
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Med., 22 July 2022
Sec. Infectious Diseases – Surveillance, Prevention and Treatment
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.906469
This article is part of the Research TopicEmerging SARS-COV-2 Variants: Genomic Variations, Transmission, Pathogenesis, Clinical Impact and InterventionsView all 65 articles
 Naniek Isnaini1
Naniek Isnaini1 Yan Mardian2
Yan Mardian2 Dewi Lokida1,2
Dewi Lokida1,2 Fajar Budiono1
Fajar Budiono1 Deni P. Butar-butar2
Deni P. Butar-butar2 Dona Arlinda2,3
Dona Arlinda2,3 Gustiani Salim2
Gustiani Salim2 Herman Kosasih2*
Herman Kosasih2* Wahyu Nawang Wulan2
Wahyu Nawang Wulan2 Jacqueline Perodin4
Jacqueline Perodin4 Aaron Neal5
Aaron Neal5 H. Clifford Lane5
H. Clifford Lane5 Muhammad Karyana2,3
Muhammad Karyana2,3Background: Reinfection with SARS-CoV-2 has been well documented, yet little is known about the degree of protection a previous infection provides against reinfection, especially against Variants of Concern (VOC).
Case presentation: Here we describe a case of an unvaccinated 49-year-old man who experienced two sequential SARS-CoV-2 infections with two different variants, as evidenced by genomic sequencing. The first episode was caused by the Pango lineage B.1.466.2 and resulted in severe COVID-19 with 5 days in an intensive care unit (ICU). The second episode occurred approximately 6 months later, during the Delta surge in Indonesia. Genomic analysis showed that the second infection was caused by the Delta variant (Pango lineage B.1.617.2) and resulted in mild disease that did not require hospitalization. No SARS-CoV-2 nucleic acid was detected between the two episodes, but both binding and neutralizing antibodies to SARS-CoV-2 were detected prior to the reinfection, with the second infection leading to an increase in the levels of antibody.
Conclusion: We confirmed that the patient experienced a reinfection instead of persistent viral shedding from the first infection based on epidemiological, clinical, serological, and genomic analyses. Our case supports the hypothesis that SARS-CoV-2 reinfection may occur once antibody titers decrease or following the emergence of a new variant. The milder presentation in the patient’s second infection deserves further investigation to provide a clear picture of the role of post-infection immunity in altering the course of subsequent disease.
According to WHO data as of June 12, 2022, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has reached over 533 million cases, with approximately 6.3 million deaths across the globe (1). Individuals who have recovered from the coronavirus disease (COVID-19) generate variable immune responses and remain at risk of reinfection (2). Several prior studies reported associations between age, comorbidities, and the severity of the acute infection with the magnitude and durability of the immune response (3, 4). This can be seen in several studies linking increasing disease severity with more robust humoral and cellular immune responses, suggesting that the immune response to natural infection is highly variable (5–7).
Reinfection with SARS-CoV-2 has been defined as a second positive RT-PCR test at least 90 days apart from the first infection, for which prolonged shedding of SARS-CoV-2 or viral RNA has been ruled out (8). Ideally, respiratory specimens from both episodes would be available to sequence to determine the different clades of SARS-CoV-2 (9). Patients who have recovered from COVID-19 have a lower risk of reinfection (8), though as the COVID-19 pandemic continues, reinfection has become increasingly common (10, 11). Since August 2020, few cases of COVID-19 reinfection have been reported with genomic data from paired episodes, thus making it challenging to establish true reinfection caused by different variants vs. persistent infection (2).
The scientific understanding of SARS-CoV-2 reinfection is still developing (12). Waning immunity from a previous infection and the emergence of new variants might negatively affect the protection provided by infection-acquired immunity, ultimately leading to the occurrence of reinfections (13). Related to waning immunity, Townsend et al. estimated that the time to reinfection by SARS-CoV-2 under endemic conditions would likely occur between 3 and 63 months after peak antibody response, with a median of 16 months (10). Since November 2021, five SARS-CoV-2 variants have been designated as Variants of Concern (VOC): Alpha (Pango Lineage B.1.1.7), Beta (Pango Lineage B.1.351), Gamma (Pango Lineage have since circulated P.1), Delta (Pango Lineage B.1.617.2), and Omicron (Pango Lineage B.1.1.529). These VOCs have since circulated worldwide (14). Among those variants, data indicate that Delta, which was first identified in India in December 2020 and rapidly spread globally, is associated with enhanced pathogenicity, can cause more severe disease, is 40–60% more transmissible than Alpha and is almost twice as transmissible as the original Wuhan strain of SARS-CoV-2 (15–17). The emergence of these variants has raised concerns about the breadth of neutralizing and cross-protective antibody responses induced by previous infection (18). To date, there have been few reports on sequential SARS-CoV-2 infections with supporting molecular evidence (2, 19), including sequence data. Here we report a confirmed reinfection case in Indonesia, with the first infection caused by Pango lineage B.1.466.2 and the second infection, 6 months later, caused by the Delta VOC.
A summary reinfection case timeline is presented in Figure 1. A 49-year-old male resident of Tangerang, Banten, Indonesia, worked as a driver and utility worker at Tangerang District Hospital. He lived alone in a private room and commuted every day to the hospital on public transportation. He had not received COVID-19 vaccination, and his only known chronic condition was hypertension, for which he received no routine treatment. On January 10, 2021, the patient developed clinical symptoms consistent with COVID-19, including fever, cough, runny nose, anosmia, headache, myalgia, and arthralgia. On January 13, 2021, the patient was admitted to the emergency department. Physical examination revealed the patient was fully conscious, with high blood pressure (175/110 mmHg), fever (38°C), respiratory rate of 22 breaths/min, heart rate of 98 beats/min, and oxygen saturation of 97%. Nasopharyngeal (NP) and oropharyngeal (OP) swabs were collected for RT-PCR testing, and the following day he was diagnosed with SARS-CoV-2 infection (Ct values: Nucleocapsid (N) gene = 23.5 and ORF1ab gene = 30.2) but remained in the emergency department due to overloaded hospital COVID-19 wards. An initial chest X-ray showed bilateral pulmonary infiltrates, and his laboratory tests showed elevated C-reactive protein (CRP) (66.3 mg/L), elevated D-dimer (241 ng/mL), leukocytosis (10.9 × 103/μL) with neutrophil-to-lymphocyte ratio (NLR) of 9.6, and mild thrombocytopenia (134 × 103/μL). The patient was admitted to the COVID-19 ward on January 15 and received favipiravir, methylprednisolone, antihypertensives, and vitamin C. A repeated RT-PCR test showed that he was still positive (Ct values: N-gene = 23.0 and ORF1ab gene = 28.4). A summary of serial blood tests performed during hospitalization is shown in Table 1.
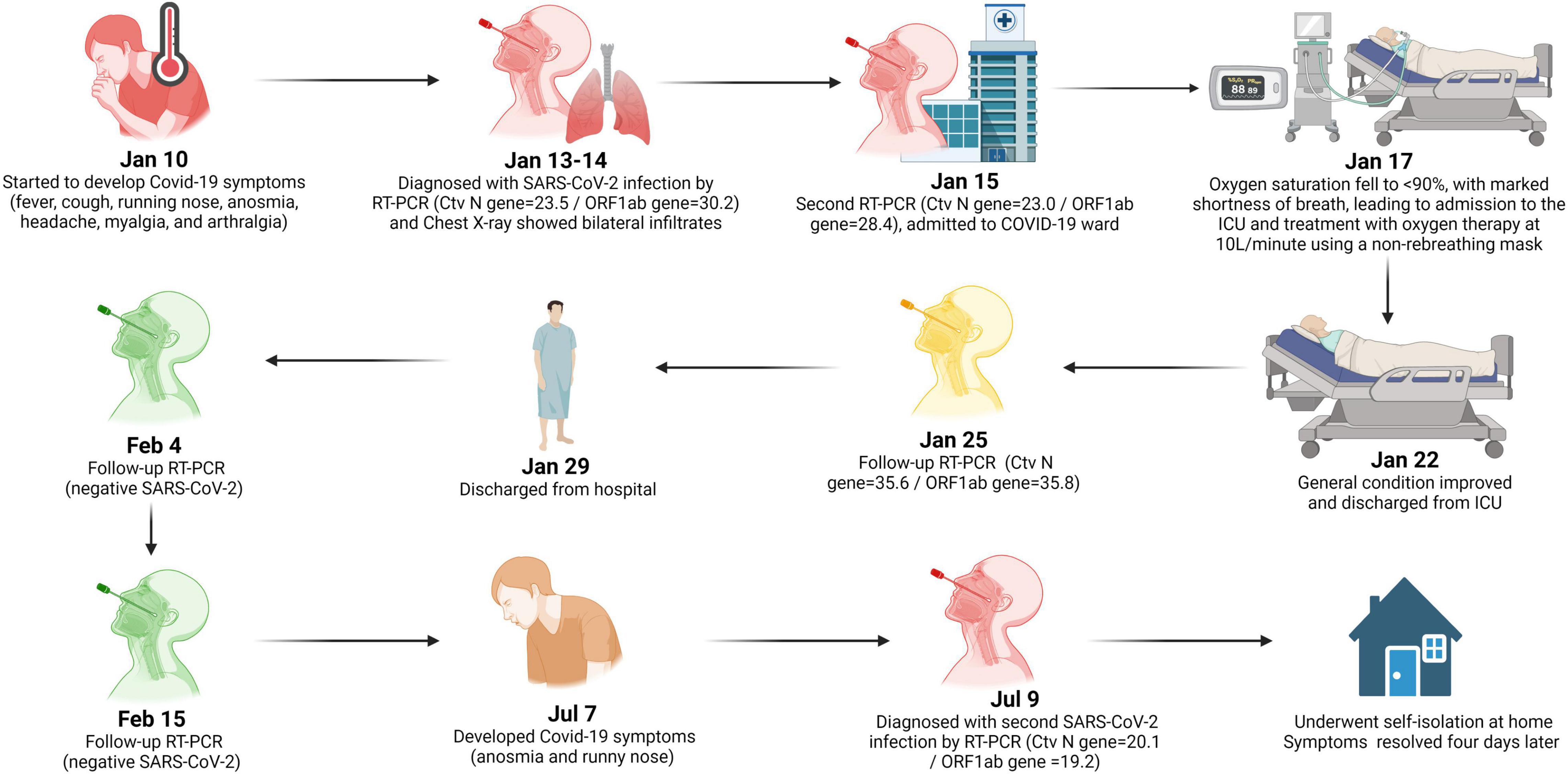
Figure 1. Timeline of reinfection case. Image was created with BioRender.com.
On January 17, the patient’s oxygen saturation fell to 88% with marked shortness of breath, leading to his admission to the ICU and treatment with oxygen therapy at 10L/minute using a non-rebreathing mask. While in the ICU, his antiviral treatment was changed to a 5-day course of remdesivir and meropenem, per the attending physician’s clinical judgment. No bacterial culture and antimicrobial susceptibility testing (AST) were performed. He also received heparin, which was then switched to fondaparinux sodium due to increased risk of bleeding, and a furosemide injection due to high blood pressure (up to 163/97 mmHg). After 3 days in the ICU, his condition improved, and his oxygen therapy was weaned to a nasal cannula. On January 22, the patient was moved to the regular COVID-19 ward, received oral levofloxacin for 5 days, and continued with oral cefixime, per clinical judgment due to an elevated leukocyte count (15.2 × 103/μL). A secondary bacterial infection was never confirmed. A follow-up RT-PCR test on January 25 detected low levels of viral RNA (Ct values: N-gene = 35.6 and ORF1ab gene = 35.8). The patient met discharge criteria [i.e., 10 days after onset of symptoms and at least three additional days without symptoms (20)] on January 29 and left the hospital. A summary of the patient’s treatments is shown in Table 2.
On January 25, 2021, serum was collected and antibodies to the SARS-CoV-2 spike protein receptor binding domain (S-RBD) were detected by chemiluminescence immunoassay (CLIA) CL-900i® SARS-CoV-2 S-RBD IgG (Mindray, Shenzhen, China), with an IgG index of 350.8 U/mL. Further testing using a surrogate virus neutralization test (sVNT) (GenScript, Piscataway, United States) resulted in a percent inhibition value of 83%. Following the patient’s discharge on January 29, follow-up RT-PCR tests on NP and OP specimens were performed on February 4 and 15, both of which were negative. Genomic sequencing of the SARS-CoV-2 Spike (S) gene from specimens collected on January 13, 2021, revealed S-D614G and S-N439K mutations. The full sequence of the S-gene (3,822-bp) was mapped and aligned to the reference genome obtained from the GISAID database (21). The sequence showed a 100% similarity with accession ID: EPI_ISL_1284133, belonging to Pango Lineage B.1.466.2 or GH clade on GISAID (22). Subsequent serological testing was performed at several timepoints after the initial COVID-19 episode (Figure 2A), with a peak S-RBD IgG index of 998.4 U/mL and a peak sVNT value of 88% on February 17. By May 24, the S-RBD IgG index declined to 230.5 U/mL and sVNT to a value of 76%.
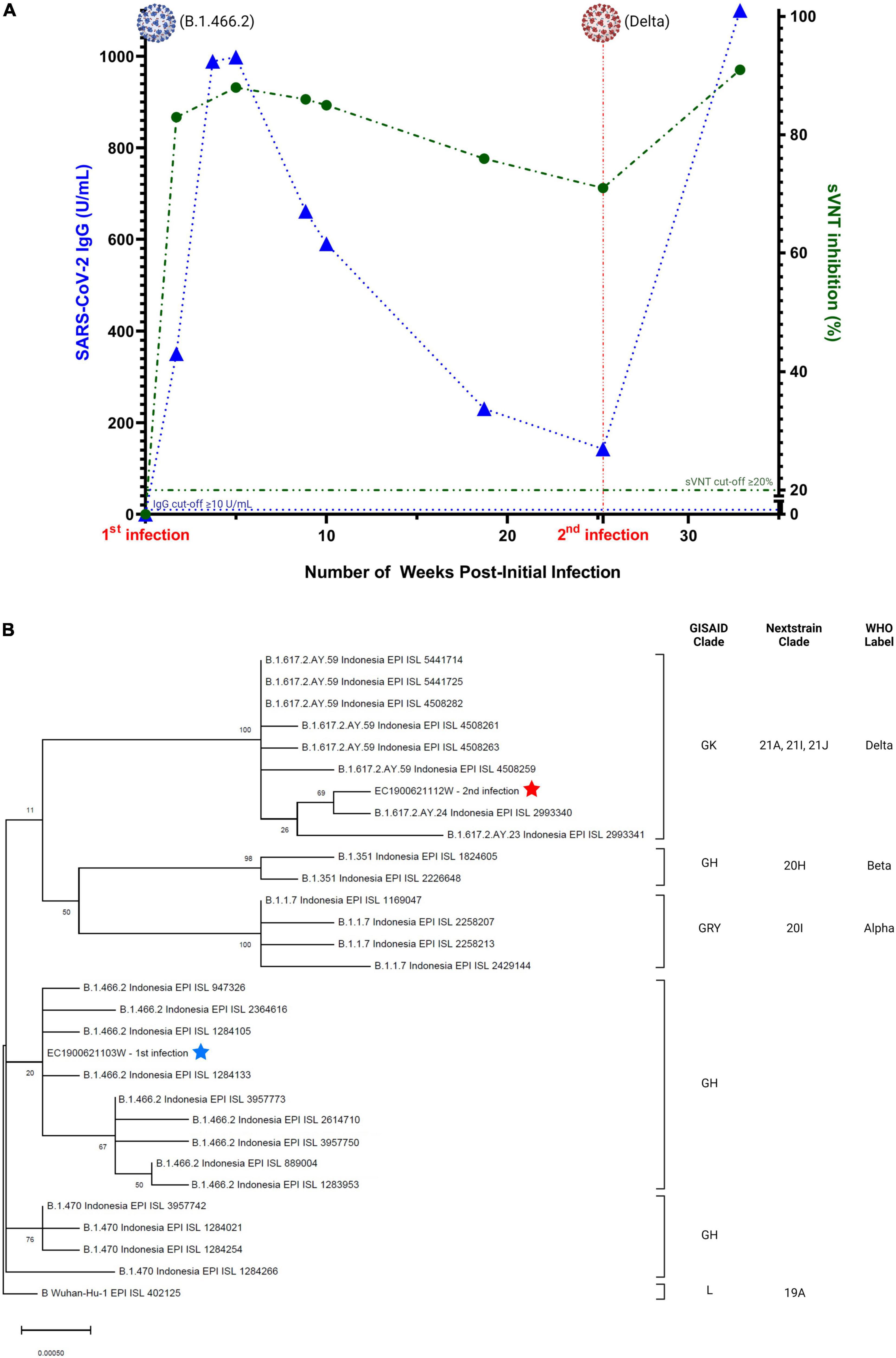
Figure 2. (A) Serial serology of SARS-CoV-2 IgG (blue) and neutralizing antibody (green). Image was created with GraphPad Prism 9.3.1 and BioRender.com, (B) phylogenetic analysis of SARS-CoV-2 genomes showing the relationship between the two distinct variants of two infection episodes. The tree was constructed by maximum likelihood method. Clade information as inferred by GISAID, Nextstrain, and Pangolin nomenclatures, are shown. The reference genome Wuhan-Hu-1 (GenBank accession number NC_045512.2) is used as the root of the tree.
The second episode of SARS-CoV-2 infection occurred around July 7, 2021, during the surge of the Delta variant in Indonesia (23). The patient developed mild symptoms consistent with COVID-19, including anosmia and runny nose. Two days later, he underwent RT-PCR testing at Tangerang District Hospital and was positive for SARS-CoV-2 (Ct values: N-gene = 20.1 and ORF1ab gene = 19.2). No routine blood test and chest imaging were performed. S-gene sequencing revealed mutations in the S gene (T19R, G142D, E156G, Del157/158, A222V, L452R, T478K, D614G, P681R, and D950N) that were consistent with the Delta variant, belonging to Pango Lineage B.1.617.2 or clade 21A on Nextstrain (22). Genomic analysis showed that the first episode viral genome belonged to a different clade/lineage from the second viral genome (Figure 2B). The patient underwent self-isolation at home and received treatment with vitamins, oseltamivir, paracetamol, and candesartan. His symptoms resolved 4 days later, with no follow-up on RT-PCR test. Serology testing from a blood specimen collected on July 9, 2021, revealed low antibody levels (S-RBD IgG = 142.7U/mL and sVNT = 71%). Subsequent testing on August 31, 2021, demonstrated boosting of humoral immunity (S-RBD IgG > 1,000 U/mL and sVNT = 91%). Details of the laboratory assays are summarized in Supplementary Table 1.
Our case report documents the first confirmed SARS-CoV-2 reinfection case in Indonesia, though it is highly likely that there have been many instances of reinfection nationwide. We observed a patient with severe COVID-19 disease caused by the B.1.466.2 variant in the first episode followed by milder disease caused by the Delta variant approximately 6 months later. Serial serology testing showed a persistent but gradual decrease of SARS-CoV-2 antibody titers after the first episode, with a boost of humoral immunity following the second infection. Although we did not perform testing on fecal specimens between episodes to rule out prolonged viral shedding in the GI tract (24), this presented case met all criteria for a confirmed diagnosis of reinfection: (1) Confirmation of a true first episode (Ct values < 35), (2) proof of a reinfection with SARS-CoV-2 RT-PCR test with Ct < 35, (3) confirmation of infection with two different/distinct phylogenetic strains/variants, and (4) negative RT-PCR tests between the first and second episodes (8). Furthermore, the epidemiological and serological analysis also confirmed that our case was a true reinfection, in which the second episode occurred during the Delta wave in Indonesia, boosting the antibody levels from the prior infection without a history of vaccine administration.
The non-sterilizing immunity generated against SARS-CoV-2 infection has made it challenging to differentiate prolonged shedding from a true reinfection (25). The US CDC maintains that the best evidence of reinfection is differing clades/lineages/variants of SARS-CoV-2, as defined in Nextstrain and GISAID, between an initial and subsequent infection (9). Based on WHO guidelines, complete or partial S-gene sequences can be used to confirm infection with a specific variant (26). In this case report, we relied on targeted S-gene sequencing to determine viral variants. The sequencing results of the first virus showed 100% similarity with the B.1.466.2 virus lineage, which was reported to be an indigenous dominant strain in Indonesia before the Delta variant surge, with an exponential growth from October 2020 to February 2021 (27, 28). While our S-gene sequence fully aligns with B.1.466.2, we cannot be certain that the whole genome contains the additional expected mutations (with mutation frequencies > 99%) of ORF1a-T1168I, ORF1a-P1640L, ORF1b-P314L, ORF3a-Q57H, and N-T205I in the B.1.466.2 variant (28). The virus from the second episode can be definitively identified as the Delta variant due to the presence of specific Delta mutations in the sequenced S-gene (L452R, T478K, and P681R) (29).
Recovery from many viral diseases is followed by a period of infection-induced immunologic protection against reinfection (30). Antibodies against SARS-CoV-2 include both binding and neutralizing antibodies (31), with the antigenic targets most frequently assessed being the S and N proteins (30). IgM, IgA, and IgG isotypes may be produced against any of these antigens and can be detected in the blood starting from 5 to 15 days following symptom onset (32, 33). Both binding and neutralizing antibody titers rise faster and reach a higher peak in persons with more severe COVID-19 (4, 30). Several studies have suggested that the waning of antibody levels occurs within several months after infection (34, 35), and the incidence of subsequent SARS-CoV-2 reinfection may be inversely associated with baseline antibody titers (36). As shown in our report, IgG and neutralizing antibodies steadily declined after the first infection episode, were at their lowest levels just prior to the second infection and increased following reinfection. There are insufficient data to determine antibody titer thresholds indicative of an individual’s protection from infection, and the level of protection may not be the same for all viral variants (30). One analysis suggested that the antibody level associated with protection against severe disease is lower than the level associated with protection against infection (37).
Despite growing evidence that immune responses remain robust and protective against reinfection with antigenically similar variants by 80–90% for at least 5–7 months after primary infection (36, 38–40), our report supports the idea that the duration of protection may be shorter with new viral variants. In laboratory studies, decreased neutralization to Delta and other variants has been demonstrated in non-Delta convalescent and post-vaccination sera (41–43). Combined in vitro experimentation and epidemiology studies have highlighted that mutations in the Spike-RBD of the Delta variant may result in increased infectivity and reduced neutralization sensitivity to sera from individuals infected with prior variants (29, 44, 45). These escape mutations are thought to be implicated in reinfection (46), but the observed reduction in effectiveness has been modest, with continued strong protection against hospitalization, severe disease, and death (30). Persistent immunity from SARS-CoV-2 specific T cells may have an important role in reducing viral replication and limiting the pathogenicity of infection (47), though we did not examine T cell responses as part of this case.
Reports on the clinical severity of reinfection compared to primary infection vary (2, 19, 48). In most cases, reinfections have appeared to be mild, with lower rates of pneumonia, heart failure, and acute kidney injury compared to primary infection episodes (48). Our clinical observations and lab results suggest that natural immunity from the patient’s initial infection reduced the severity of disease during the subsequent infection with the Delta variant. It is important to point out that this is not universally the case in the setting of reinfection (49–51). Future studies of cases of reinfection are needed to provide a clear picture of how robust the quality, quantity, and durability of protective immunity elicited by natural infection with SARS-CoV-2 is and its role in altering the course of subsequent infection. Given the rapid evolution of the virus and highly variable levels of host protection, it may be difficult to draw strong generalizations.
The patient did not receive any COVID-19 vaccines before or between his first and second episodes of infection. During that time, due to limited supplies, Indonesian national policy for receiving the COVID-19 vaccine only included individuals with no comorbidities, blood pressure less than 140/90 mmHg and no prior infection with COVID-19 (52). Thus, our patient was not eligible to receive the vaccine following his initial infection. However, the policy has since changed; COVID-19 survivors with blood pressure under 180/110 mmHg who are at least 1-month post-infection are now eligible (53, 54). Currently, the patient has received two doses of the Pfizer-BioNTech vaccine (on September 10, 2021, and October 1, 2021) and a booster dose with the Moderna vaccine (on December 31, 2021).
Several studies have shown that strong immune response, termed “hybrid immunity,” arises from a combination of natural and vaccine-generated immunity (55). Vaccination of convalescent individuals boosted the humoral immune response to well above the threshold of neutralization for multiple variants (29). Furthermore, memory B cell and CD4 + T cell components directed toward multiple spike and non-spike regions appear to be enhanced during hybrid immunity (29, 55). A recent report from Israel found that people who had been infected with SARS-CoV-2 and then received one dose of the Pfizer-BioNTech messenger RNA (mRNA) vaccine were less likely to be reinfected than previously infected individuals who were unvaccinated [hazard ratio, 0.18 (95% CI, 0.15–0.20)] (56). The substantial growing body of evidence strongly indicates that vaccination after infection significantly enhances protection and further reduces the risk of reinfection against an array of circulating viral variants, including Delta (30, 56–58).
A major strength of this report was our serial documentation of RT-PCR and serologic testing during and between both episodes of infection. This is rarely documented in routine practice. However, our study has several limitations. First, this is a case study of one individual, and therefore no broad conclusions or recommendations can be drawn. Second, we only relied on targeted S-gene sequencing to determine viral variants, although it is still acceptable within WHO guidelines (26). Whole genome sequences would be valuable in determining exact variant/Pango lineage. Third, T-cell responses were not measured in this study. Future studies in confirmed reinfection cases may provide a better picture of how the different components of the adaptive immune response can prevent or limit reinfection (39, 59). Fourth, we did not perform the gold standard virus neutralization test (VNT) or plaque reduction neutralization test (PRNT), which requires biosafety level 3 (BSL-3) facilities. However, the chemiluminescent immunoassays and surrogate neutralization assay (sVNT) used in this study have demonstrated good concordance with the gold standard PRNT for SARS-CoV-2 antibody detection in human sera (60, 61). They thus can serve as valid alternative to the VNT test in neutralizing activity measurements.
We reported a case of SARS-CoV-2 reinfection with distinct variants in Indonesia (8). The milder presentation in the second infection deserves further investigation to provide a clear picture of the role of post-infection immunity in altering the course of subsequent disease. As SARS-CoV-2 continues to circulate and mutate, vaccination following natural infection may prove to be an important strategy to increase protection against reinfection and disease.
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
NI, FB, and DL performed clinical assessments and were responsible for data collection. DB-b, GS, and WW performed laboratory assays. YM, HK, DL, JP, AN, and HL reviewed data and wrote the manuscript. All authors contributed to the manuscript development, provided critical review, and have approved the final version.
This manuscript has been funded in whole or in part with the Indonesia Ministry of Health and Federal funds from the United States National Institute of Health’s National Institute of Allergy and Infectious Diseases (contract no. 75N91019D00024), through the Indonesia Research Partnership on Infectious Disease (INA-RESPOND). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
JP was employed by Leidos Biomedical Research, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the Indonesia Research Partnership on Infectious Disease (INA-RESPOND) for their support. We are also grateful to the laboratory team (Yuanita Jayadi, and Rizki Amalia Sari) at Tangerang District Hospital, Tangerang, Indonesia. We also thank Aly Diana and Adhella Menur Naysilla for technical assistance with the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.906469/full#supplementary-material
1. World Health Organization. COVID-19 Weekly Epidemiological Update. Geneva: World Health Organization (2021). p. 1–33.
2. Wang J, Kaperak C, Sato T, Sakuraba A. COVID-19 reinfection: a rapid systematic review of case reports and case series. J Investig Med. (2021) 69:1253–5. doi: 10.1136/jim-2021-001853
3. Chvatal-Medina M, Mendez-Cortina Y, Patiño PJ, Velilla PA, Rugeles MT. Antibody responses in COVID-19: a review. Front Immunol. (2021) 12:633184. doi: 10.3389/fimmu.2021.633184
4. Mardian Y, Kosasih H, Karyana M, Neal A, Lau C-Y. Review of current COVID-19 diagnostics and opportunities for further development. Front Med. (2021) 8:562–85. doi: 10.3389/fmed.2021.615099
5. Long Q-X, Tang X-J, Shi Q-L, Li Q, Deng H-J, Yuan J, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. (2020) 26:1200–4.
6. Shrotri M, van Schalkwyk MCI, Post N, Eddy D, Huntley C, Leeman D, et al. T cell response to SARS-CoV-2 infection in humans: a systematic review. PLoS One. (2021) 16:e0245532. doi: 10.1371/journal.pone.0245532
7. Garrido C, Hurst JH, Lorang CG, Aquino JN, Rodriguez J, Pfeiffer TS, et al. Asymptomatic or mild symptomatic SARS-CoV-2 infection elicits durable neutralizing antibody responses in children and adolescents. JCI Insight. (2021) 6:e150909. doi: 10.1172/jci.insight.150909
8. Yahav D, Yelin D, Eckerle I, Eberhardt CS, Wang J, Cao B, et al. Definitions for coronavirus disease 2019 reinfection, relapse and PCR re-positivity. Clin Microbiol Infect. (2021) 27:315–8. doi: 10.1016/j.cmi.2020.11.028
9. CDC. Investigative Criteria for Suspected Cases of SARS-CoV-2 Reinfection (ICR). (2020). Available online at: https://www.cdc.gov/coronavirus/2019-ncov/php/invest-criteria.html (accessed on March 14, 2022).
10. Townsend JP, Hassler HB, Wang Z, Miura S, Singh J, Kumar S, et al. The durability of immunity against reinfection by SARS-CoV-2: a comparative evolutionary study. Lancet Microbe. (2021) 2:e666–75. doi: 10.1016/S2666-5247(21)00219-6
11. Graham F. Daily briefing: why COVID-19 reinfections are surging. Nature. (2022). [Epub ahead of print]. doi: 10.1038/d41586-022-00500-0
12. Lauring AS, Hodcroft EB. Genetic variants of SARS-CoV-2—what do they mean? JAMA. (2021) 325:529–31. doi: 10.1001/jama.2020.27124
13. Cavanaugh AM, Spicer KB, Thoroughman D, Glick C, Winter K. Reduced risk of reinfection with SARS-CoV-2 after COVID-19 vaccination—Kentucky, May–June 2021. Morb Mortal Wkly Rep. (2021) 70:1081–3. doi: 10.15585/mmwr.mm7032e1
15. Saito A, Irie T, Suzuki R, Maemura T, Nasser H, Uriu K, et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. (2022) 602:300–6. doi: 10.1038/s41586-021-04266-9
16. Yazdanpanah F, Yazdanpanah S, Rezaei N. COVID-19 Delta variation; more contagious or more pernicious? Acta Biomed. (2022) 92:e2021454.
17. Shiehzadegan S, Alaghemand N, Fox M, Venketaraman V. Analysis of the delta variant B.1.617.2 COVID-19. Clin Pract. (2021) 11:778–84.
18. Edara VV, Hudson WH, Xie X, Ahmed R, Suthar MS. Neutralizing antibodies against SARS-CoV-2 variants after infection and vaccination. JAMA. (2021) 325:1896–8. doi: 10.1001/jama.2021.4388
19. Shastri J, Parikh S, Aggarwal V, Agrawal S, Chatterjee N, Shah R, et al. Severe SARS-CoV-2 breakthrough reinfection with delta variant after recovery from breakthrough infection by alpha variant in a fully vaccinated health worker. Front Med. (2021) 8:1379. doi: 10.3389/fmed.2021.737007
20. World Health Organization. Criteria for Releasing COVID-19 Patients From Isolation: Scientific Brief, 17 June 2020. Geneva: World Health Organization (2020).
21. Khare S, Gurry C, Freitas L, Schultz MB, Bach G, Diallo A, et al. GISAID’s role in pandemic response. China CDC Wkly. (2021) 3:1049–51. doi: 10.46234/ccdcw2021.255
22. Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. (2018) 34:4121–3. doi: 10.1093/bioinformatics/bty407
23. Dyer O. Covid-19: Indonesia becomes Asia’s new pandemic epicentre as delta variant spreads. BMJ. (2021) 374:n1815. doi: 10.1136/bmj.n1815
24. Morone G, Palomba A, Iosa M, Caporaso T, De Angelis D, Venturiero V, et al. Incidence and persistence of viral shedding in COVID-19 post-acute patients with negativized pharyngeal swab: a systematic review. Front Med. (2020) 7:562–70. doi: 10.3389/fmed.2020.00562
25. Nicholson EG, Avadhanula V, Fragoso S, Stroh R, Ye X, Bond N, et al. SARS-CoV-2 re-infection versus prolonged shedding: a case series. Influenza Other Respi Viruses. (2021) 15:691–6. doi: 10.1111/irv.12879
26. European Centre for Disease Prevention and Control. Methods for the Detection and Identification of SARS-CoV-2 Variants. Geneva: World Health Organization (2021).
27. Fibriani A, Stephanie R, Alfiantie AA, Siregar ALF, Pradani GAP, Yamahoki N, et al. Analysis of SARS-CoV-2 genomes from West Java, Indonesia. Viruses. (2021) 13:2097. doi: 10.3390/v13102097
28. Zhu M, Zeng Q, Saputro BIL, Chew SP, Chew I, Frendy H, et al. Tracking the molecular evolution and transmission patterns of SARS-CoV-2 lineage B.1.466.2 in Indonesia based on genomic surveillance data. Virol J. (2022) 19:103. doi: 10.1186/s12985-022-01830-1
29. Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. (2021) 596:276–80. doi: 10.1038/s41586-021-03777-9
30. National Center for Immunization. Science Brief: SARS-CoV-2 Infection-induced and Vaccine-induced Immunity. CDC COVID-19 Science Briefs. Atlanta, GA: Centers for Disease Control and Prevention (2021). p. 1–16.
31. Wu J, Liang B, Chen C, Wang H, Fang Y, Shen S, et al. SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. Nat Commun. (2021) 12:1813. doi: 10.1038/s41467-021-22034-1
32. Röltgen K, Powell AE, Wirz OF, Stevens BA, Hogan CA, Najeeb J, et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol. (2020) 5:eabe0240. doi: 10.1126/sciimmunol.abe0240
33. Post N, Eddy D, Huntley C, van Schalkwyk MCI, Shrotri M, Leeman D, et al. Antibody response to SARS-CoV-2 infection in humans: a systematic review. PLoS One. (2020) 15:e0244126. doi: 10.1371/journal.pone.0244126
34. Arkhipova-Jenkins I, Helfand M, Armstrong C, Gean E, Anderson J, Paynter RA, et al. Antibody response after SARS-CoV-2 infection and implications for immunity: a rapid living review. Ann Intern Med. (2021) 174:811–21. doi: 10.7326/M20-7547
35. Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. (2020) 5:1598–607. doi: 10.1038/s41564-020-00813-8
36. Lumley SF, O’Donnell D, Stoesser NE, Matthews PC, Howarth A, Hatch SB, et al. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med. (2020) 384:533–40. doi: 10.1056/NEJMoa2034545
37. Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. (2021) 27:1205–11. doi: 10.1038/s41591-021-01377-8
38. Vitale J, Mumoli N, Clerici P, De Paschale M, Evangelista I, Cei M, et al. Assessment of SARS-CoV-2 reinfection 1 year after primary infection in a population in Lombardy, Italy. JAMA Intern Med. (2021) 181:1407–8. doi: 10.1001/jamainternmed.2021.2959
39. Abu-Raddad LJ, Chemaitelly H, Coyle P, Malek JA, Ahmed AA, Mohamoud YA, et al. SARS-CoV-2 antibody-positivity protects against reinfection for at least seven months with 95% efficacy. EClinicalMedicine. (2021) 35:1–12. doi: 10.1016/j.eclinm.2021.100861
40. Hansen CH, Michlmayr D, Gubbels SM, Mølbak K, Ethelberg S. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet. (2021) 397:1204–12. doi: 10.1016/S0140-6736(21)00575-4
41. Lucas C, Vogels CBF, Yildirim I, Rothman JE, Lu P, Monteiro V, et al. Impact of circulating SARS-CoV-2 variants on mRNA vaccine-induced immunity. Nature. (2021) 600:523–9. doi: 10.1038/s41586-021-04085-y
42. Edara V-V, Pinsky BA, Suthar MS, Lai L, Davis-Gardner ME, Floyd K, et al. Infection and vaccine-induced neutralizing-antibody responses to the SARS-CoV-2 B.1.617 variants. N Engl J Med. (2021) 385:664–6. doi: 10.1056/NEJMc2107799
43. Sapkal GN, Yadav PD, Sahay RR, Deshpande G, Gupta N, Nyayanit DA, et al. Neutralization of Delta variant with sera of CovishieldTM vaccinees and COVID-19-recovered vaccinated individuals. J Travel Med. (2021) 28:taab119. doi: 10.1093/jtm/taab119
44. Liu C, Ginn HM, Dejnirattisai W, Supasa P, Wang B, Tuekprakhon A, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. (2021) 184:4220–36.e13.
45. Mlcochova P, Kemp S, Dhar MS, Papa G, Meng B, Ferreira IATM, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. (2021) 599:114–9. doi: 10.1038/s41586-021-03944-y
46. Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. (2021) 29:44–57.e9. doi: 10.1016/j.chom.2020.11.007
47. Bertoletti A, Le Bert N, Qui M, Tan AT. SARS-CoV-2-specific T cells in infection and vaccination. Cell Mol Immunol. (2021) 18:2307–12. doi: 10.1038/s41423-021-00743-3
48. Qureshi AI, Baskett WI, Huang W, Lobanova I, Hasan Naqvi S, Shyu C-R. Reinfection with severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) in patients undergoing serial laboratory testing. Clin Infect Dis. (2021) 74:294–300. doi: 10.1093/cid/ciab345
49. Tillett RL, Sevinsky JR, Hartley PD, Kerwin H, Crawford N, Gorzalski A, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. (2021) 21:52–8. doi: 10.1016/S1473-3099(20)30764-7
50. Larson D, Brodniak SL, Voegtly LJ, Cer RZ, Glang LA, Malagon FJ, et al. A case of early reinfection with severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2). Clin Infect Dis. (2020) 73:e2827–8. doi: 10.1093/cid/ciaa1436
51. Shastri J, Parikh S, Agrawal S, Chatterjee N, Pathak M, Chaudhary S, et al. Clinical, serological, whole genome sequence analyses to confirm SARS-CoV-2 reinfection in patients from Mumbai, India. Front Med. (2021) 8:215. doi: 10.3389/fmed.2021.631769
52. Cabinet Secretariat of the Republic of Indonesia. Gov’t Unveils Criteria for People Unable to Receive Sinovac’s Vaccine. (2020). Available online at: https://setkab.go.id/en/govt-unveils-criteria-for-people-unable-to-receive-sinovacs-vaccine/ (accessed on June 20, 2022).
53. Yanto TA, Octavius GS, Heriyanto RS, Ienawi C, Nisa H, Pasai HE. Psychological factors affecting COVID-19 vaccine acceptance in Indonesia. Egypt J Neurol psychiatry Neurosurg. (2021) 57:177. doi: 10.1186/s41983-021-00436-8
54. Voice of Indonesia. COVID-19 Survivors Can Now Be Vaccinated After A Month Of Recovery. (2021). Available online at: https://voi.id/en/news/90156/covid-19-survivors-can-now-be-vaccinated-after-a-month-of-recovery (accessed on June 21, 2022).
56. Gazit S, Shlezinger R, Perez G, Lotan R, Peretz A, Ben-Tov A, et al. The incidence of SARS-CoV-2 reinfection in persons with naturally acquired immunity with and without subsequent receipt of a single dose of BNT162b2 vaccine. Ann Intern Med. (2022) 175:674–81. doi: 10.7326/M21-4130
57. Hall V, Foulkes S, Insalata F, Kirwan P, Saei A, Atti A, et al. Protection against SARS-CoV-2 after Covid-19 Vaccination and Previous Infection. N Engl J Med. (2022) 386:1207–20. doi: 10.1056/NEJMoa2118691
58. Hammerman A, Sergienko R, Friger M, Beckenstein T, Peretz A, Netzer D, et al. Effectiveness of the BNT162b2 vaccine after recovery from Covid-19. N Engl J Med. (2022) 386:1221–9. doi: 10.1056/NEJMoa2119497
59. Cox RJ, Brokstad KA. Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nat Rev Immunol. (2020). 20:581–582. doi: 10.1038/s41577-020-00436-4
60. Prap M, Ronald K, Tot Y, Hds C, Kmy M, Bc J, et al. Evaluation of a SARS-CoV-2 surrogate virus neutralization test for detection of antibody in human, canine, cat, and hamster sera. J Clin Microbiol. (2022) 59:e2504–20. doi: 10.1128/JCM.02504-20
61. Cristiano A, Pieri M, Sarubbi S, Pelagalli M, Calugi G, Tomassetti F, et al. Evaluation of serological anti-SARS-CoV-2 chemiluminescent immunoassays correlated to live virus neutralization test, for the detection of anti-RBD antibodies as a relevant alternative in COVID-19 large-scale neutralizing activity monitoring. Clin Immunol. (2022) 234:108918. doi: 10.1016/j.clim.2021.108918
Keywords: case report, reinfection, COVID-19, Indonesia, Delta variant
Citation: Isnaini N, Mardian Y, Lokida D, Budiono F, Butar-butar DP, Arlinda D, Salim G, Kosasih H, Wulan WN, Perodin J, Neal A, Lane HC and Karyana M (2022) Mild reinfection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Delta variant: First case report from Indonesia. Front. Med. 9:906469. doi: 10.3389/fmed.2022.906469
Received: 28 March 2022; Accepted: 27 June 2022;
Published: 22 July 2022.
Edited by:
Éric Bergeron, Centers for Disease Control and Prevention (CDC), United StatesReviewed by:
Sandra Barroso-Arévalo, Complutense University of Madrid, SpainCopyright © 2022 Isnaini, Mardian, Lokida, Budiono, Butar-butar, Arlinda, Salim, Kosasih, Wulan, Perodin, Neal, Lane and Karyana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Herman Kosasih, aGtvc2FzaWhAaW5hLXJlc3BvbmQubmV0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.