
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 23 December 2021
Sec. Precision Medicine
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.798571
This article is part of the Research Topic Personalized Medicine and Infectious Disease Management View all 11 articles
Background: Individuals infected with the COVID-19 virus present with different symptoms of varying severity. In addition, not all individuals are infected despite exposure. Risk factors such as age, sex, and comorbidities play a major role in this variability; however, genetics may also be important in driving the differences in the incidence and prognosis of the disease. An Insertion/Deletion (I/D) polymorphism in the ACE1 gene (rs1799752) may explain these genetic differences. The aims of this study were to determine the potential role of ACE1 I/D genetic polymorphism in the risk of contracting COVID-19 as well as predicting the severity of COVID-19 infection.
Methods: Three-hundred and eighty-seven non-related Lebanese subjects, 155 controls and 232 cases, who presented to the American University of Beirut Medical Center (AUBMC) for COVID-19 PCR testing were recruited. Clinical data were collected via filling a questionnaire and accessing the medical records. Peripheral blood was withdrawn for DNA isolation, and genotyping performed with standard PCR followed by band visualization on agarose gel.
Results: In our study population, previously described risk factors such as gender, age, and comorbidities were associated with increase in disease susceptibility and severity. ACE1 I was the least common allele, and there was a positive association between ACE1 I and the risk of contracting the COVID-19 disease. More specifically, the frequency of II genotype was significantly higher among cases when compared to controls (P = 0.035) with individuals with the II genotype having greater risk for contracting the COVID-19 disease: OR = 2.074, P = 0.048 in the multivariate analysis. As for disease severity, the DD genotype and D allele were associated with increased risk for developing severe symptoms (OR = 2.845, P = 0.026 and OR = 2.359, P = 0.014, respectively), and the DD genotype with necessitating hospitalization (OR = 2.307, P = 0.042). In parallel, D allele carriers showed a significantly increased risk for developing hypoxia: OR = 4.374, P = 0.045.
Conclusion: We found a positive association between ACE1 I and the risk of contracting the COVID-19 disease, and between ACE1 D and a worse outcome of the COVID-19 infection. Therefore, genotyping for ACE1 I/D polymorphism could be used to assess risk and predict severity for better prognosis and management of the disease.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive-sense single-stranded RNA virus that is responsible for the globally transmissible coronavirus disease of 2019 (COVID-19) (1). It has been observed across infected populations worldwide that symptoms are displayed with dissimilar presentations of varying severity. In addition, not all individuals are infected despite a history of exposure, including multiple direct exposures, to COVID-19. Several factors have been described in the literature for their potential role in the risk of contracting COVID-19 as well as that of developing complications. These include age, sex, blood group, smoking history, comorbidities, obesity, and intake of ACE inhibitors (ACEI) or angiotensin receptor blockers (ARBs) (2–8). In addition to these risk factors, genetics may play a contributing role in COVID-19 infection (9). With inconclusive data, few studies have highlighted the roles of transmembrane protease serine 2 (TMPRSS2), angiotensin converting enzyme 1 (ACE1), and ACE2 gene variants in the susceptibility and severity of SARS-CoV-2 infection (10–12).
TMPRSS2 expression facilitates the entry of the virus into host cells through ACE2 (13). Both ACE1 and ACE2 are endogenous proteins involved in the renin-angiotensin system (RAS), which regulates the homeostasis of blood pressure and fluid electrolyte balance (14). In lung vascular endothelium, ACE1 converts Angiotensin I into Angiotensin II that promotes vasoconstriction, inflammation, and thrombosis (14). ACE2 converts Angiotensin II into Angiotensin 1–7 that acts inversely to Angiotensin II and hence promotes vasodilation (14). When SARS-CoV-2 enters human cells by binding its spike (S) protein to ACE2, lower levels of this membrane receptor become available for the suppression of Angiotensin II (15). Consequently, the balance of the RAS can be distorted in favor of vasoconstriction, inflammation, and thrombosis, potentially complicating the outcome of COVID-19 infection (14, 15).
An Insertion/Deletion (I/D) polymorphism in the ACE1 gene (rs1799752) may explain the differences in genetic susceptibilities across variable geographic populations. The ACE1 D/D genotype correlates with a higher activity of the ACE1 enzyme, hence increasing the levels of Angiotensin II with secondary lowering of ACE2 expression (16). Despite some negative results (17), few studies showed the DD genotype to be associated with a significantly higher risk for COVID-19 morbidity and mortality (18, 19). Moreover, a higher I/D-allele frequency ratio has been associated with higher recovery rates despite an increase in infectivity (20). A comprehensive review done in 2021 regarding the association between ACE1 (I/D) polymorphism and COVID-19 symptoms is referenced for the reader (21). The data are less conclusive concerning the association between ACE1 (I/D) genetic polymorphism and risk of contracting the disease. For instance, an initial analysis by Delanghe et al. (22) of disease spread in 25 European countries with ACE1 historical genetic data showed a significant association between COVID-19 cases and higher frequency of the ACE1 I allele (22). In contrast, Yamamoto et al. (23) observed that the Europeans have a higher probability of being infected by SARS-CoV-2 compared to Asian populations who have a higher frequency of the ACE1 II genotype. Importantly, the negative correlation between COVID-19 incidence and ACE1 II genotype was weakened when they added data from the Middle East, stating that the Middle East should be considered an important factor for future studies (23). This is especially the case since, and as per Saab et al. (24), the Middle Eastern population such as the Lebanese, have a lower frequency of the ACE1 I allele when compared to the D allele.
The aims of this study were to determine the potential role of ACE1 I/D genetic polymorphism in the risk of contracting COVID-19 as well as predicting the severity of COVID-19 infection. We hypothesized that the ACE1 I allele is associated with an increased risk of contracting the SARS-CoV-2 virus, while the ACE1 D allele is associated with a worse prognosis depicted as increased severity of signs, symptoms, and sequelae following COVID-19 infection.
This study was approved by the Institutional Review Board (IRB) of the American University of Beirut (AUB). Three-hundred and eighty-seven Lebanese adult subjects were recruited given they had presented to the AUB Medical Center (AUBMC) for COVID-19 PCR testing (irrespective of result), COVID-19 hospitalization, or post-COVID-19 persistent symptoms. The recruitment process entailed a one-time participation that included informed consent process, data collection, and peripheral blood withdrawal for DNA isolation and ACE1 genotyping.
Data for this study were obtained via a questionnaire and access through medical records on the electronic heath information system EPIC. Information collected included demographics, comorbidities, medications intake, date of PCR testing, and COVID-19 disease presentation, management, and progression for each participant.
Peripheral blood was collected in EDTA containing tubes, processed into aliquots and stored at −80°C. DNA was then isolated using FlexiGene® DNA Kit by QIAGEN® (Germany) as per the manufacturer's guidelines. Isolated DNA was read using the DS-11 Spectrophotometer (DeNovix®, USA) for quantification and purity assessment and stored at −20°C. Genotyping for ACE1 insertion/deletion polymorphism was carried out by polymerase chain reaction (PCR) followed by gel visualization with primers and experimental conditions as previously described (25). Individuals homozygous for the D allele and I allele were identified by a single 190 bp fragment and a single 490 bp fragment, respectively. Heterozygous individuals were identified by the presence of both fragments.
The collected data were transcribed onto Microsoft Excel then exported to SPSS® (IBM, USA) for description and analysis. A P < 0.05 was considered statistically significant.
The ACE1 polymorphism was analyzed using four separate associations: one for the alleles (I vs. D), and the remaining three for the genotypes (II vs. DI vs. DD, D-carriers, and I-carriers). The D-carrier association was (II vs. DI + DD), and that of the I-carrier was (DD vs. DI + II). The genotype frequencies in controls were checked for Hardy Weinberg Equilibrium (HWE) using chi-square test.
Baseline characteristics included in the analysis were age, body mass index (BMI), sex, blood group (containing A or not), smoking (never, ever), comorbidities, and intake of ACEI or ARBs. Comorbidities were classified as follows: dyslipidemia, hypertension, diabetes, heart disease (coronary artery disease or heart failure), kidney disease (chronic kidney disease or end-stage renal disease), lung disease (chronic obstructive pulmonary disease or interstitial lung disease or asthma), cerebrovascular disease (stroke or carotid stenosis), coagulation disorders (hemophilia or von Willebrand disease), and cancer.
For the association of ACE1 (I/D) polymorphism with contracting COVID-19 disease, participants infected with COVID-19 (cases) were compared to those who were not (controls). For the association of ACE1 (I/D) polymorphism with severity and outcome of COVID-19 infection, three comparisons were carried out: mild vs. moderate vs. severe disease, hospitalized vs. non-hospitalized, and hypoxic (SpO2 <94%) vs. non-hypoxic (SpO2 ≥94%) upon hospitalization. Disease severity was classified according to the WHO clinical progression scale into three stages: stage I (mild), stage II (moderate), and stage III (severe) (26). Mild presentation included any combination of the following: fever and/or chills, cough, shortness of breath, sore throat, congestion and/or rhinorrhea, fatigue, myalgias, headache, nausea and/or vomiting, diarrhea, anosmia, and ageusia. The moderate disease stage included symptomatic patients who were hospitalized with evident radiographic lung inflammation and a blood oxygen saturation (SpO2) ≥94% with minimal or no oxygen therapy required (26). Severe disease included critically ill patients with marked lung infiltrates on Chest X-Ray or CT scan and hypoxia (SpO2 <94%) who required hospitalization with essential oxygen therapy by either nasal cannula, face mask, non-invasive ventilation (NIV), and/or mechanical ventilation with intubation (26).
Association analyses were carried out using Fisher's Exact test for categorical variables and independent sample t-test or one-way ANOVA with post-hoc Bonferroni for continuous variables as applicable. Binary or multinomial logistic regressions were used for the associations with ACE1 (I/D) polymorphism at both the univariate and multivariate level since these are the main focus of the study. Multivariate regression entailed adjustment for all statistically significant covariates at the univariate level. Results are presented as number (percentage) N (%), mean ± standard deviation (SD) at the univariate level, and odds ratios (OR) (adjusted and unadjusted) with 95% confidence intervals.
Additional analysis was performed to explore previously reported association(s) of the ACE1 polymorphism with comorbidities.
Three-hundred and eighty-seven non-related Lebanese subjects, 155 controls and 232 cases, who presented to AUBMC for COVID-19 PCR testing were recruited and included in this study. The three genotypes were in HWE (P = 0.281). ACE1 I allele was the least common with a frequency of 31.0% and a II genotype frequency of 7.8% in controls (Table 1). These numbers are in line with the literature stating that the I allele is least common in Caucasians and Middle Easterners, and most common in Asians [Supplementary Table 1; (24, 27)].
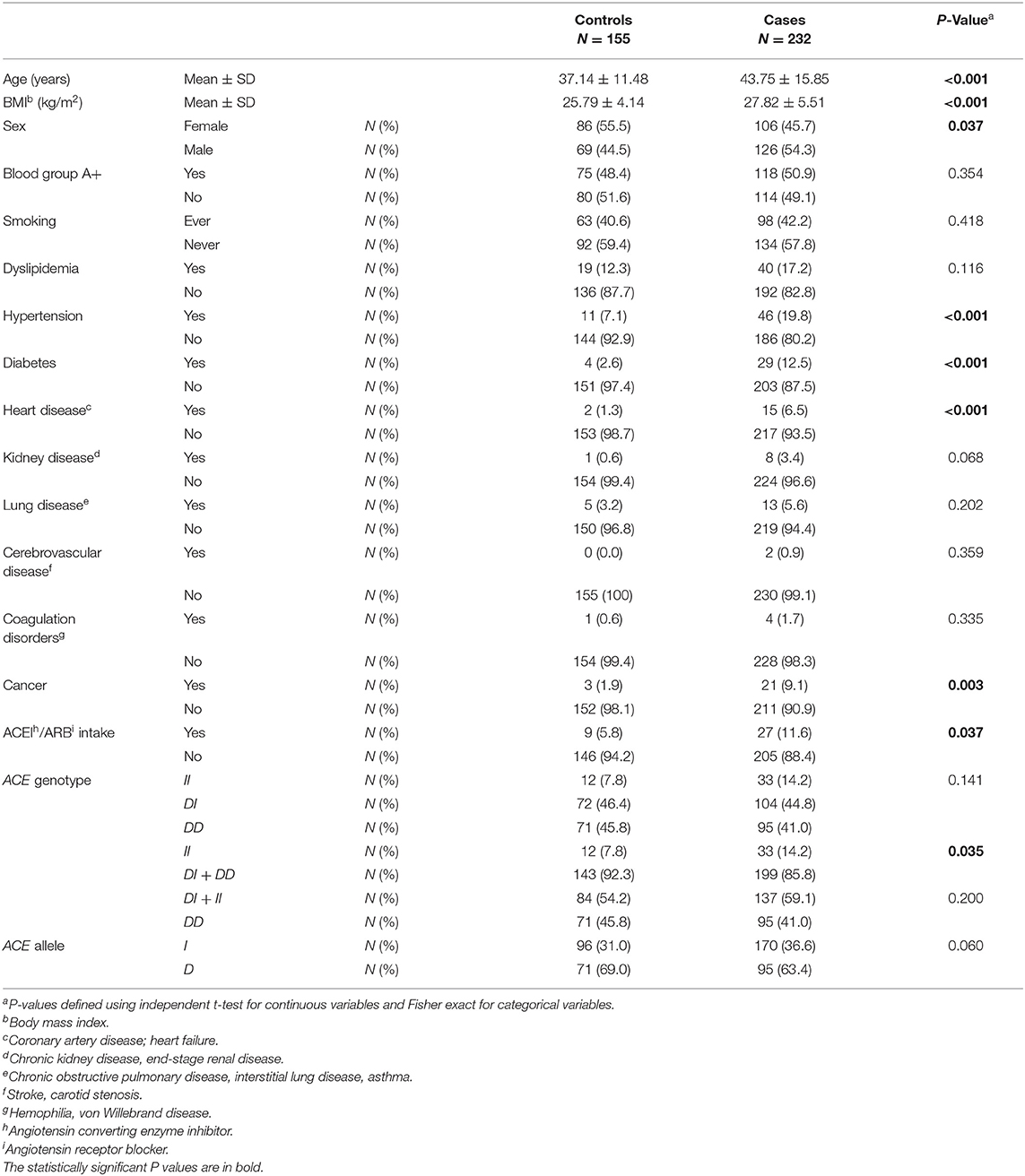
Table 1. Association between baseline characteristics and ACE1 polymorphism in COVID-19 positive cases vs. COVID-19 negative controls.
When comparing baseline characteristics to predict disease susceptibility in cases vs. controls (Table 1), the cases were both older and of higher BMI. There was a larger proportion of males in the infected group compared to that of the uninfected group. Hypertension, diabetes, heart disease, and cancer were all significant comorbid predictors for COVID-19 susceptibility. Moreover, there was a greater proportion of participants taking ACEI/ARBs among the case group when compared to controls (Table 1).
Compared to ACE1 D, the frequency of the II genotype was significantly higher among individuals infected with COVID-19 (14.2 vs. 7.8%; P = 0.035; Table 1). After adjusting for age, BMI, sex, hypertension, diabetes, heart disease, cancer, and ACEI/ARBs intake, binary logistic regression showed that, compared to D allele carriers, individuals with the II genotype were at increased risk for contracting the virus (OR = 2.074; P = 0.048; Supplementary Table 2 and Figure 1).
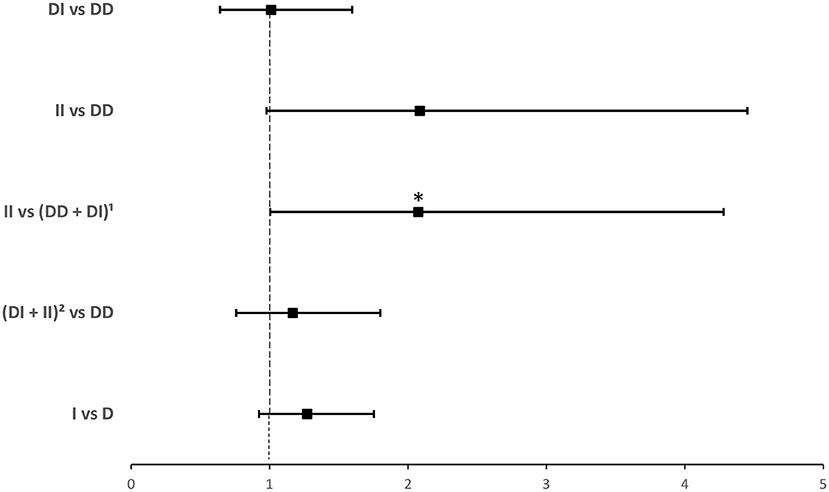
Figure 1. Forest plot showing adjusted odds ratios ± 95% confidence intervals of ACE1 polymorphism for contracting SARS-CoV-2. Multivariate analysis included variables that were statistically significant in the association analysis shown in Table 1; *P < 0.05. 1D allele carriers. 2I allele carriers.
Among the 232 cases, 223 were symptomatic: 136 (61.0%) had mild symptoms, 26 (11.7%) had moderate symptoms and 61 (27.3%) had severe symptoms. The mean ± SD of symptoms' duration was 10.14 ± 8.56 days.
As show in Table 2, compared to cases with mild infection, those with moderate and severe infection were older and of higher BMI. There were larger proportions of males among moderate and severe cases compared to mild cases. Dyslipidemia, hypertension, diabetes, heart disease, kidney disease, coagulation disorders, and cancer were significant comorbid predictors for moderate and severe disease vs. mild disease. There was also a larger proportion of ACEI/ARBs intake among moderate and severe cases when compared to controls (Table 2).
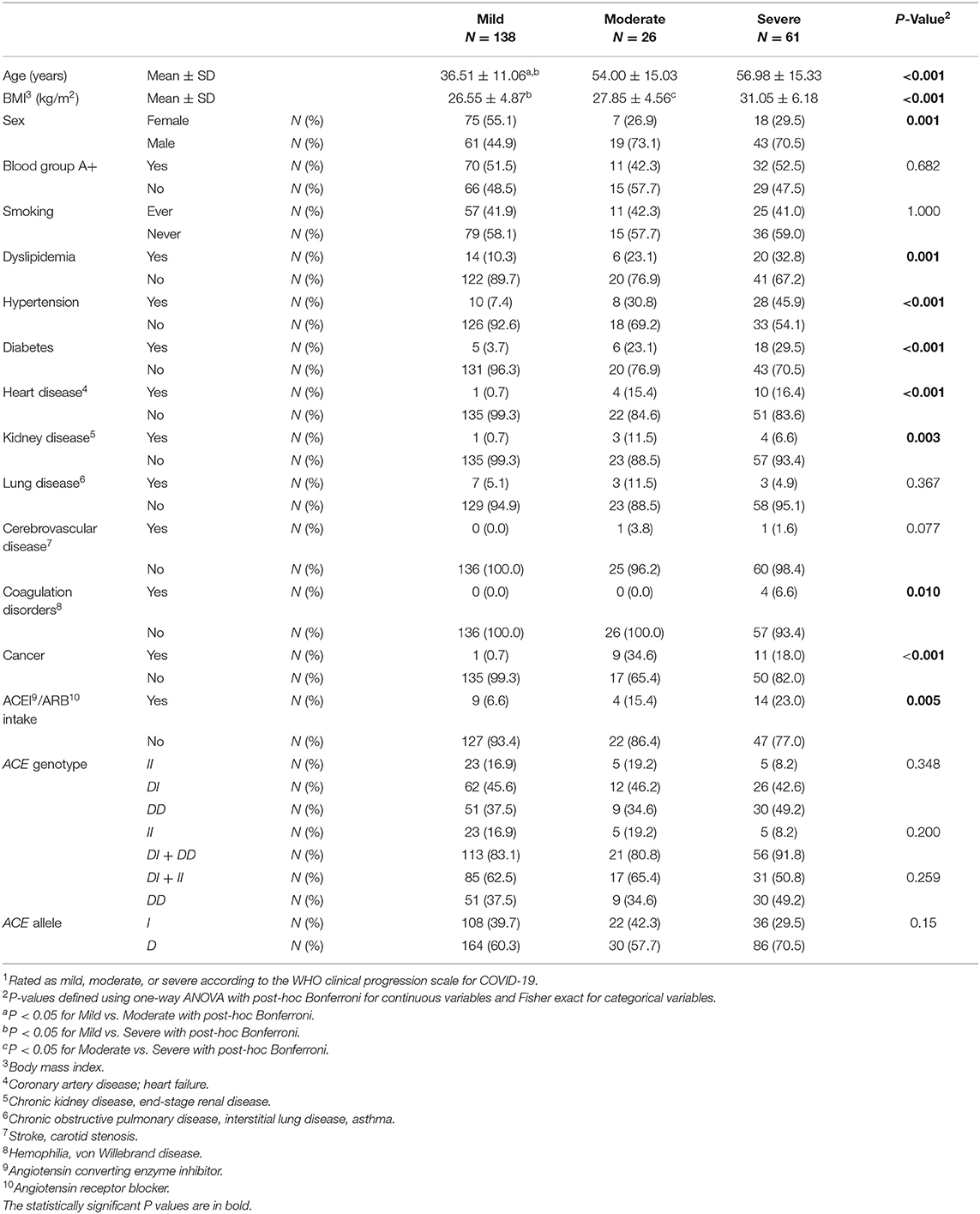
Table 2. Association between baseline characteristics and ACE1 polymorphism with disease severity1 in symptomatic COVID-19 cases.
ACE1 I/D genotype and allele frequencies were not significantly associated with disease severity although there was a trend of higher DD genotype and D allele frequencies in cases with severe symptoms of COVID-19 disease (Table 2). After adjusting for age, BMI, sex, significant comorbidities, and ACEI/ARBs intake, multinomial logistic regression showed that symptomatic cases with the DD genotype had a higher risk of developing severe disease following SARS-CoV-2 infection (OR = 5.751; P = 0.038) when compared to symptomatic II individuals. In addition, and compared to symptomatic I carriers, symptomatic cases with the DD genotype were more likely to develop severe disease following infection (OR = 2.845; P = 0.026). Similarly, the D allele was significantly associated with more severe disease presentation (OR =2.359; P = 0.014: Supplementary Table 3 and Figure 2).
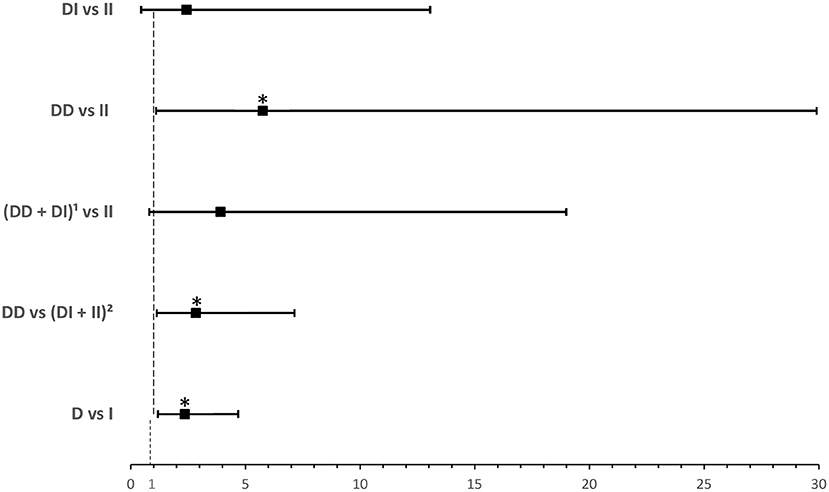
Figure 2. Forest plot showing odds ratios ± 95% confidence intervals of ACE1 polymorphism for developing severea disease in symptomatic COVID-19 cases. Multivariate analysis included variables that were statistically significant in the association analysis shown in Table 2; *P < 0.05. aRated as mild, moderate, or severe according to WHO clinical progression scale for COVID-19 with mild disease as Reference. 1D allele carriers. 2I allele carriers.
Among the 232 cases, 144 (62.1%) were non-hospitalized while 88 (37.9%) were hospitalized. The mean ± SD of length of stay was 13.45 ± 13.73 days.
It is shown in Table 3 that hospitalized patients were older and of higher BMI. There was a significantly larger proportion of hospitalized males compared to non-hospitalized males. Dyslipidemia, hypertension, diabetes, heart disease, kidney disease, coagulation disorders, and cancer were significant comorbid predictors for hospitalization. Additionally, there was a larger proportion of ACEI/ARBs intake among hospitalized cases (Table 3).
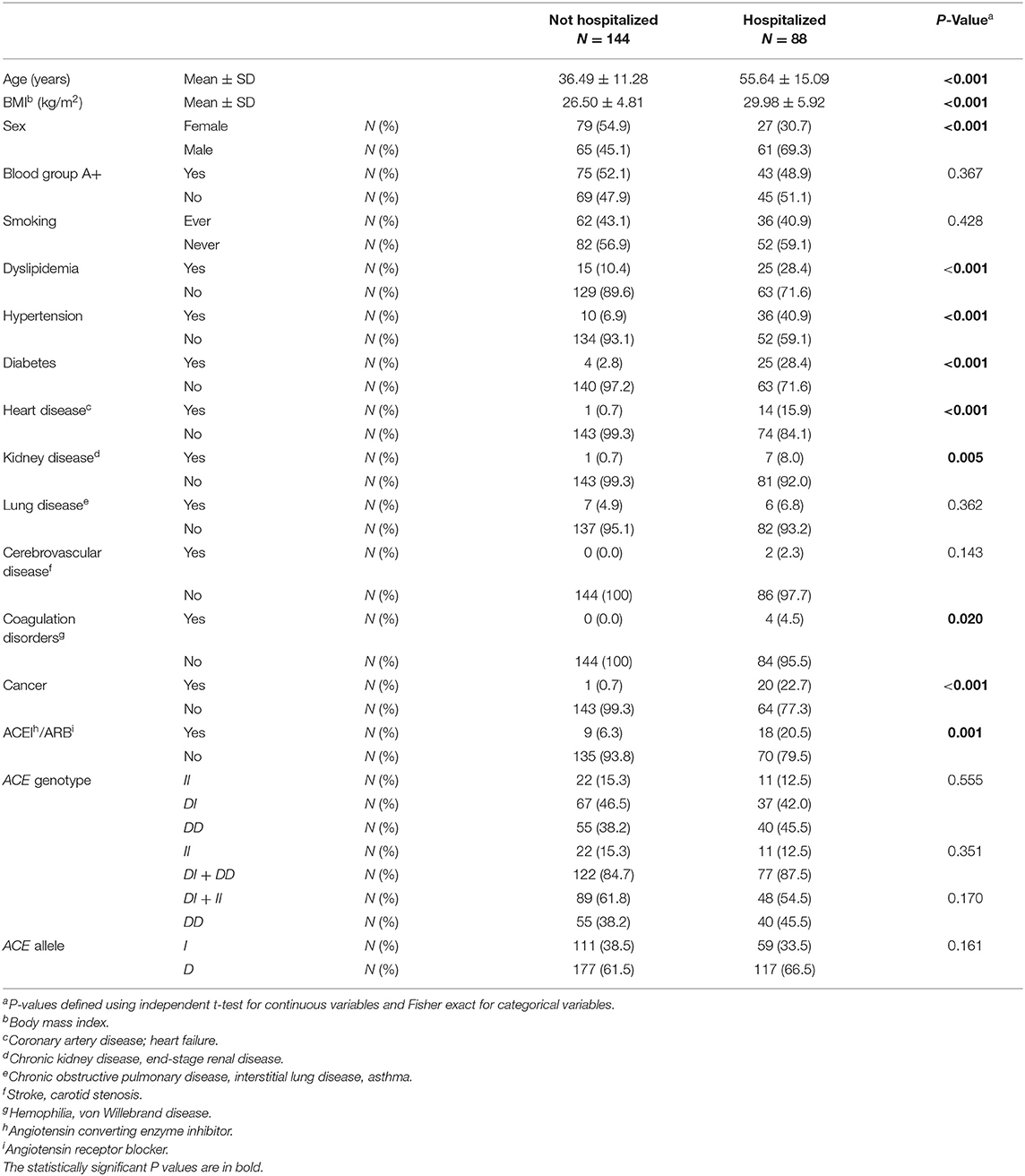
Table 3. Association between baseline characteristics and ACE1 polymorphism with hospitalized vs. non-hospitalized COVID-19 cases.
ACE1 I/D genotype and allele frequencies were not significantly associated with hospitalization although there was a trend of higher DD genotype and D allele frequencies in hospitalized cases with COVID-19 disease (Table 3). After adjusting for age, BMI, sex, significant comorbidities, and ACEI/ARBs intake, binary logistic regression showed that, compared to I carriers, individuals with the DD genotype were at higher risk for hospitalization following infection (OR = 2.307; P = 0.042; Supplementary Table 4 and Figure 3).
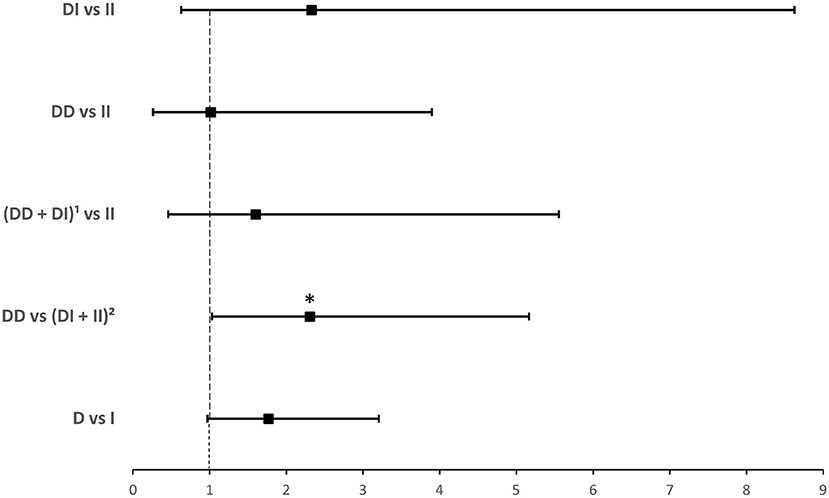
Figure 3. Forest plot showing adjusted odds ratios ± 95% confidence intervals of ACE1 polymorphism for hospitalization for COVID-19. Multivariate analysis included variables that were statistically significant in the association analysis shown in Table 3; *P < 0.05. 1D allele carriers, 2I allele carriers.
Among the 88 hospitalized patients, 26 (29.5%) were not hypoxic as opposed to 62 (70.5%) that were.
As shown in Table 4, hypoxic patients only had significantly higher BMI when compared to admitted patients without hypoxia. There was a slightly larger proportion of hypoxic males compared to non-hypoxic males, but this result was not statistically significant. There were no significant comorbid predictors for developing hypoxia; nevertheless, there was an increasing trend for dyslipidemia, hypertension, diabetes, and ACEI/ARBs intake among hypoxic patients (Table 4).

Table 4. Association between baseline characteristics and ACE1 polymorphism with hypoxic vs. non-hypoxic hospitalized COVID-19 cases.
ACE1 I/D genotype and allele frequencies were not significantly associated with hypoxia although there was a trend of higher DD genotype and D allele frequencies in hypoxic hospitalized cases with COVID-19 disease (Table 4). After adjusting for BMI, binary logistic regression showed that, compared to the II genotype, D allele carriers were at an increased risk for developing hypoxia following infection (OR = 4.374; P = 0.045; Supplementary Table 5 and Figure 4).
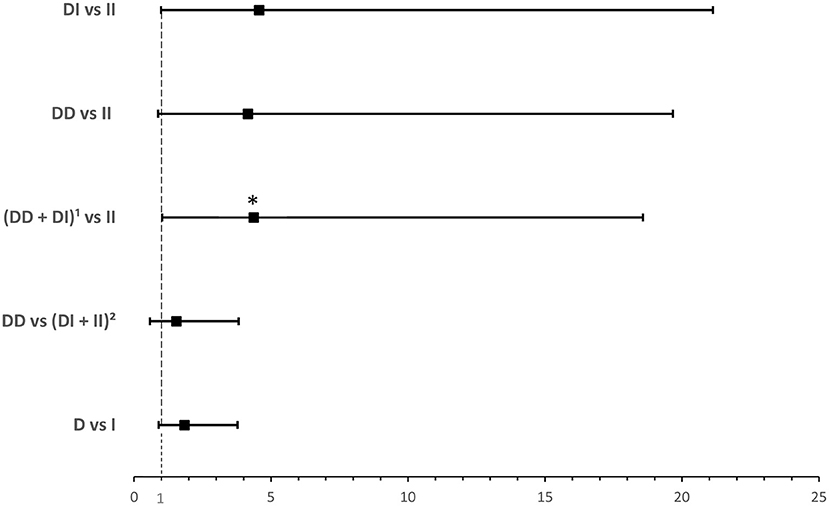
Figure 4. Forest plot showing adjusted odds ratios ± 95% confidence intervals of ACE1 polymorphism for developing hypoxia in hospitalized COVID-19 cases. Multivariate analysis adjusted for BMI; *P < 0.05. 1D allele carriers. 2I allele carriers.
Ever since the outbreak, people realized that the SARS-CoV-2 virus hits every individual differently with varying symptoms and severity. There has been a plethora of articles from different populations and ethnicities discussing the factors that are considered to be risk factors for both symptoms and severity of the COVID-19 disease, but with only few related to genetics. This study is the first to evaluate these factors in Lebanese Arabs. We show that almost all previously reported factors and comorbidities also predict disease susceptibility and severity in the Lebanese population. We also show a positive correlation between ACE1 I and the risk of contracting the COVID-19 disease, and between ACE1 D and worse COVID-19 infection. These results suggest that genotyping for ACE1 I/D polymorphism could be used to assess risk and predict severity for better prognosis and management of the disease. This is especially important for Middle Easterners in general and the Lebanese in particular who, and similarly to the results of the current study, have a higher frequency of the ACE1 D allele when compared to the I allele (24, 25).
Most of the associated demographics, health-related behaviors, and comorbidities can be explained at the physiological level. For instance for age, ACE2 receptor, being the key factor in the entry of the virus, is more highly expressed in well-differentiated ciliated epithelial cells found in adults (2). Moreover, the immunity of an older individual is weaker than the immunity of children due to immunosenescence and the presence of central memory T cells rather than naïve T cells (2). Our results agree with the literature since the mean age (in years) is significantly higher in the infected cases when compared to the non-infected controls, and it is significantly higher with disease severity. Concerning sex, ACE2 being an X-linked gene can be considered as a disadvantage for infected males, since lower ACE2 expression may correlate with lesser conversion of Angiotensin II into Angiotensin 1–7 (28). Moreover, testosterone suppresses the immune system in males, which affects the T cell responses (29). These findings are compatible with our results that show that the majority of cases and those with worse outcome are males. In our study, the mean BMI (kg/m2) was also significantly higher in the infected cases and associated with more severe disease. This can be explained by the fact that the adipose tissue expresses ACE2 receptors as much as the pulmonary tissues (7). Accordingly, obese individuals have higher levels of circulating ACE2 with secondarily higher disease susceptibility and adverse outcome (30). As for blood group, data are still non-conclusive. For example, it has been shown that carriage of blood group A was associated with a higher rate of COVID-19 infection when compared to blood group O (31). However, it is felt that individuals with blood group A also have more underlying comorbidities (29), which could be the reason behind the significance seen in infected patients. In our study, blood group did not show any significant difference with neither risk nor severity of the disease.
Concerning health-related behavior, smoking is one of the most common risk factor for many diseases. That is why smoking is expected to further complicate the symptoms of COVID-19. Smoking is shown to increase the gene expression of ACE2 in the lungs (4). Moreover, nicotine upregulates the activity of renin and ACE1 thus activating ACE/Angiotensin II/AT1R pathway, and decreases the activity of AT2R by downregulating the activity of ACE2 (32). A systematic review has shown that current smokers had a lower risk for developing severe outcome when compared to former smokers (33). However, a preliminary meta-analysis on five studies in China, and similarly to our results, has shown that active smoking is not significantly related to the severity of COVID-19 (34). Further data are needed to resolve this controversy.
To date, it is still unclear whether ACEI and/or ARBs should be kept in patients who contract COVID-19. There are currently two contradicting hypotheses in the literature that RAS inhibition could be both harmful and protective (8). In our study, ACEI/ARBs were significantly more frequently taken in the worse disease outcome group. However, it is possible that these results relate to the fact that this group of subjects has underlying comorbidities that necessitate ACEI/ARBs treatment. As a matter of fact, people with underlying comorbidities such as diabetes, hypertension (HTN), cardiovascular diseases (CVD), chronic kidney diseases (CKD), lung diseases (COPD and asthma), cerebral vascular disease, and coagulation disorders are at a higher risk of worse COVID-19 severity and outcome (35–37). Our results clearly show that comorbidities are risk factors for contracting the virus and developing a worse COVID-19 disease outcome.
In relation to disease susceptibility, available data, most of which are literature and database searches, are at times contradictory (21). For example on one hand, Yamamoto et al. (23) showed that countries with higher frequency of the ACE1 I allele had less susceptibility to COVID-19. On the other hand, Delanghe et al. (22) showed that a high frequency of ACE1 I allele increases the prevalence of COVID-19 cases. Nevertheless, when Yamamoto et al. (23) specifically looked at Middle Eastern populations, they found a weaker association with the D allele, hence the need for further investigations. To our knowledge, we are the first to evaluate such an association in patients. We confirmed Delanghe et al.'s (22) simulations by showing that the frequency of II was significantly highest in infected cases when compared to controls coupled with a significantly higher risk of contracting the COVID-19 disease after adjusting for confounders.
As for disease outcome, ACE1 DD genotype leads to higher activity of ACE1 enzyme thus lowering ACE2 causing an increase in the amount of angiotensin II left active. Although lower levels of ACE2 could mean that there is less chance for SARS-CoV-2 to bind and enter the host cell, high levels of angiotensin II would act through AT1R and further cause cardiovascular and lung pathologies (16). As a matter of fact, Gomez et al. (16) found that ACE1 DD genotype was more frequent in severe COVID-19 cases, suggesting that there is an association between ACE1 DD genotype and the severity of COVID-19. Furthermore, ACE1 DD genotype has been correlated with respiratory failure (12) and increased death rate (38) in patients infected with COVID-19. In addition, an ecologic meta-regression showed that there is a link between ACE1 I/D polymorphism and the recovery rate of COVID-19 whereby faster recovery was correlated with higher frequency ratio of the I/D allele (20). Our results are in agreement with the literature. Notably, it could be argued that the latter association is due to the known associations of the ACE1 D allele with cardiovascular comorbidities. In our cohort of infected cases however, we found no significant associations with any of the comorbidities (Supplementary Table 6).
This study has few limitations. First, the sample size is limited to a single country and institution, and is relatively small. Of note that we did not estimate needed sample size at study initiation because of lack of such data at the time and the study being exploratory. Nevertheless, our sample size for the severity outcome is very similar to two recent investigations, one with Spanish Caucasians (16) and another with Indians (39). Second, the study entailed multiple testing, the adjustment of which could lead to loss of statistical significance. In fact for the severity outcome whereby we assessed three independent outcomes, it may be relevant to set the significance level at 0.016 (0.05/3). With such adjustment, only the association between the D allele and disease severity remains statistically significant (OR =2.359; P = 0.014). Notably, disease severity was classified as mild, moderate, and severe as per the WHO progression scale scoring system (26), a scoring system that is based on a constellation of assessment tools for severity following infection that includes hospitalization status, oxygen saturation, and need for oxygen therapy. With a larger representative sample, it is possible to have independently increased risks for both hospitalization and hypoxia with the D allele after accounting for multiple testing (P < 0.016). Additional data from other institutions and populations may address these two limitations with the opportunity to perform a meta-analysis. Third, the study only evaluated the ACE1 I/D polymorphism and did not look at other possible SNPs in ACE1. Moreover, it would be relevant to look at ACE2 and TMPRSS2 variants, as these two genes are important factors in the entry of SARS-CoV-2 (40). Finally, the role of ACEI/ARBs in COVID-19 disease is still unresolved and it would be interesting to evaluate whether there is any interaction between ACE polymorphisms and these drugs in the SARS-COV2 setting (41).
To our knowledge, we are the first to evaluate the association of ACE1 genetic polymorphism with COVID-19 disease susceptibility and outcome in a Middle Eastern Arab population such as the Lebanese. Despite its limitations, results of this study suggest that genotyping for ACE1 I/D polymorphism could be used to elicit the disease risk and severity for better prognosis and management. Further studies are needed to evaluate additional genetic variants in different ethnicities and populations.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by the American University of Beirut Institutional Review Board under protocol: BIO-2020-0259. The patients/participants provided their written informed consent to participate in this study.
CS, RM, IA, and NZ contributed conception and design of the study. HS recruited study subjects and collected data. KJ performed the experiments. HS, KJ, and NZ organized the database, performed the statistical analysis, and wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by National Council for Scientific Research Lebanon: The flash call COVID-19 management in Lebanon. Diana Tamari Sabbagh Scholars Program (DTSSP)—Award for MS Graduate Students in biomedical research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.798571/full#supplementary-material
1. Cevik M, Kuppalli K, Kindrachuk J, Peiris M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ. (2020) 371:m3862. doi: 10.1136/bmj.m3862
2. Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin Immunol. (2020) 215:108427. doi: 10.1016/j.clim.2020.108427
3. Goel R, Bloch EM, Pirenne F, Al-Riyami AZ, Crowe E, Dau L, et al. ABO blood group and COVID-19: a review on behalf of the ISBT COVID-19 Working Group. Vox Sang. (2021) 116:849–61. doi: 10.1111/vox.13076
4. Cai G, Bosse Y, Xiao F, Kheradmand F, Amos CI. Tobacco smoking increases the lung gene expression of ACE2, the receptor of SARS-CoV-2. Am J Respir Crit Care Med. (2020) 201:1557–9. doi: 10.1164/rccm.202003-0693LE
5. Ejaz H, Alsrhani A, Zafar A, Javed H, Junaid K, Abdalla AE, et al. COVID-19 and comorbidities: deleterious impact on infected patients. J Infect Public Health. (2020) 13:1833–9. doi: 10.1016/j.jiph.2020.07.014
6. Froldi G, Dorigo P. Endothelial dysfunction in Coronavirus disease (2019) (COVID-19): gender and age influences. Med Hypotheses. (2020). 144:110015. doi: 10.1016/j.mehy.2020.110015
7. Sattar N, McInnes IB, McMurray JJV. Obesity is a risk factor for severe COVID-19 infection: multiple potential mechanisms. Circulation. (2020) 142:4–6. doi: 10.1161/CIRCULATIONAHA.120.047659
8. Patel AB, Verma A. COVID-19 and angiotensin-converting enzyme inhibitors and angiotensin receptor blockers: what is the evidence? JAMA. (2020) 323:1769–70. doi: 10.1001/jama.2020.4812
9. Zeberg H, Paabo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. (2020) 587:610–2. doi: 10.1038/s41586-020-2818-3
10. Russo R, Andolfo I, Lasorsa VA, Iolascon A, Capasso M. Genetic analysis of the coronavirus SARS-CoV-2 host protease TMPRSS2 in different populations. Front Genet. (2020) 11:872. doi: 10.3389/fgene.2020.00872
11. Wooster L, Nicholson CJ, Sigurslid HH, Lino Cardenas CL, Malhotra R. Polymorphisms in the ACE2 locus associate with severity of COVID-19 infection. MedRxiv. (2020). doi: 10.1101/2020.06.18.20135152
12. Annunziata A, Coppola A, Lanza M, Simioli F, Imitazione P, Pepe N, et al. ACE DD polymorphism in severe COVID-19. J Transl Sci. (2020) 7:1. doi: 10.15761/JTS.1000396
13. Hou Y, Zhao J, Martin W, Kallianpur A, Chung MK, Jehi L, et al. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. (2020) 18:216. doi: 10.1186/s12916-020-01673-z
14. D'Ardes D, Boccatonda A, Rossi I, Guagnano MT, Santilli F, Cipollone F, et al. COVID-19 and RAS: unravelling an unclear relationship. Int J Mol Sci. (2020) 21:3003. doi: 10.3390/ijms21083003
15. Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X, et al. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. (2020) 6:11. doi: 10.1038/s41421-020-0147-1
16. Gomez J, Albaiceta GM, Garcia-Clemente M, Lopez-Larrea C, Amado-Rodriguez L, Lopez-Alonso I, et al. Angiotensin-converting enzymes (ACE, ACE2) gene variants and COVID-19 outcome. Gene. (2020) 762:145102. doi: 10.1016/j.gene.2020.145102
17. Karakas CS, Cakmak GG, Piskin N, Acikgoz B, Altinsoy B, Kurucu IB, et al. Polymorphisms of ACE (I/D) and ACE2 receptor gene (Rs2106809, Rs2285666) are not related to the clinical course of COVID-19: a case study. J Med Virol. (2021) 93:5947–52. doi: 10.1002/jmv.27160
18. Zheng H, Cao JJ. Angiotensin-converting enzyme gene polymorphism and severe lung injury in patients with coronavirus disease 2019. Am J Pathol. (2020) 190:2013–7. doi: 10.1016/j.ajpath.2020.07.009
19. Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. (2020) 63:364–74. doi: 10.1007/s11427-020-1643-8
20. Hatami N, Ahi S, Sadeghinikoo A, Foroughian M, Javdani F, Kalani N, et al. Worldwide ACE (I/D) polymorphism may affect COVID-19 recovery rate: an ecological meta-regression. Endocrine. (2020) 68:479–84. doi: 10.1007/s12020-020-02381-7
21. Yamamoto N, Nishida N, Yamamoto R, Gojobori T, Shimotohno K, Mizokami M, et al. Angiotensin-Converting Enzyme (ACE) 1 gene polymorphism and phenotypic expression of COVID-19 symptoms. Genes. (2021) 12:1572. doi: 10.3390/genes12101572
22. Delanghe JR, Speeckaert MM, De Buyzere ML. The host's angiotensin-converting enzyme polymorphism may explain epidemiological findings in COVID-19 infections. Clin Chim Acta. (2020) 505:192–3. doi: 10.1016/j.cca.2020.03.031
23. Yamamoto N, Ariumi Y, Nishida N, Yamamoto R, Bauer G, Gojobori T, et al. SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene. (2020) 758:144944. doi: 10.1016/j.gene.2020.144944
24. Saab YB, Gard PR, Overall AD. The geographic distribution of the ACE II genotype: a novel finding. Genet Res. (2007) 89:259–67. doi: 10.1017/S0016672307009019
25. Akra-Ismail M, Makki RF, Chmaisse HN, Kazma A, Zgheib NK. Association between angiotensin-converting enzyme insertion/deletion genetic polymorphism and hypertension in a sample of Lebanese patients. Genet Test Mol Biomarkers. (2010) 14:787–92. doi: 10.1089/gtmb.2010.0096
26. WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis. (2020) 20:e192–7. doi: 10.1016/S1473-3099(20)30483-7
27. Han C, Han XK, Liu FC, Huang JF. Ethnic differences in the association between angiotensin-converting enzyme gene insertion/deletion polymorphism and peripheral vascular disease: a meta-analysis. Chronic Dis Transl Med. (2017) 3:230–41. doi: 10.1016/j.cdtm.2017.07.002
28. Gemmati D, Bramanti B, Serino ML, Secchiero P, Zauli G, Tisato V. COVID-19 and individual genetic susceptibility/receptivity: role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the double X-chromosome in females be protective against SARS-CoV-2 compared to the single X-chromosome in males? Int J Mol Sci. (2020) 21:3474. doi: 10.3390/ijms21103474
29. Peckham H, de Gruijter NM, Raine C, Radziszewska A, Ciurtin C, Wedderburn LR, et al. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat Commun. (2020) 11:6317. doi: 10.1038/s41467-020-19741-6
30. Emilsson V, Gudmundsson EF, Aspelund T, Jonsson BG, Gudjonsson A, Launer LJ, et al. Serum levels of ACE2 are higher in patients with obesity and diabetes. Obes Sci Pract. (2021) 7:239–43. doi: 10.1002/osp4.472
31. Li J, Wang X, Chen J, Cai Y, Deng A, Yang M. Association between ABO blood groups and risk of SARS-CoV-2 pneumonia. Br J Haematol. (2020) 190:24–7. doi: 10.1111/bjh.16797
32. Oakes JM, Fuchs RM, Gardner JD, Lazartigues E, Yue X. Nicotine and the renin-angiotensin system. Am J Physiol Regul Integr Comp Physiol. (2018) 315:R895–906. doi: 10.1152/ajpregu.00099.2018
33. Farsalinos K, Barbouni A, Poulas K, Polosa R, Caponnetto P, Niaura R. Current smoking, former smoking, and adverse outcome among hospitalized COVID-19 patients: a systematic review and meta-analysis. Ther Adv Chronic Dis. (2020) 11:2040622320935765. doi: 10.1177/2040622320935765
34. Lippi G, Henry BM. Active smoking is not associated with severity of coronavirus disease 2019. (COVID-19). Eur J Intern Med. (2020) 75:107–8. doi: 10.1016/j.ejim.2020.03.014
35. Sanyaolu A, Okorie C, Marinkovic A, Patidar R, Younis K, Desai P, et al. Comorbidity and its impact on patients with COVID-19. SN Compr Clin Med. (2020) 1–8. doi: 10.1007/s42399-020-00363-4
36. Iniguez M, Perez-Matute P, Villoslada-Blanco P, Recio-Fernandez E, Ezquerro-Perez D, Alba J, et al. ACE gene variants rise the risk of severe COVID-19 in patients with hypertension, dyslipidemia or diabetes: a spanish pilot study. Front Endocrinol. (2021) 12:688071. doi: 10.3389/fendo.2021.688071
37. Vetter P, Vu DL, L'Huillier AG, Schibler M, Kaiser L, Jacquerioz F. Clinical features of covid-19. BMJ. (2020) 369:m1470. doi: 10.1136/bmj.m1470
38. Adamzik M, Frey U, Sixt S, Knemeyer L, Beiderlinden M, Peters J, et al. ACE I/D but not AGT (-6)A/G polymorphism is a risk factor for mortality in ARDS. Eur Respir J. (2007) 29:482–8. doi: 10.1183/09031936.00046106
39. Verma S, Abbas M, Verma S, Khan FH, Raza ST, Siddiqi Z, et al. Impact of I/D polymorphism of angiotensin-converting enzyme 1 (ACE1) gene on the severity of COVID-19 patients. Infect Genet Evol. (2021) 91:104801. doi: 10.1016/j.meegid.2021.104801
40. Torre-Fuentes L, Matias-Guiu J, Hernandez-Lorenzo L, Montero-Escribano P, Pytel V, Porta-Etessam J, et al. ACE2, TMPRSS2, and furin variants and SARS-CoV-2 infection in Madrid, Spain. J Med Virol. (2021) 93:863–9. doi: 10.1002/jmv.26319
Keywords: ACE1, COVID, risk, severity, genetic polymorphism
Citation: Saad H, Jabotian K, Sakr C, Mahfouz R, Akl IB and Zgheib NK (2021) The Role of Angiotensin Converting Enzyme 1 Insertion/Deletion Genetic Polymorphism in the Risk and Severity of COVID-19 Infection. Front. Med. 8:798571. doi: 10.3389/fmed.2021.798571
Received: 20 October 2021; Accepted: 29 November 2021;
Published: 23 December 2021.
Edited by:
Hamdi Mbarek, Qatar Foundation, QatarReviewed by:
Mohamad Saad, Qatar Computing Research Institute, QatarCopyright © 2021 Saad, Jabotian, Sakr, Mahfouz, Akl and Zgheib. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Imad Bou Akl, aWIwOEBhdWIuZWR1Lmxi; Nathalie K. Zgheib, bmsxNkBhdWIuZWR1Lmxi
†These authors share first authorship
‡These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.