
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Med. , 08 November 2022
Sec. Gene and Cell Therapy
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.1026931
The polymerase δ-interacting protein (POLDIP) family is a new family that can interact with DNA polymerase δ (delta). The members of the POLDIP family include POLDIP1, POLDIP2, and POLDIP3. Screened by the two-hybrid method, POLDIP1, POLDIP2, and POLDIP3 were initially discovered and named for their ability to bind to the p50 subunit of DNA polymerase δ. Recent studies have confirmed that POLDIPs are involved in the regulation of signal transduction pathways in neurodevelopment, neuropsychiatric diseases, cardiovascular diseases, tumors, and other diseases. However, each protein participates in different signaling pathways. In this review, we elucidate upon the family in terms of their genes and protein structures, their biological functions, in addition to the pathways that they are involved in during the development of diverse diseases. Finally, to provide new insights to the scientific community, we used the TCGA database to analyze and summarize the gene expressions of POLDIP family members in various tumors, as well as the correlations between their expressions and the overall survival times of tumor patients. Our data summary will give researchers working on cancer new concepts.
Polymerase δ-interacting protein (POLDIP) is a new family that interacts with DNA polymerase (delta). POLDIP1, POLDIP2, and POLDIP3 are members of the POLDIP family. POLDIP1, POLDIP2, and POLDIP3 were initially discovered and named for their ability to bind to the p50 subunit of DNA polymerase using the two-hybrid method (1, 2). Recent research has confirmed that POLDIPs play a role in the regulation of signal transduction pathways in neurodevelopment, neuropsychiatric diseases, cardiovascular diseases, tumors, and other diseases. However, each protein participates in a different signaling pathway. In this review, we elucidate the family in terms of their genes and protein structures, biological functions, and pathways that they are involved in during the development of various diseases.
Nucleic POLDIP1 contains one functional domain and one functional motif. Its N-terminal has a BTB/POZ domain (residues 41–138) that is involved in the ubiquitination and degradation of ras homolog family member A (RhoA). The C-terminal possesses the motif QTKV-EFP (residues 249–255), a proliferating cell nuclear antigen (PCNA)-bind motif (Figure 1A) (1, 3). Co-induced with interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α), POLDIP1 can directly interact with PCNA and the small subunit (p50) of DNA polymerase δ to enhance DNA polymerase δ activity. Moreover, POLDIP1 can interact with a variety of proteins to play various biological functions (see Table 1).
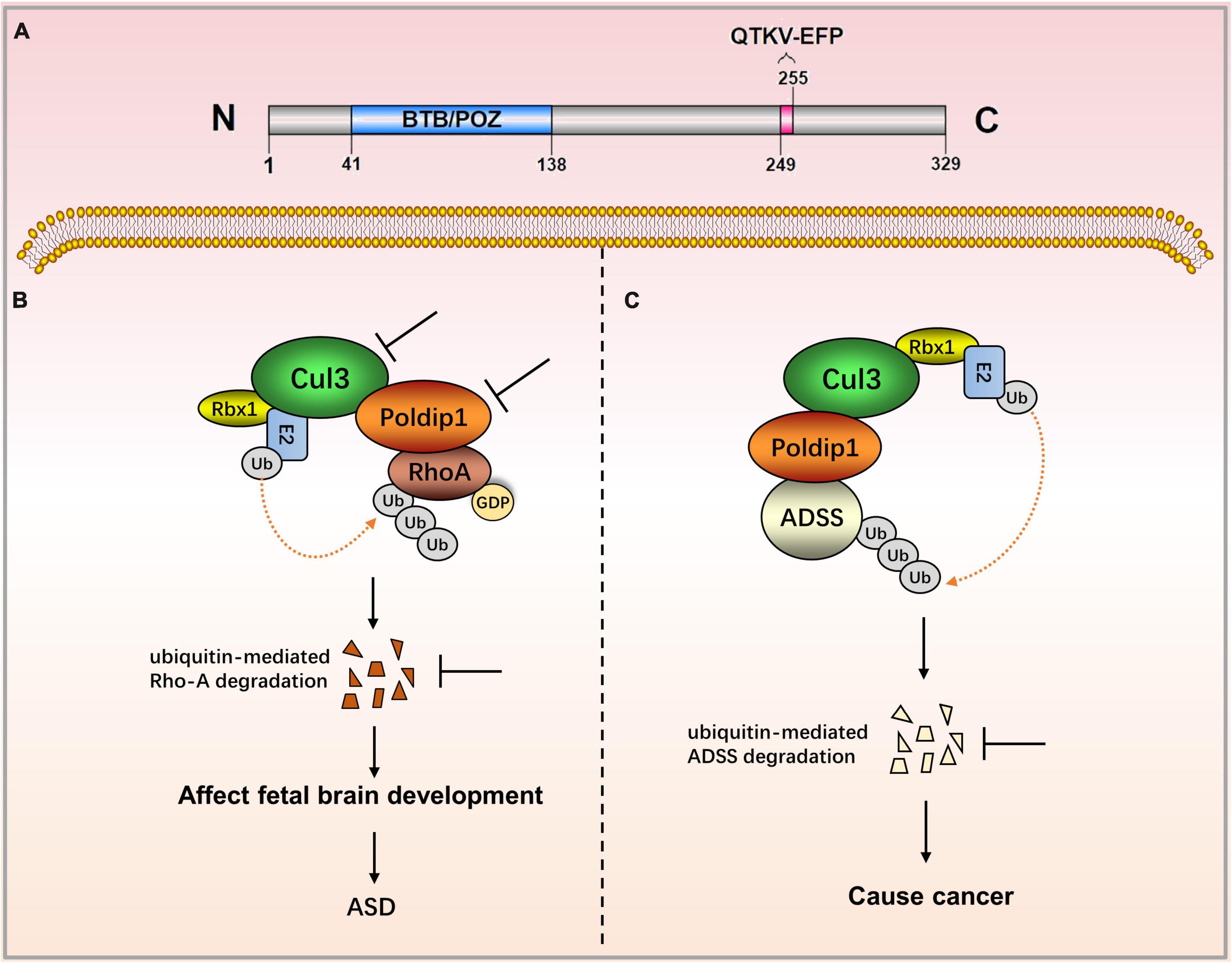
Figure 1. The role of POLDIP1 in the modulation of disease-related signaling pathways. (A) Domain map of POLDIP1. Its N-terminal has a BTB/POZ domain (residues 41–138) that is involved in the ubiquitination and degradation of RhoA. The C-terminal possesses the motif QTKV-EFP (residues 249–255), a proliferating cell nuclear antigen (PCNA)-bind motif. (B) Decreased Cul3 in ASD patients can reduce the physical interaction between POLDIP1 and Cul3, resulting in an abnormal expression of RhoA, which subsequently affects fetal brain development. (C) POLDIP1 can target adenylosuccinate synthase (ADSS), and it can accelerate its ubiquitination and degradation. POLDIP1 will promote the degradation and reduction of ADSS, and that will promote carcinogenesis.
Polymerase δ-interacting protein 2 is a mitochondrial protein with multiple subcellular localizations. POLDIP2 can be expressed in the mitochondria, nucleus, cell membrane, and mitotic spindle. POLDIP2 has two independent domains: the YccV domain at the N-terminal (residues 64–186) and the DUF525 domain at the C-terminal (residues 231–368) (Figure 2A) (4, 5). An assessment of the POLDIP2 sequence revealed three supposed PCNA binding motifs between residues 81–88, 151–158, and 193–200 (6). Ever-growing evidence has shown that, in addition to the interaction between PCNA and polymerase δ, POLDIP2 can interact with a variety of binding partners and participate in the cell cycle, focal adhesion transition, and cell migration (see Table 2).
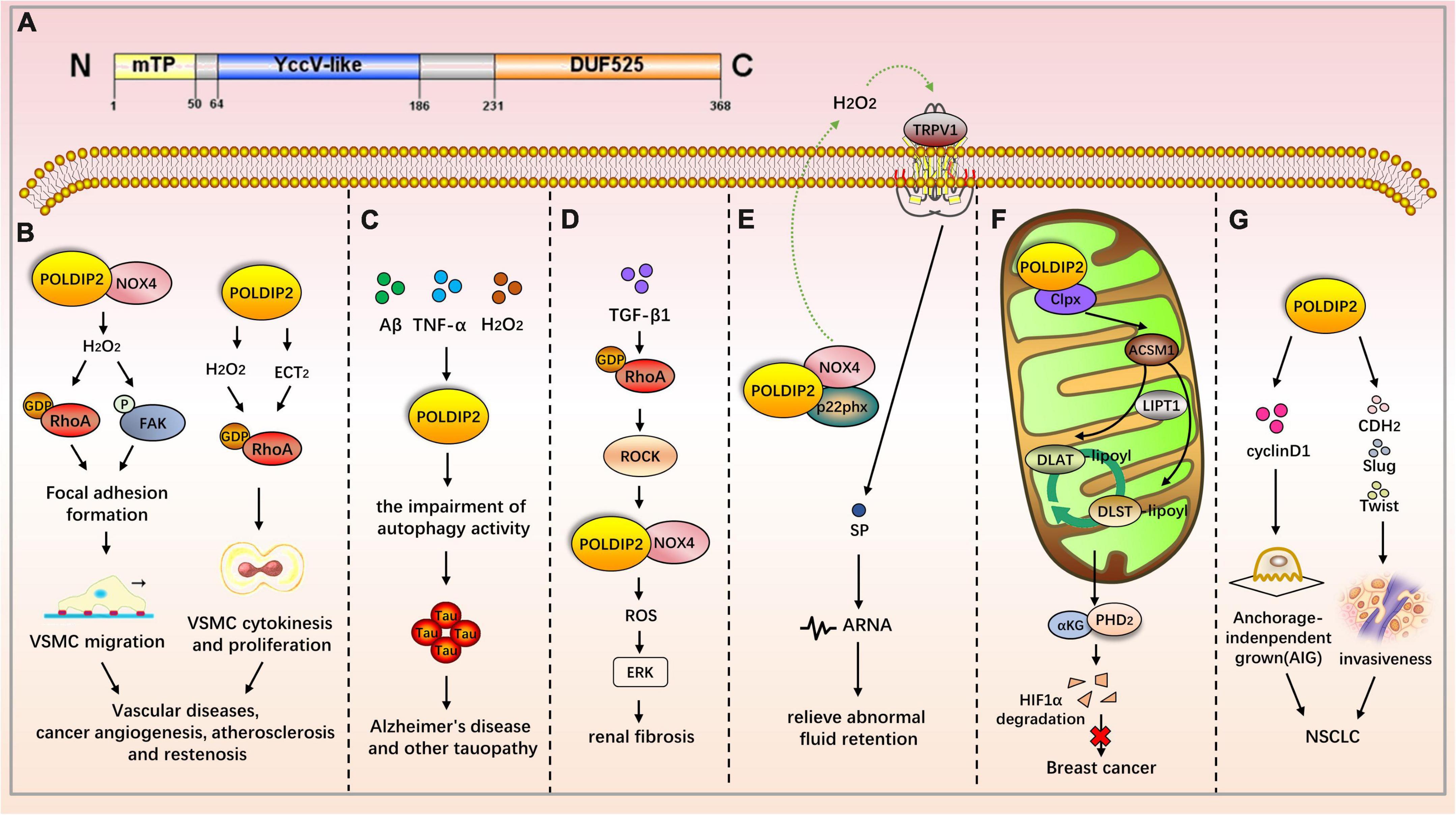
Figure 2. The role of POLDIP2 in the modulation of disease-related signaling pathways. (A) Domain map of POLDIP2. POLDIP2 contains an N-terminal mitochondrial targeting peptide (mTP) and two main functional domains: a DUF525 domain and a hemimethylated YccV-like domain. DUF525 may be involved in protein-protein interaction and cation efflux, respectively. YccV may bind to DNA and regulate its expression. (B) POLDIP2 can increase the focal adhesion turnover and cell polarization of VSMCs. POLDIP2 participates in the migration of VSMCs by promoting the NOX4/RhoA/FAK pathway. POLDIP2 is involved in the cytokinesis and proliferation of VSMCs by activating RhoGEF epithelial cell transformation sequence 2 (ECT2) and its downstream molecule RhoA. (C) POLDIP2 is a novel regulator of Tau aggregation in Alzheimer’s disease and another tauopathy. Expression of POLDIP2 can be increased in neuronal cells by the multiple stresses, including Aβ, TNF-α, and H2O2. POLDIP2 overexpression can induce impairments of autophagy activity and partially proteasome activity and subsequently result in Tau aggregation. (D) In the TGF-β-induced rat renal myofibroblast differentiation model, the activation of the RhoA/Rock/POLDIP2/NOX4/ROS pathway can induce the activation of renal myofibroblasts, which may be of great significance in the pathogenesis of renal fibrosis. (E) Lin et al. have studied the effect of NOX4/p22phox/POLDIP2 interactions on the activity of TRPV1-mediated mechanosensation. Using co-immunoprecipitation experiments, they have demonstrated that POLDIP2 can interact with NOX4 and p22phox in renal pelvis lysates. The elevation of intrapelvic pressure (IPP) stimulates POLDIP2 expression and enhances the association between POLDIP2 and p22phox, which further activates NOX4. NOX4 stimulates TRPV1 by producing H2O2, increases the release of substrate P (SP), and boosts afferent renal nerve activity (ARNA). The mechanism clarifies the cause of renal non-response to fluid retention and provides a new therapeutic strategy for relieving abnormal fluid retention. (F) POLDIP2, a mitochondrial protein, can increase mitochondrial lipoacylation, enhance cell respiration, and reduce the growth rate of cancer cells. POLDIP2 binding to CLPX can restrain lipoic acid-activating enzyme Ac-CoA synthetase medium-chain family member 1 (ACSM1) degradation. Consequently, lipoyl-AMP is produced, and lipoyltransferase 1 (LIPT1) has a substrate for the lipoylation of dihydrolipoamide S-acetyltransferase (DLAT) and dihydrolipoamide S-succinyltransferase (DLST). The lipoylation promotes the tricarboxylic acid (TCA) cycle, leading to prolyl hydroxylase-2 (PHD2) production and HIF-1α degradation. However, POLDIP2 expression is declined in breast cancer cells. (G) POLDIP2 is downregulated in NSCLC tissues, and the overexpressed POLDIP2 increases the anchorage independent growth (AIG) and proliferation of NSCLC cells. As shown in the mechanism study, POLDIP2 knockdown can significantly impair the expression of cell proliferation, cyclin D1, epithelial mesenchymal transition (EMT) markers, cdh2, and slug and twist, thereby indicating that POLDIP2 participates in regulating tumor growth and invasiveness.
The subcellular localization of POLDIP3 is mainly located in nuclear spots and at the exon junction complex (EJC). In mitotic cells, POLDIP3 is located in the cytoplasm. POLDIP3 also has two independent domains: the APIM domain (residues 53–125) and an RNA recognition motif (RRM) domain (residues 277–357) (Figure 3A) (7). POLDIP3 can interact with DNA polymerase δ and PCNA, and it plays an important role in DNA replication. POLDIP3 also has a variety of binding partners as well as many functions, several of which seem to be the result of specifically related protein interactions (see Table 3). Additionally, since POLDIP3 possesses RRM, it is speculated that POLDIP3 may participate in RNA processing, nuclear output, and stability to participate in RNA metabolism.
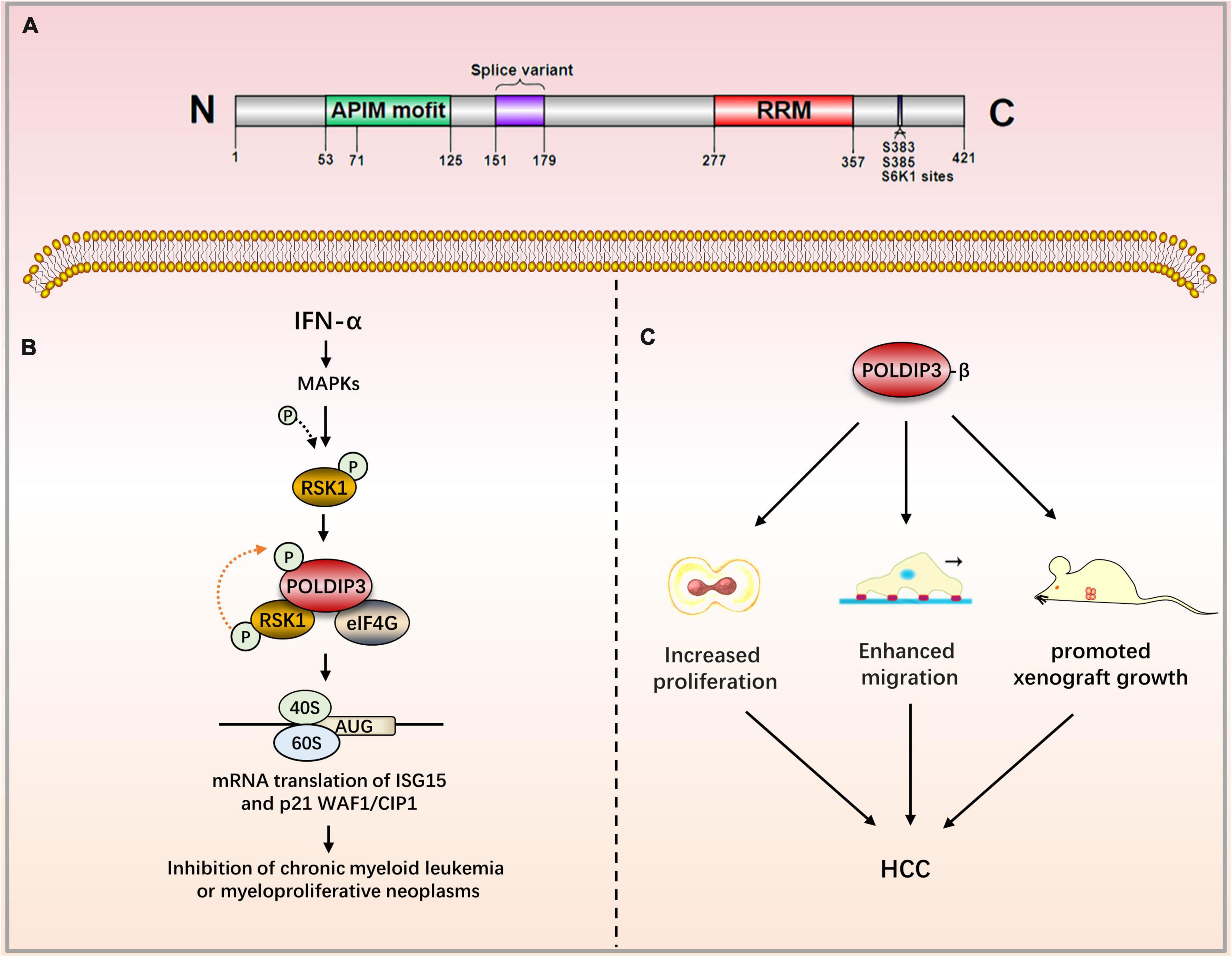
Figure 3. The role of POLDIP3 in the modulation of disease-related signaling pathways. (A) Domain map of POLDIP3. The N-terminal of POLDIP3 contains a cluster of five AlkB homolog 2 PCNA-Interacting motifs (APIMs), in which POLDIP3 can bind with PCNA and the p50 subunit of polδ at its residues 71–125 and 53–125, respectively. The C-terminal 277–357 amino acid residues of POLDIP3 possess an RRM similar to ALY/REF (RNA and export factor binding proteins) and can bind with RNA. Meanwhile, the RRM domain can bind with ribosomal protein S6 kinase 1 (S6K1). The phosphorylation of POLDIP3 locates at s383/s385. (B) Ribosomal protein S6 kinase (RSK1) can induce POLDIP3 phosphorylation in an IFN-α-dependent manner. POLDIP3 phosphorylation leads to greater interactions between POLDIP3 and eukaryotic initiation factor 4G (EIF4G) to form a unique IFN-induced RSK1-POLDIP3-EIF4G complex, thus promoting the mRNA translation of interferon-stimulated genes (ISG). (C) POLDIP3-β overexpression can increase the proliferation and migration of hepatocellular carcinoma cells and promote the growth of xenotransplantation. IFN-α, Type I interferons Alpha; RSK1, p90 ribosomal protein S6 kinase; eIF4G, eukaryotic initiation factor; ISG, IFN-stimulated genes; CML, chronic myeloid leukemia; HCC, Hepatocellular carcinoma.
Although the POLDIP family has been linked to several neurological diseases, cancers, and other diseases, there may be many undiscovered links between the POLDIP family and other diseases, particularly tumors. While summarizing the relationship between the POLDIP family and a variety of diseases, this review also examined the relationships between POLDIP family protein expression and carcinogenesis, as well as the survival time of various types of tumors, using the TCGA database.
This review summarizes the POLDIP family’s involvement in various biological functions, as well as the pathways in which it participates during the occurrence of multiple diseases. This knowledge will help scientists find novel targets and treatment options for related diseases.
DNA POLDIP1, also known as BACURD1, FKSG86, PDIP1, hBACURD1, and KCTD13, was originally discovered by He et al. Through a yeast two-hybrid system experiment in 2000, He et al. (1) found a new protein interacting with small subunit p50 of DNA polymerase δ and named it POLDIP1. The human POLDIP1 gene is located on 16p11.2. Its molecular weight is about 30 kDa. POLDIP1 is widely expressed in multiple tissues, including the heart, kidney, and brain tissues.
Examining the subcellular distribution of POLDIP1 by indirect immunofluorescence and confocal microscopy, He et al. (1) found that POLDIP1 was located in the nucleus. Further studies found that POLDIP1 and PCNA were co-located in the replication focus of S-phase MCF-7 cells. Two conserved sites were found in the protein structure of the POLDIP1 gene: a BTB/POZ domain in the C-terminal region as well as a PCNA-binding motif in the N-terminal region (1, 2).
Human POLDIP1 can interact with PCNA and p50, the small subunit of DNA polymerase δ; therefore, it can enhance the activity of DNA polymerase δ in the presence of PCNA. Its synthesis can be co-induced by TNF-α and IL-6 (1). POLDIP1, acting as a meson, can bind to MYB proto-oncogenes like 2 (MYBL2) and PCNA, thereby participating in the process of DNA repair (3). Utilizing a BTB/POZ domain with about 120 amino acid residues at its N-terminal, POLDIP1 can act as a substrate-specific adaptor protein to form a complex with cullin-3 ubiquitin ligase. POLDIP1 acts as an adaptor protein and participates in the ubiquitination and degradation of RhoA; it does so to maintain the actin cytoskeleton structure and cell morphology (4), promote the transmission of neural synapses, and to facilitate cell migration as well as the development of brain size (7–9). POLDIP1 is an interaction partner of Rnd2 and Rnd3, both of which are members of the rho family GTPase (RND) protein family. This interaction affects the long-term localization and dendritic maturation of cortical neurons (10).
Polymerase δ-interacting protein 1 plays an important role in neurocognitive impairment. The POLDIP1 gene can modulate early neural development and the head size phenotype. Copy number deletion of POLDIP1 (located at 16p11.2) led to the macrocephalic phenotype, whereas copy number duplication of POLDIP1 yielded a microcephalic phenotype in zebrafish embryos (8). The integration of zebrafish and mouse data has indicated that the microcephalic phenotype results from decreased neural progenitor cells and increased apoptosis of developing brain cells, whereas the macrocephalic phenotype is attributed to increased neural progenitor cells and no alteration in brain cell apoptosis. Some studies have also demonstrated that POLDIP1 knockout reduced the synaptic transmission in area CA1 of the hippocampus, although they have proven that POLDIP1 deletion cannot affect brain size (7). A critical 118 kb-deletion region on 16p11.2 from a three-generation autism spectrum disorder (ASD) family has been identified, and POLDIP1 is located exactly within the region (11). Meanwhile, researchers have revealed copy number variants (CNVs) in a family pedigree of autism on 16p11.2, in which they have analyzed both inherited and de novo rearrangements of POLDIP1 (8). POLDIP1 is a target gene that causes cognitive dysfunction in patients with 16p11.2 deletion (12). Decreased Cul3 in ASD patients can reduce the physical interaction between POLDIP1 and Cul3, resulting in an abnormal expression of RhoA, which subsequently affects fetal brain development (9).
Polymerase δ-interacting protein 1 has recently been implicated in biological ubiquitination and protein degradation. POLDIP1 can bind to and accelerate the ubiquitination and degradation of adenylosuccinate synthase (ADSS), an enzyme that catalyzes the synthesis of adenosine monophosphate (AMP). ADSS expression in lung adenocarcinoma tissues is significantly lower than in normal tissues, according to researchers. Based on the above findings, it is hypothesized that POLDIP1 will promote the degradation and reduction of ADSS and that it will promote carcinogenesis (13). Furthermore, data from the cosmic database show that POLDIP1 may promote the occurrence of breast tumors, as high expression of POLDIP1 has been found in 14% of breast tumor samples (14).
Liu et al. (6) identified a novel protein that can bind to the p50 subunit of DNA polymerase delta. They named it PDIP38, which is an alias for DNA POLDIP2. POLDIP2 is also known as pold4, pdip38, and p38. The human POLDIP2 gene is located at 17q11.2, and it encodes a protein with a molecular weight of about 28 kDa. POLDIP2 is generally expressed in 27 tissues, including kidney, liver, and heart tissues among others. Of note, the expression level of POLDIP2 in myeloid cells is very low (15).
Polymerase δ-interacting protein 2 has been found in multiple organelles, including mitochondria, spliceosomes, and nuclei. Its subcellular localization depends on the state of cell proliferation and its interactions with cell adhesion receptors. Cell fractionation experiments have shown that the majority of POLDIP2 can distribute in mitochondrial precipitation and that only a very small amount of it is present in the nucleus (16). However, there are disagreements about the subcellular localization of POLDIP2. POLDIP2 is dynamically localized in the cell surface or the nucleus under the influence of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1, CD66a), which acts as a cell adhesion receptor as well as an interacting protein of POLDIP2 (17). The above results indicate that POLDIP2 can regulate multiple cellular functions based on its subcellular localization and binding partners (17, 18).
Liu et al. (6) identified POLDIP2 as a binding chaperone of p50 (a small subunit of DNA polymerase δ). POLDIP2 is a multi-functional protein dealing in DNA replication and repair. In addition to its role in DNA replication and damage repair, POLDIP2 is involved in mitochondrial function modulation, extracellular matrix (ECM) regulation, cell cycle progression, focal adhesion turnover, and cell migration (19). In addition to p50, data have indicated that POLDIP2 can interact with a variety of proteins and perform its multiple functions. By binding to polymerase δ and PCNA, POLDIP2 can participate in DNA replication (6). Additionally, POLDIP2, as an intermediate, interacts with Pol η, Polζ, and Rev1, and it is involved in DNA replication and DNA translesion synthesis (TLS) (20). When 8-oxo-7,8-dihydroguanine (8-oxo-G) lesions occur during DNA replication, POLDIP2 can interact with Pol λ, which is involved in the correct bypass of 8-oxo-7,8-dihydroguanine (8-oxo-G) lesions (21). POLDIP2 can also stimulate the activity of primpol and therefore enhance primpol’s ability to bind DNA, which plays a repair role in 8-oxo-G TLS damage (22). Interacting with the E7 oncoprotein of human papillomavirus 16 and simultaneously inhibiting Pol δ activity, POLDIP2 plays an important role in Pol δ-mediated viral DNA replication (23). Binding with p22phox to stimulate NADPH oxidase 4 (NOX4), POLDIP2 controls focal adhesion turnover and affects vascular smooth muscle cell (VSMC) migration (15). POLDIP2 can interact with NOX4 or P22phox, and it can regulate the activity of the transient receptor potential vanilloid 1 (TRPV1) channel in rat kidneys (24). After stimulating NOX4, POLDIP2 expression increases the production of endogenous reactive oxygen species (ROS) catalyzed by NOX4, thereby indicating its crucial role in regulating the nuclear redox environment (15). POLDIP2 can interact with CEACAM1 (carcinoembryonic antigen-related cell adhesion molecule 1, CD66a) to manage its subcellular localization, which in turn facilitates CEACAM1-mediated cell survival, differentiation, and growth (17). Following UV-induced DNA damage, POLDIP2 translocates to the spliceosome loci of mouse double minute 2 homolog (MDM2). In doing so, it participates in UV-induced selective MDM2 transcript splicing (25).
Defective POLDIP2 can decrease the activity of NOX4 in cardiac muscle tissues. POLDIP2 knockout promotes the destruction of the aortic valve’s elastic lamina. This leads to excessive deposition of the ECM, resulting in impaired aortic contractions and reduced compliance. Further mechanism studies have indicated that defective POLDIP2 decreases NOX4 activity; the VSMCs consequently produce less ROS and secrete more type I collagen. This indicates that POLDIP2 is involved in regulating NADPH oxidase activity, thus affecting the structure and function of vessels (26). POLDIP2 can increase the focal adhesion turnover and cell polarization of VSMCs. POLDIP2 participates in the migration of VSMCs by promoting the NOX4/RhoA/FAK pathway (27). The excessive proliferation of VSMCs is one mechanism of atherosclerosis and restenosis. POLDIP2 is involved in the cytokinesis and proliferation of VSMCs by activating RhoGEF epithelial cell transformation sequence 2 (ECT2) and its downstream molecule RhoA (28).
Polymerase δ-interacting protein 2 is upregulated after ischemic stroke, which boosts cytokine TNF-α and IL-6 production as well as matrix metalloproteinase (MMP) activation, thereby mediating the increase of blood–brain barrier (BBB) permeability. Therefore, POLDIP2 is a potential drug target to reduce edema and stroke mortality (29). Neuronal degeneration caused by Tau aggregation can be found in a variety of neurological diseases, including Alzheimer’s disease and Parkinson’s disease. Studies have shown that POLDIP2 regulates Tau aggregation. POLDIP2 expression increases Tau aggregation without affecting Tau phosphorylation. It has been found that POLDIP2 overexpression can result in reduced cellular autophagy activity and partial proteasome activity, which are primarily performed by the DUF525 domain of POLDIP2 (30).
In the TGF-β-induced rat renal myofibroblast differentiation model, the activation of the RhoA/Rock/POLDIP2/NOX4/ROS pathway can induce the activation of renal myofibroblasts, which may be of great significance in the pathogenesis of renal fibrosis (31). However, POLDIP2 controls VSMC migration by activating NOX4/RhoA (27). The activation pathways mediated by POLDIP2 in the two kinds of literature are different. Lin et al. have studied the effect of NOX4/p22phox/POLDIP2 interactions on the activity of TRPV1-mediated mechanosensation. Using co-immunoprecipitation experiments, they have demonstrated that POLDIP2 can interact with NOX4 and p22phox in renal pelvis lysates. The elevation of intrapelvic pressure (IPP) stimulates POLDIP2 expression and enhances the association between POLDIP2 and p22phox, which further activates NOX4. NOX4 stimulates TRPV1 by producing H2O2, increases the release of substrate P (SP), and boosts afferent renal nerve activity (ARNA). The effect is to make the body respond to the changes in hydrostatic pressure in the renal pelvis. The mechanism clarifies the cause of renal non-response to fluid retention and provides a new therapeutic strategy for relieving abnormal fluid retention (24).
Acute respiratory distress syndrome (ARDS) is a fatal disease marked by acute hypoxia and non-cardiogenic pulmonary edema. Poldip2 is involved in the activation of β2-integrin during the inflammatory response, which mediates neutrophil-to-endothelium adhesion in ARDS (32).
Grinchuk et al. have identified a new complex sense-antisense architecture (CSAGA) on 17q11. The CSAGA contains five genes: tmem97, ift20, tnfaip1, polip2, and tmem199, all of which are termed the tnfaip1/POLDIP2 CSAGA. This CSAGA is linked to the amplification of 17q11.2 genomic regions and is associated with the expression of ERBB2 in breast cancer. The co-expression pattern of the CSAGA may correlate to the histological grade and prognosis of breast cancer (33). Downregulation of POLDIP2 and binding to CLPX allow for caseinolytic peptidase (Clp) activation and lipoic acid-activating enzyme Ac-CoA synthetase medium-chain family member 1 (ACSM1) degradation. Consequently, no lipoyl-AMP is produced, and lipoyltransferase 1 (LIPT1) has no substrate for the lipoylation of dihydrolipoamide S-acetyltransferase (DLAT) and dihydrolipoamide S-succinyltransferase (DLST). The lipoylation defect represses the tricarboxylic acid (TCA) cycle, leading to HIF-1α prolyl hydroxylase-2 (PHD2) metabolic inhibition and HIF-1α lasting expression. Overexpression of POLDIP2, a mitochondrial protein, can increase mitochondrial lipoacylation, enhance cell respiration, and reduce the growth rate of cancer cells, thereby indicating the key role of POLDIP2 in hypoxia and metabolic adaptation of breast cancer cells (34). Gene analyses from peripheral blood mononuclear cells have shown that POLDIP2 mRNA expression was negatively correlated with the risk of non-small cell lung cancer (NSCLC) (35). POLDIP2 is downregulated in NSCLC tissues, and the overexpressed POLDIP2 increases the anchorage independent growth (AIG) and proliferation of NSCLC cells. As shown in the mechanism study, POLDIP2 knockdown can significantly impair the expression of cell proliferation, cyclin D1, epithelial mesenchymal transition (EMT) markers, cdh2, and slug and twist, thereby indicating that POLDIP2 participates in regulating tumor growth and invasiveness (36). Additionally, POLDIP2 mutations have been linked to the development of tumors. A frameshift mutation in POLDIP2 was discovered in HPV-negative undifferentiated tongue sarcoma (37). Shear mutation of POLDIP2 was found in secretory breast carcinoma (SBC) (38).
Polymerase δ-interacting protein 3, also known as pdip3, pdip46, and Skar, was identified by Hernandes et al. in 2003. Through a yeast two-hybrid experiment, they found that POLDIP3, like POLDIP2, was a novel protein interacting with the p50 small subunit of human DNA polymerase δ (pol δ) (6). The human POLDIP3 gene is located on chromosome 22q13.2. The total molecular weight of POLDIP3 is about 42 kD. Human POLDIP3 is widely expressed in 27 tissues, including the spleen, ovary, and bone marrow tissues. The POLDIP3 protein is mainly located in the nucleus and at the EJC (39). In mitotic cells, POLDIP3 is located in the cytoplasm (40). The N-terminal of POLDIP3 contains a cluster of five AlkB homolog 2 PCNA-Interacting motifs (APIMs), in which POLDIP3 can bind with PCNA and the p50 subunit of pol δ at its residues 71–125 and 53–125, respectively (41). The C-terminal 277–357 amino acid residues of POLDIP3 possess an RRM similar to ALY/REF (RNA and export factor binding proteins) and can bind with RNA (39). Meanwhile, the RRM domain can bind with ribosomal protein S6 kinase 1 (S6K1), as well (41).
Since the discovery of POLDIP3, little research focused on its function has been performed. POLDIP3 can interact with DNA Pol δ and PCNA to promote cell replication and proliferation (42). POLDIP3 can directly communicate with Pol δ to activate DNA polymerase δ. Therefore, the alteration of POLDIP3 expression or its mutation may affect the function of Pol δ, thereby modulating genomic stability (41). POLDIP3 plays a role in the synthesis of leading-strand and lagging-strand DNA. During lagging strand DNA synthesis, when Polδ3 encounters secondary structure obstacles, POLDIP3 will accelerate the dissociation of Polδ3 from DNA and trigger the conversion of DNA polymerase to Polδ4/POLDIP3. Similarly, Pol ε is a principal enzyme for leading strand DNA synthesis. When Pol ε faces secondary structure obstacles during DNA synthesis, POLDIP3 can boost the depolymerization of Pol ε from DNA and ignite the switch of DNA polymerase from Pol ε to Polδ4/POLDIP3 (16). The functional interaction between telomere length 1 (RTEL1) and POLDIP3 has been recently demonstrated. An R-loop (RNA-DNA hybridization) at the genomic region where transcription and replication intersect can obstruct DNA replication. When DNA replication is blocked, POLDIP3 acts as a subunit of Pol δ; With other components of PCNA, it can also recruit RETL1 to the replication fork of the stalled transcription site to prevent a collision between the replication fork and RNA polymerase.
This in turn prevents an R-loop; and finally, this protects genome-wide replication and genome integrity (43). POLDIP3 can interact with ribosomal protein S6K1 to enhance the translation efficiency of mRNA (40). Under the stress state, the activated mTOR signaling pathway can induce POLDIP3, which is already located on the EJC. This leads to the recruitment of S6K1 into the newly synthesized mRNA, thereby facilitating the phosphorylation of several proteins in a cap-binding complex-mRNA protein (CBC mRNP) complex. The effect is to promote the translation efficiency of mRNA (40). Meanwhile, activated by the mTOR and PI3K signaling pathways, S6K1 triggers the phosphorylation of POLDIP3 at s383/s385. This phosphorylation is necessary to combine S6K1 and POLDIP3 (39). Because of its RRM, POLDIP3 has been speculated to be involved in RNA processing, nuclear export, and stability. It also takes part in RNA metabolism (39).
Polymerase δ-interacting protein 3 is the binding partner of the enhancer of the rudimentary homolog (ERH), as well. ERH is a transcription regulator that affects the expression of several genes in the cell cycle. Further studies have found that the site of interaction with ERH is located at POLDIP3’s C-terminal residues 274–421. This region is not continuous and can be further divided into two sub-regions. The larger (sub-region I) contains residues 274–368, and the smaller (sub-region II) contains residues 379–421. Since POLDIP3 is thought to affect cell size, this interaction of ERH/POLDIP3 is widespread in cell growth control (42). Recently, POLDIP3 has been identified as a novel member of the human transcription-export (TREX) complex. It is well known that the TREX complex plays an important role in mRNA nuclear export (44). Moreover, it has been demonstrated that overexpression of POLDIP3 can lead to the obstruction of enormous mRNA exportation from the nucleus (45). Ribosomal protein S6 kinase (RSK1) can induce POLDIP3 phosphorylation in an IFN-α-dependent manner. POLDIP3 phosphorylation leads to greater interactions between POLDIP3 and eukaryotic initiation factor 4G (EIF4G) to form a unique IFN-induced RSK1-POLDIP3-EIF4G complex, thus promoting the mRNA translation of interferon-stimulated genes (ISG) (46).
Studies have shown that the alternative splicing of POLDIP3 is related to amyotrophic lateral sclerosis (ALS). ALS is a neurodegenerative disease caused by the selective loss of motor neurons. In ALS motoneurons, the TDP-43 protein (43 kDa tar DNA binding protein) dislocates from the nucleus and enters the cytoplasm to form an inclusion body. Depletion of TDP-43 can result in abnormal splicing of POLDIP3 mRNA. In TDP-43 siRNA-treated cell models, wild-type POLDIP3 (variant 1) decreased, while mutated POLDIP3 (variant 2, lacking exon 3) increased. Therefore, detecting the variety law of POLDIP3/variant-2 in cerebrospinal fluid is of great significance for the diagnosis and evaluation of the progress of ALS (47).
Systemic vasculitis forms a group of heterogeneous autoimmune diseases characterized by vascular inflammation and an antibody reaction with autoantigens in the vascular wall. Using the serum of patients with clinical atypical renal vasculitis, researchers have found that POLDIP3 is a novel autoantigen, one that can be used as a diagnostic marker of autoimmune disease systemic vasculitis (48).
Kroczynska et al. have demonstrated that POLDIP3 plays an important role in type I interferons (IFN)-induced anti-leukemia and anti-tumor responses. Experiments have shown that POLDIP3 knockdown in primary leukemia CFU-GM progenitor cells from patients with chronic myeloid leukemia (CML)–or in primary malignant early erythrocyte progenitor cells (BFU-E) from patients with polycythemia vera–have reduced IFN-α-induced inhibitory effects on colony formation. Mechanism studies have illuminated how POLDIP3 knockdown results in the mRNA translation defects of key interferon stimulated genes (ISG) ISG15 and p21WAF1/CIP1 (41). Kroczynska et al. evaluated the effect of POLDIP3 knockdown on IFN-α-induced anti-colon adenocarcinoma responses. The results have shown that IFN-α treatment can inhibit the growth of HT29, a malignant colon adenocarcinoma cell; this inhibition, however, is reversed by POLDIP3 knockdown. This study established the role of POLDIP3 in IFN-α-induced colon cancer prevention (41). Lou et al. have constructed a prognosis model composed of 10 genes related to RNA processing, including POLDIP3, by using bioinformatics methods that predict the prognosis of patients with gastric cancer. They have found that their prognosis model could be used to predict the treatment response and prognosis of patients suffering from gastric cancer (49). Liu et al. also found that the expression of POLDIP3-β (POLDIP3 transcript lacking exon 3) in liver cancer tissues is significantly upregulated compared to paired adjacent non-cancerous liver tissues. POLDIP3-β overexpression can increase the proliferation and migration of hepatocellular carcinoma cells and promote the growth of xenotransplantation. POLDIP3-α (full-length containing exon 3) has much weaker effects on HCC cells. In short, POLDIP3-β will be a promising target for the treatment of liver cancer (50). Utilizing network-based microarray analysis and a visualization platform,1 researchers have found that a loss in POLDIP3 copy numbers leads to poor overall or recurrence-free survival in patients with neuroblastoma (51).
To date, the DNA POLDIP family contains three members, all of which can interact with DNA polymerase δ. It is widely known that human Pol δ (DNA polymerase delta) was first isolated from bovine bone marrow by Byrnes in 1976. The Pol δ belongs to the B family and is one of the principal DNA polymerases in eukaryotes. DNA polymerase δ is a holoenzyme composed of p125, p50, p68, and p12 subunits. It plays an important role in leading-strand and lagging-strand DNA synthesis (1, 2). To further study the role of the Pol δ, researchers in different laboratories have used yeast two-hybrid technology to find its interacting proteins. As a result, the three POLDIPs binding to the p50 subunit of Pol δ were identified by two separate laboratories. Researchers initially believed that POLDIPs were mainly involved in DNA replication and damage repair. However, recent strecent have confirmed that POLDIPs are involved in the regulation of signal transduction pathways in neurodevelopment, neuropsychiatric diseases, cardiovascular diseases, tumors, and other diseases. However, each protein participates in different signaling pathways.
Polymerase δ-interacting protein 1 research has primarily focused on neural development. POLDIP1 is the primary determinant of head size, and it can regulate early neural development (see Figure 1). POLDIP1 can be used as a substrate-specific adaptor protein to form a complex with cullin-3 ubiquitin ligase, as well as to participate in the ubiquitination and degradation of a wide range of proteins (4, 11).
So far, research on POLDIP2 is the most extensive and comprehensive, focusing mainly on cardiovascular disease, neurological disease, renal disease, and cancers (Figure 2). After interacting with NOX4, POLDIP2 participates in the migration and proliferation of VSMCs and renal myofibroblasts, thus playing an important role in vascular and renal fibrosis-related diseases (27, 31). However, the mechanism of its involvement in neurological disease and tumors need to be illuminated.
Few studies on POLDIP3 have been performed to date. Relevant studies on POLDIP3 have mainly assessed its role in the occurrence of neurological diseases, immune diseases, and cancer. However, the precise mechanisms remain unclear (Figure 3).
Last but not least, we examined and summarized the gene expressions of the POLDIP family members in a range of malignancies to offer fresh perspectives to the scientific community. Using the TCGA database, we also looked at the relationships between their expressions and overall patient survival times (summarized in Figure 4). For researchers working on cancer, our data summary can offer fresh perspectives.
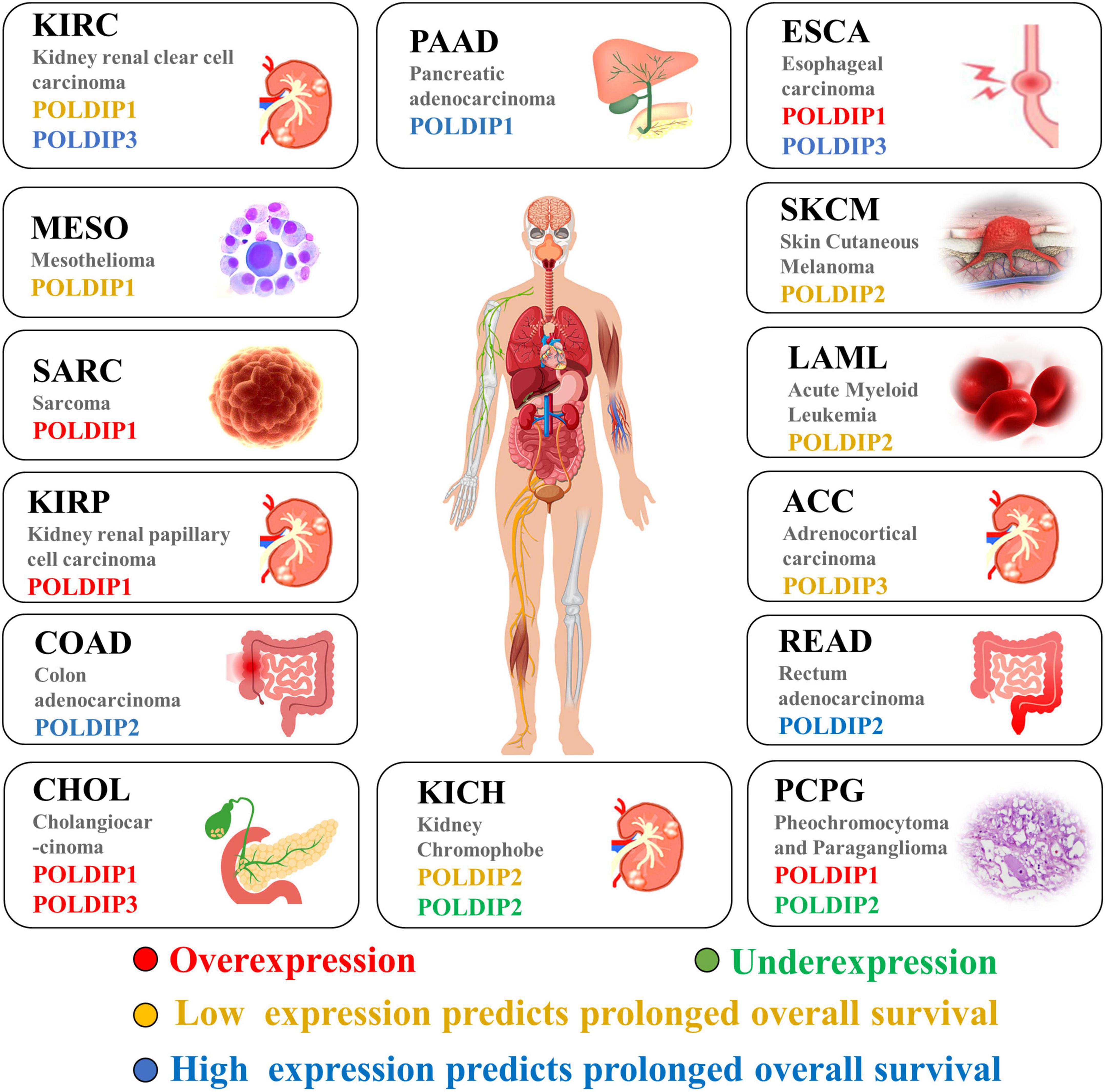
Figure 4. The POLDIP family in human cancers. Synthesis of the current knowledge on the role of POLDIP proteins in different cancer types. POLDIPs whose expression is downregulated in cancers are in green; POLDIPs whose expression is upregulated in cancers are in red; POLDIPs whose low-expression predicts prolonged overall survival are in yellow; POLDIPs whose high-expression predicts prolonged overall survival are in blue; all information is the result of data extrapolated by analysis of TCGA.
Regarding the various therapeutic targeting options for the POLDIP family in diseases, therapeutic modalities of diseases will primarily focus on several approaches described below and in Table 4: (1) POLDIPs gene therapy, for instance, adenoviral-mediated POLDIPs overexpression/inhibition, (2) targeted the py against POLDIPs interactive proteins (3) targeted therapy against critical factors involved in POLDIPs-related signaling pathways, and (4) POLDIPs mutants-related therapy, that is to develop mutant-specific POLDIPs restoration drugs. However, many aspects of POLDIPs’ function, such as the details of POLDIPs’ interactive proteins and the precise knowledge of POLDIPs mutations in diseases, have remained elusive. POLDIPs targeting therapy is still in its early stages.
JD designed the study. PH, LW, and NZ were major contributors to wrote the manuscript. JD and PH made substantial contributions to the design of the manuscript and revised it critically for important intellectual content. LW and PH created all the figures for the manuscript. NZ and HZ collected relevant data from TCGA and analyzed it. All authors have read and agreed to the published version of the manuscript.
This work was supported by Guangxi Science and Technology Project for Bases and Talents (Guike AD20297024), the Innovation Project of Guangxi Graduate Education (YCSW2021258), the Graduate Research Program of Guilin Medical University (GYYK2022009), and the National Natural Science Foundation of China (30901712, 81872221, and 82160408).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ACSM1, Ac-CoA synthetase medium-chain family member 1; ADSS, adenylosuccinate synthase; AIG, anchorage independent growth; ALS, amyotrophic lateral sclerosis; AMP, adenosine monophosphate; ASD, autism spectrum disorder; ARNA, afferent renal nerve activity; ARDS, acute respiratory distress syndrome; SBC, secretory breast carcinoma; BBB, blood–brain barrier; CSAGA, complex sense-antisense architecture; DLAT, dihydrolipoamide S-acetyltransferase; ECM, extracellular matrix; ECT2, epithelial cell transformation sequence 2; EIF4G, eukaryotic initiation factor 4G; EJC, exon junction complex; EMT, epithelial mesenchymal transition; ERH, enhancer of the rudimentary homolog; FAK, focal adhesion kinase; IL-6, interleukin-6; IPP, intrapelvic pressure; ISG, interferon-stimulated genes; LIPT1, lipoyl-AMP is produced, and lipoyltransferase 1; NOX4, NADPH oxidase 4; NSCLC, non-small cell lung cancer; MDM2, mouse double minute 2 homolog; MMP, matrix metalloproteinase; MYBL2, MYB proto-oncogene like 2; PCNA, proliferating cell nuclear antigen; PHD2, prolyl hydroxylase-2; POLDIP, polymeraseδ-interacting protein; RND, rho family GTPase; ROS, reactive oxygen species; RRM, RNA recognition motif; RSK1, ribosomal protein S6 kinase; RTEL1, telomere length 1; SP, substrate P; S6K1, ribosomal protein S6 kinase 1; TCA, tricarboxylic acid; TLS, DNA translesion synthesis; TNF-α, tumor necrosis factor-α; TREX, transcription-export; TRPV1, transient receptor potential vanilloid 1; VSMC, vascular smooth muscle cell.
1. He H, Tan CK, Downey KM, So AG. A tumor necrosis factor alpha- and interleukin 6-inducible protein that interacts with the small subunit of DNA polymerase delta and proliferating cell nuclear antigen. Proc Natl Acad Sci USA. (2001) 98:11979–84.
2. Zhou J, Hu X, Xiong X, Liu X, Liu Y, Ren K, et al. Cloning of two rat PDIP1 related genes and their interactions with proliferating cell nuclear antigen. J Exp Zool A Comp Exp Biol. (2005) 303:227–40. doi: 10.1002/jez.a.150
3. Werwein E, Schmedt T, Hoffmann H, Usadel C, Obermann N, Singer JD, et al. B-Myb promotes S-phase independently of its sequence-specific DNA binding activity and interacts with polymerase delta-interacting protein 1 (Pdip1). Cell Cycle. (2012) 11:4047–58. doi: 10.4161/cc.22386
4. Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, et al. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell. (2009) 35:841–55. doi: 10.1016/j.molcel.2009.09.004
5. Kulik AA, Maruszczak KK, Thomas DC, Nabi-Aldridge NLA, Carr M, Bingham RJ, et al. Crystal structure and molecular dynamics of human POLDIP2, a multifaceted adaptor protein in metabolism and genome stability. Protein Sci. (2021) 30:1196–209. doi: 10.1002/pro.4085
6. Liu L, Rodriguez-Belmonte EM, Mazloum N, Xie B, Lee MY. Identification of a novel protein, PDIP38, that interacts with the p50 subunit of DNA polymerase delta and proliferating cell nuclear antigen. J Biol Chem. (2003) 278:10041–7. doi: 10.1074/jbc.M208694200
7. Escamilla CO, Filonova I, Walker AK, Xuan ZX, Holehonnur R, Espinosa F, et al. Kctd13 deletion reduces synaptic transmission via increased RhoA. Nature. (2017) 551:227–31. doi: 10.1038/nature24470
8. Golzio C, Willer J, Talkowski ME, Oh EC, Taniguchi Y, Jacquemont S, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. (2012) 485:363–7. doi: 10.1038/nature11091
9. Lin GN, Corominas R, Lemmens I, Yang X, Tavernier J, Hill DE, et al. Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron. (2015) 85:742–54. doi: 10.1016/j.neuron.2015.01.010
10. Gladwyn-Ng I, Huang L, Ngo L, Li SS, Qu Z, Vanyai HK, et al. Bacurd1/Kctd13 and Bacurd2/Tnfaip1 are interacting partners to Rnd proteins which influence the long-term positioning and dendritic maturation of cerebral cortical neurons. Neural Dev. (2016) 11:7. doi: 10.1186/s13064-016-0062-1
11. Crepel A, Steyaert J, De la Marche W, De Wolf V, Fryns JP, Noens I, et al. Narrowing the critical deletion region for autism spectrum disorders on 16p11.2. Am J Med Genet B Neuropsychiatr Genet. (2011) 156:243–5. doi: 10.1002/ajmg.b.31163
12. Martin Lorenzo S, Nalesso V, Chevalier C, Birling MC, Herault Y. Targeting the RHOA pathway improves learning and memory in adult Kctd13 and 16p11.2 deletion mouse models. Mol Autism. (2021) 12:1. doi: 10.1186/s13229-020-00405-7
13. Miller JC, Blake DC Jr., Herzog CR. Adenylosuccinate synthetase 1 gene is a novel target of deletion in lung adenocarcinoma. Mol Carcinog. (2009) 48:1116–22. doi: 10.1002/mc.20563
14. Angrisani A, Di Fiore A, De Smaele E, Moretti M. The emerging role of the KCTD proteins in cancer. Cell Commun Signal. (2021) 19:56.
15. Lyle AN, Deshpande NN, Taniyama Y, Seidel-Rogol B, Pounkova L, Du P, et al. Poldip2, a novel regulator of Nox4 and cytoskeletal integrity in vascular smooth muscle cells. Circ Res. (2009) 105:249–59. doi: 10.1161/CIRCRESAHA.109.193722
16. Lee M, Wang X, Zhang S, Zhang Z, Lee EYC. Regulation and modulation of human DNA polymerase δ activity and function. Genes. (2017) 8:190. doi: 10.3390/genes8070190
17. Klaile E, Müller MM, Kannicht C, Otto W, Singer BB, Reutter W, et al. The cell adhesion receptor carcinoembryonic antigen-related cell adhesion molecule 1 regulates nucleocytoplasmic trafficking of DNA polymerase delta-interacting protein 38. J Biol Chem. (2007) 282:26629–40. doi: 10.1074/jbc.M701807200
18. Klaile E, Kukalev A, Obrink B, Müller MM. PDIP38 is a novel mitotic spindle-associated protein that affects spindle organization and chromosome segregation. Cell Cycle. (2008) 7:3180–6. doi: 10.4161/cc.7.20.6813
19. Hernandes MS, Lassègue B, Griendling KK. Polymerase δ-interacting protein 2: a multifunctional protein. J Cardiovasc Pharmacol. (2017) 69:335–42. doi: 10.1097/FJC.0000000000000465
20. Tissier A, Janel-Bintz R, Coulon S, Klaile E, Kannouche P, Fuchs RP, et al. Crosstalk between replicative and translesional DNA polymerases: PDIP38 interacts directly with Poleta. DNA Repair. (2010) 9:922–8. doi: 10.1016/j.dnarep.2010.04.010
21. Maga G, Crespan E, Markkanen E, Imhof R, Furrer A, Villani G, et al. DNA polymerase δ-interacting protein 2 is a processivity factor for DNA polymerase λ during 8-oxo-7,8-dihydroguanine bypass. Proc Natl Acad Sci USA. (2013) 110:18850–5. doi: 10.1073/pnas.1308760110
22. Guilliam TA, Bailey LJ, Brissett NC, Doherty AJ. PolDIP2 interacts with human PrimPol and enhances its DNA polymerase activities. Nucleic Acids Res. (2016) 44:3317–29.
23. Xie B, Li H, Wang Q, Xie S, Rahmeh A, Dai W, et al. Further characterization of human DNA polymerase delta interacting protein 38. J Biol Chem. (2005) 280:22375–84.
24. Lin CS, Lee SH, Huang HS, Chen YS, Ma MC. H2O2 generated by NADPH oxidase 4 contributes to transient receptor potential vanilloid 1 channel-mediated mechanosensation in the rat kidney. Am J Physiol Renal Physiol. (2015) 309:F369–76. doi: 10.1152/ajprenal.00462.2014
25. Wong A, Zhang S, Mordue D, Wu JM, Zhang Z, Darzynkiewicz Z, et al. PDIP38 is translocated to the spliceosomes/nuclear speckles in response to UV-induced DNA damage and is required for UV-induced alternative splicing of MDM2. Cell Cycle. (2013) 12:3184–93. doi: 10.4161/cc.26221
26. Sutliff RL, Hilenski LL, Amanso AM, Parastatidis I, Dikalova AE, Hansen L, et al. Polymerase delta interacting protein 2 sustains vascular structure and function. Arterioscler Thromb Vasc Biol. (2013) 33:2154–61.
27. Datla SR, McGrail DJ, Vukelic S, Huff LP, Lyle AN, Pounkova L, et al. Poldip2 controls vascular smooth muscle cell migration by regulating focal adhesion turnover and force polarization. Am J Physiol Heart Circ Physiol. (2014) 307:H945–57. doi: 10.1152/ajpheart.00918.2013
28. Huff LP, Kikuchi DS, Faidley E, Forrester SJ, Tsai MZ, Lassègue B, et al. Polymerase-δ-interacting protein 2 activates the RhoGEF epithelial cell transforming sequence 2 in vascular smooth muscle cells. Am J Physiol Cell Physiol. (2019) 316:C621–31. doi: 10.1152/ajpcell.00208.2018
29. Hernandes MS, Lassègue B, Hilenski LL, Adams J, Gao N, Kuan CY, et al. Polymerase delta-interacting protein 2 deficiency protects against blood-brain barrier permeability in the ischemic brain. J Neuroinflammation. (2018) 15:45. doi: 10.1186/s12974-017-1032-1
30. Kim Y, Park H, Nah J, Moon S, Lee W, Hong SH, et al. Essential role of POLDIP2 in Tau aggregation and neurotoxicity via autophagy/proteasome inhibition. Biochem Biophys Res Commun. (2015) 462:112–8. doi: 10.1016/j.bbrc.2015.04.084
31. Manickam N, Patel M, Griendling KK, Gorin Y, Barnes JL. RhoA/Rho kinase mediates TGF-β1-induced kidney myofibroblast activation through Poldip2/Nox4-derived reactive oxygen species. Am J Physiol Renal Physiol. (2014) 307:F159–71. doi: 10.1152/ajprenal.00546.2013
32. Ou Z, Dolmatova E, Mandavilli R, Qu H, Gafford G, White T, et al. Myeloid poldip2 contributes to the development of pulmonary inflammation by regulating neutrophil adhesion in a murine model of acute respiratory distress syndrome. J Am Heart Assoc. (2022) 11:e025181. doi: 10.1161/JAHA.121.025181
33. Grinchuk OV, Motakis E, Kuznetsov VA. Complex sense-antisense architecture of TNFAIP1/POLDIP2 on 17q11.2 represents a novel transcriptional structural-functional gene module involved in breast cancer progression. BMC Genomics. (2010) 11(Suppl 1):S9. doi: 10.1186/1471-2164-11-S1-S9
34. Paredes F, Sheldon K, Lassègue B, Williams HC, Faidley EA, Benavides GA, et al. Poldip2 is an oxygen-sensitive protein that controls PDH and αKGDH lipoylation and activation to support metabolic adaptation in hypoxia and cancer. Proc Natl Acad Sci USA. (2018) 115:1789–94. doi: 10.1073/pnas.1720693115
35. Chian CF, Hwang YT, Terng HJ, Lee SC, Chao TY, Chang H, et al. Panels of tumor-derived RNA markers in peripheral blood of patients with non-small cell lung cancer: their dependence on age, gender and clinical stages. Oncotarget. (2016) 7:50582–95. doi: 10.18632/oncotarget.10558
36. Chen YC, Kuo CC, Chian CF, Tzao C, Chang SY, Shih YL, et al. Knockdown of POLDIP2 suppresses tumor growth and invasion capacity and is linked to unfavorable transformation ability and metastatic feature in non-small cell lung cancer. Exp Cell Res. (2018) 368:42–9. doi: 10.1016/j.yexcr.2018.04.011
37. Chan JYK, Poon PHY, Zhang Y, Ng CWK, Piao WY, Ma M, et al. Case Report: exome sequencing reveals recurrent RETSAT mutations and a loss-of-function POLDIP2 mutation in a rare undifferentiated tongue sarcoma. F1000Res. (2018) 7:499. doi: 10.12688/f1000research.14383.1
38. Lei T, Deng X, Peng Y, Chen T. The genomic profile of double primary secretory breast carcinoma in one patient provides evidence for the treatment of such carcinoma: a case report. Pathol Res Pract. (2022) 236:154006. doi: 10.1016/j.prp.2022.154006
39. Richardson CJ, Bröenstrup M, Fingar DC, Jülich K, Ballif BA, Gygi S, et al. SKAR is a specific target of S6 kinase 1 in cell growth control. Curr Biol. (2004) 14:1540–9.
40. Ma XM, Yoon SO, Richardson CJ, Jülich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. (2008) 133:303–13. doi: 10.1016/j.cell.2008.02.031
41. Wang X, Zhang S, Zheng R, Yue F, Lin SH, Rahmeh AA, et al. PDIP46 (DNA polymerase δ interacting protein 46) is an activating factor for human DNA polymerase δ. Oncotarget. (2016) 7:6294–313. doi: 10.18632/oncotarget.7034
42. Smyk A, Szuminska M, Uniewicz KA, Graves LM, Kozlowski P. Human enhancer of rudimentary is a molecular partner of PDIP46/SKAR, a protein interacting with DNA polymerase delta and S6K1 and regulating cell growth. Febs J. (2006) 273:4728–41. doi: 10.1111/j.1742-4658.2006.05477.x
43. Björkman A, Johansen SL, Lin L, Schertzer M, Kanellis DC, Katsori AM, et al. Human RTEL1 associates with Poldip3 to facilitate responses to replication stress and R-loop resolution. Genes Dev. (2020) 34:1065–74. doi: 10.1101/gad.330050.119
44. Dufu K, Livingstone MJ, Seebacher J, Gygi SP, Wilson SA, Reed R. ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev. (2010) 24:2043–53. doi: 10.1101/gad.1898610
45. Folco EG, Lee CS, Dufu K, Yamazaki T, Reed R. The proteins PDIP3 and ZC11A associate with the human TREX complex in an ATP-dependent manner and function in mRNA export. PLoS One. (2012) 7:e43804. doi: 10.1371/journal.pone.0043804
46. Kroczynska B, Mehrotra S, Majchrzak-Kita B, Arslan AD, Altman JK, Stein BL, et al. Regulatory effects of SKAR in interferon α signaling and its role in the generation of type I IFN responses. Proc Natl Acad Sci USA. (2014) 111:11377–82. doi: 10.1073/pnas.1405250111
47. Shiga A, Ishihara T, Miyashita A, Kuwabara M, Kato T, Watanabe N, et al. Alteration of POLDIP3 splicing associated with loss of function of TDP-43 in tissues affected with ALS. PLoS One. (2012) 7:e43120. doi: 10.1371/journal.pone.0043120
48. Avila J, Acosta E, Machargo MD, Arteaga MF, Gallego E, Cañete H, et al. Autoantigenic nuclear proteins of a clinically atypical renal vasculitis. J Autoimmune Dis. (2008) 5:3. doi: 10.1186/1740-2557-5-3
49. Lou S, Meng F, Yin X, Zhang Y, Han B, Xue Y. Comprehensive characterization of RNA processing factors in gastric cancer identifies a prognostic signature for predicting clinical outcomes and therapeutic responses. Front Immunol. (2021) 12:719628. doi: 10.3389/fimmu.2021.719628
50. Liu XN, Yuan JH, Wang TT, Pan W, Sun SH. An alternative POLDIP3 transcript promotes hepatocellular carcinoma progression. Biomed Pharmacother. (2017) 89:276–83. doi: 10.1016/j.biopha.2017.01.139
Keywords: DNA polymerase δ, POLDIP1, POLDIP2, POLDIP3, signaling pathway
Citation: Huang P, Wu L, Zhu N, Zhao H and Du J (2022) The polymerase δ-interacting protein family and their emerging roles in diseases. Front. Med. 9:1026931. doi: 10.3389/fmed.2022.1026931
Received: 24 August 2022; Accepted: 24 October 2022;
Published: 08 November 2022.
Edited by:
Peiqing Zhao, Zibo Central Hospital, ChinaReviewed by:
Wei Zhao, Shandong University, ChinaCopyright © 2022 Huang, Wu, Zhu, Zhao and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Du, c3VubnlkdWp1YW5AZ2xtYy5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.