- Wellcome-Wolfson Institute for Experimental Medicine, School of Medicine, Dentistry and Biomedical Sciences, Queen’s University Belfast, Belfast, United Kingdom
Introduction: Acute respiratory distress syndrome (ARDS) is one of the main causes of Intensive Care Unit morbidity and mortality. Metabolic biomarkers of mitochondrial dysfunction are correlated with disease development and high mortality in many respiratory conditions, however it is not known if they can be used to assess risk of mortality in patients with ARDS.
Objectives: The aim of this systematic review was to examine the link between recorded biomarkers of mitochondrial dysfunction in ARDS and mortality.
Methods: A systematic review of CINAHL, EMBASE, MEDLINE, and Cochrane databases was performed. Studies had to include critically ill ARDS patients with reported biomarkers of mitochondrial dysfunction and mortality. Information on the levels of biomarkers reflective of energy metabolism and mitochondrial respiratory function, mitochondrial metabolites, coenzymes, and mitochondrial deoxyribonucleic acid (mtDNA) copy number was recorded. RevMan5.4 was used for meta-analysis. Biomarkers measured in the samples representative of systemic circulation were analyzed separately from the biomarkers measured in the samples representative of lung compartment. Cochrane risk of bias tool and Newcastle-Ottawa scale were used to evaluate publication bias (Prospero protocol: CRD42022288262).
Results: Twenty-five studies were included in the systematic review and nine had raw data available for follow up meta-analysis. Biomarkers of mitochondrial dysfunction included mtDNA, glutathione coupled mediators, lactate, malondialdehyde, mitochondrial genetic defects, oxidative stress associated markers. Biomarkers that were eligible for meta-analysis inclusion were: xanthine, hypoxanthine, acetone, N-pentane, isoprene and mtDNA. Levels of mitochondrial biomarkers were significantly higher in ARDS than in non-ARDS controls (P = 0.0008) in the blood-based samples, whereas in the BAL the difference did not reach statistical significance (P = 0.14). mtDNA was the most frequently measured biomarker, its levels in the blood-based samples were significantly higher in ARDS compared to non-ARDS controls (P = 0.04). Difference between mtDNA levels in ARDS non-survivors compared to ARDS survivors did not reach statistical significance (P = 0.05).
Conclusion: Increased levels of biomarkers of mitochondrial dysfunction in the blood-based samples are positively associated with ARDS. Circulating mtDNA is the most frequently measured biomarker of mitochondrial dysfunction, with significantly elevated levels in ARDS patients compared to non-ARDS controls. Its potential to predict risk of ARDS mortality requires further investigation.
Systematic review registration: [https://www.crd.york.ac.uk/prospero], identifier [CRD42022288262].
Introduction
Acute Respiratory Distress Syndrome is a principle cause of respiratory failure in critically ill patients requiring mechanical ventilation, characterized by severe pulmonary inflammation, diffuse alveolar damage and pulmonary edema (1). Due to a lack of effective treatment, ARDS results in substantial mortality of up to 30–40% (2). In addition to this, the current COVID-19 pandemic reports ARDS as one of the leading causes of ICU mortality, presenting an urgent need for advancement in ARDS research (3). ARDS pathogenesis remains nebulous; consequently, pharmacological therapies that reduce the severity of lung injury in preclinical models have not yet been translated into effective clinical treatment options. Therefore, further research into the mechanisms of ARDS pathogenesis and translational therapies is imperative.
Mitochondria are complex para-symbiotic organelles that perform a myriad of diverse yet interconnected functions, producing ATP and biosynthetic intermediates while also contributing to cellular stress responses such as autophagy and apoptosis (4). Acute inflammation can alter various mitochondrial functions, including reduced levels oxidative phosphorylation, and thus ATP production, increased mtROS production, increased apoptosis, as well as altered mitochondrial biogenesis and mitophagy (5). Dysfunctional mitochondria release multiple forms of damage-associated molecular patterns (DAMPs), such as ATP and mtDNA (6, 7). Similar to pathogenic stimuli, mitochondrial DAMPs can activate innate immunoreceptors, thus contributing to a vicious cycle of dysregulated inflammation. Other biochemical markers of mitochondrial dysfunction described in the literature include direct (lactate, pyruvate, lactate-to-pyruvate ratio, ubiquinone, alanine) and indirect markers (creatine kinase (CK), carnitine, aspartate aminotransferase (AST), alanine aminotransferase (ALT) and ammonia) (8). Clinical observational studies demonstrate that biochemical markers of mitochondrial dysfunction are associated with higher mortality and a higher risk of disease development in many respiratory conditions as well as sepsis (6, 9).
This review aims to assess the association between levels of biomarkers of mitochondrial dysfunction in any biological sample with mortality and other physiological and clinical outcomes in critically ill patients with ARDS. This will be carried out by presenting and appraising current research publications using standardized predefined assessable outcome measurements.
Methods
This systematic review was conducted in accordance to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) and Cochrane guidelines. Please see Supplementary Methods 1 for extended explanations of search criterion.
Literature search
The databases CINAHL, EMBASE, MEDLINE, and Cochrane were systematically searched using predefined search terms for headings: Mitochondria, ARDS, and patient; synonyms and analogous terms of these headlines were defined in Table 1. In order to be eligible studies must include adult (18y/o) participants with ARDS in intensive care units (ICU) (critically ill patients). The severity, cause, and duration of ARDS will not be restricted. The definition of ARDS was not a limiting factor. Covid-ARDS was not included in this systematic review due to differences in disease pathophysiology. No other exclusion criteria was applied to patients. Published relevant studies up to a March 5, 2022 were searched. Full search code, database limitation and limits applied for each database search can be found on PROSPERO (CRD42022288262).
Study selection
Following the initial procurement of studies, by McClintock and Mulholland independently, from search databases, articles were retrieved in full text and stored on Endnote software. A 97% similarity in search results was obtained upon comparison of independent searches. Endnote enabled removal of duplicate articles. Those studies initially applicable were reviewed in full and criterion assessed, those failing to meet criterion were omitted.
Data extraction
The primary outcome was to assess the association between levels of biomarkers of mitochondrial dysfunction in clinical samples and ARDS patient mortality. The secondary outcome was to assess the association between levels of biomarkers of mitochondrial dysfunction in clinical samples and; (i) disease development and (ii) aggravation of ARDS disease severity. Alongside outcome data, the following information was also extracted: patient characteristics (age and sex), year of study publication, study design, sample size, characteristics of ARDS, type of biomarker, type of clinical sample, time of sampling, methods of biomarker measurement, concentration levels of the biomarkers.
All study designs were eligible for inclusion in this systematic review. Studies lacking a comparator/control, for example in the instance of retrospective case reports, were not eligible for meta-analysis inclusion. In the case of interventional studies, the data were extracted from the non-interventional/control arm of the study; this was to ensure that the mitochondrial biomarkers were not confounded by the intervention carried out.
Where possible biomarker concentration data for meta-analysis was collected in the form of mean, standardized mean difference (SMD) and “N” study participant. In the cases where median with interquartile range (IQR) were the only data provided, they were converted to Mean ± SD using the range rule (the standard deviation of a sample is approximately equal to one-fourth of the range of the data). Any standard errors provided were converted to standard deviation for consistency. RevMan software 5.4 was used to store and analyze data, as recommend by Cochrane guidelines. The random-effects model using the inverse- variance method was used on this statistical software, as this allowed for studies with the same biomarker lacking the same units to be compared. The I2 statistics was used to analyze between-study heterogeneity, and values higher than 50% was considered as high heterogeneity. P-values less than 0.05 were considered significant.
Quality assessment
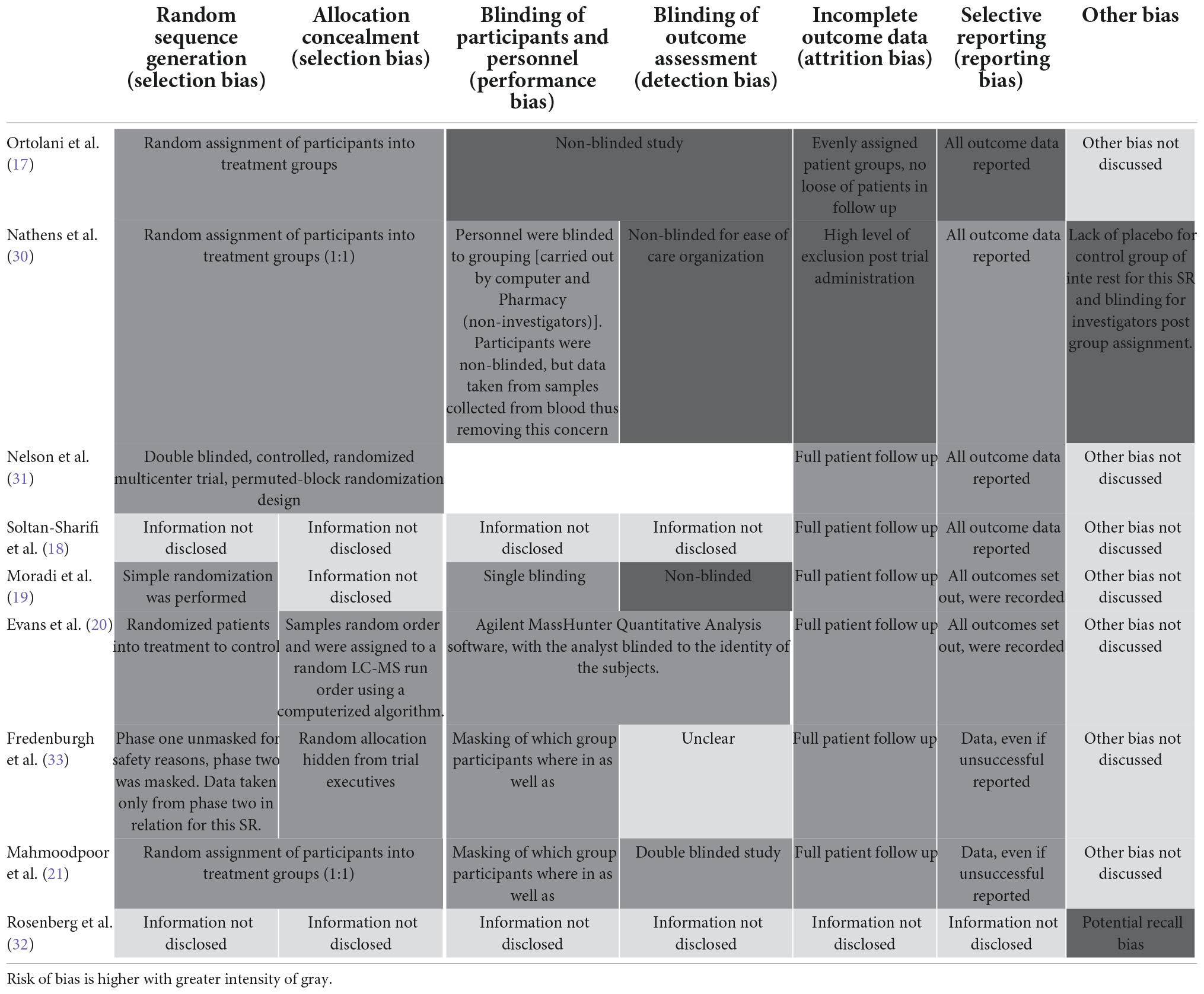
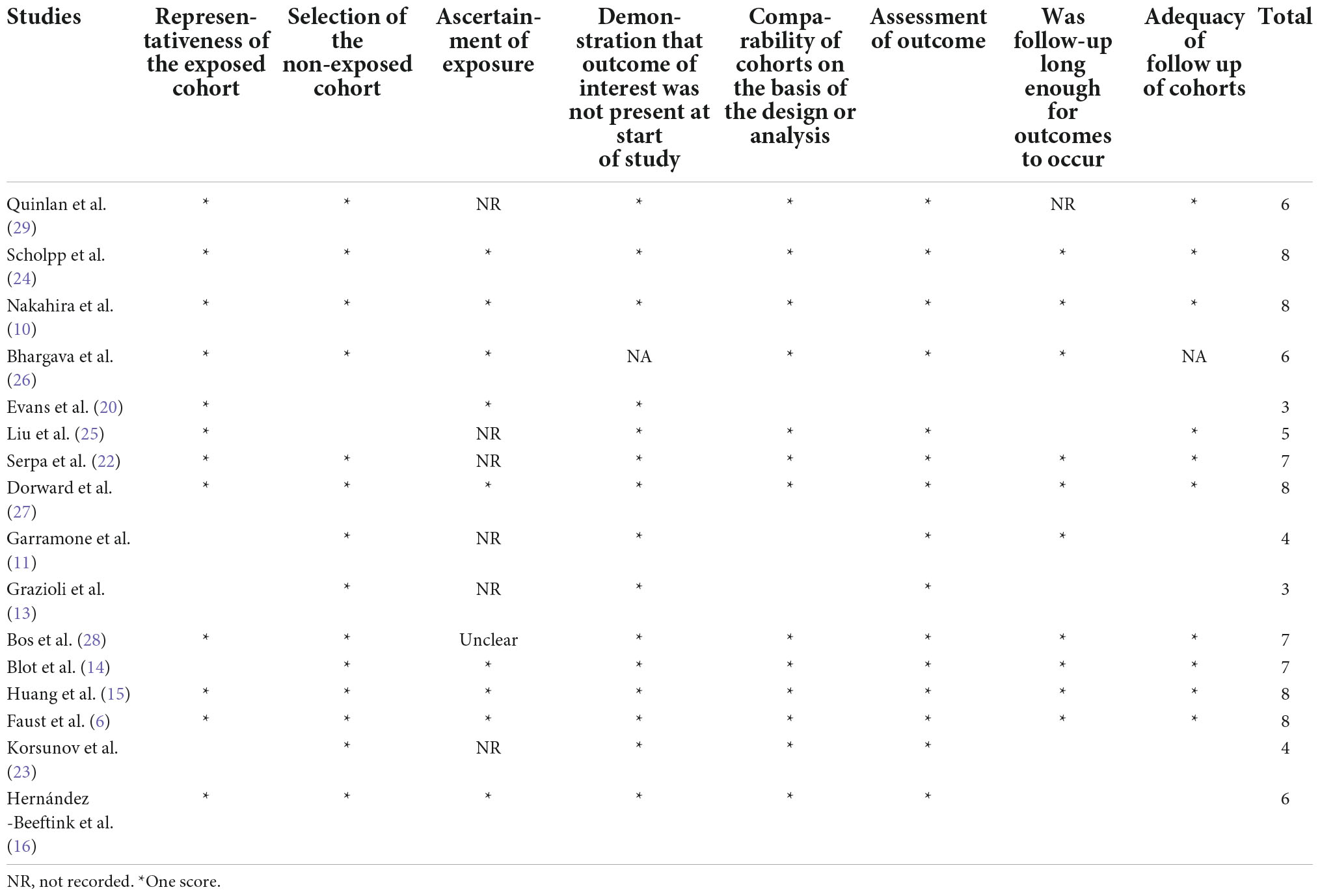
Given the inclusion of all study designs in this systematic review, the risk of bias assessment methods used to assess study quality were chosen based on applicable nature to study design. The study design was confirmed using the SIGN checklist prior to assessmen.1 Randomized Control Trials (RCT) were grouped together and assessed using the Cochrane risk of bias tool2 and non-randomized studies, were assessed using the Newcastle-Ottawa Scale (NOS).3 Studies were considered high quality if the NOS score was more than six points.
Results
Study selection
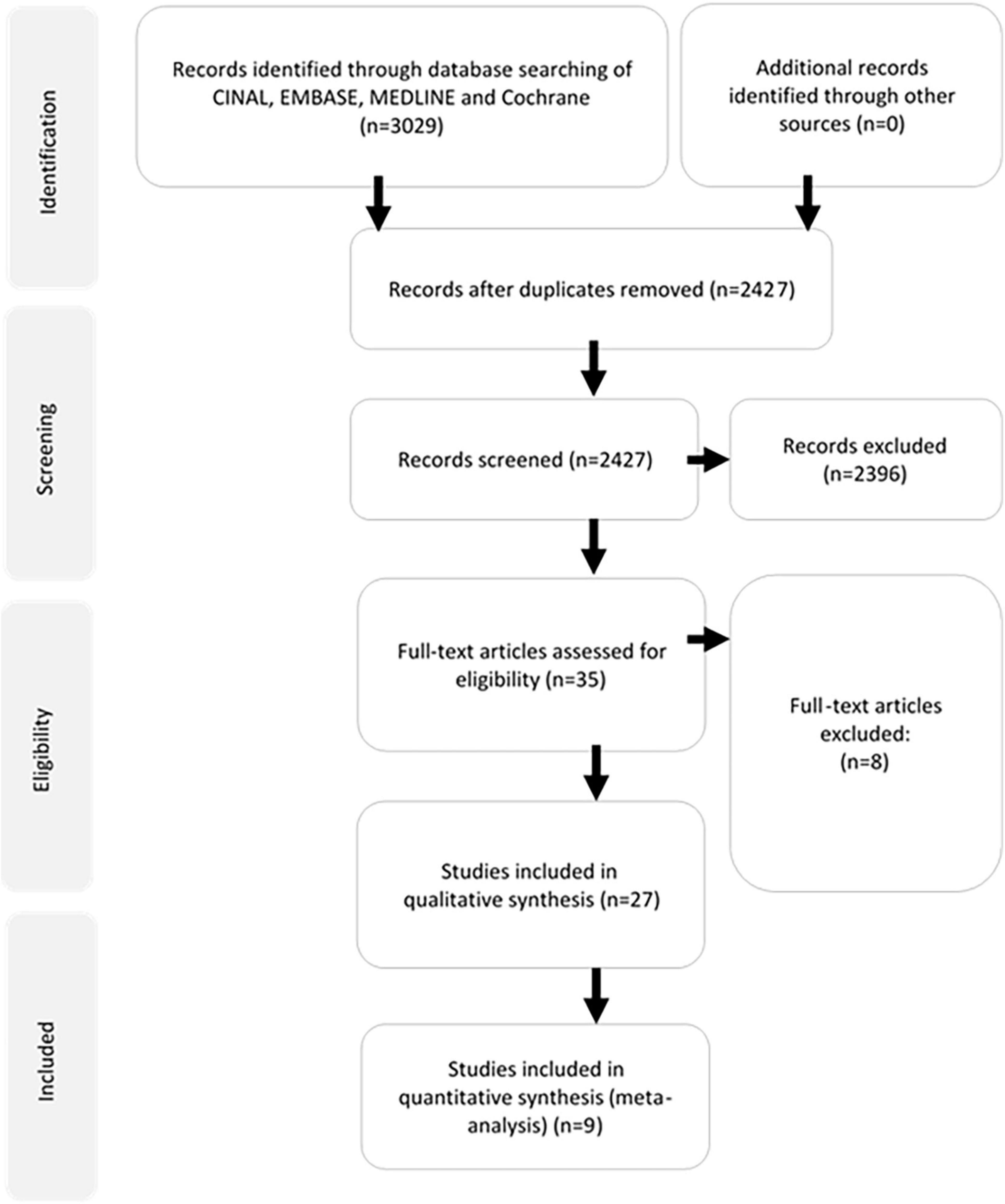
A total of 3,029 articles were identified through search of the four databases. Twenty-six articles were included in the review, nine of which contained sufficient information for meta-analysis. The selection process has been summarized according to the PRISMA guidelines in Figure 1. No potentially relevant papers were excluded from review.
Patient and study characteristics
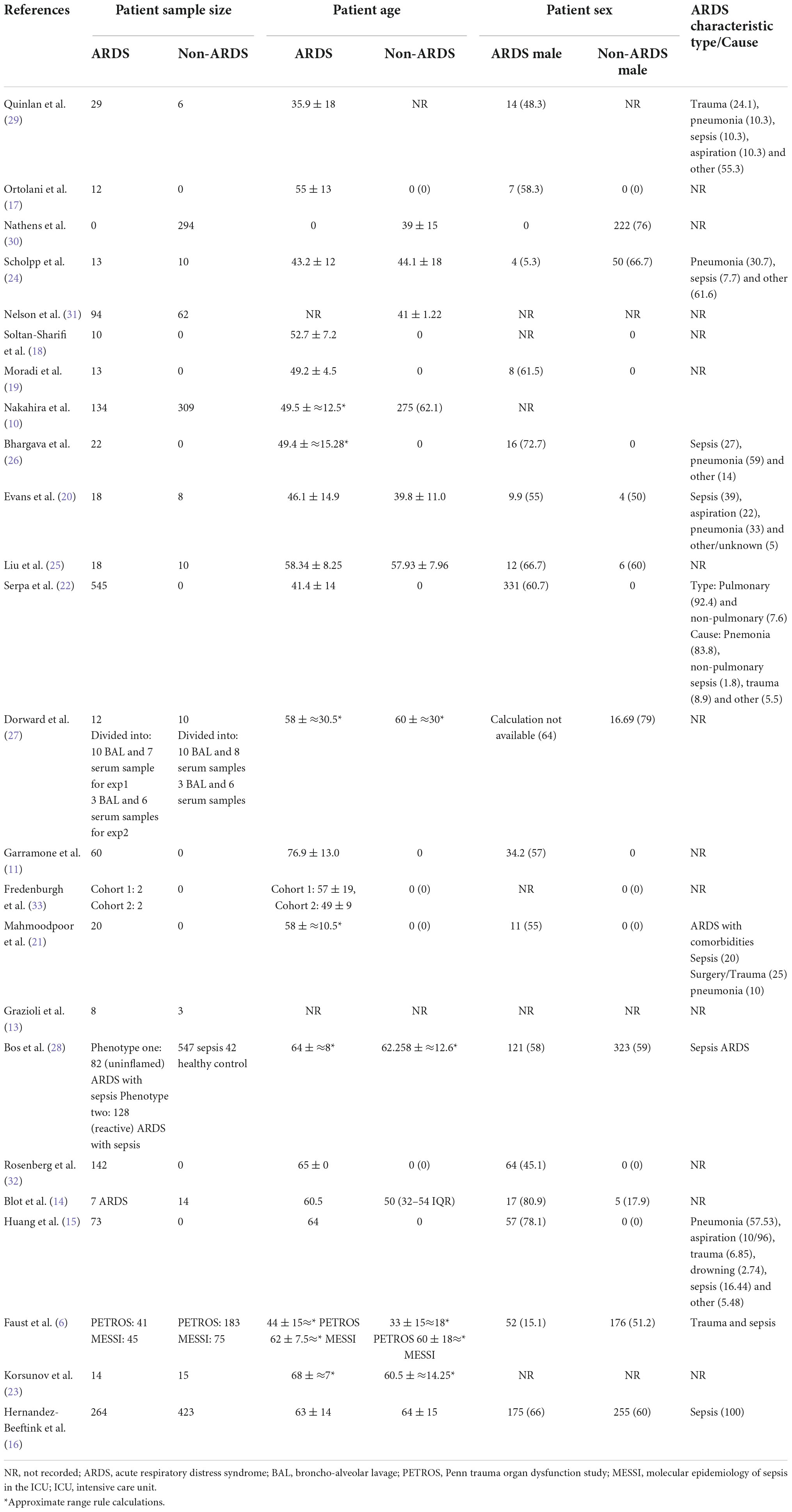
Participant characteristics are summarized in the Table 2. The mean number of ARDS patients across all included articles was 79 and the mean non-ARDS was 148. In the ARDS studies there was a greater number of male participants (53.9% male), than in the non-ARDS (52.8% male). The mean ages of the ARDS participants was 55.4 and non-ARDS 51.3. The type and cause of ARDS varied, in the studies that provided this information, pneumonia and sepsis were the most prevalent causes of ARDS. There was a degree in variation of sample collection time, the majority of samples were collected upon enrollment or day 0 (52%) and the maximal collection time was 35 days. 29 sample sources across 25 studies. Sample sources were: blood (34.4%), plasma (34.4%), BAL (27.6%) and muscle tissue (3.45%).
Biomarkers of mitochondrial dysfunction reported in the included studies were: mitochondrial DNA (mtDNA) (eight studies) (6, 10–16) glutathione coupled mediators [(referenced retrospectively, glutathione, glutanation, glutathione S-transferase (GST), L-gluatamate and glutathione perosidase)] (five studies) (17–21), lactate (three studies) (20, 22, 23), malondialdehyde (MDA) (three studies) (17, 24, 25), metabolic signaling pathways and mediators [(referenced retrospectively: Nicotinamide adenine dinucleotide (NADH), N-terminal peptide FMNPLAQ - also known as NADH2, glyceraldehyde-3-phosphate dehydrogenase-like 6 (GAPDHL6), sirtuin enrichment, xanthine and hypoxanthine)] (four studies) (26–29), oxidative stress associated markers [(hydrogen peroxide (H2O2), super oxidase dismutase (SOD), ascorbate, alpha tocopherol, beta-carotene and retinol] (three studies) (24, 30, 31) (Table 3). Notably, multiple studies reported more than one biomarker, as recorded in Tables 2–5. Methods of sample analysis varied depending on the nature of sample biomarker. Mitochondrial DNA was recurrently analyzed using PCR (based on mitochondrial copy number|), other markers were analyzed using HPLC, ELISA, enzyme immunoassay, mass spectrometry and GeneTitan Affymetrix (Table 3).
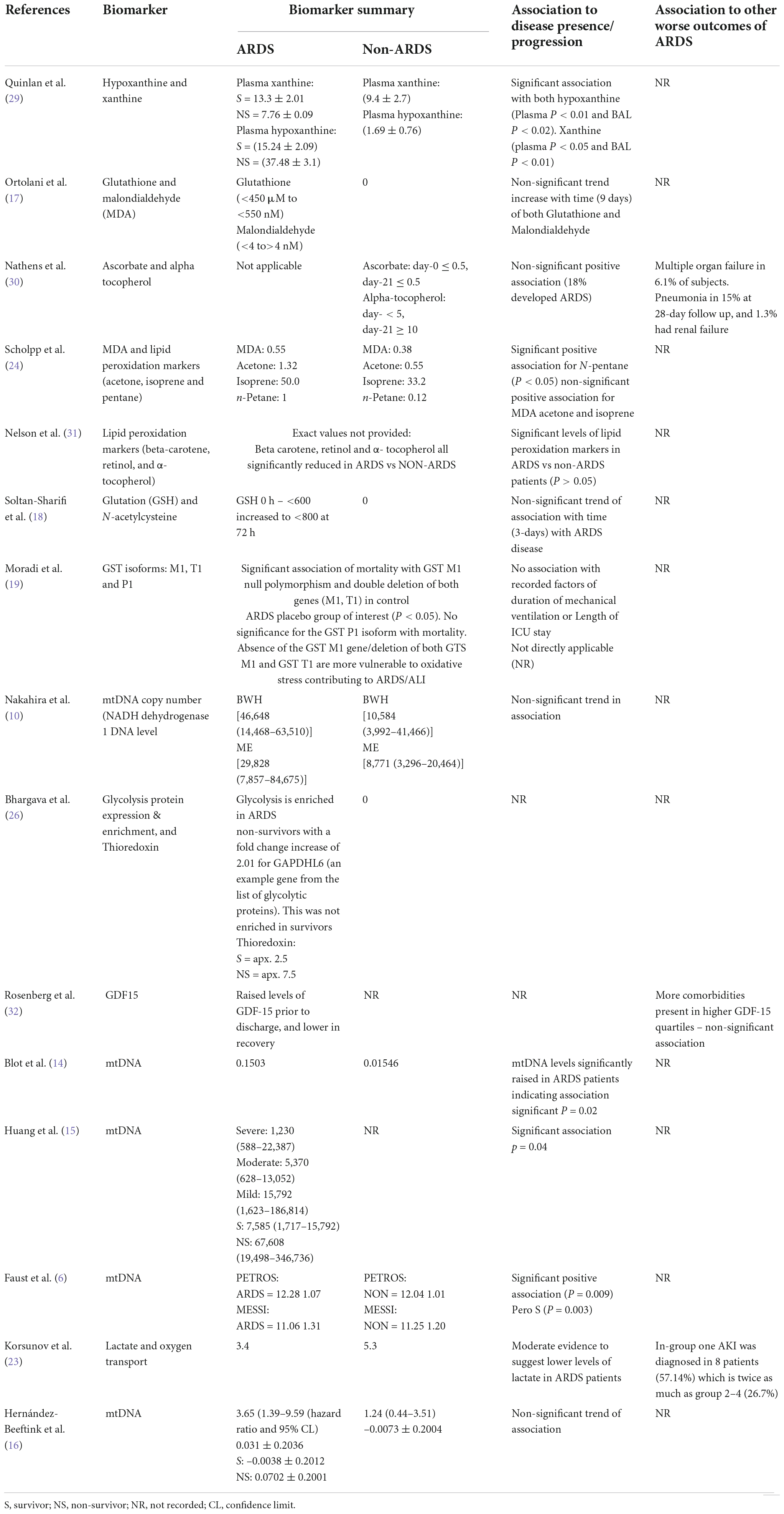
Table 5. Secondary outcome: Association between levels of mitochondrial biomarkers and risks of development of new complications of ARDS or worsening of severity or development of ARDS.
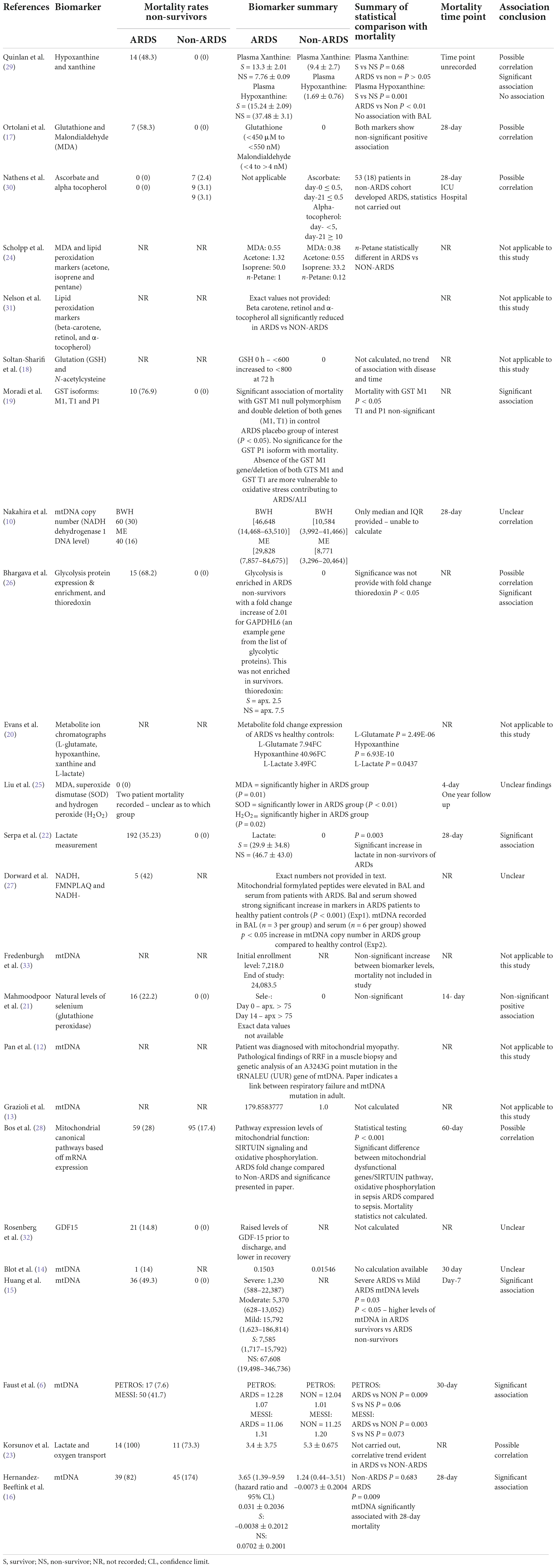
Mortality was recorded in seventeen of the twenty-five studies included in this review (Table 4). Mortality time point recording ranged from 4 to 60-day; however a number of studies failed to record a specific time point of mortality used in the analysis. Studies were examined for significant associations between raised biomarker levels and mortality outcomes, as well as non-significant trends. Due to a number of studies reporting multiple biomarkers, two studies fell into both of these categories (27, 30).
In the 17 studies which had recorded mortality, seven (41%) reported a significant association between elevated levels of mitochondrial biomarkers and higher mortality of ARDS patients (6, 15, 16, 19, 22, 27, 30). From these studies the significantly associated biomarkers were: hypoxanthine (30), GST isoform M1 (19), Thioredoxin (27), lactate (22) and mtDNA (6, 14–16). Due to the lack of provided numerical information only three studies were eligible for inclusion into the meta-analysis for association with mortality. Mitochondrial DNA was the only biomarker reported with significant association in more than one study.
Seven studies (41%) reported numerical trends toward association of higher levels of mitochondrial biomarkers with higher mortality, however the association did not reach statistical significance, or insufficient numerical information was provided (17, 23, 27, 29–32). Positively but non-significantly associated with mortality biomarkers consisted of: xanthine (30), glutathione, MDA (17), ascorbate, alpha-tocopherol (31), GAPDHL6 (27), glutathione peroxidase (selenium) (21), mediators of sirtuin signaling pathway, mediators of oxidative phosphorylation (29) and lactate (23).
The five (29%) remaining study biomarkers, from the 17 eligible for mortality association assessment, did not show any significant, nor general trend with biomarker levels and mortality (10, 14, 25, 27, 32). Of these five studies, two reported mortality of ARDS and non-ARDS groups together (10, 25), and two did not report mortality in non-ARDS group (14, 27), due to inability to draw comparisons between ARDS and non-ARDS cohorts for these four studies, no conclusions of association could be drawn. The remaining study has unclear findings. In Rosenberg et al., raised levels of GDF-19 were found in patients prior to hospital discharge, before declining in recovery. Whilst patient mortality was recorded, due to the temporary nature of increased biomarker levels no conclusive association can be drawn (33).
Development of other adverse clinical outcomes was recorded in five studies. These clinical outcomes included: multiple organ failure, renal failure, pulmonary fibrosis, atrial thrombus, hypotension and acute kidney injury. No statistical correlations of measured levels of mitochondrial biomarkers and development of other adverse clinical outcomes were performed (Table 5).
20 out of 25 studies assessed association of levels of mitochondrial biomarkers and risks of ARDS development or progression (Table 5). Eight studies (40%) reported a significant association between higher levels of mitochondrial biomarkers and the risk of developing ARDS or worsening of ARDS severity (6, 14, 15, 24, 27–29, 31). The biomarkers that indicated significant correlation with ARDS progression include; xanthine, hypoxanthine (29), N-pentane (24). Lipid peroxidation markers (31), NADH, NADH2 (27), sirtuin, mediators of oxidative phosphorylation (27), and mtDNA (6, 14, 15). Nine studies (45%) reported non-significant trend toward association between biomarker levels and risk of development or progression of ARDS (12, 13, 16–18, 20, 24, 30, 33). The biomarker are as follows: MDA (17, 24), glutathione (17), ascorbate, alpha-tocopherol (30), GSH, N-acetylcysteine (18), metabolite ion chromatograph (20), and mtDNA (12, 13, 16, 33). One study showed an opposing finding, with decreased biomarker levels in association with ARDS disease progression, reporting higher lactate levels in non-ARDS compare to ARDS (23).
Meta-analysis
Ten publications reported mean, standard deviation, and “n” number for inclusion in the meta-analysis. First, we compared the blood, plasma, broncho-alveolar lavage fluid (BAL), and lung epithelial lining fluid (ELF) levels of mitochondrial biomarkers between ARDS and non-ARDS subjects. To reflect the biological differences between biomarkers measured in the systemic circulation vs. lung compartment, peripheral blood, arterial blood and plasma were combined for comparison under the category “blood based biomarkers” and biomarkers measured in the BAL or ELF were combined under the category “BAL based biomarkers”. Eight out of ten studies were eligible for this comparison (6, 10, 13, 14, 16, 23, 24, 29). Several studies provided information on multiple biomarkers, Faust et al., and Nakahira et al., reported data from two cohorts, the data on different biomarkers and different cohorts were included in meta-analysis separately (Figure 2). Collectively, 609ARDS patients and 1,054 non-ARDS were included in the comparison, of these 743 ARDS and 1,363 non-ARDS samples were blood based and the remainder were BAL. Biomarkers that were eligible for meta-analysis inclusion were: xanthine, hypoxanthine, (29), acetone, N-pentane, isoprene (24), lactate (23) and mtDNA (6, 10, 13, 14, 16). Xanthine and hypoxanthine are mediators involved in mitochondrial redox balance (34), acetone, isoprene and N-pentane are indicators of metabolic changes in association with oxidative stress (35–37) and accumulation of lactate is a metabolic indicator of oxidative phosphorylation impairment (38).
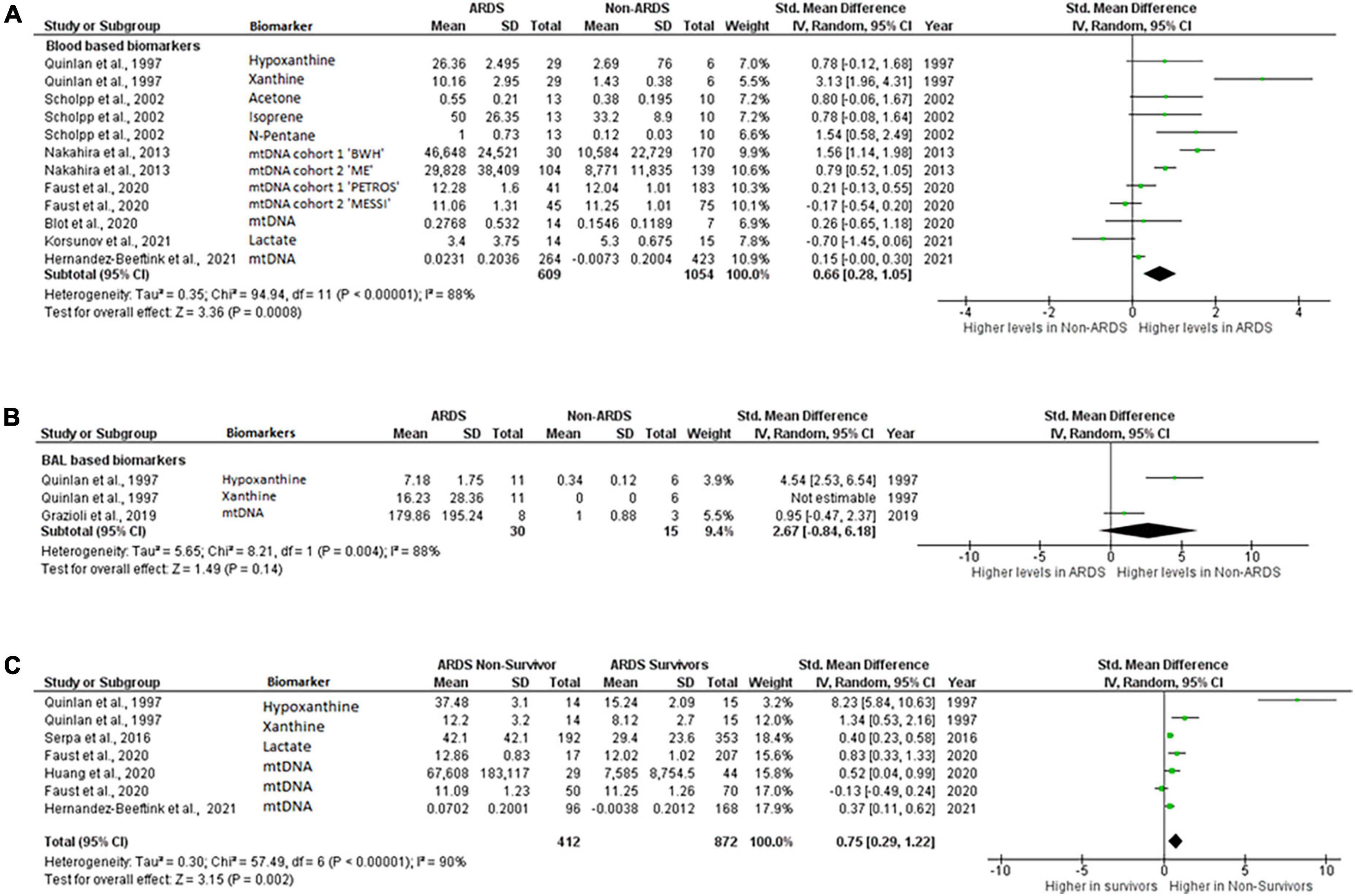
Figure 2. Levels of biomarkers of mitochondrial dysfunction. Forest plot meta-analysis of the levels of biomarkers in ARDS patients and non-ARDS controls in (A) blood samples P = 0.008 and (B) BAL samples P = 0.14 (C) ARDS survivors vs ARDS non-survivors, P = 0.002. Data analysis generated on RevMan 5.4.
Mitochondrial biomarker levels in the blood based samples were significantly higher in ARDS than in non-ARDS controls. Standardized mean difference 0.66 [0.28,1.05], overall effect Z = 3.36, P = 0.0008. Heterogeneity, I2 = 88%, P < 0.00001. I2 values show very large heterogeneity across non-ARDS and ARDS comparisons (Figure 2A).
Difference in the levels of the BAL based mitochondrial biomarkers did not reach statistical significance. Standardized mean difference 2.67 [–0.84,6.18], overall effect Z = 1.49, P = 0.14. Heterogeneity, I2 = 88%, P = 0.004. I2 values also show very large heterogeneity across non-ARDS and ARDS BAL biomarker comparisons (Figure 2B).
Next, levels of blood based mitochondrial biomarkers were compared between survivors and those who died from ARDS. The blood biomarkers eligible for this analysis were, hypoxanthine, xanthine (31), mtDNA (6, 15, 16) and lactate (22). All these biomarkers were measured within 24-h of enrollment. Mortality was recorded at 7- (15), 28- (22) or 30-days (6), Quinlan et al., did not specify the time when mortality was recorded (31) (Table 4). By meta-analysis, levels of biomarkers were significantly higher in non-survivors compared to survivors of ARDS. Standardized mean difference 0.37 [0.11,0.62], overall effect Z = 3.15, P = 0.002. Heterogeneity, I2 = 90%, P < 0.00001. I2 values show very large heterogeneity across non-ARDS and ARDS comparisons (Figure 2C).
Among the studies included in this review, mitochondrial DNA was the most frequently measured biomarker, therefore separate meta-analysis was performed on these studies. Three studies reported levels of mtDNA in plasma, serum or whole blood from ARDS and non-ARDS subjects (6, 10, 16). In the study of Nakahira et al., samples were collected upon enrollment into trial, Faust et al., collected samples upon arrival to emergency department (6, 10) and Hernández-Beeftink et al., recorded collection at 24 h after sepsis diagnosis (16). 484 ARDS patients and 990 non-ARDS patients were included in this comparison. Circulating mitochondrial DNA levels in patients with ARDS were significantly higher than in non-ARDS control groups. 0.50 [0.03,0.98], overall effect Z = 2.07, P = 0.04. Heterogeneity, I2 = 93%, P < 0.00001. Again, I2 values show very large heterogeneity across non-ARDS and ARDS comparisons (Figure 3A). Although Blot et al., and Grazioli et al., reported significant elevation of mtDNA in the BAL samples of ARDS patients compared to healthy controls in small cohorts (5 ARDS vs.3 heathy and 7 ARDS vs. 3 healthy, respectively), numerical information provided in these studies was not sufficient to carry out meta-analysis. Also, Nakahira et al., displayed graphs with significant differences in mtDNA copy numbers between patients with and without ARDS however raw values were not provided and thus could not be included in meta-analysis.
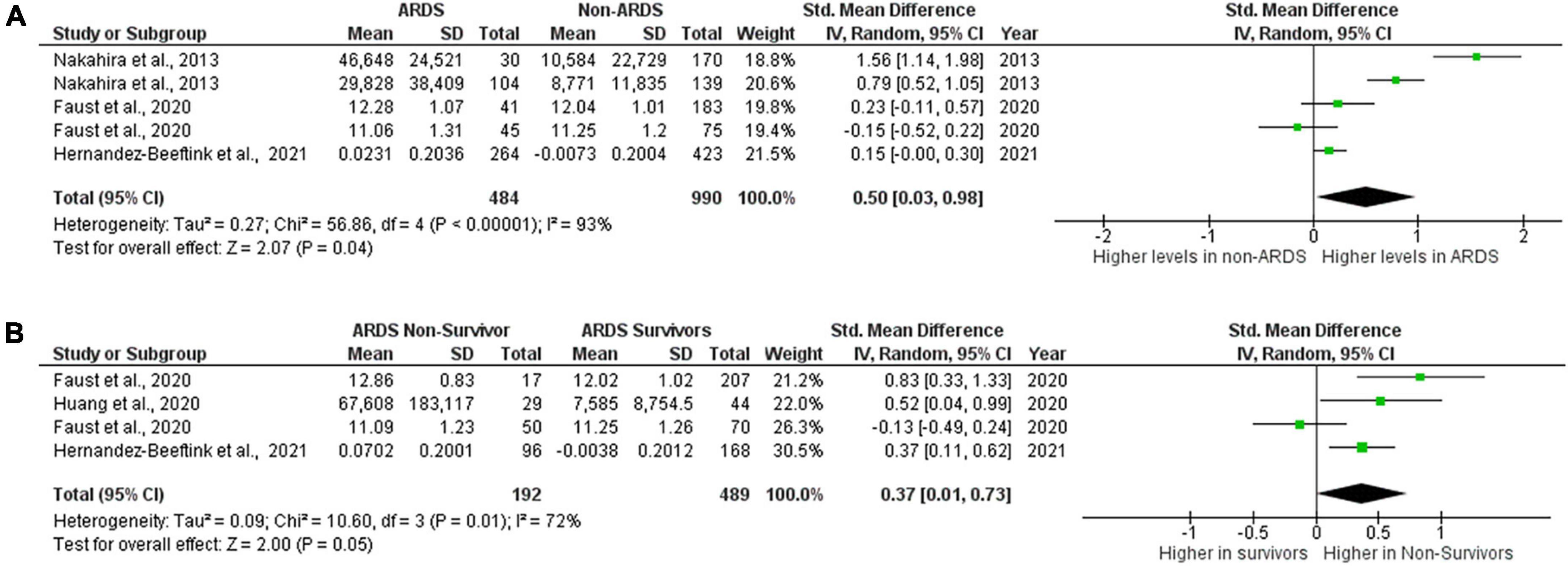
Figure 3. Levels of circulating mtDNA. Forest plot meta-analysis of the levels of circulating mtDNA in ARDS patients and non-ARDS controls, P = 0.04 (A) and in ARDS survivors vs ARDS non- survivors P = 0.05 (B). Data analysis generated on RevMan 5.4.
Three studies also reported data on the levels of circulating mtDNA in ARDS survivors and non-survivors (6, 15, 16). Both Haung et al., and Hernandez-Beeftink et al., collected samples at 24 h or 1 day after presentation (15, 16), Faust et al., collected samples at presentation (Table 3). There were 489 ARDS survivors and 192 ARDS non-survivors included in the comparison. Mortality was recorded at 30 days (Faust et al.), 28 days (Hernandez-Beeftink et al.) and 7 days (Huang et al.) (Table 4). Results of meta-analysis did not allow to draw definitive conclusions about whether or not levels of mtDNA are elevated in non-survivors as overall P value is on the border of significance, although there is a numeric trend toward higher levels in non-survivors. Standardized mean difference 0.37 [0.01,0.73], overall effect Z = 2.00, P = 0.05. Heterogeneity, I2 = 72%, P = 0.01. I2 values show large heterogeneity across ARDS survivor and non-survivor comparisons (Figure 3B).
Publication bias
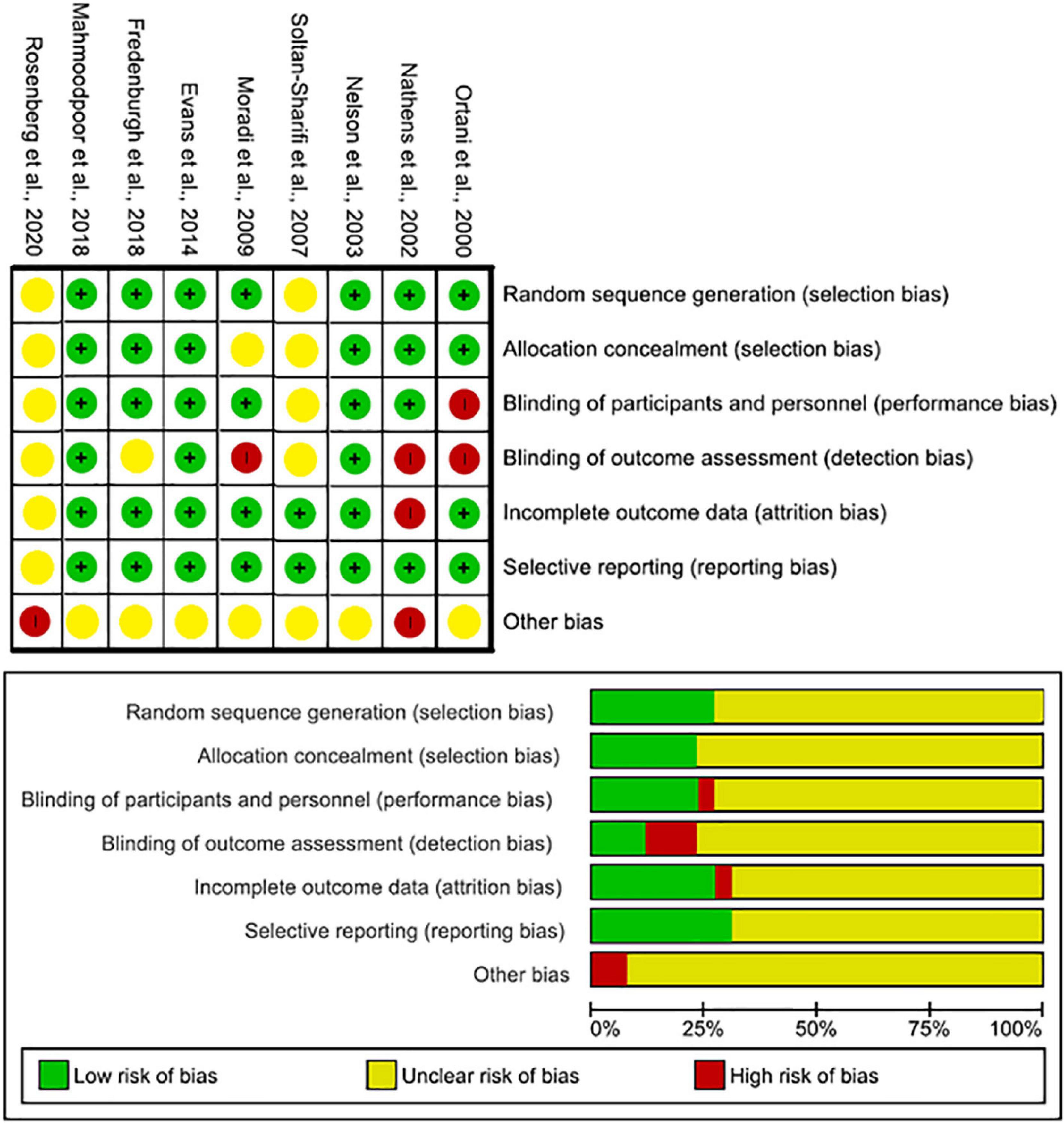
The nine RCT trials were assessed using the Cochrane risk of bias charts. Five studies were classed as low risk, and two as high risks (Figure 4 and Table 6) (17–21, 30–33). The remaining 16, cohort and case–control studies, were assessed by the Newcastle–Ottawa Scale (NOS) (10, 13–16, 20, 22–29). The mean score was 6. 12 out of 16 of studies were considered low risk (Table 7).
Discussion
Clinical and biological markers for prediction of ARDS outcomes are based upon inflammatory indicators, including IL-6, IL-8, RAGE, Ang-2, C-reactive protein and procalcitonin (39, 40). A systematic review by van der Zee et al. examined the multivariate biomarkers associated with ARDS disease, in which RAGE and Ang-2 showed significant association with the risk of ARDS development; yet none were significantly correlated to mortality (40). This likely is contingent on the heterogeneous nature of ARDS pathophysiology.
Pneumonia and sepsis were the top two the most frequent and most devastating causes of ARDS in the studies included in the review. Recently mitochondrial dysfunction, specifically ability of immune cells to switch between glycolytic and oxidative phosphorylation pathways has emerged as a mechanism of pathogenesis of sepsis (41). Patients with sepsis have been shown to have decreased expression of mitochondrial quality mitophagy markers PINK1 and PARKIN, elevated levels of mtDNA, dysfunctional mitochondrial morphology and decreased mitochondrial mass, as well as increased cell death due to calcium overload and raised levels of reactive oxygen species (42–44). The consolidated contribution of mitochondrial dysfunction with the pathogenesis of sepsis, alongside the well-established sepsis induction of ARDS; combined made it plausible to hypothesize that mitochondrial dysfunction might too contribute to ARDS (5, 45).
To our knowledge, this is the first systematic review with meta-analysis investigating association of levels of biomarkers of mitochondrial dysfunction with ARDS. Majority of studies included into this review reported positive trends toward association of elevated levels of biomarkers of mitochondria dysfunction with ARDS. These trends reached statistical significance in the cases of mtDNA, xanthine, hypoxanthine, lactate, isoprene and n-pentane in the blood based samples, however statistically significant difference is absent in BAL samples. Of note, levels of xanthine were not detectable in the BAL of non-ARDS patients which could have impacted the results of meta-analysis.
Importantly, levels of circulating hypoxanthine, xanthine, mtDNA and lactate measured at early time points after presentation were significantly elevated in those who survived ARDS compared to non-survivors, suggesting a potential role of mitochondrial dysfunction in ARDS pathogenesis.
MtDNA was the most frequently measured biomarker across the included studies. Levels of circulating cell-free mtDNA were significantly higher in ARDS patients compared to non-ARDS in six studies. This was further confirmed by meta-analysis of the three studies which have provided necessary raw values for comparison. Difference in mtDNA levels between ARDS survivors and non-survivors did not reach statistical significance by meta-analysis with overall P = 0.05, however there was strong trend toward higher levels in non-survivors. Interestingly, Faust et al., also reported mtDNA levels at 48 h after presentation which were significantly higher in non-survivors than in survivors in both cohorts (6), suggesting that later time points might be more appropriate for measurements of mtDNA as predictor of mortality in ARDS. Taken together, these data indicate an association of mitochondrial dysfunction with ARDS pathophysiology and highlight blood mtDNA as important mediator of ARDS pathogenesis with the potential to serve as a biomarker for predicting the risk of mortality.
This systematic review identified ten different biomarkers of mitochondrial dysfunction measured in ARDS patients. Initial overview of mitochondrial biomarkers in ARDS vs non-ARDS patients showed significantly higher levels in the ARDS patient groups, regardless of the cause of ARDS (Figure 2 and Tables 4, 5). However, 6 of these biomarkers were only measured in one study; the top four most frequently measured biomarkers were (i) mtDNA, (ii) glutathione, (iii) lactate, and (iv) MDA. The study weighting of the meta-analysis was largely driven by blood mtDNA as the most frequently measured biomarker. Therefore, we carried out separate meta-analysis of the studies that reported levels of mtDNA. Plasma mtDNA levels were significantly higher in ARDS vs non-ARDS at time points from 0 to 24 h from presentation. Interestingly, Bolt et al., and Grazioli et al., also reported significant elevation in mtDNA levels in the BAL samples in ARDS patients compared to non-ARDS controls, although the information provided in these studies was not sufficient to run meta-analysis. However, the sample size was small in both studies (7 heathy vs. 7 ARDS and 3 healthy vs. 5ARDS BAL samples, respectively), therefore further studies are required to investigate the significance of alveolar release of mtDNA in ARDS, as a potential biomarker of lung injury.
Mitochondrial DNA levels are currently used as prognostic biomarker in a number of diseases such as Parkinson’s disease and type two diabetes in combination with coronary heart disease (46). Hernandez-Beeftink et al., observed that mtDNA copies in the whole blood were significantly associated with 28-day survival in sepsis patients who developed ARDS (hazard ratio = 3.65, 95% confidence interval = 1.39–9.59, p = 0.009) but not in sepsis patients without ARDS. These findings support the hypothesis that cell free mtDNA copies at sepsis diagnosis could be considered an early prognostic biomarker in sepsis-associated ARDS patients. Results of this review support the potential use of mtDNA as ARDS biomarker; however, more research would be required to determine the most appropriate sample (plasma or whole blood) as well as best time points for sample collection. Additionally, studies recorded mtDNA levels in different units (e.g., copy numbers per μl, μmol, intensities); although the meta-analysis model considers this factor, there is a need for standardization of the measurement units. Circulating mtDNA levels may facilitate the stratification of patients, however, future studies are necessary to standardize the technique and to define more accurate cut-off points.
Glutathione, and its downstream mediators, hypoxanthine and xanthine, follow a similar trend to mtDNA, with elevated levels in ARDS patients (Tables 1, 2). Both hypoxanthine and xanthine are converted to uric acid through xanthine oxidase, resulting in ROS production and leading to oxidative stress (47, 48). In similar fashion, decreased glutathione reduction and increased redox imbalances are known to be associated with mitochondrial disorders (49). It is feasible that one, or both of these mediators could act as prognostic biomarkers for ARDS given the positive trends observed across the two studies (20, 21). Glutathione mediators were recorded in the blood and BAL samples, demonstrating similar trends. To further this avenue, measurement of all three ROS mediators in larger cohorts would be required.
Oxidative damage to lipids, amino acids, and DNA leads to accumulation of malondialdehyde (MDA) (Tables 4, 5). MDA inhibits mitochondrial complex I, II and V, thus impacting the functionality of present mitochondria (50). Due to lack of control comparator for mortality/other worse outcomes, it was not possible to draw any definitive conclusions in regards to MDA. One out of three studies found significantly higher levels in ARDS compared to non-ARDS (17, 51), while the other two studies showed a non-significant trend with higher levels in ARDS (24). MDA was measured in both BAL and plasma; this variation could be driving the lack of conclusive results. Interestingly, studies which investigated the levels of lactate, another metabolite indirectly representative of mitochondrial dysfunction, also reported controversial findings (52). Evans et al., observed a fold-change increase in lactate in ARDS vs non-ARDS, while Serpa et al., demonstrated a higher level of lactate in ARDS non-survivors. On the contrary, Korsunov et al., reported higher levels of lactate in non-ARDS vs ARDS. No data comparisons around lactate were significant across the three studies (20, 22, 23). As there is no consistency across all three studies, it is plausible that metabolites such as MDA and lactate are not useful as prospective biomarkers for ARDS clinical outcomes (53).
One study examined a biomarker associated to mitochondria genetic defects; GDF-15 (Tables 4, 5). GDF-15 is a secretory protein induced by mitochondrial stress, overexpressed in patients with mitochondrial point mutation syndromes (54, 55). The outcomes of this biomarker, as described in Tables 4, 5, do indicate a possible link of association of mitochondrial dysfunction with ARDS, however the lack of raw numbers and limited size of the cohort did not allow for definitive conclusions; current evidence would not support the use of this genetic biomarker in ARDS.
The quality assessment imply a minimal risk of bias across the board of studies included. Main findings were drawn from meta-analysis, of which only one study, Korsunov et al., presented with NOS score of four, due to lack of provided information.
Limitations
This review had several limitations. First, the lack of global representation across study cohorts could influence the predictive power of the examined biomarkers. Secondly, a large variance in study size resulted in high I2 values across all meta-analysis carried out; some of the smaller studies included less than 100 patients could be underpowered for significance calculations. Finally, regardless of standardized mean difference calculation weighting of studies, the inconsistency in biomarker units, as well as different methods of analysis of the same type of biomarkers could affect the statistical conclusions drawn from this small-scale meta-analysis.
Conclusion
This systematic review and meta-analysis suggest that increased levels of biomarkers of mitochondrial dysfunction are positively associated with ARDS. Blood-based biomarkers were the most appropriate for assessment of mitochondrial dysfunction. Circulating mtDNA is the most frequently measured biomarker of mitochondrial dysfunction; circulating mtDNA levels are significantly higher in ARDS patients compared to non-ARDS controls. Mitochondrial DNA is a plausible biomarker candidate for further investigation of its role in ARDS pathogenesis. Further research is required to explore the role of mitochondrial biomarkers in greater populations of ARDS patients and between ARDS subphenotypes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
AK designed the study, revised and independently checked the manuscript. CM and NM conducted the systematic searches and data extraction. CM merged the studies and wrote the manuscript. All authors approved the submitted version.
Funding
AK was supported by UKRI Medical Research Council Research (MR/R025096/1 and MR/S009426/1). CM was supported by Department for Economy Ph.D. studentship.
Acknowledgments
We are very grateful for Professor Danny McAuley and Dr. Bronwen Connolly for their guidance and professional advice during preparation of this systematic review.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.1011819/full#supplementary-material
Footnotes
- ^ https://www.sign.ac.uk/assets/study_design.pdf
- ^ https://methods.cochrane.org/bias/resources/rob-2-revised-cochrane-risk-bias-tool-randomized-trials
- ^ http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp
References
2. Matthay M, Zemans R, Zimmerman G, Arabi Y, Beitler J, Mercat A, et al. Acute respiratory distress syndrome. Nat Rev Dis Prim. (2018) 5:18. doi: 10.1038/s41572-019-0069-0
3. Fan E, Beitler J, Brochard L, Calfee C, Ferguson N, Slutsky A, et al. COVID-19-associated acute respiratory distress syndrome: is a different approach to management warranted? Lancet Respir Med. (2020) 8:816–21. doi: 10.1016/S2213-2600(20)30304-0
4. Schumacker P, Gillespie M, Nakahira K, Choi A, Crouser E, Piantadosi C, et al. Mitochondria in lung biology and pathology: more than just a powerhouse. Am J Physiol Lung Cell Mol Physiol. (2014) 306:962–74. doi: 10.1152/ajplung.00073.2014
5. Ten V, Ratner V. Mitochondrial bioenergetics and pulmonary dysfunction: current progress and future directions. Paediatr Respir Rev. (2020) 34:37–45. doi: 10.1016/j.prrv.2019.04.001
6. Faust H, Reilly J, Anderson B, Ittner C, Forker C, Zhang P, et al. Plasma mitochondrial DNA levels are associated with ARDS in trauma and sepsis patients. Chest. (2020) 157:67–76. doi: 10.1016/j.chest.2019.09.028
7. Simmons J, Lee Y, Pastukh V, Capley G, Muscat C, Muscat D, et al. Potential contribution of mitochondrial (mt) DNA damage associated molecular patterns (DAMPs) in transfusion products to the development of acute respiratory distress syndrome (ARDS) after multiple transfusions. Physiol Behav. (2017) 176:139–48. doi: 10.1097/TA.0000000000001421
8. Rossignol D, Frye R. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. (2012) 17:290–314. doi: 10.1038/mp.2010.136
9. Cloonan S, Kim K, Esteves P, Trian T, Barnes P. Mitochondrial dysfunction in lung ageing and disease. Eur Respir Rev. (2020) 29:200165.
10. Nakahira K, Kyung S, Rogers A, Gazourian L, Youn S, Massaro A, et al. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. (2013) 10:e1001577; discussione1001577. doi: 10.1371/journal.pmed.1001577
11. Garramone A, Cangemi R, Bresciani E, Carnevale R, Bartimoccia S, Fante E, et al. Early decrease of oxidative stress by non-invasive ventilation in patients with acute respiratory failure. Intern Emerg Med. (2018) 13:183–90. doi: 10.1007/s11739-017-1750-5
12. Pan X, Wang L, Fei G, Dong J, Zhong C, Lu J, et al. Acute respiratory failure is the initial manifestation in the adult-onset A3243G tRNALeu mtDNA mutation: a case report and the literature review. Front Neurol. (2019) 10:780. doi: 10.3389/fneur.2019.00780
13. Grazioli S, Dunn-Siegrist I, Pauchard L, Blot M, Charles P, Pugin J. Mitochondrial alarmins are tissue mediators of ventilator-induced lung injury and ARDS. PLoS One. (2019) 14:e0225468. doi: 10.1371/journal.pone.0225468
14. Blot M, Jacquier M, Aho Glele L, Beltramo G, Nguyen M, Bonniaud P, et al. CXCL10 could drive longer duration of mechanical ventilation during COVID-19 ARDS. Crit Care. (2020) 24:632.
15. Huang L, Chang W, Huang Y, Xu X, Yang Y, Qiu H. Prognostic value of plasma mitochondrial DNA in acute respiratory distress syndrome (ARDS): a single-center observational study. J Thorac Dis. (2020) 12:1320–8. doi: 10.21037/jtd.2020.02.49
16. Hernández-Beeftink T, Guillen-Guio B, Rodríguez-Pérez H, Marcelino-Rodríguez I, Lorenzo-Salazar J, Corrales A, et al. Whole-blood mitochondrial DNA copies are associated with the prognosis of acute respiratory distress syndrome after sepsis. Front Immunol. (2021) 12:737369. doi: 10.3389/fimmu.2021.737369
17. Ortolani O, Conti A, De Gaudio A, Masoni M, Novelli G. Protective effects of N-Acetylcysteine and rutin on lipid peroxidation of the lung epithelium during the adult respiratory distress syndrome. Shock. (2000) 13:14–8. doi: 10.1097/00024382-200013010-00003
18. Soltan-Sharifi M, Mojtahedzadeh M, Najafi A, Khajavi M, Rouini M, Moradi M, et al. Improvement by N-acetylcysteine of acute respiratory distress syndrome through increasing intracellular glutathione, and extracellular thiol molecules and anti-oxidant power: evidence for underlying toxicological mechanisms. Hum Exp Toxicol. (2007) 26:697–703. doi: 10.1177/0960327107083452
19. Moradi M, Mojtahedzadeh M, Mandegari A, Soltan-Sharifi M, Najafi A, Khajavi M, et al. The role of glutathione-S-transferase polymorphisms on clinical outcome of ALI/ARDS patient treated with N-acetylcysteine. Respir Med. (2009) 103:434–41. doi: 10.1016/j.rmed.2008.09.013
20. Evans C, Karnovsky A, Kovach M, Standiford T, Burant C, Stringer K. Untargeted LC-MS metabolomics of bronchoalveolar lavage fluid differentiates acute respiratory distress syndrome from health. J Proteome Res. (2014) 13:640–9. doi: 10.1021/pr4007624
21. Mahmoodpoor A, Hamishehkar H, Shadvar K, Ostadi Z, Sanaie S, Saghaleini S, et al. The effect of intravenous selenium on oxidative stress in critically ill patients with acute respiratory distress syndrome. Immunol Invest. (2019) 48:147–59. doi: 10.1080/08820139.2018.1496098
22. Serpa Neto A, Schmidt M, Azevedo L, Bein T, Brochard L, Beutel G, et al. Associations between ventilator settings during extracorporeal membrane oxygenation for refractory hypoxemia and outcome in patients with acute respiratory distress syndrome: a pooled individual patient data analysis: mechanical ventilation during ECMO. Intens Care Med. (2016) 42:1672–84.
23. Korsunov, V, Georgiyants M, Skoryk V. Central hemodynamics and oxygen transport in patients with acute respiratory distress syndrome caused by Covid-19 and their impact on the course and outcomes of the disease. EUREKA Heal Sci. (2021) 1:3–11.
24. Scholpp J, Schubert J, Miekisch W, Geiger K. Breath markers and soluble lipid peroxidation markers in critically III patients. Clin Chem Lab Med. (2002) 40:587–94. doi: 10.1515/CCLM.2002.101
25. Liu D, Luo G, Luo C, Wang T, Sun G, Hei Z. Changes in the concentrations of mediators of inflammation and oxidative stress in exhaled breath condensate during liver transplantation and their relations with postoperative ARDS. Respir Care. (2015) 60:679–88. doi: 10.4187/respcare.03311
26. Bhargava M, Becker T, Viken K, Jagtap P, Dey S, Steinbach M, et al. Proteomic profiles in acute respiratory distress syndrome differentiates survivors from non-survivors. PLoS One. (2014) 9:e109713. doi: 10.1371/journal.pone.0109713
27. Dorward D, Lucas C, Doherty M, Chapman G, Scholefield E, Conway Morris A, et al. Novel role for endogenous mitochondrial formylated peptide-driven formyl peptide receptor 1 signalling in acute respiratory distress syndrome. Thorax. (2017) 72:928–36. doi: 10.1136/thoraxjnl-2017-210030
28. Bos L, Scicluna B, Ong D, Cremer O, Van Der Poll T, Schultz M. Understanding heterogeneity in biologic phenotypes of acute respiratory distress syndrome by leukocyte expression profiles. Am J Respir Crit Care Med. (2019) 200:42–50.
29. Quinlan G, Lamb N, Tilley R, Evans T, Gutteridge J. Plasma hypoxanthine levels in ARDS: implications for oxidative stress, morbidity, and mortality. Am J Respir Crit Care Med. (1997) 155:479–84. doi: 10.1164/ajrccm.155.2.9032182
30. Nathens A, Neff M, Jurkovich G, Klotz P, Farver K, Ruzinski J, et al. Randomized, prospective trial of antioxidant supplementation in critically III surgical patients. Ann Surg. (2002) 236:814–22. doi: 10.2174/138955707781024526
31. Nelson J, DeMichele S, Pacht E, Wennberg A, Gadek J, Drake J, et al. Effect of enteral feeding with eicosapentaenoic acid, γ-linolenic acid, and antioxidants on antioxidant status in patients with acute respiratory distress syndrome. J Parenter Enter Nutr. (2003) 27:98–104. doi: 10.1177/014860710302700298
32. Rosenberg B, Hirano M, Quinzii C, Colantuoni E, Needham D, Lederer D, et al. Growth differentiation factor-15 as a biomarker of strength and recovery in survivors of acute respiratory failure. Thorax. (2019) 74:1099–101. doi: 10.1136/thoraxjnl-2019-213621
33. Fredenburgh L, Perrella M, Barragan-Bradford D, Hess D, Peters E, Welty-Wolf K, et al. A phase I trial of low-dose inhaled carbon monoxide in sepsis-induced ARDS. JCI Insight. (2018) 3:e124039.
34. Gladden J, Zelickson B, Wei C, Ulasova E, Zheng J, Ahmed M, et al. Novel insights into interactions between mitochondria and xanthine oxidase in acute cardiac volume overload. Free Radic Biol Med. (2011) 51:1975–84. doi: 10.1016/j.freeradbiomed.2011.08.022
35. Tanda N, Hoshikawa Y, Sato T, Takahashi N, Koseki T. Exhaled acetone and isoprene in perioperative lung cancer patients under intensive oral care: possible indicators of inflammatory responses a nd metabolic changes. Biomed Res. (2019) 40:29–36. doi: 10.2220/biomedres.40.29
36. Hall J, Crane F. Disruption of mitochondrial membrane by acetone extraction. Biochim Biophys Acta. (1971) 2:682–6.
37. Akobsson-Borin A, Aberg F, Dallner G. Lipid peroxidation of microsomal and mitochondrial membranes extracted with n-pentane and reconstituted with ubiquinol, dolichol and cholesterol. Biochim Biophys Acta. (1994) 2:159–66. doi: 10.1016/0005-2760(94)90022-1
38. Villar J, Herrán-Monge R, González-Higueras E, Prieto-González M, Ambrós A, Rodríguez-Pérez A, et al. Clinical and biological markers for predicting ARDS and outcome in septic patients. Sci Rep. (2021) 11:22702.
39. Yan J, Rao Q. Biomarkers in the diagnosis and prognostic assessment of acute respiratory distress syndrome. J Transl Intern Med. (2014) 2:160–3.
40. Van Der Zee P, Rietdijk W, Somhorst P, Endeman H, Gommers D. A systematic review of biomarkers multivariately associated with acute respiratory distress syndrome development and mortality. Crit Care. (2020) 24:243. doi: 10.1186/s13054-020-02913-7
41. Rahmel T, Marko B, Nowak H, Bergmann L, Thon P, Rump K, et al. Mitochondrial dysfunction in sepsis is associated with diminished intramitochondrial TFAM despite its increased cellular expression. Sci Rep. (2020) 10:21029. doi: 10.1038/s41598-020-78195-4
42. Knutie S, Gabor C, Kohl K, Rohr J. Mitochondrial DNA in Sepsis. Physiol Behav. (2017) 176:139–48.
43. van der Slikke E, Star B, van Meurs M, Henning R, Moser J, Bouma H. Sepsis is associated with mitochondrial DNA damage and a reduced mitochondrial mass in the kidney of patients with sepsis-AKI. Crit Care. (2021) 25:36. doi: 10.1186/s13054-020-03424-1
44. Preau S, Vodovar D, Jung B, Lancel S, Zafrani L, Flatres A, et al. Energetic dysfunction in sepsis: a narrative review. Ann Intens Care. (2021) 11:104. doi: 10.1186/s13613-021-00893-7
45. Robinson MJ, Krasnodembskaya AD. Therapeutic targeting of metabolic alterations in acute respiratory distress syndrome. Eur Respir Rev. (2020) 29:200114. doi: 10.1183/16000617.0114-2020
46. Lowes H, Pyle A, Santibanez-Koref M, Hudson G. Circulating cell-free mitochondrial DNA levels in Parkinson’s disease are influenced by treatment. Mol Neurodegener. (2020) 15:10. doi: 10.1186/s13024-020-00362-y
47. Kristal B, Vigneau-Callahan K, Moskowitz A, Matson W. Purine catabolism: links to mitochondrial respiration and antioxidant defenses? Arch Biochem Biophys. (1999) 370:22–33.
48. Vergeade A, Mulder P, Vendeville C, Ventura-Clapier R, Thuillez C, Monteil C. Xanthine oxidase contributes to mitochondrial ROS generation in an experimental model of cocaine-induced diastolic dysfunction. J Cardiovasc Pharmacol. (2012) 60:538–43. doi: 10.1097/FJC.0b013e318271223c
49. Enns G, Cowan T. Glutathione as a redox biomarker in mitochondrial disease—implications for therapy. J Clin Med. (2017) 6:50. doi: 10.3390/jcm6050050
50. Long J, Liu C, Sun L, Gao H, Liu J. Neuronal mitochondrial toxicity of malondialdehyde: inhibitory effects on respiratory function and enzyme activities in rat brain mitochondria. Neurochem Res. (2009) 34:786–94. doi: 10.1007/s11064-008-9882-7
51. Liu J, Zou Y, Tang Y, Xi M, Xie L, Zhang Q, et al. Circulating cell-free mitochondrial deoxyribonucleic acid is increased in coronary heart disease patients with diabetes mellitus. J Diabetes Investig. (2016) 7:109–14. doi: 10.1111/jdi.12366
52. Glancy B, Kane D, Kavazis A, Goodwin M, Willis W, Gladden L. Mitochondrial lactate metabolism: history and implications for exercise and disease. J Physiol. (2021) 599:863–88.
53. Metwaly S, Winston B. Systems biology ARDS research with a focus on metabolomics. Metabolites. (2020) 10:207. doi: 10.3390/metabo10050207
54. Ayuso P, Martínez C, Pastor P, Lorenzo-Betancor O, Luengo A, Jiménez-Jiménez F, et al. An association study between heme oxygenase-1 genetic variants and Parkinson’s disease. Front Cell Neurosci. (2014) 8:298. doi: 10.3389/fncel.2014.00298
Keywords: acute respiratory distress syndrome, biomarker, mitochondrial dysfunction, mitochondrial DNA, mortality, systematic review, meta-analysis, ARDS
Citation: McClintock CR, Mulholland N and Krasnodembskaya AD (2022) Biomarkers of mitochondrial dysfunction in acute respiratory distress syndrome: A systematic review and meta-analysis. Front. Med. 9:1011819. doi: 10.3389/fmed.2022.1011819
Received: 04 August 2022; Accepted: 18 November 2022;
Published: 14 December 2022.
Edited by:
Shahd Horie, University of Galway, IrelandReviewed by:
Alejandro Pablo Adam, Albany Medical College, United StatesDaniel O’Toole, University of Galway, Ireland
Copyright © 2022 McClintock, Mulholland and Krasnodembskaya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Catherine R. McClintock, Y21jY2xpbnRvY2swNUBxdWIuYWMudWs=
 Catherine R. McClintock
Catherine R. McClintock Niamh Mulholland
Niamh Mulholland Anna D. Krasnodembskaya
Anna D. Krasnodembskaya