
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med., 19 April 2022
Sec. Hepatology
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.862278
This article is part of the Research TopicNAFLD in the World StageView all 7 articles
 Wen Wang1†
Wen Wang1† Xiaojing Liu1†
Xiaojing Liu1† Peiyao Wei1
Peiyao Wei1 Feng Ye1
Feng Ye1 Yunru Chen1
Yunru Chen1 Lei Shi1
Lei Shi1 Xi Zhang1
Xi Zhang1 Jianzhou Li1
Jianzhou Li1 Shumei Lin1*
Shumei Lin1* Xueliang Yang2*
Xueliang Yang2*Background and Aims: Non-alcoholic fatty liver disease (NAFLD) is a major chronic liver disease worldwide, and non-alcoholic steatohepatitis (NASH) is one of its pathological subtypes. The pathogenesis of NASH has not yet been fully elucidated. The purpose of this study was to identify the hub genes and pathways involved in NASH using bioinformatics methods. The hub genes were confirmed in human and animal models.
Materials and Methods: Three Gene Expression Omnibus (GEO) datasets (GSE48452, GSE58979, and GSE151158) of NASH patients and healthy controls were included in the study. We used GEO2R to identify differentially expressed genes (DEGs) between NASH patients and healthy controls. Functional enrichment analyses were then performed to explore the potential functions and pathways of the DEGs. In all DEGs, only two genes were highly expressed in NASH patients throughout the three datasets; these two genes, SPP1 and CXCL9, were further studied. Serum and liver tissues from NASH patients and healthy controls were collected. Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were measured in NASH patients and healthy controls. Liver tissues were stained with hematoxylin and eosin. Immunohistochemical staining was used to evaluate the expression levels of the two genes in liver tissues. Male C57BL/6J mice were fed a methionine choline-deficient (MCD) diet for 8 weeks, after which serum ALT and AST levels were measured and liver tissues were stained.
Results: SPP1 and CXCL9 were the hub genes detected in the three datasets. “Lipid metabolism,” “inflammatory response,” and “lymphocyte activation” were the most significant biological functions in GSE48452, GSE58979, and GSE151158, respectively. Kyoto Encyclopedia of Genes and Genomes pathway analysis showed that the toll-like receptor signaling pathway was significantly enriched in NASH patients. Serum ALT and AST levels were significantly increased in NASH patients compared to healthy controls. Liver tissues had more serious steatosis, hepatocyte ballooning degeneration, and lobular inflammatory infiltration, and the expression of SPP1 and CXCL9 in liver cells was significantly upregulated in NASH patients compared to healthy controls. MCD diet mice were consistent with NASH patients.
Conclusion: SPP1 and CXCL9 may play important roles in NASH pathogenesis and could be potential therapeutic targets and biomarkers of NASH in the future. Further experimental studies are needed to confirm our results.
Non-alcoholic fatty liver disease (NAFLD) is pathologically classified as non-alcoholic fatty liver (NAFL) or non-alcoholic steatohepatitis (NASH) based on the presence of ballooned hepatocytes (1). NASH is characterized by steatosis, ballooning necrosis around steatotic hepatocytes, mild inflammation, and fibrosis (2, 3). The global prevalence of NAFLD in the general population is estimated to be 25%, whereas that of NASH is estimated to range from 3 to 5% (4). Over the past 20 years, the prevalence of NAFLD in China has doubled, and the disease is most prevalent in the urban population (27%). Globally, China is expected to have the fastest increase in NAFLD incidence due to urbanization (5). As the prevalence of NAFLD continues to rise, NASH has the potential to become the most common cause of cirrhosis and end-stage liver disease in the coming decades (6).
Currently, the pathogenesis of NASH is unclear. The “two hit hypothesis” is a classic theory that describes the development and progression of NAFLD/NASH, but this view is too simplistic to recapitulate the molecular and metabolic changes that underlie the pathogenesis of NASH (7, 8). Recently, a “multiple-hit hypothesis” has replaced the “two hit hypothesis” to explain disease pathogenesis in NASH (9). This theory suggests that multiple factors, such as insulin resistance, hormones secreted by adipose tissue, nutrition, lifestyle, gut microbiota, inflammatory genes, and genetic and epigenetic factors, work together to induce NASH in genetically predisposed subjects, providing a more accurate explanation for the pathogenesis of NASH (10–14). At present, the management of NASH primarily focuses on diet and exercise, but still fails to achieve good clinical outcomes. NASH pharmacotherapy is under study, and to date, there are no Food and Drug Administration-approved drugs to treat NASH (15). It is important to understand the precise molecular mechanisms of NASH pathogenesis and develop and identify therapeutic targets for effective diagnosis, management, and therapy. Thus, determining key genes that affect the occurrence and development of NASH can contribute to the elucidation of its molecular mechanisms and the identification of new diagnostic markers and therapeutic targets.
Therefore, we analyzed differentially expressed genes (DEGs) in the liver tissues of patients with NASH using data downloaded from the Gene Expression Omnibus (GEO) database to identify the hub genes involved in the pathogenesis of the disease. Next, the potential mechanisms of the DEGs in NASH were explored using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. Finally, we verified the hub genes in the liver tissues of NASH patients and methionine choline-deficient (MCD) diet mice; these hub genes may be candidate diagnostic or therapeutic biomarkers for NASH.
We obtained the gene expression datasets of NASH from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). We systematically searched the studies using the following terms: “fat liver,” “non-alcoholic fatty liver disease,” “non-alcoholic steatohepatitis,” and “Homo sapiens.” The gene expression profiles of GSE48452, GSE58979, and GSE151158 were downloaded. The NASH and healthy control (HC) patient sample sizes were 18 vs. 14 in GSE48452, 8 vs. 10 in GSE58979, and 17 vs. 21 in GSE151158, respectively.
The original microarray data were analyzed using the online tool GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r), which allows researchers to identify DEGs in each group. Genes with P < 0.05 and |log FC| > 1.5 were considered as DEGs. The overlapping DEGs in all three datasets were analyzed using Venn diagrams (http://bioinformatics.psb.ugent.be/webtools/Venn/).
GO provides comprehensive information on the gene function of individual genomic products based on defined features comprising molecular function, biological processes, and cellular components. KEGG is a biological information database for understanding high-level biological functions and utilities. These analyses and annotations were performed using the Database for Annotation, Visualization, and Integrated Discovery (https://david.ncifcrf.gov/), which provides functional annotation information regarding genes and proteins for researchers to explore the biological meaning of a specified gene list. Statistical significance was set at P < 0.05.
NASH liver tissues and blood samples were obtained from morbidly obese patients undergoing bariatric surgery, and normal liver tissues and blood samples were taken from adults without obesity who underwent liver resection for hemangioma. The study was conducted according to the principles of the Declaration of Helsinki, and written informed consent was obtained from all patients. The study was reviewed and approved by the Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University (ethics approval number: 2018QN-13).
Male C57BL/6J mice (6-week-old) were supplied by the Experimental Animal Center of Xi'an Jiaotong University. Mice were housed in an animal center with a light-dark cycle of 12 h and free access to food and water. They were randomly divided into two groups: standard chow diet (HC group, n = 5) and MCD diet (NASH group, n = 5). After 8 weeks of feeding, liver tissues and blood samples were collected for the subsequent experiments. All experimental procedures were approved by the Animal Ethics Committee of Xi'an Jiaotong University (ethics approval number: 2018QN-13) and followed the animal ethical guidelines.
Liver tissues were fixed, dehydrated, cleared, and immersed in paraffin wax. The paraffin-embedded tissues were sliced into 3–4 μm-thick sections. All samples from NASH patients, HCs, and mice were reviewed histologically using H&E staining. Immunohistochemistry was performed using an anti-SPP1 antibody (Abclonal, Wuhan, China) and anti-CXCL9 antibody (ThermoFisher, Waltham, MA, USA).
Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were evaluated to assess liver inflammation using an LST008 Biochemistry Analyzer (Hitachi, Ltd., Tokyo, Japan).
All data are presented as the mean ± standard deviation. Statistical analysis between groups used an unpaired two-tailed Student's t-test. Statistical significance was set at P < 0.05. All analyses were performed in GraphPad Prism 6 (GraphPad Software, San Diego, America).
Three datasets were downloaded from the GEO database: GSE48452, GSE58979, and GSE151158. A total of 295 DEGs were identified in the GSE48452 dataset, including 194 upregulated and 101 downregulated genes (Figure 1A). Additionally, 407 DEGs were identified in the GSE58979 dataset, including 178 upregulated genes and 229 downregulated genes (Figure 1B). Finally, 160 DEGs were identified in the GSE151158 dataset, including 146 upregulated genes and 14 downregulated genes (Figure 1C). Heatmaps indicated the expression patterns of the top 20 DEGs in the datasets GSE48452 (CYP7A1, PEG10, TRHDE, FADS2, ENO3, TMEM154, FMO1, NCAM2, MEP1B, GRID1, RFC1, SQLE, ME1, AKR1B10, FABP4, SPP1, P4HA1, IGFBP2, IGFBP1, CHAC1), GSE58979 (ALB, SERPINA1, IL1RN, TNMD, TRDN, IGFBP1, FPR2, S100A8, S100A9, GADD45B, LDLR, IGHM, GPAT3, MYH11, CCL21, MMRN1, CDH19, SLITRK5, SPANXB1, CBLN4), and GSE151158 (SPP1, CCL20, LAIR1, LGALS3, FCGR2A, THY1, ITGAX, CD83, CCL18, CDKN1A, IL8, CXCR4, CXCL10, CXCL9, CXCL11, IL2RG, LAMP3, CCL19, B3GAT1, ZBTB16) (Figures 1D–F). To compare the NASH and HC groups, the intersection of DEGs from these datasets was presented in a Venn diagram and two upregulated genes were subsequently identified: SPP1 and CXCL9 (Figures 1G,H).
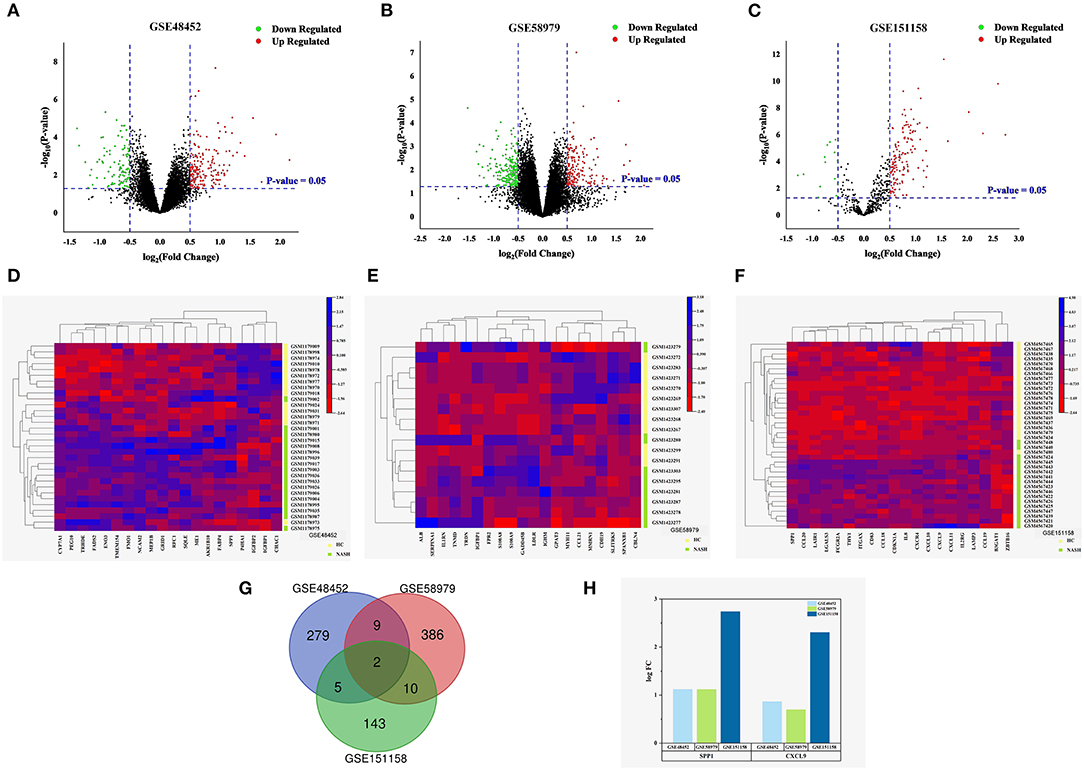
Figure 1. Identification of DEGs in NASH. (A–C) Volcano plot of the differentially expressed genes of the three datasets. Red points indicate DEGs with a logFC > 0.5 and P < 0.05, green points indicate DEGs with a logFC < −0.5 and P < 0.05. (A) GSE48452. (B) GSE58979. (C) GSE151158. (D–F) Heat map for the top 20 DEGs between NASH and normal liver tissues of the three datasets. Green lines represent NASH, and yellow lines represent HC. (D) GSE48452 (containing 18 NASH tissues and 14 normal liver tissues). (E) GSE58979 (8 NASH tissues and 10 normal liver tissues). (F) GSE151158 (17 NASH tissues and 21 normal liver tissues). (G,H) Venn diagram and the expression levels of the intersecting genes in GSE48452, GSE58979, and GSE151158. (G) Venn diagram displaying two DEGs present in both NASH and HC. (H) Expression levels of the intersecting genes (logFC = log Fold Change).
GO analysis was applied to investigate the biological functions of the identified DEGs in the three datasets. The biological process analysis in GSE48452 primarily included lipid metabolism (e.g., steroid metabolic process, peroxisome proliferator-activated receptor signaling pathway, cellular response to lipids, and fatty acid derivative metabolic process) (Figure 2A). The biological process analysis in GSE58979 mainly included inflammatory response (e.g., leukocyte migration, myeloid leukocyte activation, regulation of leukocyte activation, and response to interferon-gamma) (Figure 2B). Similarly, the biological process analysis in GSE151158 primarily included inflammatory response (e.g., lymphocyte activation, adaptive immune response, cytokine signaling in the immune system, cytokine-cytokine receptor interaction, leukocyte differentiation, and positive regulation of immune response) (Figure 2C).
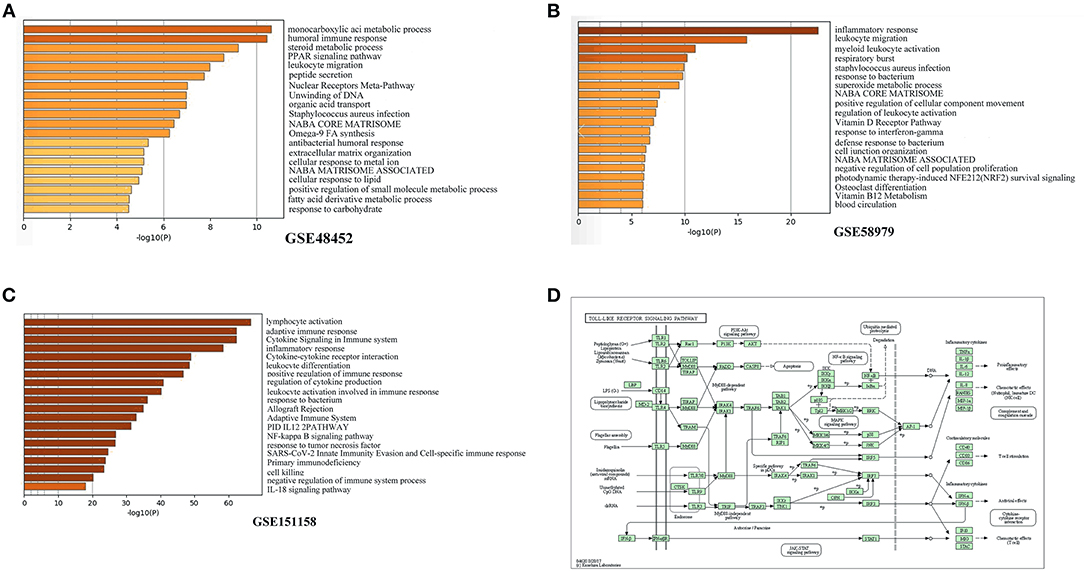
Figure 2. Functional enrichment analysis of DEGs. (A–C) Top 20 biological functions according to gene ontology (GO) analysis of the two DEGs. (A) GSE48452. (B) GSE58979. (C) GSE151158. (D) KEGG functional analysis: colored map of the toll-like receptor signaling pathway.
KEGG pathway analysis was utilized to elucidate the pathway based on the two DEGs identified previously. The toll-like receptor signaling pathway was significantly enriched in KEGG pathways in NASH patients (hsa04620; P = 0.015) (Figure 2D).
To further validate SPP1 and CXCL9, identified using bioinformatics analysis, we examined the expression of the two DEGs in NASH patients and HCs. H&E staining confirmed that NASH patients had more serious steatosis, hepatocyte ballooning degeneration, and lobular inflammatory infiltration than the HC group (Figure 3A). Serum ALT and AST levels were significantly elevated in NASH patients compared to the HC group (Figures 3B,C). Immunohistochemical results showed that the expression levels of SPP1 and CXCL9 were significantly upregulated in NASH liver tissues compared to healthy liver tissues, which is consistent with the results of the bioinformatics analysis mentioned previously (Figure 3D).
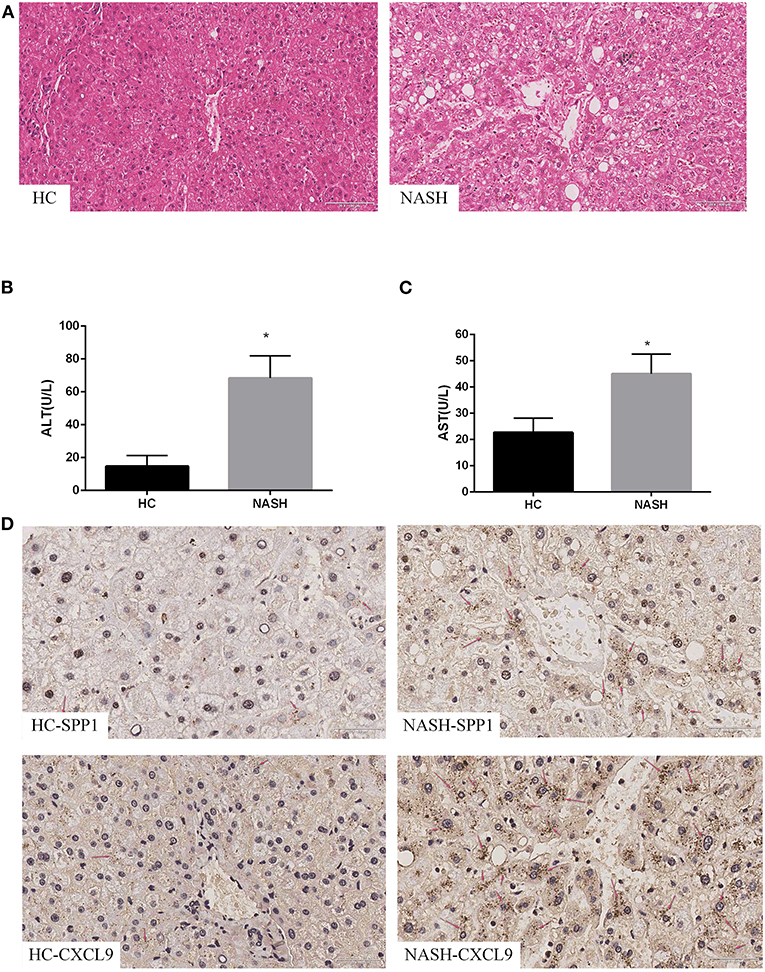
Figure 3. Expression of SPP1 and CXCL9 in NASH patients. Serum biochemical indices change, morphological analysis and immunohistochemistry images of the expression of SPP1 and CXCL9 in NASH and HC group of human liver samples. (A) The liver samples were stained with hematoxylin–eosin (20×) in NASH and HC groups. The gray arrows represent steatosis, the red arrows represent hepatocyte ballooning degeneration, and the black arrows represent lobular infiltration. (B,C) Serum content of ALT and AST changes between HC patients and NASH patients. ALT and AST concentrations were significantly higher in NASH group than those in HC group. n = 5, *P < 0.05 compared with HC group. (D) Determination of the expression levels of SPP1 and CXCL9 in NASH and HC group of human liver samples using immunohistochemical staining (40×). The red arrows represent SPP1 or CXCL9 proteins in liver cells.
Based on the results in human samples, we further verified the expression of SPP1 and CXCL9 in an MCD diet mouse model. H&E staining confirmed that MCD diet mice had more serious steatosis, hepatocyte ballooning degeneration, and lobular inflammatory infiltration than the HC group (Figure 4A). Serum ALT and AST levels in the MCD diet mice were significantly higher than those in the HC group (Figures 4B,C). Immunohistochemical results showed that the expression levels of SPP1 and CXCL9 were significantly upregulated in MCD diet mice compared to HC mice, which is consistent with the above results (Figure 4D).
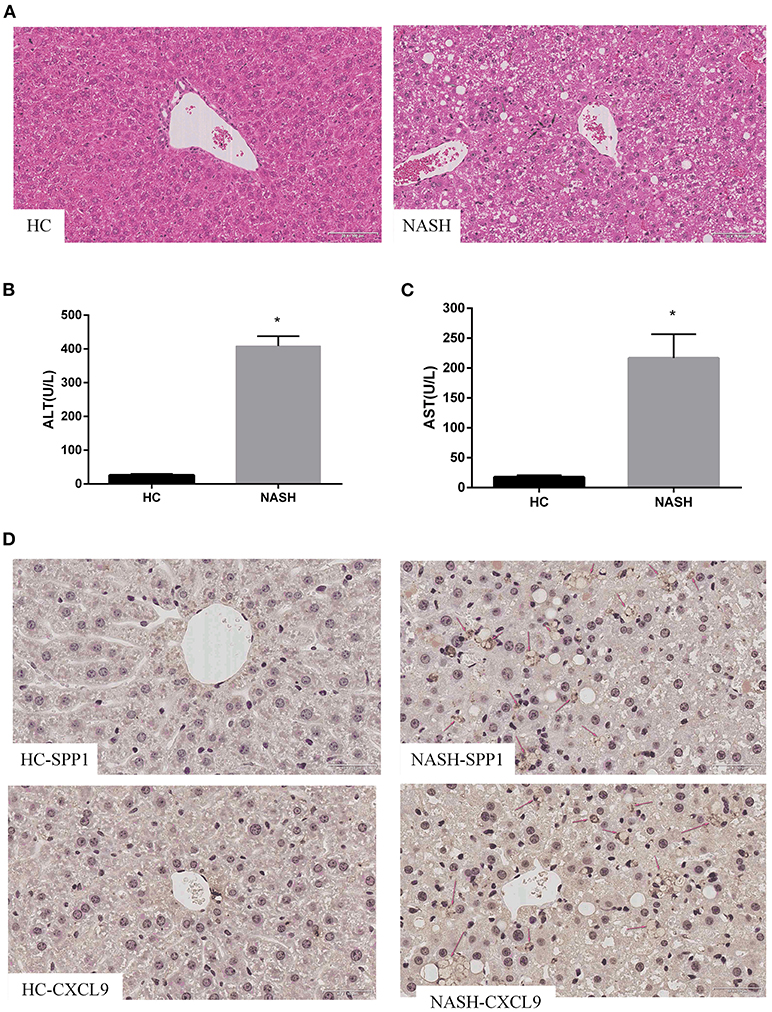
Figure 4. Expression of SPP1 and CXCL9 in MCD diet mice. Serum biochemical indices change, morphological analysis and immunohistochemistry images of the expression of SPP1 and CXCL9 in NASH and HC group of mice liver samples after MCD diet treatment for 8 weeks. (A) The liver samples were stained with hematoxylin–eosin (20×) in NASH and HC groups. The gray arrows represent steatosis, the red arrows represent hepatocyte ballooning degeneration, and the black arrows represent lobular infiltration. (B,C) Serum content of ALT and AST changes after MCD diet treatment for 8 weeks. ALT and AST concentrations were significantly higher in NASH group than those in HC group. n = 3, *P < 0.05 compared with HC group. (D) Determination of the expression levels of SPP1 and CXCL9 in NASH and HC group of mice liver samples using immunohistochemical staining (40×). The red arrows represent SPP1 or CXCL9 proteins in liver cells.
In this study, bioinformatics analysis was utilized to identify DEGs related to NASH. The potential role of DEGs in NASH was explored using GO and KEGG pathway analyses. The hub genes were verified in the liver tissues of NASH patients and HC patients. We identified two hub genes, SPP1 and CXCL9, both of which had upregulated expression levels. Functional enrichment analysis showed that DEGs were mainly enriched in lipid metabolism, inflammatory response, and lymphocyte activation in these datasets. The toll-like receptor signaling pathway was the main enrichment pathway for the hub genes. GO and KEGG pathway analyses indicated that one of the mechanisms of NASH pathogenesis involves changes that affect metabolism and stress responses (16). The upregulation of SPP1 and CXCL9 genes in NASH livers was also confirmed in patient and mice samples. These results may contribute to the understanding of the pathogenesis of NASH and identification of potential diagnostic or therapeutic biomarkers.
Secreted phosphoprotein 1 (SPP1), also known as Osteopontin (OPN), is a secreted phosphorylated glycoprotein. SPP1 is seldomly expressed in normal liver tissue and is mainly expressed in activated Kupffer cells, hepatic macrophages, hepatic stellate cells, and hepatocytes under pathological conditions (17–19). As a multifunctional protein, SPP1 is involved in multiple liver diseases by promoting inflammatory responses, cell activation, proliferation, and migration, and is closely related to the occurrence, development, and prognosis of fatty liver, liver fibrosis, and liver cancer (20, 21). Previous studies have revealed that the expression of SPP1 is upregulated in NAFLD and NASH liver tissues. In a new swine model of NASH, SPP1 gene expression was significantly positively correlated with lipid droplet area and inflammation, and when NASH was reversed, SPP1 gene expression was significantly reduced (22). A cross-sectional survey of 19 NASH patients reported that plasma SPP1 levels in NASH patients were higher than those in HC patients, whereas plasma and hepatic SPP1 levels in patients with advanced fibrosis were higher than those in patients with no or mild fibrosis, indicating that the histological characteristics of disease severity in NASH patients are associated with SPP1 (23). Serum SPP1 levels were measured in 179 patients with NAFLD and 123 HC patients and were significantly elevated in patients with NAFLD, and were independently associated with liver enzymes and portal inflammation (24). Inhibition of SPP1 can improve inflammation in adipose and liver tissues (25). Furthermore, other studies have confirmed that knocking out the SPP1 gene can aggravate the progression of fatty liver disease. SPP1 deficiency may reduce liver-related mortality, which may be associated with the promotion of a systemic pro-inflammatory milieu by SPP1. This suggests that SPP1 plays a dual role in the pathogenesis of fatty liver and that SPP1 may be involved in the internal control of excessive lipid uptake in the liver, thereby protecting the liver from lipotoxicity, apoptosis, and subsequent fibrosis and hepatocyte proliferation (26).
The expression of C-X-C motif chemokine ligand 9 (CXCL9) was also upregulated in the liver tissues of patients with NASH. The CXCL9 gene is a member of a chemokine superfamily that encodes secreted proteins involved in immunoregulatory and inflammatory processes and is thought to be involved in T cell trafficking. The encoded protein binds to C-X-C motif chemokine 3 (CXCL3) and is a chemoattractant for lymphocytes but not for neutrophils. CXCL9 has been reported to be involved in various pathological processes, including tumor development, immunity, and inflammation (27, 28). Overexpression of CXCL9 mRNA was observed in both NASH and simple steatosis mouse models, and hepatocytes and sinusoid endothelial cells secreting CXCL9 protein were localized in areas infiltrated with inflammatory cells (29). The expression of CXCL9 was upregulated in NASH without fibrosis in a high-risk cohort of adults with obesity (12). Another cohort study showed that the serum concentration of CXCL9 in patients with chronic liver disease was significantly higher than that in HC patients and was positively correlated with the severity of liver fibrosis (30). Increased expression of CXCL9 was also found in hepatocytes of patients with chronic hepatitis C virus infection, and its expression levels were associated with liver fibrosis (31). Overexpression of CXCL9 is associated with tumor progression. In mouse models, a chronically increasing trend in CXCL9 levels was associated with the progression from NAFLD to hepatocellular carcinoma in male mice (32, 33). These studies suggest that CXCL9 is an important factor in chronic liver inflammation and that its expression and role in NAFLD require further research.
Atherosclerosis is the leading cause of death in NAFLD and NASH patients. Chemokines play an important role in NAFLD and NASH with atherosclerosis. IL-17, which is released by the visceral adipose tissue, can induce Eotaxin secretion via the smooth muscle cells in atherosclerotic blood vessels. Eotaxin is another member of the CC chemokine subfamily and was able to predict carotid intima-media thickness (IMT), a marker of atherosclerosis (34). CXCL9 expression also may be regulated by IL-17. IL-17 can upregulate the expression of chemokines, such as CXCL9, in mice with hypersensitivity pneumonitis, while IL-17 gene-deficient mice had reduced levels of CXCL9 (35). Obesity and insulin resistance are central to NAFLD and hepatic steatosis. Stem cell growth factor-beta (SCGF-β) levels were positively correlated to insulin resistance and hepatic steatosis severity in a retrospective study on obese patients (36). In addition, CXCL9 and SCGF-β levels were higher in unstable plaques compared to stable plaques (37). The relationship between CXCL9 and SCGF-β in NAFLD requires further research.
In conclusion, using bioinformatics analysis of NASH patients and healthy controls, we identified two hub genes (SPP1 and CXCL9), which were significantly upregulated. The upregulation of SPP1 and CXCL9 in NASH livers was also verified using human and mouse samples. Our study confirmed that SPP1 and CXCL9, which play key roles in NASH pathogenesis, are potential targets for the prevention and treatment of NASH. However, the findings of this study need to be validated by further experimental studies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.ncbi.nlm.nih.gov/geo/.
The studies involving human participants were reviewed and approved by the Ethics Committee of The First Affiliated Hospital of Xi'an Jiaotong University. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Animal Ethics Committee of Xi 'an Jiaotong University.
WW and XL contributed to the data analysis, experiments, and manuscript writing. PW, FY, and YC contributed to the data analysis and the data interpretation. LS and XZ conducted the experiments and analyzed the data. JL wrote and revised the manuscript. SL and XY conceived and designed the study and wrote the manuscript. All authors read and approved the submitted version.
This study was supported by the National Major Scientific and Technological Project (Nos. 2017ZX10201201 and 2017ZX10202202) and Institutional Foundation of The First Affiliated Hospital of Xi'an Jiaotong University (No. 2018QN-13).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank MEDEE (medee.cn/sy) for English language polishing service.
1. Tanaka N, Kimura T, Fujimori N, Nagaya T, Komatsu M, Tanaka E. Current status, problems, and perspectives of non-alcoholic fatty liver disease research. World J Gastroenterol. (2019) 25:163–77. doi: 10.3748/wjg.v25.i2.163
2. Yki-Järvinen H. Non-alcoholic fatty liver disease as a cause and a consequence of metabolic syndrome. Lancet Diabetes Endocrinol. (2014) 2:901–10. doi: 10.1016/S2213-8587(14)70032-4
3. Bedossa P. Pathology of non-alcoholic fatty liver disease. Liver Int. (2017) 37:85–9. doi: 10.1111/liv.13301
4. Sheka AC, Adeyi O, Thompson J, Hameed B, Crawford PA, Ikramuddin S. Nonalcoholic steatohepatitis: a review. JAMA. (2020) 323:1175–83. doi: 10.1001/jama.2020.2298
5. Younossi ZM. Non-alcoholic fatty liver disease - A global public health perspective. J Hepatol. (2019) 70:531–44. doi: 10.1016/j.jhep.2018.10.033
6. Vernon G, Baranova A, Younossi ZM. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharmacol Ther. (2011) 34:274–85. doi: 10.1111/j.1365-2036.2011.04724.x
7. Ullah R, Rauf N, Nabi G, Ullah H, Shen Y, Zhou YD, et al. Role of nutrition in the pathogenesis and prevention of non-alcoholic fatty liver disease: recent updates. Int J Biol Sci. (2019) 15:265–76. doi: 10.7150/ijbs.30121
8. Wong VW, Chitturi S, Wong GL, Yu J, Chan HL, Farrell GC. Pathogenesis and novel treatment options for non-alcoholic steatohepatitis. Lancet Gastroenterol Hepatol. (2016) 1:56–67. doi: 10.1016/S2468-1253(16)30011-5
9. Hashimoto E, Taniai M, Tokushige K. Characteristics and diagnosis of NAFLD/NASH. J Gastroenterol Hepatol. (2013) 28:64–70. doi: 10.1111/jgh.12271
10. Buzzetti E, Pinzani M, Tsochatzis EA. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism. (2016) 65:1038–48. doi: 10.1016/j.metabol.2015.12.012
11. Peng C, Stewart AG, Woodman OL, Ritchie RH, Qin CX. Non-alcoholic steatohepatitis: a review of its mechanism, models and medical treatments. Front Pharmacol. (2020) 11:603926. doi: 10.3389/fphar.2020.603926
12. Subudhi S, Drescher HK, Dichtel LE, Bartsch LM, Chung RT, Hutter MM, et al. Distinct hepatic gene-expression patterns of NAFLD in patients with obesity. Hepatol Commun. (2022) 6:77–89. doi: 10.1002/hep4.1789
13. Cotter TG, Rinella M. Nonalcoholic fatty liver disease 2020: the state of the disease. Gastroenterology. (2020) 158:1851–64. doi: 10.1053/j.gastro.2020.01.052
14. Pafili K, Roden M. Nonalcoholic fatty liver disease (NAFLD) from pathogenesis to treatment concepts in humans. Mol Metab. (2021) 50:101122. doi: 10.1016/j.molmet.2020.101122
15. Roeb E, Geier A. Nonalcoholic steatohepatitis (NASH) - current treatment recommendations and future developments. Z Gastroenterol. (2019) 57:508–17. doi: 10.1055/a-0784-8827
16. Machado MV, Diehl AM. Pathogenesis of nonalcoholic steatohepatitis. Gastroenterology. (2016) 150:1769–77. doi: 10.1053/j.gastro.2016.02.066
17. Nagoshi S. Osteopontin: versatile modulator of liver diseases. Hepatol Res. (2014) 44:22–30. doi: 10.1111/hepr.12166
18. Bruha R, Vitek L, Smid V. Osteopontin - A potential biomarker of advanced liver disease. Ann Hepatol. (2020) 19:344–52. doi: 10.1016/j.aohep.2020.01.001
19. Jiang X, Zhang F, Ji X, Dong F, Yu H, Xue M, et al. Lipid-injured hepatocytes release sOPN to improve macrophage migration via CD44 engagement and pFak-NFκB signaling. Cytokine. (2021) 142:155474. doi: 10.1016/j.cyto.2021.155474
20. Wang G, Chen S, Zhao C, Li X, Zhao W, Yang J, et al. Comparative analysis of gene expression profiles of OPN signaling pathway in four kinds of liver diseases. J Genet. (2016) 95:741–50. doi: 10.1007/s12041-016-0673-7
21. Wang C, He M, Peng J, Li S, Long M, Chen W, et al. Increased plasma osteopontin levels are associated with nonalcoholic fatty liver disease in patients with type 2 diabetes mellitus. Cytokine. (2020) 125:154837. doi: 10.1016/j.cyto.2019.154837
22. Herrera-Marcos LV, Martínez-Beamonte R, Macías-Herranz M, Arnal C, Barranquero C, Puente-Lanzarote JJ, et al. Hepatic galectin-3 is associated with lipid droplet area in non-alcoholic steatohepatitis in a new swine model. Sci Rep. (2022) 12:1024. doi: 10.1038/s41598-022-04971-z
23. Arendt BM, Teterina A, Pettinelli P, Comelli EM, Ma DWL, Fung SK, et al. Cancer-related gene expression is associated with disease severity and modifiable lifestyle factors in non-alcoholic fatty liver disease. Nutrition. (2019) 62:100–07. doi: 10.1016/j.nut.2018.12.001
24. Yilmaz Y, Ozturk O, Alahdab YO, Senates E, Colak Y, Doganay HL, et al. Serum osteopontin levels as a predictor of portal inflammation in patients with nonalcoholic fatty liver disease. Dig Liver Dis. (2013) 45:58–62. doi: 10.1016/j.dld.2012.08.017
25. Kiefer FW, Zeyda M, Gollinger K, Pfau B, Neuhofer A, Weichhart T, et al. Neutralization of osteopontin inhibits obesity-induced inflammation and insulin resistance. Diabetes. (2010) 59:935–46. doi: 10.2337/db09-0404
26. Nardo AD, Grün NG, Zeyda M, Dumanic M, Oberhuber G, Rivelles E, et al. Impact of osteopontin on the development of non-alcoholic liver disease and related hepatocellular carcinoma. Liver Int. (2020) 40:1620–33. doi: 10.1111/liv.14464
27. Song X, Shen Y, Lao Y, Tao Z, Zeng J, Wang J, et al. CXCL9 regulates acetaminophen-induced liver injury via CXCR3. Exp Ther Med. (2019) 18:4845–51. doi: 10.3892/etm.2019.8122
28. Ikeda A, Aoki N, Kido M, Iwamoto S, Nishiura H, Maruoka R, et al. Progression of autoimmune hepatitis is mediated by IL-18-producing dendritic cells and hepatic CXCL9 expression in mice. Hepatology. (2014) 60:224–36. doi: 10.1002/hep.27087
29. Semba T, Nishimura M, Nishimura S, Ohara O, Ishige T, Ohno S, et al. The FLS (fatty liver Shionogi) mouse reveals local expressions of lipocalin-2, CXCL1 and CXCL9 in the liver with non-alcoholic steatohepatitis. BMC Gastroenterol. (2013) 13:120. doi: 10.1186/1471-230X-13-120
30. Tacke F, Zimmermann HW, Berres ML, Trautwein C, Wasmuth HE. Serum chemokine receptor CXCR3 ligands are associated with progression, organ dysfunction and complications of chronic liver diseases. Liver Int. (2011) 31:840–9. doi: 10.1111/j.1478-3231.2011.02504.x
31. Jiménez-Sousa M, Gómez-Moreno AZ, Pineda-Tenor D, Medrano LM, Sánchez-Ruano JJ, Fernández-Rodríguez A, et al. CXCL9-11 polymorphisms are associated with liver fibrosis in patients with chronic hepatitis C: a cross-sectional study. Clin Transl Med. (2017) 6:26. doi: 10.1186/s40169-017-0156-3
32. Mirshahi F, Aqbi HF, Cresswell K, Saneshaw M, Coleman C, Jacobs T, et al. Longitudinal studies can identify distinct inflammatory cytokines associated with the inhibition or progression of liver cancer. Liver Int. (2020) 40:468–72. doi: 10.1111/liv.14323
33. Wang YH, Huang JH, Tian ZF, Zhou YF, Yang J. The role of CXC cytokines as biomarkers and potential targets in hepatocellular carcinoma. Math Biosci Eng. (2019) 17:1381–95. doi: 10.3934/mbe.2020070
34. Tarantino G, Costantini S, Finelli C, Capone F, Guerriero E, La Sala N, et al. Is serum Interleukin-17 associated with early atherosclerosis in obese patients? J Transl Med. (2014) 12:214. doi: 10.1186/s12967-014-0214-1
35. Joshi AD, Fong DJ, Oak SR, Trujillo G, Flaherty KR, Martinez FJ, et al. Interleukin-17-mediated immunopathogenesis in experimental hypersensitivity pneumonitis. Am J Respir Crit Care Med. (2009) 179:705–16. doi: 10.1164/rccm.200811-1700OC
36. Tarantino G, Citro V, Balsano C, Capone D. Could SCGF-beta levels be associated with inflammation markers and insulin resistance in male patients suffering from obesity-related NAFLD? Diagnostics. (2020) 10:395. doi: 10.3390/diagnostics10060395
Keywords: NASH, SPP1, CXCL9, bioinformatics analysis, MCD diet
Citation: Wang W, Liu X, Wei P, Ye F, Chen Y, Shi L, Zhang X, Li J, Lin S and Yang X (2022) SPP1 and CXCL9 Promote Non-alcoholic Steatohepatitis Progression Based on Bioinformatics Analysis and Experimental Studies. Front. Med. 9:862278. doi: 10.3389/fmed.2022.862278
Received: 25 January 2022; Accepted: 24 March 2022;
Published: 19 April 2022.
Edited by:
Mark Muthiah, National University Hospital, SingaporeReviewed by:
Thomas Cotter, University of Texas Southwestern Medical Center, United StatesCopyright © 2022 Wang, Liu, Wei, Ye, Chen, Shi, Zhang, Li, Lin and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueliang Yang, amlhbmppYW5kYW5kYW4xOTg3QDEyNi5jb20=; Shumei Lin, bGluc2h1bWVpMTIzQDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.