
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 01 April 2022
Sec. Translational Medicine
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.807382
This article is part of the Research Topic Implementation of AI and Machine Learning Technologies in Medicine View all 21 articles
 Wenle Li1,2†
Wenle Li1,2† Tao Hong3†
Tao Hong3† Wencai Liu4†
Wencai Liu4† Shengtao Dong5
Shengtao Dong5 Haosheng Wang6
Haosheng Wang6 Zhi-Ri Tang7
Zhi-Ri Tang7 Wanying Li2
Wanying Li2 Bing Wang2
Bing Wang2 Zhaohui Hu8
Zhaohui Hu8 Qiang Liu1*
Qiang Liu1* Yong Qin9*
Yong Qin9* Chengliang Yin10*
Chengliang Yin10*Background: This study aimed to develop and validate machine learning (ML)-based prediction models for lung metastasis (LM) in patients with Ewing sarcoma (ES), and to deploy the best model as an open access web tool.
Methods: We retrospectively analyzed data from the Surveillance Epidemiology and End Results (SEER) Database from 2010 to 2016 and from four medical institutions to develop and validate predictive models for LM in patients with ES. Patient data from the SEER database was used as the training group (n = 929). Using demographic and clinicopathologic variables six ML-based models for predicting LM were developed, and internally validated using 10-fold cross validation. All ML-based models were subsequently externally validated using multiple data from four medical institutions (the validation group, n = 51). The predictive power of the models was evaluated by the area under receiver operating characteristic curve (AUC). The best-performing model was used to produce an online tool for use by clinicians to identify ES patients at risk from lung metastasis, to improve decision making and optimize individual treatment.
Results: The study cohort consisted of 929 patients from the SEER database and 51 patients from multiple medical centers, a total of 980 ES patients. Of these, 175 (18.8%) had lung metastasis. Multivariate logistic regression analysis was performed with survival time, T-stage, N-stage, surgery, and bone metastasis providing the independent predictive factors of LM. The AUC value of six predictive models ranged from 0.585 to 0.705. The Random Forest (RF) model (AUC = 0.705) using 4 variables was identified as the best predictive model of LM in ES patients and was employed to construct an online tool to assist clinicians in optimizing patient treatment. (https://share.streamlit.io/liuwencai123/es_lm/main/es_lm.py).
Conclusions: Machine learning were found to have utility for predicting LM in patients with Ewing sarcoma, and the RF model gave the best performance. The accessibility of the predictive model as a web-based tool offers clear opportunities for improving the personalized treatment of patients with ES.
Ewing sarcoma (ES) is an aggressive sarcoma with a high propensity for local recurrence and distant metastasis in children and adolescents (1, 2). ES is the second most common primary bone malignancy, accounting for 5% of all child and adolescent cancers (3). ES frequently involves the diaphysis region of long bones (4). Despite the development of new treatment regimens, ES has a high likelihood of tumor metastasis, leading to a worsening prognosis and resulting in a poor 5-year survival rate of only 20–45% (4, 5). In a retrospective study of 975 patients with ES, 5-year survival and 5-year relapse-free survival rates for patients with localized disease were 70 and 55%, respectively, but only 33 and 21% for those with distant metastasis disease (6).
Although diagnostic imaging techniques have improved dramatically during the past 30 years, metastatic status can only be detected in approximately 20–25% of ES patients (3), with the lung being the most common metastatic site (5, 7, 8). Computed tomography (CT) scans of the chest are usually carried out to detect lung metastasis. However, given the high cost, radiation damage, and low efficiency of detection of metastatic nodules, new strategies are urgently required to accurately predict the development of lung metastasis in patients with ES (9, 10).
Machine learning (ML) has emerged as a powerful computer-based method of data mining and analysis and has been extensively applied as a “prediction tool” in a multitude of different scientific, engineering, and medical scenarios (11–15). ML has been shown to detect more interactions between variables, and to be more accurate than conventional statistical methods (14, 16). ML algorithms have been applied to model clinical outcome and to improve cognition of tumor growth and progression (17). However, although numerous ML-based predictive models of tumor development have been reported, no study has been conducted in predicting lung metastasis associated with Ewing Sarcoma.
The Surveillance Epidemiology and End Results (SEER) database contains data for around 26% of the United States population and is commonly used to study rare diseases since it overcomes the obstacle of inadequate case numbers (18–20). We constructed several ML-based models of LM in patients with ES, using the SEER database. External validation was subsequently performed using data from multiple medical centers to predict the probability of LM with the aim of improving individualized patient management. The best model was uploaded as a web-based tool.
Data were sourced from the SEER database and four medical institutions in China: Liuzhou People's Hospital, Second Affiliated Hospital of Jilin University, Xianyang Central Hospital, and Second Affiliated Hospital of Dalian Medical University, respectively. This retrospective study did not use personal identifying information and thus did not require informed patient consent or Institutional Ethics Committee Board approval.
Patients selected from the SEER database (2010–2016) who were diagnosed with ES originating in bone, as identified by ICD-O-3/WHO 2008 morphology code 9260d, composed the “training” group. Criteria for exclusion were more than one primary tumor and incomplete clinicopathological information. The “validation” group was composed of ES patient data obtained from four hospitals in different regions of China, from 2010 to 2018. All cases featured complete clinicopathological data and follow-up information and no other primary tumors. Demographic and clinicopathological variables included in both groups were: race, age, sex, primary site, laterality, T-stage, N-stage, M-stage, surgery, radiation, chemotherapy, bone metastasis, and survival times. For consistency with SEER database records, “race” in the Chinese medical records was classified as “other”. Detailed treatments, such as surgery, radiation, and chemotherapy were classified as Yes or No, and were not recorded in the SEER database.
Using demographic and clinicopathological data, we explored the effect of variables (p < 0.05) in univariate analysis, in the multifactorial regression model, and in predictive models based on the ML algorithms. Six different ML algorithms were applied independently to develop predictive models of LM in patients with ES, as follows: Random Forest (RF), Logistic regression (LR), Extreme gradient boosting (XGB), Gradient boosting machine (GBM), Multilayer perceptron (MLP), and Decision tree (DT) (21, 22). For the training process of the ML algorithms using python (version 3.8), we employed 10-fold cross-validation to avoid overfitting (23). We also calculated the average value of the area under receiver operating characteristic curve (AUC) to evaluate the predictive power of each model.
The ML algorithms were subsequently applied to the validation group and the AUC was again calculated to evaluate the predictive performance of all models. The higher the AUC value, the better the model. Finally, the best-performing model was designed as a web-based tool for predicting the likelihood of LM in ES patients.
As a model inspection technique, permutation feature importance can be used for any fitted estimator (24–26). Thus, a total of 100 independent training simulation results were applied to assess the most important variables in each predictive model using permutation feature importance analysis. We further assessed the relative contribution of four key clinical variables to LM predictive models using spearman correlation of features analysis and plotted a correlation heat map.
All data were extracted from the SEER database via the SEER * Stat software (version 8.3.6). All analyses were performed using python (version 3.8). The baseline variables between the training group and validation group were compared using Student's t tests and Pearson chi-square test. A two-sided p < 0.05 was deemed to have statistical significance.
A total of 980 patients with ES were enrolled in this study; 929 patients originating from the SEER database were assigned to the training group; and 51 patients from four medical centers in China were assigned to the validation group (Table 1). There were significant differences between the two groups in terms of race, T-stage, and radiation (p < 0.05). In the validation group, all patients were classified under race as “others”. The proportion of radiation was significantly higher in the validation group than in the training group. In addition, more patients were diagnosed as TX in the training group. The remaining variables were not significantly different in both groups (Table 1). Lung metastasis occurred in 185 (18.9%) cases, the median age of the patients was 22.25 years (SD = 16.3), more than 85% of the patients were Caucasian and 534 (57.5%) patients were male. Comparison of the baseline data between the lung metastasis group and no lung metastasis group, revealed significant differences for the following factors: T-stage, N-stage, M-stage, surgery, bone metastasis, and survival time (p < 0.001). The demographic and clinicopathological variables of all 980 patients are summarized in Table 2.
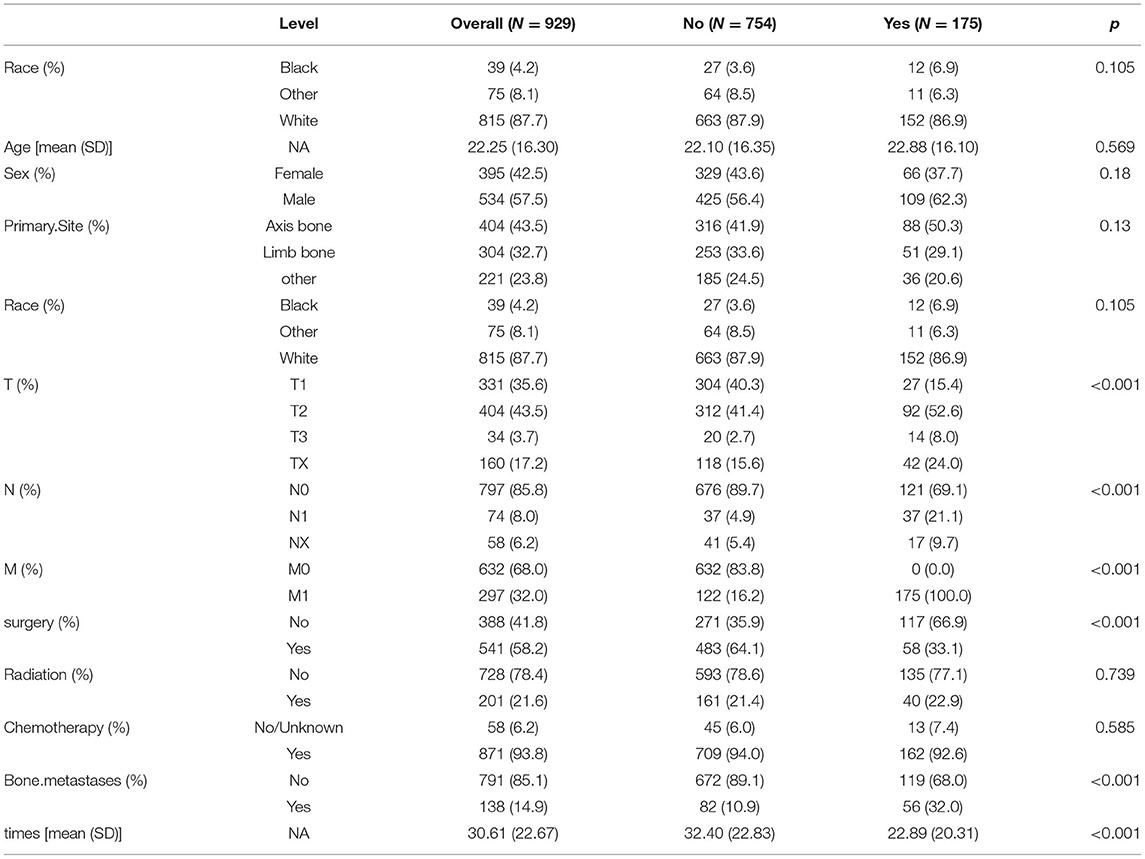
Table 2. Baseline table of patients in the Ewing sarcoma lung metastasis group vs. the no lung metastasis group.
The following variables were shown to have significant correlation with the development of LM in univariate analysis (p < 0.05): survival time, T-stage, N-stage, surgery, and bone metastasis (p < 0.001) (Table 3). Multifactorial LR analysis based on the variables (p < 0.05) in univariate analysis, demonstrated that T- stage (T2, OR = 2.7018, 95% CI = 1.690–4.317; T3, OR = 4.0378, 95% CI = 1.773–9.194; TX, OR = 3.1468, 95% CI = 1.778–5.566), N1 stage [vs. N0 stage, N1, (OR = 5.102, 95% CI = 3.048–8.540)], and bone metastasis (OR = 1.685, 95% CI = 1.090–2.605) were independent negative predictors of LM while survival time (OR = 0.988, 95% CI = 0.979–0.997) and surgery (OR = 0.451, 95% CI = 0.309–0.658) were positive predictors.
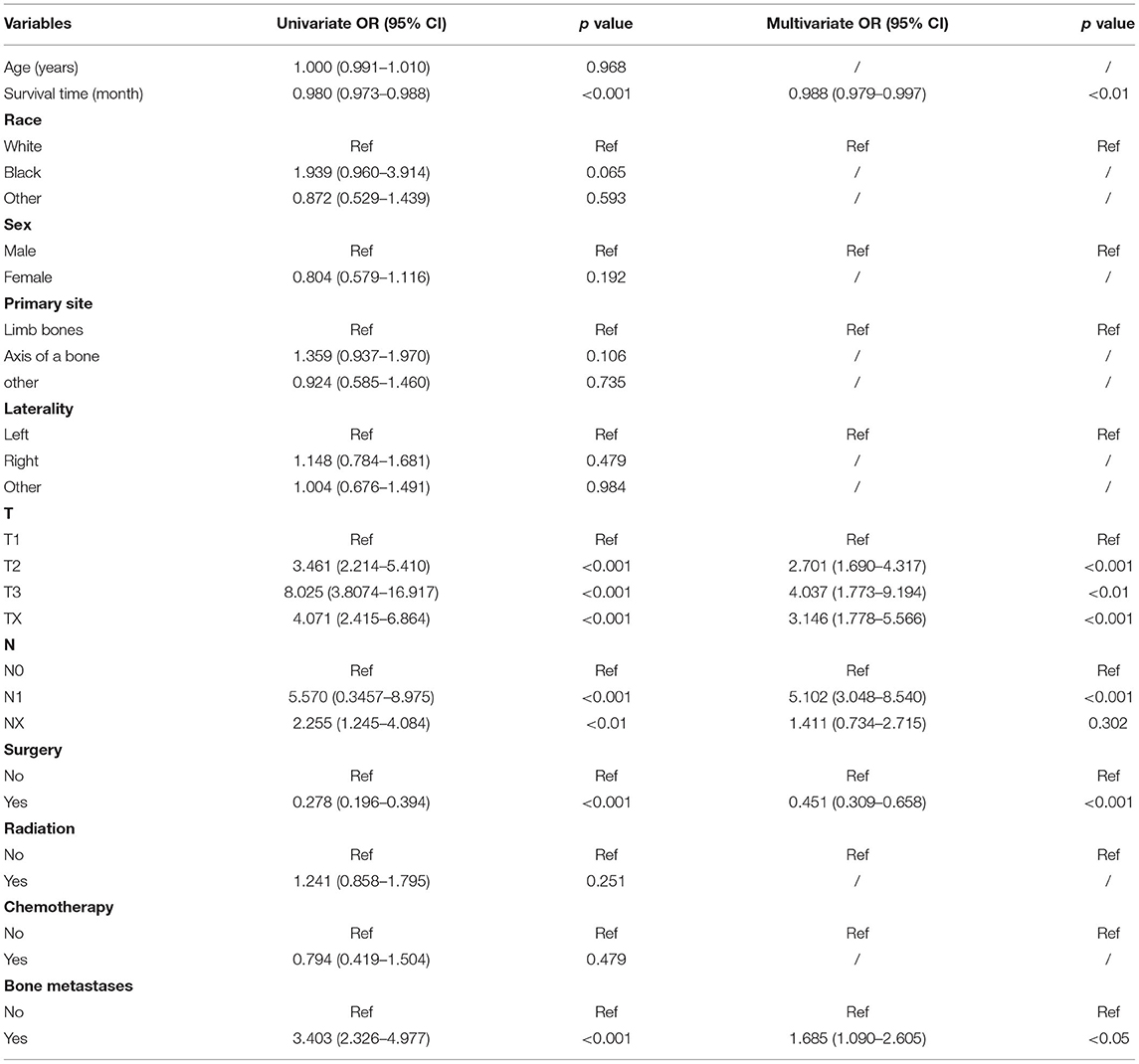
Table 3. Univariate and multifactorial logistic regression analysis of risk factors for lung metastasis in patients with Ewing sarcoma.
Six ML-based models for predicting LM in ES patients were developed based on the training group data. The average AUC of the six models determined by 10-fold cross-validation is shown in Figure 1, with the RF model achieving the best performance (AUC = 0.775). When the models established in training were subjected to external validation (Figure 2), the RF model still achieved the best performance (AUC = 0.705) in predicting LM and was accordingly selected as the design for a web-based, predictive tool.
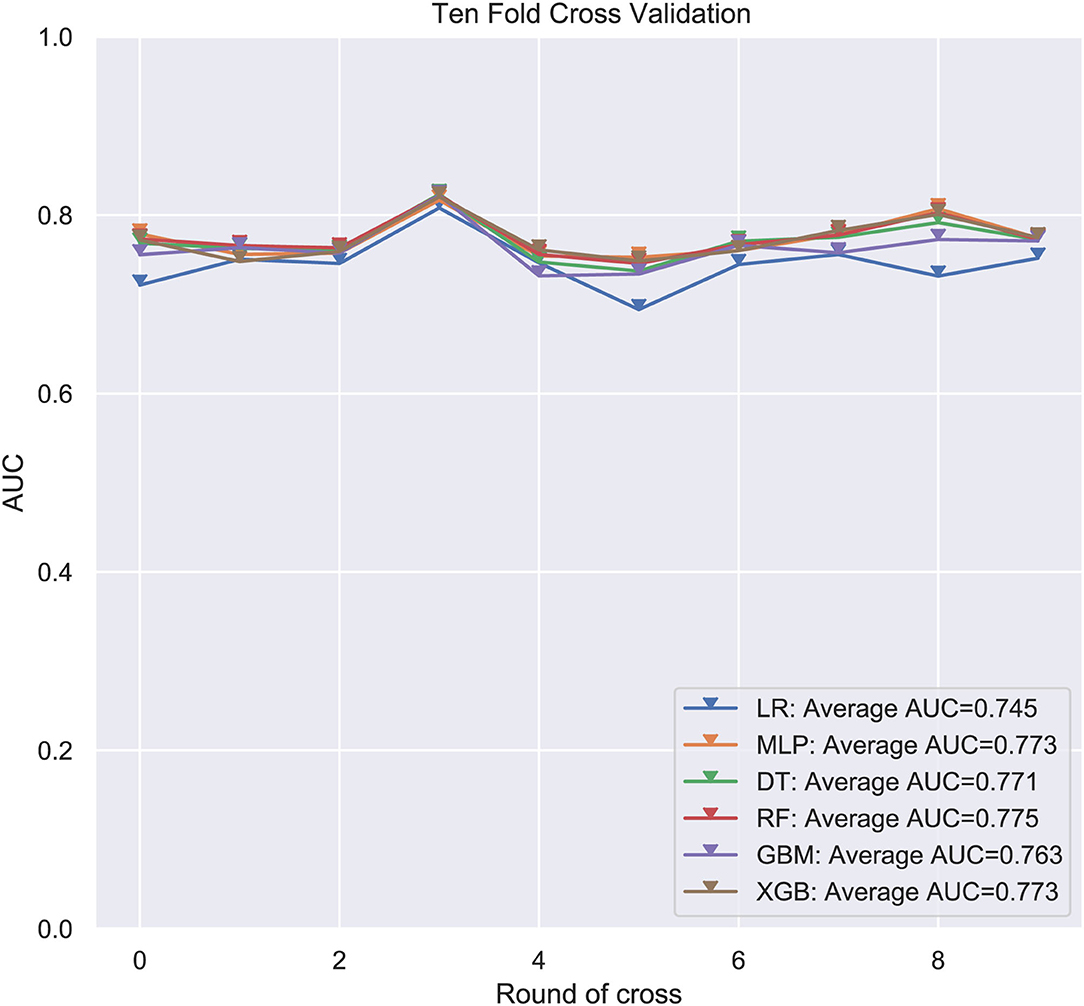
Figure 1. Average area under the curve (AUC) values of 10-fold cross-validation. RF, Random forest predictive model; DT, Decision tree; XGB, Extreme gradient boosting; GBM, Gradient boosting machine; MLP, Multilayer perceptron; LR, Logistic regression; AUC used as an indicator of performance, RF model achieved the best predictive performance while the MLP model showed the lowest.

Figure 2. External validation of machine learning algorithms. RF, Random Forest; DT, Decision tree; XGB, Extreme gradient boosting; GBM, Gradient boosting machine; MLP, Multilayer perceptron; LR, Logistic regression; AUC, area under the curve.
In consideration of clinical utility (Figure 3), we focused on four variables (T-stage, N-stage, surgery, and bone metastasis) to construct ML-based predictive models for LM in ES patients. Although there were slight differences in the importance of variables identified by each model; three factors, such as surgery, T-stage and N-stage, consistently ranked in the top three, and bone metastasis ranked fourth. The relative importance of variables in predicting LM using the RF model decreased in the order: surgery > T-stage > N-stage > bone metastasis. Analysis using spearman correlation of features approach revealed no significant positive correlation between any variable, and a negative correlation between surgery and the other three variables, indicating that all variables were independent (Figure 4).
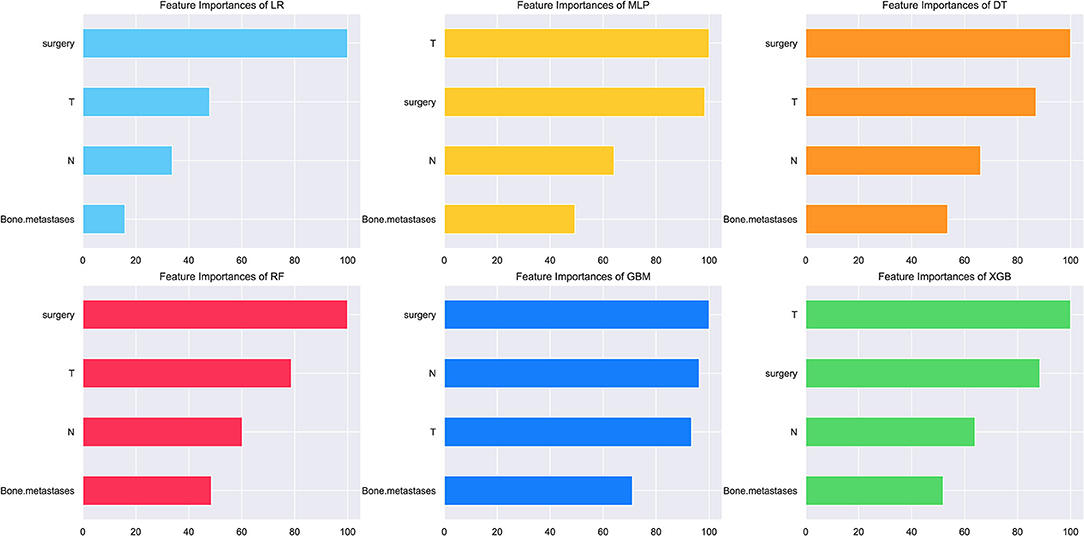
Figure 3. The relative importance of variables for the prediction of LM using ML algorithms. Surgery, T-stage and N-stage ranked in the top three in all prediction models, with bone metastasis ranked fourth.
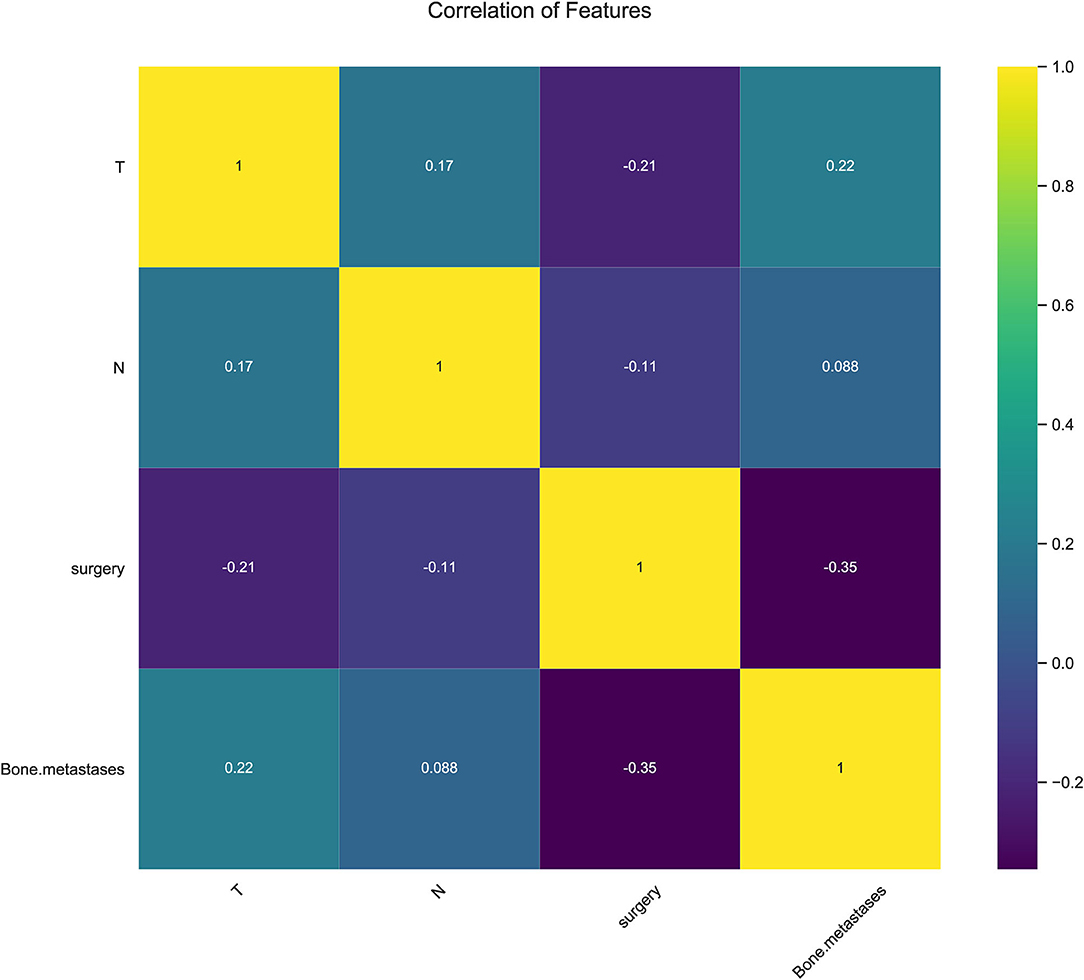
Figure 4. Results of Pearson correlation of features analysis between all variables showing no obvious correlation between every two variables.
The best-performing RF model was used to design a web-based tool to assist clinicians in predicting lung metastasis in ES patients (https://share.streamlit.io/liuwencai123/es_lm/main/es_lm.py) (Figure 5).
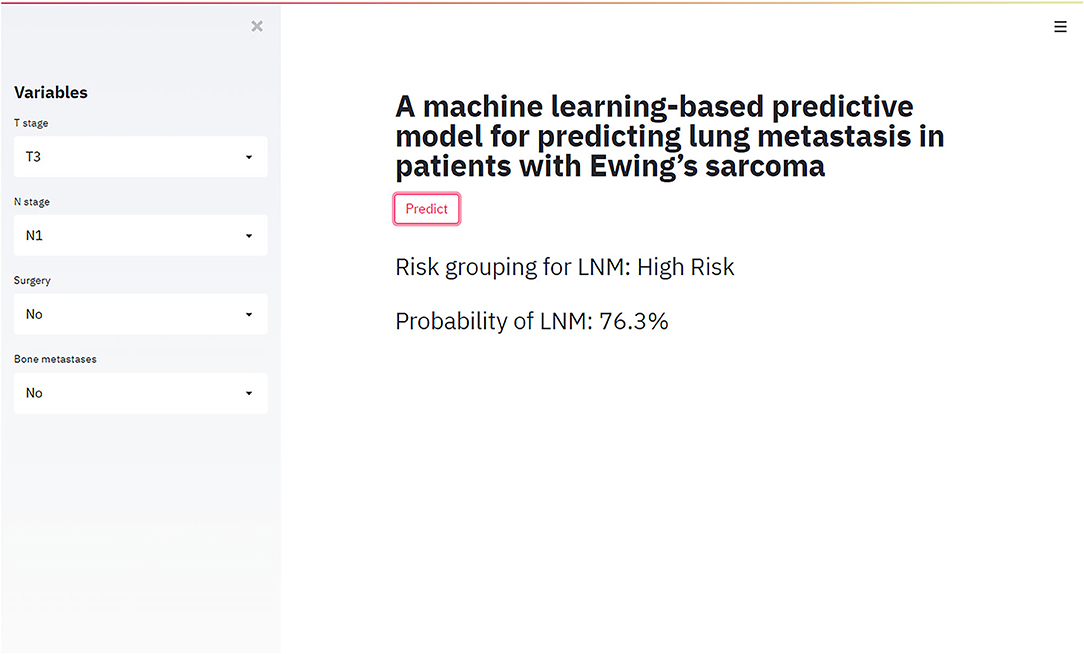
Figure 5. The web-based tool designed for predicting lung metastasis in patients with Ewing sarcoma.
Multi-modal therapy of metastatic disease based on chemotherapy, surgery, and radiation would be improved dramatically by the availability of reliable methods for predicting metastasis (27, 28). Many mathematical models of tumor malignancy employ multivariate regression or correlation analysis, which usually require the variables to be independent and linear (29–32). In addition to traditional univariate and multivariate analysis, we used multiple ML algorithms, which are widely applied in healthcare data analysis, to construct predictive models of LM in ES patients. We found that the RF model provided the best performance. RF is a commonly used ML algorithm that has a proven track record in handling large complex nonlinear datasets (33, 34). We subsequently designed a rapid web-based clinical tool, which is based on the RF model, for predicting lung metastasis in patients with ES.
Patient survival time was positively related to LM in univariate analysis. However, when considering clinical practice, survival time has no meaning for patients initially diagnosed with ES, and it is difficult to assess the survival time of a part of the patient population. Thus, survival time was not considered as a variable in ML models.
In the present study, four clinical variables: surgery, T-stage, N-stage, and bone metastasis were found to be the most important factors for predicting LM status by ML algorithms. We identified surgery as a protective factor against LM. To our knowledge, this factor has not been included previously in LM risk prediction models. Surgery is not only a vital form of treatment, but also plays a significant diagnostic role, which enables more accurate TNM staging and prognosis of ES patients. Surgery ranked first in order of importance in most of the predictive models developed in the present study, while T-stage (tumor size) ranked in the top two in all models investigated and was highly predictive of LM, similar to previous reports (35, 36). Large tumor volume indicates a longer growth cycle, resulting in a more proliferative and aggressive state, thus increasing the occurrence of lung metastasis. The correlation heat map showed that the T-stage correlated negatively with surgery since radical surgical treatment is difficult for large tumors, and lung metastasis is more likely.
Extensive investigations have consistently demonstrated that patients with regional node involvement were more prone to develop distant metastasis (37–41). Since the lung is associated with an abundance of lymphatic vessels, a tumor is more likely to metastasize to the lung when lymph nodes are positive. However, due to the scarcity of lymphatic vessels in bone tumor, it is conventionally accepted that dissemination to lymph nodes is uncommon (4, 42). Applebaum et al., for example, found that only 6.3% (91/1,452) of cases featured lymph node involvement (37). In contrast, our study revealed a much higher rate of lymph node metastasis, approximately 18.9% (185/980).
Importantly, our ML-based models revealed that bone metastasis was an important predictor of LM in ES patients, ranking fourth in importance behind surgery, T-stage and N-stage variables. Of the 138 patients in the two combined cohorts (training group and validation group) who had bone metastasis, 40.6% (56/138) also displayed lung metastasis. This figure was significantly higher than the number of patients who showed LM without bone metastasis (15%, 119/791).
Our present study of ML-based models for predicting LM in ES patients contained certain limitations which, nonetheless, serve as a guide for future improvements. Firstly, the information accessed from the SEER database was to a certain degree limited. Clinical information, such as the precise surgical treatment, surgical margin status, tumor marker, vascular invasion, radiation dosage, and chemotherapy modalities were unavailable, which limits the predictive value of the developed models. Secondly, the data from the SEER database was retrospective, which may introduce bias in data selection. However, while cognizant of these limitations, our study affirmed that ML-based prediction models can effectively identify the likelihood of LM in patients with ES by inspection of clinical factors such as surgery, N-stage, T-stage, and bone metastasis. The RF model performed best according to ROC analysis and was subsequently used to produce a web-based tool designed to help clinicians identify ES patients with lung metastasis, improve decision making and optimize individual treatment. Increased case data and multicenter studies are anticipated to lead to improvements in predictive performance.
Machine learning algorithms were applied to develop a prognostic tool for predicting the risk of LM in patients with ES. A RF model performed best and was engineered as a web-based tool for use by clinicians to improve patient diagnosis and treatment.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
CY, QL, and YQ designed the study. WL and TH collected and evaluated the data and wrote the first draft of the manuscript. All authors contributed to the interpretation of the results and the final draft of the manuscript.
This study was supported by the National Clinical Research Center for Orthopedics, Sports Medicine and Rehabilitation, and the Jiangsu China-Israel Industrial Technical Research Institute Foundation, 2021-NCRC-CXJJ-ZH-11.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn/) for providing linguistic services.
1. Khan S, Abid Z, Haider G, Bukhari N, Zehra D, Hashmi M, et al. Incidence of Ewing's sarcoma in different age groups, their associated features, and its correlation with primary care interval. Cureus. (2021) 13:e13986. doi: 10.7759/cureus.13986
2. Yu H, Ge Y, Guo L, Huang L. Potential approaches to the treatment of Ewing's sarcoma. Oncotarget. (2017) 8:5523–39. doi: 10.18632/oncotarget.12566
3. Balamuth NJ, Womer RB. Ewing's sarcoma, Lancet Oncol. (2010) 11:184–92. doi: 10.1016/S1470-2045(09)70286-4
4. Shi J, Yang J, Ma X, Wang X. Risk factors for metastasis and poor prognosis of Ewing sarcoma: a population based study. J Orthop Surg Res. (2020) 15:88. doi: 10.1186/s13018-020-01607-8
5. Gaspar N, Hawkins DS, Dirksen U, Lewis IJ, Ferrari S, Le Deley MC, et al. Ewing sarcoma: current management and future approaches through collaboration. J Clin Oncol. (2015) 33:3036–46. doi: 10.1200/JCO.2014.59.5256
6. Cotterill SJ, Ahrens S, Paulussen M, Jürgens HF, Voûte PA, Gadner H, et al. Prognostic factors in Ewing's tumor of bone: analysis of 975 patients from the European Intergroup Cooperative Ewing's sarcoma study group. J Clin Oncol. (2000) 18:3108–14. doi: 10.1200/JCO.2000.18.17.3108
7. Esiashvili N, Goodman M, Marcus RB Jr. Changes in incidence and survival of Ewing sarcoma patients over the past 3 decades: surveillance Epidemiology and End Results data. J Pediatr Hematol Oncol. (2008) 30:425–30. doi: 10.1097/MPH.0b013e31816e22f3
8. Arpaci E, Yetisyigit T, Seker M, Uncu D, Uyeturk U, Oksuzoglu B, et al. Prognostic factors and clinical outcome of patients with Ewing's sarcoma family of tumors in adults: multicentric study of the Anatolian Society of Medical Oncology. Med Oncol. (2013) 30:469. doi: 10.1007/s12032-013-0469-z
9. Völker T, Denecke T, Steffen I, Misch D, Schönberger S, Plotkin M, et al. Positron emission tomography for staging of pediatric sarcoma patients: results of a prospective multicenter trial. J Clin Oncol. (2007) 25:5435–41. doi: 10.1200/JCO.2007.12.2473
10. Mikulić D, Ilić I, Cepulić M, Giljević JS, Orlić D, Zupancić B, et al. Angiogenesis and Ewing sarcoma–relationship to pulmonary metastasis and survival. J Pediatr Surg. (2006) 41:524–9. doi: 10.1016/j.jpedsurg.2005.11.058
11. Mo X, Chen X, Ieong C, Zhang S, Li H, Li J, et al. Early prediction of clinical response to etanercept treatment in juvenile idiopathic arthritis using machine learning. Front Pharmacol. (2020) 11:1164. doi: 10.3389/fphar.2020.01164
12. Jin S, Kostka K, Posada JD, Kim Y, Seo SI, Lee DY, et al. Prediction of major depressive disorder following beta-blocker therapy in patients with cardiovascular diseases. J Pers Med. (2020) 10. doi: 10.3390/jpm10040288
13. Vey J, Kapsner LA, Fuchs M, Unberath P, Veronesi G, Kunz M. A toolbox for functional analysis and the systematic identification of diagnostic and prognostic gene expression signatures combining meta-analysis and machine learning. Cancers (Basel). (2019) 11. doi: 10.3390/cancers11101606
14. Stumpo V, Staartjes VE, Esposito G, Serra C, Regli L, Olivi A, et al. Machine learning and intracranial aneurysms: from detection to outcome prediction. Acta Neurochir Suppl. (2022) 134:319–31. doi: 10.1007/978-3-030-85292-4_36
15. Zilcha-Mano S, Roose SP, Brown PJ, Rutherford BR. A machine learning approach to identifying placebo responders in late-life depression trials. Am J Geriatr Psychiatry. (2018) 26:669–77. doi: 10.1016/j.jagp.2018.01.001
16. Frizzell JD, Liang L, Schulte PJ, Yancy CW, Heidenreich PA, Hernandez AF, et al. Prediction of 30-day all-cause readmissions in patients hospitalized for heart failure: comparison of machine learning and other statistical approaches. JAMA Cardiol. (2017) 2:204–9. doi: 10.1001/jamacardio.2016.3956
17. Zhu W, Xie L, Han J, Guo X. The application of deep learning in cancer prognosis prediction. Cancers (Basel). (2020) 12. doi: 10.3390/cancers12030603
18. Doll KM, Rademaker A, Sosa JA. Practical guide to surgical data sets: surveillance, epidemiology, and end results (SEER) database. JAMA Surg. (2018) 153:588–9. doi: 10.1001/jamasurg.2018.0501
19. Mao W, Deng F, Wang D, Gao L, Shi X. Treatment of advanced gallbladder cancer: a SEER-based study. Cancer Med. (2020) 9:141–50. doi: 10.1002/cam4.2679
20. Duggan MA, Anderson WF, Altekruse S, Penberthy L, Sherman ME. The surveillance, epidemiology, and end results (seer) program and pathology: toward strengthening the critical relationship. Am J Surg Pathol. (2016) 40:e94–94e102. doi: 10.1097/PAS.0000000000000749
21. Gonzalez GH, Tahsin T, Goodale BC, Greene AC, Greene CS. Recent advances and emerging applications in text and data mining for biomedical discovery. Brief Bioinform. (2016) 17:33–42. doi: 10.1093/bib/bbv087
22. Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery, Lancet Oncol. (2019) 20:e262–262e273. doi: 10.1016/S1470-2045(19)30149-4
23. Sturgiss EA, Rieger E, Haesler E, Ridd MJ, Douglas K, Galvin SL. Adaption and validation of the Working Alliance Inventory for General Practice: qualitative review and cross-sectional surveys. Fam Pract. (2019) 36:516–22. doi: 10.1093/fampra/cmy113
24. Altmann A, Toloşi L, Sander O, Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics. (2010) 26:1340–7. doi: 10.1093/bioinformatics/btq134
25. Mi X, Zou B, Zou F, Hu J. Permutation-based identification of important biomarkers for complex diseases via machine learning models. Nat Commun. (2021) 12:3008. doi: 10.1038/s41467-021-22756-2
26. Yang JB, Shen KQ, Ong CJ, Li XP. Feature selection for MLP neural network: the use of random permutation of probabilistic outputs. IEEE Trans Neural Netw. (2009) 20:1911–22. doi: 10.1109/TNN.2009.2032543
27. Kibiş EY, Büyüktahtakin IE. Optimizing multi-modal cancer treatment under 3D spatio-temporal tumor growth. Math Biosci. (2019) 307:53–69. doi: 10.1016/j.mbs.2018.10.010
28. Liu Z, Zhong Y, Zhou X, Huang X, Zhou J, Huang D, et al. Inherently nitric oxide containing polymersomes remotely regulated by NIR for improving multi-modal therapy on drug resistant cancer. Biomaterials. (2021) 277:121118. doi: 10.1016/j.biomaterials.2021.121118
29. Pearce O, Delaine-Smith RM, Maniati E, Nichols S, Wang J, Böhm S, et al. Deconstruction of a metastatic tumor microenvironment reveals a common matrix response in human cancers. Cancer Discov. (2018) 8:304–19. doi: 10.1158/2159-8290.CD-17-0284
30. Arefan D, Hausler RM, Sumkin JH, Sun M, Wu S. Predicting cell invasion in breast tumor microenvironment from radiological imaging phenotypes. BMC Cancer. (2021) 21:370. doi: 10.1186/s12885-021-08122-x
31. Liu Z, Mi M, Li X, Zheng X, Wu G, Zhang L. A lncRNA prognostic signature associated with immune infiltration and tumour mutation burden in breast cancer. J Cell Mol Med. (2020) 24:12444–56. doi: 10.1111/jcmm.15762
32. Madekivi V, Boström P, Karlsson A, Aaltonen R, Salminen E. Can a machine-learning model improve the prediction of nodal stage after a positive sentinel lymph node biopsy in breast cancer. Acta Oncol. (2020) 59:689–95. doi: 10.1080/0284186X.2020.1736332
33. Lebedev AV, Westman E, Van Westen GJ, Kramberger MG, Lundervold A, Aarsland D, et al. Random Forest ensembles for detection and prediction of Alzheimer's disease with a good between-cohort robustness. Neuroimage Clin. (2014) 6:115–25. doi: 10.1016/j.nicl.2014.08.023
34. Speiser JL, Miller ME, Tooze J, Ip E. A comparison of random forest variable selection methods for classification prediction modeling. Expert Syst Appl. (2019) 134:93–101. doi: 10.1016/j.eswa.2019.05.028
35. Ye C, Dai M, Zhang B. Risk factors for metastasis at initial diagnosis with ewing sarcoma. Front Oncol. (2019) 9:1043. doi: 10.3389/fonc.2019.01043
36. Ramkumar DB, Ramkumar N, Miller BJ, Henderson ER. Risk factors for detectable metastatic disease at presentation in Ewing sarcoma - An analysis of the SEER registry. Cancer Epidemiol. (2018) 57:134–9. doi: 10.1016/j.canep.2018.10.013
37. Applebaum MA, Goldsby R, Neuhaus J, DuBois SG. Clinical features and outcomes in patients with Ewing sarcoma and regional lymph node involvement. Pediatr Blood Cancer. (2012) 59:617–20. doi: 10.1002/pbc.24053
38. van der Kamp MF, Muntinghe F, Iepsma RS, Plaat B, van der Laan B, Algassab A, et al. Predictors for distant metastasis in head and neck cancer, with emphasis on age. Eur Arch Otorhinolaryngol. (2021) 278:181–90. doi: 10.1007/s00405-020-06118-0
39. Javidiparsijani S, Brickman A, Lin DM, Rohra P, Ghai R, Bitterman P, et al. Is regional lymph node metastasis of head and neck paraganglioma a sign of aggressive clinical behavior: a clinical/pathologic review. Ear Nose Throat J. (2021) 100:447–53. doi: 10.1177/0145561319863373
40. Chu PY, Chen YF Li CY, Yang JS, King YA, Chiu YJ, et al. Factors influencing locoregional recurrence and distant metastasis in Asian patients with cutaneous melanoma after surgery: a retrospective analysis in a tertiary hospital in Taiwan. J Chin Med Assoc. (2021) 84:870–6. doi: 10.1097/JCMA.0000000000000586
41. Kilic C, Kimyon Comert G, Cakir C, Yuksel D, Codal B, Kilic F, et al. Recurrence pattern and prognostic factors for survival in cervical cancer with lymph node metastasis. J Obstet Gynaecol Res. (2021) 47:2175–84. doi: 10.1111/jog.14762
Keywords: Ewing sarcoma, lung metastasis, machine learning algorithms, multicenter, web calculator
Citation: Li W, Hong T, Liu W, Dong S, Wang H, Tang Z-R, Li W, Wang B, Hu Z, Liu Q, Qin Y and Yin C (2022) Development of a Machine Learning-Based Predictive Model for Lung Metastasis in Patients With Ewing Sarcoma. Front. Med. 9:807382. doi: 10.3389/fmed.2022.807382
Received: 02 November 2021; Accepted: 07 March 2022;
Published: 01 April 2022.
Edited by:
Chris Hodge, The University of Sydney, AustraliaReviewed by:
Steven Christopher Smith, Virginia Commonwealth University Health System, United StatesCopyright © 2022 Li, Hong, Liu, Dong, Wang, Tang, Li, Wang, Hu, Liu, Qin and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengliang Yin, Y2hlbmdsaWFuZ3lpbkAxNjMuY29t; Yong Qin, cWlueW9uZzAxMjVAMTI2LmNvbQ==; Qiang Liu, bTEzOTkyMDc5NjY4QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.