- 1Department of Tropical Medicine, Medical Microbiology and Pharmacology, John A. Burns School of Medicine, University of Hawai‘i at Mānoa, Honolulu, HI, United States
- 2Pacific Center for Emerging Infectious Diseases, John A. Burns School of Medicine, University of Hawai‘i at Mānoa, Honolulu, HI, United States
- 3Department of Quantitative Health Sciences, John A. Burns School of Medicine, University of Hawai‘i at Mānoa, Honolulu, HI, United States
Uncontrolled transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to the emergence of several variants of concern (VOC). As vaccine-induced neutralizing antibodies against VOC waned over time, breakthrough infections (BTIs) have been reported primarily among healthcare workers or in long-term care facilities. Most BTIs were identified by reverse transcription-polymerase chain reaction (RT-PCR) or antigen test for individuals experiencing symptoms, known as symptomatic BTIs. In this study, we detected seroconversion of anti-nucleocapsid (N) antibody to identify both symptomatic and asymptomatic BTIs in a cohort of COVID-19-naive university employees and students following two or three doses of mRNA vaccines. We reported 4 BTIs among 85 (4.7%) participants caused by the Omicron and Delta VOC during the transition from the Delta to Omicron wave of the pandemic; three were symptomatic and confirmed by RT-PCR test and one asymptomatic. A symptomatic reinfection two and half months after a BTI was found in one participant. Two of three symptomatic BTIs and the reinfection were confirmed by whole genome sequencing. All were supported by a >4-fold increase in neutralizing antibodies against the Delta or Omicron variant. Moreover, we found both symptomatic and asymptomatic BTIs can boost neutralizing antibodies against VOC with variable degrees ranging from 2.5- to 77.4-fold increase in neutralizing antibody titers. As BTIs continue, our findings highlight the application of anti-N antibody test to ongoing studies of immunity induced by spike-based vaccine, and provide new insights into the establishment of herd immunity in the community during the post-vaccination era.
Introduction
Unconstrained transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in the emergence of several variants of concern (VOC), including previously circulating VOC, the Alpha (Pango lineage: B.1.1.7), Beta (B.1.351), Gamma (P1), and Delta (B.1.617.2) variants, and the currently circulating VOC, the Omicron (B.1.1.529) variant and Omicron subvariants under monitoring such as BA.4, BA.5, and BA.2.75 (1, 2). Genomic surveillance of variants revealed that the Delta variant rose from <1% of circulating viruses in the US in early May 2021 to >50% in June, and to >95% in all 10 regions by August 2021. Since the first US Omicron case reported on 12/1/2021, the Omicron variant increased from >1% of circulating lineages in early December, to >50% in late December, and to >99% in late January 2022 (3, 4). The proportion of circulating variants in Hawaii, belonging to region 9, followed a similar trend.
In agreement with reports that vaccine-induced neutralizing antibodies against SARS-CoV-2 reduced and waned over time, breakthrough infections (BTIs) have been well documented following two or three doses of mRNA vaccines (5–16). Studies of BTI have been primarily focused on healthcare workers or residents in long-term care facilities (5, 14, 17–20). Based on reverse transcription-polymerase chain reaction (RT-PCR) results, Rana et al. reported a BTI rate of 2.0% among 3650 healthcare workers following the second dose of vaccine (14). Similarly, Bergwerk et al. reported a BTI rate of 2.7% among 1497 fully vaccinated healthcare workers (5). The BTI rates in long-term care homes were reported to be 39.6 and 9.3% among fully vaccinated residents and staff, respectively, during an outbreak of Gamma variant, and 6.5 to 50% among fully vaccinated residents during the Beta variant outbreak (17, 19, 20). The extent and rate of BTIs among non-healthcare workers remain understudied. The most common method to identify BTIs is RT-PCR or antigen test for individuals experiencing symptoms, thereby identifying symptomatic BTIs. Detection of anti-nucleocapsid (N) protein antibody has potential to identify both symptomatic and asymptomatic BTIs among COVID-19-naive vaccinees who received a spike (S)-based vaccine such as mRNA vaccines (21). Although recent studies reported potent and broad neutralizing antibodies induced by BTIs (22–26), how a BTI boosts neutralizing antibodies against VOC in individuals remains incompletely understood. In this study, we employed a combination of anti-N and anti-S enzyme-linked immunosorbent assays (ELISAs) to investigate BTI in a cohort of university employees and students following two or three doses of mRNA vaccines and examine neutralizing antibodies before and after BTI in Hawaii.
Materials and methods
Human subjects
With the approval of Institutional Review Board of the University of Hawaii (2020-00406), coded plasma samples were obtained with informed consents from study participants (university employees and students), who were COVID-19-naïve or recovered cases at enrollment and received two or three doses of mRNA-1273 (Moderna) or BNT162b2 (Pfizer) vaccine between August 2020 and February 2022 (27, 28). The samples included COVID-19-naïve participants before vaccination (n = 28), COVID-19-naïve participants following one (n = 55), two (n = 147), or three (n = 20) doses of a mRNA vaccine, RT-PCR-confirmed cases of SARS-CoV-2 natural infection (NI) before vaccination (n = 19), and NI followed by one (n = 10) or two (n = 7) doses of a mRNA vaccine (Table 1). All participants were otherwise healthy adults based on a self-reported questionnaire.
SARS-CoV-2 ELISAs
Anti-N and anti-S antibodies were detected by SARS-CoV-2 Detect IgG ELISA (InBios) and Platelia SARS-CoV total Ab ELISA (BioRad), respectively.
Plasmids
Plasmids pNL4-3 R-E-miRFP, which contains the miRFP gene replacing the Luc gene of an env-defective HIV-1 reporter construct pNL4-3.Luc.R- E-, and D614G, which contains the S gene of the SARS-CoV-2 Wuhan-Hu-1 strain with D614G mutation and C-terminal 19-residue truncation, have been described previously (28). The S genes (Alpha, Delta) were synthesized (Integrated DNA Technologies) by two fragments [residues 1 to 461, flanked by KpnI and AflII (an introduced silent site mutation) sites, and residues 461 to 1254, flanked by AflII and NotI sites], and cloned into the plasmid D614G (with KpnI and NotI sites) by 3-fragment ligation to generate plasmid Alpha. Two-step cloning (residues 1 to 461 first, followed by residues 461 to 1254) and one-step cloning of four fragments (residues 1 to 461, residues 461 to 853, and residues 853 to 1254) by NEBuilder HiFi DNA assembly kit (New England Biolab) were performed to generate plasmids Delta and Omicron, respectively. All plasmids were confirmed by sequencing of the entire S gene insert (Supplementary Figure 1) and verified for expression by transfection and Western blot analysis (28, 29).
Generation of SARS-CoV-2 pseudovirus
To generate pseudoviruses, HEK-293T cells were seeded in 10-cm dish 1 day before transfection, co-transfected with pNL4-3 R-E-miRFP (12 μg) and S plasmid (3 μg) using lipofectamine 2000, and incubated with DMEM media containing 10% FBS (28). The supernatants were collected at 48 h post transfection, followed by low-speed centrifugation at 300 × g for 10 min, aliquoted and stored at −80°C. Previously, 1.65 × 109 RNA copies of pseudovirus per well were used for each neutralization test, resulting in miRFP signals 10 times higher than mock-infected well at 72 h post-infection (28). To titrate each pseudovirus, threefold serially diluted supernatants were inoculated to HEK-293ThACE2 cells by spin infection (28); miRFP signals were quantitated at 72 h post-infection, and the amount of pseudovirus that resulted in miRFP signals 10 times higher than the mock-infected wells was used for neutralization test. Pseudovirus D614G was referred as the wild type D614G strain, which predominated since March 2020 and contains the S gene similar to that of the USA-WA1 strain used in the mRNA vaccine with one amino acid substitution (G at residue 614) (2).
SARS-CoV-2 pseudovirus neutralization test
HEK-293T-hACE cells (2 × 104 cells/well) were seeded onto 96-well plates 1 day before infection. Pseudovirus (D614G, Alpha, Delta, or Omicron variant) was mixed with fourfold serial dilutions of plasma at 1:1 ratio, incubated at 37°C for 1 h, and added to each well for spin infection. At 72 h, the plates were scanned by Li-Cor Odyssey imager (28). The % of infection at different plasma dilutions (from 1:10 to 1:10,240 dilutions) were calculated by the formula (intensity of plasma + pseudovirus—intensity of media only)/(intensity of pseudovirus only—intensity of media only) × 100. The % neutralization = 100 −% of infection (28). NT50 titer was the plasma dilution that reached 50% neutralization using 4-parameter non-linear regression analysis (GraphPad 6.0) (Supplementary Figure 2). NT50 titer <10 was arbitrarily assigned as 5.
Processing of nasal swabs
Nasal swabs were collected between 2 and 5 days following symptom onset and stored in virus transport medium (VTM). SARS-CoV-2 infection was confirmed by quantitative RT-PCR using the CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel.
Whole genome sequencing
Total RNA was extracted from 200 μl of VTM using the MagMax™ Viral/Pathogen II kit with the Kingfisher™ Flex Purification System. RNA was reverse transcribed into cDNA using random hexamers (SuperScript™ IV first-strand synthesis system, Life Technology). PCR amplification of the SARS-CoV-2 genome was adapted from the ARTIC network1 sequencing protocol (30) using the ARTIC V4.1 primers. Purified PCR amplicons were submitted to the Advanced Studies in Genomics, Proteomics and Bioinformatics (ASGPB) core at the University of Hawai‘i at Mānoa for library preparation and sequencing. Sequencing libraries were prepared using the Nextera XT kit according to the manufacturer’s instructions and sequenced using the Illumina MiSeq V3 platform.
Whole genome sequencing and analysis
The consensus sequences from the reads were generated by the nf-core/viralrecon pipeline (31), using the options for the Illumina amplicon-based library protocol. Consensus genome sequences were submitted to GenBank after validation using VADR SARS-CoV-2 models (32). The genomes were classified into lineages using Pangolin (33) and into clades using Nextclade (34).
Statistical analysis
The two-tailed Mann–Whitney test was used to compare quantitative variables between two groups (GraphPad 6.0).
Results
To examine the feasibility of using anti-N and anti-S ELISAs to distinguish COVID-19-naïve, COVID-19-naïve vaccinees, and COVID-19-recovered cases with or without vaccination in our study which involved S-based vaccines only, we first tested sequential samples from COVID-19-naive participants (n = 27) and participants with RT-PCR-confirmed SARS-CoV-2 NI (n = 4), including pre-vaccination and ∼2 weeks following one or two doses of mRNA (Moderna or Pfizer) vaccine. As expected, neither anti-S nor anti-N antibody was detectable in the naïve group before vaccination, and anti-S but not anti-N antibody was detected following administration of one (26/27 positive) or two (27/27 positive) doses of a mRNA vaccine (Figure 1A). Both anti-S and anti-N antibodies were detected in the NI group and within the NI group after one or two doses of mRNA vaccines (Figure 1B).
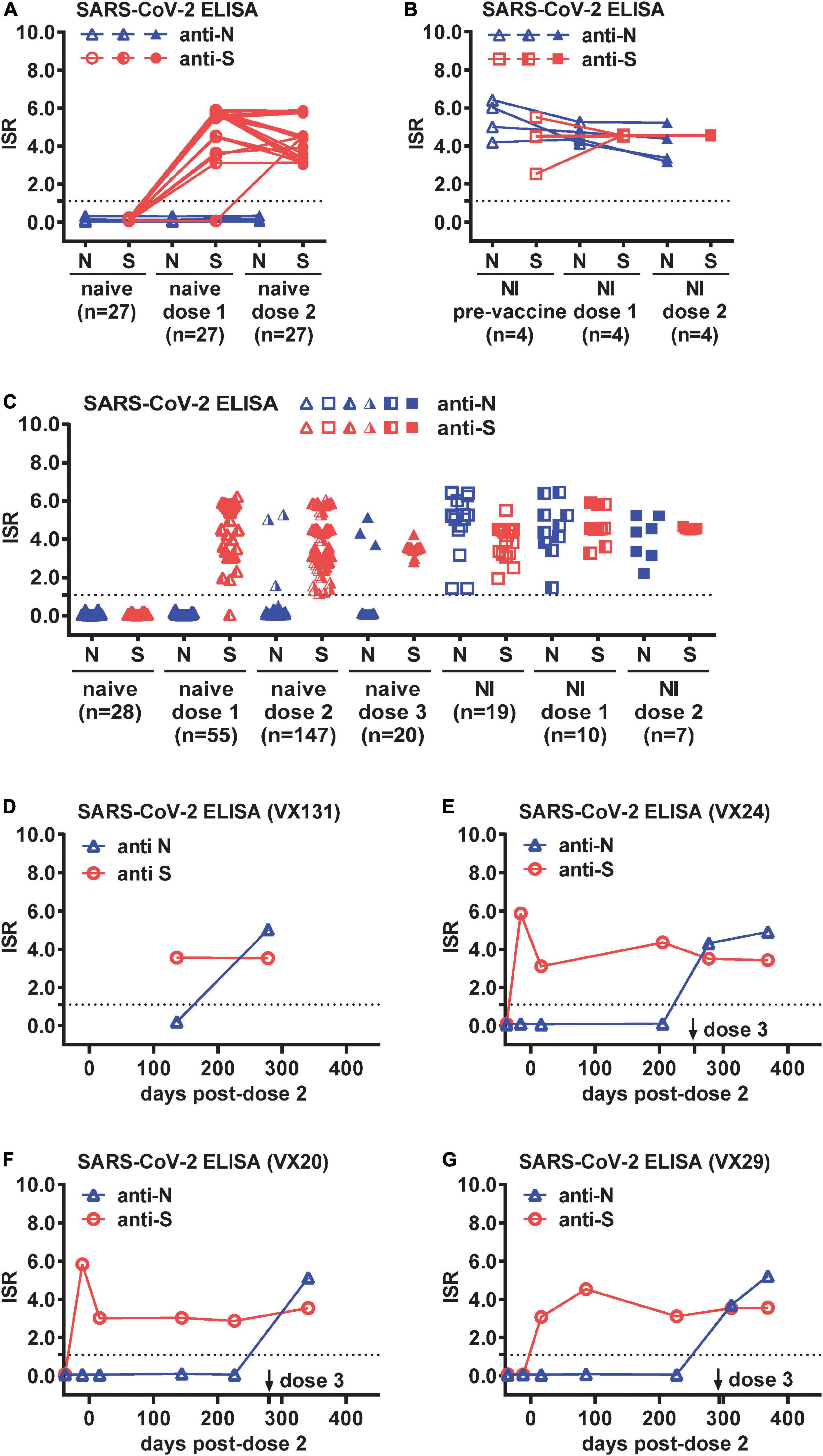
Figure 1. Breakthrough infections (BTIs) identified by anti-N and anti-S ELISAs among COVID-19-naïve vaccinees. Results of anti-N and anti-S ELISAs of sequential plasma samples from (A) COVID-19-naïve (n = 27) and (B) SARS-CoV-2 natural infection (NI) (n = 4) panels before and after one and two doses of a mRNA vaccine (Moderna or Pfizer). (C) Results of anti-N and anti-S ELISAs of single or sequential plasma samples from COVID-19-naïve participants following one (n = 55), two (n = 147), or three (n = 20) doses of a mRNA vaccine, and controls from COVID-19-naïve (n = 28), NI (n = 19), and NI followed by one (n = 10) or two (n = 7) doses of a mRNA vaccine. (D–G) Results of anti-N and anti-S ELISAs of sequential plasma samples from four COVID-19-naïve participants (VX131, VX24, VX20, VX29) with a BTI identified by anti-N antibody seroconversion following two or three doses of Moderna vaccine. Dotted lines indicate the cut-off of immunological status ratio (ISR) values for ELISAs. Data are the mean of duplicates from one experiment.
We next tested larger panels to identify BTIs, including samples from COVID-19-naïve participants following one (n = 55), two (n = 147), or three (n = 20) doses of a mRNA vaccine, as well as controls from COVID-19-naïve (n = 28), NI (n = 19), and NI followed by one (n = 10) or two (n = 7) doses of a mRNA vaccine (Table 1). The age, gender and sampling days following vaccination in each panel were comparable except that the NI post-dose 1 panel were older compared with the naïve post-dose 1 or 2 panel (p = 0.02, two-tailed Mann–Whitney test) and had shorter sampling days after vaccination compared with the naïve post-dose 1, 2, or 3 panel (p = 0.02, 0.0007, or 0.002, respectively, two-tailed Mann–Whitney test) (Table 1). Of the 222 samples from 85 COVID-19-naïve participants, anti-N antibody was detected in six participants following two or three doses of a mRNA vaccine (Figure 1C). While two of them had no earlier samples available, suggesting a possible BTI, the other four had earlier samples (anti-N antibody negative) to demonstrate anti-N antibody seroconversion, indicating a BTI (Figures 1D-G). Of the four BTIs, one was after the second dose and three after the third dose. The four BTIs occurred between November 2021 and February 2022, which was during the transition from the Delta to Omicron wave of the pandemic.
We further performed pseudovirus neutralization test to assess neutralizing antibodies against the wild type D614G strain and the Alpha, Delta and Omicron VOC before and after BTI for the four participants, and compared with time course including symptoms and RT-PCR results. Participant VX131 received the second dose of Moderna vaccine on 5/5/2021, started to have COVID-19 symptoms >7 months later with an RT-PCR positive test on 12/20/2022; the symptomatic BTI was supported by anti-N antibody seroconversion 2 months later (2/24/2022) and the NT50 titers showed 3.1 to 9.0-fold increase with a >4-fold increase against the Omicron variant (9.0-fold), the predominant VOC at the time (Figure 2A). Participant VX24 received the third dose of Moderna vaccine on 11/2/2021 and had anti-N antibody seroconversion on 11/24/2021 without apparent symptoms, suggesting an asymptomatic BTI, which was supported by a >4-fold increase in NT50 titers against the Delta variant (6.0-fold), the predominant VOC then (Figure 2B). Two and half months later, VX24 developed COVID-19 symptoms with an RT-PCR positive test (2/3/2022), suggesting a new and symptomatic reinfection; the NT50 titers 3 weeks later (2/24/2022) revealed a >4-fold increase against the predominant Omicron variant (5.1-fold) then (Figure 2B). Participant VX20, who received the third dose of Moderna vaccine on 11/27/2021, started having symptoms 1 month later with an RT-PCR positive test (1/4/2022); the symptomatic BTI was supported by anti-N antibody seroconversion 3 weeks later (1/27/2022) and a >4-fold increase in NT50 titers against the predominant Omicron variant at the time. Interestingly, the NT50 titers showed a 32.8 to 70.0-fold increase against the four variants tested (Figure 2C), suggesting stronger booster effect of BTI on neutralization titers compared with the first two cases. Participant VX29, who received the third dose of Moderna vaccine on 12/10/2021, started having symptoms 1 week later with an RT-PCR positive test (12/20/2022); the symptomatic BTI was supported by anti-N antibody seroconversion 9 days later (12/29/2022) and a >4-fold increase in NT50 titers against the predominant Omicron variant. Notably, the NT50 titers revealed a 11.0 to 77.4-fold increase against the four variants tested (Figure 2D). Follow-up NT50 titers 2 months later (2/24/2022) revealed a slightly further increase (1.3 to 1.7-fold) against the four variants.
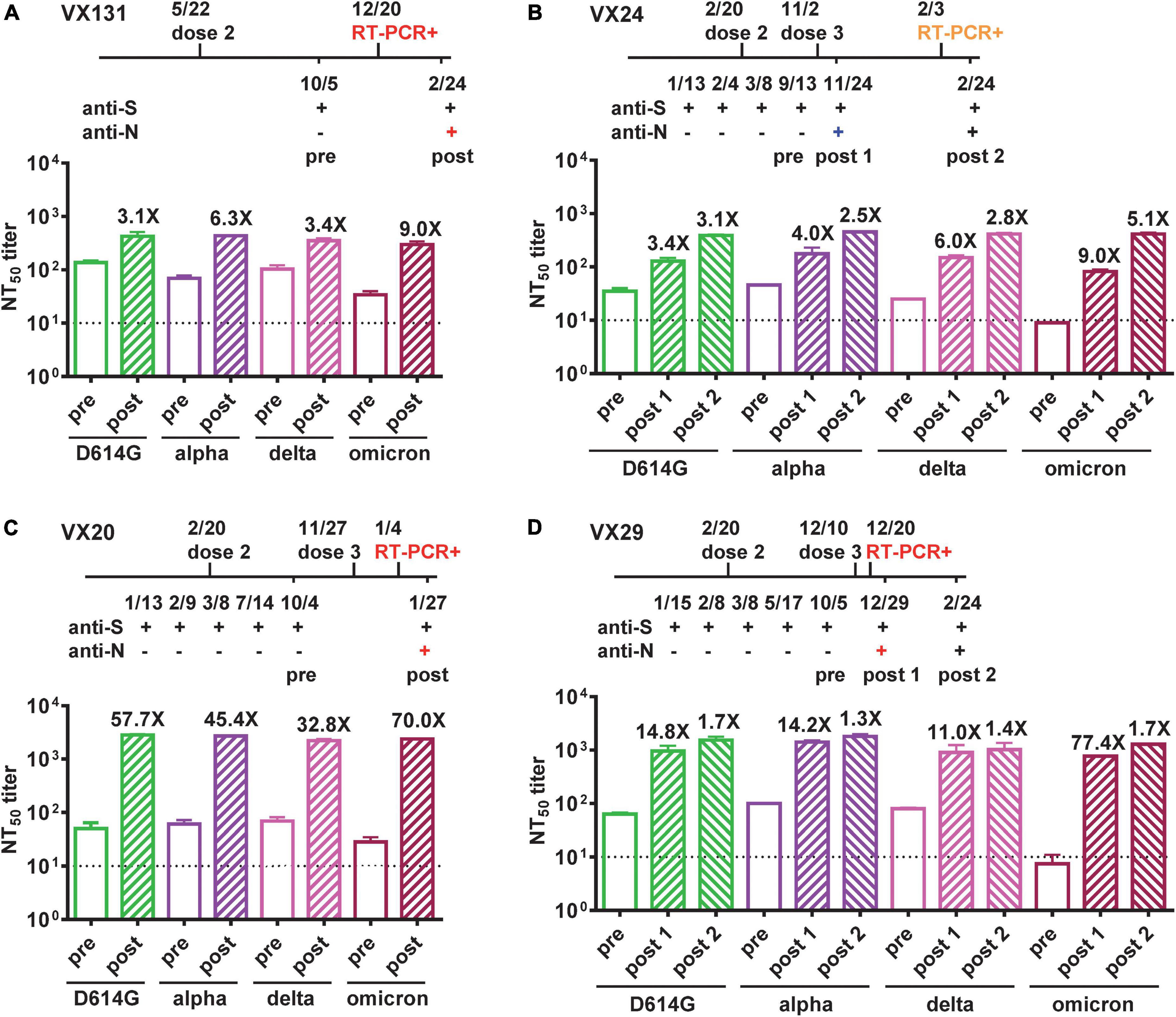
Figure 2. Neutralizing antibodies against VOC before and after BTIs. (A–D) Time course of vaccination, anti-N and anti-S ELISAs, and RT-PCR test of four COVID-19-naïve participants (VX131, VX24, VX20, VX29) who received two or three doses of Moderna vaccine with a BTI identified by anti-N antibody seroconversion. Symptomatic BTIs with positive RT-PCR test and anti-N ELISA are shown in red, asymptomatic BTI with positive anti-N ELISA in blue, and symptomatic reinfection with positive RT-PCR test in orange. NT50 titers against D614G, Alpha, Delta and Omicron variants before (pre, open bars) and after (post, hatched bars) BTIs were determined by pseudovirus neutralization test (28). Number x = fold increase in NT50 titers compared with previous time point. Dotted lines indicate NT50 titer = 10. Data are the means and standard deviations of duplicates from one experiment.
To confirm the variants responsible for BTIs and reinfection, we conducted whole genome sequencing of 2 available RT-PCR positive samples from the 4 BTI cases and one from the reinfection case. The identified variants were BA.1.1.2 (VX-131) and BA.1.1 (VX-29) for BTIs and BA.1.1 (VX-24) for reinfection (Table 2).
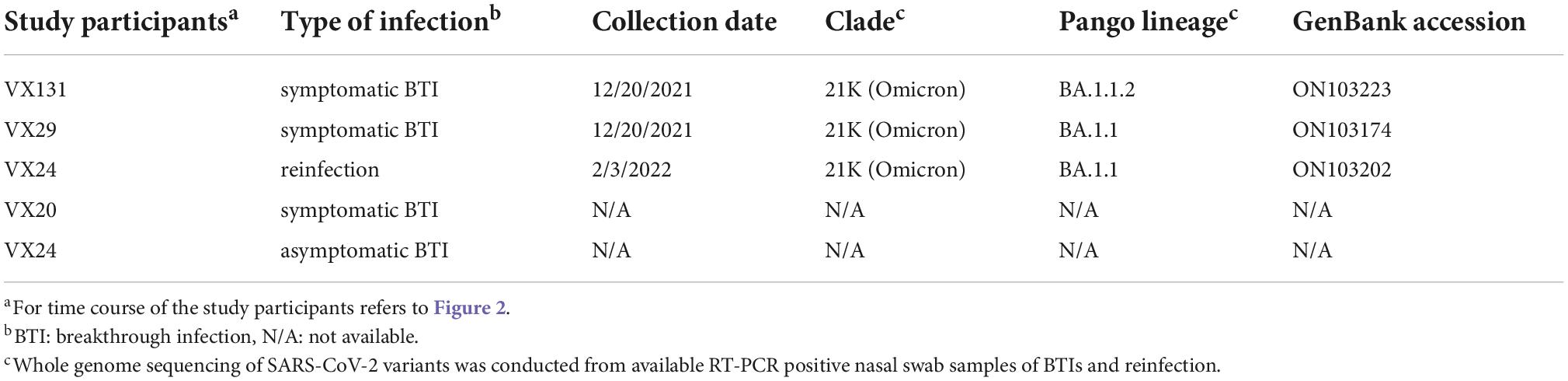
Table 2. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants confirmed by whole genome sequencing.
Discussion
In this study, we report that two simple IgG ELISAs can distinguish COVID-19-naïve (both anti-N and anti-S negative), COVID-19-naïve vaccinees (anti-S positive only), or COVID-19-recovered cases with or without vaccination (both anti-N and anti-S positive) in places where only S-based vaccines, such as mRNA, adenovirus-vectored, and recombinant S protein vaccines are implemented. With available sequential samples, anti-N antibody seroconversion can identify BTI among COVID-19-naïve vaccinees (anti-S positive and anti-N seroconversion).
Using anti-N antibody to identify BTIs, Laing et al. reported a BTI rate of 0.88% for symptomatic infection and 25% for asymptomatic infection in 227 healthcare workers (21). Others reported BTI rates of 0.6 and 0.8% among 4111 and 130 fully vaccinated healthcare workers, respectively (35, 36). Another study reported a BTI rate of 4.6 and 5.3% in patients with immune-mediated inflammatory diseases with (n = 3207) and without immunosuppressants (n = 985), respectively, and 4.0% in 822 healthy controls (37). Our finding of a BTI rate of 4.7% among 85 fully vaccinated non-healthcare workers was similar to the reported rate of healthy controls.
Interestingly, we found that BTIs have variable booster effects on neutralizing antibodies against VOC among the four participants, all young female (21 to 27 years old), healthy without immunocompromised conditions and receiving the Moderna vaccine. Of the four participants, one (VX131) had a BTI after the second dose, three had BTIs after the third dose, either at 1 month (VX20) or <3 weeks (VX24 and VX29) following the third dose. Due to the short time interval between the third dose and BTI, there was no sampling during this window to distinguish between the booster effects of the third dose of mRNA vaccine and BTI. Nonetheless, variable booster effects of BTI plus the third dose were observed; VX24 had a 3.4 to 9.0-fold increase in NT50 titers against the four variants tested, whereas VX20 and VX29 had a 32.8 to 70.0-fold and 11.0 to 77.4-fold increase in NT50 titers, respectively.
Since the variants responsible for BTIs in this study were only confirmed in two out of four by genome sequencing, they were all assessed by the predominant circulating variant and supported by neutralization test, which showed a >4-fold increase in NT50 titers against the predominant variant at the time (Figure 2). It is worth noting that despite previous studies reported the booster effect of BTI on NT50 titers was not specific to the circulating variant, a >4-fold increase in NT50 titers against the variant causing BTI was always observed (23, 38). Consistent with the reported intervals between infection and re-infection, 46 days to 6 months (39), VX24 experienced a symptomatic reinfection during the Omicron wave two and half months after the BTI, which was supported by a 5.1-fold increase in NT50 titers against the Omicron variant (Figure 2B). A recent study reported a Delta variant BTI followed by reinfection with the Omicron variant in a fully vaccinated healthcare worker (40); our study reports a similar case of BTI followed by reinfection in the community, which may reflect what was happening in the real world. Notably, the confirmation of the BA.1.1 and BA.1.1.2 subvariants in two BTIs on 12/20/2021 by whole genome sequencing was in agreement with reports of these subvariants during the initial phase of Omicron wave (2).
With the widespread transmission of the Omicron VOC and its subvariants since late 2021, it is likely that BTIs and reinfections occur commonly as supported by the rapid rise of the seroprevalence of anti-N antibody (41, 42). Given the vaccine-induced and infection-induced immunity together with the attenuated replication and pathogenicity of the Omicron VOC, most BTI and reinfections resulted in asymptomatic infection or mild disease, however, the concern of new VOC with increased virulence remains (43–46). Although an increase in anti-N antibody seroprevalence could compromise the detection of BTIs by anti-N antibody alone, future studies using anti-N antibody test (IgG and/or IgM) in combination with RT-PCR or antigen test could still identify BTIs among COVID-19-naïve vaccinees (anti-N negative during the window period before seroconversion) and distinguish them with reinfection among COVID-19-recovered cases (anti-N positive). Investigation of the extent of BTIs and reinfections in the community would provide new insights into future vaccine and booster strategies to combat the morbidity and mortality caused by next VOC and further our understanding of how herd immunity is built in the post-vaccination era. As BTIs, which has been shown to boost immunity (22–26), continue, an immediate application of our study is to use anti-N antibody test and/or RT-PCR or antigen test to identify BTIs in ongoing COVID-19 vaccine studies aiming to explicit the breadth and durability of vaccine-induced immunity.
There were several limitations. First, the sample size was small. Nonetheless, comparing with other studies using anti-N antibody to identify BTI within the same period as this study, the BTI rate (4.7%) based on 85 participants in our study was similar to the BTI rate (4.0%) based on 822 healthy controls in one study and lower than that (25%) based on 227 healthcare workers in another study (21, 37). Second, anti-N antibody test cannot be used to identify SARS-CoV-2 infection (either NI, BTI or reinfection) in countries where inactivated COVID-19 vaccines such as Covaxin, which contains the N protein, are implemented. Nonetheless, recent studies have shown anti-ORF8 and ORF3b antibodies as new serological markers of SARS-CoV-2 infection in these countries (47, 48). Third, since these BTIs occurred when Omicron, and subvariants BA.1, BA1.1, and BA.2 were circulating, which have been shown to be antigenically equidistant from the wild-type virus and shared similar neutralization profiles, neutralization test was performed against the parental Omicron VOC (B.1.1.529) only but not the BA.4 or BA.5 subvariant (49, 50). Fourth, as the peak of IgG antibody was reported to be at 2 and 3 weeks following the second dose of mRNA vaccine and infection, respectively (12, 51), we measured the NT50 titers within 3 weeks following BTIs, presumably the peak of IgG, in three cases (VX24, VX20, and VX29), and 2 months following BTI in another case (VX131).
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of the University of Hawai’i (2020-00406). The patients/participants provided their written informed consent to participate in this study.
Author contributions
WKW, VRN, YCD, YCL, LLC, ACT, and YQ contributed to study design, experiments, and data analysis. WKW, VRN, and LLC contributed to sample collection and manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants 51R01AI149502-02 (WKW) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), 5P30GM114737-05, 3P30GM114737-05S1, and 3P30GM103466-20S1 from the National Institute of General Medical Sciences, NIH, MedRes-2022-00000789 (WKW) from the Hawai’i Community Foundation, and a contract (CT-MAY-2000282) (VRN) from the City and County of Honolulu. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We thank Eileen Nakano, Renn Silve Salomon, Joanne Yeung, Jolene Siu, and Nicholas Miwa for technical assistance, sample processing, storage and retrieval, and Jennifer Saito at the ASGPB Core, University of Hawai‘i at Mānoa for assistance with whole genome sequencing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.1019490/full#supplementary-material
Footnotes
References
1. Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. (2021) 19:409–24. doi: 10.1038/s41579-021-00573-0
2. World Health Organization [WHO]. Tracking SARS-CoV-2 Variants. (2022). Available online at: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants (accessed August 2, 2022).
3. Centers for Disease Control and Prevention. COVID Data Tracker. (2022). Available online at: https://covid.cdc.gov/covid-data-tracker/#variant-proportions (accessed August 6, 2022).
4. Lambrou AS, Shirk P, Steele MK, Paul P, Paden CR, Cadwell B, et al. Genomic surveillance for SARS-CoV-2 variants: predominance of the delta (B.1.617.2) and Omicron (B.1.1.529) variants — United States, June 2021–January 2022. MMWR Morb Mortal Wkly Rep. (2022) 71:206–11. doi: 10.15585/mmwr.mm7106a4
5. Bergwerk M, Gonen T, Lustig Y, Amit S, Lipsitch M, Cohen C, et al. Covid-19 breakthrough infections in vaccinated health care workers. N Engl J Med. (2021) 385:1474–84. doi: 10.1056/NEJMoa2109072
6. Edara VV, Pinsky BA, Suthar MS, Lai L, Davis-Gardner ME, Floyd K, et al. Infection and vaccine-induced neutralizing-antibody responses to the SARS-CoV-2 B.1.617 variants. N Engl J Med. (2021) 385:664–6. doi: 10.1056/NEJMc2107799
7. Goel RR, Painter MM, Apostolidis SA, Mathew D, Meng W, Rosenfeld AM, et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. (2021) 374:abm0829. doi: 10.1126/science.abm0829
8. Hoffmann M, Arora P, Groß R, Seidel A, Hörnich BF, Hahn AS, et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. (2021) 184:2384–93. doi: 10.1016/j.cell.2021.03.036
9. Liu C, Ginn HM, Dejnirattisai W, Supasa P, Wang B, Tuekprakhon A, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. (2021) 184:4220–36. doi: 10.1016/j.cell.2021.06.020
10. Liu Y, Liu J, Xia H, Zhang X, Fontes-Garfias CR, Swanson KA, et al. Neutralizing activity of BNT162b2-elicited serum. N Engl J Med. (2021) 384:1466–8. doi: 10.1056/NEJMc2102017
11. Mlcochova P, Kemp S, Dhar MS, Papa G, Meng B, Ferreira IATM, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. (2021) 599:114–9.
12. Pegu A, O’Connell SE, Schmidt SD, O’Dell S, Talana CA, Lai L, et al. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science. (2021) 373:1372–7. doi: 10.1101/2021.05.13.444010
13. Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. (2022) 602:671–5. doi: 10.1038/s41586-021-04389-z
14. Rana K, Mohindra R, Pinnaka L. Vaccine breakthrough infections with SARS-CoV-2 variants. N Engl J Med. (2021) 385:e7. doi: 10.1056/NEJMc2107808
15. Rössler A, Riepler L, Bante D, von Laer D, Kimpel J. SARS-CoV-2 Omicron variant neutralization in serum from vaccinated and convalescent persons. N Engl J Med. (2022) 386:698–700. doi: 10.1056/NEJMc2119236
16. Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. (2021) 593:130–5. doi: 10.1038/s41586-021-03398-2
17. Bailly B, Guilpain L, Bouiller K, Chirouze C, N’Debi M, Soulier A, et al. BNT162b2 mRNA vaccination did not prevent an outbreak of SARS COV-2 variant 501Y.V2 in an elderly nursing home but reduced transmission and disease severity. Clin Infect Dis. (2022) 74:517–20. doi: 10.1093/cid/ciab446
18. Kuhlmann C, Mayer CK, Claassen M, Maponga T, Burgers WA, Keeton R, et al. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet. (2022) 399:625–6. doi: 10.1016/S0140-6736(22)00090-3
19. Vanker A, McGeer A, O’Byrne G, Snyder ECR, Salisbury DA, Guthrie JL, et al. Adverse outcomes associated with SARS-CoV-2 variant B.1.351 infection in vaccinated residents of a long term care home, Ontario, Canada. Clin Infect Dis. (2022) 74:751–2. doi: 10.1093/cid/ciab523
20. Williams C, Al-Bargash D, Macalintal C, Stuart R, Seth A, Latham J, et al. COVID-19 outbreak associated with a SARS-CoV-2 P.1 lineage in a long-term care home after implementation of a vaccination program – Ontario, April-May 2021. Clin Infect Dis. (2022) 74:1085–8. doi: 10.1093/cid/ciab617
21. Laing ED, Weiss CD, Samuels EC, Coggins SA, Wang W, Wang R, et al. Durability of antibody response and frequency of SARS-CoV-2 infection 6 months after COVID-19 vaccination in healthcare workers. Emerg Infect Dis. (2022) 28:828–32. doi: 10.3201/eid2804.212037
22. Bates TA, McBride SK, Leier HC, Guzman G, Lyski ZL, Schoen D, et al. Vaccination before or after SARS-CoV-2 infection leads to robust humoral response and antibodies that effectively neutralize variants. Sci Immunol. (2022) 7:eabn8014. doi: 10.1126/sciimmunol.abn8014
23. Lechmere T, Snell LB, Graham C, Seow J, Shalim ZA, Charalampous T, et al. Broad neutralization of SARS-CoV-2 variants, including Omicron, following breakthrough infection with Delta in COVID-19-vaccinated individuals. mBio. (2022) 17:e0379821. doi: 10.1101/2021.12.01.21266982
24. Müller L, Andrée M, Ostermann PN, Jazmati N, Flüh G, Fischer JC, et al. SARS-CoV-2 infection in fully vaccinated individuals of old age strongly boosts the humoral immune response. Front. Med. (2021) 8:746644. doi: 10.3389/fmed.2021.746644
25. Walls AC, Sprouse KR, Bowen JE, Joshi A, Franko N, Navarro MJ, et al. SARS-CoV-2 breakthrough infections elicit potent, broad, and durable neutralizing antibody responses. Cell. (2022) 185:872–80. doi: 10.1016/j.cell.2022.01.011
26. Yadav PD, Sapkal GN, Sahay RR, Patil DY, Deshpande GR, Jain R, et al. Elevated neutralization of Omicron with sera of COVID-19 recovered and breakthrough cases vaccinated with Covaxin than two dose naïve vaccinees. J Infect. (2022) 84:834–72. doi: 10.1016/j.jinf.2022.03.016
27. Ching LL, Tseng AC, Nakano E, Salomon RC, Wang WK, Shikuma C, et al. COVID-19 vaccination and booster induced authentic-virus neutralizing antibody response is superior to SARS-CoV-2 natural infection induced response. J. Clin. Virol. (2022) 152:105185. doi: 10.1016/j.jcv.2022.105185
28. Tsai WY, Ching LC, Hsieh SC, Melish ME, Nerriah VR, Wang WK. A real-time and high-throughput neutralization test based on SARS-CoV-2 pseudovirus containing monomeric infrared fluorescent protein as reporter. Emerg Microbe Infect. (2021) 10:894–905. doi: 10.1080/22221751.2021.1925163
29. V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. (2020) 28:1–16. doi: 10.1038/s41579-020-00468-6
30. Quick J. nCoV-2019 Sequencing Protocol v3 (LoCost) V.3. (2020). Available online at: https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye (accessed August 25, 2020).
31. Patel H, Sarai Varona S, Monzón S, Espinosa-Carrasco J, Heuer ML, Underwood A, et al. nf-core/Viralrecon: nf-core/Viralrecon v2.5 – Manganese Monkey. (2022) Available online at: https://zenodo.org/record/6827984#.Y292unZBzIU (accessed July 13, 2022).
32. Schäffer AA, Hatcher EL, Yankie L, Shonkwiler L, Brister JR, Karsch-Mizrachi I, et al. VADR: validation and annotation of virus sequence submissions to GenBank. BMC Bioinformatics. (2020) 21:211. doi: 10.1186/s12859-020-3537-3
33. O’Toole Á, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. (2021) 7:veab064. doi: 10.1093/ve/veab064
34. Aksamentov I, Roemer C, Hodcroft E, Neher R. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. (2021) 6:3773. doi: 10.21105/joss.03773
35. Allen N, Brady M, Ni Riain U, Conlon N, Domegan L, Carrion Martin AI, et al. Prevalence of antibodies to SARS-CoV-2 following natural infection and vaccination in Irish hospital healthcare workers: changing epidemiology as the pandemic progresses. Front Med. (2022) 8:758118. doi: 10.3389/fmed.2021.758118
36. Wang RC, Murphy CE IV, Kornblith AE, Hohenstein NA, Carter CM, Wong AHK, et al. SARS COV-2 anti-nucleocapsid and anti-spike antibodies in an emergency department healthcare worker cohort: september 2020 – April 2021. Am J Emerg Med. (2022) 54:81–6. doi: 10.1016/j.ajem.2022.01.055
37. Boekel L, Stalman EW, Wieske L, Hooijberg F, van Dam KPJ, Besten YR, et al. Breakthrough SARS-CoV-2 infections with the Delta (B.1.617.2) variant in vaccinated patients with immune-mediated inflammatory diseases using immunosuppressants: a substudy of two prospective cohort studies. Lancet Rheumatol. (2022) 4:e417–29. doi: 10.1016/S2665-9913(22)00102-3
38. Sahay RR, Patil DY, Sapkal GN, Shete AM, Yadav PD. Cases of SARS-CoV-2 reinfection with Omicron BA.2 post breakthrough infection with Delta and Kappa variants. Infect Dis. (2022) 23:1–4. doi: 10.1080/23744235.2022.2114538
39. Cohen JI, Burbelo PD. Reinfection with SARS-CoV-2: implications for vaccines. Clin Infect Dis. (2021) 73:e4223–8. doi: 10.1093/cid/ciaa1866
40. Pandit P, Bhatt P, Sahay RR, Joshi Y, Patil DY, Yadav PD. A case of breakthrough infection with SARS-CoV-2 Delta derivative and reinfection with Omicron variant in a fully vaccinated health care professional. J Infect. (2022) 85:e15–7. doi: 10.1016/j.jinf.2022.04.023
41. Centers for Disease Control and Prevention. Nationwide COVID-19 Infection-Induced Antibody Seroprevalence (Commercial laboratories). Atlanta, GA: US Department of Health and Human Services (2022).
42. Jones JM, Stone M, Sulaeman H, Fink RV, Dave H, Levy ME, et al. Estimated US infection- and vaccine-induced SARS-CoV-2 seroprevalence based on blood donations, July 2020-May 2021. JAMA. (2021) 326:1400–9.
43. Altmann DM, Boyton RJ. COVID-19 vaccination: the road ahead. Science. (2022) 375:1127–32. doi: 10.1126/science.abn1755
44. Dance A. Omicron’s lasting mysteries: four questions scientists are racing to answer. Nature. (2022) 603:22–4. doi: 10.1038/d41586-022-00428-5
45. Halfmann PJ, Iida S, Iwatsuki-Horimoto K, Maemura T, Kiso M, Scheaffer SM, et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. (2022) 603:687–92.
46. Shuai H, Chan JF, Hu B, Chai Y, Yuen TT, Yin F, et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. (2022) 603:693–9. doi: 10.1038/s41586-022-04442-5
47. Hachim A, Kavian N, Cohen CA, Chin AWH, Chu DKW, Mok CKP, et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat Immunol. (2020) 21:1293–301. doi: 10.1038/s41590-020-0773-7
48. Wang X, Lam JY, Wong WM, Yuen CK, Cai JP, Au SW, et al. Accurate diagnosis of COVID-19 by a novel immunogenic secreted SARS-CoV-2 orf8 Protein. mBio. (2020) 11:e02431-20. doi: 10.1128/mBio.02431-20
49. Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. (2022) 608:593–602. doi: 10.1038/s41586-022-04980-y
50. Iketani S, Liu L, Guo Y, Liu L, Chan JF, Huang Y, et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. (2022) 604:553–6. doi: 10.1038/s41586-022-04594-4
51. Lumley SF, Wei J, O’Donnell D, Stoesser NE, Matthews PC, Howarth A, et al. The duration, dynamics, and determinants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody responses in individual healthcare workers. Clin Infect Dis. (2021) 73:e699–709. doi: 10.1101/2020.11.02.20224824
Keywords: severe acute respiratory syndrome coronavirus 2, variants of concern, breakthrough infection, neutralizing antibodies, anti-nucleocapsid antibody, whole genome sequencing
Citation: Dai YC, Lin YC, Ching LL, Tseng AC, Qin Y, Nerurkar VR and Wang WK (2022) Identification of severe acute respiratory syndrome coronavirus 2 breakthrough infections by anti-nucleocapsid antibody among fully vaccinated non-healthcare workers during the transition from the delta to omicron wave. Front. Med. 9:1019490. doi: 10.3389/fmed.2022.1019490
Received: 15 August 2022; Accepted: 11 November 2022;
Published: 29 November 2022.
Edited by:
Sergio E. Rodriguez, Centers for Disease Control and Prevention (CDC), United StatesReviewed by:
Raphael Niklaus Sieber, Statens Serum Institut (SSI), DenmarkVeronica Ueckermann, University of Pretoria, South Africa
Copyright © 2022 Dai, Lin, Ching, Tseng, Qin, Nerurkar and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Kung Wang, d2FuZ3drQGhhd2FpaS5lZHU=
†These authors have contributed equally to this work and share first authorship
 Yu-Ching Dai
Yu-Ching Dai Yen-Chia Lin
Yen-Chia Lin Lauren L. Ching
Lauren L. Ching Alanna C. Tseng
Alanna C. Tseng Yujia Qin3
Yujia Qin3 Vivek R. Nerurkar
Vivek R. Nerurkar Wei-Kung Wang
Wei-Kung Wang