
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Med., 10 October 2022
Sec. Infectious Diseases – Surveillance, Prevention and Treatment
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.1010489
This article is part of the Research TopicEmerging SARS-COV-2 Variants: Genomic Variations, Transmission, Pathogenesis, Clinical Impact and Interventions, Volume IIView all 28 articles
In late December 2019, health authorities reported a cluster of patients with pneumonia of unknown cause that was epidemiologically linked to a seafood market in Wuhan, Hubei Province, China (1). The etiological agent was identified as a novel coronavirus, eventually named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and the respiratory illness was designated as coronavirus disease-2019 (COVID-19) by the World Health Organization (WHO) (2). Since its emergence, the rapid global spread of SARS-CoV-2 has provoked a catastrophic impact in our health and economic systems, causing a devastating pandemic and at the same time testing the resilience of the human population. As of September 5, 2022, more than 604.5 million cases of COVID-19 and 6.4 million deaths have been reported around the world. Most of the cases have been reported by the USA, followed by India, Brazil, and France (3).
The emergence of SARS-CoV-2 has been characterized by the identification of several variants of concern (VOCs) during the pandemic course: alpha (B.1.1.7), beta (B.1.351), gamma (P.1), and delta (B.1.617.2) (4, 5). Over the last 2 years these emerging variants have been associated with an abrupt increase in the number of COVID-19 cases, catalyzing several waves of the pandemic in many countries around the globe (6). More recently, the omicron (B.1.1.529) variant was first reported in Botswana and South Africa at the end of November 2021. After its emergence, this new variant initially named BA.1, spread rapidly across the world and was classified as a VOC by the WHO on 26 November 2021. The BA.1 subvariant rapidly spread around the world and outcompeted other SARS-CoV-2 variants, such as delta variant (7). In a rapidly moving field of study, a cumulative body of findings has demonstrated that the omicron variant is associated with high transmissibility and less severe illness in the human population, has resistance against most therapeutic antibodies, has robust binding to human ACE2 receptor, and may escape from neutralizing antibody responses in both convalescent and vaccinated individuals (8–11).
However, in the past few months, multiple subvariants (BA.1.1, BA.2, BA.2.12.1, BA.3, BA. 4, and BA.5) of the omicron have emerged and raised great concerns to global health (12). These omicron subvariants carry a distinctive constellation of mutations, including several that have been previously determined to be of virological importance to other previous SARS-CoV-2 variants (Figure 1) (13). Among them, BA.2 has recently spread to many countries worldwide (14). Within a short time, research groups around the world have rapidly provided relevant insights about this novel omicron subvariant. Recent progress has shown that the effective reproduction number (R0) of BA.2 was 1.4-fold higher than that of BA.1 subvariant (7). Immunological studies demonstrated that the immunity induced by most COVID-19 vaccines administered to human populations is not effective against BA.2 subvariant (7). Collectively, in vitro and in vivo experiments showed that the BA.2 spike confers high capacity to replicate in human nasal epithelial cells and is more pathogenic than the BA.1, as demonstrated in hamsters (7).
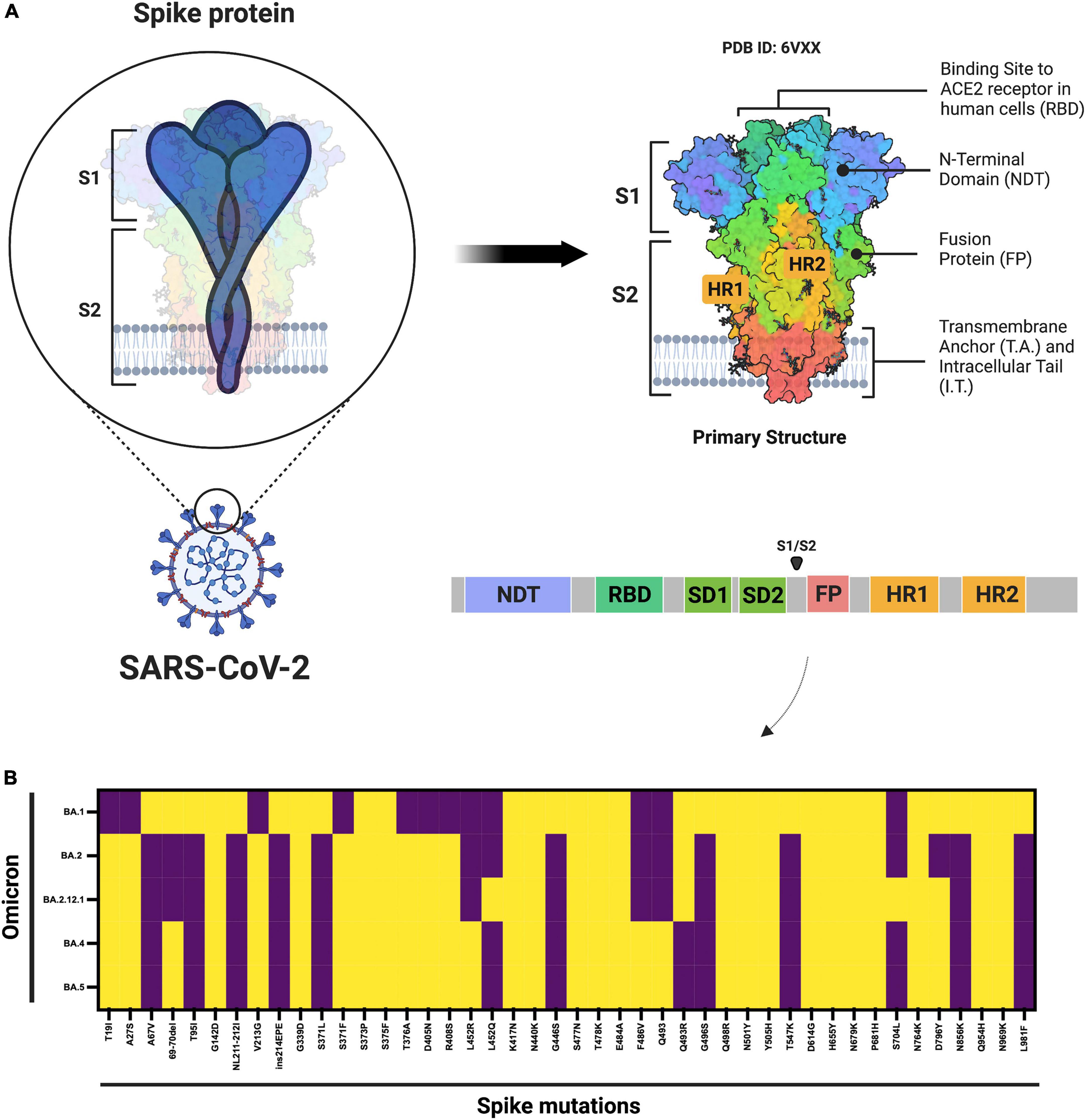
Figure 1. Schematic diagram of the SARS-CoV-2 spike protein and omicron subvariant mutations. (A) Shows the architecture of the SARS-CoV-2 spike protein. (B) Shows the mutations that have been identified in the omicron BA.1, BA.2, BA.2.12.1, and BA.4 or BA.5 subvariants. ACE2, angiotensin–converting enzyme 2; FP, fusion peptide; HR1. heptad repeat 1; HR2, heptad repeat 2; NTD, N-terminal domain; RBD, receptor-binding domain; SD1, subdomain 1; SD2, subdomain 2. Yellow denotes the presence, while purple denotes the absence of the specific mutation. The figure was created with Biorender.com.
As the COVID-19 pandemic progressed, other omicron subvariants are becoming protagonists (Table 1). BA.5 is currently the dominant subvariant in the USA, indicating that this subvariant may have selective advantages when compared to other SARS-CoV-2 omicron variants (15). Moreover, many countries around the world have documented cases associated with the circulation of these novel omicron subvariants.1 A recent report provided relevant immunological insights in terms of neutralizing antibody titers produced against the Wuhan virus along with omicron subvariants (BA.1, BA.2, BA.2.12.1, BA.4 or BA.5). In that study, Hachmann and colleagues conducted a study including 27 individuals who had been vaccinated and boosted with mRNA vaccine (BNT162b2) and 27 individuals who had been infected with the BA.1 or BA.2 subvariants (12). The results revealed that the omicron subvariants (BA.2.12.1, BA.4, and BA.5) substantially escape neutralizing antibodies induced by previous natural infection and vaccination. Interestingly, the immunological data demonstrated that the neutralizing antibody titers against the subvariants (BA.4, BA.5 or BA.2.12.1) was lower than antibody titers against the BA.1 and BA.2 omicron subvariants (12). Taken together, these findings indicate that the SARS-CoV-2 omicron variant has evolved multiple immune evasion strategies to escape from host the immune response for successful viral replication. This is exemplified due to fact that the omicron spike inefficiently utilizes the TMPRSS2 for cell entry via plasma membrane fusion. Instead, the omicron variant demonstrates a greater dependency on cell entry via the endocytic route (16).
The widespread dissemination of the SARS-CoV-2 omicron variant has been a devastating threat to pandemic control, indicating that we need to reconsider several features of the virus that had been previously thought to be established. After approximately 2 years and 5 months since the beginning of the pandemic, the emergence of new omicron subvariants introduces uncertainty about the end of the COVID-19 pandemic, at least for now. Although vaccine deployment has contributed to the reduction in the number of hospitalizations and deaths, many countries worldwide have experienced an abrupt increase in the number of COVID-19 cases in the past few months, catalyzing a new wave of the COVID-19 pandemic.
With this in mind, there are some other factors that the healthcare authorities can consider to declare the end of the COVID-19 pandemic: (i) disease severity and mortality due to new SARS-CoV-2 variants and they differ in vaccinated and non-vaccinated individuals. To address this question, recent findings suggested that alpha, beta, gamma, and delta SARS-CoV-2 variants are more serious than the Wuhan virus in terms of hospitalization, intensive care unit (ICU) admission, and mortality (17). In patients with the omicron VOC, the risk of hospitalization or death was considered lower when compared to infected patients with delta variant (18). However, it is important to consider that the high transmissibility and immune evasion properties of the omicron subvariants can lead to an increase in the rate of infection and mortality in older people with comorbidities, as we can see in the case of Japan and China after the emergence of the omicron subvariants (3, 19). While most SARS-CoV-2 variants are linked with breakthrough infections in fully vaccinated individuals, a cumulative body of data has shown that the vaccinated recipients showed a faster clearance time compared to non-vaccinated individuals (20) and reduced risk of death in patients infected with SARS-CoV-2 variants (21). Instead, unvaccinated individuals remained at the highest risk of infection, severe outcomes, and death (21). Therefore, continued efforts to increase vaccination and the establishment of booster campaigns in the human population are of paramount importance to provide protection and overcome the COVID-19 crisis. (ii) How about the authorized antiviral therapies against new omicron variants and comparative efficacy among vaccinated and non-vaccinated individuals. In this way, antiviral drugs and mass inoculations demonstrated a critical role in treating COVID-19 patients, especially to reduce the number of severe cases and deaths (22). According to the Food and Drug Administration (FDA), COVID-19-related therapies include antiviral drugs, immunomodulators, neutralizing antibody therapies, cell therapies, and gene therapies.2 Based on the ongoing and past pandemic control experiences, some antiviral drugs are questionable for the treatment of patients infected with the omicron VOC and for use in clinical practice (23). Recent studies have shown that the omicron VOC is resistant to most therapeutic monoclonal antibodies (mAbs) (13). In terms of antiviral drugs, recent reports have shown that nirmatrelvir, remdesivir, PF-0730481472, and molnupiravir are effective against infection with the omicron VOC, indicating that these options may be used for clinical practice for the treatment of patients (23, 24). More recently, multiple reports demonstrated that the PAXLOVID™ (oral tablets of nirmatrelvir and ritonavir, a SARS-CoV-2 protease inhibitor) was able to reduce the risk of hospitalization or death by 89% (within 3 days of symptom onset) and 88% (within 5 days of symptom onset) when compared to the placebo group (25). Despite efforts and recent advances, we need to revise and update frequently the therapeutic arsenal against the SARS-CoV-2 infection based on the past lessons, current experiences, and features of the omicron variant and other SARS-CoV-2 variants. As we have seen for other viruses (e.g., human immunodeficiency virus, HIV), if we create selective pressure on the virus, this can favor the emergence of mutations that help it to survive in the presence of the drug, especially in the case of use of protease inhibitor or viral life cycle inhibitor drugs.
Based on our current scenario, what should we expect from our future with SARS-CoV-2? As WHO Director General Dr. Tedros Adhanom Ghebreyesus said, “the pandemic is far from over – and it will not be over anywhere until it’s over everywhere.” Considering all these factors discussed above appears we must take a step back regarding the relaxation of COVID-19 control measures and indicate that it is not yet time to let our guard down in the face of this devastating virus, especially after the emergence of the omicron subvariants. But the good news is if we look at the current epidemiological scenario in the USA, the most affected country during the COVID-19 pandemic. Analyzing the number of confirmed COVID-19 cases, deaths, hospital admissions, and patients in ICU per million people it is evident that we can see a light at the end of the tunnel, but we still have a considerable way to go to see the end of the pandemic phase (Figure 2). What should we expect in the coming months? Different countries around the world will experience the coming phase differently based on some critical factors: vaccine coverage, availability and application of boosters in the human population, dynamics of seasonality, demographics, government policies, and implementation of strategies to reduce the transmission of the virus.
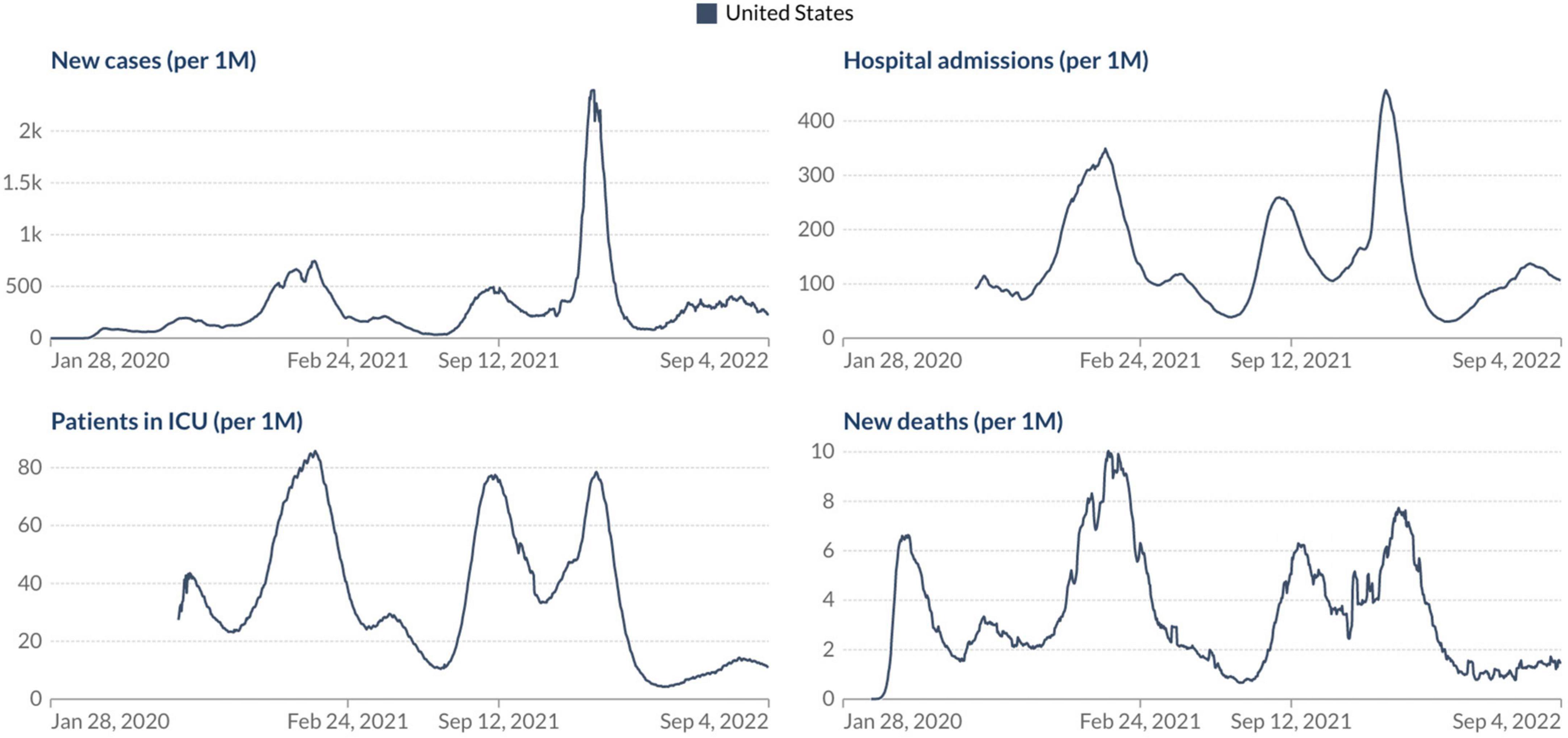
Figure 2. Epidemiological situation of the COVID-19 pandemic in the USA. Johns Hopkins University CSSE COVID-19 Data (https://ourworldindata.org/).
Notably, the SARS-CoV-2 omicron variant will probably not be the last VOC, which suggests that we should prepare for the emergence of new variants that can lead to further immune evasion and render current vaccine ineffective over time. The administration of booster shots using mRNA vaccines as an extra layer of protection will be of paramount importance to overcome the COVID-19 pandemic and combat the impact of further emerging SARS-CoV-2 variants. As with flu, it appears that we should monitor and update the composition of COVID-19 vaccines frequently, as COVID-19 may continue to be an endemic disease in the world. At the same time, we need to learn to live the new “normal” lifestyle with the hope that SARS-CoV-2 does not bring serious concerns to global health in the near future.
SS conceived the work, wrote the original draft, and reviewed the final manuscript.
SS is supported by a Postdoctoral Fellowship sponsored by the University of Toronto, Canada. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Figure 1 was created with Biorender.com under academic license.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. (2020) 382:727–33. doi: 10.1056/NEJMoa2001017
2. Rambaut A, Holmes EC, O’Toole Á, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. (2020) 5:1403–7. doi: 10.1038/s41564-020-0770-5
3. Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. (2020) 20:533–4. doi: 10.1016/S1473-3099(20)30120-1
4. da Silva SJR, de Lima SC, da Silva RC, Kohl A, Pena L. Viral load in COVID-19 patients: implications for prognosis and vaccine efficacy in the context of emerging SARS-CoV-2 variants. Front Med (Lausanne). (2021) 8:836826. doi: 10.3389/fmed.2021.836826
5. Silva SJRD, Pena L. Collapse of the public health system and the emergence of new variants during the second wave of the COVID-19 pandemic in Brazil. One Health. (2021) 13:100287. doi: 10.1016/j.onehlt.2021.100287
6. da Silva SJR, do Nascimento JCF, Germano Mendes RP, Guarines KM, Targino Alves da Silva C, da Silva PG, et al. Two years into the COVID-19 pandemic: lessons learned. ACS Infect Dis. (2022) 8:1758–814. doi: 10.1021/acsinfecdis.2c00204
7. Yamasoba D, Kimura I, Nasser H, Morioka Y, Nao N, Ito J, et al. Virological characteristics of the SARS-CoV-2 omicron BA.2 spike. Cell. (2022) 185:2103–15.e19. doi: 10.1016/j.cell.2022.04.035
8. Silva, SJRD, Kohl A, Pena L, Pardee K. Recent insights into SARS-CoV-2 omicron variant. Rev Med Virol. (2022) e2373. doi: 10.1002/rmv.2373
9. Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. (2021) 184:2372–83.e9. doi: 10.1016/j.cell.2021.03.013
10. Planas, D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, et al. Considerable escape of SARS-CoV-2 omicron to antibody neutralization. Nature. (2021). 602:671–75. doi: 10.1038/s41586-021-04389-z
11. Nyberg T, Ferguson NM, Nash SG, Webster HH, Flaxman S, Andrews N, et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet. (2022) 399:1303–12. doi: 10.1016/S0140-6736(22)00462-7
12. Hachmann NP, Miller J, Collier AY, Ventura JD, Yu J, Rowe M, et al. Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N Engl J Med. (2022) 387:86–8. doi: 10.1056/NEJMc2206576
13. Hoffmann M, Krüger N, Schulz S, Cossmann A, Rocha C, Kempf A, et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. (2021). 185:447–56. doi: 10.1016/j.cell.2021.12.032
14. Yu J, Collier AY, Rowe M, Mardas F, Ventura JD, Wan H, et al. Neutralization of the SARS-CoV-2 omicron BA.1 and BA.2 variants. N Engl J Med. (2022) 386:1579–80. doi: 10.1056/NEJMc2201849
16. Meng B, Abdullahi A, Ferreira IATM, Goonawardane N, Saito A, Kimura I, et al. Altered TMPRSS2 usage by SARS-CoV-2 omicron impacts tropism and fusogenicity. Nature. (2022) 603:706–14. doi: 10.1038/s41586-022-04474-x
17. Ong SWX, Chiew CJ, Ang LW, Mak TM, Cui L, Toh MPHS, et al. Clinical and virological features of SARS-CoV-2 variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta). Clin Infect Dis. (2021). 75:e1128–36. doi: 10.1093/cid/ciab721
18. Ulloa AC, Buchan SA, Daneman N, Brown KA. Estimates of SARS-CoV-2 omicron variant severity in Ontario, Canada. JAMA. (2022) 327:1286–8. doi: 10.1001/jama.2022.2274
19. Zhang X, Zhang W, Chen S. Shanghai’s life-saving efforts against the current omicron wave of the COVID-19 pandemic. Lancet. (2022) 399:2011–2. doi: 10.1016/S0140-6736(22)00838-8
20. Kissler SM, Fauver JR, Mack C, Tai CG, Breban MI, Watkins AE, et al. Viral dynamics of SARS-CoV-2 variants in vaccinated and unvaccinated persons. N Engl J Med. (2021) 385:2489–91. doi: 10.1056/NEJMc2102507
21. Cohn BA, Cirillo PM, Murphy CC, Krigbaum NY, Wallace AW. SARS-CoV-2 vaccine protection and deaths among US veterans during 2021. Science. (2022) 375:331–6. doi: 10.1126/science.abm0620
22. Barnette K, Gordon MS, Rodriguez D, Gary Bird T, Skolnick A, Schnaus M, et al. Oral sabizabulin for high-risk, hospitalized adults with Covid-19: interim analysis. NEJM Evid. (2022). 1:1–11.
23. Li P, Wang Y, Lavrijsen M, Lamers MM, de Vries AC, Rottier RJ, et al. SARS-CoV-2 omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination. Cell Res. (2022) 32:322–4. doi: 10.1038/s41422-022-00618-w
24. Takashita E, Kinoshita N, Yamayoshi S, Sakai-Tagawa Y, Fujisaki S, Ito M, et al. Efficacy of antibodies and antiviral drugs against Covid-19 omicron variant. N Engl J Med. (2022) 386:995–8. doi: 10.1056/NEJMc2119407
25. Hammond J, Leister-Tebbe H, Gardner A, Abreu P, Bao W, Wisemandle W, et al. Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19. N Engl J Med. (2022) 386:1397–408. doi: 10.1056/NEJMoa2118542
26. Cameroni E, Bowen JE, Rosen LE, Saliba C, Zepeda SK, Culap K, et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. (2021). 602:664–70. doi: 10.1038/s41586-021-04386-2
27. Garcia-Beltran WF, St Denis KJ, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML, et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. (2022) 185:457–66.e4. doi: 10.1016/j.cell.2021.12.033
28. Xu Y, Wu C, Cao X, Gu C, Liu H, Jiang M, et al. Structural and biochemical mechanism for increased infectivity and immune evasion of omicron BA.2 variant compared to BA.1 and their possible mouse origins. Cell Res. (2022) 32:609–20. doi: 10.1038/s41422-022-00672-4
29. Andreano E, Paciello I, Marchese S, Donnici L, Pierleoni G, Piccini G, et al. Anatomy of omicron BA.1 and BA.2 neutralizing antibodies in COVID-19 mRNA vaccinees. Nat Commun. (2022) 13:3375. doi: 10.1038/s41467-022-31115-8
30. Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by omicron infection. Nature. (2022) 608:593–602. doi: 10.1038/s41586-022-04980-y
31. Wang Q, Guo Y, Iketani S, Li Z, Mohri H, Wang M, et al. SARS-CoV-2 omicron BA.2.12.1, BA.4, and BA.5 subvariants evolved to extend antibody evasion. bioRxiv [Preprint]. (2022). doi: 10.1038/s41586-022-05053-w
Keywords: SARS-CoV-2, pandemic, variants of concern, omicron, public health
Citation: Silva SJRd (2022) The emergence of new SARS-CoV-2 omicron subvariants introduces uncertainty about the end of the COVID-19 pandemic. Front. Med. 9:1010489. doi: 10.3389/fmed.2022.1010489
Received: 03 August 2022; Accepted: 08 September 2022;
Published: 10 October 2022.
Edited by:
Pragya Dhruv Yadav, ICMR-National Institute of Virology, IndiaReviewed by:
Chaitanya Kurhade, University of Texas Medical Branch at Galveston, United StatesCopyright © 2022 Silva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Severino Jefferson Ribeiro da Silva, amVmZmVyc29uLnNpbHZhQHV0b3JvbnRvLmNh, amVmZmVyc29uYmlvdGVjdmlyb0BnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.