
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Med. , 20 June 2022
Sec. Gastroenterology
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.890127
This article is part of the Research Topic Burden of Diarrhoeal Diseases View all 11 articles
Irritable bowel syndrome (IBS) is a chronic disorder, which its causative agent is not completely clear; however, the interaction between microorganisms and gastrointestinal (GI) epithelial cells plays a critical role in the development of IBS and presenting symptoms. During recent decades, many studies have highlighted the high prevalence of Blastocystis sp. in patients with IBS and suggested a probable role for this protist in this disease. Recent studies have documented changes in the gut microbiota composition in patients with IBS regarding the presence of Blastocystis sp., but it is not clear that either disturbance of the gut during GI disorders is a favorable condition for Blastocystis sp. colonization or the presence of this protist may lead to alteration in the gut microbiota in IBS patients. In this review, we comprehensively gather and discuss scientific findings covering the role of Blastocystis sp. in IBS via gut microbiota shifting.
Irritable bowel syndrome (IBS) is a chronic disorder, which is known by abdominal pain and abnormal defection. IBS seems to be a gut-brain axis-related disease; therefore, it is also called a functional gastrointestinal (GI) disorder (1, 2). IBS is a commonly reported disorder in clinical practices, affecting 10–20% of the world's population (3).
IBS could be asymptomatic; however, this disease is characterized by symptoms such as abdominal pain, variable bowel habits, and bloating (1). IBS is a multifactorial disorder. Even though its causative agent is not clear; gut microbiota disturbance appears to play an important role in this disorder (4). Early studies have documented the role of microbial gastroenteritis [post-infectious IBS (PI-IBS)] (5, 6) and overuse of broad-spectrum antibiotics in the development of IBS (7). Studies on the gut microbiota in patients with IBS suggest that microbial dysbiosis may increase the severity and duration of IBS symptoms. Accordingly, it was documented that changes in the gut microbiota composition play a critical role in establishing, developing, and flaring the symptoms of IBS (8). Nevertheless, the manipulation of the intestinal microbiota can be considered a new treatment method for patients with IBS. For example, fecal microbiota transplantation (FMT) reduced inflammation and symptoms in people with IBS and could be regarded as a treatment strategy (9).
In addition to the critical role of bacteria in the development of IBS, the high prevalence of some protozoa, such as Blastocystis sp., has highlighted the putative role of this protist in the development of IBS. Although a bilateral correlation between the presence of Blastocystis sp. and the gut microbiota composition has been reported in many studies, it is not clear whether the gut conditions and gut microbiota perturbation lead to higher colonization of Blastocystis sp. in the gut or colonization of Blastocystis sp. may lead to dysbiosis (10, 11).
It has been suggested that microbial infections may lead to a mild inflammation through the intestine and the development of IBS. Although the results contradict (12), the emerging role of intestinal parasites in the development of IBS is now being investigated (13). Intestinal parasites can increase the gut permeability and contact of lumen antigens with the lower layers of the intestine, provoking immune responses, and developing chronic inflammation (14, 15).
Accordingly, a higher prevalence of some protozoa (Giardia, Blastocystis, and Cryptosporidium) was recorded among the patients with IBS compared to healthy controls (16). The correlation between previous infection with Giardia and an increased risk of IBS was strongly suggested by retrospective studies. In this regard, in a retrospective cohort study performed by Dormond et al. (17), which was carried out on military personnel, the risk of developing chronic GI disorders, such as IBS, was assessed in those with documented giardiasis and the findings represented an increased risk of IBS in Giardia-infected personnel (17). Nakao et al. (18), in a large retrospective study, which employed the 2006–2010 MarketScan commercial insurance database, showed that despite considering confounding factors, such as anxiety, depression, and healthcare utilization, giardiasis increased the risk of subsequent IBS (18). Although the mechanism beyond the role of giardiasis in increasing the risk of IBS is not clear, destruction of the intestinal barrier unity during colonization of G. lamblia appears to be an important factor. For example, it was demonstrated that infection by Giardia may lead to gut barrier dysfunctions via downregulation of tight junctions, such as claudin-1, and induction of apoptosis in epithelial cells throughout the intestine (19).
There is evidence speculating the role of early-age infection by Cryptosporidium in the development of IBS in adolescence, via increased jejunal sensitivity to bloating (20). In addition, the intestinal stage of a nematode worm, Trichinella spiralis, seems to provoke a mild inflammation over the gut epithelial cell, which can lead to visceral sensitivity and changes in intestinal motility (21–23). It was shown that transient mucosal inflammation in Swiss mice during the acute phase of infection by T. spiralis can lead to an alteration in neuromuscular functions, even after the resolution of Trichinella-caused inflammation (21). These findings were then supported by Venkova et al. (23), who demonstrated jejunal inflammation due to T. spiralis, which induced long-term changes in muscle contractility and enteric neurotransmission, even after recovery from mucosal inflammation (23). Eventually, it was demonstrated that during the GI phase of trichinosis, T. spiralis can induce a long-term remodeling of epithelial functions (22). Therefore, the persistent inflammation and neuromuscular dysfunctions during the intestinal stage of trichinosis, seem to increase the risk of IBS development, particularly in those who are susceptible.
Dientamoeba fragilis is an intestinal protozoan, which, together with Blastocystis sp., are the most prevalent protozoa reported in patients with IBS (24–26). The probable correlation between D. fragilis and IBS was first reported by Borody et al. (27) who showed that eradication of the protozoan led to amelioration of IBS-like symptoms. However, further investigations on the correlation between D. fragilis and IBS have reported controversial results. For example, Yakoob et al. (28) demonstrated a higher prevalence of D. fragilis in patients with IBS compared to healthy controls. Engsbro et al. (25) reported that 35–41% of patients with IBS carried out D. fragilis in the Danish population, and Ibrahim et al. (29) supported previous studies and documented a higher prevalence of D. fragilis in patients with IBS compared to control subjects. A case of PI-IBS due to D. fragilis in a patient who traveled to Mexico points out the probable role of this protozoan in the development of IBS, as well (30). However, in contrast to these studies, Engsbro et al. (31) analyzed the response to anti-D. fragilis treatment in 25 patients with IBS who carried the protozoan and showed the lack of correlation between microbiological response to treatment and clinical manifestations of patients with IBS.
Blastocystis sp. is frequently reported from patients with IBS, and numerous studies have suggested a correlation between carrying this protist and IBS; however, it is not clear whether either Blastocystis sp. leads to IBS/IBS-like symptoms or perturbed conditions of the GI tract provide a favorable niche for colonization of Blastocystis sp. One of the first studies that examined a probable parasite as a causative agent of IBS was performed by Yakoob et al. (32), who reported a statistically significant higher prevalence of Blastocystis sp. in patients with IBS [46% (44 of 95)] compared to the controls [7% (4 of 55)]. Surangsrirat et al. (33), in a case–control study in Thailand, documented a higher frequency of Blastocystis sp. in patients with IBS (16.7%) compared to the control group (10%), although this difference was not statistically significant. In addition, Das et al. (34) demonstrated that the prevalence of Blastocystis sp. was three times higher than that reported in healthy subjects.
The close frequency rate of Blastocystis sp. in patients with IBS and control groups in many studies and a greater prevalence rate of Blastocystis sp. in control groups compared to patients with IBS have obscured an established linkage between the presence of this protist and the development of IBS (35–37).
The presence of correlation between genetic lineages of Blastocystis sp. and the development of IBS has also been evaluated. Although a correlation between the presence of ST1 (38) and ST3 (39) with IBS was suggested, most of the studies failed to associate the presence of a certain subtype with IBS development. In this regard, Pena et al. (40) analyzed the subtype distribution of Blastocystis sp. in patients with IBS compared to healthy controls that the results showed the presence of ST1 and ST2 in both groups, while ST3 and ST4 were only characterized by healthy controls and patients with IBS, respectively. The lack of linkage between certain subtypes and IBS was observed in Indian subjects, where ST3 was the dominant subtype in both IBS and control groups followed by ST1 (34). Subtypes 1 and 3 were also reported to be the major genetic lineages in IBS and control subjects in other studies (41–43).
The expression of certain enzymes in Blastocystis sp. isolated from patients with IBS was also evaluated. In this regard, Nagel et al. (44) indicated the higher presence of a Blastocystis sp. protein (probably a cysteine protease) in subjects with IBS compared to healthy controls. They also claimed that some single nucleotide polymorphisms (SNPs) in IL-8 and IL-10 probably affect the relative risk of IBS development in individuals who carry Blastocystis sp. (45). Although the effects of genetic polymorphisms through the IBS-related signature regions of IL-6, IL-8, and IL-10 might be different in various ethnic groups, these SNPs could increase the risk of IBS development (46, 47). In this line, a recently published study proposed significantly higher serum levels of IL-6, IL-8, IL-10, IFN-γ, and TNF-α in patients with IBS who colonized with Blastocystis sp., proposing the critical effects of Blastocystis sp., in IBS development (48) via modulation of IBS-related cytokines (49).
Taken together, although colonization of a certain subtype of Blastocystis sp. does not seem to be correlated with IBS, it was suggested that there are significant differences in the protease activity of different subtypes of Blastocystis sp. (50). Importantly, proteases released by Blastocystis sp. can disrupt the epithelial barrier and actin filaments, increase gut permeability, and subsequently develop IBS (51–53). Moreover, a most recently published study suggested that Blastocystis sp. ST3 can modulate the expression levels of microRNAs involved in the gut barrier integrity, and claudin-7 (54). Notably, claudin-7 is categorized among pre-sealing tight junction proteins and plays a critical role in reducing the permeability of the gut (55).
The microbiota is comprised of the microbial community of the human body, which is made up of a variety of microorganisms, including bacteria, viruses, fungi, and protozoa. The gut microbiota consists of between 10 and 100 trillion cells, which are ~10 times the total cells of a human (56). The gut microbiome has the highest number of microbial communities. The number of microbial genes in the gut is estimated to be 150 times higher than the genes of human origin (57). Current documentations demonstrated four bacterial phyla, including Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria, are the core of the gut microbiota of which Firmicutes and Bacteroidetes are predominant (58–60).
The gut microbiota composition is linked to a couple of intestinal and extra-intestinal disorders (61–64). Changes in the GI microbiota (is known as dysbiosis) can affect the immune responses, metabolism, and intestinal permeability, resulting in a pre-inflammatory state. Such changes can disrupt the functions of the host's immunity and metabolic systems, which may lead to diseases, such as diabetes, obesity, GI, neurological, and autoimmune disorders (61, 63, 65, 66). A number of studies have demonstrated a link between changes in the gut microbiota and incidences of gut-related diseases, such as obesity, non-alcoholic steatohepatitis (NASH), IBS, inflammatory bowel disease (IBD), celiac disease, and GI neoplasms (61, 66–68).
The correlation between gut microbiota dysbiosis and IBS conditions is well-established (69, 70). A correlation between the severity of IBS and a gut microbiota signature has been demonstrated (71). PI-IBS, small intestine bacterial overgrowth (SIBO), stress, antibiotics, diet, and early childhood experiences shape the gut microbiota and affect the incidence rate of IBS (72).
A gut microbiota analysis suggested a doubled ratio of Firmicutes to Bacteroidetes, an increased number of Dorea, Ruminococcus, and Clostridium spp., and a decreased number of Faecalibacterium spp. in patients with IBS compared to healthy controls (69). Tap et al. (71) reported an association between the gut microbiota composition and the severity of IBS. They showed that enterotypes Prevotella and Bacteroides represented the lowest and highest proportion in patients with severe IBS, respectively. In addition, it was claimed that the highest and lowest proportion of Bacteroides were observed in IBS-D and IBS-C, respectively (71). These results were then confirmed by Zhuang et al. (73) who documented the Bacteroidetes in stool samples of patients with IBS-D, and suggested an association between the gut microbiota composition with the pathogenicity of IBS (73). IBS has four types, which are characterized based on the stool formation and the number of defecation, including IBS-C with constipation; IBS-D with diarrhea; IBS-M, which has intermittent bowel pattern with a mix of diarrhea and constipation; and IBS-U, which is not easily classified into any of the mentioned groups (74). Metagenomics studies suggest that the gut microbiota may present different patterns according to the types of IBS. The results of a quantitative real-time PCR, which was employed to amplify the 16S rRNA gene of the gut bacteria, showed that the number of bacteria, such as R. productus, C. coccoides, Villonella, Tetiotamicron, Pseudomonas aeruginosa, and Gram-negative bacteria in patients with IBS was higher, but the number of Lactobacillus was lower than healthy individuals. In addition, the number of Violinella species in patients with IBS-C and P. aeruginosa in people with IBS-C and IBS-D were higher than in healthy individuals (75).
The 16S rRNA gene sequencing revealed that although the gut microbiota diversity was similar between patients with IBS and healthy controls, the richness of bacteria in patients with IBS-D was lower than in other groups, while a significant increase in Proteobacteria and decrease in Firmicutes, Fusobacteria, and Actinobacteria were observed in patients with IBS-D (76). Apart from the changes in the diversity and richness of the gut microbiota, the overgrowth of the bacterial community throughout the small intestine is thought to be associated with IBS (4). A systematic review by Pittayanon et al. (77) on 24 studies revealed that Enterobacteriaceae (phylum Proteobacteria), family Lactobacillaceae, and genus Bacteroides were increased, and uncultured Clostridiales I, genus Faecalibacterium (e.g., F. prausnitzii), and genus Bifidobacterium were decreased in patients with IBS compared to healthy controls. Nevertheless, the presence of a microbiome signature, how IBS alters the gut microbiota compositions, and the role of diet changes on the gut microbiota alterations in patients with IBS are the main unclear issues that need to be investigated (Table 1).
There is growing evidence linking inflammation with IBS. Inflammation may lead to changes in smooth muscles and GI nerves, which are resulted in GI dysfunctions (86, 87). The presence and the number of mast cells, release of histamine and tryptase, distance of intestinal nerve to mast cells in patients with IBS characterized by Room II, and abdominal pain were compared to healthy controls that the infiltration of mast cells and release of their contents in proximity to mucosal innervation were probably correlated with IBS and abdominal pain (88).
The correlation between low level of inflammation and IBS manifestations was suggested in a study by Ohman et al. (89) who showed an increased frequency of blood T cells expressing CD69 and integrin b7/HLA-DR. They concluded that T-cell activation supports low-grade inflammation and symptom generation in patients with IBS (89). This finding was later supported by Nasser et al. (90), who showed an immune activation of CD4+ T cell derived from patients with IBS-D, while it was not correlated with physiological stress. However, they suggested that immune activation is could be a trigger of or a parallel phenomenon with IBS (90). The higher number and the activation of mucosal B lymphocytes and plasma cells, together with an increased number of mast cells in the mucosal jejunal biopsy of patients with IBS-D, probably contribute to the presentation of the disorder (91). Therefore, it seems that a mild inflammation due to enhanced humoral and innate immunity throughout the intestine could be correlated with IBS (91, 92). Importantly, amelioration of the clinical manifestations of IBS-D via activation of mast cells and modulating of the intestinal innate immunity followed by oral prescription of disodium cromoglycate (DSCG) confirmed the hypothesis of correlation between mild inflammation and IBS development (93).
The role of gut microbiota in the arrangement of the immune responses in IBS is still investigating; however, it seems that microbe-association pattern recognition plays a determinative role in orchestrating the immune responses (94). In this regard, the role of toll-like receptors (TLRs) as cross-road between gut microbiota, immune responses, and IBS, has been highlighted. Among TLRs, TLR4 seems to be more involved in the development of IBS. TLR4 interacts with lipopolysaccharide (LPS) pattern, which is the most outer surface component of almost all Gram-negative bacteria (95, 96). It was demonstrated that stimulation of TLR4 by LPS can lead to motivation of the enteric nervous system and motility of the intestine (97). This finding was further investigated and supported by the comparison of the expression of TLRs in biopsy samples of patients with IBS and healthy controls in which the results implied a significant elevation of TLR4 in patients with IBS (98). Furthermore, Belmonte et al. (99) evidenced that the levels of TLR may be different based on the IBS subtypes. In this regard, they reported a significant upregulation of TLR2 and TLR4 in patients with IBS-M together with elevation of IL-8 and IL-1β (99). Recently, Jalanka et al. (100) analyzed the correlation of the gene expression of TLR4 and correlated receptors in patients with IBS and supported the probable role of a low inflammation due to bacteria in the intestine of patients with IBS. Therefore, it seems that a change in the gut microbiota composition may arrange a chronic inflammation and subsequent IBS.
Many studies demonstrated the effects of colonization of Blastocystis sp. on the gut microbiota, in both composition and richness. It was suggested that the presence of Blastocystis sp. reduced a Firmicutes/Bacteroidetes ratio (F/B ratio) in two cohort studies, FACSA and UNEME, which were performed in healthy people and individuals with metabolic disorders in Mexico (101). The mean proportions of Faecalibacterium spp. and Ruminococcaceae in the Blastocystis sp.-positive group, and Enterococcus spp. in the Blastocystis sp.-negative group were abundant in Korean populations (102). The high richness of Faecalibacterium, Prevotella, Ruminococcaceae UCG-002, Muribaculaceae, Rikenellaceae, Acidaminococcaceae, Phascolarctobacterium, and Ruminococcaceae UCG-005 in individuals who carry Blastocystis sp., and E. hirae, E. faecalis, E. durans, Enterococcaceae, Lactobacillales, and Bacilli in subjects who were negative for Blastocystis sp. was observed (102). Importantly, this study concluded that the presence of Blastocystis sp. is an indicator of healthy gut microbiota (102). Most recently, it was shown that Blastocystis sp. ST4 inhibited the growth of B. vulgatus, which suggests the protective role of Blastocystis sp. ST4 in the gut barrier integrity from damage due to the bacteria (103). Later, an altered gut microbial composition was documented in normal healthy mice and Rag1−/− mice colonized by Blastocystis sp. ST4, mainly by an increased proportion of Clostridia vadinBB60 group and Lachnospiraceae NK4A136 group, respectively (104). These results confirmed the protective role of Blastocystis sp. ST4 in the modulation of the gut microbiota to reduce inflammation (104). Blastocystis sp. was reported to significantly increase alpha diversity in carriers. Accordingly, the presence of Blastocystis sp. was associated with enriched Firmicutes and Bacteroidetes, and the genera Prevotella, Faecalibacterium, Flavonifractor, Clostridium, Succinivibrio, and Oscillibacter, whereas Proteobacteria and the genera Escherichia, Bacteroides, Klebsiella, and Pseudomonas were enriched in Blastocystis sp.-negative group (105).
In contrast, a positive correlation between the presence of Blastocystis sp. and C. difficile was suggested in patients with IBD, which was also correlated with the number of defecation in these patients (10). Furthermore, Nourrisson et al. (106) investigated the gut microbiota in four classes of IBS and concluded that the presence of Blastocystis sp. was significantly correlated with an increase of Lactobacilli and a decrease of Bifidobacterium sp. and F. prausnitzii in male patients. Therefore, they suggested that Blastocystis sp. colonization may lead to a decrease in protective bacteria (106). To clear the role of Blastocystis sp., in shifting the gut conditions or gut microbiota composition in IBS, an experimental study was performed by Defaye et al. (107) in rats. Accordingly, they orally inoculated Blastocystis sp. ST4 in rats and evaluated the gut microbiota, inflammation, behavior, and short-chain fatty acid (SCFA). Interestingly, their findings showed that the presence of Blastocystis sp. was resulted in non-inflammatory colonic hypersensitivity with increased serine protease activity, anxiety- and depressive-like behaviors, the relative abundance of Oscillospira with a decrease in Clostridium, and lower levels of SCFAs (107), all of which highlight the role of Blastocystis sp. in the development of IBS and its symptoms, as well as microbiota shifting in patients with IBS (107).
Recently, the eukaryome and prokaryome profiles of patients with IBS-C showed that the colonization of Blastocystis sp. not only induces changes in prokaryotic microbiota of the gut, particularly Tenericutes phylum and Ruminococcaceae family, but also this protist may disturb eukaryome population, particularly fungi, in patients with IBS (108). Importantly, gut mycobiome seems to play an important role in the development, presentation of symptoms, and response to treatment in patients with GI disorders (109–111). Nevertheless, the role of Blastocystis sp. in the gut microbiota changes is controversial, and there is evidence of a neutral role of this protist in the gut microbiota composition in patients with IBS (74). The controversial correlation between the gut microbiota composition and colonization of Blastocystis sp. (112) is thought to be related to antibiotic consumption (113). Remarkably, antibiotics can alter the gut microbiota composition and change the gut lumen from a favorable niche for Blastocystis sp. colonization toward an inimical condition for the protist (Figure 1 and Table 2).
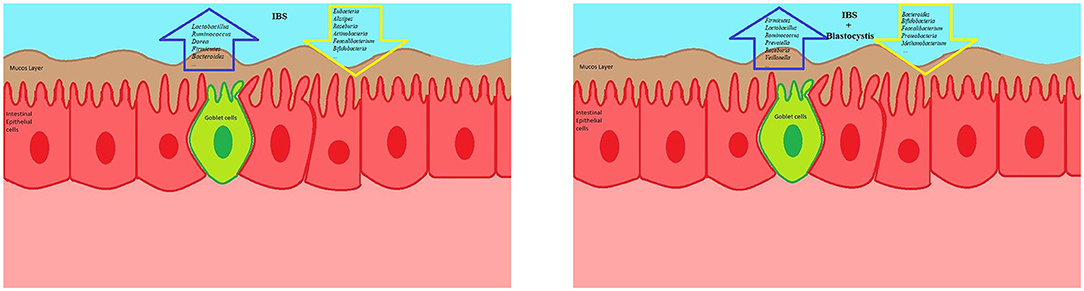
Figure 1. The schematic view presenting a correlation between Blastocystis sp. and certain microbiota signatures. The presence of Blastocystis sp. has not been linked with specific signature of gut microbiota. Although this protist seems to a healthy indicator for the gut microbiota, Blastocystis sp. can reduce a Firmicutes/Bacteroidetes ratio and protective bacteria. In addition, the low and high presence of Blastocystis sp. in IBD and IBS, respectively, might be related to the differences in the gut microbiota composition between IBD and IBS.
Blastocystis sp. is a prevalent protist, which is reported from apparently healthy subjects, as well as individuals with a variety of GI disorders. Although the presence of Blastocystis sp. has not been linked with certain symptoms or disorders, this protist is reported in high prevalence rate in some diseases, such as IBS. IBS conditions seem to provide a favorable niche for colonization of Blastocystis sp. in the intestine. It is not clear how Blastocystis sp. communicates with the gut microbiota in healthy and disease conditions, particularly in patients with IBD and IBS. Some studies suggested that Blastocystis sp. is a healthy gut indicator, while the high prevalence of this protist in IBS proposes a correlation between improper gut conditions and Blastocystis sp. colonization. Nevertheless, recent studies have indicated a correlation between Blastocystis sp. and gut microbiota. It is not clear that either Blastocystis sp. may manipulate the gut microbiota composition or an altered gut microbiota in IBS may provide favorable conditions for colonization of Blastocystis sp.
Gut permeability is a key point of IBS development. Blastocystis sp. discharges a number of proteins, particularly a broad spectrum of proteases, which affect the gut permeability and tight junctions. Although limited data, the secretion levels and types of proteases and the effects of proteases on the human cells are supposed to be different in Blastocystis sp. strains. Therefore, the study of proteases, particularly cysteine protease, derived from different isolates and subtypes on the gut permeability and tight junction proteins provides interesting data. There is no documented study investigating the role of extracellular vesicles (EVs) discharged from either Blastocystis sp. or Blastocystis sp.-affected host cell on the gut permeability. Indeed, a cross-talk between Blastocystis sp. and gut microbiota via EVs may play a role in the successful colonization of the protist or the gut microbiota composition of the gut. EVs play an important role in cross-talk between microorganisms and the study of EVs (released from Blastocystis sp., gut microbiota, and/or host cells) would be interesting. However, researches on Blastocystis sp. is at the beginning stages and more studies are needed to be performed.
This study was approved by the ethical standards (IR.IAU.SRB.REC.1400.241) released by Ethical Review Committees of Islamic Azad University Science and Research Branch, Tehran, Iran.
HM and ESM: conceived and designed. AO, HM, ESM, and AY: data gathering and literature review. AO and HM: writing the manuscript. AS: reviewing and editing the manuscript. All authors read and approved the final version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank all members of the Foodborne and Waterborne Diseases Research Center for their support.
1. Ford AC, Lacy BE, Talley NJ. Irritable bowel syndrome. N Engl J Med. (2017) 376:2566–78. doi: 10.1056/NEJMra1607547
2. Lacy BE, Pimentel M, Brenner DM, Chey WD, Keefer LA, Long MD, et al. ACG Clinical guideline: management of irritable bowel syndrome. Am J Gastroenterol. (2021) 116:17–44. doi: 10.14309/ajg.0000000000001036
3. Dale HF, Lied GA. Gut microbiota and therapeutic approaches for dysbiosis in irritable bowel syndrome: recent developments and future perspectives. Turk J Med Sci. (2020) 50:1632–41. doi: 10.3906/sag-2002-57
4. Bennet SM, Ohman L, Simren M. Gut microbiota as potential orchestrators of irritable bowel syndrome. Gut Liver. (2015) 9:318–31. doi: 10.5009/gnl14344
5. Berumen A, Lennon R, Breen-Lyles M, Griffith J, Patel R, Boxrud D, et al. Characteristics and risk factors of post-infection irritable bowel syndrome after Campylobacter enteritis. Clin Gastroenterol Hepatol. (2021) 19:1855–63.e1851. doi: 10.1016/j.cgh.2020.07.033
6. Sung J, Morales W, Kim G, Pokkunuri V, Weitsman S, Rooks E, et al. Effect of repeated Campylobacter jejuni infection on gut flora and mucosal defense in a rat model of post infectious functional and microbial bowel changes. Neurogastroenterol Motil. (2013) 25:529–37. doi: 10.1111/nmo.12118
7. Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. (2016) 65:1906–15. doi: 10.1136/gutjnl-2016-312297
8. Fagoonee S, Pellicano R. Does the microbiota play a pivotal role in the pathogenesis of irritable bowel syndrome? J Clin Med. (2019) 8:1808. doi: 10.3390/jcm8111808
9. Johnsen PH, Hilpüsch F, Valle PC, Goll R. The effect of fecal microbiota transplantation on IBS related quality of life and fatigue in moderate to severe non-constipated irritable bowel: secondary endpoints of a double blind, randomized, placebo-controlled trial. EBioMed. (2020) 51:102562. doi: 10.1016/j.ebiom.2019.11.023
10. Azimirad M, Gol SMA, Javanmard E, Mirjalali H, Yadegar A, Aghdaei HA, et al. Blastocystis and Clostridioides difficile: evidence for a synergistic role in colonization among IBD patients with emphasis on ulcerative colitis. Turk J Gastroenterol. (2021) 32:500–7. doi: 10.5152/tjg.2021.19644
11. Mirjalali H, Abbasi MR, Naderi N, Hasani Z, Mirsamadi ES, Stensvold CR, et al. Distribution and phylogenetic analysis of Blastocystis sp. subtypes isolated from IBD patients and healthy individuals in Iran. Europ J Clin Microbiol Infect Dis. (2017) 36:2335–42. doi: 10.1007/s10096-017-3065-x
12. Morgan DR, Benshoff M, Cáceres M, Becker-Dreps S, Cortes L, Martin CF, et al. Irritable bowel syndrome and gastrointestinal parasite infection in a developing nation environment. Gastroenterol Res Pract. (2012) 2012:343812. doi: 10.1155/2012/343812
13. Lee YY, Annamalai C, Rao SSC. Post-infectious irritable bowel syndrome. Curr Gastroenterol Rep. (2017) 19:56. doi: 10.1007/s11894-017-0595-4
14. Beatty JK, Bhargava A, Buret AG. Post-infectious irritable bowel syndrome: mechanistic insights into chronic disturbances following enteric infection. World J Gastroenterol. (2014) 20:3976–85. doi: 10.3748/wjg.v20.i14.3976
15. Motomura Y, Khan WI, El-Sharkawy RT, Verma-Gandhu M, Grencis RK, Collins SM. Mechanisms underlying gut dysfunction in a murine model of chronic parasitic infection. Am J Physiol Gastrointestinal Liver Physiol. (2010) 299:G1354–60. doi: 10.1152/ajpgi.00324.2010
16. Jadallah KA, Nimri LF, Ghanem RA. Protozoan parasites in irritable bowel syndrome: a case-control study. World J Gastrointestinal Pharmacol Therap. (2017) 8:201–7. doi: 10.4292/wjgpt.v8.i4.201
17. Dormond M, Gutierrez RL, Porter CK. Giardia lamblia infection increases risk of chronic gastrointestinal disorders. Trop Dis Travel Med Vaccin. (2016) 2:17. doi: 10.1186/s40794-016-0030-0
18. Nakao JH, Collier SA, Gargano JW. Giardiasis and subsequent irritable bowel syndrome: a longitudinal cohort study using health insurance data. J Infect Dis. (2017) 215:798–805. doi: 10.1093/infdis/jiw621
19. Troeger H, Epple HJ, Schneider T, Wahnschaffe U, Ullrich R, Burchard GD, et al. Effect of chronic Giardia lamblia infection on epithelial transport and barrier function in human duodenum. Gut. (2007) 56:328–35. doi: 10.1136/gut.2006.100198
20. Marion R, Baishanbo A, Gargala G, François A, Ducrotté P, Duclos C, et al. Transient neonatal Cryptosporidium parvum infection triggers long-term jejunal hypersensitivity to distension in immunocompetent rats. Infect Immun. (2006) 74:4387–9. doi: 10.1128/IAI.02055-05
21. Barbara G, Vallance BA, Collins SM. Persistent intestinal neuromuscular dysfunction after acute nematode infection in mice. Gastroenterology. (1997) 113:1224–32. doi: 10.1053/gast.1997.v113.pm9322517
22. Venkova K, Greenwood-van Meerveld B. Long-lasting changes in small intestinal transport following the recovery from Trichinella spiralis infection. Neurogastroenterol Motil. (2006) 18:234–42. doi: 10.1111/j.1365-2982.2005.00753.x
23. Venkova K, Palmer JM, Greenwood-Van Meerveld B. Nematode-induced jejunal inflammation in the ferret causes long-term changes in excitatory neuromuscular responses. J Pharmacol Exp Therap. (1999) 290:96–103.
24. Abedi SH, Fazlzadeh A, Mollalo A, Sartip B, Mahjour S, Bahadory S, et al. The neglected role of Blastocystis sp. and Giardia lamblia in development of irritable bowel syndrome: a systematic review and meta-analysis. Microbial Pathog. (2022) 162:105215. doi: 10.1016/j.micpath.2021.105215
25. Engsbro AL, Stensvold CR, Vedel Nielsen H, Bytzer P. Prevalence, incidence, and risk factors of intestinal parasites in Danish primary care patients with irritable bowel syndrome. Scand J Infect Dis. (2014) 46:204–9. doi: 10.3109/00365548.2013.861609
26. Rostami A, Riahi SM, Haghighi A, Saber V, Armon B, Seyyedtabaei SJ. The role of Blastocystis sp. and Dientamoeba fragilis in irritable bowel syndrome: a systematic review and meta-analysis. Parasitol Res. (2017) 116:2361–71. doi: 10.1007/s00436-017-5535-6
27. Borody T, Warren E, Wettstein A, Robertson G, Recabarren P, Fontella A, et al. Eradication of Dientamoeba fragilis can resolve IBS-like symptoms. J Gastroenterol Hepatol. (2002) 17:A103.
28. Yakoob J, Jafri W, Beg MA, Abbas Z, Naz S, Islam M, et al. Blastocystis hominis and Dientamoeba fragilis in patients fulfilling irritable bowel syndrome criteria. Parasitol Res. (2010) 107:679–84. doi: 10.1007/s00436-010-1918-7
29. Ibrahim AN, Al-Ashkar AM, Nazeer JT. Additional glance on the role of Dientamoeba fragilis & Blastocystis hominis in patients with irritable bowel syndrome. Iranian J Parasitol. (2018) 13:100–7.
30. Ali S, Khetpal N, Khan MT, Rasheed M, Asad-Ur-Rahman F, Echeverria-Beltran K. A mexican honeymoon marred by gastrointestinal upset: a case of Dientamoeba fragilis causing post-infectious irritable bowel syndrome. Cureus. (2017) 9:e1992. doi: 10.7759/cureus.1992
31. Engsbro AL, Stensvold CR, Nielsen HV, Bytzer P. Treatment of Dientamoeba fragilis in patients with irritable bowel syndrome. Am J Trop Med Hyg. (2012) 87:1046–52. doi: 10.4269/ajtmh.2012.11-0761
32. Yakoob J, Jafri W, Jafri N, Khan R, Islam M, Beg MA, et al. Irritable bowel syndrome: in search of an etiology: role of Blastocystis hominis. Am J Trop Med Hyg. (2004) 70:383–5. doi: 10.4269/ajtmh.2004.70.383
33. Surangsrirat S, Thamrongwittawatpong L, Piyaniran W, Naaglor T, Khoprasert C, Taamasri P, et al. Assessment of the association between Blastocystis infection and irritable bowel syndrome. J Med AssocThailand. (2010) 93(Suppl. 6):S119–24.
34. Das R, Khalil S, Mirdha BR, Makharia GK, Dattagupta S, Chaudhry R. Molecular characterization and subtyping of Blastocystis sp ecies in irritable bowel syndrome patients from north india. PLoS ONE. (2016) 11:e0147055. doi: 10.1371/journal.pone.0147055
35. Khademvatan S, Masjedizadeh R, Rahim F, Mahbodfar H, Salehi R, Yousefi-Razin E, et al. Blastocystis and irritable bowel syndrome: frequency and subtypes from Iranian patients. Parasitol Int. (2017) 66:142–5. doi: 10.1016/j.parint.2017.01.005
36. Krogsgaard LR, Engsbro AL, Stensvold CR, Nielsen HV, Bytzer P. The prevalence of intestinal parasites is not greater among individuals with irritable bowel syndrome: a population-based case-control study. Clin Gastroenterol Hepatol. (2015) 13:507–13.e502. doi: 10.1016/j.cgh.2014.07.065
37. Salvador F, Lobo B, Goterris L, Alonso-Cotoner C, Santos J, Sulleiro E, et al. Blastocystis sp. carriage and irritable bowel syndrome: is the association already established? Biology. (2021) 10:340. doi: 10.3390/biology10040340
38. Fouad SA, Basyoni MM, Fahmy RA, Kobaisi MH. The pathogenic role of different Blastocystis hominis genotypes isolated from patients with irritable bowel syndrome. Arab J Gastroenterol. (2011) 12:194–200. doi: 10.1016/j.ajg.2011.11.005
39. Unalan-Altintop T, Vahabov C, Ergunay K, Kurt O, Kav T, Akyon Y, et al. Investigation of Dientamoeba fragilis and Blastocystis in patients from Turkey with ulcerative colitis and irritable bowel syndrome: any relation with genotypes?. Acta Trop. (2022) 231:106451. doi: 10.1016/j.actatropica.2022.106451
40. Peña S, Carrasco G, Rojas P, Castillo D, Ozaki LS, Mercado R. Determination of subtypes of Blastocystis sp. in Chilean patients with and without inflammatory bowel syndrome, a preliminary report. Parasit Epidemiol Cont. (2020) 8:e00125. doi: 10.1016/j.parepi.2019.e00125
41. Dogruman-Al F, Kustimur S, Yoshikawa H, Tuncer C, Simsek Z, Tanyuksel M, et al. Blastocystis subtypes in irritable bowel syndrome and inflammatory bowel disease in Ankara, Turkey. Mem Instit Oswaldo Cruz. (2009) 104:724–7. doi: 10.1590/S0074-02762009000500011
42. Jimenez-Gonzalez DE, Martinez-Flores WA, Reyes-Gordillo J, Ramirez-Miranda ME, Arroyo-Escalante S, Romero-Valdovinos M, et al. Blastocystis infection is associated with irritable bowel syndrome in a Mexican patient population. Parasitol Res. (2012) 110:1269–75. doi: 10.1007/s00436-011-2626-7
43. Vargas-Sanchez GB, Romero-Valdovinos M, Ramirez-Guerrero C, Vargas-Hernandez I, Ramirez-Miranda ME, Martinez-Ocaña J, et al. Blastocystis isolates from patients with irritable bowel syndrome and from asymptomatic carriers exhibit similar parasitological loads, but significantly different generation times and genetic variability across multiple subtypes. PLoS ONE. (2015) 10:e0124006. doi: 10.1371/journal.pone.0124006
44. Nagel R, Traub RJ, Kwan MM, Bielefeldt-Ohmann H. Blastocystis sp ecific serum immunoglobulin in patients with irritable bowel syndrome (IBS) versus healthy controls. Parasite Vector. (2015) 8:453. doi: 10.1186/s13071-015-1069-x
45. Olivo-Diaz A, Romero-Valdovinos M, Gudiño-Ramirez A, Reyes-Gordillo J, Jimenez-Gonzalez DE, Ramirez-Miranda ME, et al. Findings related to IL-8 and IL-10 gene polymorphisms in a Mexican patient population with irritable bowel syndrome infected with Blastocystis. Parasitol Res. (2012) 111:487–91. doi: 10.1007/s00436-012-2830-0
46. Romero-Valdovinos M, Gudiño-Ramírez A, Reyes-Gordillo J, Martínez-Flores WA, Ramírez-Miranda ME, Maravilla P, et al. Interleukin-8 and−10 gene polymorphisms in irritable bowel syndrome. Mol Biol Rep. (2012) 39:8837–43. doi: 10.1007/s11033-012-1745-2
47. Xiao QY, Fang XC, Li XQ, Fei GJ. Ethnic differences in genetic polymorphism associated with irritable bowel syndrome. World J Gastroenterol. (2020) 26:2049–63. doi: 10.3748/wjg.v26.i17.2049
48. Ismail MH, Molan AL, Abbas SK. Serological levels of cytokines in irritable bowel syndrome (IBS) patients and non-IBS subjects with and without Blastocystis spp. infection. Annal Parasitol. (2022) 68:77–85. doi: 10.17420/ap6801.411
49. Vara EJ, Brokstad KA, Hausken T, Lied GA. Altered levels of cytokines in patients with irritable bowel syndrome are not correlated with fatigue. Int J General Med. (2018) 11:285–91. doi: 10.2147/IJGM.S166600
50. Karamati SA, Mirjalali H, Niyyati M, Yadegar A, Asadzadeh Aghdaei H, Haghighi A, et al. Association of Blastocystis ST6 with higher protease activity among symptomatic subjects. BMC Microbiol. (2021) 21:285. doi: 10.1186/s12866-021-02341-9
51. Mirza H, Tan KS. Blastocystis exhibits inter- and intra-subtype variation in cysteine protease activity. Parasitol Res. (2009) 104:355–61. doi: 10.1007/s00436-008-1203-1
52. Mirza H, Wu Z, Teo JD, Tan KS. Statin pleiotropy prevents rho kinase-mediated intestinal epithelial barrier compromise induced by Blastocystis cysteine proteases. Cell Microbiol. (2012) 14:1474–84. doi: 10.1111/j.1462-5822.2012.01814.x
53. Puthia MK, Lu J, Tan KS. Blastocystis ratti contains cysteine proteases that mediate interleukin-8 response from human intestinal epithelial cells in an NF-κB-dependent manner. Eukaryot Cell. (2008) 7:435–43. doi: 10.1128/EC.00371-07
54. Mohammad Rahimi H, Yadegar A, Asadzadeh Aghdaei H, Mirjalali H, Zali MR. Modulation of microRNAs and claudin-7 in Caco-2 cell line treated with Blastocystis sp., subtype 3 soluble total antigen. BMC Microbiol. (2022) 22:111. doi: 10.1186/s12866-022-02528-8
55. Kozieł MJ, Ziaja M, Piastowska-Ciesielska AW. Intestinal barrier, claudins and mycotoxins. Toxins. (2021) 13:758. doi: 10.3390/toxins13110758
56. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. (2007) 449:804–10. doi: 10.1038/nature06244
57. Zhu B, Wang X, Li L. Human gut microbiome: the second genome of human body. Protein Cell. (2010) 1:718–25. doi: 10.1007/s13238-010-0093-z
58. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. (2011) 473:174–80. doi: 10.1038/nature09944
59. Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, et al. Conducting a microbiome study. Cell. (2014) 158:250–62. doi: 10.1016/j.cell.2014.06.037
60. Siezen RJ, Kleerebezem M. The human gut microbiome: are we our enterotypes? Microb Biotechnol. (2011) 4:550–3. doi: 10.1111/j.1751-7915.2011.00290.x
61. Berbisá MÁ F, Nielsen KR, Ingham AC, Midjord J, Hammer T, et al. Similar gut bacterial composition between patients with ulcerative colitis and healthy controls in a high incidence population: a cross-sectional study of the faroe islands IBD cohort. Inflamm Bowel Dis. (2022). doi: 10.1093/ibd/izab355. [Epub ahead of print].
62. McCarthy S, Barrett M, Kirthi S, Pellanda P, Vlckova K, Tobin AM, et al. Altered skin and gut microbiome in hidradenitis suppurativa. J Invest Dermatol. (2022) 142:459–68.e415. doi: 10.1016/j.jid.2021.05.036
63. Thompson MD, Kang J, Faerber A, Hinrichs H, Özler O, Cowen J, et al. Maternal obesogenic diet regulates offspring bile acid homeostasis and hepatic lipid metabolism via the gut microbiome in mice. Am J Physiol Gastrointestinal Liver Physiol. (2022) 322:G295–309. doi: 10.1152/ajpgi.00247.2021
64. Wang L, Xian YF, Loo SKF, Ip SP, Yang W, Chan WY, et al. Baicalin ameliorates 2,4-dinitrochlorobenzene-induced atopic dermatitis-like skin lesions in mice through modulating skin barrier function, gut microbiota and JAK/STAT pathway. Bioorgan Chem. (2022) 119:105538. doi: 10.1016/j.bioorg.2021.105538
65. Janeiro MH, Ramírez MJ, Solas M. Dysbiosis and alzheimer's disease: cause or treatment opportunity? Cell Mol Neurobiol. (2022) 42:377–87. doi: 10.1007/s10571-020-01024-9
66. Sheykhsaran E, Abbasi A, Ebrahimzadeh Leylabadlo H, Sadeghi J, Mehri S, Naeimi Mazraeh F, et al. Gut microbiota and obesity: an overview of microbiota to microbial-based therapies. Postgrad Med J. (2022). doi: 10.1136/postgradmedj-2021-141311. [Epub ahead of print].
67. Passos M, Moraes-Filho JP. Intestinal microbiota in digestive diseases. Arq Gastroenterol. (2017) 54:255–62. doi: 10.1590/s0004-2803.201700000-31
68. Thibaut MM, Bindels LB. Crosstalk between bile acid-activated receptors and microbiome in entero-hepatic inflammation. Trend Mol Med. (2022) 28:223–36. doi: 10.1016/j.molmed.2021.12.006
69. Rajilić-Stojanović M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. (2011) 141:1792–801. doi: 10.1053/j.gastro.2011.07.043
70. Shankar V, Reo NV, Paliy O. Simultaneous fecal microbial and metabolite profiling enables accurate classification of pediatric irritable bowel syndrome. Microbiome. (2015) 3:73. doi: 10.1186/s40168-015-0139-9
71. Tap J, Derrien M, Törnblom H, Brazeilles R, Cools-Portier S, Doré J, et al. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology. (2017) 152:111–23.e118. doi: 10.1053/j.gastro.2016.09.049
72. Bhattarai Y, Muniz Pedrogo DA, Kashyap PC. Irritable bowel syndrome: a gut microbiota-related disorder? Am J Physiol Gastrointestinal Liver Physiol. (2017) 312:G52–62. doi: 10.1152/ajpgi.00338.2016
73. Zhuang X, Tian Z, Li L, Zeng Z, Chen M, Xiong L. Fecal microbiota alterations associated with diarrhea-predominant irritable bowel syndrome. Front Microbiol. (2018) 9:1600. doi: 10.3389/fmicb.2018.01600
74. Nagel R, Traub RJ, Allcock RJ, Kwan MM, Bielefeldt-Ohmann H. Comparison of faecal microbiota in Blastocystis-positive and Blastocystis-negative irritable bowel syndrome patients. Microbiome. (2016) 4:47. doi: 10.1186/s40168-016-0191-0
75. Shukla R, Ghoshal U, Dhole TN, Ghoshal UC. Fecal microbiota in patients with irritable bowel syndrome compared with healthy controls using real-time polymerase chain reaction: an evidence of dysbiosis. Dig Dis Sci. (2015) 60:2953–62. doi: 10.1007/s10620-015-3607-y
76. Mei L, Zhou J, Su Y, Mao K, Wu J, Zhu C, et al. Gut microbiota composition and functional prediction in diarrhea-predominant irritable bowel syndrome. BMC Gastroenterol. (2021) 21:105. doi: 10.1186/s12876-021-01693-w
77. Pittayanon R, Lau JT, Yuan Y, Leontiadis GI, Tse F, Surette M, et al. Gut microbiota in patients with irritable bowel syndrome-a systematic review. Gastroenterology. (2019) 157:97–108. doi: 10.1053/j.gastro.2019.03.049
78. Lo Presti A, Zorzi F, Del Chierico F, Altomare A, Cocca S, Avola A, et al. Fecal and mucosal microbiota profiling in irritable bowel syndrome and inflammatory bowel disease. Front Microbiol. (2019) 10:1655. doi: 10.3389/fmicb.2019.01655
79. Zhu X, Hong G, Li Y, Yang P, Cheng M, Zhang L, et al. Understanding of the site-specific microbial patterns towards accurate identification for patients with diarrhea-predominant irritable bowel syndrome. Microbiol Spect. (2021) 9:e0125521. doi: 10.1128/Spectrum.01255-21
80. Sciavilla P, Strati F, Di Paola M, Modesto M, Vitali F, Cavalieri D, et al. Gut microbiota profiles and characterization of cultivable fungal isolates in IBS patients. Appl Microbiol Biotechnol. (2021) 105:3277–88. doi: 10.1007/s00253-021-11264-4
81. Lopetuso LR, Petito V, Graziani C, Schiavoni E, Paroni Sterbini F, Poscia A, et al. Gut microbiota in health, diverticular disease, irritable bowel syndrome, and inflammatory bowel diseases: time for microbial marker of gastrointestinal disorders. Dig Dis. (2018) 36:56–65. doi: 10.1159/000477205
82. Sundin J, Rangel I, Fuentes S, Heikamp-de Jong I, Hultgren-Hörnquist E, de Vos WM, et al. Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Alim Pharmacol Therap. (2015) 41:342–51. doi: 10.1111/apt.13055
83. Giamarellos-Bourboulis E, Tang J, Pyleris E, Pistiki A, Barbatzas C, Brown J, et al. Molecular assessment of differences in the duodenal microbiome in subjects with irritable bowel syndrome. Scandinavian J Gastroenterol. (2015) 50:1076–87. doi: 10.3109/00365521.2015.1027261
84. Liu Y, Zhang L, Wang X, Wang Z, Zhang J, Jiang R, et al. Similar fecal microbiota signatures in patients with diarrhea-predominant irritable bowel syndrome and patients with depression. Clin Gastroenterol Hepatol. (2016) 14:1602–11.e1605. doi: 10.1016/j.cgh.2016.05.033
85. Carroll IM, Ringel-Kulka T, Keku TO, Chang YH, Packey CD, Sartor RB, et al. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointestinal Liver Physiol. (2011) 301:G799–807. doi: 10.1152/ajpgi.00154.2011
86. Drossman DA. Functional gastrointestinal disorders: history, pathophysiology, clinical features and rome IV. Gastroenterology. (2016) 150:1262–1279.e2. doi: 10.1053/j.gastro.2016.02.032
87. Muller PA, Koscsó B, Rajani GM, Stevanovic K, Berres ML, Hashimoto D, et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. (2014) 158:300–13. doi: 10.1016/j.cell.2014.04.050
88. Barbara G, Stanghellini V, De Giorgio R, Cremon C, Cottrell GS, Santini D, et al. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology. (2004) 126:693–702. doi: 10.1053/j.gastro.2003.11.055
89. Ohman L, Isaksson S, Lindmark AC, Posserud I, Stotzer PO, Strid H, et al. T-cell activation in patients with irritable bowel syndrome. Am J Gastroenterol. (2009) 104:1205–12. doi: 10.1038/ajg.2009.116
90. Nasser Y, Petes C, Simmers C, Basso L, Altier C, Gee K, et al. Activation of peripheral blood CD4+ T-cells in ibs is not associated with gastrointestinal or psychological symptoms. Sci Rep. (2019) 9:3710. doi: 10.1038/s41598-019-40124-5
91. Vicario M, González-Castro AM, Martínez C, Lobo B, Pigrau M, Guilarte M, et al. Increased humoral immunity in the jejunum of diarrhoea-predominant irritable bowel syndrome associated with clinical manifestations. Gut. (2015) 64:1379–88. doi: 10.1136/gutjnl-2013-306236
92. Chatoo M, Li Y, Ma Z, Coote J, Du J, Chen X. Involvement of corticotropin-releasing factor and receptors in immune cells in irritable bowel syndrome. Front Endocrinol. (2018) 9:21. doi: 10.3389/fendo.2018.00021
93. Lobo B, Ramos L, Martínez C, Guilarte M, González-Castro AM, Alonso-Cotoner C, et al. Downregulation of mucosal mast cell activation immune response in diarrhoea-irritable bowel syndrome by oral disodium cromoglycate: a pilot study. United Europ Gastroenterol J. (2017) 5:887–97. doi: 10.1177/2050640617691690
94. Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. (2007) 449:819–26. doi: 10.1038/nature06246
95. Bertani B, Ruiz N. Function and biogenesis of lipopolysaccharides. EcoSal Plus. (2018) 8:10.1128/ecosalplus.ESP-0001-2018. doi: 10.1128/ecosalplus.ESP-0001-2018
96. Park BS, Lee JO. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med. (2013) 45:e66. doi: 10.1038/emm.2013.97
97. Anitha M, Vijay-Kumar M, Sitaraman SV, Gewirtz AT, Srinivasan S. Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology. (2012) 143:1006–16.e1004. doi: 10.1053/j.gastro.2012.06.034
98. Brint EK, MacSharry J, Fanning A, Shanahan F, Quigley EM. Differential expression of toll-like receptors in patients with irritable bowel syndrome. Am J Gastroenterol. (2011) 106:329–36. doi: 10.1038/ajg.2010.438
99. Belmonte L, Beutheu Youmba S, Bertiaux-Vandaële N, Antonietti M, Lecleire S, Zalar A, et al. Role of toll like receptors in irritable bowel syndrome: differential mucosal immune activation according to the disease subtype. PLoS ONE. (2012) 7:e42777. doi: 10.1371/journal.pone.0042777
100. Jalanka J, Lam C, Bennett A, Hartikainen A, Crispie F, Finnegan LA, et al. Colonic gene expression and fecal microbiota in diarrhea-predominant irritable bowel syndrome: increased toll-like receptor 4 but minimal inflammation and no response to mesalazine. J Neurogastroenterol Motil. (2021) 27:279–91. doi: 10.5056/jnm20205
101. Yañez CM, Hernández AM, Sandoval AM, Domínguez MAM, Muñiz SAZ, Gómez JOG. Prevalence of Blastocystis and its association with Firmicutes/Bacteroidetes ratio in clinically healthy and metabolically ill subjects. BMC Microbiol. (2021) 21:339. doi: 10.1186/s12866-021-02402-z
102. Kim MJ, Lee YJ, Kim TJ, Won EJ. Gut microbiome profiles in colonizations with the enteric protozoa Blastocystis in Korean populations. Microorganisms. (2021) 10:34. doi: 10.3390/microorganisms10010034
103. Deng L, Tan K. Interactions between Blastocystis subtype ST4 and gut microbiota in vitro. Parasit Vector. (2022) 15:80. doi: 10.1186/s13071-022-05194-x
104. Deng L, Wojciech L, Png CW, Koh EY, Aung TT, Kioh D, et al. Experimental colonization with Blastocystis ST4 is associated with protective immune responses modulation of gut microbiome in a DSS-induced colitis mouse model. Cell Mol Life Sci. (2022) 79:245. doi: 10.1007/s00018-022-04271-9
105. Stensvold CR, Sørland BA, Berg R, Andersen LO, van der Giezen M, Bowtell JL, et al. Stool microbiota diversity analysis of Blastocystis-positive and Blastocystis-negative individuals. Microorganisms. (2022) 10:326. doi: 10.3390/microorganisms10020326
106. Nourrisson C, Scanzi J, Pereira B, NkoudMongo C, Wawrzyniak I, Cian A, et al. Blastocystis is associated with decrease of fecal microbiota protective bacteria: comparative analysis between patients with irritable bowel syndrome and control subjects. PLoS ONE. (2014) 9:e111868. doi: 10.1371/journal.pone.0111868
107. Defaye M, Nourrisson C, Baudu E, Lashermes A, Meynier M, Meleine M, et al. Fecal dysbiosis associated with colonic hypersensitivity and behavioral alterations in chronically Blastocystis-infected rats. Sci Rep. (2020) 10:9146. doi: 10.1038/s41598-020-66156-w
108. Nourrisson C, Scanzi J, Brunet J, Delbac F, Dapoigny M, Poirier P. Prokaryotic and eukaryotic fecal microbiota in irritable bowel syndrome patients and healthy individuals colonized with Blastocystis. Front Microbiol. (2021) 12:713347. doi: 10.3389/fmicb.2021.713347
109. Beheshti-Maal A, Shahrokh S, Ansari S, Mirsamadi ES, Yadegar A, Mirjalali H, et al. Gut mycobiome: the probable determinative role of fungi in IBD patients. Mycoses. (2021) 64:468–76. doi: 10.1111/myc.13238
110. Das A, O'Herlihy E, Shanahan F, O'Toole PW, Jeffery IB. The fecal mycobiome in patients with irritable bowel syndrome. Sci Rep. (2021) 11:124. doi: 10.1038/s41598-020-79478-6
111. Gu Y, Zhou G, Qin X, Huang S, Wang B, Cao H. The potential role of gut mycobiome in irritable bowel syndrome. Front Microbiol. (2019) 10:1894. doi: 10.3389/fmicb.2019.01894
112. Deng L, Wojciech L, Gascoigne N, Peng G, Tan K. New insights into the interactions between Blastocystis, the gut microbiota, and host immunity. PLoS Pathog. (2021) 17:e1009253. doi: 10.1371/journal.ppat.1009253
Keywords: irritable bowel syndrome, gut microbiota, Blastocystis sp., dysbiosis, post-infectious-IBS
Citation: Olyaiee A, Sadeghi A, Yadegar A, Mirsamadi ES and Mirjalali H (2022) Gut Microbiota Shifting in Irritable Bowel Syndrome: The Mysterious Role of Blastocystis sp. Front. Med. 9:890127. doi: 10.3389/fmed.2022.890127
Received: 07 March 2022; Accepted: 09 May 2022;
Published: 20 June 2022.
Edited by:
Md. Robiul Karim, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshReviewed by:
Ali Rostami, Babol University of Medical Sciences, IranCopyright © 2022 Olyaiee, Sadeghi, Yadegar, Mirsamadi and Mirjalali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hamed Mirjalali, aGFtZWRfbWlyamFsYWxpQGhvdG1haWwuY29t; aGFtZWRtaXJqYWxhbGlAc2JtdS5hYy5pcg==; Elnaz Sadat Mirsamadi, ZWxuYXpfbWlyNjJAeWFob28uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.