- 1Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran
- 2Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
- 3Laboratory of Experimental Pharmacology, Istituto Di Ricovero e Cura a Carattere Scientifico Istituto Tumori Giovanni Paolo II, Bari, Italy
- 4Medical Oncology Unit, IRCCS Istituto Tumori “Giovanni Paolo II” of Bari, Bari, Italy
- 5Medical Oncology Unit, Department of Human Pathology “G. Barresi, University of Messina, Messina, Italy
- 6Department of Immunology, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
Background: The programmed death-ligand 1 (PD-L1)/PD-1 axis is one of the well-established inhibitory axes in regulating immune responses. Besides the significance of tumor-intrinsic PD-L1 expression in immune evasion, its oncogenic role has been implicated in various malignancies, like non-small cell lung cancer (NSCLC). As small non-coding RNAs, microRNAs (miRs) have pivotal roles in cancer biology. The current study aimed to systematically review the current knowledge about the significance of PD-L1-inhibiting miRs in NSCLC inhibition and their underlying mechanisms.
Materials and methods: We conducted the current scoping review based on the PRISMA-ScR statement. We systematically searched Embase, Scopus, Web of Science, PubMed, Ovid, EBSCO, ProQuest, Cochrane Library, African Index Medicus, and Pascal-Francis up to 4 April 2021. We also performed in silico tumor bulk RNA sequencing and single-cell RNA sequencing to further the current knowledge of the non-coding RNA-mediated tumor-intrinsic PD-L1 regulation and the PD-L1/PD-1 axis in NSCLC.
Results: The ectopic expression of hsa-miR-194-5p, hsa-miR-326, hsa-miR-526b-3p, hsa-miR-34a-5p, hsa-miR-34c-5p, hsa-miR-138-5p, hsa-miR-377-3p, hsa-let-7c-5p, hsa-miR-200a-3p, hsa-miR-200b-3p, hsa-miR-200c-3p, and hsa-miR-197-3p, as PD-L1-inhibiting miR, inhibits NSCLC development. These PD-L1-inhibiting miRs can substantially regulate the cell cycle, migration, clonogenicity, invasion, apoptosis, tumor chemosensitivity, and host anti-tumoral immune responses. Based on single-cell RNA sequencing results, PD-L1 inhibition might liberate the tumor-infiltrated CD8+ T-cells and dendritic cells (DCs)-mediated anti-tumoral immune responses via disrupting the PD-L1/PD-1 axis.
Conclusion: Given the promising preclinical results of these PD-L1-inhibiting miRs in inhibiting NSCLC development, their ectopic expression might improve NSCLC patients’ prognosis; however, further studies are needed to translate this approach into clinical practice.
Introduction
Lung cancer is projected to be the leading cause of cancer death in the United States; it has been estimated that lung cancer will be responsible for 350 deaths each day in 2022. Also, lung cancer will be the second most commonly affected cancer in both males and females in 2022. Besides, the 5-year relative survival of lung cancer patients is 22% in all races and stages (1). It has been reported that approximately 85% of lung cancer cases are grouped into NSCLCs; lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) are the most common subtypes of NSCLC (2). Considering the prevalence, associated burden, and poor survival of affected patients, there is a pressing need to identify novel therapeutic approaches to improve the prognosis of the patients.
Cancer immunotherapy is a novel therapeutic approach that leverages the host immune system to reject tumors (3). The immunosuppressive tumor microenvironment shields malignant cells from anti-tumoral immune responses and reprograms the residing immune cells in a way that they contribute to tumor growth and migration (4). Growing evidence has highlighted the significance of pathological expression of inhibitory immune checkpoint molecules in suppressing anti-tumoral immune responses (5). The PD-L1/PD-1 is one of the well-studied inhibitory immune checkpoint axes that can regulate immune responses. It has been reported that the increased expression of PD-L1 is associated with decreased overall survival and recurrence-free survival of NSCLC patients (6). Following the significant role of the PD-L1/PD-1 axis in facilitating tumor growth, various monoclonal antibodies targeting this axis have been developed, investigated, and approved for NSCLC patients (7). Besides the immunosuppressive role of tumor-intrinsic PD-L1, it is implicated in stimulating oncogenic signaling pathways. For instance, increased expression of PD-L1 in NSCLC cells increases tumor proliferation and colony numbers; however, PD-L1 depletion reserves these pro-tumoral effects in NSCLC cells (8). In cisplatin-treated NSCLC cells, PD-L1 knockdown decreases cell proliferation, migration, and invasion of NSCLC cells (6). It has been shown that the mitogen-activated protein kinase (MAPK) pathway stimulates PD-L1 expression in NSCLC cells, and inhibiting the MAPK pathway decreases PD-L1 protein and mRNA expression (9). Besides, the epidermal growth factor receptor (EGFR)-mediated pathway, as an oncogenic signaling pathway, upregulates PD-L1 expression in NSCLC cells (10). In colorectal cancer cells, PD-L1 knockdown has decreased the protein and mRNA expression of phosphatidylinositol-3-kinase and protein kinase B (PI3K) and protein kinase B (Akt), which are the essential factors of the PI3K/Akt oncogenic signaling pathway (11). Therefore, PD-L1 is considerably involved in oncogenesis and tumor development. Despite the promising results of administrating monoclonal antibodies against the PD-L1/PD-1 axis for NSCLC patients, these antibodies primarily target the PD-L1/PD-1 expression on various cells. Indeed, they are not designed to inhibit the mRNA or protein expression of oncogenic signaling pathway elements in malignant cells (12).
miRs are small non-coding RNAs that have considerable roles in the post-transcriptional regulation of gene expression (13). These non-coding RNAs substantially regulate various signaling pathways, and the downregulation of tumor-suppressive miRs and the upregulation of oncomiRs have been associated with tumor development (14). Recent studies have shown that ectopic expression of specific PD-L1-targeting miRs in cancerous cells, like colorectal cancer and triple-negative breast cancer cells, can suppress tumor-intrinsic PD-L1 expression, stimulate anti-tumoral immune responses, substantially inhibit oncogenic signaling pathways, and decrease tumor development (12, 15). Accumulating studies have identified PD-L1-inhibiting miRs that some of them can suppress NSCLC development (6, 16–18). Xie et al. have shown that the ectopic expression of miR-140, as a PD-L1-inhibiting miR, is associated with decreased expression of cyclin E and suppressed cell proliferation in NSCLC cells (19). Chen et al. have reported that miR-526b-3p increased expression downregulates the expression of c-Myc and MDR1 and enhances cisplatin-cytotoxicity in NSCLC cells (16). Song et al. have shown that increased expression of hsa-miR-138-5p decreases tumor proliferation and arrests the cell cycle of NSCLC cells via downregulating CCND3, Ki67, CDC20, and MCM2 (20). However, there is no study to comprehensively collate and investigate their role in NSCLC development.
In the current study, we aimed to thoroughly and systematically search the literature to identify the current knowledge of the significance of PD-L1-inhibiting miR ectopic expression in NSCLC development. We also leveraged the cancer genome atlas (TCGA) data to study their clinical significance in the affected patients. Besides searching the literature, we leveraged in silico data to predict the circular RNA (circRNA)/miR/PD-L1 axes in NSCLC, which might translate into introducing novel therapeutic approaches. Finally, we applied single-cell RNA sequencing to study the potential results of the ectopic expression of PD-L1-inhibiting miRs on the immune cells residing in the NSCLC microenvironment.
Materials and methods
Scoping review of literature
Scoping review protocol
We applied Arksey and O’Malley’s framework (21), which was improved by Levac et al. (22). Also, the current scoping review is consistent with the preferred reporting items for systematic reviews and meta-analyses extension for scoping reviews (PRISMA-ScR) guidelines (23). The five steps of this scoping review consist of developing the research question, finding the relevant papers, selecting papers, data charting, and summarizing and reporting the obtained results.
The research question
Concerning the oncogenic role of tumor-intrinsic PD-L1 in developing colorectal cancer and triple-negative breast cancer (12, 15), the current scoping review aimed to study the current knowledge on the therapeutic potentiality of tumor-intrinsic PD-L1-inhibiting miRs to suppress NSCLC development.
Finding the published records
The Embase, Scopus, Web of Science, PubMed, Ovid, EBSCO, ProQuest, Cochrane Library, African Index Medicus, and Pascal-Francis were systematically searched to identify the published papers before 4 April 2021 without any restriction on language, country, and time. The following keywords were used to identify the relevant papers: (“programmed death-ligand 1” OR “PD-L1” OR “P.D. L1” OR “PDL1” OR “B7-H1” OR “B7 H1” OR “B7H1” OR “CD274” OR “cluster of differentiation 274” OR “CD 274” OR “cluster of differentiation274” OR “B7 homolog 1” OR “PDCD1 ligand 1” OR “PDCD1LG1” OR “PDCD1L1” OR “HPD-L1” OR “B7-H1 antigen” OR “programmed death 1 ligand 1”) and (“microRNA-” OR “microRNA” OR “micro RNA-” OR “micro RNA” OR “miRNA-” OR “miRNA” OR “miR-” OR “miR” OR “miRNA-” OR “miRNA” OR “MicroRNAs”) and (“NSCLC” OR “lung tumor” OR “lung malignancy” OR “lung” OR “non-small-cell lung carcinoma” OR “non-small-cell lung cancer” OR “non-small-cell lung carcinoma” OR “non-small cell lung carcinoma” OR “non-small cell lung carcinoma” OR “non-small-cell lung cancer” OR “non-small cell lung cancer” OR “squamous cell carcinoma of lung” OR “lung squamous cell carcinoma” OR “SCC” OR “large cell carcinoma” OR “carcinoma, non-small-cell lung” OR “lung neoplasms” OR “Adenocarcinoma of Lung” OR “Carcinoma, Large Cell” OR “Carcinoma, Squamous Cell” OR “lung cancer” OR “lung tumor” OR “non-small cell lung cancer” OR “adenocarcinoma” OR “lung adenocarcinoma” OR “squamous cell carcinoma”). The medical subject headings (MeSh) and Emtree terms were used in our systematic search to increase the sensitivity of the systematic search.
Study selection
Two independent co-authors (MS and FG) thoroughly reviewed the obtained records in two distinct phases. In the first phase, those co-authors independently screened the title and abstract of the retrieved records. In the second phase, they independently reviewed the full text of the remaining papers. The senior author, i.e., BB, was consulted to resolve any disagreements.
The included papers in the current scoping review must meet the following criteria, i.e., the included studies must be original articles published in English, and those papers must study the effect of miR ectopic expression on the expression of tumor-intrinsic PD-L1 in human NSCLC cells. The following records were excluded from the current scoping review: (I) duplicated records, (II) review articles, (III) meeting abstracts, (IV) editorial papers, (V) non-English papers, (VI) viewpoints, (VII) case reports, (VIII) book chapters, (IX) notes, (X) erratum, and (XI) the papers that did not meet the aforementioned inclusion criteria.
Data charting
We extracted the following results from the included papers: the first author, publication year, studied miR(s), studied cell line, the effect of studied miR(s) on tumor development, and the proposed cross-talk with other coding and non-coding RNAs.
Summarizing and reporting the obtained results
Besides summarizing the findings of the included studies, the current scoping review discusses the findings regarding the effect of these PD-L1-inhibiting miRs on NSCLC development that have not been presented in the included studies. Also, we included in silico investigations to present more insights regarding the significance of the PD-1/PD-L1 axis based on single-cell RNA sequencing.
In silico investigation
The significance of identified programmed death-ligand 1-inhibiting miRs on biological pathways
We used the miRPathDB v2.0 to access the WikiPathways dataset and analyze them (24). We adjusted our analyses based on a minimum of four significant miRs per pathway and strong experimental evidence.
Identifying the circRNA-miR-PD-L1 axes in non-small cell lung cancer
We also studied the circRNA/miR/PD-L1 axis in NSCLC cancer patients. We accessed the GSE158695 and GSE63805 datasets via the Gene Expression Omnibus (GEO) database1 to identify the significantly upregulated circRNAs and downregulated miRs in NSCLCs, respectively. Then, we used the data of microT-CDS to predict the potential miRs that can target PD-L1. The common miRs between these are selected for further analyses. After that, we accessed the circMine2 to identify the targets of the identified significantly upregulated circRNA in NSCLC tissues. The | log2FC| ≥ 1 and adjusted P-value < 0.05 were the cut-offs for considering significantly differentially expressed miRs and circRNAs.
The prognostic significance of identified programmed death-ligand 1-inhibiting miRs in patients with lung adenocarcinoma
We accessed the TCGA-LUAD and TCGA-LUSC data via the UCSC Cancer Browser3. In our survival analyses, we only included primary tumor tissues and considered the upper quartile as “high” expression and the lower quartile as “low” expression.
Single-cell RNA sequencing
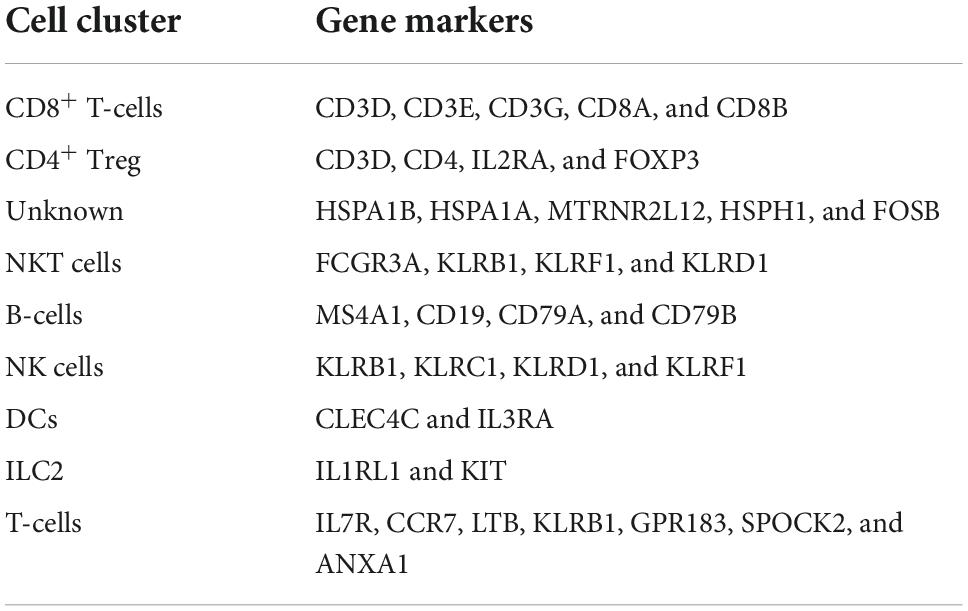
After highlighting the significance of PD-L1-inhibiting miRs on NSCLC development and biological pathways, we aimed to investigate which cells might be potentially liberated from the immunosuppressive PD-L1/PD-1 axis in the NSCLC microenvironment. For this purpose, we accessed to GSE144945 dataset via Gene Expression Omnibus (GEO) database (see text footnote 1). This dataset applied single-cell RNA sequencing on 10 tissues for NSCLC patients using GPL16791 Illumina HiSeq 2500 (Homo sapiens) (25). After downloading the dataset, we used Seurat (version 4.1.0) R package to analyze the raw data (24). Cells with mitochondrial gene expression of < 10% and expressed genes of above 500 were included in our analyses. We scaled them and ran the principal component analysis after normalizing data, identifying variable genes, and integrating data from multiple samples. Then, the uniform manifold approximation and projection (UMAP) clustering algorithms with a resolution of 0.11 were used to cluster the cells into a two-dimensional figure. Afterward, the PanglaoDB dataset and the information demonstrated in the original study were used to annotate the identified clusters according to the pertained gene markers (25, 26). The applied gene markers are demonstrated in Table 1.
Results
Included studies
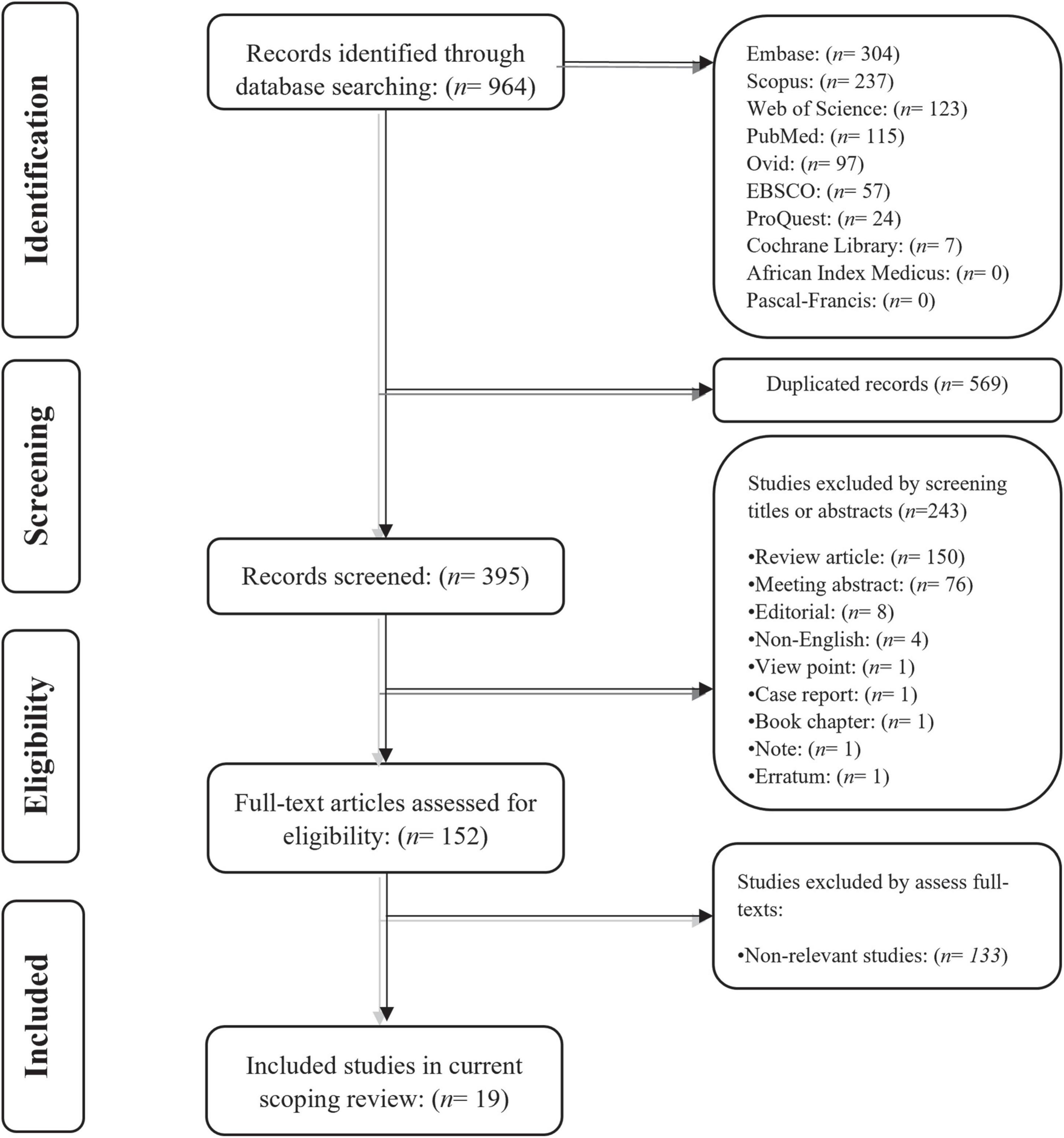
Our systematic search on Embase, Scopus, Web of Science, PubMed, Ovid, EBSCO, ProQuest, Cochrane library, African Index Medicus, and Pascal-Francis databases retrieved 964 potentially relevant records for the current scoping review. Following removing the duplicated records, the title and abstract of the remaining 395 papers were screened independently by two co-authors. After removing the records that were not suitable for the scoping review, those co-authors independently reviewed the full text of the remaining papers for consideration to be included in the study. Finally, 19 original studies were included in this scoping review. The literature identification flowchart is shown in Figure 1.
The characteristics of the included studies
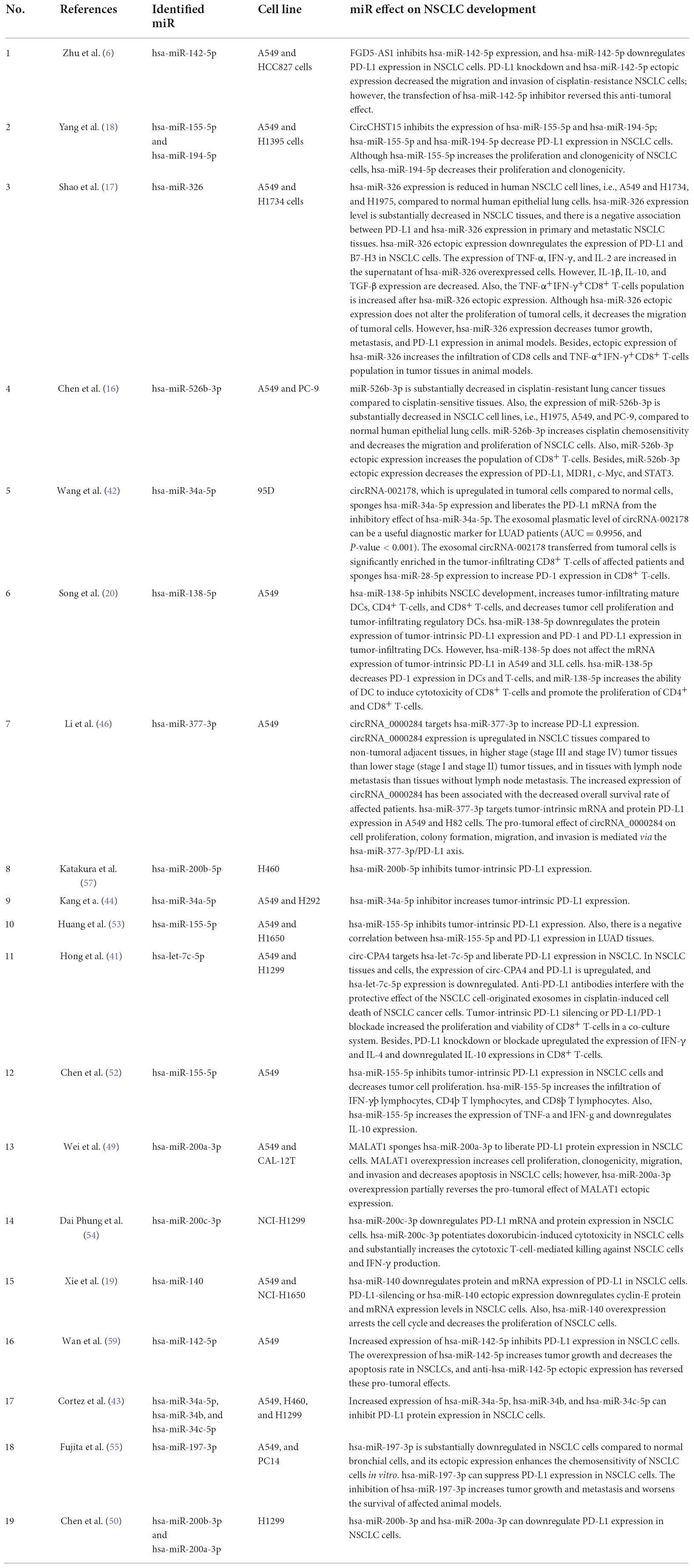
The 19 included studies were published between 2014 and 2021. A549 is the most studied NSCLC cell line in the included studies (Table 2). Our results have indicated that the increased expression of hsa-miR-142-5p, hsa-miR-155-5p, hsa-miR-194-5p, hsa-miR-326, hsa-miR-526b-3p, hsa-miR-138-5p, hsa-miR-377-3p, hsa-miR-200b-5p, hsa-miR-34a-5p, hsa-let-7c-5p, hsa-miR-200a-3p, hsa-miR-200c-3p, hsa-miR-140, hsa-miR-34b, hsa-miR-34c-5p, hsa-miR-197-3p, and hsa-miR-200b-3p inhibit tumor-intrinsic PD-L1 in NSCLC cells (Table 2).
The significance of the identified programmed death-ligand 1-inhibiting miRs on biological pathways
We used the WikiPathways dataset to analyze the significance of the identified PD-L1-inhibiting miRs on biological pathways. Our results have indicated that hsa-miR-34a-5p, hsa-miR-377-3p, hsa-miR-200a-3p, and hsa-miR-34c-5p are significantly enriched for regulating the cell cycle (Figure 2). Also, our results have indicated that hsa-miR-34a-5p, hsa-let-7c-5p, hsa-miR-200c-3p, and hsa-miR-34c-5p are significantly enriched for regulating the PI3K/Akt signaling pathway (Figure 2).
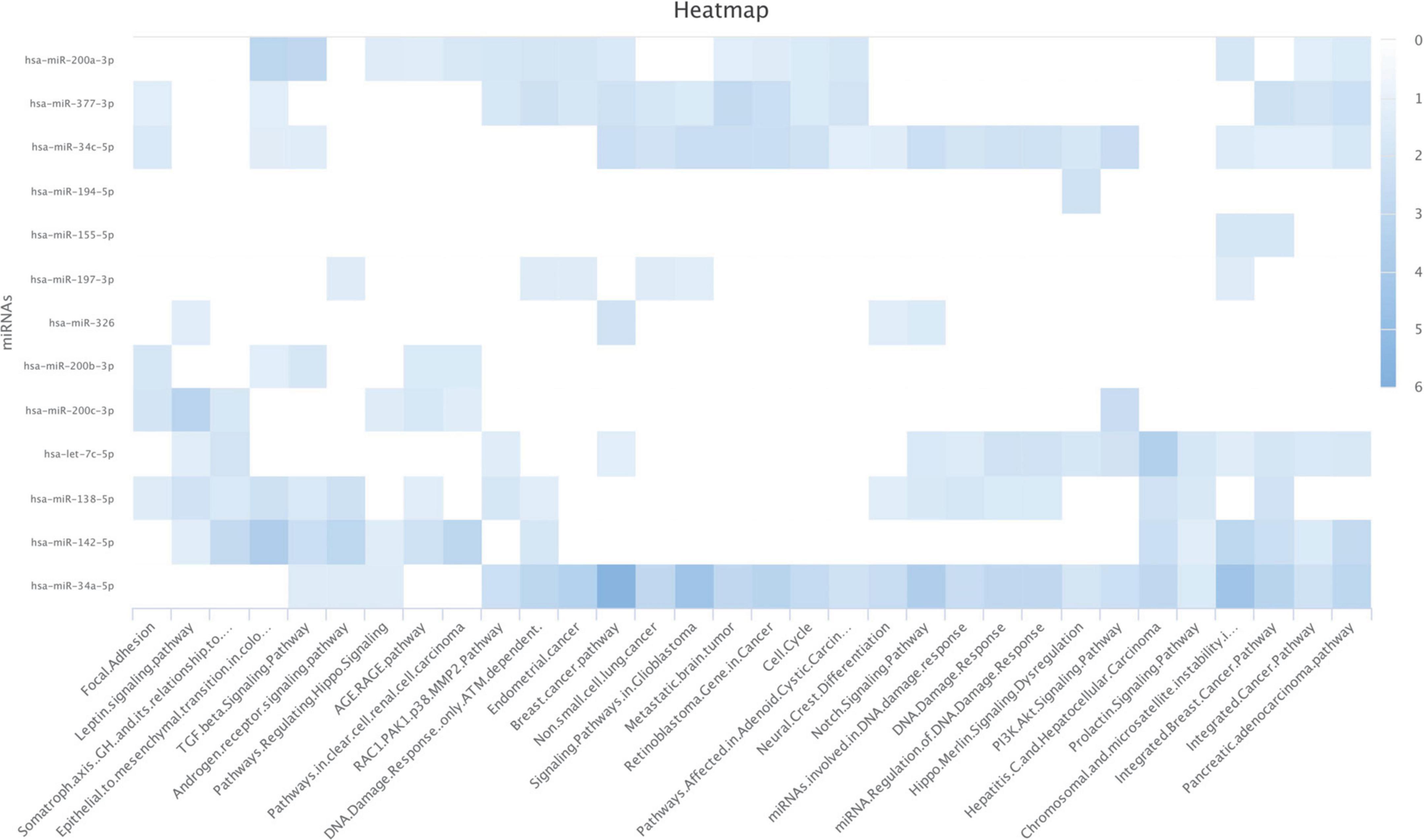
Figure 2. Enrichment analyses on the identified programmed death-ligand 1 (PD-L1)-inhibiting miRs based on the WikiPathways. The darker color shows a more significant one.
The prognostic values of the identified programmed death-ligand 1-inhibiting miRs in patients with lung adenocarcinoma and lung squamous cell carcinoma
Our results have indicated that high expression of hsa-miR-200a-3p, as a PD-L1-inhibiting miR, is significantly associated with improved disease-specific survival of patients with LUAD (P-value = 0.01844) (Figure 3A). Also, high expression of hsa-let-7c-5p, as a PD-L1-inhibiting miR, is significantly associated with enhanced disease-specific survival of patients with LUSC (P-value = 0.001504) (Figure 3B).
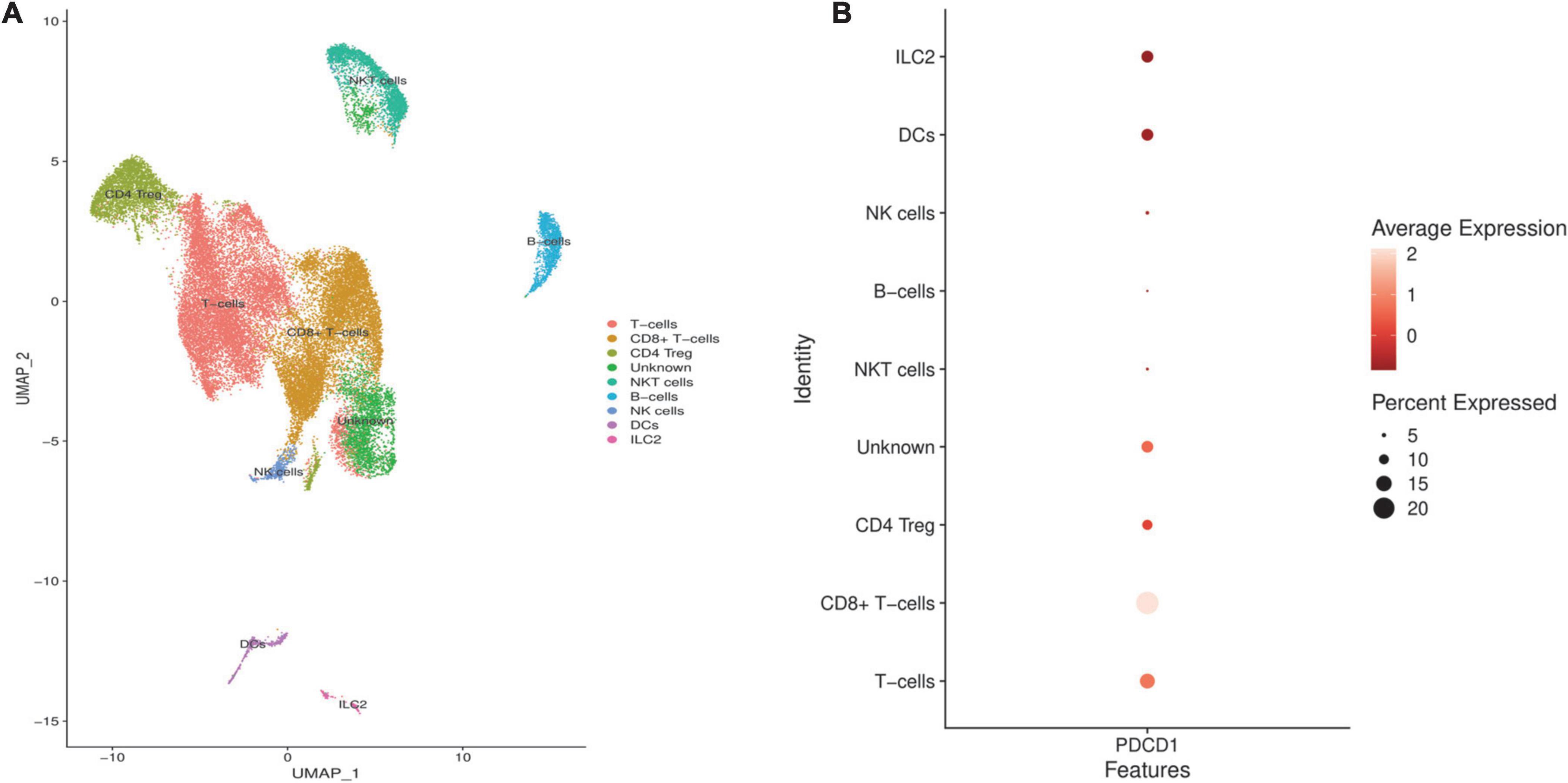
Figure 3. The prognostic values of hsa-miR-200a-3p and hsa-let-7c-5p, as programmed death-ligand 1 (PD-L1)-inhibiting miRs, based on TCGA-LUAD and TCGA-LUSC databases. (A) High expression of hsa-miR-200a-3p is associated with enhanced disease-specific survival of LUAD patients. (B) High expression of hsa-let-7c-5p is associated with improved disease-specific survival of patients with lung squamous cell carcinoma (LUSC).
The circRNA/miR/PD-L1 axes in non-small cell lung cancer
We analyzed the GSE158695 dataset to identify significantly upregulated circRNAs in NSCLC tissues compared to normal lung tissues. For this aim, we normalized the expression values of 3 NSCLC and 3 non-tumoral tissues (Figure 4A). Our results have shown that the expression profile of the included tissues is consistent with two distinct patterns (Figure 4B). We have identified 12 upregulated circRNAs in NSCLC tissues (Figure 4C).
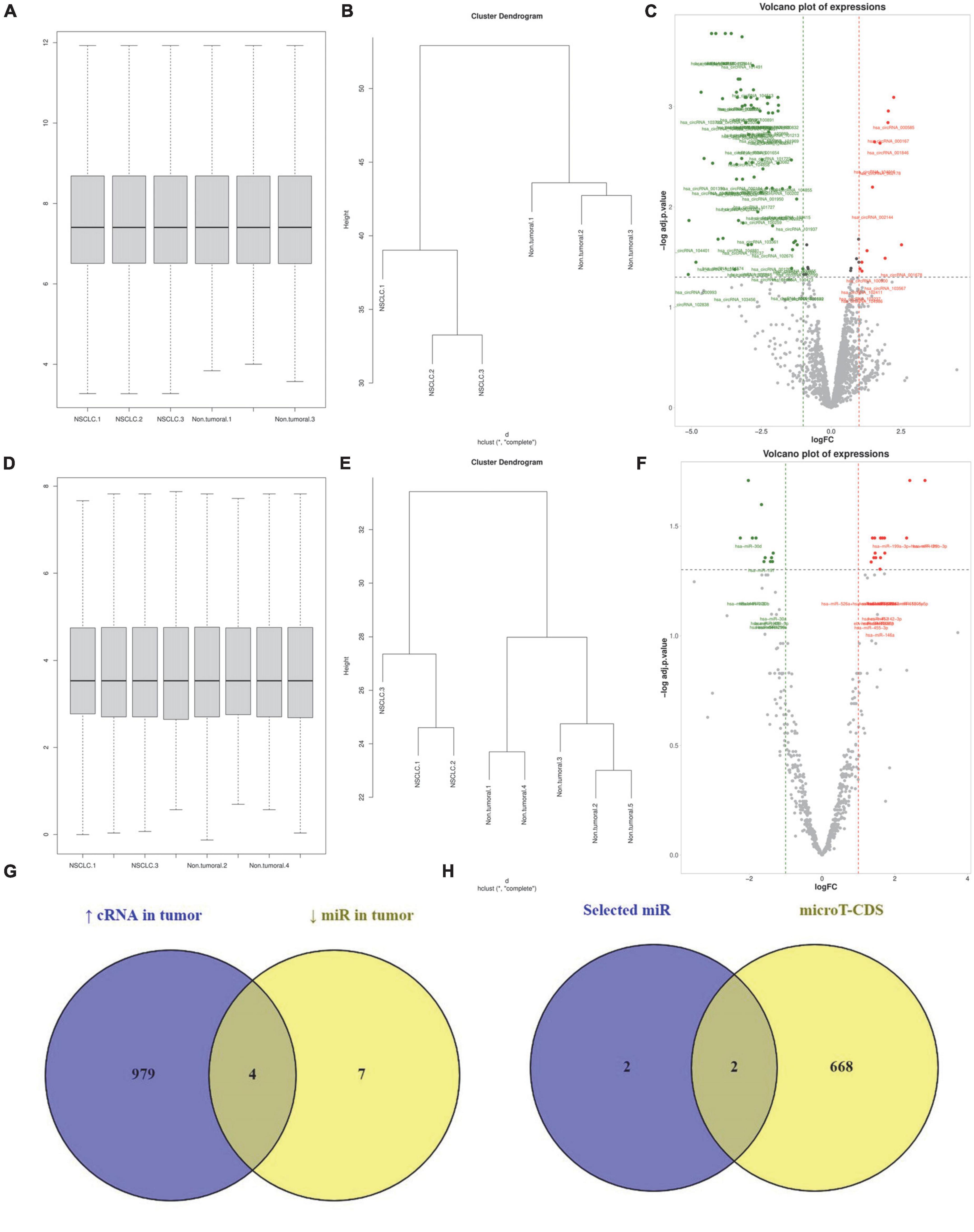
Figure 4. has_circ_0002476/hsa-miR-216a-5p/PD-L1 and hsa_circ_0063526/hsa-miR-1299/PD-L1 might increase programmed death-ligand 1 (PD-L1) expression in non-small cell lung cancer (NSCLC). (A–C) Normalizing, clustering, and identifying differentially expressed circRNAs from the GSE158695 dataset. (D–F) Normalizing, clustering, and identifying differentially expressed circRNAs from the GSE63805 dataset. (G) Identifying the commonality between the miR targets of upregulated circRNAs in NSCLC tissues between the downregulated miRs in NSCLC tissues. (H) Identifying which of those miRs can target PD-L1 based on the microT-CDS database.
In the same manner, we analyzed the GSE63805 dataset to identify significantly downregulated miRs in NSCLC tissues. After data normalization and clustering, our results have demonstrated 11 significantly downregulated miRs in NSCLC tissues (Figures 4D–F, respectively). We have demonstrated that 4 miR-targets of those 12 upregulated circRNAs are among the 11 significantly downregulated miRs; these miRs are hsa-miR-216a-5p, hsa-miR-144-3p, hsa-miR-1299, and hsa-miR-1909-3p (Figure 4G). Based on microT-CDS, hsa-miR-216a-5p and hsa-miR-1299 might directly target PD-L1 expression (Figure 4H). Therefore, our in silico results suggested that has_circ_0002476/hsa-miR-216a-5p/PD-L1 and hsa_circ_0063526/hsa-miR-1299/PD-L1 might be two axes in NSCLC that might increase PD-L1 expression in NSCLC tissues.
The expression pattern of PD-1 in non-small cell lung cancer microenvironment
We analyzed the GSE144945 dataset to identify the expression pattern of PD-1 in the NSCLC microenvironment and highlight which cell populations might be liberated from the tumor-intrinsic PD-L1 inhibition in the NSCLC microenvironment. After excluding low-quality cells, we included 32,116 cells from 10 NSCLC tissues. Based on the abovementioned markers, we identified 9 cell populations residing in the included NSCLC microenvironment (Figure 5A). Our results have indicated that PD-1 might be considerably expressed in the T-cells, CD8+ T-cells, CD4 regulatory T-cells (Tregs), DCs, and ILC2 in the included tumor microenvironment of NSCLCs (Figure 5B).
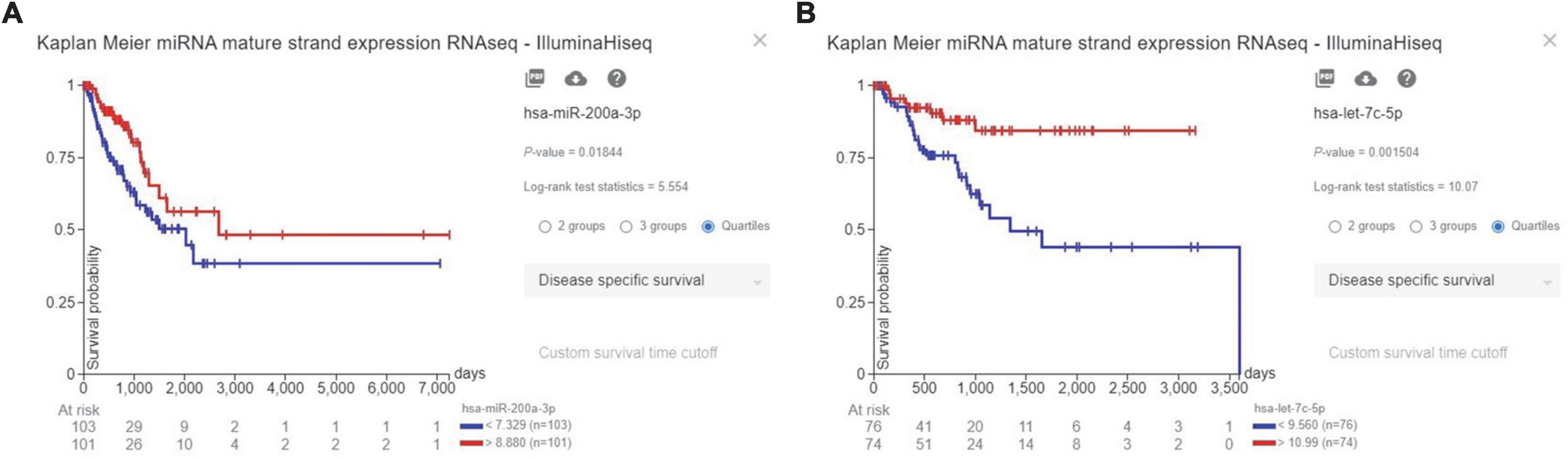
Figure 5. Identifying programmed death-ligand 1 (PD-L1) expression pattern in the included non-small cell lung cancers (NSCLCs) microenvironment. (A) The clustering and cell annotation displayed nine cell clusters in the included tumor microenvironment of NSCLCs. (B) PD-1 is considerably expressed in the T-cells, CD8+ T-cells, CD4 Treg, DCs, and ILC2 in the included NSCLCs microenvironment.
Discussion
In 2022, lung cancer will be the second most commonly diagnosed and the most common cause of cancer-related death in men and women in the United States (1). NSCLC is responsible for the majority of lung cancer cases, and most patients are diagnosed when the disease is advanced (27). Although there have been notable advances in treating NSCLCs, disease progression and resistance development have been considerable burdens for affected patients (28). In this regard, a better understanding of NSCLC biology and its microenvironment can pave the way for developing novel treatments for affected patients.
As small non-coding RNAs, miRs have pivotal roles in the post-transcriptional regulation of genes. It is well-established that miRs can bind to the 3’ untranslated region (UTR) of target mRNAs and facilitate translation inhibition; therefore, they are involved in regulating various biological processes (29). Growing evidence has highlighted that specific miRs can pave the way for tumor development, and other miRs can inhibit tumor development and metastasis (12, 15). Tumor-suppressive miRs target the expression of oncogenes, while oncomiRs target the expression of tumor-suppressive genes. In this regard, the ectopic expression of tumor-suppressive miRs and suppressing the expression of oncomiRs can inhibit tumor development and migration (30). Since miRs can regulate a vast number of gene expressions, both directly and indirectly, they carry valuable therapeutic potential for cancer treatment (31).
Recently, circRNAs have emerged as a new class of non-coding of RNAs that considerably regulate gene expression (32). It has been reported that circRNAs are involved with miR sponges, protein/peptide translators, RNA-binding protein sponges, and regulating gene splicing and transcription (31). One of the most studied circRNA-mediated interactions is the cross-talk between circRNA and miRs; circRNAs have multiple sites for binding miRs that can “sponge” them and therefore regulate the expression of various genes (33). Thus, identifying the circRNA/miR/mRNA axes that have significant roles in cancer development/inhibition can be a milestone in cancer treatment.
Immunotherapy is one of the novel and promising treatments for a subset of cancer patients. Stimulating the host’s anti-tumoral immune responses is the cornerstone of immunotherapeutic approaches (3). Effective rejection of tumors can also eradicate cancerous cells and prevent tumor relapse; however, the immunosuppressive tumor microenvironment has been implicated in tumor development and shielding tumoral cells from anti-tumoral immune responses (34). Inhibitory immune checkpoint axes considerably attenuate anti-tumoral immune responses and contribute to tumor growth (3). The PD-1/PD-L1 axis is one of these inhibitory axes that can maintain this pathological immune tolerance against neoplastic cells (3). In this regard, multiple clinical studies have demonstrated the beneficial effect of administrating monoclonal antibodies that inhibit the PD-1/PD-L1 axes in NSCLC patients. Following the promising results, the FDA has approved pembrolizumab (anti-PD-1), nivolumab (anti-PD-1), atezolizumab (anti-PD-L1), and durvalumab (anti-PD-L1) for patients with NSCLC. Thus, targeting this inhibitory axis is pivotal in treating NSCLC (7).
Besides inhibiting anti-tumoral immune responses, recent findings have indicated that tumor-intrinsic inhibitory immune checkpoint molecules have oncogenic effects in various cancers, like triple-negative breast cancer, NSCLC, and melanoma (35–38). In TNBC cells, PD-L1 knockdown has substantially decreased cell viability, stemness, cell cycle progression, and migration. Also, tumor-intrinsic PD-L1 silencing has stimulated apoptosis of TNBC cells and decreased the CD25+ forkhead box P3 (Foxp3)+ Tregs (36). Qiu et al. have reported that PD-L1 overexpression has substantially increased cell proliferation and migration in glioblastoma cells via the mitogen-activated extracellular signal-regulated kinase (MEK)/extracellular signal-related kinase (Erk) pathway both in vitro and in vivo (39). Wei et al. have reported that increased expression of PD-L1 increases the self-renewal of colorectal cancer cells and PD-L1 knockdown decreases colorectal cancer stemness (40). Yu et al. have demonstrated that PD-L1 knockdown has substantially decreased cell viability and reduced tumor clonogenicity and migration in NSCLC cells (35). Hong et al. have shown that tumor-intrinsic PD-L1 silencing or PD-L1/PD-1 blockade can increase the proliferation and viability of CD8+ T-cells, upregulate the expression of IFN-γ and IL-4, and downregulate IL-10 expression in a co-culture system with NSCLC cells (41). Therefore, inhibiting tumor-intrinsic PD-L1 can stimulate anti-tumoral immune responses and suppress oncogenic signaling pathways, leading to suppressed tumor growth.
Considering the inhibitory effect of tumor-intrinsic PD-L1 on anti-tumoral immune responses, the established oncogenic effect of tumor-intrinsic PD-L1 on tumor development, and the multi-target and considerable effect of miRs on mRNAs expression, identifying PD-L1-inhibiting miRs and studying the overall effect of their ectopic expression on tumoral cells can pave the way for the development of novel therapeutic approaches. It has been reported that the ectopic expression of hsa-miR-497-5p, hsa-miR-424-5p, hsa-miR-34a-5p, hsa-miR-200c-3p, hsa-miR-138-5p, hsa-miR-383-5p, hsa-miR-195-5p, hsa-miR-570-3p, and hsa-miR-3609 can considerably inhibit tumor proliferation, migration, stemness, tumor-intrinsic PD-L1 expression in TNBC cells. Besides contributing to the transformation of the tumor microenvironment from immunosuppressive to pro-inflammatory, their ectopic expression has been associated with enhanced chemosensitivity in TNBC cells (15). Consistent with this, it has been shown that ectopic expression of hsa-miR-140-3p, hsa-miR-382-3p, hsa-miR-148a-3p, hsa-miR-93-5p, hsa-miR-200a-3p, hsa-miR-200c-3p, hsa-miR-138–5p, and hsa-miR-15b-5p can suppress PD-L1 expression and inhibit tumor proliferation and migration in colorectal cancer cells; these PD-L1-inhibiting miRs substantially transform the tumor microenvironment from immunosuppressive to pro-inflammatory and improve the chemosensitivity of colorectal cancer cells regardless of the microsatellite state (12). In the current study, we aimed to identify the effect of PD-L1-inhibiting miRs on NSCLC development and investigate the circRNA/miR/PD-L1 axes based on the literature and in silico data.
Zhu et al. have identified the FGD5-AS1/hsa-miR-142-5p/PD-L1 axis in NSCLC cells. They have reported that FGD5-AS1 inhibits hsa-miR-142-5p expression, and hsa-miR-142-5p ectopic expression downregulates PD-L1 expression and decreases the migration and invasion of cisplatin-resistant NSCLC cells (6). Yang et al. have identified the circCHST15/hsa-miR-194-5p/PD-L1 axis in NSCLC cells; they have shown that circCHST15 inhibits the expression of hsa-miR-194-5p and hsa-miR-194-5p ectopic expression decreases the proliferation and clonogenicity of NSCLC cells (18). Wang et al. have identified the circRNA-002178/hsa-miR-34a-5p/PD-L1 axis in NSCLC cells; circRNA-002178 inhibits the expression of hsa-miR-34a-5p, leading to the liberation of PD-L1 expression. Of interest, it has been reported that the exosomal plasmatic level of circRNA-002178 has a significant diagnostic value in LUAD patients (AUC = 0.9956, and P-value < 0.001) (42). Consistent with this, increased expression of hsa-miR-34a-5p mimic and hsa-miR-34a-5p inhibitor have decreased and increased PD-L1 expression in NSCLC cells (43, 44). It has been reported that hsa-miR-34a-5p overexpression inhibits the proliferation, migration, and invasion of LUAD cells. Also, hsa-miR-34a-5p overexpression stimulates apoptosis of LUAD cells (45). We have shown that hsa-miR-34a-5p is significantly enriched for regulating the PI3K/Akt pathway and cell cycle. Li et al. have highlighted the circRNA_0000284/hsa-miR-377-3p/PD-L1 axis in NSCLC cells, and the protumoral effect of circRNA_0000284 on cell proliferation, clonogenicity, migration, and invasion of NSCLC cells is mediated via the hsa-miR-377-3p/PD-L1 axis. They have shown that the expression level of circRNA_0000284 is substantially upregulated in NSCLC tissues compared to non-tumoral tissues, in higher stage NSCLC tissues compared to lower stage NSCLC tissues, in tissues with lymph node metastasis compared with the tissues without lymph node metastasis. Of interest, circRNA_0000284 expression level has been associated with the inferior overall survival of affected patients (46). Besides, it has been shown that hsa-miR-377-3p decreases the cell viability, migration, and invasion of NSCLC cells (47). We have shown that hsa-miR-377-3p is significantly enriched for regulating the cell cycle. Hong et al. have shed light on the circ-CPA4/hsa-let-7c-5p/PD-L1 axis in NSCLC cells; they have shown that circ-CPA4 targets hsa-let-7c-5p and liberate PD-L1 expression in NSCLC (41). Also, it has been reported that hsa-let-7c-5p suppresses the cell viability of NSCLC cells (48). Our results have indicated that increased expression of hsa-let-7c-5p is associated with improved disease-specific survival of LUSC patients. Also, we have demonstrated that hsa-let-7c-5p is significantly enriched for regulating the PI3K/Akt pathway. Wei et al. have highlighted the MALAT1/hsa-miR-200a-3p/PD-L1 axis in NSCLC cells; they have shown that MALAT1 sponges hsa-miR-200a-3p to liberate PD-L1 protein expression in NSCLC cells and MALAT1 overexpression increases clonogenicity, cell migration, proliferation, and invasion and decrease apoptosis in NSCLC cells. However, hsa-miR-200a-3p ectopic expression partially reverses the pro-tumoral effect of MALAT1 in NSCLC cells (49). Chen et al. have reported that hsa-miR-200a-3p suppresses tumor-intrinsic PD-L1 expression in NSCLC cells (50). Tan et al. have reported that ectopic expression of hsa-miR-200a-3p decreases the proliferation, invasion, and migration of NSCLC cells (51). Our results have shown that increased expression of hsa-miR-200a-3p is associated with enhanced disease-specific survival of LUAD patients. Besides, we have shown that hsa-miR-200a-3p is significantly enriched for regulating the cell cycle. Yang et al. have highlighted the CircCHST15/hsa-miR-155-5p/PD-L1 axis in NSCLC cells; circCHST15 suppresses hsa-miR-155-5p expression and liberates PD-L1 expression in NSCLC cells. Yang et al. have reported that hsa-miR-155-5p ectopic expression increases the proliferation and clonogenicity in H1395 and A549 cells (18). Consistent with this, Chen et al. have reported that hsa-miR-155-5p transfection decreases PD-L1 expression; however, Chen et al. have reported that hsa-miR-155-5p transfection suppresses the proliferation of A549 cells. They have shown that hsa-miR-155-5p increases the infiltration of IFN-γþ lymphocytes, CD4þ T lymphocytes, and CD8þ T lymphocytes, upregulates the expression of TNF-α and IFN-g, and downregulates IL-10 expression (52). In line with these, Huang et al. have demonstrated that hsa-miR-155-5p targets mRNA and protein PD-L1 expression in A549 and H1650 cells. Also, they have shown a negative correlation between hsa-miR-155-5p expression and PD-L1 expression in LUAD tissues (53).
It has been reported that hsa-miR-326 ectopic expression downregulates PD-L1 and B7-H3 expression in NSCLC cells. Besides, there have been upregulation in TNF-α, IFN-γ, and IL-2 expression and downregulation in IL-1β, IL-10, and TGF-β expression in the supernatant of hsa-miR-326 overexpressed cells. The overexpression of hsa-miR-326 has substantially decreased tumor growth, metastasis, and PD-L1 expression in animal models and increased the infiltration of CD8+ cells and TNF-α+ IFN-γ+ CD8+ T-cells population in tumor tissues in animal models of NSCLC (17). Chen et al. have demonstrated that miR-526b-3p can enhance cisplatin chemosensitivity, decrease the migration and proliferation of NSCLCs, and increase the population of CD8+ T-cells (16). Song et al. have reported that hsa-miR-138-5p inhibits NSCLC proliferation, decreases PD-L1 protein expression, and increases tumor-infiltrating mature DCs, CD4+ T cells, and CD8+ T-cells. Also, hsa-miR-138-5p downregulates PD-1 and PD-L1 expression in tumor-infiltrating DCs and increases the ability of DCs to induce cytotoxicity of CD8+ T-cells (20). It has been shown that hsa-miR-200c-3p downregulates PD-L1 mRNA and protein expression in NSCLC cells, and hsa-miR-200c-3p potentiates doxorubicin-induced cytotoxicity in NSCLC cells. Also, hsa-miR-200c-3p considerably enhances IFN-γ expression and the cytotoxic T-cell-mediated anti-tumoral effect against NSCLC cells (54). We have shown that hsa-miR-200c-3p is significantly enriched for regulating the PI3K/Akt pathway. Xie et al. have shown that miR-140 downregulates the protein and mRNA expression of PD-L1, arrests the cell cycle, and decreases the proliferation of NSCLC cells (19). Fujita et al. have demonstrated that the increased expression of hsa-miR-197-3p suppresses PD-L1 expression and enhances the chemosensitivity of NSCLC cells; hsa-miR-197-3p inhibition increases tumor growth and its migration and decreases the survival of affected animals (55). Cortez et al. have shown that hsa-miR-34c-5p and hsa-miR-34b suppress PD-L1 expression in NSCLC cells (43). It has been reported that hsa-miR-34c-5p increased expression decreases the migration and invasion of NSCLC cells (56). Our results have indicated that hsa-miR-34c-5p is significantly enriched for regulating the PI3K/Akt pathway and the cell cycle. It has been reported that hsa-miR-200b-3p and hsa-miR-200b-5p downregulate tumor-intrinsic PD-L1 expression in NSCLC cells (50, 57). The ectopic expression of hsa-miR-200b-3p has substantially decreased the migration and invasion of NSCLC cells (58). However, further studies are needed to clarify the overall effect of the hsa-miR-200b-5p ectopic expression on NSCLC cells. Despite the inhibitory role of hsa-miR-142-5p on tumor-intrinsic NSCLC cells, the overexpression of hsa-miR-142-5p increases tumor growth and decreases the apoptosis rate in NSCLC cells (59). In contrast, Jiang et al. have reported that hsa-miR-142-5p overexpression decreases cell proliferation, migration, and invasion and increases the apoptosis of NSCLC cells (60). Therefore, further studies are needed to evaluate the overall effect of the hsa-miR-142-5p ectopic expression on NSCLC development. Table 3 summarizes the tumor-suppressive PD-L1-inhibiting miRs in NSCLC.
System biology approaches have provided valuable insights into the expression profile of various human conditions (61, 62). Following systematically reviewing the literature, we aimed to leverage the data obtained from system biology to predict the potential circRNA/PD-L1-targeting miRs in NSCLC tissues. Our in silico results have highlighted the hsa_circ_0002476/hsa-miR-216a-5p/PD-L1 and hsa_circ_0063526/hsa-miR-1299/PD-L1 in NSCLC. Based on our in silico analyses, hsa-miR-216a-5p and hsa-miR-1299 can be significantly downregulated in NSCLCs and can suppress PD-L1 expression. Also, hsa_circ_0002476 and hsa_circ_0063526 can be significantly upregulated in NSCLC tissues, and hsa_circ_0002476 and hsa_circ_0063526 can inhibit hsa-miR-216a-5p and hsa-miR-1299 expression. Therefore, hsa_circ_0002476 and hsa_circ_0063526 might sponge hsa-miR-216a-5p and hsa-miR-1299, respectively, liberating PD-L1 expression in NSCLC. Consistent with our in silico investigation, Pang et al. have shown that hsa-miR-216a-5p is substantially decreased in NSCLC cells and tissues compared to normal cells and tissues, respectively. Also, the ectopic expression of hsa-miR-216a-5p has substantially decreased the proliferation, invasion, and migration and stimulated apoptosis in NSCLC cells (63). Recently, Chai et al. have demonstrated that miR-1299 expression has substantially decreased in NSCLC tissues compared to normal tissues (64). In line with these, Cao et al. have also reported that miR-1299 expression is substantially decreased in NSCLC cells and tissues compared to non-tumoral cells and tissues, respectively. Also, miR-1299 ectopic expression suppresses proliferation, migration, invasion, and epithelial-mesenchymal transition processes and stimulates apoptosis in NSCLC cells by regulating the PI3K/Akt pathway (65).
Single-cell RNA sequencing can be considered a milestone in understanding various cells residing in the tumor microenvironment and their expression profile (66). This novel approach enables us to dissect each cell of tumor tissue, thoroughly examine its expression profile, categorize them into groups, and investigate the expression of a specific gene in these cell groups (67). Following identifying the significance of ectopic expression of PD-L1-inhibiting miRs in NSCLC development and discussing the underlying mechanisms through systematic literature review and tumor bulk analysis, we aimed to answer the question of which cells residing in the tumor microenvironment of NSCLC might be liberated following ectopic expression of PD-L1-inhibiting miRs in NSCLC cells. For this aim, we analyzed 32,116 cells from 10 NSCLC tissues to identify which cells express PD-1 in the tumor microenvironment of those NSCLC tissues. Based on our analyses, tumor-infiltrated T-cells, CD8+ T-cells, CD4 Treg, DCs, and ILC2 might considerably express PD-1; therefore, ectopic expression of hsa-miR-194-5p, hsa-miR-326, hsa-miR-526b-3p, hsa-miR-34a-5p, hsa-miR-34c-5p, hsa-miR-138-5p, hsa-miR-377-3p, hsa-let-7c-5p, hsa-miR-200a-3p, hsa-miR-200b-3p, hsa-miR-200c-3p, and hsa-miR-197-3p might stimulate CD8+ T-cells and DCs-mediated anti-tumoral immune responses via inhibiting tumor-intrinsic PD-L1 expression in NSCLC.
The current study has several limitations that should be noted. First, we only included papers published in English before the mentioned date. The second limitation stems from the intrinsic limitation of single-cell RNA sequencing approaches, i.e., the count of sequenced cells in our single-cell RNA sequencing was limited compared to the count of sequenced cells in tumor bulk analysis. Nevertheless, this study has strengths as well. First, we systematically searched ten databases, i.e., Embase, Scopus, Web of Science, PubMed, Ovid, EBSCO, ProQuest, Cochrane library, African Index Medicus, and Pascal-Francis, to retrieve available papers published before the mentioned data. Second, we included in silico tumor bulk and single-cell RNA sequencing to enrich this study. Third, we predicted circRNA/miR/PD-L1 axes based on bioinformatic analyses and discussed their significance based on a thorough literature review of NSCLC development.
Conclusion
Overall, the ectopic expression of hsa-miR-194-5p, hsa-miR-326, hsa-miR-526b-3p, hsa-miR-34a-5p, hsa-miR-34c-5p, hsa-miR-138-5p, hsa-miR-377-3p, hsa-let-7c-5p, hsa-miR-200a-3p, hsa-miR-200b-3p, hsa-miR-200c-3p, and hsa-miR-197-3p, as PD-L1-inhibiting miR, can inhibit NSCLC development and stimulate anti-tumoral immune responses. Based on the single-cell RNA sequencing analyses, PD-1 might be considerably expressed on CD8+ T-cells and DCs; therefore, the ectopic expression of the abovementioned PD-L1-inhibiting miRs might stimulate the CD8+ T-cells and DCs-mediated anti-tumoral immune responses. Considering the multi-target nature of circRNA and miRs, identifying tumor-suppressive circRNA and miRs can provide a promising opportunity to overcome tumor growth, metastasis, and therapy resistance. Nevertheless, further studies are needed to identify, validate, and translate them into clinical practice.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: geo databases, GSE158695, GSE63805, and GSE144945; and the TGCA database.
Author contributions
MS: developing the research question, performing the in silico and single-cell RNA sequencing analyses, searching the databases, selecting the studies from the initial systematic search, and writing the manuscript. FG and HH: selecting the studies from the initial systematic search. NA and NH: extracting the results from selected studies. AD and NSh: searching the literature. OB: reviewing and editing the manuscript. NSi and BB: reviewing and editing the manuscript and supervising the study. All authors contributed to the article and approved the submitted version.
Acknowledgments
The research protocol was approved and supported by Student Research Committee, Tabriz University of Medical Sciences (IR.TBZMED.VCR.REC.1401.211 and grant number: 70568).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/geo/
- ^ http://www.biomedical-web.com/circmine/home
- ^ https://xenabrowser.net/
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. (2022) 72:7–33.
2. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. (2018) 553:446–54.
3. Fasano R, Shadbad MA, Brunetti O, Argentiero A, Calabrese A, Nardulli P, et al. Immunotherapy for hepatocellular carcinoma: New prospects for the cancer therapy. Life. (2021) 11:1355.
4. Ghorbaninezhad F, Asadzadeh Z, Masoumi J, Mokhtarzadeh A, Kazemi T, Aghebati-Maleki L, et al. Dendritic cell-based cancer immunotherapy in the era of immune checkpoint inhibitors: From bench to bedside. Life Sci. (2022) 297:120466. doi: 10.1016/j.lfs.2022.120466
5. Shadbad MA, Asadzadeh Z, Hosseinkhani N, Derakhshani A, Alizadeh N, Brunetti O, et al. A systematic review of the tumor-infiltrating CD8+ T-Cells/PD-L1 axis in high-grade glial tumors: Toward personalized immuno-oncology. Front Immunol. (2021) 12:734956. doi: 10.3389/fimmu.2021.734956
6. Zhu F, Niu R, Shao X, Shao X. FGD5-AS1 promotes cisplatin resistance of human lung adenocarcinoma cell via the miR-142-5p/PD-L1 axis. Int J Mol Med. (2021) 47:523–32. doi: 10.3892/ijmm.2020.4816
7. Vaddepally RK, Kharel P, Pandey R, Garje R, Chandra AB. Review of indications of FDA-approved immune checkpoint inhibitors per NCCN guidelines with the level of evidence. Cancers. (2020) 12:738. doi: 10.3390/cancers12030738
8. Qiao X, Zhao F. Long non-coding RNA Opa interacting protein 5-antisense RNA 1 binds to micorRNA-34a to upregulate oncogenic PD-L1 in non-small cell lung cancer. Bioengineered. (2022) 13:9264–73. doi: 10.1080/21655979.2022.2036904
9. Stutvoet TS, Kol A, De Vries EG, De Bruyn M, Fehrmann RS, Terwisscha Van Scheltinga AG, et al. MAPK pathway activity plays a key role in PD-L1 expression of lung adenocarcinoma cells. J Pathol. (2019) 249:52–64. doi: 10.1002/path.5280
10. Chen N, Fang W, Zhan J, Hong S, Tang Y, Kang S, et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-driven NSCLC: Implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J Thorac Oncol. (2015) 10:910–23. doi: 10.1097/JTO.0000000000000500
11. Chen Y, Huang Y, Lu X, Wang G, Chi P. Antitumor effects of the silencing of programmed cell death ligand 1 in colorectal cancer via immunoregulation. Oncol Rep. (2018) 40:3370–80. doi: 10.3892/or.2018.6738
12. Shadbad MA, Asadzadeh Z, Derakhshani A, Hosseinkhani N, Mokhtarzadeh A, Baghbanzadeh A, et al. A scoping review on the potentiality of PD-L1-inhibiting microRNAs in treating colorectal cancer: Toward single-cell sequencing-guided biocompatible-based delivery. Biomed Pharmacother. (2021) 143:112213. doi: 10.1016/j.biopha.2021.112213
13. Dastmalchi N, Safaralizadeh R, Banan Khojasteh SM, Sam MR, Latifi-Navid S, Hussen BM, et al. An updated review of the cross-talk between microRNAs and epigenetic factors in cancers. Curr Med Chem. (2021) 28:8722–32.
14. Shahverdi M, Hajiasgharzadeh K, Sorkhabi AD, Jafarlou M, Shojaee M, Tabrizi NJ, et al. The regulatory role of autophagy-related miRNAs in lung cancer drug resistance. Biomed Pharmacother. (2022) 148:112735.
15. Shadbad MA, Safaei S, Brunetti O, Derakhshani A, Lotfinejad P, Mokhtarzadeh A, et al. A systematic review on the therapeutic potentiality of PD-L1-inhibiting microRNAs for triple-negative breast cancer: Toward single-cell sequencing-guided biomimetic delivery. Genes. (2021) 12:1206. doi: 10.3390/genes12081206
16. Chen K-B, Yang W, Xuan Y, Lin A-J. miR-526b-3p inhibits lung cancer cisplatin-resistance and metastasis by inhibiting STAT3-promoted PD-L1. Cell Death Dis. (2021) 12:1–10. doi: 10.1038/s41419-021-04033-8
17. Shao L, He Q, Wang J, He F, Lin S, Wu L, et al. MicroRNA-326 attenuates immune escape and prevents metastasis in lung adenocarcinoma by targeting PD-L1 and B7-H3. Cell Death Discov. (2021) 7:1–10. doi: 10.1038/s41420-021-00527-8
18. Yang J, Jia Y, Wang B, Yang S, Du K, Luo Y, et al. Circular RNA CHST15 sponges miR-155-5p and miR-194-5p to promote the immune escape of lung cancer cells mediated by PD-L1. Front Oncol. (2021) 11:364. doi: 10.3389/fonc.2021.595609
19. Xie W-B, Liang L-H, Wu K-G, Wang L-X, He X, Song C, et al. MiR-140 expression regulates cell proliferation and targets PD-L1 in NSCLC. Cell Physiol Biochem. (2018) 46:654–63. doi: 10.1159/000488634
20. Song N, Li P, Song P, Li Y, Zhou S, Su Q, et al. MicroRNA-138-5p suppresses non-small cell lung cancer cells by targeting PD-L1/PD-1 to regulate tumor microenvironment. Front Cell Dev Biol. (2020) 8:540. doi: 10.3389/fcell.2020.00540
21. Arksey H, O’malley L. Scoping studies: Towards a methodological framework. Int J Soc Res Methodol. (2005) 8:19–32.
22. Levac D, Colquhoun H, O’brien KK. Scoping studies: Advancing the methodology. Implement Sci. (2010) 5:1–9.
23. Tricco AC, Lillie E, Zarin W, O’brien KK, Colquhoun H, Levac D, et al. PRISMA extension for scoping reviews (PRISMA-ScR): Checklist and explanation. Ann Intern Med. (2018) 169:467–73.
24. Kehl T, Kern F, Backes C, Fehlmann T, Stöckel D, Meese E, et al. miRPathDB 2.0: A novel release of the miRNA pathway dictionary database. Nucleic Acids Res. (2020) 48:D142–7. doi: 10.1093/nar/gkz1022
25. Cho J-W, Park S, Kim G, Han H, Shim HS, Shin S, et al. Dysregulation of TFH-B-TRM lymphocyte cooperation is associated with unfavorable anti-PD-1 responses in EGFR-mutant lung cancer. Nat Commun. (2021) 12:1–16. doi: 10.1038/s41467-021-26362-0
26. Franzén O, Gan L-M, Björkegren JL. PanglaoDB: A web server for exploration of mouse and human single-cell RNA sequencing data. Database. (2019) 2019:baz046. doi: 10.1093/database/baz046
27. Majeed U, Manochakian R, Zhao Y, Lou Y. Targeted therapy in advanced non-small cell lung cancer: current advances and future trends. J Hematol Oncol. (2021) 14:1–20.
28. Lin JJ, Shaw AT. Resisting resistance: Targeted therapies in lung cancer. Trends Cancer. (2016) 2:350–64.
29. O’brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. (2018) 9:402. doi: 10.3389/fendo.2018.00402
30. Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics. (2019) 11:1–24.
31. Szczepanek J, Skorupa M, Tretyn A. MicroRNA as a potential therapeutic molecule in cancer. Cells. (2022) 11:1008.
32. Khan S, Jha A, Panda AC, Dixit A. Cancer-associated circRNA–miRNA–mRNA regulatory networks: A meta-analysis. Front Mol Biosci. (2021) 8:671309. doi: 10.3389/fmolb.2021.671309
33. Yu C-Y, Kuo H-C. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. (2019) 26:1–12.
34. Hosseinkhani N, Derakhshani A, Kooshkaki O, Abdoli Shadbad M, Hajiasgharzadeh K, Baghbanzadeh A, et al. Immune checkpoints and CAR-T cells: The pioneers in future cancer therapies? Int J Mol Sci. (2020) 21:8305. doi: 10.3390/ijms21218305
35. Yu W, Hua Y, Qiu H, Hao J, Zou K, Li Z, et al. PD-L1 promotes tumor growth and progression by activating WIP and β-catenin signaling pathways and predicts poor prognosis in lung cancer. Cell Death Dis. (2020) 11:1–16. doi: 10.1038/s41419-020-2701-z
36. Lotfinejad P, Kazemi T, Safaei S, Amini M, Baghbani E, Shotorbani SS, et al. PD-L1 silencing inhibits triple-negative breast cancer development and upregulates T-cell-induced pro-inflammatory cytokines. Biomed Pharmacother. (2021) 138:111436. doi: 10.1016/j.biopha.2021.111436
37. Shadbad MA, Hajiasgharzadeh K, Derakhshani A, Silvestris N, Baghbanzadeh A, Racanelli V, et al. From melanoma development to RNA-modified dendritic cell vaccines: Highlighting the lessons from the past. Front Immunol. (2021) 12:623639. doi: 10.3389/fimmu.2021.623639
38. Baghbani E, Noorolyai S, Rahmani S, Shanehbandi D, Shadbad MA, Aghebati-Maleki L, et al. Silencing tumor-intrinsic CD73 enhances the chemosensitivity of NSCLC and potentiates the anti-tumoral effects of cisplatin: An in vitro study. Biomed Pharmacother. (2022) 145:112370. doi: 10.1016/j.biopha.2021.112370
39. Qiu XY, Hu DX, Chen WQ, Chen RQ, Qian SR, Li CY, et al. PD-L1 confers glioblastoma multiforme malignancy via Ras binding and Ras/Erk/EMT activation. Biochim Biophys Acta Mol Basis Dis. (2018) 1864:1754–69. doi: 10.1016/j.bbadis.2018.03.002
40. Wei F, Zhang T, Deng S-C, Wei J-C, Yang P, Wang Q, et al. PD-L1 promotes colorectal cancer stem cell expansion by activating HMGA1-dependent signaling pathways. Cancer Lett. (2019) 450:1–13. doi: 10.1016/j.canlet.2019.02.022
41. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J Exp Clin Cancer Res. (2020) 39:149. doi: 10.1186/s13046-020-01648-1
42. Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. (2020) 11:1–11. doi: 10.1038/s41419-020-2230-9
43. Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, et al. PDL1 regulation by p53 via miR-34. J Natl Cancer Inst. (2016) 108:djv303. doi: 10.1093/jnci/djv303
44. Kang DY, Sp N, Jo ES, Rugamba A, Hong DY, Lee HG, et al. The inhibitory mechanisms of tumor PD-L1 expression by natural bioactive gallic acid in non-small-cell lung cancer (NSCLC) cells. Cancers. (2020) 12:727. doi: 10.3390/cancers12030727
45. Li W, Pan T, Jiang W, Zhao H. HCG18/miR-34a-5p/HMMR axis accelerates the progression of lung adenocarcinoma. Biomed Pharmacother. (2020) 129:110217. doi: 10.1016/j.biopha.2020.110217
46. Li L, Zhang Q, Lian K. Circular RNA circ_0000284 plays an oncogenic role in the progression of non-small cell lung cancer through the miR-377-3p-mediated PD-L1 promotion. Cancer Cell Int. (2020) 20:1–11. doi: 10.1186/s12935-020-01310-y
47. Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y, et al. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. (2016) 7:51784–814.
48. Lv X, Fang Z, Qi W, Xu Y, Chen W. Long non-coding RNA HOXA11-AS facilitates proliferation of lung adenocarcinoma cells via targeting the let-7c-5p/IGF2BP1 axis. Front Genet. (2022) 13:831397. doi: 10.3389/fgene.2022.831397
49. Wei S, Wang K, Huang X, Zhao Z, Zhao Z. LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis. Int J Immunopathol Pharmacol. (2019) 33:2058738419859699. doi: 10.1177/2058738419859699
50. Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn Y-H, Byers LA, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat commun. (2014) 5:1–12. doi: 10.1038/ncomms6241
51. Tan T, Xu X, Lu X, Wang X. MiRNA-200a-3p suppresses the proliferation, migration and invasion of non-small cell lung cancer through targeting IRS2. Eur Rev Med Pharmacol Sci. (2020) 24:712–20. doi: 10.26355/eurrev_202001_20050
52. Chen H, Wang Z, Wang W, Liu Y, He C, Chen G. MIR-155-5P promote tumor immunity by regulating PD-L1 expression in lung cancer cells. Acta Med Mediterranea. (2020) 36:2385–90.
53. Huang J, Weng Q, Shi Y, Mao W, Zhao Z, Wu R, et al. MicroRNA-155-5p suppresses PD-L1 expression in lung adenocarcinoma. FEBS Open Bio. (2020) 10:1065–71. doi: 10.1002/2211-5463.12853
54. Dai Phung C, Nguyen HT, Choi JY, Pham TT, Acharya S, Timilshina M, et al. Reprogramming the T cell response to cancer by simultaneous, nanoparticle-mediated PD-L1 inhibition and immunogenic cell death. J Control Release. (2019) 315:126–38. doi: 10.1016/j.jconrel.2019.10.047
55. Fujita Y, Yagishita S, Hagiwara K, Yoshioka Y, Kosaka N, Takeshita F, et al. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol Ther. (2015) 23:717–27. doi: 10.1038/mt.2015.10
56. Shen Z, Sun S. CircPTCH1 promotes migration in lung cancer by regulating MYCN expression through miR-34c-5p. Onco Targets Ther. (2021) 14:4779–89. doi: 10.2147/OTT.S324015
57. Katakura S, Kobayashi N, Hashimoto H, Kamimaki C, Tanaka K, Kubo S, et al. MicroRNA-200b is a potential biomarker of the expression of PD-L1 in patients with lung cancer. Thoracic Cancer. (2020) 11:2975–82. doi: 10.1111/1759-7714.13653
58. Tang Q, Li M, Chen L, Bi F, Xia H. miR-200b/c targets the expression of RhoE and inhibits the proliferation and invasion of non-small cell lung cancer cells. Int J Oncol. (2018) 53:1732–42.
59. Wan J, Ling X, Peng B, Ding G. miR-142-5p regulates CD4+ T cells in human non-small cell lung cancer through PD-L1 expression via the PTEN pathway. Oncol Rep. (2018) 40:272–82. doi: 10.3892/or.2018.6439
60. Jiang Q, Xing W, Cheng J, Yu Y. Knockdown of lncRNA XIST suppresses cell tumorigenicity in human non-small cell lung cancer by regulating miR-142-5p/PAX6 axis. OncoTargets Ther. (2020) 13:4919. doi: 10.2147/OTT.S238808
61. Derakhshani A, Javadrashid D, Hemmat N, Dufour A, Solimando AG, Shadbad MA, et al. Identification of common and distinct pathways in inflammatory bowel disease and colorectal cancer: A hypothesis based on weighted gene co-expression network analysis. Front Genet. (2022) 13:848646. doi: 10.3389/fgene.2022.848646
62. Nomiri S, Karami H, Baradaran B, Javadrashid D, Derakhshani A, Nourbakhsh NS, et al. Exploiting systems biology to investigate the gene modules and drugs in ovarian cancer: A hypothesis based on the weighted gene co-expression network analysis. Biomed Pharmacother. (2022) 146:112537. doi: 10.1016/j.biopha.2021.112537
63. Pang L, Zhang Q, Wu Y, Yang Q, Zhang J, Liu Y, et al. Long non-coding RNA CCAT1 promotes non-small cell lung cancer progression by regulating the miR-216a-5p/RAP2B axis. Exp Biol Med. (2021) 246:142–52. doi: 10.1177/1535370220961013
64. Chai J, Qin L, Zhang G, Hua P, Jin C. Long non-coding MELTF antisense RNA 1 promotes and prognosis the progression of non-small cell lung cancer by targeting miR-1299. Bioengineered. (2022) 13:10594–604. doi: 10.1080/21655979.2022.2063563
65. Cao S, Li L, Li J, Zhao H. MiR-1299 impedes the progression of non-small-cell lung cancer through EGFR/PI3K/AKT signaling pathway. Onco Targets Ther. (2020) 13:7493–502. doi: 10.2147/OTT.S250396
66. Derakhshani A, Rostami Z, Safarpour H, Shadbad MA, Nourbakhsh NS, Argentiero A, et al. From oncogenic signaling pathways to single-cell sequencing of immune cells: Changing the landscape of cancer immunotherapy. Molecules. (2021) 26:2278. doi: 10.3390/molecules26082278
Keywords: non-small-cell lung carcinoma, PD-L1, microRNAs, circular RNA, single-cell RNA sequencing
Citation: Shadbad MA, Ghorbaninezhad F, Hassanian H, Ahangar NK, Hosseinkhani N, Derakhshani A, Shekari N, Brunetti O, Silvestris N and Baradaran B (2022) A scoping review on the significance of programmed death-ligand 1-inhibiting microRNAs in non-small cell lung treatment: A single-cell RNA sequencing-based study. Front. Med. 9:1027758. doi: 10.3389/fmed.2022.1027758
Received: 25 August 2022; Accepted: 10 October 2022;
Published: 31 October 2022.
Edited by:
Somchai Chutipongtanate, University of Cincinnati, United StatesReviewed by:
Behrouz Shamsaei, University of Cincinnati, United StatesAlbino Eccher, Integrated University Hospital Verona, Italy
Copyright © 2022 Shadbad, Ghorbaninezhad, Hassanian, Ahangar, Hosseinkhani, Derakhshani, Shekari, Brunetti, Silvestris and Baradaran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicola Silvestris, bnNpbHZlc3RyaXNAdW5pbWUuaXQ=; Behzad Baradaran, YmFyYWRhcmFuYkB0YnptZWQuYWMuaXI=
†These authors have contributed equally to this work
‡These authors share last authorship
 Mahdi Abdoli Shadbad
Mahdi Abdoli Shadbad Farid Ghorbaninezhad
Farid Ghorbaninezhad Hamidreza Hassanian
Hamidreza Hassanian Noora Karim Ahangar
Noora Karim Ahangar Negar Hosseinkhani
Negar Hosseinkhani Afshin Derakhshani
Afshin Derakhshani Najibeh Shekari2
Najibeh Shekari2 Oronzo Brunetti
Oronzo Brunetti Nicola Silvestris
Nicola Silvestris Behzad Baradaran
Behzad Baradaran