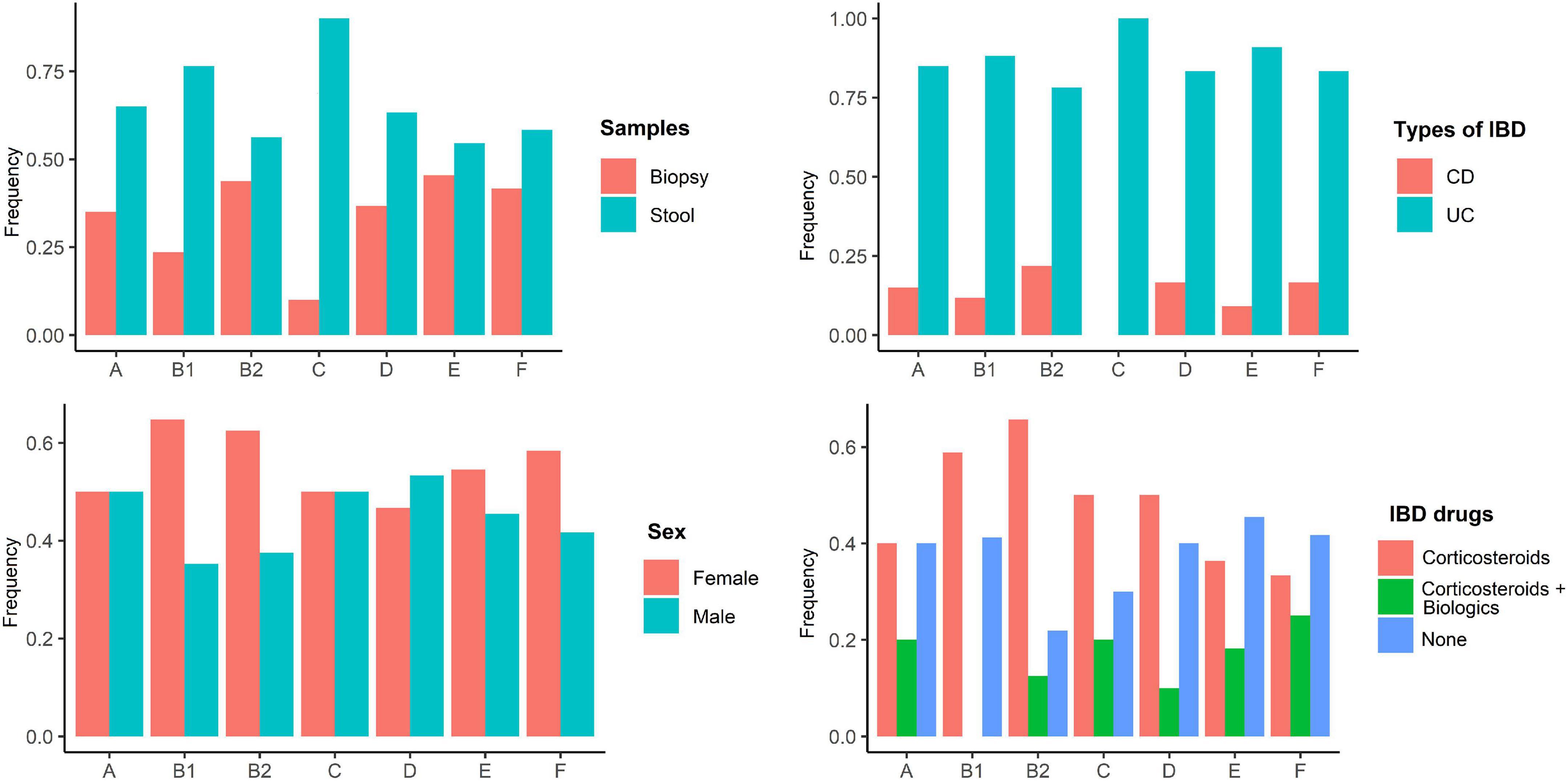
Figure 3. Frequency of phylogenetic groups according to the gender of patients, type of samples, type of disease, and the used biologics and anti-inflammatory drugs.
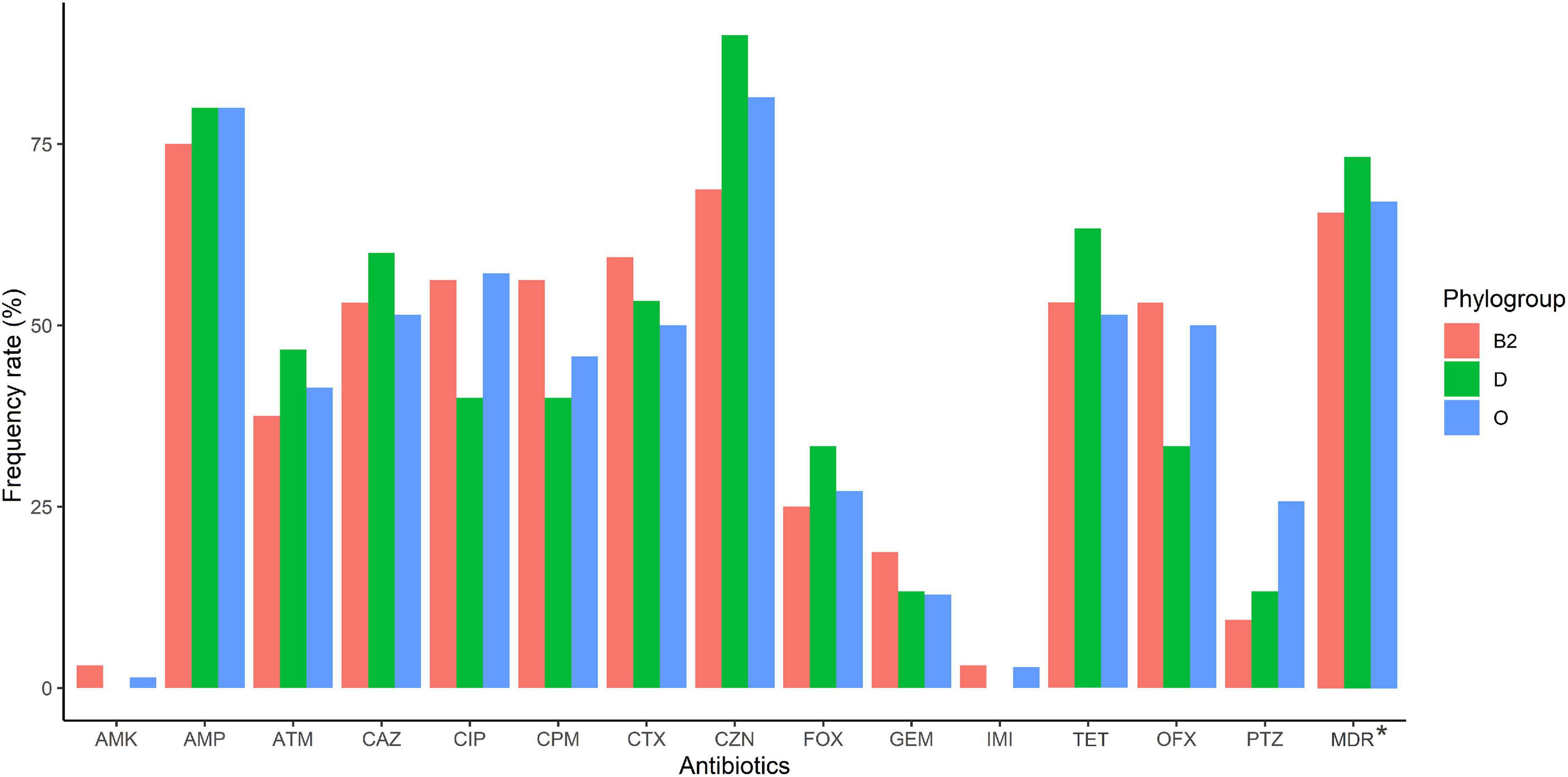
Figure 4. Antimicrobial resistance of IBD-associated E. coli isolates according to phylogroup (B2, D, and other than B2 and D [O]). Amikacin (AMK), ampicillin (AMP), cefotaxime (CTX), piperacillin/tazobactam (PTZ), cefazolin (CZN), cefoxitin (FOX), ceftazidime (CAZ), cefepime (CPM), imipenem (IMI), aztreonam (ATM), ciprofloxacin (CIP), ofloxacin (OFX), gentamicin (GEM) and tetracycline (TET).
Discussion
Several independent investigations have suggested a considerable increase in a specific population of E. coli belonging to phylogroup B2 among patients with IBD (14). However, some animal studies have described overgrowth of E. coli after DSS-induced colitis, which suggest that E. coli enrichment in the gut can be as a consequence of inflammation (19). Any answer concerning the role of E. coli in the pathogenesis of IBD needs isolation and characterization of this bacterium for further in-depth analysis of host-microbe interactions. Moreover, much of the research on these E. coli strains, as well as their potential impact in IBD, has been performed in Western countries (Europe, North America, and Australia), however there is insufficient data regarding the characterization of E. coli isolated from CD and UC patients in the Middle East. It is currently not well comprehended whether the overgrowth of such putative pathogens described among IBD populations is more widespread in the patients in regions with increasing IBD incidence, especially in the Middle East countries and more specifically in Iran. In the current study, we performed phylogenetic analysis and our findings demonstrated that most E. coli strains were classified into phylogenetic groups B2 (24.2%) and D (22.7%), which are reported to be genetically similar to ExPEC strains (49). The strains belonged to phylogruops B2 and D harbor chuA gene, which is a heme iron acquisition gene and involves in heme utilization. The strains that carry chuA are able to survive and persist inside macrophages, which can be suggested as a major contributor to the multiplication of E. coli strains in the inflamed human intestines. Importantly, the upregulating of the chuA stimulates the release of TNF-α, which may promote dysbiosis and microbe-driven intestinal inflammation (50). E. coli strains related to phylogroups B2 and D carry more virulence-associated genes compared to E. coli related to the other phylogroups. Furthermore, B2 and D phylogroups usually possess certain virulence factors, which can lead to extraintestinal infections and also give them the ability to persist within the human gastrointestinal tract (51). Dadi et al. reported that E. coli strains which are engaged in extraintestinal infections, including urinary tract infections (UTIs) are most likely to belong to phylogroup B2 or, to a lesser degree, to phylogroup D (52). Moreover, IBD patients with anorectal complications, which include perianal abscesses and anal fistulas, appear to have an increased risk for UTIs. It could be explained that in these patients anorectal complications could lead to bacterial translocation from the perineum to the bladder (53, 54). Accordingly, the overgrowth of E. coli strains with extraintestinal virulence capacity in IBD patients could act as a risk factor for UTI development. Martinez-Medina et al. (55) noticed that IBD-associated E. coli strains, which have biofilm-producing capacity, mainly belong to B2 phylogroup. Clinical observations revealed that the density of the mucosal biofilm on intestinal tissues was higher in IBD patients than in patients with irritable bowel syndrome (IBS) or controls (56, 57). Biofilm formation by E. coli strains especially AIEC in human gut gives the strains an advantage for intestinal colonization and consequently increases their chance to invade the intestinal epithelium and further results in mucosal inflammation (55). Although several studies have reported increased colonization by strains belonging to phylogroups B2 and D in IBD patients (58–60), some authors indicated a similar distribution of phylogroups among IBD and healthy cohorts (6, 61).
In the current study, PCR pathotyping analysis revealed that ETEC and DAEC were the prevalent pathotypes, where lt gene was detected in 21 (15.9%) E. coli isolates, and daaD was identified in 11 (8.3%) isolates. An important insight of our study is a relatively higher prevalence of ETEC pathotype than that reported by others; in which Meheissen et al. reported the detection of lt in two out of 60 (3.3%) and Kmetova et al found five lt out of 437 (1.2%) samples tested. Previously, Brubaker et al. described that ETEC infection significantly induces intestinal and systemic inflammation among subjects who remained asymptomatic (62). Notably, an important insight of the mentioned study was the extent to which asymptomatic ETEC infection could cause significant intestinal inflammation that may lead to further inflammation-mediated GI disorders such as IBD. More according to a large-scale study by Eybpoosh et al. ETEC was the second most frequent E.coli pathotype in Iran and was detected in all investigated provinces including twenty-nine cities (63). We suggest that the high prevalence of ETEC among IBD patients in our study could support the potential role of ETEC strains in inducing intestinal inflammation and their impact on IBD progression in such endemic regions.
Though still controversial, some preliminary evidence suggests the link between DAEC and UC. In, Burke and Axon reported that a majority of isolated E. coli strains from the stool of UC patients were DAEC pathotypes with both enteropathogenic and enterotoxigenic properties, on the contrary to the isolates from healthy subjects (64). DAEC strains harboring Afa/Dr have been indicated to adhere to the colonic epithelium of UC patients and to induce proinflammatory cytokines TNF-α and IL-8 via the interaction of their fimbriae with membrane-bound host receptors (65). Additionally, some research on the pathogenesis of DAEC indicates that Afa/Dr fully differentiated epithelial cells, inducing the rearrangement of brush border-associated cytoskeletal proteins F-actin followed by the loss of the epithelial cell microvilli (66). In the present work, EIEC and STEC were not detected in the collected samples from CD and UC patients. Importantly, however, there is no epidemiological evidence directly linking the colonization of DAEC to the development and progression of UC. Accordingly, more large-scale case-control studies are needed to clarify the potential role of E. coli pathotypes in the pathogenesis of IBD.
In the present study, we found that approximately half of the cultured E. coli strains from UC and CD patients were non-susceptible to three or more antimicrobial categories and would be categorized as MDR. To our knowledge, only a very small fraction of studies have reported the antimicrobial resistance patterns of Enterobacteriaceae strains among IBD patients. In a study conducted by Martinez-Medina et al., it was reported that mucosa-associated E. coli isolates from CD patients were more frequently resistant to β-lactams than those isolated from the intestine of control subjects (67). Dogan et al. reported that CD-associated E. coli frequently manifests resistance to commonly used antimicrobials, particularly, resistance to ciprofloxacin, rifaximin/rifampin, and trimethoprim/sulfamethoxazole (68). They also found that most of the E. coli isolates from CD patients were MDR compared to those from healthy subjects.
More importantly, 69% of E. coli-colonized patients with IBD harbored strains that were resistant to ciprofloxacin, which is commonly used as empirical antibiotic therapy to treat IBD flare. This rate of ciprofloxacin-resistant E. coli among Iranian IBD patients is significantly higher than those found in the study performed by Dogan et al., in which 29% of E. coli-colonized ileal CD patients harbored E. coli with resistance to ciprofloxacin (68). Interestingly, we found that ciprofloxacin-resistant E. coli were more common in the IBD patients with prior use of ciprofloxacin as antibiotic therapy for IBD flare (73.3%) compared to those patients without adjunct ciprofloxacin therapy (47.5%). Ciprofloxacin is used in combination with metronidazole in the treatment of IBD patients, especially in severe cases or during flare-ups (69). Some previous studies have suggested ciprofloxacin is an adjunct to infliximab (73% vs. 39%, P = 0.12) or adalimumab (71% vs. 47%, P = 0.047), and had a higher response than these two biologics alone (70, 71). In addition to recent studies demonstrating the association of AIEC pathotype with CD, and the ability of ciprofloxacin to eradicate bacteria within macrophages, several investigators suggest that ciprofloxacin may be particularly useful for treating IBD (69). Moreover, patients with ileal or ileal with right-sided CD and circulating antibodies directed against E. coli have been shown to have a higher response rate to budesonide plus ciprofloxacin than the group with no antibody predominant profiles (72). However, our findings demonstrate substantial implications for this strategy. Hence, we suggest that antimicrobial resistance along with non-targeted antimicrobial selection could contribute to IBD flare-ups related to overgrowth of ciprofloxacin-resistant E. coli in those who have received ciprofloxacin for the long term.
Conclusion
In the present study, we found that E. coli strains colonize the gut of Iranian patients with IBD most frequently belonged to phylogenetic groups B2 and D. We also conclude that E.coli isolates from IBD patients have been revealed to be resistant to commonly used antibiotics, in which most of them harbored strains that would be categorized as MDR. We also consider the possibility that the colonization with ciprofloxacin-resistant E. coli strains may arise due to the empirical therapy. We suggest that antimicrobial therapy in IBD patients should be informed by knowledge of antimicrobial susceptibility of the gut resident Enterobacteriaceae. However, there are some limitations in this study. First, a relatively low sample size of CD patients and the single center recruitment, could be technical impediments that can adversely influence the statistical power of the results. In addition, the lack of healthy control group and the lack of follow-up of patients are other limitations of the present work. Further research should be conducted to investigate the characteristics of mucosa-associated E. coli especially AIEC pathotype and their contribution to the inflammatory process in Iranian patients with IBD.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by the Institutional Ethical Review Committee of Research Institute for Gastroenterology and Liver Diseases (RIGLD) at Shahid Beheshti University of Medical Sciences (Project No. IR.SBMU.RIGLD.REC.1399.052). The patients/participants provided their written informed consent to participate in this study.
Author contributions
BFN and BN performed the sample collection and microbiological experiments. BFN and HH wrote the manuscript draft. AY contributed to the study design, conceptualization, and methodology. SS and MA contributed to the colonoscopy procedures and provided clinical consultations. AY and GE provided the important intellectual content and critically revised the manuscript. All authors read and approved the final version of the manuscript and authorship.
Funding
This study was supported by a research grant (Project No. 1139) from the Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Acknowledgments
We thank the Taleghani Hospital Endoscopy Department for their cooperation and help in performing this study. We are grateful to all laboratory staff of the Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran. We also thank the staff of the Department of Microbiology and Microbial Biotechnology, Faculty of Life Sciences and Biotechnology, Shahid Beheshti University, Tehran, Iran.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.985300/full#supplementary-material
Abbreviations
AIEC, adherent-invasive E. coli; BHI, brain heart infusion; CD, Crohn’s disease; CDAI, Crohn’s disease activity index; DEC, Diarrheagenic E. coli; DAEC, diffusely adherent E. coli; EAEC, enteroaggregative E. coli; EIEC, enteroinvasive E. coli; EPEC, enteropathogenic E. coli; ERIC-PCR, enterobacterial repetitive intergenic consensus sequence polymerase chain reaction; ETEC, enterotoxigenic E. coli; ExPEC, extraintestinal pathogenic E. coli; IBD, inflammatory bowel disease; MDR, multidrug-resistant; MLST, multilocus sequence typing; PBS, phosphate buffer solution; PFGE, pulsed-field gel electrophoresis; RAPD, randomly amplified polymorphic DNA; STEC, Shiga toxin-producing E. coli; UC, ulcerative colitis; UTIs, urinary tract infections.
References
1. Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. (2017) 14:573. doi: 10.1038/nrgastro.2017.88
2. Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. (2007) 369:1627–40. doi: 10.1016/S0140-6736(07)60750-8
3. Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. (2013) 62:1505–10. doi: 10.1136/gutjnl-2012-303954
4. Zuo T, Ng SC. The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front Microbiol. (2018) 9:2247. doi: 10.3389/fmicb.2018.02247
5. Torres J, Hu J, Seki A, Eisele C, Nair N, Huang R, et al. Infants born to mothers with IBD present with altered gut microbiome that transfers abnormalities of the adaptive immune system to germ-free mice. Gut. (2020) 69:42–51. doi: 10.1136/gutjnl-2018-317855
6. Elliott TR, Hudspith BN, Wu G, Cooley M, Parkes G, Quiñones B, et al. Quantification and characterization of mucosa-associated and intracellular Escherichia coli in inflammatory bowel disease. Inflamm Bowel Dis. (2013) 19:2326–38. doi: 10.1097/MIB.0b013e3182a38a92
7. Axelrad JE, Joelson A, Nobel YR, Lawlor G, Green PH, Lichtiger S, et al. Enteric infection in relapse of inflammatory bowel disease: the utility of stool microbial PCR testing. Inflamm Bowel Dis. (2017) 23:1034–9. doi: 10.1097/MIB.0000000000001097
8. Martinez-Medina M, Garcia-Gil LJ. Escherichia coli in chronic inflammatory bowel diseases: an update on adherent invasive Escherichia coli pathogenicity. World J Gastrointest Pathophysiol. (2014) 5:213. doi: 10.4291/wjgp.v5.i3.213
9. Russo TA, Johnson JR. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis. (2000) 181:1753–4. doi: 10.1086/315418
10. Mirsepasi-Lauridsen HC, Vrankx K, Engberg J, Friis-Møller A, Brynskov J, Nordgaard-Lassen I, et al. Disease-specific enteric microbiome dysbiosis in inflammatory bowel disease. Front Med. (2018) 5:304. doi: 10.3389/fmed.2018.00304
11. Vejborg RM, Hancock V, Petersen AM, Krogfelt KA, Klemm P. Comparative genomics of Escherichia coli isolated from patients with inflammatory bowel disease. BMC Genomics. (2011) 12:316. doi: 10.1186/1471-2164-12-316
12. Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+ D phylogenetic group in inflammatory bowel disease. Gut. (2007) 56:669–75. doi: 10.1136/gut.2006.099796
13. Yang H, Mirsepasi-Lauridsen HC, Struve C, Allaire JM, Sivignon A, Vogl W, et al. Ulcerative colitis-associated E. coli pathobionts potentiate colitis in susceptible hosts. Gut Microbes. (2020) 12:1847976. doi: 10.1080/19490976.2020.1847976
14. Mirsepasi-Lauridsen HC, Vallance BA, Krogfelt KA, Petersen AM. Escherichia coli pathobionts associated with inflammatory bowel disease. Clin Microbiol Rev. (2019) 32:e60–18. doi: 10.1128/CMR.00060-18
15. Mazzarella G, Perna A, Marano A, Lucariello A, Rotondi Aufiero V, Sorrentino A, et al. Pathogenic role of associated adherent-invasive Escherichia coli in Crohn’ disease. J Cell Physiol. (2017) 232:2860–8. doi: 10.1002/jcp.25717
16. Shaler CR, Elhenawy W, Coombes BK. The unique lifestyle of Crohn’s disease-associated adherent-invasive Escherichia coli. J Mol Biol. (2019) 431:2970–81. doi: 10.1016/j.jmb.2019.04.023
17. Nadalian B, Yadegar A, Houri H, Olfatifar M, Shahrokh S, Asadzadeh Aghdaei H, et al. Prevalence of the pathobiont adherent-invasive Escherichia coli and inflammatory bowel disease: a systematic review and meta-analysis. Eur J Gastroenterol Hepatol. (2021) 36:852–63. doi: 10.1111/jgh.15260
18. Lee JG, Han DS, Jo SV, Lee AR, Park CH, Eun CS, et al. Characteristics and pathogenic role of adherent-invasive Escherichia coli in inflammatory bowel disease: potential impact on clinical outcomes. PLoS One. (2019) 14:e0216165. doi: 10.1371/journal.pone.0216165
19. Palmela C, Chevarin C, Xu Z, Torres J, Sevrin G, Hirten R, et al. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut. (2018) 67:574–87. doi: 10.1136/gutjnl-2017-314903
20. Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. (2000) 66:4555–8. doi: 10.1128/AEM.66.10.4555-4558.2000
21. Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. (2010) 8:207–17. doi: 10.1038/nrmicro2298
22. Gorgannezhad L, Sreejith KR, Christie M, Jin J, Ooi CH, Katouli M, et al. Core-shell beads as microreactors for phylogrouping of E. coli strains. Micromachines. (2020) 11:761. doi: 10.3390/mi11080761
23. Stoppe NC, Silva JS, Carlos C, Sato MIZ, Saraiva AM, Ottoboni LMM, et al. Worldwide phylogenetic group patterns of Escherichia coli from commensal human and wastewater treatment plant isolates. Front Microbiol. (2017) 8:2512. doi: 10.3389/fmicb.2017.02512
24. Carlos C, Pires MM, Stoppe NC, Hachich EM, Sato MI, Gomes TA, et al. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol. (2010) 10:161. doi: 10.1186/1471-2180-10-161
25. Giaffer M, Holdsworth C, Duerden B. Virulence properties of Escherichia coli strains isolated from patients with inflammatory bowel disease. Gut. (1992) 33:646–50. doi: 10.1136/gut.33.5.646
26. Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. (1998) 115:1405–13. doi: 10.1016/S0016-5085(98)70019-8
27. Micenková L, Frankovièová L, Jaborníková I, Bosák J, Dítì P, Šmarda J, et al. Escherichia coli isolates from patients with inflammatory bowel disease: ExPEC virulence- and colicin-determinants are more frequent compared to healthy controls. Int J Med Microbiol. (2018) 308:498–504. doi: 10.1016/j.ijmm.2018.04.008
28. Cooke EM, Ewins SP, Hywel-Jones J, Lennard-Jones J. Properties of strains of Escherichia coli carried in different phases of ulcerative colitis. Gut. (1974) 15:143–6. doi: 10.1136/gut.15.2.143
29. Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser A-L, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. (2004) 127:412–21. doi: 10.1053/j.gastro.2004.04.061
30. Asiabar AS, Aghdaei HA, Sabokbar A, Zali MR, Feizabadi MM. Prevalence of adherent-invasive Escherichia coli with fimH gene isolated from Iranian patients with ulcerative colitis. Jundishapur J Microbiol. (2017) 10:e13858. doi: 10.5812/jjm.13858
31. Asiabar AS, Aghdaei HA, Sabokbar A, Zali MR, Feizabadi MM. Investigation of adherent-invasive E. coli in patients with Crohn’s disease. Med J Islam Repub Iran. (2018) 32:11. doi: 10.14196/mjiri.32.11
32. Nitzan O, Elias M, Peretz A, Saliba W. Role of antibiotics for treatment of inflammatory bowel disease. World J Gastroenterol. (2016) 22:1078–87. doi: 10.3748/wjg.v22.i3.1078
33. Magro F, Langner C, Driessen A, Ensari A, Geboes K, Mantzaris G, et al. European consensus on the histopathology of inflammatory bowel disease. J Crohns Colitis. (2013) 7:827–51. doi: 10.1016/j.crohns.2013.06.001
34. Peyrin-Biroulet L, Panés J, Sandborn WJ, Vermeire S, Danese S, Feagan BG, et al. Defining disease severity in inflammatory bowel diseases: current and future directions. Clin Gastroenterol Hepatol. (2016) 14:348–354.e17. doi: 10.1016/j.cgh.2015.06.001
35. Martinez-Medina M, Aldeguer X, Lopez-Siles M, González-Huix F, López-Oliu C, Dahbi G, et al. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn’s disease. Inflamm Bowel Dis. (2009) 15:872–82. doi: 10.1002/ibd.20860
36. Iyadorai T, Mariappan V, Vellasamy KM, Wanyiri JW, Roslani AC, Lee GK, et al. Prevalence and association of pks+ Escherichia coli with colorectal cancer in patients at the university Malaya medical centre, Malaysia. PLoS One. (2020) 15:e0228217. doi: 10.1371/journal.pone.0228217
37. Meheissen M, Header D, Abdelaty K. Phylogenetic and pathotype analysis of Escherichia coli stool isolates from Egyptian patients with inflammatory bowel disease. Germs. (2019) 9:172. doi: 10.18683/germs.2019.1173
38. Tille P. Bailey & Scott’s Diagnostic Microbiology-e-Book. Amsterdam: Elsevier Health Sciences (2015).
39. Sabat G, Rose P, Hickey W, Harkin J. Selective and sensitive method for PCR amplification of Escherichia coli 16S rRNA genes in soil. Appl Environ Microbiol. (2000) 66:844–9. doi: 10.1128/AEM.66.2.844-849.2000
40. CLSI.Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. Wayne, PA: Clinical and Laboratory Standards Institute (2020).
41. European Committee on Antimicrobial Susceptibility Testing.Breakpoint Tables for Interpretation of MICs and Zone Diameters. Växjö: EUCAST (2020).
42. Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas M, Giske C, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
43. Rajendran P, Ajjampur SSR, Chidambaram D, Chandrabose G, Thangaraj B, Sarkar R, et al. Pathotypes of diarrheagenic Escherichia coli in children attending a tertiary care hospital in South India. Diagn Microbiol Infect Dis. (2010) 68:117–22. doi: 10.1016/j.diagmicrobio.2010.06.003
44. Nguyen TV, Le Van P, Le Huy C, Gia KN, Weintraub A. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J Clin Microbiol. (2005) 43:755–60. doi: 10.1128/JCM.43.2.755-760.2005
45. Saka HK, Dabo NT, Muhammad B, García-Soto S, Ugarte-Ruiz M, Alvarez J. Diarrheagenic Escherichia coli pathotypes from children younger than 5 years in Kano State, Nigeria. Front Public Health. (2019) 7:348. doi: 10.3389/fpubh.2019.00348
46. Clermont O, Christenson JK, Denamur E, Gordon DM. The C lermont E scherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. (2013) 5:58–65. doi: 10.1111/1758-2229.12019
47. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. (2006) 60:1136–51. doi: 10.1111/j.1365-2958.2006.05172.x
48. Clermont O, Gordon D, Denamur E. Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology. (2015) 161:980–8. doi: 10.1099/mic.0.000063
49. Fang X, Monk JM, Mih N, Du B, Sastry AV, Kavvas E, et al. Escherichia coli B2 strains prevalent in inflammatory bowel disease patients have distinct metabolic capabilities that enable colonization of intestinal mucosa. BMC Syst Biol. (2018) 12:66. doi: 10.1186/s12918-018-0587-5
50. Dogan B, Suzuki H, Herlekar D, Sartor RB, Campbell BJ, Roberts CL, et al. Inflammation-associated adherent-invasive Escherichia coli are enriched in pathways for use of propanediol and iron and M-cell translocation. Inflamm Bowel Dis. (2014) 20:1919–32. doi: 10.1097/MIB.0000000000000183
51. Nowrouzian FL, Adlerberth I, Wold AE. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect. (2006) 8:834–40. doi: 10.1016/j.micinf.2005.10.011
52. Dadi BR, Abebe T, Zhang L, Mihret A, Abebe W, Amogne W. Distribution of virulence genes and phylogenetics of uropathogenic Escherichia coli among urinary tract infection patients in Addis Ababa, Ethiopia. BMC Infect Dis. (2020) 20:108. doi: 10.1186/s12879-020-4844-z
53. Herbert J, Teeter E, Burstiner LS, Doka R, Royer A, Owings AH, et al. Urinary manifestations in African American and Caucasian inflammatory bowel disease patients: a retrospective cohort study. BMC Urol. (2022) 22:1. doi: 10.1186/s12894-021-00951-z
54. Peyrin-Biroulet L, Pillot C, Oussalah A, Billioud V, Aissa N, Balde M, et al. Urinary tract infections in hospitalized inflammatory bowel disease patients: a 10-year experience. Inflamm Bowel Dis. (2012) 18:697–702. doi: 10.1002/ibd.21777
55. Martinez-Medina M, Naves P, Blanco J, Aldeguer X, Blanco JE, Blanco M, et al. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC). BMC Microbiol. (2009) 9:202. doi: 10.1186/1471-2180-9-202
56. Srivastava A, Gupta J, Kumar S, Kumar A. Gut biofilm forming bacteria in inflammatory bowel disease. Microb Pathog. (2017) 112:5–14. doi: 10.1016/j.micpath.2017.09.041
57. Chassaing B, Darfeuille-Michaud A. The σE pathway is involved in biofilm formation by Crohn’s disease-associated adherent-invasive Escherichia coli. J Bacteriol. (2013) 195:76–84. doi: 10.1128/JB.01079-12
58. Rhodes JM. The role of Escherichia coli in inflammatory bowel disease. Gut. (2007) 56:610–2. doi: 10.1136/gut.2006.111872
59. Dreux N, Denizot J, Martinez-Medina M, Mellmann A, Billig M, Kisiela D, et al. Point mutations in FimH adhesin of Crohn’s disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog. (2013) 9:e1003141. doi: 10.1371/journal.ppat.1003141
60. Costa RF, Ferrari ML, Bringer M-A, Darfeuille-Michaud A, Martins FS, Barnich N. Characterization of mucosa-associated Escherichia coli strains isolated from Crohn’s disease patients in Brazil. BMC Microbiol. (2020) 20:178. doi: 10.1186/s12866-020-01856-x
61. Schippa S, Conte MP, Borrelli O, Iebba V, Aleandri M, Seganti L, et al. Dominant genotypes in mucosa-associated Escherichia coli strains from pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. (2009) 15:661–72. doi: 10.1002/ibd.20818
62. Brubaker J, Zhang X, Bourgeois AL, Harro C, Sack DA, Chakraborty S. Intestinal and systemic inflammation induced by symptomatic and asymptomatic enterotoxigenic E. coli infection and impact on intestinal colonization and ETEC specific immune responses in an experimental human challenge model. Gut Microbes. (2021) 13:1891852. doi: 10.1080/19490976.2021.1891852
63. Eybpoosh S, Mostaan S, Gouya MM, Masoumi-Asl H, Owlia P, Eshrati B, et al. Frequency of five Escherichia Coli pathotypes in Iranian adults and children with acute diarrhea. PLoS One. (2021) 16:e0245470. doi: 10.1371/journal.pone.0245470
64. Burke D, Axon A. Ulcerative colitis and Escherichia coli with adhesive properties. J Clin Pathol. (1987) 40:782–6. doi: 10.1136/jcp.40.7.782
65. Servin AL. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin Microbiol Rev. (2005) 18:264–92. doi: 10.1128/CMR.18.2.264-292.2005
66. Le Bouguénec C, Servin AL. Diffusely adherent Escherichia coli strains expressing Afa/Dr adhesins (Afa/Dr DAEC): hitherto unrecognized pathogens. FEMS Microbiol Lett. (2006) 256:185–94. doi: 10.1111/j.1574-6968.2006.00144.x
67. Martinez-Medina M, Strozzi F, Ruiz Del Castillo B, Serrano-Morillas N, Ferrer Bustins N, Martínez-Martínez L. Antimicrobial resistance profiles of adherent invasive Escherichia coli show increased resistance to β-lactams. Antibiotics. (2020) 9:251. doi: 10.3390/antibiotics9050251
68. Dogan B, Scherl E, Bosworth B, Yantiss R, Altier C, McDonough PL, et al. Multidrug resistance is common in Escherichia coli associated with ileal Crohn’s disease. Inflamm Bowel Dis. (2012) 19:141–50. doi: 10.1002/ibd.22971
69. Arnold GL, Beaves MR, Pryjdun VO, Mook WJ. Preliminary study of ciprofloxacin in active Crohn’s disease. Inflamm Bowel Dis. (2002) 8:10–5. doi: 10.1097/00054725-200201000-00002
70. West R, Van der Woude C, Hansen B, Felt-Bersma R, Van Tilburg A, Drapers J, et al. Clinical and endosonographic effect of ciprofloxacin on the treatment of perianal fistulae in Crohn’s disease with infliximab: a double-blind placebo-controlled study. Aliment Pharmacol Ther. (2004) 20:1329–36. doi: 10.1111/j.1365-2036.2004.02247.x
71. Dewint P, Hansen BE, Verhey E, Oldenburg B, Hommes DW, Pierik M, et al. Adalimumab combined with ciprofloxacin is superior to adalimumab monotherapy in perianal fistula closure in Crohn’s disease: a randomised, double-blind, placebo controlled trial (ADAFI). Gut. (2014) 63:292–9. doi: 10.1136/gutjnl-2013-304488
Keywords: Escherichia coli, inflammatory bowel disease, antimicrobial resistance, phylogrouping, pathotypes, multidrug-resistant phenotype
Citation: Nadalian B, Nadalian B, Houri H, Shahrokh S, Abdehagh M, Yadegar A and Ebrahimipour G (2022) Phylogrouping and characterization of Escherichia coli isolated from colonic biopsies and fecal samples of patients with flare of inflammatory bowel disease in Iran. Front. Med. 9:985300. doi: 10.3389/fmed.2022.985300
Received: 03 July 2022; Accepted: 05 August 2022;
Published: 29 August 2022.
Edited by:
Xiumin Wang, Feed Research Institute (CAAS), ChinaReviewed by:
Babatunde Odetoyin, Obafemi Awolowo University, NigeriaLiset Candelaria Pérez-Vázquez, Benemérita Universidad Autónoma de Puebla, Mexico
Copyright © 2022 Nadalian, Nadalian, Houri, Shahrokh, Abdehagh, Yadegar and Ebrahimipour. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abbas Yadegar, YS55YWRlZ2FyQHNibXUuYWMuaXI=; YmFiYWtfeTE5ODNAeWFob28uY29t; Gholamhossein Ebrahimipour, Zy1lYnJhaGltaUBzYnUuYWMuaXI=
†ORCID: Abbas Yadegar, orcid.org/0000-0002-2135-7581
 Banafsheh Nadalian
Banafsheh Nadalian Bahareh Nadalian2
Bahareh Nadalian2 Hamidreza Houri
Hamidreza Houri Shabnam Shahrokh
Shabnam Shahrokh Abbas Yadegar
Abbas Yadegar