- 1State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Guangdong Provincial Key Laboratory of Ophthalmology and Visual Science, Guangdong Provincial Clinical Research Center for Ocular Diseases, Sun Yat-sen University, Guangzhou, China
- 2Guizhou Provincial People’s Hospital, Guizhou University, Guiyang, China
- 3Ophthalmology Department of Zhuhai Integrated Traditional Chinese and Western Medicine Hospital, Zhuhai, China
Retinitis pigmentosa (RP), characterized by the gradual loss of rod and cone photoreceptors that eventually leads to blindness, is the most common inherited retinal disorder, affecting more than 2.5 million people worldwide. However, the underlying pathogenesis of RP remains unclear and there is no effective cure for RP. Mutations in the Mer receptor tyrosine kinase (MERTK) gene induce the phagocytic dysfunction of retinal pigment epithelium (RPE) cells, leading to RP. Studies have indicated that filamentous actin (F-actin)—which is regulated by chaperonin-containing TCP1 subunit 5 (CCT5)—plays a vital role in phagocytosis in RPE cells. However, whether CCT5/F-actin signaling is involved in MERTK-associated RP remains largely unknown. In the present study, we specifically knocked down MERTK and CCT5 through siRNA transfection and examined the expression of CCT5 and F-actin in human primary RPE (HsRPE) cells. We found that MERTK downregulation inhibited cell proliferation, migration, and phagocytic function; significantly decreased the expression of F-actin; and disrupted the regular arrangement of F-actin. Importantly, our findings firstly indicate that CCT5 interacts with F-actin and is inhibited by MERTK siRNA in HsRPE cells. Upregulating CCT5 using CCT5-specific lentiviral vectors (CCT5-Le) rescued the cell proliferation, migration, and phagocytic function of HsRPE cells under the MERTK knockdown condition by increasing the expression of F-actin and restoring its regular arrangement via the LIMK1/cofilin, but not the SSH1/cofilin, pathway. In conclusion, CCT5 protects against the effect of MERTK knockdown in HsRPE cells and demonstrates the potential for effective treatment of MERTK-associated RP.
Introduction
Retinitis pigmentosa (RP) is the most common inherited retinal disease that affected tens of millions of people worldwide and is caused by a series of gene mutations that eventually lead to progressive retinal degeneration (1). As the most common inherited retinal disease, RP affects more than 2.5 million people worldwide (2, 3). However, the pathogenesis of RP remains unclear, and there is currently no effective treatment (4). Studies have shown that the Mer receptor tyrosine kinase (MERTK) gene mutation can lead to RP in humans (5). In the Royal College of Surgeons (RCS) rat, which is a typical animal model of human autosomal recessive inherited RP, the disease was found to be caused by the Mertk gene mutation (6). The MERTK gene encodes the Mer receptor tyrosine kinase, which belongs to the TAM receptor kinase family and participates in the phagocytic process (5). Retinal pigment epithelium (RPE) can constantly phagocytize the shed photoreceptor outer segment (POS), which plays a vital role in maintaining retinal homeostasis, as POS renewal is essential for visual function (7). The phagocytic dysfunction of RPE is an important aspect of the pathogenesis of RP (8). According to previous studies, a mutation in the MERTK gene leads to the phagocytic dysfunction of RPE cells, which is responsible for MERTK-associated RP (5). However, the mechanism underlying the MERTK gene mutation leading to phagocytic dysfunction remains largely unknown.
The process of POS phagocytosis is based on the rigorous control of the distribution and expression of the actin cytoskeleton (9). Actin, as a major component of the cytoskeleton, plays a vital role in the POS phagocytosis process. A previous study showed alphavbeta5 binding POS which is the first step of the POS phagocytosis process is required actin (10). However, nearly decades after the phenomenon was first described, much remains unknown regarding the role of the actin cytoskeleton in this process. Recent studies have emphasized that the rearrangement of RPE cytoskeletal filamentous actin (F-actin) is essential for POS internalization (11). The formation of phagocytic cups via the early recruitment of F-actin lays the foundation for phagocytosis, which means any factor that obstructs the expression of F-actin or disrupts the arrangement of F-actin will lead to phagocytic dysfunction, as the phagocytic cup formed by the orderly aggregation of F-actin combined with POS is the key to phagocytosis initiation (12). Previous studies demonstrated that the F-actin arrangement is abnormal in RPE cells with low phagocytotic activity (13). Recent studies also indicated that phagocytes formed by the recruitment of F-actin and the binding of POS must be combined with MERTK (14). In other words, the MERTK gene mutation may impair the recruitment of F-actin. However, the involvement of abnormal epithelial F-actin cytoskeleton in MERTK-associated RP remains largely unknown.
The TCP-1 cyclic complex, also known as the chaperonin-containing TCP-1 (CCT), is composed of eight parallel subunits (CCT1–8) (15–17). Assisted by ATP binding and hydrolysis, the main function of CCT is to help refold misfolded or unfolded proteins. Between 5% and 10% of the proteins in mammalian cells interact with CCT (18). In S. cerevisiae, CCT deficiency mutations lead to death, demonstrating the importance of CCT function (19). Actin is one of the main substrates of CCT; however, the exact mechanism by which CCT promotes actin folding is not yet understood (20, 21). A recent study found that CCT was required for efficient actin myofilament assembly (22) and that CCT5-specific ATP binding was required for efficient actin folding (23). Additionally, CCT5 controls lysosome biogenesis via the actin cytoskeleton (24). The actin/CCT5 pathway is implicated in multiple diseases—including hereditary sensory neuropathies (23), legionella pneumophila infection (25), muscle atrophy (26), and Alzheimer’s disease (24)—suggesting that CCT5 is associated with various cellular physiological processes in different tissues. Nonetheless, whether the F-actin/CCT5 pathway plays a vital role in MERTK-associated RP remains to be elucidated. Therefore, the present study explored the relationship between CCT5, F-actin, and RP, and the potential molecular mechanism underlying this disease.
Results
Mer Receptor Tyrosine Kinase siRNA Inhibited Cell Proliferation and Induced Morphological Changes and Phagocytic Dysfunction in Human Primary RPE Cells
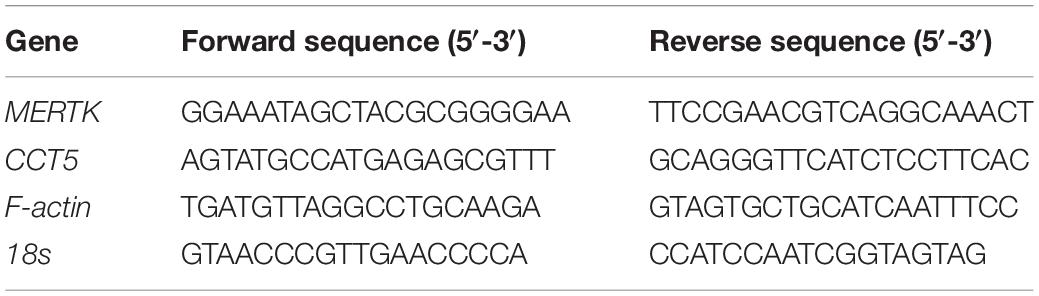
On the 7 days after the HsRPE cells were extracted and cultured in DMEM medium, we observed cultured cells with an inverted phase-contrast microscope, we found each cell contains abundant brownish-yellow pigments which is the characterization of the HsRPE cell. This result suggests that we’re extracting the HsRPE cells, not other nerve cells, fibroblasts, blood vessel cells, etc (Figure 1A). To further verify that the cells we extracted were HsRPE cells, we stained the cultured cells with RPE65, an RPE cell-specific protein. The result showed every cultured cell is stained with RPE65 (green) (Figure 1B). These results indicate that we have successfully extracted and cultured HsRPE cells. To establish a model of MERTK-associated RP in vitro, MERTK siRNA was applied to HsRPE cells. To test the target-specific efficacy of the MERTK siRNA, two types of MERTK siRNA (MERTK siRNA1 and MERTK siRNA2) were used. Both the mRNA and protein expression of MERTK in the siRNA groups were significantly reduced compared with those in the negative control (NC) siRNA groups (Figures 1C,D). Since MERTK siRNA2 more effectively downregulated MERTK than MERTK siRNA1, we applied MERTK siRNA2 in the subsequent experiments. A Cell Counting Kit-8 (CCK8) assay was used to determine the viability of HsRPE cells. Compared with the NC siRNA group, the group with knocked down MERTK exhibited suppressed cell proliferation, as the optical density (OD) value of the NC siRNA group was higher than that of the MERTK siRNA group (Figure 1E). We then observed the changes in cell morphology using an inverted microscope and found that the cellular morphologies of the HsRPE cells were very different between the NC siRNA group and the MERTK siRNA group. The cells of the NC siRNA group had a typical polygonal appearance; however, the MERTK siRNA-transfected cells became elongated in shape (Figure 1F). Wound-healing assays and Transwell migration assays were performed to assess the wound-healing ability and migration of HsRPE cells. The results demonstrated that the number of migrated HsRPE cells and their migration distance was significantly decreased in the MERTK siRNA group compared with the NC siRNA group (Figures 1G,H). We performed the phagocytosis assay combined with the immunofluorescence assay to evaluate the effect of MERTK siRNA on the phagocytic function of HsRPE cells. For the phagocytosis assay, Nile red-labeled particles and MERTK immunofluorescent staining were used. In the NC siRNA group, the expression of MerTK (green) is significantly higher than MERTK siRNA group, and massive red particles were phagocytized into the HsRPE cells, while the number of red particles that were phagocytized into the HsRPE cells was significantly decreased in the MERTK siRNA group (Figure 1I). These results indicate that MERTK-specific siRNA inhibited cell proliferation and induced morphological changes and phagocytic dysfunction in HsRPE cells.
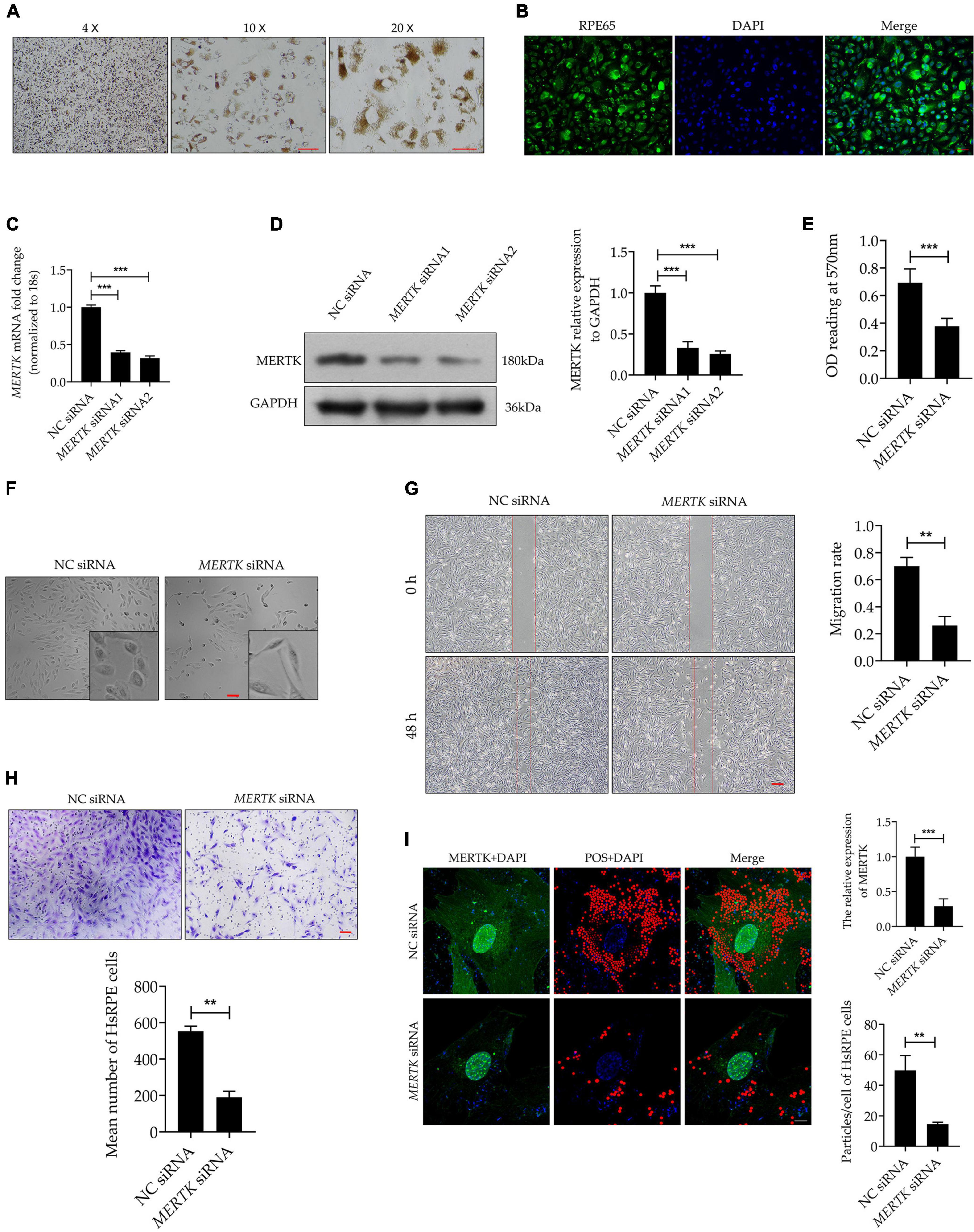
Figure 1. MERTK siRNA inhibited cell proliferation and induced morphological changes and phagocytic dysfunction in HsRPE cells. (A) The primary cultured HsRPE cells were observed with an inverted microscope at different magnifications (4X, 10X, and 20X). Scale bars: white bar 100 μm; red bars 10 μm. (B) The HsRPE cells were identified by immunofluorescence staining with RPE65 antibody. Scale bars: 50 μm. (C) HsRPE cells were treated with NC, MERTK siRNA1, or MERTK siRNA2 (20 nM) for 24 h, and the expression of MERTK was analyzed by RT-PCR. (D) HsRPE cells were treated with NC, MERTK siRNA1, or MERTK siRNA2 (20 nM) for 48 h, and MERTK expression was determined by Western blotting. (E) HsRPE cells were treated with NC or MERTK siRNA2 (MERTK siRNA) for 48 h, and cell proliferation was analyzed by CCK8. (F) HsRPE cells were treated the same as in (E), and cell morphology was observed with an inverted microscope. Scale bars: 100 μm. (G) HsRPE cells were treated the same as in (E), and wound-healing assays were performed to assess the wound-healing capabilities of HsRPE cells. Scale bars: 200 μm. (H) HsRPE cells were treated the same as in (E), and Transwell assays were performed to evaluate the migration activity of HsRPE cells (purple). Scale bars: 100 μm. (I) HsRPE cells were treated the same as in (E), and their phagocytic ability was examined using phagocytosis assays. The red dots represent the particles and the green dye represents the MERTK protein. Scale bars: 5 μm. **P < 0.01; *** P < 0.001.
Mer Receptor Tyrosine Kinase siRNA Downregulated the Expression of Chaperonin-Containing TCP1 Subunit 5, Which Interacted and Co-localized With Filamentous Actin in Human Primary RPE Cells
Filamentous actin plays a vital role in phagocytosis. To determine whether F-actin was involved in MERTK-associated RP, we detected F-actin by quantitative real-time PCR (RT-PCR) and Western blot assays separately. Both the RT-PCR and Western blot assays demonstrated that MERTK siRNA significantly inhibited F-actin at both the mRNA and protein levels, as compared with the NC group (Figures 2A,B). Furthermore, to detect the distribution and arrangement of F-actin, we performed the immunofluorescence analysis. The results indicated that F-actin demonstrated a highly regular radial distribution of filamentous order in the NC group. However, in the MERTK siRNA group, the F-actin clumped together in a disorderly arrangement around the nucleus (Figure 2C). CCT5 plays an important role in actin regulation; therefore, to determine whether CCT5 was involved in the regulation of F-actin in MERTK-associated RP, CCT5 expression at the mRNA and protein levels was assessed. The RT-PCR results indicated that the downregulation of MERTK significantly suppressed the expression of CCT5 at the mRNA level, compared with that in the NC group, in HsRPE cells (Figure 2D). In line with the mRNA-level results, the Western blotting results demonstrated that CCT5 expression was significantly suppressed by MERTK siRNA in HsRPE cells (Figure 2E). To analyze whether CCT5 and F-actin directly interacted, we performed immunolocalization and co-immunoprecipitation (Co-IP) assays. Immunofluorescence analysis demonstrated that CCT5 and F-actin co-localized in the cytoplasm of HsRPE cells (Figure 2F), and Co-IP analysis demonstrated that CCT5 proteins could directly bind with F-actin proteins, but could not directly bind with MERTK proteins in HsRPE cells (Figures 2G,H). These results indicate that CCT5 interacts and co-localizes with F-actin but not MERTK and that MERTK siRNA can decrease F-actin expression via the regulation of CCT5 expression in HsRPE cells.
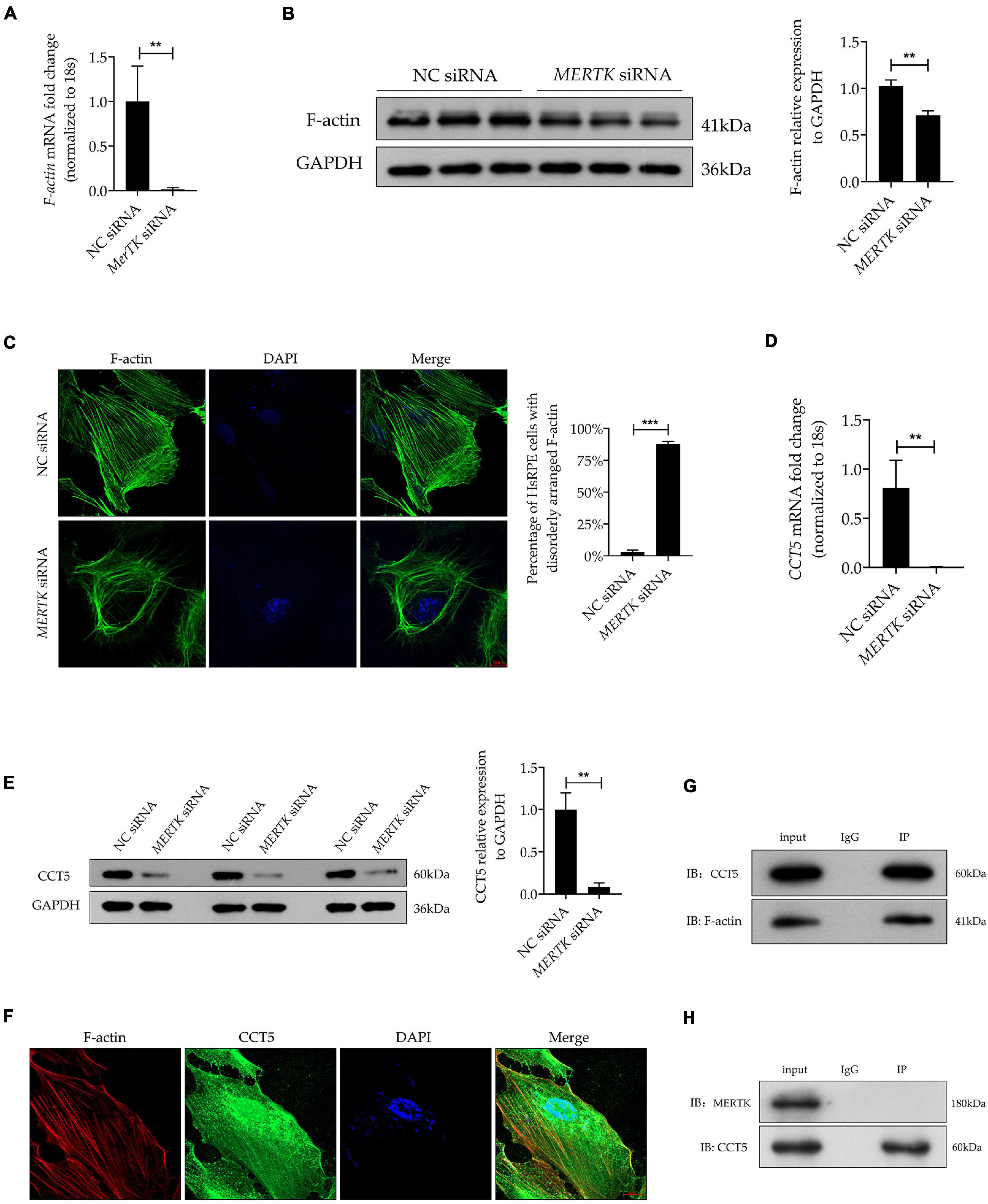
Figure 2. MERTK siRNA downregulated the expression of CCT5, which interacted and co-localized with F-actin in HsRPE cells. (A) HsRPE cells were treated with NC or MERTK siRNA for 24 h, and the expression of F-actin was analyzed by RT-PCR. (B) HsRPE cells were treated with NC or MERTK siRNA for 48 h, and the expression of F-actin was analyzed by Western blotting. (C) HsRPE cells were treated the same as in (B), and the distribution and organization of F-actin (green) were detected by immunofluorescence analysis. Scale bars: 10 μm. (D) HsRPE cells were treated the same as in (A), andCCT5 expression was analyzed by RT-PCR. (E) HsRPE cells were treated the same as in (B), and CCT5 expression was analyzed by Western blotting. (F) The co-localization (yellow) of F-actin (red) and CCT5 (green) in HsRPE cells was assessed by immunolocalization analysis. Scale bars: 10 μm. (G) The binding between F-actin and CCT5 in HsRPE cells was verified by co-immunoprecipitation. (H) The binding between MERTK and CCT5 in HsRPE cells was verified by co-immunoprecipitation. **P < 0.01; *** P < 0.001.
Upregulation of Chaperonin-Containing TCP1 Subunit 5 Expression Recovered the Morphology and Migration Function Destroyed by Mer Receptor Tyrosine Kinase siRNA in Human Primary RPE Cells
For a better insight into the involvement of CCT5 in RP, CCT5-specific siRNA and lentiviral vectors were used to downregulate and upregulate CCT5 expression separately. We tested two siRNAs (CCT5 siRNA1 and CCT5 siRNA2) and selected CCT5 siRNA2, showing the best downregulation efficiency, for the subsequent experiments (Figure 3A). After the CCT5 siRNA was transfected into HsRPE cells, the expression of CCT5 was significantly downregulated. On the other hand, treatment with CCT5-specific lentiviral vectors (CCT5-Le) significantly upregulated the expression of CCT5, compared with that in the MERTK siRNA group, at both the mRNA and protein levels (Figures 3B,C). CCK8 assays were used to detect the viability of HsRPE cells. Compared with the MERTK siRNA group, the MERTK siRNA + CCT5 siRNA group demonstrated significant further suppression of cell proliferation, as the optical density (OD) value of the MERTK siRNA + CCT5 siRNA group was lower than that of the MERTK siRNA groups. However, the viability of HsRPE cells in the MERTK siRNA + CCT5-Le group was significantly higher than that seen in the MERTK siRNA group (Figure 3D). These results demonstrate that the upregulation of CCT5 expression enhanced the viability of HsRPE cells, which was inhibited by MERTK siRNA. Next, we observed changes in cell morphology using an inverted microscope. In the MERTK siRNA + CCT5 siRNA group, the shape of the HsRPE cells became more elongated than in the MERTK siRNA group. However, in the MERTK siRNA + CCT5-Le group, the shape of the HsRPE cells became typically polygonal, as in the NC group (Figure 3E). These results indicate that upregulating CCT5 expression restored the morphological dysfunction induced by MERTK siRNA. Wound-healing and Transwell-migration assays demonstrated that the migration distance and the number of migrated HsRPE cells were significantly suppressed in the MERTK siRNA group compared with the control group and that CCT5 siRNA further suppressed these parameters. However, CCT5-Le significantly increased the migration distance and the number of migrated HsRPE cells, which were suppressed by MERTK siRNA (Figures 3F,G). These results indicate that upregulation of CCT5 expression restored the morphology and migration function, which were disrupted in HsRPE cells with MERTK siRNA.
Figure 3. Upregulation of CCT5 expression recovered the morphology and migration function disrupted by MERTK siRNA in HsRPE cells. (A) HsRPE cells were treated with NC, CCT5 siRNA1, or CCT5 siRNA2 (20 nM) for 24 h, and CCT5 expression was analyzed by RT-PCR. (B) HsRPE cells were treated with or without CCT5 siRNA or CCT5-specific lentiviral vectors (CCT5-Le) after exposure to NC or MERTK siRNA for 24 h, and the expression of CCT5 was analyzed by RT-PCR. (C) HsRPE cells were treated with or without CCT5 siRNA or CCT5-specific lentiviral vectors (CCT5-Le) after exposure to NC or MERTK siRNA for 48 h, and CCT5 expression was analyzed by Western blotting. (D) HsRPE cells were treated the same as in (C), and cell proliferation was determined by CCK8 assay. (E) HsRPE cells were treated the same as in (C), and morphological changes were observed using an inverted microscope. Scale bars: 100 μm. (F) HsRPE cells were treated the same as in (C), and wound-healing assays were performed to assess the wound-healing capabilities of HsRPE cells. Scale bars: 200 μm. (G) HsRPE cells were treated the same as in (C), and Transwell assays were performed to evaluate the migration activity of HsRPE cells (purple). Scale bars: 100 μm. # P > 0.05; *P < 0.05; **P < 0.01; *** P < 0.001.
Upregulating Chaperonin-Containing TCP1 Subunit 5 Expression Rescued the Phagocytic Function Disrupted by Mer Receptor Tyrosine Kinase siRNA by Restoring the Expression and Organization of Filamentous Actin in Human Primary RPE Cells
To investigate the function of CCT5-Le in the phagocytic process of HsRPE cells, we performed the phagocytosis assay combined with the immunofluorescence assay. The MERTK siRNA group exhibited a significantly decreased expression of CCT5 (green) and a significantly decreased quantity of red particles that were phagocytized into the HsRPE cells, as compared with the NC siRNA group. In the MERTK siRNA + CCT5 siRNA group, the expression of CCT5 was further decreased and the number of red particles phagocytized into HsRPE cells was further reduced compared with that in the MERTK siRNA group. By contrast, the MERTK siRNA + CCT5-Le group exhibited a significantly increased CCT5 expression and quantity of red particles phagocytized into HsRPE cells compared with the MERTK siRNA group (Figure 4A). To investigate whether CCT5-Le rescued the phagocytic function via the regulation of F-actin, the expression of F-actin was then examined. Our results indicate that MERTK siRNA significantly decreased the expression of F-actin at both the mRNA and protein levels, as compared with that in the NC group. In the MERTK siRNA + CCT5 siRNA group, the expression of F-actin was decreased further compared with that in the MERTK siRNA group. However, in the MERTK siRNA + CCT5-Le group, F-actin expression was significantly increased, compared with that in the MERTK siRNA and MERTK siRNA + CCT5 siRNA groups (Figures 4B,C). Furthermore, to detect whether CCT5-Le restored the distribution and arrangement of F-actin, we performed the immunofluorescence analysis. In the MERTK siRNA + CCT5 siRNA group, the distribution and arrangement of F-actin became more irregular than that in the MERTK siRNA group. However, in the MERTK siRNA + CCT5-Le group, the distribution and arrangement of F-actin were recovered, as seen in the NC group (Figure 4D). These results indicate that the upregulation of CCT5 expression rescued the phagocytic function, which was disrupted by MERTK siRNA, by restoring the expression and organization of F-actin in HsRPE cells.
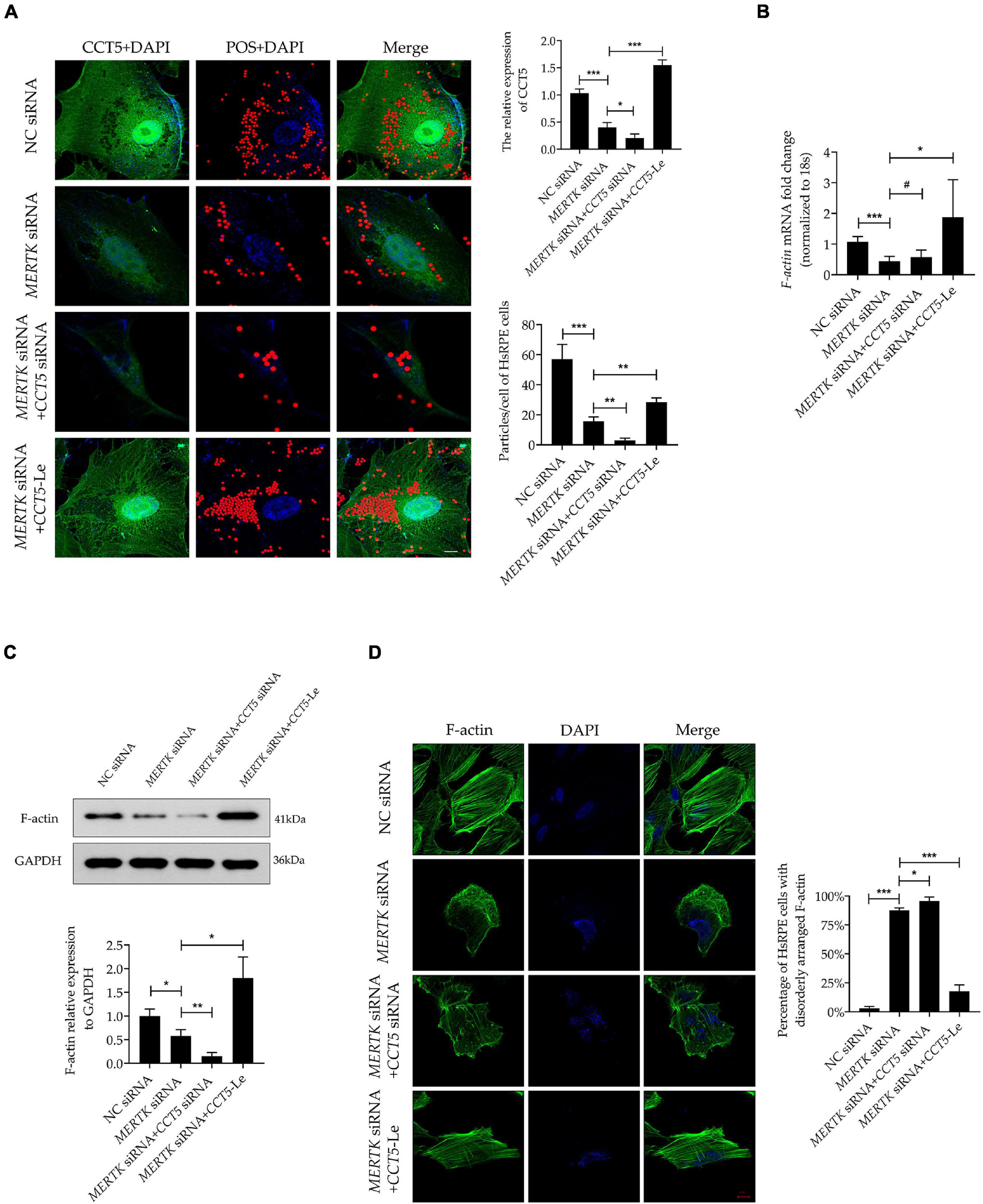
Figure 4. CCT5 rescued the phagocytic function disrupted by MERTK siRNA by restoring the expression and organization of F-actin in HsRPE cells. (A) HsRPE cells were treated with or without CCT5 siRNA or CCT5-Le after exposure to NC or MERTK siRNA for 48 h, and the phagocytic ability of HsRPE cells was examined by phagocytosis assay. The red dots represent the particles and the green dye represents the CCT5 protein. Scale bars: 5 μm. (B) HsRPE cells were treated with or without CCT5 siRNA or CCT5-Le after exposure to NC or MERTK siRNA for 24 h, and F-actin expression was analyzed by RT-PCR. (C) HsRPE cells were treated the same as in (A), and F-actin expression was analyzed by Western blotting. (D) HsRPE cells were treated the same as in (A), and the distribution and organization of F-actin (green) were detected by immunofluorescence analysis. Scale bars: 10 μm. # P > 0.05; *P < 0.05; **P < 0.01; *** P < 0.001.
Chaperonin-Containing TCP1 Subunit 5 Rescued Human Primary RPE Cells With Mer Receptor Tyrosine Kinase-Associated Retinitis Pigmentosa via the LIM-Kinase 1/Cofilin, but Not the SSH1/Cofilin, Pathway
Phosphorylated cofilin (p-cofilin) and cofilin are essential regulators of F-actin dynamics, regulating the polymerization and depolymerization of F-actin in the processes of phagocytosis and migration. To determine whether cofilin and p-cofilin were involved in CCT5-Le’s rescue of the phagocytosis and migration function of HsRPE cells with MERTK-associated RP, we next determined their levels of expression. Notably, we found that the expression of both cofilin and p-cofilin was suppressed in the MERTK siRNA group compared with that in the NC group. In the MERTK siRNA + CCT5 siRNA group, the expression of both cofilin and p-cofilin was decreased further when compared with that in the MERTK siRNA group. However, in the MERTK siRNA + CCT5-Le group, the expression of both cofilin and p-cofilin was increased compared with that in the MERTK siRNA and MERTK siRNA + CCT5 siRNA groups (Figure 5A). Cofilin is inactivated via phosphorylation by LIM-kinase 1 (LIMK1) and testicular protein kinase 1 (TESK1) and reactivated via dephosphorylation by slingshot 1 (SSH1). Therefore, to investigate whether LIMK1, TESK1, and SSH1 regulated the activity of cofilin in the process of CCT5-Le’s rescue of the phagocytosis and migration function of HsRPE cells under MERTK-associated RP, we detected the expression of LIMK1, TESK1, and SSH1. The results indicate that the expression of LIMK1, TESK1, and SSH1 was decreased in the MERTK siRNA group, compared with that in the NC group. However, in the MERTK siRNA + CCT5 siRNA group, only the expression of LIMK1 and TESK1, and not SSH1, was further decreased compared with that in the MERTK siRNA group. In the MERTK siRNA + CCT5-Le group, the expression of LIMK1 and TESK1 was increased, but the expression of SSH1 again demonstrated no significant change compared with that in the MERTK siRNA and MERTK siRNA + CCT5 siRNA groups (Figure 5B). These results indicate that CCT5-Le rescued the phagocytosis and migration function which were destroyed by MERTK siRNA via the LIMK1/cofilin, but not the SSH1/cofilin, pathway.
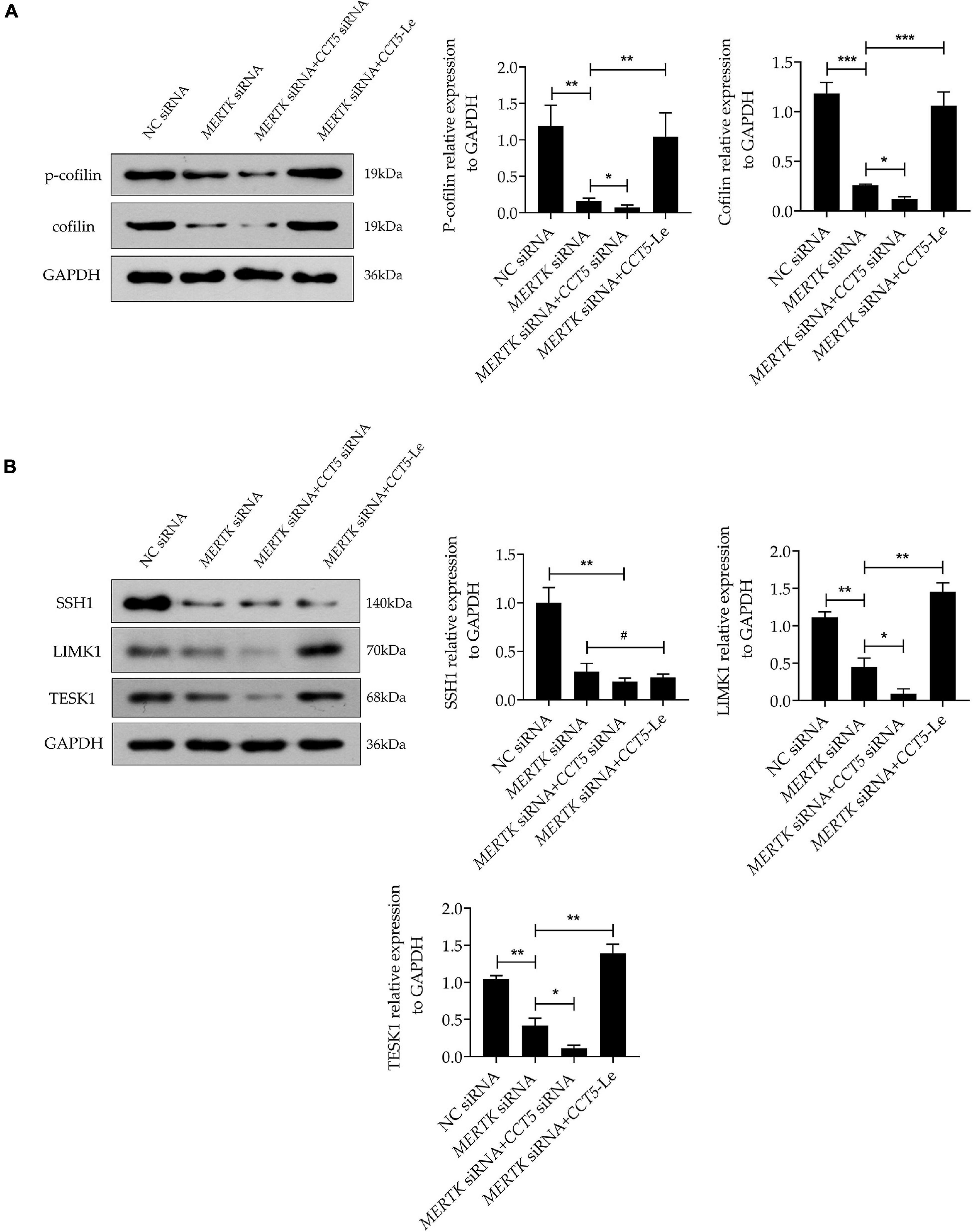
Figure 5. CCT5 rescued HsRPE cells with MERTK-associated RP via the LIMK1/cofilin, but not the SSH1/cofilin, pathway. (A) HsRPE cells were treated with or without CCT5 siRNA or CCT5-Le after exposure to NC or MERTK siRNA for 48 h, and the cofilin and p-cofilin expression was analyzed by Western blotting. (B) HsRPE cells were treated the same as in (A), and the expression of LIMK1, TESK1, and SSH1 was analyzed by Western blotting. *P < 0.05; **P < 0.01; *** P < 0.001.
Discussion
Retinitis pigmentosa is caused by multiple molecular interactions, of which the phagocytic dysfunction induced by a MERTK mutation in RPE cells is one of the most vital (1). As a cytoskeleton actin protein, F-actin plays an important role in cellular motility and phagocytosis (27). CCT5 acts as a chaperone protein and plays a vital role in maintaining the normal function and structure of actin proteins (28). Recently, numerous studies have demonstrated the role that F-actin plays in the phagocytosis of RPE cells (29). However, to date, studies have failed to determine whether CCT5 is associated with F-actin regulation or whether it plays an essential role in the progression of RP. Therefore, in this study, we focused on the role of CCT5’s regulation of F-actin in the pathogenesis of RP and the molecular mechanism of such.
We found that downregulating MERTK inhibited cell proliferation, migration, and phagocytic function. Additionally, knocking down MERTK significantly inhibited F-actin’s expression and disrupted its regular arrangement. Importantly, our study revealed, for the first time, that CCT5 interacts with F-actin and that CCT5 expression is inhibited by MERTK siRNA in HsRPE cells. Upregulating CCT5 rescued the cell proliferation, migration, and phagocytic function which were destroyed by MERTK siRNA via an increase in the expression of F-actin, as well as the remodeling of its regular arrangement. Moreover, we discovered that CCT5 promoted the remodeling of F-actin’s arrangement via the LIMK1/cofilin, but not the SSH1/cofilin, pathway (Figure 6).
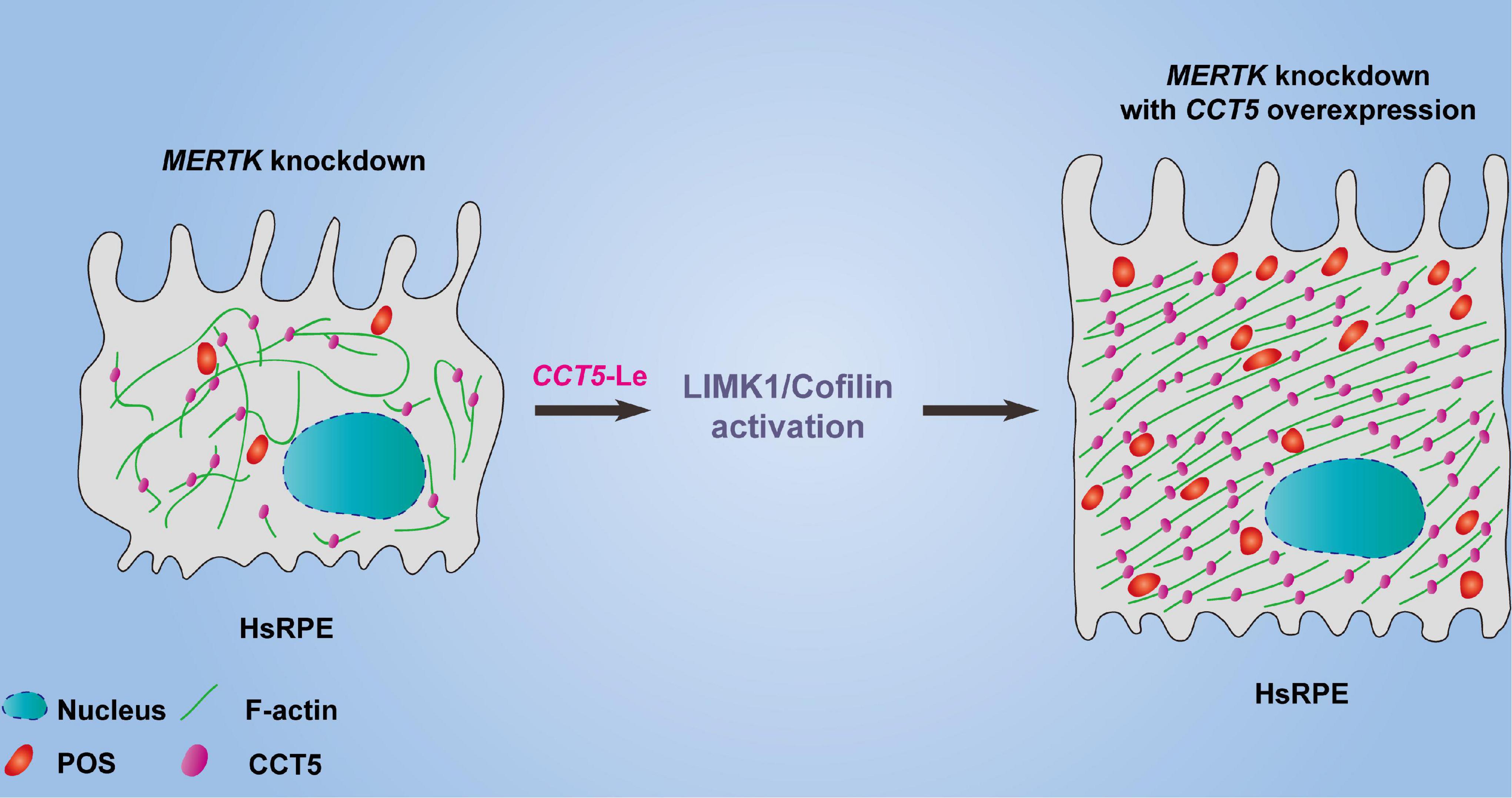
Figure 6. The model diagram of CCT5-specific lentiviral vectors (CCT5-Le) rescued the cell phagocytic function of HsRPE cells under MERTK siRNA condition by increasing the expression of F-actin and restoring its regular arrangement via the LIMK1/cofilin pathway.
Phagocytosis in RPE cells is a highly conserved, complex process that has evolved to counter the constant shedding of POS (30). To maintain the healthy photoreceptor–RPE interface of the retina, RPE cells constantly phagocytize the POS; however, the mutation of MERTK results in phagocytic dysfunction in RPE cells, leading to RP. However, which part of the phagocytic process is disrupted in RPE cells with the MERTK mutation remains to be elucidated. Ensheathment is required for POS fragmentation before internalization. The MERTK ligands Gas6 and Protein S contribute to the initiation of POS ensheathed by RPE (31). Abnormalities in the ensheathment, fragmentation, and internalization process in POS phagocytosis were found to occur in MERTK-mutated RPE while restoring MERTK expression in the RPE cells of RP patients reversed these abnormalities (32). The ensheathment, fragmentation, and internalization process require an appropriate F-actin distribution and arrangement (33). Therefore, the appropriate expression and correct spatial organization of F-actin filaments are essential for normal proliferation, migration, and phagocytic function (34–36). A recent study reported that, due to the lack of Mertk, the RPE cells of RCS rats could not aggregate F-actin and bind POS, even in the presence of protein S (14). Additionally, a previous study revealed that hypophagocytic RPE cells contain F-actin stress fibers but lack adjacent lateral circumferential F-actin (13). Together, these studies indicate that F-actin is involved in MERTK-associated RP. In line with these findings, our MERTK-associated RP model in HsRPE cells demonstrated that knocking down MERTK significantly inhibited the expression of F-actin and disrupted its regular arrangement. Moreover, we found that downregulating MERTK expression inhibited proliferation, migration, and phagocytic function.
Filamentous actin is regulated by various actin-binding proteins (37). Cofilin is a ubiquitous actin-binding protein that binds to F-actin only in its dephosphorylated state. When dephosphorylated cofilin binds to actin, it depolymerizes F-actin; however, phosphorylation at Ser 3 inactivates cofilin which then separates from, and thereby repolymerizes, F-actin (38). The recombination of the actin cytoskeleton via polymerization and depolymerization of F-actin by cofilin is indispensable for various cellular activities, such as phagocytosis, cytokinesis, and cell migration (39). The balance between F-actin and cofilin is vital for preserving the normal physiological function of cells. Once this balance is disturbed, many diseases can result—including Alzheimer’s disease (40, 41), mitotic disorder (42), cancers (43), optic nerve injury (44), migration disorder (35), immune dysfunction (45), and more. However, whether cofilin takes part in MERTK-associated RP retains a veil of mystery. First, we revealed that knocking down MERTK significantly inhibited the expression of both cofilin and p-cofilin in HsRPE cells. These results are consistent with the abnormal expression and arrangement of F-actin in our MERTK-associated RP model in HsRPE cells. The phosphorylation of cofilin is controlled by the LIMK1/TESK1 pathway (46–48). LIMK1 and TESK1 are, therefore, two key components of the signal transduction pathways that link extracellular stimulation to changes in cytoskeletal structure (49, 50). It was reported that the N-terminal kinase of TESK1 may be a putative candidate accounting for LIMK-independent cofilin phosphorylation, as it shows approximately 50% amino-acid identity with LIMK (51, 52). On the other hand, cofilin is activated by dephosphorylation by the phosphatase SSH1. SSH1’s activity is strongly increased by its binding to F-actin (53). According to previous studies, the overexpression of LIMK1 and TESK1 in cultured cells results in the accumulation of F-actin, and the overexpression of SSH1 leads to the depolymerization of F-actin (49, 50, 53). However, no currently available research indicates a regulatory relationship between LIMK1, TESK1, SSH1, cofilin, and F-actin in RP. In our study, we revealed that the expression of LIMK1, TESK1, and SSH1 was inhibited after MERTK expression was downregulated. These results explain why knocking down MERTK significantly inhibited the expression of both cofilin and p-cofilin in HsRPE cells.
As a chaperonin, CCT5 assists in the folding of the unfolding or misfolded proteins, and actin is reported to be a natural substrate for CCT5 (21). It is reported that the effect of CCT5 on the z-disk of sarcomere cells is required for the efficient assembly of actin filaments, and CCT5-specific ATP binding is required for the efficient folding of actin in vivo. In addition, the mutant A-actin subtype that causes linear myopathy in patients acquires its pathogenic conformation through this function of CCT5 (22). Julie et al. found that CCT depletion did not affect actin-polypeptide synthesis but led to a decrease in natural actin levels and a disturbance of actin-based cell motility (54). McCormack et al. found that actin appeared to fold in association with chaperonin and that actin site II—located at the apex of actin subdomain 4—was the main binding site for CCT binding (55). Llorca et al. produced a three-dimensional reconstruction of the CCT and α-actin complex, which demonstrated that α-actin interacts with the apical domain of one of the two CCT subunits. Furthermore, they found that actin’s binding to CCT is subunit-specific and geometrically dependent (21). CCT5’s interactions in HsRPE cells with the MERTK mutation have not yet been reported. In our study, we discovered that CCT5 interacted with F-actin and that CCT5’s expression was inhibited by MERTK siRNA in HsRPE cells. To verify that F-actin was regulated by CCT5, we downregulated and upregulated CCT5 separately in HsRPE cells. We found that upregulating the expression of CCT5 using CCT5-Le increased the expression of F-actin and remodeled its distribution and arrangement in our MERTK-associated RP model in HsRPE cells. When the expression of CCT5 was decreased using CCT siRNA, the effect was exactly opposite to that of CCT5-Le. In addition to its effect on actin, CCT5 has been reported to be connected to multiple cellular processes including proliferation, migration, and apoptosis via its relation with numerous proteins such as cyclin D1, polymerase basic protein, cell-division cycle protein 20, and P53 (56–59). In our study, we found that CCT5 regulated the expression of LIMK1 and TESK1 but not SSH1. Moreover, upregulating CCT5 rescued the cell proliferation, migration, and phagocytic function of HsRPE cells under MERTK-associated RP. These results indicate that CCT5 protects against MERTK-associated RP in RPE cells through interactions with F-actin and the activation of the LIMK1/cofilin pathway.
However, there are several limitations to our study. First, we only performed in vitro experiments in an RPE cell model established using MERTK siRNA. In vivo experiments should be performed in RCS rats to verify the role of CCT5 in MERTK-associated RP. Many questions remain to be answered in RCS rats, including whether the expression of CCT5 is abnormal in the RPE cells of RCS rats and whether regulating the expression of CCT5 can rescue the RP. Second, although CCT consists of eight subunits, we only detected the function of CCT5 in HsRPE cells. Thus, whether other paralogous subunits are involved in RP requires further exploration. Third, we found that the expression of cofilin, p-cofilin, LIMK1, and TESK1 was upregulated by CCT5-Le, but how CCT5 increases their expression—whether by increasing their transcription or by decreasing their degradation—remains to be determined. Although LIMK1 and TESK1 control the phosphorylation of cofilin, no studies have demonstrated that they control the expression of cofilin. Therefore, whether CCT5 directly increases total cofilin expression or indirectly increases cofilin levels via other signals requires further study. We will attempt to find answers to these questions in future research.
In summary, despite the abovementioned limitations, our results demonstrate, for the first time, that the cell proliferation, migration, and phagocytic function of RPE cells are inhibited by MERTK-associated RP due to the abnormal expression of F-actin and disruption of its regular arrangement. Importantly, our findings reveal that CCT5 interacts with F-actin and regulates its expression and arrangement. Upregulating CCT5 protects against the effect of MERTK knockdown in RPE cells through its interactions with F-actin and activation of the LIMK1/cofilin pathway. These findings provide a new perspective for research into the mechanisms underlying MERTK-associated RP and provide a new direction for future studies of the molecular mechanisms of RP pathogenesis.
Materials and Methods
Cell Culture
Human donor eyes were obtained from the Guangdong Eye Bank in accordance with the 2013 Declaration of Helsinki. Written consent was obtained from the donor or the donor’s family for the eye to be used in medical research. The study was approved by the Ethics Committee of Zhongshan Eye Center, Sun Yat-sen University. The human primary RPE (HsRPE) cells were separated from human donor eyes as previously described (60). All the HsRPE cells were cultured in DMEM containing 4.5 g/L glucose, which was supplemented with 1% penicillin/streptomycin and 10% FBS, in a humidified incubator with a 5% CO2 atmosphere at 37°C.
siRNA Transfections
The siRNAs for MERTK and CCT5 were designed and synthesized by RiboBio Inc. (Guangzhou, China). The sequences of the MERTK siRNA and CCT5 siRNA are listed in Table 1. Transient transfections of the siRNAs for human MERTK (20 nM) and CCT5 (20 nM) were conducted using Lipofectamine RNAiMAX (Invitrogen, New York, NY, United States), according to the manufacturer’s protocol. Morphological changes in the cells were investigated using an inverted microscope (Carle ZEISS Axio Observer 7, Oberkohen, Baden-wurttemberg, Germany), 48 h after siRNA transfection.
Real-Time PCR
The RNA of the HsRPE samples was extracted using Trizol reagent (Invitrogen, New York, NY, United States). After cDNA was synthesized by reverse transcribing the RNA, the RT-PCR assay was performed using a LightCycler 480 SYBR Green I Master (Roche, Indianapolis, IN, United States). The values for each gene were normalized to the levels of tubulin or 18S mRNA. The sequences of the primers used for RT-PCR are listed in Table 2.
Western Blot Assay
Cell lysis buffer was used to extract the total protein of the HsRPE cell samples before it was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The total protein was then transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, United States). The PVDF membranes were incubated with different primary antibodies: anti-MERTK (Cell Signaling Technology, 4319S), anti-F-actin (Abcam, ab130935), anti-CCT5 (Proteintech, 11603-1-AP), anti-cofilin (Abcam, ab54532), anti-phosphorylated cofilin (p-cofilin, Abcam, ab12866), anti-LIMK1 (Affinity, AF6345), anti-TESK1 (Affinity, DF4012), anti-SSH1 (Abcam, ab76943), and anti-GAPDH (Abcam, ab8245). The membranes were incubated with the corresponding secondary antibody for 1 h at room temperature. The binding of specific antibodies was visualized using Chemiluminescent fluid (Millipore, WBKLS0500).
Cell Counting Kit-8 Assay
A CCK8 kit (Beyotime, Shanghai, China) was used to determine the cell viability. The HsRPE cells were cultured at a density of 7 × 103 cells/well in 96-well plates, after exposure to MERTK siRNA with or without the interference of CCT5 expression for 48 h. The CCT8 assay was performed as previously described in Zhou et al. (61), and the absorption values at a wavelength of 570 nm were evaluated.
Wound-Healing and Transwell-Migration Assays
Wound-healing assays were performed as previously described in Qi et al. (62). Briefly, HsRPE cells from all the groups were seeded into 24-well plates until 100% confluence was attained. Wounds were scratched in the confluent cell layers using 200 uL pipette tips, and the samples were cultured for a further 24 h. The Transwell assays were performed in 24-well plates with a chamber insert (8 μm pore size) (Corning, 3422, Corning, NY, United States). After 24 h of incubation with transfected MERTK siRNAs, CCT5 siRNAs, or CCT5 lentiviral vectors (CCT5-Le), the underlying HsRPE cells were stained with crystal violet before the number of migrated cells was determined using an inverted microscope (Carle ZEISS Axio Observer 7, Oberkohen, Baden-wurttemberg, German).
Phagocytosis Assay
Phagocytosis assays were performed as previously described in Irschick et al. (63). To assess the phagocytosis of HsRPE cells, Nile red-labeled Fluospheres (Sigma-Aldrich, F8825) were used. HsRPE cells from all the groups were cultured until the cell confluence reached 80%. Then, the medium was replaced and 10 μL/mL of diluted 2 μm-diameter particles were added. HsRPE cells were phagocytized with microspheres in a 5% CO2 atmosphere at 37°C for 6 h and then washed with PBS three times to remove the unphagocytized particles before being stained with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min. Finally, the number of microspheres was quantified by measuring the total fluorescence present using a confocal microscope (Carle ZEISS Axio Imager Z2, Oberkohen, Germany).
Immunofluorescence
The immunofluorescence assays were performed as previously described in Feng et al. (64). HsRPE cells were fixed with 4% paraformaldehyde for 15 min, followed by blocking with 5% BSA at room temperature for 1 h. The cell samples were incubated with RPE65 (Abcam, ab235950), F-actin (Abcam, ab130935), CCT5 (Proteintech, 11603-1-AP), and MERTK (Cell Signaling Technology, 4319S) antibodies. Cell samples were then incubated with secondary antibodies for 1 h at room temperature. Subsequently, the nuclei of the cells were stained using DAPI for 7 min. All the images were taken using a confocal microscope (Carle ZEISS LSM 980, Oberkohen, Germany).
Co-immunoprecipitation
The total protein of the HsRPE cell samples was extracted using cell lysis buffer, leaving 100 μL of cell lysate as the input group. Two 1.5 EP tubes were filled with 400 μL of HsPRE cell lysis solution, and 100 uL of RIPA lysis buffer was added to each. One tube was incubated under rotation with 5 μg of anti-CCT5 (Proteintech, 11603-1-AP), while the other tube was incubated under rotation with 5 μg of rabbit anti-IgG (Cell Signaling Technology, 2729) at 4°C overnight. Fifty microliters of Protein A/G Magnetic Beads (Bimake Ferromagnetic Bead, B23202) were added to two additional 1.5 mL EP tubes. The magnetic beads were washed twice with 1 ml of precooled PBS and centrifuged at 3000 rpm for 1 min. Next, the magnetic beads were rotated and blocked with 3% BSA at 4°C for 30 min, followed by centrifugation at 3000 rpm for 1 min, absorbing the blocking solution, adding the antibody-protein complexes into the magnetic beads, and incubating under rotation at room temperature for 1 h. After incubation, the magnetic beads were washed with washing buffer. After the centrifugation of the proteins at 12,000 rpm and 4°C for 1 min, the supernatant was discarded carefully. Next, the proteins were separated by SDS-PAGE and transferred onto PVDF membranes. The membranes were probed with anti-F-actin (Abcam, ab130935) and anti-CCT5 (Proteintech, 11603-1-AP), and their corresponding secondary antibodies. Specific antibody binding was visualized using chemiluminescent fluid (Millipore, WBKLS0500).
Transfection and Expression of Chaperonin-Containing TCP1 Subunit 5-Le
Lentiviral vectors were used to upregulate the expression of CCT5. The recombined plasmid, contained pLV-CMV.hCCT5.EF1.CopGFP-T2A-Puro.Wpre, was used to produce the lentivirus. The 293 T cells were then transfected with the lentivirus. After 72 h, the 293 T cells were collected, the concentrated virus was extracted by centrifugation, and the virus titer was detected. The HsRPE cells were then inoculated in 6-well plates and incubated until their confluence was about 30%, and 50 μL of virus was added (titer: 2 × 108 TU/mL). After another 48 h of culture, the expression of the target gene in the lentivirus was observed by measuring the green fluorescent protein (GFP), and the mRNA and protein levels were detected. The sequences of the lentiviral vectors of CCT5 are listed in Table 3.
Statistical Analysis
Statistical analysis was performed using SPSS 26.0. An unpaired t-test and one-way ANOVA were used to analyze and compare the differences between groups. The data are expressed as the means ± standard deviations (SD) of at least three independent trials. A p-value less than 0.05 was considered statistically significant.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Zhongshan Eye Center, Sun Yat-sen University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
LF designed the experiments and wrote the manuscript. HL performed the experiments. YD performed the literature search and provided experimental support. TZ, YtZ, ZdL, LZ, and XW collected the data. GW, LbZ, ZJ, and ZL analyzed the data. ZO and YW performed the extraction of HsRPE cells. YhZ supervised the project and reviewed the manuscript. LF, HL, and YD have made equal contributions to the study and should therefore be regarded as equal authors. All authors have read and approved the published version of the manuscript.
Funding
Our study was funded by the Science and Technology Program of Guangzhou, China (202102010216); the National Key Research and Development Project of China (2020YFA0112701); the National Natural Science Foundation of China (82171057).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Caibin Deng for his helpful suggestions, to Jing Chen for her help in the culture of the HsRPE cells, and to Jiangna Chen for her help regarding the Western blot analysis. We thank the members of the Zhongshan Ophthalmic Center at Sun Yat-sen University for their helpful comments and suggestions during this study.
Abbreviations
CCK8, Cell Counting Kit-8; CCT, chaperonin-containing TCP-1; CCT5, chaperonin-containing TCP1 subunit 5; CCT5-Le, CCT5-specific lentiviral vectors; Co-IP, co-immunoprecipitation; F-actin, filamentous actin; HsRPE, human primary RPE; LIMK1, LIM-kinase 1; MERTK, Mer receptor tyrosine kinase; NC, negative control; OD, optical density; p-cofilin, Phosphorylated cofilin; POS, photoreceptor outer segment; RCS, Royal College of Surgeons; RP, retinitis pigmentosa; RPE, retinal pigment epithelium; RT-PCR, quantitative real-time PCR; SSH1, slingshot 1; TESK1, testicular protein kinase 1.
References
1. Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. (2006) 368:1795–809. doi: 10.1016/s0140-6736(06)69740-7
2. Humphries P, Kenna P, Farrar GJ. On the molecular genetics of retinitis pigmentosa. Science. (1992) 256:804–8. doi: 10.1126/science.1589761
3. Dias MF, Joo K, Kemp JA, Fialho S, Cunha ADS, Woo SJ, et al. Molecular genetics and emerging therapies for retinitis pigmentosa: basic research and clinical perspectives. Prog Retin Eye Res. (2018) 63:107–31. doi: 10.1016/j.preteyeres.2017.10.004
4. Ducloyer J-B, Le Meur G, Cronin T, Adjali O, Weber M. Gene therapy for retinitis pigmentosa. Med Sci. (2020) 36:607–15. doi: 10.1051/medsci/2020095
5. Gal A, Li Y, Thompson D, Weir J, Orth U, Jacobson S, et al. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat Genet. (2000) 26:270–1. doi: 10.1038/81555
6. D’Cruz PM, Yasumura D, Weir J, Matthes MT, Abderrahim H, Lavail MM, et al. Mutation of the receptor tyrosine kinase gene MERTK in the retinal dystrophic RCS rat. Hum Mol Genet. (2000) 9:645–51. doi: 10.1093/hmg/9.4.645
7. Kwon W, Freeman SA. Phagocytosis by the retinal pigment epithelium: recognition, resolution, recycling. Front Immunol. (2020) 11:604205. doi: 10.3389/fimmu.2020.604205
8. Essner E, Pino RM, Griewski RA. Distribution of anionic sites on the surface of retinal pigment epithelial and rod photo-receptor cells. Curr Eye Res. (1981) 1:381–9. doi: 10.3109/02713688109019975
9. Bulloj A, Duan W, Finnemann SC. PI 3-kinase independent role for AKT in F-actin regulation during outer segment phagocytosis by RPE cells. Exp Eye Res. (2013) 113:9–18. doi: 10.1016/j.exer.2013.05.002
10. Finnemann SC, Rodriguez-Boulan E. Macrophage and retinal pigment epithelium phagocytosis: apoptotic cells and photoreceptors compete for alphavbeta3 and alphavbeta5 integrins, and protein kinase C regulates alphavbeta5 binding and cytoskeletal linkage. J Exp Med. (1999) 190:861–74. doi: 10.1084/jem.190.6.861
11. Bulloj A, Maminishkis A, Mizui M, Finnemann SC. Semaphorin4D-PlexinB1 Signaling Attenuates Photoreceptor Outer Segment Phagocytosis by Reducing Rac1 Activity of RPE Cells. Mol Neurobiol. (2018) 55:4320–32. doi: 10.1007/s12035-017-0649-5
12. Freeman SA, Goyette J, Furuya W, Woods EC, Bertozzi CR, Bergmeier W, et al. Integrins form an expanding diffusional barrier that coordinates phagocytosis. Cell. (2016) 164:128–40. doi: 10.1016/j.cell.2015.11.048
13. Müller C, Charniga C, Temple S, Finnemann SC. Quantified F-actin morphology is predictive of phagocytic capacity of stem cell-derived retinal pigment epithelium. Stem Cell Rep. (2018) 10:1075–87. doi: 10.1016/j.stemcr.2018.01.017
14. Mao Y, Finnemann S. Acute RhoA/Rho kinase inhibition is sufficient to restore phagocytic capacity to retinal pigment epithelium lacking the engulfment receptor MERTK. Cells. (2021) 10:1927. doi: 10.3390/cells10081927
15. Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. Embo J. (1992) 11:4767–78. doi: 10.1002/j.1460-2075.1992.tb05582.x
16. Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, et al. 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc Natl Acad Sci USA. (2010) 107:4967–72. doi: 10.1073/pnas.0913774107
17. Rommelaere H, Van Troys M, Gao Y, Melki R, Cowan NJ, Vandekerckhove J, et al. Eukaryotic cytosolic chaperonin contains t-complex polypeptide 1 and seven related subunits. Proc Natl Acad Sci USA. (1993) 90:11975–9. doi: 10.1073/pnas.90.24.11975
18. Frydman J, Hartl FU. Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms. Science. (1996) 272:1497–502. doi: 10.1126/science.272.5267.1497
19. Dekker C, Stirling P, McCormack EA, Filmore H, Paul A, Brost RL, et al. The interaction network of the chaperonin CCT. Embo J. (2008) 27:1827–39. doi: 10.1038/emboj.2008.108
20. Llorca O, Martín-Benito J, Grantham J, Ritco-Vonsovici M, Willison KR, Carrascosa JL, et al. The ‘sequential allosteric ring’ mechanism in the eukaryotic chaperonin-assisted folding of actin and tubulin. Embo J. (2001) 20:4065–75. doi: 10.1093/emboj/20.15.4065
21. Llorca O, McCormack EA, Hynes GM, Grantham JJ, Cordell J, Carrascosa JL, et al. Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature. (1999) 402:693–6. doi: 10.1038/45294
22. Berger J, Berger S, Li M, Jacoby AS, Arner A, Bavi N, et al. In vivo function of the chaperonin TRiC in α-actin folding during sarcomere assembly. Cell Rep. (2018) 22:313–22. doi: 10.1016/j.celrep.2017.12.069
23. Sergeeva O, Tran MT, Haase-Pettingell C, King JA. Biochemical characterization of mutants in chaperonin proteins CCT4 and CCT5 associated with hereditary sensory neuropathy. J Biol Chem. (2014) 289:27470–80. doi: 10.1074/jbc.m114.576033
24. Pavel M, Imarisio S, Menzies FM, Jimenez-Sanchez M, Siddiqi FH, Wu X, et al. CCT complex restricts neuropathogenic protein aggregation via autophagy. Nat Commun. (2016) 7:13821. doi: 10.1038/ncomms13821
25. Levanova N, Steinemann M, Böhmer KE, Schneider S, Belyi Y, Schlosser A, et al. Characterization of the glucosyltransferase activity of Legionella pneumophila effector SetA. Naunyn-Schmiedeberg Arch Pharmacol. (2018) 392:69–79. doi: 10.1007/s00210-018-1562-9
26. Ogneva IV, Biryukov NS. Lecithin prevents cortical cytoskeleton reorganization in rat soleus muscle fibers under short-term gravitational disuse. PLoS One. (2016) 11:e0153650. doi: 10.1371/journal.pone.0153650
27. Gruenheid S, Finlay BB. Microbial pathogenesis and cytoskeletal function. Nature. (2003) 422:775–81. doi: 10.1038/nature01603
28. Mádi A, Mikkat S, Ringel B, Thiesen HJ, Glocker MO. Profiling stage-dependent changes of protein expression in Caenorhabditis elegans by mass spectrometric proteome analysis leads to the identification of stage-specific marker proteins. Electrophoresis. (2003) 24:1809–17. doi: 10.1002/elps.200305390
29. Czuczman MA, Fattouh R, van Rijn J, Canadien V, Osborne S, Muise AM, et al. Listeria monocytogenes exploits efferocytosis to promote cell-to-cell spread. Nature. (2014) 509:230–4. doi: 10.1038/nature13168
30. Kim J-Y, Zhao H, Martinez J, Doggett TA, Kolesnikov AV, Tang PH, et al. Noncanonical Autophagy Promotes the Visual Cycle. Cell. (2013) 154:365–76. doi: 10.1016/j.cell.2013.06.012
31. Nandrot EF. Opposite roles of MERTK ligands Gas6 and protein s during retinal phagocytosis. Adv Exp Med Biol. (2018) 1074:577–83. doi: 10.1007/978-3-319-75402-4_70
32. Almedawar S, Vafia K, Schreiter S, Neumann K, Khattak S, Kurth T, et al. MERTK-dependent ensheathment of photoreceptor outer segments by human pluripotent stem cell-derived retinal pigment epithelium. Stem Cell Rep. (2020) 14:374–89. doi: 10.1016/j.stemcr.2020.02.004
33. Hayes MJ, Shao D, Bailly M, Moss SE. Regulation of actin dynamics by annexin 2. Embo J. (2006) 25:1816–26. doi: 10.1038/sj.emboj.7601078
34. Haralambiev L, Nitsch A, Jacoby JM, Strakeljahn S, Bekeschus S, Mustea A, et al. Cold atmospheric plasma treatment of chondrosarcoma cells affects proliferation and cell membrane permeability. Int J Mol Sci. (2020) 21:2291. doi: 10.3390/ijms21072291
35. Bisaria A, Hayer A, Garbett D, Cohen D, Meyer T. Membrane-proximal F-actin restricts local membrane protrusions and directs cell migration. Science. (2020) 368:1205–10. doi: 10.1126/science.aay7794
36. Kitano M, Nakaya M, Nakamura T, Nagata S, Matsuda M. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature. (2008) 453:241–5. doi: 10.1038/nature06857
37. McGough A. F-actin-binding proteins. Curr Opin Struct Biol. (1998) 8:166–76. doi: 10.1016/s0959-440x(98)80034-1
38. Hirayama A, Adachi R, Otani S, Kasahara T, Suzuki K. Cofilin plays a critical role in IL-8-dependent chemotaxis of neutrophilic HL-60 cells through changes in phosphorylation. J Leukoc Biol. (2006) 81:720–8. doi: 10.1189/jlb.0506314
39. Shiozaki N, Nakano K, Kushida Y, Noguchi TQP, Uyeda TQP, Wloga D, et al. ADF/Cofilin Is Not Essential but Is Critically Important for Actin Activities during Phagocytosis in Tetrahymena thermophila. Eukaryot Cell. (2013) 12:1080–6. doi: 10.1128/ec.00074-13
40. Bamburg JR, Bernstein BW. Actin dynamics and cofilin-actin rods in alzheimer disease. Cytoskeleton. (2016) 73:477–97. doi: 10.1002/cm.21282
41. Kang DE, Woo JA. Cofilin, a master node regulating cytoskeletal pathogenesis in alzheimer’s disease. J Alzheimer’s Dis. (2019) 72:S131–44. doi: 10.3233/JAD-190585
42. Jin Z, Yao X, Wen L, Hao G, Kwon J, Hao J, et al. AIP1 and Cofilin control the actin dynamics to modulate the asymmetric division and cytokinesis in mouse oocytes. Faseb J. (2020) 34:11292–306. doi: 10.1096/fj.202000093r
43. Chang C-Y, Leu J-D, Lee Y-J. The actin depolymerizing factor (ADF)/cofilin signaling pathway and DNA damage responses in cancer. Int J Mol Sci. (2015) 16:4095–120. doi: 10.3390/ijms16024095
44. Hao Q, Zhang Y, Li X, Liang L, Shi H, Cui Z, et al. Upregulated neuregulin-1 protects against optic nerve injury by regulating the RhoA/cofilin/F-actin axis. Life Sci. (2020) 264:118283. doi: 10.1016/j.lfs.2020.118283
45. Burkhardt JK, Carrizosa E, Shaffer MH. The Actin Cytoskeleton in T Cell Activation. Annu Rev Immunol. (2008) 26:233–59. doi: 10.1146/annurev.immunol.26.021607.090347
46. Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, et al. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. (1998) 393:809–12. doi: 10.1038/31735
47. Sakurai K, Talukdar I, Patil VS, Dang J, Li Z, Chang K-Y, et al. Kinome-wide Functional Analysis Highlights the Role of Cytoskeletal Remodeling in Somatic Cell Reprogramming. Cell Stem Cell. (2014) 14:523–34. doi: 10.1016/j.stem.2014.03.001
48. Røsok O, Pedeutour F, Ree AH, Aasheim H-C. Identification and Characterization of TESK2, a Novel Member of the LIMK/TESK Family of Protein Kinases, Predominantly Expressed in Testis. Genomics. (1999) 61:44–54. doi: 10.1006/geno.1999.5922
49. Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon C, Bernard O, et al. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nat Cell Biol. (1998) 393:805–9. doi: 10.1038/31729
50. Wang L, Buckley AF, Spurney RF. Regulation of cofilin phosphorylation in glomerular podocytes by testis specific kinase 1 (TESK1). Sci Rep. (2018) 8:12286. doi: 10.1038/s41598-018-30115-3
51. Sarmiere PD, Bamburg JR. Regulation of the neuronal actin cytoskeleton by ADF/cofilin. J Neurobiol. (2003) 58:103–17. doi: 10.1002/neu.10267
52. Yu Q, Wu C, Chen Y, Li B, Wang R, Huang R, et al. Inhibition of LIM kinase reduces contraction and proliferation in bladder smooth muscle. Acta Pharm Sin B. (2021) 11:1914–30. doi: 10.1016/j.apsb.2021.01.005
53. Eiseler T, Döppler H, Yan IK, Kitatani K, Mizuno K, Storz P. Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat Cell Biol. (2009) 11:545–56. doi: 10.1038/ncb1861
54. Grantham J, Brackley KI, Willison KR. Substantial CCT activity is required for cell cycle progression and cytoskeletal organi-zation in mammalian cells. Exp Cell Res. (2006) 312:2309–24. doi: 10.1016/j.yexcr.2006.03.028
55. McCormack EA, Rohman MJ, Willison KR. Mutational screen identifies critical amino acid residues of beta-actin mediating interaction between its folding intermediates and eukaryotic cytosolic chaperonin CCT. J Struct Biol. (2001) 135:185–97. doi: 10.1006/jsbi.2001.4389
56. Meng Y, Yang L, Wei X, Luo H, Hu Y, Tao X, et al. CCT5 interacts with cyclin D1 promoting lung adenocarcinoma cell migration and invasion. Biochem Biophys Res Commun. (2021) 567:222–9. doi: 10.1016/j.bbrc.2021.04.105
57. Zhang X, Lin X, Qin C, Huang K, Sun X, Zhao L, et al. Avian Chaperonin Containing TCP1 Subunit 5 Supports Influenza A Virus Replication by Interacting With Viral Nucleoprotein, PB1, and PB2 Proteins. Front Microbiol. (2020) 11:538355. doi: 10.3389/fmicb.2020.538355
58. Ooe A, Kato K, Noguchi S. Possible involvement of CCT5, RGS3, and YKT6 genes up-regulated in p53-mutated tumors in resistance to docetaxel in human breast cancers. Breast Cancer Res Treat. (2006) 101:305–15. doi: 10.1007/s10549-006-9293-x
59. Wang Q, Huang W-R, Chih W-Y, Chuang K-P, Chang C-D, Wu Y, et al. Cdc20 and molecular chaperone CCT2 and CCT5 are required for the Muscovy duck reovirus p10.8-induced cell cycle arrest and apoptosis. Veter Microbiol. (2019) 235:151–63. doi: 10.1016/j.vetmic.2019.06.017
60. Terluk MR, Ebeling MC, Fisher CR, Kapphahn RJ, Yuan C, Kartha RV, et al. N-Acetyl-L-cysteine Protects Human Retinal Pigment Epithelial Cells from Oxidative Damage: Implications for Age-Related Macular Degeneration. Oxidative Med Cell Longev. (2019) 2019:1–14. doi: 10.1155/2019/5174957
61. Zhou J, Jiang Y, Chen H, Wu Y, Zhang L. Tanshinone I attenuates the malignant biological properties of ovarian cancer by inducing apoptosis and autophagy via the inactivation of PI3K/AKT/mTOR pathway. Cell Prolif. (2019) 53:e12739. doi: 10.1111/cpr.12739
62. Qi T, Jing R, Wen C, Hu C, Wang Y, Pei C, et al. Interleukin-6 promotes migration and extracellular matrix synthesis in retinal pigment epithelial cells. Histochem Cell Biol. (2020) 154:629–38. doi: 10.1007/s00418-020-01923-4
63. Irschick EU, Sgonc R, Böck G, Wolf H, Fuchs D, Nussbaumer W, et al. Retinal pigment epithelial phagocytosis and metabolism differ from those of macrophages. Ophthalmic Res. (2004) 36:200–10. doi: 10.1159/000078778
Keywords: retinitis pigmentosa, MERTK, CCT5, f-actin, phagocytosis
Citation: Feng L, Li H, Du Y, Zhang T, Zhu Y, Li Z, Zhao L, Wang X, Wang G, Zhou L, Jiang Z, Liu Z, Ou Z, Wen Y and Zhuo Y (2022) Chaperonin-Containing TCP1 Subunit 5 Protects Against the Effect of Mer Receptor Tyrosine Kinase Knockdown in Retinal Pigment Epithelial Cells by Interacting With Filamentous Actin and Activating the LIM-Kinase 1/Cofilin Pathway. Front. Med. 9:861371. doi: 10.3389/fmed.2022.861371
Received: 24 January 2022; Accepted: 09 March 2022;
Published: 13 April 2022.
Edited by:
Hetian Lei, Shenzhen Eye Hospital, ChinaReviewed by:
Tsz Kin Ng, Shantou University and the Chinese University of Hong Kong, ChinaGuoxing Xu, First Affiliated Hospital of Fujian Medical University, China
Copyright © 2022 Feng, Li, Du, Zhang, Zhu, Li, Zhao, Wang, Wang, Zhou, Jiang, Liu, Ou, Wen and Zhuo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yehong Zhuo, emh1b3loQG1haWwuc3lzdS5lZHUuY24=
†These authors have contributed equally to this work
 Lujia Feng
Lujia Feng Haichun Li1†
Haichun Li1† Yuwen Wen
Yuwen Wen Yehong Zhuo
Yehong Zhuo