
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 28 June 2022
Sec. Nephrology
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.881322
This article is part of the Research Topic The Role of Immune Microenvironment in the Kidney Pathogenesis View all 6 articles
Background: IgA nephropathy (IgAN) is the most common primary glomerular disease and the leading cause of the end-stage renal disease in the world. The pathogenesis of IgAN has not been well elucidated, and yet treatment is limited. High-throughput microarray has been applied for elucidating molecular biomarkers and potential mechanisms involved in IgAN. This study aimed to identify the potential key genes and therapeutics associated with IgAN using integrative bioinformatics and transcriptome-based computational drug repurposing approach.
Methods: Three datasets of mRNA expression profile were obtained from the gene expression omnibus database and differentially expressed genes (DEGs) between IgAN glomeruli and normal tissue were identified by integrated analysis. Gene ontology and pathway enrichment analyses of the DEGs were performed by R software, and protein-protein interaction networks were constructed using the STRING online search tool. External dataset and immunohistochemical assessment of kidney biopsy specimens were used for hub gene validation. Potential compounds for IgAN therapy were obtained by Connectivity Map (CMap) analysis and preliminarily verified in vitro. Stimulated human mesangial cells were collected for cell proliferation and cell cycle analysis using cell counting kit 8 and flow cytometry, respectively.
Results: 134 DEGs genes were differentially expressed across kidney transcriptomic data from IgAN patients and healthy living donors. Enrichment analysis showed that the glomerular compartments underwent a wide range of interesting pathological changes during kidney injury, focused on anion transmembrane transporter activity and protein digestion and absorption mostly. Hub genes (ITGB2, FCER1G, CSF1R) were identified and verified to be significantly upregulated in IgAN patients, and associated with severity of renal lesions. Computational drug repurposing with the CMap identified tetrandrine as a candidate treatment to reverse IgAN hub gene expression. Tetrandrine administration significantly reversed mesangial cell proliferation and cell cycle transition.
Conclusion: The identification of DEGs and related therapeutic strategies of IgAN through this integrated bioinformatics analysis provides a valuable resource of therapeutic targets and agents of IgAN. Especially, our findings suggest that tetrandrine might be beneficial for IgAN, which deserves future research.
IgA nephropathy (IgAN) is the most common primary glomerular disease, with an incidence of about 25 patients per 1 million people in the population and the highest incidence in the Asia Pacific region (1, 2). It is a frequent cause of end-stage renal disease (ESRD), progressive glomerular and interstitial sclerosis leads to end-stage kidney failure in 20–40% of patients within 20 years after diagnosis, which brings heavy disease and economic burden to the individual and the whole society (3). A kidney biopsy is required for IgAN diagnosis which is characterized by mesangial immune deposits of IgA1/galactose-deficient IgA1 (Gd-IgA1) with C3 and occasionally IgG or IgM (4, 5). To date, multiethnic genome-wide association studies involving over 20,000 individuals have identified risk loci predisposing to IgAN, highlighting the importance of innate and adaptive immunity in this disorder (6). Published gene chips and high-throughput sequencing also give clues to the pathogenesis of IgAN, while there is still a lack of clear understanding of the pathogenesis of IgAN and short of effective therapeutics for disease progression. In-depth exploration of the public datasets, combined with laboratory validation, may reveal associated risk genes.
Connectivity Map (CMap) is one of the first publicly available tools for drug repurposing, utilizing transcriptomics to form unbiased connections between diseases, genes, and drugs (7). These connections can identify drugs that reverse large numbers of genes disturbed in disease (8). In this study based on bioinformatics, three IgAN datasets were integrated from the GEO database and performed differentially expressed genes (DEGs), gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG) pathway analyses. The protein-protein interaction (PPI) network was performed, and hub genes with high connectivity in network that are expected to play an important role in understanding the biological mechanisms of IgAN were identified, and were further validated with external and laboratory data. We also interrogated the CMap with disease-related DEGs and identified tetrandrine as a reversal agent, and examined the impact of tetrandrine on human mesangial cells. Overall, our output suggest potential key genes and novel therapeutic agents for IgAN.
The microarray data were downloaded from the gene expression omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) using the keywords “IgA nephropathy,” “Homo sapiens” and “Expression profiling by array” were included in the next round of screening. The gene expression microarray datasets GSE104948 (9), GSE93798 (10) GSE37463 (11) were selected and downloaded for DEG analysis. Probes were matched to the gene symbols using the annotation files provided by the manufacturer. DEGs were identified by sva/limma R package (12) (version 4.1.0) after consolidation and batch normalization, with threshold |log2FC| >1 and adjusted p < 0.05. Heatmap (https://CRAN.R-project.org/package=pheatmap) and volcano plot were subsequently performed using R software to visualize the DEGs.
GO enrichment and KEGG pathway analyses were performed to explore the potential biological processes and utilities, cellular components, and molecular functions of DEGs using colorspace (13) and stringi (https://stringi.gagolewski.com/) R packages. Significantly relevant signal pathways were identified via clusterProfiler package (14) in R software with p <0.05 and q < 0.05. The STRING database (https://string-db.org) (15) was used to explore potential protein-protein interactions (PPI) interactions between DEGs, and a network interaction matrix was built. An interaction with a combined score > 0.7 was set as the cut-off value. Densely connected clusters in the PPI network were visualized by R.
To validate the expression of hub genes in other glomerulonephritis types and IgAN, we use GSE116626 (16) as an independent validate cohort, comprising 52 IgAN patients, 22 non-IgAN glomerulonephrites (non-IgAN GN), and 7 kidney living donors (LD). The relative mRNA expression levels of hub genes in each patient were extracted from the raw data, which were then analyzed by Graphpad Prism 6. Statistical differences were analyzed by Student's t-test, and p < 0.05 was considered statistically significant. Analyses were conducted with GraphPad Prism 6.0 statistical software.
Formaldehyde-fixed and paraffin-embedded tissue sections (4 mm thick) were used for immunohistochemistry as previously described (17). After dehydration, slides in citrate solution were subjected to a microwave antigen retrieval process. Primary antibodies were employed against the integrin beta 2 (ITGB2) (Santa Cruz Biotechnology, sc-8420), fc fragment of IgE receptor Ig (FCER1G) (Santa Cruz Biotechnology, sc-390222), and colony-stimulating factor 1 receptor (CSF1R) (Santa Cruz Biotechnology, sc-46662), periodic acid-Schiff (PAS) was used instead of primary antibodies as negative controls for all staining. Finally, horseradish peroxidase (HRP)-conjugated polymer (Abcam, USA) was used for the visualized detection under light microscopy (Nikon Tokyo, Japan). OD values were analyzed by ImageJ software (National Institute of Health, USA). The use of renal specimens of IgAN patients was approved by the Ethics Committee of the Second Xiangya Hospital of Central South University (2019SNK1222000) according to the Declaration of Helsinki.
The CMap (https://portals.broadinstitute.org/cmap) is an open resource that links disease, genes, and drugs by similar or opposite gene expression profiles (18), and was used to explore potential drugs targeting IgAN. Upregulation and the downregulation DEGs were converted to probe sets in the chip type to query the CMap. A list of CMap instances predicted to reverse IgAN DEGs was obtained, with connectivity score and p-value.
The human mesangial cells (HMCs) collected from (CBR130735, Cellbio, China) were cultivated in Dulbecco's Modified Eagle's Medium (DMEM)/F-12 medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS) under 5% CO2 at 37 °C. Monomeric human IgA1 (Abcam, ab91020) was heated and aggregated at 65°C for 150 min on a dry plate heater to obtain aggregated IgA1 (aIgA1) as previously described (19). After cooling at room temperature, the proteins were centrifuged at 11,000g for 5 min to remove insoluble precipitant. The supernatants were then used as heat-aggregated IgA. Mesangial cells were treated with or without IgA aggregates at a final concentration of 25 μg/ml for 24 h. The dose of IgA aggregates was selected based on our previous study in mesangial cells (17). Tetrandrine (10 mM in 1 ml dimethyl sulfoxide) was purchased from ApexBio Technology (Houston, USA). Cells were incubated with aIgA1 alone or with a combination of tetrandrine (0–10 μM) concentrations for 24 h.
Cell proliferation was accessed by cell counting kit-8 (CCK8) assay (Dojindo, Japan) following the manufacturer's recommended procedure. 10 μl of CCK8 was added to treated cells in 96-wells and incubated at 37 °C. The absorbance was detected at 450 nm using spectrophotometer.
Cell cycle was measured by cell cycle and apoptosis kit (Wellbiology, China), and the treated cells were fixed overnight with 70% cold ethanol at 4°C, then stained with a mixture of propidium iodide and RNase A at 37°C for 30 min. The distribution of the cell cycle phase was measured by FlowJo software.
This study was conducted according to the flow chart shown in Figure 1. The microarray data archives for glomeruli of IgAN were searched and downloaded from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) using the “IgA nephropathy,” with “Homo sapiens” and “Expression profiling by array” filters in the next round of screening. Three relevant microarray expression profiles (GSE104948, GSE93798, GSE37463) were selected between IgAN and normal glomeruli compartment. A total of 144 human glomerular compartment tissue samples were screened, of which 74 were IgAN patients and 70 were healthy donors. Details of the GEO datasets in this study were shown in Table 1. A total of 134 DEGs were identified after consolidation and batch normalization (Figure 2A; Supplementary Table S1), including 54 upregulated and 80 downregulated genes. The volcano plot (Figure 2B) showed the distribution of DEGs between IgAN and normal controls.
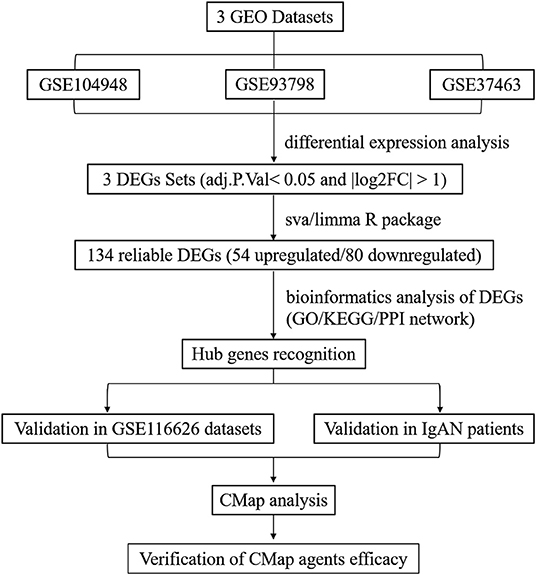
Figure 1. Flowchart presenting the process of identification of potential hub genes and therapeutic agents in IgAN.
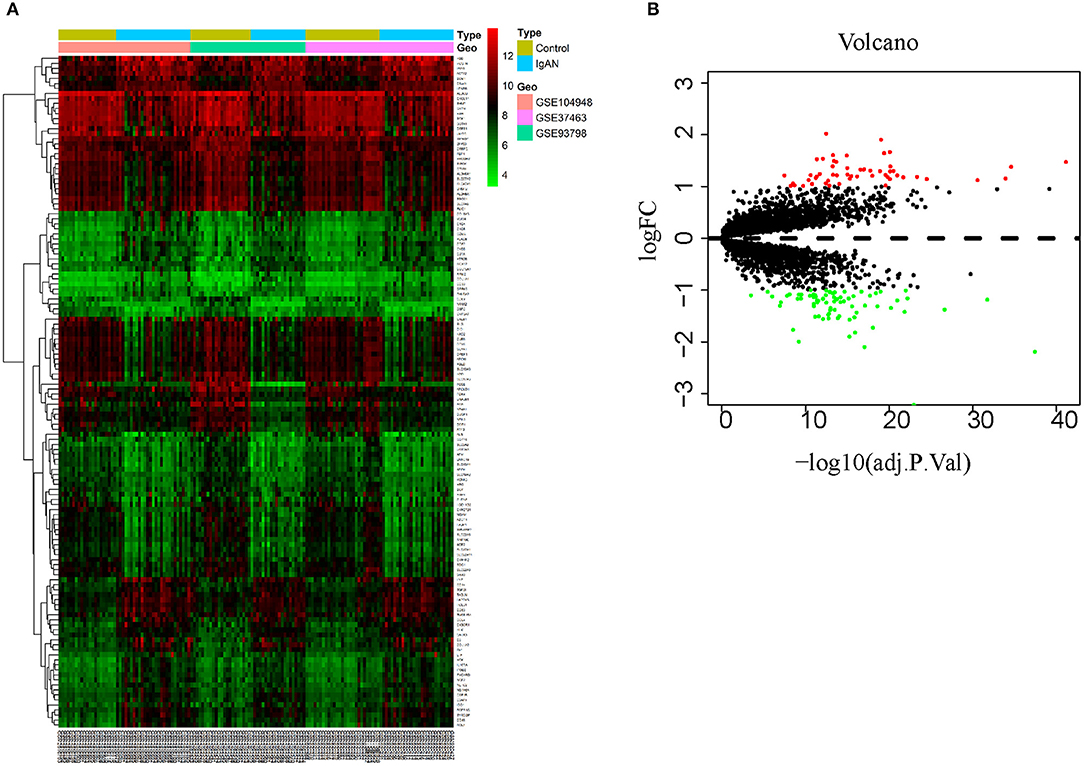
Figure 2. Identification of DEGs between IgAN and normal tissues. (A) Heat map of 134 DEGs identified by integrated analysis of the GEO datasets. Red areas represent highly expressed genes and green areas represent lowly expressed genes in glomeruli from IgAN subjects compared with healthy living donors. (B) Volcano plot analysis identifies DEGs. Red dots represent 54 upregulated genes and green dots represent 80 downregulated genes in glomeruli from IgAN subjects compared with healthy living donors.
To identify comprehensive information on biological processes, cellular components, and molecular functions gene function, GO and KEGG pathway analyses of DEGs were performed. The top twelve GO terms with the highest degree of enrichment were shown in Figure 3A. GO terms mainly concentrated on anion transmembrane transporter activity, extracellular matrix structural constituent, and organic anion transmembrane transporter activity. Figure 3B presented seven enriched KEGG pathways of the DEGs, protein digestion and absorption, pertussis, complement, and coagulation cascades were identified as the top three significantly relevant signal pathways.
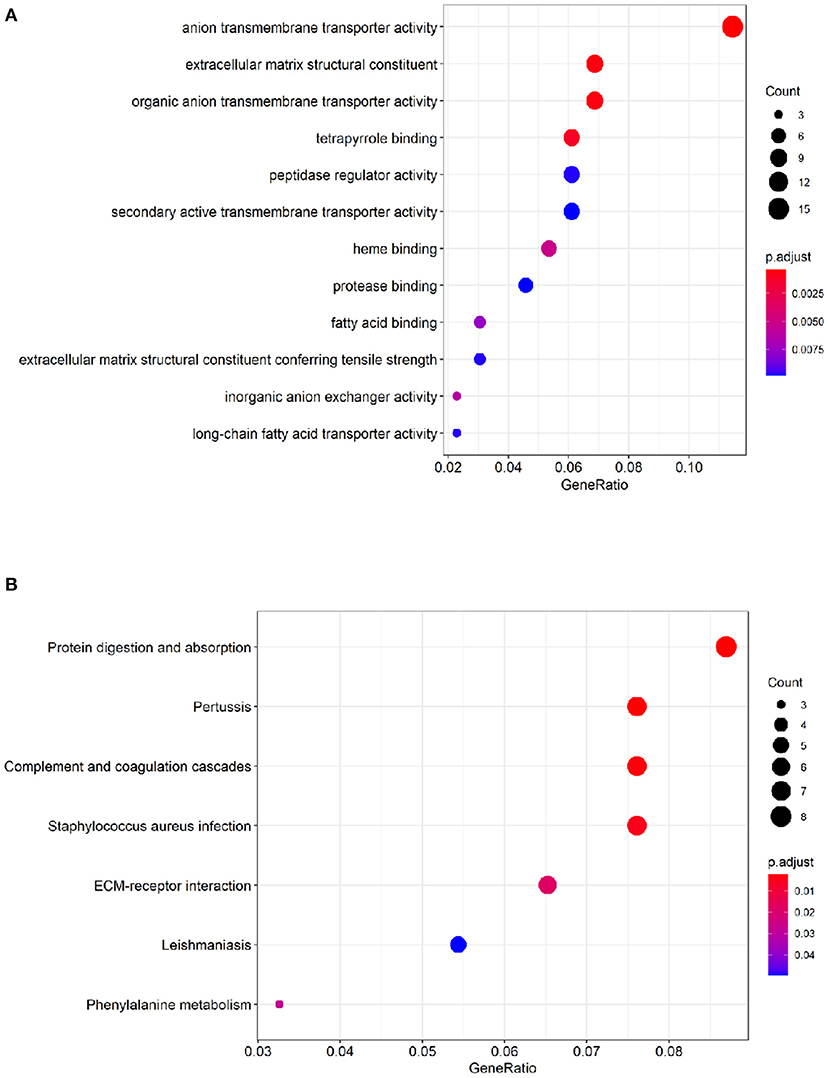
Figure 3. Functional enrichment analysis of the DEGs. (A) Top 12 GO terms of the DEGs. (B) 7 enriched KEGG pathways of the DEGs. The color depth of the nodes refers to the adj P-value. The size of the nodes refers to the number of genes.
The STRING database (https://string-db.org) was used to explore potential interactions between DEGs. High confidence score>0.7 serves as the cut-off point for the construction of protein-protein interaction (PPI) network, visualization of the PPI network was shown in Figure 4A. A total of 91 nodes and 194 interactions were calculated by R software, the top 30 candidate hub genes which may play a central role in this network were identified in Figure 4B. Five genes with the highest score were obtained as ITGB2, FCER1G, complement C3a receptor 1 (C3AR1), CSF1R, transmembrane immune signaling adaptor TYROBP (TYROBP).
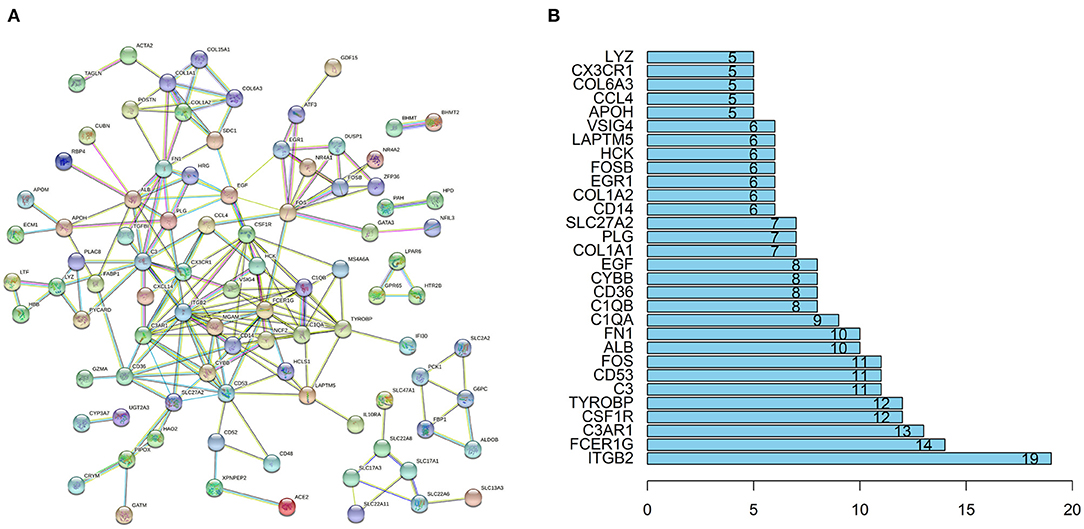
Figure 4. PPI network and the hub genes of DEGs. (A) The PPI network of DEGs was constructed using String with high confidence. (B) The top 30 key genes were obtained from the PPI network. The numbers on the bar represent the number of related nodes for each gene.
To verify the reliability of the data and the link between key genes and IgAN, an external dataset GSE116626 (16), which was uploaded by Cox et al. in 2018 and updated in 2020, was used to validate the top 5 hub genes. ITGB2, FCER1G, C3AR1, CSF1R, TYROBP expression profiles were screened in archival FFPE biopsy samples of 52 IgAN patients, 22 non-IgAN glomerulonephrites (non-IgAN GN), and 7 kidney living donors (LD). The transcriptional levels of ITGB2, FCER1G, and CSF1R were significant higher in IgAN compared to either healthy donors or non-IgAN (Figure 5A), while C3AR1 and TYROBP transcriptional levels were not significant (data not shown). We further investigated the ITGB2, FCER1G, and CSF1R transcriptional and protein levels in IgAN with different composite renal lesions, patients were stratified into 4 groups based on the E, C,and T (but not M or S) scores: minimal (E0, C0, T0); active (E1 and/or C1, C2, T0); chronic (T1 or T2, E0, C0);mixed group composed of active and chronic lesions (E1 and/or C1, C2, T1 or T2) as previously described (16). Compared with minimal lesions or active lesions, these three genes mRNA expression were significantly increased in the mixed lesions group. Also, FCER1G mRNA level was significantly upregulated in the mixed lesion group compared to chronic lesions (Figure 5B). Moreover, biopsy specimens of minimal change disease (MCD) and IgAN were collected in our hospital. Through immunohistochemistry, ITGB2, FCER1G, and CSF1R were significantly elevated in IgAN patients with different renal pathology compared with MCD (Figure 6).
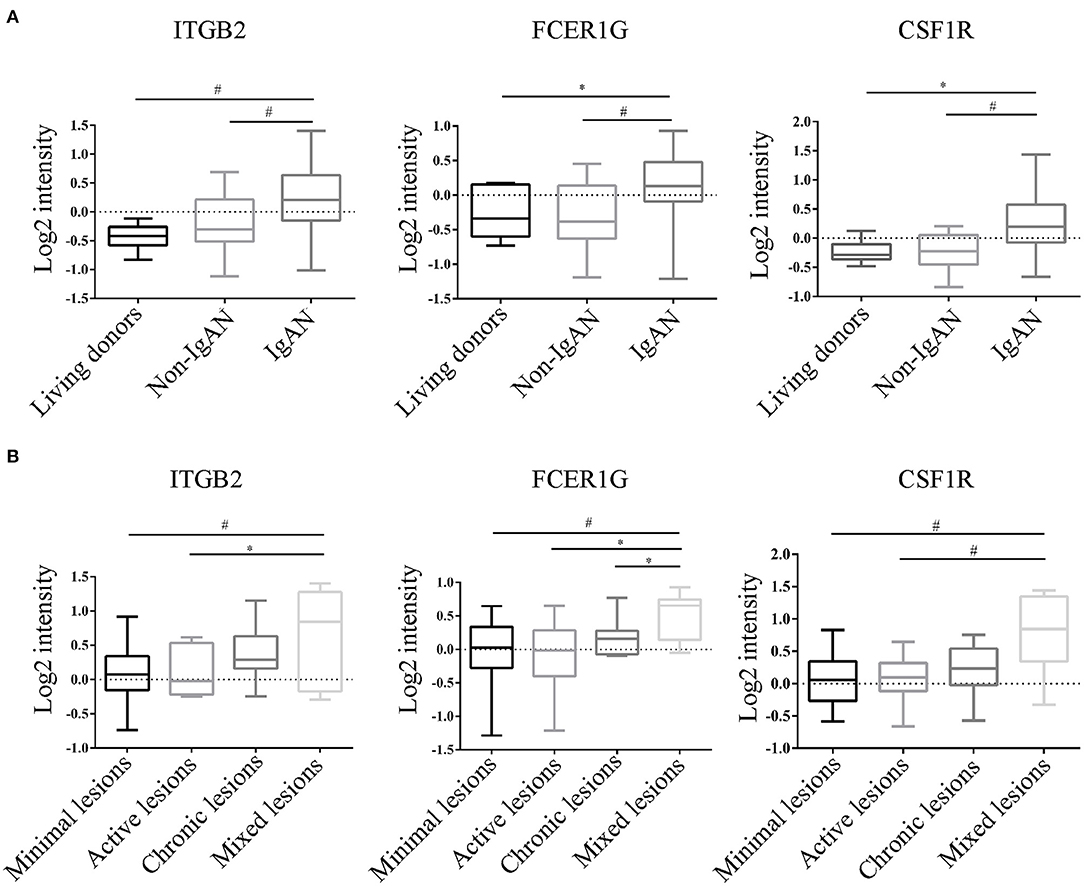
Figure 5. Transcriptional levels were analyzed in kidney biopsy specimens of living donors, non-IgAN, and IgAN patients in the GSE116626 dataset. (A) Levels of ITGB2, FCER1G, CSF1R in living donors, non-IgAN, and IgAN patients. (B) Levels of ITGB2, FCER1G, CSF1R with minimal, active, chronic, and mixed renal lesions in IgAN. *p < 0.05, #p < 0.01.
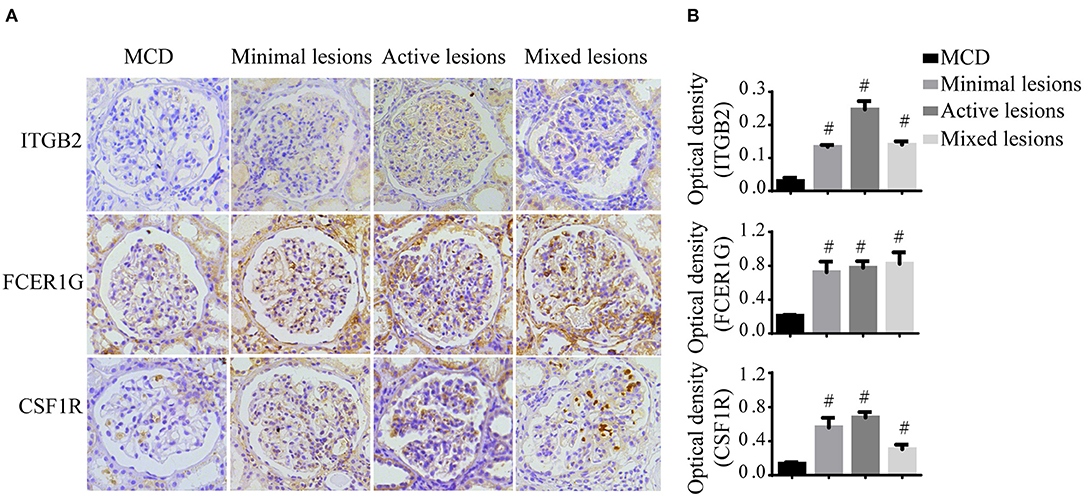
Figure 6. Immunohistochemistry of ITGB2, FCER1G, and CSF1R in IgAN and MCD kidneys. (A) Representative images of ITGB2, FCER1G, and CSF1R glomerular immunohistostaining from MCD and IgAN patients with the minimal lesion, active lesion, mixed lesions. (B) The average optical density of ITGB2, FCER1G, and CSF1R was analyzed by ImageJ software. Data were presented as mean ± SD. Magnification 40×. #p < 0.01 vs. MCD. MCD, minimal change disease.
To screen potential therapeutic small molecular compounds for IgAN, we performed CMap (http://portals.broadinstitute.org/cmap/) analysis and obtained a list of drug candidates predicted to reverse altered expression of DEGs (Figure 7A). Tetrandrine, an antirheumatic and analgesic agent, was the top-ranked candidate with the highest negative connectivity score (Figure 7B) and was selected for subsequent validation experiments.
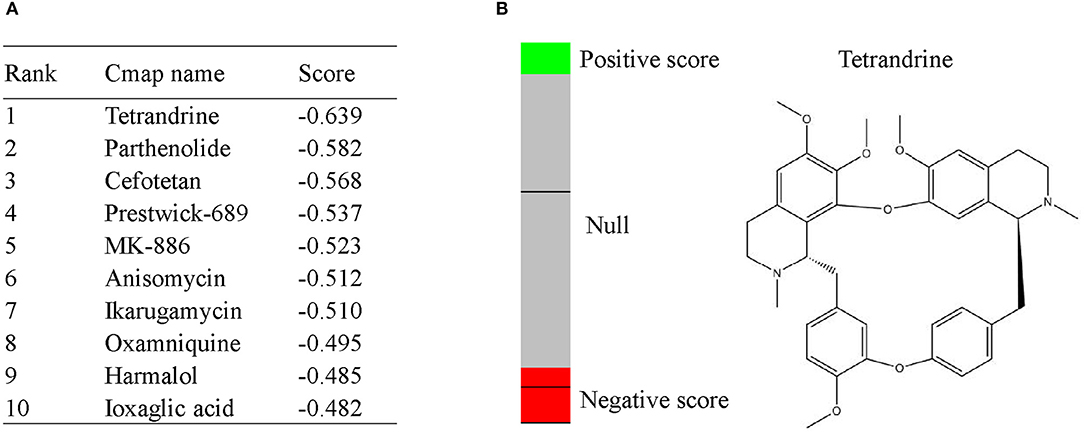
Figure 7. CMap analysis indicated potential treatment options for IgAN. (A) Top10 CMap compounds predicted to reverse altered DEGs expression. (B) The bar graph represents the connectivity score data for tetrandrine. The black horizontal lines represent each instance performed with the respective compound. Instances in the red area indicate negative correlation scores, and instances in the green area indicate positive ones. No correlation could be detected for instances in the gray area. The chemical structure of tetrandrine was shown.
Considering that mesangial cell proliferation is a prominent pathological feature of IgAN, to further investigate the renal protective effect of tetrandrine, we focused its role on the proliferation of human mesangial cells (HMCs). 25 μg/ml aIgA1-stimulated HMCs was used to mimic the IgAN HMC model in vitro, as previously described (17). The CCK8 assay showed that tetrandrine at concentrations of 2.5–10 μM for 24 h had strong inhibitory effects on aIgA1-induced mesangial proliferation, and did not alter the viability of HMCs (Figure 8A). The representative light microscopy was shown in Figure 8B. To further confirm the effect of tetrandrine on the proliferation of HMCs, cell cycle analysis was carried out by flow cytometry. After 24 h incubation with aIgA1, the proportion of cells in S and G2/M phases increased, while 5 μM tetrandrine retained more cells in G0/G1 phase (Figure 8C).
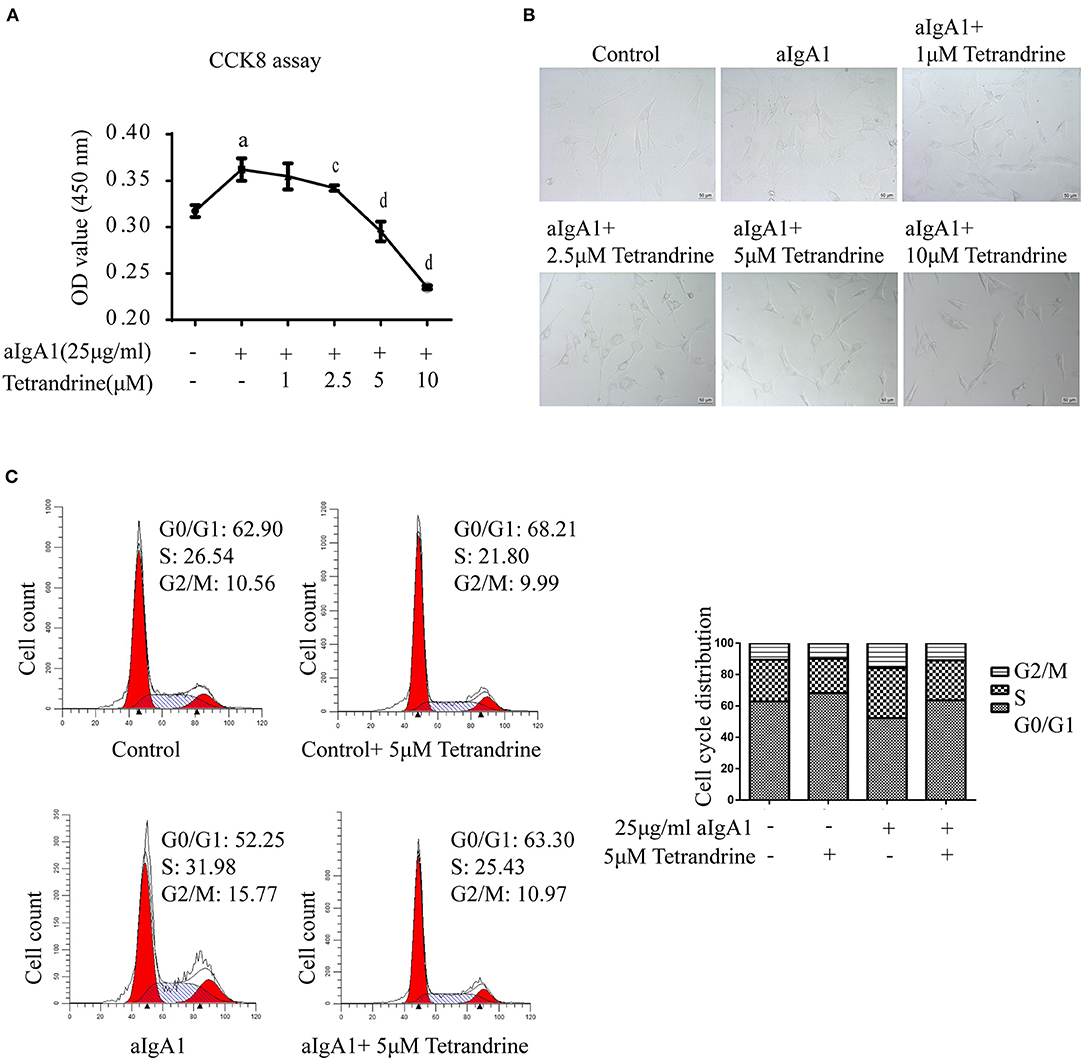
Figure 8. Effect of tetrandrine on the proliferation of HMCs. (A) Cell proliferation of aIgA1-induced HMCs under 1–10 μM tetrandrine treatment was detected by CCK8 assay. Data were presented as mean ± SD (n = 3 in each group). (B) Representative light microscopy images were presented. (C) The cell cycle of HMCs treated with 25 μg/ml aIgA1 and 5 μM tetrandrine was detected by flow cytometry. ap < 0.05 vs. control; cp < 0.05, dp < 0.01 vs. aIgA1 group.
IgAN was recognized as a separate category of glomerulonephritis when Berger & Hinglais applied the immunofluorescence staining technique to renal biopsy in 1968. Much progress has occurred in understanding immunologic and biochemical defects underlying IgAN, but there is a paucity of effective treatments for IgAN. Here, we presented a transcriptome-based drug repurposing approach identifying potential treatments for IgAN.
First, we described IgAN DEGs using combined analyses of three glomeruli microarray data, since glomerular injury is better associated with a progressive decline of kidney function than tubule cell activation and injury. Interestingly, extracellular matrix structural constituent and complement and coagulation cascades were observed in functional enrichment analyses, represented genes were fibronectin (FN1) which was revealed to aid in IgA deposition (20), and C3, a verified complement colocalized with IgA deposition (21, 22). Our previous report also showed that coagulation parameters prothrombin time (PT) and the activated partial thromboplastin time (APTT) were associated with the prognosis of IgAN (23).
Second, we screened three hub genes-ITGB2, FCER1G, and CSF1R and validated them in an additional published dataset and our patients. These genes were reported to be enhanced in immune cells (macrophage) and demonstrated a prominent inflammatory signature, reflecting increased infiltrating local inflammation in IgAN. ITGB2 encodes the integrin beta chain, which combines with multiple different alpha chains to form different integrin heterodimers. It participates in cell surface-mediated signaling as well as immune response, and defects in this gene cause leukocyte adhesion deficiency (24). ITGB2 has also been reported to be negatively correlated with eGFR in patients with CKD (25) and involved in cell adhesion and extracellular matrix remodeling in renal cancer (26). FCER1G is located on chromosome 1q23.3 and encodes the cytoplasmic Fc receptor gamma chain (FcRg) of immunoglobulin. It is a signal-transducing subunit containing an immunoreceptor tyrosine-based activation motif (ITAM) that transduces activation signals from various immunoreceptors that play a critical role in allergic reactions, immune cell activation, and chronic inflammatory programs (27, 28). It has been illustrated that FCER1G participated in various kidney diseases, such as diabetic kidney disease and clear cell renal cell carcinoma (29, 30). Recently FCER1G was identified as a marker of inflammatory dendritic cells and abundantly expressed in the periglomerular region of the lupus nephritis kidney (31). CSF1R encodes a tyrosine-protein kinase that acts as a cell-surface receptor for CSF1 and IL-34 and regulates the production and differentiation of most circulating/tissue-resident macrophages. Activation of CSF1R in the kidney could induce monocyte recruitment from the blood and proliferation and survival of tissue-resident macrophages, as well as skew macrophages toward an M2 phenotype (32), CSF1R knockout mice significantly reduced macrophages (33). Another bioinformatics analysis has also predicted that ITGB2, FCER1G and CSF1R were involved in the development and progression of IgAN (34), but our study is the first to verify this in biopsy specimens and explore related therapeutics. In addition, our results indicated that ITGB2, FCER1G and CSF1R were highly expressed in IgAN glomeruli and in mesangial cells of frozen IgAN sections, which may contribute to recruit monocytes/macrophages and further amplifying inflammatory responses. It has been reported that galactose deficient-IgA1 isolated from IgAN patient serum could initiate podocyte macrophage transdifferentiation (35), we speculate that mesangial cells may also have similar transformation characteristics and need further exploration.
Third, we applied our disease characteristics to CMap, which has the advantage that most of the recorded compounds have already been approved by regulators, so the benefits demonstrated in experimental models could be quickly translated into human disease (7, 36). After screening more than 1,000 compounds through CMap pattern-matching algorithms linked IgAN hub genes to drugs that could likely reverse them based on empirical evidence, tetrandrine was identified as the top candidate. With molecular formula C38H42N2O6 and weight 622.3, tetrandrine has been used in the treatment of T cell lymphoma, rheumatism, and silicosis in China for several decades (37, 38). Tetrandrine has been reported to possess extensive pharmacological properties including anticancer (39), anti-inflammation (40, 41), immunomodulatory effects (42), and regulates several important biological processes in the pathogenesis of kidney diseases. Tetrandrine could alleviate podocyte injury induced by TRPC6 overexpression (43), decrease 24 h urine protein and glomerular cell proliferation in the membranous glomerulopathy (44), and restrain diabetic process and renal damage in diabetic nephropathy (45). Early studies have shown that tetrandrine could inhibit the activation of mesangial cells by down-regulating the ERK/ NF-κB signaling pathway (46), suggesting a protective effect on the pathological proliferated mesangial cells mediated renal damage. Here, consistent with other reports of cell cycle regulation (47–50), tetrandrine was observed to significantly inhibit cell cycle transition at G1/S boundary as well as aIgA1-induced mesangial cells proliferation. Further comprehensive investigation of the therapeutic effect and mechanism of tetrandrine on IgA nephropathy is warranted.
In conclusion, we identified differential expression genes between kidneys of IgAN and healthy controls, and validated the hub genes ITGB2, FCER1G, and CSF1R closely associated with IgAN renal damage. CMap analysis provided tetrandrine as a potential therapeutic compound to reverse the disease signature, possibly due in part to its renal protective effects by inhibiting cell cycle transition and proliferation of HMC. Together, our outputs could empower the novel potential targets and treatment to IgAN therapy.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by Ethics Committee of the Second Xiangya Hospital of Central South University. The patients/participants provided their written informed consent to participate in this study.
MX contributed to study design, experiments, and draft the manuscript. DL, HYL, and LP contributed to data collection and statistical analysis/interpretation. DY contributed to pathological experimental techniques. CT, GC, and YL contributed to review and editing. HL gave the final approval for the article to be published. All authors have read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (82070737), Hunan Provincial Natural Science Foundation of China (2021JJ30940), Hunan Provincial Innovation Foundation for Postgraduate (CX20210368), and Fundamental Research Funds for the Central Universities of Central South University (2021zzts0366).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We sincerely appreciate the time and effort of all who contributed to this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.881322/full#supplementary-material
1. McGrogan A, Franssen C, de Vries C. The incidence of primary glomerulonephritis worldwide: a systematic review of the literature. Nephrol Dial Transplant. (2011) 26:414–30. doi: 10.1093/ndt/gfq665
2. Lai KN, Tang SC, Schena FP, Novak J, Tomino Y, Fogo AB, et al. IgA nephropathy. Nat Rev Dis Primers. (2016) 2:16001. doi: 10.1038/nrdp.2016.1
3. Barbour S, Coppo R, Zhang H, Liu Z, Suzuki Y, Matsuzaki K, et al. Evaluating a new international risk-prediction tool in IgA nephropathy. JAMA Intern Med. (2019) 179:942–52. doi: 10.1001/jamainternmed.2019.0600
4. Jennette J. The immunohistology of IgA nephropathy. Am J Kidney Dis. (1988) 12:348–52. doi: 10.1016/S0272-6386(88)80022-2
5. Suzuki H, Yasutake J, Makita Y, Tanbo Y, Yamasaki K, Sofue T, et al. IgA nephropathy and IgA vasculitis with nephritis have a shared feature involving galactose-deficient IgA1-oriented pathogenesis. Kidney Int. (2018) 93:700–5. doi: 10.1016/j.kint.2017.10.019
6. Kiryluk K, Li Y, Moldoveanu Z, Suzuki H, Reily C, Hou P, et al. GWAS for serum galactose-deficient IgA1 implicates critical genes of the O-glycosylation pathway. PLoS Genet. (2017) 13:e1006609. doi: 10.1371/journal.pgen.1006609
7. Lamb J, Crawford E, Peck D, Modell J, Blat I, Wrobel M, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. (2006) 313:1929–35. doi: 10.1126/science.1132939
8. Williams V, Konvalinka A, Song X, Zhou X, John R, Pei Y, et al. Connectivity mapping of a chronic kidney disease progression signature identified lysine deacetylases as novel therapeutic targets. Kidney Int. (2020) 98:116–32. doi: 10.1016/j.kint.2020.01.029
9. Grayson P, Eddy S, Taroni J, Lightfoot Y, Mariani L, Parikh H, et al. Metabolic pathways and immunometabolism in rare kidney diseases. Ann Rheum Dis. (2018) 77:1226–33. doi: 10.1136/annrheumdis-2017-212935
10. Liu P, Lassén E, Nair V, Berthier C, Suguro M, Sihlbom C, et al. Transcriptomic and proteomic profiling provides insight into mesangial cell function in IgA nephropathy. J Am Soc Nephrol. (2017) 28:2961–72. doi: 10.1681/ASN.2016101103
11. Berthier C, Bethunaickan R, Gonzalez-Rivera T, Nair V, Ramanujam M, Zhang W, et al. Cross-species transcriptional network analysis defines shared inflammatory responses in murine and human lupus nephritis. J Immunol. (2012) 189:988–1001. doi: 10.4049/jimmunol.1103031
12. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47. doi: 10.1093/nar/gkv007
13. Zeileis A, Fisher JC, Hornik K, Ihaka R, McWhite CD, Murrell P, et al. colorspace: a toolbox for manipulating and assessing colors and palettes. J Stat Softw. (2020 96:1–49. doi: 10.18637/jss.v096.i01
14. Yu G, Wang L, Han Y, He Q. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. (2012) 16:284–7. doi: 10.1089/omi.2011.0118
15. Szklarczyk D, Gable A, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. (2019) 47:D607–13. doi: 10.1093/nar/gky1131
16. Cox SN, Chiurlia S, Divella C, Rossini M, Serino G, Bonomini M, et al. Formalin-fixed paraffin-embedded renal biopsy tissues: an underexploited biospecimen resource for gene expression profiling in IgA nephropathy. Sci Rep. (2020) 10:15164. doi: 10.1038/s41598-020-72026-2
17. Xia M, Liu D, Tang X, Liu Y, Liu H, Liu Y, et al. Dihydroartemisinin inhibits the proliferation of IgAN mesangial cells through the mTOR signaling pathway. Int Immunopharmacol. (2020) 80:106125. doi: 10.1016/j.intimp.2019.106125
18. Subramanian A, Narayan R, Corsello S, Peck D, Natoli T, Lu X, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. (2017) 171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049
19. Hyun YY, Kim IO, Kim MH, Nam DH, Lee MH, Kim JE, et al. Adipose-derived stem cells improve renal function in a mouse model of IgA nephropathy. Cell Transplant. (2012) 21:2425–39. doi: 10.3727/096368912X639008
20. Zheng Y, Lu P, Deng Y, Wen L, Wang Y, Ma X, et al. Single-cell transcriptomics reveal immune mechanisms of the onset and progression of IgA nephropathy. Cell Rep. (2020) 33:108525. doi: 10.1016/j.celrep.2020.108525
21. Rizk D, Maillard N, Julian B, Knoppova B, Green T, Novak J, et al. The emerging role of complement proteins as a target for therapy of IgA nephropathy. Front Immunol. (2019) 10:504. doi: 10.3389/fimmu.2019.00504
22. Chen P, Yu G, Zhang X, Xie X, Wang J, Shi S, et al. Plasma galactose-deficient IgA1 and C3 and CKD progression in IgA nephropathy. Clin J Am Soc Nephrol. (2019) 14:1458–65. doi: 10.2215/CJN.13711118
23. Xia M, Liu D, Peng L, Li Y, Liu H, Wu L, et al. Coagulation parameters are associated with the prognosis of immunoglobulin a nephropathy: a retrospective study. BMC Nephrol. (2020) 21:447. doi: 10.1186/s12882-020-02111-1
24. Yaz I, Ozbek B, Bildik H, Tan C, Oskay Halacli S, Soyak Aytekin E, et al. Clinical and laboratory findings in patients with leukocyte adhesion deficiency type I: a multicenter study in turkey. Clin Exp Immunol. (2021) 206:47–55. doi: 10.1111/cei.13645
25. Wang W, Shen J, Qi C, Pu J, Chen H, Zuo Z. The key candidate genes in tubulointerstitial injury of chronic kidney diseases patients as determined by bioinformatic analysis. Cell Biochem Funct. (2020) 38:761–72. doi: 10.1002/cbf.3545
26. Boguslawska J, Kedzierska H, Poplawski P, Rybicka B, Tanski Z, Piekielko-Witkowska A. Expression of genes involved in cellular adhesion and extracellular matrix remodeling correlates with poor survival of patients with renal cancer. J Urol. (2016) 195:1892–902. doi: 10.1016/j.juro.2015.11.050
27. Andreu P, Johansson M, Affara N, Pucci F, Tan T, Junankar S, et al. FcRgamma activation regulates inflammation-associated squamous carcinogenesis. Cancer Cell. (2010) 17:121–34. doi: 10.1016/j.ccr.2009.12.019
28. Liang Y, Wang P, Zhao M, Liang G, Yin H, Zhang G, et al. Demethylation of the FCER1G promoter leads to FcεRI overexpression on monocytes of patients with atopic dermatitis. Allergy. (2012) 67:424–30. doi: 10.1111/j.1398-9995.2011.02760.x
29. Liu S, Wang C, Yang H, Zhu T, Jiang H, Chen J. Weighted gene co-expression network analysis identifies FCER1G as a key gene associated with diabetic kidney disease. Ann Transl Med. (2020) 8:1427. doi: 10.21037/atm-20-1087
30. Chen L, Yuan L, Wang Y, Wang G, Zhu Y, Cao R, et al. Co-expression network analysis identified FCER1G in association with progression and prognosis in human clear cell renal cell carcinoma. Int J Biol Sci. (2017) 13:1361–72. doi: 10.7150/ijbs.21657
31. Parikh S, Malvar A, Shapiro J, Turman J, Song H, Alberton V, et al. A novel inflammatory dendritic cell that is abundant and contiguous to T cells in the kidneys of patients with lupus nephritis. Front Immunol. (2021) 12:621039. doi: 10.3389/fimmu.2021.621039
32. Perry H, Okusa M. Driving change: kidney proximal tubule CSF-1 polarizes macrophages. Kidney Int. (2015) 88:1219–21. doi: 10.1038/ki.2015.324
33. Lin W, Xu D, Austin C, Caplazi P, Senger K, Sun Y, et al. Function of CSF1 and IL34 in macrophage homeostasis, inflammation, and cancer. Front Immunol. (2019) 10:2019. doi: 10.3389/fimmu.2019.02019
34. Chen X, Sun M. Identification of key genes, pathways and potential therapeutic agents for IgA nephropathy using an integrated bioinformatics analysis. J Renin Angiotensin Aldosterone Syst. (2020) 21:1470320320919635. doi: 10.1177/1470320320919635
35. Peng W, Pei G, Tang Y, Tan L, Qin W. IgA1 deposition may induce NLRP3 expression and macrophage transdifferentiation of podocyte in IgA nephropathy. J Transl Med. (2019) 17:406. doi: 10.1186/s12967-019-02157-2
36. Lamb J. The connectivity map: a new tool for biomedical research. Nat Rev Cancer. (2007) 7:54–60. doi: 10.1038/nrc2044
37. Xu W, Chen S, Wang X, Tanaka S, Onda K, Sugiyama K, et al. Molecular mechanisms and therapeutic implications of tetrandrine and cepharanthine in T cell acute lymphoblastic leukemia and autoimmune diseases. Pharmacol Ther. (2021) 217:107659. doi: 10.1016/j.pharmthera.2020.107659
38. Song M, Wang J, Sun Y, Han Z, Zhou Y, Liu Y, et al. Tetrandrine alleviates silicosis by inhibiting canonical and non-canonical NLRP3 inflammasome activation in lung macrophages. Acta Pharmacol Sin. (2021) 43:1274–84. doi: 10.1038/s41401-021-00693-6
39. Luan F, He X, Zeng N. Tetrandrine: a review of its anticancer potentials, clinical settings, pharmacokinetics and drug delivery systems. J Pharm Pharmacol. (2020) 72:1491–512. doi: 10.1111/jphp.13339
40. Gao L, Feng Q, Zhang X, Wang Q, Cui Y. Tetrandrine suppresses articular inflammatory response by inhibiting pro-inflammatory factors via NF-κB inactivation. J Orthop Res. (2016) 34:1557–68. doi: 10.1002/jor.23155
41. Zhao H, Kong L, Shen J, Ma Y, Wu Z, Li H, et al. Tetrandrine inhibits the occurrence and development of frozen shoulder by inhibiting inflammation, angiogenesis, and fibrosis. Biomed Pharmacother. (2021) 140:111700. doi: 10.1016/j.biopha.2021.111700
42. Ho L, Lai J. Chinese herbs as immunomodulators and potential disease-modifying antirheumatic drugs in autoimmune disorders. Curr Drug Metab. (2004) 5:181–92. doi: 10.2174/1389200043489081
43. Yu J, Zhu C, Yin J, Yu D, Wan F, Tang X, et al. Tetrandrine Suppresses Transient Receptor Potential Cation Channel Protein 6 Overexpression- Induced Podocyte Damage via Blockage of RhoA/ROCK1 Signaling. Drug Des Devel Ther. (2020) 14:361–70. doi: 10.2147/DDDT.S234262
44. Yin J, Lin J, Yu J, Wei X, Zhu B, Zhu C. Tetrandrine may treat membranous glomerulopathy via P13K/Akt signalling pathway regulation: Therapeutic mechanism validation using Heymann nephritis rat model. Bioengineered. (2021) 12:6499–515. doi: 10.1080/21655979.2021.1973862
45. Su L, Cao P, Wang H. Tetrandrine mediates renal function and redox homeostasis in a streptozotocin-induced diabetic nephropathy rat model through Nrf2/HO-1 reactivation. Ann Transl Med. (2020) 8:990. doi: 10.21037/atm-20-5548
46. Wu C, Wang Y, Lin C, Chen H, Chen Y. Tetrandrine down-regulates ERK/NF-κB signaling and inhibits activation of mesangial cells. Toxicol In Vitro. (2011) 25:1834–40. doi: 10.1016/j.tiv.2011.09.024
47. Chen T, Ji B, Chen Y. Tetrandrine triggers apoptosis and cell cycle arrest in human renal cell carcinoma cells. J Nat Med. (2014) 68:46–52. doi: 10.1007/s11418-013-0765-0
48. Chen S, Liu W, Wang K, Fan Y, Chen J, Ma J, et al. Tetrandrine inhibits migration and invasion of human renal cell carcinoma by regulating Akt/NF-κB/MMP-9 signaling. PLoS ONE. (2017) 12:e0173725. doi: 10.1371/journal.pone.0173725
49. Meng L, Zhang H, Hayward L, Takemura H, Shao R, Pommier Y. Tetrandrine induces early G1 arrest in human colon carcinoma cells by down-regulating the activity and inducing the degradation of G1-S-specific cyclin-dependent kinases and by inducing p53 and p21Cip1. Cancer Res. (2004) 64:9086–92. doi: 10.1158/0008-5472.CAN-04-0313
Keywords: IgA nephropathy, hub gene, bioinformatics, tetrandrine, mesangial cell
Citation: Xia M, Liu D, Liu H, Peng L, Yang D, Tang C, Chen G, Liu Y and Liu H (2022) Identification of Hub Genes and Therapeutic Agents for IgA Nephropathy Through Bioinformatics Analysis and Experimental Validation. Front. Med. 9:881322. doi: 10.3389/fmed.2022.881322
Received: 22 February 2022; Accepted: 09 May 2022;
Published: 28 June 2022.
Edited by:
Ratnakar Tiwari, Northwestern University, United StatesReviewed by:
Kongning Li, Hainan Medical University, ChinaCopyright © 2022 Xia, Liu, Liu, Peng, Yang, Tang, Chen, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Liu, bGl1aG9uZzYxOEBjc3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.