
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 16 May 2022
Sec. Pulmonary Medicine
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.889020
This article is part of the Research Topic Chronic Airway Diseases, Lung Cancer, and Their Interaction View all 11 articles
 Ping Yang1,2
Ping Yang1,2 Zhenchao Wu2
Zhenchao Wu2 Chao Liu3
Chao Liu3 Jiajia Zheng4
Jiajia Zheng4 Nan Wu2
Nan Wu2 Zhangli Wu1,2
Zhangli Wu1,2 Juan Yi1
Juan Yi1 Ming Lu2,3*†
Ming Lu2,3*† Ning Shen1,2,3*†
Ning Shen1,2,3*†Background: Sequence type 11 (ST11) Klebsiella pneumoniae (Kp) is highly prevalent in China and is a typical sequence type among KPC-producing isolates. This study aimed to evaluate the clinical outcomes and microbiological features of ST11 Kp infections.
Methods: A retrospective cohort study was conducted at Peking University Third Hospital from January 2017 to March 2021. Clinical data were collected from medical records. Antimicrobial susceptibility testing and string tests were performed. Whole-genome sequencing was used to analyze the capsular serotypes, detect virulence-associated genes, and perform multilocus sequence typing. The risk of all-cause mortality in ST11 Kp-infected patients was compared to that in non-ST11 Kp-infected patients.
Results: From 139 patients infected with Kp, 49 ST11 Kp (35.3%) strains were isolated. The Charlson comorbidity index in the ST11 group was higher than that in the non-ST11 group (3.94 ± 1.59 vs. 2.41 ± 1.54, P = 0.001). A greater number of ST11 Kp-infected patients required ICU admission (46.9 vs. 16.7%, P < 0.001) and mechanical ventilation (28.6 vs. 10.0%, P = 0.005). All ST11 isolates presented a multidrug-resistant (MDR) phenotype, and twenty-nine (59.2%) hypervirulent Kp (hvKp) were identified. Twenty-four ST11 strains presented with hypermucoviscosity. The presence of capsular types K47 and K64 was frequent in the ST11 Kp strains (P < 0.001). The key virulence-associated genes rmpA, rmpA2, iucA, iroB, and peg344 were present in 26.5, 42.9, 59.2, 0, and 26.5% of the isolates, respectively, in the ST11 group. Twenty-one ST11 isolates harbored the combination of iucA+rmpA2. The 30-day mortality rate and sequential organ failure assessment (SOFA) score were significantly higher in ST11 Kp-infected patients than in non-ST11 Kp-infected patients (P < 0.01). ST11 Kp infection appeared to be an independent risk factor for mortality in ST11 Kp-infected patients.
Conclusions: A high prevalence of the ST11 clone was found in the hospital, which accounted for elevated antimicrobial resistance and exhibited great molecularly inferred virulence. Patients with ST11 Kp infection had a tendency toward increased 30-day mortality and SOFA scores. ST11 Kp infection was an independent risk factor for mortality, suggesting that enhanced surveillance and management are essential.
Klebsiella pneumoniae (Kp), an important community-acquired and nosocomial gram-negative bacterial pathogen, causes fatal infections, including pneumonia, urinary tract infection, pyogenic liver abscess, bacteremia and so on (1). Two distinct pathotypes are currently circulating: hypervirulent Klebsiella pneumoniae (hvKp) and classical Klebsiella pneumoniae (cKp), each of which poses great challenges in clinical practice (2–5). cKp infection mainly occurs in immunocompromised hosts and patients with physiological barrier breakdown in the medical environment. cKp has the ability to acquire various antibiotic resistance genes and rapidly become resistant to all available antibiotics, which is a serious threat to public health. The worldwide spread of multidrug-resistant (MDR) cKp is mainly driven by a special clone termed sequence type 11 (ST11) (6, 7). In China, ST11 Kp is endemic, and most KPC-producing isolates are typically of this sequence type (8). In stark contrast, hvKp infection is mainly acquired by healthy individuals of any age in the community and is highly associated with aggressive invasive infections in hospitals, such as bacteremia and pyogenic liver abscess (4, 9, 10). The previous definition of hvKp was determined by the hypermucoviscosity phenotype (string test showing a positive result) (11).
In fact, not all hvKp isolates possess the hypermucoviscosity phenotype, as confirmed by in vitro and in vivo studies (3, 5, 12–14). Studies have demonstrated that genetic traits perform better than hypermucoviscosity as a marker for differentiating hvKp and cKp (14, 15). Five key virulence-associated genes, iucA, iroB, peg-344, rmpA and rmpA2, showed higher diagnostic accuracy for defining hvKp than the hypermucoviscosity phenotype and other virulence-associated genes (15).
ST11, previously identified as cKp, commonly presents with lower virulence (16, 17). However, Gu et al. (18) first demonstrated a fatal outbreak caused by ST11 carbapenem-resistant (CR) hvKp in the ICU. The acquisition of pVir-CR-hvKp4 contributed to the convergence of carbapenem resistance and hypervirulence (18). In addition, the emergence of hybrid conjugative virulence plasmids triggered MDR hvKp formation (19, 20).
Contrary to common belief, the phenomenon of convergence of carbapenem resistance and hypervirulence in Kp is increasing in frequency, which may lead to worse clinical outcomes (18, 19). Few studies have focused on the outcomes and microbiological characteristics of ST11 Kp strains. Liu et al. (21) revealed that the 3-year survival rate of ST11 Kp-infected patients was 73.68%. Another study reported that the in-hospital mortality in the ST11 group was 36.7% (22). However, these previous studies on the mortality of ST11 Kp infection compared to that of non-ST11 infection were controversial. Therefore, we conducted a retrospective study to analyze the clinical outcomes, antibiotic resistance, virulence, and all-cause mortality risk of ST11 Kp infection. We found that ST11 Kp isolates accounted for elevated antimicrobial resistance and exhibited great molecularly inferred virulence. ST11 Kp infection was an independent risk factor for mortality and an increased sequential organ failure assessment (SOFA) score.
A retrospective cohort study was conducted on Kp culture-positive patients enrolled from January 2017 to March 2021 at Peking University Third Hospital. Clinical data and patient information were obtained from medical records, which included basic demographics, medical history, usage of invasive devices, infection site, infection type, blood examination results, ICU admission, and mechanical ventilation after Kp infection. The Charlson comorbidity index (CCI) was calculated based on the medical history.
The main inclusion criteria were as follows: (1) age ≥ 18 years old and (2) Kp was cultured positively and was associated with clinical infectious manifestations at the same time. The exclusion criteria included the following: (1) the bacterial strain not viable after storage and (2) duplicate isolates from the same patient within 3 months. The primary outcome was survival and all-cause mortality at 30 days after Kp infection, and the secondary outcome was the SOFA score.
Hospital-acquired infection and community-acquired infection were defined as previously described (4). The definition of metastatic infection was based on the clinical diagnosis (the presence of > 1 infection site) in the same patient (23).
This study protocol was approved by the Peking University Third Hospital Medical Science Research Ethics Committee (M2021545).
These specimens were from the respiratory system, urine, blood, drainage fluid, and other body sites. The standardized isolation, culture, and identification were conducted in the Department of Clinical Microbiology. All strains were stored at −80°C.
Strain identification and antimicrobial susceptibility testing (AST) were performed by a Vitek 2 system (bioMérieux, Marcy-l'Étoile, France). If necessary, we also applied the disk diffusion method. The AST results were interpreted according to the 2020 Clinical and Laboratory Standards Institute (CLSI) guidelines (24). The following antimicrobial agents were tested: piperacillin/tazobactam, cefoperazone/sulbactam, ceftazidime, cefepime, imipenem, meropenem, levofloxacin, amikacin, minocycline, and trimethoprim/sulfamethoxazole. The definition of an MDR strain was resistance to three or more different antimicrobial categories (25). Carbapenem-resistant Klebsiella pneumoniae (CRKP) was defined based on resistance to imipenem or meropenem.
The DNA of all the isolated Kp strains was extracted by using a GenePure Pro Automatic Nucleic Acid Purification System (NPA-32P, Bioer Technology, Hangzhou, Zhejiang, China) and MagaBio Bacterium DNA Fast Purification Kit (BSC45S1E, Bioer Technology, Hangzhou, Zhejiang, China). Each Kp sample was fully mixed with 180 μL of TET buffer (with added lysozyme) and incubated at 37°C for 30~60 min. Two microliters of RNase A (RT405-02, TIANGEN, Beijing, China) was added, and the samples were shaken for 15 s and incubated at 15–25°C for 5 min. Next, each prepared sample and 20 μl of Proteinase K were transferred into kit columns 1 and 7, and then plugged in 8-strip tip and ran the machine program. DNA concentration and purity were evaluated by a NanoDrop (ThermoFisher, Waltham, America).
All strains were sequenced using the Illumina HiSeq 2500 platform by constructing paired-end libraries to obtain 150 bp reads. The clean data were obtained using FastQC and assembled using SPAdes (v.3.13) with the default parameters.
Whole-genome sequencing was used to analyze the capsular serotypes, identify virulence-associated genes, and perform multilocus sequence typing (MLST). Raw data were filtered to remove low-quality reads and then assembled using SPAdes (v.3.13). The definition of ST11 Kp was based on MLST using MLST (v.2.0) (Center for Genomic Epidemiology). Capsular types were analyzed using Kleborate software (v.0.3.0). Resistance and virulence genes were annotated by comparison with relevant databases (ResFinder, Virulence Factor Database) using BLAST software (v.2.2.18). HvKp is defined based on the combination of peg-344, iroB, iucA, rmpA, or rmpA2 positivity (15).
Sequencing reads were mapped to the K. pneumoniae HS11286 using bowtie 2 v2.2.8 and single nucleotide polymorphisms (SNPs) were identified by using Samtools v1.9 and combined according to the reference genome (SGH-10) using the iSNV-calling pipeline (https://github.com/generality/iSNV-calling). High-quality SNPs (more than 5 reads of mapping quality > 20) were retained. Regions of recombination were detected by Gubbins (26), and the polymorphic sites located in recombination regions were removed. The concatenated sequences of filtered polymorphic sites conserved in all genomes (core genome SNPs, cgSNPs) were used to perform phylogenetic analysis using the maximum likelihood method by iqTree2.1.2 (27).
The hypermucoviscous phenotype was determined by the string test as described previously (2). All isolates were inoculated onto Columbia agar with sheep blood (PB0123A, OXOID, Beijing, China) and incubated at 37°C overnight. The string test was considered positive when a bacteriology inoculation loop was able to generate a viscous string > 5 mm in length by touching and pulling a single colony upward.
Data analysis was performed using SPSS software (v.25.0). Measurement data were assessed as the mean ± standard deviation, and count data are reported as percentages. We used the T test and Wilcoxon test for the analysis of continuous variables. We performed the χ2 or Fisher's exact test for categorical variables. All tests were 2-tailed. A P-value < 0.05 was considered statistically significant. The all-cause mortality within 30 days between the two groups was estimated using a Kaplan–Meier curve and log-rank tests. Univariate logistic regression analyses were performed to identify the risk factors associated with death. A multivariable logistic regression analysis was conducted for independent risk factors for death (the variables with P < 0.05 were included). Univariate linear regression analyses were performed to identify the factors associated with death. A multivariable linear regression analysis was conducted for independent risk factors for elevated SOFA scores (the variables with P < 0.05 were included).
Isolates from 139 Kp infection cases were collected from January 2017 to March 2021. Among these isolates, 49 strains (35.3%, 49/139) were identified as ST11, which was the most prevalent sequence type in this study. Non-ST11 strains (64.7%, 90/139) mainly included ST23 (18.9%, 17/90), ST15 (8.9%, 8/90), ST86 (6.7%, 6/90) and so on. The median age of patients with ST11 Kp infections was 80.04 ± 12.41 years, and 29 patients (59.2%) were male. Most ST11 Kp strains were isolated from patients in the emergency department and ICU (55.1 and 26.5%, respectively), followed by the geriatric department (10.2%) (Supplementary Figure 1). Compared with those in the non-ST11 group, more patients in the ST11 group had cardiovascular disease (93.9 vs. 64.4%, P < 0.001), cerebrovascular disease (63.3 vs. 36.7%, P = 0.003), and urinary disease (51.0 vs. 28.9%, P = 0.010). Furthermore, the CCI was higher in the ST11 group (3.94 ± 1.59 vs. 2.41 ± 1.54, P = 0.001). In addition, the ST11 group showed significantly more antibiotic exposure within the previous 90 days (100.0 vs. 60.0%, P < 0.001). A significant number of patients with invasive catheters were infected by ST11 Kp isolates (100.0 vs. 55.6%, P < 0.001), which included central intravenous catheters (67.3 vs. 26.0%, P < 0.001), urinary catheters (91.8 vs. 70.0%, P = 0.006), endotracheal tubes (32.7 vs. 14.0%, P = 0.028) and gastrostomy tubes (89.8 vs. 60.0%, P = 0.001). In this study, the most common ST11 Kp infection sites were the respiratory system (65.3%), followed by urine (16.3%). Fewer patients suffered from bloodstream infection in the ST11 group than in the non-ST11 group (6.1 vs. 18.9%, P = 0.040), whereas there was no significant difference among other sites between the two groups. Compared to non-ST11 Kp strains, ST11 Kp strains were more closely related to hospital-acquired infections (100.0 vs. 74.4%, P < 0.001). In contrast, more community-acquired infections occurred in non-ST11 isolates (0 vs. 25.6%, P < 0.001). Furthermore, blood testing indicators (red blood cell count, hemoglobin, and albumin) of patients with ST11 Kp infection were significantly lower than those of the non-ST11 group (P < 0.01). However, patients with ST11 Kp infection had a higher hematocrit (P = 0.013). In addition, a significant number of patients with ST11 Kp infection required ICU admission (46.9 vs. 16.7%, P < 0.001) and mechanical ventilation after Kp detection (28.6 vs. 10.0%, P = 0.005) (Table 1).
Subgroup analysis was performed according to cKp and hvKp infection. The clinical characteristics of ST11 Kp infection in the hvKp and cKp subgroups were mostly similar to the above results. Notably, more metastatic infection occurred with ST11 Kp strains of the hvKp subgroup (34.5 vs. 10.9%, P = 0.013) (Supplementary Table 2). In the cKp subgroup, more patients suffered from respiratory system infection in the ST11 group than in the non-ST11 group (75.0 vs. 47.7%, P = 0.041) (Supplementary Table 2). Additionally, in the ST11 group, the ICU admission rate was higher in patients with cKp infection than in those with hvKp infection (34.5 vs. 65.0%, P = 0.035), while the other clinical characteristics showed no significant differences (Supplementary Table 3).
All the ST11 Kp isolates (100.0%) were MDR, while only 28 of 90 non-ST11 (31.1%) Kp strains were MDR. Moreover, CRKP in the ST11 group and non-ST11 group accounted for 100 and 14.4%, respectively. Notably, all 49 ST11 Kp strains (100.0%) were resistant to piperacillin/tazobactam, cefoperazone/sulbactam, ceftazidime, cefepime, imipenem, and meropenem. Significantly, the rates of resistance to all antibiotics in the non-ST11 group were <50%. However, except for minocycline and trimethoprim/sulfamethoxazole, the rates of resistance to most antibiotics were approximately 100% in ST11 Kp isolates. Additionally, for the antibiotics commonly used in clinical practice, such as β-lactamase inhibitors and carbapenems, the resistance rate of the ST11 group was significantly higher than that of the non-ST11 group (P < 0.001) (Table 2, Supplementary Table 1).
In terms of resistance genes, all the ST11 Kp strains harbored the beta-lactamase gene blaKPC−2. Additionally, a large number of ST11 isolates presented the beta-lactamase gene blaTEM−1D (93.9%) and the fosfomycin resistance gene fosA3 (91.8%). Over half of the ST11 isolates possessed the aminoglycoside resistance gene aadA2 and the beta-lactamase genes blaCTX−M−65 and blaSHV−11, which were significantly more frequently detected than in the non-ST11 group (P < 0.001, P < 0.001 and P = 0.022, respectively). There was a strong tendency for the presence of K47 (53.1 vs. 1.1%, P < 0.001) and K64 (42.9 vs. 1.1%, P < 0.001) in ST11 Kp strains. In contrast, there was a tendency for the presence of K1 (0 vs. 20.0%, P < 0.001) and K2 (0 vs. 15.6%, P = 0.002) in non-ST11 Kp isolates. Twenty-four ST11 strains showed a hypermucoviscous phenotype, which was a significantly lower number than that in the non-ST11 group (49.0 vs. 68.9%, P = 0.021). The virulence-associated genes rmpA, rmpA2, iucA, iroB, and peg344 were present in 26.5, 42.9, 59.2, 0, and 26.5% of the isolates in the ST11 group, respectively. Notably, all the ST11 Kp isolates harbored the siderophore yersiniabactin genes. Additionally, the ybt-ICEKp3 cluster was highly clustered in the ST11 group (100.0 vs. 6.7%, P < 0.001). peg589, which is related to a poor prognosis in an animal model, was highly associated with the ST11 group (61.2 vs. 41.1%, P = 0.023). Importantly, twenty-nine (59.2%) ST11 Kp strains were MDR hvKp. Additionally, twenty-one ST11 isolates harbored the combination of iucA+rmpA2, which showed no significant difference in both groups (P = 0.649) (Figure 1, Supplementary Table 4).
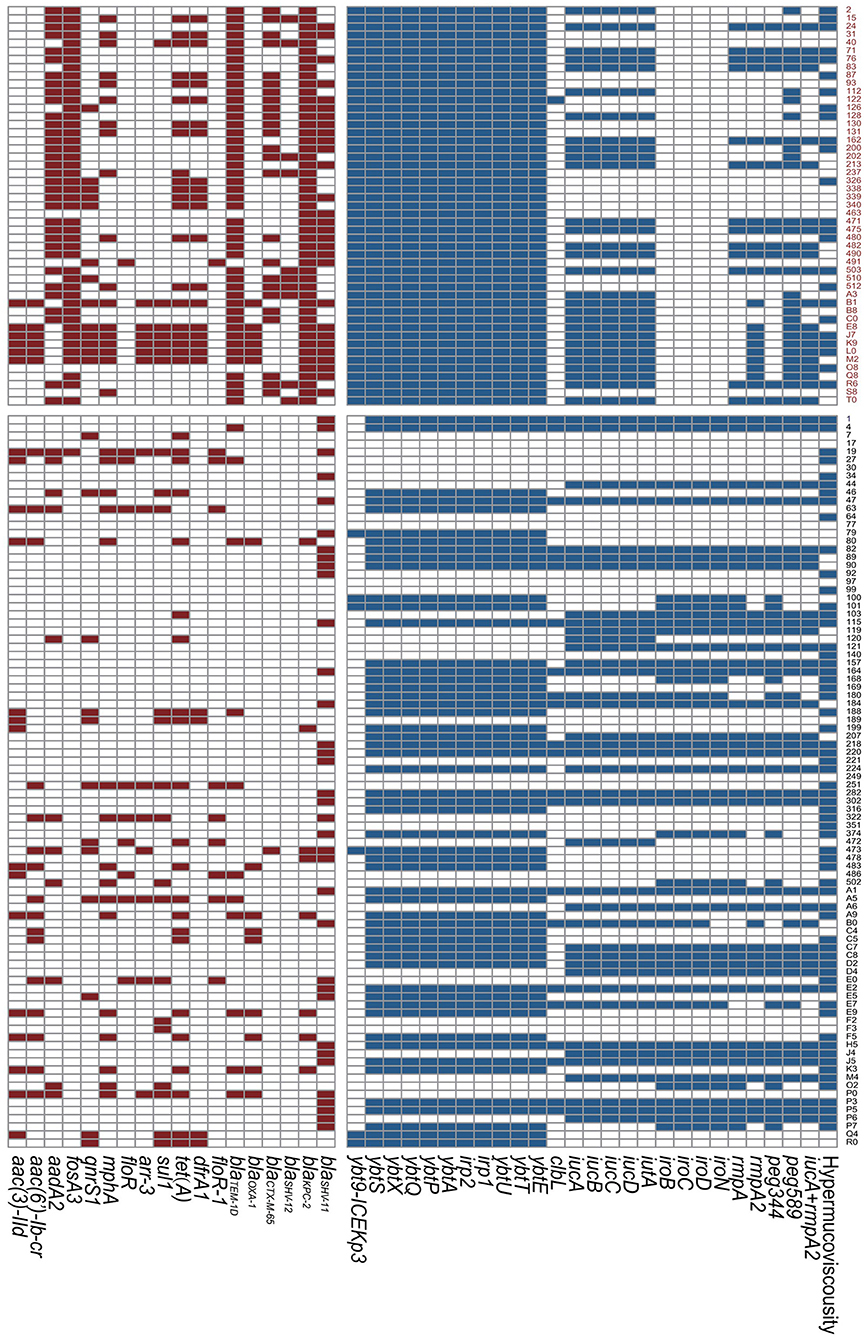
Figure 1. Resistance genes and virulence genes of Klebsiella pneumoniae strains. The colored blocks represent existence of genes. Red, ST11 group; Black, Non-ST11 group.
Among all the ST11 Kp strains, two distinguished clades were identified by the phylogenetic analysis. Clade a contained 23 strains, and clade b comprised 26 strains. Notably, a great number of hvKp belonged to the clade a (21/29). Besides, two clades had the same 30-day mortality (Figure 2).
The survival curve revealed that the 30-day mortality rate in patients with ST11 Kp infection was significantly higher than that in patients without ST11 Kp infection (38.3 vs. 12.5%, P = 0.001) (Figure 3). Similar results were observed in the hvKp (35.7 vs. 8.9%, P = 0.005) and cKp subgroups (42.1 vs. 16.3%, P = 0.029) (Supplementary Figure 2). In the ST11 group, the mortality among patients with cKp and hvKp infections was similar (35.7 vs. 42.1%, P = 0.644) (Supplementary Figure 3).
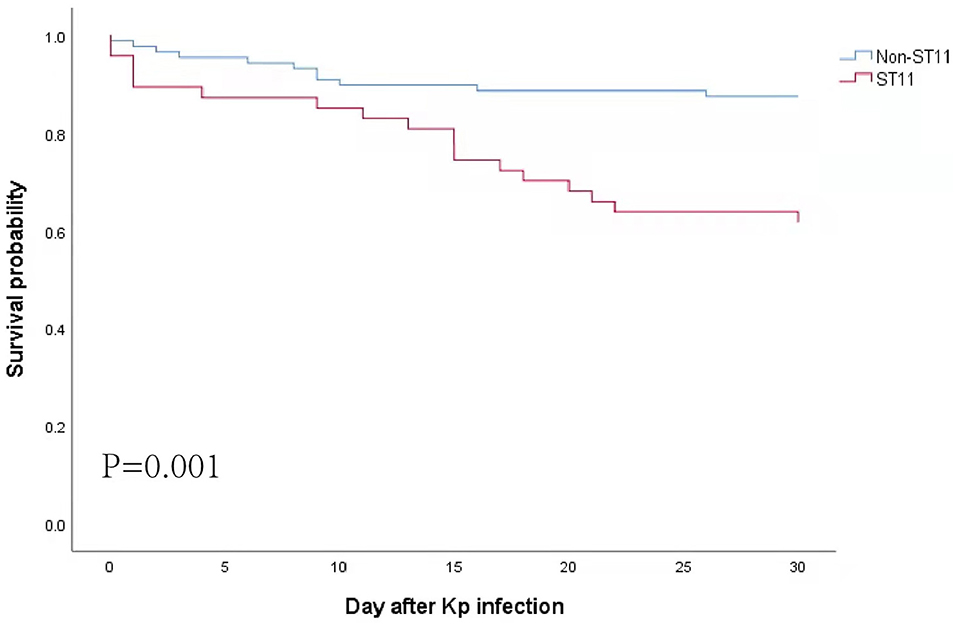
Figure 3. Kaplan–Meier curves for all-cause 30-day mortality. Statistical significance was determined by the log-rank test.
Univariate regression analysis showed that ST11 Kp infection [odds ratio (OR) = 4.345] and the CCI (OR = 1.598) were statistically significant risk factors associated with 30-day mortality. In addition, multivariate analysis revealed that ST11 Kp infection appeared to be an independent risk factor for ST11 Kp infection (OR = 2.786) (Table 3).
The SOFA score was significantly higher in patients with ST11 infection than in the non-ST11 infection patients (6.31 ± 6.04 vs. 2.47 ± 2.97, P < 0.001) (Table 1, Supplementary Figure 4). The results of univariate linear analysis revealed that the risk factors for an elevated SOFA score included ST11 Kp infection [risk ratio (RR) = 3.839]) and increased CCI (RR = 0.597). Multivariate analysis showed that compared to non-ST11 infection, ST11 Kp infection caused an increase in SOFA score (RR = 3.579) (Table 4).
The prevalent sequence types of Kp are diverse worldwide. In China, previous studies revealed that ST11 Kp is the main endemic clone, typically presenting as CRKP (7, 28, 29). Similarly, we found that ST11 Kp was highly prevalent in our study. We found that the CCI was significantly higher in patients with ST11 Kp infection than in the non-ST11 infection patients. In addition, our data revealed that the ST11 group was highly associated with catheter usage, ICU admission and mechanical ventilation after Kp detection. It seemed that ST11 Kp-infected patients might have a complicated status. Importantly, the results suggested that all ST11 Kp strains were MDR. Surprisingly, the majority of MDR ST11 isolates were hvKp. The emergence of MDR ST11 hvKp strains threatens the viability of the current therapeutic approach, increasing the severity of infection. The nosocomial dissemination of MDR ST11 hvKp isolates is alarming, and medical staff need to enhance the infection control and management of ST11 strains with “superbug” characteristics and high virulence.
A previous study demonstrated that ST11 Kp was a common class of cKp and was notorious for acquiring various antibiotic resistance genes (30). Notably, the ratio of MDR strains has increased rapidly among ST11 isolates (7, 31). Previous studies reported that ST11 isolates had higher levels of resistance to aztreonam, fosfomycin, amikacin and meropenem than non-ST11 strains (7, 32). Our results also revealed that all ST11 isolates presented with an MDR phenotype, while the drug resistance rate in the non-ST11 group was generally low. Importantly, most MDR-ST11 Kp acquired virulence-associated genes and then evolved into MDR ST11 hvKp strains (18). In terms of the virulence gene spectrum, our data showed that the prevalence of rmpA2 (42.9%) and iucA (59.2%) was very high, and most of the MDR ST11 hvKp possessed iucA+rmpA2, similar to previous reports, suggesting that they might carry pVir-CR-hvKP4-like virulence plasmids (18, 33, 34). Studies have reported that these strains tested positive on the string test and presented hypervirulence in Galleria mellonella infection and human neutrophil models (18, 35). Compared with pLVPK, a 41,231 bp depletion occurred in pVir-CR-hvKP4 that resulted in the loss of the virulence genes rmpA and iro, but the iuc and rmpA2 genes were retained (18, 34). Further investigations are required to determine whether genetic deletion has a potential effect on the hypervirulence phenotype. Previous studies demonstrated that the K1 and/or K2 capsule serotypes were commonly associated with enhanced virulence (36, 37). However, none of the MDR hvKp strains possessed the K1/K2 serotype in this study. The K47 and K64 serotypes were common within the ST11 group, which is similar to previous studies (38, 39). This result suggested that other genetic elements may play key roles in virulence. A previous study showed that ST11-KL47 was replaced by ST11-KL64 as the endemic subclone (40), which should be further confirmed in a large study. ICEKp represents a key virulence element that exerts a strong influence on the pathogenicity of Kp isolates. ICEKp is responsible for scavenging iron from host transport proteins, thereby enhancing survival and replication within the host (41, 42). Lam et al. (41) reported that ybt 9 and ybt 10 were predominant within the ST11 group, and the ICEKp3 element was highly associated with ybt 8 and ybt 9. In this study, ybt9-ICEKp3 was dominant in ST11 isolates, which was similar to a previous study with longitudinal genomic surveillance (43).
As described above, ST11 Kp strains presented all kinds of virulence determinants and showed both MDR and hypervirulent phenotypes, indicating that the prognosis of the patients infected with MDR ST11 hvKp was poor. Gu et al. (18) first reported an outbreak caused by ST11 CR-hvKp in the ICU, and all the patients presented with a poor prognosis. Compared with that of non-ST11 Kp-infected patients (18/20, 90.00%), the 3-year survival rate (28/38, 73.68%) was lower in a report from Liu et al. (21). However, another study reported no significant difference in in-hospital mortality between the ST11 and non-ST11 groups (P = 0.795) (22). Remarkably, previous studies have set different endpoints that lead to contradictory conclusions. Too long or too short of a study period results in confounding factors that affect the conclusion. Some studies have indicated that 30-day mortality is a better indicator to analyze the clinical outcomes of infected patients. Of note, the previous study enrolled patients with hospital-acquired pneumonia caused by CRKP, while we enrolled patients with all kinds of Kp infections. Our study highlighted that ST11 Kp infection was significantly associated with higher 30-day mortality than non-ST11 infection. Interestingly, our subgroup analysis revealed that the mortality among patients with cKp and hvKp infections was similar in patients with ST11 strain infection. Notably, the ST11 isolates themselves, not the cKp or hvKp, might be responsible for the poor prognosis. Increased attention should be given to the prevention and control of ST11 Kp infections.
A previous study reported that CRKP appeared to be an independent risk factor for 1-year postoperative mortality in patients after kidney transplantation (44). Additionally, some studies demonstrated that CRKP infection was one of the independent risk factors for death from Kp bloodstream infection (45, 46). However, these studies did not distinguish the specific sequence types that might exert different influences on the mortality of Kp-infected patients. Li et al. observed a high percentage (20/35, 57.1%) of KPC-producing isolates among hvKp strains, in which ST11 strains were dominant (17/35, 48.6%). They found that the KPC-producing isolates were an independent predictor for 30-day mortality of Kp bacteremia patients (47). Similarly, another previous study revealed that ST11 was the most prevalent (66.7%), nearly all of the ST11 isolates were blaKPC positive, and blaKPC was an independent risk factor for 14-day mortality. It is believed that ST11 strains are the dominant Kp clone in CRKP strains in China, typically carrying blaKPC and producing carbapenemase (8, 31). Similarly, all the CRKP isolates were ST11 in our study. In this study, ST11 Kp infection was independently associated with 30-day mortality in Kp-infected patients, indicating that close attention should be given to ST11 strains, not only CRKP.
The SOFA score, a universally recognized indicator to evaluate sepsis, was significantly associated with 30-day mortality in patients with KPC-producing Kp and CRKP infection (48–51). Our study showed that patients with ST11 Kp infection had an elevated SOFA score (RR = 3.579). Moreover, multivariable linear regression revealed that ST11 Kp infection could lead to an increase in the SOFA score, indicating that ST11 Kp strains could cause more serious infections, a higher risk of sepsis and a worse prognosis than non-ST11 Kp strains.
The main limitation of our study is the selection bias and small sample size because it was a retrospective study conducted at a single center. Therefore, further prospective multicenter studies are desirable. Additionally, apart from virulence-associated genes detected by whole-genome sequencing, identification of Kp virulence in in vitro and in vivo models by using objective evidence, such as Galleria mellonella, mouse, or human neutrophil assays, is needed.
In summary, ST11 Kp infection was an independent risk factor for mortality and an elevated SOFA score. Our research demonstrated the notable conclusion that a high prevalence of ST11 Kp strains might be the main cause of high 30-day mortality and SOFA scores in Kp-infected patients. All the ST11 strains presented an MDR phenotype and exhibited great molecularly inferred virulence. For this superbug, it is of great importance to enhance clinical awareness, control and management of ST11 Kp infections.
The genome sequences in this study were deposited into the China National Center for Bioinformation under BioProject accession no. PRJCA007641. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
The studies involving human participants were reviewed and approved by Peking University Third Hospital Medical Science Research Ethics Committee. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
NS and ML contributed to the study design. NW, ZheW, and JY collected the clinical data. JZ and PY collected the laboratory data and performed the tests. CL and ZhaW analyzed and interpreted the data. ZheW and PY drafted the manuscript. CL revised the manuscript. All authors have reviewed and approved the final version of the manuscript.
This study was supported by Beijing Key Clinical Specialty Funding (010071) and by the Clinical Cohort Construction Program of Peking University Third Hospital (BYSYDL2019007).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Peking University Third Hospital for providing a laboratory for the experiments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.889020/full#supplementary-material
1. Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol Mol Biol Rev. (2016) 80:629–61. doi: 10.1128/MMBR.00078-15
2. Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. (2013) 4:107–18. doi: 10.4161/viru.22718
3. Catalan-Najera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence. (2017) 8:1111–23. doi: 10.1080/21505594.2017.1317412
4. Li W, Sun G, Yu Y, Li N, Chen M, Jin R, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. (2014) 58:225–32. doi: 10.1093/cid/cit675
5. Liu C, Shi J, Guo J. High prevalence of hypervirulent Klebsiella pneumoniae infection in the genetic background of elderly patients in two teaching hospitals in China. Infect Drug Resist. (2018) 11:1031–41. doi: 10.2147/IDR.S161075
6. Tóth A, Damjanova I, Puskás E, Jánvári L, Farkas M, Dobák A, et al. Emergence of a colistin-resistant KPC-2-producing Klebsiella pneumoniae ST258 clone in Hungary. Eur J Clin Microbiol Infect Dis. (2010) 29:765–9. doi: 10.1007/s10096-010-0921-3
7. Wang Q, Wang X, Wang J, Ouyang P, Jin C, Wang R, et al. Phenotypic and genotypic characterization of carbapenem-resistant enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012-2016). Clin Infect Dis. (2018) 67:S196–205. doi: 10.1093/cid/ciy660
8. Liu Y, Zhang X, Cai L, Zong Z. Enhanced survival of ST-11 carbapenem-resistant Klebsiella pneumoniae in the intensive care unit. Infect Control Hosp Epidemiol. (2020) 41:740–2. doi: 10.1017/ice.2020.68
9. Pomakova DK, Hsiao CB, Beanan JM, Olson R, MacDonald U, Keynan Y, et al. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: an emerging and under-recognized pathogenic variant. Eur J Clin Microbiol Infect Dis. (2012) 31:981–9. doi: 10.1007/s10096-011-1396-6
10. Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. (2012) 12:881–7. doi: 10.1016/S1473-3099(12)70205-0
11. Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. (2004) 199:697–705. doi: 10.1084/jem.20030857
12. Zhang Y, Zhao C, Wang Q, Wang X, Chen H, Li H, et al. High prevalence of hypervirulent klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. (2016) 60:6115–20. doi: 10.1128/AAC.01127-16
13. Lin YC, Lu MC, Tang HL, Liu HC, Chen CH, Liu KS, et al. Assessment of hypermucoviscosity as a virulence factor for experimental Klebsiella pneumoniae infections: comparative virulence analysis with hypermucoviscosity-negative strain. BMC Microbiol. (2011) 11:50. doi: 10.1186/1471-2180-11-50
14. Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun. (2015) 83:3325–33. doi: 10.1128/IAI.00430-15
15. Russo TA, Olson R, Fang CT, Stoesser N, Miller M, MacDonald U, et al. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J Clin Microbiol. (2018) 56:e00776–18. doi: 10.1128/JCM.00776-18
16. Chiang TT, Yang YS, Yeh KM, Chiu SK, Wang NC, Lin TY, et al. Quantification and comparison of virulence and characteristics of different variants of carbapenemase-producing klebsiella pneumoniae clinical isolates from Taiwan and the United States. J Microbiol Immunol Infect. (2016) 49:83–90. doi: 10.1016/j.jmii.2015.08.011
17. Liu Y, Liu PP, Wang LH, Wei DD, Wan LG, Zhang W. Capsular polysaccharide types and virulence-related traits of epidemic KPC-producing klebsiella pneumoniae isolates in a Chinese university hospital. Microb Drug Resist. (2017) 23:901–7. doi: 10.1089/mdr.2016.0222
18. Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. (2018) 18:37–46. doi: 10.1016/S1473-3099(17)30489-9
19. Dong N, Yang X, Zhang R, Chan EW, Chen S. Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerg Microbes Infect. (2018) 7:146. doi: 10.1038/s41426-018-0146-6
20. Yang X, Wai-Chi Chan E, Zhang R, Chen S. A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat Microbiol. (2019) 4:2039–43. doi: 10.1038/s41564-019-0566-7
21. Liu S, Wang X, Ge J, Wu X, Zhao Q, Li YM, et al. Analysis of carbapenemase-resistant genotypes of highly virulent klebsiella pneumoniae and clinical infection characteristics of different MLST types. Evid Based Complement Alternat Med. (2021) 2021:3455121. doi: 10.1155/2021/3455121
22. Zuo Y, Zhao D, Song G, Li J, Xu Y, Wang Z. Risk factors, molecular epidemiology, and outcomes of carbapenem-resistant klebsiella pneumoniae infection for hospital-acquired pneumonia: a matched case-control study in eastern China during 2015-2017. Microb Drug Resist. (2021) 27:204–11. doi: 10.1089/mdr.2020.0162
23. Wang JH, Liu YC, Lee SS, Yen MY, Chen YS, Wang JH, et al. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin Infect Dis. (1998) 26:1434–8. doi: 10.1086/516369
24. Institute CaLS. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed. Wayne, PA: CLSI supplement M100 CLSI (2020).
25. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
26. Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. (2015) 43:e15. doi: 10.1093/nar/gku1196
27. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ, IQ-TREE a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. (2015) 32:268–74. doi: 10.1093/molbev/msu300
28. Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. (2016) 7:895. doi: 10.3389/fmicb.2016.00895
29. Li H, Zhang J, Liu Y, Zheng R, Chen H, Wang X, et al. Molecular characteristics of carbapenemase-producing Enterobacteriaceae in China from 2008 to 2011: predominance of KPC-2 enzyme. Diagn Microbiol Infect Dis. (2014) 78:63–5. doi: 10.1016/j.diagmicrobio.2013.10.002
30. Kuehn BM. “Nightmare” bacteria on the rise in US hospitals, long-term care facilities. Jama. (2013) 309:1573–4. doi: 10.1001/jama.2013.2922
31. Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11 the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. (2011) 66:307–12. doi: 10.1093/jac/dkq431
32. Zhou M, Yang Q, Lomovskaya O, Sun D, Kudinha T, Xu Z, et al. In vitro activity of meropenem combined with vaborbactam against KPC-producing Enterobacteriaceae in China. J Antimicrob Chemother. (2018) 73:2789–96. doi: 10.1093/jac/dky251
33. Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. (2019) 32:e00001–19. doi: 10.1128/CMR.00001-19
34. Yang X, Dong N, Chan EW, Zhang R, Chen S. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in klebsiella pneumoniae. Trends Microbiol. (2021) 29:65–83. doi: 10.1016/j.tim.2020.04.012
35. Zhang Y, Jin L, Ouyang P, Wang Q, Wang R, Wang J, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. (2020) 75:327–36. doi: 10.1093/jac/dkz446
36. Yeh KM, Kurup A, Siu LK, Koh YL, Fung CP, Lin JC, et al. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J Clin Microbiol. (2007) 45:466–71. doi: 10.1128/JCM.01150-06
37. Wang TC, Lin JC, Chang JC, Hiaso YW, Wang CH, Chiu SK, et al. Virulence among different types of hypervirulent Klebsiella pneumoniae with multi-locus sequence type (MLST)-11, Serotype K1 or K2 strains. Gut Pathog. (2021) 13:40. doi: 10.1186/s13099-021-00439-z
38. Dong N, Zhang R, Liu L, Li R, Lin D, Chan EW, et al. Genome analysis of clinical multilocus sequence Type 11 Klebsiella pneumoniae from China. Microb Genom. (2018) 4:e000149. doi: 10.1099/mgen.0.000149
39. Liu C, Du P, Xiao N, Ji F, Russo TA, Guo J. Hypervirulent Klebsiella pneumoniae is emerging as an increasingly prevalent K. pneumoniae pathotype responsible for nosocomial and healthcare-associated infections in Beijing, China. Virulence. (2020) 11:1215–24. doi: 10.1080/21505594.2020.1809322
40. Zhou K, Xiao T, David S, Wang Q, Zhou Y, Guo L, et al. Novel subclone of carbapenem-resistant klebsiella pneumoniae sequence type 11 with enhanced virulence and transmissibility, China. Emerg Infect Dis. (2020) 26:289–97. doi: 10.3201/eid2602.190594
41. Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom. (2018) 4:e000196. doi: 10.1099/mgen.0.000196
42. Holden VI, Bachman MA. Diverging roles of bacterial siderophores during infection. Metallomics. (2015) 7:986–95. doi: 10.1039/C4MT00333K
43. Zhu C, Li C, Lai CKC, Ng R, Chau KY, Wong KT, et al. Longitudinal genomic characterization of carbapenemase-producing enterobacteriaceae (CPE) reveals changing pattern of CPE isolated in Hong Kong hospitals. Int J Antimicrob Agents. (2021) 58:106430. doi: 10.1016/j.ijantimicag.2021.106430
44. Zhang F, Zhong J, Ding H, Pan J, Yang J, Lan T, et al. Analysis of risk factors for carbapenem-resistant klebsiella pneumoniae infection and its effect on the outcome of early infection after kidney transplantation. Front Cell Infect Microbiol. (2021) 11:726282. doi: 10.3389/fcimb.2021.726282
45. Xiao T, Zhu Y, Zhang S, Wang Y, Shen P, Zhou Y, et al. A retrospective analysis of risk factors and outcomes of carbapenem-resistant klebsiella pneumoniae bacteremia in nontransplant patients. J Infect Dis. (2020) 221:S174–s83. doi: 10.1093/infdis/jiz559
46. Chang H, Wei J, Zhou W, Yan X, Cao X, Zuo L, et al. Risk factors and mortality for patients with Bloodstream infections of klebsiella pneumoniae during 2014-2018: clinical impact of carbapenem resistance in a large tertiary hospital of China. J Infect Public Health. (2020) 13:784–90. doi: 10.1016/j.jiph.2019.11.014
47. Li J, Ren J, Wang W, Wang G, Gu G, Wu X, et al. Risk factors and clinical outcomes of hypervirulent Klebsiella pneumoniae induced bloodstream infections. Eur J Clin Microbiol Infect Dis. (2018) 37:679–89. doi: 10.1007/s10096-017-3160-z
48. Falcone M, Bassetti M, Tiseo G, Giordano C, Nencini E, Russo A, et al. Time to appropriate antibiotic therapy is a predictor of outcome in patients with bloodstream infection caused by KPC-producing Klebsiella pneumoniae. Crit Care. (2020) 24:29. doi: 10.1186/s13054-020-2742-9
49. Freire MP, Abdala E, Moura ML, de Paula FJ, Spadão F, Caiaffa-Filho HH, et al. Risk factors and outcome of infections with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae in kidney transplant recipients. Infection. (2015) 43:315–23. doi: 10.1007/s15010-015-0743-4
50. Wu D, Xiao J, Ding J, Jia Y, Guo Z, Liu H, et al. Predictors of mortality and drug resistance among carbapenem-resistant enterobacteriaceae-infected pancreatic necrosis patients. Infect Dis Ther. (2021) 10:1665–76. doi: 10.1007/s40121-021-00489-5
Keywords: Klebsiella pneumoniae, ST11, risk factor, virulence, multidrug resistance
Citation: Yang P, Wu Z, Liu C, Zheng J, Wu N, Wu Z, Yi J, Lu M and Shen N (2022) Clinical Outcomes and Microbiological Characteristics of Sequence Type 11 Klebsiella pneumoniae Infection. Front. Med. 9:889020. doi: 10.3389/fmed.2022.889020
Received: 03 March 2022; Accepted: 19 April 2022;
Published: 16 May 2022.
Edited by:
Yi Liu, Shandong Provincial Hospital, ChinaReviewed by:
Farzad Badmasti, Pasteur Institute of Iran, IranCopyright © 2022 Yang, Wu, Liu, Zheng, Wu, Wu, Yi, Lu and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Lu, bHVtaW5ncHVoM0AxNjMuY29t; Ning Shen, c2hlbm5pbmdwdXRoQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.