
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
HYPOTHESIS AND THEORY article
Front. Med. , 12 April 2022
Sec. Infectious Diseases – Surveillance, Prevention and Treatment
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.852998
This article is part of the Research Topic Emerging SARS-COV-2 Variants: Genomic Variations, Transmission, Pathogenesis, Clinical Impact and Interventions View all 65 articles
The ongoing pandemic Coronavirus Disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a matter of global concern in terms of public health Within the symptoms secondary to SARS-CoV-2 infection, hyposmia and anosmia have emerged as characteristic symptoms during the onset of the pandemic. Although many researchers have investigated the etiopathogenesis of this phenomenon, the main cause is not clear. The appearance of the new variant of concern Omicron has meant a breakthrough in the chronology of this pandemic, presenting greater transmissibility and less severity, according to the first reports. We have been impressed by the decrease in anosmia reported with this new variant and in patients reinfected or who had received vaccination before becoming infected. Based on the literature published to date, this review proposes different hypotheses to explain this possible lesser affectation of smell. On the one hand, modifications in the SARS-CoV-2 spike protein could produce changes in cell tropism and interaction with proteins that promote virus uptake (ACE-2, TMPRSS2, and TMEM16F). These proteins can be found in the sustentacular cells and glandular cells of the olfactory epithelium. Second, due to the characteristics of the virus or previous immunity (infection or vaccination), there could be less systemic or local inflammation that would generate less cell damage in the olfactory epithelium and/or in the central nervous system.
Neurological symptoms such as loss of taste and, specifically, olfactory dysfunction have been consistently reported within the different SARS-CoV-2 variants (1, 2). Olfactory dysfunction, defined as the reduction (hyposmia) or total loss (anosmia) of smell during sniffing or eating, better predicts the disease than other well-known symptoms such as fever and cough (3, 4). Despite the high incidence of these symptoms, underlying mechanisms have been unclear (5). Hyposmia was not initially recognized to be linked to COVID-19 and was only mentioned to affect about 5% of COVID-19 patients in one of the first studies from China (1). However, a much higher prevalence was reported in subsequent studies from Europe, the Middle East, and North America (6–9). A recent systematic review and meta-analysis with 23,353 patients diagnosed with COVID-19 (10) pointed out that prevalence of smell loss and taste loss were 38 and 30.35%, respectively, with significant differences between Western countries and East Asian ones. Although there is a great plethora of factors related to the appearance of this phenomenon, we must not forget the important advances that are being made with regard to host genetic factors (11) and their implication in susceptibility to infection, immune response, and even symptomatology accompanying SARS-CoV-2 infection.
One of the possible rationales could implicate that Caucasians have more often an Angiotensin-converting enzyme 2 (ACE2) variant expressed in the olfactory epithelium (12, 13) (presumably in the sustentacular cells of the olfactory epithelium). Because this protein is one of those used by the receptor binding domain (RBD) of the SARS-CoV-2 Spike Protein to infect the cell (14), genetic variability and the mutation rate within the RBD domain is of particular interest in the context of population differences in the prevalence of hyposmia.
Recently, the Norwegian Institute of Public Health reported the demographics of 117 cases infected with Omicron variant (15). Strikingly, loss of smell and taste was reported in 12% (median duration 2 days) and 23% (median duration 2.5 days), respectively (15). This feature has also been supported by US State Department of Health (16) and by others studies (17, 18).
Vihta et al. (17) showed loss of smell and taste was found to be less common among Omicron compared to Delta cases (13% of Omicron cases, 34% of Delta cases, odds ratio 0.22, 95% CI: 0.21–0.23). Moreover, Boscolo-Rizzo et al. (18), in a pre-printed version, presented data on a prospective study on mild-to-moderate symptomatic adult patients and reported that 24.6% of patients had smell alterations during the proxy Omicron period compared to 62.6% during the comparator period mainly driven by the delta variant (p < 0.001). Finally, a recent report led by Maisa et al. collecting 468 Omicron cases in different regions of France reported an 8.3% of anosmia (19).
The Omicron (B.1.1.529) COVID-19 variation of concern (VOC) was first identified in South Africa on November 9, 2021 (20). This variant was accounted for to the World Health Organization (WHO) by South African experts on November 24, 2021, after which the WHO's Technical Advisory Group on SARS-CoV-2 Virus Evolution (TAG-VE) was reconvened on November 26, 2021, which prompted B.1.1.529 to being meant as a VOC (21).
The omicron variant seems to be highly transmissible, presenting many substitutions in the spike glycoprotein and appeared at a time when a large part of the world population had received SARS-CoV-2 vaccination.
We hypothesize that Omicron variant features or previously acquired immunity, either due to previous infection or vaccination, could explain the lower incidence of olfactory disturbances. Omicron could produce less hyposmia due to differences in cell tropism, mechanism of entry into cells and by producing less inflammatory dysregulation (Figure 1).
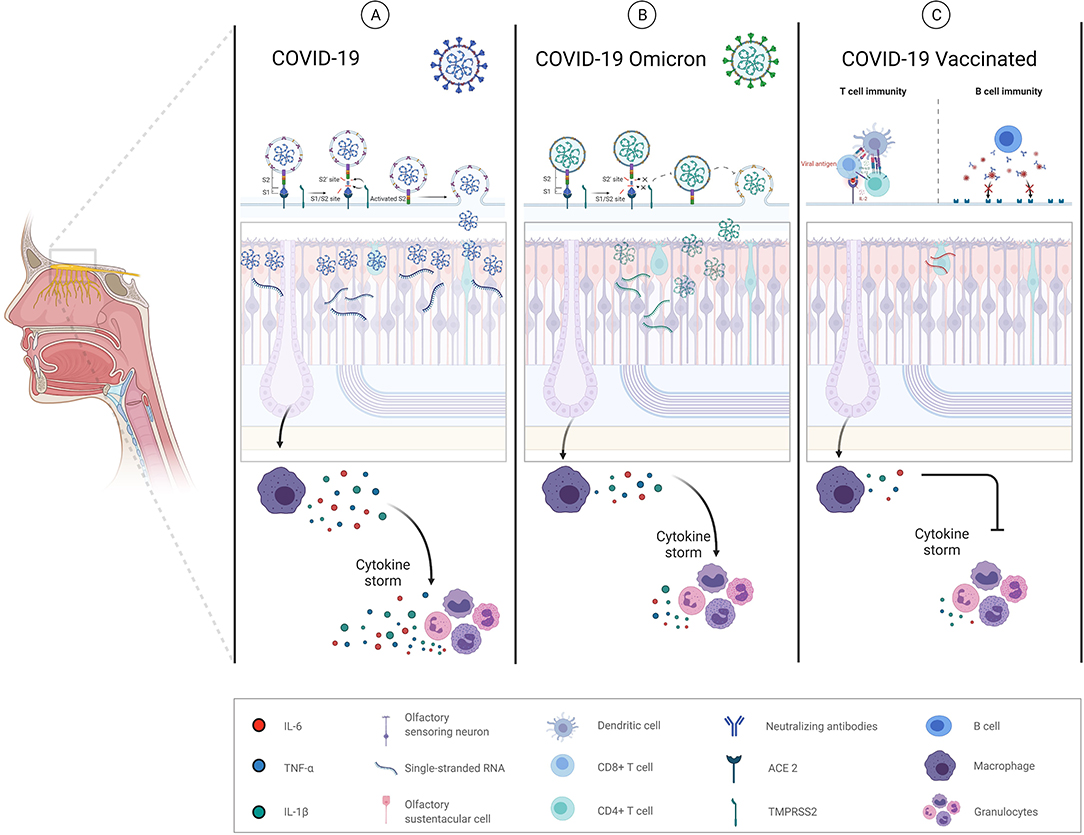
Figure 1. Schematic mechanism proposal of anosmia/hyposmia in COVID-19. (A) SARS-CoV-2 alpha variant. (1) Infection and destruction of olfactory epithelial supporting cells (olfactory sustentacular cells), resulting in inflammation, and abnormalities in local homeostasis; (2) infection or immune-mediated damage of surrounding cells (vascular cells) resulting in hypoperfusion, inflammatory cell recruitment, cytokine release, and the production of chemical neurotoxins. (B) SARS-CoV-2 Omicron variant. Omicron mechanism of cell entry based on a TMPRS22 dependent and non-dependent manner may involve a more heterogenous tropism with less replication and the consequent minor local inflammation produced. (C) SARS-CoV-2 vaccines may not alter the levels of replicating virus in the olfactory epithelium once viral infection establishes but may instead protect through accelerated clearance or enhanced neutralization of infectious viral particles.
Due to its significant prevalence, hyposmia has been one of the neurological symptoms that has aroused the greatest interest in the scientific community. A wide-variety of scenarios such as olfactory cleft syndrome (22), systemic and local inflammation in the olfactory epithelium, apoptosis of the olfactory receptor neurons and sustentacular cells, changes in the olfactory cilia and odor transmission, injury of microglial cells, effect on the olfactory bulb, epithelial olfactory injury (e.g., Sustentacular cells), as well as damage of olfactory receptor neurons and olfactory stem cells neurons have been described as possible mechanisms (23). Damage to these olfactory stem cell neurons may be the cause for persistent anosmia after COVID-19 recovery despite having acquired specific immunity.
The omicron variant seems to be highly transmissible, presenting many substitutions in the spike glycoprotein. Several mutations in the RBD and S2 region of the spike protein are predicted to impact transmissibility and affinity for the ACE-2 receptor (24). Brann et al. found that the gene encoding the ACE2 receptor protein, which SARS-CoV-2 utilizes to enter human cells, is not expressed in olfactory sensory neurons (OSNs) (25). Nevertheless, two specific cell types in the olfactory epithelium expressed ACE2 at similar levels to what has been observed in the lower respiratory tract cells. These included sustentacular cells, which embraced sensory neurons and are thought to provide structural and metabolic support, and basal cells, which act as stem cells that regenerate the olfactory epithelium after damage. ACE2 receptors are also found in glial cells, neurons and capillary endothelium and the interaction with the virus could be one of the mechanisms that allows it to enter into the central nervous system (26).
The key to understand such differences in infectivity of SARS-CoV-2 may lie in the frequency of variants in the virus entry proteins, ACE2, and Transmembrane serine protease 2 (TMPRSS2), which may depend on cell type and racial or ethnic groups (27, 28).
Recently, Bentley et al. studied Omicron features using an established K18-hACE2 mouse model of SARS-CoV-2 (29). The infection of hACE2 mice with Omicron variant led to less viral load in oral swabs and nasal tissue and to less severe clinical signs and less severe pneumonia compared with Delta variant. However, more studies are needed because McMahan et al. suggested that Omicron infection may lead to increased upper respiratory tract disease but reduced lower respiratory tract disease (30) compared with WA1/2020 variant. These results should be taken cautiously since no animal model can predict with absolute certainty the consequences of infection in humans. Meanwhile, Hong Kong investigators studied Alpha, Beta, Delta and Omicron variants in ex vivo explant cultures of human bronchus and lung. They suggested that B.1.1.529 replicates faster in the human bronchus and less in lung cells, which may explain its greater transmissibility and putative lower disease severity (31). At the same time, they propose a less-dependent TMPRSS2 activation in Omicron variant compared with Delta, suggesting that Omicron may have a broader spectrum of target cells compared to Delta (32). This last proposal was also supported by Thomas et al., showing that Omicron achieved this rapid replication rate by becoming less specialized in its cellular tropism, entering cells in both a TMPRSS2-dependent and –independent manner (33). Recently it was proposed by Meng et al. that TMPRSS2 usage may be impacted by ACE2 levels after studies in organoid systems and human nasal epithelial cultures (34).
While the switch in passage pathway has expanded its intrinsic contagiousness, the less effective utilization of cell surface TMPRSS2 by Omicron Spike protein has also brought about a diminishing affinity for syncytia arrangement (34). This may explain the decreased disease severity impression since syncytia have been reported in the autopsy of COVID cases and the efficient cleavage at the furin site that underlies syncytia formation has been associated with enhanced disease severity in animal models (35).
Lastly, another essential aspect that should be considered is the fusogenic capacity of this new SARS-CoV-2 VOC. Recently, Meng et al. showed Omicron spike is relatively poorly cleaved and impaired in mediating cell-cell fusion and syncytia formation (34). This observation is of interest because, for syncytia formation, Transmembrane 16F (TMEM16F) protein is one of the proteins that are activated by the SARS-COV-2 spike (35). This calcium-activated chloride channel is also located on the cilia of olfactory sensory neurons and appears to be involved in olfactory signal transduction (35, 36). Perhaps Omicron variant does not alter these proteins, and this could explain, in part, the lower incidence of olfactory alterations.
In summary, the different data from ex vivo, mouse model or in vitro studies suggest that the Omicron variant has a particular cellular entry mechanism that may explain the lower incidence of olfactory impairment. Still such interpretations need to be qualified because the semiotics of COVID-19 is determined not only by virus replication but also by dysregulated innate immune responses.
Olfaction is a complex process comprising multiple components, including receptors, nerves, and structures of the brain. Cell-signaling processes are critical in any complex sensory system such as olfaction, where a myriad selection of cytokines (37) or even intermediate metabolic substances such as zinc play an essential role (38).
Before the COVID-19 breakthrough, IL-6 was already considered a possible causal factor for initiation of hyposmia reflective of local or systemic immunological/inflammatory changes in blood, saliva, or nasal mucus (39). This hypothesis is consistent with finding smell loss among patients with inflammatory rheumatoid arthritis (40). Elevated IL-6 level has been previously reported in nasal lavage fluid from patients with naturally acquired parvovirus (41), in addition, other viruses have been found in turbinate epithelial cells of patients with post-viral olfactory dysfunction (42).
Sensorineural olfactory loss could occur due to destruction of the olfactory neuroepithelium by toxic inflammatory factors (such as TNF-α, IL-6, and IL-1β) (43). Cazzolla et al. observed in 67 COVID patients that the resolution of olfactory alterations is accompanied by a progressive decrease of IL-6 levels to normal values (44). Thus, a new approach is opened in which the decrease of IL-6 from the beginning to the disappearance of the symptoms would have greater clinical relevance than the initial peak values of IL-6. Elevated IL-6 level could act as an endogenous substance regulating olfactory neuronal activity because it has been shown to regulate neuronal (45) and glial cell activity (46). For instance, Neuropoietin, an IL-6–related cytokine that affects signaling through ciliary neurotrophic factor receptor (47), could directly inhibit smell function because inhibition of several ciliary factors has induced smell loss in patients with other syndromic disorders (48, 49).
In contrast to these findings, Bax et al. failed to associate different inflammatory markers (CRP, IL-6) with the presence or absence of anosmia in 93 COVID patients (50). However, it is possible that any nasal IL-6 production within the olfactory epithelium is insufficient to contribute significantly to serum IL-6 levels.
Finally, Torabi et al. analyzed TNF-α and IL-1β levels in olfactory epithelial biopsies from patients with confirmed COVID-19 and uninfected controls (51). In this study, the authors proposed that the inflammatory infiltration that occurs in response to local TNF-α expression may lead to a considerable expansion of the olfactory submucosa and causes damage to OSN (52).
A plausible scenario would be one in which the Omicron variant would not markedly produce such a pro-inflammatory environment in the olfactory epithelium and/or central nervous system, providing additional explanation for the low rate of anosmia and hyposmia reported with this variant.
About 50 mutations have been detected in the Omicron variant (30 in the spike protein) (20). These mutations, especially those of the spike protein, make the virus capable of evading, at least in part, the neutralizing antibodies generated by vaccination or previous infections.
However, beyond generating a specific humoral response, both vaccination and previous infections produce a longer-lasting cellular response mediated by CD4 and CD8 T-cells. This response seems to be maintained at 70–80% against the Omicron variant (53–55).
In addition to the intrinsic characteristics of the Omicron variant, this previous specific immunity could explain why the new variant generates less severe infection and causes a lesser and less lasting cytokine storm.
The fact of generating a lower inflammatory response in the olfactory epithelium and a faster elimination of the virus could explain the lower incidence of hyposmia. It has already been shown that vaccinated patients (especially with two doses), who are re-infected by SARS-CoV-2 (before the appearance of the Omicron variant) had a lower incidence of smell alterations and duration of symptoms was shorter (56, 57). However, the only data we have found from the smell loss registry in symptomatic COVID-19 patients after fully vaccination suggest that anosmia/hyposmia may be an existing finding in these subjects (58). Further studies involving close and personalized symptom monitoring together with a correct genome characterization of SARS-CoV-2 are necessary to shed light on this topic.
Since the advent of the Omicron variant, the general impression in the medical community is that there is a lower incidence of hyposmia in infected people. Despite the lack of studies with large sample size, preliminary data from Norway have reported only 12% alterations in smell compared to 38% in the other variants (15).
Among the causes that may explain this lower incidence are the characteristics of the new variant and its interaction with the organism and its response to infection, and, finally, the role of previously acquired immunity (by previous infection or vaccination).
The Omicron variant presents a series of mutations in the spike protein that affect the affinity for the ACE2 receptor, generate a less specialized cell tropism (cell entry based on a TMPRS22 dependent and non-dependent manner) and, finally, a lower capacity for cell fusion. On the other hand, because the new variant appears less pathogenic, it may generate a lower viral load and less local or systemic inflammation (e.g., IL-6, TNF-α, IL-1β, etc.).
Finally, one of the most plausible explanations is that the previous immunity generated by vaccines or past infections causes less local or systemic inflammation for less time and faster viral elimination.
However, we must not ignore that the different possible explanations that we propose in this manuscript should be taken cautiously. Several elucidations have been suggested (55); nevertheless; we are aware that some of them may not be verified in the future and others are based on unreviewed preprinted articles which will be needing future validation. In order to delve into this phenomenon, more studies are required to establish the exact mechanisms of how SARS-CoV-2 elicits olfactory system damage.
In conclusion, less direct damage by the virus and less inflammation—either due to virus characteristics or previous immunity—could explain less damage to the olfactory epithelium and/or central nervous system. Larger sample size studies are needed to establish the incidence of olfactory alterations in patients with Omicron, to be able to associate them with certain risk factors and to better understand the etiopathogenesis.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
JR-S: study concept and design and figure editing. JR-S and BB: methodology, literature review, data collection, analysis, interpretation, writing-original draft manuscript, and review-original final manuscript. All authors have read and agreed to the published version of the manuscript.
This work has received support from FEDER funds and the FIS project form Instituto de Carlos III (PI19/00019).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We apologize to authors of several high-profile articles whose work was not included owing to space limitations.
1. Mao L, Jin H, Wang M, Hu Y, Chen S, He Q, et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. (2020) 77:683–90. doi: 10.1001/jamaneurol.2020.1127
2. Lechien JR, Chiesa-Estomba CM, De Siati DR, Horoi M, Le Bon SD, Rodriguez A, et al. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur Arch Otorhinolaryngol. (2020) 277:2251–61. doi: 10.1007/s00405-020-05965-1
3. Callejon-Leblic MA, Moreno-Luna R, Del Cuvillo A, Reyes-Tejero IM, Garcia-Villaran MA, Santos-Peña M, et al. Loss of smell and taste can accurately predict COVID-19 infection: a machine-learning approach. J Clin Med. (2021) 10:570. doi: 10.3390/jcm10040570
4. Liou J-M, Chen M-J, Hong T-C, Wu M.-S. Alteration of taste or smell as a predictor of COVID-19. Gut. (2021) 70:806–7. doi: 10.1136/gutjnl-2020-322125
5. Meng X, Deng Y, Dai Z, Meng Z. COVID-19 and anosmia: a review based on up-to-date knowledge. Am J Otolaryngol. (2020) 41:102581. doi: 10.1016/j.amjoto.2020.102581
6. Spinato G, Fabbris C, Polesel J, Cazzador D, Borsetto D, Hopkins C, et al. Alterations in smell or taste in mildly symptomatic outpatients with SARS-CoV-2 infection. JAMA. (2020) 323:2089–90. doi: 10.1001/jama.2020.6771
7. Yan CH, Faraji F, Prajapati DP, Boone CE, DeConde AS. Association of chemosensory dysfunction and COVID-19 in patients presenting with influenza-like symptoms. Int Forum Allergy Rhinol. (2020) 10:806–13. doi: 10.1002/alr.22579
8. Bagheri SH, Asghari A, Farhadi M, Shamshiri AR, Kabir A, Kamrava SK, et al. Coincidence of COVID-19 epidemic and olfactory dysfunction outbreak in Iran. Med J Islam Repub Iran. (2020) 34:62. doi: 10.34171/mjiri.34.62
9. Giacomelli A, Pezzati L, Conti F, Bernacchia D, Siano M, Oreni L, et al. Self-reported olfactory and taste disorders in patients with severe acute respiratory coronavirus 2 infection: a cross-sectional study. Clin Infect Dis. (2020) 71:889–90. doi: 10.1093/cid/ciaa330
10. von Bartheld CS, Hagen MM, Butowt R. Prevalence of chemosensory dysfunction in COVID-19 patients: a systematic review and meta-analysis reveals significant ethnic differences. ACS Chem Neurosci. (2020) 11:2944–61. doi: 10.1021/acschemneuro.0c00460
11. Niemi MEK, Karjalainen J, Liao RG, Neale BM, Daly M, Ganna A, et al. Mapping the human genetic architecture of COVID-19. Nature. (2021) 600:472–7. doi: 10.1038/s41586-021-03767-x
12. Bilinska K, Jakubowska P, Von Bartheld CS, Butowt R. Expression of the SARS-CoV-2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory epithelium: identification of cell types and trends with age. ACS Chem Neurosci. (2020) 11:1555–62. doi: 10.1021/acschemneuro.0c00210
13. Butowt R, von Bartheld CS. Anosmia in COVID-19: underlying mechanisms and assessment of an olfactory route to brain infection. Neuroscientist. (2021) 27:582–603. doi: 10.1177/1073858420956905
14. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. (2020) 581:221–4. doi: 10.1038/s41586-020-2179-y
15. Brandal LT, MacDonald E, Veneti L, Ravlo T, Lange H, Naseer U, et al. Outbreak caused by the SARS-CoV-2 Omicron variant in Norway, November to December 2021. Eurosurveillance. (2021) 26:2101147. doi: 10.2807/1560-7917.ES.2021.26.50.2101147
16. Washington State Department of Health. SARS-CoV-2 Sequencing and Variants in Washington State (2022).
17. Pouwels KB, Pritchard E, Matthews PC, Stoesser N, Eyre DW, Vihta KD, et al. Effect of Delta variant on viral burden and vaccine effectiveness against new SARS-CoV-2 infections in the UK. Nat Med. (2021) 27:2127–35. doi: 10.1038/s41591-021-01548-7
18. Boscolo-Rizzo P, Tirelli G, Meloni P, Hopkins C, Madeddu G, De Vito A, et al. COVID-19-related smell and taste impairment with widespread diffusion of SARS-CoV-2 Omicron variant. Int Forum Allergy Rhinol. (2022) 1–9. doi: 10.1101/2022.02.17.22271116
19. Maisa A, Spaccaferri G, Fournier L, Schaeffer J, Deniau J, Rolland P, et al. First cases of Omicron in France are exhibiting mild symptoms, November 2021-January 2022. Infect Dis Now. (2022) 12:S2666–9919(22)00036-7. doi: 10.1016/j.idnow.2022.02.003
20. World Health Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern (2021).
22. Altundag A, Yildirim D, Tekcan Sanli DE, Cayonu M, Kandemirli SG, Sanli AN, et al. Olfactory cleft measurements and COVID-19-related anosmia. Otolaryngol Head Neck Surgery. (2021) 164:1337–44. doi: 10.1177/0194599820965920
23. Bilinska K, Butowt R. Anosmia in COVID-19: a bumpy road to establishing a cellular mechanism. ACS Chem Neurosci. (2020) 11:2152–5. doi: 10.1021/acschemneuro.0c00406
24. Dejnirattisai W, Huo J, Zhou D, Zahradník J, Supasa P, Liu C, et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. (2022) 185:467–84.e15. doi: 10.1016/j.cell.2021.12.046
25. Brann DH, Tsukahara T, Weinreb C, Lipovsek M, Berge KVD, Gong B, et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci Adv. (2020) 6:eabc5801. doi: 10.1126/sciadv.abc5801
26. Bhola S, Trisal J, Thakur V, Kaur P, Kulshrestha S, Bhatia SK, et al. Neurological toll of COVID-19. Neurol Sci. (2022) 43:2171–86. doi: 10.1007/s10072-022-05875-6
27. Meng X, Pan Y. COVID-19 and anosmia: the story so far. Ear Nose Throat J. (2021) 29:1455613211048998. doi: 10.1177/01455613211048998
28. Najafloo R, Majidi J, Asghari A, Aleemardani M, Kamrava SK, Simorgh S, et al. Mechanism of anosmia caused by symptoms of COVID-19 and emerging treatments. ACS Chem Neurosci. (2021) 12:3795–805. doi: 10.1021/acschemneuro.1c00477
29. Bentley EG, Kirby A, Sharma P, Kipar A, Mega DF, Bramwell C, et al. SARS-CoV-2 Omicron-B.1.1.529 variant leads to less severe disease than Pango B and Delta variants strains in a mouse model of severe COVID-19. bioRxiv. (2021) 2021.12.26.474085. doi: 10.1101/2021.12.26.474085
30. McMahan K, Giffin V, Tostanoski L, Chung B, Siamatu M, Suthar M, et al. Reduced pathogenicity of the SARS-CoV-2 Omicron variant in hamsters. bioRxiv. (2022) 2022.01.02.474743. doi: 10.1101/2022.01.02.474743
31. Chan M, Hui K, Ho J, Cheung M-C, Ng K-C, Ching R, et al. SARS-CoV-2 Omicron variant replication in human respiratory tract ex vivo. Nature. (2021) 603:715–20. doi: 10.1038/s41586-022-04479-6
32. Willett BJ, Grove J, MacLean OA, Wilkie C, Logan N, De Lorenzo G, et al. The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. medRxiv. (2021) 2022.01.03.21268111. doi: 10.1101/2022.01.03.21268111
33. Peacock TP, Brown JC, Zhou J, Thakur N, Newman J, Kugathasan R, et al. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv. (2021) 2021.12.31.474653. doi: 10.1101/2021.12.31.474653
34. Meng B, Abdullahi A, Ferreira IATM, Goonawardane N, Saito A, Kimura I, et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. (2022). 603:706–14. doi: 10.1038/s41586-022-04474-x
35. Braga L, Ali H, Secco I, Chiavacci E, Neves G, Goldhill D, et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature. (2021) 594:88–93. doi: 10.1038/s41586-021-03491-6
36. Henkel B, Drose DR, Ackels T, Oberland S, Spehr M, Neuhaus EM. Co-expression of anoctamins in cilia of olfactory sensory neurons. Chem Senses. (2015) 40:73–87. doi: 10.1093/chemse/bju061
37. Rothwell NJ, Hopkins SJ. Cytokines and the nervous system II: actions and mechanisms of action. Trends Neurosci. (1995) 18:130–6. doi: 10.1016/0166-2236(95)93890-A
38. Atkin-Thor E, Goddard BW, O'Nion J, Stephen RL, Kolff WJ. Hypogeusia and zinc depletion in chronic dialysis patients. Am J Clin Nutr. (1978) 31:1948–51. doi: 10.1093/ajcn/31.10.1948
39. Henkin RI, Schmidt L, Velicu I. Interleukin 6 in hyposmia. JAMA Otolaryngol Head Neck Surg. (2013) 139:728–34. doi: 10.1001/jamaoto.2013.3392
40. Heckmann JG, Heckmann SM, Lang CJ, Hummel T. Neurological aspects of taste disorders. Archiv Neurol. (2003) 60:667–71. doi: 10.1001/archneur.60.5.667
41. Wang JH, Kwon HJ, Jang YJ. Detection of parainfluenza virus 3 in turbinate epithelial cells of postviral olfactory dysfunction patients. Laryngoscope. (2007) 117:1445–9. doi: 10.1097/MLG.0b013e318063e878
42. Suzuki M, Saito K, Min WP, Vladau C, Toida K, Itoh H, et al. Identification of viruses in patients with postviral olfactory dysfunction. Laryngoscope. (2007) 117:272–7. doi: 10.1097/01.mlg.0000249922.37381.1e
43. Yee KK, Pribitkin EA, Cowart BJ, Vainius AA, Klock CT, Rosen D, et al. Neuropathology of the olfactory mucosa in chronic rhinosinusitis. Am J Rhinol Allergy. (2010) 24:110–20. doi: 10.2500/ajra.2010.24.3435
44. Cazzolla AP, Lovero R, Lo Muzio L, Testa NF, Schirinzi A, Palmieri G, et al. Taste and smell disorders in COVID-19 patients: role of interleukin-6. ACS Chem Neurosci. (2020) 11:2774–81. doi: 10.1021/acschemneuro.0c00447
45. Jüttler E, Tarabin V, Schwaninger M. Interleukin-6 (IL-6): a possible neuromodulator induced by neuronal activity. Neuroscientist. (2002) 8:268–75. doi: 10.1177/1073858402008003012
46. Galiano M, Liu ZQ, Kalla R, Bohatschek M, Koppius A, Gschwendtner A, et al. Interleukin-6 (IL6) and cellular response to facial nerve injury: effects on lymphocyte recruitment, early microglial activation and axonal outgrowth in IL6-deficient mice. Eur J Neurosci. (2001) 14:327–41. doi: 10.1046/j.0953-816x.2001.01647.x
47. Derouet D, Rousseau F, Alfonsi F, Froger J, Hermann J, Barbier F, et al. Neuropoietin, a new IL-6-related cytokine signaling through the ciliary neurotrophic factor receptor. Proc Natl Acad Sci USA. (2004) 101:4827–32. doi: 10.1073/pnas.0306178101
48. Afzelius BA. A human syndrome caused by immotile cilia. Science. (1976) 193:317–9. doi: 10.1126/science.1084576
49. Zaghloul NA, Katsanis N. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J Clin Investig. (2009) 119:428–37. doi: 10.1172/JCI37041
50. Bax F, Tascini C, Valente M, Marini A, Surcinelli A, Pellitteri G, et al. Hyposmia and dysgeusia in COVID-19: indication to swab test and clue of CNS involvement. Neurol Clin Pract. (2021) 11:e92–6. doi: 10.1212/CPJ.0000000000001029
51. Torabi A, Mohammadbagheri E, Akbari Dilmaghani N, Bayat A-H, Fathi M, Vakili K, et al. Proinflammatory cytokines in the olfactory mucosa result in COVID-19 induced anosmia. ACS Chem Neurosci. (2020) 11:1909–13. doi: 10.1021/acschemneuro.0c00249
52. Lane AP, Turner J, May L, Reed R. A genetic model of chronic rhinosinusitis-associated olfactory inflammation reveals reversible functional impairment and dramatic neuroepithelial reorganization. J Neurosci. (2010) 30:2324–9. doi: 10.1523/JNEUROSCI.4507-09.2010
53. Liu Y, Zeng Q, Deng C, Li M, Li L, Liu D, et al. Robust induction of B cell and T cell responses by a third dose of inactivated SARS-CoV-2 vaccine. Cell Discov. (2022) 8:10. doi: 10.1038/s41421-022-00373-7
54. Keeton R, Tincho MB, Ngomti A, Baguma R, Benede N, Suzuki A, et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature. (2022) 603:488–92. doi: 10.1038/s41586-022-04460-3
55. Tarke A, Coelho CH, Zhang Z, Dan JM, Yu ED, Methot N, et al. SARS-CoV-2 vaccination induces immunological memory able to cross-recognize variants from alpha to Omicron. bioRxiv. (2021) 2021.12.28.474333. doi: 10.1101/2021.12.28.474333
56. Antonelli M, Penfold R, Merino J, Sudre C, Molteni E, Berry S, et al. Risk factors and disease profile of post-vaccination SARS-CoV-2 infection in UK users of the COVID Symptom Study app: a prospective, community-based, nested, case-control study. Lancet Infect Dis. (2021) 22:43–55. doi: 10.1016/S1473-3099(21)00460-6
57. Malhotra S, Mani K, Lodha R, Bakhshi S, Mathur VP, Gupta P, et al. SARS-CoV-2 reinfection rate and estimated effectiveness of the inactivated whole virion vaccine BBV152 against reinfection among health care workers in New Delhi, India. JAMA Netw Open. (2022) 5:e2142210. doi: 10.1001/jamanetworkopen.2021.42210
Keywords: COVID-19, anosmia, Omicron variant, inflammation, vaccines
Citation: Rodriguez-Sevilla JJ, Güerri-Fernádez R and Bertran Recasens B (2022) Is There Less Alteration of Smell Sensation in Patients With Omicron SARS-CoV-2 Variant Infection? Front. Med. 9:852998. doi: 10.3389/fmed.2022.852998
Received: 20 January 2022; Accepted: 23 March 2022;
Published: 12 April 2022.
Edited by:
Renan Pedra de Souza, Universidade Federal de Minas Gerais, BrazilReviewed by:
Paolo Boscolo-Rizzo, University of Trieste, ItalyCopyright © 2022 Rodriguez-Sevilla, Güerri-Fernádez and Bertran Recasens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Jose Rodriguez-Sevilla, anJvZHJpZ3VlenNldmlsbGFAcHNtYXIuY2F0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.